Graphical abstract
Keywords: Pinus elliottii needles, Ultrasound-Assisted Extraction, Response Surface Methodology, Kinetic Model, Antioxidant activity, UHPLC-Q-Exactive Orbitrap MS/MS
Highlights
-
•
Ultrasound-assisted extraction of PNś polyphenolic compounds was optimized by RSM.
-
•
UAE showed higher efficient extraction for polyphenols than that of CSE.
-
•
PNś extracts exhibited strong antioxidant activity.
-
•
37 kinds of polyphenolic compounds were identified by UHPLC-Q-Exactive Orbitrap MS/MS.
-
•
Kinetic model suggested the extraction process of PNs is diffusion-controlled.
Abstract
Extracting polyphenolic bioactive compounds from Pinus elliottii needles, a forestry residue, promises economic and environmental benefits, however, relevant experimental data are lacking. Herein, a comprehensive investigation of the polyphenolic composition of pine needles (PNs) was carried out. Ultrasound-Assisted Extraction (UAE) was applied to extract the polyphenolic compounds of pine needles. The optimal conditions of extracts were determined by Response Surface Methodology (RSM). The maximum total phenolic content (TPC) of 40.37 mg GAE/g PNs was achieved with solid–liquid ratio of 1:20, 60 % ethanol, and 350 W for 25 min at 45 °C. Polyphenolic extracts showed antioxidant activity in scavenging free radicals and reducing power (DPPH, IC50 41.05 μg/mL; FRAP 1.09 mM Fe2+/g PNs; ABTS, IC50 214.07 μg/mL). Furthermore, the second-order kinetic model was also constructed to describe the mechanism of the UAE process, with the extraction activation energy estimated at 12.26 kJ/mol. In addition, 37 compounds in PNs were first identified by UHPLC-Q-Exactive Orbitrap MS/MS, including flavonoids and phenolic acids. The results suggest that Ultrasound-Assisted is an effective method for the extraction of natural polyphenolic compounds from pine needles and this study could serve as a foundation for utilizing phenolics derived from PNs in the food and pharmaceutical industries.
1. Introduction
Pinus genus consists of approximately one hundred and twenty species, growing in a wide range of Asia, Europe, and North America, many of which are quite harsh and extreme [1]. Pinus elliottii is a highly economically valuable species within the Pinus genus, characterized by its rapid growth, high resin production, and resilience [2], [3]. Its wood is used in both the timber and pulp industries, and the exuded turpentine from pine is an important raw material for food, pharmaceuticals, and chemicals. Pine needles (PNs) the leaves of Pinaceae plants from the main residue of pine trees are fast-growing, naturally stocked, widely distributed, and harvested throughout the year. In 2018, the yearly net yield of pine needles in the Western Himalayan region alone was 67.99 million tons. However, most of the pine needles are discarded directly, and highly flammable waste PNs are mostly responsible for forest fires, which cause significant harm to flora, fauna, and forest land and emit greenhouse gas into the atmosphere [4]. Currently, only a small percentage have been processed into manufactured products, such as pine needle powders added to food, pine needle tea, and pine needle beverages. The low utilization rate leads to a significant waste of resources. Numerous studies have shown that pine needles are of high nutritional and medicinal value with abundant protein, vitamins, minerals, and essential oils [5], [6], [7], [8], [9]. However, previous studies have primarily examined the PNs' volatile components, with no published report specifically addressing the analysis, identification, antioxidant activity, and TPC of polyphenolic compounds in PNs.
Polyphenolic compound is a type of natural active substance extracted from plants including Pinus genus, which belong to the main products of secondary metabolites [10]. It is present in the leaves, the bark and the fruit and exhibits valuable biological properties with potential health benefits effects and bioactivity as high value-added compounds, such as antioxidant, anti-inflammatory, and antimicrobial activities [11]. So it is a beneficial natural antioxidant source for use in food additives, cosmetic ingredients, or as precursors for the pharmaceutical industry [12].
The traditional method for polyphenolic compound extraction is Conventional Solvent Extraction (CSE), which often intensifies mass transfer by heating, causing the polyphenolic compound to be released into the solvent water [13]. Nonetheless, Ultrasound-Assisted Extraction (UAE) is an eco-friendly method that reduces the time of extraction, solvent volume, and energy input compared to CSE [14]. UAE primarily facilitates the release of soluble compounds from the plant by inducing cavitation that damages the cell walls, improves mass transfer, and enables the solvent to access cell contents [15]. However, the application of UAE involves the interaction of multiple factors. So, it is important to optimize the extraction conditions by Response Surface Methodology (RSM) increasingly utilized to design multivariate experiments to reduce the number of analyses required in the optimization process, resulting in richer and more precise results than traditional full factorial experiments [16], [17], [18]. In addition, physical or empirical kinetic models have been used to describe the mechanism of the solid–liquid extraction process [19].
The extraction of PNs’ polyphenolic compounds, a forestry residue, has many economic and environmental benefits and good prospects in high-end bioactive utilization. Salzano de Luna et al. reported the results of three extraction methods (conventional maceration, microwave-assisted extraction, UAE) for Pi̅nǔs pi̅nea needle extracts [20]. However, much of the relevant data is currently lacking, including the types of polyphenolic compounds found in pine needles, the extraction methods and efficiencies, and the kinetic model of the extraction, which is essential for the industrial exploitation of the extraction process.
The objective of this work was a comprehensive study of pine needles (pinus elliottii). The extraction conditions of polyphenolic compounds from pine needles (pinus elliottii) were optimized by RSM. A second-order extraction dynamics model was created to analyze the impact of temperature on the extraction process. The antioxidant capacity of PNs extraction was evaluated by DPPH, FRAP, and ABTS. The polyphenolic compounds were identified by UHPLC-Q-Exactive Orbitrap MS/MS. The research results are intended to provide a foundation for future applications of polyphenolic compounds from pine needles (Pinus elliottii) in the field of food and pharmaceuticals.
2. Materials and methods
2.1. Raw material and solvents
Fresh pine needles (Pinus elliottii) were purchased from Wuzhou, Guangxi Zhuang Autonomous Region, China. Fresh pine needles were frozen at −4 °C. For use, they were first cut into approximately 2 cm long pieces. Then it was placed in a blast drying oven (DHG-9030A) at 45 °C for 24 h. Finally, placed in a pulverizer (FW100, Taisite Instrument Co., Tianjin, China) for 5 min. They were pulverized into powder and sieved by a 60-mesh screen.
Gallic acid, Rutin, DPPH (2,2-diphenyl-1-picrylhydrazyl), Vitamin C (L-Ascorbic acid), Folin-Ciocalteu’s phenol reagent, Pyrogallol, TPTZ (2,4,6-Tri(2-pyridyl)-s-triazine), ABTS (2,2′-azino-bis(3ethylbenzothiazoline-6-sulfonic acid)) were purchased from Macklin Company (Shanghai, China). The other chemical reagents were of analytical grade.
2.2. Ultrasound-assisted extraction of polyphenolic extract
A sample of crushed pine needles (1 g) is placed in a centrifuge tube (polypropylene, 50 mL, round bottom) and mixed with an appropriate volume of aqueous ethanol solution. Centrifuge tubes containing samples are sonicated in a sonication unit at different power, time, and temperature. All samples were placed in the fixed position of the ultrasound bath (KQ-500DB, China). Each sample was repeated three times. Parameters were adjusted according to the parameter settings of Single-factor and RSM experiments. After UAE, the mixture was placed in a high-speed centrifuge (TG16K-Ⅱ, DONGWANG INSTRUMENT, China) and spun at 10,270 g for 10 min. The resultant supernatant was collected, diluted to 100 mL, and then stored at a temperature of −4 °C.
2.3. Single-factor experimental and RSM based on CCD experimental design
Single-factor experiments were conducted using various power levels (250, 300, 350, 400, 450 W), temperatures (25, 35, 45, 55, 65 ℃), ethanol concentrations (40, 50, 60, 70, 80 %), solid–liquid ratios (1:X, X = 5, 10, 15, 20, 25), and times (10, 15, 20, 25, 30 min) (Table 1).
Table 1.
Single dependent variables of extraction conditions experimental design.
| Single factor | Power (W) | Temperature (℃) | Ethanol Concentration (%) | Solid-liquid ratio (1:X) | Time (min) |
|---|---|---|---|---|---|
| Power | 250,300,350, 400,450 | 45 | 60 | 15 | 20 |
| Temperature | 350 | 25,35,45, 55,65 | 60 | 15 | 20 |
| Ethanol concentration | 350 | 45 | 40,50,60, 70,80 | 15 | 20 |
| Solid-liquid ratio | 350 | 45 | 60 | 5,1015, 20,25 | 20 |
| Time | 350 | 45 | 60 | 20 | 10,15,20, 25,30 |
Design-Expert 13 was used to establish Response Surface Methodology (RSM) based on Central Composite Design (CCD) for investigating the Ultrasound-Assisted Extraction process of PNs’ polyphenolic compounds. Table 2 displays the outcome of the RSM process. RSM was designed to explore four independent factors with TPC as the response variable.
Table 2.
