Abstract
Rationale & Objective:
The current classification system for focal segmental glomerulosclerosis (FSGS) and minimal change disease (MCD) does not fully capture the complex structural changes in kidney biopsies, nor the clinical and molecular heterogeneity of these diseases.
Study Design:
Prospective observational cohort study.
Setting & Participants:
N=221 MCD and FSGS patients enrolled in the Nephrotic Syndrome Study Network (NEPTUNE).
Exposures:
The NEPTUNE Digital Pathology Scoring System (NDPSS) was applied to generate scores for 37 glomerular descriptors.
Outcomes:
Time from biopsy to complete proteinuria remission, time from biopsy to kidney disease progression (40% eGFR decline or kidney failure), and eGFR over time.
Analytical Approach:
Cluster analysis was used to group patients with similar morphologic characteristics. Glomerular descriptors and patient clusters were assessed for associations with outcomes using adjusted Cox models and linear mixed models. Messenger RNA from glomerular tissue was used to assess differentially expressed genes between clusters and identify genes associated with individual descriptors driving cluster membership.
Results:
Three clusters were identified: X (N=56), Y (N=68), and Z (N=97). Clusters Y and Z had higher probabilities of proteinuria remission (HR [95% CI]= 1.95 [0.99, 3.85] and 3.29 [1.52, 7.13], respectively), lower hazards of disease progression 0.22 [0.08, 0.57] and 0.11 [0.03, 0.45], respectively), and lower loss of eGFR over time compared with X. Cluster X had 1920 DEGs compared to Y+Z, which reflected activation of pathways of immune response and inflammation. Six individual descriptors driving the clusters individually correlated with clinical outcomes and gene expression.
Limitations:
Low prevalence of some descriptors and biopsy from a single time point.
Conclusions:
The NDPSS allows for characterization of FSGS/MCD patients into clinically and biologically relevant subgroups and uncovers histologic parameters associated with clinical outcomes and molecular signatures not included in current classification systems.
Keywords: nephrotic syndrome, proteinuria, glomerular disease, renal pathology, morphologic features, clinical outcomes, transcriptomics, gene expression, molecular analysis
INTRODUCTION
The current classification system of focal segmental glomerulosclerosis (FSGS) and minimal change disease (MCD)1 is based on a few selected morphologic parameters to derive a diagnosis, therefore it does not capture complex combinations of discrete structural changes in the renal parenchyma nor the clinical and molecular heterogeneity of these diseases. As a result, it has limited utility in predicting disease progression, response to therapy, or guiding discovery of new therapeutic approaches. New methodologies are necessary to extract useful information from the kidney structural changes that can be integrated with clinical and biomarker data to identify patient-specific therapeutic targets and ultimately improve patient care.2,3
The Nephrotic Syndrome Study Network (NEPTUNE) Digital Pathology Scoring System (NDPSS) was developed to capture the morphologic complexity of glomerular diseases agnostic to conventional categories.4 The NDPSS applies quantitative and semi-quantitative metrics to evaluate the presence and extent of discrete structural changes (“descriptors”) in the glomeruli, tubulointerstitium, and vessels.4,5 Because of its granularity, this approach is well-suited for integration of this information with other data using systems biology approaches.
To test the hypothesis that quantification of the discrete glomerular structural changes uncovers clinically and biologically relevant information not available using conventional approaches, we applied the NDPSS to kidney biopsies from MDC/FSGS patients. The resulting glomerular descriptor-based profiles were used to identify subgroups of participants with morphologic similarities and test associations of these subgroups and individual descriptors with clinical outcomes and molecular phenotypes.
METHODS
Study Sample
NEPTUNE is a prospective cohort of children and adults enrolled at the time of first clinically indicated biopsy with proteinuria >0.5 mg/mg in phase 1 and >1.5mg/mg in phase 2.5 NEPTUNE participants with a kidney biopsy in the Digital Pathology Repository (DPR) and a diagnosis of MCD or FSGS were included in this study. FSGS included patients with at least one glomerular lesion of segmental sclerosis.1 MCD was further classified into MCD (defined as ≥75% effacement with age-expected global sclerosis) or MCD-Like (defined as <75% effacement or ≥75% effacement with global sclerosis exceeding that expected for age).6 According to the NEPTUNE protocol, kidney biopsies are not further characterized based on etiology (i.e., primary, secondary and adaptive). Clinical demographics, medical history, kidney failure status, medication exposures and local laboratory data, were collected at each study visit (Supplementary Methods). All NEPTUNE participants provided written informed consent (adults) or assent with parental written consent (children). The study was approved by the University of Michigan Institutional Review Board (HUM00158219).
NEPTUNE Digital Pathology Scoring System (NDPSS) and Patient Clustering by Morphologic Profiles
A reference manual illustrating the comprehensive list of glomerular structural and cellular abnormalities (37 descriptors) and an electronic scoring sheet were first created (Table S1). Glomeruli were then annotated (enumerated) across all levels (Figure S1 A–B) and each glomerulus was scored by study pathologists using all levels and stains, (NDPSS) (Table S1 & Figure S1C).4,7 4,8 The percentage of glomeruli with each descriptor was calculated for each biopsy. Ward’s hierarchical clustering algorithm was applied to cluster participants into subgroups (Item S1). The final cluster solution was compared with conventional diagnosis (MCD/MCD-Like and FSGS subtypes) 1,9. To identify the strongest individual descriptors driving cluster membership, we used elastic net penalized multinomial regression with a penalization mixing parameter of α=0.01, and assessed whether a subset of descriptors could predict cluster membership by repeating the model with only the top 5, 10, 15, 20, 25 and 30 descriptors and comparing prediction accuracy across models.
Clinical Outcomes Analysis
NEPTUNE participants are followed prospectively with study visits three times per year in the first year and twice yearly thereafter, up to 5 years. Clinical outcomes of interest included (1) time from biopsy to complete remission of proteinuria, defined as urine protein creatinine ratio (UPCR) <0.3 mg/mg, (2) time from biopsy to a composite disease progression outcome of at least 40% decline in estimated glomerular filtration rate (eGFR) 10 with eGFR <90 mL/min/1.73m2 or kidney failure, and (3) longitudinal eGFR over time. For the two time-to-event outcomes, we used Cox proportional hazards models. Participants who did not experience a complete proteinuria remission or the disease progression outcome were censored at the time of their last UPCR measurement or at the time of last eGFR measurement or kidney failure ascertainment date respectively. For longitudinal eGFR, we used a linear mixed model with random intercept and random slope for each participant to account for repeated measures. Interactions between time from biopsy and cluster groups were tested in the longitudinal eGFR model to assess whether eGFR slope differed across clusters.
We performed both sequentially adjusted and unadjusted models between cluster groups and outcomes. For adjusted models, we first adjusted for demographics and clinical characteristics. If outcomes had sufficient numbers of events or observations to avoid overfitting, we adjusted for age, sex, self-reported black race, Hispanic ethnicity, eGFR at biopsy, UPCR at biopsy, kidney disease duration as of biopsy, and any immunosuppression use at or within 30 days prior to biopsy; if there were not enough events or observations to avoid overfitting, we used backward selection to choose adjustment covariates. Then, we additionally adjusted for participant disease diagnosis (MCD/MCD-Like vs. FSGS). To assess whether differences in post-biopsy immunosuppressive treatment between groups mediated the association between cluster groups and outcomes, we conducted post-hoc analyses that included immunosuppressant use after biopsy as a binary time-varying indicator in models. Finally, we used unadjusted models to assess associations between each individual glomerular descriptor and each outcome. P-values were controlled for false discovery rate (FDR) using the Benjamini and Hochberg linear step-up method.11
Molecular Data and Analysis
Messenger ribonucleic acid (mRNA) expression from RNA sequencing of microdissected glomeruli was available for 126 of the MCD/MCD-Like and FSGS participants (N=56 and N=70, respectively). (Item S1). Glomerular gene expression from cluster X was compared to Y+Z using Significance Analysis of Microarray (SAM). Differentially expressed genes (DEGs) were defined as fold change greater than 1.3 and false discovery rate (FDR)-corrected q-value less than 5%. Pearson correlation coefficient was calculated between each glomerular gene expression value and log10-transformed descriptor percentage score (descriptors with 0% were transformed to 1% to avoid undefined values), with adjustment for multiple testing (q-value less than 5%). Significant gene lists were analyzed for enrichment of canonical pathways and upstream regulators using the Ingenuity Pathway Analysis Software Suite.12 An activation z-score was derived from the direction of gene regulation observed to the expected direction of gene regulation to infer the activation state of a putative upstream regulator,12 with adjustment for multiple testing using Bonferroni. Upstream regulators were identified using both enrichment as well as direction of expression change with known cause and effect relationships.
In addition, a heatmap of cell type specific transcripts was generated using single-cell RNA sequencing data from reference nephrectomies (see Item S1). The expression values of the subset of cell-specific genes were extracted from the glomerular compartment bulk transcriptome data and were labeled according to cell type.14
RESULTS
Cluster Analysis Results
Three descriptor-based clusters of participants, X, Y and Z, were found (Figure 1A & Figure S2). Cluster X (56 participants) had the most frequent representation of structural changes, followed by Cluster Y (68 participants), and Cluster Z (97 participants). When we conducted a sensitivity analysis using unscaled data the cluster results remained unchanged.
Figure 1: Hierarchical clustering of 221 NEPTUNE participants with FSGS and MCD based on 39 glomerular descriptors.
