Abstract
MET alterations, including MET exon 14 skipping variants, MET amplification, MET overexpression, and MET fusion, play pivotal roles in primary tumorigenesis and acquired resistance to targeted therapies, especially EGFR tyrosine kinase inhibitors. They represent important diagnostic, prognostic, and predictive biomarkers in many solid tumor types. However, the detection of MET alterations is challenging due to the complexity of MET alterations and the diversity of platform technologies. Therefore, techniques with high sensitivity, specificity, and reliable molecular detection accuracy are needed to overcome such hindrances and aid in biomarker-guided therapies. The current review emphasizes the role of MET alterations as oncogenic drivers in a variety of cancers and their involvement in the development of resistance to targeted therapies. Moreover, our review provides an overview of and recommendations on the selection of various cross-platform technologies for the detection of MET exon 14 skipping variants, MET amplification, MET overexpression, and MET fusion. Furthermore, challenges and hurdles underlying these common detection platforms are discussed.
Keywords: detection platform, exon 14 skipping, MET alterations, MET amplification, MET fusion, MET overexpression, targeted therapies
Introduction
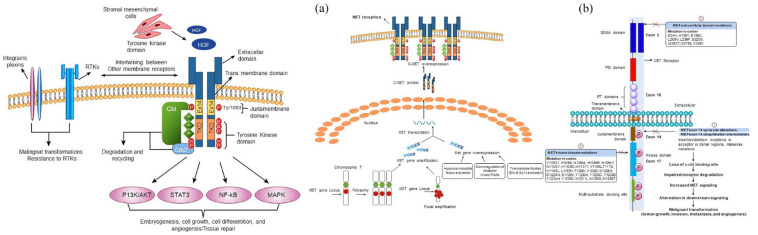
MET (mesenchymal–epithelial transition factor) is a widely expressed tyrosine kinase receptor that binds with the natural ligand hepatocyte growth factor (HGF) and plays a vital role in embryogenesis, cell growth, cell differentiation, and angiogenesis. 1 MET activation negatively affects tyrosine kinase inhibitors (TKIs) effectiveness due to the intertwining between the MET and receptor tyrosine kinase (RTK) [epidermal growth factor receptor (EGFR)] signaling pathways. 2 Dysregulation of the HGF-MET axis potentially arises by a variety of mechanisms, including mutational activation, such as exon 14 splice site alteration, exon 14 ubiquitination site mutation, kinase domain mutation, extracellular domain mutation, and amplification of the MET proto-oncogene or gene copy number (GCN) gain due to polysomy or focal amplification 3 or by overexpression that occurs either due to alteration in transcription factors [erythroblast Transformation specific (Ets) and specificity protein 1 (Sp1)] or by transcriptional upregulation due to hypoxia-inducible factor activation and downregulation of repressor microRNAs (miR-1, miR-34, and miR-449a). 4 These dysregulations lead to malignant transformation (tumor growth, invasion, metastasis, and angiogenesis) through alterations in downstream cellular signaling pathways (Ras, PI3K/Akt, STAT3, and NF-κB). 5 A schematic illustration of the normal MET signaling pathway and various mechanisms underlying aberrant MET signaling pathways is represented in Figure 1.
Figure 1.
(1) Normal MET signaling pathway (left): The binding of hepatocyte growth factor (HGF) leads to receptor dimerization that activates the receptor and subsequently activates downstream signaling pathways including Ras, PI3K/Akt, STAT3, and NF-κB. This signaling basically instigates embryogenesis, cell growth, cell differentiation, angiogenesis, and tissue repair. Furthermore, downregulation of MET receptor is initiated by CBL and ubiquitin-mediated degradation, extracellular shedding as well as proteolytic cleavage. Intertwining between MET and other membrane receptors which includes plexins, integrins, EGFR, and other RTKs promotes malignant transformations and drug resistance. (2) Dysregulation of MET signaling pathway (right). Major mechanisms include (a) MET amplification (polysomy and focal amplification) and overexpression. (b) MET exon 14 skipping variants. (1) MET exon 14 splice site variants result in exon 14 exclusion, thereby lacking ubiquitin-binding site in the juxtamembrane domain and ultimately impairing MET degradation and increasing MET signaling. Furthermore, Missense mutations in the juxtamembrane domain prevent spliceosome binding and modify the Y1003 ubiquitylation site in the MET protein. (2) Mutations in the kinase domain lead to increased activation of the MET kinase and can be associated with conformational changes that favor the inactive conformation state. (3) The implications of extracellular mutations that include HGF-binding site (SEMA domain) are currently unclear. All these induce alterations in downstream signaling and ultimately induce mutagenic transformations.
CBL, Casitas B lineage lymphoma proto-oncogene; Ets, erythroblast transformation specific; HGF, hepatocyte growth factor; IPT, immunoglobulin-like plexins and transcription factor; NF-κB, nuclear factor kappa-light-chain-Zkinase; PSI domain, plexin–semaphorin–integrin; SEMA, semaphorin; Sp1, specificity protein 1; STAT3, signal transducer and activator of transcription 3.
In view of this pivotal role of MET in tissue remodeling and morphogenesis, scholars in several studies have reported that MET alterations, particularly MET exon 14 skipping variants, MET amplification, MET overexpression, and MET fusion, play a key role in pathogenesis and alteration in sensitivity to targeted therapies and contribute to the development of acquired tumor cell resistance to treatment with EGFR-targeting TKIs in different cancer types. 6 A few anti-MET-TKIs have been developed for MET-directed targeted therapies, such as tepotinib and capmatinib, which have been approved in the United States and Japan, respectively, and savolitinib, which was approved by the National Medical Products Administration of China in June 2021,7–9 as well as glumetinib, which has been approved in China in March 2023. In view of the importance of MET gene alterations in cancer pathogenesis and therapy, the detection of MET abnormities has become increasingly important in clinical practice to guide patient selection for targeted therapy, and testing for MET exon 14 skipping variants and amplifications has already been recommended by the NCCN guidelines in treatment-naive and TKI-resistant non-small-cell lung cancer (NSCLC) patients, respectively. 10 This review will summarize the present situation and future of MET alteration detection technology, especially exon 14 skipping, amplification testing, and overexpression testing, to provide a landscape of MET alteration-associated detection and targeted therapy updates.
MET variation
MET exon 14 skipping variants in cancers
A diverse range of variations involving the kinase domain, exon 14, intronic splice site, and SEMA domain can occur within MET (Figure 1). Furthermore, a splicing variant in MET leading to loss of MET exon 14 emerged as a biomarker and offers a potential therapeutic target in several cancer types. 11 Therefore, robust approaches for the detection of such skipping events in MET exon 14 are critical in the clinical management of cancers, specifically NSCLCs, and other cancers harboring MET exon 14 skipping variants. 3 The prevalence of MET exon 14-skipping variants is widely reported in lung cancer, with a frequency of 0.9–4%. 12 Among all cancers, the prevalence of MET exon 14 skipping variants is widely reported in NSCLCs, and the most widely used platform for the detection of this variant includes next-generation sequencing (NGS) followed by reverse transcription polymerase chain reaction (RT-PCR). A summarization of prevalence and prognosis is presented in Table 1.
Table 1.
Summarization on prevalence and prognosis of MET exon14 skipping variants.
| Study group | Type of cancer | Disease stage | Race | Sample type | Method | Study population (N) | MET Exon 14 skipping variants detection rate (%) | Main findings determining the prognostic value |
|---|---|---|---|---|---|---|---|---|
| Li et al. 13 | NSCLC | Stage I–IV | Asian (Chinese) | Tissue | DNA- and RNA-based NGS | 77 | 20.8 (PSCs) | – |
| Yu et al. 14 | – | – | Tissue | NGS | 46 | 9 | – | |
| Vuong et al. 15 | – | Asian and Western | – | NGS, RT-qPCR, Sanger Sequencing | 18,464 | 3.0 (Western patients) 2.0 (Asian patients) |
Exon 14 skipping mutations were significantly associated with shorter OS (HR 1.82 (1.04–3.19), p = 0 0.04 | |
| Awad et al. 16 | Stage I–IV |
White, Non-Hispanic |
Tissue | NGS | 933 | 3.0 | – | |
| Lung et al. 17 | Stage I–IV |
– | Tissue | RT-qPCR | 196 | 1.02 | – | |
| Okuda et al. 18 | Stage I–IV |
– | Tissue | RT-PCR | 178 | 1.7 | – | |
| Pruis et al. 19 | Stage IIIA–IV | – | Tissue | DNA-based NGS | 1497 | 2.0 | – | |
| Heist et al. 20 | Stage I–IV |
– | Tissue | NGS | 54 | 19 | – | |
| Xu et al. 21 | Stage I–IV/ unknown |
Asian (Chinese) |
Tissue | RNA-based NGS | 951 | 1.7 | – | |
| Dubbink et al. 22 | – | – | Tissue | DNA-based NGS | 676 | 2.7 | – | |
| Frampton et al. 6 | – | – | Tissue | NGS | 38,028 | 0.6 | – | |
| Lee et al. 23 | – | – | Tissue | RT-PCR | 51 | 9.8 | – | |
| Hu et al. 24 | Glioblastoma | Grade IV | – | – | RNA-based NGS | 78 | 14 | MET ex14 showed significantly poorer OS compared to those without MET ex14 (p=3.52×10-3) |
| Lee et al. 23 | Gastric cancer | – | – | Tissue | RT-PCR | 42 | 7.1 | – |
| Colon cancer | – | – | Tissue | RT-PCR | 43 | 9.3 | – | |
| Rectal cancer | – | – | Tissue | RT-PCR | 23 | 0 | – | |
| Hepatocellular carcinoma | – | – | Tissue | RT-PCR | 15 | 0 | – | |
| Sarcoma | – | – | Tissue | RT-PCR | 9 | 0 | – | |
| Pancreatic cancer | – | – | Tissue | RT-PCR | 5 | 0 | – | |
| Cholangiocarcinoma | – | – | Tissue | RT-PCR | 6 | 0 | – | |
| Melanoma | – | – | Tissue | RT-PCR | 5 | 0 | – | |
| Esophageal squamous carcinoma | – | – | Tissue | RT-PCR | 1 | 0 | – | |
| Renal cell carcinoma | – | – | Tissue | RT-PCR | 1 | 0 | – |
‘–’, Not reported; AJCC, American Joint Committee on Cancer; HR, hazard ratio; NGS, next-generation sequencing; NSCLC, non-small-cell lung cancer; OS, overall survival; PSCs, pulmonary sarcomastoid carcinomas; RT-PCR, reverse transcription polymerase chain reaction; RT-qPCR, quantitative reverse transcription polymerase chain reaction.
Detection of MET exon 14 skipping variants
Genome-wide sequencing revealed heterogenic forms of MET exon 14 variants at the DNA level, thereby making it challenging to detect either by amplification refractory mutation system (ARMS)-PCR or by DNA NGS panels; instead, RNA-based testing can further improve testing accuracy. 16 Furthermore, limited reports are available on the comparison of these detection platforms.
