Abstract
Introduction
Diarrhea is still a significant global public health problem. There are currently no systematic evaluation of the modeling areas and approaches to predict diarrheal illness outcomes. This paper reviews existing research efforts in predictive modeling of infectious diarrheal illness in pediatric populations.
Methods
We conducted a systematic review via a PubMed search for the period 1990–2021. A comprehensive search query was developed through an iterative process and literature on predictive modeling of diarrhea was retrieved. The following filters were applied to the search results: human subjects, English language, and children (birth to 18 years). We carried out a narrative synthesis of the included publications.
Results
Our literature search returned 2671 articles. After manual evaluation, 38 of these articles were included in this review. The most common research topic among the studies were disease forecasts 14 (36.8%), vaccine‐related predictions 9 (23.7%), and disease/pathogen detection 5 (13.2%). Majority of these studies were published between 2011 and 2020, 28 (73.7%). The most common technique used in the modeling was machine learning 12 (31.6%) with various algorithms used for the prediction tasks. With change in the landscape of diarrheal etiology after rotavirus vaccine introduction, many open areas (disease forecasts, disease detection, and strain dynamics) remain for pathogen‐specific predictive models among etiological agents that have emerged as important. Additionally, the outcomes of diarrheal illness remain under researched. We also observed lack of consistency in the reporting of results of prediction models despite the available guidelines highlighting the need for common data standards and adherence to guidelines on reporting of predictive models for biomedical research.
Conclusions
Our review identified knowledge gaps and opportunities in predictive modeling for diarrheal illness, and limitations in existing attempts whilst advancing some precursory thoughts on how to address them, aiming to invigorate future research efforts in this sphere.
Keywords: diarrhea, machine learning, pediatric, predictive modeling
1. INTRODUCTION
Diarrhea is a global public health problem causing approximately 1.7 billion episodes in children annually, 1 , 2 with a significant proportion of diarrheal morbidity and mortality occurring in low‐income countries due to resource and infrastructural challenges to the health system. 3 , 4 Diarrheal disease, if not well managed, has a plethora of poor outcomes; malnutrition, longer duration diarrhea, dehydration, and even death. 5 , 6 Diarrhea is the second leading cause of mortality in children aged <5 years globally, causing about 1.5 million deaths a year, translating into nearly one in every nine child deaths. 7 , 8
The goal of predictive modeling in healthcare is to identify the likelihood of health events in patients or the population and such efforts guide healthcare providers and policy makers in making preventive strategies and local interventions, which ultimately reduce morbidity and mortality as well as the associated economic and social burden. 9 , 10 The early identification of diarrheal disease and its progression, in the backdrop of limited diagnostic capabilities in the developing world and budgetary constraints, is a key step in assessing and treating diarrheal illness in a cost‐effective manner, while improving quality of care and averting poor outcomes. Predictive models that are data‐driven can synthesize clinical, administrative, and socio‐economic data and complement clinician judgment while providing additional insights. Additionally, the prediction of disease incidence, seasonality, and outbreaks could help public health authorities to understand transmission dynamics and seasonal patterns in advance and aid them in planning and selecting the main response actions thereby enhancing the efficacy and timeliness of local interventions and preventive strategies translating to reduced morbidity and mortality. 11 , 12 , 13
Despite the advancement of predictive modeling in healthcare, there is currently no systematic evaluation, to the best of our knowledge, of the modeling areas and approaches in diarrheal disease. This paper, therefore, focuses on giving a synopsis of existing attempts to predict infectious diarrheal disease in children aged <18 years and their shortcomings. Additionally, we seek to identify topical and methodological gaps in knowledge that can be built upon to ameliorate predictive modeling for infectious diarrheal disease in pediatric populations.
2. QUESTION(S) OF INTEREST
What are the topical and methodological knowledge gaps in in predictive modeling for diarrheal disease in pediatric populations?
3. METHODS
Our study was conducted in line with the Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA) guidelines. 14 The systematic review protocol for this study was developed and iteratively refined with inputs from all study co‐authors. The protocol was registered with the international prospective register of systematic reviews (PROSPERO) (registration number: CRD42021241479): an online repository of systematic review protocols, maintained by the Center for Reviews and Dissemination at the University of York.
We developed a search strategy, limited to PubMed, to retrieve publications on predictive modeling of diarrheal illness between January 1, 1990 and December 31, 2021. The initial search strategy used was: Diarrhea AND (Predict OR Forecast). Two independent reviewers (Billy Ogwel and Gabriel Otieno) evaluated the title and abstract of each retrieved publication to determine the probable pertinence. Pertinence was assessed based on pre‐defined inclusion criteria ensuring that the publication's primary focus addressed some aspect of predictive modeling of diarrheal illness. We included all primary epidemiological study designs conducted in any country. We excluded literature that did not undergo a peer review: gray literature, conference proceedings, dissertations, and posters. The full texts of potentially relevant articles were further examined to make a decision on their inclusion.
Citations of included publications were also reviewed and if a citation was found pertinent but missing from the original search results, the search query was expanded by adding a keyword phrase extracted from the citation so as to include the relevant citation and other similar articles. Additionally, specific keyword phrases were used to narrow our search results by excluding irrelevant articles. The above process was iterated until a comprehensive search was developed.
The final search query used was: (Diarrh* OR gastroenteritis) AND (Predict* OR Forecast OR “risk scoring”) NOT (necrotizing OR appendicitis OR cancer OR ulcer OR IBD OR “Inflammatory bowel disease” OR colitis OR “Eosinophilic Esophagitis” OR Crohn). The following filters were applied to the search results: human subjects, English language, and children (birth to 18 years). The final literature review was done on articles that met the pre‐defined inclusion criteria (prediction of any aspect of infectious diarrhea in pediatric populations). Risk of bias and quality of the studies were not evaluated as this paper was descriptive in nature and no inferences were made based on the validity of the estimates of the performance metrics of the included studies. Difference of opinion about inclusion of articles were resolved by discussions among the two reviewers and where necessary a third reviewer (Bryan O. Nyawanda). One reviewer (Billy Ogwel) extracted the following information from the included articles: aspect of diarrheal illness, country, data used in modeling, modeling technique, time period, and performance of the predictive models. In terms of data analysis, we carried out a narrative synthesis of included publications.
4. RESULTS
A total of 2671 articles from the PubMed search were reviewed. Fifty nine of these appeared to be pertinent after review of titles and abstracts, and underwent full‐text review, where after 38 were established to be relevant and are discussed in this paper (Figure 1). Majority of the studies were conducted in Asia 17 (44.7%) 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 with 8 (47.1%) of them conducted in Bangladesh alone. Eight studies (21.1%), 4 (10.5%), and 2 (5.3%) reported data from Africa, 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 Americas, 40 , 41 , 42 , 43 and Europe, 44 , 45 respectively. Seven studies (17.4%) were multi‐site 46 , 47 , 48 , 49 , 50 , 51 , 52 of which 4 (57.1%) were reported from the Global Enteric Multicenter Study (GEMS). 53 The most common research topics among the studies were disease forecasts 14 (36.8%), vaccine‐related 9 (23.7%), and disease/pathogen detection 5 (13.2%). Majority of these studies were published between 2011 and 2020, 28 (73.7%). The most common technique used in the modeling was machine learning 12 (31.6%) with various algorithms used for the prediction tasks (Table 1).
FIGURE 1.
