Abstract
TRP channels are important pharmacological targets in physiopathology. TRPV2 plays distinct roles in cardiac and neuromuscular function, immunity, and metabolism, and is associated with pathologies like muscular dystrophy and cancer. However, TRPV2 pharmacology is unspecific and scarce at best. Using in silico similarity-based chemoinformatics we obtained a set of 270 potential hits for TRPV2 categorized into families based on chemical nature and similarity. Docking the compounds on available rat TRPV2 structures allowed the clustering of drug families in specific ligand binding sites. Starting from a probenecid docking pose in the piperlongumine binding site and using a Gaussian accelerated molecular dynamics approach we have assigned a putative probenecid binding site. In parallel, we measured the EC50 of 7 probenecid derivatives on TRPV2 expressed in Pichia pastoris using a novel medium-throughput Ca2+ influx assay in yeast membranes together with an unbiased and unsupervised data analysis method. We found that 4-(piperidine-1-sulfonyl)-benzoic acid had a better EC50 than probenecid, which is one of the most specific TRPV2 agonists to date. Exploring the TRPV2-dependent anti-hypertensive potential in vivo, we found that 4-(piperidine-1-sulfonyl)-benzoic acid shows a sex-biased vasodilator effect producing larger vascular relaxations in female mice. Overall, this study expands the pharmacological toolbox for TRPV2, a widely expressed membrane protein and orphan drug target.
Keywords: Ion channels, TRP channels, TRPV2, Drug discovery, Membrane proteins, Biophysics, Docking, Cardiovascular, Computational biology, Structural biology, Pharmacology
Graphical Abstract

1. Introduction
The Transient receptor potential (TRP) ion channel family is second largest after the potassium channel family. Many TRPs are somatosensory channels that respond to physicochemical stimuli, including temperature, mechanical stress, or osmotic changes [1]. Thus, TRPs are key therapeutic targets in physiopathology, including cancer [2], and bacterial and viral infectivity [3], [4]. TRP vanilloid 1 (TRPV1) has a vast pharmacological toolbox related to acute and chronic pain management [5]. The closest TRPV1 homolog, TRPV2, is widely expressed in humans and important in cardiac structure[6], immunity[3], and nervous system development and physiopathology [7], [8], [9]. TRPV2 is a noxious heat and mechanical sensor, and so far the endogenous activation of this channel is related to oxidative stress through methionine (Met) oxidation [2] and phosphatidylinositol (PIP2) lipid binding [10]. The sparse TRPV2 pharmacology includes cannabidiol (CBD) derivatives, lipids like PIP2 and lysophosphatidylcholine (LPC), and few mid-specific modulators, like agonist probenecid (PBCD) and antagonist tranilast [11]. PBCD and tranilast are already in the clinic for the treatment of gout and allergy [12]. Tranilast has been repurposed as a TRPV2-targeted drug in heart failure [13]. On the other hand, PBCD has been repurposed as a Pannexin-1-targeted inhibitor in hypertension, but also as heart failure drug in a recent clinical trial [14], [15], [16].
Structures of rabbit and rat TRPV2 were determined by cryogenic electron microscopy (cryo-EM) and X-ray crystallography [17], [18], [19], [20], along with structures of other TRPVs [21]. Specifically, TRPV2-bound ligands that have been resolved so far are CBD [22], 2-aminoethoxydiphenyl borate (2-APB) [23], [24], piperlongumine (PLG) [25], and Δ9-tetrahydrocannabiorcol (C16) [26]. The PBCD binding site remains elusive although it has been shown to induce specific conformational changes in TRPV2 and to act synergistically with C16, indicating a different binding site [26]. In sum, TRPV2 is an important target with an increasing number of physiopathological implications and its pharmacology is being extensively explored, although thus far with limited success.
Here we present a multidisciplinary study combining chemoinformatics, computational structural biophysics, a new in vitro activity assay with new pharmacological data analysis to find a TRPV2 ligand, 4-(Piperidine-1-sulfonyl)-benzoic acid (PSBA) with slightly higher efficacy than PBCD in vitro. Comparison of the activity of PBCD and PSBA in physiological conditions using an animal ex vivo model of vascular reactivity [27], shows that PSBA modulates vascular relaxation in a sex-biased fashion. In sum, our results provide an interesting pharmacological toolbox to explore the pharmacophysiology of TRPV2, and a pipeline that can be easily translated to other ion channels.
2. Results and discussion
2.1. Chemoinformatics similarity search for putative TRPV2 modulators
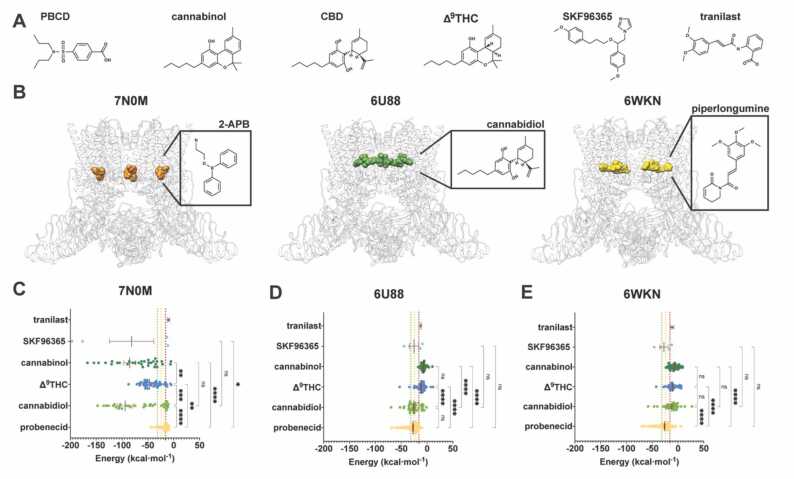
To expand the pharmacological toolbox for TRPV2, we used PBCD, CBD, ∆9-tetrahydrocannabinol (∆9THC), cannabinol, and SKF96365 (Fig. 1A), described as the TRPV2 pharmacology by Vriens et al. [5], as seeds in a similarity-based chemoinformatics search in ZINC [28], CHEMBL [29], DrugBank [30], and TCM [31] databases to find compounds active on TRPV2 with minimal activity on TRPV1 (see Supporting Information). This search yielded a set of 270 compounds (grouped in five families of 148, 46, 38, 34, and 4 hits related to PBCD, ∆9THC, CBD, cannabinol, and SKF96365, respectively) ranked using the Tanimoto difference coefficient [32], with diversity in the number of rings, partition coefficient, polar surface area, hydrogen-bond donor and acceptors (Supporting Dataset 1).
Fig. 1.