CCD with responses of the dependent variables to extraction conditions.
| Run | Power A (W) | Temperature B (℃) | Solid-liquid C (1:X) | Time D (min) | TPC (mg GAE/g PNs) |
|---|---|---|---|---|---|
| 1 | 1(4 0 0) | −1(35) | 1(20) | 1(25) | 36.98 |
| 2 | −2(2 5 0) | 0(45) | 0(15) | 0(20) | 32.72 |
| 3 | −1(3 0 0) | −1 | 1 | −1(15) | 34.72 |
| 4 | −1 | −1 | 1 | 1 | 37.02 |
| 5 | 0(3 5 0) | 0 | 0 | 0 | 40.37 |
| 6 | −1 | −1 | −1(10) | −1 | 31.98 |
| 7 | 0 | 2(65) | 0 | 0 | 36.44 |
| 8 | 1 | −1 | −1 | 1 | 34.86 |
| 9 | −1 | 1 | 1 | −1 | 35.78 |
| 10 | 1 | 1 | 1 | 1 | 37.66 |
| 11 | 0 | −2(25) | 0 | 0 | 33.8 |
| 12 | 1 | 1 | −1 | −1 | 32.53 |
| 13 | −1 | 1 | −1 | −1 | 33.27 |
| 14 | 0 | 0 | 0 | −2(10) | 32.09 |
| 15 | 0 | 0 | 0 | 0 | 40.1 |
| 16 | 0 | 0 | 0 | 0 | 40.83 |
| 17 | 1 | −1 | −1 | −1 | 30.92 |
| 18 | 0 | 0 | 0 | 2(30) | 37.95 |
| 19 | 0 | 0 | 0 | 0 | 38.73 |
| 20 | 0 | 0 | −2(5) | 0 | 35.61 |
| 21 | −1 | −1 | −1 | 1 | 34.08 |
| 22 | 1 | −1 | 1 | −1 | 32.84 |
| 23 | −1 | 1 | −1 | 1 | 34.66 |
| 24 | 2(4 5 0) | 0 | 0 | 0 | 31.95 |
| 25 | −1 | 1 | 1 | 1 | 38.07 |
| 26 | 1 | 1 | −1 | 1 | 35.76 |
| 27 | 0 | 0 | 0 | 0 | 39.16 |
| 28 | 0 | 0 | 2(25) | 0 | 40.66 |
| 29 | 0 | 0 | 0 | 0 | 39.45 |
| 30 | 1 | 1 | 1 | −1 | 33.81 |
2.4. Total phenolic and flavonoid contents of PNs extracts
The total phenolic content (TPC) within the supernatant of the PNs extracts was assessed using the Folin-Ciocalteu method in accordance with the adjustments of Fatima et al. [21]. Initially, 20 μL supernatant was diluted to 1 mL and 1 mL of Folin-Ciocalteu solution was then added. Following a 3-minute incubation period, 3 mL of 10 % Na2CO3 solution was added. The combination was then kept away from light and left to stand for 40 min before measurement. The content and standard curve were calculated by measuring the absorbance at 765 nm with a UV–visible spectrophotometer (UV-2600, SHIMADZU, Japan). The results were presented as milligrams of gallic acid equivalents (GAE) per gram of pine needles (mg GAE/g PNs).
The total flavonoid content (TFC) was determined in the PNs extracts (supernatant) using Wang et al.'s method with little adjustments [22], [23]. The 50 μL sample solution was diluted to 2 mL and then mixed with 0.15 mL 5 % NaNO2 solution and 2 mL of deionized water. The mixture was shaken thoroughly and left for 6 min after adding 0.15 mL 10 % Al(NO3)3 solution. Then, 2.0 mL of 4 % NaOH solution was added, and the mixture was shaken thoroughly and left to stand for 15 min, protected from light. The absorbance was measured at a wavelength of 510 nm with 60 % ethanol employed as a blank control, and subsequent calculations were made for both content and standard curve. The results were presented as milligrams of rutin equivalent (RE) per gram of pine needles (mg RE/g PNs).
2.5. Kinetic modeling
The extraction kinetic model refers to the second-order extraction kinetic model [24], [25], [26]. Assume that the extraction process is as follows.
| (1) |
And assume that (1) the extractable fraction distributes evenly within the solid (2) the solid is well distributed in the extraction solvent, and (3) a constant diffusion coefficient for the extractable fraction.
The leaching rate of polyphenols from the pine needle can be expressed as Eq (2):
| (2) |
Ct (mg GAE/mL) is the concentration of PNs’ polyphenolic at t (min).
Cs (mg GAE/mL) is the concentration of polyphenolic in the supernatant at saturation.
k (mL/mg·min) represents the constant for the rate of second-order extraction.
The extracted kinetic constants were calculated using the formula Eq (2), integrating under the boundary conditions (Ct = 0-t and t = 0-t), and expressed as either a formal equation Eq (3) or a linear equation Eq (4).
| (3) |
| (4) |
h (mL/mg·min) is the starting extraction rate at which Ct and t tend to 0.
TPC at any given moment can be expressed as Eq (5) after rearrangement.
| (5) |
when plotting with 't' as the x-axis coordinate and '' as the y-axis coordinate, the slope of the line is given by , and the intercept by . This provides the second-order extraction rate constant.
According to Arrhenius Equation Eq (6), the rate constant rises as the temperature increases.
| (6) |
k (mL/mg·min) is the extraction rate constant.
k0 (mL/mg·min) is the thermal independent factor.
Ea (J/mol) is extraction activation energy.
R (8.314 J·mol−1·K) is the gas constant.
T (K) is the absolute liquid phase extraction temperature.
Taking lnk as the x-axis coordinate and 1/T as the y-axis coordinate, the resulting equation can be linearized Eq (7), always resulting in the thermal independent factor (k0) and the activation energy (Ea).
| (7) |
To determine the agreement between predicted and measured data, the R-squared (R2) was utilized. The R2 equation is shown in Eq (8).
| (8) |
Here n corresponds to the count of samples. yi denotes the actual measured data of the sample. refers to the model-fitted data of the sample, and y represents the average of all experimental data. The standardized R2 falls between 0 and 1 with higher values indicating a stronger correlation between the measured and the model-fit data.
2.6. Antioxidant activity of PNs extracts
2.6.1. DPPH radical scavenging capacity assay
The method of Zhang et al. was used to assess the scavenging capacity of PNs extracts (supernatant) for DPPH radicals with minor modifications [27]. The supernatant was initially diluted 20 times. Afterwards, the DPPH reagent was combined with the diluted samples (3 mL + 1 mL) at room temperature, then the combination was incubated in the dark for 30 min. The free radical scavenging assay was checked at 520 nm wavelength while utilizing Vitamin C and Trolox as positive control. The formula Eq (9) was used to calculate the DPPH radical scavenging activity:
| (9) |
A0 represents the DPPH absorbance value when no sample is present. Ai denotes the solution's absorbance value to DPPH. Ai0 denotes the solution's absorbance value when DPPH is absent.
For the Single-factor experiment, standard curves were established using different concentrations of vitamin C or Trolox solutions instead of samples. The obtained results were quantified in grams of Vitamin C equivalency per gram of pine needles (mg VC/g PNs).
2.6.2. Ferric reducing antioxidant power assay (FRAP)
The PNs extracts' ability to remove Fe3+ was assessed by the FRAP method of Martínez-Patiño et al. with minor modifications [28]. The FRAP solution (2.5 mL, preheated to 37 °C) was combined with the supernatant (0.5 mL) and immersed in a water bath(37 °C, 30 min), and then measuring the absorbance of FeSO4, which was used as a reference compound, at 593 nm. Quantifying the antioxidant activity by following a standard curve and expressing it as mM Fe2+/g pine needles. As a positive control, Vitamin C and Trolox were utilized.
2.6.3. ABTS assay
The PNs extracts' ability to reduce ABTS•+ was assessed using the ABTS method outlined by Sridhar & Charle but with certain modifications [29]. ABTS working solution was prepared by combining equal amounts of K2S2O8 solution (2.6 mM) and ABTS (7.4 mM) and maintaining the mixture with no exposure to light for 12–16 h at room temperature. The absorbance of ABTS working solution should reach 0.68 ± 0.02 by diluting with Ethanol. Next, combine 2.5 mL ABTS working solution (diluted) with 0.5 mL either various concentrations of the sample, ascorbic acid or Trolox solution. Incubate the mixture in the dark for 6 min at RT and the photometrics were taken at 750 nm. The ABTS radical scavenging activity was calculated by Eq (10):
| (10) |
A0 denotes the absorbance value of ABTS without the PNs extracts. Ai represents the absorbance value of the PNs extracts to ABTS.
2.7. UHPLC-Q-Exactive Orbitrap MS/MS analysis of polyphenolic compounds
2.7.1. Chromatographic conditions
Chromatographic conditions Refer to V. González de Peredo et al. with appropriate modifications [30]. Column: ACQUITY UPLCBEH C18 column (50 mm × 2.1 mm, 1.7 μm); column temperature: 30 ℃; mobile phase: 0.1 % formic acid in water (A)-methanol (B); gradient elution: 0 ∼ 2 min, 5 % B; 2 ∼ 13 min, 5 % B-95 % B; 13 ∼ 14 min, 95 % B-100 % B; 14 ∼ 16 min, 100 % B; 16 ∼ 16.1 min, 100 % B-5 % B; 16.1 ∼ 19 min, 5 % B; flow rate was 0.3 mL/min; injection volume was 2 μL.
2.7.2. Mass spectrometry conditions
Mass spectrometry was performed using Q-Exactive Orbitrap MS/MS (Thermo Fisher Scientific, America). The ion source was heated electrospray (HESI, 350 ℃), and the samples were acquired in Full MS/dd-MS/MS scanning mode, with a primary scanning resolution of 70,000, a secondary scanning resolution of 17,500, and a scanning range of m/z 50–1500; the positive and negative ion detection modes were used, with a transfer capillary temperature of 320 ℃; the sheath gas was 35 psi, and the flow rate of auxiliary gas was 10 psi spray voltage of 3.0 kV.
2.8. Statistical analysis
The experiments were replicated three times, and the average value, accompanied by the standard deviation (SD), was documented for trial. ANOVA was utilized to analyze the data, and significance was determined at a P-value below 0.05. Design Expert 13 was used to perform the statistical analyses, while Compounds Discovery 3.1 was employed to interpret the mass spectral outcomes. The graphs were plotted using Origin 2021.
3. Results and discussion
3.1. Single-factor experiments for optimization of extraction conditions
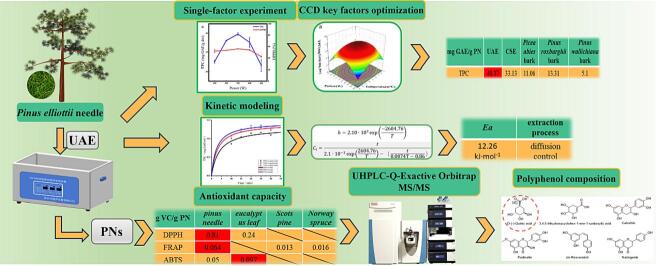
The study conducted single-factor experiments to determine the effects of various influencing factors on TPC and DPPH. Results can be seen in Fig. 1.
