Abstract
The relationship between cancer stem cells (CSCs) in oral squamous cell carcinoma (OSCC) and programmed cell death ligand 1 (PD-L1)/programmed cell death 1 (PD-1) remains unclear. Therefore, the present study aimed to clarify the association between the CD44v3high/CD24low immunophenotype of CSCs in OSCC and PD-L1/PD-1 co-expression, and to assess the prognostic effect of CSCs in terms of immune checkpoint molecules. Formalin-fixed, paraffin-embedded tissue samples and clinicopathological data from 168 patients with OSCC were retrospectively retrieved. Immunohistochemical staining and reverse transcription quantitative polymerase chain reaction were applied to a tissue microarray of the invasive front of each case. Semi-automated cell counting was used to assess CD44v3, CD24, PD-L1 and PD-1 expression by immunohistochemistry (IHC) using a digital image analysis program. Associations between immunological markers and clinicopathological variables were estimated. Patients with the CSC immunophenotype CD44v3high/CD24low, and patients with a high PD-L1/PD-1-positive cell density in the tumor parenchyma and stroma had significantly lower survival rates. Furthermore, patients with the CSC immunophenotype (CD44v3high/CD24low) and high PD-L1/PD-1 co-expression had even lower survival rates (P<0.01, log-rank test). Notably, there was a positive correlation between CD44v3 and PD-L1 expression (τ=0.1096, P=0.0366, Kendall rank correlation coefficient) and a negative correlation between CD24 and PD-1 expression (τ=−0.1387, P=0.0089, Kendall rank correlation coefficient). Additionally, the high CD44v3 expression group, as determined by IHC, exhibited significantly decreased expression of U2 small nuclear RNA auxiliary factor 1 (U2AF1) at the mRNA level compared with that in the low CD44v3 expression group (P<0.001, Mann-Whitney U test), and U2AF1 and PD-L1 mRNA expression exhibited a significant negative correlation (τ=−0.3948, P<0.001, Kendall rank correlation coefficient). In conclusion, CSCs in OSCC may evade host immune mechanisms and maintain CSC stemness via PD-L1/PD-1 co-expression, resulting in unfavorable clinical outcomes.
Keywords: CSC, OSCC, CD44v3, CD24, programmed cell death ligand 1, programmed cell death 1, immune checkpoint inhibitors
Introduction
Oral cancer is the eighth most common cancer worldwide with high morbidity and low survival rates, the majority of which is oral squamous cell carcinoma (OSCC) (1). Despite recent advances in diagnostic accuracy, treatment modalities, and combinations of various chemotherapeutic agents that have improved quality of life, mortality from this disease is high because of local recurrence and distant metastasis. Such treatment failures are due to a small population of cancer stem cells (CSCs) responsible for tumorigenesis and resistance to conventional therapies such as pharmacotherapy and radiotherapy (2,3).
Therefore, attempts have been made to identify CSCs using various cell surface markers, such as CD44, its variant form CD44v3, and CD24, in various solid tumor types including oral and breast cancers (4–9). However, most stem cell markers currently used to identify CSCs do not have sufficient specificity, and a single marker is not sufficient as direct therapeutic targets (8,10,11). These markers are routinely used in combination for the isolation of pure CSCs. In OSCC, CD44 or CD44v3 combined with CD24 is used for CSC isolation. CD44v3 is an alternatively spliced variant of CD44, a multifunctional transmembrane glycoprotein expressed in many cancer types (12,13). CD44v3 is regulated by U2 small nuclear RNA auxiliary factor 1 (U2AF1), an essential spliceosome component, which enhances cancer cell proliferation and promotes stemness of CSCs (14). On the other hand, CD24 is a glycosylphosphatidylinositol anchor (15) that binds to the extracellular matrix and cell membrane (16). CD24 expression in cancer is reduced by Twist, one of the well-known Epithelial-mesenchymal transition (EMT) factors (17), and it is speculated that CD24 depletion is a prerequisite for EMT induction (8). We previously reported that CD44v3high/CD24low cells in OSCC have CSC characteristics, namely a self-renewal capacity and drug resistance, and that patients with more CSCs have a less favorable prognosis (7). On the other hand, dysfunction of the immune system, which is crucial for carcinogenesis and evolution of various solid tumor types (18–21), may also be involved in treatment failure of OSCC. In particular, immune checkpoint molecules, such as programmed cell death ligand 1 (PD-L1) and programmed cell death 1 (PD-1), play an essential role in immune evasion of tumors (22). They are also associated with the degree of tumor malignancy (19–21). When tumor cells overexpress PD-L1, this molecule binds to the PD-1 receptor of T cells around the tumor, activating the PD-L1/PD-1 checkpoint pathway to attenuate immune responses (23). PD-L1/PD-1 co-expression is an independent poor prognostic for OSCC (24,25). Recent studies have shown that PD-L1/PD-1 checkpoint inhibition is effective for treating head and neck squamous cell carcinomas (HNSCCs), including OSCC, and is beginning to be applied clinically (26,27). The only clinical immune checkpoint inhibitor used for OSCC targets the PD-L1/PD-1 axis, the key molecule in OSCC.
Recently, in HNSCC, there have been a few reports of a potential association between PD-L1 and CSCs. CD44-positive CSCs increase PD-L1 expression and promote T cell-mediated immunosuppression in HNSCC (28), and tumors expressing PD-L1 mediate immunosuppression, EMT, and CSC phenotype (29,30). In OSCC, PD-L1 knockdown has also been shown to suppress the induction of Akt phosphorylation and Stat3 phosphorylation (31), suggesting that PD-L1 may contribute to the maintenance of CSC stemness through Akt phosphorylation. However, the relationship between CSCs and the PD-L1/PD-1 axis in OSCC is still unclear, with only a few reports discussing the association. Therefore, we performed a clinicopathological study to clarify the relationship between cell surface markers CD44v3 and CD24 and immune checkpoint molecules PD-L1 and PD-1.
Materials and methods
Study design and patient cohort
This study was a retrospective analysis of 168 untreated OSCC patients (mean age: 69.307 years; age range: 28–95 years) who visited the Kurume University Hospital Dental and Oral Medical Center from January 2013 to December 2015. Eligible patients had biopsy- or resection-proven OSCC, no prior treatment for oral cancer, and a sufficient sample volume to prepare a tissue microarray (TMA) for immunohistochemical analysis. The design and methods of this study complied with the ethical guidelines of the Declaration of Helsinki and the guidelines for research involving human subjects of the Ethical Review Committee of the Clinical Research Center of Kurume University Hospital. Institutional review and approval were obtained prior to commencing the study (approval number: 22217). Written informed consent was obtained before patient participation in the study, and clinical specimens were obtained from each patient in accordance with the approved guidelines.
Postoperative recurrence was defined as lymph node (LN) metastasis and/or local recurrence. In this study, 41 of 168 patients had postoperative recurrence. Of these patients, 16 were LN metastases, 9 were local recurrences, and 16 were LN metastases and local recurrences. Sixteen of 168 patients died during the observation period, and were insufficient to assess patient prognosis by overall survival. As the primary treatment, surgery was performed in 158 cases, of which 58 underwent cervical neck dissection. Postoperative therapy, including radiotherapy and/or chemotherapy, was conducted for 40 of the 168 patients. Thirty-eight of the 41 patients with postoperative recurrence underwent cervical neck dissection and/or reoperation. The other three patients received systemic chemotherapy because of distant metastases. The cases and clinicopathological features were shown in Table I.
Table I.
