Abstract
OBJECTIVES:
We sought to assess whether genetic associations with metabolite concentrations in septic shock patients could be used to identify pathways of potential importance for understanding sepsis pathophysiology.
DESIGN:
Retrospective multicenter cohort studies of septic shock patients.
SETTING:
All participants who were admitted to 27 participating hospital sites in three countries (Australia, New Zealand, and the United Kingdom) were eligible for inclusion.
PATIENTS:
Adult, critically ill, mechanically ventilated patients with septic shock (n = 230) who were a subset of the Adjunctive Corticosteroid Treatment in Critically Ill Patients with Septic Shock trial (ClinicalTrials.gov number: NCT01448109).
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
A genome-wide association study was conducted for a range of serum metabolite levels for participants. Genome-wide significant associations (p ≤ 5 × 10–8) were found for the two major ketone bodies (3-hydroxybutyrate [rs2456680] and acetoacetate [rs2213037] and creatinine (rs6851961). One of these single-nucleotide polymorphisms (SNPs) (rs2213037) was located in the alcohol dehydrogenase cluster of genes, which code for enzymes related to the metabolism of acetoacetate and, therefore, presents a plausible association for this metabolite. None of the three SNPs showed strong associations with risk of sepsis, 28- or 90-day mortality, or Acute Physiology and Chronic Health Evaluation score (a measure of sepsis severity).
CONCLUSIONS:
We suggest that the genetic associations with metabolites may reflect a starvation response rather than processes involved in sepsis pathophysiology. However, our results require further investigation and replication in both healthy and diseased cohorts including those of different ancestry.
Keywords: genetics, metabolomics, molecular biology, sepsis
KEY POINTS
Question: The genetic etiology of circulating metabolite levels in a septic shock cohort.
Findings: Genome-wide association studies were conducted with measured metabolite levels as the phenotype of interest in a septic shock cohort. Significant results were reported for two ketone bodies (3-hydroxybutyrate and acetoacetate) and creatinine.
Meanings: The findings suggest that these metabolite changes are either an association with sepsis pathophysiology or a reflection of a starvation response.
Metabolites are small molecules that reflect biological processes that have long been used as markers for diagnosis, prognosis, and evaluation of treatment efficacy within clinical medicine (1, 2). Circulating blood metabolites typically exhibit significant heritabilities (3), although substantial variability exists in estimates depending upon the particular metabolite assayed (2, 4–6). Genome-wide association studies (GWAS) have identified hundreds of quantitative trait loci robustly associated with serum/plasma levels of a wide range of metabolites (1, 2, 6–15). Perhaps unsurprisingly, the genetic variants reported in these studies have often mapped to genes that encode enzymes, metabolite transporters, and regulators of metabolism (6). These have included variants that have been previously identified in GWAS of common complex diseases, providing a potential upstream mechanism for (or downstream consequence of) the underlying disease associations (6, 8). Given that the genetic variants associated with metabolomic phenotypes typically display much larger effect sizes than those associated with common complex diseases (6), it has been proposed that large-scale GWAS of metabolites could provide a useful adjunct and powerful discovery tool for improved understanding of the molecular pathways underlying common diseases.
Previous GWAS of metabolite concentrations have predominantly been performed in large (healthy) population-based cohorts (1, 2, 6–15). However, it is likely that the genetic etiology of metabolite concentrations differs according to the health status of participants. For example, a range of complex diseases exhibit metabolite patterns that are vastly different from those in healthy populations (16). Identifying genetic variants of relatively large effect that are associated with dysregulated metabolite levels may therefore provide a complementary strategy for identifying genetic loci and biological pathways important in disease pathogenesis.
In the present study, the genetic basis of serum metabolite levels in a sample of individuals with septic shock was investigated. Sepsis is an abnormal host response to infection that results in tissue and organ damage (17). To date, there have only been a small number of sepsis GWAS (18–22). These have yielded a handful of genome-wide significant loci that have not replicated upon subsequent investigation (18). Septic shock represents the most severe form of sepsis, with a significantly higher mortality rate due to circulatory and metabolic pathologies. This study was designed to assess whether the genetic etiology of metabolite concentrations in septic shock patients differed from the genetic etiology of metabolites in population-based samples, and further whether focusing on genetic variation associated with dysregulated metabolite concentrations could be used to identify pathways that may be important in terms of sepsis pathophysiology.
