Abstract
Primary open-angle glaucoma (POAG) is a complex disease with a strong hereditably component. Several genetic variants have recently been associated with POAG, partially due to technological improvements such as next-generation sequencing (NGS). The aim of this study was to genetically analyze patients with POAG to determine the contribution of rare variants and hypomorphic alleles associated with glaucoma as a future method of diagnosis and early treatment. Seventy-two genes potentially associated with adult glaucoma were studied in 61 patients with POAG. Additionally, we sequenced the coding sequence of CYP1B1 gene in 13 independent patients to deep analyze the potential association of hypomorphic CYP1B1 alleles in the pathogenesis of POAG. We detected nine rare variants in 16% of POAG patients studied by NGS. Those rare variants are located in CYP1B1, SIX6, CARD10, MFN1, OPTC, OPTN, and WDR36 glaucoma-related genes. Hypomorphic variants in CYP1B1 and SIX6 genes have been identified in 8% of the total POAG patient assessed. Our findings suggest that NGS could be a valuable tool to clarify the impact of genetic component on adult glaucoma. However, in order to demonstrate the contribution of these rare variants and hypomorphic alleles to glaucoma, segregation and functional studies would be necessary. The identification of new variants and hypomorphic alleles in glaucoma patients will help to configure the genetic identity of these patients, in order to make an early and precise molecular diagnosis.
Introduction
It is estimated that almost 112 million people worldwide will have glaucoma in 2040, which will continue to be the second leading cause of blindness worldwide [1]. It is known that half of all glaucoma cases go undiagnosed, and therefore efficient screening methods for detecting glaucoma are essential. Genetic studies have enormously progressed in recent years with the advent of the Human Genome Project [2]. Increasingly, molecular genetics is seen as a diagnostic tool in daily clinical practice. Molecular genetic analysis could be used to detect the disease in its early stage (pre-symptomatic relatives), to perform phenotypic-genotypic correlations, to personalize treatment, to refine the prognosis of the disease and to offer genetic counselling [3].
Current next-generation sequencing (NGS) allows millions of DNA sequences to be produced in a single reaction, avoiding the limitations of the Sanger method of gene-to-gene and exon-to-exon sequencing [4]. In this way, through NGS, a large amount of data is obtained and a large number of variants will be observed in each individual. However, the classification of the gene variants is the challenge posed in current genetic molecular diagnosis.
Through linkage analysis, 23 loci and four genes (MYOC/TIGR, CYP1B1, OPTN, and WDR36) had already been associated with glaucoma in the past years. Early-onset glaucoma (<40 years) is more likely to be inherited according to a classic Mendelian pattern involving single genes, mainly CYP1B1 in primary congenital glaucoma (PCG) and MYOC in juvenile open-angle glaucoma (JOAG), whereas glaucoma in adults tends to be more complex due to its multifactorial inheritance [5, 6]. Family grouping is a known risk factor for glaucoma in the adult, with a risk 1–10 times greater than the observed in general population among the first-degree relatives of an affected individual [7, 8]. Therefore, the surveillance of these individuals is indicated for early detection and treatment [9, 10].
However, these disease-causing genes mentioned above account for <10% of primary open angle glaucoma (POAG) cases in the general population [11]. Therefore, in the past decade, efforts have been made to elucidate the genetic causes of adult glaucoma. The application of advanced genetic technology has increased the list of candidate genes [6]. Recent Big Data studies using genome-wide association study (GWAS) have identified over 100 loci related with oxidative stress, DNA repair mechanisms, mitochondrial DNA genes, sex hormones, and phenotypic traits associated with glaucoma [12]. Quantitative endophenotypic agents that act as risk factors (pachymetry, axial length, anterior chamber depth…) and glaucomatous risk stratification have been identified [13, 14]. Therefore, genetic screening appears increasingly promising not only for PCG and JOAG diagnosis, but also for POAG.
In 2006, our research group created the Spanish Multicentre Glaucoma Group (Estudio Multicéntrico Español de Investigación Genética del Glaucoma, EMEIGG), including 18 Eye Hospitals throughout Spain. We performed an initial study to identify pathogenic variants in the MYOC and CYP1B1 genes, which were the only fully-described glaucoma-causing genes at that time [15]. The aim of this study is to test a custom gene panel that gathered recently described candidate genes associated with POAG using NGS and to evaluate the impact of the implementation of NGS as a future method for diagnosis and early treatment of adult glaucoma. And, additionally, based on the NGS results, we set out to evaluate the impact of hypomorphic alleles of CYP1B1 in a new cohort of patients with POAG.
Materials and methods
Patients
A total of 74 patients signed written informed consent to participate in this study and were asked to fill in a questionnaire including personal, biographic, demographic, family, and clinical data. This study was carried out in accordance with the Declaration of Helsinki and approved by the Research Ethics Committee of the Hospital Clínic of Barcelona.
All participating ophthalmologists from the EMEIGG completed a standardized questionnaire to homogenize the data collection. A full history was taken, including systemic and ophthalmologic disease, family members affected by glaucoma (number and type of relative) and family history of consanguinity. POAG was diagnosed in the presence of compatible perimetric lesions correlated with typical glaucomatous changes of the optic nerve in patients over 40 years and absence of a cause for secondary glaucoma diagnosis. Patients affected with low-tension glaucoma (LTG) which is a form of POAG in which there is a glaucomatous optic neuropathy in the presence of intraocular pressures (IOP) lower than 20 mmHg were also included in the study. Patients with early-onset glaucomas who were diagnosed with PCG or JOAG were excluded.
Each referring ophthalmologist graded the disease stage, based on the ophthalmologic exams, the aspect of the optic disc, and visual field-testing results, and classified each case as initial, moderate, or severe glaucoma according to the Hodapp-Parrish-Anderson grading scale, which refers to visual field mean defect (MD) (initial if MD < −6 dB, moderate if MD between −6 and −12 dB, and advanced if MD > −12 dB). Data about the number of eye surgeries and current ocular hypotensive medication were recorded. The presence of thin pachymetry was also annotated as being an endophenotypic trait related to glaucoma conversion and severity.
Study of glaucoma-related genes using NGS
Sixty-one patients (all unrelated, except two siblings) with POAG (or LTG) (Table 1) were included in the NGS study, with a mean age at diagnosis of 51 ± 12 years (range: 18–76 years). Although the study included adult glaucoma cases, four patients were younger than 40 years at the time of diagnosis (18–27 years), but did not display the typical features of JOAG, and were considered as early-onset POAG.