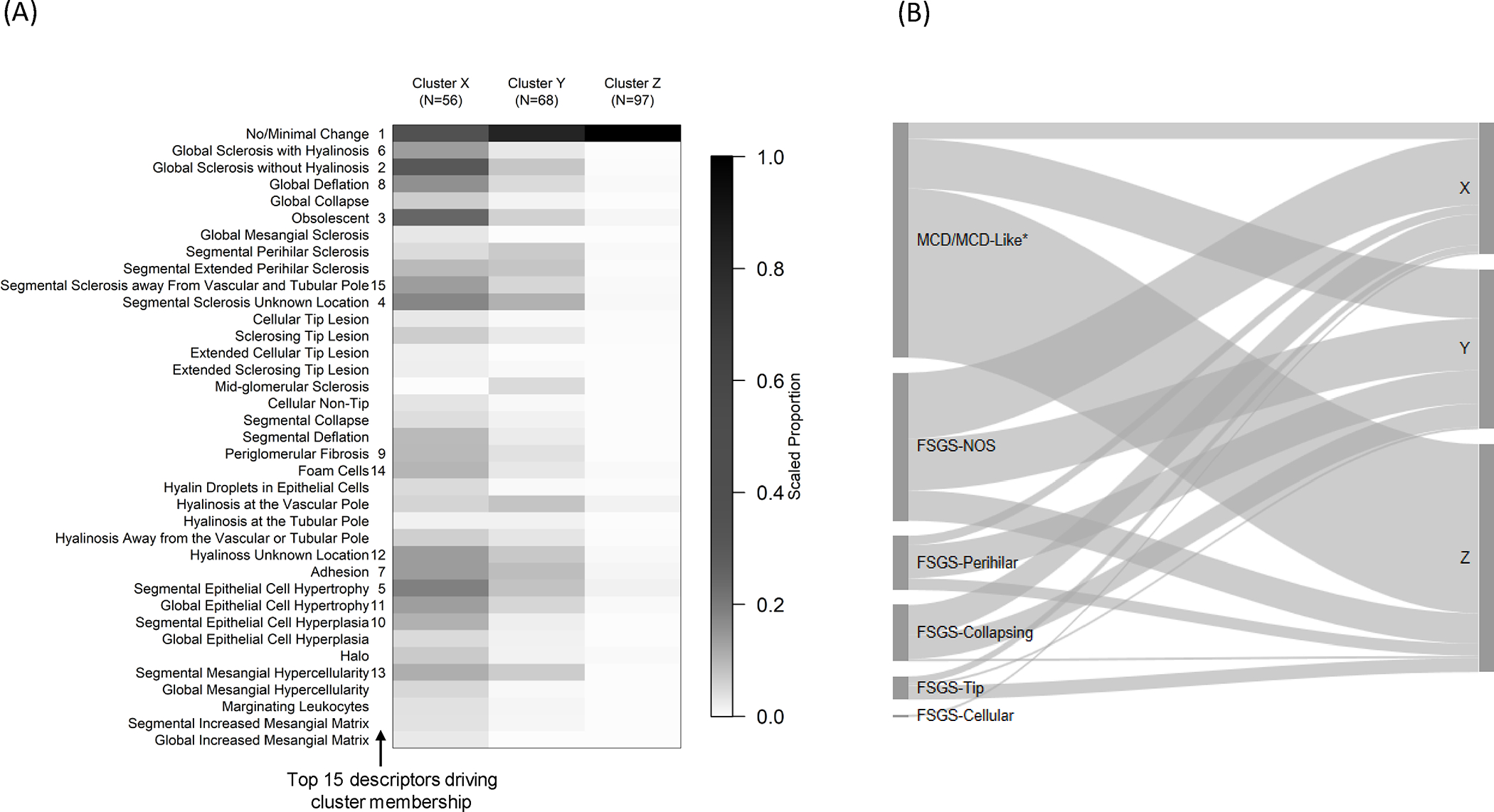
The heat map (A) shows the mean value of each descriptor by cluster after scaling each descriptor to 0–1 based on the observed range (i.e. the descriptor’s observed minimum is scaled to 0 and maximum to 1). The darker cells therefore represent greater percentages of glomeruli with each descriptor relative to others in the sample. The numbers 1–15 to the right side of descriptor labels indicate variable importance in driving cluster membership, obtained from variable entry order in a penalized multinomial regression model. The Sankey diagram (B) shows relationships between conventional classification categories (left) and cluster membership (right). The width of each ribbon-shaped band is proportional to the number of patients in that band. *MCD refers to cases with ≥75% effacement with no global sclerosis or global sclerosis expected for age, and MCD-Like refers to cases with <75% effacement with or without presence of global sclerosis exceeding that expected for age or ≥75% effacement with global sclerosis exceeding that expected for age.
The top descriptors driving cluster membership were identified using penalized multinomial regression and the top 15 shown in Figure 1A. The accuracy of cluster membership using fewer than the full set of descriptors was assessed based on a hierarchy of these top descriptors. Using only the top 5 descriptors yielded 81.4% accuracy, the top 10 83.3%, the top 15 89.1%, the top 20 88.2%, the top 25 89.6%, and the top 30 88.7%. Compared with the prediction accuracy of 88.2% using all 37 glomerular descriptors, the prediction accuracy began to level off with 15 descriptors.
Participants in Clusters X and Y had higher proportions of adults (77% and 66%, respectively) and FSGS participants (89% and 69%, respectively), whereas Cluster Z participants had more pediatric participants (69%) and MCD/MCD-Like participants (74%) (Table 1 A&B). Cluster X had the highest percentage of African American participants (48%). Cluster Z participants had the highest mean eGFR and UPCR and had a greater proportion on immunosuppressive medications at biopsy. Each cluster contained participants with MCD/MCD-Like and FSGS subtypes. While over half of Cluster Z patients had MCD/MCD-Like, there was little other concordance between cluster groups and FSGS subtypes (Figure 1B).
Table 1A.
Patient characteristics at biopsy overall and by glomerular cluster
| Total (n=221) | Cluster X (n=56) | Cluster Y (n=68) | Cluster Z (n=97) | |
|---|---|---|---|---|
|
| ||||
| Age (years) | 28.6 (22.0) | 40.5 (22.0) | 34.8 (21.4) | 17.3 (16.5) |
| Pediatric patients | 47% (103) | 23% (13) | 34% (23) | 69% (67) |
| Adult patients | 53% (118) | 77% (43) | 66% (45) | 31% (30) |
| Male | 62% (136) | 61% (34) | 60% (41) | 63% (61) |
| African American b | 36% (78) | 48% (27) | 30% (20) | 33% (31) |
| Hispanic a | 23% (51) | 22% (12) | 28% (19) | 21% (20) |
| Disease Cohort | ||||
| MCD/MCD-Like | 45% (99) | 11% (6) | 31% (21) | 74% (72) |
| FSGS | 55% (122) | 89% (50) | 69% (47) | 26% (25) |
| eGFR at biopsy c | 85.6 (41.3) | 54.0 (31.4) | 80.2 (32.8) | 108.5 (38.5) |
| UPCR at biopsy b | 5.38 (7.92) | 4.36 (4.31) | 4.47 (4.63) | 6.69 (10.88) |
| BMI b | ||||
| Underweight | 3% (7) | 6% (3) | 2% (1) | 3% (3) |
| Normal weight | 30% (65) | 20% (11) | 30% (20) | 36% (34) |
| Overweight | 27% (58) | 28% (15) | 30% (20) | 24% (23) |
| Obese | 40% (85) | 46% (25) | 38% (25) | 37% (35) |
| Serum Albumin, g/dL d | 3.16 (1.08) | 3.28 (1.12) | 3.25 (1.04) | 3.02 (1.08) |
| Any edema present b | 42% (91) | 44% (24) | 39% (26) | 43% (41) |
| On Immunosuppressive medications at biopsy | 31% (69) | 16% (9) | 19% (13) | 48% (47) |
| Duration of kidney disease as of biopsy (months) | 4 (1, 25) | 5 (1, 52) | 3 (1, 25) | 6 (1, 18) |
| RNA-sequencing data available | 126 | 27 | 38 | 61 |
Table values are mean (standard deviation), median (Q1, Q3), percent (frequency), rate, or frequency.
MCD, minimal change disease; FSGS, focal segmental glomerulosclerosis; eGFR, estimated glomerular filtration rate; UPCR, urinary protein/creatinine ratio.
Missing <1%;
Missing <5%;
Missing <10%;
Missing n=44
Table 1B.
Patient follow-up characteristics overall and by glomerular cluster
| Total (n=221) | Cluster X (n=56) | Cluster Y (n=68) | Cluster Z (n=97) | |
|---|---|---|---|---|
|
| ||||
| Follow-up time (years), median (Q1, Q3) | 4.17 (2.25, 4.67) | 3.08 (2.04, 4.54) | 4.50 (2.63, 4.75) | 4.17 (2.33, 4.58) |
| Number of complete remission events | 107 | 18 | 38 | 51 |
| Rate of complete remission (# of events per 100 person-years) a | 34.8 | 17.8 | 30.8 | 61.1 |
| Number of disease progression (≥40% decline in eGFR with eGFR<90 or kidney failure) events | 30 | 19 | 8 | 3 |
| Rate of disease progression (# of events per 100 person-years) b | 4.8 | 15.9 | 3.6 | 1.1 |
Q1, first quartile; Q3, third quartile
n=181 (n=25 had complete remission at biopsy; n=9 missing UPCR at biopsy; n=6 had no follow-up UPCR)
n=193 (n=22 missing eGFR at biopsy; n=6 had no follow-up eGFR)
Clinical outcomes
Cluster X demonstrated both the lowest probability of proteinuria remission and highest probability of disease progression (Figure 2). While Clusters Y and Z had similarly low probabilities of disease progression, Cluster Z had higher probabilities of remission of proteinuria. Cox models gave similar results, both unadjusted and after adjusting for baseline demographics and clinical characteristics (Table 2). In adjusted models, Clusters Y and Z had higher probabilities of proteinuria remission compared to Cluster X (HR [95% CI] = 1.95 [0.99, 3.85] and 3.29 [1.52, 7.13], respectively) and lower probabilities of disease progression (HR [95% CI] = 0.22 [0.08, 0.57] and 0.11 [0.03, 0.45], respectively).