DNA-based NGS
A detailed methodology of NGS is represented in Figure 2(c). Generally, two types of NGS-based assays, namely, amplicon-based and hybrid capture-based NGS platforms differing in DNA enrichment methods, are used in clinical settings. 25 The major limitation associated with routinely used commercially available amplicon-based NGS panels for detection of MET exon 14 skipping variants includes the frequent emergence of new regions to be covered that lead to allelic dropouts and sequencing errors. 26 Earlier, Davies et al. compared amplicon-mediated DNA-based NGS versus RNA-based NGS in NSCLC tumor samples for MET exon 14 skipping variants and reported that among 286 samples tested by both assays, RNA-based testing detected 10 positive samples, 6 of which were not detected by the DNA-based assay. Further examination revealed that genomic deletion involving primer binding sequences was the likely cause of false negatives reported and led to the further conclusion that amplicon DNA-based NGS misses the detection of a substantial fraction of MET exon 14 alterations as they are located outside the amplified regions. 27 However, amplicon-based NGS has the advantage of improved capture of targets and sequencing of difficult regions with shorter turnaround time when compared to hybrid-based NGS. 3 The pitfalls associated with amplicon-based NGS can be addressed with the adoption of hybrid-based NGS, where it not only allows the identification of hotspot mutations but also interrogates entire coding sequences of oncogenes, tumor suppressor genes, and introns of selected genes that are involved in gene fusions and further allows the assessment of copy number alterations. 28 Furthermore, if the designed algorithm and probe/bait sufficiently cover the region of interest, DNA-based assays using hybrid capture-mediated target enrichment are less likely to produce false-negative results. This is exemplified by the studies from Frampton et al. and Awad et al., where a wide variety of MET exon 14 skipping variants, including large deletions, were successfully detected by employing hybrid-based NGS assays.6,16 Furthermore, a hybrid-based approach enables corrections for some of the sequencing bias and allele dropout issues associated with amplicon-based NGS assay. 29 Although the depth of coverage of genes of interest in both platforms was high, hybrid-based NGS, with its noteworthy advantage, outperforms amplicon-based NGS. 3
Figure 2.
Schematic illustrations of workflow of various MET aberration detection techniques: (a) polymerase chain reaction, quantitative reverse transcription polymerase chain reaction (RT-qPCR) is based on conventional PCR which utilizes a fluorescent readout to measure the amount of PCR product after each round of amplification 30 ; (b) fluorescence in situ hybridization (FISH) assay involves three steps, that is, sample fixation and denaturing the sample that involves conversion of double-stranded DNA into single-stranded DNA and subsequent hybridization where denatured single-stranded DNA was tagged with fluorescent-labeled single-stranded DNA probes and followed by visualization of the hybridized probe-target DNA complexes under fluorescent microscope. 31 (c) Next-generation sequencing (NGS) is a high-throughput technology that utilizes parallel sequencing of multiple fragments to determine the sequence. It is a complicated process with multiple steps and high requirements of quality control, different NGS platforms adopt their own specific protocol in the sequencing methods. An overview of NGS workflow (both DNA based and RNA based) was represented. (d) Immunohistochemistry: It involves the utilization of anti-MET antibodies [monoclonal antibodies (SP44, cMET, and MET4) or polyclonal antibodies (MET AF276)].
NGS, next-generation sequencing; RT-qPCR, reverse transcription polymerase chain reaction.
RNA-based NGS or RT-PCR
MET exon 14 variants can also be detected at the RNA level. Li et al. conducted a study that involved the comparison of DNA- and RNA-based NGS for the detection of MET exon 14 skipping variants in pulmonary sarcomatoid carcinomas (PSCs) and reported a concordance of 96.1% between these platforms and concluded that RNA-based sequencing was the most accurate because some somatic variants not covering MET exon 14 splice sites might also induce skipping. 13 Furthermore, DNA sequencing cannot confirm the absence of the exon, as modifications such as splicing occur post-translationally. 27 At this juncture, RNA-based NGS platforms have the potential to complement DNA-based NGS platforms where RNA sequencing permits the direct recognition of the loss of exon 14 transcription. 27 In a study, Jurkiewicz et al. compared the performance of DNA versus RNA-based NGS assays for the detection of MET-ex14 skipping variants in 644 lung adenocarcinoma samples and concluded that DNA-based NGS panels can potentially miss MET-ex14 skipping when the primers do not target both the 3′ and 5′ splice sites of introns 13 and 14, respectively. 32 Furthermore, due to the diverse nature of MET exon 14 splice site alterations, interpreting variants that truly result in exon 14 skipping is challenging, and this becomes more strenuous when these alterations are located in deeper introns; since RNA-based platform involves sequencing mRNA that is devoid of introns, this kind of challenge can be avoided. 33
RNA-based RT-PCR can also be used for MET-ex14 skipping detection and shows good concordance with RNA-based NGS. 12 Although NGS is rapid in comparison to traditional Sanger sequencing, it is still too expensive to be affordable by small laboratories or an individual. 34 By contrast, RT-PCR is easy to perform, more widespread, and has a shorter turnaround time. RT-qPCR is usually designed in the MET exon 13 and 15 region primers for the detection of specific amplification products. This method has a high accuracy in detecting MET 14 variants but it can miss some special and rare forms of MET variations. Meanwhile, RNA-based analysis is highly reliant on the quality of RNA, which can be suboptimal in some clinical samples. 27 RNA-based testing is not part of the routine workflow in many molecular detection laboratories, as acquiring sufficient RNA material remains a large obstacle in contrast to DNA acquisition. Therefore, when RNA quality is at risk, an alternative is DNA-based NGS, where DNA is less difficult to obtain and less vulnerable to degradation.
Targeted therapies for MET exon 14 skipping variants
The constant discovery of actionable activating variants has led to targeted therapies with new-generation TKIs and improved overall survival and time to progress. 26 Among all reviewed literature, NSCLC lung cancer accounts for almost all. Earlier, a phase I PROFILE 1001 study reported the efficacy of crizotinib [median progression-free survival (mPFS) 7.3 months and overall response rate (ORR) of 32%] in advanced-stage NSCLC patients harboring MET exon 14 alterations, and these results led to the inclusion of crizotinib in the NCCN guidelines. With this breakthrough, several highly selective MET inhibitors, such as tepotinib, capmatinib, savolitinib, and glumetinib, have been tested in patients with MET exon 14-altered NSCLC and found to be more potent than crizotinib. 35
Tepotinib was developed for the treatment of solid tumors and demonstrated promising clinical efficacy and safety profiles in patients with advanced NSCLC with a confirmed MET exon 14 skipping variant in the multinational phase II VISION study. 36 In March 2020, it was approved for use in Japan for this indication and was subsequently approved by the FDA on 3 February 2021. Capmatinib was developed for the treatment of lung cancer, inhibiting cancer cell growth driven by the mutant MET variant, including exon 14 skipping. In May 2020, capmatinib received its first global approval for the treatment of adults with metastatic NSCLC with MET exon 14-skipping variants as detected by an FDA-approved test. 8 Savolitinib was developed for the treatment of NSCLC, gastric cancer, colorectal cancer, and papillary and clear cell renal cell carcinoma. Based on the results of a pivotal phase II trial, savolitinib yielded promising activity and had an acceptable safety profile in patients with NSCLC/pulmonary sarcomatoid carcinoma and was recently approved in June 2021 in China for the treatment of metastatic NSCLC with MET exon 14-skipping alterations in patients who have progressed after or who are unable to tolerate platinum-based chemotherapy, conditional on the results of a phase III trial. 37 Glumetinib was developed for the treatment of lung cancer and showed durable antitumor activity with manageable toxicity in patients with locally advanced or metastatic MET ex14-positive NSCLC in the phase II GLORY study. 38 In March 2023, it was approved in China for this indication.
Furthermore, reports from several clinical studies cannot be compared directly due to different populations and inclusion criteria in the clinical efficacy of several MET-TKI inhibitors, including tepotinib 39 [independent review committee (IRC): ORR 57.3% (treatment naive), 45.0% (pretreated), 40 capmatinib (ORR: 44% (pretreated) and ORR: 68.3% (treatment naive) 35 , mPFS 12.5 m (treatment naive), 5.5 m (pretreated); mOS 25.5 m (treatment naive), not reported (pretreated))], 41 and savolitinib [phase IIIb study, IRC: ORR 58.6% (treatment naive), mPFS 13.8 m (treatment naive) 42 ; phase II study(36% PSC and 21% CNS metastases): TRES set: ORR 49.2%, FAS set: mPFS 6.9 m, mOS 12.5 m; in PSC, mPFS 5.5 m, mOS 10.6 m; other NSCLC (non-PSC), mPFS 7.0 m, mOS 17.3m 43 ] and glumetinib 38 (BIRC: overall ORR 66%, mPFS 8.5 m, mOS 17.3 m; in treatment-naïve patients: ORR 71%, mPFS 11.7 m) in patients harboring MET exon 14 skipping variants in NSCLC. A summarization of MET exon 14 skipping variant targeted therapy outcomes in advanced NSCLC is represented in Table 2.
Table 2.
MET exon 14 skipping variants targeted therapy outcomes in advanced NSCLC.
| Study group | Type of cancer | Drug | Stage | Study phase | No. of patients | Assay used | ORR | Median PFS in months (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Drilon et al. 44 | NSCLC | Crizotinib (9% PSCs) | Advanced | I | 69 | NGS, RT-PCR | Overall: 32% (21/65) ORR by alteration type: Indels: 25% (5/20). Point mutations: 36% (12/33). Splice acceptor site mutations: 31% (5/16). Splice donor site mutations: 32% (12/37). |
7.3 (5.4–9.1). |
| Moro-Sibilot et al. 45 | Crizotinib (PSCs% unknown) | Advanced | II | 28 | NGS and Sanger sequencing | ORR: 10.7% | 3.6 (1.6–7.1) | |
| Wolf et al. 35 | Capmatinib (INC280) (none-PSCs) |
Advanced | II | Pretreated: 100 treatment naive: 60 | RT-PCR, NGS | Pretreated: 44%, treatment naive: 68.3% | Pretreated: 5.5 (4.2–8.1) Treatment naive: 12.5 (8.3–18.0) |
|
| Mazieres et al. 39 | Tepotinib (PSCs% unknown) | Advanced | II | Overall: 313 Pretreated: 149 Treatment naive: 164 |
RNA-based NGS | Overall: 51.4% Pretreated: 45%, Treatment naive: 57.3% |
Overall: 11.2 (9.5–13.8) Pretreated: 11.0 (8.2–13.7), Treatment naive: 12.6 (9.7–17.7) |
|
| Lu et al. 46 | PSCs and other NSCLCs | Savolitinib (36% PSCs) | Advanced | II | PSCs: 25, non-PSCs: 45 | Sanger DNA sequencing or DNA-based NGS | TRES (tumor response evaluable set): ORR: 53.2% PSCs: 50%, other NSCLCs Non-PSCs: 55% |
Overall: 6.9 (4.6–8.3) PSCs: 5.5 (2.6–6.9), Other NSCLCs (non-PSCs): 7.0 (5.5–13.7) |
| Yu et al. 38 | NSCLC | Glumetinib (6% PSCs) | Advanced | II | Overall: 84 Pretreated: 38 Treatment-naive: 46 |
RNA-based RT-PCR | Overall: 66% Pretreated: 60% Treatment-naive: 71% |
Overall: 8.5 (7.6–9.7) Pretreated: 7.6 (4.1–9.6), Treatment-naive: 11.7 (7.6–21.9) |
| Lu et al. 42 | Savolitinib (8% PSCs) |
Advanced | IIIB | 87 (Treatment naive) |
NA | ORR: 58.6% | 13.8 (9.7, NR) |
mPFS, median progression free survival; NGS, next-generation sequencing; NR, not reached; NSCLC, non-small-cell lung cancer; ORR, overall response rate; PFS, progression-free survival; PSCs, pulmonary sarcomatoid carcinomas; RT-PCR, reverse transcription polymerase chain reaction.
