Graphical Abstract
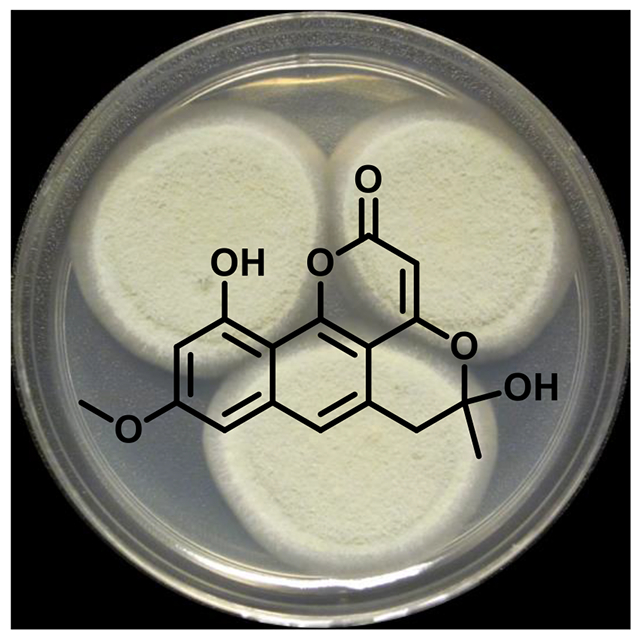
Pannorin B (1), a new naphthopyrone derivative, was obtained from an endophytic fungal culture of Penicillium sp. (G324), which was isolated from the leaves of milk thistle (Silybum marianum). Four known compounds, including O-methyldihydrogladiolic acid (2), 10,20-dehydro[12,13-dehydroprolyl-2-(1,1-dimethylallyltryptophyl)diketopiperazine] (3), 12,13-dehydroprolyl-2-(1,1-dimethylallyltryptophyl)diketopiperazine (4), and deoxybrevianamide E (5) were also encountered. The structures of 1–5 were determined via analysis of a suite of NMR and MS data.
Keywords: NMR, 1H NMR, 13C NMR, Fungal endophyte, Secondary metabolites, Milk thistle, Silybum marianum
Introduction
The herbal remedy, milk thistle (Silybum marianum), has been used in traditional medicine for various liver, kidney, and gall bladder ailments. For over a decade, our research group has been investigating the flavonolignans obtained from this medicinal herb for cancer chemoprevention and hepatoprotection.[1–6] Recently, we extended our studies towards examining the diversity as well as distribution patterns of fungal endophytes in leaves, stem, and roots of milk thistle.[7] These fungi inhabit the internal living tissues of the host plants asymptomatically, though they may also cause disease over time.[8] In addition to the phylogenetic profiling of these endophytes, a series of fungal extracts were also examined for chemical composition. Although the plant-endophyte relationship may or may not be mutualistic, the compounds produced by some endophytes could play a role in the growth and survival of the host. In a previous study, Penicillium restrictum, isolated from milk thistle, yielded promising secondary metabolites.[9] Hence, in pursuit of interesting chemistry, a related monoverticillate endophytic Penicillium sp. was explored.
Results and Discussion
Natural products chemistry studies of an extract from solid-substrate fermentation cultures of G324, which was isolated from leaves of milk thistle and identified as a Penicillium sp. (see Supporting Information and Figs. S6 and S7), afforded one new naphthopyrone, which we have termed pannorin B (1), and four previously known metabolites [O-methyldihydrogladiolic acid (2), 10,20-dehydro[12,13-dehydroprolyl-2-(1,1-dimethylallyltryptophyl)diketopiperazine] (3), 12,13-dehydroprolyl-2-(1,1-dimethylallyltryptophyl)diketopiperazine (4), and deoxybrevianamide E (5)](Fig. 1).[10–12] The known compounds were identified by favorable comparisons of their NMR and MS data to the literature.
Figure 1.