TRPV2 molecular docking results. (A) Chemical structures of PBCD (PBCD), cannabinol, CBD, Δ9THC, SKF96365 and tranilast. Hydrogens are omitted for clarity. (B) TRPV2 structure and binding site of 2-APB (orange) in 7N0M, CBD (green) in 6U88, and PLG (yellow) in 6WKN. TRPV2 is shown in cartoon, colored in white. Ligands are shown as spheres. (C-E) Predicted binding energy of the 270 compounds, including tranilast, at the 2-APB binding site in 7N0M (C), CBD binding site in 6U88 (D), and PLG binding site in 6WKN (E). Ligands are categorized into families based on seed compounds. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by one-way ANOVA.
2.2. Ligand-based virtual screening and PBCD binding site assignment
Taking advantage of the TRPV2 structural biology to date, we have tried to assign a putative binding site for PBCD, using docking. To define the best docking strategy, we tried two alternative protocols. For the first approach, we used TRPV2 apo (pdb id 6U84) and ligand-bound (pdb id 6U88 and 6WKN) structures to perform a global unbiased docking approach for PBCD, cannabidiol, and tranilast using a box for the overall transmembrane domain (TMD) of TRPV2. We obtained scattered docking poses throughout the TMD not converging in the resolved binding site (Fig. S1 and Table S1), thus remaining inconclusive to assign a putative PBCD binding site.
For the second approach we tried a local docking strategy using as targets the binding sites for CBD, PLG, and 2-APB in their respectives cryo-EM structures of TRPV2 (PDB codes 6U88, 6WKN, and 7N0M, respectively, Fig. 1B). As a benchmark, we determined the binding energies of the original ligands, obtaining values of − 31.5 kcal mol-1, − 24.3 kcal mol-1, and − 15.8 kcal mol-1 for CBD in 6U88, PLG in 6WKN, and 2-APB in 7N0M, respectively (dashed lines in Fig. 1C-E). We also docked tranilast (Fig. 1A) as a reference molecule, obtaining binding energies in the − 9 to − 12 kcal mol-1 range. Then, to define a putative PBCD binding site, we performed a family-based docking approach using the 270 compounds identified by chemoinformatics. Fig. 1 C-E show the binding energy values for docking the 270 compounds in all three structures, grouped into their families, expecting to assess which TRPV2 structure allowed us to discriminate between the PBCD family and the others. Docking results in 7N0M are highly biased to negative energy binding values (average binding energies for all families are lower than −15.8 kcal mol-1) but with high dispersion in binding energy within the members of the same family (Fig. 1C), arguing for the lack of chemical specificity. The PBCD family showed the largest energy mean value, indicating that the 2-APB site was not the most favorable.
In the CBD binding site in 6U88, docking of the CBD family compounds docking yielded very negative and narrowly distributed binding energy values (25.6 ± 2.2 kcal mol-1; Fig. 1D). The CBD family mean binding energies (−25.6 kcal mol-1) values, represent better scores than for the ∆9THC and cannabinol families, but not as low as the resolved CBD (−31.5 kcal mol-1), however it allows to discriminate accurately between CBD and other cannabis derivatives. Intriguingly, PBCD-family compounds bind with similar affinity to the CBD binding site as CBD-derived compounds showing no significant differences in binding energies (Fig. 1D) indicating that PBCD may occupy the CBD binding site. However, a recent study defining the C16 binding site in TRPV2 pretreated with PBCD suggested that the CBD binding site is unlikely to be occupied by PBCD [26].
Docking the PBCD family in the PLG binding site (6WKN) shows the highest affinity, i.e., the lowest mean energy binding for PBCD family (−26.1 kcal mol-1), significantly lower binding energy compared to cannabis-derived subfamilies (Fig. 1E), arguing for the PLG binding site as the most suitable binding site for PBCD-family compounds using a local docking approach.
In fact, in the original study describing TRPV2 activation by CBD [22], cells were pretreated with a high concentration of PBCD (2.5 mM versus PBCD TRPV2 EC50 = 25 µM), because this PBCD treatment has been routinely applied to inhibit organic anion transporters in calcium imaging assays, and the role of PBCD as a TRPV2 activator was just emerging [33]. The resulting CBD EC50 of 3.7 µM for rat TRPV2 [22] may therefore have been affected by PBCD sensitization as demonstrated in a recent study of TRPV2 incubated with the cannabinoid C16 and PBCD [26]. As discussed below, this same situation where cells were pretreated with PBCD happened with the discovery of 2-APB as an activator for TRPV2 [34].
C16 occupies a binding site between the CBD and PLG binding sites but the PBCD binding site remains unknown [26], yet another recent TRPV2 structure solved in the presence of CBD and PIP2 shows that CBD binding site is slightly shifted because the presence of the lipid [35], arguing for an effect of the lipid annulus of TRPV2 as a ligand-binding modulator. Superimposing the different TRPV2 binding sites resolved so far defines a continuous binding region for ligands such as CBD (PDB code 6U88), PLG (PDB code 6WKN), and 2-APB (PDB codes 7N0M, 7YEP), as depicted in Fig. 2A. Thus, TRPV2 structural biology points to a broad and continuous ligand binding region ranging from the CBD binding site to the TM4-TM5 linker (Fig. 2A). As mentioned above, PLG binding site is the most favorable for PBCD-family binding in our virtual docking. This also holds for the PBCD binding energies, which correspond to − 29.5 ± 1.3, − 28.0 ± 0.3, and − 28.0 ± 1.3 kcal mol-1 for 6WKN, 6U88, and 7N0M, respectively.
Fig. 2.
(A) Combined representation of the binding sites of CBD (6U88), PLG (6WKN), and 2-APB (rat 7N0M, mouse 7YEP). Protein is depicted in cartoon and color-coded by chains: A (light pink), B (light blue), C (light green), D (light yellow). Ligands are shown as sticks and surface is also displayed: CBD in red, PLG in blue, 2-APB in 7N0M in purple, 2-APB in 7YEP in yellow. (B) Clustering by Principal Component Analysis (PCA) of the 200 ns Ligand Gaussian Accelerated molecular dynamics (LiGaMD) simulation of PBCD (PBCD) initially bound to the PLG site in TRPV2 (pdb id 6WKN). Cluster 1 and cluster 2 are the left and right, respectively. (C) Representation of TRPV2 embedded in the membrane. Representative structure of PBCD clusters is shown as: purple (cluster 1) and green (cluster 2). Ligands are shown as spheres. (D) TRPV2 residues and lipids involved in the binding of PBCD (PBCD distance < 4.0 Å) in both clusters. Residues are shown as sticks and the chain is indicated as a subindex and amino acid carbon atoms are colored according to the respective subunit.