MET amplification
MET amplification in cancers
MET GCN gain occurs either by polysomy or focal amplification (Figure 1).3,47 Tumors harboring de novo MET amplifications (high level) are primarily dependent on the MET signaling pathway for growth and are found across a wide variety of solid tumors, 3 while tumors harboring acquired MET amplifications rely on other oncogene alterations (such as EGFR mutation) and develop a secondary dependence on the MET signaling pathway as a mechanism of resistance to targeted EGFR therapies. 48 Reports from several studies have indicated that MET gene amplification drives treatment resistance, particularly therapies targeting EGFR.49–53
MET amplification occurs in a broad type of cancer as protooncogenes or resistance mechanisms. A summarization of the prevalence and prognosis of MET amplification in various cancers is presented in Table 3.
Table 3.
Summarization on prevalence and prognosis of MET amplification in various cancers.
| Study group | Type of cancer | Disease stage | Race | Sample type | Method | MET amplification type | Treatment | Study population (N) | MET amplification detection rate N (%) | Prognostic value |
|---|---|---|---|---|---|---|---|---|---|---|
| Ahn et al. 10 | NSCLC | Stage IV | Asian and non-Asian | Tissue | FISH | Acquired | EGFR-TKI + MET-TKI | 108 | 32 (29.6) | FISH10+ has better survival than FISH5+ |
| Tong et al. 54 | NSCLC | Stage I–IV | - | Tissue | FISH | – | – | 687 | 29 (4.2) | – |
| Go et al. 55 | Stage I–IV | Asian (Korean) |
Tissue | FISH | De novo | EGFR-TKI naive | 180 | 7 (3.9) | High MET GCN leads to shorter survival | |
| Okamoto et al., 56 | Stage IIIB/IV | – | Tissue | FISH | – | – | 229 | 9 (3.9) | – | |
| Schildhaus et al. 49 | – | – | Tissue | FISH | De novo | Untreated | 693 | 20 (3) | – | |
| Suryavanshi et al. 57 | Stage IV | Tissue | FISH | – | – | 76 | 5 (6.57) | |||
| Guo et al. 58 | – | Asian and Non-Asian | Tissue | RT-PCR, FISH, and SISH | – | – | 5516 | 4–18 | High GCN was significantly associated with shorter OS (HR 1.90 [1.35–2.68], p = 0.001) | |
| Onozato et al. 59 | Stage I–IV | – | Tissue | RT-PCR | – | – | 148 | 2 (1.4) | – | |
| Bean et al. 60 | – | – | Tissue | RT-PCR | Acquired | EGFR-TKI | 43 | 9 (21%) | MET amplification occurs independently of EGFRT790 M mutation and clinically relevant to gefitinib and erlotinib resistance | |
| De novo | Untreated | 62 | 2 (3%) | |||||||
| Xia et al. 61 | Stage I–IV | Han Chinese | Tissue | RT-PCR | – | – | 110 | 6 (5.5) | – | |
| Xu et al. 62 | – | – | Tissue | fluorescent RT-PCR | De novo | – | 368 | 33 (8.97%) | MET gene amplification in a primary tumor helps in guiding MET-targeted therapies | |
| Chen et al. 63 | – | – | Tissue | RT-PCR | – | – | 257 | 29 (11.28) | – | |
| Blood | – | – | 318 | 31 (9.75) | ||||||
| Beau-Faller et al. 64 | Stage I–IV |
Non-Asian | Tissue | RT-PCR | – | – | 106 | 22 (21) | – | |
| Okuda et al. 18 | Stage I–IV |
– | Tissue | RT-PCR | – | – | 213 | 12 (5.6) | – | |
| Tsuta et al. 65 | Stage I–IV |
– | Tissue | BISH | – | – | 844 | 92 (10.9) | – | |
| Zhang et al. 66 | Colorectal adeno- carcinoma |
AJCC Stage I–IV |
– | Tissue | FISH | De novo | – | 294 | 13 (4.4) | – |
| Jardim et al. 67 | Metastasis CRC | Advanced (metastasis) | Asian, Black, Hispanic, White |
Tissue/needle aspiration biopsies | FISH | – | – | 208 | 4 (1.9) | – |
| Raghav et al. 68 | – | – | Tissue | FISH (MET/CEP7 ratio > 2) | De novo | Untreated | 208 | 4 (1.9) | – | |
| – | – | ctDNA | NGS (GCN > 4 copies) | Acquired | EGFR-TKI therapy | 53 | 12 (22.6%) | – | ||
| – | – | Tissue | NGS (GCN > 4 copies) | – | – | 279 | 6 (2.2%) | – | ||
| Yu et al. 69 | GC | – | Asian and Western | – | FISH, and SISH | – | – | 2210 | 8–82 |
MET amplification was significantly associated with shorter OS (HR 2.112 [1.622–2.748]) |
| Graziano et al. 70 | – | – | Tissue | FISH | De novo | – | 230 | 1–5 | – | |
| de Melo Gagliato et al. 71 | Breast cancer | Stage IV | Asian, American, Indian, Black, Hispanic, White | Tissue | FISH | – | – | 107 | 63 (4.7) | – |
| Jardim et al. 67 | Advanced (metastasis) | Asian, Black, Hispanic, White |
Tissue/needle aspiration biopsies | FISH | – | – | 64 | 3 (4.6) | – | |
| Jardim et al. 67 | Ovarian cancer | Advanced (metastasis) | Asian, Black, Hispanic, White |
Tissue/needle aspiration biopsies | FISH | – | – | 110 | 4 (3.6%) | – |
| Gastroesophageal | Advanced (metastasis) |
Asian, Black, Hispanic, White |
Tissue/needle aspiration biopsies | – | – | 77 | 5 (6.4) | – | ||
| Melanoma | Advanced (metastasis) | Asian, Black, Hispanic, White |
Tissue/ needle aspiration biopsies | – | – | 61 | 2 (3.2) | – | ||
| Renal cancer | Advanced (metastasis) | Asian, Black, Hispanic, White | Tissue/needle aspiration biopsies | – | – | 28 | 4 (14.28) | – | ||
| Pal et al. 72 | PRCC | Advanced | – | Tissue | FISH | De novo | – | 169 | 13% (Type 1 PRCC) and 3% (type 2 PRCC) | – |
| Kato et al. 73 | Esophageal carcinoma | Stage I–III | – | Tissue | FISH | De novo | – | 196 | 1 (2) | – |
‘–’, Not reported; AJCC, American joint committee on cancer; cfDNA, circulating cell-free DNA; GC, gastric carcinoma; HR, hazard ratio; mCRC, metastatic colorectal cancer; NSCLC, non-small-cell lung cancer; PRCC, papillary renal cell carcinoma; RT-PCR, reverse transcription polymerase chain reaction; RT-qPCR, reverse transcription polymerase chain reaction; FISH, fluorescence in situ hybridization.
Regarding polysomy, there is controversial evidence regarding whether MET polysomy is the driver of gene alteration for tumorigenesis. Lai et al. found that although up to 26% of TKI-naive EGFR mutant-positive NSCLC harbor high MET GCN by FISH, this did not significantly affect response to TKIs, except in patients identified as MET-amplified but not polysomy. 74 In the phase II Acse study, advanced NSCLC patients with MET polysomy and low amplification (MET/CET7 ratio between 1.8 and 2.2) did not respond well to crizotinib, with a mPFS of 3.2 months and a mOS of 7.7 months. 45 However, in the phase 1b/2 TATTON study, osimertinib and savolitinib showed encouraging antitumor activity in both MET amplification and polysomy patients recruited based on FISH positivity, with an ORR of 30% in part B1 (after prior 3G TKIs). 12 The subgroup analysis results showed that the response of focal amplification was better than that of polysomy (ORR 31% versus 28%) but more research is needed for verification. Therefore, MET amplification may be considered as a biomarker for MET-directed therapies, while polysomy still needs to be studied more.
Detection of MET amplification
MET amplification (copy number gain) can be detected by several techniques, such as FISH, NGS, and ddPCR. 3 As the magnitude of MET amplification is a continuous variable, determining the cutoff for MET positivity is more challenging. 3 At present, no consensus exists on the most appropriate diagnostic cutoff point for MET amplification. 75 Different detection platforms for MET amplification testing exist and have ineligible disparities in terms of sensitivity and specificity.
Fluorescence in situ hybridization (FISH)
FISH is a cytogenetic technique used for obtaining spatial genomic and quantification of nucleic acids in the cellular environment and has emerged as the gold standard technique for the detection of chromosomal abnormalities. 76
In the FISH assay, MET copy number increases can be defined either by Cappuzzo criteria or by University of Colorado Cancer Center (UCCC) criteria, which calculate the ratio of MET to chromosome enumerating probe against chromosome 7 (CEP7). The Cappuzzo et al. criteria define MET amplification as a mean of five or more copies of MET per cell (MET gene copy number (GCN) ⩾5). 77 Furthermore, other alternate definitions suggest a MET GCN of ⩾6 78 and a MET GCN of ⩾15. 79 However, the determination of GCN cannot differentiate between MET focal amplification and polysomy, and this limitation is overcome by the UCCC approach involving the calculation of the MET-to-CEP7 ratio that adjusts the number of chromosomes present, thereby differentiating selective MET focal amplifications and chromosomal duplication. In general, a MET to CEP7 ratio ⩾ 2.0 defines MET focal amplification. 73 However, several other studies categorized the degree of MET focal amplification into low (⩾1.8–⩽2.2), intermediate (>2.2–<5), and high (⩾5). 75
The FISH technique is advantageous in the detection of MET amplification in light of its comparable performance characteristics and potential for cost-effectiveness and limited complexity in testing when compared with NGS. Furthermore, FISH offers direct visualization of the tested samples, which was not possible by NGS, and another added benefit of this technique is that it represents a suitable technology for the detection of intratumoral heterogeneity within tissue samples. Although FISH is the current gold standard for MET amplification testing, the prevalence of MET amplification detected by FISH is variable in the literature, which is likely attributable to a lack of standardization of technique and/or patient selection criteria and different cutoffs for defining MET positivity. 63 In addition, the main limitation of FISH is that it can only be applied to tissue samples, while tissue feasibility is low in advanced NSCLC patients, especially patients who progressed on previous TKIs.