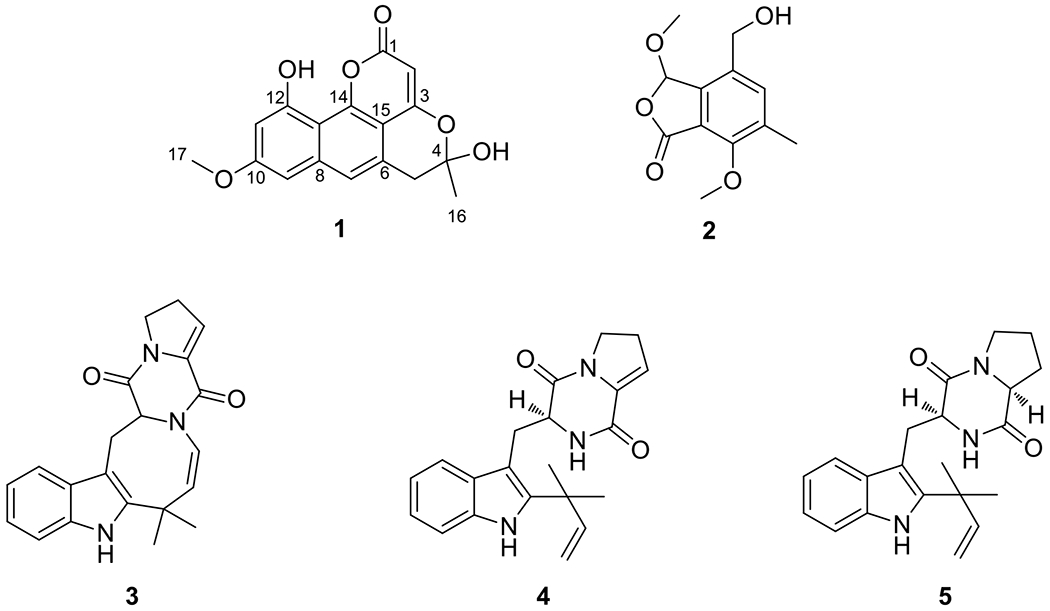
Structures of compounds 1–5.
The molecular formula of pannorin B (1) was determined to be C17H14O6 (index of hydrogen deficiency of 11) on the basis of HRESIMS data. The 1H NMR spectrum (Table 1 and Fig. S1) displayed three aromatic signals, an olefin signal, a methoxy singlet, one methyl singlet, and a pair of doublets at δH 3.17 and δH 3.26 (J = 16.8 Hz); the latter were characteristic of isolated diastereotopic methylene protons. Two singlets (δH 7.45 and δH 10.34), corresponding to exchangeable protons (4-OH and 12-OH), were also observed. In addition to the signals expected for the structural features discussed above, the 13C NMR data (Table 1 and Fig. S2) revealed the presence of one quaternary sp3 carbon (δC 101.4; C-4), one ester group (δC 164.5; C-1), and eight non-protonated sp2 carbons, suggesting a highly conjugated ring system as supported by the UV data (λmax 363, 319, and 271 nm). The NMR chemical shifts for meta-coupled aromatic protons (δH 6.85, δC 98.8; C-9 and δH 6.61, δC 101.9; C-11) indicated the presence of oxygenated substituents on carbons adjacent to these positions (δC 160.3 for C-10 and δC 156.7 for C-12). Key HMBC correlations (Table 1 and Fig. S4) from H-9 to C-10, C-11, and C-13 (δC 107.7), as well as from H-11 to C-9, C-10, C-12, and C-13 supported a tetra-substituted aromatic ring. A three-bond correlation from H3-17 to C-10 confirmed the position of the methoxy group. HMBC correlations from H-9 to C-7 (δC 121.0) and from H-7 (δH 7.37) to C-13 and C-15 (δC 104.6) suggested the presence of a highly substituted naphthalene-type ring system. The methyl singlet (H3-16) showed HMBC correlations to methylene C-5 (δC 38.1) and a doubly-oxygenated quaternary C-4 (δC 101.4). The position of the hydroxy group at C-4 was confirmed by HMBC correlations from 4-OH (δH 7.45) to C-4 and C-5. Additional HMBC correlations from H2-5 to C-4, C-6 (δC 127.9), C-7, and C-15 connected this partial structure to the fused bicyclic ring system. The remaining olefinic proton, H-2 (δH 5.66), showed HMBC correlations to carbonyl C-1, C-3 (δC 162.2), and C-15, suggesting a pyrone ring via linking the oxygen atom to C-14. Finally, the ether linkage between C-3 and hemiacetal C-4 was supported by chemical shifts for the respective carbons and accounted for the last remaining unsaturation, thus completing the gross structure of 1 (Fig. 1). Unfortunately, the absolute configuration of 1 could not be determined since the sample exhibited no optical activity, suggesting the presence of a racemic mixture.