Fig. 1.
Single dependent variables of extraction conditions experimental design on tpc and dpph: power (a), temperature (b), ethanol concentration (c), solid–liquid ratio (d), and time (e).
3.1.1. Influence of sonication power on the extraction of polyphenolic compounds from PNs
As illustrated in Fig. 1a, as ultrasound power (250–350 W) increased, TPC and DPPH also increased gradually but tapered off when the power exceeded 350 W. This phenomenon was noted in the extraction of phenolic compounds from okra [31]. Nipornram et al. mentioned in their paper that low-power sonication treatment accelerates the delivery, spread, and dissolution of intracellular polyphenolic compounds, resulting in increased extraction rates [32]. Additionally, they indicated that higher power can cause the collapse of cavitation vesicles, generating higher pressure that strengthens the degradation of polyphenolic compounds.
3.1.2. Influence of sonication temperature on the extraction of polyphenolic compounds from PNs
The results depicted in Fig. 1b indicate that sonication temperature has a positive effect in the range of 25–45 °C and total phenolic content decreased sharply above 45 °C. Heating can enhance extraction rates by potentially disrupting the cell structure, resulting in fragmentation of both the cell wall and membrane. Meanwhile, as the temperature increases, the solvent's viscosity and surface tension decrease. And high temperatures impact the strength of cavitation, resulting in reduced extraction efficiency [33]. Furthermore, a study by Setyaningsih et al. showed that cinnamic acid, catechins, stilbenes, and flavonols were less stable in ultrasound-assisted extraction and started to degrade after 50 °C [34]. However, the extraction temperature did not significantly impact DPPH radical scavenging activity in this study. This may be because raising the extraction temperature beyond a certain threshold can cause the polyphenolic compounds extracted at lower temperatures to degrade, as well as decompose residual polyphenolic compounds that remain in the plant matrix [35].
3.1.3. Influence of ethanol concentration on the extraction of polyphenolic compounds from PNs
Ultrasound-assisted extraction in ethanol solution promotes efficient diffusion of polyphenolic compounds [36]. Therefore, the concentration of ethanol used greatly affects the extraction process. Fig. 1c shows that the TPC generally increases with the percentage of ethanol in water, up to a concentration range of 40–60 %. However, beyond 60 %, the TPC decreases sharply. The ethanol and water blend proved more effective than the single solvent system for extracting polyphenolic compounds. In the extraction of polyphenolic compounds from coconut skin, Yang et al. used ultrasound-assisted extraction and compared the effect of different solvents (50 % acetone, 50 % ethanol, acetone, ethanol, and water) on the extraction [37]. Luo et al. also reported that the extraction rate of polyphenolic compounds tended to increase and then decrease with increasing ethanol concentration, reaching a peak at 50 % ethanol concentration [38]. Whereas, DPPH did not change significantly at 40–70 % and decreased extremely rapidly after further increasing the ethanol concentration. The above phenomena indicate that the polyphenolic compounds extracted from different plants exhibit different polarities. Ibukunoluwa Fola et al. suggested that aqueous ethanol had higher extraction efficiency attributed to the following two reasons: the presence of ethanol creates a concentration gradient depending on the amount of ethanol, which promotes the solvent's diffusion into the solute and thus enhances mass transfer; water helps to swell the solute while ethanol disrupts the bonding between the solutes and plant matrix [39]. Therefore, 60 % ethanol was selected for extraction.
3.1.4. Influence of solid–liquid ratio on the extraction of polyphenolic compounds from PNs
It appears that an increase in the solid–liquid ratio(1:X) from 5 to 20 increased TPC. However, TPC decreased slightly with higher solid–liquid ratios. Increasing the solid–liquid ratio can enhance the yield of the active compound by increasing the concentration difference between the outside and inside of the cell, thus improving the mass transfer rate [40]. However, at higher solid–liquid ratios, this effect weakens because mass transfer depends mostly on the feedstock quantity [41]. Changes in DPPH could be attributed to variations in the polyphenolic compounds content, given that the trend of DDPH parallels that of TPC.
3.1.5. Influence of sonication time on the extraction of polyphenolic compounds from PNs
From Fig. 1e, it is observed that increasing the extraction time from 10 to 25 min leads to an increase in TPC, while a further increase in time to over 25 min causes a decrease in TPC. Sun et al. in Kudingcha extraction of polyphenols found a gradual increase in phenolic content from 20 to 60 min and after 60 min the phenolic content started to decrease [42]. However, the activity of DPPH did not change significantly with extraction time.
3.2. Results and analysis of RSM models for UAE
Since UAE results from the interaction of several factors, it is crucial to study the interaction and the strength of each factor's effects. To further optimize the extraction effect, power, temperature, time, and solid–liquid ratio were selected for RSM on TPC under a fixed ethanol concentration of 60 %.
3.2.1. Optimization of extraction parameters by RSM
The significance of the second-order multinomial equation fitted to the experimental data was estimated by Analysis of Variance (ANOVA). The F-test revealed a model F-value (57.24), with a P-value (P < 0.0001), indicating that the model was highly significant. Additionally, the coefficient of determination R2 (0.982) was nearly 1, indicating a strong correlation between the predicted and actual values.
The model's lack of fit test resulted in an F-value (0.243) and a p-value (0.973). P > 0.05, indicating the insignificance of the misfit term and affirming the accuracy of the mathematical model for calculating TPC. With an R2 (0.982), the model accuracy is considered good. Additionally, the corrected coefficient of determination () indicates a satisfactory correlation between the predicted and actual values values. The coefficient of variation (CV < 10 %), indicates high reproducibility and precision of the model.
3.2.2. Analysis of 3D response surface
According to the F-values, it's apparent that the extraction conditions affecting TPC are in the order of D (time) > C (solid–liquid ratio) > B (temperature) > A (power). This indicates that temperature time, and solid–liquid ratio are the primary factors influencing UAE extraction. According to Table 3, the significant factors in multinomial equation are the linear terms B, C, and D, as well as the quadratic term of AD.
Table 3.
ANOVA for fitted second-order multinomial models.
| Source | Sum of Squares | df | Mean Square | F-value | P-value | |
|---|---|---|---|---|---|---|
| Model | 249.96 | 14 | 17.85 | 57.24 | < 0.0001 | significant |
| A-Power | 1.38 | 1 | 1.38 | 4.43 | 0.0525 | |
| B-Temperature | 7.50 | 1 | 7.50 | 24.06 | 0.0002 | |
| C- solid–liquid ratio | 34.85 | 1 | 34.85 | 111.72 | < 0.0001 | |
| D-Time | 50.93 | 1 | 50.93 | 163.27 | < 0.0001 | |
| AB | 0.0020 | 1 | 0.0020 | 0.0065 | 0.9368 | |
| AC | 1.20 | 1 | 1.20 | 3.84 | 0.0688 | |
| AD | 3.13 | 1 | 3.13 | 10.04 | 0.0064 | |
| BC | 0.0240 | 1 | 0.0240 | 0.0770 | 0.7852 | |
| BD | 0.1849 | 1 | 0.1849 | 0.5928 | 0.4533 | |
| CD | 0.2304 | 1 | 0.2304 | 0.7387 | 0.4036 | |
| A2 | 102.99 | 1 | 102.99 | 330.17 | < 0.0001 | |
| B2 | 42.27 | 1 | 42.27 | 135.53 | < 0.0001 | |
| C2 | 6.52 | 1 | 6.52 | 20.92 | 0.0004 | |
| D2 | 43.99 | 1 | 43.99 | 141.04 | < 0.0001 | |
| Residual | 4.68 | 15 | 0.3119 | |||
| Lack of Fit | 1.53 | 10 | 0.1530 | 0.2430 | 0.9727 | not significant |
| Pure Error | 3.15 | 5 | 0.6297 | |||
| Cor Total | 254.64 | 29 | R2 | 0.9816 | ||
| Std. Dev. | 0.5585 | Adjusted R2 | 0.9645 | |||
| Mean | 35.83 | 29 | Predicted R2 | 0.9476 | ||
| C.V. % | 1.56 | Adeq Precision | 22.9406 |
The second-order multinomial equation expression was as follows:
| (11) |
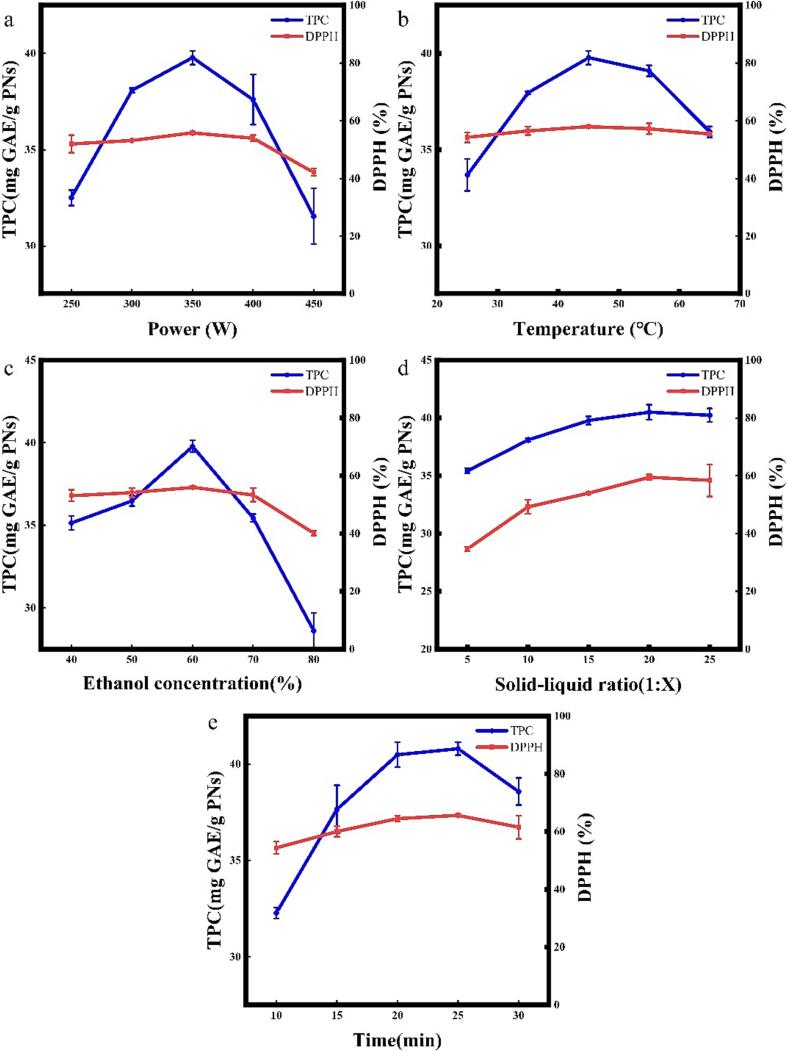
To determine the effect of independent variables on the TPC of PNs extracts, two variables were altered within the experimental range, and the third variable was kept consistent. This approach was followed to obtain a 3D plot of the interaction (Fig. 2). The obtained response surface plots tool shows significant changes, indicating that the factor changes have a significant effect on TPC. The contours of the response surface plots for power and time were significantly elliptical and the P-value (<0.05) indicates that the interaction between time and power has a significant impact on TPC extraction.
