Summary
While fruit flies (Drosophila melanogaster) and humans exhibit immune system dysfunction in space, studies examining their immune systems’ interactions with natural parasites in space are lacking. Drosophila parasitoid wasps modify blood cell function to suppress host immunity. In this study, naive and parasitized ground and space flies from a tumor-free control and a blood tumor-bearing mutant strain were examined. Inflammation-related genes were activated in space in both fly strains. Whereas control flies did not develop tumors, tumor burden increased in the space-returned tumor-bearing mutants. Surprisingly, control flies were more sensitive to spaceflight than mutant flies; many of their essential genes were downregulated. Parasitoids appeared more resilient than fly hosts, and spaceflight did not significantly impact wasp survival or the expression of their virulence genes. Previously undocumented mutant wasps with novel wing color and wing shape were isolated post-flight and will be invaluable for host-parasite studies on Earth.
Subject areas: Parasitology, Biological sciences, Entomology
Graphical abstract
Highlights
-
•
Spaceflight enhances immune gene expression and tumor development in fruit flies
-
•
Drosophila parasitoid wasps can develop in space without losing virulence
-
•
Parasitoid mutants with visible alterations were obtained
Parasitology; Biological sciences; Entomology
Introduction
As humans become a space-faring species, they must confront the dual and long-term challenges of microgravity and radiation. Robust immune physiology and intact genomes are vital to the success of manned space exploration efforts. Animals and plants harbor a diverse array of microbial and metazoan pathogens and parasites. While the effects of the space environment in low Earth orbit are documented on many host species, with a focus on humans,1 the effects of space on the accompanying pathogens and parasites are not known. This question is especially important as immune-compromised humans (and other animal hosts), or those with dysfunctional immunity, venture into space. Experiments with the Drosophila model organism and studies on astronauts over the last decade have revealed how highly conserved innate immune functions and mechanisms are altered in space.2,3 These and earlier studies (reviewed by Iyer et al.4) also demonstrated that fruit flies can successfully develop in space, opening the system to further inquiry. In this study, we examined the effects of spaceflight on D. melanogaster infested with their natural parasitoid wasps that journeyed to and back from the International Space Station (ISS), aboard SpaceX-14, on a 34-day mission.
Parasitoid wasps represent a large class of obligate parasitic insects, and many are used in the biocontrol of agricultural pests. The term endoparasitoid refers to the wasp whose preimaginal stages develop within the host larva and pupa. The developing parasitoid (or parasite) eats the host as both continue to develop; the host builds a puparium that a parasite ultimately occupies and emerges from. Drosophila larvae and pupae serve as hosts to more than 60 species of such parasitic wasps.5 The female wasp’s sharp ovipositor pierces the first line of host defense—the larval exoskeleton, made of cuticle—to introduce an egg and venom. This step triggers an encapsulation response in which larval blood cells (hemocytes) of the hosts’ innate immune system (macrophage-like plasmatocytes—called macrophages here—and their derivatives, lamellocytes) cooperate to recognize, surround, melanize, and destroy the wasp egg.6 This melanotic capsule itself, composed of dozens of macrophages and lamellocytes, shields the host from the dead parasite as it continues to become an adult fly. Encapsulation, an innate immune response, is conserved across many multicellular animals.6,7
Encapsulation of the parasite egg by Drosophila larvae is not always fatal to the parasite, as active and passive means to avoid encapsulation have evolved to assure that at least some percentage of parasitic wasp infections are successful. Leptopilina heterotoma (Lh) is highly successful on many Drosophila species and is considered to be a generalist parasite.8 Its venom contains spiked extracellular vesicle (EV)-like structures with immune-suppressive activities.9,10 Lh EVs are surrounded by a phospholipid bilayer, and their protein composition is similar to that of mammalian EVs.11,12 L. boulardi (Lb) is a specialist parasite and is successful on a narrower host range than Lh.8,13 Lb EVs also bear spikes, are immune-suppressive, and have a protein profile similar to Lh EVs.13,14,15,16 There is no evidence that either Lb or Lh EV-like structures possess a genome or that they replicate in either the wasp or the host. The protein-coding genes of Lh EVs are present in the wasp genome.12
While the venom EVs from both Lb and Lh are immune suppressive, their effects on hemocytes differ greatly. Lb EVs enter lamellocytes and alter lamellocyte shape to suppress encapsulation without killing them.17 Lh EVs, on the other hand, lyse lamellocytes within a few hours after infection.18,19 Lh EVs also kill circulating macrophages as well as hemocyte progenitors housed within a small hematopoietic organ called the lymph gland.18,20 Thus, despite differing in the ways they attack host hemocytes, both wasps effectively create immune-deficient hosts, making it conducive for wasp development. In 20–25 days, one adult wasp emerges per fly pupa; male and female wasps mate as free-living adults, and females then deposit eggs into fly larvae to initiate another generation.8,13,21
In this Fruit Fly Lab-03 (FFL-03) spaceflight experiment, we compared the development of two fly strains: one healthy and another suffering from chronic inflammation (CI). The latter strain carries a dominant germline mutation in the Janus kinase (JAK) gene to activate the transcription factor called Signal Transducer and Activator of Transcription (STAT). The mutation constitutively activates the fly’s innate immune JAK-STAT and Toll signaling pathways.22,23,24 Components of both pathways are highly conserved in evolution, including humans. Both immune pathways also control hematopoietic development. Their genetic activation in the fly leads to the overgrowth of hematopoietic progenitors. As a result, small granuloma-like tumors form melanized structures that are observed in the larval hemolymph.23,24,25,26 In their physical properties, these small tumors are analogous to the hemocyte capsules formed around wasp eggs and are considered to be autoimmune-like reactions to self-tissue.27,28 Tumor growth, inflammatory gene expression, and metabolic inflammation in hopTum-l larvae are sensitive to aspirin administration.24 Thus, the hopTum-l background is a “stand-in” disease model for animal leukemia and CI.22,23,24 In humans, aberrant JAK-STAT and Toll-nuclear factor κB (NF-κB) signaling is linked to malignancies, immune deficiency, inflammation, autoimmunity, and cancer and is therefore the focus of therapeutic measures.29,30
The aims of this study were to (a) compare how conditions in the ISS affect healthy control and CI fly hosts; (b) examine if the development of endoparasites occurs normally in the ISS; and (c) study if parasite virulence is affected in space. We report that fewer than expected tumor-free control fruit fly strain returned to Earth, but, unexpectedly, the tumor-bearing CI strain succeeded in space just as well as ground controls. In both strains, genes encoding extracellular matrix (ECM) proteins or ECM-associated proteins (together, matrisome proteins) that contribute to cuticle and chorion structure and function were strongly affected in space, but genes essential for life were downregulated only in the control fruit fly strain. Orthologs of many of these genes have disease relevance in humans. However, inflammation-related genes were activated in both strains, and, while the control flies remained free of tumors, the tumor burden in the space-returned CI strain increased. The effects of spaceflight on parasite emergence were minimal, if any, and changes in the expression of virulence genes were modest, at best. The venom activity of Lh from space wasps was comparable to that of ground wasps, and their virulence EV particles were normally distributed within the hosts. Visible mutations affecting wing shape, color, and ovipositor integrity were identified in the progeny of space L. heterotoma.
Pathogens and parasites pose a threat to astronaut’s health during spaceflight. Bacteria reared under microgravity conditions exhibit an increase in virulence.31,32 Thus, in addition to assessing changes in host immune mechanisms, it is important to evaluate changes in pathogen or parasite virulence in space in the context of their natural hosts. Our studies show that metazoan parasites can develop normally in space, retaining their infection strategies. Continued analysis of such systems will be important to help establish multigenerational studies and to obtain a fine-grained understanding of the long-term effects of space on diverse organisms.
Results
Spaceflight compromises y w but not hopTum-l emergence
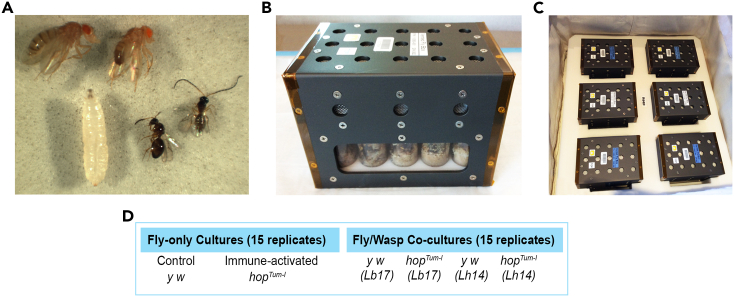
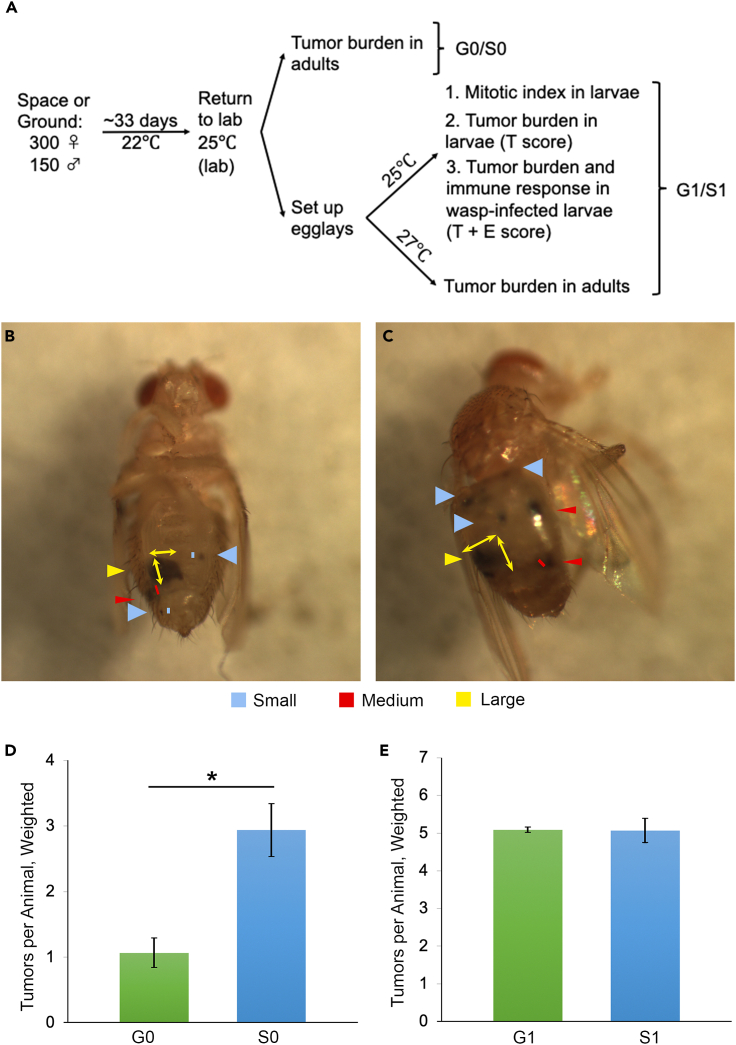
Lb and Lh females attack their larval hosts, introducing one or more egg and venom into the host’s body cavity. Depending on the host’s ability to defend itself and the wasp’s ability to overcome host defenses, one of the two insects emerges alive from the host’s puparium (Figure 1A). To study the effects of spaceflight on this host-parasite scuffle, naive hosts or Lb17-/Lh14-infected hosts were held in ventilated fly boxes (VFBs); these fly boxes were then placed into a cargo transfer bag (CTB) aboard the ISS or at the ground facility tracking ISS conditions (Figures 1B and 1C). The FFL-03 experimental design consisted of two fly-only cultures (fly strains y w and y w, hopTum-l) and four fly/parasite co-cultures on each host strain (Figure 1D). Survival of animals was scored post-flight (see STAR Methods).
Figure 1.
The FFL-03 experimental design
(A) D. melanogaster adults (top left) and larva (bottom left) imaged together with parasitoid wasps (bottom right). Female wasps have shorter antennae than males (right).
(B) Vented fly box (VFB) with 15 holding slots for fly vials.
(C) Six VFBs in one cargo transfer bag (CTB). Samples in the CTB were placed in the ISS by the crew. The ISS flight conditions were mirrored at the Kennedy Space Center laboratory, where ground control samples were housed for the duration of the mission.
(D) The FFL-03 experimental design consisted of two fly-only cultures and four fly/wasp co-cultures, as shown. See STAR methods for more details.
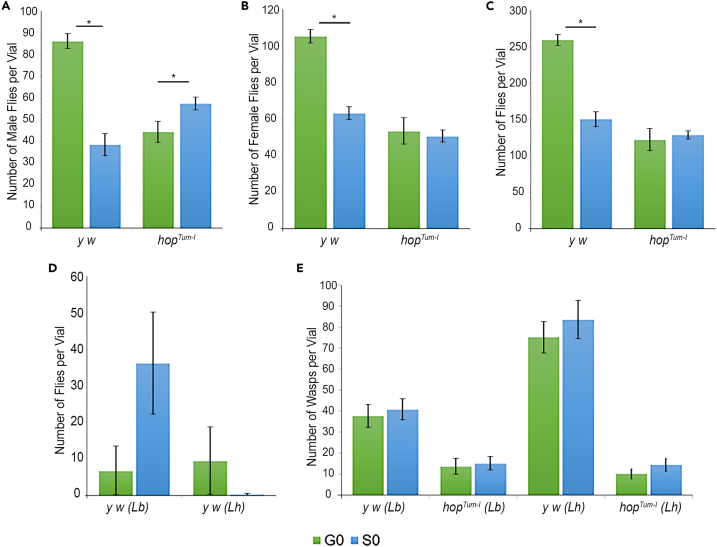
We found that the overall yield of both male and female y w flies in each vial was roughly half of that of the ground-reared flies (Figures 2A–2C; FFL-03 ground control and space samples retrieved post-flight are referred to as G0 and S0, respectively). The trend was somewhat different for the hopTum-l animals that did not appear to suffer from the effects of spaceflight; while spaceflight actually favored male viability slightly but significantly (average 44 versus 57 males/vial), female development remained unaffected (Figures 2A–2C). Thus, under the FFL-03 experimental protocol, animals of the two genetic backgrounds responded differently to spaceflight, underscoring the complex interplay of these factors. There was no significant difference in the yields of flies and wasps from the fly/wasp co-culture vials (Figures 2D and 2E).
Figure 2.
Survival of flies and wasps in ground control (G0) and space (S0) samples
(A–C) The average numbers of adult male (A), adult female (B), and all adult (C) flies per vial in naive y w and hopTum-l cultures. The number of animals, averaged from 14 vials, is shown below. Error bars show standard error. y w males per vial in G0 and S0 samples = 86 ± 3 and 39 ± 5; p ≤ 2.23 × 10−8. hopTum-l males per vial in G0 and S0 samples = 44 ± 5 and 57 ± 3; p = 0.03. y w females per vial in G0 and S0 samples = 105 ± 4 and 63 ± 4; p = 1.03 × 10−8. hopTum-l females per vial in G0 and S0 samples = 53 ± 7 and 51 ± 3; p = 0.74. Total y w adults per vial in G0 and S0 samples = 260 ± 8 and 151 ± 10; p = 5.38 × 10−10. Total hopTum-l adults per vial in G0 and S0 samples = 122 ± 15 and 129 ± 6; p = 0.38.