Table 1.
1H (400 MHz) and 13C (100 MHz) NMR data of pannorin B (1) in (CD3)2SO.
| # | δH (mult., J) | δ C | HMBC (H→ #C) |
|---|---|---|---|
| 1* | 164.5 | ||
| 2 | 5.66 ( s) | 91.4 | 1, 3, 15 |
| 3* | 162.2 | ||
| 4 | 101.4 | ||
| 5 | 3.26 (d, 16.8) | 38.1 | 4, 6, 7 (wk), 15 (wk) |
| 3.17 (d, 16.8) | 4, 6, 7, 15 | ||
| 6 | 127.9 | ||
| 7 | 7.37 (br s) | 121.0 | 5, 8, 9, 13, 15 |
| 8 | 138.9 | ||
| 9 | 6.85 (d, 2.4) | 98.8 | 7, 10 (wk), 11, 13 |
| 10 | 160.3 | ||
| 11 | 6.61 (d, 2.4) | 101.9 | 9, 10, 12, 13 |
| 12 | 156.7 | ||
| 13 | 107.7 | ||
| 14 | 151.8 | ||
| 15 | 104.6 | ||
| 16 | 1.67 (s) | 27.2 | 4, 5 |
| 17 | 3.85 (s) | 55.3 | 10 |
| 4-OH | 7.45 (s) | 4, 5, 16 (wk) | |
| 12-OH | 10.34 (s) | 11, 12, 13 |
These assignments may be interchanged.
Biogenetically, pannorin B (1) appears to be polyketide-derived and possesses a partial structure similar to another fungal natural product, pannorin.[13] Other than the methylation of one of the phenolic hydroxyl groups and the presence of an additional acetyl unit undergoing cyclization to form the fourth ring, compound 1 could be biosynthesized in a manner analogous to the construction of pannorin.[14]
Some of our previous research on compounds from milk thistle has been directed toward prostate cancer chemoprevention.[1–3] Thus, the pure compounds isolated from milk thistle endophytic fungal extracts were examined for cytotoxicity against human prostate carcinoma (PC3) cells, exactly as described in detail previously.[7] Compounds 1, 3, and 5 were tested in this assay but were inactive at 25 μM concentration. Compounds 3, 4, and 5 have not been evaluated in the literature for biological activity. However, O-methyldihydrogladiolic acid (2) has been reported as phytotoxic to the germination and growth of lettuce,[10] which was interesting given its biosynthesis by a plant endophyte;[7] it was not tested for cytotoxicity in our assays due to degradation over time.
Experimental
General experimental procedures
Optical rotation data were acquired on a Rudolph Research Autopol III polarimeter (Rudolph Research Analytical, Flanders, NJ, USA). UV data were obtained using a Varian Cary 100 Bio UV-vis spectrophotometer (Varian Medical Systems, Palo Alto, CA, USA). HRESIMS data were collected using an electrospray ionization (ESI) source coupled to a LTQ Orbitrap XL system (Thermo Fisher Scientific, San Jose, CA, USA) in positive and negative ionization modes via a liquid chromatography/autosampler system comprised of an Acquity UPLC system (Waters Corp., Milford, MA, USA). A CombiFlash Rf system using a 12 g RediSep Rf Si-gel Gold column (both from Teledyne-Isco, Lincoln, NE, USA) was employed for normal-phase flash column chromatography. High-performance liquid chromatography (HPLC) separations were performed utilizing Varian ProStar HPLC systems equipped with ProStar 210 pumps and a ProStar 335 photodiode array detector, using Galaxie Chromatography Workstation software (version 1.9.3.2, Varian Inc.). YMC ODS-A (Waters Corp.; 5μm; 250 × 10 mm column; semi-preparative HPLC) or Kinetex C18 (Phenomenex, Torrance, CA, USA; 5μm; columns of dimensions 250 × 21.2 mm; preparative HPLC and 250 × 4.6 mm; analytical HPLC) were used for HPLC. For UPLC analysis, a BEH C18 (Waters Corp.; 1.7 μm; 50 × 2.1 mm) column was used with data collected and analyzed using Empower 3 software. The solvents were obtained from Fisher Scientific.