Fig. 2.
3D response surface and contour plots of TPC.(a)A × B; (b)A × C; (c)A × D; (d)B × C; (e)B × D; (f)C × D;
Different factors affected the TPC with varying intensity. At constant solid–liquid ratio and time (Fig. 2a), too low or too high power and temperature resulted in a decrease in TPC extraction. The value progressively increased as power and temperature rose, peaking at 350 W and 45 °C respectively. Then TPC started to decrease. This may be due to the fact that power and temperature extraction produce the same effect; increasing power and increasing temperature increases the rate of mass transfer, but high power and high temperature in turn lead to polyphenol decomposition. At constant temperature and time (Fig. 2b), TPC first increases and then decreases with increasing power. For the solid-to-liquid ratio, TPC always reaches a maximum value at 1:20 regardless of the power, and TPC remains essentially constant as the solid–liquid ratio increases. This could be because the concentration difference effect on mass transfer is no longer significant when the solid–liquid ratio reaches 1:20. Mass transfer at this point is determined by the raw material amount. In the case of constant power and time (Fig. 2d), the same phenomenon is observed.
When examining the power and temperature over time (Fig. 2c, e), it becomes clear that they have comparable response surface plots. This similarity suggests that the effects resulting from their interactions are alike at that particular time. The RSM plots (Fig. 2f) showing the variation of TPC concerning the solid–liquid ratio and time indicate a constant TPC value for solid–liquid ratios greater than 1:20 and that increasing the time at the same solid–liquid ratio results in an increase followed by a decrease in TPC.
The model expression of TPC was designed to obtain the optimal extraction conditions, power 350 W, 45 °C, solid–liquid ratio 1:20, 25 min, and the maximum TPC under the optimal extraction conditions was 40.37 0.14 mg GAE/g PNs. This method enhanced 21 % compared to the conventional extraction method (33.13 ± 0.39 mg GAE/g PNs). Chmelová et al. reported a TPC of 11.06 ± 0.00 mg GAE/g dry matter from Picea abies bark [43]. Maimoona et al. reported the number of polyphenols in Pinus roxburghii bark and Pinus wallichiana bark as 1331 ± 0.24 mg GAE/100 g dry matter and 510 ± 1.81 mg GAE/100 g dry matter [44]. Compared with the polyphenols extracted from Picea abies bark, Pinus roxburghii bark, and Pinus wallichiana bark, PNs possessed excellent phenolic content, suggesting the pine needle is a good source of natural polyphenol.
3.3. Kinetic model
The obtained data underwent processing and were plotted according to the second-order kinetic model. Mathematical regression analysis of the data at each temperature (Table 4) was utilized to calculate various kinetic parameters, such as the saturation extraction concentration, the extraction rate constant, and the initial extraction rate. The accuracy of the secondary kinetic model's fit for all experimental data was determined by its mathematical regression coefficient (R2 > 0.95). The correlation between the temperature and the kinetic parameters' fit is evident.
Table 4.
Parameters of the secondary kinetic model.
| Temperature (K) | A = 1/Cs | B = 1/h | Cs = 1/B (mg/g) | h = 1/A | k = h/(Cs)2 (g/(mg•min)) | R2 |
|---|---|---|---|---|---|---|
| 298 | 3.04 | 0.77 | 1.3 | 0.33 | 0.19 | 0.970 |
| 308 | 2.13 | 0.71 | 1.41 | 0.47 | 0.2 | 0.981 |
| 318 | 1.76 | 0.69 | 1.45 | 0.57 | 0.27 | 0.982 |
3.3.1. Kinetic analysis
The obtained data from UAE were analyzed and the second-order kinetic model was plotted. Mathematical regression analysis of the data at each temperature identified kinetic parameters including Cs, k, and h. The mathematical regression coefficients (R2 > 0.96) allowed the fit of the secondary kinetic model to be determined for all the experimental data. Temperature strongly influences the kinetic parameters.
Eq (4) implies that k and h are bounded solely by (Cs2). Thus, the relationship between 1/T and lnh was established for the extraction of total polyphenols using 60 % ethanol, as shown in Eq (12):
| (12) |
The concentration–time relationship can be determined by plotting saturation concentration (Cs) against time using the equation given in Eq (13):
| (13) |
The equations show nearly identical slopes, suggesting that the concentration at saturation in the solvent rises at the same rate as temperature. When extracting a 60 % v/v aqueous ethanol solution, applying h to Eq (5) results in the following relationship of Ct with temperature and time: Eq (14):
| (14) |
3.3.2. Activation energy coefficient
According to the linearized Arrhenius equation, when plotting lnk against 1/T, the rate constant rises as the temperature increases. With knowledge of k0 and k, Arrhenius' law can be used to determine the activation energy Ea. Eq. (15) gives the total polyphenol when the ethanol concentration is at 60 % v/v.
| (15) |
The positive activation energy value (12.26 kJ·mol−1) achieved for the uae process of pine needles polyphenols indicates that the process is a heat-absorbing reaction and that diffusion controls the extraction process. [45], [46]. The value obtained falls within the documented range of activation energies (Ea) for extracting bioactive compounds from biomass via solid–liquid extraction.
3.3.3. Validity of model development
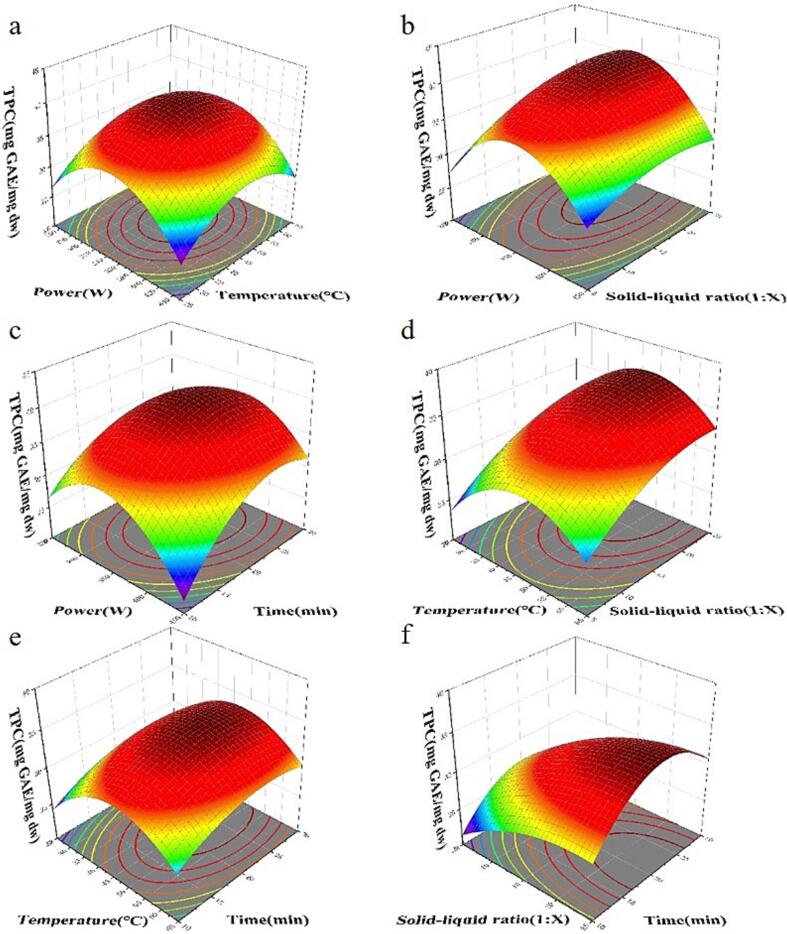
The total concentration of phenolic compounds was determined using Eq (14). Comparison was made between the experimental findings and those validated by a second-order model developed within a temperature range of 298–328 K (Fig. 3). The model accurately accounted for both experimental and calculated datasets, validating the relationship and its suitability for the intended purpose.
Fig. 3.
Comparison of calculated and experimental values of TPC concentrations in pine needles by UAE.
3.4. Antioxidant activity of pine needle extraction from Pinus elliottii needles
Because Pinus elliottii needles are rich in polyphenols, this means that as a food or nutrient, the determination of their antioxidant activity is crucial. A single assessment of antioxidant activity has limitations, so three assessment methods (DPPH, FRAP, ABTS) were utilized to assess the antioxidant activity of polyphenolic extracts from pine needles.
DPPH radical scavenging assays have been widely used to evaluate antioxidants due to their good stability and reproducibility. In the DPPH assay, polyphenol compounds can provide hydrogen that reduces the DPPH radical to DPPH-H, so they are used to evaluate antioxidant scavenging activity. As depicted in Fig. 1, the solid–liquid ratio significantly impacted the antioxidant activity of pine needles, while power, temperature, time, and ethanol concentration had a lesser effect. In terms of IC50 for DPPH, PNs was 41.05 ± 0.48 μg/mL, while VC was 33.41 ± 0.12 μg/mL and Trolox was 23.91 ± 0.13 μg/mL. Li et al. reported an IC50 of 0.139 mg/mL for the DPPH scavenging capacity of the eucalyptus leaf [47]. Compared with VC and the polyphenols extracted from the eucalyptus leaf, the antioxidant capacity of the polyphenols extracted from pines needles is just below VC and much higher than eucalyptus, suggesting the pines needles have great potential in DPPH scavenging capacity.