Associations of the CSC immunophenotype with various clinicopathological characteristics and PD-L1/PD-1 expression.
| CSC immunophenotype | ||||
|---|---|---|---|---|
|
|
||||
| Characteristic | Cases (%) | CSC | non-CSC | P-value |
| Age, years | NS | |||
| >60 | 40 (23.8) | 14 | 26 | |
| ≤60 | 128 (76.2) | 32 | 96 | |
| Sex | NS | |||
| Male | 87 (51.8) | 25 | 62 | |
| Female | 81 (48.2) | 21 | 60 | |
| Location | NS | |||
| Tongue | 76 (45.2) | 23 | 53 | |
| Gingiva | 63 (37.5) | 12 | 51 | |
| Floor of mouth | 11 (6.5) | 4 | 7 | |
| Buccal mucosa | 13 (7.7) | 6 | 7 | |
| Palate | 3 (1.8) | 1 | 2 | |
| Lip | 2 (1.2) | 0 | 2 | |
| Histological gradea | NS | |||
| Grade 1 | 141 (83.9) | 42 | 99 | |
| Grade 2/3 | 27 (16.1) | 4 | 23 | |
| Lymphatic vessel invasion | 0.063 | |||
| Yes | 48 (28.6) | 18 | 30 | |
| No | 120 (71.4) | 28 | 92 | |
| pT classification | NS | |||
| T1/T2 | 122 (72.6) | 38 | 84 | |
| T3/T4 | 46 (27.4) | 8 | 38 | |
| Nodal statusb | 0.02 | |||
| N(+) | 64 (38.1) | 24 | 40 | |
| N(−) | 104 (61.9) | 22 | 82 | |
| Stage | NS | |||
| I/II | 101 (60.1) | 30 | 71 | |
| III/IV | 67 (39.9) | 16 | 51 | |
| Mode of invasion (YK classification)c | NS | |||
| Grade 1/2 | 66 (39.3) | 16 | 50 | |
| Grade 3 | 58 (34.5) | 15 | 43 | |
| Grade 4C/4D | 44 (26.2) | 15 | 29 | |
| Post-operative local recurrence | 0.043 | |||
| Yes | 25 (14.9) | 11 | 14 | |
| No | 143 (85.1) | 35 | 108 | |
| Post-operative lymph node metastasis | 0.006 | |||
| Yes | 32 (19.0) | 15 | 17 | |
| No | 136 (81.0) | 31 | 105 | |
| Post-operative recurrenced | 0.002 | |||
| Yes | 41 (24.4) | 19 | 22 | |
| No | 127 (75.6) | 27 | 100 | |
| PD-L1/PD-1 | 0.002 | |||
| Group Ae | 45 (26.8) | 8 | 37 | |
| Group Be | 76 (45.2) | 16 | 60 | |
| Group Ce | 47 (28.0) | 22 | 25 | |
The histological grade was determined by the World Health Organization classification (2017). G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated.
Nodal status indicates the presence of lymph node metastasis at the time of the first visit to our department.
Mode of tumor invasion was determined by Yamamoto-Kohama criteria (20).
Postoperative recurrence was defined as lymph node metastasis and/or local recurrence.
Group A, PD-L1low/PD-1low; group B, PD-L1high/PD-1low or PD-L1low/PD-1high; group C, PD-L1high/PD-1high. P<0.05 was considered significant. CSC, cancer stem cell; PD-1, programmed cell death 1; PD-L1, PD ligand 1; NS, not significant; YK, Yamamoto-Kohama.
TMA preparation
A TMA was used for multiple histological analyses of tissue specimens. TMA blocks were prepared using an arraying device (Azumaya, Tokyo, Japan). Depending on the amount of tissue collected, two or three cores of 5 mm in diameter were punched from the invasive front of the tumor portion of the Formalin-fixed, paraffin-embedded (FFPE) tissue block of each patient and placed onto the recipient TMA block. Each TMA block contained 11 cores.
Histopathological analysis
Biopsy or resected specimens were used in study. Specimens were prepared as 4 µm-thick sections from TMA FFPE blocks and stained with hematoxylin-eosin to confirm the diagnosis and histological grade by experienced oral pathologists (KT and YA.). The histopathological tumor-node-metastasis (TNM) stage was defined by the Union for International Cancer Control TNM Classification of Malignant Tumors, 8th edition. The mode of tumor invasion was defined by Yamamoto-Kohama (YK) criteria as follows: grade 1, well-defined borderline; grade 2, cords and a less marked borderline; grade 3, a group of cells and no distinct borderline; grade 4C, diffuse invasion of the cord-like type; grade 4D, diffuse invasion of the widespread type (32). Patient and tumor characteristics were shown in Table I.
Immunohistochemistry (IHC)
FFPE TMA block samples were cut at 4 µm thicknesses, examined on a coated glass slide, and labeled with antibodies using the Bond-III autostainer (Leica Microsystems, Newcastle, UK). Primary antibodies were against CD44v3 (mouse anti-human monoclonal antibody, cat. no. BMS144, 1:200 dilution, clone VFF-327 variant 3, Bender MedSystems, Vienna, Austria), CD24 (mouse anti-human monoclonal antibody, cat. No. NB100-64861, 1:100 dilution, clone SN3, Novus Biologics, Briarwood, USA), PD-L1 (rabbit anti-human monoclonal antibody, 1:50 dilution, clone 28–8, cat. no. ab205921, Abcam, Cambridge, MA, USA), and PD-1 (mouse anti-human monoclonal antibody, 1:100 dilution, clone NAT105, cat. no. ab52587, Abcam, Cambridge, MA, USA). Briefly, antigen retrieval was performed by heat treatment in epitope retrieval solution 1 (pH 6) for 30 min, followed by attenuating non-specific protein binding by incubation for 30 min with 10% goat serum. Anti-CD44v3 and CD24 antibodies were incubated for 30 min. Anti-PD-L1 and -PD-1 antibodies were treated by heat in epitope retrieval solution 2 (pH 9) for 30 min and then incubated for 30 min. The automated system used a Refine polymer detection system (Leica Microsystems, Newcastle, UK) with horseradish peroxidase polymer as the secondary antibody and 3,3′ diaminobenzidine as the chromogen. Negative controls included PBS instead of primary antibody. Expression analysis was performed to measure the positive expression area in all cases using Win ROOF software (version 7.4.5, Mitani Corporation, Osaka, Japan). Images of positive cells were selected for clarity in five high-power fields of the invasive front from each IHC specimen using a CCD digital camera (DXM1200, Nikon, Tokyo, Japan) with light microscope (BX43, Olympus, Tokyo, Japan). The digitized data of the positive expression area (µm2) were measured and averaged. The labeling index for each case was calculated as follows: for CD44v3, CD24, and PD-L1, the denominator was all the area of all cancer cells in the region of interest (the invasive front in the TMA core), and the numerator was all the area of cancer cells stained by DAB for each antibody in the same region of interest. For PD-1, the denominator is all the area of inflammatory cells in the region of interest around the tumor (the invasive front in the TMA core). The numerator is all the area of inflammatory cells stained by DAB (CD24-positive cells) in the same region of interest. The percentage of positive cells, or labeling index, for each antibody, was calculated as a percentage by dividing the numerator by the denominator mentioned above. Finally, the median value was calculated by univariate analysis (n=168) for the labeling index for each antibody. The calculation of the labeling index was based on the previous report (24).
mRNA extraction from TMA sample
For the extraction of RNA, each TMA block from 168 OSCC cases was used. The TMA blocks were sectioned to a thickness of 10 µm using a Leica RM2245 microtome (Leica Microsystems K.K., Tokyo, Japan) with an RNase-free water-treated blade. For the preparation of RNA from TMA blocks, the RNeasy FFPE kit (QIAGEN GmbH, Hilden, Germany) was used according to manufacturer's recommendations. Depending on each TMA core, 1 to 4 sections of 10 µm thickness were used per preparation of RNA. For RNA preparation from RNAlater stabilized samples, RNeasy plus kits were used. RNA isolation was carried out in an RNase-free environment. RNA yields were determined by Nanodrop ND 1000 (Thermo Fisher Scientific, Waltham, MA, USA).
Complementary DNA (cDNA) preparation followed by reverse transcription quantitative polymerase chain reaction (RT-qPCR) for gene expression assay
cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI, USA) according to the manufacturer's instructions. RT-qPCR was performed to examine the expression of U2AF1, CD24, PD-L1, and PD-1 with ABI PRISM 7500 (Applied Biosystems, Foster City, CA, USA). Gene expression assays and primer and probe mixes were used for U2AF1, CD24, PD-L1, and PD-1, and β-actin [assay IDs (Hs00739599_m1, Hs04405694_m1, Hs00204257_m1, Hs01550088_m1, and Hs99999903_m1, respectively; Thermo Fisher Scientific, Waltham, MA, USA)], and thermal cycler conditions were as follows: initial incubation at 95°C for 10 min, then 40 cycles alternating in turn with 95°C for 10 sec, 60°C for 20 sec, and 72°C for 15 sec, and then maintained at 72°C for 10 min. Comparative gene expression analysis was performed using the 2(−ΔΔCq) method (33) with normalization to the level of internal control gene, β-actin.