We used the PLG site as the starting docking pose for putative PBCD binding (Fig. 2B), and instead of classical molecular dynamics (MD) simulations [36] to refine the PBCD binding site we used a Gaussian accelerated molecular dynamics simulations (3 replicates, 500 ns of simulation time) to explore further the docking space and the binding energies (LiGaMD [37]) of PBCD in TRPV2 in a lipid bilayer. The LiGaMD results show two distinguishable PBCD binding clusters with and average binding energy of − 88.9 ± 0.5 kcal mol-1 (Fig. 2B and C and Supporting Information movie S1). PBCD in cluster 1 is mainly stabilized within the same TRPV2 subunit by lipids (cholesterol and POPC) hydrogen bonds with residues Tyr471 (TM3) and Asn511 (TM4), and by van der Waals interactions with Phe472/Leu475 (TM3), Leu510 (TM4), and Gln530/Leu534 (TM4/TM5 linker) of one subunit and Ala628, Leu631 and Leu632 (TM6) of the next subunit (Fig. 2D). PBCD in cluster 2 is stabilized by ligand-lipid interactions with cholesterol, POPC and POPG, and ligand-protein hydrophobic interactions mainly by Leu478 (TM3) of one TRPV2 subunit and Leu624 (TM6) of the adjacent one (Fig. 2D). Both clusters show a phosphatidylcholine molecule participating in the binding of PBCD (Fig. 2 C and D), which has also been observed in previous experimental work with CBD-bound TRPV2 structures, where lipid-like densities interact and likely stabilize the binding of CBD [35]. Overall, this study highlights the importance of including the lipid bilayer in computational models of membrane proteins to account for these potentially crucial lipid-ligand interactions.
Supplementary material related to this article can be found online at doi:10.1016/j.csbj.2023.12.028.
The following is the Supplementary material related to this article Video S1..
Compared with other ligands from resolved structures, both PBCD clusters mainly occupy the 2-APB binding site in mTRPV2 (PDB code 7YEP, Supporting Information movie S1), located near the vanilloid pocket, surrounded mainly by hydrophobic residues and lipid molecules, highlighting the relevance of ligand-lipid and protein-lipid interactions in the binding of PBCD.
2.3. Design of a Ca2+ assay to test TRPV2 activity
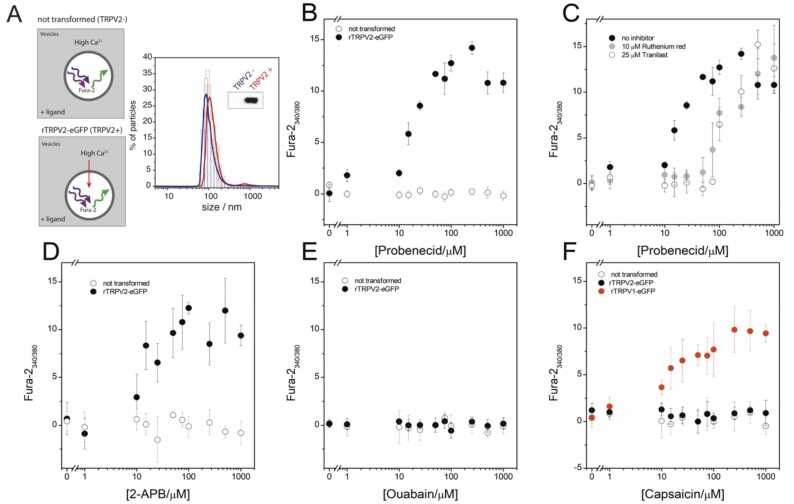
In a drug discovery approach, we selected and purchased seven commercially available PBCD derivatives contained in our dataset (Table 1), to find new, and hopefully more potent, TRPV2 activators compared to PBCD. We designed a functional assay using heterologously expressed TRPV2 to test for TRPV2 activation by these compounds. Our assay uses purified membranes of cells expressing TRPV2, and thus measures drug responses directly due to TRPV2 and no other intracellular signaling pathways, such as G-protein coupled receptor pathways triggered by cannabis derivatives, or organic transporter inhibition derived by PBCD. First, we produced large amounts of TRPV2 in the eukaryote yeast Pichia pastoris [38]. To monitor protein expression, we fused enhanced green fluorescent protein (eGFP) to human, mouse, or rat TRPV2 (TRPV2-eGFP). Rat TRPV2 yielded the most monodisperse protein in fluorescence size exclusion (FSEC) thermal stability experiments (Fig. S2). We thus purified dodecylmaltoside (DDM) solubilized rat TRPV2-eGFP (Fig. S3); its ATR-FTIR spectrum agrees at the secondary structure level with that expected from 3D structures (Fig. S4 and Table S2). Rat TRPV2-eGFP was active in planar lipid membranes (Fig. S5); the first demonstration, to the best of our knowledge, of an active TRP channel expressed in and purified from P. pastoris, which opens biotechnological perspectives in term of production scalability for pharmacological, and structural and molecular biology processes.
Table 1.
Summary of physicochemical data on Compounds 1 to 8, ZINC database identifier, chemical structure, chemical name, ΔTanimoto, molecular weight (MW), n-octanol and water partition coefficient log(coctanol cwater-1) (cLogP).
| Compound | ZINC number | Chemical structure | Name | ΔTanimoto | MW [Da] | cLogP |
|---|---|---|---|---|---|---|
| 1 | 00001982 | 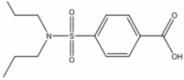 |
4-[(Dipropylamino)sulfonyl]-benzoic acid (PBCD) | 0.775 | 284.4 | 1.94 |
| 2 | 00001323 | 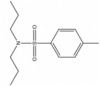 |
4-Methyl-N,N-dipropyl-benzenesulfonamide | 0.564 | 255.4 | 3.88 |
| 3, 4 | 00033305 00033306 |
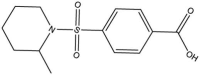 |
4-[(2-Methylpiperidin-1-yl)-sulfonyl]-benzoic acid | 0.584 | 282.4 | 1.63 |
| 5 | 00112604 | 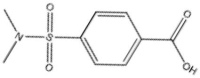 |
4-[(Dimethylamino)sulfonyl]-benzoic acid | 0.564 | 228.2 | 0.38 |
| 6 | 00190172 |  |
4-(Piperidine-1-sulfonyl)-benzoic acid (PSBA) | 0.630 | 268.3 | 1.24 |
| 7 | 00874371 | 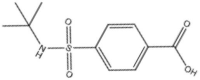 |
4-[(Tert-butylamino)sulfonyl]-benzoic acid | 0.652 | 256.3 | 1. 60 |
| 8 | 01681377 | 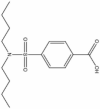 |
4-Dibutylsulfonyl-benzoic acid | 0.671 | 312.4 | 2.72 |
We next developed a higher throughput fluorescence-based Ca2+ influx assay. Briefly, P. pastoris was lysed and membranes from TRPV2-eGFP-expressing or control strains were extruded to 100 nm in the presence of the ratiometric Ca2+ sensor Fura-2 to generate Fura-2-loaded vesicles.