The FRAP assay measures the antioxidant capacity of PNs extracts through their ability to reduce Fe3+. The measured FRAPs for PNs’ extracts, VC, and Trolox were 1.09 ± 0.01 mM Fe2+/g PNs, 16.98 ± 0.37 mM Fe2+/g VC, and 7.77 ± 0.66 mM Fe2+/g Trolox, respectively. Agarwal et al. reported a FRAP of 13.71 ± 0.75 mg VC/g dw for Scots pine and 16.16 ± 2.40 mg VC/g dw for Norway spruce [48]. After calculation, the antioxidant capacity of PNs was 64.19 mg VC/g PNs, four times that of Scots pine and five times that of Norway spruce.
ABTS·radicals are positively charged and can obtain electrons from antioxidant molecules to form stable neutral molecules. The ABTS diammonium salt reacts with K2S2O8 resulting in the creation of stabilized ABTS·, with a maximum absorption at 750 nm, so the antioxidant activity can be assessed by measuring the scavenging ability of the ABTS·radicals. The IC50 value for PNs ' ABTS radical scavenging ability was 214.07 ± 0.26 ug/mL, Whereas the IC50 values of antioxidant activity of VC and Trolox were 10.38 ± 0.82 µg/mL, 22.1 ± 0.39 µg/mL, respectively. This may be related to the content of polyphenols in pine needles. Li et al. reported that the eucalyptus leaf has the ABTS radical scavenging capacity with IC50 of 0.111 mg/mL [47]. Compared to the eucalyptus leaf, the ABTS radical scavenging capacity of pine needles is only half that of eucalyptus leaves, because The free radical scavenging ability was related to polyphenols [49].
By evaluating the antioxidant capacity of PNs and comparing it with the results of VC and other studies, the study revealed that the polyphenolic extract of PNs exhibits potential as a natural antioxidant.
3.5. Qualitative analysis of polyphenolic extracts by UHPLC-Q-Exactive Orbitrap MS
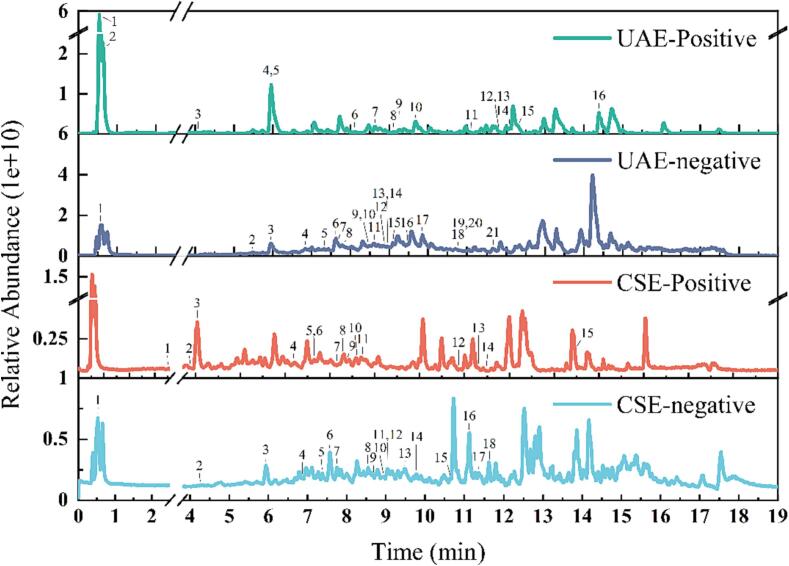
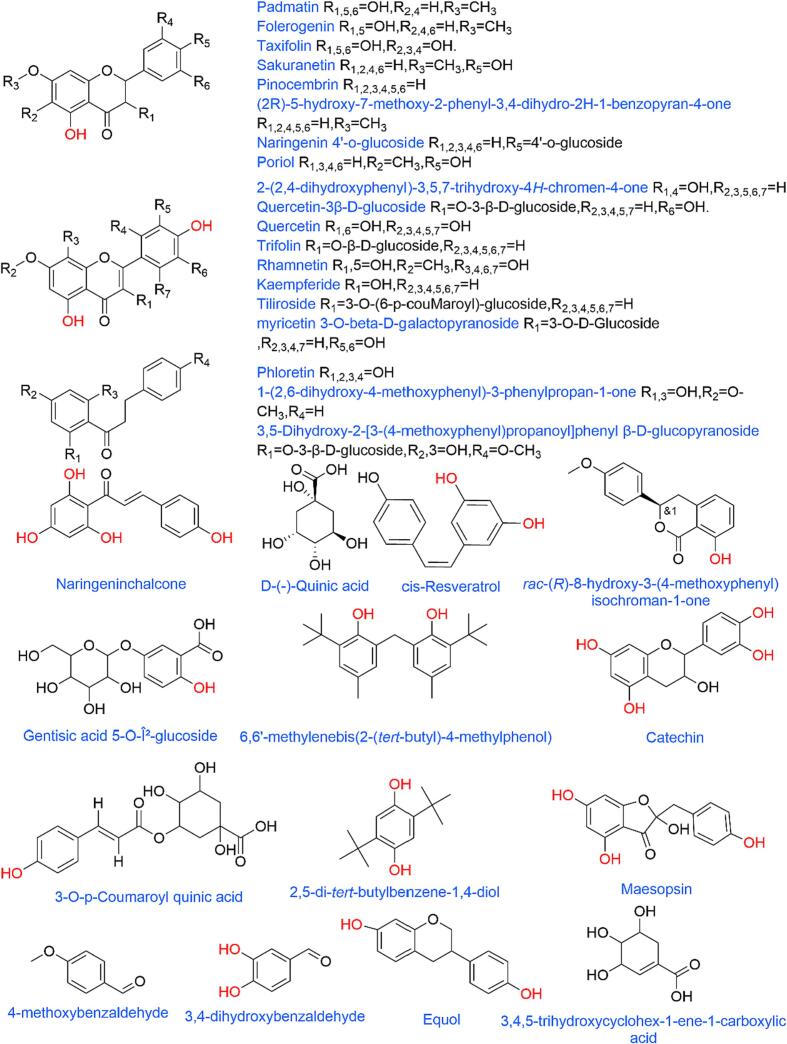
UHPLC-Q-Exactive Orbitrap MS/MS was utilized for characterizing the primary polyphenolic compounds in pine needles. Fig. 4 shows the UHPLC-Q-Exactive Orbitrap MS/MS analysis of the extracted compounds (CSE, UAE) using positive and negative ionic modes of the total ion current diagram (TIC) of negative and positive ionic modes. Compounds were identified by the resolution of Full MS/dd-MS/MS mass spectra and the determination of the exact mass number of molecular ions and fragments by UHPLC-Q-Exactive Orbitrap MS/MS, and the obtained data were analyzed by comparison with the McCloud database. 37 polyphenolic compounds form were identified in UAE extract (Table 5) and 32 polyphenolic compounds form were identified in CSE extract (Table S1). Furthermore, Fig. 5 illustrates the elaborate structures of the compounds.
Fig. 4.
Full ion chromatography analysis of PNs exacts (UAE and CSE) in negative and positive ionic mode.
Table 5.
Characterization of polyphenolic compounds (UAE) from pine needles by UHPLC-Q-Exactive Orbitrap MS/MS in positive and negative ionic mode.