Next-generation sequencing
Similar to the FISH assay, there is no consensus on a single definition of MET amplification, and the cutoff for MET amplification varies across different NGS platforms. 80 Hybrid capture-based NGS is known to be more accurate in assessing copy number variation in MET and other genes, as it interrogates broader regions of the genome and removes sequence replicates. By contrast, amplicon-based NGS covers a limited genomic territory, and sequence replicates cannot be removed, affecting sequence coverage depth. 3 NGS can be used to analyze both tissue and plasma ctDNA or other bodily fluids, which will facilitate biomarker testing extensively. However, certain technical limitations, including sample selection, low tumor cell fraction, and low DNA quality of tumor samples can increase background noise, hindering the accurate analysis of copy number gain/loss. 80 As reported in the TATTON study, where osimertinib (3rd EGFR-TKI) and savolitinib (MET-TKI) combined therapy in NSCLC patients with MET-amplified EGFR-TKI resistance demonstrated encouraging antitumor activity, FISH positivity was defined centrally as either focal amplification (MET:CEP7 ratio ⩾ 2) or polysomy (gene copy number ⩾ 5 if MET:CEP7 <2), while tissue NGS from Foundation Medicine showed higher positive-percent agreement (PPA, also known as sensitivity) for FISH focal amplification (88%) but lower PPA for FISH polysomy (4%). Similarly, a comparison of ctDNA NGS with FISH yielded negative-percent agreement (NPA) of 90% and a modest PPA of only 25%. 45 Various factors affect the sensitivity of ctDNA detection by NGS, such as sequencing depth, threshold of bioinformatic analysis, and techniques to enrich tumor-derived signals and reduce background noise. The high specificity and low sensitivity of the currently available NGS assay indicate that copy number variation (CNV) detection by NGS needs to be optimized, and retesting with FISH should be considered to avoid missing MET amplification; while NGS may serve as an alternative selection for MET polysomy with higher sensitivity and specificity and guide the clinical treatment. 81
Although plasma ctDNA testing of MET amplification is challenging, it is a direction for future development to fulfill the clinical needs given that plasma specimens are dominant in late-phase patients. In addition, plasma harvested from peripheral blood is less invasive and can reflect the disease progression dynamically while avoiding tumor heterogenicity in single surgery or biopsy samples. 82
Polymerase chain reaction
MET amplifications can be detected by employing a PCR technique specifically (RT-PCR). Similar to FISH and NGS, the cutoff for defining MET amplification positivity was not standardized. 62 A comparison of the basic principle and procedure involved in different PCR techniques is represented in Figure 2(a). The major limitation associated with RT-PCR is that the success relies on the RNA quality in the specimen tested. Inadequate fixation or prolonged ischemia of tissue leads to false-negative results. These limitations were outweighed by the introduction of droplet digital PCR (ddPCR), which has high sensitivity and accuracy levels for absolute representation of a given nucleic acid sequence. Droplet digital PCR enables the absolute quantification of nucleic acids present in the sample by partitioning the sample into independent PCR subreactions where each partition contains a few or no target sequences and is subjected to PCR wherein the fraction of amplification positive partitions is used to quantify the concentration of the target sequence with a statistically defined accuracy using Poisson’s statistics. 83
The performance of ddPCR for MET amplification testing is not well characterized compared to FISH and NGS. 84 However, the consistency between ddPCR and FISH is reported in some small sample studies for assay analytical validation of developed ddPCR methodology; the sensitivity and specificity of tissue ddPCR and FISH are both 100%, while the sensitivity and specificity of tissue and blood ddPCR are 66.67% and 98.86%, respectively. 85 Given that insufficient tissue can be retrieved after resistance to EGFR-TKIs, further studies to confirm the testing capability of blood ddPCR as an alternative detection tool for MET amplification is needed in the future.
Targeted therapies for MET amplification
The prevalence of acquired MET amplification is enriched gradually after lines of EGFR-TKI treatment. In lung cancer, MET amplification occurred in 1−4% of treatment-naive patients, while the prevalence increased to 5−22% after first- and second-generation TKI treatment and 5−50% after third-generation TKI osimertinib treatment. 86 The amplification of MET occurred independently of the EGFR T790M mutation and was clinically relevant to gefitinib and erlotinib resistance. 60
There are several MET-TKIs in development for MET amplification-positive NSCLC patients, such as crizotinib, tepotinib, capmatinib, and savolitinib, and criteria for patient inclusion are mainly based on FISH and immunohistochemistry (IHC). The cutoff for MET amplification positivity varies among studies 83 Furthermore, it is evident from the literature that patients harboring higher MET GCN achieve better clinical outcomes from the targeted therapy.
For de novo MET amplification NSCLC, in a phase I PROFILE 1001 study in which NSCLC patients were stratified based on the degree of MET amplification and the activity of crizotinib examined in relation to the level of MET amplification, there was a high amplification group (MET-to-CEP7 ratio ⩾ 4) with reported ORR of 38.1% and median PFS of 6.7 months compared with a low amplification group (MET-to-CEP7 ratio ⩾ 1.8 to ⩽2.2) with ORR of 33.3% and median PFS of 1.8 months and an intermediate amplification group (MET-to-CEP7 ratio > 2.2 to <4) with ORR of 14.3% and median PFS of 1.9 months. 87 Earlier, the results of the GEOMETRY mono-1 study demonstrated the efficacy and safety of capmatinib in patients with high-level (GCN) ⩾10 compared with low-(GCN < 4) or mid-level (GCN 4–5 or 6–9) MET-amplified advanced NSCLC. Patients with GCN ⩾ 10 treatment-naive and/or receiving 1 or 2 lines of therapy exhibited better ORRs of 40% and 29%, respectively. 35 It seems that high amplification status is associated with better response to MET-TKIs compared to low amplification status given the current evidence. Recently, the results from the VIKTORY umbrella trial showed that savolitinib monotherapy exhibited an ORR of 50% (10/20) in a subset of gastric cancer patients harboring MET amplifications, and further genomic analysis revealed that patients with MET GCN > 10 (by tissue NGS) had high response rates to savolitinib [ORR 70% (7/10)] and concluded that the subset of patients with MET amplifications achieved the largest absolute decrease in tumor burden. 83 Furthermore, despite there existing evidence of MET inhibition by foretinib, Shah et al. reported disappointing results for foretinib, which might be due to disparities in the selection of the study population, study design, or drug itself. 88 Furthermore, a summarization of MET amplification targeted therapy outcomes by MET copy number status across different cancer types is represented in Table 4.
Table 4.
MET amplification targeted therapy outcomes.
| Study group | Cancer type | Drug | Study phase | Study population | Assay used | Amplification criteria | MET subgroup | Subgroup (N) | ORR | Median PFS (months) |
|---|---|---|---|---|---|---|---|---|---|---|
| Camidge et al. 89 | NSCLCs | Crizotinib | Phase I | 38 | FISH | MET-to-CEP7 ratio ⩾ 1.8 | MET-to-CEP7 ratio ⩾ 1.8–⩽2.2 | 3 | 33% (1/3) | 1.8 |
| MET-to-CEP7 ratio > 2.2 to <4.0 | 14 | 14% (2/14) | 1.9 | |||||||
| MET-to-CEP7 ratio ⩾ 4.0 | 20 | 38% (8/21) | 6.7 | |||||||
| Moro-Sibilot et al. 45 | Crizotinib | Phase II Acsé study |
25 | FISH | MET GCN ⩾ 6 and IHC 2 +/ 3+ | All patients | – | 16% (4/25) | 3.2 | |
| Schuler et al. 90 | Capmatinib | Phase I | 44 | FISH | MET-to-CEP7 ratio ⩾ 2.0 or MET GCN ⩾ 5, H-score ⩾ 150, IHC 2 +/ 3+ | MET GCN ⩾ 5 | 8 | 63% (5/8) | – | |
| Wu et al. 91 | Capmatinib + gefitinib | Phase II | 100 | FISH and IHC | MET GCN ⩾ 4 | All patients | – | 29% (29/100) | ||
| MET GCN ⩾ 6 | 36 | 47% (17/36) | ||||||||
| Yu et al. 92 | Savolitinib + osimertinib | Phase II ORCHARD study | 20 | NGS | - | MET amplification | 17 | 41% (7/17) | – | |
| Hartmaier et al. 93 | Savolitinib + osimertinib | Phase Ib TATTON study | 42 | FISH | MET GCN ⩾ 5 or MET-to-CEP7 ratio ⩾ 2.0 | Part D all patients | – | 62% | 9.0 | |
| Ahn et al. 10 | Savolitinib + osimertinib | Phase II SAVANNAH study | 196 | FISH and IHC | MET GCN ⩾ 5 and/or MET-to-CEP7 ratio ⩾ 2.0, IHC ⩾ 50% tumor cell 3+ | FISH10+ and/or IHC90+ | 108 | 49% | 7.1 | |
| Liam et al. 94 | Tepotinib + gefitinib | Phase II INSIGHT study | 31 | FISH IHC | IHC ⩾ 50% tumor cell 2 + 3+ MET GCN ⩾ 5 or MET-to-CEP7 ratio ⩾ 2.0 |
All patients | – | 45.2% | 4.9 | |
| IHC 3+ | 19 | 68.4% | 8.3 | |||||||
| MET amplification | 12 | 66.7% | 16.6 | |||||||
| Kim et al. 95 | Tepotinib + osimertinib | Phase II INSIGHT 2 study | 128 | FISH LBx NGS |
MET GCN ⩾ 5 and/or MET-to-CEP7⩾2 LBx NGS,GCN ⩾ 2.3 |
TBx FISH LBx NGS |
98(TBx FISH) | 50.0% | 5.6 | |
| Yu et al. 96 | Glumetinib + osimertinib | Phase Ib/II | 30 | FISH | GCN ⩾ 5 and/or MET-to-CEP7⩾2 | MET amplification | 11(Post 3G EGFR-TKI) | 60% (36.4% in post 3G EGFR-TKI) |
6.9 | |
| Qin et al. 97 | Hepatocellular carcinoma | Capmatinib | Phase II | 30 | FISH | MET H-score ⩾ 50 or MET-to-CEP7 ratio ⩾ 2.0 or MET GCN ⩾ 5 | All patients | – | 10% (3/30) | – |
| MET IHC score 3 + or 2 + in ⩾ 50% of tumor cells and MET GCN ⩾ 5 | 10 | 30% (3/10) | – | |||||||
| Lee et al. 83 | Gastroesophageal cancers | Savolitinib | Phase II | 715 (25 MET amplified) | NGS | – | MET GCN > 10 | 20 | 70% (7/10) | – |
| Shah et al. 88 | Foretinib | Phase II | 74 | FISH | MET-to-CEP7 ratio ⩾ 2.0 | All patients | – | 0% (0/71) | – | |
|
MET- amplified |
3 | 0% (0/3) | – | |||||||
| Bang et al. 98 | Solid tumors | Capmatinib | Phase I | 38 | FISH | – | MET GCN < 4 | 22 | 0% (0/22) | – |
| MET GCN ⩾ 4 to <6 | 6 | 0% (0/6) | ||||||||
| MET GCN ⩾ 6 | 3 | 0% (0/3) | ||||||||
| Angevin et al. 99 | SAR125844 | Phase I | 72 | FISH | MET-to-CEP7 ratio ⩾ 2.0 and MET GCN > 4 | All patients | – | 17% (5/29) | – |
‘–’, not reported; CEP 7, chromosome 7; FISH, fluorescence in situ hybridization; GCN, gene copy number; IHC, immunohistochemistry; NSCLC, non-small-cell lung cancer; PRCC, papillary renal cell carcinoma; NGS, next-generation sequencing; PFS, progression-free survival; ORR, overall response rate.
As shown in Table 4, the commonly used platform for MET amplification testing and patient screening is FISH, combined with the IHC platform for some studies. The criteria for MET amplification varied across studies, and a unified cutoff has not yet been established. Both the MET-to-CEP7 ratio and MET GCN number were used separately or combined. It seems that a better response to MET-TKIs or combined therapy occurs in patients with higher MET amplification status.