MATERIALS AND METHODS
Study Participants and Sampling
The Adjunctive Corticosteroid Treatment in Critically Ill Patients with Septic Shock (ADRENAL) trial is an international randomized double-blinded placebo-controlled trial designed to assess the utility of IV hydrocortisone treatment compared with placebo in critically ill, mechanically ventilated patients with septic shock (ClinicalTrials.gov number: NCT01448109). The ADRENAL-Gene Expression Study (GEPS) is a prespecified substudy of the ADRENAL trial designed to investigate the genetics and genomics of septic shock (23). The ADRENAL-GEPS study was approved by institutional review board (Board Name: University of Queensland Human Research Ethics Committee B; Approval Number: 2015000018; Approval Date: March 11, 2017; Study Title: Gene expression profiling in critically ill patients with septic shock: The ADRENAL-GEPS study). Previous written informed consent or written consent to consent to continue was obtained for all participants, according to the legal requirements in each jurisdiction, concurrently with consent for the ADRENAL trial. The study was conducted in accordance with the Helsinki Declaration of 1975. All ADRENAL participants who were admitted to 27 participating hospital sites in three countries (Australia, New Zealand, and the United Kingdom) were eligible for inclusion in ADRENAL-GEPS. The inclusion/exclusion criteria are summarized in the Supplementary Methods S1 (http://links.lww.com/CCX/B290) (18, 23). Blood samples from ADRENAL-GEPS participants (n = 578) were collected at the time of randomization before administration of corticosteroids/placebo. Blood was collected into 2 × 2.5 mL EDTA, 2 × 2.5 mL serum blood collection vacuettes (Cat. No. 455071; Interpath, Somerset, Australia), and 1 × 2.5 mL PAXgene RNA Vacutainer (Cat. No. 762165; Becton Dickinson, Franklin Lakes, New Jersey). The present study describes 230 fully genotyped individuals of White European ancestry who were randomly selected from the ADRENAL-GEPS substudy to have their serum nuclear magnetic resonance (NMR) metabolome profiles measured. The exclusion of individuals of different ancestry from analyses is standard procedure in GWAS because allelic frequency differences between populations can produce spurious associations, and linkage disequilibrium patterns can vary across ancestries and obscure/complicate the interpretation of genetic signals.
Genotyping
DNA extractions were performed on 200 µL of whole blood using a QIAsymphony (QIAGEN, Venlo, The Netherlands) SP instrument according to the manufacturer’s protocol. Genomic DNA was eluted in 100 µL of buffer ATE and quantified using the Trinean Dropsense 96. Samples were genotyped on the Illumina Infinium Global Screening Array (GSA)-24+ v1.0 (Illumina, San Diego, California). The arrays were scanned on an Illumina iScan system, and the raw fluorescence intensity data were normalized and clustered for each sample using Illumina Genome Studio (v 2.0.3) (Illumina, San Diego, California). Genotypes were called using the standard Illumina GSA-24v1-0_A6 Cluster File (Illumina, San Diego, California).
The PLINK v1.90b3.31 software package (https://pngu.mgh.harvard.edu/purcell/plink/) was used to carry out a number of standard quality control (QC) procedures on the entire ADRENAL-GEPS cohort (24). Pre-imputation QC procedures included excluding one of each pair of cryptically related individuals (genome-wide proportion of alleles identical by descent > 0.185), high autosomal heterozygosity (F > 0.2), sex inconsistencies, low minor allele frequency (MAF < 1%), low genotyping rate (< 95%), and departure from Hardy-Weinberg equilibrium (p < 10–6). Imputation was performed against the HRCr1.1 panel using the Michigan Imputation Server (https://imputationserver.sph.umich.edu) that implements Eagle v2 for phasing and IMPUTE2 for imputation (25). Post-imputation QC was performed using the same thresholds. Additionally, single-nucleotide polymorphisms (SNPs) with low imputation quality (R2 < 0.2), ambiguous SNPs that may result in strand mix-ups (e.g., A/T and C/G SNPs), triallelic SNPs, and SNPs with MAF that differed by greater than 10% from HRCr1.1 allele frequencies were removed. For further details regarding QC procedures refer to D’Urso et al (18) and Supplementary Methods S2 (http://links.lww.com/CCX/B290).
NMR Metabolomic Spectra Collection
Serum samples were thawed at room temperature and left on ice for sample preparation. For each serum sample, a total volume of 200 μL for each sample was prepared consisting of a 100 μL aliquot of serum sample, 80 μL 63.75 mM sodium phosphate buffer (pH 7.4), and 20 μL D2o. The prepared serum samples (200 μL) were transferred to 3 mm NMR tubes. 1H NMR spectra were acquired on a Bruker AV900 NMR spectrometer operating at a 1H frequency of 900.13 MHz and equipped with a 5-mm self-shielded z-gradient triple resonance probe.
The sera 1H NMR spectra were collected at a temperature of 310 K. Both a 1D Carr-Purcell-Meiboom-Gill (1D CPMG) and a 1D 1H-Nuclear Overhauser Effect Spectroscopy (1D NOESY) spectrum were acquired for each of the serum samples included in the ADRENAL cohort. 1D CPMG spectra were acquired with the standard cpmgpr1d pulse sequence . A fixed spin-spin relaxation delay 2nτ of 80 ms (τ = 500 μs) was used to eliminate the broad signals from high molecular weight analytes. Water suppression irradiation was applied during the relaxation delay of 4.0 seconds. For sera 1D CPMG spectra 65536 data points with a spectral width of 20 ppm were collected into 56 transients and 16 dummy scans.
In addition to standard 1D CPMG spectra collection for serum samples 1D NOESY were also collected for potential investigation of lipid changes. For all ADRENAL serum samples a 1D NOESY spectrum was acquired with the standard noesypr1d pulse sequence . The water signal was suppressed by a continuous wave irradiation during both the relaxation delay 4.0 seconds and the mixing time (τm) of 100 ms. For serum 1D NOESY spectra 65536 data points with a spectral width of 20 ppm were collected into 32 transients and eight dummy scans.
For one of the pooled quality controls (PQCs) 2D spectrum were collected in the form of 1H-13C heteronuclear single quantum coherence (HSQC) and 1H-1H Total Correlation Spectroscopy (TOCSY). Both HSQC and TOCSY spectrum were used as an aid in metabolite identification.