Statistical analysis
Statistical analyses were performed using JMP software version 16 (SAS Institute Inc., Cary, NC, USA). The Pearson χ2 test and Fisher's exact test were used to assess the significance of between-group differences in clinicopathological characteristics. Kendall rank correlation coefficient was used to analyze the correlation between CD44v3 and PD-L1 expression in IHC, PD-1 and CD24 expression in IHC, and PD-L1 and U2AF1 expression at mRNA level. Mann-Whitney U test was used to analyze the correlation between the immunophenotype of CD44v3 and U2AF1 at mRNA level, immunophenotype and genotype of CD24, immunophenotype and genotype of PD-L1, and immunophenotype and genotype of PD-1. Univariate and multivariate analyses using a Cox proportional hazards regression model were applied to examine the effect of clinicopathological characteristics, CSC markers (CD44v3 and CD24), and PD-L1/PD-1 co-expression on postoperative recurrence. Survival curves were generated by the Kaplan-Meier method, and the log-rank test was used to calculate P-values. P<0.05 was considered statistically significant.
Results
Immunohistochemical evaluation of CSC markers and PD-L1/PD-1
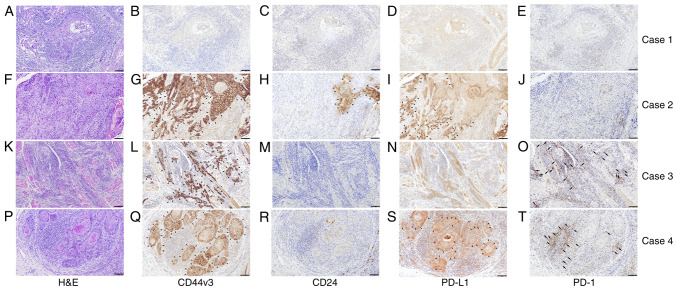
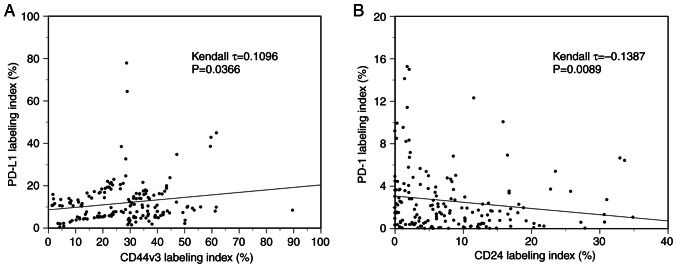
Immunohistochemical analysis was performed to examine the expression of CD44v3, CD24, PD-L1, and PD-1 in 168 OSCC patients. Various expression levels of CD44v3 (Fig. 1B, G, L and Q), CD24 (Fig. 1C, H, M and R), and PD-L1 (Fig. 1D, I, N and S) were observed in tumor cell membranes at the invasive front among the cases. Various levels of PD-1 expression were also analyzed in inflammatory cells around the invasive front of tumors (Fig. 1E, J, O and T). The median labeling indices of CD44v3, CD24, PD-1, and PD-L1 were 29.1, 6.1, 1.6, and 6.1%, respectively. Next, all OSCC patients were classified into high and low expression groups for each molecule using the median value as the cutoff. All OSCC patients were then classified into two subgroups by the CSC immunophenotype. Specifically, CD44v3highCD24low was designated as the CSC group, and CD44v3low/CD24low and CD44v3high/CD24high as the non-CSC group. Additionally, all OSCC patients were classified into three subgroups by the mode of PD-L1/PD-1 co-expression as follows: group A, PD-L1low/PD-1low; group B, PD-Lhigh1/PD-1low or PD-L1low/PD-1high; group C, PD-L1high/PD-1high. Based on these classifications, four different cases with different expressions of each cell surface marker were shown in Fig. 1, and the clinical characteristics of each were as follows: Fig. 1A-E (Case 1, 61 age, male, tongue), Fig. 1F-J (Case 2, 91 age, female, lower gingiva), Fig. 1K-O (Case 3, 39 age, female, tongue), and Fig. 1P-T (Case 4, 54 age, male, tongue), respectively. Then, the immunophenotypes of each case are as follows: Case 1; CD44v3low/CD24low (non-CSC pattern) and PD-L1low/PD-1low (group A), Case 2; CD44v3high/CD24high (non-CSC pattern) and PD-L1high/PD-1low (group B), Case 3; CD44v3high/CD24low (CSC pattern) and PD-L1low/PD-1high (group B). Case 4; CD44v3high/CD24low (CSC pattern) and PD-L1high/PD-1high (group C). The CD44v3-positive rate was significantly positively correlated to the PD-L1 labeling index (τ=0.1096, P=0.0366, Kendall rank correlation coefficient) (Fig. 2A). On the other hand, the CD24-positive rate was significantly negatively correlated to the PD-1 labeling index (τ=−0.1387, P=0.0089, Kendall rank correlation coefficient) (Fig. 2B). However, as the τ-values were <0.3/-0.3, these correlations are considered to be very weak.
Figure 1.
Representative micrographs of H&E staining and immunohistochemical classification in accordance with CD44v3, CD24, PD-L1, and PD-1 expression patterns. Four different cases with different expressions of each cell surface marker are shown. (A-E) Photomicrographs of the same case (Case 1, 61 age, male, tongue). (A) H&E staining. (B) CD44v3 and (C) CD24 immunoreactivity. (D) PD-L1 and (E) PD-1 immunoreactivity. This case is classified as CD44v3low/CD24low (non-CSC pattern) and PD-L1low/PD-1low (group A). (F-J) Photomicrographs of the same case (Case 2, 91 age, female, lower gingiva). (F) H&E staining. (G) CD44v3 and (H) CD24 immunoreactivity. Both CD44v3 and CD24 are expressed on the cancer cell membrane. (I) PD-L1 and (J) PD-1 immunoreactivity. PD-L1 is expressed on the cancer cell membrane. This case is classified as CD44v3high/CD24high (non-CSC pattern) and PD-L1high/PD-1low (group B). (K-O) Photomicrographs of the same case (Case 3, 39 age, female, tongue). (K) H&E staining. (L) CD44v3 and (M) CD24 immunoreactivity. CD44v3 is expressed on the cancer cell membrane. (N) PD-L1 and (O) PD-1 immunoreactivity. PD-1 expression is observed in lymphocytes around cancer nests. This case is classified as CD44v3high/CD24low (CSC pattern) and PD-L1low/PD-1high (group B). (P-T) Photomicrographs of the same case (Case 4, 54 age, male, tongue). (P) H&E staining. (Q) CD44v3 and (R) CD24 immunoreactivity. CD44v3 is expressed on the cancer cell membrane. (S) PD-L1 and (T) PD-1 immunoreactivity, respectively. PD-L1 is expressed on the cancer cell membrane. PD-1 expression is observed in lymphocytes around cancer nests. This case is classified as CD44v3high/CD24low (CSC pattern) and PD-L1high/PD-1high (group C). Arrowheads indicate regions where CD44v3, CD24, or PD-L1 expression is highlighted. Arrows indicate regions where PD-1 expression is highlighted. Scale bars: 100 µm. H&E, hematoxylin and eosin; PD-1, programmed cell death 1; PD-L1, programmed cell death ligand 1.
Figure 2.
Correlations between CD44v3 positivity with PD-L1 expression patterns and CD24 positivity with PD-1 expression patterns. (A) A significant positive correlation is observed between the CD44v3-positive rate and PD-L1 labeling index (τ=0.1096, P=0.0366). (B) A significant negative correlation is observed between the CD24-positive rate and PD-1 labeling index (τ=−0.1387, P=0.0089). The Kendall rank correlation coefficient was used for statistical analysis. PD-1, programmed cell death 1; PD-L1, programmed cell death ligand 1.