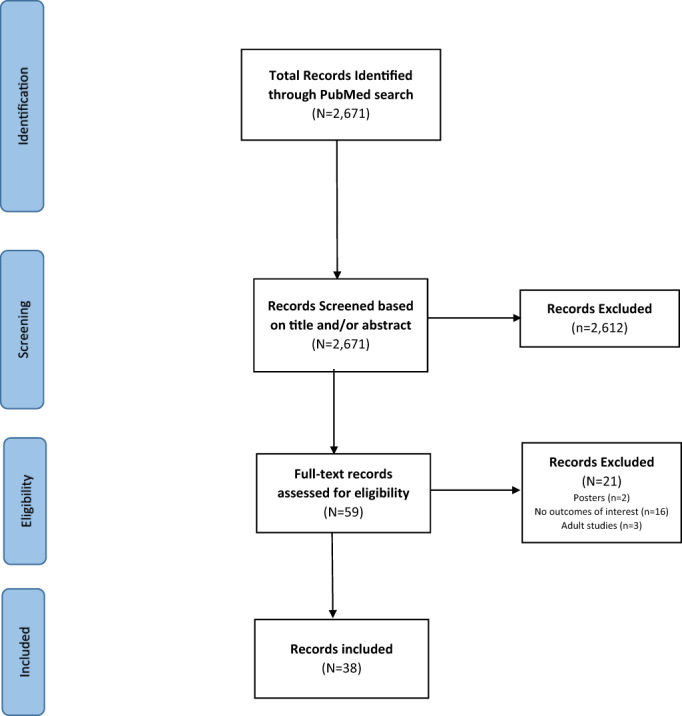
Flowchart of included studies of predictive modeling in diarrheal disease.
TABLE 1.
Characteristics of included studies in predictive modeling for diarrheal disease.
| N = 38 | |
|---|---|
| Characteristics | n (%) |
| Region | |
| Asia | 17 (44.7) |
| Africa | 8 (21.1) |
| Americas | 4 (10.5) |
| Europe | 2 (5.3) |
| Multi‐site | 7 (18.4) |
| Topics | |
| Disease forecasts | 14 (36.8) |
| Vaccine‐related | 9 (23.7) |
| Disease/pathogen detection | 5 (13.2) |
| Outcomes | 4 (10.5) |
| Strain dynamics | 3 (7.9) |
| Determinants of diarrheal disease burden | 2 (5.3) |
| Seasonality | 1 (2.6) |
| Year of publication | |
| 1990–2000 | 1 (2.6) |
| 2001–2010 | 7 (18.4) |
| 2011–2020 | 28 (73.7) |
| 2021 | 2 (5.3) |
| Modeling techniques | |
| Machine learning algorithms | 12 (31.6) |
| SIS‐/SIRS‐like compartmental models a | 6 (15.8) |
| Dynamic, deterministic compartmental models | 5 (13.2) |
| Logistic regression | 4 (10.5) |
| Auto regression Model | 3 (7.9) |
| Multiplicative Holt‐Winters method | 1 (2.6) |
| Linear regression | 1 (2.6) |
| Fourier analysis | 1 (2.6) |
| Fitness models | 1 (2.6) |
| Data assimilation: ensemble Kalfman filter | 1 (2.6) |
| Gravity models | 1 (2.6) |
| Multiple regression models | 1 (2.6) |
| Spatially‐explicit stochastic model | 1 (2.6) |
SIS‐ (susceptible‐infectious‐susceptible)/SIRS‐like (susceptible‐infectious‐recovered‐susceptible) compartmental models.
4.1. Predicting disease forecasts
Much of the existing work on predictive modeling of diarrhea (14 [36.8%]) is focused on predicting diarrheal disease forecasts with five studies focusing on all‐cause diarrhea, 15 , 16 , 17 , 32 , 33 while nine were specific to cholera. 23 , 24 , 25 , 29 , 30 , 31 , 40 , 41 , 42 The prediction of rotavirus‐specific incidence have been incorporated in vaccine impact predictions discussed under the sub‐topic vaccine‐related predictions.
The studies that predicted forecasts of all‐cause diarrhea used data collected from China, 15 , 17 Indonesia, 16 Mali, 32 and Botswana. 33 The modeling techniques used in these predictions were Machine learning (Multiple Linear Regression; Random Forest [RF] Regression; Support Vector (SVM) Regression; Gradient Boosting Regression; Convolutional Neural Network; Neural Network (NN) Regression), Auto regression models (autoregressive integrated moving average [ARIMA/X]; seasonal‐auto‐regressive‐integrated‐moving‐average [SARIMA/X]), compartmental susceptible‐infected‐recovered‐susceptible (SIRS) model, multiplicative Holt‐Winters method, and parsimonious model (PM; Table 2). The data used in the above modeling included: morbidity, meteorological/climate and search indices data. The results from the studies varied: Fang et al. 15 reported that the RF model outperformed the ARIMA/X models with a mean absolute percentage error (MAPE) of approximately 20.0%; Medina et al. 32 realized a MAPE circa 25.0% using the multiplicative Holt‐Winters method; Pangestu et al. 16 reported an accuracy of 78.6% on the SARIMA model; Wang et al. 17 found the PM model to outperform all the other models in three metrics; the SIRS model built by Heaney et al. 33 had a mean Root Mean Square Error (RMSE) and correlation of 0.79 and 0.99, respectively between the observations and simulations across all wet season outbreaks, while across dry season outbreaks, the mean RMSE and correlation were 1.33 and 0.99, respectively (Table 3).
TABLE 2.
Categorization and current status of research topics on predictive modeling for diarrheal disease.
| Primary Category | Sub‐category | Modeling methods for existing predictive models |
|---|---|---|
| Disease forecast | All‐cause diarrhea | Random Forest, autoregressive integrated moving average (ARIMA/X), seasonal‐auto‐regressive‐integrated‐moving‐average (SARIMA/X), multiplicative Holt‐Winters method, compartmental susceptible‐infected‐recovered‐susceptible (SIRS) model, Parsimony Model, gravity models, Multiple Linear Regression, Random Forest Regression, Support Vector Regression, Gradient Boosting Regression, Extreme Gradient Boosting Regression, Convolutional Neural Network, Neural Network Regression |
| Cholera | SIRS‐like models, data assimilation: ensemble Kalfman filter, individual‐based spatially‐explicit stochastic model, logistic regression, SARIMA, model, auto regression model, multiple regression models | |
| Disease/pathogen detection | All‐cause diarrhea | Naïve Bayes, linear discriminant analysis, quadratic discriminant analysis, support vector machine, Artificial Neural Network |
| Viral etiology | Random Forest, logistic regression | |
| Bacterial etiology | Random Forest, logistic regression | |
| Rotavirus | Classification trees | |
| Strain dynamics | Rotavirus | Fourier analysis |
| Norovirus | Fitness models | |
| Shigella | Logistic regression, Neural Network, support vector machines | |
| Outcomes | Dehydration | Logistic regression/recursive partitioning model |
| Malnutrition | Linear regression | |
| Hospitalization | None | |
| Prolonged/persistent diarrhea | None | |
| Mortality | None | |
| Seasonality | Principal‐Component Analysis, K‐means clustering, classification and regression trees | |
| Vaccine | Vaccine impact | SIS‐ (susceptible‐infectious‐susceptible), SIRS‐like compartmental models, ensemble models, dynamic, deterministic compartmental model, periodic regression models, age‐structured compartmental mode |
| Vaccine cost‐effectiveness | Dynamic model | |
| Vaccine hesitancy | Logistic regression, Random Forest, and Neural Networks | |
| Determinants of diarrheal disease burden | Classification and Regression Trees (CART) | |
TABLE 3.
