Abstract
Mutation signatures associated with apolipoprotein B mRNA editing catalytic polypeptide-like 3A/B (APOBEC3A/B) cytidine deaminases are prevalent across cancers, implying their roles as mutagenic drivers during tumorigenesis and tumor evolution. APOBEC3A (A3A) expression induces DNA replication stress and increases the cellular dependency on the ataxia telangiectasia and Rad3-related (ATR) kinase for survival. Nonetheless, how A3A induces DNA replication stress remains unclear. We show that A3A induces replication stress without slowing replication forks. We find that A3A induces single-stranded DNA (ssDNA) gaps through PrimPol-mediated repriming. A3A-induced ssDNA gaps are repaired by multiple pathways involving ATR, RAD51, and translesion synthesis. Both ATR inhibition and trapping of poly(ADP-ribose) polymerase (PARP) on DNA by PARP inhibitor impair the repair of A3A-induced gaps, preferentially killing A3A-expressing cells. When used in combination, PARP and ATR inhibitors selectively kill A3A-expressing cells synergistically in a manner dependent on PrimPol-generated gaps. Thus, A3A-induced replication stress arises from PrimPol-generated ssDNA gaps, which confer a therapeutic vulnerability to gap-targeted DNA repair inhibitors.
APOBEC3A induces DNA gaps and offers therapeutic opportunities.
INTRODUCTION
Apolipoprotein B mRNA editing catalytic polypeptide-like 3A (APOBEC3A) belongs to the APOBEC family of cytidine deaminases. APOBEC3A (A3A) catalyzes the deamination of cytosine to uracil in single-stranded DNA (ssDNA) or RNA (1–3). Recent sequencing studies on cancer genomes have identified A3A and A3B enzymes as key drivers of mutations that promote tumor heterogeneity, tumor evolution, and drug resistance (1, 4). A3A/B mutation signature, namely, single-base substitution (SBS2) and SBS13, has been identified in several different types of cancers, including breast, lung, head-and-neck, cervical, esophageal, and bladder cancers that abnormally express A3A/B (5–7). A3A expression is associated with poor overall survival in pancreatic ductal adenocarcinoma and drives chromosomal instability and pancreatic cancer metastasis (8). A3A also induces PD-L1 expression in cancer and, thus, simultaneously promotes mutagenesis and tumor immune evasion (9). Overall, A3A expression provides cancer cells with a selective growth and survival advantage.
DNA replication is constantly challenged by obstacles. Upon encountering these obstacles, cells slow down or stall ongoing replication in a transient manner. This stalling or slowing down and activation of corresponding response pathways is often termed a replication stress response. Cells have evolved several DNA damage tolerance (DDT) mechanisms to cope with replication stress that overcome obstacles and allow continued DNA synthesis. Human cells broadly use three distinct DDT mechanisms, (i) PrimPol-dependent repriming, (ii) translesion synthesis (TLS), and (iii) RAD51-dependent template switching (TS) (10, 11). Replication forks facing obstacles can also undergo regression on DNA through a RAD51-mediated process called fork reversal, generating four-way DNA structures that stabilize stalled forks and promote their recovery (12–14).
Repriming of DNA synthesis by PrimPol overcomes replication obstacles and allows replication fork progression (15). Upon fork stalling, repriming by PrimPol allows reinitiation of DNA synthesis downstream of the lesion, leaving behind ssDNA gaps that need to be filled post-replicatively. PrimPol-generated ssDNA gaps arise in various contexts and can act as potential problematic lesions that undermine genome integrity by serving as both a cause and consequence of replication stress. Fork reversal and PrimPol-dependent repriming have been shown to be in direct competition to respond to replication stress and loss of fork reversal factors engages PrimPol-dependent repriming (16, 17). Inhibition of poly(ADP-ribose) polymerase (PARP) leads to RECQ1 activation and promotes the formation of PrimPol-dependent ssDNA gaps by inhibiting fork reversal (18, 19). Moreover, certain PARP inhibitors can also trap PARP at ssDNA gaps and prevent their repair/ fill-in post-replicatively leading to gap persistence (20). Last, cancers with inactivation of tumor suppressors, such as BRCA1/2-mutant cancers, or with expression of oncogenes such as Cyclin E1 harbor PrimPol-generated ssDNA gaps (21).
Although several types of replication stress can lead to ssDNA gaps, cells use various pathways to limit the persistence of gaps by using post-replicative gap repair/fill-in mechanisms. RAD51 is a key factor in the repair of ssDNA gaps post-replicatively (17, 22, 23). Studies in yeast have suggested a role for RAD51 in post-replicative gap repair in response to alkylating agents (24). RAD51-dependent ssDNA gap repair in the G2 phase of the cell cycle was observed in avian DT-40 cells where the loss of RAD51 led to the accumulation of RPA-bound ssDNA specifically in the G2 phase (23). In human cells, RAD51 was shown to be involved in gap repair in the S phase (17). In response to bulky lesions, PrimPol-generated gaps are primarily repaired by RAD51 post-replicatively (25). In addition to RAD51-dependent repair, TLS and POLQ-dependent pathways have also been shown to be important for gap repair or gap suppression (17, 26, 27). Overall, there seems to be a balance between the accumulation of gaps and their repair, and cellular changes tilting this balance toward gap accumulation can lead to replication stress-induced genomic instability.
Previously, we have demonstrated that A3A expression induces replication stress in cancer cells by increased deamination of cytosine into uracil, which is subsequently excised leaving abasic sites (28). In the absence of ataxia telangiectasia and Rad3-related (ATR) activity, A3A-expressing cells accumulate ssDNA leading to increased levels of double-stranded breaks (DSBs). As a result, A3A-expressing cells are exquisitely sensitive to ATR inhibition (ATRi) (28, 29). Other replication inhibitors do not sensitize A3A-expressing cells as much as ATRi, suggesting that ATR has a unique function in protecting cells against A3A-induced replication stress. These findings raise important questions about the nature of replication stress induced by A3A, how A3A expression leads to ssDNA accumulation and increased DSBs, and how ATR protects the genome against A3A-induced replication stress.