Analysis of CSC markers-related gene and PD-L1/PD-1 gene expression by RT-qPCR
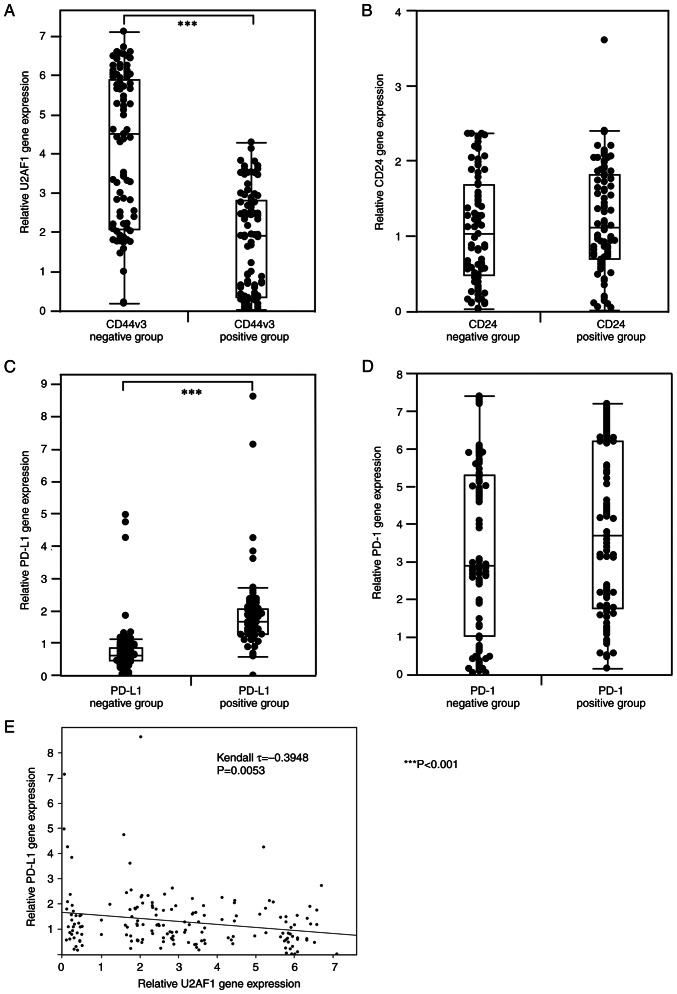
RT-qPCR was performed to examine the expression of CSC-related molecules (U2AF1 and CD24) and PD-L1/PD-1 at mRNA level in 168 OSCC patients. For CD24 and PD-1, no significant correlation was found between the immunophenotype and the genotype. On the other hand, a significant negative correlation was found between the expression of CD44v3 in IHC and U2AF1 at mRNA level (P<0.001, Mann-Whitney U test) (Fig. 3A). Moreover, a significant positive correlation was found between the immunophenotype and genotype of PD-L1 (P<0.001, Mann-Whitney U test) (Fig. 3B). In addition, U2AF1 expression was significantly negatively correlated to the PD-L1 expression at mRNA level (τ=−0.3948, P=0.0053, Kendall rank correlation coefficient) (Fig. 3E). On the other hand, for PD-1 and CD24, no significant correlation was observed between the immunophenotype in IHC and the genotype at the mRNA level, respectively (Fig. 3C and D).
Figure 3.
The expression of cancer stem cell-related molecules (U2AF1 and CD24) and PD-L1/PD-1 at mRNA level and its correlation with protein expression in IHC of each molecule (CD44v3, CD24, PD-L1, and PD-1). (A) CD44v3-negative group by IHC expresses higher mRNA levels of U2AF1 than the CD44v3-positive group (P<0.001). (B) The PD-L1-positive group in IHC expresses higher mRNA levels of PD-L1 than the PD-L1 negative group in IHC. (P<0.001). (C) No significant correlation is found in the mRNA expression level of CD24 between the CD24-positive and negative groups in IHC. (D) No significant correlation is found in the mRNA expression level of PD-1 between the PD-1-positive and negative groups in IHC. (E) U2AF1 expression was significantly negatively correlated to the PD-L1 expression at mRNA level (τ=−0.3948, P0.0053). The Mann-Whitney U test is used for statistical analysis for (A-D). For (E), the Kendall rank correlation coefficient is used for statistical analysis. ***P<0.001. PD-1, programmed cell death 1; PD-L1, programmed cell death ligand 1; U2AF1, U2 small nuclear RNA auxiliary factor 1.
Associations of the CSC immunophenotype with various clinicopathological factors and PD-L1/PD-1 expression
As shown in Table I, the associations of CSC surface markers with clinicopathological factors were investigated in all 168 OSCC patients. The nodal status (prevalence rates of cervical LN metastasis), postoperative local recurrence, and postoperative LN metastasis were significantly higher in the CSC group than in the non-CSC group (Table I). Moreover, PD-L1/PD-1 co-expression was significantly associated with the CSC immunophenotyped (Table I). Conversely, other clinical factors, including age, gender, location, histological grade, lymphatic vessel invasion, pT classification, stage, and mode of tumor invasion, did not differ significantly between CSC and non-CSC groups.
As shown in Table II, the associations between postoperative recurrence-free survival (RFS) and individual clinicopathological factors, including CSC markers and PD-L1/PD-1 co-expression, were assessed by univariate and multivariate analyses. In univariate analysis of postoperative RFS, nodal status [hazard ratio (HR) 2.12, 95% confidence interval (CI) 1.14–3.92], mode of invasion (HR 3.01, 95% CI 1.38–6.55), CSC immunophenotype (HR 3.90, 95% CI 2.05–7.43), PD-L1/PD-1 expression (HR 4.43, 95% CI 1.78–11.06), and CSC immunophenotype combined with PD-L1/PD-1 co-expression (HR 9.61, 95% CI 4.58–20.16) were significant predictors of postoperative recurrence. Further multivariate analysis showed that the CSC immunophenotype (HR 2.89, 95% CI 1.41–5.95) and PD-L1/PD-1 co-expression (HR 3.01, 95% CI 1.38–6.55) were independent poor prognostic factors for postoperative recurrence. No significant association was found regarding age, gender, location, histological grade, or pT classification in univariate and multivariate analyses.
Table II.
Univariate and multivariate analyses of postoperative recurrence-free survival of individual characteristics.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
|
|
|||
| Characteristic | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years | ||||
| >60 | Reference | Reference | ||
| ≤60 | 2.52 (0.99–6.43) | 0.053 | 2.16 (0.19–4.23) | 0.364 |
| Sex | ||||
| Male | Reference | Reference | ||
| Female | 1.01 (0.54–1.85) | 0.980 | 1.45 (0.72–2.91) | 0.301 |
| Location | ||||
| Tongue | Reference | Reference | ||
| Gingiva | 1.45 (0.72–2.91) | 0.300 | 1.05 (0.42–2.64) | 0.913 |
| Floor of mouth | 1.53 (0.44–5.34) | 0.500 | 1.09 (0.28–4.16) | 0.904 |
| Buccal mucosa | 1.18 (0.34–4.12) | 0.790 | 0.49 (0.13–1.91) | 0.308 |
| Palate | 3.29 (0.75–14.55) | 0.200 | 3.87 (0.82–18.24) | 0.095 |
| Lip | 3.74 (0.48–28.79) | 0.120 | 4.82 (1.24–32.79) | 0.180 |
| Histological gradea | ||||
| Grade 1 | Reference | Reference | ||
| Grade 2/3 | 1.55 (1.75–3.16) | 0.230 | 3.23 (1.44–7.26) | 0.410 |
| pT classification | ||||
| T1/T2 | Reference | Reference | ||
| T3/T4 | 1.58 (0.84–2.98) | 0.160 | 1.98 (0.77–5.09) | 0.156 |
| Nodal statusb | ||||
| N(−) | Reference | Reference | ||
| N(+) | 2.12 (1.14–3.92) | 0.016 | 1.49 (0.73–3.02) | 0.273 |
| Mode of invasion (YK classification)c | ||||
| Grade 1/2 | Reference | Reference | ||
| Grade 3 | 1.22 (0.57–2.60) | 0.610 | 1.13 (0.53–2.42) | 0.753 |
| Grade 4C/4D | 3.01 (1.38–6.55) | 0.006 | 1.39 (1.02–5.61) | 0.342 |
| Immunophenotype of CSC | ||||
| Non-CSC | Reference | Reference | ||
| CSC | 3.90 (2.05–7.43) | <0.0001 | 2.89 (1.41–5.95) | 0.039 |
| PD-L1/PD-1 | ||||
| Group Ad | Reference | Reference | ||
| Group Bd | 1.80 (0.69–4.62) | 0.220 | 1.59 (0.61–4.17) | 0.434 |
| Group Cd | 4.43 (1.78–11.06) | 0.001 | 3.54 (1.37–9.12) | 0.028 |
| Immunophenotype of CSC combined with PD-L1/PD-1 | ||||
| CSC and Group Ad, or non-CSC | Reference | |||
| CSC and Group Bd | 2.62 (0.98–6.97) | 0.054 | ||
| CSC and Group Cd | 9.61 (4.58–20.16) | <0.0001 | ||
The histological grade was determined by the World Health Organization classification (2017). G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated.