Comprehensive list of included studies in predictive modeling for diarrheal disease.
| Authors | Year of publication | Time period of the study | Country | Aspect of diarrheal disease | Technique used for predictive modeling | Data used (studied variables) | Performance of model |
|---|---|---|---|---|---|---|---|
| Pascual et al. | 2008 | 1966–2005 | Bangladesh | Cholera incidence/outbreaks | TSIR or TSIRS model | Laboratory‐confirmed cholera infection data, year and month of infection, climate data | The authors observed that lack of extreme events between 2001 and 2005 would have been anticipated with 75% confidence half a year ahead with a model fitted to data up to 2000. |
| Pasetto et al. | 2017 | 2010 | Haiti | Cholera incidence/outbreaks | Data assimilation: ensemble Kalfman filter | Laboratory‐confirmed cholera infection data, year and month of infection, rainfall data | The authors showed that the assimilation procedure with the sequential update of the parameters outperformed calibration schemes based on Markov chain Monte Carlo. Moreover, in a forecasting mode, the model predicted the spatial incidence of cholera at least 1 month ahead. |
| Bertuzzo et al. | 2016 | 2010–2017 | Haiti | Cholera incidence/outbreaks | Individual‐based spatially‐explicit stochastic model | Epidemiological dynamics and health‐care practice data | The model captured the timing and the magnitude of the peaks correctly (Nash–Sutcliffe index = 0.79). The authors showed that the probability that the epidemic would go extinct before the end of 2016 was of the order of 1%. |
| Jutla et al. | 2015 | 2002–2010 | Bangladesh | Cholera incidence/outbreaks | Logistic regression | River discharge data, terrestrial water storage (TWS) data, cholera prevalence data | The authors observed that TWS representing had an asymmetrical, strong association with cholera prevalence in the spring (τ = −0.53; P < 0.001) and autumn (τ = 0.45; P < 0.001) up to 6 months in advance. |
| Daisy et al. | 2020 | 2000–2013 | Bangladesh | Cholera incidence/outbreaks | Seasonal‐auto‐regressive‐integrated‐moving‐average (SARIMA) model | Cholera incidence and climatic variables | Root Mean Square Error (RMSE) = 14.7; mean absolute error (MAE) = 11. |
| Bengtsson et al. | 2015 | 2010 | Haiti | Cholera incidence/outbreaks | Gravity models | Case data, mobility data | Area under the Curve (AUC) = 0.79 for mobile phone‐based model |
| Matsuda et al. | 2008 | 1983–2002 | Bangladesh | Cholera incidence/outbreaks | Auto‐regression model | Cholera patient data, climate data | The authors reported a Pearson's correlation coefficient of 0.95 between the monthly number of patients predicted by the model and the actual monthly number of patients. |
| Koepke et al. | 2016 | 2000–2007; 2010–2013 | Bangladesh | Cholera incidence/outbreaks | SIRS model | Cholera case data, environmental variables | The authors showed that their model successfully predicted an increase in the number of infected individuals in the population weeks before the observed number of cholera cases increased. |
| Jutla et al. | 2013 | 1998–2010 | Bangladesh | Cholera incidence/outbreaks | Multiple regression models | Cholera incidence, sewage discharge data, satellite environmental determinants | Accuracy = 75% |
| Levine et al. | 2015 | 2014 | Bangladesh | Dehydration | Logistic regression/recursive partitioning model | Historical, demographic, clinical, and nutritional data | The authors reported an AUC of 0.79 (95% confidence interval [CI] = 0.74–0.84) for severe dehydration and 0.78 (95% CI = 0.74–0.81) for some (any) dehydration for the new DHAKA Dehydration Score. Additionally, their score had a 90% agreement between independent raters, with a Cohen's Kappa of 0.75 (95% CI = 0.66–0.85) among children with a repeat clinical exam. |
| Levine et al. | 2013 | 2010–2012 | Rwanda | Dehydration | Logistic regression | Demographic and clinical data | The authors reported AUCs of 0.72 (95% CI = 0.60–0.85), 0.73 (95% CI = 0.62–0.84), and 0.80 (95% CI = 0.71–0.89) for the WHO severe dehydration scale, CDC scale, and Clinical Dehydration Scale, respectively, in the full cohort. They also showed that only the Clinical Dehydration Scale was a significant predictor of severe disease when used in infants, with an AUC of 0.77 (95% CI = 0.61–0.93). |
| Zodpey et al. | 1999 | 1996–1997 | India | Dehydration | Logistic regression | Demographic and clinical data | The authors reported sensitivity, specificity, positive predictive value, Cohen's kappa, and overall predictive accuracy of 0.81, 0.81, 0.81, 0.61, and 0.86, respectively. |
| Alexander and Blackburn | 2013 | 2006–2009 | Botswana | Determinants of diarrheal disease burden | Cluster analysis/classification and regression trees | Hospital surveillance data | |
| Green et al. | 2009 | 200–2007 | Global | Determinants of diarrheal disease burden | Classification and Regression Trees (CART) | WASH, government spending, literacy levels | Mean squared prediction error (MSE) of 0.225 |
| Fang et al. | 2020 | 2012–2016 | China | Diarrhea incidence | Random Forest, autoregressive integrated moving average (ARIMA/X) | Morbidity and meteorological data | 20% mean absolute percentage error (MAPE) with actual values; 30% MAPE between ARIMAX and ARIMA Model |
| Pangestu et al. | 2020 | 2010–2019 | Indonesia | Diarrhea incidence | Seasonal‐auto‐regressive‐integrated‐moving‐average (SARIMA/X) | Burden of disease estimates and climate data | Accuracy = 78.6% |
| Wang et al. | 2020 | 2012–2016 | China | Diarrhea Incidence | Parsimony Model (PM)/Multiple Linear Regression/Random Forest Regression/Support Vector Regression/Gradient Boosting Regression/Extreme Gradient Boosting Regression/Convolutional Neural Network/Neural Network Regression | Historical outpatient visit counts, meteorological factors (MF) and Baidu search indices (BSI) | The authors observed that the PM model obtained the best performance in terms of three metrics benefiting from MF and BSI data. |
| Medina et al. | 2007 | 1996–2004 | Mali | Diarrhea incidence/seasonality | Multiplicative Holt‐Winters method | Clinical data and climate data | MAPE circa 25%. |
| Heaney et al. | 2020 | 2007–2017 | Botswana | Diarrheal incidence/outbreaks | Compartmental susceptible‐infected‐recovered‐susceptible (SIRS) model | Hospital surveillance data | The authors reported that the average RMSE and correlation between the observations and simulations across all wet season outbreaks was 0.79 and 0.99, respectively. Similarly, they reported an average RMSE and correlation across dry season outbreaks as 1.33 and 0.99, respectively. |
| Maniruzzaman et al. | 2020 | 2014 | Bangladesh | Diarrheal infection | naïve Bayes/linear discriminant analysis/quadratic discriminant analysis/support vector machine | Demographic health survey | Support Vector Machine (SVM) with radial basis kernel yielded 65.61% accuracy, 66.27% sensitivity, and 52.28% specificity. |
| Abubakar and Olatunji | 2019 | 2013 | Nigeria | Diarrheal infection | Artificial Neural Network | Demographic and health survey data | High accuracy of 95.78 and 95.63% during training and testing phases |
| Brander et al. | 2019 | 2007–2011 | The Gambia, Mali, Mozambique, Kenya, Pakistan, India, Bangladesh | Malnutrition | Linear regression | Clinical, historical, anthropometric | AUC of 0.67 (95% CI = 0.64–0.69) |
| Suzuki et al. | 2016 | 2006–2015 | Japan | Norovirus strain dynamics | Fitness models | Sequence data, year and month of isolation | The authors showed that their model predicted GII.3 and GII.4 would contract, whereas GII.17 would expand and predominate in the 2015–2016 season. |
| Garbern et al. | 2021 | Mali and Bangladesh | Pathogen detection/clinical profiles/viral etiology | Random Forest/logistic regression | Clinical, historical, anthropometric and microbiologic data | AUC of 0.754 (0.665–0.843) | |