MET overexpression
MET overexpression in cancers
MET overexpression can be caused by gene amplification, gene mutation, transcriptional enhancement [activation of specificity protein 1 (Sp1), erythroblast transformation specific (Ets)], or by post-transcriptional mechanisms that lead to malignant transformations (Figure 1). 4
The clinicopathological impacts of MET overexpression in various cancers have been investigated in several studies. The prevalence of MET overexpression was reported to be 39.8%, 100 33.7%, 101 and 58.8% 102 in cases of gallbladder carcinoma, triple-negative breast cancer, and NSCLC, respectively. However, the potential correlation of MET overexpression with patient outcome is inconsistent across tumors. Some of the studies indicated that MET overexpression was significantly related to shorter OS or PFS in bladder cancer 103 and glioblastoma multiforme, 104 while some indicated no correlation to prognosis in NSCLC 102 or lung adenocarcinoma 105 Thus, a detailed understanding of the relationship between MET overexpression and prognosis is still needed.
Detection of MET overexpression
MET overexpression can be analyzed using immunohistochemistry (IHC), which provides a semiquantitative information on MET expression. 3 The prevalence rate of MET overexpression is approximately 13.7−63.7% in all NSCLCs. Among them, the prevalence rate of MET overexpression is approximately 30.4−37% in advanced NSCLC after EGFR-TKI treatment. Different scoring systems were used to quantify the level of MET expression by IHC. 106 In clinical trial settings, the level of expression is quantified as a clinical score (on a scale of 0–3+). The H-score (range from 0–300) is another scoring system that involves multiplying the percent of cells with staining scores of 1+, 2+, and 3+ by their staining intensity score. 107 In general, an H-score ⩾ 200 denotes overexpression, and the specific cutoff range varies among studies. 108 Recently, a Chinese expert consensus on MET immunohistochemistry detection and interpretation standards for NSCLC has been proposed, aiming to improve the quality of detecion and interpretation to further guide the clinical treatment and studies. 109
Some MET IHC antibodies for MET overexpression testing are shown in Table 5, including SP44, D1C1, and 3077. At present, many domestic and foreign antibodies for detecting MET amplification have obtained domestic medical device product status (recorded in the National Medical Products Administration), involving multiple clone numbers. The staining performance of different antibodies varies, and the interpretation criteria of current clinical research combine the expression intensity and percentage of relevant antibodies in tumor cells at the same time.
Table 5.
MET overexpression targeted therapy outcomes.
| Study | Treatment | Study phase | Study population | Assay used | Antibody | Interpretation standards | Cutoff | MET subgroup | Subgroup (N) | ORR | Median PFS (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCT03539536 110 | Teliso-v | 2 | Non-squamous (EGFR-WT): 58 Squamous: 28 |
IHC | SP44 | Clinical scoring | Non-squamous: ⩾25% tumor cell 3+; Squamous: ⩾75% tumor cell 1+ |
Non-squamous (total) | 52 | 36.5% | – |
| Non-squamous (25–50% tumor cell 3+) | 29 | 24.1% | – | ||||||||
| Non-squamous (⩾50% tumor cell 3+) | 23 | 52.2% | – | ||||||||
| Squamous | 27 | 11.1% | – | ||||||||
| TATTON 93 | Savolitinib + osimertinib | 1b | 180 | FISH, NGS or IHC | SP44 | Clinical scoring | FISH: GCN ⩾ 5 or MET/CEP7 ⩾ 2 NGS: GCN ⩾ 5 IHC: ⩾50% tumor cell 3+ |
Overall (B2/B3/D) | 111 | 62–67% | 9.0–11.1 |
| IHC ⩾ 50% tumor cell 3+ | B1: 13 B2: 4 |
B1: 46% | – | ||||||||
| SAVANNAH Ahn et al. 10 |
Savolitinib + osimertinib | 2 | 196 | FISH or IHC | SP44 | Clinical scoring | FISH: GCN ⩾ 5 or MET/CEP7 ⩾ 2 IHC: ⩾50% tumor cell 3+ |
Total | 193 | 32% | 5.3 |
| FISH10+ and/or IHC90+ | 108 | 49% | 7.1 | ||||||||
| NCT01610336 91 | Capmatinib + gefitinib | 1b/2 | 100 | FISH or IHC | 3077 | Clinical scoring | FISH: GCN ⩾ 4 IHC: ⩾50% tumor cell 2 +/ 3+ |
Total | 100 | 29% | – |
| IHC ⩾ 50% tumor cell 2+ | 16 | 19% | – | ||||||||
| IHC ⩾ 50% tumor cell 3+ | 78 | 32% | 5.45 | ||||||||
| NCT04077463 111 (CHRYSALIS-2) | Amivantamab + lazertinib | 1/1b | 108 | IHC | SP44 | Clinical scoring | IHC: ⩾25% tumor cell 3+ | MET+: ⩾25% tumor cell 3+ MET-: others |
28 (MET+) | 61% | 12.2 |
| NCT02099058 112 | Teliso-V + osimertinib | 1/1b | 25 | IHC | SP44 | Clinical scoring | IHC: ⩾25% tumor cell 3+ | Total | 18 | 58% | |
| High ⩾ 50% tumor cell 3+ | 10 | 50% | |||||||||
| Intermediate 25–49% tumor cell 3+ | 8 | 63% |
‘–’, not reported; CEP 7, Chromosome 7; FISH, fluorescence in situ hybridization; GCN, gene copy number; IHC, immuohistochemistry; NGS, next-generation sequencing; WT, wild type; PFS, progression-free survival; ORR, overall response rate.
Targeted therapies for MET overexpression
MET can be overexpressed in certain cancers harboring primary and/or secondary MET exon 14 alterations or MET amplifications, 3 and many studies have indicated that MET overexpression and gene amplification are prognostic survival factors for many cancers, including gastric carcinomas. 113 At this juncture, the results from several clinical trials involving monotherapy with anti-MET antibodies (onartuzumab, emibetuzumab), anti-HGF antibodies (ficlatuzumab, rilotumumab), TKIs (crizotinib, tivantinib, cabozantinib), and other therapeutic agents suggest that the overall activity of these monotherapies in MET-overexpressing cancers is low, indicating that MET overexpression is not consistently predictive of benefit from MET-directed therapies. 114
By contrast, combination with EGFR-directed therapies is effective. Wu et al. conducted an INSIGHT trial aiming to evaluate the efficacy and safety of tepotinib plus gefitinib in NSCLC patients harboring MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitors and concluded that patients with MET IHC3+ or MET amplifications showed a better response, where PFS and OS were longer with tepotinib plus gefitinib (PFS 8.3 months, OS 29.1 months) than with chemotherapy (PFS 4.4 months, OS 17.9 months) in patients with IHC3+ (⩾50% tumor cells with strong intensity) MET overexpression, while the IHC2+ (⩾50% tumor cells with moderate intensity) subgroup showed a poor response to combination therapy with a MET inhibitor and EGFR inhibitor. 115 Overall, IHC is the only detection method for MET expression in clinical trials. The cutoff for MET expression varies across studies, and 50% of tumor cells 2+/3+ are usually used as criteria. Among the studies in Table 5, subgroup analysis suggested that a higher percentage of tumor cell 3+ or the same percentage with a higher staining score generally improved clinical outcomes. Therefore, given the limited clinical studies and a small number of patients, higher MET overexpression can reflect better outcomes for MET-TKIs plus EGFR-TKIs treatment in EGFR-resistant advanced NSCLC patients harboring MET overexpression. More studies and specific diagnostic criteria for MET overexpression are required to identify patients who can benefit more from combination therapy with MET-TKIs and EGFR-TKIs in this setting.
Furthermore, MET overexpression is not a reliable indicator of MET exon 14 alterations or MET amplifications, and reports from several studies have indicated the same where MET overexpression determined by IHC does not correlate with MET amplification. 116 This difference in correlation might be due to the inclusion of samples (featuring a low level of amplification) that do not result in considerable protein expression or protein expression modulation by posttranscriptional and posttranslational factors. 3 Therefore, patients harboring activating alterations in MET can be investigated for the presence of MET overexpression, but unfortunately, MET overexpression is not a reliable indicator of MET amplifications/MET exon 14 skipping variants. 3 Furthermore, a growing number of clinical trials are in the pipeline to explore the relationship between MET expression and MET amplification; however, unifying guidelines for standard scoring systems for IHC are required to obtain consensus among different trials.
MET fusion
MET fusion in cancers
The MET fusion was first found in chemically transformed osteosarcoma cell lines, which was the TPR-MET fusion. 3 Thereafter, MET fusions were identified in a variety of tumors over the years, such as gastric cancers, lung adenocarcinoma, thyroid carcinoma, hepatocellular carcinoma, and glioma. 3 Beyond TPR-MET, multiple other fusions have been identified, including PTPRZ1-MET, CLIP2-MET CAPZA2-MET, ST7-MET, TRIM24-MET, KIF5B-MET, RBPMS-MET, and EML4-MET, most of which have been reported in case reports.117–122 The exact frequency of MET fusion in these cancers is poorly defined; of them, glioma had the highest proportion at 15%. 123 As a result, MET fusions and their therapeutic implications have been largely ignored. MET fusions have rarely been described in NSCLC, with an overall frequency of approximately 0.29%, and half of the fusion types are intragenic fusions.122,124 A large real-world multicenter study for the Chinese population detected putative MET fusions with a prevalence of 0.15% in 79,803 solid tumors, while the majority of them were lung cancer patients (75.4%). 125 It is worth considering that some patients with MET fusion can benefit from MET-TKI therapy.
Detection of MET fusion
Numerous assays can detect MET fusions, including FISH, RT-PCR, and NGS. However, complex/novel rearrangements may result in inadequate FISH for detecting many MET fusions. 3 Therefore, NGS has become the increasingly preferred assay in the clinic.
NGS with traditional amplicon-based enrichment is less accurate in identifying gene fusions with unknown partners, while a technique termed anchored multiplex PCR (AMP) possibly addresses this limitation. 126 In AMP, a ‘half-functional’ NGS adapter is ligated to cDNA fragments that are derived from input RNA, and then the amplification between gene-specific primers and a primer to the adapter leads to target enrichment. As a result, the gene fusions of interest, even if they involve a novel fusion partner, should be detected. 126 Currently, targeted RNA-based NGS (tRNA-seq) is increasingly being applied in molecular detection for gene fusion in solid tumors, which is efficient for the simultaneous detection of actionable gene fusions, splice variants, single nucleotide variants (SNVs), and indels. 127 RNA-seq is not only a well-validated tool for detecting gene fusions in fresh-frozen tumors but also in formalin-fixed, paraffin-embedded (FFPE) tumor samples. It showed a sensitivity of 83.3% in clinical FFPE specimens, with a negative prediction value of 94.3%, and was regarded as a complement DNA-based NGS assay. 128
In NSCLC, sequentially combining DNA NGS and RNA NGS was shown to be one of the most efficient strategies for fusion detection; it was feasible on small tissue samples and could drastically reduce the complexity and cost of molecular workup. 129 Song et al. 130 developed a convenient single-tube, dual-template assay, and an integrated bioinformatics pipeline for relevant variant calling, in which RNA was used for fusion detection, whereas DNA was used for SNVs and insertion and deletions (indels). This method was considered to benefit not only most patients carrying target fusion but also those with rare variations. 130 Wei et al. 131 designed a lung-cancer-specific targeted all-in-one transcriptome-based assay based on single primed enrichment technology which covered gene loci that are related to selecting optimal targeted therapy in advanced NSCLC and could simultaneously identify mutations, gene fusions, and exon skipping events. This assay was shown to identify all the expected mutations at the transcriptome level and to reach an accuracy of close to 100%. 131
Targeted therapies for MET fusion
Precisely targeted therapy has been incredibly underexplored in MET fusion-positive cancers. Nevertheless, in recent years, there have been many clinical case reports presenting the potential for MET-TKI therapy. Among these, crizotinib (monotherapy or combination therapy) has been described as having surprising clinical responses in patients with a variety of MET fusion-positive glioblastoma and lung adenocarcinomas, including the gene fusion types CUX1-MET, HLA-DRB1-MET, CAV1-MET, ARL1-MET, PRKAR1A-MET, bringing substantial tumor shrinkage and associated relief of symptoms.132–137 Blanc-Durand et al. 138 reported a patient with NSCLC with brain metastasis harboring an HLA-DRB1-MET gene fusion who successively received crizotinib and cabozantinib and the selective inhibitor tepotinib and experienced rapid responses associated with a tremendous improvement in physical function during each treatment cycle. The potential role of capmatinib was also reported in a patient with chemotherapy-refractory metastatic cholangiocarcinoma harboring a CAPZA-2-MET fusion. 139 Kang et al. 140 attempted to explain the potential resistance mechanisms of MET inhibitors in patients with de novo MET fusions and found that secondary mutations D1228H/N or D1246N are worth further exploration. Multiple clinical trials are ongoing to evaluate the efficacy of MET-TKIs in tumor patients with MET fusions (NCT03993873, NCT02978261, NCT01639508, and CTR20181664 141 ).
