SUMMARY
Neuroblastoma (NB), the most common cancer in the first year of life, presents almost exclusively in the trunk. To understand why an early-onset cancer would have such a specific localization, we xenotransplanted human NB cells into discrete neural crest (NC) streams in zebrafish embryos. Here, we demonstrate that human NB cells remain in an undifferentiated, tumorigenic state when comigrating posteriorly with NC cells but, upon comigration into the head, differentiate into neurons and exhibit decreased survival. Furthermore, we demonstrate that this in vivo differentiation requires retinoic acid and brain-derived neurotrophic factor signaling from the microenvironment, as well as cell-autonomous intersectin-1-dependent phosphoinositide 3-kinase-mediated signaling, likely via Akt kinase activation. Our findings suggest a microenvironment-driven explanation for NB’s trunk-biased localization and highlight the potential for induced differentiation to promote NB resolution in vivo.
In brief
In this study, Treffy et al. perform targeted xenotransplantation of human neuroblastoma cancer cells into vertebrate embryos and identify signaling factors that promote region-specific differentiation of malignant cells into neurons that die. These findings may have implications for our understanding of neuroblastoma’s origins and localization in pediatric patients.
Graphical abstract
INTRODUCTION
Neuroblastoma (NB) has traditionally been thought to arise from sympathoadrenal cells in the trunk (Matthay et al., 2016; Ratner et al., 2016), where this malignancy is predominantly localized (Vo et al., 2014). Sympathoadrenal cells, in turn, are derived from neural crest cells (NCCs), highly migratory stem cells that contribute to a variety of cell types during vertebrate embryonic development and that share striking genetic and phenotypic similarities with metastatic cancer cells (Gallik et al., 2017; Jiang et al., 2011). Given NB’s presentation in early childhood and in utero (Gigliotti et al., 2009), as well as its appropriation of developmental signaling pathways, including those responsible for NCCs’ proliferative and migratory capabilities, NB has often been described as a “developmental cancer” (Gallik et al., 2017; Jiang et al., 2011; Marshall et al., 2014; Nakagawara, 1998; Ratner et al., 2016).
Although early childhood NB may arise from differentiated cells (Ratner et al., 2016; Reiff et al., 2010), this transformation would necessitate a number of malignancy-inducing mutations. Meanwhile, previous studies have shown that a small number of genetic modifications are sufficient to transform primary cultured and immortalized NCCs into NB in vitro (Olsen et al., 2017; Schulte et al., 2013). Therefore, we hypothesized that NB might be derived directly from aberrant NCCs in vivo. However, given the neural crest’s (NC’s) contributions to multiple lineages throughout the developing embryo, the concept of NB derivation directly from the NC raises immediate questions as to why this cancer with a low genetic threshold for malignancy would localize almost exclusively to the trunk of young patients.
One potential contributor to localization is the microenvironment, but it remains unclear as to how an embryonic or infant microenvironment might affect the migration and behavior of malignant NB. Building on previous work in zebrafish (Casey and Stewart, 2018; Kaufman et al., 2016; Kendall et al., 2018; Topczewska et al., 2006; Zhu et al., 2012), we devised a high-resolution model system to inject, track, and analyze the in vivo behavior of human NB cells, including patient-derived xenografts (PDXs), alongside endogenous NCCs (Figure 1A) in an embryonic microenvironment similar to that encountered by malignant NB cells in utero and in infants. Here, we present evidence in zebrafish and mouse embryos suggesting that NB’s anatomical localization is due to permissive versus differentiating microenvironments. Furthermore, we identify two microenvironmental factors, retinoic acid (RA) and brain-derived neurotrophic factor (BDNF), that are required for region-specific differentiation in vivo. Finally, we pinpoint two downstream cell-autonomous drivers of differentiation, the scaffold protein intersectin-1 (ITSN1) and phosphoinositide 3-kinase (PI3K)/Akt signaling.
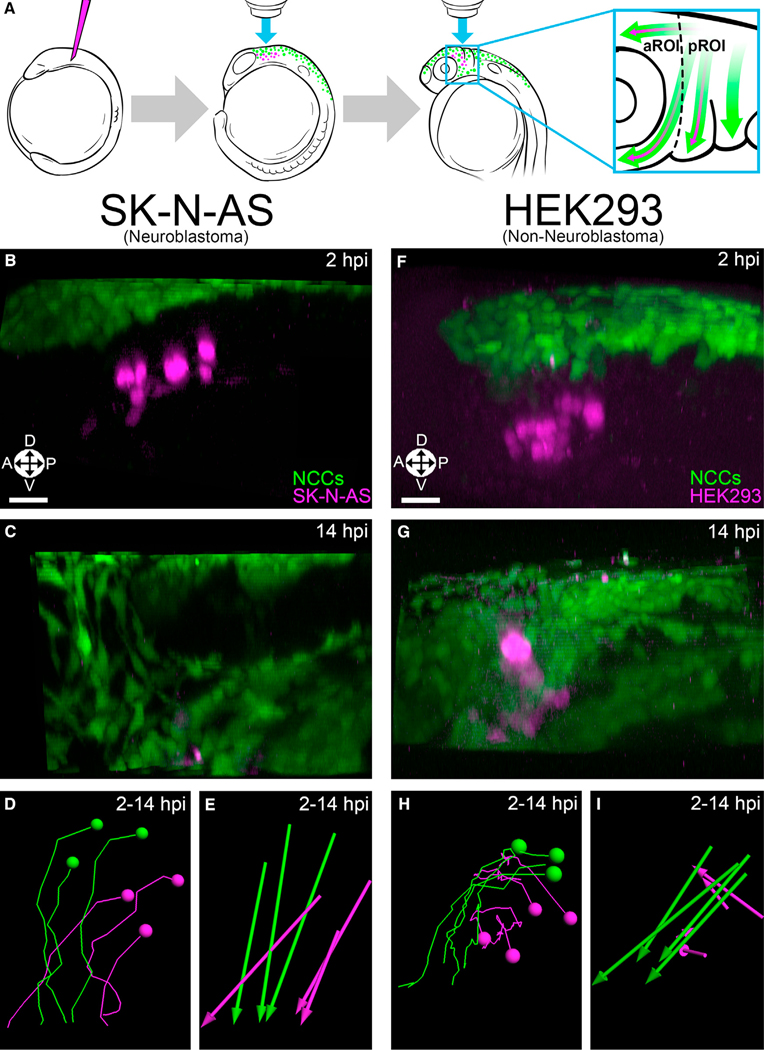
Figure 1. NB cells, but not control cells, comigrate with cranial NCCs.
(A) Schematic representation of the procedure used to study NB in live zebrafish embryos. Dotted line divides the aROI and pROI, and arrows indicate NC (green) and NB (purple) migration.
(B and C) 3D projections of Hoechst 34580-labeled (nuclear) injected SK-N-AS NB cells (purple; n = 8 embryos) alongside NCCs (green) at 2 hpi (premigratory) (B) and 14 hpi (C) in zebrafish embryos.
(D and E) SK-N-AS cell tracks (D) and displacements (E) at 2–14 hpi.
(F and G) 3D projections of Hoechst 34580-labeled HEK293 cells (n = 5 embryos) alongside NCCs at 2 hpi (F) and 14 hpi (G). HEK293 cell tracks (H) and dis- placements (I) at 2–14 hpi. Orientation: A: anterior; P, posterior; L, lateral; D, dorsal; V, ventral. Scale bar (B–I), 30 μm.
RESULTS
NB cells comigrate with NCCs into distinct microenvironments
We first injected human KELLY NB cells into the zebrafish trunk NC, which gives rise to the sympathoadrenal lineage, and observed ventrolateral comigration with NCCs along endogenous migration pathways (Video S1), consistent with previous work in chicken embryos (Delloye-Bourgeois et al., 2017). Next, we examined the behavior of NB cells in unexplored microenvironments outside of those normally seen by the sympathoadrenal lineage. To do so, we took advantage of a branch point of the anterior-most cranial NC stream in zebrafish from which some NCCs migrate further into an anterior region of interest (aROI) to form several cell types, including neurons, whereas other NCCs instead invade the first pharyngeal arch to form cartilage (defined here as our posterior region of interest (pROI)) (Theveneau and Mayor, 2012) (Figure 1A). This natural “fork in the migratory road” facilitated anatomically precise injections of human NB cells that then migrated in one of two directions leading to diverse microenvironments. Injections of three different human NB cell lines (KELLY, SK-N-AS, and IMR-5) into this location yielded comigration with NCCs similar to that seen in the trunk (Figures 1B–1E, S1A–S1C′, and S1G; Video S2). To rule out non-specific cellular movement, we injected the embryonic kidney cell line HEK293, ovarian cancer cell line Ovcar-8, and melanoma cell line A375 and found that none of them comigrated with cranial NCCs (Figures 1F–1I and S1D–S1G; Video S3).
NB cells undergo region-specific differentiation and death
Although several NB cell lines can be stimulated to differentiate into neurons in vitro (Singh et al., 2011; Voigt et al., 2000), this is not the case for SK-N-AS cells (Guglielmi et al., 2014; Singh et al., 2011) (Figures S2A–S2C). However, upon injecting SK-N-AS cells into the branch point of the anterior-most cranial NC stream in zebrafish embryos and tracking migration into the aROI or pROI, we discovered cells that had ceased migrating and adopted a neuron-like morphology, as defined by an elongated cell body, rapidly extending unipolar or bipolar projections of typical axonal diameter with growth cones, and temporally stable projections (Figures 2A–2F; Video S4 versus S5; Video S6). Time-lapse imaging revealed that these newly differentiated neurons underwent apoptosis within a few hours (Figures 2E and 2F; Video S7).
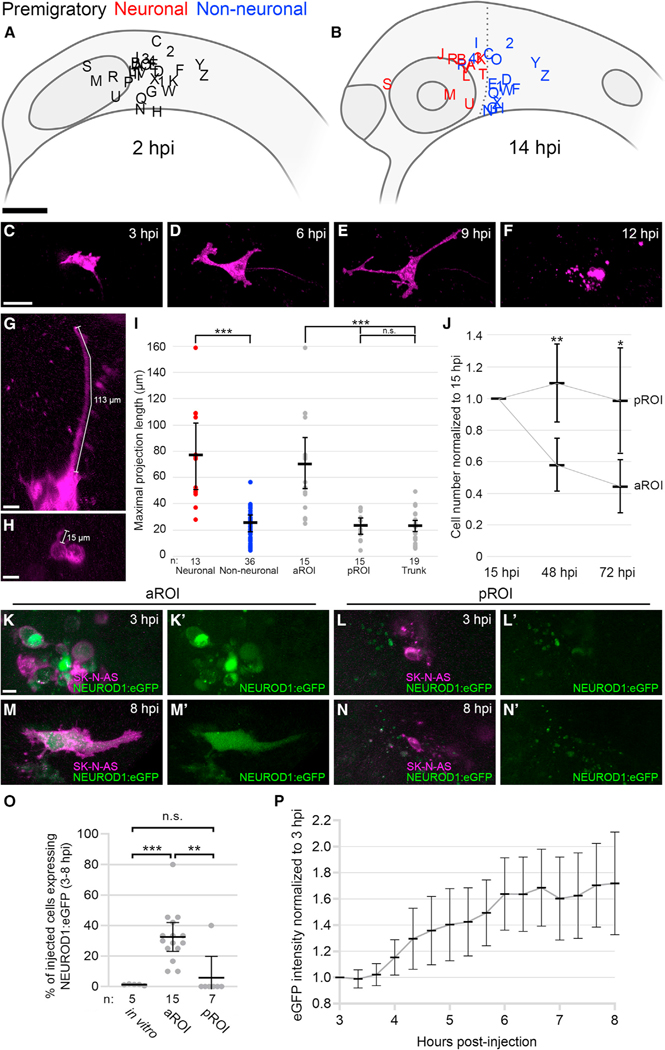
Figure 2. The neurogenic microenvironment of the developing zebrafish head induces NB differentiation into neurons.