Signal processing of 1H NMR spectra was performed in TopSpin, Version 3.6.2 (Bruker BioSpin, Rheinstetten, Germany). The free induction delays (FIDs) of the serum 1D CPMG spectra were multiplied by a sine window function shifted by π/2 before Fourier transformation. The serum 1D CPMG spectra were aligned to the anomeric proton of glucose (δ = 5.22 ppm). Phase and baseline correction were performed manually in TopSpin.
Metabolite Identification and Quantification
Metabolites in the sera 1D CPMG spectra were identified using Chenomx NMR Suite, Version 8.5.1 (Chenomx, Edmonton, AB, Canada) and the human metabolome database (26). The PQC HSQC spectrum was used to help confirm metabolite assignment. Quantification of identified metabolites was performed manually in Chenomx NMR Suite. For a list of identified metabolites with resonance assignments refer to Supplementary Table S1 (http://links.lww.com/CCX/B290).
No internal chemical standard was included in the serum samples, instead serum quantification was a relative measure, using the integrated region of formate as a reference in Chenomx Processor. The formate concentration Chenomx Processor referenced was a ratio of integrated regions of . Some metabolites could not be quantified in all collected 1H NMR samples due either to signal overlap or relative low concentration.
Metabolite GWAS
Outlying metabolite values that had a z score greater than three () were removed before analyses and distributions of metabolites were inverse normal transformed. For a list of metabolites and initial sample size with number of excluded outliers refer to Supplementary Table S2 (http://links.lww.com/CCX/B290). Ordinary least squares linear regression analysis was performed across the genome assuming an underlying additive genetic model as implemented in PLINK (24). The first five principal components (PCs) from a PC analysis of the cleaned ADRENAL-GEPS GWAS dataset were used as covariates as well as age and sex (18). PCs were calculated for the entire ADRENAL-GEPS cohort including individuals of non-European descent as well as individuals for whom metabolite data was not collected (n = 576) (18). Genome-wide significant SNPs from the current metabolite GWAS were looked up in a previously published GWAS of 28- and 90-day mortality, sepsis risk, and Acute Physiology and Chronic Health Evaluation (APACHE) score (a measure of disease severity) involving the entire ADRENAL-GEPS cohort (18). We also looked up our top genome-wide significant SNPs in PHENOSCANNER (http://www.phenoscanner.medschl.cam.ac.uk/) (27, 28) and the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI) GWAS catalog (https://www.ebi.ac.uk/gwas/) (29). Finally, genome-wide significant variants from the NHGRI-EBI GWAS catalog for 3-hydroxybutyrate, acetoacetate, and creatinine (i.e., the three metabolites that displayed genome-wide significant results) were checked for replication in our scan.
Gene- and Pathway-Based Analyses
Gene-based and gene-set enrichment analyses were performed using Multi-marker Analysis of GenoMic Annotation (MAGMA) as implemented by the Functional Mapping and Annotation of GWAS platform (fuma.ctglab.nl) (30). Gene-based tests were performed to estimate the degree of association between the total genetic variation in each gene and each metabolite phenotype (31). The significance threshold for MAGMA gene-based tests was set at p ≤ 2.655 × 10–6 given SNPs were mapped to 18830 protein-coding genes. The output of the gene-based tests, the gene p values, and gene correlation matrix were then used to conduct gene-set analyses. MAGMA implements a competitive gene-set analysis that uses the full distribution of SNP p values to determine whether genes in a particular gene set are more strongly associated with the phenotype of interest than other genes in the genome (31). MAGMA by default uses 10678 gene sets (curated gene sets: 4761, GO terms: 5917) from MsigDB v6.2. All MAGMA analyses were performed on summary level statistics. MAGMA gene-set analysis considers results significant for Bonferroni corrected p values (p ≤ 4.682 × 10–6).
Metabolites and Clinical Variables Analysis
Targeted metabolite analysis with select clinical variables as outcomes was performed. Outcomes of interest were those related to mortality of septic shock, 28- and 90-day mortality, and those related to renal function, prior renal replacement therapy (RRT), RRT post-randomization, and dialysis use, as a number of metabolites measured are known to be associated with renal function. Each metabolite was used as an explanatory variable for individual logistic regression models. Subsequent logistic regression models included similar clinical covariates as presented in the original ADRENAL trial (23). Clinical covariates that were included in addition to individual metabolite levels in the subsequent logistic regression models were patient sex, age, APACHE II, admission type (medical or surgical), trial site, and the use of RRT in the 24 hours before randomization.
Statistical Power Analyses
We estimated the statistical power (two-tailed α = 0.05) to detect an association between genome-wide significant genetic variants related to metabolite concentrations (i.e., variants for creatinine, 3-hydroxybutyrate, and acetone) and sepsis outcomes (i.e., APACHE II score and susceptibility to sepsis). For continuous outcomes (APACHE II score), we estimated statistical power using the Genetic Effects Power Calculator (https://zzz.bwh.harvard.edu/gpc/) (32). We assumed that each genome-wide significant variant explained the same proportion of variance in the relevant metabolite that was empirically estimated in our study, and then derived the expected variance explained in APACHE score given possible sizes of the causal relationship between metabolite level and APACHE score.