Fungal strain
Whole plants and seeds of Silybum marianum (L.) Gaertn. (Asteraceae) were obtained from Horizon Herbs, LLC (Williams, OR, USA), and a voucher specimen was deposited in the University of North Carolina Herbarium (NCU602014).[7] Four different collections of whole plants and seeds were obtained for the study in 2011 (Lot # 6490; Lot # 6510), 2012 (Lot # 12348), and 2013 (Lot # 6462). A strain (G324) of the Penicillium sp. was isolated from the leaves of milk thistle (Lot # 12348). The fungal culture is maintained at the University of North Carolina at Greensboro, Department of Chemistry and Biochemistry Fungal Culture Collection. Details for the isolation, molecular identification, and phylogenetic analysis of the fungal strain can be found in the supporting information and Figures S6 and S7.
Fermentation, extraction, and isolation
The cultures of G324 were subsequently grown on 2% MEA, Potato Dextrose Agar (PDA, Difco), and 2% soy peptone, 2% dextrose, and 1% yeast extract (YESD). For chemical extractions, fungal cultures were grown on rice.[15] To make seed cultures for inoculating rice, a piece of a fresh culture grown in MEA media was excised from the leading edge of the colony and transferred to a liquid medium containing 2% soy peptone, 2% dextrose and 1% yeast extract (YESD media). Following incubation (7 days) at 22 °C with agitation, the culture was used to inoculate 50 mL of rice media prepared using rice and twice the volume of rice with H2O in a 250 mL Erlenmeyer flask. This was incubated at 22 °C until the cultures showed good growth (14 – 21 days) depending on the growth of individual species to generate the screener culture. For large-scale production of fungal cultures for chemical extractions, a seed culture was grown in 50 mL YESD media in a 250 mL Erlenmeyer flask for 7 days at 22 °C with agitation. After the seed culture was grown for 7 days, about 10 mL of liquid culture was used to inoculate each of the 4 flasks of rice, which was prepared in the same manner as the screener scale culture.
To a screener solid-substrate fermentation culture (G324) grown on rice, 60 mL of 1:1 CH3OH–CHCl3 were added. The culture was chopped into small pieces with a spatula and shaken overnight (∼125 rpm at room temperature) using a rotary shaker. The sample was vacuum filtered, and the remaining residues were washed with small volumes of 1:1 CH3OH–CHCl3. To the filtrate, 90 mL each of CHCl3 and H2O were added, followed by stirring for 30 min. The organic layer was collected and evaporated to dryness under reduced pressure and partitioned between 100 mL of 1:1 CH3OH–CH3CN and 100 mL of hexanes. The CH3OH–CH3CN layer was evaporated to dryness under vacuum to yield 70 mg of extract. Scaled-up cultures were extracted by parallel processing of four such flasks using the above protocol to yield a combined 499 mg of crude extract.
The CH3OH /CH3CN extract (499 mg) was adsorbed on a minimal amount of Celite 545 (Acros Organics, Geel, Belgium). The sample was dried before loading on a cartridge. This cartridge of adsorbed material was placed atop of a silica gel column and subjected to normal-phase silica gel flash column chromatography employing a step gradient elution with hexanes, CHCl3, and CH3OH (30 mL/min flow rate and 61.0 column volumes). The resulting fractions were combined according to the UV and ELSD data to afford four pools. Pool 4 (96 mg) was subjected to preparative RP-HPLC (gradient elution using CH3CN in H2O: 40–80% over 40 min and 80-100% CH3CN over 10 min ; λ = 210 and 254 nm; flow rate = 21.2 mL/min) affording 2 (7.5 mg, tR 10.0 min), 3 (3.2 mg, tR 12.5 min), 1 (6.9 mg, tR 15.0 min), an impure sample of 4 (5.9 mg, tR 16.0 min), and 5 (1.7 mg, tR 17.0 min). Compound 4 could not be recovered in sufficient purity. Purity of 1 was determined by UPLC using a gradient elution of 20% CH3CN in H2O to 100% CH3CN over 4.5 min (Fig. S5). Gradual degradation of compound 2 was observed over time, even upon dry storage. All of the previously known compounds encountered in this extract (2–5) were identified by comparison of their 1H NMR, 13C NMR, and/or MS data with literature values.[10–12]
Pannorin B (1): Light yellow powder; [α]25D 0.0 (c 0.20, CH3OH); UV (CH3OH) λmax (log ε) 363 (3.6), 319 (3.4), 271 (3.5) nm; 1H NMR data [(CD3)2SO, 400 MHz] and 13C NMR [(CD3)2SO, 100 MHz] data, see Table 1 and Figs. S1 and S2; HRESIMS obsd. m/z 313.0703 [M−H]− (calcd. for C17H13O6, 313.0718).