In this study, we further investigated the nature of replication stress induced by A3A. Using single-molecule DNA fiber assays, we found that A3A expression induces replication stress without slowing down replication forks. Instead, A3A induces ssDNA gaps in nascent DNA during DNA replication in a manner dependent on its deaminase activity. These ssDNA gaps are generated by PrimPol, which is recruited to replication forks encountering A3A-generated uracils and subsequent base excision repair (BER) intermediates. We further show that A3A-induced ssDNA gaps are repaired by post-replicative repair/ fill-in mechanisms dependent on ATR, but not on its downstream effector Chk1. Depletion of RAD51 or inhibition of RAD51 with a small-molecule inhibitor also blocks gap repair in a manner epistatic with ATRi, suggesting that ATR and RAD51 function in concert with gap repair. Notably, A3A-expressing cells are sensitive to talazoparib, a PARP inhibitor (PARPi) that efficiently traps PARP at ssDNA gaps, and to loss of REV1, which is critical for TLS. Furthermore, the combination of ATRi and PARPi displays a strong synergy in killing A3A-expressing cells, and TLS inhibitor (TLSi) also enhances the killing of A3A-expressing cells by ATRi. Together, our findings provide valuable insights into the nature of A3A-induced replication stress and the DNA repair pathways responding to it, suggesting that therapies combining ATRi with PARPi or TLSi may be effective strategies to eliminate A3A-expressing cells in tumors.
RESULTS
A3A induces replication stress without slowing replication forks
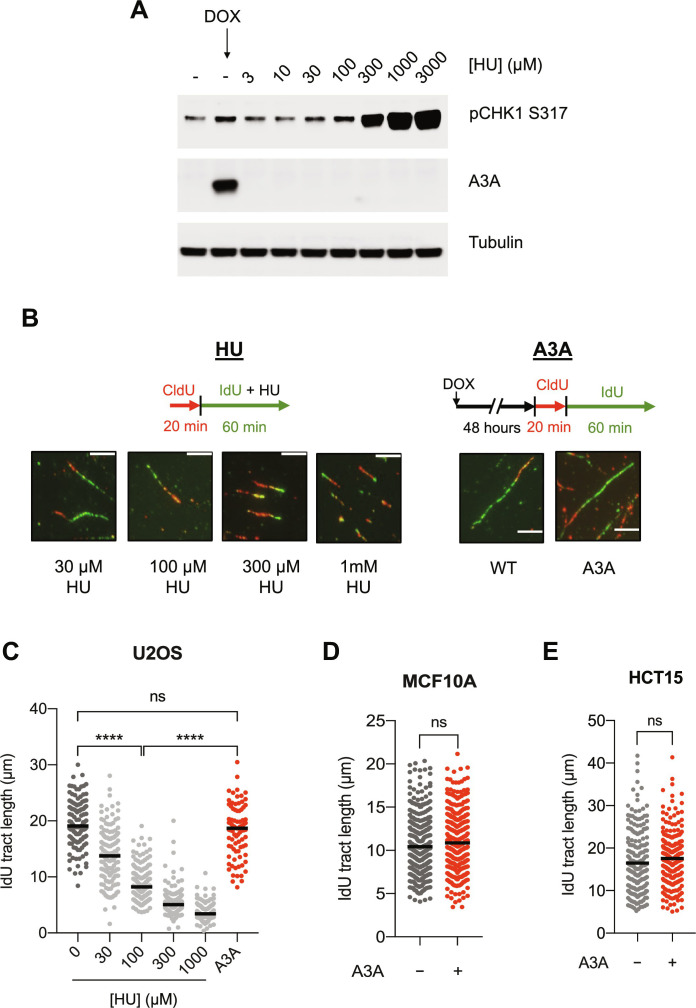
We and others recently showed that A3A-mediated cytidine deamination induces replication stress (28, 29). To understand how A3A induces replication stress, we conditionally expressed A3A in the human osteosarcoma cell line U2OS and used phosphorylated Chk1 (p-Chk1S317), a substrate of ATR, as a surrogate to detect replication stress. Consistent with our previous report, A3A expression led to an increase of p-Chk1S317 compared to control cells (28). To quantitatively assess the level of A3A-induced replication stress, we compared the level of A3A-induced p-Chk1S317 with that induced by hydroxyurea (HU), an inhibitor of DNA replication. Treatment of cells with increasing concentrations of HU led to an expected dose-dependent increase of pChk1S317 (Fig. 1A). Notably, the level of p-Chk1S317 in A3A-expressing cells was comparable to that in cells treated with 100 μM HU, but much lower than that in cells treated with higher concentrations of HU (Fig. 1A). Thus, A3A expression in the inducible cell line generates a detectable but relatively modest level of replication stress.
Fig. 1. A3A expression induces replication stress but does not slow down replication forks.
(A) U2OS-A3A cells were treated with either indicated concentrations of HU for 1 hour or with doxycycline (DOX; 200 ng/ml) to induce A3A for 48 hours before processing samples for Western blot analysis. (B) Top: Scheme for DNA fiber analysis of samples treated with HU (left) or A3A (right). Bottom: Representative images of DNA fibers of samples treated with appropriate concentrations of HU or with A3A expression. All scale bars are 40 pixels in width. (C) Quantification of 5-iodo-2′-deoxyuridine (IdU) tract lengths in U2OS cells for the respective samples shown in (B). 0 μM HU sample was the same as wild type (WT) in (B). (D and E) Quantification of IdU tract lengths of control and A3A-expressing MCF10A and HCT15 cells. For (C) to (E), the black horizontal line indicates the median IdU tract length from at least 125 fibers quantified from two independent experiments (n > 125). Statistical significance was determined by one-way analysis of variance (ANOVA) using Tukey’s multiple comparisons test in (C) or using an unpaired two-tailed t test in (D) and (E).