| Brintz et al. | 2020 | 2007–2011 | The Gambia, Mali, Mozambique, Kenya, Pakistan, India, Bangladesh | Pathogen detection/clinical profiles/viral etiology/Bacterial etiology | Random Forest/logistic regression | Clinical, historical, anthropometric and microbiologic data | AUC = 0.825; specificity = 0.85; sensitivity = 0.59, negative predictive value (NPV) = 0.82; positive predictive value (PPV) = 0.64 |
| Ayers et al. | 2016 | 2007–2011 | Kenya | Pathogen detection/clinical profiles/Rotavirus | Classification trees | Clinical, historical, anthropometric and microbiologic data | AUC = 0.816 on training: AUC = 0.6125 on test data |
| Pitzer et al. | 2011 | 1985–2009* | Italy, Hungary, Spain, Japan, United States, Australia | Rotavirus strain dynamics | Fourier analysis | Laboratory‐confirmed rotavirus infection data, sequence data, vaccination | The authors showed that their model explained the coexistence and cyclical pattern in the distribution of genotypes observed in most developed countries: predominant rotavirus strains cycle with periods (T) ranging from 3 to 11 years |
| Chao et al. | 2019 | 2007–2011 | The Gambia, Mali, Mozambique, Kenya, Pakistan, India, Bangladesh | Seasonality | Principal‐Component Analysis/K‐means clustering | Microbiological and weather data | The authors observed that rotavirus was most prevalent during the drier “winter” months and out of phase with bacterial pathogens, which peaked during hotter and rainier times of year corresponding to “monsoon,” “rainy,” or “summer” seasons. |
| Adamker et al. | 2018 | 2002–2015 | Israel | Shigella species/Outcomes | Logistic Regression (LR), Neural Network (NN), and Support Vector Machines (SVM) | National Shigella data as collated by the Ministry of Health (MoH) Division of Epidemiology | Accuracy of 93.2% (Shigella species) and 94.9% (hospitalization) |
| Freiesleben de Blasio et al. | 2014 | Kazakhstan | Vaccine cost‐effectiveness | Dynamic model | The authors reported that a vaccination program with 90% coverage would prevent ≈880 rotavirus deaths and save an average of 54,784 life‐years for children <5 years of age. They also showed that Indirect protection accounted for 40% and 60% reduction in severe and mild rotavirus gastroenteritis, respectively | ||
| Bar‐Lev et al. | 2021 | 2014–2018 | Israel | Vaccine hesitancy | Logistic regression, Random Forest and Neural Networks | Demographic, clinical, socio‐economic data, vaccination, social media traffic | The authors observed that the performance of models for Rotavirus, Hepatitis A and Hepatitis B, were close to random (accuracy <0.63 and F1 < 0.65). Additionally, they reported a negative association between on‐line discussions and vaccination. |
| de Blasio et al. | 2010 | 2005–2008 | Kyrgyzstan | Vaccine impact | Deterministic age‐structured dynamic model | Key features of rotavirus epidemiology, rotavirus associated events (death, hospitalization, outpatient visits), vaccination | The authors reported that a routine rotavirus vaccination program at 95% coverage and 54% effectiveness against severe infection was estimated to lead to a 56% reduction in rotavirus‐associated deaths and a 50% reduction in hospital admissions, while outpatient visits and homecare episodes would decrease by 52% compared to baseline levels after 5 years of intervention. |
| Atchison et al. | 2010 | 1998–2007 | England and Wales | Vaccine impact/seasonality | Deterministic age‐structured dynamic model | Key features of rotavirus epidemiology, vaccination | The authors showed that their model reproduced the strong seasonal pattern and age distribution of rotavirus disease observed in England and Wales. Furthermore, they observed that their model predicted that vaccination would provide both direct and indirect protection within the population resulting in 61% reduction of rotavirus disease incidence. |
| Park et al. | 2017 | 2009–2012 | Niger | Vaccine Impact/transmission dynamics | Susceptible‐infected‐recovered (SIR)‐like compartmental models/Ensemble models | Clinic admissions data and healthcare seeking data | The authors reported that their model predicted the current burden of severe rotavirus disease to be 2.6%–3.7% of the population each year and that a two‐dose vaccine schedule achieving 70% coverage could reduce burden by 39%–42%. |
| Pitzer et al. | 2012 | 1999–2009 | England and Wales | Vaccine impact/transmission dynamics | SIS‐ (susceptible‐infectious‐susceptible)/SIRS‐like (susceptible‐infectious‐recovered‐susceptible) compartmental models | Laboratory‐confirmed rotavirus infection data | The authors showed that their models predicted that during the initial year after vaccine introduction, incidence of severe Rotavirus gastroenteritis (RVGE) would be reduced 1.8–2.9 times more than expected from the direct effects of the vaccine alone (28%–50% at 90% coverage), but over a 5‐year period following vaccine introduction severe RVGE would be reduced only by 1.1–1.7 times more than expected from the direct effects (54%–90% at 90% coverage). They also reported that projections for the long‐term reduction of severe RVGE ranged from a 55% reduction at full coverage to elimination with at least 80% coverage. |
| Effelterre et al. | 2009 | France, Germany, Italy, Spain and the United Kingdom | Vaccine impact/transmission dynamics | Dynamic, deterministic compartmental model | Burden of disease estimates: hospitalizations, emergency‐room visits and primary‐care visits | The authors reported that with vaccination coverage rates of 70%, 90%, and 95% their model predicted that, in addition to the direct effect of vaccination, herd protection induced a reduction in RV‐related gastroenteritis (GE) incidence of 25%, 22%, and 20%, respectively, for RV‐GE of any severity, and of 19%, 15%, and 13%, respectively, for moderate‐to‐severe RV‐GE, 5 years after implementation of a vaccination program. | |
| Asare et al. | 2020 | 2007–2015 | Ghana | Vaccine impact/transmission dynamics | SIRS‐like model | Epidemiological data and vaccination data | The authors showed that their model captured the spatio‐temporal variations in rotavirus incidence across the three sites and showed good agreement with the age distribution of observed cases |
| Olson et al. | 2020 | 2002–2016 | United States | Vaccine impact/transmission dynamics | Periodic regression models/ age‐structured compartmental mode | Case data, Emergency department (ED) visits data, hospitalization data, vaccination data | The authors reported that their published mechanistic model qualitatively predicted patterns more than 2 years in advance. |
The studies that focused on the prediction of cholera forecasts utilized data from two countries: six from Bangladesh 23 , 24 , 25 , 29 , 30 , 31 and three from Haiti. 40 , 41 , 42 The modeling techniques employed by these studies were SIRS models, auto regression models, logistic regression (LR), multiple regression, gravity models, spatially‐explicit stochastic model, and data assimilation: ensemble Kalfman filter. The modeling was based on the following data: morbidity, climate, environmental, mobility, sewage discharge, terrestrial water storage, and satellite data. The results were diverse: Jutla et al. 31 reported an accuracy of 75.0% on the multiple regression model; Bengtsson et al. 42 reported an Area Under the Curve (AUC) of 79.0% on the mobile phone‐based model; Pascual et al. 23 got an accuracy of 75.0% on their SIRs model; Pasetto et al. 40 reported that the assimilation procedure outperformed other calibration schemes and it was able to forecast the spatial incidence of cholera at least 1 month ahead; Bertuzzo et al. 41 using a spatially‐explicit stochastic model observed an overall good agreement with the data, capturing the timing and the magnitude of the peaks correctly (Nash–Sutcliffe index = 0.79); Daisy et al. 25 achieved the best fit in the model using climate data (RMSE = 14.7, Mean of Absolute value of Errors [MAE] = 11); Matsuda et al. 29 achieved a correlation of 0.95 between simulations and actual number of disease.