O-Methyldihydrogladiolic acid (2): HRESIMS obsd. m/z 207.0643 [M−CH3OH+H]+ (calcd. for C11H11O4, 207.0652); 1H NMR data were fully consistent with those reported in literature; the structure was also confirmed by analysis of 2D NMR data (Spectral assignments, Table S1 and Fig. S8).[10]
10,20-Dehydro[12,13-dehydroprolyl-2-(1,1-dimethylallyltryptophyl)diketopiperazine] (3): HRESIMS obsd. m/z 348.1691 [M+H]+ (calcd. for C21H22N3O2, 348.1707); 1H NMR data were consistent with those reported in literature; the structure was also confirmed by analysis of 2D NMR data (Spectral assignments, Table S2 and Fig. S9).[11]
12,13-Dehydroprolyl-2-(1,1-dimethylallyltryptophyl)diketopiperazine (4): HRESIMS obsd. m/z 350.1849 [M+H]+ (calcd. for C21H24N3O2, 350.1863); 1H NMR data were consistent with those reported in literature; the structure was also confirmed by analysis of 2D NMR data (Spectral assignments, Table S2 and Fig. S10).[11]
Deoxybrevianamide E (5): HRESIMS obsd. m/z 352.2004 [M+H]+ (calcd. for C21H26N3O2, 352.2020) ; 1H NMR data (Fig. S11) were fully consistent with those reported in literature.[11, 12]
NMR data
NMR spectra (1H, 13C, 1H-13C HSQC, and 1H-13C HMBC) were recorded at 25 °C in (CD3)2SO on a JEOL ECS-400 NMR spectrometer (399.78 MHz for 1H and 100.53 MHz for 13C; JEOL Ltd., Tokyo, Japan) equipped with auto tune 5 mm field gradient tunable Royal probe (NM-03810RO5/UPG). The 1H and 13C chemical shifts were referenced to the residual solvent peak of (CD3)2SO at 2.05 ppm and 39.5 ppm, for 1H and 13C, respectively. The 1H sweep width was set at 5997 Hz for all experiments with a 90° pulse for 1H of 6.4 μs and 13C sweep width 25131 Hz with a 90° pulse for 13C of 11.6 μs. The digital resolution of 1H NMR was 0.37 Hz and that of 13C NMR was 0.77 Hz. The edited-gradient 1H-13C HSQC was acquired with 13C sweep width of 16084 Hz and 256 t1 increments. Each increment was acquired with 16 transients. One-bond coupling constant delay was set using 145 Hz and MPF8 decoupling was applied during acquisition. The gradient 1H-13C HMBC was acquired using 64 transients per increment with 256 t1 increments. A sweep width of 20105 Hz was used for the 13C dimension. One-bond coupling constant of 145 Hz and long-range coupling constant of 8 Hz were used to set the delays in the pulse sequence.
Supplementary Material
Figure 2.
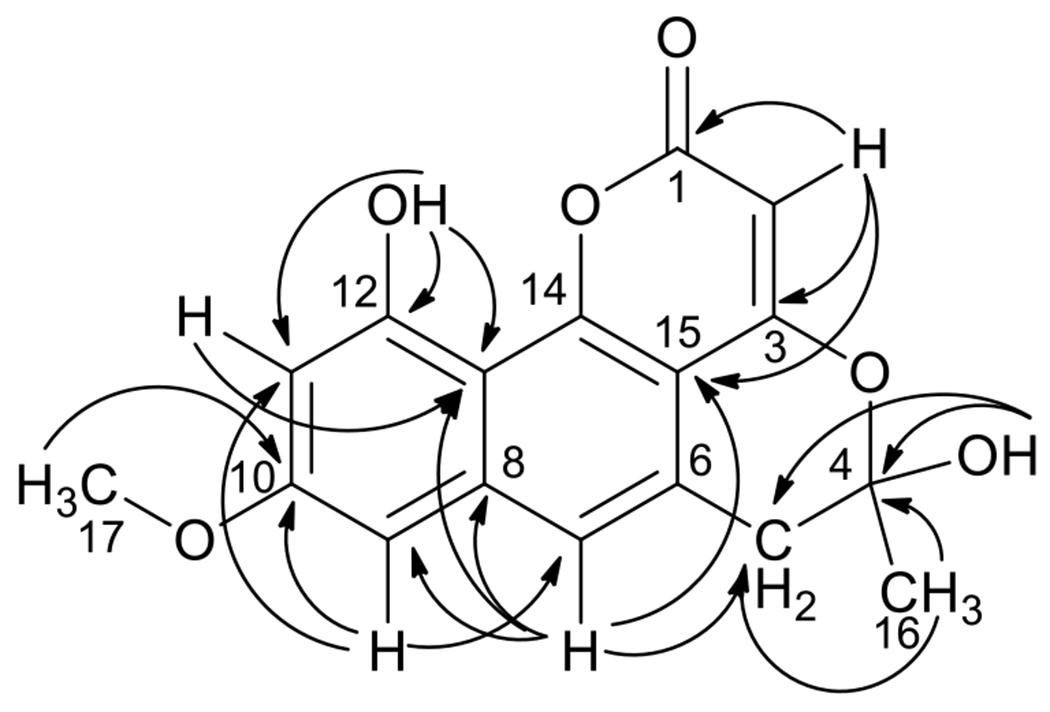
Key HMBC correlations for pannorin B (1).