Figure 2: Kaplan-Meier curves for each of the three clusters showing cumulative probability of complete proteinuria remission (left) and the survival probability of ≥40% decline in eGFR with eGFR <90 or kidney failure (right) by clusters.
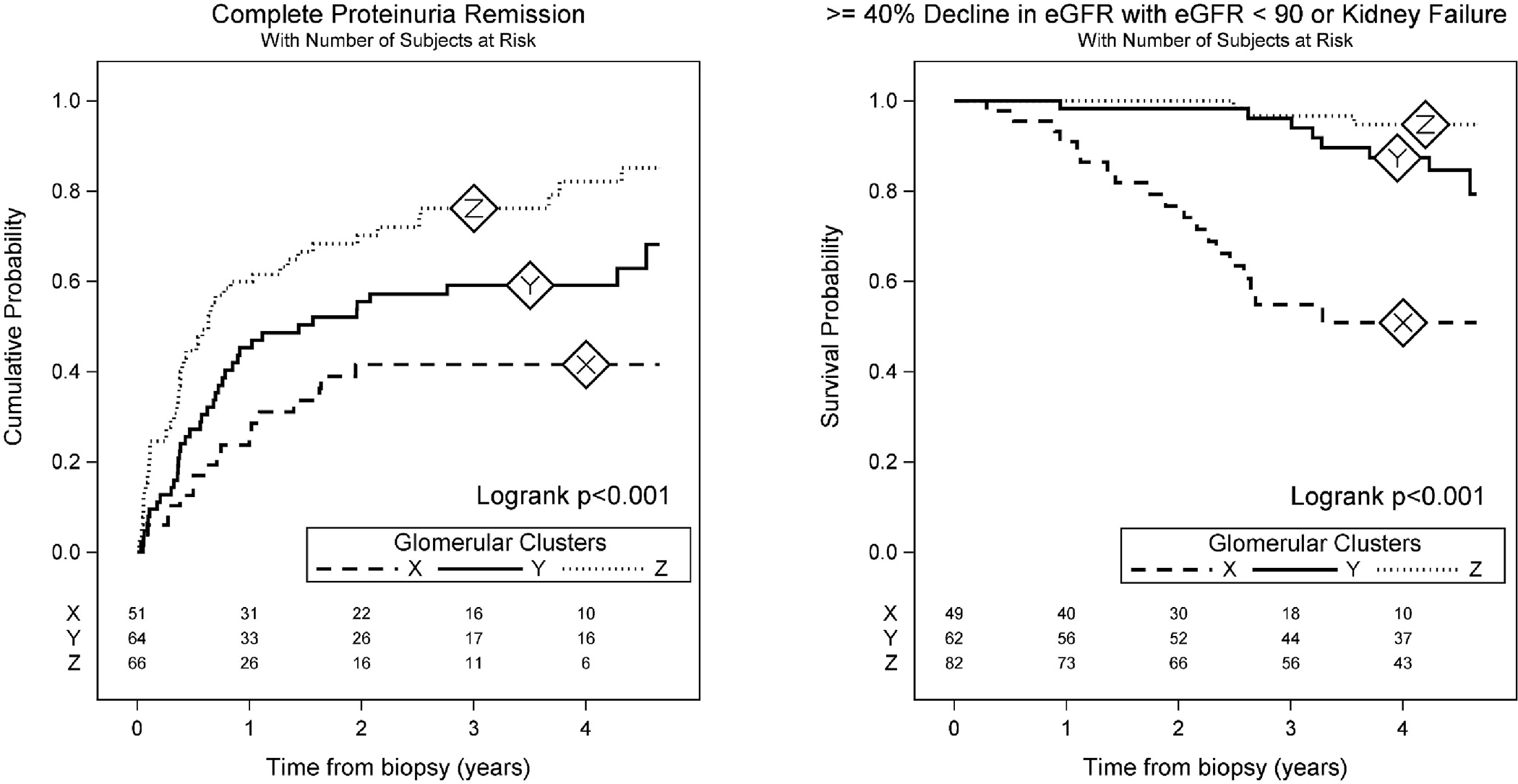
Cluster X had the lowest (worst) probability of proteinuria remission and highest (worst) probability of disease progression, while Cluster Z had the highest proteinuria remission probabilities and Clusters Y and Z had similar disease progression probabilities.
Table 2:
Associations between glomerular clusters and time-to-event outcomes from Cox models, unadjusted and adjusted for demographics, clinical characteristics, and disease diagnosis.
| A. Complete remission of proteinuria | ||||||
|---|---|---|---|---|---|---|
| Unadjusted | Adjusted for demographics + clinical characteristics a | Adjusted for disease diagnosis + demographics + clinical characteristics a | ||||
|
| ||||||
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Glomerular Clusters | <0.001 | 0.01 | 0.3 | |||
| X (reference) | ||||||
| Y | 1.82 (1.03, 3.23) | 0.04 | 1.95 (0.99, 3.85) | 0.05 | 1.60 (0.80, 3.18) | 0.2 |
| Z | 3.10 (1.79, 5.37) | <.001 | 3.29 (1.52, 7.13) | 0.003 | 2.02 (0.86, 4.74) | 0.1 |
|
| ||||||
| B. Composite of ≥40% decline in eGFR with eGFR < 90 or ESRD | ||||||
| Unadjusted | Adjusted for selected demographics + clinical characteristics a | Adjusted for disease diagnosis + selected demographics + clinical characteristics b | ||||
|
| ||||||
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Glomerular Clusters | <0.001 | <0.001 | 0.002 | |||
| X (reference) | ||||||
| Y | 0.19 (0.08, 0.45) | <0.001 | 0.22 (0.08, 0.57) | 0.002 | 0.22 (0.09, 0.54) | <0.001 |
| Z | 0.06 (0.02, 0.20) | <.001 | 0.11 (0.03, 0.45) | 0.002 | 0.29 (0.07, 1.12) | 0.07 |
Adjusted for demographics (patient age, sex, black race, Hispanic ethnicity) + clinical characteristics included patient age, sex, black race, Hispanic ethnicity, eGFR and UPCR at biopsy, kidney disease duration as of biopsy and immunosuppressant use at biopsy. Additional adjustment for disease diagnosis included MCD/MCD-Like vs. FSGS.
Due to a limited number of events of the composite outcome, adjustment covariates were selected based on backward elimination:
adjusted demographics + clinical characteristics included kidney disease duration as of biopsy, and eGFR and UPCR at biopsy;
adjusted demographics + clinical characteristics included kidney disease duration as of biopsy and UPCR at biopsy. Additional adjustment for disease diagnosis included MCD/MCD-Like vs. FSGS.
For the longitudinal eGFR outcome, after adjusting for baseline demographics and clinical characteristics, Cluster X had the steepest decline in eGFR (average slope of −5.5 mL/min/1.73m2 per year with 95% CI of −7.9 to −3.2, Figure 3). Clusters Y and Z had slower eGFR decline rates, with average eGFR slopes (95% CI) that were 3.07 (0.17 to 5.97) and 4.01 (1.19 to 6.85) greater than Cluster X, respectively (Table S2). After additionally adjusting for disease diagnosis, cluster groups were still significantly associated with disease progression (overall p-value=0.002, Table 2) and eGFR change over time (overall p-value for the interaction between cluster groups and time from biopsy=0.02, Table S2). The magnitude of the association between cluster groups and eGFR change over time was similar after adjustment for disease diagnosis versus before, while effect estimates for cluster groups on time-to-event outcomes were slightly dampened. Post-hoc analyses that included immunosuppressant use after biopsy showed little difference in results, implying little evidence that the effects of clusters on outcomes are explained by variation in subsequent immunosuppressant treatment. Unadjusted associations between individual descriptors and outcomes for the top 15 glomerular descriptors that drive cluster membership are shown in Table 3 and Table S3).
Figure 3: Estimated eGFR trajectories by cluster from longitudinal eGFR models.