Since replication stress is commonly associated with slowing or stalling of replication forks, we next tested whether A3A expression reduces replication fork speed using the DNA fiber assay. We sequentially labeled nascent DNA with thymidine analogs 5-chloro-2′-deoxyuridine (CldU) and 5-iodo-2′-deoxyuridine (IdU), and then analyzed the length of labeled replication tracts in DNA fibers. To compare the effects of A3A and HU on fork speed, cells were either treated with doxycycline (DOX) to induce A3A expression for 48 hours before labeling or treated with HU for 1 hour during the IdU labeling. In both A3A-expressing and HU-treated cells, DNA replication forks encounter replication stress during IdU labeling, allowing us to examine the effects on fork speed by measuring the length of IdU-labeled tracts. As expected, the treatment of cells with HU led to a dose-dependent slowing of replication forks (Fig. 1, B and C). In particular, cells treated with 100 μM HU showed an approximately twofold reduction in fork speed. Unexpectedly, however, although cells induced to express A3A and cells treated with 100 μM HU displayed similar levels of replication stress as indicated by p-Chk1S317, A3A expression did not slow down replication forks as 100 μM HU did (Fig. 1, B and C). To validate the results from U2OS cells, we inducibly expressed A3A in human colon adenocarcinoma HCT15 and human mammary epithelial MCF10A cell lines (fig. S1, A and B). Consistent with our results from U2OS, A3A expression did not lead to a slowdown of forks but increased p-Chk1S317 levels (fig. S1, A and B). These results suggest that A3A expression induces replication stress without slowing replication forks.
A3A induces PrimPol-generated ssDNA gaps through cytosine deamination
When replication forks encounter various types of impediments on DNA, multiple DDT pathways are activated to help forks overcome impediments and continue DNA synthesis (Fig. 2A). One of the DDT pathways is driven by PrimPol-mediated repriming, which allows replication forks to resume DNA synthesis ahead of stalled DNA polymerases but leaves behind ssDNA gaps. Another DDT pathway is mediated by fork reversal, which is driven by fork-remodeling factors including SMARCAL1, ZRANB3, and HLTF. Notably, whereas fork reversal tends to slow down replication forks, PrimPol-mediated priming does not (16, 30). Our observation that A3A induces replication stress without slowing replication forks raised the possibility that PrimPol-mediated repriming is activated in response to A3A expression.
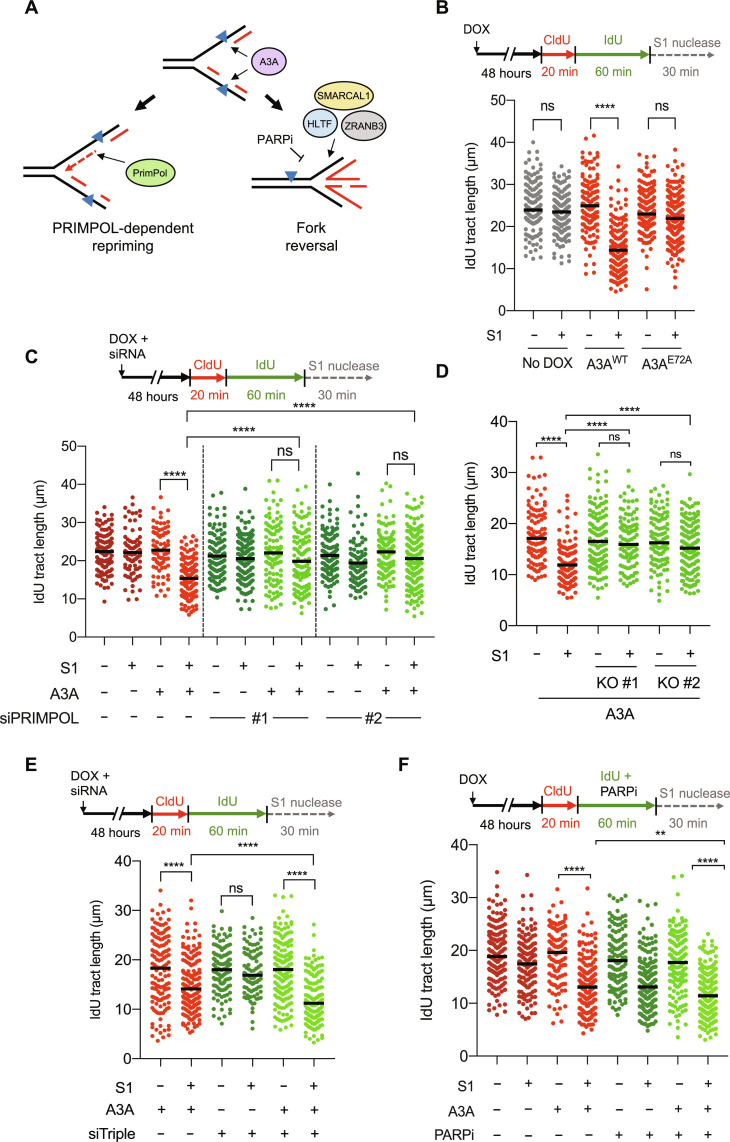
Fig. 2. A3A expression induces ssDNA gaps in a PRIMPOL-dependent manner.
(A) Model depicting a competition between PrimPol-dependent repriming and fork reversal when replication forks encounter A3A-induced DNA lesions. Fork reversal is inhibited in the presence of PARPi or by loss of fork reversal factors HLTF, SMARCAL1, and ZRANB3. (B) A3A deaminase activity generates ssDNA gaps. (C and D) A3A induces ssDNA gaps in a PrimPol-dependent manner. U2OS-A3A cells depleted of PrimPol with 10 nM small interfering RNAs (siRNAs) in (C) or two independent U2OS-A3A PrimPol KO clones in (D) were used to detect IdU-positive replication tracts. (E and F) U2OS-A3A cells were depleted of all three fork reversal factors SMARCAL1, HLTF, and ZRANB3 with 5 nM siRNA each in (E) or were treated with 100 nM talazoparib during the IdU labeling period in (F) and processed for DNA fiber analysis. The black horizontal line indicates the median IdU tract length from more than 125 IdU-positive replication tracts having a CldU-IdU double-positive signal from three independent experiments (n > 125). Statistical significance was determined by one-way ANOVA using Tukey’s multiple comparisons test.