In the case of binary traits (i.e., susceptibility to sepsis), for each individual we simulated a SNP, a metabolite associated with the SNP (with variance explained equal to that which was empirically estimated in our study), and then a (standard normal) latent variable that represented liability to sepsis and was a function of the simulated metabolite. Individuals whose liability score exceeded a threshold based on a lifetime risk of sepsis (in this case, a threshold in the upper right tail of the distribution corresponding to 1.16% of the total area under the curve) (33) were coded as affected, while the remainder of simulated individuals were coded as controls. We performed rejection sampling until the same number of cases and controls as in the present study was obtained. We then performed logistic regression on each replicate. For all power calculations/simulations, we assumed perfect linkage disequilibrium between trait and marker loci and the same allele frequencies as in the article. We performed 1000 simulated replicates for each condition.
RESULTS
Genome-Wide Association Studies
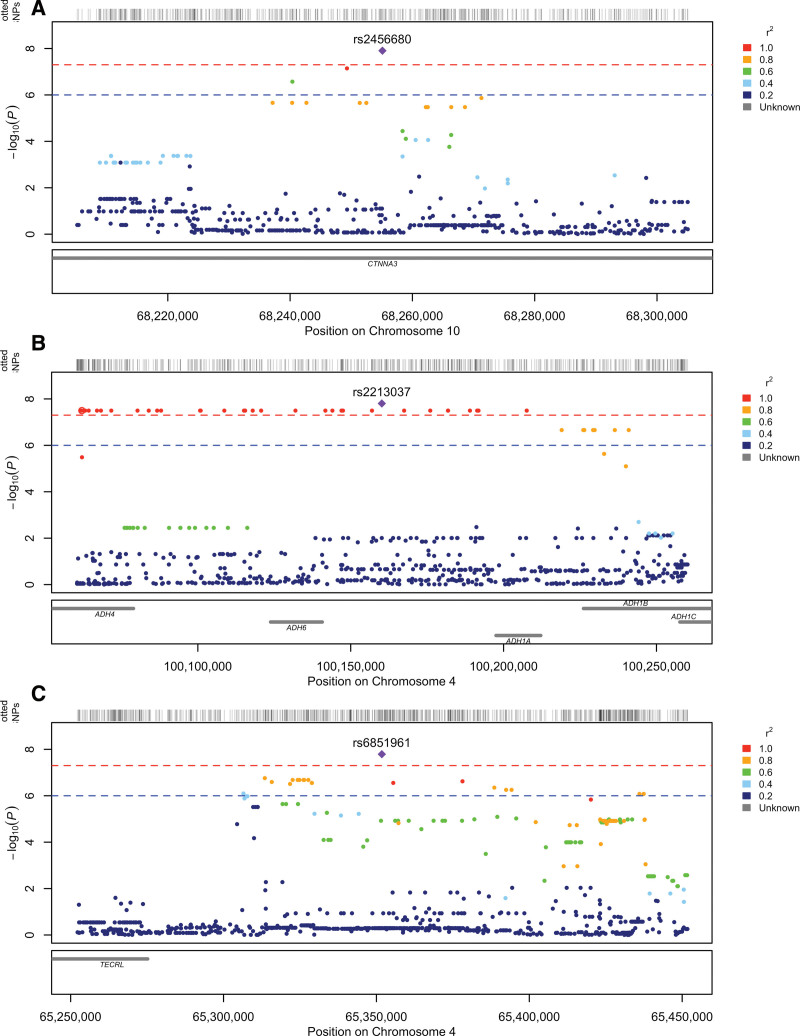
GWAS for 20 measured metabolites were conducted. Quantile-quantile plots (Supplementary Figs. S1–S20, http://links.lww.com/CCX/B290) and genomic inflation factors (Supplementary Table S3, http://links.lww.com/CCX/B290), which ranged from 0.985 to 1.007, were consistent with our GWAS being well controlled for population stratification and other possible biases. Three metabolite phenotypes, 3-hydroxybutyrate, acetoacetate, and creatinine, contained SNPs that passed the criterion for genome-wide significance (p ≤ 5 × 10–8). Manhattan plots for these three metabolites are presented in Supplementary Figures S21–S23 (http://links.lww.com/CCX/B290) with regional plots of significant SNPs presented in Figure 1. For Manhattan plots of the other metabolite, GWAS with no genome-wide significant SNPs refer to Supplementary Figures S24–S40 (http://links.lww.com/CCX/B290). The lead SNPs and their associated metabolite are shown in Table 1; and Supplementary Table S4 (http://links.lww.com/CCX/B290). For lead SNPs that did not reach genome-wide significance but reached the threshold for suggestive significance (p ≤ 1 × 10–5), refer to Supplementary Table S5 (http://links.lww.com/CCX/B290).
Figure 1.
Regional plots for metabolite GWAS with genome-wide significant SNPs. A, rs2456680 with association to 3-hydroxybutyrate levels. B, rs2213037 with association to acetoacetate levels. C, rs6851961 with association to creatinine levels.
TABLE 1.
Genome-Wide Significant Single-Nucleotide Polymorphisms With a p ≤ 5 × 10–8 From the Different Metabolite Genome-Wide Association Studies
| Genome-Wide Association Studies Phenotype | Single-Nucleotide Polymorphism | Chromosome: Base Pair Positiona | Variant Type | Minor Allele Frequency | Effect Allele/Other Allele | Beta (se) | p |
|---|---|---|---|---|---|---|---|
| 3-Hydroxybutyrate | rs2456680 | 10:68255097 | Intronic CTNNA3 | 0.48 | G/A | 0.60 (0.10) | 1.24 × 10–8 |
| Acetoacetate | rs2213037 | 4:100160177 | Intronic LOC100507053 | 0.97 | C/T | 1.72 (0.29) | 1.57 × 10–8 |
| Acetoacetate | rs10014222 | 4:100062064 | Intronic ADH4, Intronic LOC100507053 | 0.97 | G/A | 1.76 (0.030) | 3.19 × 10–8 |
| Creatinine | rs6851961 | 4:65351771 | Intergenic | 0.49 | G/A | 0.54 (0.09) | 1.62 × 10–8 |
The hg37 human genome build was used.