TRPV2 presence or absence was confirmed by immunodetection (Fig. 3A). We then exposed these vesicles, devoid of cytosolic signaling machinery, to compounds in a high Ca2+ buffer and measured the Fura-2 fluorescence ratio at 340/380 nm excitation wavelengths. First, we tested the drug dose response depending on TRPV2 orhtologs, so we tested PBCD, 2-APB and CBD against human, mouse and rat TRPV2 (Fig. S6). We selected the rTRPV2 for future measurements because it was the isoform showing higher fluorescence intensity in drug dose-dependent measurements. To validate signal specificity, we measured PBCD dose responses in the absence (PBCD EC50 ∼ 25 μM Fig. 3B) or presence of inhibitors such as tranilast and ruthenium red (PBCD EC50 ∼ 200 μM; Fig. 3 C). Non-selective agonist 2-APB also activated TRPV2 (EC50 ∼ 20 μM, Fig. 3D). At this point, it is key to realize that PBCD has been related to TRPV2’s pharmacology, unknowingly from the beginning. First evidence of 2-APB modulation on TRPV2, yielded an EC50 of 129 µM in HEK293, but those measurements were taken in the presence of 2 mM of PBCD [34], similarly to the first studies with CBD [22], arguing for drug-sensitization effects on the activation of TRPV2, as shown for 2-APB [34], CBD [22], [35], and C16 [26]. In contrast, neither ouabain (Fig. 3E and Supporting Information Fig. S15), a modulator of Na/K-ATPase and IP3 receptors, nor RN1747, a TRPV4 inhibitor (Supporting Information Fig. S16), changed the fluorescence of rat TRPV2-GFP vesicles. Also, we observed a capsaicin response with TRPV1-GFP-containing vesicles (EC50 ∼ 20 μM), but not TRPV2-GFP vesicles (Fig. 3 F). Although the capsaicin values for TRPV1, the closest homolog to TRPV2, are not comparable to those in the literature (nM range) [39], capsaicin, still behaves as a negative control for TRPV2. All these controls are key to validate our assay to measure Ca2+ influx into TRPV2-GFP-containing vesicles.
Fig. 3.
(A) Illustration of the Ca2+ flux yeast membrane-based functional assay. Particle size distribution of the Fura-2 loaded vesicles from cells expressing TRPV2 or not, as detected by immunoblotting. (B-F) Dose response curves showing increase in Fura-2 fluorescence ratio by Ca2+ influx into P. pastoris membrane vesicles containing rat TRPV2 (rTRPV2-eGFP) loaded with Fura-2 Ca2+ sensitive dye in the presence of increasing concentrations of PBCD (B); PBCD in the presence/absence of inhibitors (ruthenium red and tranilast at 10 μM and 25 μM, respectively) (C); 2-APB (D); ouabain (E) as a negative control; capsaicin in the presence of rTRPV1-eGFP or rTRPV2-eGFP Fura-2 loaded vesicles (F). Each plot represents the average ratio of Fura-2 fluorescence at excitation wavelengths 340 and 380 nm (average ± S.E.M; n = 4 independent experiments) as a function of fluorescence intensity.
2.4. EC50 estimation and data analysis
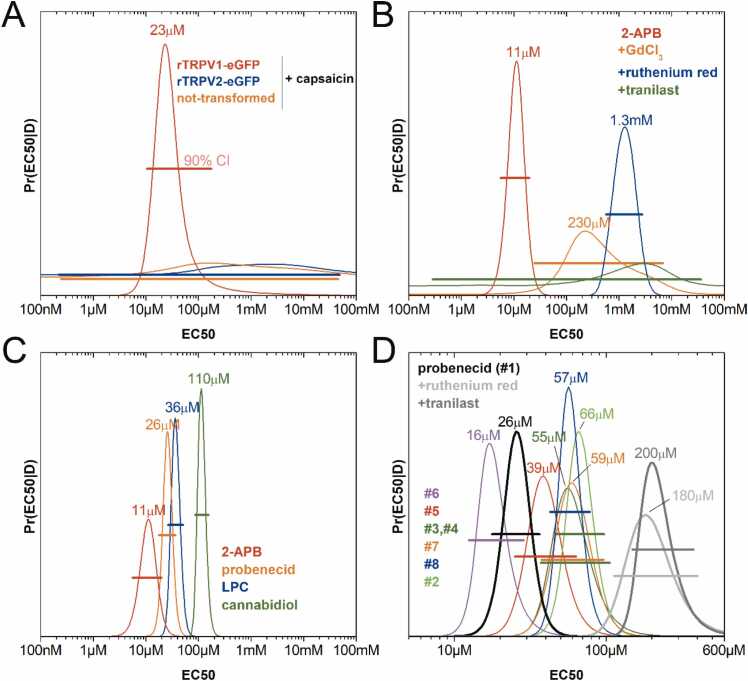
One open question in the analysis of activity data to estimate EC50 values, particularly with noisy titration curves, is how to use all the experimental data available to avoid selection bias, while at the same time preventing the inclusion of noisier/more disperse data sets in the analysis that might corrupt the estimation of EC50 values. Another important issue is how to obtain, besides the best possible estimate for EC50, reliable confidence intervals for its value that account for all possible uncertainties, to be able to meaningfully rank EC50 values from different compounds. We implemented an unbiased and unsupervised global parameter estimation method using Bayesian inference (Supporting Information Methods and Figs. S6 to S8 for technical details). Briefly, Bayesian analysis was applied to four individual datasets for a given compound to obtain four probability distributions for EC50 given only the data (other parameters were integrated out) with the distribution broadness reflecting how informative was an individual dataset on the value of EC50. Following the rules of probability, a global probability distribution for EC50 was obtained by the multiplication of all four individual probabilities, a step that automatically makes narrower probability distributions to contribute more than broader distribution to the final probability distribution for EC50. This allowed us to make use of all the available information in the data about EC50, even when datasets had apparent differences in quality. Finally, from the (posterior) probability distribution for EC50 we could obtain realistic confidence levels (see Supporting Information on how we integrated out nuisance parameters to obtain probabilities for EC50).