To test whether A3A expression leads to ssDNA gaps in nascent DNA, we used the S1 nuclease, which specifically cleaves ssDNA, into the DNA fiber assay (31). Upon sequential labeling of nascent DNA with CldU and IdU, DNA fibers were digested with the S1 nuclease. If ssDNA gaps were present in nascent DNA, then the length of IdU+ tracts would be shortened by S1. In cells induced to express wild-type A3A (A3AWT), IdU+ tract length was shortened compared to that in uninduced control cells (Fig. 2B, lanes 3 and 4), showing the induction of ssDNA gaps by A3AWT. In contrast to A3AWT, the catalytically dead A3AE72A mutant did not induce ssDNA gaps (Fig. 2B, lanes 5 and 6; and fig. S2, A to D). In HCT15 and MCF10A cells conditionally expressing A3AWT, A3AWT also induced ssDNA gaps, confirming this effect of A3A in different cancer and noncancerous cells (fig. S2, E and F). Thus, the cytidine deaminase activity of A3A induces ssDNA gaps in nascent DNA.
Next, we investigated whether A3A-induced ssDNA gaps were generated by PrimPol. The knockdown of PrimPol using two independent small interfering RNAs (siRNAs) in A3A-expressing cells almost completely reversed the shortening of replication tracts by S1 (Fig. 2C and fig. S2G), showing the presence of PrimPol-generated ssDNA gaps. To confirm this result, we generated U2OS PrimPol knockout (KO) cell lines and tested the effects of A3A expression on ssDNA gaps with the S1 fiber assay. Using two independent PrimPol KO clones, we confirmed the presence of PrimPol-generated ssDNA gaps in A3A-expressing cells (Fig. 2D and fig. S2H). These results suggest that A3A induces PrimPol-generated ssDNA gaps. Consistent with the induction of ssDNA gaps by A3A, we observed an increase in phospho-Chk1 (p-Chk1) levels in A3A-expressing cells (fig. S2I), which may reflect the activation of ATR by ssDNA gaps. However, the knockdown of PrimPol did not substantially reduce p-Chk1 levels (fig. S2I), suggesting that A3A can induce ssDNA gaps through PrimPol-independent mechanisms. It is possible that both PrimPol and DNA polymerase α (Pol α)-primase contribute to the A3A-induced repriming and formation of ssDNA gaps. Loss of PrimPol may prevent gap formation on the leading strand and the cleavage of replication tracts by the S1 nuclease, but A3A-induced gaps can still form on the lagging strand by Pol a-primase.
The induction of ssDNA gaps by A3A is influenced by the choice of DDT pathways
Previous studies by us and others suggested that fork reversal and PrimPol-mediated repriming are competing DDT pathways at stressed forks (16, 17, 19, 20). When both fork reversal and repriming are activated in cells, loss of fork reversal is expected to increase repriming (Fig. 2A). Upon the simultaneous knockdown of SMARCAL1, HLTF, and ZRANB3 in A3A-expressing cells (fig. S2J), the level of A3A-induced ssDNA gaps, as revealed by the S1-dependent shortening of replication tracts, was increased (Fig. 2E, lanes 1, 2, 5, and 6). This result suggests that both fork reversal and repriming are activated by A3A, and the choice between these two DDT pathways influences the induction of ssDNA gaps by A3A.
PARP1 promotes the accumulation of reversed forks by inhibiting the RECQ1 helicase, which resolves reversed forks (18). Therefore, PARP inhibitor (PARPi) is expected to decrease fork reversal in favor of repriming (Fig. 2A). Consistent with our recent studies (20), PARPi increased ssDNA gaps in the absence of A3A (Fig. 2F, lanes 1, 2, 5, and 6). In A3A-expressing cells, PARPi enhanced the shortening of replication tracts by S1 (Fig. 2F, lanes 4 and 8), suggesting an increase of ssDNA gaps. These results lend further support to the notion that promoting PrimPol-mediated repriming in A3A-expressing cells increases ssDNA gaps.
Recent studies have shown a synthetic lethal relationship between loss of HMCES and A3A expression (32). We tested whether HMCES plays a role in suppressing A3A-induced ssDNA gaps. Consistent with previous results (33), we observed a slowing of replication forks in A3A-expressing cells after loss of HMCES suggesting that HMCES helps maintain replication fork speed in A3A-expressing cells (fig. S3). Loss of HMCES itself led to a shortening of replication tracts after S1 treatment indicative of ssDNA gaps. However, depletion of HMCES in A3A-expressing cells did not lead to a further reduction in IdU tract lengths compared to A3A-expressing cells proficient in HMCES.
A3A triggers PrimPol through UNG-generated abasic sites
A3A-mediated cytidine deamination generates uracil in genomic DNA, which is excised by uracil glycosylases to give rise to abasic sites during BER. To understand whether PrimPol-mediated repriming is triggered by A3A-induced abasic sites, we knocked down the uracil glycosylase UNG, the primary glycosylase removing genomic uracil (34), in A3A-expressing cells and tested its effects on replication tracts. Depletion of UNG in A3A-expressing cells reduced replication fork speed (fig. S4A, lanes 1 and 3). In addition, loss of UNG in A3A-expressing cells reduced the shortening of replication tracts by S1, which is evident by the reduced ratio of tract length between untreated and S1-treated fibers (fig. S4, A and B). Why replication forks slowdown in the absence of UNG still requires further investigation. Nonetheless, the reduction in S1-dependent shortening of replication tracts in UNG-depleted cells suggests that UNG contributes to the formation of ssDNA gaps by PrimPol.
SMUG1 is another glycosylase that may process genomic uracil as a backup for UNG and contribute to the formation of PrimPol-dependent ssDNA gaps (26). To determine whether SMUG1 contributes to the formation of ssDNA gaps in A3A-expressing cells, we depleted SMUG1 in A3A-expressing cells and analyzed its effects on replication tracts. In contrast to the loss of UNG, the depletion of SMUG1 did not slow replication forks or alter the level of ssDNA gaps (fig. S4, A and B). These results suggest that A3A-generated genomic uracil is primarily processed by UNG, giving rise to abasic sites that impede DNA polymerases and trigger PrimPol-mediated repriming.
PARP and TLS are involved in the response to A3A-induced ssDNA gaps
While PrimPol-mediated repriming allows replication forks to continue DNA synthesis when encountering certain types of impediments, PrimPol-generated ssDNA gaps could be a source of genomic instability if not repaired properly (Fig. 3A). To understand how A3A-induced gaps are repaired, we sought to establish an assay to follow the removal of gaps after they are formed during DNA replication. We pulse-labeled nascent DNA in A3A-expressing cells and then used the S1 fiber assay to measure the levels of ssDNA gaps in labeled replication tracts over time (Fig. 3B). The levels of A3A-induced gaps, as indicated by the S1-dependent shortening of replication tracts, were gradually reduced over time. Notably, A3A-induced ssDNA gaps were largely removed 16 hours after they were generated in nascent DNA. Thus, we established an assay to follow the repair of A3A-induced gaps, allowing us to test the effects of various DDR proteins or drugs on gap repair in A3A-expressing cells.