The only genetic variant that was significantly associated with serum levels of 3-hydroxybutyrate was the imputed SNP rs2456680. This SNP is located in an intronic region of a protein-coding gene CTNNA3 (Fig. 1A) and the effect allele “G” was associated with elevated levels of 3-hydroxybutyrate (p = 1.24 × 10–8; Table 1).
Acetoacetate had one lead SNP, rs2213037, that reached genome-wide significance. The effect allele, the C-allele, of the genotype SNP rs2213037 was associated with increased levels of acetoacetate (p = 1.57 × 10–8; Table 1). The rs2213037 SNP lies in an intronic region of a noncoding RNA gene LOC100507053 located among the alcohol dehydrogenase (ADH) cluster of genes (Fig. 1B). An additional SNP, rs10014222, which is in linkage disequilibrium with the leading acetoacetate GWAS SNP (r2 = 0.59), also reached genome-wide significance (p = 3.19 × 10–8) (Table 1). The rs10014222 SNP falls within overlapping gene region with both the intronic region of the protein-coding gene ADH4 and the intronic region of the noncoding RNA gene LOC100507053. It is of note that a number of SNPs associated with acetoacetate reach genome-wide significance with these SNPs falling across a number of genes, including several ADH genes (ADH1A, ADH1B, ADH4, ADH5, and ADH6).
The genotyped SNP rs6851961 was the only genetic variant that was significantly associated with levels of creatinine (p = 1.62 × 10–8; Table 1). This SNP did not fall in a reported gene region (Fig. 1C), the closest protein-coding gene being TECRL about 100 kb centromeric to rs6851961. Given that creatinine has a known association with renal function a second creatinine GWAS was performed with additional covariates (prior RRT, RRT post-randomization, and dialysis use). The same signal was still present after correction for renal function (results not shown).
Look-Up of Genome-Wide Significant Variants
The top SNPs at the three genome-wide significant loci were not associated with sepsis-related outcomes in the wider ADRENAL-GEPS cohort (p > 0.05) (Supplementary Table S6, http://links.lww.com/CCX/B290). Nor were they associated with phenotypes in the PHENOSCANNER (p > 1 × 10–5) or NHGRI-EBI GWAS databases (p > 5 × 10–8).
A list of SNPs that have shown evidence of association (p ≤ 1 × 10–5) with 3-hydroxybutyrate, acetoacetate, and creatinine was generated using the GWAS catalog and queried for replication (p < 0.05) in our corresponding metabolite GWAS summary statistics (Supplementary Table S7 and S8, http://links.lww.com/CCX/B290). Of these three metabolites, creatinine was the only one to have SNPs of previous studies replicated in our study. Creatinine had seven SNPs which replicated for a nominal p value (p < 0.05) taking into consideration direction of effect allele for both studies. However, given the large number of SNPs queried no replicated SNPs passed multiple testing correction (p < 8.36 × 10–5).
Gene-Based Test and Gene-Set Analysis (MAGMA)
None of the metabolite phenotypes produced significant results with gene-based tests implemented in MAGMA. The significant results from the competitive gene-set analysis, which was performed in MAGMA, are presented in Table 2. The only metabolite phenotype which reported a significant gene set that passed multiple testing corrections was 3-hydroxybutyrate. The significant gene set related to circulating levels of 3-hydroxybutyrate was a gene set related to G1/S-specific transcription as recorded in the Reactome Pathway Database (https://reactome.org) (34).
TABLE 2.
Gene-Sets Reported by Multi-Marker Analysis of GenoMic Annotation Competitive Gene-Set Analysis
| Gene Set | No. of Genes | Beta (se) | p | Bonferonni Corrected p |
|---|---|---|---|---|
| G1/S-specific transcription | 29 | 0.68 (0.14) | 2.74 × 10–7 | 4.24 × 10–3 |
Metabolite and Clinical Variable Analysis
Logistic regression analysis without covariates revealed a few metabolites associated with mortality, 28-day mortality (creatine, myo-inositol, pyruvate, and tyrosine), and 90-day mortality (pyruvate) (Supplementary Table S9, http://links.lww.com/CCX/B290). However, many metabolites were associated with renal function, RRT prior randomization (myo-inositol, pyruvate, and tyrosine), and RRT post-randomization (creatine, myo-inositol, pyruvate, and tyrosine). Logistic regression analysis with the inclusion of covariates showed pyruvate was associated with 28-day mortality (p = 0.0356) (Supplementary Table S10, http://links.lww.com/CCX/B290). In these analyses, pyruvate was not associated with markers of renal function. Of note, creatinine was consistently associated with markers of renal function both with and without the inclusion of covariates and did not show an association with sepsis mortality.
Statistical Power Analyses
Statistical power analyses suggested that our study did not have sufficient power to detect a relationship between a metabolite associated variant and sepsis-related outcomes—assuming a variant of relatively small effect—as would be expected for a complex trait like sepsis (Supplementary Tables S11 and S112, http://links.lww.com/CCX/B290).