(D and E) The number of G0 adult flies (D) and wasps (E) per vial in fly-wasp co-cultures. Error bars show standard error. G0 y w from Lb infections = 7 ± 7; S0 y w from Lb infections = 36 ± 14; p = 0.07. G0 y w from Lh infections = 10 ± 9; S0 y w from Lh infections = 0.36 ± 0.17; p = 0.33. G0 Lb on y w hosts = 38 ± 5; S0 Lb on y w hosts = 41 ± 5; p = 0.31. G0 Lb on hopTum-l hosts = 14 ± 4; S0 Lb on hopTum-l hosts = 15 ± 3; p = 0.78. G0 Lh on y w hosts = 75 ± 7; S0 Lh on y w hosts = 84 ± 9; p = 0.48. G0 Lh on hopTum-l hosts = 10 ± 2; S0 Lh on hopTum-l hosts = 14 ± 3; p = 0.26. See also Figure S9.
Effects of spaceflight on gene expression in adult flies
To understand the molecular basis of the effects of spaceflight on flies, we analyzed bulk RNA sequencing (RNA-Seq) gene expression results in naive adult flies. Differentially expressed genes (DEGs) were defined as adjusted p < 0.05 and |log2Fold Change (FC)| > 1. The close clustering of the four replicates and high variation among the four sample types in the principal-component analysis (PCA) show that the experiment was well controlled (Figure S1A). Overall, the y w strain was more sensitive to spaceflight than hopTum-l, with 13.22% versus 6.78% DEGs (spaceflight versus ground control samples), respectively (Table S1), a difference that is also reflected in the PCA plot (Figure S1A). A separate RNA-Seq analysis of white-eyed (w1118) flies, also on the SpaceX-14 mission, reported 6.49% and 3.25% DEGs in S0 versus G0 female and male fly heads, respectively (false discovery rate [FDR]-adjusted p values < 0.05; total number of genes expressed was 13,99133). Although the sample types in the two studies are different, these results suggest that the y w strain is more sensitive to space conditions than the y w hopTum-l and the w1118 strains. On Earth, the hopTum-l strain showed 9.69% DEGs relative to y w, with more than two-thirds of DEGs upregulated and the remaining DEGs downregulated. In space, this differential expression was enhanced (14.03% of genes affected), but the proportion of upregulated versus downregulated genes was roughly equal (Table S1).
Volcano plots of the four comparisons revealed distinct gene identity profiles in each comparison (Figures S1B–S1E). This distinction between the strains is also clearly evident from Gene Ontology-based Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showing active Toll/Imd (immune deficiency) signaling not only in naive G0 hopTum-l compared to y w flies but, surprisingly, also in S0 y w and hopTum-l flies, relative to their G0 counterparts (Figure S2A). Furthermore, for these same three comparisons, the top 20 significantly enriched (adjusted p value < 0.05) Gene Ontology terms in Biological Process enrichment analyses included host defense terms such as response to bacterium, response to other organism, and response to biotic stimulus (Figure S2B). These four comparisons allowed us to decouple the effects of genetic strain differences in the S0/G0 culture conditions. We next examined changes in expression patterns of the most significantly DEGs (Top DEGs), genes essential to fly viability (Essential DEGs), and immune genes in the JAK/STAT and Toll/Imd pathways (Pathway DEGs), as summarized in Figure S1F.
Top DEGs in space include members of protease, endopeptidase, and matrisome gene families
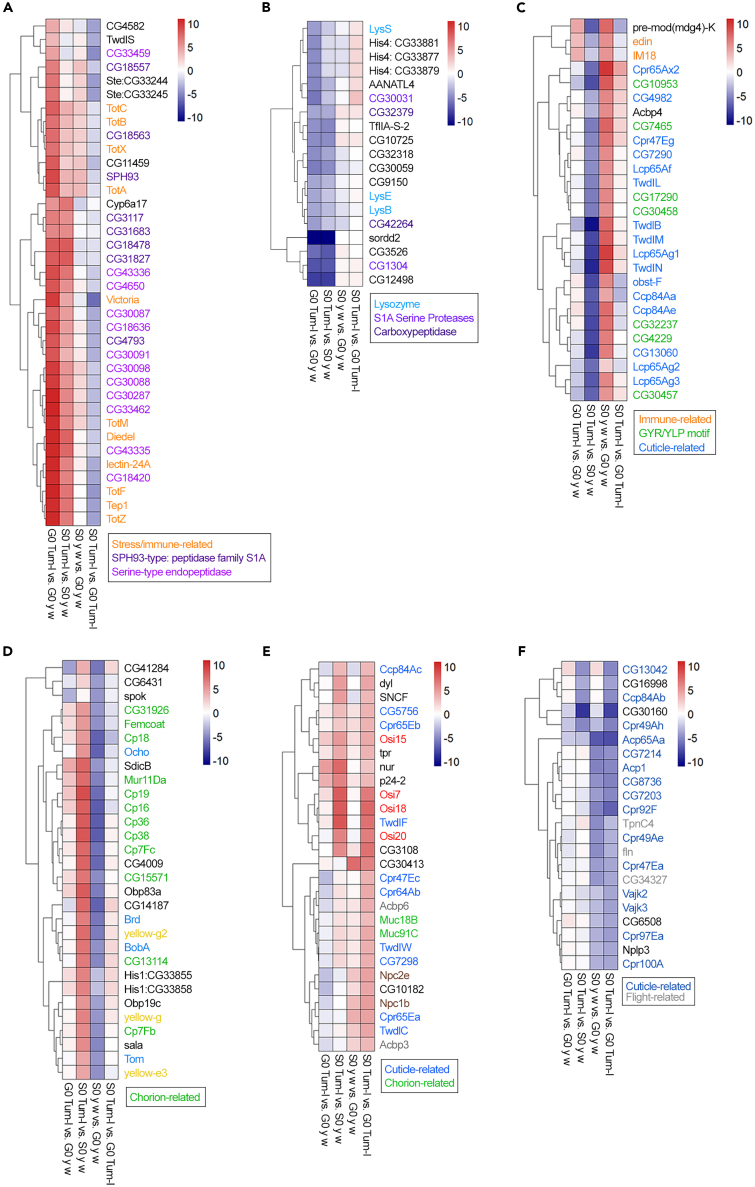
We analyzed 172 annotated Top DEGs (see STAR methods for selection of Top DEGs) and identified six profiles described here in two broad categories (Figures 3A–3F). In the first category (Figures 3A and 3B), we consider the Top DEGs showing differential expression in the two fly strains as follows. (a) Top DEGs upregulated in G0 hopTum-l versus G0 y w that maintain their expression patterns in space (S0 hopTum-l versus S0 y w) include genes annotated for stress- and immune-related peptides (Tot and Tep family members), proteases, and endopeptidases (Figure 3A). (b) The Top DEGs downregulated in G0 hopTum-l versus G0 y w that maintain their expression patterns in space (S0 hopTum-l versus S0 y w) include members of the lysozyme gene family, histone H4 genes, serine proteases, carboxypeptidases, and other enzymes (Figure 3B). Exceptions to both of these trends were also observed (Figures 3A and 3B). These results define strong differences in DEG patterns in hopTum-l and y w flies. They also show that the molecular gene expression differences between two genetic strains, for many but not all genes, are maintained in spaceflight conditions.
Figure 3.
Differential expression of Top DEGs in S0/G0 adult flies
(A and B) Top DEGs upregulated (A) or downregulated (B) in G0 hopTum-l vs. G0 y w. Most genes in panel A that maintain differential expression in space are Toll pathway target genes34,35 (TotM and Tep1 family members, SPH93, and CG18563). Lectin-24A is predicted to bind galactose, and its human homolog is associated with the 3MC syndrome 3.36 Notable DEGs in panel (B) with disease-relevant human orthologs are CG32379, CG32318, and CG30059.37
(C and D) Comparisons showing differential expression of upregulated (C) and downregulated (D) genes in S0 y w versus G0 y w. Cuticle-related and chorion-family genes are included in these profiles. The fly histone H1s (orthologous to the human H1 gene, implicated in acute lymphoblastic leukemia38) and CG6431, a triacylglycerol hydrolase (orthologous to several human disease genes implicated in cardiovascular disease and type-2 diabetes), are downregulated (D).
(E) Genes in hopTum-l whose expression is activated in space. Cuticle-related and chorion-family genes are included in this profile. CG3108 is predicted to enable metallocarboxypeptidase activity, whose human orthologs are implicated in familial febrile seizures 11 and familial temporal lobe epilepsy 5.37
(F) Genes in y w and hopTum-l whose expression is downregulated in space. Cuticle-related and flight-related genes are included. Flight-related genes are CG34327 (associated with abnormal flight), flightin (fln), and Troponin C isoform 4 (TpnC4). See also Figure S1.
In the second broad category, we consider the effects of spaceflight on Top DEGs in individual strains as follows (Figures 3C–3F). (a) Top DEGs upregulated in S0 y w, when compared to the respective ground control group (i.e., G0 y w), encode (i) structural proteins for cuticle (e.g., chitin-binding domain-containing proteins of the Twdl, CPLCA, and Cpr families) and (ii) proteins with the GYR/YLP domain or immune response-related proteins (Figure 3C). (b) Top DEGs downregulated in S0 y w flies but not in S0 hopTum-l flies, when compared to respective ground control groups, include members of the yellow gene family (yellow-g, yellow-g2, yellow-e3), bearded gene family (Brd, Tom, BobA), Histone H1s, and several genes for chorion constituents and chorion assembly (e.g., Cp7Fb, Cp7Fc, Cp16, Cp19, Cp36, Cp38, Mur11Da; Figure 3D). These Cp genes are clustered on the X and third chromosomes, and the gene clusters are selectively amplified in the follicle cells of the ovary.39 (c) Top DEGs upregulated in S0 hopTum-l, when compared to the respective ground control group (G0 hopTum-l), include Cpr family genes, Osiris family genes, tracheal-prostasin (tpr), a protein important for sleep regulation (nemuri, nur), and CG3108 (Figure 3E). (d) Top downregulated DEGs in S0 versus G0 flies of both genetic backgrounds include several cuticle-related (CPLCA, CPLCP) families and flight-related genes (e.g., flightin [fln] and Troponin C isoform 4 [TpnC4]; Figure 3F). Chitin supports the epidermal and tracheal cuticles and the peritrophic matrices that line the gut while the chorion makes up the egg shell. These comparative RNA-Seq results suggest that matrisome genes of many multigene families that encode proteins of the Drosophila cuticle40 are sensitive to spaceflight.
Essential genes are downregulated in y w flies exposed to spaceflight
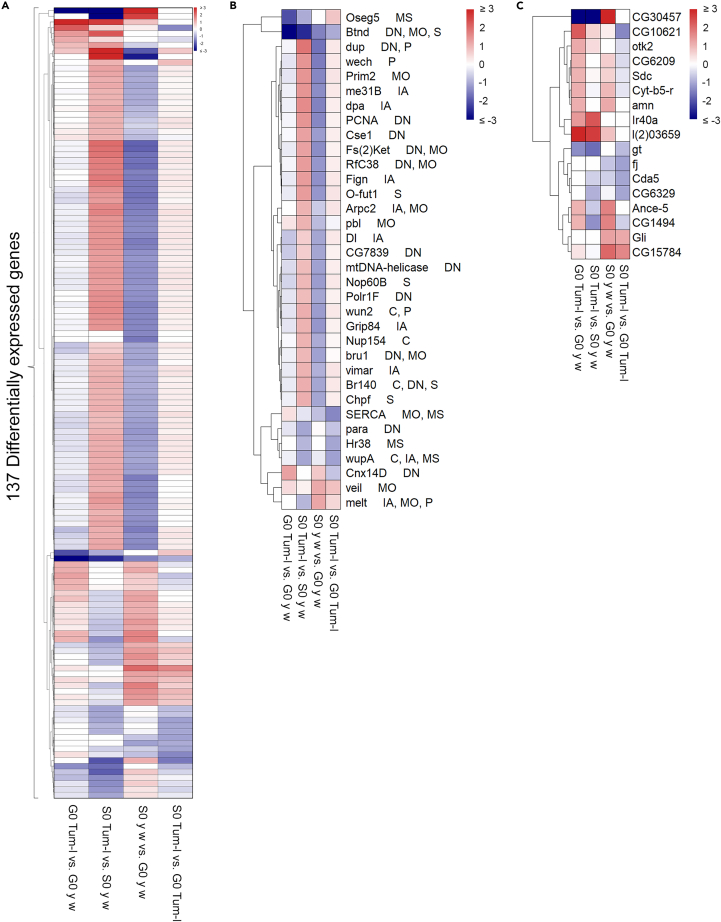
To further understand the differential effects of spaceflight on fly survival, we examined the expression of 1,024 essential genes that have been identified in genetic experiments as indispensable for life (see STAR methods). Across the four comparisons, we found 137 essential genes to be differentially expressed (adjusted p < 0.05 and |log2FC| > 1). Strikingly, the space environment had a profound effect on the differential expression of these genes between the two genetic backgrounds. For example, in G0 hopTum-l versus G0 y w samples, only 15 differentially expressed essential genes were either upregulated (11 genes) or downregulated (4 genes), whereas spaceflight conditions affected the transcription of 95 genes in the S0 hopTum-l versus S0 y w comparison (81 genes upregulated, 14 genes downregulated, Figure 4A).
Figure 4.
Differential expression of essential genes in S0/G0 adult flies
(A) The expression of 137 genes essential to fly viability that were differentially expressed (adjusted p < 0.05 and |log2FC| > 1) in one or more comparisons is shown.
(B) Expression of 34 genes pertinent to human health. The DIOPT category and score-relevant human disease or trait linked to the human ortholog were determined using the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool.41 The disease/trait category for the fly gene refers to at least one human disease or trait linked to one or more human ortholog. MS, musculoskeletal; DN, development/neurological; MO, metabolic-/obesity-related; S, skin-related; P, pulmonary; IA, infection and/or autoimmune; C, coronary-/cardiac-related.
(C) The differential expression of essential genes with no human ortholog or none-to-low disease relevance. See also Figure S1.
The most striking effects of spaceflight were seen in y w flies: 96 genes were either upregulated (24 genes) or downregulated (72 genes) in y w space-flown samples compared to the respective ground control samples. The most strongly upregulated DEGs of these 96 DEGs, with log2-(FC) > 2, include Cpr65Ec (cuticular protein), CG15784, and CG30457 (both latter genes lack annotation). The most highly downregulated genes are S1A serine protease Notopleural and histone H1 His1:CG31617, with log2-(FC) < −1.9. Remarkably, gene expression changes were milder in space-flown hopTum-l samples, with the expression of only 10 genes significantly affected; either 2 genes were upregulated or 8 genes were downregulated, compared to respective ground control samples (Figure 4A). These results likely explain why y w flies were more sensitive to spaceflight than hopTum-l flies.
Differentially expressed essential genes reveal association with human disease conditions
To assess why S0 y w flies were sensitive to spaceflight and if they have relevance to human conditions, we filtered the 137 essential DEGs for a DIOPT (Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool) score of at least 12 and identified 34 high-scoring DEGs that we grouped into broad categories. Strikingly, we observed 22 DEGs that were significantly downregulated in S0 y w versus G0 y w flies. Oseg5 and Biotinidase expression was downregulated in G0 hopTum-l versus G0 y w flies; for both genes, this downregulation was maintained in space. Only two genes, veil and melt, showed significant upregulation (Figure 4B). This analysis revealed that human orthologs of these 34 fly genes affect many aspects of human physiology (see Figure 4B for details). The remaining 17 essential DEGs, with a low DIOPT value (i.e., < 12), were annotated with enzymatic, developmental, or unknown functions. The most significantly upregulated genes in hopTum-l versus y w flies were l(2)03659 (enables organic anion transporter activity) and Ir40a (involved in response to humidity changes),42 both maintaining differential expression in space. In addition, six DEGs, each in S0 y w flies and S0 hopTum-l flies, were either upregulated or downregulated, relative to their corresponding ground controls (Figure 4C).