Nodal status indicates the presence of lymph node metastasis at the time of the first visit to our department.
Mode of tumor invasion was determined by Yamamoto-Kohama (YK) criteria (20).
Group A, PD-L1low/PD-1low; group B, PD-L1high/PD-1low or PD-L1low/PD-1high; group C, PD-L1high/PD-1high. P<0.05 was considered significant. CSC, cancer stem cell; PD-1, programmed cell death 1; PD-L1, PD ligand 1; NS, not significant; YK, Yamamoto-Kohama.
Prognostic effect of CSC markers and PD-L1/PD-1 co-expression in OSCC patients
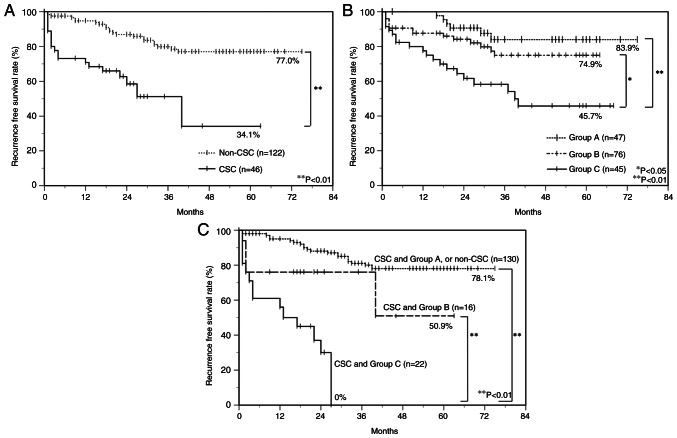
To estimate the relationship between immunoreactivity for CSC markers and PD-L1/PD-1 and the prognosis of OSCC patients, postoperative RFS rates were calculated using the Kaplan-Meier method. There was a significant difference in the postoperative RFS rate between the CSC group (34.1%) and the non-CSC group (77.0%) (log-rank test, P<0.01) (Fig. 4A). Furthermore, all OSCC patients were classified into three subgroups by the mode of PD-L1/PD-1 expression as follows: group A, PD-L1low/PD-1low; group B, PD-L1high/PD-1low or PD-L1low/PD-1high; group C, PD-L1high/PD-1high, and there was a significant difference in the postoperative RFS rate between group C (45.7%) and group B (74.9%) (log-rank test, P<0.05), and between group C and group A (83.9%) (log-rank test, P<0.01) (Fig. 4B). Moreover, group C with a combined CSC immunophenotype had a significantly poorer prognosis than group B with the combined CSC immunophenotype and group A with the combined CSC immunophenotype or non-CSC group. RFS rates in group C with the combined CSC immunophenotype, group B with the combined CSC immunophenotype, and group A with the combined CSC or non-CSC immunophenotype were 0, 50.6, and 78.1%, respectively (Fig. 4C).
Figure 4.
Postoperative RFS curves of OSCC patients. Cumulative survival curves of OSCC patients classified by the expression patterns of (A) CSC markers, (B) PD-L1/PD-1, and (C) combined CSC markers and PD-L1/PD-1. (A) CSC cases have poor prognoses, and there is a significant difference between CSC and non-CSC cases. (B) The time to postoperative RFS is significantly shorter in group C. (C) The time to postoperative RFS is significantly shorter in combined CSC and group C. Based on the PD-L1/PD-1 immunophenotypes, group A, B, and C are characterized as follows: Group A, PD-L1low/PD-1low; group B, PD-L1high/PD-1low or PD-L1low/PD-1high; group C, PD-L1high/PD-1high. The Kaplan-Meier method was used to construct survival curves, and the log-rank test was used to calculate P-value. *P<0.05 and **P<0.01. CSC, cancer stem cell; OSCC, oral squamous cell carcinoma; PD-1, programmed cell death 1; PD-L1, programmed cell death ligand 1; RFS, recurrence-free survival.
Discussion
There have been only a few reports on the relationship between immune checkpoint molecules PD-L1/PD-1 and CSCs in HNSCCs including OSCC (28–30). To our knowledge, this is the first study to address the prognostic effect of CD44v3 and CD24, cell surface markers of CSCs, in terms of PD-L1 and PD-1 co-expression in OSCC. In this study, 168 OSCC specimens were analyzed and grouped according to the expression results of the CSC markers, i.e., CD44v3 and CD24, and the immune checkpoint molecules, i.e., PD-1 and its ligand PD-L1 in IHC.
We believe that CSCs in OSCC have a critical unfavorable prognostic effect in terms of PD-L1/PD-1 co-expression for several reasons. First, we identified the CSC immunophenotype CD44v3high/CD24low in OSCC as a critical unfavorable prognostic factor. Although our results may revealed that the phenotype of CSCs was not correlate with histological morphology, such as histological differentiation or infiltration pattern (YK classification), it is even more critical to identify CSCs using cell surface markers such as CD44v3 and CD24. We found that the patient population with the CSC immunophenotype was significantly correlated to unfavorable clinical outcomes (postoperative LN metastasis and/or local recurrence) compared with the patient population with the non-CSC immunophenotype (CD44v3high/CD24high and CD44v3low/CD24high or low), which was similar to our previous study (7).
The second reason for the CSC prognostic effect in terms of PD-L1/PD-1 co-expression was that univariate analysis revealed that the PD-L1/PD-1 co-expression group was significantly associated with unfavorable clinical outcomes compared with groups expressing either PD-L1, PD-1, or neither. Furthermore, multivariate analysis revealed that PD-L1/PD-1 co-expression was an independent poor prognostic factor. These findings were consistent with previous studies (24,25). Cancer cells stimulate PD-1 receptors on T cell membranes via PD-L1, and these signals act as so-called T cell brakes and inhibit cancer cell attack by T cells (19–21,23). These findings suggest that PD-L1 expression in the tumor parenchyma and PD-1 expression in inflammatory cells in the stroma play an essential role in immune evasion of OSCC.