Fig. 4 shows resulting probability distributions of EC50 values for control and test compounds (with a 90% confidence interval in brackets). Capsaicin activated TRPV1-GFP with EC50 = 23 [11−175] μM but induced no measurable increase of fluorescence signal for TRPV2-GFP (Fig. 4A and Fig. S11), leading to a featureless probability distribution for EC50. 2-APB activated TRPV2-GFP with EC50 = 11 [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20] μM (Fig. 4B). The presence of the inhibitor ruthenium red (10 μM) did not fully prevent the activation of TRPV2 by 2-APB but increased its EC50 to 1.3 [0.6–2.7] mM. On the other hand, the addition of the inhibitors GdCl3 (100 μM) and tranilast (25 μM) almost completely prevented the activation of TRPV2 by 2-APB (Fig. S10). The resulting small signals decreased the precision of the 2-APB EC50 estimates; for GdCl3 it increased to 233 [25−7000] μM, while for tranilast the probability distribution of the 2-APB EC50 was so broad as to be undefined (Fig. 4B and Fig. S9). Other known TRPV2 agonists yielded activating signals with EC50 values of 26 [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36] μM for PBCD, 36 [27–51] μM for LPC, and 110 [87–155] μM for CBD (Fig. 4C and Figs. S6-S8, S10, and S12). Thus, our approach of a membrane-based in vitro activity assay and our EC50 data analysis allowed us to robustly measure and compare drug’s activity on isolated cell membranes. Previously published TRPV2 results from assays in mammalian cells do not rule out indirect effects, and showed irreproducibility or EC50 differences among labs [12]. For instance, CBD is the most active non-specific TRPV2 agonist in some studies but seems to show species dependence with reported EC50 values ranging from 10-8 to 10-6 M for mouse or rat TRPV2, and no activation of human TRPV2 [40]. The CBD EC50 we obtained for rat TRPV2 is 110 μM (10-4 M), a lower efficacy compared to values reported in the literature. This difference may be due to one or a combination of i) altered accessibility of the CBD binding site in our system, ii) the lack of downstream amplification from signaling in a cell-based assay, and iii) the lack of PBCD pretreatment [26], [33], [41].
Fig. 4.
Posterior probability distributions of EC50 for different compounds, estimated from a dose-dependent fluorescence assay sensitive to TRPV2-mediated Ca2+ transport. Note that conditions lacking dose-dependent fluorescence changes provide extremely broad (uncertain) probability distributions. (A) EC50 of capsaicin for TRPV1, TRPV2, and control. (B) EC50 of 2-ABP for TRPV2 in the absence and presence of different inhibitors. (C) EC50 of known TRPV2 activators. (D) EC50 of PBCD and PBCD derivatives for TRPV2. Horizontal bars for each EC50 indicate the 90% confidence interval (CI) for each probability distribution.
2.5. Agonist-mode determination of PBCD-derivatives activity
Finally, we tested the agonist activity of seven commercially available PBCD derivatives using our Ca2+ influx fluorescence assay (Fig. 4D and Fig. S13). Ranking compounds by efficiency (lower to higher EC50) we found: 6 > PBCD (1) > 5 > 3,4 > 8 > 7 > 2. Thus, 6 (subsequently named PSBA), replacing the two N-linked propyl groups with a piperidine ring, can activate TRPV2 at a lower concentration than PBCD. Using a Bayesian hypothesis test to the EC50 probability distributions for PSBA and PBCD (Fig. S21), we find an 84% confidence that the EC50 is indeed lower for PSBA than PBCD (Figs. S16 and S17).
2.6. Assessing vascular responses to TRPV2 agonists
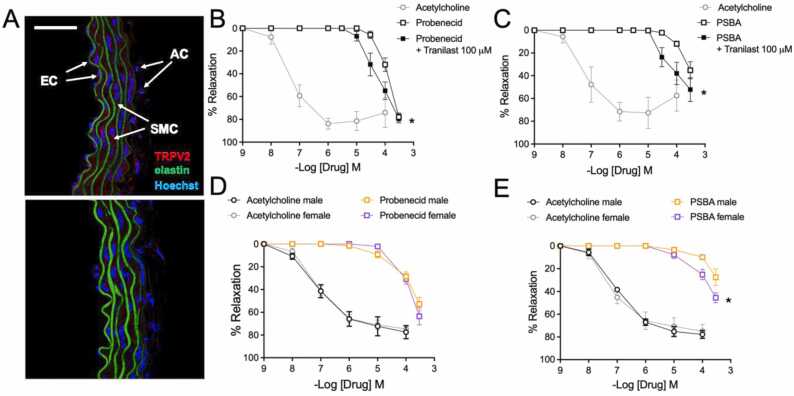
To evaluate whether the higher efficiency of PSBA as compared to PBCD for activation of TRPV2 in silico/in vitro has physiological relevance, we measured vascular relaxations in an ex vivo model of vascular reactivity. We first examined TRPV2 expression in aortas from young (3- to 4-months-old) male Oncine France 1 mice using immunocytochemistry (Fig. 5A). TRPV2 was expressed in all three layers of the thoracic aorta wall. We next compared the effects of PBCD and PSBA on vascular function. Neither compound was able to induce aortic contractions. Previous studies have evidenced the vasodilator properties of PBCD in the rat aorta [16] and in the human forearm and the leg vasculature [42], and we have recently show the vasodilator properties of PBCD over TRPV2 [27]. Consistently, both compounds induced concentration-dependent relaxations in U46619-precontracted arteries that were lower than relaxations to acetylcholine, an endothelium-dependent muscarinic receptor agonist that was used as a relaxation control (Fig. 5B and C). PSBA evoked significantly lower relaxations than probenecid (Fig. 5B vs. C). Interestingly, blockade with the TRPV2 inhibitor tranilast (100 µM) led to a similar potentiation of probenecid and PSBA relaxations (Fig. 5B and C). The lower relaxations of PSBA suggest a lesser capacity of this compound to activate endothelial TRPV2, while the similar potentiation effect of tranilast on both compound relaxations suggests similar activation of smooth muscle TRPV2.
Fig. 5.