Spaceflight activates the expression of immune genes in S0 adults and increases inflammatory tumor burden in S0 hopTum-l
A comparison of the Gene Ontology-based pathway and Biological Process enrichment terms associated with the y w versus hopTum-l transcriptional profiles provided a “big picture” view of the differential effects of space on the two strains. Both plots show that, while most GO terms and almost all biological processes are affected in y w flies, only a few parameters change in both strains. Most notable of these are the Toll/Imd pathway and lysosomal pathway terms that are significantly enriched in the space samples for both genotypes, suggesting the activation of inflammation-related genes in space (Figures S2A and S2B).
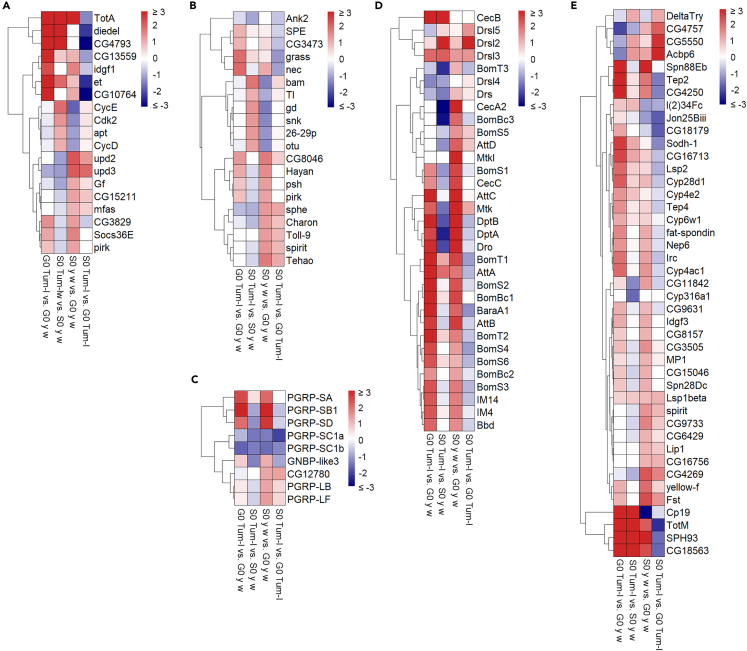
Since the hopTum-l mutant fly strain carries a constitutively active JAK enzyme,43 the transcriptional activation of known JAK-STAT target genes is expected in hopTum-l flies relative to y w flies without this mutation. Consistent with this expectation we found many target genes (Idgf1, SOCS36E, TotA, CG3829, CG4793, CG13559, and CG10764) to be differentially upregulated in G0 hopTum-l versus G0 y w flies. High levels of JAK-STAT pathway genes TotA, diedel, et, and CG4793 in hopTum-l versus y w were maintained in space (Figure 5A). Similarly, several Toll pathway components (PGRPs SA, SB1, and SD, grass, SPE) and target genes were significantly upregulated in G0 hopTum-l versus G0 y w flies (Figures 5B–5E).
Figure 5.
Differentially expressed immune signaling genes in S0/G0 adult flies
(A) JAK-STAT pathway components and target genes are shown based on fold induction. Genes were grouped based on their annotation in FlyBase37 as follows: upd2 and upd3 are ligands; CycD, CycE, and Cdk2 are positive regulators; diedel, Socs36E, and et are negative regulators. The target genes are TotA, Idgf1, pirk, mfas, CG3829, CG4793, CG13559, CG10764, CG15211, and Gf.
(B–E) Differentially expressed Toll pathway components: core components (B), pathogen recognition receptors (C), antimicrobial peptides (D), and other targets (E). Only significantly affected DEGs (adjusted p < 0.05 and |log2FC| > 1, in one or more comparisons) are shown. See also Figures S1 and S2.
Whereas many immune genes are constitutively active in hopTum-l flies, remarkably, spaceflight activated the expression of a majority of these immune genes in y w flies (Figures 5B–5E). The JAK-STAT pathway ligands (upd2 and upd3), several core components (SOCS36E), and target genes (TotA, Gf, mfas, and CG15211) were activated in S0 y w versus G0 y w flies (Figure 5A). A clear signature for Toll/Imd pathway activation was also observed in S0 y w adults. Elevated expression of pathway components (CG8046, Toll-9, Figure 5B), pathogen recognition receptors (PRRs, PGRPs SA, SB1, SD, Figure 5C), several antimicrobial peptide (AMP) genes (Figure 5D), and other Toll/Imd pathway target genes (Figure 5E) was observed in S0 y w versus G0 y w flies. Notably, some Toll/Imd pathway target genes (e.g., Drsl2, 3, 5) were also activated in S0 hopTum-l versus G0 hopTum-l flies (Figure 5D).
To examine if the immune gene expression changes identified in space animals might have influenced tumor development in S0 hopTum-l adults (Figure 6A), we scored tumor number and size (small, medium, large tumors, T score; see STAR methods and Figures 6B and 6C). In 25–30 adults from three independent G0 and S0 cultures, the average weighted T score was 1.07 ± 0.22 in G0 adults and 2.94 ± 0.40 in S0 flies (Figure 6D), although this difference is lost in the adult G1 and S1 progeny of these G0/S0 animals (Figure 6E). Tumors were not detected in S0 y w animals, even though they showed evidence of high immune signaling.
Figure 6.
Tumor burden in naive G0/S0 hopTum-l fly adults
(A) Experimental design for space and ground hopTum-l flies for post-flight experiments. The progeny of 300 females and 150 adult males were raised either in the ISS or at KSC (see STAR methods) at 22°C, and then at 25°C, once returned to the laboratory. Tumors were scored in larval and adult samples.
(B and C) Representative G1 hopTum-l adult female (B) and male (C), showing tumor size classes: small, medium, and large.
(D and E) Weighted average number of tumors per animal (tumor burden) in adult fly samples retrieved from ground (G0) and space (S0) (D) and values from their ground (G1) and space (S1) adult progeny (E). The weighted tumor numbers per animal in G0 and S0 adults were 1.07 ± 0.22 and 2.94 ± 0.40, respectively (mean ± standard error; p = 0.015, Student’s t test; ∗ indicates p < 0.05). The weighted average tumor numbers per animal in G1 and S1 hopTum-l adult males were 5.09 ± 0.08 and 5.07 ± 0.33, respectively (mean ± standard error; p = 0.95, Student’s t test). See also Figures S1 and S2.
High immune gene expression persists in naive S1 larvae
As in the mutant hopTum-l adults, high gene expression levels of the JAK-STAT and Toll/Imd pathway genes encoding ligands, cytoplasmic mediators, and targets were recorded in naive G1 and S1 hopTum-l larvae when compared to G1 y w and S1 y w larvae, respectively (Figures S3A–S3E). Thus, in hopTum-l larvae, the immune effects of spaceflight are evaluated in the background of constitutive immune signaling.
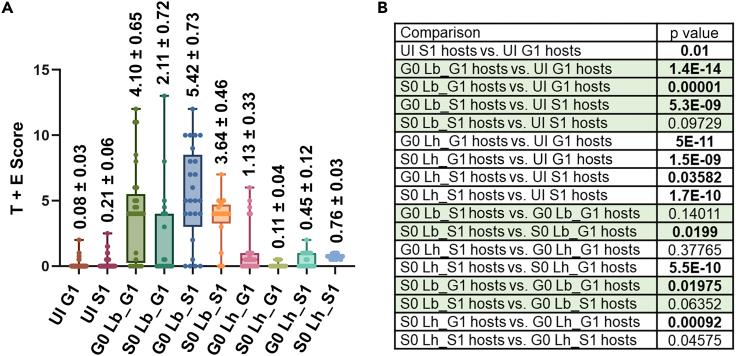
We assessed if naive S1 y w or hopTum-l larvae showed any lingering effects of space observed in their S0 parents. Spaceflight-responsive differentially expressed immune genes in naive S1 y w larvae included CG10764, LUBEL, PGRP-SB1, -SC1a, -SC1b, and AMP genes, CecB, AttC, Drs, and Drsl5 (Figures S3A–S3D). Similarly, naive S1 hopTum-l larvae showed elevated expression of immune targets DptA, DptB, CecA2, Drsl3, Drsl5, and CecA1 relative to their G1 counterparts (Figure S3D). Thus, in both genetic backgrounds, immune signaling was elevated in S1 larvae, even though the affected target genes were different. Naive S1 versus G1 hopTum-l larvae maintained the difference in inflammatory tumor burden observed in their parents (0.08 ± 0.03 and 0.21 ± 0.06, respectively, p = 0.010; Figures 7A and 7B), although the mitotic index in the underlying hematopoietic tissue was not significantly different (2.74 ± 0.93% of G1 and 1.95 ± 0.66% of S1 macrophages were phospho-histone H3 positive; p = 0.53).
Figure 7.
Cellular immunity in G1 and S1 hopTum-l larvae, post parasitization
(A) Tumor (T) scores of uninfected (UI) and tumor and encapsulation (T + E) scores of G0/S0 Lb- or G0/S0 Lh-infected G1/S1 hopTum-l larvae. Value above each bar indicates the mean ± standard error.
(B) Summary of the statistical analysis of pairwise comparisons of T or T + E scores (see STAR methods). Lb17 wasp infection rows are shaded for clarity. p values < 0.05 (Mann-Whitney U test for unpaired samples) were considered significant (bold). See also Figures S3–S7.
Tumor-encapsulation scores are higher in S1 than in G1 hopTum-l hosts
Previous studies with hopTum-l larvae have shown that Lb17 attack provokes stronger cellular immune reactions than Lh14 does. While Lb17 attack elicits encapsulation of the wasp egg due to an overabundance of circulating hemocytes (not recruited into the melanized tumors), Lh14-infected hopTum-l larvae instead develop small melanized specks8 (see Figure S4 for examples of immune reactions). To assess if naive S1 versus G1 hopTum-l larval hosts differ in their ability to mount an encapsulation response, we exposed them to G0/S0 Lb17 or Lh14 wasps and computed a composite tumor-encapsulation (T + E) score for these animals, which measures their self- and wasp egg-induced encapsulation response. The experimental design consisted of scoring T + E scores from all G/S and host/parasite combinations. The following findings emerged from our comparisons of naive versus Lb17- or Lh14-infected hopTum-l hosts.
First, naive versus G0 or S0 Lb-infected G1 or S1 hopTum-l hosts showed a ∼17- to 52-fold increase in T + E scores, and scores from three of the four combinations differed significantly (Figures 7A and 7B). The T + E score in naive versus G0/S0 Lh-infected G1 or S1 hopTum-l hosts in all four combinations similarly increased from 1.4- to 14-fold, and the increases were significantly different across all four comparisons (Figures 7A and 7B). It is noteworthy that the canonical Toll pathway target Drs is strongly upregulated in S1 hopTum-l hosts after Lb17 infection, but its expression remains unchanged after Lh14 infection (Drs, Figures S3F and S3G). This trend is characteristic of these wasps’ effects on wild-type hosts8 and suggests that the overall differences in wasp virulence were maintained in spaceflight.
Second, S1 compared to G1 hosts, infected by S0 (but not G0) wasps of both species (S0 Lb/Lh_S1 hosts versus S0 Lb/Lh_G1 hosts), were more competent at mounting an immune response (3.64 ± 0.46 versus 2.11 ± 0.72 [p = 0.0199] for Lb17 and 0.76 ± 0.03 versus 0.11 ± 0.04 [p = 5.55 e−10] for Lh14; Figures 7A and 7B). Thus, not only is the tumor score higher in naive S1 compared to G1 hopTum-l larvae but S1 hosts are also more immune reactive than G1 hosts.
Third, on ground hosts, space wasps of both species differed from ground wasps in their ability to elicit encapsulation. T + E scores in G1 hopTum-l hosts subjected to S0 or G0 parasites of the same species (S0 Lb/Lh_G1 hosts versus G0 Lb/Lh_G1 hosts) showed that, for both parasites, the T + E values for S0 wasp infections were significantly lower than the corresponding values for G0 parasites (2.11 ± 0.72 versus 4.10 ± 0.65 for Lb17 [p = 0.0199] and 0.11 ± 0.04 versus 1.13 ± 0.33 [p = 0.000915] for Lh14). The difference was not significant in space hosts (Figures 7A and 7B). These results suggest that, while S1 larval progeny may be adapting to normal gravity and other terrestrial conditions, they retain altered immune physiology of their space parents, albeit transiently. S0 wasps largely maintain their inherent virulent strategies, although we could detect subtle differences in their abilities to affect host immunity relative to G0 wasps.
Lymph gland morphologies of naive and infected G0 and S0 hosts
Upon Lb17 attack, hemocytes in wild-type lymph gland lobes divide and differentiate into lamellocytes and disperse into the hemocoel to surround the wasp egg.44,45,46 Hemocytes in the hopTum-l lymph gland actively divide and differentiate in the absence of infection.43 Because of the high inflammation state of space animals, we examined differences in lymph gland lobes from third-instar G0 and S0 animals. A normal lymph gland has multiple, paired lobes, consisting of hemocytes, positioned along the tubular dorsal vessel that continues into a pulsating heart, which directs the flow of the hemolymph and circulating hemocytes into the body cavity (Figures S5A–S5D). The anterior-most lobes are the largest, with hemocytes at varying differentiation stages. The posterior lobes are smaller in size and harbor fewer differentiated cells than the anterior lobes. The margins of all lobes from naive y w ground and space hosts were continuous with none-to-little dispersal. Like lobes from G0 naive y w animals, lobes from S0 naive y w animals did not exhibit any hemocyte loss or any other obvious morphological change (Figures S5A–S5H).
In contrast to naive y w animals, all anterior lobes of naive G0 and S0 hopTum-l larvae were dispersed due to the effect of the mutation (Figures S5I–S5P). The mutation also results in some lobes detaching from the dorsal vessel, and many GFP-positive lamellocytes were present in G0 and S0 hopTum-l samples. Some tumors were observed as small aggregates or as free-floating structures in the hemolymph made of macrophages and lamellocytes (Figures S5J and S5N). No clear difference in the posterior lobes was detected due to spaceflight in either fly strain.
G1 and S1 y w host lymph glands were unresponsive after Lb parasitization (Figures S6A–S6F). In naive G1 hopTum-l lymph glands, GFP-expressing cells were absent (Figures S6G and S6H), whereas in S1 hosts lamellocytes were clearly observed regardless of infection status (Figures S6I–S6P). In some samples, lamellocyte-rich tumors were associated with the anterior and posterior lobes (e.g., S1 sample, Figures S6K and S5L). In only one S1 host, the anterior lobes exhibited a strong anti-wasp response (Figure S6O). Thus, even though the T + E scores rise in response to Lb infection, this difference was not reflected in the lymph gland morphologies of the animals we examined.
Lh14 infection results in the gradual loss of hemocytes in all lobes. This effect is attributed to the presence of EVs in the entire lymph gland.20 EVs were present in hematopoietic progenitor cells and dorsal vessels of G0 and S0 Lh-infected G1 and S1 y w hosts, and this association was accompanied by the loss of hematopoietic progenitors (Figures S7A–S7F). Similarly, in G1 and S1 hopTum-l hosts, the progenitor population was reduced, and EVs were present in the lymph glands of hosts after S0 Lh infection. (G0 Lh wasps were not available for this experiment.) Lymph glands from uninfected G1 or S1 hosts do not possess EVs (Figures S7G–S7N). Thus, spaceflight does not affect the entry or distribution of Lh EVs in the hemolymphatic system.