Table 1. Clinical data of patients studied by NGS and Sanger sequencing.
| Patients NGS N = 61 | Patients Sanger N = 13 | |||
|---|---|---|---|---|
| Gender | Female | 33 | 8 | |
| Male | 28 | 5 | ||
| Diagnosis | POAG | Initial | 9 | 2 |
| Moderate | 14 | 8 | ||
| Severe | 31 | 3 | ||
| LTG | Severe | 3 | - | |
| Early-onset POAG | Moderate | 1 | - | |
| Severe | 3 | - | ||
| Family history | Yes | 50 | 11 | |
| No | 11 | 2 | ||
| Surgery | Yes | 38 | 6 | |
| No | 23 | 7 | ||
| Ocular hypotensive medication | Yes | 15 | 1 | |
| No | 46 | 12 | ||
LTG: Low-Tension Glaucoma; POAG: Primary Open-Angle Glaucoma.
Thirty-nine genes potentially associated with POAG were studied (Table 2). After a thorough search in OMIM, Orphanet and Human Gene Mutation Database (HGMD), we selected the classic known glaucoma-causing genes, genes associated with elevated IOP or POAG risk, anterior segment dysgenesis, glaucoma, glaucoma-related endophenotypic traits, optic nerve pathology or augmented susceptibility and retinal vessels anomalies and, finally, syndromic glaucoma-causing genes. Besides, additional genes have been included because a potential association with glaucoma has been suggested in some scientific literature studies [16–25].
Table 2. List of genes included in the custom glaucoma panel designed.
| ADRB1 | ERCC2 | OCLM |
|---|---|---|
| ADRB2 | GALC | OLFM2 |
| AGTR2 | GAS7 | OPA1 |
| ATOH7 | GSTM1 | OPTC |
| BCAS3 | HK2 | OPTN |
| BMP4 | LMX1B | PAX6 |
| CARD10 | MFN1 | PON1 |
| CAV1 | MFN2 | SIX1 |
| CAV2 | MTHFR | SIX6 |
| CDC7 | MYOC | TGFBR3 |
| CDKN2B | NCK2 | TMCO1 |
| COL8A2 | NOS3 | WDR36 |
| CYP1B1 | NTF4 | XRCC1 |
Libraries were generated through gene capture by hybridization with the SeqCap EZ system (NimbleGene, custom panel “cl1_GM1”) and subsequent massive parallel sequencing was carried out with a HiSeq™ 2000 platform sequencer (Illumina) at Sistemas Genómicos, S.L. Sequence reads were aligned with the reference genome GRCh38/hg38. Alignment was made using the BWA (Burrows-Wheeler Aligner) tool and scripts designed by Sistemas Genómicos, S.L. Genetic variants in coding and flanking sequences of genes with frequencies <1% were annotated. Annotated sequencing data was investigated using the filtering system for nonsense, missense, synonym and intronic variants located up to +10 bp in the flanking sequences. All exons and intronic regions up to +10 bp from the exons showed coverages greater than 20x. UTR regions were not analyzed. SNV annotation was made using ANNOVAR [26].
Sanger sequencing of CYP1B1 in a new cohort of POAG patients
After observing the results of the NGS study, we included 13 additional patients for the study of the CYP1B1 gene using Sanger sequencing. These 13 patients were not studied by NGS. The coding sequence of the CYP1B1 gene was directly sequenced, following the same selection criteria as before (Table 1). The mean age at diagnosis was 55 ± 10 years (range: 40–79 years).
Specific oligonucleotides were designed to amplify the two coding exons of CYP1B1 (NM_000104.4; S1 Table).
Interpretation and classification of variants
In silico prediction algorithms included in Varsome (https://varsome.com/) were used to classify variants. The in silico tools used were: CADD, Polyphen2, DEOGEN2, MutPred, FATHMM-XF, Mutation assessor, MVP, PROVEAN, EIGEN, LRT, SIFT, BLOSUM, DANN, LIST-S2, M-CAP, MutationTaster and PrimateAI. ClinVar database was also consulted to check the classification reported by other subscribers.
General population frequencies and the highest frequencies in a population were obtained from gnomAD v3.1.2. Final variant interpretation was performed according to the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) recommendations [27]. In this way, we considered rare variants classified/described as pathogenic, likely pathogenic or variant of uncertain significance (VUS) with low frequencies in the general population (<1%), as well as variants that have been described as hypomorphic variants.
Results
Rare genetic variants detected in POAG patients by NGS
We detected nine rare or hypomorphic variants (Table 3) in 10 patients (16%) by NGS (Table 4). These variants were found in seven of the glaucoma-related genes. All variants were detected in heterozygosity and classified as VUS. The allelic frequency of these variants in general population was very low (<1%) (Table 3). Most patients described with variants showed a severe POAG phenotype (70%, Table 4). Additionally, all patients with a rare genetic variant had a family history of glaucoma and most of them had required surgery or maximal ocular medication (Table 4). The presence of thin pachymetry readings was detected in two patients (patient 1 and patient 8).
Table 3. Description and classification of variants detected in this study.
All variants were detected in heterozygosity.
| Gene | Transcript | Variant (cDNA) | Variant (protein) | ACMG criteria | ClinVar classification (number of times) | ACMG classification | AF in general population | Highest AF and population |
|---|---|---|---|---|---|---|---|---|
| CYP1B1 | NM_000104.4 | c.241T>A | p.Y81N | PS3, PP3, BS1, BS2 | VUS (2); LB (1); B (2) | VUS (Hypomorphic) | 0.35% | 1.36% European (Finnish) |
| CARD10 | NM_014550.4 | c.1307C>T | p.T436M | PM2, BP4 | Not reported | VUS | 0.01% | 0.03% Ashkenazi Jewish |
| CARD10 | NM_014550.4 | c.2952C>A | p.C984X | PM2, BP4 | Not reported | VUS | Not found | Not found |
| MFN1 | NM_033540.3 | c.1601G>A | p.R534Q | PM2, PP3 | Not reported | VUS | Not found | Not found |
| MFN1 | NM_033540.3 | c.2101G>T | p.E701X | PM2 | Not reported | VUS | Not found | Not found |
| OPTC | NM_014359.4 | c.893G>A | p.R298H | PM2 | Not reported | VUS | 0.03% | 0.21% Latino/Admixed American |
| OPTN | NM_001008212.2 | c.1552C>T | p.Q518X | PM2 | Not reported | VUS | Not found | Not found |
| SIX6 | NM_007374.3 | c.635C>T | p.T212M | PM2, PP3 | VUS (2) | VUS (Hypomorphic) | 0.02% | 0.03% European (non-Finnish) |
| WDR36 | NM_139281.3 | c.892G>A | p.E298N | PM2, PP3 | Not reported | VUS | 0.01% | 0.11% Ashkenazi Jewish |
| CYP1B1 ¶ | NM_000104.4 | c.83C>G | p.S28W | PM2, PP5, PS1 | Not reported | VUS (Hypomorphic) | 0.004% | 0.12% Ashkenazi Jewish |
¶Variant detected only in the study by Sanger sequencing. ACMG: American College of Medical Genetics; AF: allelic frequency; B: benign; LB: likely benign; NGS: next-generation sequencing; VUS: variant of uncertain significance.
