Abstract
Three-dimensional (3D) shape lies at the core of understanding the physical objects that surround us. In the biomedical field, shape analysis has been shown to be powerful in quantifying how anatomy changes with time and disease. The Shape AnaLysis Toolbox (SALT) was created as a vehicle for disseminating advanced shape methodology as an open source, free, and comprehensive software tool. We present new developments in our shape analysis software package, including easy-to-interpret statistical methods to better leverage the quantitative information contained in SALT’s shape representations. We also show SlicerPipelines, a module to improve the usability of SALT by facilitating the analysis of large-scale data sets, automating workflows for non-expert users, and allowing the distribution of reproducible workflows.
Keywords: Shape analysis, Statistics, Open-source software
1. Introduction
Statistical shape analysis is an essential area of research in computer science and mathematics, with application areas as diverse as biology, anatomy, agriculture, or paleontology. Shape represents critical morphometric features not encoded in simple derived measurements such as volume or linear distances. Over the past three decades, our research team and others have been developing shape analysis methodologies that are widely used within their own labs. However, these methods have not fully transitioned into clinical applications due to requiring the users to be experts in computer science or statistics.
SlicerSALT was created with the vision to integrate recent methodological advancements into a user-friendly toolkit. SlicerSALT meets the critical need of the broad scientific community to have access to technologies otherwise only available to specific computer science research labs, all while encouraging reproducibility and transparency. It benefits from a large international user community by making use of the 3D Slicer ecosystem as well as Kitware’s open source libraries such as the Insight Toolkit (ITK) [10] and the Visualization Toolkit (VTK) [12]. SALT follows strict quality standards for software development and testing. These factors position SlicerSALT as one of the leaders in shape analysis software development and dissemination and create conventions and quality standards for future work.
SlicerSALT fills a niche for researchers interested in a broad range of biomedical shape analysis tasks. Most other packages focus on tasks such as image segmentation (itkSNAP [16]) or specific neuroimaging pipelines (FreeSurfer [5], FSL [6]). There are only a few freely distributed toolkits for generic shape analysis tasks, including ShapeWorks [3], Deformetrica [4] or Statismo [1]. However, those packages each have a relatively narrow focus. Deformetrica deals with deformations of the 2D or 3D ambient space while ShapeWorks and Statismo are only designed for point distribution models (PDMs) or principal component analysis (PCA). SALT, on the other hand, provides users with a wide variety of tools for both creating different shape representations and performing statistical analysis to accomodate a wider range of applications.
This paper is intended to present the new features in SlicerSALT v4.0, as compared with previous versions [15]. Over the past six years SlicerSALT has provided biomedical researchers with access to sophisticated shape analysis methodology otherwise reserved for computer science experts. In this new release, we intend to expand this toolkit towards novel and interpretable shape statistics, in order to maximize the potential benefits of the geometric information contained in medical data and expand its use in order to support impactful biomedical research.
2. Shape Representations and Correspondence
In addition to standard PDM models computed using SPHARM-PDM, SlicerSALT 4.0 has improved support for skeletal shape models and producing correspondence via registration. We have also updated various visualization tools for examining shape populations for quality control.
2.1. Skeletal Representations
The functionality for creating, displaying, analyzing, and storing skeletal representations (s-reps) was completely re-engineered for SlicerSALT 4.0. These updates were necessary to make the tools easier to use as well as allowing reuse of the code in other modules.
We have developed a new SRep module that allows users to see basic information about s-reps as well as modify their display properties. The user can easily change spoke and skeletal grid thicknesses, colors, opacity, and visibility for different parts of the s-rep.
This module also adds a new Medical Reality Modeling Language (MRML) data node for s-reps. This allows modules in this and other extensions to easily use, manipulate, and display s-rep data as it would with other data types native to 3D Slicer. Additionally, this makes it easier to save, load and share s-rep files as they can be read and written via the standard 3D Slicer Save and Load buttons as well as loaded via drag-and-drop. The new JSON-based file format is also more human readable and allows the data to be read using standard JSON parsing libraries.
S-reps are fit to objects using the SRepCreator and SRepRefiner modules. First, an input surface is given to the SRepCreator which generates an s-rep using a method based on surface curvature flow to estimate a best-fitting ellipsoid [8]. An s-rep is analytically computed from the ellipsoid and propagated back to the original surface via thin-plate splines. The SRepRefiner takes this initial s-rep and uses an optimization process to improve its fit to the original object surface as well as geometric properties such as ensuring that s-rep spokes are orthogonal to the object boundary.