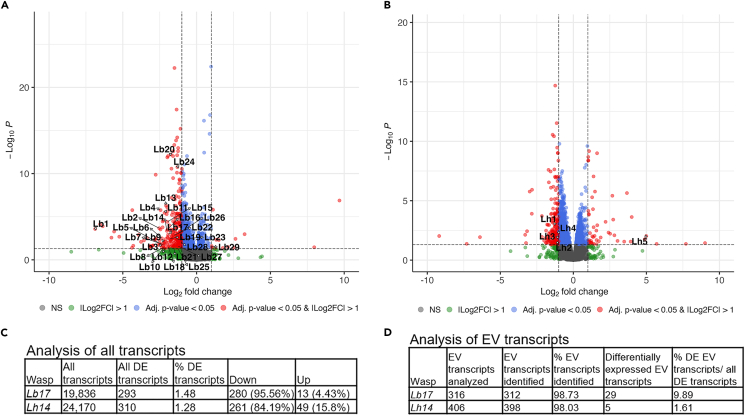
Perturbation of Lb and Lh EV gene expression
The venom fluid containing the EVs is injected into the host during oviposition. Due to their close association with and direct effects on target hemocytes,17,20 our definition of virulence proteins in this study is restricted to those that make up the EVs. Since standard annotations of wasp transcripts are not available, a custom pipeline was developed to analyze gene expression changes against corresponding transcriptomes (Figure S8A). Overall, in adult male and female wasps, the effects of spaceflight were detected (Figures S8B and S8C), although they were modest, and less than 1.5% of all transcripts identified showed significant change when compared to respective ground controls (293 and 310 transcripts, for Lb17 and Lh14, respectively). A majority of the transcripts were downregulated in both wasps (Figures 8A–8C; Tables S2 and S3). Furthermore, the intensity of gene expression change was also mild. Of the downregulated DEGs (adjusted p < 0.05), more than 80% showed log (2)-FC between −1.00 and −2.00 (228/280 in Lb17 and 250/261 in Lh14). The remaining downregulated DEGs showed log (2)-FC between −2.00 and −9.18. Only 6 of all DEGs showed a log (2)-FC > 2 for Lb17; this number was 16 for Lh14. These DEGs do not appear to belong to a specific functional class, and many lack clear annotations.
Figure 8.
Differential expression of EV genes
(A and B) Enhanced volcano plots for DEGs in Lb17 (A) and Lh14 (B) S0 vs. G0 wasp samples. Numerical identifiers correspond to EV genes in Tables 1 and 2, with their corresponding accession numbers.
(C) Number of transcripts from adult male and female wasps, showing fluctuation in space. The term “all transcripts” refers to the number of transcript sequences in the Lb17 (GAJA00000000) and Lh14 (GAJC00000000) transcriptomes that aligned with the corresponding RNA-Seq reads.47 The term “all DE transcripts” refers to the number of differentially expressed transcripts in spaceflight vs. ground control samples (Tables S2 and S3). A majority of transcripts were downregulated. See STAR methods and Figure S8 for more details.
(D) Transcripts coding for EV proteins in the RNA-Seq samples show that 312 Lb17 EV and 398 Lh14 EV transcripts were identified for each wasp. Of these, only 29 Lb17 and 5 Lh14 transcripts were differentially expressed in S0 vs. G0 samples. Their identities, fold-change values, and conserved protein domains are shown in Tables 1 and 2. See also Figures S8, S10, and S11.
The identities of the proteins constituting L. boulardi EVs were recently published.14 Our comparative analysis indicated that Lb EV proteins show an overall profile similar to the Lh EV profile; both are enriched in mammalian EV proteins.15 A majority (> 98%) of the EV transcripts were identified in our RNA-Seq analysis (312/316 for Lb17 and 398/406 for Lh14). In Lb17, 29/312 (9.89%) EV genes were significantly differentially expressed (Figure 8D). Of these, 28 were downregulated (Table 1). Two mildly downregulated genes are predicted to encode RhoGAP superfamily proteins (GAJA01018902.1, log (2)-FC = −1.27, and GAJA01006288.1, log (2)-FC = −1.09). A RhoGAP superfamily protein called LbGAP in Lb EVs is implicated in wasp virulence.48,49 Of the two downregulated wasp RhoGAP superfamily proteins, only one (GAJA01018902.1) showed homology to LbGAP: 69.7% nucleotide sequence identity (68% query coverage; 8e-55). For Lh14, only 5/398 (1.61%) transcripts were significantly differentially expressed. Of these, only one gene, lacking annotation, was about 5-fold upregulated (Table 2). Of the downregulated genes, three lack annotations, and one is predicted to encode a protein with a chitin-binding domain.
Table 1.
Lb17 EV transcripts whose expression is significantly altered (adjusted p < 0.05 and |log2FC| > 1) in S0 wasps compared to G0 wasps
| ID | Accession | Conserved domain | LOG (2)-FC S0 vs. G0 | Adjusted p value S0 vs. G0 |
|---|---|---|---|---|
| 1 | GAJA01001411.1 | CYP4 cd20628 (0E+00) | −6.87725 | 0.00026306 |
| 2 | GAJA01017988.1 | ycf1 superfamily cl42951 (1.59E-04) | −2.53668 | 0.00021388 |
| 3 | GAJA01015791.1 | None | −2.45226 | 0.01972584 |
| 4 | GAJA01018823.1 | PLN02872 superfamily cl28691 (2.42E-38) | −2.44316 | 1.71E-06 |
| 5 | GAJA01012169.1 | None | −2.43751 | 0.01366223 |
| 6 | GAJA01009829.1 | LRR_8 pfam13855 (4.09E-08) | −2.35265 | 0.00318438 |
| 7 | GAJA01006376.1 | ZnMc superfamily cl00064 (1.72E-28) | −2.23343 | 0.01726026 |
| 8 | GAJA01020203.1 | TIL cd19941 (3.57E-03) | −2.22991 | 0.0362285 |
| 9 | GAJA01000968.1 | None | −2.22716 | 0.00809954 |
| 10 | GAJA01006002.1 | None | −2.2178 | 0.03823238 |
| 11 | GAJA01012568.1 | None | −2.17458 | 3.62E-06 |
| 12 | GAJA01020069.1 | None | −2.12765 | 0.04720412 |
| 13 | GAJA01018222.1 | PBP_GOBP pfam01395 (8.21E-20) | −2.07279 | 5.15E-07 |
| 14 | GAJA01008705.1 | CAP_euk cd05380 (4.17E-39) | −2.01981 | 0.00043155 |
| 15 | GAJA01016048.1 | None | −1.98327 | 3.90E-05 |
| 16 | GAJA01014376.1 | None | −1.95966 | 0.00030382 |
| 17 | GAJA01015285.1 | None | −1.93316 | 0.00119667 |
| 18 | GAJA01020187.1 | serpin42Da-like cd19601 (5.03E-116) | −1.93082 | 0.03067199 |
| 19 | GAJA01019354.1 | Amino_oxidase pfam01593 (1.15E-64) | −1.82038 | 0.0035116 |
| 20 | GAJA01005947.1 | nuc_hydro superfamily cl0026 (8.92E-81) | −1.74632 | 6.09E-13 |
| 21 | GAJA01008032.1 | NADB_Rossman superfamily (1.00E-130) | −1.54859 | 0.03714499 |
| 22 | GAJA01011758.1 | None | −1.54458 | 0.00149792 |
| 23 | GAJA01019353.1 | Amino_oxidase pfam01593 (1.15E-64) | −1.32183 | 0.00449535 |
| 24 | GAJA01019883.1 | MYSc_Myh1_insects_crustaceans cd14909 (0E+00) | −1.30444 | 1.74E-11 |
| 25 | GAJA01018902.1 | RhoGAP superfamily cl02570 (1.14E-51) | −1.27277 | 0.04705272 |
| 26 | GAJA01008333.1 | PTZ00184 superfamily cl33172 (2.73E-22) | −1.26373 | 0.0008629 |
| 27 | GAJA01016214.1 | RT_like superfamily cl02808 (7.14E-03) | −1.26184 | 0.02510846 |
| 28 | GAJA01006288.1 | RhoGAP superfamily cl02570 (1.11E-22) | −1.09051 | 0.00805544 |
| 29 | GAJA01013892.1 | S10_plectin pfam03501 (1.28E-64) | 1.020586 | 0.0253683 |
Conserved domains and motifs were detected via the Conserved Domain Database.50 GenBank accession numbers and E-values of the domain or protein family are shown. Transcripts are organized by log (2)-FC in their expression. ID refers to the Lb identifier for each transcript labeled in the volcano plot in Figure 8A. The master record for Lb17 transcriptome in the Transcriptome Shotgun Assembly (TSA) sequence database is GenBank: GAJA00000000.
Table 2.
Lh14 EV transcripts whose expression is significantly altered in S0 wasps compared to G0 wasps (adjusted p < 0.05 and |log2FC| > 1)
| ID | Accession | Conserved domain | LOG (2)-FC S0 vs. G0 |
Adjusted p value S0 vs. G0 |
|---|---|---|---|---|
| 1 | GAJC01017287.1 | None | −1.23674 | 0.00096682 |
| 2 | GAJC01017061.1 | Chitin_bind_4 pfam00379 (3.07E-19) | −1.11582 | 0.03116203 |
| 3 | GAJC01000522.1 | None | −1.09305 | 0.01189165 |
| 4 | GAJC01000520.1 | None | −1.02435 | 0.00556429 |
| 5 | GAJC01012741.1 | None | 4.993669 | 0.00937726 |
Transcripts are organized by log (2)-FC in gene expression. Conserved domain(s) predicted from the Conserved Domain Database50 search results are also shown. ID refers to the Lh identifier for each transcript labeled in the Volcano Plot in Figure 8B. The master record for the Lh14 TSA sequence database is GenBank: GAJC00000000.
Spaceflight does not significantly affect wasp emergence
Spaceflight did not significantly affect the overall development of wasps. The yields per vial of each wasp species reared in space were comparable to those of ground control samples (Figure 2E). The relative success of parasite versus host in individual vials suggested that each Leptopilina species successfully disarmed both y w and hopTum-l hosts in space and also on Earth (Figures 2D and 2E). However, while no hopTum-l hosts escaped parasitization by Lb or Lh in either ground or space cultures, some y w flies emerged in the co-culture vials (Figure 2D), likely because these hosts overcame parasite infection or remained uninfected by the available parasites. When total wasps were scored relative to all insects (% wasps/total insects in each vial; Figure S9A), we found no significant difference in space- versus ground-reared wasp samples. Both wasp species raised on either fly host showed equal success in flight and ground conditions (Figure S9B), reinforcing this conclusion.
EV morphology and Lh14 virulence
We next investigated whether the morphology of EV particles produced by both wasp species is affected by spaceflight. In the scanning electron micrographs of gold-coated samples of G0 and S0 Lb17 wasp venom, we observed no gross morphological difference (Figures S10A–S10G). The overall shapes and morphologies reported previously9,16 were maintained in both samples. In addition to the regularly shaped, spiked particles, there were many particles with heterogeneous shapes, although most had spike-like processes. We observed small membranous protrusions on the particles from both G0 and S0 EV samples that were previously not reported. The G0 and S0 Lh14 EV morphologies were also indistinguishable (and similar to previously reported structures9), in scanning electron micrographs of gold-coated samples (Figures S10H–S10S). Overall, the Lh14 particle morphologies were uniform, and the particles were spiked, with variable spike length.
An ex vivo assay was used to detect changes in G0 versus S0 L. heterotoma virulence. Lamellocytes from laboratory-reared hopTum-l larvae were exposed to venom fluid from G0 or S0 wasps. Venom from both wasp types promoted changes in lamellocyte shape compared to the buffer control (6.50% ± 0.76% G0; 6.12% ± 0.71% S0; 2.99% ± 0.06% buffer control), but there was no significant difference in the S0 versus G0 Lh14 venom activity (Figures S11A–S11C). These results are consistent with the unremarkable difference in the overall L. heterotoma success (emergence) of S0 versus G0 Lh14 wasps.
Mutant Lh14 strains
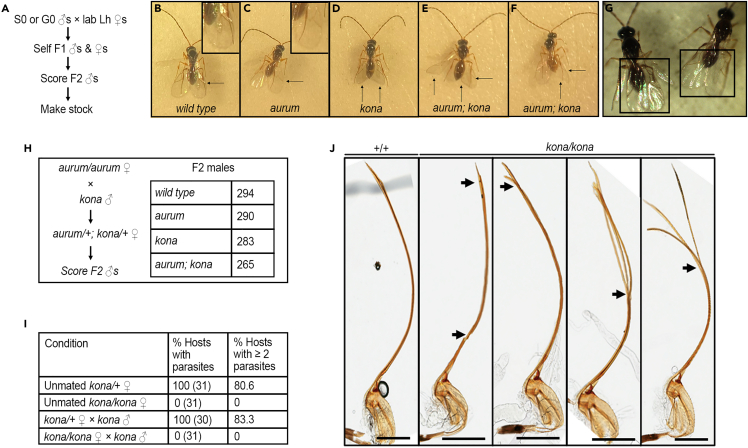
Space-radiation-induced mutations in D. melanogaster have previously been reported.51,52 We hypothesized that parasitoid wasps are similarly sensitive to high levels of radiation in the ISS. We therefore examined the grandsons of space-returned males (Figure 9A). While no heritable defects were observed for any Lb17 or ground-raised Lh14, we obtained two recessive mutations in Lh14 (Figures 9B–9G). In the aurum1 mutant, melanization of the wings is affected, making the normal wings appear golden (horizontal arrows; Figures 9B, 9C, 9E, and 9F; side-by-side view in panel G). This mutant is homozygous viable, and a pure-breeding stock could be established (see STAR methods). A second phenotypic alteration was identified in this pure-breeding aurum1 stock, where the posterior wing margin in affected individuals is angular as opposed to the wild-type round shape (vertical arrows; Figures 9D–9F). In rare cases, this kona1 mutation was incompletely penetrant, where only one wing was affected (Figure 9F). To our knowledge, these are the only live mutant animals in this class of wasps.
Figure 9.
Mutant strains of L heterotoma
(A) Steps employed for the isolation of mutant wasps.
(B–F) Phenotypes of mutant wasps (C–F), identified from space samples compared to wild-type (B) Lh wasps. Males of wild type, aurum1 (pigmentation of the wing blade and of wing veins affected), and kona1 (wing shape affected) wasps are shown. Horizontal arrows point to differences in melanization in wild-type versus aurum1 mutants. The color difference is subtle but consistent. Vertical arrows point to the angular wing shape of the kona1 mutant as compared to the rounded wild-type wing blade. Double mutants in panels (E) and (F) possess both mutant traits. The shape of the left wing in panel (F) is wild-type in appearance, while the right one (arrow) is angular. Samples in panels B–F were photographed at the same time.
(G) A side-by-side comparison of wild type (left) and aurum1 (right) wings shows a clear difference in the melanization of wing veins.
(H) The aurum and kona loci appear to be unlinked, as F2 males from unmated heterozygous females were scored in roughly equal proportions.
(I) Homozygous kona1 females are unable to oviposit based on observations of more than 30 hosts dissected. (The number of hosts scored is indicated in parentheses).
(J) Representative ovipositors from wild-type and mutant wasps. Homozygous kona1 females with defective ovipositors show areas of compromised integrity or have branched ends (arrows) compared to the continuous ovipositors with sharp ends from wild-type control wasps (+/+). Posterior is to the top. Scale bar = 300 μm. See also Figure S12.