The third reason, which was most important for the critical unfavorable prognostic effect of CSCs in terms of PD-L1/PD-1 co-expression, was that the CSC immunophenotype and PD-L1/PD-1 co-expression were independent unfavorable prognostic factors for OSCC, they influenced each other, and CSC marker and PD-L1/PD-1 co-expression had a more substantial prognostic effect. Furthermore, our results revealed that PD-L1/PD-1 co-expression was significantly associated with the CSC immunophenotype, suggesting that PD-L1 and PD-1 play an essential role in maintaining CSC stemness in OSCC. Indeed, previous reports have shown that PD-L1 plays a crucial role in CSC expansion in addition to its role as an immune checkpoint (34,35). Moreover, in breast cancer, where CD44high/CD24low has been reported to be a CSC immunophenotype, the PD-L1/PD-1 axis activates Phosphoinositide 3-kinase/Akt and Extracellular signal-regulated kinase 1/2 signaling pathways to promote cancer stemness and drug resistance (36–38). To investigate the relationship between the Akt pathway and PD-L1/PD-1 axis in CSCs in OSCC, we focused on U2FA1, which has been reported to be a splicing factor of CD44v3 and a down regulator of the Akt pathway upstream of this pathway (14). Furthermore, U2AF1 has been reported to suppress PD-L1 expression (39). In the present study, the CD44v3-positive group showed significantly lower expression of U2AF1 at mRNA level than that of the CD44v3-negative group. Furthermore, a negatively significant correlation was observed between U2AF1 and PD-L1 expression at mRNA level. These results suggest that the downregulation of U2AF1, which acts in a suppressive manner to activate the Akt pathway, contributes to the promotion of CD44v3 splicing, the maintenance of CSC stemness, and the increase in PD-L1 expression. However, the role of U2AF1 in CSCs of OSCC remains unclear, with only one report indicating that U2AF1 is involved in carcinogenesis from oral precancerous lesions (40). Also, another report on the relationship between U2AF1 and PD-L1 found no correlation between their expression (41). On the other hand, Geum et al (31) support our study. They reported that PD-L1 knockdown suppressed the induction of Akt phosphorylation. In other words, PD-L1 activates Akt and may be involved in maintaining OSCC stemness. However, the involvement of U2AF1 in PD-L1 to Akt is still unknown with only a few reports. Thus, further validation is needed to elucidate the role of U2AF1 in CSCs in OSCC. To confirm the further relationship between CSCs and PD-L1/PD-1, we examined the expression correlation between CD44v3 and PD-L1, and CD24 and PD-1. Regarding the relationship between CD44v3 and PD-L1, our results revealed a significant positive correlation between CD44v3 and PD-L1. Although these correlations are considered to be very weak due to the low τ-value of <0.3, our results may suggest that CSCs in OSCC express PD-L1 on the tumor cell membrane surface and play a role in immune evasion. Previous studies support our findings by reporting that CD44v3high exosomes have higher levels of immunosuppressive proteins such as PD-L1 than CD44v3low exosomes, correlating to a higher stage and LN metastasis in OSCC patients (42).
Regarding the relationship between CD24 and PD-1, a significant negative correlation was found between CD24 in the tumor parenchyma and PD-1 expression in surrounding inflammatory cells. Although these correlations are considered to be very weak due to the low τ-value of <-0.3, our results may suggest that downregulation of CD24 plays a role in evading host immune responses via high PD-1 expression on the stroma of CSCs. A previous report supports the results of our study. Kim et al (43) reported that CD24 bound to T cells via intracytoplasmic high-mobility group box 1, an inflammation-associated protein, and promoted activation of CD8+ effector T cells (43). In other words, CD24low cancer cells reduce CD8+ T cell activity and attenuated host immune responses compared with CD24high cancer cells. Another study also supports our findings. Xiao et al (44) reported that high expression of PD-1 correlated to decreased CD24 expression and cytokine production in B cells infiltrating hepatocellular carcinoma (44). Furthermore, in another previous report of breast cancer stem cells, in which CD24low was considered to be an immunophenotype of CSCs, increased expression of PD-L1 was observed in CD24low cancer cells (36). Although this study does not directly address the negative correlation between CD24 and PD-1, it can be inferred that PD-1 expression is elevated along with PD-L1 in CD24low breast cancer (36). On the other hand, Mirhashemi et al (45) reported contradictory results to our study. They performed RT-qPCR analysis of clinical specimens from 15 oral epithelial dysplasia and 45 cases of OSCC. They reported that as histologic grade increased from oral epithelial dysplasia to low-grade OSCC and from low-grade OSCC to high-grade OSCC, the expression of both CD44 and CD24 increased and correlated more intensely with the expression of these two cell surface markers. However, they also mentioned that there were conflicting reports, including our previous report (7), on the CD24 phenotype of CSCs in OSCC, and further verification was needed to assert that CD44high and CD24high were the phenotypes of CSCs in OSCC. On the contrary, Ghuwalewala et al (9) support our results. They report that MiR-146a, which directly targets IRAK1, traf6, and numb genes in OSCC and confers tumorigenicity, contributes to the enrichment of CSCs in OSCC through increased expression of stem cell markers, including CD44 and decreased CD24 levels. In addition, a previous study also supports our results. It reveals an increase in CD44highCD24low cells in IHC from low in normal tissues to relatively high in OSCC (8). Thus, more data are needed before definitive conclusions can be drawn because of the conflicting results of previous studies on the interaction of CD24 with PD-1 and the possible tumor specificity of CD24 functions. At least concerning CSCs in OSCC, attenuated expression of CD24 appears to play a role in evasion of CSCs from host immunity by enhancing PD-1 expression in inflammatory cells in the tumor stroma.
Although there is no doubt that CSCs are a significant unfavorable prognostic factor in terms of PD-L1/PD-1 co-expression, the following five points need to be considered to ensure the validity of this study: 1. the reasons for focusing on the invasive front of the tumor as a region of interest. 2. the limitation of the ability to profile mRNA expression extracted from FFPE, 3. the validity of the cutoff values used in the IHC analysis of the four antibodies (CD44v3, CD24, PD-L1, PD-1) used in this study, 4. the heterogeneity of expression of each antibody within the tumor tissue, and 5. the impact of core selection in TMA. 1. The reason for focusing on the invasive front of the tumor was that these areas are suitable for observing cross-talk between the parenchyma and stroma via the PD-L1/PD-1 axis, as described in a previous report (24) 2. Regarding the limitation of the ability to profile mRNA expression extracted from FFPE, the expression of CD44v3 in IHC was significantly negatively correlated with the expression of the molecules U2AF1, and the expression of PD-L1 in IHC was significantly positively correlated with the expression of PD-L1 at mRNA level. On the other hand, for CD24 and PD-1, no significant difference was found between the immunophenotype and the genotype. This discrepancy between the immunophenotype and genotype may be due to the effect of chemical modification to mRNA during FFPE specimen preparation and the degradation of nucleic acids during mRNA extraction process (46). 3. Regarding the cutoff values, the validity of the labeling indices (median values) obtained in this study was compared with the cutoff values described in previous literatures on IHC studies in OSCC for each molecule. No reports were found mentioning a cutoff value for CD44v3 in IHC, but as far as the CD44 standard isoform is concerned, a range of 10–50% was reported, and no uniform value existed (47,48). As far as CD24 is concerned, only one reference mentioned a cutoff value in IHC, which was reported as 37.4% (47). As far as PD-L1 is concerned, the range of cutoff values in IHC was reported to be 1–20% (24,25,49). As far as PD-1 is concerned, only one reference mentions a cutoff value in IHC, which was reported as 30% (24). The previous reports listed above regarding cutoff values in IHC for CD44 and PD-L1 include review articles (25,48,49), indicating no uniform cutoff value in the IHC study. Oliveira et al (50) mentioned that the cutoff values differ among studies of the same antibody because of the different antibodies used and staining conditions in IHC. Therefore, as is done in many IHC clinical studies, cutoff value was established for each antibody, using the median value in this study. In particular, we mention the criteria of PD-L1 positivity in this study, concerning the immunohistochemical analysis of PD-L1. For PD-L1 expression analysis, the antibody clone used in this study (clone 28-8) was the same clone used in the companion diagnosis of nivolumab for HNSCC treatment. In clinical practice, a cutoff value of 1% is considered positive. However, this clinical cutoff value is lower than the cutoff value of 6.1% adopted in this study. The difference in cutoff values between clinical practice and the present study may be attributed to multiple factors, including the different staining conditions and analysis methods used in the companion diagnosis. Another factor contributing to the discrepancy is that only the area of the invasive portion was selected as the stained area for PD-L1 in this study, rather than the entire tumor. Indeed, PD-L1 expression is spatiotemporally heterogeneous, and there is heterogeneity in PD-L1 expression rates within tumors and at different treatment points (20,51,52). Thus, PD-L1 expression levels are not well understood by single treatment point or single time sampling. 4. Regarding the heterogeneity of expression of each antibody in tumor tissue and 5. the impact of core selection in TMA, it has been reported that the expression of CD44(v), CD24, PD-L1, and PD-1 are all heterogeneous in OSCC in the same tissue (48,53–56). Boxberg et al (54) reported that this heterogeneity in the expression of each antibody had the risk of missing heterogeneous tumor regions when using TMA for immunohistochemical analysis while noting that increasing the number of cores collected per case may allow for a close match between tissue cores and staining results on the entire slide (56). In this study, by sampling at least two to three cores per case, we reduced the possibility of missing heterogeneous expression regions and, as much as possible, ensured the validity of each molecule's expression in IHC from TMA. In addition, we add one point of the limitations of this study. In this study, for CD44v3, we could not perform direct RT-qPCR due to the lack of commercially available primer.