Table 4. Clinical characteristics of patients and variants detected in this study.
All variants were detected in heterozygosity.
| Patient | Gender | Age at diagnosis | Diagnosis | Features | Family history of glaucoma | Consanguinity | Surgery | Ocular hypotensive medication | Gene | Transcript | Variant (protein) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Next-generation sequencing | |||||||||||
| Patient 1 | Female | 55 | POAG | Initial glaucoma, with PDS and myopia | Yes (mother, sister) | No | No | No | CYP1B1 | NM_000104.4 | p.Y81N |
| Patient 2 | Male | 76 | POAG | Severe glaucoma | No | No | Yes | Yes | CYP1B1 | NM_000104.4 | p.Y81N |
| Patient 3 | Female | 55 | POAG | Moderate glaucoma | Yes (daughter) | No | Yes | No | SIX6 | NM_007374.3 | p.T212M |
| Patient 4¶ | Female | 42 | POAG | Severe glaucoma, with retinal venous and arterial occlusions | Yes (brother) | No | Yes | No | CARD10 | NM_014550.4 | p.T436M |
| Patient 5¶ | Male | 30 | POAG | Severe glaucoma, with retinal venous and arterial occlusions | Yes (sister) | No | Yes | No | CARD10 | NM_014550.4 | p.T436M |
| Patient 6 | Female | 65 | POAG | Severe glaucoma | Yes (son, 2 siblings) | Yes | No | Yes | CARD10 | NM_014550.4 | p.C984X |
| MFN1 | NM_033540.3 | p.R534Q | |||||||||
| Patient 7 | Female | 65 | POAG | Moderate glaucoma | Yes (4 siblings, nephew) | No | No | Yes | OPTC | NM_014359.4 | p.R298H |
| Patient 8 | Male | 25 | Early-onset POAG | Severe glaucoma | Yes (father, brother) | No | Yes | No | OPTN | NM_001008212.2 | p.Q518X |
| Patient 9 | Male | 60 | LTG | Severe glaucoma | Yes (daughter) | No | Yes | No | WDR36 | NM_139281.3 | p.E298N |
| Patient 10 | Female | 68 | POAG | Severe glaucoma | Yes (father, brother) | No | Yes | No | MFN1 | NM_033540.3 | p.E701X |
| Sanger sequencing of CYP1B1 | |||||||||||
| Patient 11 | Male | 58 | POAG | Moderate glaucoma | No | No | Yes | No | CYP1B1 | NM_000104.4 | p.Y81N |
| Patient 12 | Female | 60 | POAG | Initial glaucoma | Yes (maternal grandmother) | No | No | No | CYP1B1 | NM_000104.4 | p.Y81N |
| Patient 13 | Male | 60 | POAG | Moderate glaucoma and myopia | Yes (mother, 2 siblings) | No | No | No | CYP1B1 | NM_000104.4 | p.S28W |
¶Siblings. LTG: low-tension glaucoma; NGS: next-generation sequencing; PDS: pigment dispersion syndrome; POAG: primary open-angle glaucoma.
The only variant detected in two unrelated patients was the variant p.Y81N in the CYP1B1 gene (Table 4). Although different in silico algorithms and functional studies predict a possible pathogenicity of this variant, it has been identified in 528 individuals from genomes and exomes available in gnomAD, even in three of them in a homozygosis state. Nevertheless, the comparison of residues between organisms showed marked conservation of Y81 in different organisms (Fig 1) and its association with POAG has been reported several times [15, 28–31], being described as a hypomorphic variant in some cases [32–35]. Therefore, this variant was classified as a VUS.
Fig 1.
Comparison of the evolutionary conservation of p.S28W (left) and p.Y81N (right) in the CYP1B1 gene in the reference genome of some organisms.
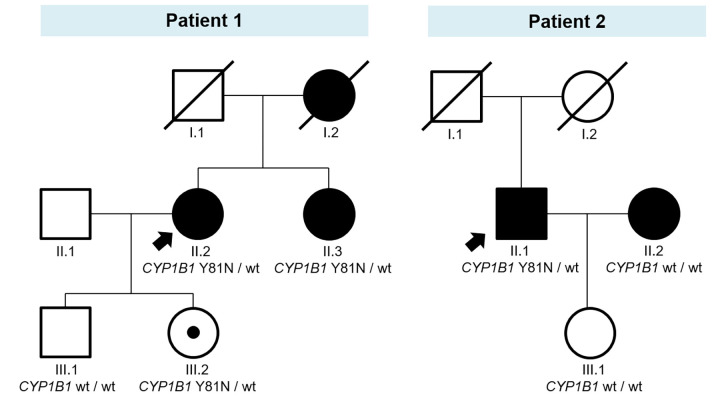
Although familial studies of VUS are not recommended in clinical practice [36], the segregation study of the p.Y81N variant in the CYP1B1 gene was performed in both families for research purposes (Fig 2). The variant segregated with the disease in the family of patient 1. The patient’s sister, who was also diagnosed with POAG, carried the p.Y81N variant in CYP1B1 in heterozygosity. The variant was also detected in the patient’s daughter, who had not signs of glaucoma, but has ocular hypertension. The family of patient 2 was smaller and not informative. The segregation study showed that the patient’s daughter, who had no signs of glaucoma or ocular hypertension, did not have the variant. His wife also had glaucoma, but she also underwent NGS testing, and no variant was detected.
Fig 2.
Pedigrees of patients in whom the p.Y81N variant in the CYP1B1 gene was detected in the NGS study: patient 1 (left) and patient 2 (right). Arrows indicate the index case. Black symbols indicate glaucoma phenotypes, and carriers of the variant are indicated by black dots in the symbols. wt: wild-type allele.
We also observed two siblings with the variant p.T436M in the CARD10 gene. Both siblings (a 45-year-old female and a 43-year-old male) had a very aggressive POAG phenotype (Fig 3), with very high IOP and pigment dispersion syndrome (pigment dots scattered around the corneal endothelium and pigmented trabecular meshword), although other characteristic features of classic pigmentary glaucoma (typical Krukenberg spindle and iris atrophy) were missing. Both also developed retinal venous and arterial occlusions. Aside from the siblings, we detected another variant in the CARD10 gene (p.C984X) in a 65-year-old female, along with a second variant in the MFN1 gene (p.R534Q). This patient had a severe POAG phenotype, and was the only patient in the study with a family history of consanguinity (grandparents were first degree cousins). Regarding the MFN1 gene, we also detected another variant (p.E701X) in a 68-year-old patient with severe glaucoma. Both variants in the CARD10 gene and in the MFN1 gene have not been reported in ClinVar and most of them do not appear in gnomAD, except for the variant p.T436M in the CARD10 gene that was identified in 12 individuals in heterozygosity. All of them have one or two deleterious predictors and should therefore be classified as VUS.