(A and B) Lateral view schematics of zebrafish embryos injected with membrane-localized mCherry-expressing SK-N-AS NB cells (empty or scrambled vector; n = 30 embryos) at 2 hpi (A) and 14 hpi (B), for aROI and pROI assays only. Black letters/numbers indicate sites of injection; red and blue letters/numbers indicate final locations of neuronal and non-neuronal cells, respectively.
(C–F) Still images from Video S7 of an injected SK-N-AS cell undergoing differentiation and apoptosis in the aROI.
(G and H) Representative 3D projections of neuronal (G) or non-neuronal (H) SK-N-AS cells (purple) with length measurements.
(I) Scatter plot of neuronal (red) and non-neuronal (blue) maximal projection lengths and their segregation into aROI, pROI, and trunk groups (gray).
(J) The ratio of the number of SK-N-AS cells at 48 and 72 hpi in comparison with 15 hpi (n = 21 embryos for aROI and n = 9 embryos for pROI).
(K–N′) 3D projections of injected NEUROD1:eGFP-expressing SK-N-AS cells present in the aROI or pROI at 3 hpi (K–L′) and 8 hpi (M–N′).
(O) The percentage of SK-N-AS cells expressing NEUROD1:eGFP in vitro (cultured cells) or in vivo in the aROI or pROI.
(P) Normalized eGFP intensity of single NEUROD1:eGFP-expressing cells in the aROI at 3–8 hpi (n = 10 cells, 1 representative cell/embryo). Horizontal bars (I, J, O, and P) denote mean values, with 95% confidence intervals indicated. n.s. p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar: (A and B) 100 μm; (C–F) 40 μm; (G, H, and K–N′) 15 μm.
To determine what might cause this surprising differentiation in a subset of injected embryos, we plotted the initial and final locations of all SK-N-AS cell groups and binned them according to morphology. Spatiotemporal analysis revealed that all SK-N-AS-derived neuron-like cells were found exclusively in the aROI, whereas SK-N-AS cell migration into the pROI did not yield neuron-like cells (Figure 2B). To quantitate the distribution of neuronal morphology, an in vitro NB differentiation assay measuring maximal neuronal projections (Guglielmi et al., 2014) was modified for in vivo use by measuring the longest projections over the course of our time-lapse imaging (Figures 2G and 2H). Compared across regions, injected cells that migrated into the aROI demonstrated maximal projection lengths significantly longer than those that had migrated into the pROI (Figure 2I). We also injected SK-N-AS cells directly into the aROI or pROI and observed similar results, suggesting that comigration with NCCs (Figure 1) is transporting NB cells into differentiating or permissive microenvironments. Meanwhile, injections of SK-N-AS cells into the trunk NC did not yield differentiation (Figure 2I). Evaluating projection lengths across the breadth of our 2–14 h post-injection (hpi) time-lapses, we observed the peak occurrence of fully differentiated neurons at 6–8 hpi (Figure S2J). To quantitate NB cell survival post-differentiation, we followed SK-N-AS cells in vivo and observed significantly decreased cell numbers at 48 and 72 hpi in the differentiation-inducing aROI, whereas cell numbers in the non-differentiating pROI and trunk stayed the same or increased (Figures 2J and S2K). Cumulatively, these findings speak to distinct, region-specific microenvironments that drive the differentiation and subsequent death of NB cells in vivo.
To further confirm the NB differentiation phenotype and clarify its timeline, we generated and validated a live reporter of early neurogenesis in which the human NEUROD1 promoter region drives eGFP expression (Figure S3). Injection and tracking of NEUROD1:eGFP-expressing SK-N-AS cells in vivo revealed stark differences in the number of eGFP-expressing cells in the aROI versus the pROI at 3–8 hpi (Figures 2K–2O; Video S8). eGFP levels increased gradually over time prior to the appearance of neuronal morphology (Figure 2P), consistent with the morphology-based identification of peak differentiation at 6–8 hpi (Figure S2J). Finally, we confirmed the presence of differentiated neurons via immunofluorescence staining for neurofilament (NF). NF staining was rarely seen in vitro in SK-N-AS cells (Figures S2A–S2C), but in vivo, NF-positive, SK-N-AS-derived cells were present in the aROI at 6, 14, and 24 hpi (Figures S2D, S2F, and S2H), whereas all assayed cells in the pROI at the same time points remained NF negative (Figures S2E, S2G, and S2I).
Taken together, our morphology, projection length, and molecular analyses suggest that a neurogenic microenvironment encountered by anteriorly migrating cranial NC streams in the developing zebrafish head can induce comigrating NB cells to differentiate and die, whereas NB cells outside of that microenvironment retain their migratory, undifferentiated state.
N-MYC amplification and/or the ALK mutation F1174L are present in significant subsets of NB (Barone et al., 2013), but SK-N-AS cells express wild-type levels of N-MYC and do not have ALK mutations. To determine whether region-specific differentiation could occur in the context of N-MYC amplification and/or the ALK mutation F1174L, we repeated our zebrafish experiments with N-MYC-amplified IMR-5 and KELLY NB cells, with KELLY also harboring the F1174L mutation (Table 1). Results for both cell lines yielded region-specific differentiation similar to that observed with SK-N-AS cells (Figure S4). In contrast, mesenchymal-like GIMEN NB cells (van Nes et al., 2013) and A375 melanoma cells were unable to differentiate in vivo (Figure S4). These results suggest that microenvironmentally mediated differentiation is independent of N-MYC amplification and ALK mutation but is unable to differentiate some mesenchymal-like cells that may not retain the capacity for neurogenesis.
Table 1.
Cell line characteristics
| Cell line | N-MYC | ALK mutation |
|---|---|---|
| SK-N-AS | non-amplified | wild type |
| IMR-5 | amplified | wild type |
| KELLY | amplified | F1174L |
| GIMEN | non-amplified | wild type |
Expression level of N-MYC and presence/absence of F1174L mutation in ALK in NB cell lines SK-N-AS, IMR-5, KELLY and GIMEN.
Although SK-N-AS, IMR-5, and KELLY cells are derived from high-risk NB, cell lines are far removed from their source tumors. Therefore, we also tested whether genetically and phenotypically heterogeneous, high-risk PDXs behaved similarly in our zebrafish model system. Upon injection, N-MYC amplified, stage 4 primary tumor-derived NB PDXs exhibited robust region-specific differentiation (Figure S4), suggesting that the aROI’s microenvironment can differentiate even high-risk human NB tumors in vivo.
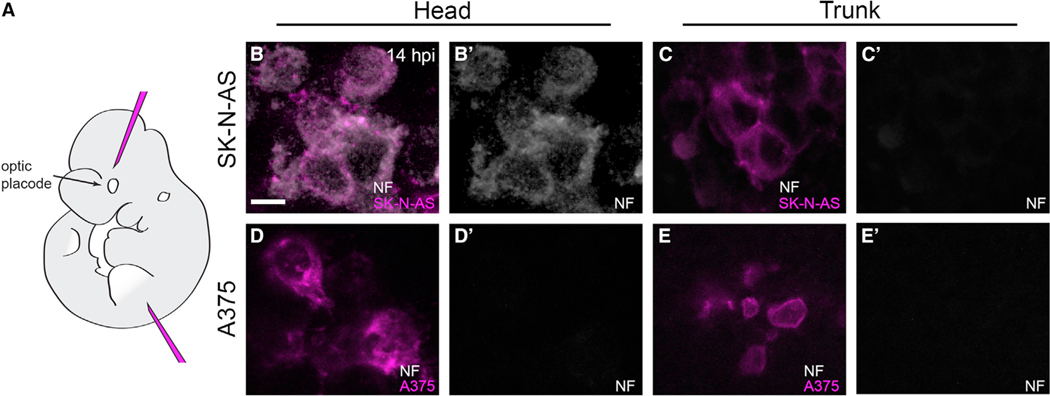
Finally, we asked whether the differentiation phenotype seen in zebrafish embryos could manifest in other vertebrates. Zebrafish and mouse cranial development are highly conserved (Bronner and LeDouarin, 2012); therefore, we injected SK-N-AS cells into mouse embryo heads at a developmentally analogous time point (E10.5; Figure 3A), cultured injected embryos ex vivo, and assayed for NF immunofluorescence. In line with our zebrafish findings, NF-positive SK-N-AS cells were found at 14 hpi in the murine head but not in the trunk, whereas A375 melanoma cells were negative for NF staining in both regions (Figures 3B–3E′). These mammalian results lend further support to the hypothesis that the vertebrate cranial microenvironment adjacent to NC streams is not conducive to NB progression and instead promotes neuronal differentiation.
Figure 3. SK-N-AS cells in the developing murine head undergo neuronal differentiation.
(A) Lateral view schematic of a mouse E10.5 embryo indicating the locations of head and trunk xenotransplantation.
(B–E′)5 μm optical slices of whole-mount immunolabeling for anti-neurofilament (NF; white) of representative SK-N-AS NB (B–C′) or A375 melanoma (D–E′) cells at 14 hpi in E10.5 mouse embryos. Head injections targeted a region populated by cranial neural crest cells, analogous to that targeted in zebrafish. 71% of assayed embryos (n = 12/17 embryos) with SK-N-AS injections into the head harbored NF-positive cells, whereas 0% of assayed embryos (n = 0/11 embryos) with SK-N-AS injections into the trunk harbored NF-positive cells. No embryos with A375 melanoma injections into the head (n = 0/16 embryos) or trunk (n = 0/14 embryos) harbored NF-positive cells. Scale bar, 10 μm.