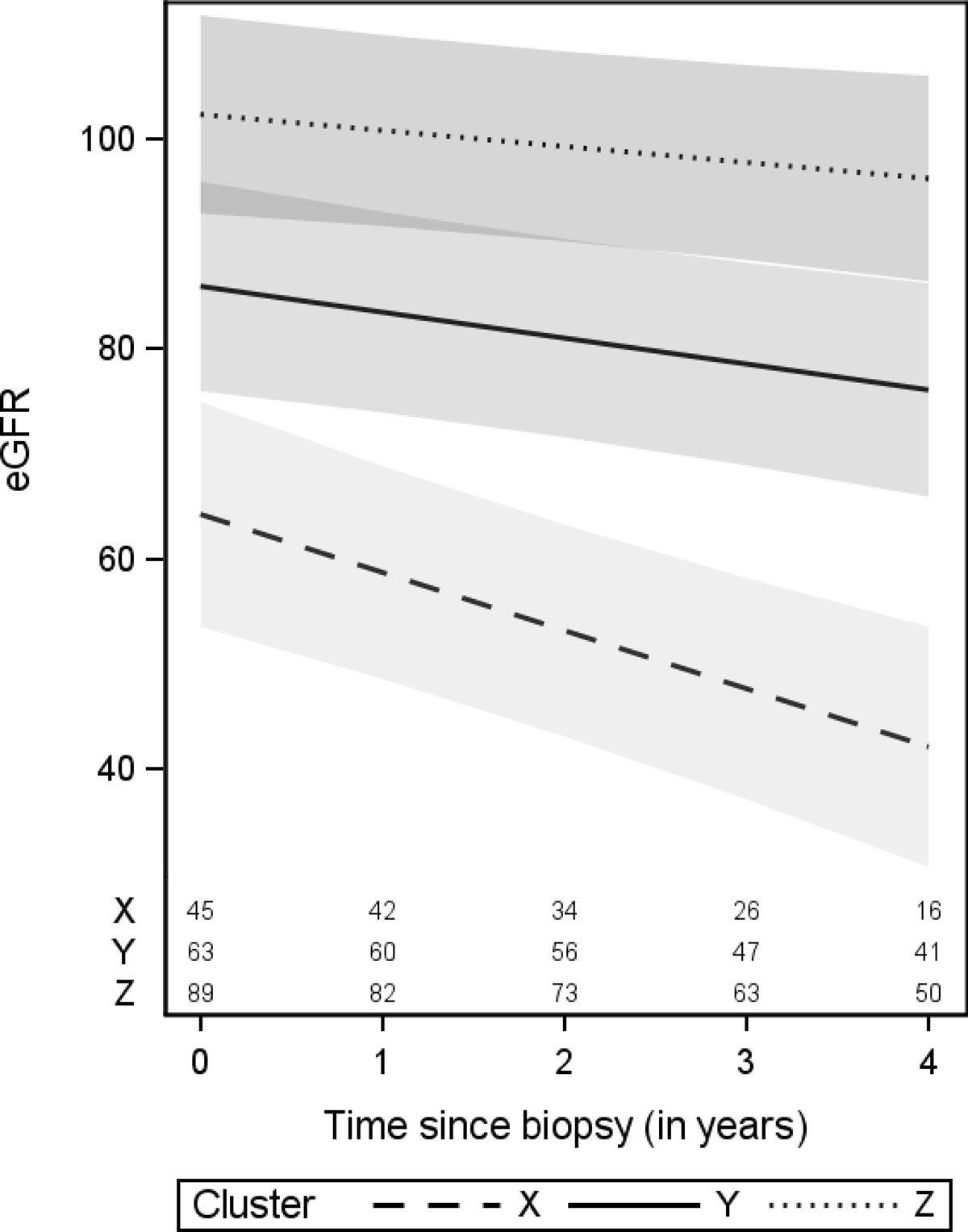
eGFR trajectories in this figure were estimated from the model adjusted for demographics and clinical characteristics. Cluster-specific averages were used for eGFR at biopsy (57, 79, and 109 mL/min/1.73m2 for Clusters X, Y, and Z, respectively). Confidence bands are based on 95% pointwise confidence intervals. Numbers at the bottom of the graph represent the number of patients available for analysis at each time point.
Table 3.
Unadjusted associations of the top 15 individual glomerular descriptor and outcomes. Effect estimates presented are for a one-tenth increase in the observed range (observed max – observed min) of a descriptor.
| % of Glomeruli with Descriptor | Complete Remission | Composite Progression | Longitudinal eGFR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| %N with 0 glomeruli with descriptor | Median (IQR) among non-zero values* | Mean | Min | Max | HR (95% CI) | P-value** | HR (95% CI) | P-value** | Estimate (95% CI) | P-value** | |||
|
| |||||||||||||
| No/Minimal Changes | 1 | 0% | 90.9 (68.4 – 100.0) | 78.6 | 6.3 | 100 | 1.25 (1.14, 1.37) | <0.001 | 0.71 (0.64, 0.80) | <.001 | 8.0 (6.7, 9.4) | <.001 | |
| Global Sclerosis without Hyalinosis | 2 | 59% | 12.2 (6.1 – 23.9) | 6.7 | 0 | 66.7 | 0.82 (0.71, 0.95) | 0.02 | 1.30 (1.14, 1.48) | <0.001 | −10.3 (−12.6, −8.0) | <.001 | |
| Obsolescent | 3 | 62% | 7.7 (3.7 – 21.1) | 6.2 | 0 | 73.3 | 0.69 (0.56, 0.85) | 0.003 | 1.40 (1.24, 1.59) | <0.001 | −9.0 (−11.6, −6.5) | <.001 | |
| Segmental Sclerosis Unknown Location | 4 | 59% | 9.1 (4.8 – 14.9) | 4.9 | 0 | 61.5 | 0.64 (0.51, 0.80) | <0.001 | 1.25 (1.09, 1.42) | 0.004 | −7.3 (−10.3, −4.3) | <.001 | |
| Segmental Visceral Epithelial Cell Hypertrophy | 5 | 55% | 6.7 (3.9 – 15.8) | 5.6 | 0 | 70.6 | 0.88 (0.75, 1.03) | 0.2 | 1.17 (1.00, 1.36) | 0.1 # | −4.3 (−7.3, −1.4) | 0.01 | |
| Global Sclerosis with Hyalinosis | 6 | 81% | 6.7 (4.0 – 14.3) | 2.1 | 0 | 50 | 0.71 (0.52, 0.97) | 0.1 # | 1.37 (1.17, 1.61) | <0.001 | −10.3 (−13.8, −6.8) | <.001 | |
| Adhesion | 7 | 61% | 6.7 (4.2 – 10.5) | 3.8 | 0 | 57.9 | 0.69 (0.55, 0.87) | 0.006 | 1.37 (1.20, 1.55) | <.001 | −7.5 (−10.6, −4.4) | <.001 | |
| Global Deflation | 8 | 76% | 6.0 (3.2 – 14.3) | 2.5 | 0 | 43.8 | 0.77 (0.62, 0.94) | 0.03 | 1.29 (1.11, 1.50) | 0.003 | −9.8 (−13.0, −6.6) | <.001 | |
| Periglomerular Fibrosis | 9 | 75% | 7.7 (3.9 – 14.3) | 2.7 | 0 | 75 | 0.51 (0.34, 0.78) | 0.006 | 1.22 (1.01, 1.47) | 0.09 # | −14.1 (−18.6, −9.6) | <.001 | |
| Segmental Visceral Epithelial Cell Hyperplasia | 10 | 84% | 7.0 (3.5 – 12.8) | 1.7 | 0 | 52.9 | 0.90 (0.72, 1.14) | 0.5 | 1.36 (1.06, 1.74) | 0.04 | −8.3 (−13.1, −3.5) | 0.003 | |
| Global Visceral Epithelial Cell Hypertrophy | 11 | 79% | 6.2 (3.2 – 13.5) | 1.9 | 0 | 35.3 | 0.99 (0.87, 1.13) | 0.9 | 1.19 (1.00, 1.43) | 0.1 | −6.3 (−9.8, −2.9) | 0.002 | |
| Hyalinosis Unknown Location | 12 | 77% | 4.7 (3.3 – 9.1) | 1.6 | 0 | 27.3 | 0.66 (0.51, 0.84) | 0.004 | 1.23 (1.06, 1.44) | 0.02 | −5.5 (−8.7, −2.3) | 0.003 | |
| Segmental Mesangial Hypercellularity | 13 | 80% | 9.1 (4.8 – 18.2) | 2.9 | 0 | 60 | 1.03 (0.91, 1.16) | 0.8 | 1.13 (0.94, 1.34) | 0.3 | 2.0 (−1.2, 5.2) | 0.3 | |
| Foam Cells | 14 | 77% | 6.7 (3.6 – 12.8) | 2.7 | 0 | 75 | 0.92 (0.77, 1.09) | 0.4 | 1.13 (0.94, 1.35) | 0.3 | −3.6 (−7.4, 0.2) | 0.1 | |
| Segmental Sclerosis away From Vascular and Tubular Pole | 15 | 87% | 3.6 (2.4 – 6.5) | 0.6 | 0 | 10.5 | 0.83 (0.71, 0.97) | 0.05 | 1.26 (1.11, 1.44) | 0.002 | −4.2 (−6.9, −1.5) | 0.009 | |
Median (IQR) among non-zero values were based on only a few participants for descriptors with >95% zero values.
P-values were controlled for false discovery rate (FDR) using the Benjamini and Hochberg linear step-up method.
Covariates where 95% CI indicated statistically significant at level 0.05, but FDR adjusted p-values were statistically insignificant.
Molecular Analysis Results
A total of 1920 genes were differentially expressed when cluster X (worst clinical outcomes) was compared to Y+Z combined (Table S4). For insight into the biological and molecular mechanisms, we subjected the DEGs to canonical pathway and upstream regulator enrichment analysis and identified the top 12 canonical pathways predicted to be enriched based on DEGs (Table 4A and Table S5A). Notable are the several pathways important for inflammation and immune responses in the kidney, such as dendritic cell maturation, B cell receptor signaling, communication between innate and adaptive immune cells, and role of pattern recognition receptors.15 Table 4B and Table S5B) shows the top 12 upstream regulators predicted to be causally upstream of the associated DEGs from X vs Y+Z. The list includes several cytokines also known to be associated with kidney disease progression, such as tumor necrosis factor (TNF), IFN-Gamma, and interleukin-1-beta (IL1B), and growth factors such as transforming growth factor beta (TGFB).16–18 Differential regulation of 15 genes chosen from those that predict the activation of these top 4 upstream regulators was validated by the Affymetrix ST 2.1 platform for 77 cases of cluster X relative to Y+Z (14 vs 63 respectively) (Table S6 and Item S1). All 15 DEGs match in direction of fold change of glomerular expression between RNA sequencing and Affymetrix platforms, and differential expression was significant to FDR < 0.05 for 13 genes, with the remaining at FDR of 0.052 and 0.054.