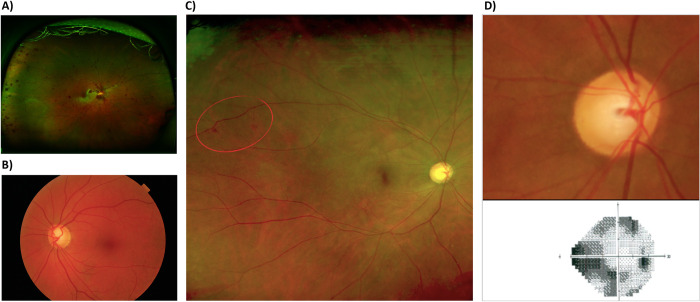
Fig 3. Ophthalmologic studies in patients 4 and 5 (siblings) with the variant p.T436M in the CARD10 gene.
A: Optomap image of ocular fundus of the patient 4 showing macular whitening and a myriad of splinter retinal hemorrhages and venous tortuosity secondary to central retinal vein occlusion with cilioretinal artery occlusion in the right eye. B: Funduscopic image of the left eye of the patient 4 showing severe optic disk cupping in the context of advanced glaucoma with pigment dispersion syndrome. C: Optomap image of the right eye fundus of the patient 5 showing mild macular whitening with scant peripheral hemorrhages one month after non-ischemic central retinal vein occlusion associated with cilioretinal artery occlusion in the right eye. D: Retinography of the patient 5 (up) showing severe right optic disk cupping causing advanced visual field constriction seen on perimetry (below).
The other genetic variants were identified in OPTC (p.p.R298H), OPTN (p.Q518X), SIX6 (p.T212M), and WDR36 (p.E298N). As shown in Table 3, all these variants have very low allele frequencies in general population and most of them have not been described in ClinVar. It is interesting to note that one of these variants, p.T212M in the SIX6 gene, has also been reported in the literature as hypomorphic variant [37].
Analysis of hypomorphic CYP1B1 variants
It has been reported that CYP1B1 may play a role in the pathogenesis of POAG in a significant proportion of cases [30, 31, 34]. For this purpose, we sequenced the coding region of this gene in a new cohort of POAG patients. Heterozygous missense variants were detected in three POAG patients (23%, Table 4). None of these patients had severe glaucoma. The aforementioned variant p.Y81N in the CYP1B1 gene was also detected in two patients of these three patients (Table 4).
The variant c.83C>G (p.S28W) in the CYP1B1 gene was identified in a 66-year-old male patient, diagnosed with myopia magna and moderate glaucoma at the age of 60 (Fig 4). His mother and two siblings were also affected by glaucoma. He was taking ocular hypotensive medication, but his IOP remained very high despite maximal treatment. Finally, surgery was necessary in both eyes. To the best of our knowledge, this variant has never been reported in ClinVar and has been detected at a very low frequency in the general population (0.004%). According to the ACMG/AMP recommendations, the variant was classified as VUS following criteria: PM2 (absent from controls in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium) and PP5 (reputable source recently reports variant as pathogenic, but not functional experimental evidence). Similarly to the variant p.Y81N, the comparison of residues between different organisms showed a marked conservation of S28 in mammals and other organisms (Fig 1).
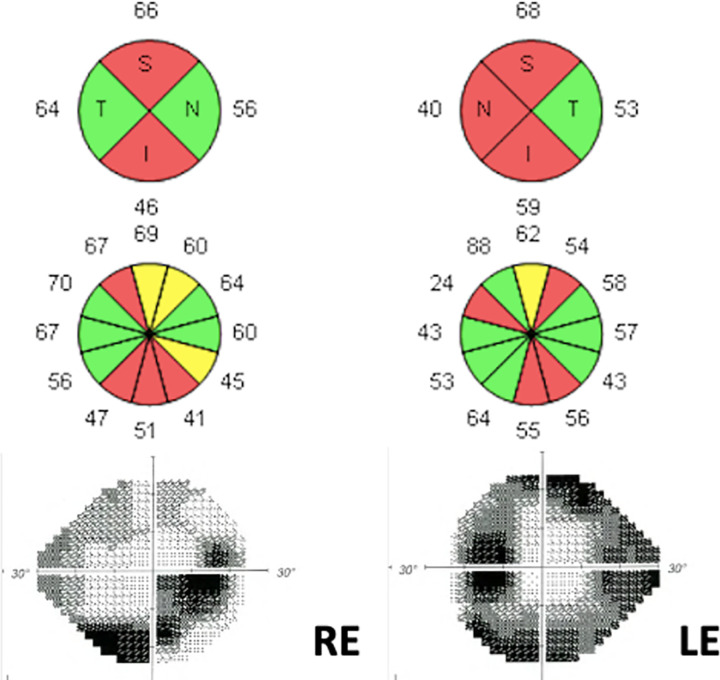
Fig 4. Ancillary tests of patient 13.
This figure shows the structural and functional damage of the right (RE) and left eye (LE). The optical coherence tomography shows severe peripapillary retinal nerve fiber layer thinning that correlates with the visual field findings that depicts a marked inferior arcuate scotoma for the RE and tubular island of vision for the LE.
Discussion
According to Lander and Schork [38], the genetic study of adult glaucoma is complex since its transmission mechanisms are sometimes unclear and have variable penetrance and late onset. Adult glaucoma is a multifactorial genetic disease whose outcome is also influenced by a number of environmental factors, many of them unknown. In addition, there are several ethnic and geographic disparities between populations, which further complicate the genetic study of glaucoma [39]. Thanks to recent technological improvements, such as NGS, the list of candidate genes possibly associated with glaucoma has increased considerably since the initial Sanger direct sequencing studies, when only four genes showed a strong association with glaucoma [6].
In this study, we applied NGS to study a cohort of patients with POAG. Hypomorphic alleles and rare variants that were not previously described in the literature or with a very low population frequency have been detected in 10 out of 65 patients assessed. The variant p.Y81N in CYP1B1 gene was the most frequent variant without considering related patients by NGS (two patients). Given the intimate relationship between CYP1B1 and glaucoma and the results observed in the first group, we sequenced the CYP1B1 gene in a different cohort of patients, detecting the variant p.Y81N in two more POAG patients. Thus, the variant p.Y81N was detected in four unrelated patients (two patients from the NGS study and two patients from the Sanger study).