RA is required for NB differentiation and may prime NB to respond to other differentiating signals
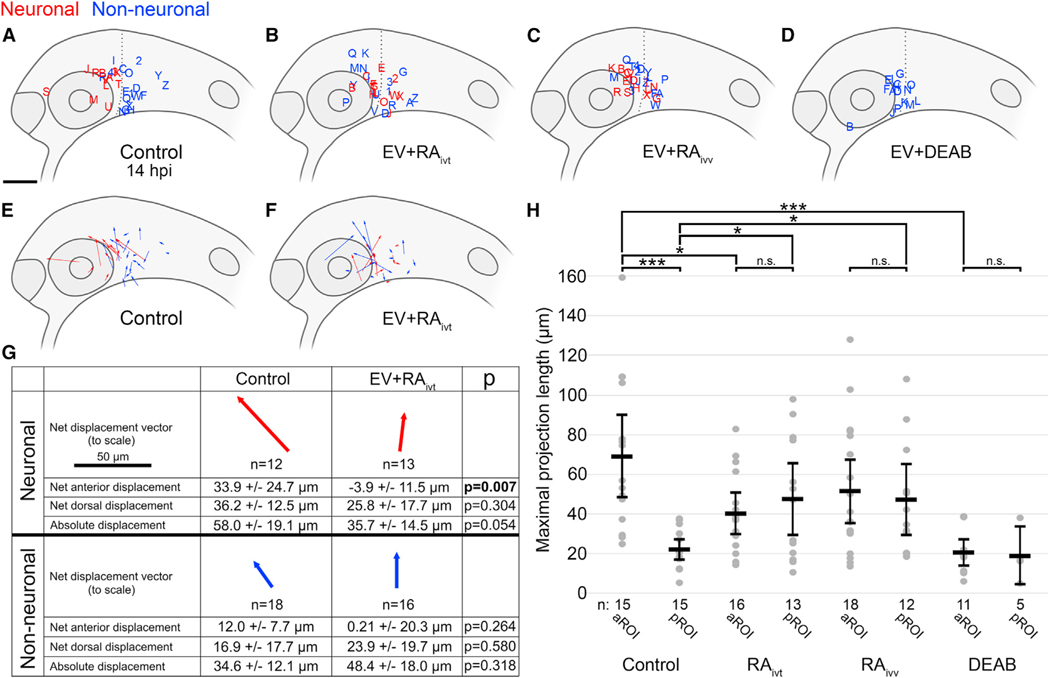
The current treatment protocol for advanced stage NB often relies on non-specific therapies with many side effects, and although there have been significant advancements in treatment, survival remains poor in comparison with that for other childhood malignancies (Pinto et al., 2015; Santiago et al., 2017). RA is an established but poorly understood therapeutic agent that induces in vitro differentiation, growth inhibition, and apoptosis in many NB cell lines (Matthay et al., 1999; Reynolds et al., 2003; Singh et al., 2011; Voigt et al., 2000), but its effects remain largely uncharacterized in vivo. As previously noted, RA does not stimulate differentiation of SK-N-AS cells in vitro (Guglielmi et al., 2014; Singh et al., 2011) (Figures S2A–S2C). At time points when differentiation occurs in zebrafish, RA and its synthesizing enzyme raldh2 have been shown to be differentially expressed in our regions of interest, with high expression in the aROI, particularly in the developing eye, versus low expression in the pROI (Shimozono et al., 2013). To ascertain RA’s possible effects, we treated SK-N-AS cells with RA for 24 h prior to injection into zebrafish embryos, followed by time-lapse imaging as before. In contrast to the stark anterior/posterior segregation observed with untreated SK-N-AS cells (Figure 4A), RA-treated SK-N-AS cells differentiated in embryos in both the aROI and pROI (Figures 4B, S5A, and S5B), with no significant difference seen between the lengths of maximal projections in those regions (Figure 4H). Next, we injected untreated SK-N-AS cells into host embryos that received RA treatment from 2 to 14 hpi, recapitulating these findings (Figures 4C, 4H, S5C, and S5D). To understand the mitigation of the previously observed segregated differentiation, pre- and post-migration coordinates of injected cells were used to determine total directional migration. This quantitation revealed that RA-treated SK-N-AS cells migrated anteriorly significantly less than untreated SK-N-AS cells did (Figures 4E–4G), suggesting that RA-treated cells may be more responsive to other differentiating signals in the microenvironment and that the addition of exogenous RA is overpowering RA’s endogenous differential expression (Shimozono et al., 2013). Finally, treatment of embryos with the retinaldehyde dehydrogenase inhibitor diethylaminobenzaldehyde (DEAB), which suppresses RA production (Perz-Edwards et al., 2001), resulted in a complete loss of differentiation (Figures 4D, 4H, S5E, and S5F), suggesting that microenvironment-derived RA signaling is required for NB differentiation in vivo.
Figure 4. Microenvironmental RA signaling is required for NB differentiation.
(A–D) Lateral view schematics of zebrafish embryos at 14 hpi that were injected with membrane-localized mCherry-expressing SK-N-AS NB cells that either differentiated into neurons (red) or not (blue) during time-lapse imaging. (A) Empty vector (EV) or scrambled vector (n = 30 embryos), duplicated from Figure 2B for clarity; (B) EV + 5 μM RA on cultured cells for 24 h pre-injection (n = 29 embryos); (C) EV + 1 μM RA on host embryos post-injection through time-lapse (n = 30 embryos); (D) EV + 50 μM DEAB on host embryos 2 h pre-injection through time-lapse (n = 16 embryos).
(E and F) Displacement vectors for untreated SK-N-AS cells (n = 30 embryos) (E) and RA-treated SK-N-AS cells (n = 29 embryos) (F). Red and blue vectors are for neuronal and non-neuronal cells, respectively.
(G) Aggregates of displacements for untreated and RA-treated SK-N-AS cells divided into neuronal and non-neuronal categories. Arrows are scaled, and their orientations correspond to orientations in (E) and (F). Net anterior and dorsal displacements, as well as absolute displacements across all axes, are shown along with p values. Displacements are affected by embryo growth such that dorsal values are skewed positively despite most cells migrating ventrally; therefore, comparisons are made only relative to appropriate controls.
(H) Scatter plot of maximal projection lengths and their segregation into aROI and pROI groups stratified by tested conditions. Control aROI and pROI columns are duplicated from Figure 2I for clarity and are composed of scrambled and empty vector data points. Horizontal bars denote mean values, with 95% confidence intervals indicated. n.s. p > 0.05; *p < 0.05; ***p < 0.001. ivt, in vitro; ivv, in vivo. Scale bar (A–F), 100 μm. Scale bar (G), 50 μm.
BDNF is expressed in the aROI and is required for NB differentiation
Since RA treatment alone is insufficient to induce differentiation of SK-N-AS cells in vitro (Guglielmi et al., 2014; Singh et al., 2011) (Figures S2A–S2C), we hypothesized that differentiation in vivo may require additional microenvironment-derived signals. The neurotrophic factors nerve growth factor (NGF) and BDNF, along with their receptors TrkA and TrkB, respectively, are known to influence NB differentiation in vitro (Brodeur et al., 2009; Hoehner et al., 1995). Although NGF is expressed near our region of interest in zebrafish embryos (Thisse et al., 2004), treatment of SK-N-AS cells and injected zebrafish embryos with the TrkA antagonist K252a did not impact differentiation (Figure S6A). BDNF, meanwhile, is expressed broadly near NC migration pathways (Lum et al., 2001; Thisse et al., 2004) and has been suggested to have diverse, often conflicting influences on NB, including a variety of possible effects on metastasis and differentiation (Brodeur et al., 2009; Hoehner et al., 1995; Hua et al., 2016), leaving its role in a complex in vivo microenvironment unclear. While RA expression has been shown to increase in the developing zebrafish eye (Shimozono et al., 2013) directly prior to our differentiation phenotype peak (Figure S2J), BDNF expression has been less well characterized at these early time points. Therefore, we performed in situ hybridization chain reaction (HCR) to detect zebrafish BDNF mRNA. In contrast to broader expression patterns later in embryonic development (Lum et al., 2001; Thisse et al., 2004), BDNF mRNA was highly localized at 19 h post-fertilization (hpf) in the developing olfactory placode, i.e., the future epithelium of the nose (Figures 5A and 5B). Thus, anteriorly migrating NC streams and comigrating NB cells are brought in proximity to the developing zebrafish eye and nose and encounter high levels of RA and BDNF, respectively, followed by increasing levels of NEUROD1:eGFP expression in NB cells (Figure 2P) and neuronal differentiation (Figure S2J).
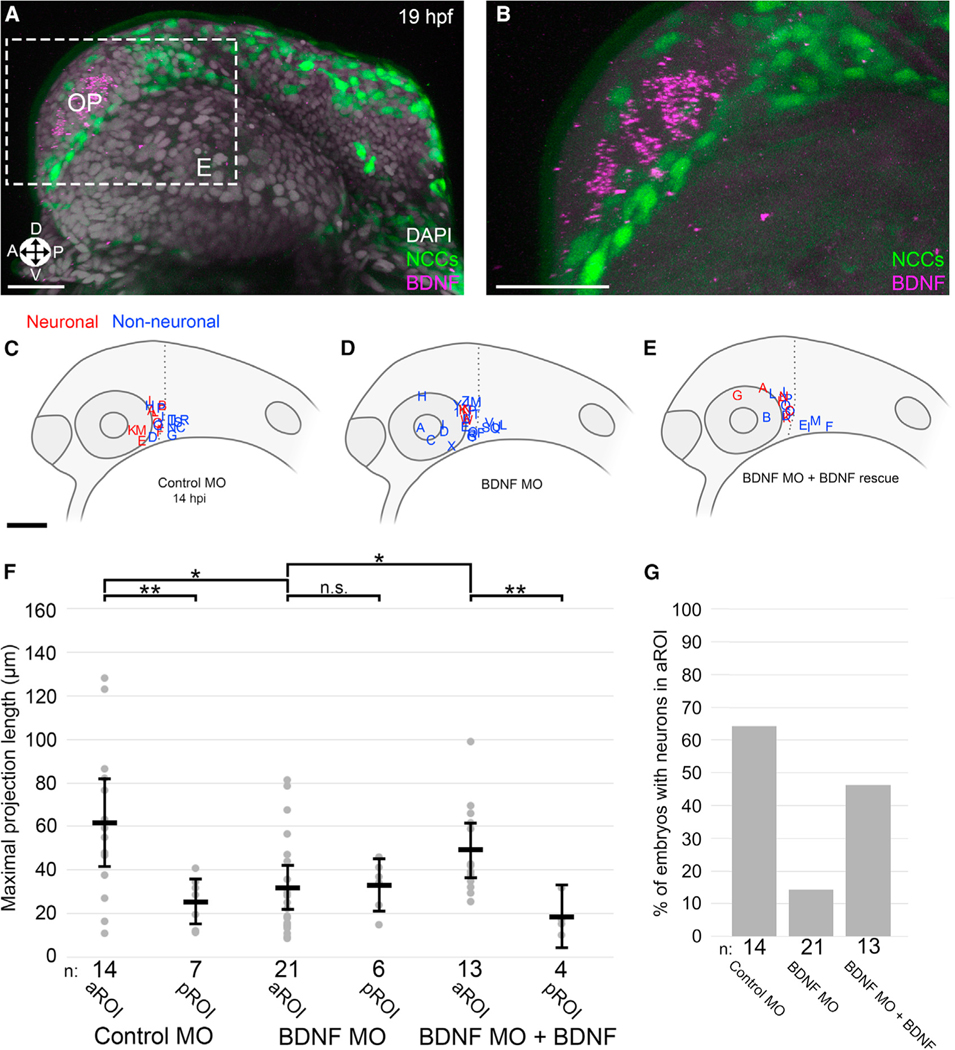
Figure 5. Microenvironmental BDNF signaling is required for NB differentiation.
(A and B) Representative 3D projection of BDNF HCR (purple), NCCs (green), and DAPI nuclear labeling (gray) (A) with inset (B) (gray) in 19 hpf zebrafish embryos, corresponding developmentally to 5 hpi. E, eye; OP, olfactory placode; Orientation: A, anterior; P, posterior; D, dorsal; V, ventral.