4.2. Predicting vaccine‐related topics
Rotavirus is a leading cause of diarrheal morbidity and mortality. 1 , 54 Consequently, rotavirus vaccines have been incorporated into national immunization programs following recommendations by the World Health Organization (WHO). 55 Nine studies focused on predictions around rotavirus vaccine with seven studies focusing on vaccine impact, 28 , 35 , 38 , 43 , 44 , 45 , 47 one on vaccine hesitancy, 27 and one on vaccine cost‐effectiveness 19 (Table 3).
The seven studies on vaccine impact used data from Niger, 35 Ghana, 38 United States, 43 Kyrgyzstan, 28 England and Wales, 44 , 45 and a multi‐site study (France, Germany, Italy, Spain, and the United Kingdom). 47 These studies built predictive models using SIRs models, periodic regression models and deterministic age‐structured dynamic model (Table 2). The models used the following data: Key features of rotavirus epidemiology, rotavirus‐associated events (death, hospitalization, outpatient visits), vaccination, and healthcare seeking data. The results were comparable: Park et al. 35 predicted a 39%–42% reduction on burden at a 70.0% vaccine coverage; Pitzer et al. 44 predicted that incidence of severe disease would be reduced 1.1‐1.7 times more than expected from the direct effects (54%–90.0% at 90.0% coverage) 5 years after roll‐out; Effelterre et al. 47 reported that with vaccination coverage rates of 70.0%, 90.0%, and 95.0% the model predicted that, in addition to the direct effect of vaccination, herd protection induced a reduction in disease incidence (any severity) of 25.0%, 22.0%, and 20.0%, respectively, and of 19.0%, 15.0%, and 13.0%, respectively, for moderate‐to‐severe disease, 5 years after roll‐out; De Blasio et al. 28 predicted a reduction of 56.0%, 50.0%,52.0% for deaths, admissions, and outpatient visits, respectively, 5 years after vaccine roll‐out at a 95.0% coverage and 54.0% effectiveness; Atchison et al. 45 predicted a 61.0% reduction in incidence (Table 3).
Freiesleben de Blasio et al. 19 predicted cost‐effectiveness of rotavirus vaccine in Kazakhstan using a dynamic model. They reported that at a 90.0% coverage, ≈880 rotavirus deaths would be averted and an average of 54,784 life‐years for children <5 years of age would be saved in a 20‐year period. Additionally, they found 40.0% and 60.0% reduction in severe and mild rotavirus gastroenteritis due to indirect protection, respectively. Bar‐Lev et al. 27 predicted vaccine hesitancy in Israel using machine learning algorithms (LR, RF, and NN). The performances of all algorithms were close to random in this study.
4.3. Predicting disease/pathogen detection
Five studies focused on disease/pathogen detection: two all‐cause diarrhea 18 , 39 ; one viral‐only etiology 49 ; one viral/bacterial etiology 48 ; one rotavirus. 37 Other than the rotavirus model, there are no other existing pathogen‐specific prediction models. Two studies focused on the prediction of diarrheal infection. Both studies used machine learning algorithms in the modeling of demographic health survey data in Bangladesh 18 and Nigeria, 39 respectively. Maniruzzaman et al. 18 reported the SVM model with radial basis kernel outperformed other models yielding 65.6% accuracy, 66.3% sensitivity, and 52.3% specificity. Abubakar and Olatunji 39 reported an accuracy of 95.6% on their artificial neural network model.
Brintz et al. 48 used data from the GEMS study to predict both viral and bacterial etiologies in diarrhea utilizing RF and LR models with the following results: AUC = 82.5%; specificity = 85.0%; sensitivity = 59.0%; Negative Predictive Value (NPV) = 82.0%; Positive Predictive Value (PPV) = 64.0%. Garbern et al. 49 predicted viral‐only etiology deploying machine learning algorithms (RF and LR regression) on data from Mali and Bangladesh yielding 75.4% AUC. Ayers 37 used classification trees on the GEMS Kenya data to predict rotavirus infection realizing 61.3% AUC.
4.4. Predicting diarrheal outcomes
There were only four studies focused on diarrheal outcomes; three on dehydration 20 , 21 , 36 and one on linear growth faltering. 51 Despite risk factors for death, 56 , 57 , 58 , 59 , 60 hospitalization, 61 , 62 , 63 , 64 and prolonged/persistent diarrhea 65 , 66 , 67 , 68 being documented, there were no predictive models developed on these outcomes. The three publications on dehydration utilized data from Bangladesh, 20 Rwanda, 36 and India. 21 The primary modeling technique in all the studies was LR with historical, demographic, clinical, and nutritional data being used. The new DHAKA Dehydration Score developed by Levine et al. 20 yielded AUC of 79.0% and 78.0%, for severe and some dehydration, respectively. Levine et al. 36 also reported AUC of 72.0%, 73.0%, and 80.0% for the WHO severe dehydration scale, Centers for Disease Control and Prevention (CDC) scale, and Clinical Dehydration Scale, respectively. In the model developed by Zodpey et al., 21 the sensitivity, specificity, PPV, Cohen's kappa, and overall predictive accuracy were 81.0%, 81.0%, 81.0%, 61.0%, and 86.0%, respectively. Brander et al. 51 used the GEMS data to predict linear growth faltering based on a LR model that yielded 67.0% AUC.
4.5. Other research topics
Strain dynamics was addressed in three studies with rotavirus, 52 norovirus, 22 and Shigella 26 being the only pathogens investigated. Pitzer et al. 52 used Fourier analysis to predict rotavirus strain dynamics based on multi‐site data (Italy, Hungary, Spain, Japan, United States, Australia). Their model explained the coexistence and cyclical patterns in the distribution of genotypes observed in most developed countries: predominant rotavirus strains cycle with periods ranging from 3 to 11 years. Suzuki et al. 22 predicted norovirus strain dynamics in Japan using fitness models. They reported that the model was effective in predicting the direction of change in the proportions of genotypes, it predicted that GII.3 and GII.4 would contract, whereas GII.17 would expand and predominate in the 2015‐2016 season. Adamker et al. 26 predicted Shigella species among Shigellosis patients in Israel using machine learning algorithms (LR, NN, and SVM) achieving an accuracy of 93.2%.
There were two studies on determinants of diarrheal disease. Alexander and Burn 34 used cluster analysis and Classification and Regression Tree (CART) models to evaluate patient attributes by outbreak in Botswana. They identified two main clusters associated with patient age while neither village nor outbreak had an influence on cluster. Furthermore, CART identified sex and hospitalization as predictors of diarrhea. Additionally, water shortages and water quality deficiencies were identified in both outbreaks. Green et al. 50 also used a CART model to establish determinants of diarrheal illness at a country‐level. Improvements in rural sanitation was identified as the most important predictor for reducing diarrhea. They predicted that a 65% global reduction in unmet rural sanitation would save 1.2 million lives annually.
Finally, Chao et al. 46 used machine learning algorithms (Principal‐Component Analysis and K‐means clustering) to characterize seasonality of different pathogens from GEMS data. The key findings from the study was that rotavirus was most prevalent during the dry “winter” months and out of phase with bacterial pathogens, which peaked during hotter and rainier times of year corresponding to “monsoon,” “rainy,” or “summer” seasons.
5. DISCUSSION
Notable effort has been made in predictive modeling for infectious diarrhea in pediatric populations. The key findings from our review are: (1) Diarrheal outcomes remain under researched; (2) with the shift in the landscape of diarrheal pathogens post rotavirus vaccine introduction, many pathogen‐specific areas remain open for exploration including disease forecasts, disease detection strain dynamics and vaccine‐related predictions; (3) the need for model ensembles to gauge and mitigate structural uncertainties in predictions problems; (4) need for more diverse, neoteric, and pertinent data to improve the precision and robustness of prediction models; (5) the lack of consistency on reporting of predictive models despite available guidelines highlighting the need for common data standards need for common data standards and adherence to guidelines on reporting of predictive models for biomedical research.