To assess if aurum and kona genes are unlinked, we scored over 1,000 sons of unmated double mutant heterozygous females. Roughly equal numbers of parental and recombinant classes were observed (chi-squared test, p = 0.63; Figure 9H). Even though the kona1 strain was homozygous viable, we were unable to make a pure-breeding stock. Moreover, we did not find wasp eggs in dissected hosts, regardless of whether the infecting kona1/kona1 females were unmated or mated. This result suggested that homozygous kona1 females were unable to oviposit while their heterozygous counterparts were successful (Figure 9I). The ovipositors of kona1 homozygotes showed structural defects, including branched termini instead of the sharp, needle-like ends of the wild-type Lh ovipositor (Figure 9J). Abdominal dissection of kona1/kona1 females revealed ovaries and eggs that are morphologically indistinguishable from wild-type or heterozygous females (Figure S12). Thus, it appears that oogenesis is likely unaffected in kona1 mutants, and they are sterile due to their inability to oviposit.
Discussion
American scientists first launched fruit flies into space in 1947, and since then more than 20 spaceflight experiments with fruit flies have been conducted.4 Even parasitic wasps have been to space: in 1967, Biosatellite II carried Habrobracon juglandis into space.53 H. juglandis is a small ectoparasite that attacks moths. The Biosatellite II mission reported multiple effects of spaceflight alone, or that of radiation in conjunction with spaceflight, on D. melanogaster and H. juglandis.53,54 D. melanogaster, however, continues to be an obvious choice for space travel studies for all the advantages the model organism offers researchers on Earth. Among the most cost-effective model systems, its high fecundity and small size facilitate the study of large numbers of individual flies, which is necessary for statistically relevant experimental protocols. These previous “fruit flies in space” studies have tackled a variety of questions, including development, aging, mating, cardiac, and neural functions, that have implications for astronaut health.4,55 Despite suffering from cardiac and neurological deficits,33,56 Drosophila born in space can largely survive the dual challenges of short-term exposure to microgravity and radiation.
Strain-specific effects on survival and transcriptional readouts in Drosophila are induced by spaceflight stressors
Spaceflight conditions exerted unexpected and differential effects on our experimental Drosophila strains. While the hopTum-l flies with CI survived just as well in space as on the ground, the y w strain, free of tumors and CI, was surprisingly more vulnerable to the spaceflight stressors, and fewer y w flies emerged from the flight compared to ground cultures. Molecular analysis revealed a complex interplay between genetic and environmental conditions. Three trend lines are noteworthy, which are discussed in the following. First, specific expression profiles of Top DEGs attributed to genetic differences were maintained in space, suggesting that the gene regulatory circuits underlying these differences may be sufficiently robust and may therefore remain refractory to spaceflight.
Second, the Top DEGs include genes encoding structural proteins for the cuticle and the chorion, an observation that was also reported in a previous multigenerational study on wild-type flies.55 Some of the cuticle-related DEGs were upregulated in one of the two S0 genetic strains, but not in the other S0 strain, while other DEGs were downregulated in S0 flies of both genetic backgrounds. This result suggests that transcriptional co-regulation of promoters among Top DEG family members may be adaptive to either or both genetic and environmental variables. Chitin and its associated proteins are part of the fly’s exoskeletal matrisome, and together they support the cells they surround structurally and functionally.57,58 A highly coordinated chitin synthesis and degradation program underlies insect growth. It would be interesting to assess if the cuticle or the chorion of S0 flies differs from that of G0 flies.
Third, numerous essential genes were downregulated in space in y w, but not in hopTum-l animals, suggesting that the cumulative effect of their reduced transcription may have led to their reduced viability. Thus, while increased tumor burden and alterations in gene expression in S0 hopTum-l adults did not compromise their overall survival, y w flies exhibited increased immune gene expression, remained tumor-free in space, and exhibited reduced survival. Future experiments will help clarify the differential effects of space on the expression of essential genes in these genetic strains. Many of these essential fly genes are conserved, and their human orthologs are linked to disease conditions. These results not only defied our expectation that animals with chronic disease would be more vulnerable to spaceflight but also suggested that, if extrapolated to other animals, spaceflight’s effects on individuals of distinct genetic backgrounds may not necessarily be straightforward to predict.
Elevated immune signaling in space hosts
Our previous molecular-genetic analysis of hopTum-l larvae showed that hyperactive Toll signaling in larval lamellocytes contributes to CI phenotypes.24 A plethora of target genes of both the JAK-STAT and Toll pathways is constitutively active in hopTum-l mutants. Strikingly, many of the same pathway components and target genes that are constitutively active in hopTum-l mutants are also upregulated in S0 y w flies, even though the S0 y w flies remained tumor free. A few AMP genes (e.g., Drs, Drsl, CecA, etc.) are further upregulated in S0 hopTum-l flies, suggesting that the increased tumor burden in S0 hopTum-l adults is fueled by ectopic JAK-STAT and Toll signaling induced in space.
Unlike their parents, however, naive S1 y w larvae did not exhibit a clear and strong inflammatory gene expression readout (see S0/G0 y w column in Figure 5D and S1/G1 y w column in Figure S3D, where constitutive immune signaling does not mask the effects of space). In contrast, like their parents, many immune genes in naive G1/S1 larvae were active in hopTum-l larvae. Even though changes in mitotic indices of host hemocytes or their lymph gland morphologies were not detected in these S1 versus G1 hopTum-l larvae, a significantly enhanced T score was observed in naive S1 versus G1 hopTum-l larvae. This increased immune reactivity was also reflected in the enhanced T + E scores in infected S1 hopTum-l larvae, relative to their G1 counterparts. The increased tumor burden detected in naive S1 hopTum-l larvae did not persist in S1 hopTum-l adults. Collectively, these observations suggest that, with re-adaptation to normal gravity, the inflammatory effects of spaceflight are transient and reversible. The highly inflamed state of hopTum-l animals provided physiological insights into the modest changes in wasp virulence due to spaceflight.
Wasp success, wasp virulence, and gene expression changes
We hypothesized that stress from spaceflight could compromise the development of either of the two partners: the fly or the parasite. Because of the complete dependence of the parasite on the host, either situation would be detrimental to parasite development. In an alternative scenario, if the effects of spaceflight on fly hosts are modest, then endoparasite development should not be affected. Wasps of both species developed successfully in the ISS environment; the more virulent Lh14 species was equally successful in both conditions, while Lb17 survival was marginally compromised in space. This difference in Lb17 versus Lh14 success may have to do with their dissimilar virulence strategies, with Lb17 being less successful on D. melanogaster than Lh14.8 Global gene expression changes in space-raised parasites were remarkably mild, and less than 1.5% of the adult parasite transcripts were affected. Consistent with parasite emergence results, the transcription of many more genes was affected in S0 Lb17 than in S0 Lh14. Thus, even though radiation in space led to germline mutations in one of the two parasites, the overall detrimental effects of microgravity were not sufficiently strong to block parasite development. We speculate that the notable overall success of both parasites in space may be due to their endoparasitoid life histories: developing internally, the preimaginal stages remain protected from the environmental challenges in space. Parasites have varied life histories and distinct survival strategies in the context of host physiology. With increased travel to space, it is inevitable that parasites will unintentionally hitch a ride with their hosts. With plans for the colonization of other planets, multigenerational experiments on “short life cycle” organisms such as D. melanogaster and their associated flora and fauna will become important. Innovative and sophisticated hardware designed for multigenerational studies will be helpful in this regard.
The virulence properties of both parasites also appeared to be largely maintained in space. This conclusion is based on (a) comparable yields of parasites in space versus ground cultures, (b) relatively stronger effects of Lb17 on the T + E score compared to Lh14, and (c) stronger transcriptional activation of Drosomycin (Drs) in S1 hosts after Lb17 infection but not after Lh14 infection (Figures S3F and S3G). It is intriguing that S0 wasps of both species were less effective at increasing the T + E score in both G1 and S1 hopTum-l hosts. This result suggests that the space environment did affect wasp physiology or virulence; the latter interpretation being consistent with reported increases in microbial virulence in space.31,32
A clear molecular explanation for differences in S0 versus G0 wasps was not apparent from the molecular analysis of wasp EV gene transcription. Transcript levels of only 29/312 Lb17 EV genes were significantly affected with all but one gene being downregulated. Only one of the two predicted RhoGAP superfamily genes shares structural similarity with the known LbGAP virulence protein,49 and even its expression was weakly downregulated in space. The transcription of only 5/398 Lh14 EV transcripts was differentially affected, with only one gene (lacking annotation) showing ∼ 5-fold upregulation. None of these genes is currently implicated in Lh14 virulence. Furthermore, in an ex vivo assay, Lh14 venom from S0 wasps was no different in its ability to distort lamellocytes than Lh14 venom from G0 wasps. The abundance and distribution of G0 and S0 Lh14 EVs in the dorsal vessel and lymph gland lobes of infected animals further suggest that the integrity of S0 Lh14 EV is comparable to that of controls. It is possible that the cumulative effects of differential gene expression changes contributed to differences between S0 and G0 wasps. Alternatively, S0 wasps may differ in their oviposition behavior, affecting the quantity of venom introduced into the host.
Mutant wasps
Haplodiploidy in Hymenoptera makes it convenient to conduct mutational studies in space, as haploid sons (developing from unfertilized eggs of mutated females) would exhibit dominant or recessive mutant phenotypes. In our study, male wasps were used to evaluate the effects of radiation on their germline, while female wasps were used in virulence assays. Screening grandsons of S0 Lh14 males developed in space resulted in the isolation of two visible mutants, one of which is homozygous viable, while the other is homozygous female-sterile. Dosimetry measurements on SpaceX-14 recorded an average absorbed dose rate of 0.3 mGy/day, or 9.6 mGy for the 32-day duration in the ISS. Being in low Earth orbit, the ISS is protected by the Earth’s magnetosphere.59 Yet, chronic exposure to these radiation levels likely resulted in DNA damage in the germline of the parasites reared in space. Despite efforts to shield the space station from the damaging consequences of radiation,60 our results support previous studies showing that animals in the ISS are at risk for DNA damage and physiological dysfunction.1 Given the success of this mission, systematic studies designed to compare the effects of radiation on Drosophila and Leptopilina may be particularly insightful, as both species yield high numbers of progeny and mutant phenotypes can be scored in the F1 generation. The risks of radiation increase considerably with the planned manned lunar missions and other deep space missions for long-term colonization.1,61 Future model organism studies can help assess the hazardous effects of radiation on soma versus germline genomes and inspire ways to avert these risks with innovative shielding designs. The isolation of mutant wasp lines will advance our understanding of wasp genomics, genetics, and host-parasite biology. The space environment is likely to have as-yet unknown impacts on animal immune function; refining our understanding of host-parasite systems with new genetic tools on Earth will broaden our understanding of immune function in space. Studies of other animal and plant hosts and their natural parasites in modeled microgravity and/or radiation will provide insights into their effects on host defense and parasite virulence.
Limitations of the study
Spaceflight experiments with live organisms are constrained by the weather, the logistics of the launch and retrieval protocols, the duration of the mission, the physical conditions within the ISS module, and available crew time. This study faced all these constraints. Although the 34-day mission duration aligned well with the overall goals of our experiment, by the end of the mission, the number of G0/S0 larvae was insufficient to set up infections with G0/S0 wasps. (Available G0/S0 larvae were dissected to examine lymph gland hemocyte morphologies [Figure S5].) It is quite likely that the effects of infection on the T + E values in G0/S0 hosts (infected with G0/S0 wasps) would be stronger than what we observed in G1/S1 hosts. The availability of fresh fly media for culturing naive G0/S0 adult flies, born in space or on ground, would have ensured sufficient numbers of larval hosts for these infection experiments, a strong consideration for designing future experiments.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-phospho-histone H3 antibody | Millipore Sigma | Catalog # 06-570 |
| mouse anti-p40 antibody | Chiu et al., 20069 | PMID: 16432035 |
| Chemicals, peptides, and recombinant proteins | ||
| 5-bromo 4-chloro 3-indoyl phosphate (BCIP) | Promega | Catalog #S381C |
| nitroblue tetrazolium (NBT) | Promega | Catalog #S380C |
| paraformaldehyde (crystalline) | Sigma | Catalog #P6148 |
| rhodamine-labeled phalloidin | Life Technologies | Catalog #R415 |
| vectashield mounting medium | Vector Labs | Catalog #H100 |
| Hoechst 33258 pentahydrate | Invitrogen | Catalog # H-1398 |
| 4% glutaraldehyde in 0.1M sodium cacodylate buffer, pH 7.4 | Electron Microscopy Sciences | Catalog # 16539-06 |
| 2% osmium tetroxide | Electron Microscopy Sciences | Catalog # 19150 |
| Biological samples | ||
| G0 and S0 adult y w and hopTum-l flies (genotypes below) | This study | N/A |
| G0 and S0 y w and hopTum-l larvae | This study | N/A |
| G1 and S1 y w and hopTum-l larvae | This study | N/A |
| G0 and S0 Leptopilina boulardi 17 adult wasps | This study | N/A |
| G0 and S0 Leptopilina heterotoma 14 adult wasps | This study | N/A |
| Critical commercial assays | ||
| RNA extraction RNA/Protein Purification plus kit | Norgen | Catalog # 48200 |
| Non-stranded NEBNext Ultra RNA Library Prep Kit for RNA-Seq sequencing libraries | New England Biolabs | Catalog #E7775 |
| Deposited data | ||
| Drosophila RNA-Seq data | NASA Open Science Data Repository | GeneLab ID: GLDS-583; OSDR ID: OSD-588; https://osdr.nasa.gov/bio/repo/data/studies/OSD-588; https://doi.org/10.26030/v9rh-5a70 |
| Leptopilina boulardi RNA-Seq data | NASA Open Science Data Repository | GeneLab ID: GLDS-587; OSDR ID: OSD-610; https://osdr.nasa.gov/bio/repo/data/studies/OSD-610; https://doi.org/10.26030/9ee4-6s36 |
| Leptopilina heterotoma RNA-Seq data | NASA Open Science Data Repository | GeneLab ID: GLDS-586; OSDR ID: OSD-609; https://osdr.nasa.gov/bio/repo/data/studies/OSD-609; https://doi.org/10.26030/5rjq-a347 |
| Experimental models: Organisms/strains | ||
| y1 w1 | Bloomington Stock Center | BDSC:1495 |
| y w hopTum-lmsn-Gal4; UAS-mCD8-GFP (hopTum-lmsn > GFP | Panettieri et al., 201929 | PMID: 31562189 |
| L. boulardi strain 17 (Lb17) | Schlenke et al., 20078 | PMID: 34051038 |
| L. heterotoma strain 14 (Lh14) | Schlenke et al., 20078 | PMID: 34051038 |
| Wing color mutant: L. heterotoma14 aurum1 | This study | Lh14 aurum1 |
| Wing shape and ovipositor mutant: L. heterotoma14 kona1 |
This study | Lh14 kona1 |
| Software and algorithms | ||
| Adobe Photoshop Creative Cloud v20-v24 | Adobe | https://www.adobe.com/ |
| Pipeline for Drosophila RNA-Seq data | NASA GeneLab: Open Science for Life in Space | GL-DPPD-7101-D; https://github.com/nasa/GeneLab_Data_Processing/blob/master/RNAseq/Pipeline_GL-DPPD-7101_Versions/GL-DPPD-7101-D.md |
| Slurm and R scripts to process and analyze Leptopilina boulardi RNA-Seq data and Leptopilina heterotoma RNA-Seq data | This study, supplement | Data S1 |
| Other | ||
| 25 mm polycarbonate Whatman membrane filter (0.1 μm pore size) | Whatman | Catalog # 110605 |
| SEM specimen mount stubs | Electron Microscopy Sciences | Catalog # 75110 |
| Illumina HiSeq 4000 (Genewiz) | Illumina, San Diego | https://www.Illumina.com |
| Illumina NovaSeq 6000 sequencer using S1 Reagent Kit v1.5 (GeneLab) | Illumina, San Diego | https://www.Illumina.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shubha Govind (sgovind@ccny.cuny.edu).