(C–E) Lateral view schematics of zebrafish embryos at 14 hpi that were injected with membrane-localized mCherry-expressing SK-N-AS NB cells after injection at the 1-cell stage with (C) 5 ng control morpholino (MO), (D) 2 ng BDNF translation-blocking MO, or (E) 2 ng BDNF translation-blocking MO + 200 ng/mL human BDNF starting at 4 hpf. Red and blue labels indicate final locations of neuronal and non-neuronal cells, respectively.
(F) Scatter plot of maximal projection lengths segregated into aROI and pROI. Horizontal bars denote mean values, with 95% confidence intervals indicated. n.s. p > 0.05, *p < 0.05, **p < 0.01.
(G) Percentage of embryos in which SK-N-AS cells differentiated into neurons in the aROI. Scale bar (A and B), 20 μm. Scale bar (C–E), 100 μm.
Given this intriguing spatiotemporal expression profile, we explored a functional role for BDNF and its receptor TrkB. In vitro, treatment of SK-N-AS cells with RA did not alter levels of human TrkB (NTRK2) expression (Figures S6E and S6F′), and treatment with both RA and BDNF was not sufficient to induce differentiation (Figures S6G and S6H′). However, in vivo treatment with the TrkB antagonist ANA-12 resulted in a near-complete loss of differentiation in the zebrafish aROI post-injection (Figures S5G, S5H, and S6B–S6D), and attenuation of BDNF production in the zebrafish microenvironment with a translation-blocking morpholino (Diekmann et al., 2009) recapitulated the ANA-12-induced phenotype (Figures 5C, 5D, 5F, 5G, S5I, and S5J). Finally, treatment of injected embryos with recombinant human BDNF protein rescued the morpholino-mediated phenotype, selectively restoring differentiation only in the aROI (Figures 5E–5G). Taken together, these data suggest that region-specific NB differentiation requires a precisely timed combination of endogenous RA and BDNF-TrkB signaling in the context of the aROI’s NC-adjacent microenvironment.
ITSN1-dependent PI3K signaling is required for NB differentiation
Having identified the necessity of two microenvironmental factors, RA and BDNF, we next evaluated the potential role of a cell-autonomous target, the scaffold protein intersectin-1 (ITSN1), which is highly expressed in NB cells (Russo and O’Bryan, 2012; Russo et al., 2015). ITSN1 is required for NB tumorigenesis (Russo and O’Bryan, 2012; Russo et al., 2015) and, of interest, is also necessary for the differentiation and survival of cortical neurons in vitro (Das et al., 2007). Given these seemingly paradoxical roles of ITSN1 in tumorigenesis and differentiation, we attempted to clarify its function in our model system. The injection of ITSN1-silenced SK-N-AS cells into zebrafish embryos (Figures 6A and 6B) yielded decreased cell numbers at 48 and 72 hpi in comparison with control SK-N-AS cells (Figures 6F and 6G), consistent with ITSN1’s known role in tumor cell survival, and yielded an almost complete loss of differentiation specifically in the aROI (Figures 6B, S5K, S5L, S7A, and S7B), with maximal projection lengths significantly shorter than in embryos injected with control (empty or scrambled vector) SK-N-AS cells (Figure 6J). To confirm the specificity of ITSN1 silencing, we expressed murine ITSN1 in ITSN1-silenced SK-N-AS cells, which rescued injected cell differentiation in the aROI (Figures S7C–S7E). These results suggest that ITSN1 is required for SK-N-AS cells to differentiate in vivo.
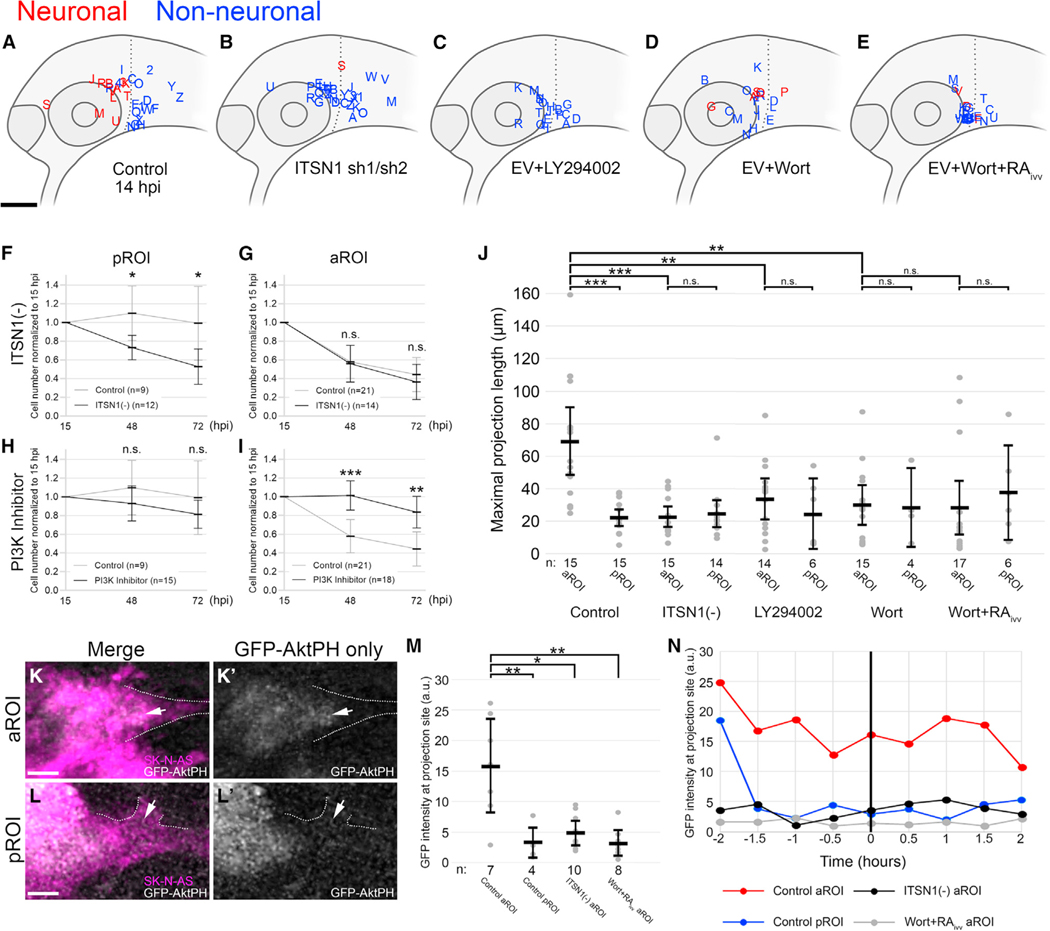
Figure 6. Cell-autonomous ITSN1-dependent PI3K signaling is required for NB differentiation.
(A–E) Lateral view schematics of zebrafish embryos at 14 hpi that were injected with membrane-localized mCherry-expressing SK-N-AS NB cells that either differentiated into neurons (red) or not (blue) during time-lapse imaging. (A) Empty vector (EV) or scrambled vector (n = 30 embryos), duplicated from Figure 2B for clarity; (B) ITSN1 short hairpin 1 (sh1) or short hairpin 2 (sh2) (n = 29 embryos combined) (C) EV + 100 μM LY294002, a reversible PI3K inhibitor, on cultured cells for 2 h pre-injection (n = 20 embryos). (D) EV + 100 nM wortmannin, an irreversible PI3K inhibitor, on cultured cells for 24 h pre-injection (n = 19 embryos). (E) EV + 1 μM wortmannin on cultured cells for 24 h pre-injection and 1 μM RA on host embryos post-injection through time-lapse (n = 23 embryos).
(F–I) Cell numbers at 48 and 72 hpi normalized to 15 hpi and their segregation into pROI or aROI groups in tested conditions compared with control SK-N-AS cells: ITSN1-silenced (sh2) SK-N-AS cells (F and G), PI3K inhibitor (either LY294002 or wortmannin)-treated SK-N-AS cells (H and I). Control pROI and aROI values are duplicated from Figure 2J for clarity.
(J) Scatter plot of maximal projection lengths and their segregation into aROI and pROI groups stratified by tested conditions. Control aROI and pROI columns are duplicated from Figure 2I for clarity and are composed of scrambled and empty vector data points.
(K–L′) 9 μm thick optical slices of representative SK-N-AS cells expressing the GFP-AktPH construct in the aROI (K and K′) or pROI (L and L′). Dashed lines denote membrane projections and arrows indicate the base of projections.
(M) Scatter plot of GFP intensity in a 2 μm diameter sphere adjacent to the cell membrane when the maximal projection first forms.
(N) GFP intensity (2 μm diameter sphere) of single representative cells over time in the aROI (red; K and K′), pROI (blue; L and L’), aROI with ITSN1 sh2 (black), or aROI post-treatment with 1 μM wortmannin and 1 μM RA (gray). Horizontal bars denote mean values, with 95% confidence intervals indicated. n.s. p > 0.05;
*p < 0.05; **p < 0.01; ***p < 0.001. ivt, in vitro; ivv, in vivo. Scale bar (A–E), 100 μm. Scale bar (K and L′), 5 μm.
Since ITSN1 has been shown to modulate PI3K signaling (Russo and O’Bryan, 2012; Russo et al., 2015), we treated SK-N-AS cells with the PI3K inhibitors LY294002 and wortmannin and found that, while PI3K inhibition had no direct effect on injected cell survival at 48 and 72 hpi (Figure 6H), PI3K-mediated signaling is required for differentiation and subsequent cell death in the aROI, including in the presence of exogenous RA treatment (Figures 6C–6E, 6I, 6J, and S5M–S5R). PI3Ks have been described as functioning through both Akt-dependent and -independent mechanisms (Bruhn et al., 2013; Janku et al., 2018). To determine the influence of Akt-dependent signaling and pinpoint its spatiotemporal dynamics during NB differentiation, we used the biosensor GFP-AktPH (Haugh et al., 2000) to quantitatively measure changes in the subcellular localization of Akt-activating PIP3 in injected SK-N-AS cells in both the aROI and pROI. GFP fluorescence quantitation revealed significantly greater GFP-AktPH localization adjacent to cell membrane sites of projections in the aROI in comparison with that in the pROI (Figures 6K–6M). Furthermore, quantitation across multiple time points demonstrated that GFP-AktPH accumulated locally several hours prior to the appearance of projections in the aROI but not in the pROI (Figure 6N). These sites of accumulation were markedly absent in ITSN1-deficient cells and in PI3K-inhibitor-treated cells, even after in vivo treatment with RA (Figures 6M and 6N). Cumulatively, these data suggest that ITSN1-dependent, spatiotemporally-specific PI3K/Akt signaling promotes NB differentiation in vivo.