Table 4A.
Top 12 canonical pathways predicted from cluster X vs Y+Z differentially expressed genes
| Ingenuity Canonical Pathways | Activation z-score | −log (p-value) |
|---|---|---|
|
| ||
| Phagosome Formation | 14.1 | |
| Hepatic Fibrosis / Hepatic Stellate Cell Activation | 12.7 | |
| Dendritic Cell Maturation | 5.47 | 9.78 |
| GP6 Signaling Pathway | 4.70 | 8.46 |
| Granulocyte Adhesion and Diapedesis | 8.08 | |
| Neuroinflammation Signaling Pathway | 4.90 | 7.38 |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 7.35 | |
| B Cell Receptor Signaling | 4.27 | 7.21 |
| Communication between Innate and Adaptive Immune Cells | 7.18 | |
| Th2 Pathway | 2.13 | 7.17 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 3.84 | 7.07 |
| Altered T Cell and B Cell Signaling in Rheumatoid Arthritis | 7.04 | |
Top 12 based on p-value. For reference, −log(0.05/1000)=4.30, so all p-values shown in the table are statistically significant at level 0.05 even after a conservative Bonferroni correction for 1000 tests. The Ingenuity Pathway Analysis database lacked enough data to calculate activation z-scores for some canonical pathways. These are left blank.
Table 4B.
Top 12 upstream regulators predicted from cluster X vs Y+Z differentially expressed genes
| Upstream Regulator (Gene ID) | Expression Fold Change | Molecule Type | Activation z-score | p-value of overlap |
|---|---|---|---|---|
|
| ||||
| TNF | 1.518 | cytokine | 10.128 | 6.47E-55 |
| IFNG | cytokine | 8.61 | 5.47E-56 | |
| IL1B | 1.667 | cytokine | 8.125 | 1.15E-31 |
| TGFB1 | 1.321 | growth factor | 7.435 | 1.71E-59 |
| IL6 | cytokine | 6.537 | 9.25E-36 | |
| ERBB2 | −1.035 | kinase | 6.053 | 6.39E-37 |
| IL2 | cytokine | 6.279 | 3.33E-20 | |
| PTGER2 | 1.314 | g-protein coupled receptor | 4.816 | 2.83E-22 |
| SPI1 | 1.664 | transcription regulator | 4.295 | 1.17E-28 |
| FOXO1 | −1.076 | transcription regulator | 4.362 | 2.48E-19 |
| STAT3 | 1.121 | transcription regulator | 3.713 | 8.16E-22 |
| CSF3 | cytokine | 2.925 | 1.36E-19 | |
Top 12 based on p-value and shown in order of descending activation z-scores. The Ingenuity Pathway Analysis database lacked enough data to calculate activation z-scores for some canonical pathways. These are left blank.
The emerging pattern of the heatmap of cell-specific gene expression values (Figure 4), with participants grouped by pathology descriptor cluster and genes grouped by cell type, indicates that the glomerular differential gene expression occurs in a cell type specific fashion. For example, podocyte enriched genes were downregulated in Cluster X relative to Y+Z whereas monocyte and macrophage enriched genes were upregulated. Furthermore, an influx of monocyte/macrophage cells is seen in cluster X compared to the others, consistent with the observed higher proportion of foam cells (Figure 1A).
Figure 4: Cell-type specific gene expression across clusters.
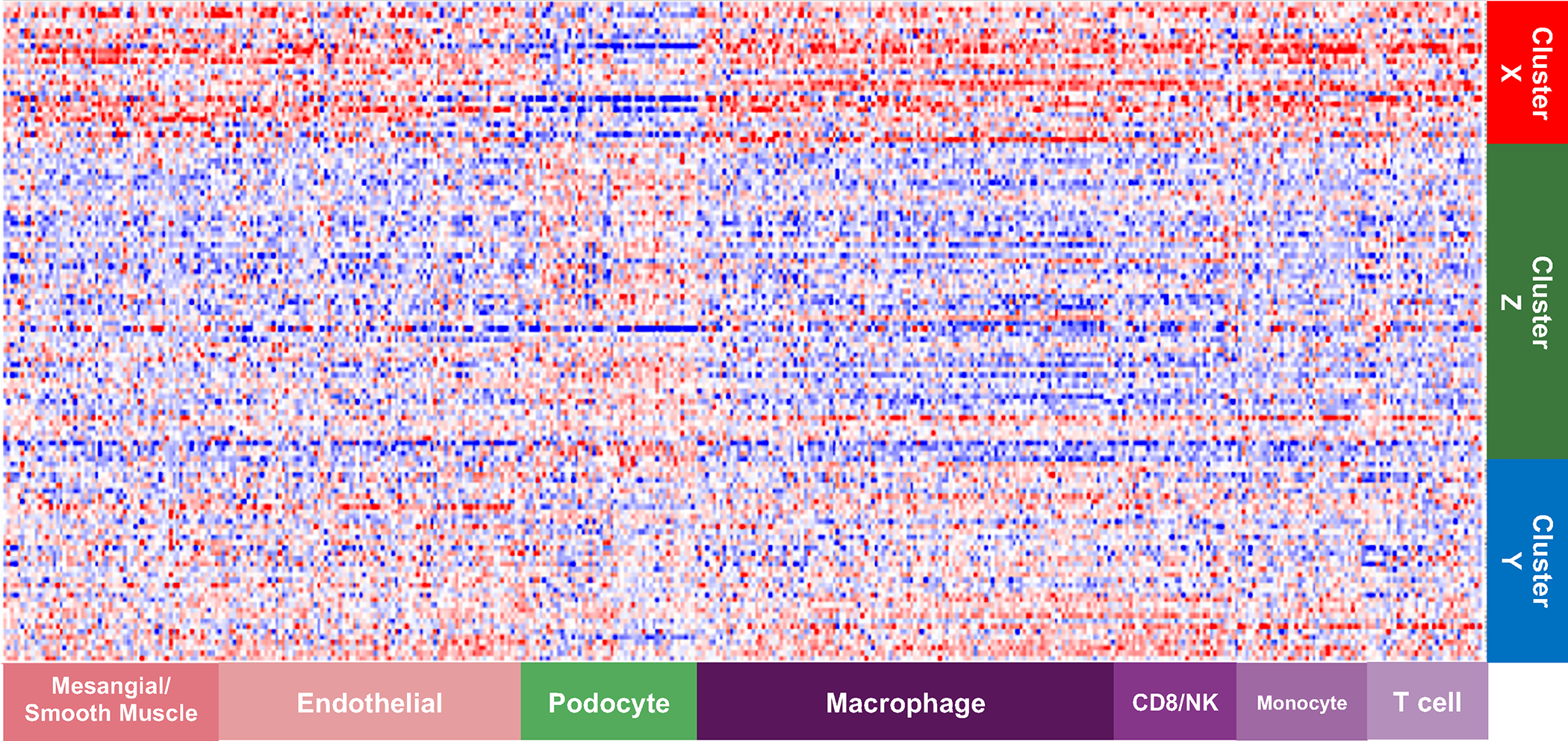
Cell-type selective gene expression profiles were overlaid onto clustered gene expression data. The expression data were row normalized as Z-scores where blue indicates low expression and red indicates high expression. The emerging pattern indicates that cell-selective gene profiles show evidence of differential gene expression across pathology defined clusters. For example, podocyte-selective gene expression is reduced in Cluster X compared to Clusters Y and Z, whereas macrophage-selective gene expression is increased. This observation strengthens confidence that descriptor clustering is identifying underlying biological mechanism.
Each of the top 15 glomerular descriptors shown to drive cluster membership (Table 3) were assessed for correlations with glomerular gene expression (Table S7). Table S8 shows the top canonical pathways and upstream regulators enriched for the seven glomerular descriptors with >700 significant positively and negatively correlated genes (full list in Table S9). Many of the top canonical pathways associated with these gene lists were similar to those associated with Cluster X and enriched for fibrosis, inflammation, and innate immunity pathways. Likewise, top predicted upstream regulators also included cytokines and growth factors TNF, IFNG, IL1B, and TGFB1. The notable exception in Table 7 are the 1401 genes that correlate with the descriptor ‘Segmental Visceral Epithelial Cell Hyperplasia’ (podocytes). Predicted canonical pathways and upstream regulators include those known to be involved with cell growth and regulation of the cell cycle.
DISCUSSION
The NDPSS, previously tested for reproducibility,18,19 capitalizes on digital imaging technology employed in NEPTUNE and facilitates a detailed, quantitative assessment of the renal biopsy.4,19,20 This granular and quantitative approach, which is agnostic to interpretative diagnoses, offers the opportunity to better explore the relationship between kidney structure, function, and molecular endophenotypes.