Other modules in SlicerSALT that work on s-reps have been modified to support this new format. These modifications will be detailed in the following relevant sections.
2.2. Registration Based Correspondence
Common techniques for creating shape representations, such as SPHARM-PDM, attempt to create models that already have inherent correspondence across a population. This works well for objects with simple topologies. However, for objects with more complicated geometries or topologies, other approaches are required. A variety of techniques have been developed to address this problem, but many of them do not have implementations that are available for wide use and do not come with groups of associated tools for analyzing the resulting representations.
SlicerSALT 4.0 now includes a module for creating corresponding representations of objects with complex geometry or non-spherical topology [14]. It takes a mesh of a representative object as a template and deforms this template to match other objects of the population via registration to create correspondence. We use ITK’s registration framework with the option of either a b-spline or diffeomorphic demons [13] registration to register a template surface mesh to each of the target objects. For this, we first create level-set representations of the template and target meshes by computing distance functions from each object. The template distance function is then registered to the target using the diffeomorphic demons registration algorithm. The resulting transform is then applied to the original template mesh, yielding a mesh that conforms to the target object’s boundary but has the topology of the template for all of the population. Figure 2 shows correspondences generated using this module.
Fig. 2.
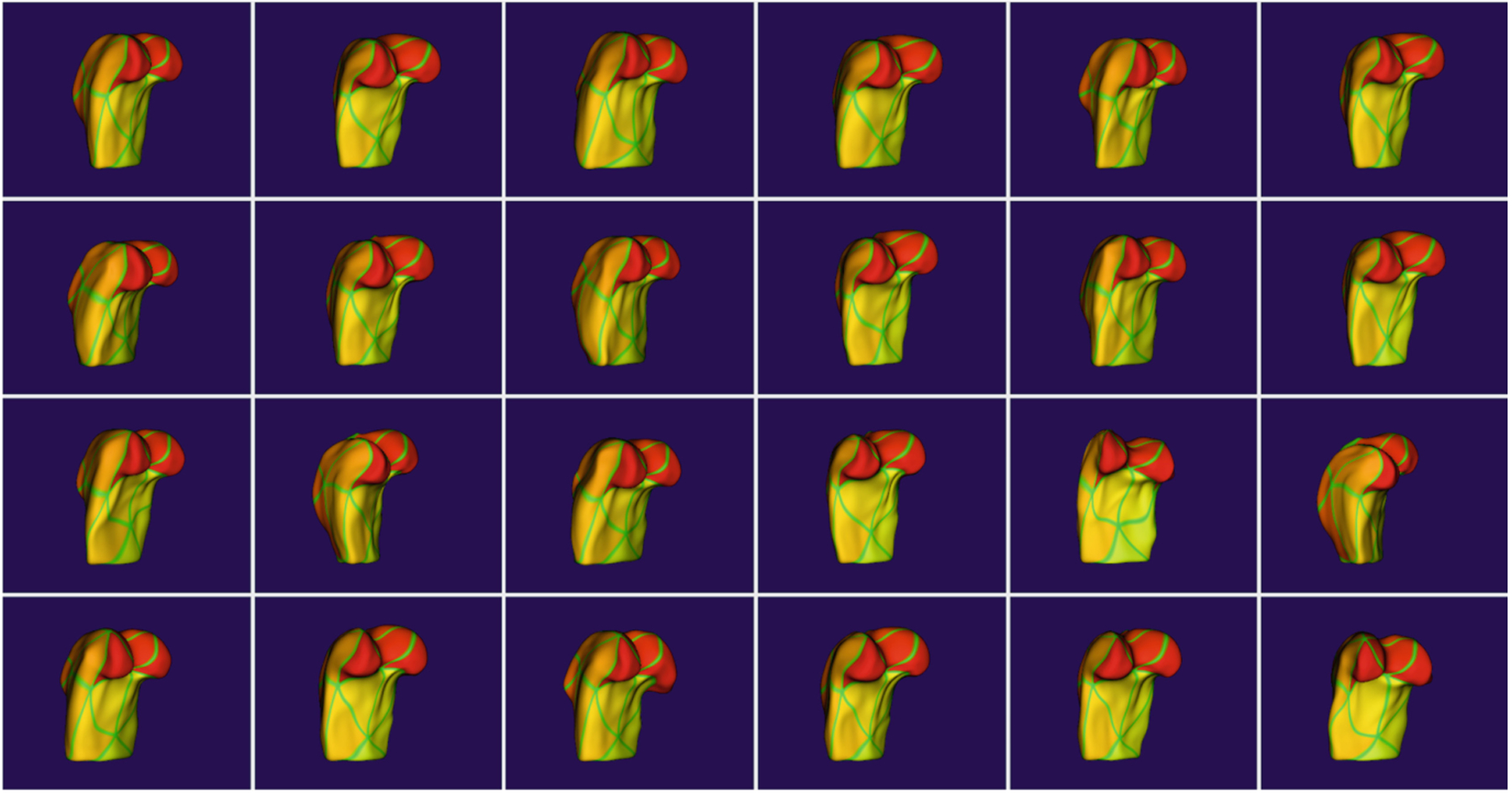
Corresponding femur models computed via registration-based correspondence. Similar coloring at similar anatomical locations show the quality of the correspondence.