Materials availability
This study generated two mutant strains of Leptopilina heterotoma. Mutant strains will be shared by the lead contact upon request.
Data and code availability
-
•
Original transcriptomic data (raw read counts and FASTQ files) from bulk RNA-Seq experiments on hosts and parasites have been deposited in and can be accessed from NASA’s publicly available Open Science Data Repository (https://osdr.nasa.gov/bio/repo). Accession numbers are listed in the key resources table.
-
•
Version D of NASA GeneLab’s bioinformatics pipeline, available on GitHub, was used to process Drosophila RNA-Seq reads. A DOI is listed in the key resources table.
-
•
A modified version of the Drosophila pipeline was used to analyze L. boulardi and L. heterotoma reads. Those scripts are available with the supplemental material Data S1 (L.boulardi_ L.heterotoma_RNAseq_processing_scripts.zip).
-
•
Microscopy data and any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request.
Experimental model and study participant details
Experimental design and sample preparation
D. melanogaster strains y1 w1 and y w hopTum-l msn-GAL4; UAS-mCD8-GFP (hopTum-l msn > GFP),24 and wasp species L. boulardi strain 17 (Lb17) and L. heterotoma strain 14 (Lh14),8 were included in the ground control and flight experiments. The hopTum-l strain carries a dominant point mutation in the human Janus kinase (JAK) homolog, hopscotch.22 This mutation confers constitutive JAK-STAT signaling, which drives hematopoietic proliferation and differentiation in larval stages; small, melanized tumors form that can be scored in adult flies. In this strain, the misshapen (msn) promoter directs Gal4 expression to lamellocytes.62 Expression of the UAS-mCD8-GFP transgene helps visualize lamellocytes, which make up the bulk of the tumors.
Extensive pre-flight testing was done to estimate the diet volume and insect numbers for optimal growth during the 34-day flight period. Six conditions (two fly-only cultures, four fly/wasp co-cultures) were distributed among six VFBs (Figure 1). The fly-only cultures were reared in polystyrene vials containing 12 mL ± 1 mL fly food, while the co-culture vials contained 10 mL ± 1 mL fly food. The difference in fly food volume accommodated the respective life histories of the two insect species; unlike flies that depend on fly food medium, wasps consume the growing fly larvae and pupae. This volume of fly food supported insect growth for 34 days while keeping carbon dioxide production to a minimum.
Prior to launch, y w fly-only cultures were initiated with 8 female and 5 male flies. The hopTum-l cultures were similarly initiated with 20 female and 10 male flies. Fly-wasp co-cultures were set up simultaneously by first adding the flies to vials for three days, whereupon the flies were removed and adult wasps introduced. For the y w co-cultures, 30 adult females and 15 adult males were added to each polystyrene vial for egg-laying. For hopTum-l egg-lays, 35 females and fifteen males were used. Fifteen egg-lays were made for each genotype, and 15 female and 15 male wasps were added to each egg-lay a day prior to launch.
Mission and hardware details
Six VFBs, each with 15 vials, were placed inside a CTB before the launch of the SpaceX-14 mission to the ISS (Figure 1). The mission was launched at 20:30 UTC (Coordinated Universal Time) on April 2nd, 2018. Wasp infection modifies fly development; it takes 20–25 days for wasps to develop into adults, while naive flies complete their development in 10–15 days. Weather-related issues delayed the original mission time from 31 days to 34 days. The ISS crew installed the samples into the Columbus Module endcone with the CTB lid open to promote air exchange between the VFBs and cabin air, where they remained until approximately two days prior to their return to Earth. Samples remained at ambient temperature (∼22°C). Temperature and humidity data in each VFB were collected throughout the flight mission. The radiation dosimeter on the ISS was the COL1A2 Radiation Assessment Detector, ISS-RAD, that detects charged particles.63 Insects remained in the VFBs throughout the mission until their return on the Dragon capsule.
A full-scale, near-synchronous ground control was conducted that was staggered 48 h from launch, using the environmental data streamed to a Space Station Processing Facility Environmental Simulator at the Kennedy Space Center (KSC). The ground control samples were manually set to recapitulate the temperature and humidity conditions of the space samples. While the temperatures between space and ground samples were similar for the duration of the experiment, the relative humidity of space samples was higher by 20–25% than that of ground samples. This was attributed to reduced convection in the microgravity environment, making it more difficult for moisture to evaporate quickly. The Dragon capsule was unberthed at 13:22 UTC on May 5th, 2018, and splashdown in the Pacific Ocean occurred on the same day at 20:00, off the coast of Long Beach, CA, from where samples were flown to New York. Ground control samples were similarly collected from KSC and flown to New York.
Condition of insect cultures
Visual inspection of the cultures indicated that most of the fly food in all vials was consumed, and there was no apparent evidence of mold or other microbial infection in any culture vial. G0 and S0 fly-only vials of both genetic backgrounds did not contain sufficient early instar larvae to set up wasp infections. (G1/S1 larval hosts were used instead for infection experiments.) Almost all adult wasps retrieved from G0/S0 cultures developed under corresponding conditions, while adult fly samples contained individuals of mixed age and from 2 to 3 generations. For all studies, appropriate numbers of animals were randomly selected from different vials.
Method details
Survival studies
Animals in each vial were sorted and scored under a dissecting microscope immediately upon arrival. For fly-only cultures, statistical analysis was performed on counts after excluding counts from vials where sample counts were incomplete. For co-cultures, statistical analysis was performed on the number of flies and wasps after excluding vials where either no insects developed or where sample counts were incomplete. The percentage of female wasps was obtained by dividing the number of female wasps by the total number of wasps in each vial.
Dissections, staining, and imaging
Depending on sample availability, 5–15 animals were dissected to examine lymph glands. Methods for larval lymph gland dissections, staining, and imaging were previously published.64,65,66,67 Lymph glands were counter-stained with rhodamine-labeled phalloidin (R415 Life Technologies) and Hoechst 33258 pentahydrate (Invitrogen). For Lh-infected G1 or S1 larvae, primary mouse anti-p40 antibody9,20 (1:1000) was used. Fixed and stained samples were imaged on the Laser Scanning Zeiss 710 or 800 confocal microscopes controlled with Zeiss Zen imaging software in the CCNY core facilities. Whole insects were imaged with a Leica MZFLIII attached to an Optronics camera. Images were collected using the Magnafire-SP software.
Mitotic index
Three-day egg-lays of G0/S0 hopTum-l adult flies (described above) were set up at 25°C. Three to five wandering third-instar larvae were used per hemocyte smear for each replicate; three biological replicates were performed. Samples were incubated with rabbit anti-phospho-histone H3 antibody (1:200, Millipore Sigma), which was visualized with goat anti-rabbit alkaline phosphatase-linked secondary antibody (1:5000, Thermo Scientific). Antibody binding was visualized by alkaline phosphatase staining (125 μg/mL BCIP and 250 μg/mL NBT, from Promega). The percentage of phospho-histone H3-positive macrophages was calculated by dividing the number of phospho-histone H3-positive macrophages by the total number of macrophages over six visual fields. Cells were scored using a Zeiss Axioplan light microscope.
Adult tumorigenesis
Tumors in randomly chosen G0 and S0 hopTum-l adults (20 males and 5–10 females, ∼22°C) were scored 6.5 days (25°C) after their return. The G1 and S1 progeny of these G0 and S0 hopTum-l flies was reared at 27°C, and their abdominal tumors were similarly scored 18.5 days post egg-lay.24 Using a dissecting microscope, abdominal tumors were categorized as follows: small (< 0.5 body segment), medium (0.5–1 body segment), and large (> 1 body segment). Small-, medium-, and large-sized tumors were arbitrarily weighed as 1, 2, and 3, respectively, and all tumors in each animal were recorded. The average number of tumor structures per animal was determined by dividing the sum of all tumors in each replicate by the total number of animals scored. Three biological replicates were performed for both S0/G0 and S1/G1 comparisons. A Student’s t test was used to determine if the averages of two datasets differed significantly from each other.
Infection and larval host immunity assay
Egg-lays of G0/S0 hopTum-l flies were set up with the same number of flies used in flight. Larval progeny, raised at 25°C for 3 days, were exposed to 10 male and 10 female G0 and S0 Lb17 or Lh14 wasps for 8 h. For each condition, hosts of the same strain were taken from more than one vial to randomize samples and ensure robust egg-laying. Similarly, wasps of the same species from different vials were combined for infections. Each experiment was replicated 3–4 times.
Infected larvae were randomly selected and examined under a dissecting microscope 3–5 days later for the presence of either melanotic tumors or melanized parasite-induced encapsulation reactions. hopTum-l tumors and parasite capsules differ in shape; tumors are globular, while capsules are typically sickle-shaped and surround a wasp egg or wasp larva. These structures can fragment into smaller, melanized pieces, making it difficult to distinguish them. Regardless of their identity, they were classified based on size: (a) specks (similar in appearance to melanized crystal cells), (b) small melanized structures, or (c) large melanized structures. To quantify spaceflight’s effects on these reactions, an arbitrary score was assigned as follows: 1–3 specks, 4–8 specks, and > 8 specks were assigned a value of 0.5, 0.75, or 1, respectively. Small- and large-melanized structures were assigned a value of 1 and 4, respectively. Where clear encapsulation reactions (with evidence of a wasp egg) were observed, the structure was also assigned a value of 4. A composite tumor and encapsulation (T + E) score was calculated from 16 to 29 G1/S1 Lb17-or Lh14-infected hopTum-l larvae. Uninfected G1/S1 hopTum-l larvae (70–89 animals) served as controls. Pairwise comparisons of the average T + E scores were made using the Mann-Whitney U test for unpaired samples.
Bulk RNA-Seq sample preparation methods
For RNA-Seq experiments (technique reviewed in68), samples were flash-frozen in dry ice and stored at −80°C. Bulk RNA-Seq analysis was performed on RNA prepared from four replicates of (a) ground control (G0) and space-flown (S0) adult flies; (b) G0 and S0 adult wasps; and (c) G1 and S1 larvae. The RNA/protein purification plus kit from Norgen Biotek Corp. was used for RNA preparation, and the extracted total RNA was stored in Norgen RNA isolation kit elution buffer.
For adult flies, three males and females each, of (a) G0 y w; (b) S0 y w; (c) G0 hopTum-l; (d) S0 hopTum-l were used for extraction. RNA preparations of G0 and S0 Lb17 and Lh14 wasps were made from three females and three males. Larval RNA was isolated from G1 and S1 y w animals. In addition, RNA was also extracted from hopTum-l larvae as follows: (a) uninfected G1; (b) uninfected S1; (c) G0 Lb-infected G1 hosts; (d) S0 Lb-infected G1 hosts; (e) G0 Lb-infected S1 hosts; (f) S0 Lb-infected S1 hosts; (g) G0 Lh-infected G1 hosts; (h) S0 Lh-infected G1 hosts; (i) G0 Lh-infected S1 hosts; and (j) S0 Lh-infected S1 hosts.
The RNA from G0 and S0 adult flies and wasps was processed at Genewiz, Inc. as follows: After poly (A)+ extraction, libraries for adult flies and wasps were prepared using the non-stranded NEBNext Ultra RNA Library Prep Kit E7775, following the manufacturer’s instructions (NEB, Ipswich, MA, USA). The libraries were validated using the Agilent TapeStation. Libraries were sequenced on the Illumina HiSeq 4000 machine for HiSeq 2 × 150 bp, single index, at Genewiz. Data output was ∼300–350 million raw paired-end reads per lane.
The total RNA prepared from G1 and S1 larval samples (stored at −80°C) was processed as follows at NASA’s GeneLab Samples Processing Laboratory. The RNA concentration was determined using a Qubit 4.0 Fluorometer, and 400 ng of each sample was used for poly (A)+ extraction. RNA-Seq sequencing libraries were prepared using the E7775 NEBNext Ultra RNA Library Prep Kit, as above. The libraries were validated by using Agilent TapeStation D1000 tape. Quantification was performed using the Qubit 4.0 Fluorometer. Libraries were pooled and sequenced on the Illumina iSeq 100 sequencer to confirm library balancing and screen for ribosomal RNA. The final library pool was sequenced on Illumina NovaSeq 6000 sequencer using S1 Reagent Kit v1.5 (300 cycles, 2 lanes).
Processing of Drosophila RNA-Seq data
The reads from adult fly and larval fly RNA were analyzed using version D of NASA GeneLab’s RNA-Seq processing pipeline (https://github.com/nasa/GeneLab_Data_Processing/blob/master/RNAseq/Pipeline_GL-DPPD-7101_Versions/GL-DPPD-7101-D.md). For this, the raw fastq files of the adult fly RNA-Seq reads obtained from Genewiz were transferred to GeneLab. Raw fastq files of both sample sets were assessed for percent rRNA (0.30–0.42% rRNA in adults; 0.20–2.73% rRNA in larvae) using HTStream SeqScreener (version 1.3.2) and filtered using Trim Galore! (version 0.6.7) powered by Cutadapt (version 2.6). Raw and trimmed fastq file quality was evaluated with FastQC (version 0.11.9), and MultiQC (version 1.11) was used to generate MultiQC reports.
D. melanogaster STAR and RSEM references were built using STAR (version 2.7.8a) and RSEM (version 1.3.1), respectively, Ensembl release 101, genome version BDGP6.28 (Drosophila_melanogaster.BDGP6.28.dna.toplevel.fa) and the following gtf annotation file: Drosophila_melanogaster.BDGP6.28.101.gtf. Trimmed reads were aligned to the Drosophila melanogaster STAR reference with STAR (version 2.7.8a). A majority (90.08%–94.47%) of trimmed reads mapped uniquely to the fly reference genome. Aligned reads were assessed for strandedness using the RSeQC Infer Experiment (version 4.0.0) and determined to be unstranded. Then aligned reads from all samples were quantified using RSEM (version 1.3.1), with strandedness set to none.
RSEM raw gene counts were imported to R (version 4.0.3) with tximport (version 1.18.0) and normalized with DESeq2 (version 1.30.0)69 median of ratios method. Differential expression analysis was performed in R (version 4.0.3) using DESeq2 (version 1.30.0); all groups were compared pairwise using the Wald test, and the likelihood ratio test was used to generate the F statistic p value. False discovery rate (adjusted p value) corrections were performed using Benjamini-Hochberg multiple testing adjustment. Gene annotations were assigned using the following Bioconductor and annotation packages: STRINGdb (v2.2.0), PANTHER.db (v1.0.10), and org.Dm.e.g.,.db (v3.12.0). Genes were considered differentially expressed if adjusted p < 0.05 and |log2FC| > 1.
Analysis of Drosophila RNA-Seq results
Global analyses
Principal Component Analysis (PCA) was performed in R (version 4.0.3) using log (2)-transformed count data from unnormalized and normalized counts. PCA plots were generated for each set of count data using ggplot2 (version 3.3.3). Volcano plots were created using EnhancedVolcano (version 1.8.0), with adjusted p < 0.05 and |log2FC| > 1 cutoff values specified.