Conclusion
The pivotal role of MET aberrations as a predictive biomarker of drug response has been reported in several clinical trials. Furthermore, several MET inhibitors demonstrated clinically meaningful efficacy in different cancers harboring MET alterations. Therefore, MET exon 14 skipping variant testing has gained prominence and has already been recommended in guidelines where capmatinib, tepotinib, and savolitinib have been approved for the treatment of NSCLCs. Furthermore, other small-molecule inhibitors, including cabozantinib and crizotinib, are in the pipeline. The literature suggests that assays such as NGS (DNA based and RNA based) could be a potential testing method in terms of sensitivity and operational procedures for the detection of MET alterations, specifically MET exon 14 skipping variants, in both tissue samples and plasma ctDNA, but may have limitations for CNV testing. In addition, the FISH assay remains a robust technique for MET amplification detection. However, it can be used only for single-gene tests and tissue samples, while NGS represents the future trend of testing choice in multialteration (MET exon 14 skipping variant, amplification, and fusion) multigene analysis and in situations of limitation to plasma samples. NGS seems to be a promising testing option. ddPCR is being developed for MET amplification testing, especially in blood. MET amplification and MET overexpression are continuous variables, so clinically meaningful cutoff points need to be standardized, particularly the cutoff for MET overexpression. MET overexpression is an emerging biomarker for MET-TKI treatment since an increasing amount of clinical data have been released to guide the treatment. Furthermore, prospective studies involving a wide range of cancer types and larger sample sizes are required in this direction for definite conclusions and to extend the spectrum of MET-targeted therapy.
Acknowledgments
None.
Footnotes
ORCID iD: Pei Yuan  https://orcid.org/0000-0002-4663-7731
https://orcid.org/0000-0002-4663-7731
Contributor Information
Pei Yuan, Department of Pathology, State Key Laboratory of Molecular Oncology, National Cancer Center/National Clinical Research Center for Cancer/ Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
Xuemin Xue, Department of Pathology, State Key Laboratory of Molecular Oncology, National Cancer Center/National Clinical Research Center for Cancer/ Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
Tian Qiu, Department of Pathology, State Key Laboratory of Molecular Oncology, National Cancer Center/National Clinical Research Center for Cancer/ Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China.
Jianming Ying, Department of Pathology, State Key Laboratory of Molecular Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, No.17, Panjiayuan Nanli, Chaoyang District, Beijing 100021, China.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Pei Yuan: Conceptualization; Data curation; Formal analysis; Writing – original draft.
Xuemin Xue: Data curation; Methodology; Writing – review & editing.
Tian Qiu: Data curation; Methodology; Writing – review & editing.
Jianming Ying: Conceptualization; Data curation; Formal analysis; Funding acquisition; Supervision; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Key Research and Development Program (2022YFC2409902), CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-1-I2M-012, 2022-I2M-C&T-B-078), and Beijing Hope Run Special Fund of Cancer Foundation of the People’s Republic of China (LC2019L04).
The authors declare that there is no conflict of interest.
Availability of data and materials: Main data are shown in this article and additional data about this study could be obtained from the corresponding author on reasonable request.
References
- 1. Cho YA, Kim EK, Heo SJ, et al. Alteration status and prognostic value of MET in head and neck squamous cell carcinoma. Cancer 2016; 7: 2197–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guo A, Villén J, Kornhauser J, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA 2008; 105: 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo R, Luo J, Chang J, et al. MET-dependent solid tumours - molecular diagnosis and targeted therapy. Nat Rev Clin Oncol 2020; 17: 569–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moosavi F, Giovannetti E, Saso L, et al. HGF/MET pathway aberrations as diagnostic, prognostic, and predictive biomarkers in human cancers. Crit Rev Clin Lab Sci 2019; 56: 533–566. [DOI] [PubMed] [Google Scholar]
- 5. Tovar EA, Graveel CR. MET in human cancer: germline and somatic mutations. Ann Transl Med 2017; 5: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015; 5: 850–859. [DOI] [PubMed] [Google Scholar]
- 7. Markham A. Tepotinib: First Approval. Drugs 2020; 80: 829–833. [DOI] [PubMed] [Google Scholar]
- 8. Dhillon S. Capmatinib: First Approval. Drugs 2020; 80: 1125–1131. [DOI] [PubMed] [Google Scholar]
- 9. Markham A. Savolitinib: First Approval. Drugs 2021; 81: 1665–1670. [DOI] [PubMed] [Google Scholar]
- 10. Ahn MJ, De Marinis F, Bonanno L, et al. EP08.02-140 MET biomarker-based preliminary efficacy analysis in SAVANNAH: savolitinib+osimertinib in EGFRm NSCLC post-osimertinib. J Thorac Oncol 2022; 17: S469–S470. [Google Scholar]
- 11. Pilotto S, Gkountakos A, Carbognin L, et al. MET exon 14 juxtamembrane splicing mutations: clinical and therapeutical perspectives for cancer therapy. Ann Transl Med 2017; 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sequist LV, Han JY, Ahn MJ, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020; 21: 373–386. [DOI] [PubMed] [Google Scholar]
- 13. Li Y, Gao L, Ma D, et al. Identification of MET exon14 skipping by targeted DNA- and RNA-based next-generation sequencing in pulmonary sarcomatoid carcinomas. Lung Cancer 2018; 122: 113–119. [DOI] [PubMed] [Google Scholar]
- 14. Yu Y, Zhang Q, Zhang J, et al. Prevalence of MET exon 14 skipping mutation in pulmonary sarcomatoid carcinoma patients without common targetable mutations: a single-institute study. J Cancer Res Ther 2019; 15: 909–913. [DOI] [PubMed] [Google Scholar]
- 15. Vuong HG, Ho ATN, Altibi AMA, et al. Clinicopathological implications of MET exon 14 mutations in non-small cell lung cancer – a systematic review and meta-analysis. Lung Cancer 2018; 123: 76–82. [DOI] [PubMed] [Google Scholar]
- 16. Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 2016; 34: 721–730. [DOI] [PubMed] [Google Scholar]
- 17. Lung J, Hung MS, Lin YC, et al. MET exon 14 skipping mutations and gene amplification in a Taiwanese lung cancer population. PLoS One 2019; 14: e0220670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okuda K, Sasaki H, Yukiue H, et al. Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer Sci 2008; 99: 2280–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pruis MA, Geurts-Giele WRR, von der TJH, et al. Highly accurate DNA-based detection and treatment results of MET exon 14 skipping mutations in lung cancer. Lung Cancer 2020; 140: 46–54. [DOI] [PubMed] [Google Scholar]
- 20. Heist RS, Shim HS, Gingipally S, et al. MET exon 14 skipping in non-small cell lung cancer. Oncologist 2016; 21: 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu Z, Li H, Dong Y, et al. Incidence and PD-L1 expression of MET 14 skipping in Chinese population: A Non-Selective NSCLC cohort study using RNA-Based sequencing. Onco Targets Ther 2020; 13: 6245–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dubbink H, Geurts-Giele W, Meijssen I, et al. One year experience of MET gene exon 14 skipping analysis in lung cancer: identification of 18 cases by NGS. J Clin Oncol 2017; 35: e20055–e20055. [Google Scholar]
- 23. Lee J, Ou SH, Lee JM, et al. Gastrointestinal malignancies harbor actionable MET exon 14 deletions. Oncotarget 2015; 6: 28211–28222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu H, Mu Q, Bao Z. Mutational landscape of secondary glioblastoma guides MET-Targeted trial in brain tumor. Cell 2018; 175: 1665–1678.e1618. [DOI] [PubMed] [Google Scholar]
- 25. McCombie WR, McPherson JD, Mardis ER. Next-generation sequencing technologies. Cold Spring Harb Perspect Med 2019; 9: a036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirot B, Doucet L, Benhenda S, et al. MET exon 14 alterations and new resistance mutations to tyrosine kinase inhibitors: risk of inadequate detection with current amplicon-based NGS panels. J Thorac Oncol 2017; 12: 1582–1587. [DOI] [PubMed] [Google Scholar]
- 27. Davies KD, Lomboy A, Lawrence CA, et al. DNA-based versus RNA-based detection of MET exon 14 skipping events in lung cancer. J Thorac Oncol 2019; 14: 737–741. [DOI] [PubMed] [Google Scholar]
- 28. Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture-based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clin Cancer Res 2015; 21: 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Samorodnitsky E, Jewell BM, Hagopian R, et al. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat 2015; 36: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009; 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 31. Huber D, Voith von Voithenberg L, Kaigala GV, et al. Fluorescence in situ hybridization (FISH): history, limitations and what to expect from micro-scale fish? Micro Nano Eng 2018; 1: 15–24. [Google Scholar]
- 32. Jurkiewicz M, Saqi A, Mansukhani M, et al. Efficacy of DNA versus RNA NGS-based methods in MET Exon 14 skipping mutation detection. J Clin Oncol 2020; 38: 9036-9036. [Google Scholar]
- 33. Farragher SM, Tanney A, Kennedy RD, et al. RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem Cell Biol 2008; 130: 435–445. [DOI] [PubMed] [Google Scholar]
- 34. O'Brien O, Wright MC, O'Brien C, et al. Cost-efficient and easy to perform PCR-Based assay to identify met exon 14 skipping in formalin-fixed paraffin-embedded (FFPE) Non-Small cell lung cancer (NSCLC) samples. Diagnostics 2019; 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolf J, Seto T, Han J-Y, et al. Capmatinib in METExon 14–mutated orMET-Amplified non-small-Cell lung cancer. New Engl J Med 2020; 383: 944–957. [DOI] [PubMed] [Google Scholar]
- 36. Paik P, Felip E, Veillon R, et al. Tepotinib in non-small-cell lung cancer with MET exon 14 skipping mutations. N Engl J Med 2020; 383: 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu S, Fang J, Li X, et al. Once-daily savolitinib in Chinese patients with pulmonary sarcomatoid carcinomas and other non-small-cell lung cancers harbouring MET exon 14 skipping alterations: a multicentre, single-arm, open-label, phase 2 study. Lancet Respir Med 2021; 9: 1154–1164. [DOI] [PubMed] [Google Scholar]
- 38. Yu Y, Zhou J, Li X, et al. Gumarontinib in patients with non-small-cell lung cancer harbouring MET exon 14 skipping mutations: a multicentre, single-arm, open-label, phase 1b/2 trial. EClinicalMedicine 2023; 59: 101952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mazieres J, Paik PK, Garassino MC, et al. Tepotinib treatment in patients with MET exon 14-Skipping Non-Small cell lung cancer: long-term follow-up of the VISION Phase 2 nonrandomized clinical trial. JAMA Oncol 2023; 9: 1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazieres J, Paik PK, Felip E, et al. 1283P tepotinib in patients (pts) with advanced NSCLC with MET exon 14 (METex14) skipping: overall efficacy results from VISION cohort A. Ann Oncol 2020; 31: S828–S829. [Google Scholar]
- 41. Vansteenkiste JF, Demedts I, Pons-Tostivint E, et al. 1176P open-label, dose escalation, phase I/II study to assess safety, tolerability, immunogenicity and preliminary clinical activity of the therapeutic cancer vaccine PDC*lung01 with or without anti-programmed death-1 (PD-1) treatment in patients with non-small cell lung cancer (NSCLC). Ann Oncol 2022; 33: S1086. [Google Scholar]
- 42. Lu S, Yu Y, Guo Q, et al. OA21.03 A phase 3b study of 1L savolitinib in patients with locally advanced or metastatic NSCLC Harboring MET exon 14 mutation. J Thorac Oncol 2023; 18: S92–S93. [Google Scholar]
- 43. Lu S, Fang J, Li X, et al. Phase II study of savolitinib in patients (pts) with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations (METex14+). J Clin Oncol 2020; 38: 9519–9519. [Google Scholar]
- 44. Drilon A, Clark JW, Weiss J, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med 2020; 26: 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moro-Sibilot D, Cozic N, Pérol M, et al. Crizotinib in c-MET- or ROS1-positive NSCLC: results of the AcSé phase II trial. Ann Oncol 2019; 30: 1985–1991. [DOI] [PubMed] [Google Scholar]