Despite the advances that have been made in predictive modeling, a number of topical areas remain open for further research. Notwithstanding the wide array of poor diarrheal outcomes and their public health impact, only four studies exist focusing on dehydration and linear growth faltering. Death, prolonged/persistent diarrhea, and malnutrition still remain uninvestigated. Furthermore, with the change in the landscape of diarrheal etiologies after rotavirus vaccine introduction, 69 , 70 other pathogens (Escherichia coli, Cryptosporidium, and Shigella) have emerged as important etiological agents of moderate‐to‐severe diarrhea in these settings 2 potentially creating the need for pathogen‐specific models in the areas of disease forecasts, disease detection, and strain dynamics. Additionally, with pipeline vaccines advancing for other enteric pathogens like cholera, Shigella, Enterotoxigenic E. coli, 71 , 72 vaccine‐related predictions would be useful for these vaccines in understanding the comprehensive epidemiological impact of vaccination and providing insights into the prospective cost‐effectiveness of vaccination by forecasting vaccine‐induced changes in the epidemiology of diarrheal disease in the population over time. 73
Although diverse and dynamic modeling approaches have been used so far, a number of methodological propositions could help to further improve the predictive modeling for diarrheal disease. A diversity of model structures should be considered in order to gauge structural uncertainties in prediction/forecasting. To enhance the characterization of forecasting uncertainty, model ensembles that aggregate a range of model structures and their individual uncertainty should be increasingly embraced. 74 In addition, since model parameters in forecasting may be subject to uncertainties, approaches such as Bayesian methodologies, which combine uncertainties and expert knowledge through choice of prior probabilities, have become prominent and could be utilized. 75 Furthermore, with the complexity and rise of data in healthcare, 76 , 77 we propose the increased adoption of machine learning algorithms in prediction tasks as they have been shown to provide additional information and understanding of the data beyond using standard statistical approaches. 37 , 78 Finally, different models reported different metrics for the same prediction task as well as incompleteness in the reporting of elements of the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) statement and there is need to define common data standards and adherence to developed guidelines on reporting of predictive models in biomedical research 79 to make comparisons across multiple models possible.
With regards to data sources for disease forecasts, adding more diverse, recent, and pertinent data will improve the precision and robustness of such models. To better predict new diarrheal events and to inform spatial spread, incorporating population density and spatial data into the modeling is vital. 80 This can gain ascendancy with the availability of novel universal geospatial datasets such as WorldPop and LandScan. Furthermore, understanding spread of diarrhea could be better understood by integrating remote sensing and satellite data, healthcare capacity, and human mobility data in the models. Additionally, we recommend use of data from multi‐site studies, where available, as they are larger, richer, and have geographical variance that could help to mitigate algorithmic bias that arises from sample and prejudice biases. 81 More recent data are also needed in the prediction of seasonality of pathogens since rotavirus vaccine has been shown to affect seasonality of rotavirus gastroenteritis 82 and climate change caused by increased global warming has also been shown to alter the distribution of infectious disease vectors and the seasonal distribution of some allergenic pollen species. 83 , 84 Finally, post‐vaccination data from diverse countries is needed to improve projections of long‐term impact of rotavirus vaccination by conducting further validation. 44
6. LIMITATION
There were several limitations with this review. First, by limiting this systematic review to PubMed, we may have missed pertinent articles not indexed within PubMed. Second, by filtering‐out articles not written in English, other predictive models for diarrheal illness may have been missed. Additionally, this review did not conduct risk of bias and quality assessment of the studies, hence bias in primary studies may have been reported leading to inaccuracy of findings. However, we did not draw any inference based on the validity of the estimates of the performance metrics of the included studies. Furthermore, the comparison of statistical measures of predictions could not be directly made across models as the outcomes being predicted were different.
7. CONCLUSION
As the first systematic review on predictive modeling for diarrheal illness, we observed substantial effort to predict various aspects of diarrheal disease. However, many topical and methodological problems remain open and there is significant scope for improvement in the predictive modeling of diarrhea. Future research in predictive modeling for diarrheal illness should seek to address them to realize more comprehensive, robust, and precise models.
AUTHOR CONTRIBUTIONS
Billy Ogwel and Bryan O. Nyawanda conceived the study. Billy Ogwel, Bryan O. Nyawanda, Richard Omore, Gabriel Otieno, and Vincent Mzazi contributed to study design and implementation. Billy Ogwel conducted literature search and review, and drafted the manuscript, and all authors critically reviewed the manuscript for intellectual content and approved the final manuscript. Bryan O. Nyawanda and Gabriel Otieno also participated in literature search. All authors read and approved the final manuscript.
FUNDING INFORMATION
The authors used their own resources.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
Not applicable.
ACKNOWLEDGMENTS
The authors acknowledge Alex Ondeng for his support of this project.
Ogwel B, Mzazi V, Nyawanda BO, Otieno G, Omore R. Predictive modeling for infectious diarrheal disease in pediatric populations: A systematic review. Learn Health Sys. 2024;8(1):e10382. doi: 10.1002/lrh2.10382
REFERENCES
- 1. Troeger C, Blacker BF, Khalil IA, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1211‐1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. WHO . Diarrhoeal Disease. 2017. https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease. Accessed November 28, 2020.
- 3. Mills A. Health care systems in low‐ and middle‐income countries. N Engl J Med. 2014;370(6):552‐557. doi: 10.1056/NEJMra1110897 [DOI] [PubMed] [Google Scholar]
- 4. Mokomane M, Kasvosve I, de Melo E, Pernica JM, Goldfarb DM. The global problem of childhood diarrhoeal diseases: emerging strategies in prevention and management. Ther Adv Infect Dis. 2018;5(1):29‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pavlinac PB, Brander RL, Atlas HE, John‐Stewart GC, Denno DM, Walson JL. Interventions to reduce post‐acute consequences of diarrheal disease in children: a systematic review. BMC Public Health. 2018;18(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitra AK, Khan MR, Alam AN. Complications and outcome of disease in patients admitted to the intensive care unit of a diarrhoeal diseases hospital in Bangladesh. Trans R Soc Trop Med Hyg. 1991;85(5):685‐687. [DOI] [PubMed] [Google Scholar]
- 7. CDC . Global Diarrhea Burden|Global Water, Sanitation and Hygiene|Healthy Water|CDC. 2018. https://www.cdc.gov/healthywater/global/diarrhea-burden.html. Accessed November 25, 2020
- 8. Wardlaw T, Salama P, Brocklehurst C, Chopra M, Mason E. Diarrhoea: why children are still dying and what can be done. Lancet. 2010;375(9718):870‐872. [DOI] [PubMed] [Google Scholar]
- 9. Javaid M, Haleem A, Pratap Singh R, Suman R, Rab S. Significance of machine learning in healthcare: Features, pillars and applications. Int J Intell Netw. 2022;3:58‐73. [Google Scholar]