| Compound no. | RT (min) | Formula | Adduct (m/z) | m/z | Title |
|---|---|---|---|---|---|
| Flvonoid and its derivtives | |||||
| 5− | 5.958 | C15H14O6 | [M−H]−1 (289.07187) | 290.0791 | Catechin |
| 9− | 9.249 | C21H20O11 | [M−H]−1 (447.09354) | 448.1008 | Trifolin |
| 8− | 8.609 | C21H22O10 | [M−H]−1 (433.11414) | 434.1214 | 7-Hydroxy-2-(4-hydroxyphenyl)-4-oxo-3,4-dihydro-2H-chromen-5-yl β-D-glucopyranoside |
| 11− | 11.127 | C16H14O5 | [M−H]−1 (285.0769) | 286.0842 | Poriol |
| 16− | 14.439 | C23H32O2 | [M−H]−1 (339.23254) | 340.2398 | 2,2′-Methylenebis(4-methyl-6-tert-butylphenol) |
| Compound no. | RT (min) | Formula | Adduct (m/z) | m/z | Title |
|---|---|---|---|---|---|
| 10− | 9.693 | C22H26O10 | [M−H]−1 (449.1457) | 450.1529 | 3,5-Dihydroxy-2-[3-(4-methoxyphenyl)propanoyl]phenyl β-D-glucopyranoside |
| 7− | 8.584 | C15H12O6 | [M−H]−1 (287.05618) | 288.0635 | 2,4,6-Trihydroxy-2-(4-hydroxybenzyl)-1-benzofuran-3(2H)-one |
| 13− | 11.863 | C16H16O4 | [M−H]−1 (241.08669) | 242.0941 | Equol |
| 4+ | 6.832 | C16 H14 O7 | [M+H]+1 (319.08162) | 318.0743 | Padmatin |
| 5+ | 7.342 | C16 H14 O6 | [M+H]+1 (303.08679) | 302.0795 | (2R,3R)-3,5-dihydroxy-2-(4-hydroxyphenyl)-7-methoxy-3,4-dihydro-2H-1-benzopyran-4-one |
| 7+ | 7.664 | C15 H12 O7 | [M+H]+1 (305.06589) | 304.0586 | Taxifolin |
| 9+ | 8.518 | C15H10 O7 | [M+H]+1 (303.05014) | 302.0428 | 2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one |
| 12+ | 8.922 | C15H10 O7 | [M+H]+1 (303.05014) | 302.0428 | Quercetin |
| 14+ | 8.991 | C21 H20 O11 | [M+H]+1 (449.10853) | 448.1012 | Trifolin |
| 15+ | 9.054 | C16 H12 O7 | [M+H]+1 (317.06592) | 316.0586 | Rhamnetin |
| 17+ | 9.99 | C30 H26 O13 | [M+H]+1 (595.1452) | 594.1379 | Tiliroside |
| 19+ | 10.972 | C16 H12 O6 | [M+H]+1 (301.0705) | 300.0632 | Kaempferide |
| Compound no. | RT (min) | Formula | Adduct (m/z) | m/z | Title |
|---|---|---|---|---|---|
| 8+ | 7.681 | C15 H12 O5 | [M+H]+1 (273.07605) | 272.0687 | Naringeninchalcone |
| 11+ | 8.663 | C15 H14 O5 | [M+H]+1 (275.09155) | 274.0842 | Phloretin |
| 18+ | 10.782 | C16 H14 O5 | [M+H]+1 (287.09164) | 286.0843 | Sakuranetin |
| 20+ | 10.988 | C15 H12 O4 | [M+H]+1 (257.08094) | 256.0736 | Pinocembrin |
| 21+ | 11.659 | C16 H14 O4 | [M+H]+1 (271.09676) | 270.0894 | (2R)-5-hydroxy-7-methoxy-2-phenyl-3,4-dihydro-2H-1-benzopyran-4-one |
| Other phenols | |||||
| 1− | 0.54 | C7H12O6 | [M−H]−1 (191.05515) | 192.0625 | D-(-)-Quinic acid |
| 3− | 4.062 | C13H16O9 | [M−H]−1 (315.07239) | 316.0797 | Gentisic acid 5-O-Î2-glucoside |
| 4− | 5.894 | C16H18O8 | [M−H]−1 (337.09296) | 338.1002 | 3-O-p-Coumaroyl quinic acid |
| 12− | 11.812 | C14H22O2 | [M−H]−1 (221.15417) | 222.1614 | 2,5-di-tert-Butylhydroquinone |
| 1+ | 0.545 | C7 H12 O6 | [M+H]+1 (193.07085) | 192.0635 | D-(-)-Quinic acid |
| 2+ | 5.475 | C8 H8 O2 | [M+H]+1 (137.05994) | 136.0526 | 4-Methoxybenzaldehyde |
| Compound no. | RT (min) | Formula | Adduct (m/z) | m/z | Title |
|---|---|---|---|---|---|
| 3+ | 5.877 | C7 H6 O3 | [M+H]+1 (139.03915) | 138.0318 | 3,4-Dihydroxybenzaldehyde |
| 6+ | 7.598 | C14 H12 O3 | [M+H]+1 (229.08612) | 228.0788 | cis-Resveratrol |
+indicates positive ion scan result, − indicates negative ion scanning results.
Fig. 5.
Structures of major polyphenolic compounds extracted from pine needles.
Only the identification process of UAE extracts is described here. Sixteen polyphenolic compounds were obtained and analyzed in the negative ion mode (Table 5). Compound 1 was found to have ionic fragments [M−H]−1 m/z 191.055, [M−H20] −1 m/z 157.01, [M−2H + K]−1 228.99, [2 M−H]−1 383.12, [2 M + FA-H]−1 429.13 at its RT 0.54 min. [M−H]−1 m/z 191.055 were analyzed by performing MS2 and comparing with the McCloud database, the characteristic fragments [M−COOH]−1 m/z 147.03, [M−2*H20]−1 m/z 157.01, [M−COOH−H2O]−1 m/z 129.02 were analyzed to coincide, the rest of the ionic fragments were judged to be D-(-)-Quinic acid by comparing with the spectra in the McCloud library. Compound 2 was found to be [M−H]−1 m/z 173.044, [M + Cl]−1 209.02, and [2 M−H]−1347.098 fragments by full MS at RT 0.64 min, and was analyzed by MS2 for [M−H]−1 m/z 173.044, and compared with the mzcloud database and analyzing the matched characteristic fragments m/z 155.03 ([M−H20]−1), 143.09, 137.02 ([M−2*H20]−1), 129.01 ([M−COOH]−1), 111.04 ([M−COOH−H2O]−1), 99.01, 83.04 and 73.02, therefore the compound was determined to be 3,4,5-trihydroxycyclohex−1-ene−1-carboxylic acid. Compound 3 was found to be [M−H]−1 m/z 315.07 by full MS at RT 4.062 min, and was analyzed and compared to MS2 by performing MS2 on the [M−H]−1 m/z 315.07 McCloud database and analyzed the matching characteristic fragments m/z 315.07, 153.01, 152.01, 109.02, 108.02, 71.01, so the compound was judged to be Gentisic acid 5-O-Î2-glucoside. Compound 4 was found to be a Gentisic acid 5-O-Î2-glucoside by full MS at RT for 5.894 min. [M−H]−1 m/z 337.093 was analyzed by MS2 of [M−H]−1 m/z 315.07 comparing with MzCloud database and analyzed the matching characteristic fragments m/z 191.05, 163.04, 119.04, 59.01 so the compound was judged as 3-O-p-Coumaroyl quinic acid.
Compound 5 was found to have ionic fragments [M−H]−1 m/z 289.07, [M−H−H2O]−1 271.06, [M + FA-H]−1 335.08, [M + Cl]−1 325.05, [M−H + TFA]−1403.06, [2 M−H]−1 579.15 by full MS at RT 5.958 min. [M−H]−1 m/z 315.07 was analyzed by MS2 comparing with MzCloud database, and analyzed the matching characteristic fragments m/z 289.07, 271.06, 245.08, 205.05, 203.07, 179.03, 151.03, 137.02, 125.02, 123.04, 109.02, so the compound 5 was judged to be Catechin. Compound 6 was found at RT 8.155 min by full MS and its [M−H]−1 m/z 479.08 was analyzed by MS2. Compared with the McCloud database, the analyzed matches were characterized by fragments m/z 317.03 ([Myricetin]), 316.02 ([Myricetin-H]−1), 178.99, 137.02, 50.01, 161,04 ([Galactose-H]−1), so the compound 6 was judged to be myricetin 3-O-beta-D-galactopyranoside. Similarly, compound 9 was judged to be Trifolin. Compound 8 was found to be [M−H]−1 m/z 433.11 by full MS at RT 8.609 min. Performing MS2 on [M−H]−1 m/z 433.11 to compare with the McCloud database, and analyzing the matched feature fragments m/z 313.07, 271.06 ([Naringenin-H]−1),151.00, etc., so the compound 8 was judged to be Naringenin 4′-o-glucoside.Similarly, other flavanones11,16 were identified as Poriol, 2,2′-Methylenebis(4-methyl-6-tert- butyl phenol).
Compound 10 was found to have ionic fragments [M−H]−1 m/z 449.14, [M + Cl]−1 m/z 485.12 by full MS at RT 9.693 min, and analyzed by MS2 for [M−H]−1 m/z 49.14 comparing to the McCloud database and analyzing the coincident characteristic fragments m/z 287.09 ([M- glucopyranoside-H]−1),193.04,181.05,93.03([Phenol-H]−1), so the compound 10 was judged to be 1-(2,6-Dihydroxy-4-methoxyphenyl)-3-phenylpropan−1-one. Synonymously compound 7 was judged to be Maesopsin.
The results in the positive ion mode were analyzed and a total of 21 polyphenolic compounds were obtained (Table 5). Compound 1 was D-(-)-Quinic acid and was identified similarly to the negative ion. Compound 2 was found to have ionic fragments [M + H]+1 m/z 137.05, [2 M + H]+1 m/z 273.11 by full MS at RT 5.474 min, and was analyzed by MS2 for [M + H]+1 m/z 137.05 comparing with the McCloud database, and analyzed for the coincident characteristic fragments m/z 122.03 ([M−CH2]+1), 109.06 ([M−CO]+1), so the compound 2 was determined to be 4-Methoxybenzaldehyde. Compound 3 was found to have ionic fragments [M + H]+1 m/z 139.03 by full MS at RT 5.877 min, and was analyzed by MS2. [M + H]+1 m/z 139.03 was analyzed to compare the McCloud database and found the matching characteristic fragments m/z 111.04 ([M−CO]+1), 93.03 ([M−CO−OH]+1), so the compound 3 was judged to be 3,4-Dihydroxybenzaldehyde.
Compound 6 was found to have ionic fragments [M + H]+1 m/z 229.08 by full MS at RT 7.598 min, and was analyzed by MS2. [M + H]+1 m/z 229.0 was analyzed comparing to the MzCloud database and analyzing the matching characteristic fragments m/z 211.07 ([M−OH]), 135.04 ([M + H-C6H5-OH]+1), 119.04 ([[M + H-C6H5-2*OH]+1]), 107.04 ([M- C6H3-2*OH-C]), 91.05 ([M−C6H3−2*OH-2*C]), so the compound 6 was judged to be cis-Resveratrol.
Compounds 4,5,7 were judged to be dihydroflavonols. The difference between compounds 4 and 7 is that the hydroxyl group of compound 4 has an H replaced by CH3. The MS2 mass spectra of compound 7 showed fragments of m/z 287.05 and 259.06, while the MS2 mass spectra of compound 4 showed fragments of m/z 301.07 and 273.01, and by comparing with the database, compound 4 was judged to be Padmatin, and compound 7 was judged to be Taxifolin, and the difference between compound 5 and compound 4 was that compound 5 had one less benzene ring, which matches with the MS2 fragments m/z 285.07 and 257.08, so compound 5 was judged to be Folerogenin.
Compounds 9,10,12,14,15,17,19 were judged to be flavonols because the fragments of m/z 303.04 were found in MS2, and they were judged to have the same body structure. Among them, compounds 10 and 12 both had similar fragments, while compound 10 had a fragment of glucoside residue with m/z 145.09, and finally compared with the McCloud database, compound 9 was judged to be 2-(2,4-dihydroxy phenyl)-3,5,7-trihydroxy-4H-chromen-4-one, compound 10 was Quercetin-3β-D-glucoside, compound 12 was Quercetin, compound 14 was Trifolin, compound 15 was Rhamnetin, compound 17 was Tiliroside, and compound 19 was Kaempferide. Compounds 8,11 were judged to be chalcogenides. Compounds 8,11 were judged to be chalcones, and similar fragments m/z 273.07 (chalcone body fragments) and m/z 107.04 (methylphenol fragments) were found in the MS2 plot, and it was finally judged that Compound 8 was Naringeninchalcone and Compound 11 was Phloretin.