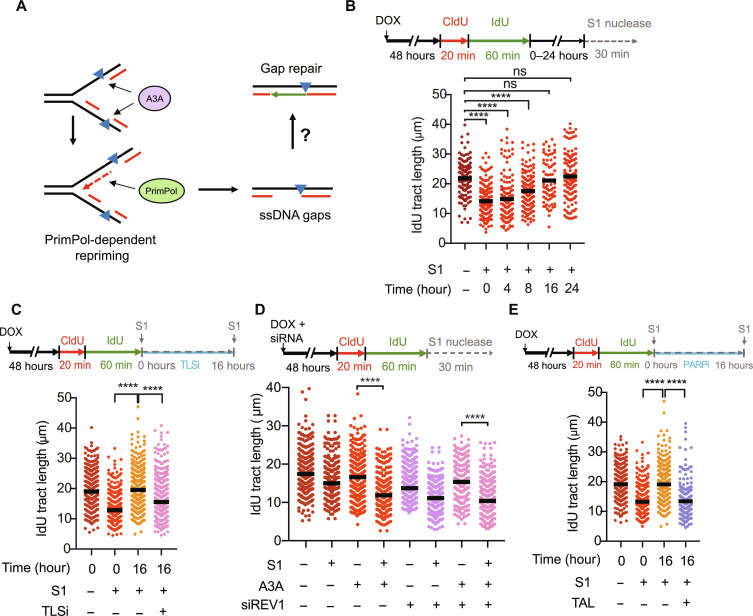
Fig. 3. ssDNA gaps induced upon A3A expression are repaired over time and can be made persistent by trapping PARP.
(A) Model depicting the hypothesis for the mechanism of repair of A3A-induced PrimPol-generated ssDNA gaps. (B) Top: Experimental scheme. After CldU and IdU labeling, A3A-expressing cells were allowed to recover in media for 0, 4, 8, 16, or 24 hours followed by treatment with S1 nuclease. Bottom: Quantification of IdU tract lengths showing repair of A3A-induced ssDNA gaps (C) Experimental scheme for testing the role of TLSi in gap repair. After CldU and IdU labeling, A3A-expressing cells were either treated with S1 immediately or were allowed to recover in media containing 10 μM TLSi (JH-RE-06) for 16 hours before S1 treatment. (D) Experimental scheme for testing the role of TLS in gap formation. U2OS-A3A cells were depleted of REV1 and processed for DNA fiber analysis. (E) Cells were recovered in media containing 100 nM PARPi (talazoparib) for 16 hours before S1 treatment. The black horizontal line indicates the median IdU tract length from more than 150 IdU-positive replication tracts having a CldU-IdU double-positive signal from three independent experiments (n > 150). Statistical significance was determined by one-way ANOVA using Tukey’s multiple comparisons test.
TLS is a DDT pathway that allows replication forks to bypass certain types of DNA lesions. Notably, TLS was recently shown to repair ssDNA gaps in BRCA1/2-deficient cells (17, 35). Furthermore, an A3A-induced mutation signature in cancer cells is dependent on REV1, a key player in TLS (6). To test whether TLS is involved in the repair of A3A-induced gaps, we pulse-labeled nascent DNA in A3A-expressing cells, and then followed gap repair in the presence or absence of a REV1 inhibitor (TLSi), JH-RE-06. In the presence of TLSi, the removal of gaps at 16 hours after labeling was modestly reduced (Fig. 3C), suggesting that TLS is a contributor to gap repair. The knockdown of REV1 with siRNA did not affect the formation of A3A-induced gaps right after nascent DNA labeling and slightly increased the S1-dependent shortening of replication tracts (Fig. 3D, lanes 3, 4, 7, and 8), supporting the idea that TLS is involved in the repair but not the formation of A3A-induced gaps. REV1 knockdown reduced fork speed and increased ssDNA gaps even in the absence of A3A (Fig. 3D, lanes 1, 2, 5, and 6), suggesting that TLS may also respond to spontaneous gaps during replication.
We recently showed that PARPi induces PrimPol-generated ssDNA gaps in nascent DNA and the trapping of PARP1/2 by PARPi on DNA prevents gap repair, giving rise to persistent ssDNA gaps that are converted to DSBs in a cell cycle–dependent manner (20). The similarities between PARPi- and A3A-induced ssDNA gaps raise the possibility that PARPi may also affect the repair of A3A-induced gaps. Talazoparib, a PARPi that traps PARP1/2 efficiently, reduced the repair of A3A-induced gaps at 16 hours after labeling (Fig. 3E). Thus, PARP is engaged in the response to A3A-induced ssDNA gaps and the trapping of PARP by PARPi blocks gap repair.
ATR and RAD51 are critical for the repair of A3A-generated ssDNA gaps
We previously showed that ATR inhibition led to ssDNA accumulation in A3A-expressing cells (28), raising the possibility that ATR is involved in gap repair. To test this possibility, we pulse-labeled nascent DNA and then followed gap repair in the presence and absence of the ATR inhibitor (ATRi) VE-821. ATRi markedly reduced the removal of gaps at 16 hours after labeling (Fig. 4A), showing that ATR is a critical regulator of the repair of A3A-induced gaps. Notably, when cells were exposed to ATRi during IdU labeling (fig. S5A, lanes 5 and 6), ATRi reduced fork speed and increased ssDNA gaps even in the absence of A3A. The effect of ATRi on fork speed, which is likely a result of increased origin firing upon ATR inhibition, is consistent with previous studies (36, 37). The induction of ssDNA gaps by ATRi suggests that ATR is involved in the response to spontaneous gaps during replication. In A3A-expressing cells, ATRi also slowed replication forks and slightly reduced IdU+ tract length after S1 treatment (fig. S5A, lanes 3, 4, 7, and 8), indicating the accumulation of both A3A-induced and spontaneous gaps. These results support the notion that ATRi does not affect the formation of A3A-induced gaps but blocks gap repair.