- 10. Leisman DE, Harhay MO, Lederer DJ, et al. Development and reporting of prediction models: guidance for authors from editors of respiratory, sleep, and critical care journals. Crit Care Med. 2020;48(5):623‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang W, Zhang J, Ma R. The prediction of infectious diseases: a bibliometric analysis. Int J Environ Res Public Health. 2020;17(17):6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yadav SK, Akhter Y. Statistical modeling for the prediction of infectious disease dissemination with special reference to COVID‐19 spread. Front Public Health. 2021;9:645405. https://www.frontiersin.org/article/10.3389/fpubh.2021.645405. Accessed May 21, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Houlihan CF, Whitworth JA. Outbreak science: recent progress in the detection and response to outbreaks of infectious diseases. Clin Med. 2019;19(2):140‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;29(372):n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fang X, Liu W, Ai J, et al. Forecasting incidence of infectious diarrhea using random forest in Jiangsu Province, China. BMC Infect Dis. 2020;20(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pangestu CJ, Piantari E, Munir M. Prediction of diarrhea sufferers in Bandung with seasonal autoregressive integrated moving average (SARIMA). J Comput Soc. 2020;1(1):61‐79. [Google Scholar]
- 17. Wang Z, Huang Y, He B, Luo T, Wang Y, Fu Y. Short‐term infectious diarrhea prediction using weather and search data in Xiamen, China. Sci Program. 2020;2020:e8814222. [Google Scholar]
- 18. Md M, Islam S, Abedin M, Md A, Hussain S. Prediction of childhood diarrhea in Bangladesh using machine learning approach. Insights Biomed Res. 2020;4(1):111‐116. https://www.scholars.direct/Articles/biomedical‐research/ibr‐4‐021.php?jid=biomedical‐research. Accessed May 7, 2022 [Google Scholar]
- 19. Freiesleben de Blasio B, Flem E, Latipov R, Kuatbaeva A, Kristiansen IS. Dynamic modeling of cost‐effectiveness of rotavirus vaccination, Kazakhstan. Emerg Infect Dis. 2014;20(1):29‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levine AC, Glavis‐Bloom J, Modi P, et al. Empirically derived dehydration scoring and decision tree models for children with diarrhea: assessment and internal validation in a prospective cohort study in Dhaka, Bangladesh. Glob Health Sci Pract. 2015;3(3):405‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zodpey S, Deshpande S, Ughade S, Kulkarni S, Shrikhande S, Hinge A. A prediction model for moderate or severe dehydration in children with diarrhoea. J Diarrhoeal Dis Res. 1999;1(17):10‐16. [PubMed] [Google Scholar]
- 22. Suzuki Y, Doan YH, Kimura H, Shinomiya H, Shirabe K, Katayama K. Predicting genotype compositions in norovirus seasons in Japan. Microbiol Immunol. 2016;60(6):418‐426. [DOI] [PubMed] [Google Scholar]
- 23. Pascual M, Chaves LF, Cash B, Rodó X, Yunus M. Predicting endemic cholera: the role of climate variability and disease dynamics. Climate Res. 2008;36(2):131‐140. [Google Scholar]
- 24. Jutla A, Akanda A, Unnikrishnan A, Huq A, Colwell R. Predictive time series analysis linking Bengal cholera with terrestrial water storage measured from gravity recovery and climate experiment sensors. Am J Trop Med Hyg. 2015;93(6):1179‐1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daisy SS, Saiful Islam AKM, Akanda AS, Faruque ASG, Amin N, Jensen PKM. Developing a forecasting model for cholera incidence in Dhaka megacity through time series climate data. J Water Health. 2020;18(2):207‐223. [DOI] [PubMed] [Google Scholar]
- 26. Adamker G, Holzer T, Karakis I, et al. Prediction of shigellosis outcomes in Israel using machine learning classifiers. Epidemiol Infect. 2018;146(11):1445‐1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bar‐Lev S, Reichman S, Barnett‐Itzhaki Z. Prediction of vaccine hesitancy based on social media traffic among Israeli parents using machine learning strategies. Isr J Health Policy Res. 2021;10(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Blasio BF, Kasymbekova K, Flem E. Dynamic model of rotavirus transmission and the impact of rotavirus vaccination in Kyrgyzstan. Vaccine. 2010;28(50):7923‐7932. [DOI] [PubMed] [Google Scholar]
- 29. Matsuda F, Ishimura S, Wagatsuma Y, et al. Prediction of epidemic cholera due to vibrio cholerae O1 in children younger than 10 years using climate data in Bangladesh. Epidemiol Infect. 2008;136(1):73‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koepke AA, Longini IM Jr, Halloran ME, Wakefield J, Minin VN. Predictive modeling of cholera outbreaks in Bangladesh. Ann Appl Stat. 2016;10(2):575‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jutla AS, Akanda AS, Islam S. A framework for predicting endemic cholera using satellite derived environmental determinants. Environ Model Software. 2013;47:148‐158. [Google Scholar]
- 32. Medina DC, Findley SE, Guindo B, Doumbia S. Forecasting non‐stationary diarrhea, acute respiratory infection, and malaria time‐series in Niono, Mali. PloS One. 2007;2(11):e1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heaney A, Alexander KA, Shaman J. Ensemble forecast and parameter inference of childhood diarrhea in Chobe District, Botswana. Epidemics. 2020;30:100372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexander K, Blackburn J. Overcoming barriers in evaluating outbreaks of diarrheal disease in resource poor settings: assessment of recurrent outbreaks in Chobe District, Botswana. BMC Public Health. 2013;13(1):775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park J, Goldstein J, Haran M, Ferrari M. An ensemble approach to predicting the impact of vaccination on rotavirus disease in Niger. Vaccine. 2017;35(43):5835‐5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levine AC, Munyaneza RM, Glavis‐Bloom J, et al. Prediction of severe disease in children with diarrhea in a resource‐limited setting. PloS One. 2013;8(12):e82386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ayers TL. Machine learning approaches for assessing moderate‐to‐severe diarrhea in children < 5 years of age, rural western Kenya 2008–2012. 2016. https://scholarworks.gsu.edu/cgi/viewcontent.cgi?article=1008&context=sph_diss
- 38. Asare EO, Al‐Mamun MA, Armah GE, et al. Modeling of rotavirus transmission dynamics and impact of vaccination in Ghana. Vaccine. 2020;38(31):4820‐4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abubakar IR, Olatunji SO. Computational intelligence‐based model for diarrhea prediction using demographic and health survey data. Soft Comput. 2020;24(7):5357‐5866. [Google Scholar]
- 40. Pasetto D, Finger F, Rinaldo A, Bertuzzo E. Real‐time projections of cholera outbreaks through data assimilation and rainfall forecasting. Adv Water Resour. 2017;108:345‐356. [Google Scholar]
- 41. Bertuzzo E, Finger F, Mari L, Gatto M, Rinaldo A. On the probability of extinction of the Haiti cholera epidemic. Stoch Environ Res Risk Assess. 2016;30(8):2043‐2455. [Google Scholar]
- 42. Bengtsson L, Gaudart J, Lu X, et al. Using mobile phone data to predict the spatial spread of cholera. Sci Rep. 2015;5(1):8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olson DR, Lopman BA, Konty KJ, et al. Surveillance data confirm multiyear predictions of rotavirus dynamics in New York City. Sci Adv. 2020;6(9):eaax0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pitzer VE, Atkins KE, de Blasio BF, et al. Direct and indirect effects of rotavirus vaccination: comparing predictions from transmission dynamic models. PloS One. 2012;7(8):e42320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Atchison C, Lopman B, Edmunds WJ. Modelling the seasonality of rotavirus disease and the impact of vaccination in England and Wales. Vaccine. 2010;28(18):3118‐3126. [DOI] [PubMed] [Google Scholar]
- 46. Chao DL, Roose A, Roh M, Kotloff KL, Proctor JL. The seasonality of diarrheal pathogens: a retrospective study of seven sites over three years. PLoS Negl Trop Dis. 2019;13(8):e0007211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Effelterre TV, Soriano‐Gabarró M, Debrus S, Newbern EC, Gray J. A mathematical model of the indirect effects of rotavirus vaccination. Epidemiol Infect. 2010;138(6):884‐897. [DOI] [PubMed] [Google Scholar]