DISCUSSION
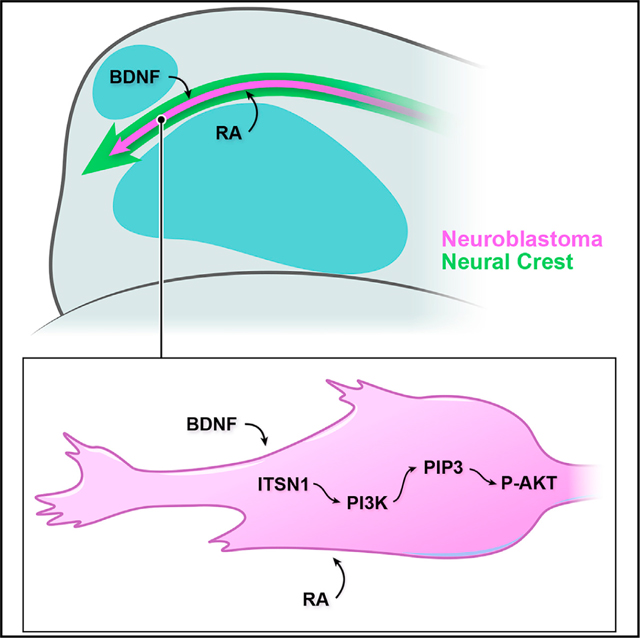
Here, we have developed a quantitative model system that probes the in vivo cell biology of NB migration, differentiation, and survival in response to microenvironmental signaling and models the response of migratory NCCs that might acquire malignancy-inducing mutations during embryogenesis. By injecting NB cells, including those not previously considered capable of differentiation, at the branch point of the anterior-most NC stream and randomizing their directional comigration with NCCs into the aROI or pROI, we have imaged and quantitated NB’s spatiotemporally distinct differentiation and death. Our findings implicate sensory organ-secreted RA and BDNF, cell-autonomous ITSN1, and PI3K-Akt signaling as critical determinants of NB differentiation in vivo along cranial NC migration routes (graphical abstract).
NCCs respond to myriad signaling factors and differentiate into a variety of derivatives that include peripheral nervous system neurons (Kasemeier-Kulesa et al., 2005; Saxena et al., 2013; Teillet et al., 1987). BDNF promotes a sensory neuron fate in NCCs (Liebl et al., 2000; Sieber-Blum, 1991; Sieber-Blum et al., 1993), and RA may also have some ability to contribute to NC differentiation (Henion and Weston, 1994; Ito and Morita, 1995). Thus, we hypothesize that mutated NCCs transformed into malignant NB cells in human embryos retain some ability to respond to differentiating signals. Our spatiotemporal tracking and differentiation analyses suggest that neurogenic structures located adjacent to cranial NC streams play critical roles in this region-specific differentiation. The developing zebrafish eye and nose express RA and BDNF, respectively, at high levels just as NCCs, along with injected NB cells, stream nearby at 4–5 hpi, which coincides with increased expression of NEUROD1:eGFP in NB cells and is followed by peak NB differentiation at 6–8 hpi. Meanwhile, directional tracking after the addition of exogenous RA points to an increased responsiveness to differentiating signals. Thus, the timely sequence of neurogenic factor upregulation in sensory organs establishes a highly differentiating microenvironment for migrating NC and NB cells, with RA likely priming NB cells to differentiate in response more readily to BDNF. Of interest, previous in vitro findings have shown that RA induces the expression of the full-length BDNF receptor TrkB in NB cell lines that readily differentiate but not in NB cells that do not respond to RA (Edsjö et al., 2003; Kaplan et al., 1993), consistent with our in vitro results in canonically non-differentiating SK-N-AS cells. Thus, the in vivo differentiation of SK-N-AS cells in the aROI may rely on higher effective concentrations of BDNF stimulating TrkB, unknown amplifiers of signaling via TrkB, and/or additional microenvironmental factors that cooperatively drive differentiation along with RA and BDNF. We attempted to demonstrate the contextual sufficiency, not just necessity, of RA and BDNF to induce differentiation in vivo by assaying their effects on NB cells in the pROI, but those experiments proved inconclusive. It is likely that there are additional microenvironmental influences that promote and/or amplify RA/BDNF-mediated differentiation in a finely tuned spatiotemporal sequence of signaling gradients that interact with streaming NCCs as they enter the aROI. Further work will be necessary to discover a combination of signaling that is sufficient to induce NB differentiation in any microenvironment.
The near absence of cranial NB tumors in young patients (Vo et al., 2014) correlates with our findings that malignant, cranially migrating NB cells, including those harboring N-MYC amplification and/or ALK mutations, respond to in vivo cues that lead to differentiation and apoptosis. Rather than being restricted to the sympathoadrenal cell lineage, we postulate that NB may also be derived directly from mutated NCCs and found predominantly in the trunk in young patients due to a permissive microenvironment. Consistent with this idea, factors secreted from the adrenal cortex, where diagnosed NB is primarily localized, are known to inhibit neuronal fate determination (Unsicker et al., 1978) and thus may help maintain NB cells in an undifferentiated state. Meanwhile, anteriorly migrating NC-derived NB cells may experience a loss of malignancy, yielding an anatomical distribution dependent on distinct microenvironments rather than fate-restricted lineages. It may well be that these early tumors are not always detected and thus not fully accounted for in known categories of risk and progression.
Uncovering the mechanisms underlying RA’s effects in patients is critical to understanding its variable therapeutic results and to pursuing therapies that target downstream or parallel pathways with greater specificity and fewer detrimental side effects. Furthermore, the paucity of in vivo knowledge regarding NB’s rapid metastasis, unpredictable differentiation, and occasional spontaneous remission speaks to the value of a versatile model system. NB heterogeneity, in particular, has long proved a challenge for treatment strategies (Esposito et al., 2017). Our ability to differentiate both genetically distinct human NB cell lines and heterogeneous stage 4 PDXs illustrates the relevance of microenvironment-induced differentiation even to high-risk human NB. Moving forward, this model system will offer a unique platform for screening candidate drugs that could promote NB differentiation and death in vivo. Comparative studies of samples derived from different patients or a single patient’s primary tumor versus a recurrent tumor, correlated with individuals’ genetic profiles, could uncover valuable information regarding patient subpopulations that are more or less inclined to respond to differentiation-promoting therapies.
Limitations of the study
These experiments were performed in zebrafish and mouse embryo experimental systems, whose spatiotemporally dynamic microenvironments will differ to some extent from those found in humans. Furthermore, although several human cell lines and patient-derived xenograft samples were used, these experimental models are inherently limited in reflecting the vast heterogeneity and diversity of tumors observed in NB patients.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Ankur Saxena (saxenaa@uic.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact.
Data and Code Availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish model
AB Zebrafish were treated and cared for in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory animals. All experiments were approved by the University of Illinois at Chicago Institutional Animal Care Committee. Zebrafish were housed in a secure aquatic facility in 3.5 L tanks with 10–20 adults per tank, daily health monitoring, and a diet of Skretting’s GEMMA Micro 75, 150, or 300, depending on age/size. Adults were not used for experiments, only as a source of fertilized eggs. Zebrafish embryos were grown, staged, and harvested as previously described (Kimmel et al., 1995; Westerfield, 2000) in egg water (15 mM NaCl, 8.3 mM CaSO4, methylene blue) at 28.5°C. The transgenic zebrafish line Tg(−4.9sox10:eGFP) was used in the AB wild-type background, and its abbreviation in the text is Sox10:eGFP (Wada et al., 2005). The sex of embryos was not determined; only embryos were used for experiments, and therefore, sex distribution was random and roughly equal in all experimental groups and treatments.
Mouse model
All experiments were approved by the University of Illinois at Chicago Institutional Animal Care Committee and comply with the USPHS Policy on Humane Care and Use of Laboratory Animals. Mice used in this manuscript were wildtype C57BL6/J mice from Jackson Laboratories (strain 000664). Mice were maintained as breeding trios in a barrier facility under standard housing conditions with daily health monitoring, weekly cage and bedding changes, and chewable nestlet enrichment. Mating mice were maintained on standard chow (Teklad). Virgin females were placed in mating cages and checked for plugs daily until mating had occurred, followed by removal to a second cage. The day mating plugs were observed was considered E0.5. At E10.5, mice were euthanized by CO2 inhalation followed by confirmatory cervical dislocation, and embryos were removed. The sex of embryos was not determined; only embryos were used for experiments, and therefore, sex distribution was random and roughly equal in all experimental groups and treatments.
Cell culture
Cells were cultured at 37°C with 5% CO2 in the following media (cell line in parentheses): DMEM with 10% FBS (HEK293 and GIMEN); DMEM with 10% FBS, 500 μM G418, and 2 μM oligomycin (SK-N-AS); RPMI with 10% FBS (KELLY); F12/DMEM with 10% FBS and 1x penicillin-streptomycin (SH-SY5Y); RPMI with 10% FBS, 250μM G418, and 1μM oligomycin (IMR-5); DMEM with 10% FBS and 800 μM G418 (A375); DMEM with 10% FBS and 1x penicillin-streptomycin (OVCAR-8). SK-N-AS cell lines (empty vector, ITSN1 scrambled vector, ITSN1 short hairpin 1 (sh1), ITSN1 short hairpin 2 (sh2)) and IMR-5 empty vector cell line were previously generated and validated by Russo et al. (Russo and O’Bryan, 2012). SK-N-AS and IMR-5 cell lines were stably transfected with mCherry-Far- nesyl-5 using Lipofectamine 2000 reagent according to manufacturer’s protocol. mCherry-Farnesyl-5 was a gift from Michael David- son (Addgene plasmid # 55045). Cells were selected using 500 mg/ml G418 (Geneticin). Following selection, colonies were pooled to generate a polyclonal cell line used for all subsequent analyses. Consistent with previous findings (Russo and O’Bryan, 2012; Russo et al., 2015), there was no observable difference in neuronal differentiation or projection lengths between cells with empty vector or scrambled vector nor between cells with either of the two short hairpins. PDX cells were thawed and plated in IMDM with 20% FBS, 4 mM L-glutamine, and 1x ITS (Sigma-Aldrich) for 2 to 7 days and injected once adherence was observed.
METHOD DETAILS
In vitro treatment and transfection
For ITSN1 rescue, SK-N-AS sh2 cells were stably transfected with an expression vector encoding CFP-tagged mouse ITSN1s (short isoform) and selected in G418. Following selection, colonies were pooled to generate a polyclonal cell line used for all subsequent analyses. Rescue was confirmed with an anti-ITSN1 polyclonal antibody (Russo and O’Bryan, 2012) (Figure S7F). All transient transfections of mCherry-Farnesyl-5 (Addgene plasmid #55045) and mCherry-Lifeact-7 (Addgene plasmid # 54491) for cell visualization and/or −2.2NEUROD1:eGFP (−2.2kb human NEUROD1 promoter driving eGFP cloned into a pcDNA3.1 vector by GenScript) for detection of early neurogenesis used Lipofectamine 3000 reagent according to manufacturer’s protocol. For RA treatment of plated cells, media on the plate was exchanged with complete media supplemented with 5 μM all-trans RA in 0.0375% DMSO 24 hours prior to cell collection, consistent with previous work (Guglielmi et al., 2014; Singh et al., 2011). For ANA-12 treatment, cells in suspension were treated for 30 minutes with 10 μM of ANA-12 in 1% DMSO in complete media followed by addition to the embryos and imaging cassette post-injection, consistent with previous work (Liang et al., 2019). For K252a treatment, cells were treated for 3 hours in culture at 1 μM in 1% DMSO in complete media followed by addition to the embryos at a concentration of 1 μM in 1% DMSO in complete egg water and to the imaging cassette post-injection at a concentration of 1 μM in 1% DMSO in imaging mold media (see below for details), consistent with previous work (Buck and Winter, 1996). For treatment with LY294002, a reversible inhibitor of PI3K, 100 μM in 1% DMSO was added 2 hours to complete media prior to cell collection, consistent with previous work (Fishwick et al., 2010). For treatment of plated cells with wortmannin, an irreversible inhibitor of PI3K, 100 nM or 1 μM (the latter when host embryos were treated with RA) in 1% DMSO was added 24 hours prior to cell collection in complete media, consistent with previous work (Kim et al., 2004; Misawa et al., 2003).