Vascular responses to TRPV2 agonists. (A) Representative photomicrograph of TRPV2 immunofluorescence (red) of confocal microscopic Oncins France 1 mice thoracic aortas (n = 3). Natural autofluorescence of elastin (green) and nuclear staining with Hoechst 33342 (10 μg/ml) (blue) are also shown. AC, adventitial cells; SMC, smooth muscle cells; EC, endothelial cells. Scale bar, 50 µm. Concentration-response relaxation curves to PBCD (B) and PSBA (C), as compared with acetylcholine, in U46619-precontracted ascending aorta rings from 3- to 4-month-old male and female Oncine France 1 mice in the presence and absence of Tranilast (100 µM). Results are mean ± SEM from n = 7 (PBCD), 6 (PSBA), 3–5 (acetylcholine) mice. Influence of sex on the TRPV2 agonists’ vascular responses; concentration-response relaxation curves to PBCD (D) and PSBA (E), as compared with acetylcholine, in U46619-precontracted aortic (thoracic + abdominal) rings from 3- to 4-month-old male and female C57BL/6 mice. Results are mean ± SEM from n = 4–5 (male) or n = 5 (female) mice. *P < 0.05 by two-way repeated measures ANOVA.
2.7. Influence of sex in TRPV2 agonists’ vascular responses – the case of PSBA
Sex-specific personalized therapies are a priority of cardiovascular disease treatment considering both the efficacy and the side-effects of treatments [43]. We subsequently studied the potential sex-dependent effects of TRPV2 agonists’. We examined aortic function in young (3- to 4-months-old) male and female C57BL/6 mice. Fig. 4 shows concentration-dependent relaxations in response to PBCD, PSBA, or acetylcholine as a function of sex. As expected, both compounds induced lower relaxations than acetylcholine. Comparing the PBCD and PSBA responses, similar relaxations were observed in females, whereas in males, relaxations to PSBA were significantly lower (P < 0.05) than PBCD (Fig. 5D vs. E). No significant differences by sex were observed in PBCD relaxations (Fig. 5D). Notably, PSBA relaxations were significantly higher in females (Fig. 5E). Previous studies demonstrated that estrogens potentiate TRPV2 activation by 2-APB [23]. The present results suggest that estrogens may also potentiate PSBA, but not probenecid, induced aortic relaxations. Altogether, relaxations induced by PSBA show sexual dimorphism because of higher responses in females. These findings hold promise to develop novel sex-dependent personalized therapies based on TRPV2 for treating vasodilator dysfunction and/or reducing cardiovascular side effects of these drugs.
2.8. Putative binding site for PSBA
Taking advantage of the computational biophysics approach, we performed the LiGaMD approximation (3 replicates, 500 ns of simulation time) to identify potential PSBA binding sites on TRPV2 using the PLG binding site (PBD 6WKN) as starting point (Fig. 6). In this case the LiGaMD approach argues for 2 overlapping clusters (converging in 10 out 11 interactions) with average binding energy of − 89.8 ± 0.07 kcal mol-1 (Fig. 6A and B). PSBA is stabilized mainly by hydrogen bonds with Thr522 (TM4/TM5 linker) of one subunit and Arg535/539 in the adjacent one; and by van der Waals interactions with His521 and Tyr525 (TM4/TM5 linker) of one subunit and Leu538, Leu542, and Val543 (TM5) of the next subunit (Fig. 6C).
Fig. 6.
(A) PCA clustering of the 200 ns LiGaMD simulation of PSBA initially bound to the PLG site in TRPV2 (pdb id 6WKN). Cluster 1 and cluster 2 are the left and right, respectively. (B). Binding site of the 2 overlapping clusters in the TRPV2 structure. Ligands are shown as spheres (C). Depiction of TRPV2 residues and lipids involved in clusters 1 and 2 (PSBA distance < 4.0 Å). Residues are shown as sticks and the chain is indicated as a subindex and amino acid carbon atoms are colored according to the respective subunit.
Interestingly, PSBA occupies a location similar to the structurally determined binding site of 2-APB in rTRPV2 (PDB codes 7T37 and 7N0M, Movie S1), between the N terminus of the TM4-TM5 linker of one subunit and TM5 of the adjacent one [24]. PSBA shares with the 2-APB binding site interactions with Thr522, Tyr525, His521, and Arg539, the last two being necessary for the 2-APB activity on rTRPV2 [24]. It has been suggested that the binding of 2-APB disrupts the cation-π interactions between these four residues, disrupting the stabilization between TM5 and the TM4-TM5 linker [24]. PSBA could be exerting its agonist effecting using a similar mechanism. PSBA interacts with cholesterol and POPC lipid tails like the ones described in the PBCD simulations above (Fig. 2). Computational analysis shows that both PBCD and PSBA explore the large binding site groove shown in Fig. 2A showing different binding modes (cluster) along this important TRPV2 region (Supporting Information movieS1). Further analysis of this dynamic exploration, indicates a crucial role of lipids in the bilayer for ligand binding, acting both as drug anchoring and/or drug accommodating molecules (Movie S1), pointing to lipids as part of the drug-binding site in TRPV2, and not merely bilayer structural plays, likely to happen in most integral membrane proteins.
3. Conclusions
As summary, our study presents a biotechnological combination of computational and experimental approaches to unveil new drug leads for TRPV2, an important physiopathological target. Using similarity-based chemoinformatics we identified 270 potential TRPV2 hits (Supporting Dataset 1) derived from known TRPV2 agonists (Fig. 1A). Using local docking, we have assigned the PBCD binding site in the vicinity of the PLG binding site to further refine it using a Gaussian accelerated molecular dynamics in a protein system embedded in the bilayer to assign the putative PBCD binding site on the cytosolic side of the transmembrane domain between TM4 and TM5, closer to the mTRPV2 2-APB binding site. To test the agonist potential of the PBCD derivatives, we have taken a biotechnological path designing a scalable screening method using vesicles derived from TRPV2-expressing P. pastoris membranes in a fluorescence-based Ca2+ influx assay, which together with a robust and unbiased data analysis allow us to determine EC50 values for different PBCD-derivatives as potential agonists. Of note, our method could be also used to measure antagonist IC50 values. We identified a novel PBCD-based molecule, namely PSBA, as a novel TRPV2 activator with both lower experimental EC50 and calculated binding free energy than PBCD, which has higher affinity for the rTRPV2 2-APB binding site. At this point it is important to highlight the role of lipids for ligand binding, and the use of computational methods to assess the dynamics of ligand binding. Finally, based on the previously described vasodilator properties of PBCD we show that both PBCD and PSBA induce concentration-dependent aortic relaxations targeting TRPV2, while exerting a smooth muscle-derived negative feedback on these responses. Importantly, we demonstrate that PSBA relaxes the mouse aorta in a sex-biased fashion, opening the window for sex-based vasodilator pharmacology to treat several vascular disorders such as hypertensive or diabetes vasculopathy. Thus, we propose a combination of computational and experimental biophysics-based strategies presented here to improve the future perspective for ligand-based drug discovery strategies for TRPV2.
Author statement
Each named author has substantially contributed to conducting the underlying research and drafting this manuscript. Additionally, the named authors have declared no conflict of interest, financial or otherwise. All authors approved the submission to CSBJ. The manuscript has not been submitted to another journal prior to this submission.