CYP1B1 is an enzyme involved in drug, fatty acid and steroid metabolism located in the endoplasmic reticulum membrane, peripheral membrane protein and microsome membrane. CYP1B1 participates in the metabolism of an as-yet-unknown biologically-active molecule that participates in eye development. Genetic variants in this gene had always been associated with PCG, an autosomal recessive inherited trait [40, 41]. However, in recent years it has been observed that CYP1B1 also could play an important role in the development of adult glaucoma [30, 31]. Hypomorphic alleles pose a challenge in the interpretation of genomic variants [42]. A hypomorphic allele results in a partial loss of the normal (wild-type) gene function, often characterized by reduced expression of the gene product (protein or RNA), although this reduction does not reach a 100% reduction in normal gene function. In the literature we found few articles about hypomorphic alleles in POAG, and most of them focus on genetic variants in CYP1B1 [31–33, 43, 44] and SIX6 [37]. In the current study, hypomorphic variants in CYP1B1 and SIX6 genes have been identified in 8% of the total POAG patient assessed. As commented before, the genetic variant p.Y81N has been identified in heterozygous state in four unrelated patients. This genetic variant has been reported at a low frequency (0.35%) in the general population and has been described as a heterozygous hypomorphic variant for POAG [32–35], with a significantly reduced enzymatic activity and reduced protein stability (18–40% of the wild-type activity) [32]. In one of the families, the segregation study revealed the presence of a variant in an individual who had not yet shown signs of glaucoma, but who had ocular hypertension. These results suggest that it might be interesting to propose familial studies in patients with CYP1B1 hypomorphic alleles to identify asymptomatic carriers of the variant for close ophthalmological follow-up.Besides, we also identified for the first time the variant c.83C>G (p.S28W) in the CYP1B1 gene in a patient with a moderate glaucoma and myopia. Interestingly, a different genetic variant but at the same position (c.83C>T) which produces the same amino acid change as the variant found in our study (p.S28W) has been described as a hypomorphic variant and reported in patients with POAG [45] and PCG [46].
Both variants identified in our study in the CYP1B1 gene caused amino acid changes and affected conserved residues located in structural domains, having the potential to modify enzyme activity by incorrect insertion of the protein into the endoplasmic reticulum membrane (p.S28W) and modification of substrate binding (p.Y81N) [45].
Additionally, we detected the variant p.T212M in the SIX6 gene in a patient with a moderate glaucoma. This variant was also described as a hypomorphic variant. SIX6 is a transcription factor with a known function in the retinal progenitor cell proliferation during the eye development [47]. In vivo analyses in zebrafish have been useful to study the effect of this variant in the eye. Modified animal models with this variant have been found to have a smaller eye size, hypothesizing that the presence of hypomorphic alleles in SIX6 could reduce the number of retinal ganglion cells and increase the risk of glaucoma [37].
Regarding the rare variants detected in other genes, we have observed variants in CARD10, MFN1, OPTC, OPTN and WDR36. Most of them have not yet been associated with glaucoma. Additional studies, including functional and segregation analyses, are imperative to confirm whether these genes and variants are indeed associated with the disease. Interestingly, we found two siblings with the variant p.T436M in the CARD10 gene. This variant has not been reported in the general population, but in silico tools predict a benign impact. Nevertheless, the variant is still considered a VUS. CARD10 encodes for a caspase recruitment domain-containing protein, which is a signaling protein in the regulation of the NF-κB (nuclear factor kappa B) pathway. Since NF-κB is involved in the regulation of cellular apoptosis, it is likely that there is a relationship between CARD10 and cell apoptosis, especially retinal ganglion cell apoptosis, producing higher optic nerve susceptibility to IOP elevations and POAG [48, 49]. In fact, Zhou et al. [18] observed a higher frequency of variants in this gene (4.28%) in patients with severe POAG compared to the control population (0.27%).
Besides, we found one patient with severe glaucoma and two VUS in CARD10 and MFN1. In these cases, the use of polygenic risk scores may be of interest. These scores have been reported in recent years as promising for stratifying individual risk and prognosis of POAG [50, 51], although further studies are still needed.
The association of POAG with some of the rare variants identified in this study should be treated with caution. For example, we detected the nonsense variant p.Q518X in the OPTN gene in one patient with POAG. Recent studies have reported that the missense variant p.E50K is the only known variant in the OPTN gene associated with glaucoma, whereas loss-of-function variants in this gene are associated with amyotrophic lateral sclerosis (ALS) [52]. However, there are cases where ALS and glaucoma coexist, as seen in one of the patients reported by Maruyama et al. [53]. Therefore, variants in the OPTN gene may contribute to some additional risk of glaucoma in certain patient populations.
Similarly, contradictory results are reported for the association of pathogenic variants in the WDR36 gene with glaucoma. Whereas several studies have indicated that genetic variants in WDR36 gene are contributing risk factors for glaucoma progression and severity [54–58], recent clinical trials and meta-analyses have suggested a lack of effect [59–61]. However, it is known that this gene has a connection to glaucoma susceptibility and even to retinal homeostasis [62, 63]. Monemi et al. demonstrated WDR36 gene expression in the lens, iris, ciliary muscles, ciliary body, trabecular meshwork, retina, and optic nerve by RT-PCR with four pathogenic variants (p.N355S, p.A449T, p.R529Q and p.D658G) associated with adult-onset POAG with implications for both high- and low-pressure glaucoma [64]. In our study, we detected the variant p.E298N in the WDR36 gene in a patient with severe LTG, whose phenotype was very similar to another reported LTG patient with a variant (p.N355S) in the WDR36 gene [58].
Some of the patients carrying rare variants (patient 1 and patient 8) presented with thin pachymetry, which is a known endophenotypic trait that increases the degree of severity of the glaucoma cases, whether this trait has been directly linked to the variant described or is a consequence of other genetic/epigenetic influences still has to be elucidated. Further studies are warranted to explore this relationship more comprehensively.
Certainly, addressing the limitations of our study is crucial. While NGS is a powerful tool for detecting genetic variations, it may have limitations in detecting copy number variations (CNV), including deletions or duplications. We acknowledge that our study focused primarily on single nucleotide variations and small indels, and we did not specifically investigate CNV. This limitation is important to consider, especially in cases where CNV could be contributing to the disease [65]. Future research efforts could explore complementary techniques or arrays designed for CNV detection to provide a more comprehensive genetic assessment of glaucoma-related variations.
Our findings suggest that NGS could be a valuable tool for the genetic assessment of glaucoma. However, to irrefutably show the contribution of these rare variants and hypomorphic alleles to glaucoma, additional studies, including functional evidence, will be necessary. The progressive identification of new rare variants and hypomorphic alleles in patients clinically diagnosed with glaucoma will help to configure the genetic identity of these patients, in order to make an early and precise molecular diagnosis. And as a clinical application of these findings, the presence of hypomorphic alleles in asymptomatic relatives of our glaucoma patients acts, in our opinion, as a red flag that suggests close monitoring of these patients and early treatment decision in case of glaucoma suspicion.