The NDPSS is inclusive of a comprehensive list of parameters, including those employed in current classification systems, that can be seen in nephrotic syndrome biopsies with a broad range of disease severity. Although some of the individual NDPSS parameters that are not included in conventional classification systems are recognized and occasionally reported by pathologists, their clinical and biologic relevance was only rarely tested. For example, the value of quantifying global sclerosis, independently from the underlying conventional diagnosis, was previously demonstrated in kidney donor and native biopsies.21,22 Similarly, descriptors reflecting podocyte hypertrophy and hyperplasia were recognized as cellular injury responses in NS,23,24 confirming previous morphometric studies demonstrating that changes in podocyte size and density predict outcome.25 In this study, both global obliteration (global sclerosis with and without hyalinosis, global deflation, and obsolescence) and podocyte hypertrophy and hyperplasia were identified within the top 15 glomerular descriptors driving cluster membership and were most prevalent in the cluster with worst outcomes (X). The importance of a comprehensive analysis to identify previously unrecognized clinically and biologically relevant parameters was emphasized by a previous study applying the NDPSS to ultrastructural analysis.26 Royal et al.26 showed that in MCD/FSGS, although some podocyte descriptors were associated with complete remission, groups of patients with the most prominent endothelial cell injury were associated with disease progression and activation of pathways reflecting endothelial cell injury.
The comprehensive collection of observational data to agnostically define subgroups of patients with morphological similarities at the glomerular level allowed for exploring whether structural signatures have clinical relevance. The descriptor-based morphologic signatures and patient clusters were associated with complete remission of proteinuria and loss of kidney function over time, even after adjusting for potential confounding factors and conventional diagnoses. Notably, there was less than full concordance between the descriptor-based clusters and traditional diagnoses of MCD and FSGS variants, highlighting that this systematic and quantifiable approach not only captured the small set of features used as criteria for conventional diagnostics, but also a variety of structural and cellular abnormalities clinically relevant when used to categorize patients with MCD/FSGS. Previous studies focused on the comparison of findings with the conventional disease categories, inferring validity of the results if the findings matched the current classification scheme. This study rather challenges this approach and opens the way for reframing the classification of NS.
Our study also links structure and function in FSGS/MCD by revealing associations between descriptor-based clusters and individual descriptors with glomerular gene expression. Previous studies on glomerular specific transcriptomic data from NEPTUNE participants have provided insights into disease molecular mechanisms by highlighting immune activation and Jak-Stat signaling in FSGS,27 characterizing expression quantitative trait loci (eQTL),14 and integrating with single-cell gene expression to define cellular expression of disease remission in FSGS.13 In our analysis we focused on Cluster X to gain insight into the biological and molecular mechanisms associated with the poorest clinical outcomes and demonstrated activation of several pathways important for inflammation and immune responses. This is congruent with recent insights into innate immunity activation in the podocyte,28 injury to which is key to FSGS and MCD pathogenesis, and current treatment strategies targeting those pathways.29 Top predicted causal upstream regulators included TNF, IFNG, IL1B, and TGFB1, which are known to be associated with inflammation and fibrosis.15–18
Using single-cell RNA sequencing data, we found gene expression patterns consistent with decreased podocyte specific gene expression and an increase in monocyte/macrophage specific gene expression in the cluster with the worst clinical outcomes. This is consistent with previous observations of podocyte loss or downregulation of podocyte genes with progressive glomerular disease25 and the recognition of macrophages and foam cells in FSGS.30
By correlating glomerular gene expression with the top glomerular descriptors driving the clusters, we identified individual parameters that may be a surrogate biomarker of mechanism and ultimately a target of novel therapeutic approaches. Seven descriptors correlated significantly with a list of >700 glomerular genes. Genes correlating with ‘Any lesion Seen’ and top descriptors capturing global obliteration and periglomerular fibrosis demonstrated enrichment for canonical pathways and predicted upstream regulators similar to Cluster X vs Y+Z. Intriguingly, the glomerular descriptor ‘Segmental Visceral Epithelial Cell Hyperplasia’ correlated with the expression of 1401 glomerular genes enriched for canonical pathways known to be associated with cell hyperplasia, growth, and cell cycle regulation, such as ‘EIF2 Signaling’, ‘mTOR Signaling’, and ‘G2/M Checkpoint Regulation’,31 and predicted upstream regulators with key roles in cell growth and gene transcription, including C-Myc, a transcription factor that promotes expression of growth related genes,32,33 and β-catenin, a modulator of cell-cell adhesion and gene transcription.34
Our findings support the biological validity of using detailed visual assessment of glomerular pathology because this approach demonstrated correlation with underlying transcriptional programs known to be active in kidney disease. These findings are not definitive; rather they suggest plausible candidate pathways that are potential targets for therapeutic intervention. A final advantage of this approach is the quantifiable data generated by the NDPSS, allowing integration with other omics datasets to understand the biologic relevance of the individual descriptors and patient clusters. These new discoveries can then be studied in animal models or cell cultures to confirm mechanisms and guide development of new treatments for clinical trials.
There are, however, some limitations to the approach. Although we were able to score biopsies from many of participants, the low prevalence of some descriptors may mask their predictive capability. For example, neither glomerular tuft collapse or tip lesions (cellular or sclerosing) were among the top descriptors driving cluster membership, even though they are recognized as features that have poor and good prognoses respectively.35 Further, morphologic features seen in cross-section might represent temporal stages of a disease process rather than distinct disease processes. The analysis of follow up biopsies in future studies might yield additional insight into the evolution of glomerular morphology with disease progression. The morphologic and molecular phenotype of pediatric patients who do not undergo biopsy remain unclear; however, this work is paving the way for future identification of blood and urine biomarkers that could be studied in a non-biopsied cohort. Relatedly, we do not have a treatment naïve or other control group for comparison of results; however, the participants included in this study would best reflect nephrotic syndrome patients encountered in the real world at the time of their first biopsy. Lastly, the current study focused only on histology glomerular descriptors; however, the renal manifestation of glomerular disease processes affects other kidney tissue compartments (tubules, interstitium, and vasculature), which all may contribute to predict outcomes. Previous studies on NEPTUNE datasets, have already shown the predictive ability of scoring for interstitial fibrosis 36 and EM descriptors26. It is expected that a comprehensive analysis integrating all histologic and ultrastructural descriptors may provide a more complete morphology profile of each biopsy that can better inform clinical outcome prediction.
The present results serve as foundational evidence that comprehensive evaluation of multiple discreet morphologic features from kidney tissue can be used to identify clinically and biologically meaningful glomerular disease categories. Additional work will be necessary to further explore and validate or refine this approach on independent glomerular disease cohorts before implementation in routine clinical practice. Given the rapid evolution of computer-aided image analysis, we anticipate that automated detection, segmentation and quantification of kidney morphologic parameters will be employed toward this goal as an aiding tool to pathologists. Successful development of deep learning models for the segmentation of histologic primitives in the NEPTUNE kidney biopsies have already been demonstrated,3,37–51 and additional computer-assisted image analysis approaches are being developed. Completion of these future studies could lead to change the current approach for morphologic analysis in clinical practice by implementing interactive human-machine protocols, and data-driven identification of reportable parameters and patient’s categorization, enhancing our ability to better predict outcome and select tailored treatments.
Supplementary Material
Figure S1: The NEPTUNE Digital Pathology Protocol (NDPP) and Scoring System (NDPSS)
Figure S2: Dendogram of hierarchical clustering of 221 NEPTUNE participants with FSGS and MCD based on 39 glomerular descriptors.
Item S1. Supplementary Methods
Table S1. 37 glomerular descriptors with definitions
Table S2: Associations between glomerular clusters and longitudinal eGFR from linear mixed models, unadjusted and adjusted for demographics, clinical characteristics, and disease diagnosis.
Table S3. Unadjusted associations between each individual glomerular descriptor (n=37) and outcomes. Effect estimates presented are for a one-tenth increase in the observed range (observed max – observed min) of a descriptor.
Table S4. Differentially expressed genes from comparison cluster X vs Y+Z
Table S5, A and B. Full list of Canonical Pathways and Upstream Regulators predicted from X vs Y+Z differentially expressed genes.
Table S6. List of 15 differentially expressed genes from comparison cluster X vs Y+Z from RNA sequencing and Affymetrix ST 2.1 platforms
Table S7, A-M. Correlation between glomerular gene expression and descriptor frequency.
Table S8, A and B. Top (a.) canonical pathways and (b.) upstream regulators predicted from the significant correlation of glomerular RNAseq-derived gene expression with log10-transformed descriptor scores
Table S9, A-M. Full list of Canonical Pathways and Upstream Regulators predicted from correlation between glomerular gene expression and descriptor frequency.
PLAIN LANGUAGE SUMMARY.
Glomerular features are associated with outcomes and transcriptomics in nephrotic syndrome.
Focal segmental glomerulosclerosis and minimal change disease are heterogeneous diseases that manifest with a variety of structural changes in the kidney that are often not captured by conventional classification systems. This study shows that a detailed morphologic analysis and quantification of these changes allows for better representation of the structural abnormalities within each patient and for grouping patients with similar morphologic profiles. The resulting clusters of patients are clinically and biologically relevant since they are associated with kidney disease progression, complete remission of proteinuria, and kidney tissue transcriptomics differences. This study serves as a foundation for eventual sub-categorization of nephrotic syndrome that can better predict outcome and select tailored treatments.