Declarations of interest
None.
Acknowledgements
Authors acknowledge financial support by the Spanish Government MCIN/AEI/ 10.13039/501100011033 (Project PID2020–120222GB-I00 to A.P.-M.), Ministerio de Universidades Margarita Salas Award (MGSD2021-10 to M.L.-M.), Universitat Autònoma De Barcelona Predoctoral Fellowship (B21P0033 to E.C.-H.), the Royal Society of Chemistry for Financial Support through a RS International Exchanges award (IES\R3\193089 to C.D.), and by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through the collaborative research center 1507 “Membrane-associated Protein Assemblies, Machineries, and Supercomplexes” – Project ID 450648163 (to UAH) and the Cluster of Excellence “Balance of the Microverse” EXC 2051 – Project-ID 390713860 (to UAH).
Material and methods
A detailed description is provided in the Supplementary Material.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.12.028.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Ramsey I.S., Delling M., Clapham D.E. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/ANNUREV.PHYSIOL.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 2.Smani T., Shapovalov G., Skryma R., Prevarskaya N., Rosado J.A. Functional and physiopathological implications of TRP channels. Biochim Et Biophys Acta (BBA) - Mol Cell Res. 2015;1853:1772–1782. doi: 10.1016/J.BBAMCR.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Link T.M., Park U., Vonakis B.M., Raben D.M., Soloski M.J., Caterina M.J. TRPV2 has a pivotal role in macrophage particle binding and phagocytosis. Nat Immunol. 2010;11:232–239. doi: 10.1038/ni.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doñate-Macián P., Jungfleisch J., Pérez-Vilaró G., Rubio-Moscardo F., Perálvarez-Marín A., Diez J., Valverde M.A. The TRPV4 channel links calcium influx to DDX3X activity and viral infectivity. Nat Commun. 2018;9:1–13. doi: 10.1038/s41467-018-04776-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vriens J., Appendino G., Nilius B. Pharmacology of Vanilloid Transient Receptor Potential Cation Channels. Mol Pharm. 2009;75:1262–1279. doi: 10.1124/MOL.109.055624. [DOI] [PubMed] [Google Scholar]
- 6.Katanosaka Y., Iwasaki K., Ujihara Y., Takatsu S., Nishitsuji K., Kanagawa M., Sudo A., Toda T., Katanosaka K., Mohri S., Naruse K. TRPV2 is critical for the maintenance of cardiac structure and function in mice. Nat Commun. 2014;5:1–14. doi: 10.1038/ncomms4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen M.R., Johnson W.M., Pilat J.M., Kiselar J., DeFrancesco-Lisowitz A., Zigmond R.E., Moiseenkova-Bell V.Y. Nerve growth factor regulates transient receptor potential vanilloid 2 via extracellular signal-regulated kinase signaling to enhance neurite outgrowth in developing neurons. Mol Cell Biol. 2015;35:4238–4252. doi: 10.1128/MCB.00549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shibasaki K., Murayama N., Ono K., Ishizaki Y., Tominaga M. TRPV2 enhances axon outgrowth through its activation by membrane stretch in developing sensory and motor neurons. J Neurosci. 2010;30:4601–4612. doi: 10.1523/JNEUROSCI.5830-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doñate-Macián P., Gómez A., Dégano I.R., Perálvarez-Marín A., Doñate-Macián P., Gómez A., Dégano I.R., Perálvarez-Marín A. A TRPV2 interactome-based signature for prognosis in glioblastoma patients. Oncotarget. 2018;9:18400–18409. doi: 10.18632/ONCOTARGET.24843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercado J., Gordon-Shaag A., Zagotta W.N., Gordon S.E. Ca2+-dependent desensitization of TRPV2 channels is mediated by hydrolysis of phosphatidylinositol 4,5-bisphosphate. J Neurosci. 2010;30:13338–13347. doi: 10.1523/JNEUROSCI.2108-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 12.Perálvarez-Marín A., Doñate-Macian P., Gaudet R. What do we know about the transient receptor potential vanilloid 2 (TRPV2) ion channel? FEBS J. 2013;280:5471–5487. doi: 10.1111/febs.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumura T., Hashimoto H., Sekimizu M., Saito A.M., Motoyoshi Y., Nakamura A., Kuru S., Fukudome T., Segawa K., Takahashi T., Tamura T., Komori T., Watanabe C., Asakura M., Kimura K., Iwata Y. Tranilast for advanced heart failure in patients with muscular dystrophy: a single-arm, open-label, multicenter study. Orphanet J Rare Dis. 2022;17 doi: 10.1186/s13023-022-02352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. Rubinstein, N. Robbins, K. Evans, G. Foster, K. Mcconeghy, T. Onadeko, J. Bunke, M. Parent, X. Luo, J. Joseph, W.-C. Wu, Repurposing Probenecid for the Treatment of Heart Failure (Re-Prosper-HF): a study protocol for a randomized placebo-controlled clinical trial, (2022). 10.1186/s13063-022-06214-y. [DOI] [PMC free article] [PubMed]
- 15.Peitzman S.J., Fernandez P.C., Bodison W., Ellis I. Ticrynafen and probenecid in hyperuricemic, hypertensive men. Clin Pharm Ther. 1979;26:205–208. doi: 10.1002/cpt1979262205. [DOI] [PubMed] [Google Scholar]
- 16.Park J.B., Kim S.-J. Anti-hypertensive effects of probenecid via inhibition of the α-adrenergic receptor. Pharm Rep. 2011;63:1145–1150. doi: 10.1016/s1734-1140(11)70633-8. [DOI] [PubMed] [Google Scholar]
- 17.Jin X., Touhey J., Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J Biol Chem. 2006;281:25006–25010. doi: 10.1074/JBC.C600153200. [DOI] [PubMed] [Google Scholar]
- 18.Huynh K.W., Cohen M.R., Jiang J., Samanta A., Lodowski D.T., Zhou Z.H., Moiseenkova-Bell V.Y. Structure of the full-length TRPV2 channel by cryo-EM. Nat Commun. 2016;7 doi: 10.1038/NCOMMS11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zubcevic L., Herzik M.A., Chung B.C., Liu Z., Lander G.C., Lee S.Y. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat Struct Mol Biol. 2016;23:180–186. doi: 10.1038/NSMB.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zubcevic L., Le S., Yang H., Lee S.Y. Conformational plasticity in the selectivity filter of the TRPV2 ion channel. Nat Struct Mol Biol. 2018;25:405–415. doi: 10.1038/S41594-018-0059-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madej M.G., Ziegler C.M. Dawning of a new era in TRP channel structural biology by cryo-electron microscopy. Pflug Arch. 2018;470:213–225. doi: 10.1007/S00424-018-2107-2/METRICS. [DOI] [PubMed] [Google Scholar]