Zebrafish embryo injections and live confocal imaging
Zebrafish embryos were staged as described in Kimmel et al. (Kimmel et al., 1995), and embryos at 11–12 hours post-fertilization (hpf), i.e. at 3–6 somite stages, were selected for injection, dechorionated with 0.3 mg/mL pronase in egg water, embedded in 0.9% low melting agarose (LMA) in 30% Danieau, and submerged in egg water. Cells at ~75% confluency were collected from culture dishes (for cells to be labeled with Hoechst 34580, a 1–1000 dilution in media was prepared and incubated at 37°C for 30 minutes prior to collection in complete media; PDX cells were also labeled with CellTracker CM-DiI (Invitrogen) at 3 ug/ml in media without FBS), suspended in media without antibiotics, allowed to settle, aspirated, placed into a new petri dish, and loaded into glass capillaries on a Nanoject III microinjector (Drummond Scientific). 0.7–1 nL (~5–10 cells) was injected into each zebrafish embryo at 12 hpf. Cells xenotransplanted for analysis in the future aROI and pROI were injected posterior to the developing eye where the NC stream splits (Figure 1A), while cells xenotransplanted into the trunk were injected into the presumptive 7th and 8th somites, consistent with the origin of the sympathoadrenal NC lineage (Corallo et al., 2016). Embryos were extracted from LMA and placed in egg water at 28.5°C for 1 hour to recover. Healthy embryos with injected cells in the desired location and desired cell density/number were held in position overnight using a protocol combining Kaufmann et al.’s approach (0.3% LMA in 30% Danieau with 0.014% tricaine) (Kaufmann et al., 2012) with Saxena et al.’s imaging molds and cassette (Saxena et al., 2013): The inverse shape of a zebrafish embryo was used as a template to create multi-position imaging molds from 0.9% LMA in 30% Danieau. Embryos were suspended in 0.3% LMA in 30% Danieau with 0.014% tricaine, positioned in the mold, and placed briefly at 4°C to solidify the 0.3% LMA. Once solidified, a 2% methylcellulose solution in 30% Danieau with 0.014% tricaine was gently placed on top. For embryos treated with RA, 1 μM of RA and 0.1% DMSO was added to the imaging cassette (both in the LMA and the methylcellulose) (2–14 hpi) while embryos treated with DEAB had 50 μM of DEAB and 0.5% DMSO added to egg water beginning at 10 hpf (2 hours pre-injection) and continuing until 14 hpi. Embryos were warmed to 32°C for 30 minutes before imaging and were kept at 32°C while imaged every 30 minutes for 11–14 hours on a Zeiss LSM 800 confocal microscope with a 40X/1.1 W objective (used for all imaging described below). Time-lapse images were rendered and analyzed using Bitplane’s Imaris software.
Mouse embryo injections and culture
E10.5 mouse embryos were collected from wild-type C57BL/6 mice and placed in culture media containing KnockOut DMEM (Invitrogen), KnockOut Serum Replacement (10%, Invitrogen), N-2 Supplement (1x, Invitrogen), Bovine Serum Albumin (2%), Penicillin (50 IU/ml), and Streptomycin (50 μg/ml), as described previously (Kalaskar and Lauderdale, 2014), but without Amphotericin-B. Embryos were externalized from the visceral yolk sac and amnion, which remained attached, and injected with 5–25 SK-N-AS cells in a 1–5 nL volume. Injected embryos were cultured for 14 hours on a platform rocker at 10 rpm in a 37°C, humidified incubator with 5% CO2.
In situ and in vitro hybridization chain reaction (HCR)
In situ: Sox10:eGFP+ zebrafish embryos were fixed at 19 hpf (equivalent to 5 hpi; stage-matched using somites to account for temperature differences yielding different growth rates), and the HCR v3.0 protocol was used as previously described (Choi et al., 2018), with custom-designed probes targeting zebrafish BDNF mRNA (Molecular Instruments, Inc.) In vitro: Untreated SH-SY5Y and SK-N-AS cells and SK-N-AS cells treated with either DMSO or 10 μM RA were fixed and NTRK2 mRNA was detected with custom-designed probes from Molecular Instruments Inc. as previously described (Choi et al., 2018). For both in situ and in vitro HCR, cells were stained with DAPI (1:2000) to label nuclei. 2 pmol HCR probes were hybridized at 37°C overnight in hybridization buffer, followed by washes at 37°C. Probes were detected by annealing of fluorescent hairpins in amplification buffer overnight at room temperature, followed by washes with 5X SSC with 0.1% tween-20. DNA probes, fluorescent hairpins and buffers were purchased from Molecular Instruments Inc.
BDNF morpholino
Morpholinos were obtained from Gene Tools, LLC. A previously used translation-blocking antisense morpholino against BDNF (CCAGTCGTAAAGGAGACCATTCAGC) (Diekmann et al., 2009; Nozawa et al., 2017) was injected into one-cell stage embryos at a concentration of 2 ng per embryo alongside injections into clutchmates of the control morpholino (CCTCTTACCTCAGTTACAATT TATA). For rescue experiments, embryos previously injected at the one-cell stage with 2 ng BDNF morpholino were treated with 200 ng/ml of human BDNF from 4 hpf onwards.
Immunolabeling
14–15 hpi zebrafish embryos were fixed and immunostained as previously described (Rajan et al., 2018). Briefly, embryos were fixed in 4% PFA overnight at 4° C, rinsed repeatedly with 1x TBS, subjected to antigen retrieval with 150 mM Tris buffer at 70°C for 25 min, rinsed repeatedly in TBST (1x TBS with 0.1% tween-20), incubated in Blocking Solution (1x TBS with 1% tween, 1% BSA, 5% DMSO, 10% donkey serum) at room temperature for 3 hours, placed in primary solution (primary antibody in.1x Blocking Solution) overnight at 4° C, washed in TBST, placed in secondary solution (secondary antibody in.1x Blocking Solution) overnight at 4° C, washed in TBST, and prepared for imaging. Anti-neurofilament heavy chain (1–200; Genetex; GTX110065) primary antibody and donkey-anti-rabbit 647 (1:1000; ThermoFisher) or donkey-anti-rabbit 488 (1:1000; ThermoFisher) secondary antibodies were used. NB cells retained their mCherry fluorescence. For 6 and 24 hpi embryos, a similar technique was used but without antigen retrieval and with an additional methanol dehydration series and acetone permeabilization prior to block: After fixation, embryos were placed in sequentially increasing methanol solutions for 5 min each (25%, 50%, 75%), placed at −20°C in 100% methanol for at least 20 minutes, rehydrated in sequentially decreasing methanol solutions for 5 min each (75%, 50%, and 25%), washed in 1X TBS, rinsed in DI water, placed in acetone for 20 minutes, rinsed in DI water, and rinsed in TBS followed by immunostaining as described above. Cell culture immunostaining was done similarly on cell culture slides following fixation at room temperature for 12 minutes with block, primary, secondary solutions at room temperature for an hour, and each wash for 5 minutes. 14 hpi mouse embryos were fixed with 4% paraformaldehyde or methanol/DMSO solution (4:1) and immunostained similar to above but without acetone and were cleared with a rapid clearing method based on triethanolamine and formamide (RTF) as described previously (Yu et al., 2018). Briefly, embryos were incubated in RTF-R1 solution (30% triethanolamine (TEA), 40% formamide (F), 30% water (W)) for 30 minutes and RTF-R2 solution (60%TEA/25%F/15%W) for 30 minutes. Anti-neurofilament heavy chain (1:500; Abcam; ab8135) and anti-RFP (1:500; Rockland; 200–101-379) primary antibodies and donkey-anti-rabbit 488 (1:1000; ThermoFisher) and donkey-anti-goat 647 (1:1000; ThermoFisher) were used. Both anti-neurofilament heavy chain primary antibodies were tested in vitro using SHSY-5Y NB cells as a positive control and secondary antibody only (no primary antibody) as a negative control.
Morphological and quantitative analysis of neuroblastoma migration, differentiation, and survival
Cell migration, behavior, and morphology were observed over the course of time-lapses using Imaris. Cell group locations were placed on scale diagrams of embryos using multiple landmarks such as NC streams and local anatomy, with reference to Kimmel’s standard descriptions (Kimmel et al., 1995), at the beginning (2 hpi) and end of each time-lapse (14 hpi). Groups of cells were binned as neuronal or non-neuronal based on neuronal morphology (elongated cell body, rapidly extending unipolar or bipolar projections of typical axonal diameter with evident growth cones, stability of projections) and lack of migration. Observed projections from cells binned as neuronal often lasted several hours, in contrast to shorter, transient projections in non-neuronal, migratory cells that persisted for minutes to less than an hour. Previously described projection length measurement methodology (Guglielmi et al., 2014) was adapted for in vivo use: Maximum length projections per embryo were identified in time-lapses, measured, and backtracked and followed to determine the time at which projections formed and the amount of time they remained. Projections that originated from cells pulling apart from each other were discarded. Maximal projection measurements were analyzed independently of morphological binning for all cell lines and PDX samples. Of note, no significant difference was observed between experimental conditions with SK-N-AS cells in any of the following: number of cells xenotransplanted, the time a neurite was initially observed, the total time a neurite remained visible, or the length of projections from neuronally classified cells. Projection measurements were compared using a two-tailed T-test.
For cell migration analysis, Imaris was used to manually track injected cells via nuclear labeling alongside cytoplasmically-labeled NCCs by annotating individual nuclei and connecting them to form migration tracks. Displacement vectors for NCCs were generated, and the Imaris reference tool’s z-plane was aligned with them. Average displacement over each time frame in the direction of net NC displacement was determined and summed for both injected cells and NCCs, and ratios of migration distance were derived and analyzed using two-tailed T-tests. Hoechst 34580-labeled nuclei that were not injected or NCCs were used as landmarks for drift correction using Imaris’s registration algorithm.
To assay cell survival, embryos with injected membrane-localized mCherry-expressing SK-N-AS cells, ITSN1-silenced (sh2) SK-N-AS cells, or SK-N-AS cells treated with LY294002 or wortmannin were surveyed and all cells were imaged and counted manually at 15 hpi, 48 hpi, and 72 hpi. Embryos were incubated at 32°C throughout the experiment in egg water when not imaged and in 0.9% LMA molds with 2–3% methylcellulose with 0.014% tricaine when imaged. Changes in cell number were calculated as a ratio in comparison to numbers at 15 hpi and evaluated using two-tailed T-test. Numbers of cells at 15 hpi were not significantly different between aROI and pROI.