Compounds 13,16 were judged to be flavonoids, where Compound 16 was the product of the substitution of Compound 13, and Compound 19 was Kaempferide. Compounds 8,11 were judged to be Chalcones, with Compound 16 being the product of the substitution of Compound 13. 13 was substituted, and after comparison with the McCloud database, compound 13 was judged to be Luteolin, and compound 16 was 2-Hydroxy-3-(5-hydroxy-7,8-dimethoxy-4-oxo-4H-chromen-2-yl)phenyl β-D- glucopyranoside. Compounds 18,20, and 21 were judged to be flavanones because of similar fragments in the MS2 spectra then compared with the McCloud database, compound 18 was judged to be Sakuranetin, compound 20 was Pinocembrin, and compound 21 was (2R)-5-hydroxy -7-methoxy-2-phenyl-3,4-dihydro-2H-1-benzopyran-4-one.
All of the above results, verified by Compounds Discovery 3.1 (trial version), match the McCloud database by more than 85 %.
Upon comparison of the polyphenol compounds in the CSE and UAE extracts, the CSE extracts had fewer species and lower content than the UAE extracts. A majority of the polyphenol compounds between CSE and UAE were similar, with only a few differences. This result could be attributed to ultrasound-assisted extraction, as it was found to be more effective in releasing polyphenol compounds from the cells. Also in the case of ultrasound-assisted, some polyphenol compounds degraded. This is the explanation for the variance in polyphenol compounds present in the extracts. The advantage of UAE over CSE is that a better extraction in terms of content and species can be achieved at the same time.
4. Conclusion
This study systematically examined the best optimal to extract polyphenols from pine needles, the identification of 37 polyphenolic components, and the antioxidant activity, and established an extraction kinetic model. Compared with CSE, ultrasound-assisted extraction effectively enhanced the extraction efficiency and extraction effect. The antioxidant activity of the extracts was determined to show that pine needles have pine needle polyphenol extracts with potential as natural antioxidants. Second-order kinetic modelling of temperature and time on phenolic content suggests that the UAE process is diffusion-controlled. With these results, we can have a comprehensive understanding of the whole process of polyphenolic extraction from pine needles and the related properties of the extracts, which provides a new way for the utilization of pine needles as forestry residues, and also provides a theoretical foundation for further exploration into the nutrition and consumption of pine needles.
CRediT authorship contribution statement
Siheng Zhang: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. Hongzhao Xie: Software. Jie Huang: Conceptualization. Qiumei Chen: Conceptualization. Xin Li: Conceptualization. Xiaopeng Chen: Conceptualization. Jiezhen Liang: Resources. Linlin Wang: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32160349,22368006), Guangxi Key Laboratory of Petrochemical Resource Processing and Process. Intensification Technology (Grant No. 2022Z002).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ultsonch.2023.106742.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Rana A.K., Guleria S., Gupta V.K., Thakur V.K. Cellulosic pine needles-based biorefinery for a circular bioeconomy. Bioresour. Technol. 2023;367 doi: 10.1016/j.biortech.2022.128255. [DOI] [PubMed] [Google Scholar]
- 2.Dias M.C., Ferreira de Oliveira J.M.P., Marum L., Pereira V., Almeida T., Nunes S., Araújo M., Moutinho-Pereira J., Correia C.M., Santos C. Pinus elliottii and P. elliottii x P. caribaea hybrid differently cope with combined drought and heat episodes. Ind. Crops Prod. 2022;176 doi: 10.1016/j.indcrop.2021.114428. [DOI] [Google Scholar]
- 3.Lai M., Zhang L., Lei L., Liu S., Jia T., Yi M. Inheritance of resin yield and main resin components in Pinus elliottii Engelm. at three locations in southern China. Ind. Crops Prod. 2020;144 doi: 10.1016/j.indcrop.2019.112065. [DOI] [Google Scholar]
- 4.Sharma V., Sharma R.K. Pine Needle Energy Potential in Conifer Forest of Western Himalayan, Environ. Nat. Resour. J. 2020;18:55–65. doi: 10.32526/ennrj.18.1.2020.06. [DOI] [Google Scholar]
- 5.Koutsaviti A., Toutoungy S., Saliba R., Loupassaki S., Tzakou O., Roussis V., Ioannou E. Antioxidant Potential of Pine Needles: A Systematic Study on the Essential Oils and Extracts of 46 Species of the Genus Pinus. Foods. 2021;10:142. doi: 10.3390/foods10010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng W.-C., Zhang Z., Gao H., Jia L.-R., He Q. Chemical Composition, Antioxidant, and Antimicrobial Activities of Essential Oil from Pine Needle (Cedrus deodara) J. Food Sci. 2012;77:C824–C829. doi: 10.1111/j.1750-3841.2012.02767.x. [DOI] [PubMed] [Google Scholar]
- 7.Juranović Cindrić I., Zeiner M., Starčević A., Stingeder G. Metals in pine needles: characterisation of bio-indicators depending on species. Int. J. Environ. Sci. Technol. 2019;16:4339–4346. doi: 10.1007/s13762-018-2096-x. [DOI] [Google Scholar]
- 8.Lučinskaitė I., Laužikė K., Žiauka J., Baliuckas V., Čėsna V., Sirgedaitė-Šėžienė V. Assessment of biologically active compounds, organic acids and antioxidant activity in needle extracts of different Norway spruce (Picea abies (L.) H. Karst) half-sib families. Wood Sci. Technol. 2021;55:1221–1235. doi: 10.1007/s00226-021-01322-5. [DOI] [Google Scholar]
- 9.Jyske T., Järvenpää E., Kunnas S., Sarjala T., Raitanen J.-E., Mäki M., Pastell H., Korpinen R., Kaseva J., Tupasela T. Sprouts and Needles of Norway Spruce (Picea abies (L.) Karst.) as Nordic Specialty—Consumer Acceptance, Stability of Nutrients, and Bioactivities during Storage. Molecules. 2020;25:4187. doi: 10.3390/molecules25184187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metsämuuronen S., Sirén H. Bioactive phenolic compounds, metabolism and properties: a review on valuable chemical compounds in Scots pine and Norway spruce. Phytochem. Rev. 2019;18:623–664. doi: 10.1007/s11101-019-09630-2. [DOI] [Google Scholar]
- 11.Esmeeta A., Adhikary S., Dharshnaa V., Swarnamughi P., Ummul Maqsummiya Z., Banerjee A., Pathak S., Duttaroy A.K. Plant-derived bioactive compounds in colon cancer treatment: An updated review. Biomed. Pharmacother. 2022;153 doi: 10.1016/j.biopha.2022.113384. [DOI] [PubMed] [Google Scholar]
- 12.Chamorro F., Carpena M., Fraga-Corral M., Echave J., Riaz Rajoka M.S., Barba F.J., Cao H., Xiao J., Prieto M.A., Simal-Gandara J. Valorization of kiwi agricultural waste and industry by-products by recovering bioactive compounds and applications as food additives: A circular economy model. Food Chem. 2022;370 doi: 10.1016/j.foodchem.2021.131315. [DOI] [PubMed] [Google Scholar]
- 13.Tang Z., Huang G., Huang H. Ultrasonic/cellulase-assisted extraction of polysaccharide from Garcinia mangostana rinds and its carboxymethylated derivative. Ultrason. Sonochem. 2023;99 doi: 10.1016/j.ultsonch.2023.106571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ojeda G.A., Sgroppo S.C., Sánchez-Moreno C., de Ancos B. Mango ‘criollo’ by-products as a source of polyphenols with antioxidant capacity. Ultrasound Assisted Extraction Evaluated by Response Surface Methodology and HPLC-ESI-QTOF-MS/MS Characterization, Food Chem. 2022;396 doi: 10.1016/j.foodchem.2022.133738. [DOI] [PubMed] [Google Scholar]
- 15.Chemat F., Rombaut N., Sicaire A.-G., Meullemiestre A., Fabiano-Tixier A.-S., Abert-Vian M. Ultrasound assisted extraction of food and natural products. Mechanisms, techniques, combinations, protocols and applications. A review. Ultrason. Sonochem. 2017;34:540–560. doi: 10.1016/j.ultsonch.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 16.Gómez-Mejía E., Rosales-Conrado N., León-González M.E., Madrid Y. Citrus peels waste as a source of value-added compounds: Extraction and quantification of bioactive polyphenols. Food Chem. 2019;295:289–299. doi: 10.1016/j.foodchem.2019.05.136. [DOI] [PubMed] [Google Scholar]
- 17.Grosso C., Ferreres F., Gil-Izquierdo A., Valentão P., Sampaio M., Lima J., Andrade P.B. Box-Behnken factorial design to obtain a phenolic-rich extract from the aerial parts of Chelidonium majus L. Talanta. 2014;130:128–136. doi: 10.1016/j.talanta.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 18.Heleno S.A., Diz P., Prieto M.A., Barros L., Rodrigues A., Barreiro M.F., Ferreira I.C.F.R. Optimization of ultrasound-assisted extraction to obtain mycosterols from Agaricus bisporus L. by response surface methodology and comparison with conventional Soxhlet extraction. Food Chem. 2016;197:1054–1063. doi: 10.1016/j.foodchem.2015.11.108. [DOI] [PubMed] [Google Scholar]