Support:
This study was supported by a grant from the National Institute of Diabetes, Digestive, and Kidney Diseases to L.B., J.B.H., B.W.G., L.B.H., L.H.M., and J.Z. (NIDDK R01-DK118431). L.H.M. is supported through funding from NIH/NIDDK, K08 DK115891–01. The Nephrotic Syndrome Rare Disease Clinical Research Network III (NEPTUNE) is part of the Rare Diseases Clinical Research Network (RDCRN), which is funded by the National Institutes of Health (NIH) and led by the National Center for Advancing Translational Sciences (NCATS) through its Office of Rare Diseases Research (ORDR). NEPTUNE is funded under grant number U54DK083912 as a collaboration between NCATS and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Additional funding and/or programmatic support for this project has also been provided by the University of Michigan, NephCure Kidney International and the Halpin Foundation. All RDCRN consortia are supported by the network’s Data Management and Coordinating Center (DMCC) (U2CTR002818). Funding support for the DMCC is provided by NCATS and the National Institute of Neurological Disorders and Stroke (NINDS). Funders did not have a role in study design, data collection, analysis, reporting, or the decision to submit for publication.
NEPTUNE Enrolling Centers
Cleveland Clinic, Cleveland, OH: K Dell*, J Sedor**, M Schachere#, J Negrey#
Children’s Hospital, Los Angeles, CA: K Lemley*, E Lim#
Children’s Mercy Hospital, Kansas City, MO: T Srivastava*, A Garrett#
Cohen Children’s Hospital, New Hyde Park, NY: C Sethna*, K Laurent #
Columbia University, New York, NY: P Canetta*, A Pradhan#
Emory University, Atlanta, GA: L Greenbaum*, C Wang**, C Kang#
Harbor-University of California Los Angeles Medical Center: S Adler*, J LaPage#
John H. Stroger Jr. Hospital of Cook County, Chicago, IL: A Athavale*, M Itteera
Johns Hopkins Medicine, Baltimore, MD: M Atkinson*, S Boynton#
Mayo Clinic, Rochester, MN: F Fervenza*, M Hogan**, J Lieske*, V Chernitskiy#
Montefiore Medical Center, Bronx, NY: F Kaskel*, M Ross*, P Flynn#
NIDDK Intramural, Bethesda MD: J Kopp*, J Blake#
New York University Medical Center, New York, NY: H Trachtman*, O Zhdanova**, F Modersitzki#, S Vento#
Stanford University, Stanford, CA: R Lafayette*, K Mehta#
Temple University, Philadelphia, PA: C Gadegbeku*, S Quinn-Boyle#
University Health Network Toronto: M Hladunewich**, H Reich**, P Ling#, M Romano#
University of Miami, Miami, FL: A Fornoni*, C Bidot#
University of Michigan, Ann Arbor, MI: M Kretzler*, D Gipson*, A Williams#, J LaVigne#
University of North Carolina, Chapel Hill, NC: V Derebail*, K Gibson*, E Cole#, J Ormond-Foster#
University of Pennsylvania, Philadelphia, PA: L Holzman*, K Meyers**, K Kallem#, A Swenson#
University of Texas Southwestern, Dallas, TX: K Sambandam*, Z Wang#, M Rogers#
University of Washington, Seattle, WA: A Jefferson*, S Hingorani**, K Tuttle**§, M Bray #, M Kelton#, A Cooper#§
Wake Forest University Baptist Health, Winston-Salem, NC: JJ Lin*, Stefanie Baker#
Data Analysis and Coordinating Center: M Kretzler, L Barisoni, J Bixler, H Desmond, S Eddy, D Fermin, C Gadegbeku, B Gillespie, D Gipson, L Holzman, V Kurtz, M Larkina, J Lavigne, S Li, S Li, CC Lienczewski, J Liu, T Mainieri, L Mariani, M Sampson, J Sedor, A Smith, A Williams, J Zee.
Digital Pathology Committee: Carmen Avila-Casado (University Health Network, Toronto), Serena Bagnasco (Johns Hopkins University), Joseph Gaut (Washington University in St Louis), Stephen Hewitt (National Cancer Institute), Jeff Hodgin (University of Michigan), Kevin Lemley (Children’s Hospital of Los Angeles), Laura Mariani (University of Michigan), Matthew Palmer (University of Pennsylvania), Avi Rosenberg (Johns Hopkins University), Virginie Royal (University of Montreal), David Thomas (University of Miami), Jarcy Zee (University of Pennsylvania) Co-Chairs: Laura Barisoni (Duke University) and Cynthia Nast (Cedar Sinai).
*Principal Investigator; **Co-investigator; #Study Coordinator
§Providence Medical Research Center, Spokane, WA
Footnotes
Disclosures:
Dr. Barisoni reports grants from NIH/NIDDK, during the conduct of the study; personal fees from Moderna, personal fees from Protalix, personal fees from Sangamo, personal fees from Vertex, outside the submitted work. Dr. Hodgin reports grants from NIH/NIDDK, during the conduct of the study; personal fees from Astrazeneca, personal fees from Gilead, personal fees from Moderna, personal fees from NovoNordisc, personal fees from Eli Lilly, personal fees from Jansen, outside the submitted work. Dr. Holzman reports grants from NIH/NIDDK during the conduct of the study. Dr. Mariani reports grants from NIH/NIDDK during the conduct of the study, personal fees from Reata, Calliditas Therapeutics and Travere Therapeutics, outside the submitted work. Dr. Zee reports grants from NIH/NIDDK during the conduct of the study. The remaining authors declare that they have no relevant financial interests.
References
- 1.D’Agati VD, Fogo AB, Bruijn JA, Jennette JC. Pathologic classification of focal segmental glomerulosclerosis: a working proposal. Am J Kidney Dis. 2004:368–82. vol. 2. [DOI] [PubMed] [Google Scholar]
- 2.Barisoni L, Hodgin JB. Digital pathology in nephrology clinical trials, research, and pathology practice. Curr Opin Nephrol Hypertens. 11 2017;26(6):450–459. doi: 10.1097/MNH.0000000000000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. Nov 2020;16(11):669–685. doi: 10.1038/s41581-020-0321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barisoni L, Nast CC, Jennette JC, et al. Digital pathology evaluation in the multicenter Nephrotic Syndrome Study Network (NEPTUNE). Clin J Am Soc Nephrol. Aug 2013;8(8):1449–59. doi: 10.2215/cjn.08370812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gadegbeku CA, Gipson DS, Holzman LB, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. Apr 2013;83(4):749–56. doi: 10.1038/ki.2012.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nast CC, Lemley KV, Hodgin JB, et al. Morphology in the Digital Age: Integrating High-Resolution Description of Structural Alterations With Phenotypes and Genotypes. Semin Nephrol. May 2015;35(3):266–78. doi: 10.1016/j.semnephrol.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barisoni L, Gimpel C, Kain R, et al. Digital pathology imaging as a novel platform for standardization and globalization of quantitative nephropathology. Clin Kidney J. Apr 2017;10(2):176–187. doi: 10.1093/ckj/sfw129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg AZ, Palmer M, Merlino L, et al. The Application of Digital Pathology to Improve Accuracy in Glomerular Enumeration in Renal Biopsies. PLoS One. 2016;11(6):e0156441. doi: 10.1371/journal.pone.0156441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stokes MB, D’Agati VD. Morphologic variants of focal segmental glomerulosclerosis and their significance. Adv Chronic Kidney Dis. Sep 2014;21(5):400–7. doi: 10.1053/j.ackd.2014.02.010 [DOI] [PubMed] [Google Scholar]
- 10.Zee J, Mansfield S, Mariani LH, Gillespie BW. Using All Longitudinal Data to Define Time to Specified Percentages of Estimated GFR Decline: A Simulation Study. Am J Kidney Dis. Jan 2019;73(1):82–89. doi: 10.1053/j.ajkd.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57(1):289–300. [Google Scholar]
- 12.Krämer A, Green J, Pollard J., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. Feb 15 2014;30(4):523–30. doi: 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menon R, Otto EA, Hoover P, et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. Mar 26 2020;5(6)doi: 10.1172/jci.insight.133267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillies CE, Putler R, Menon R, et al. An eQTL Landscape of Kidney Tissue in Human Nephrotic Syndrome. Am J Hum Genet. Aug 2 2018;103(2):232–244. doi: 10.1016/j.ajhg.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang SCW, Yiu WH. Innate immunity in diabetic kidney disease. Nat Rev Nephrol. Apr 2020;16(4):206–222. doi: 10.1038/s41581-019-0234-4 [DOI] [PubMed] [Google Scholar]
- 16.Cao Z, Cooper ME. Role of angiotensin II in tubulointerstitial injury. Semin Nephrol. Nov 2001;21(6):554–62. doi: 10.1053/snep.2001.26794 [DOI] [PubMed] [Google Scholar]
- 17.Lv W, Booz GW, Wang Y, Fan F, Roman RJ. Inflammation and renal fibrosis: Recent developments on key signaling molecules as potential therapeutic targets. Eur J Pharmacol. Feb 5 2018;820:65–76. doi: 10.1016/j.ejphar.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misseri R, Meldrum KK. Mediators of fibrosis and apoptosis in obstructive uropathies. Curr Urol Rep. Mar 2005;6(2):140–5. doi: 10.1007/s11934-005-0083-5 [DOI] [PubMed] [Google Scholar]