Supporting information
(DOCX)
Acknowledgments
We would like to thank the rest of members of the EMEIGG group (Estudio Multicéntrico Español de Investigación Genética del Glaucoma): Esperanza Gutiérrez and Marta Montero (Hospital Doce de Octubre, Madrid); Cristina Vendrell (Hospital de Viladecans, Barcelona); José Abreu (Hospital Universitario de Canarias); Carmen Cabarga (Hospital Ramón y Cajal, Madrid); Soledad Jiménez (Hospital Universitario Puerta del Mar, Cádiz); Miguel Ángel Almela (Hospital Lluis Alcanyís, Xàtiva); Jordi Loscos (Hospital Universitari Germans Trias i Pujol, Badalona); Carlos Martínez Bello (Hospital Dos de Maig, Barcelona); Sergio Torregrosa (Hospital Punta de Europa, Cádiz); Rosa Martínez (Hospital Infanta Leonor, Madrid); Tiburcio Ibáñez (Hospital San Agustín, Linares); Lourdes Iglesias (Hospital Universitario La Princesa, Madrid); Pere Viñallonga (Institut Oftalmològic, Menorca); Lluís Soler (Hospital de Manresa, Barcelona); and Carmen Carrasco (Hospital de Alcorcón, Madrid).
Data Availability
The minimal dataset is contained within the paper and its Supporting Information file.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–90. doi: 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 2.Green ED, Watson JD, Collins FS. Human Genome Project: Twenty-five years of big biology. Nature. 2015;526(7571):29–31. doi: 10.1038/526029a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagenkord J, Funke B, Qian E, Hegde M, Jacobs KB, Ferber M, et al. Design and Reporting Considerations for Genetic Screening Tests. J Mol Diagnostics. 2020;22(5):599–609. doi: 10.1016/j.jmoldx.2020.01.014 [DOI] [PubMed] [Google Scholar]
- 4.Qin D. Next-generation sequencing and its clinical application. Cancer Biol Med. 2019;16(1):4–10. doi: 10.20892/j.issn.2095-3941.2018.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiggs JL, Pasquale LR. Genetics of glaucoma. Hum Mol Genet. 2017;26(R1):R21–7. doi: 10.1093/hmg/ddx184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar S, Malik MA, Goswami S, Sihota R, Kaur J. Candidate genes involved in the susceptibility of primary open angle glaucoma. Gene. 2016;577(2):119–31. doi: 10.1016/j.gene.2015.11.032 [DOI] [PubMed] [Google Scholar]
- 7.Le A, Mukesh BN, McCarty CA, Taylor HR. Risk factors associated with the incidence of open-angle glaucoma: The visual impairment project. Investig Ophthalmol Vis Sci. 2003;44(9):3783–9. doi: 10.1167/iovs.03-0077 [DOI] [PubMed] [Google Scholar]
- 8.Green CM, Kearns LS, Wu J, Barbour JM, Wilkinson RM, Ring MA, et al. How significant is a family history of glaucoma? Experience from the Glaucoma Inheritance Study in Tasmania. Clin Exp Ophthalmol. 2007;35(9):793–9. doi: 10.1111/j.1442-9071.2007.01612.x [DOI] [PubMed] [Google Scholar]
- 9.Milla E, Gamundi MJ, Duch S, Rios J, Carballo M. Phenotypic Description of the Spanish Multicentre Genetic Glaucoma Group Cohort. J Ophthalmol. 2017;2017:1907454. doi: 10.1155/2017/1907454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong G, Kosoko-Lasaki S, Haynatzki G, Lynch HT, Lynch JA, Wilson MR. Inherited, familial and sporadic primary open-angle glaucoma. J Natl Med Assoc. 2007;99(5):559–63. [PMC free article] [PubMed] [Google Scholar]
- 11.Rao KN, Nagireddy S, Chakrabarti S. Complex genetic mechanisms in glaucoma: An overview. Indian J Ophthalmol. 2011;59(Suppl. 1):S31–42. doi: 10.4103/0301-4738.73685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X, Gharahkhani P, Hamel AR, Ong JS, Rentería ME, Mehta P, et al. Large-scale multitrait genome-wide association analyses identify hundreds of glaucoma risk loci. Nat Genet. 2023;55(7):1116–25. doi: 10.1038/s41588-023-01428-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnemaijer PWM, Leeuwen EM va., Iglesias AI, Gharahkhani P, Vitart V, Khawaja AP, et al. Multi-trait genome-wide association study identifies new loci associated with optic disc parameters. Commun Biol. 2019;2(1):1–12. doi: 10.1038/s42003-019-0634-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asefa NG, Neustaeter A, Jansonius NM, Snieder H. Heritability of glaucoma and glaucoma-related endophenotypes: Systematic review and meta-analysis. Surv Ophthalmol. 2019;64(6):835–51. doi: 10.1016/j.survophthal.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 15.Milla E, Mañé B, Duch S, Hernan I, Borràs E, Planas E, et al. Survey of familial glaucoma shows a high incidence of cytochrome P450, family 1, subfamily B, polypeptide 1 (CYP1B1) mutations in non-consanguineous congenital forms in a Spanish population. Mol Vis. 2013;19(July 2012):1707–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Can Demirdöğen B, Koçan Akçin C, Göksoy E, Yakar G, Öztepe T, Demirkaya-Budak S, et al. Paraoxonase 1 (PON1) promoter (−107T/C) and coding region (192Q/R and 55L/M) genetic variations in pseudoexfoliation syndrome and pseudoexfoliative glaucoma risk. Graefe’s Arch Clin Exp Ophthalmol. 2019;257(10):2257–70. doi: 10.1007/s00417-019-04408-w [DOI] [PubMed] [Google Scholar]
- 17.Gohari M, Mirjalili Seyed A, Akbarian-Bafghi MJ, Jarahzadeh MH, Zare-Shehneh M, Neamatzadeh H. Association of MTHFR C677T and A1298C Polymorphisms with Glaucoma Risk: a Systematic Review Meta-Analysis based 42 Case-Control Studies. Rom J Ophthalmol. 2019;63(2):107–18. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Souzeau E, Sharma S, Siggs OM, Goldberg I, Healey PR, et al. Rare variants in optic disc area gene CARD10 enriched in primary open-angle glaucoma. Mol Genet Genomic Med. 2016;4(6):624–33. doi: 10.1002/mgg3.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi D, Funayama T, Mashima Y, Takano Y, Shimizu A, Yamamoto K, et al. Association of HK2 and NCK2 with Normal Tension Glaucoma in the Japanese Population. PLoS One. 2013;8(1). doi: 10.1371/journal.pone.0054115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramdas WD, van Koolwijk LME, Lemij HG, Pasutto F, Cree AJ, Thorleifsson G, et al. Common genetic variants associated with open-angle glaucoma. Hum Mol Genet. 2011;20(12):2464–71. doi: 10.1093/hmg/ddr120 [DOI] [PubMed] [Google Scholar]