2.3. Shape Population Viewer
Shape Population Viewer is a module that allows the user to quickly visualize an entire population of objects simultaneously. This allows the user to quickly assess population-wide correspondences such as in Fig. 2 or verify there are no ill-formed meshes. This module has been extended to support s-reps as seen in Fig. 3.
Fig. 3.
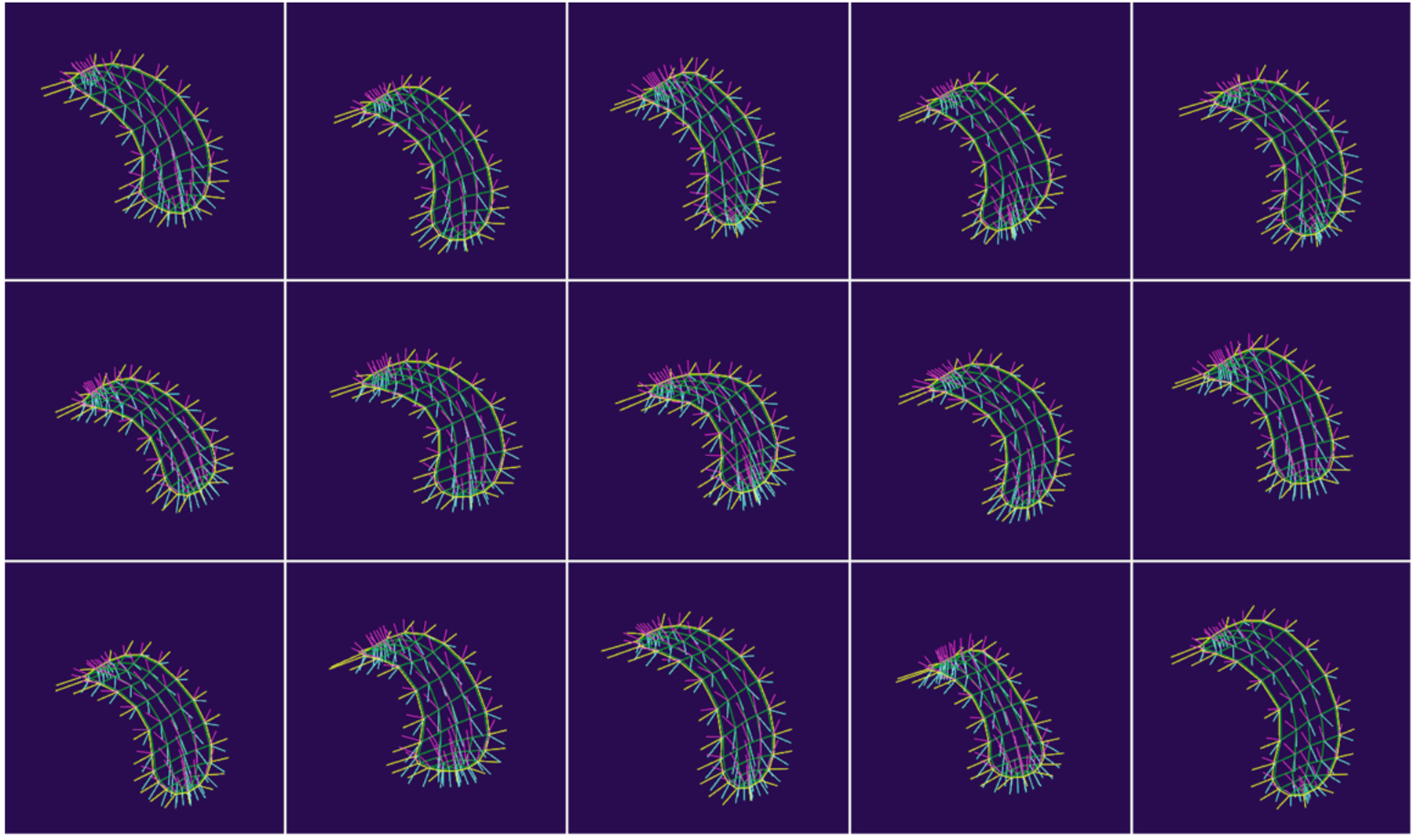
ShapePopulationViewer displaying a set of 15 s-reps.