In summary, this study revealed that CSCs in OSCC evade host immune mechanisms and maintain CSC stemness via PD-L1/PD-1 expression, resulting in unfavorable clinical outcomes, suggesting that CSCs are a potential therapeutic target for immune checkpoint inhibitors.
Acknowledgements
The authors would like to thank Ms. Kumiko Tsubone (Kurume University School of Medicine, Dental and Oral Medical Center) for their excellent assistance in the experiments.
Glossary
Abbreviations
- CSC
cancer stem cell
- OSCC
oral squamous cell carcinoma
- FFPE
formalin-fixed, paraffin-embedded
- IHC
immunohistochemistry
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- U2AF1
U2 small nuclear RNA auxiliary factor 1
- HNSCC
head and neck squamous cell carcinoma
- LN
lymph node
- YK
Yamamoto-Kohama
- EMT
epithelial-mesenchymal transition
- TMA
tissue microarray
Funding Statement
This study was supported by a Grant-in-Aid for Scientific Research (grant no. 20K18714) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contribution
KT and YN conceived and designed the study. KT, AK, MN and NS performed the experiments and acquired data, and assessed the authenticity of all the raw data to ensure its legitimacy. KT, YA, KM, JK and HN analyzed the data. KT, YN and MN drafted the manuscript. KT and JK confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The design and methods of this study complied with the ethical guidelines of the Declaration of Helsinki and the guidelines for research involving human subjects of the Ethical Review Committee of the Clinical Research Center of Kurume University Hospital. Institutional review and approval were obtained prior to commencing the study (approval no. 22217). Written informed consent was obtained before patient participation in the study, and clinical specimens were obtained from each patient in accordance with the approved guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Petersen PE. Oral cancer prevention and control-the approach of the world health organization. Oral Oncol. 2009;45:454–460. doi: 10.1016/j.oraloncology.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 2.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidwai F, Costea DE, Hutchison I, Mackenzie I. The effects of CD44 down-regulation on stem cell properties of head and neck cancer cell lines. J Oral Pathol Med. 2013;42:682–690. doi: 10.1111/jop.12076. [DOI] [PubMed] [Google Scholar]
- 4.Kalish ED, Iida N, Moffat FL, Bourguignon LY. A new CD44v3-containing isoform is involved in tumor cell growth and migration during human breast carcinoma progression. Front Biosci. 1999;4:A1–A8. doi: 10.2741/A399. [DOI] [PubMed] [Google Scholar]
- 5.Hurt EM, Kawasaki BT, Klarmann GJ, Thomas SB, Farrar WL. CD44+CD24- prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma F, Li H, Wang H, Shi X, Fan Y, Ding X, Lin C, Zhan Q, Qian H, Xu B. Enriched CD44+/CD24- population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC) Cancer Lett. 2014;353:153–159. doi: 10.1016/j.canlet.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Todoroki K, Ogasawara S, Akiba J, Nakayama M, Naito Y, Seki N, Kusukawa J, Yano H. CD44v3+/CD24- cells possess cancer stem cell-like properties in human oral squamous cell carcinoma. Int J Oncol. 2015;48:99–109. doi: 10.3892/ijo.2015.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, Panda CK, Roychoudhury S. CD44highCD24low molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Res. 2016;16:405–417. doi: 10.1016/j.scr.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 9.Ghuwalewala S, Ghatak D, Das S, Roy S, Das P, Butti R, Gorain M, Nath S, Kundu GC, Roychoudhury S. MiRNA-146a/AKT/β-catenin activation regulates cancer stem cell phenotype in oral squamous cell carcinoma by targeting CD24. Front Oncol. 2021;11:651692. doi: 10.3389/fonc.2021.651692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, Wicha MS, Prince ME. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baillie R, Tan ST, Itinteang T. Cancer stem cells in oral cavity squamous cell carcinoma: A review. Front Oncol. 2017;7:112. doi: 10.3389/fonc.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang SJ, Wong G, de Heer AM, Xia W, Bourguignon LYW. CD44 variant isoforms in head and neck squamous cell carcinoma progression. Laryngoscope. 2009;119:1518–1530. doi: 10.1002/lary.20506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourguignon LYW, Wong G, Earle C, Chen L. Hyaluronan-CD44v3 interaction with Oct4-Sox2-nanog promotes miR-302 expression leading to self-renewal, clonal formation, and cisplatin resistance in cancer stem cells from head and neck squamous cell carcinoma. J Biol Chem. 2012;287:32800–32824. doi: 10.1074/jbc.M111.308528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Zhou W, Wan Y, Lu J, Ge K, Jia C. CD44V3, an alternatively spliced form of CD44, promotes pancreatic cancer progression. Int J Mol Sci. 2022;23:12061. doi: 10.3390/ijms232012061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirruccello SJ, LeBien TW. The human B cell-associated antigen CD24 is a single chain sialoglycoprotein. J Immunol. 1986;136:3779–3784. doi: 10.4049/jimmunol.136.10.3779. [DOI] [PubMed] [Google Scholar]
- 16.Fischer GF, Majdic O, Gadd S, Knapp W. Signal transduction in lymphocytic and myeloid cells via CD24, a new member of phosphoinositol-anchored membrane molecules. J Immunol. 1990;144:638–641. doi: 10.4049/jimmunol.144.2.638. [DOI] [PubMed] [Google Scholar]
- 17.Vesuna F, Lisok A, Kimble B, Raman V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia. 2009;11:1318–1328. doi: 10.1593/neo.91084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 19.Economopoulou P, Agelaki S, Perisanidis C, Giotakis EI, Psyrri A. The promise of immunotherapy in head and neck squamous cell carcinoma. Ann Oncol. 2016;27:1675–1685. doi: 10.1093/annonc/mdw226. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Bai X, Shan F. The progress and confusion of anti-PD1/PD-L1 immunotherapy for patients with advanced non-small cell lung cancer. Int Immunopharmacol. 2020;80:106247. doi: 10.1016/j.intimp.2020.106247. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Chen Z, Li Y, Zhao W, Wu J, Zhang Z. PD-1/PD-L1 checkpoint inhibitors in tumor immunotherapy. Front Pharmacol. 2021;12:731798. doi: 10.3389/fphar.2021.731798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blank C, Gajewski TF, Mackensen A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: Implications for tumor immunotherapy. Cancer Immunol Immunother. 2005;54:307–314. doi: 10.1007/s00262-004-0593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol. 2015;33:3293–3304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maruse Y, Kawano S, Jinno T, Matsubara R, Goto Y, Kaneko N, Sakamoto T, Hashiguchi Y, Moriyama M, Toyoshima T, et al. Significant association of increased PD-L1 and PD-1 expression with nodal metastasis and a poor prognosis in oral squamous cell carcinoma. Int J Oral Max Surg. 2018;47:836–845. doi: 10.1016/j.ijom.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Lenouvel D, González-Moles MÁ, Ruiz-Ávila I, Gonzalez-Ruiz L, Gonzalez-Ruiz I, Ramos-García P. Prognostic and clinicopathological significance of PD-L1 overexpression in oral squamous cell carcinoma: A systematic review and comprehensive meta-analysis. Oral Oncol. 2020;106:104722. doi: 10.1016/j.oraloncology.2020.104722. [DOI] [PubMed] [Google Scholar]