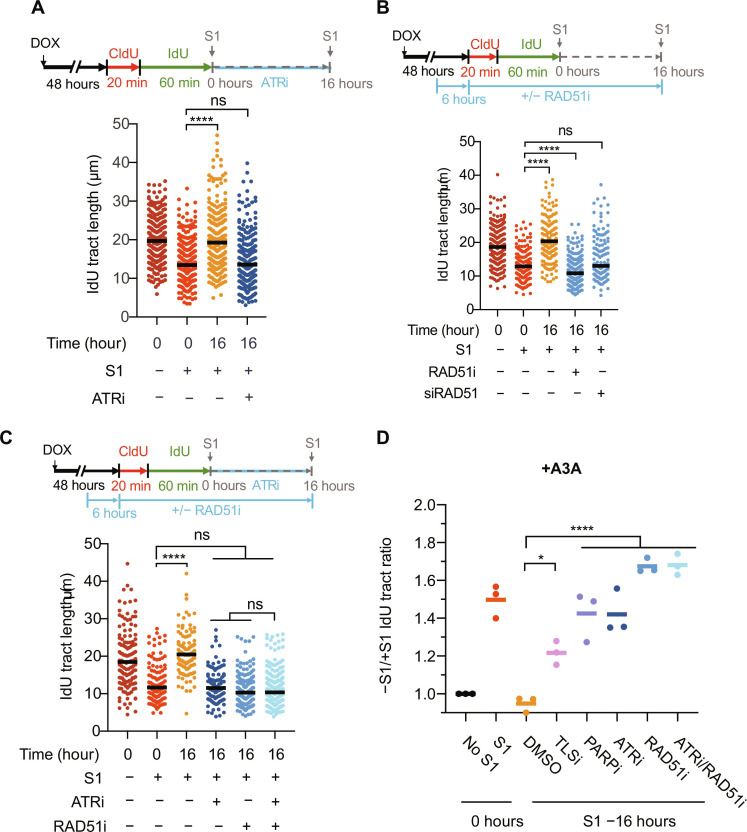
Fig. 4. ATR and RAD51 fill in A3A-induced ssDNA gaps in an epistatic manner.
(A) After CldU and IdU labeling, A3A-expressing cells were either permeabilized and treated with S1 immediately or were allowed to recover in media containing 10 μM ATRi (VE-821) for 16 hours before S1 treatment. (B) Experimental scheme to test the effect of RAD51 on gap repair. A3A-expressing cells were treated with 25 μM RAD51i (B02) 6 hours before labeling cells with CldU and IdU. The inhibitor was maintained throughout the labeling period and was present during the 16-hour recovery period. For siRAD51, cells were transfected with 10 nM siRAD51 at the same time as DOX treatment (i.e., 48 hours before CldU labeling). (C) Experimental scheme to test whether ATR and RAD51 are epistatic for repair of A3A-induced ssDNA gaps. For (A) to (C), the black horizontal line indicates the median IdU tract length from more than 150 IdU-positive replication tracts having a CldU-IdU double-positive signal (n > 150) from three independent experiments. (D) −S1/+S1 IdU tract ratios were calculated by dividing the median IdU tract length of samples treated with buffer by the median IdU tract lengths of the labeled samples treated with S1 nuclease. All samples were from A3A-expressing cells. Statistical significance was determined by one-way ANOVA using Tukey’s multiple comparisons test.
Chk1 is the key effector kinase of ATR in DNA repair and the replication checkpoint response (38). However, in contrast to ATRi, the Chk1 inhibitor (Chk1i) only modestly reduced the removal of A3A-induced gaps at 16 hours after labeling (fig. S5B). This result suggests that the function of ATR in the repair of A3A-induced ssDNA gaps is only partially mediated by Chk1. ATR regulates the loading of RAD51 at DNA breaks and stalled replication forks to promote break repair through homologous recombination (HR) and fork protection (39, 40). To investigate whether ATR promotes gap repair through RAD51, we first tested whether RAD51 is required for the repair of A3A-induced gaps. Similar to ATRi, the RAD51 inhibitor (RAD51i) B02 markedly reduced the removal of gaps at 16 hours after labeling (Fig. 4C), showing that RAD51 is critical for gap repair. The inhibitory effects of ATRi and RAD51i on gap repair were similar, and the combination of ATRi and RAD51i blocked gap repair identically to RAD51i alone (Fig. 4, C and D), suggesting that ATR promotes gap repair by regulating RAD51. When A3A-expressing cells were exposed to RAD51i during nascent DNA labeling, RAD51i did not affect the formation of A3A-induced ssDNA gaps (fig. S5C). Together, these results suggest that ATR and RAD51 act through the same pathway to promote gap repair. The requirement of RAD51 for the repair of A3A-induced gaps prompted us to test whether the HR pathway is involved. Unexpectedly, the knockdown of BRCA2, which plays a key role in loading RAD51 onto ssDNA in the canonical HR pathway, did not affect the repair of A3A-induced gaps (fig. S5D). Furthermore, in the presence of RAD51i, BRCA2 knockdown did not further alter gap repair. Thus, the BRCA2-mediated canonical HR pathway is not responsible for the repair of A3A-induced gaps, raising the possibility that A3A-induced gaps are repaired by a RAD51-dependent and HR-related pathway.