3. Shape Analysis Methods
SlicerSALT 4.0 introduces changes to several existing shape analysis modules as well as introducing several new ones.
3.1. Shape Variation Analyzer
The Shape Variation Analyzer (SVA) module is designed to take in a set of corresponding PDMs or s-reps and use principal component analysis (PCA) or composite principal nested spheres (CPNS) [11] to compute a mean shape and its major modes of variation. The user can then visualize the mean shape as well as how moving along each principal component changes the shape of the object. This module allows for a quantitative comparison of the generated shape space to those created by other methods by examining the percent of variation explained by the modes of variation as well as computing generalization, specificity, and compactness measures on the distribution.
PCA.
For Euclidean shape representations such as PDMs, SVA allows the computation of PCA directly on the point positions of the shape representation. SVA also allows the user to view the mean shape and explore the modes of variation of the PCA shape space and to evaluate the quality of the generated models as seen in Fig. 4.
Fig. 4.
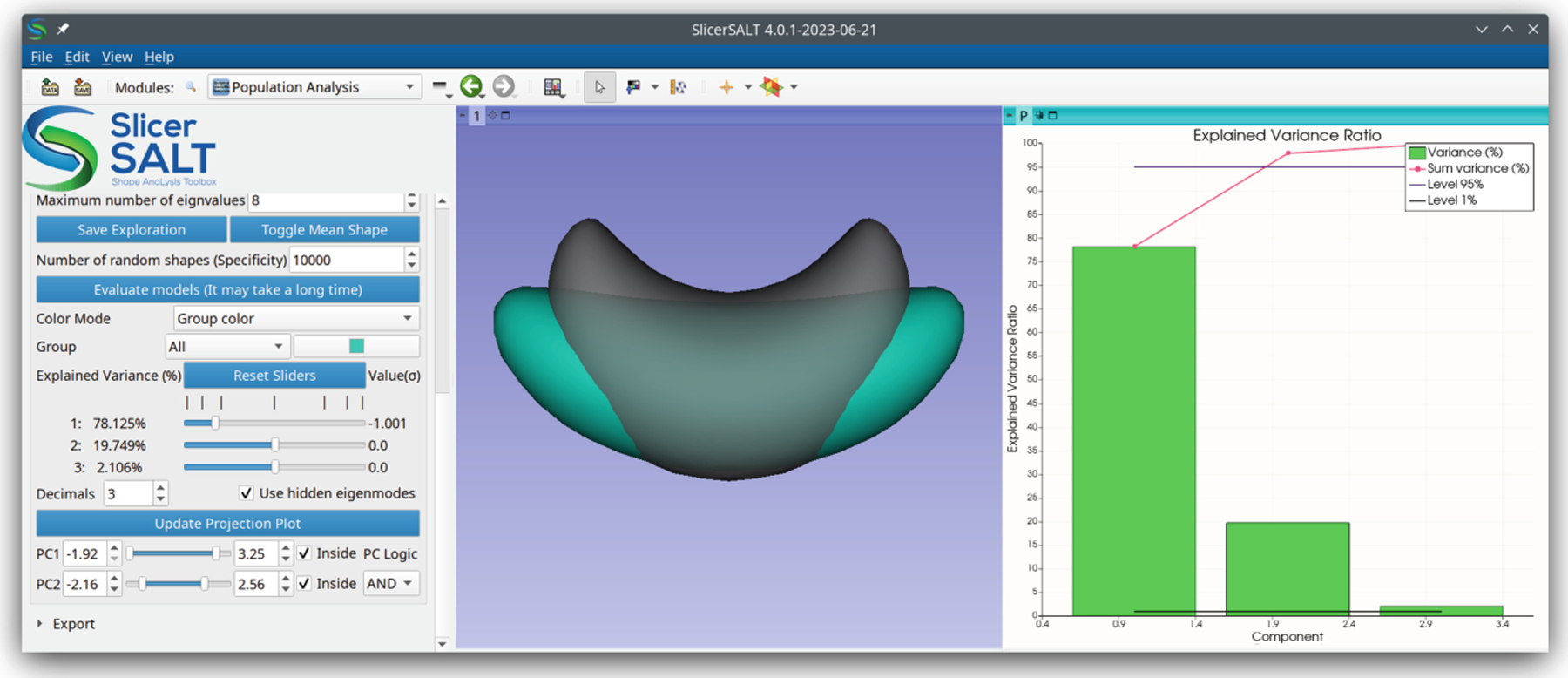
A screenshot of the SVA module in SlicerSALT. The left panel shows controls, the middle shows a mean shape (gray, transparent) alongside a shape (blue) deformed along its first principal component, and the right shows a PCA scree plot. (Color figure online)
The inputs are a list of VTK mesh files that are in correspondence. PCA results can be saved using the “Save Exploration” button and loaded using the “JSON File” entry.