- 21.Sharma S, Burdon KP, Chidlow G, Klebe S, Crawford A, Dimasi DP, et al. Association of genetic variants in the TMCO1 gene with clinical parameters related to glaucoma and characterization of the protein in the eye. Investig Ophthalmol Vis Sci. 2012;53(8):4917–25. doi: 10.1167/iovs.11-9047 [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Luo H, Yu M, Yang C, Shu Y, Gong B, et al. Association of polymorphism rs11656696 in GAS7 with primary open-Angle Glaucoma in a Chinese Population. Ophthalmic Genet [Internet]. 2019;40(3):237–41. Available from: doi: 10.1080/13816810.2019.1627465 [DOI] [PubMed] [Google Scholar]
- 23.Loomis SJ, Kang JH, Weinreb RN, Yaspan BL, Cooke Bailey JN, Gaasterland D, et al. Association of CAV1/CAV2 genomic variants with primary open-angle glaucoma overall and by gender and pattern of visual field loss. Ophthalmology. 2014;121(2):508–16. doi: 10.1016/j.ophtha.2013.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inagaki Y, Mashima Y, Fuse N, Funayama T, Ohtake Y, Yasuda N, et al. Polymorphism of β-adrenergic receptors and susceptibility of open-angle glaucoma. Mol Vis. 2006;12(January):673–80. [PubMed] [Google Scholar]
- 25.Hashizume K, Mashima Y, Fumayama T, Ohtake Y, Kimura I, Yoshida K, et al. Genetic polymorphisms in the angiotensin II receptor gene and their association with open-angle glaucoma in a Japanese population. Investig Ophthalmol Vis Sci. 2005;46(6):1993–2001. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):1–7. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou T, Souzeau E, Siggs OM, Landers J, Mills R, Goldberg I, et al. Contribution of mutations in known mendelian glaucoma genes to advanced early-onset primary open-angle glaucoma. Investig Ophthalmol Vis Sci. 2017;58(3):1537–44. doi: 10.1167/iovs.16-21049 [DOI] [PubMed] [Google Scholar]
- 29.Reis LM, Tyler RC, Weh E, Hendee KE, Kariminejad A, Abdul-Rahman O, et al. Analysis of CYP1B1 in pediatric and adult glaucoma and other ocular phenotypes. 2016;1(October):1229–38. [PMC free article] [PubMed] [Google Scholar]
- 30.Melki R, Colomb E, Lefort N, Brézin AP, Garchon HJ. CYP1B1 mutations in French patients with early-onset primary open-angle glaucoma. J Med Genet. 2004;41(9):647–51. doi: 10.1136/jmg.2004.020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel HY, Richards AJ, De Karolyi B, Best SJ, Danesh-Meyer H V., Vincent AL. Screening glaucoma genes in adult glaucoma suggests a multiallelic contribution of CYP1B1 to open-angle glaucoma phenotypes. Clin Exp Ophthalmol. 2012;40(4):208–17. doi: 10.1111/j.1442-9071.2011.02714.x [DOI] [PubMed] [Google Scholar]
- 32.López-Garrido MP, Blanco-Marchite C, Sánchez-Sánchez F, López-Sánchez E, Chaqués-Alepuz V, Campos-Mollo E, et al. Functional analysis of CYP1B1 mutations and association of heterozygous hypomorphic alleles with primary open-angle glaucoma. Clin Genet. 2010;77(1):70–8. doi: 10.1111/j.1399-0004.2009.01284.x [DOI] [PubMed] [Google Scholar]
- 33.Campos-Mollo E, López-Garrido MP, Blanco-Marchite C, Garcia-Feijoo J, Peralta J, Belmonte-Martínez J, et al. CYP1B1 mutations in Spanish patients with primary congenital glaucoma: Phenotypic and functional variability. Mol Vis. 2009;15(November 2008):417–31. [PMC free article] [PubMed] [Google Scholar]
- 34.Pasutto F, Chavarria-Soley G, Mardin CY, Michels-Rautenstrauss K, Ingelman-Sundberg M, Fernández-Martínez L, et al. Heterozygous loss-of-function variants in CYP1B1 predispose to primary open-angle glaucoma. Investig Ophthalmol Vis Sci. 2010;51(1):249–54. doi: 10.1167/iovs.09-3880 [DOI] [PubMed] [Google Scholar]
- 35.Banerjee A, Chakraborty S, Chakraborty A, Chakrabarti S, Ray K. Functional and structural analyses of CYP1B1 variants linked to congenital and adult-onset glaucoma to investigate the molecular basis of these diseases. PLoS One. 2016;11(5):1–30. doi: 10.1371/journal.pone.0156252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard S, Sun S, Regier DA. Balancing uncertainty with patient autonomy in precision medicine. Nat Rev Genet. 2019;20(5):251–2. doi: 10.1038/s41576-019-0111-9 [DOI] [PubMed] [Google Scholar]
- 37.Carnes MU, Liu YP, Allingham RR, Whigham BT, Havens S, Garrett ME, et al. Discovery and Functional Annotation of SIX6 Variants in Primary Open-Angle Glaucoma. PLoS Genet. 2014;10(5):1–10. doi: 10.1371/journal.pgen.1004372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lander ES, Schork NJ. Genetic dissection of complex traits. Science (80-). 1994;265(5181):2037–48. doi: 10.1126/science.8091226 [DOI] [PubMed] [Google Scholar]
- 39.Zukerman R, Harris A, Vercellin AV, Siesky B, Pasquale LR, Ciulla TA. Molecular genetics of glaucoma: Subtype and ethnicity considerations. Genes (Basel). 2021;12(1):1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leysen L, Cassiman C, Vermeer S, Casteels I, Balikova I. Genetics in primary congenital glaucoma: Implications in disease management and counseling. Eur J Med Genet. 2022;65(1):104378. doi: 10.1016/j.ejmg.2021.104378 [DOI] [PubMed] [Google Scholar]
- 41.Haddad A, Ait Boujmia OK, El Maaloum L, Dehbi H. Meta-analysis of CYP1B1 gene mutations in primary congenital glaucoma patients. Eur J Ophthalmol. 2021;31(6):2796–807. doi: 10.1177/11206721211016308 [DOI] [PubMed] [Google Scholar]