Blocking gap repair selectively kills A3A-expressing cells
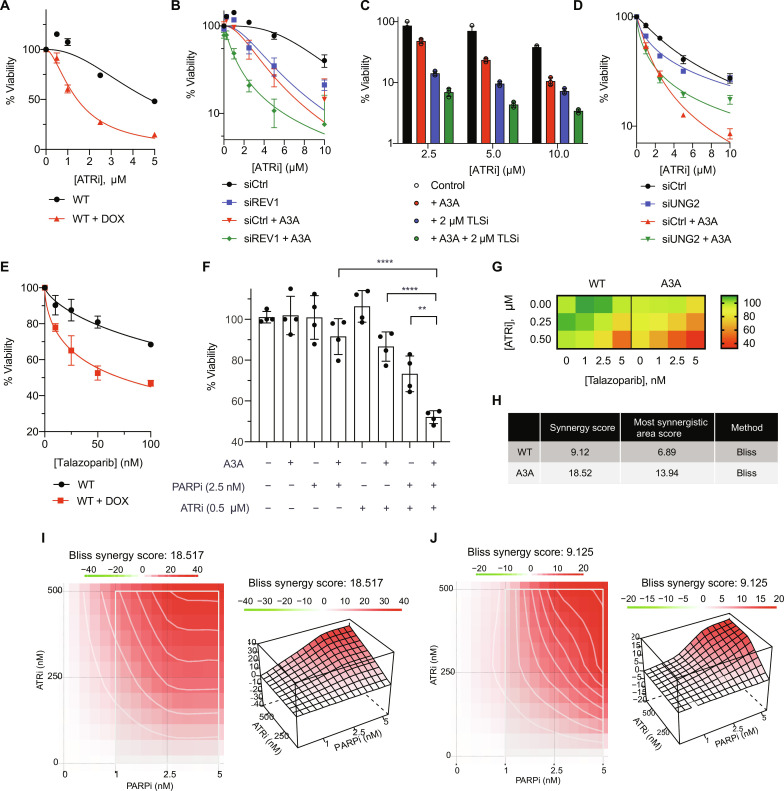
While PrimPol-generated ssDNA gaps do not kill cells immediately, these gaps, if not repaired properly, can be converted into DSBs in the next cell cycle during the S phase when the gaps collide with replication forks, leading to cell death in a cell cycle–dependent manner (20). Thus, we asked whether increasing gap formation and/or blocking gap repair in A3A-expressing cells results in synthetic lethality. As shown in our previous studies, induction of A3A expression increased the ATRi sensitivity of cells (Fig. 5A) (28), supporting the idea that blocking gap repair in A3A-expressing cells is synthetic-lethal. Because TLS allows replication forks to bypass impediments, loss of TLS is expected to increase stalling of DNA polymerase, which elevates repriming, gap formation, and ATRi sensitivity. Consistent with this possibility, the knockdown of REV1 increased ATRi sensitivity even in the absence of A3A (Fig. 5B). In A3A-expressing cells, the knockdown of REV1 further increased ATRi sensitivity (Fig. 5B). Similar observations were made when A3A-expressing cells were treated with ATRi and TLSi (Fig. 5C). In contrast to TLS inhibition, loss of UNG is expected to reduce abasic sites, which decreases polymerase stalling, repriming, gap formation, and ATRi sensitivity. The knockdown of UNG reduced the ATRi sensitivity of A3A-expressing cells (Fig. 5D). Additional SMUG1 depletion did not alter ATRi sensitivity further (fig. S6). Together, these results suggest that the sensitivity of A3A-expressing cells to ATRi can be modulated by repair pathways affecting gap levels. The ability of PARPi to induce gaps and block gap repair prompted us to test if A3A-expressing cells are sensitive to PARPi. We observed that expression of A3A increased the sensitivity of cells to talazoparib (Fig. 5E).
Fig. 5. ATRi and PARPi treatment is synergistic in killing A3A-expressing cells.
(A) U2OS-A3A cells were treated with increasing concentrations of ATRi (VE-821). Viability was measured using CellTiter-Glo 5 days after ATRi treatment and normalized to the untreated control. Data represent means with SD (n = 3). (B) siREV1 (10 nM) was added 48 hours before treatment with ATRi. Data represent means with SD (n = 3). (C) Similar to (A) except 2 μM TLSi (JH-RE-06) was added along with ATRi. Data represent means with SD (n = 3). (D) siUNG (10 nM) was added 48 hours before treatment with ATRi (n = 3). (E) Similar to (A) except cells were treated with talazoparib. Data represent means with SD (n = 3). (F) Viability of WT or A3A-expressing cells in the presence of 2.5 nM talazoparib, 0.5 μM VE-821, and combination. Data represent means with SD (n = 4). (G) Heatmap of the viability of WT or A3A-expressing cells for all concentrations tested for ATRi and PARPi. (H to J) Synergy scores calculated for ATRi and PARPi treatment for A3A-expressing cells in (I) and control cells in (J) with the Bliss method using SynergyFinder 3.0. Statistical significance was determined by one-way ANOVA using Tukey’s multiple comparisons test. Viability curves were generated using a curve fit nonlinear regression model.
The increased sensitivity of A3A-expressing cells to talazoparib prompted us to investigate whether high levels of APOBEC mutational signatures correlate with high talazoparib sensitivity in cancer cells. To test this, we analyzed whole exome sequencing data from cell lines in the Cancer Cell Line Encyclopedia (CCLE) database. We used non-negative matrix factorization (NMF) to decompose APOBEC mutational signatures and selected the cell lines with at least 10% of their mutations assigned to A3A/B (referred to as APOBEC+ cell lines) (2, 39). We observed a strong inverse correlation between the APOBEC mutational signature and the IC50 (median inhibitory concentration) of talazoparib across 39 cell lines of head-and-neck cancer, a tumor type that generally harbors high APOBEC activity (fig. S7A). However, this trend was not readily observed in other tumor types (fig. S7, B to D). Thus, APOBEC mutational signatures correlate with talazoparib sensitivity in some but not all APOBEC+ cancers.
The lack of correlation between APOBEC mutational signatures and talazoparib sensitivity in many APOBEC+ cancers raises the possibility that talazoparib alone is not sufficient to fully exploit A3A-induced gaps. This idea prompted us to investigate whether the combination of ATRi and PARPi can eliminate A3A-expressing cells more effectively than ATRi and PARPi alone. We treated A3A-expressing cells with both ATRi and PARPi and measured cell viability. These cells displayed a marked hypersensitivity when they were treated with both ATRi and PARPi, compared to uninduced cells or treated with ATRi or PARPi alone (Fig. 5, F and G). To determine whether the effects of ATRi and PARPi are synergistic, we used the Bliss method to calculate the synergy score for ATRi and PARPi (41). ATRi and PARPi were synergistic even in uninduced cells, but the synergy was further increased in A3A-expressing cells (Fig. 5, H to J). Thus, ATRi and PARPi display a stronger synergy in the presence of A3A, providing a strategy to preferentially kill A3A-expressing cells.