- 19.Liao J., Guo Z., Yu G. Process intensification and kinetic studies of ultrasound-assisted extraction of flavonoids from peanut shells. Ultrason. Sonochem. 2021;76 doi: 10.1016/j.ultsonch.2021.105661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzano de Luna M., Vetrone G., Viggiano S., Panzella L., Marotta A., Filippone G., Ambrogi V. Pine Needles as a Biomass Resource for Phenolic Compounds: Trade-Off between Efficiency and Sustainability of the Extraction Methods by Life Cycle Assessment. ACS Sustain. Chem. Eng. 2023;11:4670–4677. doi: 10.1021/acssuschemeng.2c06698. [DOI] [Google Scholar]
- 21.Fatima P., Nadeem M., Hussain A., Kausar T., Rehman A., Siddique T., Kabir K., Noreen S., Nisar R., Fatima H., Korma S.A., Simal-Gandara J. Synergistic effect of microwave heating and thermosonication on the physicochemical and nutritional quality of muskmelon and sugarcane juice blend. Food Chem. 2023;425 doi: 10.1016/j.foodchem.2023.136489. [DOI] [PubMed] [Google Scholar]
- 22.Hou J., Liang L., Su M., Yang T., Mao X., Wang Y. Variations in phenolic acids and antioxidant activity of navel orange at different growth stages. Food Chem. 2021;360 doi: 10.1016/j.foodchem.2021.129980. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y.-C., Chuang Y.-C., Hsu H.-W. The flavonoid, carotenoid and pectin content in peels of citrus cultivated in Taiwan. Food Chem. 2008;106:277–284. doi: 10.1016/j.foodchem.2007.05.086. [DOI] [Google Scholar]
- 24.Lazar L., Talmaciu A.I., Volf I., Popa V.I. Kinetic modeling of the ultrasound-assisted extraction of polyphenols from Picea abies bark. Ultrason. Sonochem. 2016;32:191–197. doi: 10.1016/j.ultsonch.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Cheung Y.-C., Siu K.-C., Wu J.-Y. Kinetic Models for Ultrasound-Assisted Extraction of Water-Soluble Components and Polysaccharides from Medicinal Fungi. Food Bioprocess Technol. 2013;6:2659–2665. doi: 10.1007/s11947-012-0929-z. [DOI] [Google Scholar]
- 26.Ho Y.-S., Harouna-Oumarou H.A., Fauduet H., Porte C. Kinetics and model building of leaching of water-soluble compounds of Tilia sapwood. Sep. Purif. Technol. 2005;45:169–173. doi: 10.1016/j.seppur.2005.03.007. [DOI] [Google Scholar]
- 27.Qiu L., Zhang M., Mujumdar A.S., Chang L. Convenient use of near-infrared spectroscopy to indirectly predict the antioxidant activitiy of edible rose (Rose chinensis Jacq “Crimsin Glory” H.T.) petals during infrared drying. Food Chem. 2022;369 doi: 10.1016/j.foodchem.2021.130951. [DOI] [PubMed] [Google Scholar]
- 28.Martínez-Patiño J.C., Gullón B., Romero I., Ruiz E., Brnčić M., Žlabur J.Š., Castro E. Optimization of ultrasound-assisted extraction of biomass from olive trees using response surface methodology. Ultrason. Sonochem. 2019;51:487–495. doi: 10.1016/j.ultsonch.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 29.Sridhar K., Charles A.L. In vitro antioxidant activity of Kyoho grape extracts in DPPH and ABTS assays: Estimation methods for EC50 using advanced statistical programs. Food Chem. 2019;275:41–49. doi: 10.1016/j.foodchem.2018.09.040. [DOI] [PubMed] [Google Scholar]
- 30.González de Peredo A.V., Vázquez-Espinosa M., Piñeiro Z., Espada-Bellido E., Ferreiro-González M., Barbero G.F., Palma M. Development of a rapid and accurate UHPLC-PDA-FL method for the quantification of phenolic compounds in grapes. Food Chem. 2021;334 doi: 10.1016/j.foodchem.2020.127569. [DOI] [PubMed] [Google Scholar]
- 31.Wang X., Liu X., Shi N., Zhang Z., Chen Y., Yan M., Li Y. Response surface methodology optimization and HPLC-ESI-QTOF-MS/MS analysis on ultrasonic-assisted extraction of phenolic compounds from okra (Abelmoschus esculentus) and their antioxidant activity. Food Chem. 2023;405 doi: 10.1016/j.foodchem.2022.134966. [DOI] [PubMed] [Google Scholar]
- 32.Nipornram S., Tochampa W., Rattanatraiwong P., Singanusong R. Optimization of low power ultrasound-assisted extraction of phenolic compounds from mandarin (Citrus reticulata Blanco cv. Sainampueng) peel. Food Chem. 2018;241:338–345. doi: 10.1016/j.foodchem.2017.08.114. [DOI] [PubMed] [Google Scholar]
- 33.Almusallam I.A., Mohamed Ahmed I.A., Babiker E.E., Al Juhaimi F.Y., Fadimu G.J., Osman M.A., Al Maiman S.A., Ghafoor K., Alqah H.A.S. Optimization of ultrasound-assisted extraction of bioactive properties from date palm (Phoenix dactylifera L.) spikelets using response surface methodology. LWT. 2021;140 doi: 10.1016/j.lwt.2020.110816. [DOI] [Google Scholar]
- 34.W. Setyaningsih, I.E. Saputro, M. Palma, C.G. Barroso, Stability of 40 phenolic compounds during ultrasound-assisted extractions (UAE), in: New York, NY USA, 2016: p. 080009. https://doi.org/10.1063/1.4958517.
- 35.Mokrani A., Madani K. Effect of solvent, time and temperature on the extraction of phenolic compounds and antioxidant capacity of peach (Prunus persica L.) fruit. Sep. Purif. Technol. 2016;162:68–76. doi: 10.1016/j.seppur.2016.01.043. [DOI] [Google Scholar]
- 36.Gil-Martín E., Forbes-Hernández T., Romero A., Cianciosi D., Giampieri F., Battino M. Influence of the extraction method on the recovery of bioactive phenolic compounds from food industry by-products. Food Chem. 2022;378 doi: 10.1016/j.foodchem.2021.131918. [DOI] [PubMed] [Google Scholar]
- 37.Yang J., Li N., Wang C., Chang T., Jiang H. Ultrasound-homogenization-assisted extraction of polyphenols from coconut mesocarp: Optimization study. Ultrason. Sonochem. 2021;78 doi: 10.1016/j.ultsonch.2021.105739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo X., Cui J., Zhang H., Duan Y., Zhang D., Cai M., Chen G. Ultrasound assisted extraction of polyphenolic compounds from red sorghum (Sorghum bicolor L.) bran and their biological activities and polyphenolic compositions. Ind. Crops Prod. 2018;112:296–304. doi: 10.1016/j.indcrop.2017.12.019. [DOI] [Google Scholar]
- 39.Olawuyi I.F., Park J.J., Lee W.Y. Effect of extraction conditions on ultrasonic-assisted extraction of polyphenolic compounds from okra (Abelmoschus esculentus L.) leaves. Korean J. Food Preserv. 2020;27:476–486. doi: 10.11002/kjfp.2020.27.4.476. [DOI] [Google Scholar]
- 40.Prakash Maran J., Manikandan S., Vigna Nivetha C., Dinesh R. Ultrasound assisted extraction of bioactive compounds from Nephelium lappaceum L. fruit peel using central composite face centered response surface design. Arab. J. Chem. 2017;10:S1145–S1157. doi: 10.1016/j.arabjc.2013.02.007. [DOI] [Google Scholar]
- 41.Yan Z., Zhang H., Dzah C.S., Zhang J., Diao C., Ma H., Duan Y. Subcritical water extraction, identification, antioxidant and antiproliferative activity of polyphenols from lotus seedpod. Sep. Purif. Technol. 2020;236 doi: 10.1016/j.seppur.2019.116217. [DOI] [Google Scholar]
- 42.Sun Y., Xu W., Zhang W., Hu Q., Zeng X. Optimizing the extraction of phenolic antioxidants from kudingcha made frrom Ilex kudingcha C.J. Tseng by using response surface methodology. Sep. Purif. Technol. 2011;78:311–320. doi: 10.1016/j.seppur.2011.01.038. [DOI] [Google Scholar]
- 43.Chmelová D., Škulcová D., Legerská B., Horník M., Ondrejovič M. Ultrasonic-assisted extraction of polyphenols and antioxidants from Picea abies bark. J. Biotechnol. 2020;314–315:25–33. doi: 10.1016/j.jbiotec.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Maimoona A., Naeem I., Saddiqe Z., Ali N., Ahmed G., Shah I. Analysis of total flavonoids and phenolics in different fractions of bark and needle extracts of Pinus roxburghii and Pinus wallichiana. J. Med. Plants Res. 2011;5:2724–2728. doi: 10.5897/JMPR.9000083. [DOI] [Google Scholar]
- 45.Wang Y.G., Yue S.T., Li D.Q., Jin M.J., Li C.Z. Kinetics and Mechanism of Y(iii) Extraction with Ca-100 Using a Constant Interfacial Cell with Laminar Flow. Solvent Extr. Ion Exch. 2002;20:345–358. doi: 10.1081/SEI-120004809. [DOI] [Google Scholar]
- 46.González-Centeno M.R., Comas-Serra F., Femenia A., Rosselló C., Simal S. Effect of power ultrasound application on aqueous extraction of phenolic compounds and antioxidant capacity from grape pomace (Vitis vinifera L.): experimental kinetics and modeling. Ultrason. Sonochem. 2015;22:506–514. doi: 10.1016/j.ultsonch.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 47.Li W., Zhang X., He Z., Chen Y., Li Z., Meng T., Li Y., Cao Y. In vitro and in vivo antioxidant activity of eucalyptus leaf polyphenols extract and its effect on chicken meat quality and cecum microbiota. Food Res. Int. 2020;136 doi: 10.1016/j.foodres.2020.109302. [DOI] [PubMed] [Google Scholar]
- 48.Agarwal C., Hofmann T., Visi-Rajczi E., Pásztory Z. Low-frequency, green sonoextraction of antioxidants from tree barks of Hungarian woodlands for potential food applications. Chem. Eng. Process. - Process Intensif. 2021;159 doi: 10.1016/j.cep.2020.108221. [DOI] [Google Scholar]
- 49.Wang M., Ding Y., Wang Q., Wang P., Han Y., Gu Z., Yang R. NaCl treatment on physio-biochemical metabolism and phenolics accumulation in barley seedlings. Food Chem. 2020;331 doi: 10.1016/j.foodchem.2020.127282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.