DISCUSSION
The current study represents the first GWAS of metabolite levels to have been performed in a cohort of individuals with septic shock. Previous GWAS of sepsis have primarily focused on mortality (i.e., 28- or 90-d mortality) as an outcome. These GWAS have reported a limited number of genome-wide significant loci (18–22) which have subsequently failed to replicate in later studies (18). In contrast, GWAS have been far more successful in identifying hundreds of genetic variants robustly associated with metabolite levels, including many of large effect (2, 4–6, 11–15). We therefore reasoned that performing a GWAS of metabolite levels in individuals with septic shock (i.e., thereby using dysregulated metabolite levels as an intermediate phenotype for sepsis) might serve as a useful complementary strategy for identifying genetic variants important in the etiology of sepsis.
The current study found genome-wide significant associations for the two major ketone bodies (3-hydroxybutyrate and acetoacetate). Both of these metabolites play a role in energy metabolism, acting as alternative energy sources during starvation. Sepsis and starvation have been previously linked, either due to decreased food intake (35, 36) or an energy deficit created by the elevated energy demands associated with an ongoing immune response (37). The typical starvation response is generally altered in septic patients, impacting disease severity and lethality. It has been suggested that sepsis mortality in relation to the starvation response is impacted by two key transcription factors, the glucocorticoid receptor (GR) and peroxisome proliferator-activated receptor alpha (PPARα). GR has well-known anti-inflammatory effects but additionally is linked to controlling gluconeogenesis, with mice GR knockouts being associated with diminished gluconeogenesis. PPARα is a nuclear receptor that among the many genes it controls includes those responsible for fatty acid β-oxidation. Fatty acid β-oxidation in hepatocytes produces acetyl-CoA, the primary substrate of ketogenesis.
Ketogenesis, the synthesis of ketone bodies, occurs in the liver mitochondria (38), and uses the mitochondrial acetyl-CoA pool to synthesize 3-hydroxybutyrate and acetoacetate (39). Both of these ketone bodies then diffuse through the blood stream to metabolically active tissues such as the brain and heart, which are then able to use them as alternative energy sources when glucose is limited (40). The genetic signals for 3-hydroxybutyrate and acetoacetate represent novel genetic associations that have not been reported in previously published metabolite GWAS. However, the top SNPs at these loci were not associated with risk of sepsis, sepsis severity, or mortality. This potentially suggests that these signals do not represent genetic variation important in the etiology of sepsis. However, it is also likely that this study is underpowered to detect an association with metabolite genetic variants with sepsis outcomes (Supplementary Tables S11 and S12, http://links.lww.com/CCX/B290). It is also possible that these results may reflect the fact that ketone bodies are dysregulated in sepsis patients where elevated energy demands can trigger a starvation response where the body shifts from using readily available energy sources to consuming energy stores (36), and consequently that ketone body associated quantitative trait loci are easier to detect in a cohort of septic shock patients than in cohorts of healthy individuals (i.e., who tend to comprise the majority of metabolomic GWAS).
Lower 3-hydroxybutyrate levels in sepsis are associated with higher observed mortality (41), and metabolic screening of sepsis patients has identified fatty acid beta-oxidation pathways as being significantly different between survivors and nonsurvivors of sepsis (42). Additionally, 3-hydroxybutyrate has been shown to have a wide variety of signaling effects, acting as an inhibitor of protein deacetylases and as a ligand on some receptors (40). The current study reports a novel genome-wide significant SNP (rs2456680) for 3-hydroxybutyrate, which mapped to an intronic region of the CTNNA3 gene. Previous studies have shown that mutations in this gene are associated with arrhythmogenic right ventricular cardiomyopathy (43) and that levels of circulating 3-hydroxybutyate are associated with disease progression potentially due to the altered energy requirements of diseased hearts (44). Sepsis, also involves alterations in energy requirements and the normal starvation response, although it is unclear exactly how CTNNA3 would play a role in this process.
There were also a number of SNPs in the ADH gene cluster that were associated with circulating acetoacetate levels at genome-wide levels of significance. However, the high degree of linkage disequilibrium in this part of the genome means that it is difficult to determine which gene is likely to be responsible for the primary association (45). Sepsis has been shown to lead to an accumulation of free fatty acids (FFAs). Increased FFAs typically results in the up-regulation of PPAR-α expression, which leads to increased ketone body production. However in sepsis, PPAR-α is downregulated, which alters β-oxidation of fatty acids and decreases ketone body production (37). Altered fatty acid transport and β-oxidation have been shown to differ between sepsis survival and nonsurvival patients (46). ADH genes are generally involved in ethanol metabolism, which produces acetyl-CoA (47), with ketogenesis using the mitochondrial acetyl-CoA pool to synthesize the two main ketone bodies (3-hydroxybutyrate and acetoacetate) (39). However, perhaps more relevant is the reported role of the ADH4 gene in fatty acid omega-oxidation, a precursor of the beta-oxidation process (48). Similar to 3-hydroxybutyrate, SNPs in this region of the genome were only borderline genome-wide significant (i.e., p = 1.57 × 10–8). Potential relationships between ADH gene variation and circulating acetoacetate levels warrant further investigation and replication in fasting cohorts.