Total displacements for each cell group were determined using the scaled schematics shown at 2 hpi and 14 hpi, which compensated for drift and embryonic growth. 2 hpi schematics were overlaid onto 14 hpi schematics using the posterior aspect of the developing eye as the anterior-posterior reference point and the edge of the yolk as the dorsal-ventral reference point. Arrows were drawn from the center point of cell groups in the 2 hpi schematics to the center point of cell groups in the 14 hpi schematics. Total displacement, net anterior displacement, and net dorsal displacement were measured. Measurements were aggregated to form vector diagrams and compared using two-tailed T-test.
NEUROD1:eGFP expression analysis
In vitro: NEUROD1:eGFP-transfected KELLY and SK-N-AS cells were treated with either 0.0375% DMSO as the control group or 10 μM RA/DMSO for 96 hours. 51.2 μm optical slices of 6 views of each cell line’s condition were captured. In vivo: NEUROD1:eGFP-transfected SK-N-AS cells were injected into wild-type zebrafish and imaged every 30 minutes for 14 hours as before. Fluorescence intensity quantitation was done for one representative cell from each of 10 embryos during 3–8 hpi using Imaris’s surface function.
PI3K/Akt activity biosensor
The PI3K biosensor GFP-AktPH (Haugh et al., 2000) was used to assay PI3K/Akt activity. Time-lapses were performed with cells transfected with GFP-AktPH/cytoplasmic mCherry and injected as described above. As before, the longest projection during the time-lapse was identified and tracked back to its point of initiation. Imaris’s Spots feature was used to create a 2 μm sphere directly adjacent to that point, and mean GFP intensity within that sphere was compared across experimental conditions using two-tailed T-tests. As an internal control for expression levels, the mean intensity of cytoplasmic mCherry was also compared using two-tailed T-tests; no significant difference was found across all experimental conditions. To measure GFP intensity over time, representative samples were chosen and GFP near the membrane where the eventual projection would form was measured in 2 μm spheres every 30 minutes from 2 hours prior to projection initiation until 2 hours after the projection first formed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of cell displacements, projection lengths and fluorescence intensity were performed with Bitplane’s Imaris software. All statistical analyses for significance were performed using two-tailed T-test for two-sample comparisons with Microsoft Excel or GraphPad Prism 9.1. n.s. p>0.05; * p<0.05; ** p<0.01; *** p<0.001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-neurofilament heavy chain | Genetex | Cat# GTX110065; RRID: AB_1950985 |
| Rabbit polyclonal anti-neurofilament heavy polypeptide | Abcam | Cat# ab8135; RRID: AB_306298 |
| Goat polyclonal anti-RFP | Rockland | Cat# 200-101-379; RRID: AB_2744552 |
| Rabbit polyclonal anti-ITSN1 antibody | Russo and O’Bryan, 2012; Laboratory of John P. O’Bryan, Medical University of South Carolina | N/A |
| Mouse monoclonal anti-β-tubulin antibody | Sigma-Aldrich | Cat# T4026; RRID: AB_477577 |
| Alexa Fluor 488 donkey anti-mouse IgG (H+L) | ThermoFisher | REF# A21202; RRID: AB_141607 |
| Alexa Fluor 488 donkey anti-rabbit IgG (H+L) | ThermoFisher | REF# A21206; RRID: AB_2535792 |
| Alexa Fluor 647 donkey anti-goat IgG (H+L) | ThermoFisher | REF# A21447; RRID: AB_2535864 |
| Alexa Fluor 647 donkey anti-mouse IgG (H+L) | ThermoFisher | REF# A31571; RRID: AB_162542 |
| Alexa Fluor 647 donkey anti-rabbit IgG (H+L) | ThermoFisher | REF# A31573; RRID: AB_2536183 |
|
| ||
| Biological samples | ||
|
| ||
| Patient-derived xenografts (PDXs) | Children’s Oncology Group Childhood Cancer Repository | COG557x; COG636x |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Lipofectamine 3000 Transfection Reagent | Invitrogen | Cat# L3000001 |
| Retinoic acid | Sigma-Aldrich | Cat# R2625 |
| ANA-12 | Sigma-Aldrich | Cat# SML0209 |
| K252a | Sigma-Aldrich | Cat# K1639 |
| LY294002 | Sigma-Aldrich | Cat# 440202 |
| Wortmannin | Sigma-Aldrich | Cat# W1628 |
| Agarose, low gelling temperature | Sigma-Aldrich | Cat# A0701 |
| Tricaine/MS-222 | Sigma-Aldrich | Cat# A5040 |
| Methyl cellulose | Sigma-Aldrich | Cat# M0512 |
| BDNF human | Sigma-Aldrich | Cat# SRP3014 |
| Triethanolamine | Sigma-Aldrich | Cat# 90279 |
| Formamide | Sigma-Aldrich | Cat# F7503 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Hoechst 34580 | Invitrogen | Cat# H21486 |
| CellTracker CM-DiI Dye | Invitrogen | Cat# C7001 |
| KnockOut DMEM | Invitrogen | Cat# 10829018 |
| KnockOut Serum Replacement | Invitrogen | Cat# 10828028 |
| N-2 Supplement | Invitrogen | Cat# 17502048 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Hybridization Chain Reaction Probes and Hairpins Version 3 | Molecular Instruments | N/A |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293 | Laboratory of John P. O’Bryan, Medical University of South Carolina | N/A |
| SK-N-AS | Laboratory of John P. O’Bryan, Medical University of South Carolina | N/A |
| KELLY | Laboratory of John P. O’Bryan, Medical University of South Carolina | N/A |
| SH-SY5Y | Laboratory of Gerardo Morfini, University of Illinois at Chicago | N/A |
| IMR-5 | Laboratory of John P. O’Bryan, Medical University of South Carolina | N/A |
| A375 | American Type Culture Collection | Cat# CRL-1619 |
| OVCAR-8 | Laboratory of Joanna E Burdette, University of Illinois at Chicago | N/A |
| GIMEN | Biohippo | Cat# 300179 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Zebrafish: AB wild-type | ZIRC | Cat# ZL1 |
| Zebrafish: Tg(−4.9sox10:eGFP) | Wada et al., 2005 | ZFIN ID: ZDB-FISH-150901-15643 |
| Mouse: C57BL/6J | Jackson Labs | Cat# 000664 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Translation-blocking morpholino against BDNF: CCAGTCGTAAAGGAGACCATTCAGC | Diekmann et al., 2009 | N/A |
| Control morpholino: CCTCTTACCTCAGTTACAATTTATA | Gene Tools | SKU:PCO-StandardControl-100 |
|
| ||
| Recombinant DNA | ||
|
| ||
| mCherry-Farnesyl-5 | Laboratory of Michael Davidson, The Florida State University | Addgene plasmid # 55045 |
| mCherry-Lifeact-7 | Laboratory of Michael Davidson, The Florida State University | Addgene plasmid # 54491 |
| GFP-AktPH | Laboratory of Tobias Meyer, Stanford University (Haugh et al., 2000) | N/A |
| −2.2NEUROD1:eGFP | This paper | N/A |
| murine ITSN1s-CFP | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Imaris | Bitplane | https://imaris.oxinst.com/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
Neuroblastoma cells comigrate with the neural crest into divergent microenvironments
Neuroblastoma cells can undergo region-specific neuronal differentiation and death
Microenvironment-derived retinoic acid and BDNF are required for differentiation
Cell-autonomous ITSN1-PI3K signaling is required for differentiation, likely via Akt
ACKNOWLEDGMENTS
We thank Dr. Robert Kelsh for the Tg(−4.9sox10:eGFP) zebrafish line, Dr. Tobias Meyer for the GFP-AktPH construct, Dr. Michael Davidson for the mCherry-Lifeact-7 and mCherry-Farnesyl-5 constructs, the Children’s Oncology Group Childhood Cancer Repository for PDX samples, the Childhood Solid Tumor Network (St. Jude Children’s Research Hospital), and Dr. Cé line Cluzeau (UIC) for valuable technical feedback and insights, and Dr. Deepak Srivastava and Dr. Abigail Green-Saxena for feedback on the manuscript. Research funding was provided by University of Illinois Lab Startup Funds, and publication costs were defrayed in part by a University of Illinois Cancer Center Mini-Grant.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.09.014.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Barone G, Anderson J, Pearson AD, Petrie K, and Chesler L. (2013). New strategies in neuroblastoma: therapeutic targeting of MYCN and ALK. Clin. Cancer Res. 19, 5814–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM, Minturn JE, Ho R, Simpson AM, Iyer R, Varela CR, Light JE, Kolla V, and Evans AE (2009). Trk receptor expression and inhibition in neuroblastomas. Clin. Cancer Res. 15, 3244–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner ME, and LeDouarin NM (2012). Development and evolution of the neural crest: an overview. Dev. Biol. 366, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn MA, Pearson RB, Hannan RD, and Sheppard KE (2013). AKT-independent PI3-K signaling in cancer - emerging role for SGK3. Cancer Manag. Res. 5, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck H, and Winter J. (1996). K252a modulates the expression of nerve growth factor-dependent capsaicin sensitivity and substance P levels in cultured adult rat dorsal root ganglion neurones. J. Neurochem. 67, 345–351. [DOI] [PubMed] [Google Scholar]
- Casey MJ, and Stewart RA (2018). Zebrafish as a model to study neuroblastoma development. Cell Tissue Res 372, 223–232. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corallo D, Candiani S, Ori M, Aveic S, and Tonini GP (2016). The zebrafish as a model for studying neuroblastoma. Cancer Cell Int 16, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Scappini E, Martin NP, Wong KA, Dunn S, Chen YJ, Miller SL, Domin J, and O’Bryan JP (2007). Regulation of neuron survival through an intersectin-phosphoinositide 3′-kinase C2beta-AKT pathway. Mol. Cell. Biol. 27, 7906–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delloye-Bourgeois C, Bertin L, Thoinet K, Jarrosson L, Kindbeiter K, Buffet T, Tauszig-Delamasure S, Bozon M, Marabelle A, Combaret V, et al. (2017). Microenvironment-driven shift of cohesion/detachment balance within tumors induces a switch toward metastasis in neuroblastoma. Cancer Cell 32, 427–443, e8. [DOI] [PubMed] [Google Scholar]
- Diekmann H, Anichtchik O, Fleming A, Futter M, Goldsmith P, Roach A, and Rubinsztein DC (2009). Decreased BDNF levels are a major contributor to the embryonic phenotype of huntingtin knockdown zebrafish. J. Neurosci. 29, 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsjö A, Lavenius E, Nilsson H, Hoehner JC, Simonsson P, Culp LA, Martinsson T, Larsson C, and På hlman S. (2003). Expression of trkB in human neuroblastoma in relation to MYCN expression and retinoic acid treatment. Lab. Invest. 83, 813–823. [DOI] [PubMed] [Google Scholar]
- Esposito MR, Aveic S, Seydel A, and Tonini GP (2017). Neuroblastoma treatment in the post-genomic era. J. Biomed. Sci. 24, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishwick KJ, Li RA, Halley P, Deng P, and Storey KG (2010). Initiation of neuronal differentiation requires PI3-kinase/TOR signalling in the vertebrate neural tube. Dev. Biol. 338, 215–225. [DOI] [PubMed] [Google Scholar]
- Gallik KL, Treffy RW, Nacke LM, Ahsan K, Rocha M, Green-Saxena A, and Saxena A. (2017). Neural crest and cancer: divergent travelers on similar paths. Mech. Dev. 148, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti AR, Di Cataldo A, Sorrentino S, Parodi S, Rizzo A, Buffa P, Granata C, Sementa AR, Fagnani AM, Provenzi M, et al. (2009). Neuroblastoma in the newborn. A study of the italian neuroblastoma registry. Eur. J. Cancer 45, 3220–3227. [DOI] [PubMed] [Google Scholar]