- 48. Brintz BJ, Howard JI, Haaland B, et al. Clinical predictors for etiology of acute diarrhea in children in resource‐limited settings. PLoS Negl Trop Dis. 2020;14(10):e0008677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garbern SC, Nelson EJ, Nasrin S, et al. External validation of a mobile clinical decision support system for diarrhea etiology prediction in children: a multicenter study in Bangladesh and Mali. eLife. 2022;11:e72294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Green ST, Small MJ, Casman EA. Determinants of national diarrheal disease burden. Environ Sci Technol. 2009;43(4):993‐999. [DOI] [PubMed] [Google Scholar]
- 51. Brander RL, Pavlinac PB, Walson JL, et al. Determinants of linear growth faltering among children with moderate‐to‐severe diarrhea in the global enteric multicenter study. BMC Med. 2019;17(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pitzer VE, Patel MM, Lopman BA, Grenfell BT, Viboud C, Parashar UD. Modeling rotavirus strain dynamics in developed countries to understand the potential impact of vaccination on genotype distributions. PNAS. 2011;108:19353‐19358. https://www.pnas.org/doi/abs/10.1073/pnas.1110507108. Accessed May 17, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Levine MM, Kotloff KL, Nataro JP, Muhsen K. The global enteric multicenter study (GEMS): impetus, rationale, and genesis. Clin Infect Dis. 2012;55(Suppl 4):S215‐S224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aliabadi N, Antoni S, Mwenda JM, et al. Global impact of rotavirus vaccine introduction on rotavirus hospitalisations among children under 5 years of age, 2008–16: findings from the global rotavirus surveillance network. Lancet Glob Health. 2019;7(7):e893‐e903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. World Health Organization . Rotavirus vaccines: an update. 2009. https://apps.who.int/iris/bitstream/handle/10665/241486/WER8451_52_533-537.PDF. Accessed May 26, 2022
- 56. Levine MM, Nasrin D, Acácio S, et al. Diarrhoeal disease and subsequent risk of death in infants and children residing in low‐income and middle‐income countries: analysis of the GEMS case‐control study and 12‐month GEMS‐1A follow‐on study. Lancet Glob Health. 2020;8(2):e204‐e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Reilly CE, Jaron P, Ochieng B, et al. Risk factors for death among children less than 5 years old hospitalized with diarrhea in rural western Kenya, 2005‐2007: a cohort study. PLoS Med. 2012;9(7):e1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Acácio S, Mandomando I, Nhampossa T, et al. Risk factors for death among children 0–59 months of age with moderate‐to‐severe diarrhea in Manhiça district, southern Mozambique. BMC Infect Dis. 2019;19(1):322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Griffin PM, Ryan CA, Nyaphisi M, Hargrett‐Bean N, Waldman RJ, Blake PA. Risk factors for fatal diarrhea: a case‐control study of African children. Am J Epidemiol. 1988;128(6):1322‐1329. [DOI] [PubMed] [Google Scholar]
- 60. Uysal G, Sökmen A, Vidinlisan S. Clinical risk factors for fatal diarrhea in hospitalized children. Indian J Pediatr. 2000;67(5):329‐333. [DOI] [PubMed] [Google Scholar]
- 61. Tornheim JA, Manya AS, Oyando N, et al. The epidemiology of hospitalization with diarrhea in rural Kenya: the utility of existing health facility data in developing countries. Int J Infect Dis. 2010;14(6):e499‐e505. [DOI] [PubMed] [Google Scholar]
- 62. Colombara DV, Faruque AS, Cowgill KD, Mayer JD. Risk factors for diarrhea hospitalization in Bangladesh, 2000–2008: a case‐case study of cholera and shigellosis. BMC Infect Dis. 2014;14:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. de Moraes Vanderlei LC, da Silva GAP, Braga JU. Risk factors for hospitalization due to acute diarrhea in children under two years old: a case‐control study. Cad Saude Publica. 2003;19(2):455‐463. [DOI] [PubMed] [Google Scholar]
- 64. Khalili B, Shahabi G, Khalili M, Mardani M, Cuevas L. Risk factors for hospitalization of children with diarrhea in Shahrekord, Iran. Arch Clin Infect Dis. 2006;1:131‐136. [Google Scholar]
- 65. Lima AAM, Guerrant RL. Persistent diarrhea in children: epidemiology, risk factors, pathophysiology, nutritional impact, and management. Epidemiol Rev. 1992;14(1):222‐242. [DOI] [PubMed] [Google Scholar]
- 66. Strand TA, Sharma PR, Gjessing HK, et al. Risk factors for extended duration of acute diarrhea in young children. PLoS One. 2012;7(5):e36436. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3348155/. Accessed November 27, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shahid NS, Sack DA, Rahman M, Alam AN, Rahman N. Risk factors for persistent diarrhoea. BMJ. 1988;297(6655):1036‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Umamaheswari B, Biswal N, Adhisivam B, Parija SC, Srinivasan S. Persistent diarrhea: risk factors and outcome. Indian J Pediatr. 2010;77(8):885‐888. [DOI] [PubMed] [Google Scholar]
- 69. Halasa N, Piya B, Stewart LS, et al. The changing landscape of pediatric viral enteropathogens in the post‐rotavirus vaccine era. Clin Infect Dis. 2021;72(4):576‐585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ballard SB, Requena D, Mayta H, Sánchez G. Enteropathogen changes after rotavirus vaccine scale‐Up. 2021. https://www.researchgate.net/publication/357130009_Enteropathogen_Changes_After_Rotavirus_Vaccine_Scale-up. Accessed May 28, 2022 [DOI] [PMC free article] [PubMed]
- 71. Das JK, Tripathi A, Ali A, Hassan A, Dojosoeandy C, Bhutta ZA. Vaccines for the prevention of diarrhea due to cholera, shigella, ETEC and rotavirus. BMC Public Health. 2013;13(3):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. PATH . Shigella and ETEC vaccine pipeline advances despite pandemic slowdown. DefeatDD. 2022. https://www.defeatdd.org/blog/shigella‐and‐etec‐vaccine‐pipeline‐advances‐despite‐pandemic‐slowdown. Accessed May 30, 2022
- 73. Rodrigues CMC, Plotkin SA. Impact of vaccines; health, economic and social perspectives. Front Microbiol. 2020;11:1526. https://www.frontiersin.org/article/10.3389/fmicb.2020.01526. Accessed May 21, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Viboud C, Sun K, Gaffey R, et al. The RAPIDD ebola forecasting challenge: synthesis and lessons learnt. Epidemics. 2018;1(22):13‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Asher J. Forecasting Ebola with a regression transmission model. Epidemics. 2018;22:50‐55. [DOI] [PubMed] [Google Scholar]
- 76. Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc J. 2019;6(2):94‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Toh C, Brody JP. Applications of machine learning in healthcare [Internet]. Smart manufacturing—when artificial intelligence meets the internet of things. IntechOpen. 2021. 10.5772/intechopen.92297 [DOI] [Google Scholar]
- 78. Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning. Nat Methods. 2018;15(4):233‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18(12):e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Desai AN, Kraemer MUG, Bhatia S, et al. Real‐time epidemic forecasting: challenges and opportunities. Health Secur. 2019;17(4):268‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Junqué de Fortuny E, Martens D, Provost F. Predictive modeling with big data: is bigger really better? Big Data. 2013;1(4):215‐226. [DOI] [PubMed] [Google Scholar]
- 82. Otieno GP, Bottomley C, Khagayi S, et al. Impact of the introduction of rotavirus vaccine on hospital admissions for diarrhea among children in Kenya: a controlled interrupted time‐series analysis. Clin Infect Dis. 2020;70(11):2306‐2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu X, Lu Y, Zhou S, Chen L, Xu B. Impact of climate change on human infectious diseases: empirical evidence and human adaptation. Environ Int. 2016;86:14‐23. [DOI] [PubMed] [Google Scholar]
- 84. Kurane I. The effect of global warming on infectious diseases. Osong Public Health Res Perspect. 2010;1(1):4‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]