Finally, a genome-wide significant SNP (rs6851961) showed association with circulating creatinine, a metabolite which is predominantly synthesized in the liver and the kidney and that accumulates in skeletal and heart muscle in order to provide a high-energy phosphate buffering system. Serum creatinine is currently used as a diagnostic measure in the Sequential (Sepsis-related) Organ Failure Assessment (17), as well as a marker of renal function for both acute kidney injury (AKI) and chronic kidney disease (CKD) (49). AKI is a frequent and serious complication of sepsis in ICU patients (50) being associated with increased mortality as well as increased risk of later CKD and end-stage kidney disease (51). The rs6851961 SNP lies in an intergenic region roughly 100 kb telomeric to TECRL the nearest protein-coding gene. TECRL is an endoplasmic reticulum protein expressed in the heart and skeletal muscle. While pathologic variants in TECRL are known to cause life-threatening arrhythmias (52), the gene is not known to be involved in sepsis pathology.
While we have reported novel genetic loci for three metabolites, these may represent type 1 errors. In particular, our genome-wide significant loci are not reported in recent metabolomic GWAS; likewise, previously reported genome-wide significant loci for these metabolites (11, 13–15) are not significant in our current scan. Nevertheless, the current study, to the best of our knowledge, is the first to investigate genetic associations with metabolites in a cohort of individuals with septic shock. As discussed, sepsis is known to disrupt metabolite levels and these changes may not be commonly observed in otherwise healthy populations. It is therefore possible that our genetic loci reflect true findings in states that have not been analyzed in large-scale GWAS to date.
CONCLUSIONS
The current study is the first to have investigated the genetic basis of circulating metabolites in a cohort of individuals with septic shock. While three genetic variants significantly associated with three different circulating metabolites were identified, these associations may reflect a generalized starvation response rather than an effect specific to sepsis. These results require further investigation and replication in both healthy and diseased cohorts including those of different ancestry.
Supplementary Material
Footnotes
This work was supported by an National Health and Medical Research Council (NHMRC) project grant (GNT2009203). Nuclear magnetic resonance (NMR) spectra were acquired on instruments of the Queensland NMR Network, which was established with funding from the Queensland State government.
Drs. Finfer, Myburgh, Cohen, Venkatesh, and Evans received grant from National Health and Medical Research Council (NHMRC) project (GNT10855159). Dr. Finfer is supported by an NHMRC Practitioner Fellowship (1117230). Dr. Evans is supported by an NHMRC investigator grant (APP2017942). The remaining authors have disclosed that they do not have any potential conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
REFERENCES
- 1.Kettunen J, Tukiainen T, Sarin A-P, et al. : Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet 2012; 44:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lotta LA, Pietzner M, Stewart ID, et al. ; MacTel Consortium: A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet 2021; 53:54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worley B, Powers R: Multivariate analysis in metabolomics. Curr Metabolomics 2013; 1:92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagenbeek FA, Pool R, van Dongen J, et al. ; BBMRI Metabolomics Consortium: Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat Commun 2020; 11:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long T, Hicks M, Yu H-C, et al. : Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet 2017; 49:568–578 [DOI] [PubMed] [Google Scholar]
- 6.Shin S-Y, Fauman EB, Petersen A-K, et al. ; Multiple Tissue Human Expression Resource (MuTHER) Consortium: An atlas of genetic influences on human blood metabolites. Nat Genet 2014; 46:543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirkan A, Henneman P, Verhoeven A, et al. : Insight in genome-wide association of metabolite quantitative traits by exome sequence analyses. PLoS Genet 2015; 11:e1004835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kettunen J, Demirkan A, Würtz P, et al. : Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 2016; 7:11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raffler J, Römisch-Margl W, Petersen A-K, et al. : Identification and MS-assisted interpretation of genetically influenced NMR signals in human plasma. Genome Med 2013; 5:13–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teslovich TM, Kim DS, Yin X, et al. ; Genetics of Obesity-Related Liver Disease Consortium (GOLD), The Alzheimer's Disease Genetics Consortium (ADGC), The DIAbetes Genetics Replication And Meta-analysis (DIAGRAM): Identification of seven novel loci associated with amino acid levels using single-variant and gene-based tests in 8545 Finnish men from the METSIM study. Hum Mol Genet 2018; 27:1664–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davyson E, Shen X, Gadd DA, et al. : Metabolomic investigation of major depressive disorder identifies a potentially causal association with polyunsaturated fatty acids. Biol Psychiatry 2023; 94:630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li-Gao R, Hughes DA, van Klinken JB, et al. : Genetic studies of metabolomics change after a liquid meal illuminate novel pathways for glucose and lipid metabolism. Diabetes 2021; 70:2932–2946 [DOI] [PubMed] [Google Scholar]
- 13.Richardson TG, Leyden GM, Wang Q, et al. : Characterising metabolomic signatures of lipid-modifying therapies through drug target mendelian randomisation. PLoS Biol 2022; 20:e3001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CJ, Sinnott-Armstrong N, Cichońska A, et al. : Integrative analysis of metabolite GWAS illuminates the molecular basis of pleiotropy and genetic correlation. Elife 2022; 11:e79348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin X, Chan LS, Bose D, et al. ; FinnGen: Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat Commun 2022; 13:1644–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MY, Hu T: Computational methods for the discovery of metabolic markers of complex traits. Metabolites 2019; 9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singer M, Deutschman CS, Seymour CW, et al. : The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Urso S, Rajbhandari D, Peach E, et al. ; CHARGE Inflammation Working Group: Septic shock: A genomewide association study and polygenic risk score analysis. Twin Research Human Genet 2020; 23:204–213 [DOI] [PubMed] [Google Scholar]