- Guglielmi L, Cinnella C, Nardella M, Maresca G, Valentini A, Mercanti D, Felsani A, and D’Agnano I. (2014). MYCN gene expression is required for the onset of the differentiation programme in neuroblastoma cells. Cell Death Dis 5, e1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugh JM, Codazzi F, Teruel M, and Meyer T. (2000). Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J. Cell Biol. 151, 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henion PD, and Weston JA (1994). Retinoic acid selectively promotes the survival and proliferation of neurogenic precursors in cultured neural crest cell populations. Dev. Biol. 161, 243–250. [DOI] [PubMed] [Google Scholar]
- Hoehner JC, Olsen L, Sandstedt B, Kaplan DR, and Påhlman S. (1995). Association of neurotrophin receptor expression and differentiation in human neuroblastoma. Am. J. Pathol. 147, 102–113. [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Gu X, Dong Y, Tan F, Liu Z, Thiele CJ, and Li Z. (2016). PI3K and MAPK pathways mediate the BDNF/TrkB-increased metastasis in neuroblastoma. Tumour Biol 37, 16227–16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, and Morita T. (1995). Role of retinoic acid in mouse neural crest cell development in vitro. Dev. Dyn. 204, 211–218. [DOI] [PubMed] [Google Scholar]
- Janku F, Yap TA, and Meric-Bernstam F. (2018). Targeting the PI3K pathway in cancer: are we making headway? Nat. Rev. Clin. Oncol. 15, 273–291. [DOI] [PubMed] [Google Scholar]
- Jiang M, Stanke J, and Lahti JM (2011). The connections between neural crest development and neuroblastoma. Curr. Top. Dev. Biol. 94, 77–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaskar VK, and Lauderdale JD (2014). Mouse embryonic development in a serum-free whole embryo culture system. J. Vis. Exp. 85, 50803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Matsumoto K, Lucarelli E, and Thiele CJ (1993). Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Eukaryotic Signal Transduction Group. Neuron 11, 321–331. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Kulesa PM, and Lefcort F. (2005). Imaging neural crest cell dynamics during formation of dorsal root ganglia and sympathetic ganglia. Development 132, 235–245. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann A, Mickoleit M, Weber M, and Huisken J. (2012). Multilayer mounting enables long-term imaging of zebrafish development in a light sheet microscope. Development 139, 3242–3247. [DOI] [PubMed] [Google Scholar]
- Kendall GC, Watson S, Xu L, LaVigne CA, Murchison W, Rakheja D, Skapek SX, Tirode F, Delattre O, and Amatruda JF (2018). PAX3-FOXO1 transgenic zebrafish models identify HES3 as a mediator of rhabdomyosarcoma tumorigenesis. eLife 7, e33800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang J, Qiao J, Thomas RP, Evers BM, and Chung DH (2004). Phosphatidylinositol 3-kinase inhibition down-regulates survivin and facilitates TRAIL-mediated apoptosis in neuroblastomas. J. Pediatr. Surg. 39, 516–521. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, and Schilling TF (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Liang J, Deng G, and Huang H. (2019). The activation of BDNF reduced inflammation in a spinal cord injury model by TrkB/p38 MAPK signaling. Exp. Ther. Med. 17, 1688–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl DJ, Klesse LJ, Tessarollo L, Wohlman T, and Parada LF (2000). Loss of brain-derived neurotrophic factor-dependent neural crest-derived sensory neurons in neurotrophin-4 mutant mice. Proc. Natl. Acad. Sci. USA 97, 2297–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum T, Huynh G, and Heinrich G. (2001). Brain-derived neurotrophic factor and TrkB tyrosine kinase receptor gene expression in zebrafish embryo and larva. Int. J. Dev. Neurosci. 19, 569–587. [DOI] [PubMed] [Google Scholar]
- Marshall GM, Carter DR, Cheung BB, Liu T, Mateos MK, Meyerowitz JG, and Weiss WA (2014). The prenatal origins of cancer. Nat. Rev. Cancer 14, 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, and Weiss WA (2016). Neuroblastoma. Nat. Rev. Dis. Primers 2, 16078. [DOI] [PubMed] [Google Scholar]
- Matthay KK, Villablanca JG, Seeger RC, Stram DO, Harris RE, Ramsay NK, Swift P, Shimada H, Black CT, Brodeur GM, et al. (1999). Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N. Engl. J. Med. 341, 1165–1173. [DOI] [PubMed] [Google Scholar]
- Misawa A, Hosoi H, Tsuchiya K, and Sugimoto T. (2003). Rapamycin inhibits proliferation of human neuroblastoma cells without suppression of MycN. Int. J. Cancer 104, 233–237. [DOI] [PubMed] [Google Scholar]
- Nakagawara A. (1998). The NGF story and neuroblastoma. Med. Pediatr. Oncol. 31, 113–115. [DOI] [PubMed] [Google Scholar]
- Nozawa K, Lin Y, Kubodera R, Shimizu Y, Tanaka H, and Ohshima T. (2017). Zebrafish Mecp2 is required for proper axonal elongation of motor neurons and synapse formation. Dev. Neurobiol. 77, 1101–1113. [DOI] [PubMed] [Google Scholar]
- Olsen RR, Otero JH, Garcıá-Ló pez J, Wallace K, Finkelstein D, Rehg JE, Yin Z, Wang YD, and Freeman KW (2017). MYCN induces neuroblastoma in primary neural crest cells. Oncogene 36, 5075–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz-Edwards A, Hardison NL, and Linney E. (2001). Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev. Biol. 229, 89–101. [DOI] [PubMed] [Google Scholar]
- Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR, et al. (2015). Advances in risk classification and treatment strategies for neuroblastoma. J. Clin. Oncol. 33, 3008–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan SG, Gallik KL, Monaghan JR, Uribe RA, Bronner ME, and Saxena A. (2018). Tracking neural crest cell cycle progression in vivo. Genesis 56, e23214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Brodeur GM, Dale RC, and Schor NF (2016). The “neuro” of neuroblastoma: neuroblastoma as a neurodevelopmental disorder. Ann. Neurol. 80, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff T, Tsarovina K, Majdazari A, Schmidt M, del Pino I, and Rohrer H. (2010). Neuroblastoma phox2b variants stimulate proliferation and dedifferentiation of immature sympathetic neurons. J. Neurosci. 30, 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CP, Matthay KK, Villablanca JG, and Maurer BJ (2003). Retinoid therapy of high-risk neuroblastoma. Cancer Lett 197, 185–192. [DOI] [PubMed] [Google Scholar]
- Russo A, and O’Bryan JP (2012). Intersectin 1 is required for neuroblastoma tumorigenesis. Oncogene 31, 4828–4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Okur MN, Bosland M, and O’Bryan JP (2015). Phosphatidylinositol 3-kinase, class 2 beta (PI3KC2beta) isoform contributes to neuroblastoma tumorigenesis. Cancer Lett 359, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago R, Vairy S, Sinnett D, Krajinovic M, and Bittencourt H. (2017). Novel therapy for childhood acute lymphoblastic leukemia. Expert Opin. Pharmacother. 18, 1081–1099. [DOI] [PubMed] [Google Scholar]
- Saxena A, Peng BN, and Bronner ME (2013). Sox10-dependent neural crest origin of olfactory microvillous neurons in zebrafish. eLife 2, e00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte JH, Lindner S, Bohrer A, Maurer J, De Preter K, Lefever S, Heukamp L, Schulte S, Molenaar J, Versteeg R, et al. (2013). MYCN and ALKF1174L are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene 32, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Shimozono S, Iimura T, Kitaguchi T, Higashijima S, and Miyawaki A. (2013). Visualization of an endogenous retinoic acid gradient across embryonic development. Nature 496, 363–366. [DOI] [PubMed] [Google Scholar]
- Sieber-Blum M. (1991). Role of the neurotrophic factors BDNF and NGF in the commitment of pluripotent neural crest cells. Neuron 6, 949–955. [DOI] [PubMed] [Google Scholar]
- Sieber-Blum M, Ito K, Richardson MK, Langtimm CJ, and Duff RS (1993). Distribution of pluripotent neural crest cells in the embryo and the role of brain-derived neurotrophic factor in the commitment to the primary sensory neuron lineage. J. Neurobiol. 24, 173–184. [DOI] [PubMed] [Google Scholar]
- Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G, Stewart J, Zage P, and Gopalakrishnan V. (2011). Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(beta-TrCP) in neuroblastoma cells. Cancer 117, 5189–5202. [DOI] [PubMed] [Google Scholar]
- Teillet MA, Kalcheim C, and Le Douarin NM (1987). Formation of the dorsal root ganglia in the avian embryo: segmental origin and migratory behavior of neural crest progenitor cells. Dev. Biol. 120, 329–347. [DOI] [PubMed] [Google Scholar]
- Theveneau E, and Mayor R. (2012). Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 366, 34–54. [DOI] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, and Thisse C. (2004). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol 77, 505–519. [DOI] [PubMed] [Google Scholar]
- Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, and Hendrix MJ (2006). Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat. Med. 12, 925–932. [DOI] [PubMed] [Google Scholar]
- Unsicker K, Krisch B, Otten U, and Thoenen H. (1978). Nerve growth factor-induced fiber outgrowth from isolated rat adrenal chromaffin cells: impairment by glucocorticoids. Proc. Natl. Acad. Sci. USA 75, 3498–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nes J, Chan A, van Groningen T, van Sluis P, Koster J, and Versteeg R. (2013). A notch3 transcriptional module induces cell motility in neuroblastoma. Clin. Cancer Res. 19, 3485–3494. [DOI] [PubMed] [Google Scholar]
- Vo KT, Matthay KK, Neuhaus J, London WB, Hero B, Ambros PF, Nakagawara A, Miniati D, Wheeler K, Pearson AD, et al. (2014). Clinical, biologic, and prognostic differences on the basis of primary tumor site in neuroblastoma: a report from the international neuroblastoma risk group project. J. Clin. Oncol. 32, 3169–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Hartmann P, and Zintl F. (2000). Differentiation, proliferation and adhesion of human neuroblastoma cells after treatment with retinoic acid. Cell Adhes. Commun. 7, 423–440. [DOI] [PubMed] [Google Scholar]
- Wada N, Javidan Y, Nelson S, Carney TJ, Kelsh RN, and Schilling TF (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977–3988. [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), (Fourth Edition) (University of Oregon Press; ). [Google Scholar]
- Yu T, Zhu J, Li Y, Ma Y, Wang J, Cheng X, Jin S, Sun Q, Li X, Gong H, et al. (2018). RTF: a rapid and versatile tissue optical clearing method. Sci. Rep. 8, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL, Rodig SJ, Neuberg DS, Helman D, Feng H, et al. (2012). Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 21, 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.