Principal Nested Spheres.
Because s-reps are non-Euclidean, the s-rep data must first be Euclideanized using principal nested spheres (PNS) [7] before PCA can be computed. This approach is known as composite principal nested spheres (CPNS) and has been shown to produce more compact representations with more meaningful modes of variations for s-reps. After CPNS is computed the user can interact with the mean s-rep and its modes of variation similarly to the PCA tools for PDMs. The module automatically chooses to use CPNS when s-reps are provided so that the user can not accidentally use the wrong analysis tools.
3.2. Distance Weighted Discrimination
Distance Weighted Discrimination (DWD) is a binary classification method designed to address shortcomings with support vector machine (SVM) performance when applied to high-dimension, low-sample-size (HDLSS) data [9]. We have created a module in SlicerSALT that allows the user to perform PDM classification based on one categorical variable. Similarly to SVM, DWD performs classification by computing the distance of each sample to a separating hyperplane, with samples lying on the same side of the hyperplane being classified together. It improves over SVM by considering the effects of all of the data on the separating hyperplane rather than just a limited set of support vectors.
3.3. Deep Learning for Geometry: FlyBy CNN
Building upon recent work to start bringing powerful deep learning methodologies to 3D Slicer, SlicerSALT now includes a projection-based convolutional learner called FlyBy CNN [2]. Specifically, 2D projection snapshots are acquired at locations along a predefined path on an encompassing sphere. The collection of snapshots forms an ordered 2.5D dataset that is analyzed via existing time series CNN approaches. An implementation via LSTM (Long Short-Term Memory) networks has been developed and applied to condylar surfaces. The results showed improved disease diagnosis performance when compared to a traditional dense network. The main advantage of this approach is that it can be applied to any general surface without the need for an existing correspondence or a consistent topology across surfaces. The disadvantage is that highly folded surfaces are not well represented with a projection approach, although shape-aware snapshot pathways are a planned extension to address this.
4. Infrastructure
In addition to new methods for creating and analyzing shapes, SlicerSALT 4.0 introduces key new application and infrastructure features to further support community research efforts.
4.1. Pipelines
SlicerPipelines is a new extension that allows the creation and use of simple modules in 3D Slicer without coding. This extension was developed as part of SlicerSALT and has been added to the Slicer Extensions Index for the broader research community to use.
Pipelines are pieces of logic that take a single MRML node as input (a model, a volume, etc.) and returns a single MRML node as output. Pipelines are created by stringing together individual existing 3D Slicer modules to produce a new derived workflow. Some examples of modules that could become parts of a pipeline are converting a model to a segmentation, using the 3D Slicer Segment Editor’s hollow, margin, and thresholding effects, and methods like decimation and smoothing from the 3D Slicer Surface Toolbox.
Because they currently can only take a single node as input and return a single node as output, Pipelines are best for simple, repetitive workflows (e.g. threshold an image, apply smoothing, convert to a model, repeat for the next image). For more complicated workflows that depend on multiple MRML inputs or user interaction, manually coding a new module is currently still required, though integration with the automated pipeline creation is planned for the future.
4.2. Sample Data and Tutorials
To aid new users in understanding how to use the methods deployed in SlicerSALT, we have integrated tutorials and example data for many modules directly into the SlicerSALT interface. Tutorials are given as links to slides on Google Drive which give the user a brief introduction to the method deployed in a module as well as walking them through its typical use. For sample data, data is automatically downloaded and verified. The user is able to inspect the data to better understand the input formats required for the module. The module UI is automatically populated with paths to the downloaded data so that the user can understand how to load their own data into the module. The user can run then run the module and inspect the sample output. An example of the Tutorials panel showing sample data being downloaded and paths automatically set is shown in Fig. 5.