Acknowledgments
This research was supported in part by a grant from the North Carolina Biotechnology Center (2011-BRG-1206). The researchers in Colorado were also supported by a grant from the National Cancer Institute/National Institutes of Health (R01 CA102514). Sequence data were generated at the mycology laboratory of Dr. A. N. Miller, Illinois Natural History Survey, University of Illinois at Urbana–Champaign. Assistance from the staff of the NMR (Dr. Franklin J. Moy) and Mass Spectrometry Facilities (Dr. Brandie M. Ehrmann) at University of North Carolina at Greensboro is gratefully acknowledged.
References
- [1].Davis-Searles PR, Nakanishi Y, Kim N-C, Graf TN, Oberlies NH, Wani MC, Wall ME, Agarwal R, Kroll DJ Cancer res. 2005, 65, 4448. [DOI] [PubMed] [Google Scholar]
- [2].Deep G, Raina K, Singh RP, Oberlies NH, Kroll DJ, Agarwal R Int. J. Cancer 2008, 123, 2750. [DOI] [PubMed] [Google Scholar]
- [3].Deep G, Gangar SC, Rajamanickam S, Raina K, Gu M, Agarwal C, Oberlies NH, Agarwal R PloS one 2012, 7, e34630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Polyak SJ, Morishima C, Lohmann V, Pal S, Lee DY, Liu Y, Graf TN, Oberlies NH P. Natl. Acad. Sci. USA 2010, 107, 5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sy-Cordero AA, Day CS, Oberlies NH J. Nat. Prod 2012, 75, 1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Graf TN, Wani MC, Agarwal R, Kroll DJ, Oberlies NH Planta Med. 2007, 73, 1495. [DOI] [PubMed] [Google Scholar]
- [7].Raja HA, Kaur A, El-Elimat T, Figueroa M, Kumar R, Deep G, Agarwal R, Faeth SH, Cech NB, Oberlies NH Mycology 2015, 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wilson D. Oikos 1995, 274. [Google Scholar]
- [9].Figueroa M, Jarmusch AK, Raja HA, El-Elimat T, Kavanaugh JS, Horswill AR, Cooks RG, Cech NB, Oberlies NH J. Nat. Prod 2014, 77, 1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ichihara A, Sawamura S, Kawakami Y, Sakamura S Agric. Biol. Chem 1985, 49, 1891. [Google Scholar]
- [11].Steyn P. Tetrahedron 1973, 29, 107. [Google Scholar]
- [12].Schkeryantz JM, Woo JC, Siliphaivanh P, Depew KM, Danishefsky SJ J. Am. Chem. Soc 1999, 121, 11964. [Google Scholar]
- [13].Ogawa H, Hasumi K, Sakai K, Murakawa S, Endo A J. Antibiot 1991, 44, 762. [DOI] [PubMed] [Google Scholar]
- [14].Vagstad AL, Newman AG, Storm PA, Belecki K, Crawford JM, Townsend CA Angew. Chem. Int. Ed 2013, 52, 1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].VanderMolen KM, Raja HA, El-Elimat T, Oberlies NH AMB Express 2013, 3, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