- 46. Lu S, Fang J, Li X, et al. Long-term efficacy, safety, and subgroup analysis of savolitinib in Chinese patients with NSCLCs harboring MET exon 14 skipping alterations. JTO Clin Res Rep 2022; 3: 100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reams AB, Roth JR. Mechanisms of gene duplication and amplification. Cold Spring Harb Perspect Biol 2015; 7: a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–1043. [DOI] [PubMed] [Google Scholar]
- 49. Schildhaus HU, Schultheis AM, Rüschoff J, et al. MET amplification status in therapy-naïve adeno- and squamous cell carcinomas of the lung. Clin Cancer Res 2015; 21: 907–915. [DOI] [PubMed] [Google Scholar]
- 50. Peng Z, Zhu Y, Wang Q, et al. Prognostic significance of MET amplification and expression in gastric cancer: a systematic review with meta-analysis. PLoS One 2014; 9: e84502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robinson KW, Sandler AB. The role of MET receptor tyrosine kinase in non-small cell lung cancer and clinical development of targeted anti-MET agents. Oncologist 2013; 18: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamashita Y, Akatsuka S, Shinjo K, et al. Met is the most frequently amplified gene in endometriosis-associated ovarian clear cell adenocarcinoma and correlates with worsened prognosis. PLoS One 2013; 8: e57724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013; 3: 658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tong JH, Yeung SF, Chan AW, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of Non-Small cell lung carcinoma with poor prognosis. Clin Cancer Res 2016; 22: 3048–3056. [DOI] [PubMed] [Google Scholar]
- 55. Go H, Jeon YK, Park HJ, et al. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol 2010; 5: 305–313. [DOI] [PubMed] [Google Scholar]
- 56. Okamoto I, Sakai K, Morita S, et al. Multiplex genomic profiling of non-small cell lung cancers from the LETS phase III trial of first-line S-1/carboplatin versus paclitaxel/carboplatin: results of a West Japan Oncology Group study. Oncotarget 2014; 5: 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Suryavanshi M, Shah A, Kumar D, et al. MET amplification and response to MET inhibitors in stage IV lung adenocarcinoma. Oncol Res Treat 2017; 40: 198–202. [DOI] [PubMed] [Google Scholar]
- 58. Guo B, Cen H, Tan X, et al. Prognostic value of MET gene copy number and protein expression in patients with surgically resected non-small cell lung cancer: a meta-analysis of published literatures. PLoS One 2014; 9: e99399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009; 4: 5–11. [DOI] [PubMed] [Google Scholar]
- 60. Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA 2007; 104: 20932–20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xia N, An J, Jiang QQ, et al. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res 2013; 39: 328–335. [DOI] [PubMed] [Google Scholar]
- 62. Xu CW, Wang WX, Wu MJ, et al. Comparison of the c-MET gene amplification between primary tumor and metastatic lymph nodes in non-small cell lung cancer. Thorac Cancer 2017; 8: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen D, Xu C, Wu J, et al. A comparison of consistency of detecting c-MET gene amplification in peripheral blood and tumor tissue of nonsmall cell lung cancer patients. J Cancer Res Ther 2015; 11 Suppl 1: C63–C67. [DOI] [PubMed] [Google Scholar]
- 64. Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naïve cohort. J Thorac Oncol 2008; 3: 331–339. [DOI] [PubMed] [Google Scholar]
- 65. Tsuta K, Kozu Y, Mimae T, et al. c-MET/phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol 2012; 7: 331–339. [DOI] [PubMed] [Google Scholar]
- 66. Zhang M, Li G, Sun X, et al. MET amplification, expression, and exon 14 mutations in colorectal adenocarcinoma. Hum Pathol 2018; 77: 108–115. [DOI] [PubMed] [Google Scholar]
- 67. Jardim DLF, Tang C, Gagliato DDM, et al. Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I clinic. Clin Cancer Res 2014; 20: 6336–6345. [DOI] [PubMed] [Google Scholar]
- 68. Raghav K, Morris V, Tang C, et al. MET amplification in metastatic colorectal cancer: an acquired response to EGFR inhibition, not a de novo phenomenon. Oncotarget 2016; 7: 54627–54631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu S, Yu Y, Zhao N, et al. C-Met as a prognostic marker in gastric cancer: a systematic review and meta-analysis. PLoS One 2013; 8: e79137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Graziano F, Galluccio N, Lorenzini P, et al. Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. J Clin Oncol 2011; 29: 4789–4795. [DOI] [PubMed] [Google Scholar]
- 71. de Melo Gagliato D, Jardim DL, Falchook G, et al. Analysis of MET genetic aberrations in patients with breast cancer at MD Anderson Phase I unit. Clin Breast Cancer 2014; 14: 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pal SK, Ali SM, Yakirevich E, et al. Characterization of clinical cases of advanced papillary renal cell carcinoma via comprehensive genomic profiling. Eur Urol 2018; 73: 71–78. [DOI] [PubMed] [Google Scholar]
- 73. Kato H, Arao T, Matsumoto K, et al. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. Int J Oncol 2013; 42: 1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lai GGY, Lim TH, Lim J, et al. Clonal MET amplification as a determinant of tyrosine kinase inhibitor resistance in epidermal growth factor receptor–mutant Non–Small-Cell lung cancer. J Clin Oncol 2019; 37: 876–884. [DOI] [PubMed] [Google Scholar]
- 75. Noonan SA, Berry L, Lu X, et al. Identifying the appropriate FISH criteria for defining MET copy number-driven lung adenocarcinoma through oncogene overlap analysis. J Thorac Oncol 2016; 11: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J Cell Sci 2003; 116: 2833–2838. [DOI] [PubMed] [Google Scholar]
- 77. Cappuzzo F, Jänne PA, Skokan M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 2009; 20: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yin X, Zhang T, Su X, et al. Relationships between chromosome 7 gain, MET Gene copy number increase and MET protein overexpression in Chinese papillary renal cell carcinoma patients. PLoS One 2015; 10: e0143468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lee HE, Kim MA, Lee HS, et al. MET in gastric carcinomas: comparison between protein expression and gene copy number and impact on clinical outcome. Br J Cancer 2012; 107: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med 2017; 141: 751–758. [DOI] [PubMed] [Google Scholar]
- 81. Sun B, Qiu T, Zeng X, et al. Detection of MET Polysomy by next-generation sequencing and its clinical relevance for MET inhibitors. Cancer Res Commun 2023; 3: 532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wu Z, Yang Z, Dai Y, et al. Update on liquid biopsy in clinical management of non-small cell lung cancer. Onco Targets Ther 2019; 12: 5097–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee J, Kim ST, Kim K, et al. Tumor genomic profiling guides patients with metastatic gastric cancer to targeted treatment: the VIKTORY Umbrella trial. Cancer Discov 2019; 9: 1388–1405. [DOI] [PubMed] [Google Scholar]
- 84. Heydt C, Becher AK, Wagener-Ryczek S, et al. Comparison of in situ and extraction-based methods for the detection of MET amplifications in solid tumors. Comput Struct Biotechnol J 2019; 17: 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fan Y, Sun R, Wang Z, et al. Detection of MET amplification by droplet digital PCR in peripheral blood samples of non-small cell lung cancer. J Cancer Res Clin Oncol 2023; 149: 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fujino T, Suda K, Mitsudomi T. Emerging MET tyrosine kinase inhibitors for the treatment of non-small cell lung cancer. Expert Opin Emerg Drugs 2020; 25: 229–249. [DOI] [PubMed] [Google Scholar]
- 87. Camidge D, Otterson G, Clark J, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. J Thorac Oncol 2018; 36: 9062–9062. [Google Scholar]
- 88. Shah MA, Wainberg ZA, Catenacci DV, et al. Phase II study evaluating 2 dosing schedules of oral foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with metastatic gastric cancer. PLoS One 2013; 8: e54014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Camidge DR, Otterson GA, Clark JW, et al. Crizotinib in patients with MET-amplified NSCLC. J Thorac Oncol 2021; 16: 1017–1029. [DOI] [PubMed] [Google Scholar]
- 90. Schuler M, Berardi R, Lim WT, et al. Phase (Ph) I study of the safety and efficacy of the cMET inhibitor capmatinib (INC280) in patients (pts) with advanced cMET+ non-small cell lung cancer (NSCLC). J Clin Oncol 2016; 34: 9067–9067. [Google Scholar]
- 91. Wu YL, Zhang L, Kim DW, et al. Phase Ib/II study of capmatinib (INC280) plus gefitinib after failure of epidermal growth factor receptor (EGFR) inhibitor therapy in patients with EGFR-mutated, MET factor-dysregulated non-small-cell lung cancer. J Clin Oncol 2018; 36: 3101–3109. [DOI] [PubMed] [Google Scholar]
- 92. Yu HA, Ambrose H, Baik C, et al. 1239P ORCHARD osimertinib + savolitinib interim analysis: A biomarker-directed phase II platform study in patients (pts) with advanced non-small cell lung cancer (NSCLC) whose disease has progressed on first-line (1L) osimertinib. Ann Oncol 2021; 32: S978–S979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hartmaier RJ, Markovets AA, Ahn MJ, et al. Osimertinib + savolitinib to overcome acquired MET-Mediated resistance in epidermal growth factor Receptor-Mutated, MET-amplified Non-Small cell lung cancer: TATTON. Cancer Discov 2023; 13: 98–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liam CK, Ahmad AR, Hsia TC, et al. Randomized trial of tepotinib plus gefitinib versus chemotherapy in EGFR-Mutant NSCLC with EGFR Inhibitor Resistance due to MET amplification: INSIGHT Final analysis. Clin Cancer Res 2023; 29: 1879–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kim TM, Guarneri V, Jye VP, et al. OA21.05 tepotinib + Osimertinib in EGFR-mutant NSCLC with MET amplification following 1L osimertinib: INSIGHT 2 primary analysis. J Thorac Oncol 2023; 18: S94. [Google Scholar]
- 96. Yu Y, Yang N, Zhang Y, et al. 305MO SCC244 plus osimertinib in patients with stage IIIB/IIIC or IV, EGFR TKI resistant EGFR-mutant NSCLC harboring MET amplification. Ann Oncol 2022; 33: S1553. [Google Scholar]
- 97. Qin S, Chan SL, Sukeepaisarnjaroen W. Erratum to a phase II study of the efficacy and safety of the MET inhibitor capmatinib (INC280) in patients with advanced hepatocellular carcinoma. Ther Adv Med Oncol 2020; 12: 1758835920913426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bang YJ, Su WC, Schuler M, et al. Phase 1 study of capmatinib in MET-positive solid tumor patients: dose escalation and expansion of selected cohorts. Cancer Sci 2020; 111: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Angevin E, Spitaleri G, Rodon J, et al. A first-in-human phase I study of SAR125844, a selective MET tyrosine kinase inhibitor, in patients with advanced solid tumours with MET amplification. Eur J Cancer 2017; 87: 131–139. [DOI] [PubMed] [Google Scholar]
- 100. Kim Y, Bang SS, Jee S, et al. Prevalence and clinicopathological significance of MET overexpression and gene amplification in patients with gallbladder carcinoma. Cancer Res Treat 2020; 52: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Weng TH, Yao MY, Xu XM, et al. RON and MET Co-overexpression are significant pathological characteristics of poor survival and therapeutic targets of tyrosine kinase inhibitors in Triple-negative breast cancer. Cancer Res Treat 2020; 52: 973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ding C, Qiu Y, Zhang J, et al. Clinicopathological characteristics of Non-Small cell lung cancer (NSCLC) patients with c-MET exon 14 skipping mutation, MET overexpression and amplification. BMC Pulm Med 2023; 23: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xu X, Zhang G, He L, et al. Clinicopathological impacts of c-Met overexpression in bladder cancer: evidence from 1,336 cases. Onco Targets Ther 2019; 12: 2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Anton J, Sudibio S, Handoko H, et al. Overexpression of c-Met is associated with poor prognosis in glioblastoma multiforme: a systematic review and meta-analyses. Asian Pac J Cancer Prev 2021; 22: 3075–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yin W, Guo M, Tang Z, et al. MET expression level in lung adenocarcinoma loosely correlates with MET copy number gain/amplification and is a poor predictor of patient outcome. Cancers 2022; 14: 2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2013; 31: 4105–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. score MS, C, ASCO Post-Calculating H-Score, https://www.ascopost.com/issues/april-10-2015/calculating-h-score/ (2015).