- 22.Pumroy R.A., Samanta A., Liu Y., Hughes T.E.T., Zhao S., Yudin Y., Rohacs T., Han S., Moiseenkova-Bell V.Y. Molecular mechanism of TRPV2 channel modulation by cannabidiol. Elife. 2019;8 doi: 10.7554/eLife.48792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su N., Zhen W., Zhang H., Xu L., Jin Y., Chen X., Zhao C., Wang Q., Wang X., Li S., Wen H., Yang W., Guo J., Yang F. Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands. Nat Chem Biol. 2023;19:72–80. doi: 10.1038/s41589-022-01139-8. [DOI] [PubMed] [Google Scholar]
- 24.Pumroy R.A., Protopopova A.D., Fricke T.C., Lange I.U., Haug F.M., Nguyen P.T., Gallo P.N., Sousa B.B., Bernardes G.J.L., Yarov-Yarovoy V., Leffler A., Moiseenkova-Bell V.Y. Structural insights into TRPV2 activation by small molecules. Nat Commun. 2022;13 doi: 10.1038/s41467-022-30083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conde J., Pumroy R.A., Baker C., Rodrigues T., Guerreiro A., Sousa B.B., Marques M.C., de Almeida B.P., Lee S., Leites E.P., Picard D., Samanta A., Vaz S.H., Sieglitz F., Langini M., Remke M., Roque R., Weiss T., Weller M., Liu Y., Han S., Corzana F., Morais V.A., Faria C.C., Carvalho T., Filippakopoulos P., Snijder B., Barbosa-Morais N.L., Moiseenkova-Bell V.Y., Bernardes G.J.L. Allosteric antagonist modulation of TRPV2 by piperlongumine impairs glioblastoma progression. ACS Cent Sci. 2021;7:868–881. doi: 10.1021/acscentsci.1c00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L., Simonsen C., Zimova L., Wang K., Moparthi L., Gaudet R., Ekoff M., Nilsson G., Hellmich U.A., Vlachova V., Gourdon P., Zygmunt P.M. Cannabinoid Non-Cannabidiol Site Modulation of TRPV2 Structure and Function. Nat Commun. 2022;13 doi: 10.1038/s41467-022-35163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perálvarez-Marín A., Solé M., Serrano J., Taddeucci A., Pérez B., Penas C., Manich G., Jiménez M., D’Ocon P., Jiménez-Altayó F. Evidence for the involvement of TRPV2 channels in the modulation of vascular tone in the mouse aorta. Life Sci. 2023;336 doi: 10.1016/J.LFS.2023.122286. [DOI] [PubMed] [Google Scholar]
- 28.Irwin J.J., Shoichet B.K. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. (+) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J.P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V., Djoumbou Y., Eisner R., Guo A.C., Wishart D.S. DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He M., Yan X., Zhou J., Xie G. Traditional Chinese Medicine Database and Application on the Web†. J Chem Inf Comput Sci. 2001;41:273–277. doi: 10.1021/CI0003101. [DOI] [PubMed] [Google Scholar]
- 32.Bajusz D., Rácz A., Héberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Chemin. 2015;7:1–13. doi: 10.1186/S13321-015-0069-3/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bang S., Kim K.Y., Yoo S., Lee S.H., Hwang S.W. Transient receptor potential V2 expressed in sensory neurons is activated by probenecid. Neurosci Lett. 2007;425:120–125. doi: 10.1016/J.NEULET.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 34.Hu H.Z., Gu Q., Wang C., Colton C.K., Tang J., Kinoshita-Kawada M., Lee L.Y., Wood J.D., Zhu M.X. 2-aminoethoxydiphenyl borate is a common activator of TRPV1, TRPV2, and TRPV3. J Biol Chem. 2004;279:35741–35748. doi: 10.1074/JBC.M404164200. [DOI] [PubMed] [Google Scholar]
- 35.Gochman A., Tan X.-F., Bae C., Chen H., Swartz K.J., Jara-Oseguera A. Cannabidiol sensitizes TRPV2 channels to activation by 2-APB. Elife. 2023;12 doi: 10.7554/eLife.86166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng S., Pumroy R.A., Protopopova A.D., Moiseenkova-Bell V.Y., Im W. Modulation of TRPV2 by endogenous and exogenous ligands: a computational study. Protein Sci. 2023;32 doi: 10.1002/PRO.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miao Y., Bhattarai A., Wang J. Ligand Gaussian accelerated molecular dynamics (LiGaMD): characterization of ligand binding thermodynamics and kinetics. J Chem Theory Comput. 2020;16:5526–5547. doi: 10.1021/ACS.JCTC.0C00395/SUPPL_FILE/CT0C00395_SI_006.MP4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suades A., Alcaraz A., Cruz E., Álvarez-Marimon E., Whitelegge J.P., Manyosa J., Cladera J., Perálvarez-Marín A. Structural biology workflow for the expression and characterization of functional human sodium glucose transporter type 1 in Pichia pastoris. Sci Rep. 2019;9:1–11. doi: 10.1038/s41598-018-37445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers B.R., Bohlen C.J., Julius D. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 2008;58:362–373. doi: 10.1016/J.NEURON.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neeper M.P., Liu Y., Hutchinson T.L., Wang Y., Flores C.M., Qin N. Activation properties of heterologously expressed mammalian TRPV2: evidence for species dependence. J Biol Chem. 2007;282:15894–15902. doi: 10.1074/jbc.M608287200. [DOI] [PubMed] [Google Scholar]
- 41.Qin N., Neeper M.P., Liu Y., Hutchinson T.L., Lou Lubin M., Flores C.M. TRPV2 is activated by cannabidiol and mediates CGRP release in cultured rat dorsal root ganglion neurons. J Neurosci. 2008;28:6231–6238. doi: 10.1523/JNEUROSCI.0504-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyberg M., Piil P., Kiehn O.T., Maagaard C., Jørgensen T.S., Egelund J., Isakson B.E., Nielsen M.S., Gliemann L., Hellsten Y. Probenecid Inhibits μ-Adrenergic Receptor-Mediated Vasoconstriction in the Human Leg Vasculature. Hypertension. 2018;71:151–159. doi: 10.1161/HYPERTENSIONAHA.117.10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalibala J., Pechère-Bertschi A., Desmeules J. Gender differences in cardiovascular pharmacotherapy-the example of hypertension: a mini review. Front Pharm. 2020 doi: 10.3389/fphar.2020.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material