- 19.Barisoni L, Troost JP, Nast C, et al. Reproducibility of the NEPTUNE descriptor-based scoring system on whole-slide images and histologic and ultrastructural digital images. Mod Pathol. July 2016;29(7):671–84. doi: 10.1038/modpathol.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zee J, Hodgin JB, Mariani LH, et al. Reproducibility and Feasibility of Strategies for Morphologic Assessment of Renal Biopsies Using the Nephrotic Syndrome Study Network Digital Pathology Scoring System. Arch Pathol Lab Med. May 2018;142(5):613–625. doi: 10.5858/arpa.2017-0181-OA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hommos MS, Zeng C, Liu Z, et al. Global glomerulosclerosis with nephrotic syndrome; the clinical importance of age adjustment. Kidney Int. 05 2018;93(5):1175–1182. doi: 10.1016/j.kint.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremers WK, Denic A, Lieske JC, et al. Distinguishing age-related from disease-related glomerulosclerosis on kidney biopsy: the Aging Kidney Anatomy study. Nephrol Dial Transplant. Dec 2015;30(12):2034–9. doi: 10.1093/ndt/gfv072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kriz W, Hähnel B, Hosser H, Rösener S, Waldherr R. Structural analysis of how podocytes detach from the glomerular basement membrane under hypertrophic stress. Front Endocrinol (Lausanne). 2014;5:207. doi: 10.3389/fendo.2014.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwimmer JA, Markowitz GS, Valeri A, Appel GB. Collapsing glomerulopathy. Semin Nephrol. Mar 2003;23(2):209–18. doi: 10.1053/snep.2003.50019 [DOI] [PubMed] [Google Scholar]
- 25.Kikuchi M, Wickman L, Hodgin JB, Wiggins RC. Podometrics as a Potential Clinical Tool for Glomerular Disease Management. Semin Nephrol. May 2015;35(3):245–55. doi: 10.1016/j.semnephrol.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royal V, Zee J, Liu Q, et al. Ultrastructural Characterization of Proteinuric Patients Predicts Clinical Outcomes. J Am Soc Nephrol. Apr 2020;31(4):841–854. doi: 10.1681/asn.2019080825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao J, Mariani L, Eddy S, et al. JAK-STAT signaling is activated in the kidney and peripheral blood cells of patients with focal segmental glomerulosclerosis. Kidney Int. Oct 2018;94(4):795–808. doi: 10.1016/j.kint.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong W, Meng XF, Zhang C. Inflammasome activation in podocytes: a new mechanism of glomerular diseases. Inflamm Res. Aug 2020;69(8):731–743. doi: 10.1007/s00011-020-01354-w [DOI] [PubMed] [Google Scholar]
- 29.Malaga-Dieguez L, Bouhassira D, Gipson D, Trachtman H. Novel therapies for FSGS: preclinical and clinical studies. Adv Chronic Kidney Dis. Mar 2015;22(2):e1–6. doi: 10.1053/j.ackd.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eom M, Hudkins KL, Alpers CE. Foam cells and the pathogenesis of kidney disease. Curr Opin Nephrol Hypertens. May 2015;24(3):245–51. doi: 10.1097/mnh.0000000000000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wengrod JC, Gardner LB. Cellular adaptation to nutrient deprivation: crosstalk between the mTORC1 and eIF2α signaling pathways and implications for autophagy. Cell Cycle. 2015;14(16):2571–7. doi: 10.1080/15384101.2015.1056947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly K, Siebenlist U. The role of c-myc in the proliferation of normal and neoplastic cells. J Clin Immunol. Mar 1985;5(2):65–77. doi: 10.1007/bf00915003 [DOI] [PubMed] [Google Scholar]
- 33.Schuhmacher M, Eick D. Dose-dependent regulation of target gene expression and cell proliferation by c-Myc levels. Transcription. Jul-Aug 2013;4(4):192–7. doi: 10.4161/trns.25907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou L, Liu Y. Wnt/β-catenin signalling and podocyte dysfunction in proteinuric kidney disease. Nat Rev Nephrol. Sep 2015;11(9):535–45. doi: 10.1038/nrneph.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Agati VD, Kaskel FJ, Falk RJ. Focal segmental glomerulosclerosis. N Engl J Med. Dec 22 2011;365(25):2398–411. doi: 10.1056/NEJMra1106556 [DOI] [PubMed] [Google Scholar]
- 36.Mariani LH, Martini S, Barisoni L, et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant. Feb 1 2018;33(2):310–318. doi: 10.1093/ndt/gfw443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayapandian CP, Chen Y, Janowczyk AR, et al. Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int. 01 2021;99(1):86–101. doi: 10.1016/j.kint.2020.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukowy JD, Dayton A, Cloutier D, et al. Region-Based Convolutional Neural Nets for Localization of Glomeruli in Trichrome-Stained Whole Kidney Sections. J Am Soc Nephrol. 08 2018;29(8):2081–2088. doi: 10.1681/ASN.2017111210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon O, Yacoub R, Jain S, Tomaszewski JE, Sarder P. Multi-radial LBP Features as a Tool for Rapid Glomerular Detection and Assessment in Whole Slide Histopathology Images. Sci Rep. 02 January 2018;8(1):2032. doi: 10.1038/s41598-018-20453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kannan S, Morgan LA, Liang B, et al. Segmentation of Glomeruli Within Trichrome Images Using Deep Learning. Kidney Int Rep. Jul 2019;4(7):955–962. doi: 10.1016/j.ekir.2019.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bueno G, Fernandez-Carrobles MM, Gonzalez-Lopez L, Deniz O. Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput Methods Programs Biomed. Feb 2020;184:105273. doi: 10.1016/j.cmpb.2019.105273 [DOI] [PubMed] [Google Scholar]
- 42.Gadermayr M, Dombrowski AK, Klinkhammer BM, Boor P, Merhof D. CNN cascades for segmenting sparse objects in gigapixel whole slide images. Comput Med Imaging Graph. 01 2019;71:40–48. doi: 10.1016/j.compmedimag.2018.11.002 [DOI] [PubMed] [Google Scholar]
- 43.Sheehan SM, Korstanje R. Automatic glomerular identification and quantification of histological phenotypes using image analysis and machine learning. Am J Physiol Renal Physiol. December 01 2018;315(6):F1644–F1651. doi: 10.1152/ajprenal.00629.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chagas P, Souza L, Araújo I, et al. Classification of glomerular hypercellularity using convolutional features and support vector machine. Artif Intell Med. March 2020;103:101808. doi: 10.1016/j.artmed.2020.101808 [DOI] [PubMed] [Google Scholar]
- 45.Murali LK, Lutnick B, Ginley B, Tomaszewski JE, Sarder P. Generative modeling for renal microanatomy. Proc SPIE Int Soc Opt Eng. Feb 2020;11320doi: 10.1117/12.2549891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouteldja N, Klinkhammer BM, Bülow RD, et al. Deep Learning-Based Segmentation and Quantification in Experimental Kidney Histopathology. J Am Soc Nephrol. January 2021;32(1):52–68. doi: 10.1681/ASN.2020050597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi R, Kawazoe Y, Shimamoto K, et al. Glomerular Classification Using Convolutional Neural Networks Based on Defined Annotation Criteria and Concordance Evaluation Among Clinicians. Kidney Int Rep. Mar 2021;6(3):716–726. doi: 10.1016/j.ekir.2020.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ginley B, Jen KY, Han SS, et al. Automated Computational Detection of Interstitial Fibrosis, Tubular Atrophy, and Glomerulosclerosis. J Am Soc Nephrol. Feb 23 2021;doi: 10.1681/ASN.2020050652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marsh JN, Liu TC, Wilson PC, Swamidass SJ, Gaut JP. Development and Validation of a Deep Learning Model to Quantify Glomerulosclerosis in Kidney Biopsy Specimens. JAMA Netw Open. January 04 2021;4(1):e2030939. doi: 10.1001/jamanetworkopen.2020.30939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilbur DC, Pettus JR, Smith ML, et al. Using Image Registration and Machine Learning to Develop a Workstation Tool for Rapid Analysis of Glomeruli in Medical Renal Biopsies. J Pathol Inform. 2020;11:37. doi: 10.4103/jpi.jpi_49_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kato T, Relator R, Ngouv H, et al. Segmental HOG: new descriptor for glomerulus detection in kidney microscopy image. BMC Bioinformatics. Sep 30 2015;16:316. doi: 10.1186/s12859-015-0739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The NEPTUNE Digital Pathology Protocol (NDPP) and Scoring System (NDPSS)
Figure S2: Dendogram of hierarchical clustering of 221 NEPTUNE participants with FSGS and MCD based on 39 glomerular descriptors.
Item S1. Supplementary Methods
Table S1. 37 glomerular descriptors with definitions
Table S2: Associations between glomerular clusters and longitudinal eGFR from linear mixed models, unadjusted and adjusted for demographics, clinical characteristics, and disease diagnosis.
Table S3. Unadjusted associations between each individual glomerular descriptor (n=37) and outcomes. Effect estimates presented are for a one-tenth increase in the observed range (observed max – observed min) of a descriptor.
Table S4. Differentially expressed genes from comparison cluster X vs Y+Z
Table S5, A and B. Full list of Canonical Pathways and Upstream Regulators predicted from X vs Y+Z differentially expressed genes.
Table S6. List of 15 differentially expressed genes from comparison cluster X vs Y+Z from RNA sequencing and Affymetrix ST 2.1 platforms
Table S7, A-M. Correlation between glomerular gene expression and descriptor frequency.
Table S8, A and B. Top (a.) canonical pathways and (b.) upstream regulators predicted from the significant correlation of glomerular RNAseq-derived gene expression with log10-transformed descriptor scores
Table S9, A-M. Full list of Canonical Pathways and Upstream Regulators predicted from correlation between glomerular gene expression and descriptor frequency.