- 42.Nolan DK, Chaudhari B, Franklin SJ, Wijeratne S, Pfau R, Mihalic Mosher T, et al. Hypomorphic alleles pose challenges in rare disease genomic variant interpretation. Clin Genet. 2021;100(6):775–6. doi: 10.1111/cge.14052 [DOI] [PubMed] [Google Scholar]
- 43.Berraho A, Serrou A, Fritez N, Annas A El, Bencherifa F, Gaboun F, et al. Genotype-phenotype correlation in Moroccan patients with primary congenital glaucoma. J Glaucoma. 2015;24(4):297–305. doi: 10.1097/IJG.0b013e31829f99b7 [DOI] [PubMed] [Google Scholar]
- 44.Chavarria-Soley G, Sticht H, Aklillu E, Ingelman-Sundberg M, Pasutto F, Reis A, et al. Mutations in CYP1B1 cause primary congenital glaucoma by reduction of either activity or abundance of the enzyme. Hum Mutat. 2008;29(9):1147–53. doi: 10.1002/humu.20786 [DOI] [PubMed] [Google Scholar]
- 45.López-Garrido MP, Sánchez-Sánchez F, López-Martínez F, Aroca-Aguilar JD, Blanco-Marchite C, Coca-Prados M, et al. Heterozygous CYP1B1 gene mutations in Spanish patients primary open-angle glaucoma. Mol Vis. 2006;12(March):748–55. [PubMed] [Google Scholar]
- 46.Reis LM, Tyler RC, Zori R, Burgesst J, Mueller J, Semina E V. A Case of 22q11.2 Deletion Syndrome with Peters Anomaly, Congenital Glaucoma, and Heterozygous Mutation in CYP1B1. Ophthalmic Genet. 2005;36(1):92–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science (80-). 2002;297(5584):1180–3. doi: 10.1126/science.1073263 [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Guo Y, Huang WJ, Ke X, Poyet JL, Manji GA, et al. CARD10 Is a Novel Caspase Recruitment Domain/Membrane-associated Guanylate Kinase Family Member That Interacts with BCL10 and Activates NF-κB. J Biol Chem. 2001;276(24):21405–9. [DOI] [PubMed] [Google Scholar]
- 49.Grabiner BC, Blonska M, Lin P, You Y, Wang D, Sun J, et al. CARMA3 deficiency abrogates G protein-coupled receptor-induced NF-κB activation. Genes Dev. 2007;21(8):984–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollitt GL, Siggs OM, Ridge B, Keane MC, Mackey DA, MacGregor S, et al. Attitudes Towards Polygenic Risk Testing in Individuals with Glaucoma. Ophthalmol Glaucoma. 2022;5(4):436–46. doi: 10.1016/j.ogla.2021.11.002 [DOI] [PubMed] [Google Scholar]
- 51.Craig JE, Han X, Qassim A, Hassall M, Cooke Bailey JN, Kinzy TG, et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. 2020;52(2):160–6. doi: 10.1038/s41588-019-0556-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fox AR, Fingert JH. Familial normal tension glaucoma genetics. Prog Retin Eye Res. 2023;96(February). doi: 10.1016/j.preteyeres.2023.101191 [DOI] [PubMed] [Google Scholar]
- 53.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295):223–6. doi: 10.1038/nature08971 [DOI] [PubMed] [Google Scholar]
- 54.Footz TK, Johnson JL, Dubois S, Boivin N, Raymond V, Walter MA. Glaucoma-associated WDR36 variants encode functional defects in a yeast model system. Hum Mol Genet. 2009;18(7):1276–87. doi: 10.1093/hmg/ddp027 [DOI] [PubMed] [Google Scholar]
- 55.Blanco-Marchite C, Sánchez-Sánchez F, López-Garrido M-P, Iñigez-de-Onzoño M, López-Martínez F, López-Sánchez E, et al. WDR36 and P53 gene variants and susceptibility to primary open-angle glaucoma: Analysis of gene-gene interactions. Investig Ophthalmol Vis Sci. 2011;52(11):8467–78. doi: 10.1167/iovs.11-7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X, Li M, Guo X, Li S, Xiao X, Jia X, et al. Mutation analysis of seven known glaucoma-associated genes in Chinese patients with glaucoma. Investig Ophthalmol Vis Sci. 2014;55(6):3594–602. doi: 10.1167/iovs.14-13927 [DOI] [PubMed] [Google Scholar]
- 57.Mookherjee S, Chakraborty S, Vishal M, Banerjee D, Sen A, Ray K. WDR36 variants in East Indian primary open-angle glaucoma patients. Mol Vis. 2011;17(September):2618–27. [PMC free article] [PubMed] [Google Scholar]
- 58.Meer E, Aleman TS, Ross AG. WDR36-Associated Neurodegeneration: A Case Report Highlights Possible Mechanisms of Normal Tension Glaucoma. Genes (Basel). 2021;12(10). doi: 10.3390/genes12101624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kramer PL, Samples JR, Monemi S, Sykes R, Sarfarazi M, Wirtz MK. The role of the WDR36 gene on chromosome 5q22.1 in a large family with primary open-angle glaucoma mapped to this region. Arch Ophthalmol. 2006;124(9):1328–31. doi: 10.1001/archopht.124.9.1328 [DOI] [PubMed] [Google Scholar]
- 60.Frezzotti P, Pescucci C, Papa FT, Iester M, Mittica V, Motolese I, et al. Association between primary open-angle glaucoma (POAG) and WDR36 sequence variance in Italian families affected by POAG. Br J Ophthalmol. 2011;95(5):624–6. doi: 10.1136/bjo.2009.167494 [DOI] [PubMed] [Google Scholar]
- 61.Liu K, He W, Zhao J, Zeng Y, Cheng H. Association of WDR36 polymorphisms with primary open angle glaucoma: A systematic review and meta-analysis. Med (United States). 2017;96(26):1–7. doi: 10.1097/MD.0000000000007291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skarie JM, Link BA. The Primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum Mol Genet. 2008;17(16):2474–85. doi: 10.1093/hmg/ddn147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chi ZL, Yasumoto F, Sergeev Y, Minami M, Obazawa M, Kimura I, et al. Mutant WDR36 directly affects axon growth of retinal ganglion cells leading to progressive retinal degeneration in mice. Hum Mol Genet. 2010;19(19):3806–15. doi: 10.1093/hmg/ddq299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, et al. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14(6):725–33. doi: 10.1093/hmg/ddi068 [DOI] [PubMed] [Google Scholar]
- 65.Davis LK, Meyer KJ, Schindler EI, Beck JS, Rudd DS, Jason Grundstad A, et al. Copy number variations and primary open-angle glaucoma. Investig Ophthalmol Vis Sci. 2011;52(10):7122–33. doi: 10.1167/iovs.10-5606 [DOI] [PMC free article] [PubMed] [Google Scholar]