KEGG enrichment analyses for adult samples
DEGs in adult fly samples (adjusted p value < 0.05 and |log2FC| > 1) were used for analysis. Gene ontology (GO) and KEGG enrichment analyses were performed using the Bioconductor package ClusterProfiler v 3.18.070 and the Drosophila database (org.Dm.e.g.,.db version 3.13). EnrichGO and EnrichKEGG functions were used to determine functionally enriched GO categories for KEGG pathways and molecular function.71 Results were visualized by R package.72
Analysis of top, essential, and pathway DEGs
The most highly modulated Top DEGs in the adult fly samples were identified by sorting log (2)-FC for each comparison and then filtering genes that met the adjusted p value significance p < 0.05 and |log2FC| > 1. From this list, the top 50 upregulated or downregulated genes were identified for each comparison, yielding ∼280 genes. Annotations and other functional information for these ∼280 genes were examined in FlyBase37 and genes with clear orthologs or paralogs and known or predicted functions (172 genes) were retained. Clustering of these 172 Top DEGs revealed six major expression profiles. The magnitude of log (2)-FC for these genes, in both directions, ranged from ∼3- to 12-fold.
For Essential DEGs, a list of 1,024 genes that were identified experimentally in P element insertion screens was compiled.73,74,75,76 Of these 1,024 genes, 137 were differentially expressed in at least one of four comparisons. The DIOPT-DIST tool,41 which predicts human orthologs of fly genes and diseases or traits associated with the human orthologs identified through GWAS or OMIM databases, was used to determine the disease relevance of these essential genes. Genes in the JAK-STAT, Toll, and Imd pathways were compiled from KEGG and/or FlyBase. Additional pathway target genes were added from the primary literature.34,77,78
For larval samples, gene set enrichment analysis of larval RNA-Seq results did not highlight immune changes, and hence the results of this analysis were not included here. Gene expression changes were examined in the context of signaling pathways as was done for the adult RNA-Seq results above.
Processing of parasite RNA-Seq reads
Raw fastq files were filtered using Trim Galore! (version 0.6.7) powered by Cutadapt (version 3.7). Raw and trimmed fastq file quality was evaluated with FastQC (version 0.11.9), and MultiQC (version 1.12) was used to generate MultiQC reports. L. heterotoma Bowtie2 and RSEM indices were built using Bowtie2 (version 2.5.0) and RSEM (version 1.3.1), respectively, and the Lh14 GAJC contig transcriptome reference from NCBI (https://sra-download.ncbi.nlm.nih.gov/traces/wgs03/wgs_aux/GA/JC/GAJC01/GAJC01.1.fsa_nt.gz). L. boulardi Bowtie2 and RSEM indices were built using Bowtie2 (version 2.5.0) and RSEM (version 1.3.1), respectively, with Lb17 GAJA contig transcriptome reference from NCBI (https://sra-download.ncbi.nlm.nih.gov/traces/wgs03/wgs_aux/GA/JA/GAJA01/GAJA01.1.fsa_nt.gz).
Trimmed reads were aligned to the respective wasp parasite species Bowtie2 transcriptome reference with Bowtie2 (version 2.5.0), and aligned reads from all samples were quantified using RSEM (version 1.3.1), with strandedness set to none. Quantification data was imported to R (version 4.1.2) with tximport (version 1.22.0) and normalized with DESeq2 (version 1.34.0) using the median of ratios method. Normalized transcript counts were subject to differential expression analysis in R (version 4.1.2) using DESeq2 (version 1.34.0); all groups were compared using the Wald test, and the likelihood ratio test was used to generate the F statistic p value. The numbers of significantly differentially expressed transcripts (adjusted p < 0.05 and |log2FC| > 1) for Lb17 and Lh14 were 293 and 310, respectively. The presence of conserved protein domains or motifs in these differentially expressed Lb17 and Lh14 DEGs was performed via CDD searches (database version CDD v3.20 - 59693 PSSMs; and expect value threshold 0.01) (Tables S2 and S3).
Analysis of parasite RNA-Seq results
Global analyses
Principal Component Analysis was performed in R (version 4.1.2) using log (2)-transformed count data from unnormalized and normalized counts. PCA plots were generated for each set of count data using ggplot2 (version 3.3.5). Volcano plots were created using EnhancedVolcano (version 1.12.0) with an adjusted p value < 0.05 and |log2FC| > 1 cutoff values specified.
Identification and analysis of parasite EV transcripts
Proteins from purified Lb17 EV have not been characterized, but the composition of purified EVs from the Lb Gotheron (LbG) strain is published.14 To identify differentially expressed Lb17 EV transcripts in space corresponding to these previously characterized 383 LbG EV proteins, we queried the LbG EV protein sequences against all 293 differentially expressed Lb17 transcripts in space (Table S2; LbG EV query sequences were kindly provided by J. Varaldi, University of Lyon). For this, NCBI’s tblastn tool was used at default settings (BLOSUM62; gap costs existence extension 1, filtered for low complexity regions, January 2023). Results where E-value ≤1 e−30, query coverage ≥50%, and percent identity of ≥50% were scored as high confidence homologs and included 27 sequences. Two sequences did not meet these criteria but were added to our results (Table 1), either due to their high log (2)-FC value (GAJA01001411.1; E = 3.00 e−100, query coverage 96%, and % identity 32.49), or due to interesting homology (GAJA01009829.1; E = 4.00 e−30, query coverage 57%, and % identity 32.69).
An estimate of the overall Lb17 EV proteome size was obtained as follows: the 383 LbG sequences were used to tblastn-search the GAJA00000000 sequences.47 This search identified 316 non-redundant EV protein-encoding GAJA sequences (E ≤ 1 e−50, query coverage ≥70%, percent identity ≥70%; NCBI BLAST+ (v 2.7.1).15,79,80,81 Some homologs were not identified due to strain differences; nevertheless, the analysis revealed that most of the LbG EV proteins are expressed in the Lb17 female abdomen. Since the Lh14 EV proteome is already characterized,11,12 tblastn analyses were not necessary, and EV genes were filtered from the 310 Lh14 DEGs (Table 2).
SEM preparation of venom EVs and imaging
Thirty venom glands of G0 and S0 wasps were dissected in 100 μL of ice-cold PBS and fixed in 100 μL of 4% glutaraldehyde in 0.1M sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences). Venom glands were processed for scanning electron microscopy imaging as follows: Ten drops of 2% osmium tetroxide aqueous solution (Electron Microscopy Sciences) were added to the sample on ice for 20 min. The sample was filtered on a wet 25-mm polycarbonate Whatman membrane filter (0.1 μm pore size), covered with a second wet filter, and suctioned with the sample in between the filters. The filters were clamped between O-rings and quickly immersed in ice-cold distilled water. The filters were then rinsed in distilled water five times, 3 min for each rinse. This was followed by dehydration in a graded (10%, 30%, 50%, 70%, 80%, 90%, 95%, 100%) series of ethanol for 5 min each. The filters were then critical point dried (Balzers, CPD 030) in liquid carbon dioxide five times, 3–5 min each. Dehydrated and dried samples were mounted on aluminum SEM specimen mount stubs (Electron Microscopy Sciences) using conductive carbon adhesive tabs (Electron Microscopy Sciences) and stored in a 60°C oven for 2 h or longer. Samples on the filter were coated with gold/palladium (Leica, EM ACE600) right before being observed on the Zeiss SUPRA 55VP scanning electron microscope. These procedures were carried out at the CCNY and CUNY ASRC core facilities.
Virulence assay for Lh14 venom
Forty hopTum-l larvae were bled in 200 μL of 7% bovine serum albumin. 50 μL of this hemolymph preparation was aliquoted into individual chambers of a 4-chamber slide and allowed to incubate at room temperature for 1 h. Venom was extracted from G0 and S0 Lh14 wasps, and protein was quantified using the Bradford method. An appropriate volume of the venom extract containing 30 μg of venom protein was added to the slide chamber with hopTum-l hemocytes. Phosphate-buffered saline (20 mM, pH 7.4) was used as a control. After incubation for 4 h at 25°C, excess liquid was removed by aspiration, hemocytes were air-dried at room temperature and fixed with 4% paraformaldehyde for 10 min. After washing, cells were stained successively with rhodamine-labeled phalloidin (R415 Life Technologies) and Hoechst 33258 pentahydrate (Invitrogen) for 15 min each at room temperature. Samples were washed three times with PBS, and stained cells were mounted in Vectashield for confocal microscopy. Lamellocytes with bipolar morphology (i.e., spindle-shaped GFP-positive lamellocytes with two pointed ends) were manually scored. Experiments were replicated three times and the Student’s t test statistic was applied to determine the significance.
Isolation and characterization of L. heterotoma mutants
G0/S0 Lb17 and Lh14 male wasps were mated with females from lab cultures, and their grandsons were examined for viable mutations affecting wing color, wing venation, wing morphology, eye color, eye morphology, and antennal morphology. Mutant grandsons were not obtained from the G0/S0 Lb17 or G0 Lh14 screens. One Lh14 grandson with golden wings (instead of the normal gray wings) was identified. This aurum1 male was mated with “lab” females; the progeny was “selfed” to obtain additional mutant males. For a pure-breeding stock, putative heterozygous unmated females were collected by placing pupae into Falcon 3072 96-well plates (Becton Dickinson) 1–3 days prior to their emergence. Strips of Scotch Tape (0.9 cm wide) were used to seal the wells. The aurum1 strain was homozygous viable, and a pure-breeding stock was established.
A second mutant phenotype was observed in the aurum1 wasp culture. The posterior wing margins of this aurum1 wasp were angular instead of the normal round ends. This mutant, kona1, assorted from the aurum1 mutation, and was similarly homozygous viable but a pure-breeding stock could not be established as homozygous kona1 females could not infect hosts. To establish this, 14 unmated, or 11 kona1 male-mated, heterozygous or homozygous kona1 females, were introduced to y w hosts. Ten males were used for the matings. Larval hosts were dissected to assess the presence of eggs.
To examine if aurum1 and kona1 mutations assort independently, unmated homozygous aurum1 females were mated with kona1 males. Thirty-six unmated double heterozygous females were used for two rounds of y w infections. Over 1,000 male progeny of the four expected genotypes were scored.
Ovaries from wild type, heterozygous, or homozygous kona1 females were fixed in ethanol, mounted in 50% glycerol, and imaged on a Nikon Eclipse TE2000-U microscope attached to a Diagnostic Instruments Inc. camera and NIS Elements Imaging Software 64.0 bit 3.22.14. Ovipositors of 10 wild type and 30 homozygous kona1 females were gently pulled out using sharp forceps, briefly dipped in 70% ethanol and PBS, fixed in 4% paraformaldehyde, and washed in PBS, before mounting in Vectashield. Images were acquired with a Leica Aperio CS2 microscope.
Quantification and statistical analysis
Sample size, experimental replication, and data analysis of results are also described in the corresponding sections above and/or the corresponding figure legend. For Figures 2 and S9, each experimental vial was considered a replicate, and statistical analysis of insect survival in space relative to ground after their return to the lab was performed in Microsoft Excel 2016. The mean numbers (± standard error) of flies and/or wasps from 14 vials were calculated in Microsoft Excel (2016). Student’s t test statistic was applied, and the difference between the corresponding space versus ground samples was significant if p < 0.05. For the data in Figures 6D and 6E, the weighted average number of tumors per animal in S0 versus G0 flies, or S1 versus G1 flies, was calculated in Microsoft Excel. The mean ± standard error was calculated, and a Student’s t test was used to determine biological significance. For Figure 7B, the non-parametric Mann-Whitney U test for unpaired samples was performed to compare all conditions pairwise. p values < 0.05 were considered significant. For Figure 9H, more than 1,000 F2 males were scored from unmated dihybrid females, and the raw numbers obtained are shown. A standard chi-square test was applied to accept the null hypothesis. For Figure 9I, at least 30 larvae per condition were dissected and scored for signs of infection. For Figure S11, the means of the proportion of bipolar cells in the L. heterotoma virulence assay were compared in experimental versus control samples using Excel and a Student’s t test. Between 129 and 1,146 lamellocytes were scored per replicate. For differential gene expression analysis in R using DESeq2, all groups were compared using the Wald test. To generate the F statistic p value and false discovery rate (adjusted p value), the likelihood ratio test was used. Corrections for multiple testing adjustment were performed using Benjamini-Hochberg. Genes were considered differentially expressed if adjusted p < 0.05 and |log2FC| > 1.
Acknowledgments
We are grateful to Edina Blazevic, Jackson Chen, Amy Gresser, Ye He, Waleed Khalid, Kevin Martin, Leo Mazur, Jorge Morales, and Zubaidul Razzak for assistance with experiments or analysis. Kristen Peach and Jack Miller provided the radiation data, and Amy Gresser designed the FFL-03 emblem in the Graphical Abstract. S. A. Tasnim Ahmed and Asif Siddiq helped with assembling figures. San-huei Lai Polo helped with data curation on OSDR. Research was funded by NASA (NNX15AB42G), the National Science Foundation (1121817 and 2022235), the National Institutes of Health (1F31GM111052-01A1), and PSC-CUNY (63816-00 51 and 66619-00 54). The sponsors or funders did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conceptualization, methodology, investigation, formal analysis, visualization, resources, writing – review and editing: all authors; software: A.M.S.-B.; data curation: A.M.S.-B., J.C., S.G.; supervision, writing – original draft: S.G.; funding acquisition, supervision: S.B., S.G.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: December 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108759.
Supplemental information
All 293 Lb17 transcripts, differentially expressed (adjusted p < 0.05 and |log2FC| > 1) in space are listed. The GAJA number is the NCBI accession number of the L. boulardi transcript. The conserved protein domain (top hit), its accession ID and E-value are shown next. Values in bold identify genes included in Table 1.
All 310 Lh14 transcripts, differentially expressed (adjusted p < 0.05 and |log2FC| > 1) in space are listed. The GAJC number is the NCBI accession number of the L. heterotoma transcript. The conserved protein domain (top hit), its accession ID and E-value are shown next. Values in bold identify genes included in Table 2.