Fig. 5.
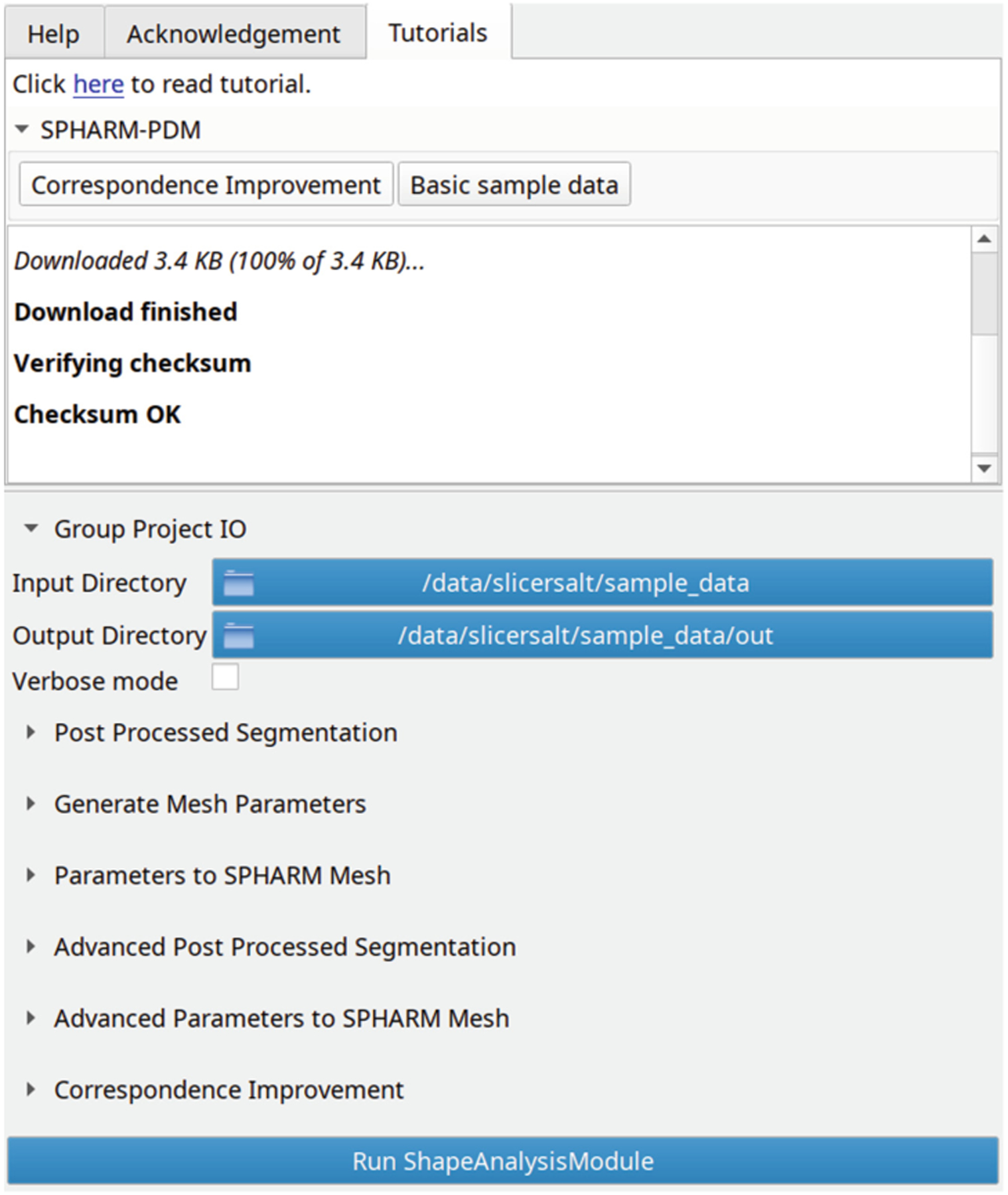
The Tutorials panel showing a link to an online tutorial as well as sample data download buttons.