- 19.Rautanen AD, Mills TCD, Gordon ACMD, et al. : Genome-wide association study of survival from sepsis due to pneumonia: An observational cohort study. Lancet Respir Med 2015; 3:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherag A, Schöneweck F, Kesselmeier M, et al. : Genetic factors of the disease course after sepsis: A genome-wide study for 28 day mortality. EBioMedicine 2016; 12:239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan L, Page G, Kirpalani H, et al. ; Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network: Genome-wide association study of sepsis in extremely premature infants. Arch Dis Child Fetal Neonatal Ed 2017; 102:F439–F445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong HR: Genetics and genomics in pediatric septic shock. Crit Care Med 2012; 40:1618–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesh B, Finfer S, Cohen J, et al. ; ADRENAL Trial Investigators and the Australian–New Zealand Intensive Care Society Clinical Trials Group: Adjunctive glucocorticoid therapy in patients with septic shock. N Engl J Med 2018; 378:797–808 [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, et al. : PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S, Forer L, Schönherr S, et al. : Next-generation genotype imputation service and methods. Nat Genet 2016; 48:1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS, Tzur D, Knox C, et al. : HMDB: The human metabolome database. Nucleic Acids Res 2007; 35:D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staley JR, Blackshaw J, Kamat MA, et al. : PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016; 32:3207–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamat MA, Blackshaw JA, Young R, et al. : PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019; 35:4851–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sollis E, Mosaku A, Abid A, et al. : The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res 2023; 51:D977–D985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K, Taskesen E, van Bochoven A, et al. : Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8:1811–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leeuw CA, Mooij JM, Heskes T, et al. : MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003; 19:149–150 [DOI] [PubMed] [Google Scholar]
- 33.Li L, Sunderland N, Rathnayake K, et al. : Epidemiology of sepsis in Australian public hospitals. Australian Commission on Safety and Quality in Health Care, 2020 [Google Scholar]
- 34.Gillespie M, Jassal B, Stephan R, et al. : The reactome pathway knowledgebase. Nucleic Acids Res 2022; 50:D687–D692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson SJ, Tsai AA, Scala CM, et al. : Adequacy of oral intake in critically ill patients 1 week after extubation. Dysphagia 2010; 25:347. [DOI] [PubMed] [Google Scholar]
- 36.Vandewalle J, Libert C: Sepsis: A failing starvation response. Trends Endocrinol Metab 2022; 33:292–304 [DOI] [PubMed] [Google Scholar]
- 37.Van Wyngene L, Vandewalle J, Libert C: Reprogramming of basic metabolic pathways in microbial sepsis: Therapeutic targets at last? EMBO Mol Med 2018; 10:e8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukao T, Mitchell G, Sass JO, et al. : Ketone body metabolism and its defects. J Inherit Metab Dis 2014; 37:541–551 [DOI] [PubMed] [Google Scholar]
- 39.Shi L, Tu BP: Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr Opin Cell Biol 2015; 33:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman JC, Verdin E: β-Hydroxybutyrate: A signaling metabolite. Annu Rev Nutr 2017; 37:51–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramazan A, Ismail E, Gurhan T, et al. : Association between beta-hydroxybutyrate levels and survival in sepsis patients. Eurasian J Med Invest 2021; 5:39 [Google Scholar]
- 42.Wang Y-H, Liu C-L, Chiu W-C, et al. : HMGCS2 mediates ketone production and regulates the proliferation and metastasis of hepatocellular carcinoma. Cancers (Basel) 2019; 11:1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Hengel J, Calore M, Bauce B, et al. : Mutations in the area composita protein αT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. Eur Heart J 2013; 34:201–210 [DOI] [PubMed] [Google Scholar]
- 44.Song J-P, Chen L, Chen X, et al. : Elevated plasma β-hydroxybutyrate predicts adverse outcomes and disease progression in patients with arrhythmogenic cardiomyopathy. Sci Transl Med 2020; 12:eaay8329. [DOI] [PubMed] [Google Scholar]
- 45.Edenberg HJ: The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health 2007; 30:5–13 [PMC free article] [PubMed] [Google Scholar]
- 46.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. : An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013; 5:195–ra195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edenberg HJ, Jerome RE, Li M: Polymorphism of the human alcohol dehydrogenase 4 (ADH4) promoter affects gene expression. Pharmacogenetics 1999; 9:25–30 [DOI] [PubMed] [Google Scholar]
- 48.Collins XH, Harmon SD, Kaduce TL, et al. : ω-Oxidation of 20-hydroxyeicosatetraenoic acid (20-HETE) in cerebral microvascular smooth muscle and endothelium by alcohol dehydrogenase 4*. J Biol Chem 2005; 280:33157–33164 [DOI] [PubMed] [Google Scholar]
- 49.Doi K: Role of kidney injury in sepsis. J Intensive Care 2016; 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Slikke EC, Star BS, van Meurs M, et al. : Sepsis is associated with mitochondrial DNA damage and a reduced mitochondrial mass in the kidney of patients with sepsis-AKI. Crit Care 2021; 25:36–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao X, Chen C, Luo W, et al. : Combining renal cell arrest and damage biomarkers to predict progressive AKI in patient with sepsis. BMC Nephrol 2021; 22:415–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devalla HD, Gélinas R, Aburawi EH, et al. : TECRL, a new life-threatening inherited arrhythmia gene associated with overlapping clinical features of both LQTS and CPVT. EMBO Mol Med 2016; 8:1390–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.