- 26.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. New Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, Jr, Psyrri A, Basté N, Neupane P, Bratland Å, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y, Shin JH, Longmire M, Wang H, Kohrt HE, Chang HY, Sunwoo JB. CD44+ cells in head and neck squamous cell carcinoma suppress t-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin Cancer Res. 2016;22:3571–3581. doi: 10.1158/1078-0432.CCR-15-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, Xiao Y, Bu LL, Zhang WF, Sun ZJ. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res. 2020;8:179–191. doi: 10.1158/2326-6066.CIR-19-0394. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, Li Y, Jia L, Kim JK, Li J, Deng P, Zhang W, Krebsbach PH, Wang CY. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell. 2021;28:1597–1613.e7. doi: 10.1016/j.stem.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geum DH, Hwang DS, Lee CH, Cho SD, Jang MA, Ryu MH, Kim UK. PD-L1 expression correlated with clinicopathological factors and akt/stat3 pathway in oral SCC. Life (Basel) 2022;12:238. doi: 10.3390/life12020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto E, Kohama G, Sunakawa H, Iwai M, Hiratsuka H. Mode of invasion, bleomycin sensitivity, and clinical course in squamous cell carcinoma of the oral cavity. Cancer. 1983;51:2175–2180. doi: 10.1002/1097-0142(19830615)51:12<2175::AID-CNCR2820511205>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, Al-Mohanna F, Al-Alwan M, Ghebeh H. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141:1402–1412. doi: 10.1002/ijc.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei F, Zhang T, Deng SC, Wei JC, Yang P, Wang Q, Chen ZP, Li WL, Chen HC, Hu H, Cao J. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 2019;450:1–13. doi: 10.1016/j.canlet.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, Dutta P, Clayton S, McCloud A, Vadgama JV. Elevated baseline serum PD-L1 level may predict poor outcomes from breast cancer in African-American and hispanic women. J Clin Med. 2022;11:283. doi: 10.3390/jcm11020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao L, Guo Q, Li X, Yang X, Ni H, Wang T, Zhao Q, Liu H, Xing Y, Xi T, Zheng L. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine. 2019;41:395–407. doi: 10.1016/j.ebiom.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha JH, Chen CT, Liao HW, Kuo CW, Khoo KH, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Judd J, Karim NA, Khan H, Naqash AR, Baca Y, Xiu J, VanderWalde AM, Mamdani H, Raez LE, Nagasaka M, et al. Characterization of KRAS mutation subtypes in non-small cell lung cancer. Mol Cancer Ther. 2021;20:2577–2584. doi: 10.1158/1535-7163.MCT-21-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin F, Wang J, Zhao K, Xin C, Shi Y, Zeng X, Xu H, Li J, Chen Q. The significance of PA28γ and U2AF1 in oral mucosal carcinogenesis. Oral Dis. 2020;26:53–61. doi: 10.1111/odi.13213. [DOI] [PubMed] [Google Scholar]
- 41.Goldschmid H, Kluck K, Ball M, Kirchner M, Allgäuer M, Winter H, Herth F, Heußel CP, Pullamsetti SS, Savai R, et al. Spatial profiling of the microenvironment reveals low intratumoral heterogeneity and STK11-associated immune evasion in therapy-naïve lung adenocarcinomas. Lung Cancer. 2023;180:107212. doi: 10.1016/j.lungcan.2023.107212. [DOI] [PubMed] [Google Scholar]
- 42.Theodoraki MN, Matsumoto A, Beccard I, Hoffmann TK, Whiteside TL. CD44v3 protein-carrying tumor-derived exosomes in HNSCC patients' plasma as potential noninvasive biomarkers of disease activity. Oncoimmunology. 2020;9:1747732. doi: 10.1080/2162402X.2020.1747732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim TS, Gorski SA, Hahn S, Murphy KM, Braciale TJ. Distinct dendritic cell subsets dictate the fate decision between effector and memory CD8+ T cell differentiation by a CD24-dependent mechanism. Immunity. 2014;40:400–413. doi: 10.1016/j.immuni.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao X, Lao XM, Chen MM, Liu RX, Wei Y, Ouyang FZ, Chen DP, Zhao XY, Zhao Q, Li XF, et al. PD-1hi identifies a novel regulatory B-cell population in human hepatoma that promotes disease progression. Cancer Discov. 2016;6:546–559. doi: 10.1158/2159-8290.CD-15-1408. [DOI] [PubMed] [Google Scholar]
- 45.Mirhashemi M, Ghazi N, Saghravanian N, Taghipour A, Mohajertehran F. Evaluation of CD24 and CD44 as cancer stem cell markers in squamous cell carcinoma and epithelial dysplasia of the oral cavity by q- RT-PCR. Dent Res J (Isfahan) 2020;17:208–212. doi: 10.4103/1735-3327.284727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall JS, Taylor J, Valentine HR, Irlam JJ, Eustace A, Hoskin PJ, Miller CJ, West CM. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. Br J Cancer. 2012;107:684–694. doi: 10.1038/bjc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Moraes FP, Lourenço SV, Ianez RCF, de Sousa EA, Silva MM, Damascena AS, Kowalski LP, Soares FA, Coutinho-Camillo CM. Expression of stem cell markers in oral cavity and oropharynx squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2017;123:113–122. doi: 10.1016/j.oooo.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Mirhashemi M, Sadeghi M, Ghazi N, Saghravanian N, Dehghani M, Aminian A. Prognostic value of CD44 expression in oral squamous cell carcinoma: A meta-analysis. Ann Diagn Pathol. 2023;67:152213. doi: 10.1016/j.anndiagpath.2023.152213. [DOI] [PubMed] [Google Scholar]
- 49.Nocini R, Vianini M, Girolami I, Calabrese L, Scarpa A, Martini M, Morbini P, Marletta S, Brunelli M, Molteni G, et al. PD-L1 in oral squamous cell carcinoma: A key biomarker from the laboratory to the bedside. Clin Exp Dent Res. 2022;8:690–698. doi: 10.1002/cre2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira LR, Oliveira-Costa JP, Araujo IM, Soave DF, Zanetti JS, Soares FA, Zucoloto S, Ribeiro-Silva A. Cancer stem cell immunophenotypes in oral squamous cell carcinoma. J Oral Pathol Med. 2011;40:135–142. doi: 10.1111/j.1600-0714.2010.00967.x. [DOI] [PubMed] [Google Scholar]
- 51.McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, Velcheti V, Herbst R, LoRusso P, Rimm DL. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2015;2:1–9. doi: 10.1001/jamaoncol.2015.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Dong Z, Jiang T, Hou L, Wu F, Gao G, He Y, Zhao J, Li X, Zhao C, et al. Heterogeneity of PD-L1 expression among the different histological components and metastatic lymph nodes in patients with resected lung adenosquamous carcinoma. Clin Lung Cancer. 2018;19:e421–e430. doi: 10.1016/j.cllc.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Athanassiou-Papaefthymiou M, Shkeir O, Kim D, Divi V, Matossian M, Owen JH, Czerwinski MJ, Papagerakis P, McHugh J, Bradford CR, et al. Evaluation of CD44 variant expression in oral, head and neck squamous cell carcinomas using a triple approach and its clinical significance. Int J Immunopathol Pharmacol. 2014;27:337–349. doi: 10.1177/039463201402700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boxberg M, Götz C, Haidari S, Dorfner C, Jesinghaus M, Drecoll E, Boskov M, Wolff KD, Weichert W, Haller B, Kolk A. Immunohistochemical expression of CD44 in oral squamous cell carcinoma in relation to histomorphological parameters and clinicopathological factors. Histopathology. 2018;73:559–572. doi: 10.1111/his.13496. [DOI] [PubMed] [Google Scholar]
- 55.Dave K, Ali A, Magalhaes M. Increased expression of PD-1 and PD-L1 in oral lesions progressing to oral squamous cell carcinoma: A pilot study. Sci Rep. 2020;10:9705. doi: 10.1038/s41598-020-66257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Straub M, Drecoll E, Pfarr N, Weichert W, Langer R, Hapfelmeier A, Götz C, Wolff KD, Kolk A, Specht K. CD274/PD-L1 gene amplification and PD-L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget. 2016;7:12024–12034. doi: 10.18632/oncotarget.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.