5. Discussion
The structures captured in 3D images coming from the biomedical fields have brought shape processing methods into the spotlight. The complex and timevarying phenomena contained in geometric structures require higher sensitivity to local variations relative to traditional markers, such as the volume of structures. In this new project period, SlicerSALT will continue providing biomedical researchers with access to sophisticated shape analysis methodology otherwise reserved for computer science experts. In addition, with the expansion towards novel and interpretable shape statistics, we will maximize the potential benefits of the geometric information contained in medical data and expand its use in order to support impactful biomedical research.
SlicerSALT development is ongoing and there are several plans for future improvements. As deep learning becomes increasingly important in the medical imaging and shape analysis communities, we plan to build upon the work described in Sect. 3.3 to introduce improved support for creating and analyzing shapes using deep learning. Our ultimate goal is to create blueprint modules for both training deep learning models and using existing models to perform inference. This will allow users to rapidly create new modules by giving them a starting point which they can customize rather than requiring module development from scratch.
To demonstrate the power of the SlicerPipelines extension for creating reusable and reproducible workflows, we plan to create example pipelines using public datasets for carrying out end-to-end shape analysis tasks. One example could be to start from a set of segmentations, create a population of PDMs using SPHARM-PDM, compute and visualize a shape space using SVA, and finally classify the inputs into two groups using DWD. These pipelines will help non-expert users quickly try various analysis methods on their own data and give more advanced researchers a way to distribute their workflows in a way that they can be easily resused.
Fig. 1.
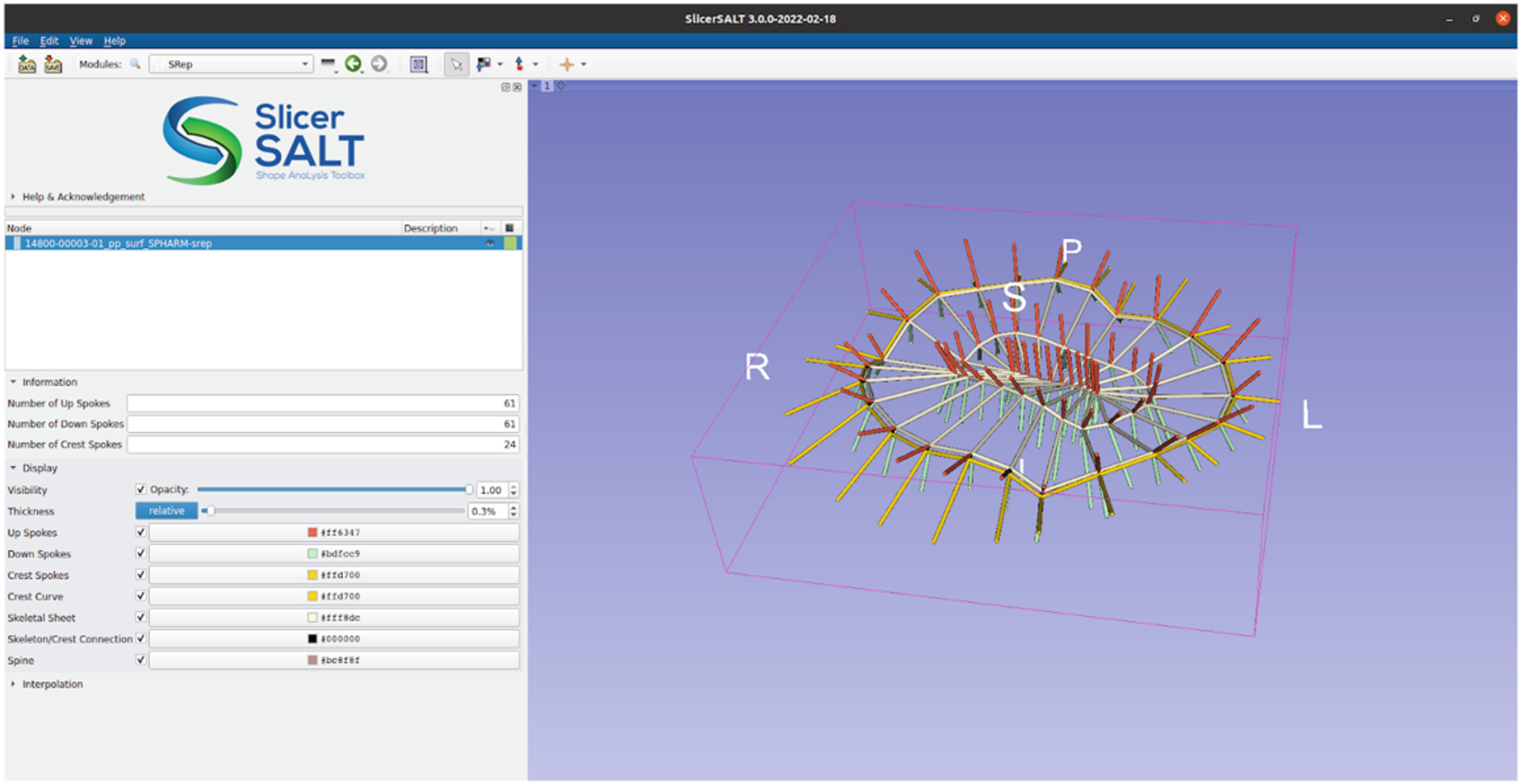
Skeletal representations module in SlicerSALT with an s-rep visualization.