References
- 1.Afshinnekoo E., Scott R.T., MacKay M.J., Pariset E., Cekanaviciute E., Barker R., Gilroy S., Hassane D., Smith S.M., Zwart S.R., et al. Fundamental Biological Features of Spaceflight: Advancing the Field to Enable Deep-Space Exploration. Cell. 2020;183:1162–1184. doi: 10.1016/j.cell.2020.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcu O., Lera M.P., Sanchez M.E., Levic E., Higgins L.A., Shmygelska A., Fahlen T.F., Nichol H., Bhattacharya S. Innate immune responses of Drosophila melanogaster are altered by spaceflight. PLoS One. 2011;6 doi: 10.1371/journal.pone.0015361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor K., Kleinhesselink K., George M.D., Morgan R., Smallwood T., Hammonds A.S., Fuller P.M., Saelao P., Alley J., Gibbs A.G., et al. Toll mediated infection response is altered by gravity and spaceflight in Drosophila. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer J., Mhatre S.D., Gilbert R., Bhattacharya S. Multi-system responses to altered gravity and spaceflight: Insights from Drosophila melanogaster. Neurosci. Biobehav. Rev. 2022;142 doi: 10.1016/j.neubiorev.2022.104880. [DOI] [PubMed] [Google Scholar]
- 5.Lue C.H., Buffington M.L., Scheffer S., Lewis M., Elliott T.A., Lindsey A.R.I., Driskell A., Jandova A., Kimura M.T., Carton Y., et al. DROP: Molecular voucher database for identification of Drosophila parasitoids. Mol. Ecol. Resour. 2021;21:2437–2454. doi: 10.1111/1755-0998.13435. [DOI] [PubMed] [Google Scholar]
- 6.Kim-Jo C., Gatti J.L., Poirié M. Drosophila Cellular Immunity Against Parasitoid Wasps: A Complex and Time-Dependent Process. Front. Physiol. 2019;10:603. doi: 10.3389/fphys.2019.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehlers S., Schaible U.E. The granuloma in tuberculosis: dynamics of a host-pathogen collusion. Front. Immunol. 2012;3:411. doi: 10.3389/fimmu.2012.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlenke T.A., Morales J., Govind S., Clark A.G. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 2007;3:1486–1501. doi: 10.1371/journal.ppat.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu H., Morales J., Govind S. Identification and immuno-electron microscopy localization of p40, a protein component of immunosuppressive virus-like particles from Leptopilina heterotoma, a virulent parasitoid wasp of Drosophila. J. Gen. Virol. 2006;87:461–470. doi: 10.1099/vir.0.81474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizki R.M., Rizki T.M. Selective destruction of a host blood cell type by a parasitoid wasp. Proc. Natl. Acad. Sci. USA. 1984;81:6154–6158. doi: 10.1073/pnas.81.19.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heavner M.E., Ramroop J., Gueguen G., Ramrattan G., Dolios G., Scarpati M., Kwiat J., Bhattacharya S., Wang R., Singh S., Govind S. Novel Organelles with Elements of Bacterial and Eukaryotic Secretion Systems Weaponize Parasites of Drosophila. Curr. Biol. 2017;27:2869–2877.e6. doi: 10.1016/j.cub.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wey B., Heavner M.E., Wittmeyer K.T., Briese T., Hopper K.R., Govind S. Immune Suppressive Extracellular Vesicle Proteins of Leptopilina heterotoma Are Encoded in the Wasp Genome. G3 (Bethesda) 2020;10:1–12. doi: 10.1534/g3.119.400349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colinet D., Schmitz A., Cazes D., Gatti J.-L., Poirié M. The origin of intraspecific variation of virulence in an eukaryotic immune suppressive parasite. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Giovanni D., Lepetit D., Guinet B., Bennetot B., Boulesteix M., Couté Y., Bouchez O., Ravallec M., Varaldi J. A behavior-manipulating virus relative as a source of adaptive genes for Drosophila parasitoids. Mol. Biol. Evol. 2020;37:2791–2807. doi: 10.1093/molbev/msaa030. [DOI] [PubMed] [Google Scholar]
- 15.Wey B. 2021. PhD Thesis: Insights into Leptopilina Spp. Immune-Suppressive Strategies Using Mixed-Omics and Molecular Approaches.https://academicworks.cuny.edu/gc_etds/4162 [Google Scholar]
- 16.Gueguen G., Rajwani R., Paddibhatla I., Morales J., Govind S. VLPs of Leptopilina boulardi share biogenesis and overall stellate morphology with VLPs of the heterotoma clade. Virus Res. 2011;160:159–165. doi: 10.1016/j.virusres.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan B., Poirié M., Gatti J.L. Parasitoid wasp venom vesicles (venosomes) enter Drosophila melanogaster lamellocytes through a flotillin/lipid raft-dependent endocytic pathway. Virulence. 2020;11:1512–1521. doi: 10.1080/21505594.2020.1838116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu H., Govind S. Natural infection of D. melanogaster by virulent parasitic wasps induces apoptotic depletion of hematopoietic precursors. Cell Death Differ. 2002;9:1379–1381. doi: 10.1038/sj.cdd.4401134. [DOI] [PubMed] [Google Scholar]
- 19.Rizki T.M., Rizki R.M., Carton Y. Leptopilina heterotoma and L. boulardi: strategies to avoid cellular defense responses of Drosophila melanogaster. Exp. Parasitol. 1990;70:466–475. doi: 10.1016/0014-4894(90)90131-u. [DOI] [PubMed] [Google Scholar]
- 20.Ramroop J.R., Heavner M.E., Razzak Z.H., Govind S. A parasitoid wasp of Drosophila employs preemptive and reactive strategies to deplete its host's blood cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizki T.M., Rizki R.M. Parasitoid-induced cellular immune deficiency in Drosophila. Ann. N. Y. Acad. Sci. 1994;712:178–194. doi: 10.1111/j.1749-6632.1994.tb33572.x. [DOI] [PubMed] [Google Scholar]
- 22.Luo H., Hanratty W.P., Dearolf C.R. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison D.A., Binari R., Nahreini T.S., Gilman M., Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panettieri S., Paddibhatla I., Chou J., Rajwani R., Moore R.S., Goncharuk T., John G., Govind S. Discovery of aspirin-triggered eicosanoid-like mediators in a Drosophila metainflammation blood tumor model. J. Cell Sci. 2019;133 doi: 10.1242/jcs.236141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu P., Pan P.C., Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 26.Govind S. Rel signalling pathway and the melanotic tumour phenotype of Drosophila. Biochem. Soc. Trans. 1996;24:39–44. doi: 10.1042/bst0240039. [DOI] [PubMed] [Google Scholar]
- 27.Hanratty W.P., Ryerse J.S. A genetic melanotic neoplasm of Drosophila melanogaster. Dev. Biol. 1981;83:238–249. doi: 10.1016/0012-1606(81)90470-x. [DOI] [PubMed] [Google Scholar]
- 28.Rizki R.M., Rizki T.M. Basement membrane abnormalities in melanotic tumor formation of Drosophila. Experientia. 1974;30:543–546. doi: 10.1007/BF01926343. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee S., Biehl A., Gadina M., Hasni S., Schwartz D.M. JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs. 2017;77:521–546. doi: 10.1007/s40265-017-0701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnabei L., Laplantine E., Mbongo W., Rieux-Laucat F., Weil R. NF-kappaB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.716469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickerson C.A., Ott C.M., Mister S.J., Morrow B.J., Burns-Keliher L., Pierson D.L. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2000;68:3147–3152. doi: 10.1128/iai.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilbert R., Torres M., Clemens R., Hateley S., Hosamani R., Wade W., Bhattacharya S. Spaceflight and simulated microgravity conditions increase virulence of Serratia marcescens in the Drosophila melanogaster infection model. NPJ Microgravity. 2020;6:4. doi: 10.1038/s41526-019-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mhatre S.D., Iyer J., Petereit J., Dolling-Boreham R.M., Tyryshkina A., Paul A.M., Gilbert R., Jensen M., Woolsey R.J., Anand S., et al. Artificial gravity partially protects space-induced neurological deficits in Drosophila melanogaster. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Gregorio E., Spellman P.T., Tzou P., Rubin G.M., Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchon N., Broderick N.A., Poidevin M., Pradervand S., Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Rooryck C., Diaz-Font A., Osborn D.P.S., Chabchoub E., Hernandez-Hernandez V., Shamseldin H., Kenny J., Waters A., Jenkins D., Kaissi A.A., et al. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat. Genet. 2011;43:197–203. doi: 10.1038/ng.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gramates L.S., Agapite J., Attrill H., Calvi B.R., Crosby M.A., Dos Santos G., Goodman J.L., Goutte-Gattat D., Jenkins V.K., Kaufman T., et al. Fly Base: a guided tour of highlighted features. Genetics. 2022;220 doi: 10.1093/genetics/iyac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis C.F., Dorak M.T. An extensive analysis of the hereditary hemochromatosis gene HFE and neighboring histone genes: associations with childhood leukemia. Ann. Hematol. 2010;89:375–384. doi: 10.1007/s00277-009-0839-y. [DOI] [PubMed] [Google Scholar]
- 39.de Cicco D.V., Spradling A.C. Localization of a cis-acting element responsible for the developmentally regulated amplification of Drosophila chorion genes. Cell. 1984;38:45–54. doi: 10.1016/0092-8674(84)90525-7. [DOI] [PubMed] [Google Scholar]
- 40.Cornman R.S. Molecular evolution of Drosophila cuticular protein genes. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enjin A., Zaharieva E.E., Frank D.D., Mansourian S., Suh G.S.B., Gallio M., Stensmyr M.C. Humidity Sensing in Drosophila. Curr. Biol. 2016;26:1352–1358. doi: 10.1016/j.cub.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanratty W.P., Dearolf C.R. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol. Gen. Genet. 1993;238:33–37. doi: 10.1007/BF00279527. [DOI] [PubMed] [Google Scholar]
- 44.Lanot R., Zachary D., Holder F., Meister M. Postembryonic hematopoiesis in Drosophila. Dev. Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 45.Louradour I., Sharma A., Morin-Poulard I., Letourneau M., Vincent A., Crozatier M., Vanzo N. Reactive oxygen species-dependent Toll/NF-kappaB activation in the Drosophila hematopoietic niche confers resistance to wasp parasitism. Elife. 2017;6 doi: 10.7554/eLife.25496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorrentino R.P., Carton Y., Govind S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 2002;243:65–80. doi: 10.1006/dbio.2001.0542. [DOI] [PubMed] [Google Scholar]
- 47.Goecks J., Mortimer N.T., Mobley J.A., Bowersock G.J., Taylor J., Schlenke T.A. Integrative approach reveals composition of endoparasitoid wasp venoms. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colinet D., Schmitz A., Depoix D., Crochard D., Poirié M. Convergent use of RhoGAP toxins by eukaryotic parasites and bacterial pathogens. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labrosse C., Stasiak K., Lesobre J., Grangeia A., Huguet E. A RhoGAP protein as a main immune suppressive factor in the Leptopilina boulari (Hymenoptera, Figitidae)-Drosophila melanogaster interaction. Insect Biochem. Mol. Biol. 2005;35:92–103. doi: 10.1016/j.ibmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 50.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ikenaga M., Yoshikawa I., Kojo M., Ayaki T., Ryo H., Ishizaki K., Kato T., Yamamoto H., Hara R. Mutations induced in Drosophila during space flight. Biol. Sci. Space. 1997;11:346–350. doi: 10.2187/bss.11.346. [DOI] [PubMed] [Google Scholar]
- 52.Filatova L.P., Vaulina E.N., Grozdova T., Prudhommeau C., Proust J. Some results of the effect of space flight factors on Drosophila melanogaster. Adv. Space Res. 1983;3:143–146. doi: 10.1016/0273-1177(83)90184-9. [DOI] [PubMed] [Google Scholar]
- 53.Buckhold B. Biosatellite II--physiological and somatic effects on insects. Life Sci. Space Res. 1969;7:77–83. [PubMed] [Google Scholar]
- 54.Thimann K.V. Biosatellite II experiments: preliminary results. Proc. Natl. Acad. Sci. USA. 1968;60:347–361. doi: 10.1073/pnas.60.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogneva I.V., Belyakin S.N., Sarantseva S.V. The Development Of Drosophila Melanogaster under Different Duration Space Flight and Subsequent Adaptation to Earth Gravity. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walls S., Diop S., Birse R., Elmen L., Gan Z., Kalvakuri S., Pineda S., Reddy C., Taylor E., Trinh B., et al. Prolonged Exposure to Microgravity Reduces Cardiac Contractility and Initiates Remodeling in Drosophila. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merzendorfer H., Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003;206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 58.Davis M.N., Horne-Badovinac S., Naba A. In-silico definition of the Drosophila melanogaster matrisome. Matrix Biol. 2019;4 doi: 10.1016/j.mbplus.2019.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassler D.M., Zeitlin C., Wimmer-Schweingruber R.F., Ehresmann B., Rafkin S., Eigenbrode J.L., Brinza D.E., Weigle G., Böttcher S., Böhm E., et al. Mars' surface radiation environment measured with the Mars Science Laboratory's Curiosity rover. Science. 2014;343 doi: 10.1126/science.1244797. [DOI] [PubMed] [Google Scholar]
- 60.Hellweg C.E., Baumstark-Khan C. Getting ready for the manned mission to Mars: the astronauts' risk from space radiation. Naturwissenschaften. 2007;94:517–526. doi: 10.1007/s00114-006-0204-0. [DOI] [PubMed] [Google Scholar]
- 61.Cucinotta F.A., To K., Cacao E. Predictions of space radiation fatality risk for exploration missions. Life Sci. Space Res. 2017;13:1–11. doi: 10.1016/j.lssr.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 62.Tokusumi T., Sorrentino R.P., Russell M., Ferrarese R., Govind S., Schulz R.A. Characterization of a lamellocyte transcriptional enhancer located within the misshapen gene of Drosophila melanogaster. PLoS One. 2009;4:e6429. doi: 10.1371/journal.pone.0006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeitlin C C.A., Beard K.B., Abdelmelek M., Hayes B.M., Johnson A.S., Stoffle N., Rios R.R. Life Sciences in Space Research; 2023. Results from the Radiation Assessment Detector on the International Space Station: Part 1, the Charged Particle Detector. [DOI] [PubMed] [Google Scholar]
- 64.Gueguen G., Kalamarz M.E., Ramroop J., Uribe J., Govind S. Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Small C., Ramroop J., Otazo M., Huang L.H., Saleque S., Govind S. An unexpected link between notch signaling and ROS in restricting the differentiation of hematopoietic progenitors in Drosophila. Genetics. 2014;197:471–483. doi: 10.1534/genetics.113.159210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalamarz M.E., Paddibhatla I., Nadar C., Govind S. Sumoylation is tumor-suppressive and confers proliferative quiescence to hematopoietic progenitors in Drosophila melanogaster larvae. Biol. Open. 2012;1:161–172. doi: 10.1242/bio.2012043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paddibhatla I., Lee M.J., Kalamarz M.E., Ferrarese R., Govind S. Role for sumoylation in systemic inflammation and immune homeostasis in Drosophila larvae. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stark R., Grzelak M., Hadfield J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 69.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wickham H. 2nd ed. Springer International Publishing; 2016. ggplot2 : Elegant Graphics for Data Analysis. Use R! [Google Scholar]
- 73.Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., Rubin G.M. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salzberg A., Prokopenko S.N., He Y., Tsai P., Pál M., Maróy P., Glover D.M., Deák P., Bellen H.J. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: mutations affecting embryonic PNS development. Genetics. 1997;147:1723–1741. doi: 10.1093/genetics/147.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bourbon H.M., Gonzy-Treboul G., Peronnet F., Alin M.F., Ardourel C., Benassayag C., Cribbs D., Deutsch J., Ferrer P., Haenlin M., et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 76.Oh S.W., Kingsley T., Shin H.H., Zheng Z., Chen H.W., Chen X., Wang H., Ruan P., Moody M., Hou S.X. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics. 2003;163:195–201. doi: 10.1093/genetics/163.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanson M.A., Lemaitre B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020;62:22–30. doi: 10.1016/j.coi.2019.11.008. [DOI] [PubMed] [Google Scholar]
- 78.Bina S., Wright V.M., Fisher K.H., Milo M., Zeidler M.P. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 2010;11:201–207. doi: 10.1038/embor.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 80.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn201. W5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All 293 Lb17 transcripts, differentially expressed (adjusted p < 0.05 and |log2FC| > 1) in space are listed. The GAJA number is the NCBI accession number of the L. boulardi transcript. The conserved protein domain (top hit), its accession ID and E-value are shown next. Values in bold identify genes included in Table 1.
All 310 Lh14 transcripts, differentially expressed (adjusted p < 0.05 and |log2FC| > 1) in space are listed. The GAJC number is the NCBI accession number of the L. heterotoma transcript. The conserved protein domain (top hit), its accession ID and E-value are shown next. Values in bold identify genes included in Table 2.
Data Availability Statement
-
•
Original transcriptomic data (raw read counts and FASTQ files) from bulk RNA-Seq experiments on hosts and parasites have been deposited in and can be accessed from NASA’s publicly available Open Science Data Repository (https://osdr.nasa.gov/bio/repo). Accession numbers are listed in the key resources table.
-
•
Version D of NASA GeneLab’s bioinformatics pipeline, available on GitHub, was used to process Drosophila RNA-Seq reads. A DOI is listed in the key resources table.
-
•
A modified version of the Drosophila pipeline was used to analyze L. boulardi and L. heterotoma reads. Those scripts are available with the supplemental material Data S1 (L.boulardi_ L.heterotoma_RNAseq_processing_scripts.zip).
-
•
Microscopy data and any additional information required to reanalyze the data reported in this paper are available from the lead contact upon request.