- 108. Mignard X, Ruppert AM, Antoine M, et al. C-MET overexpression as a poor predictor of MET amplifications or exon 14 mutations in lung sarcomatoid carcinomas. J Thorac Oncol 2018; 13: 1962–1967. [DOI] [PubMed] [Google Scholar]
- 109. Standardization Interpretation Expert Committee of MET immunohistochemistry; Molecular Pathology Collaboration Group of Tumor Pathology Committee of Chinese Anti-Cancer Association. Chinese expert consensus on MET immunohistochemistry detection and interpretation standards for non-small cell lung cancer(2023 version). Zhonghua Bing Li Xue Za Zhi 2023; 52: 1090–1097. [DOI] [PubMed] [Google Scholar]
- 110. Camidge D, Bar J, Horinouchi H, et al. Telisotuzumab vedotin (Teliso-V) monotherapy in patients (pts) with previously treated c-Met–overexpressing (OE) advanced non-small cell lung cancer (NSCLC). J Clin Oncol 2022; 40: 9016–9016. [Google Scholar]
- 111. Besse B, Baik C, Marmarelis M, et al. Predictive biomarkers for treatment with amivantamab plus lazertinib among EGFR-mutated NSCLC in the post-osimertinib setting: Analysis of tissue IHC and ctDNA NGS. J Clin Oncol. 2023; 41: 9013–9013. [Google Scholar]
- 112. Goldman J, Horinouchi H, Cho B, et al. Phase 1/1b study of telisotuzumab vedotin (Teliso-V) + osimertinib (Osi), after failure on prior Osi, in patients with advanced, c-Met overexpressing, EGFR-mutated non-small cell lung cancer (NSCLC). J Clin Oncol. 2022; 40: 9013–9013. [Google Scholar]
- 113. Zhang J, Guo L, Liu X, et al. MET overexpression, gene amplification and relevant clinicopathological features in gastric adenocarcinoma. Oncotarget 2017; 8: 10264–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Goyal L, Zheng H, Yurgelun MB, et al. A phase 2 and biomarker study of cabozantinib in patients with advanced cholangiocarcinoma. Cancer 2017; 123: 1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wu YL, Cheng Y, Zhou J, et al.; INSIGHT Investigators. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med 2020; 8: 1132–1143. [DOI] [PubMed] [Google Scholar]
- 116. Park S, Koh J, Kim DW, et al. MET amplification, protein expression, and mutations in pulmonary adenocarcinoma. Lung Cancer 2015; 90: 381–387. [DOI] [PubMed] [Google Scholar]
- 117. Chowdhury T, Lee Y, Kim S, et al. A glioneuronal tumor with CLIP2-MET fusion. NPJ Genomic Med 2020; 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chai R, Liu X, Pang B, et al. Recurrent PTPRZ1-MET fusion and a high occurrence rate of MET exon 14 skipping in brain metastases. Cancer Sci 2022; 113: 796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hiemenz MC, Skrypek MM, Cotter JA, et al. Novel TRIM24-MET Fusion in a neonatal brain tumor. JCO Precis Oncol 2019; 3: 1–6. [DOI] [PubMed] [Google Scholar]
- 120. Wu ZW, Sha Y, Chen Q, et al. Novel intergenic KIF5B-MET fusion variant in a patient with gastric cancer: A case report. World J Clin Cases 2021; 9: 3350–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gupta A, Belsky JA, Schieffer KM, et al. Infantile fibrosarcoma-like tumor driven by novel RBPMS-MET fusion consolidated with cabozantinib. Cold Spring Harb Mol Case Stud 2020; 6: a005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sun D, Wu W, Wang L, et al. Identification of MET fusions as novel therapeutic targets sensitive to MET inhibitors in lung cancer. J Transl Med 2023; 21: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zeng AL, Yan W, Liu YW, et al. Tumour exosomes from cells harbouring PTPRZ1-MET fusion contribute to a malignant phenotype and temozolomide chemoresistance in glioblastoma. Oncogene 2017; 36: 5369–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Riedel R, Fassunke J, Scheel AH, et al. MET fusions in NSCLC: Clinicopathologic features and response to MET inhibition. J Thorac Oncol 2024; 19: 160–165. [DOI] [PubMed] [Google Scholar]
- 125. Xia H, Zhang J, Chen T, et al. Molecular characterization of MET fusions from a large real-world Chinese population: a multicenter study. Cancer Med 2023; 12: 14015–14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Seager M, Aisner DL, Davies KD. Oncogenic gene fusion detection using anchored multiplex polymerase chain reaction followed by next generation sequencing. J Vis Exp 2019; 149: e59895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Claerhout S, Lehnert S, Vander Borght S, et al. Targeted RNA sequencing for upfront analysis of actionable driver alterations in non-small cell lung cancer. Lung Cancer 2022; 166: 242–249. [DOI] [PubMed] [Google Scholar]
- 128. Peng H, Huang R, Wang K, et al. Development and validation of an RNA sequencing assay for gene fusion detection in formalin-fixed, paraffin-embedded tumors. J Mol Diagn 2021; 23: 223–233. [DOI] [PubMed] [Google Scholar]
- 129. Cohen D, Hondelink LM, Solleveld-Westerink N, et al. Optimizing mutation and fusion detection in NSCLC by sequential DNA and RNA sequencing. J Thorac Oncol 2020; 15: 1000–1014. [DOI] [PubMed] [Google Scholar]
- 130. Song Z, Xu C, He Y, et al. Simultaneous detection of gene fusions and base mutations in cancer tissue biopsies by sequencing dual nucleic acid templates in unified reaction. Clin Chem 2020; 66: 178–187. [DOI] [PubMed] [Google Scholar]
- 131. Wei J, Rybczynska AA, Meng P, et al. An all-in-one transcriptome-based assay to identify therapy-guiding genomic aberrations in nonsmall cell lung cancer patients. Cancers 2020; 12: 2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. International Cancer Genome Consortium PedBrain Tumor Project. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med 2016; 22: 1314–1320. [DOI] [PubMed] [Google Scholar]
- 133. Ou L, Tang Y, Deng Y, et al. Case report: durable partial response to icotinib plus crizotinib in a lung adenocarcinoma patient with double uncommon EGFR G719D/L861Q mutations and an acquired novel CUX1-MET fusion. Front Oncol 2022; 12: 2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Davies KD, Ng TL, Estrada-Bernal A, et al. Dramatic response to crizotinib in a patient with lung cancer positive for an HLA-DRB1-MET Gene Fusion. JCO Precis Oncol 2017; 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Liu J, Li X, Peng J. A novel CAV1-MET fusion in SCLC transformation responds to crizotinib and osimertinib treatment. J Thorac Oncol 2019; 14: e126–e128. [DOI] [PubMed] [Google Scholar]
- 136. Ma Q, Kong L, Zhong D. Case report: dramatic response to crizotinib in a patient with Non-Small cell lung cancer positive for a novel ARL1-MET Fusion. Front Oncol 2022; 12: 804330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Yang Y, Zhang Y, Zhao D, et al. A novel PRKAR1A::MET Fusion dramatic response to crizotinib in a patient with unresectable lung cancer. Clin Lung Cancer 2023; 24: e50–e54. [DOI] [PubMed] [Google Scholar]
- 138. Blanc-Durand F, Alameddine R, Iafrate AJ, et al. Tepotinib efficacy in a patient with Non-Small cell lung cancer with brain metastasis harboring an HLA-DRB1-MET Gene Fusion. Oncologist 2020; 25: 916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Turpin A, Descarpentries C, Grégoire V, et al. Response to capmatinib in a MET fusion-positive cholangiocarcinoma. Oncologist 2023; 28: 80–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kang J, Deng QM, Feng W, et al. Response and acquired resistance to MET inhibitors in de novo MET fusion-positive advanced non-small cell lung cancer. Lung Cancer 2023; 178: 66–74. [DOI] [PubMed] [Google Scholar]
- 141. Bao Z, Li S, Wang L, et al. PTPRZ1-METFUsion GENe (ZM-FUGEN) trial: study protocol for a multicentric, randomized, open-label phase II/III trial. Chin Neurosurg J 2023; 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]