Acknowledgements.
SlicerSALT development has been funded by NIH NIBIB awards R01EB021391 and R56EB021391 as well as NHLBI award R01HL153166.
References
- 1.Albrecht T, et al. : Statismo - a framework for PCA based statistical models. Insight J. pp. 1–18 (2012) [Google Scholar]
- 2.Boubolo L, et al. : FlyBy CNN: a 3D surface segmentation framework. In: Medical Imaging 2021: Image Processing, vol. 11596, pp. 627–632. SPIE; (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cates J, Elhabian S, Whitaker R: ShapeWorks: particle-based shape correspondence and visualization software. Statistical Shape and Deformation Analysis: Methods, Implementation and Applications, pp. 257–298 (2017). 10.1016/B978-0-12-810493-4.00012-2 [DOI] [Google Scholar]
- 4.Durrleman S: Deformetrica (2013). http://www.deformetrica.org
- 5.Fischl B: FreeSurfer. Neuroimage 62(2), 774–781 (2012). 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM: FSL. NeuroImage 62, 782–90 (2012). 10.1016/j.neuroimage.2011.09.015, http://www.ncbi.nlm.nih.gov/pubmed/21979382 [DOI] [PubMed] [Google Scholar]
- 7.Jung S, Dryden IL, Marron JS: Analysis of principal nested spheres. Biometrika 99(3), 551–568 (2012). 10.1093/biomet/ass022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Hong J, Vicory J, Damon JN, Pizer SM: Fitting unbranching skeletal structures to objects. Med. Image Anal 70, 102020 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marron JS, Todd MJ, Ahn J: Distance-weighted discrimination. J. Am.Stat. Assoc 102, 1267–1271 (2007). 10.1198/016214507000001120, http://www.tandfonline.com/doi/abs/10.1198/016214507000001120 [DOI] [Google Scholar]
- 10.Mccormick M, Liu X, Jomier J, Marion C, Ibanez L: ITK: enabling reproducible research and open science. Front. Neuroinformatics 8 (2014). 10.3389/FNINF.2014.00013, https://pubmed.ncbi.nlm.nih.gov/24600387/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pizer SM, et al. : Nested sphere statistics of skeletal models. In: Breuß M, Bruckstein A, Maragos P (eds.) Innovations for Shape Analysis: Models and Algorithms, pp. 93–115. Springer, Berlin, Heidelberg: (2013). 10.1007/978-3-642-34141-05 [DOI] [Google Scholar]
- 12.Schroeder W, Martin K, Lorensen B: The Visualization Toolkit: An Object-oriented Approach to 3D Graphics. Kitware Inc; (2006) [Google Scholar]
- 13.Vercauteren T, Pennec X, Perchant A, Ayache N: Diffeomorphic demons: efficient non-parametric image registration. Neuroimage 45(1), S61–S72 (2009) [DOI] [PubMed] [Google Scholar]
- 14.Vicory J, Allemang D, Zukic D, Prothero J, McCormick M, Paniagua B: An open-source solution for shape modeling and analysis of objects of challenging topologies. In: Medical Imaging 2021: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 11600, pp. 137–142. SPIE; (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vicory J, et al. : SlicerSALT: shape analysis toolbox. In: Reuter M, Wachinger C, Lombaert H, Paniagua B, Lüthi M, Egger B (eds.) ShapeMI 2018. LNCS, vol. 11167, pp. 65–72. Springer, Cham: (2018). 10.1007/978-3-030-04747-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yushkevich PA, et al. : User-guided segmentation of multi-modality medical imaging datasets with ITK-SNAP. Neuroinformatics 17(1), 83–102 (2018). 10.1007/s12021-018-9385-x [DOI] [PMC free article] [PubMed] [Google Scholar]