To further probe this, we tested whether endogenous A3A/B activities are associated with the sensitivity of cancer cells to ATRi and PARPi. We tested a panel of cancer cell lines known to have different levels of endogenous A3A/B activities against ATRi and PARPi. TOV21G and MDA-MB-453 are two cell lines with high A3A/B activities, whereas SKBR3 is a cell line with low A3A/B activities. We find that TOV21G and MDA-MB-453 are much more sensitive to ATRi and PARPi than SKBR3 (fig. S8A). Furthermore, ATRi and PARPi displayed a strong synergy in killing TOV21G and MDA-MB-452 cells, but not SKBR3 cells (fig. S8A). Thus, the sensitivity of cancer cells to ATRi and PARPi, as well as the combination of ATRi and PARPi, correlates with endogenous A3A/B activities.
We then asked whether endogenous A3A expression in cancer cells is sufficient to render them susceptible to ATRi and talazoparib. Using quantitative polymerase chain reaction (qPCR), we confirmed that NCI-H2347 is a cell line that expresses endogenous A3A, whereas A3A expression is undetectable in SKBR3 (fig. S8B). We treated these cell lines with ATRi and PARPi and then analyzed the levels of γH2AX, a marker of DNA damage. In NCI-H2347, we observed an increase in γH2AX after treatment with ATRi, PARPi, or both, showing that ATRi and PARPi induce DNA damage efficiently in A3A-expressing cells (fig. S8C). Moreover, the induction of γH2AX by ATRi and PARPi in NCI-H2347 cells was suppressed by the knockdown of endogenous A3A (fig. S8C), confirming that endogenous A3A is responsible for the DNA damage induction by ATRi and PARPi. In contrast to NCI-H2347, treatment of SKBR3 with A3A siRNA did not affect ATRi and PARPi induced γH2AX (fig. S8C). Thus, as observed in NCI-H2347 cells, the endogenous A3A in cancer cells can render cancer cells suspectable to ATRi and PARPi treatments.
The sensitivity of A3A-expressing cells to ATRi and PARPi is dependent on gaps
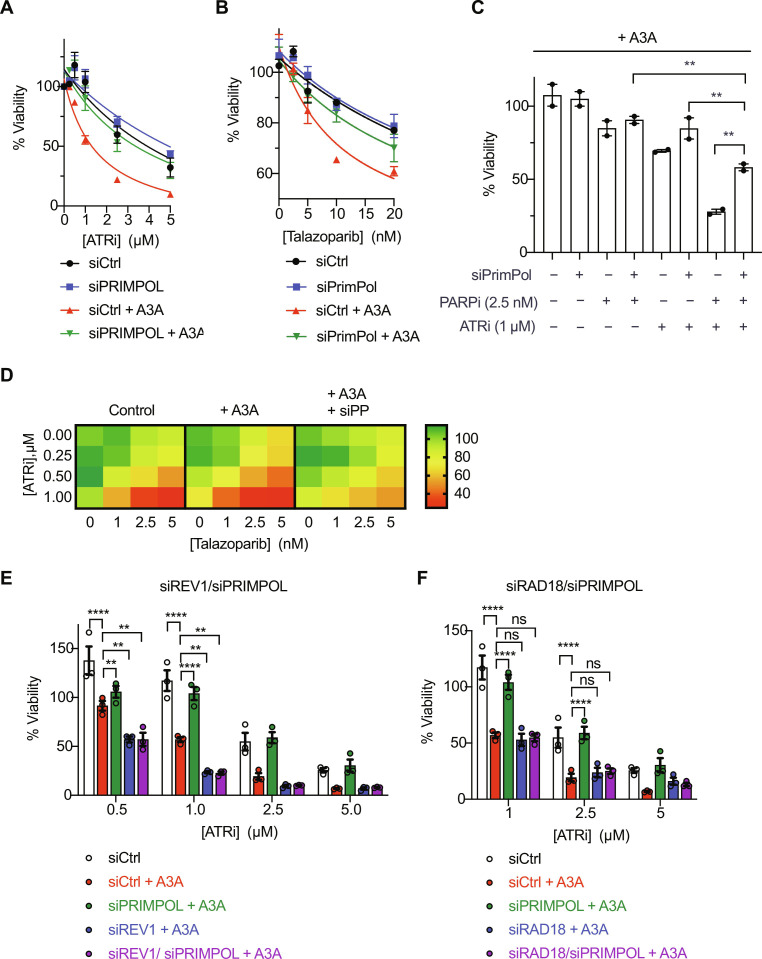
If PrimPol-generated ssDNA gaps are the cause of ATRi and PARPi sensitivities in A3A-expressing cells, one would predict that loss of PrimPol should render cells resistant to ATRi and PARPi. The knockdown of PrimPol reduced the sensitivity of A3A-expressing cells to ATRi and PARPi (Fig. 6, A and B). Moreover, PrimPol depletion also reduced the sensitivity of A3A-expressing cells to combined ATRi and PARPi treatment (Fig. 6, C and D). These results support the idea that PrimPol-generated gaps confer ATRi and PARPi sensitivity in A3A-expressing cells.
Fig. 6. Loss of PrimPol renders A3A-expressing cells resistant to ATRi and PARPi dual treatment.
(A and B) U2OS-A3A cells were treated with 10 nM PrimPol siRNA to deplete PrimPol. Forty-eight hours later, cells were treated with increasing concentrations of ATRi (VE-821) in (A) or PARPi (talazoparib) in (B). Viability was measured 5 days after ATRi/PARPi treatment and normalized to the untreated control. Data represent means with SD (n = 3). (C) Viability of WT and A3A-expressing cells in the presence of 2.5 nM talazoparib, 1 μM VE-821, or both. Data represent means with SD (n = 2). (D) Heatmap showing viability for all ATRi and PARPi concentrations. (E and F) U2OS-A3A cells were treated with 10 nM each siPrimPol and siREV1 in (E) or siRAD18 in (F) for 48 hours before ATRi or PARPi treatment. Viability was measured 5 days after ATRi/PARPi treatment and normalized to the untreated control. Data represent means with SD (n = 3). Statistical significance was determined by one-way ANOVA using Tukey’s multiple comparisons test. Viability curves were generated using a curve fit nonlinear regression model.
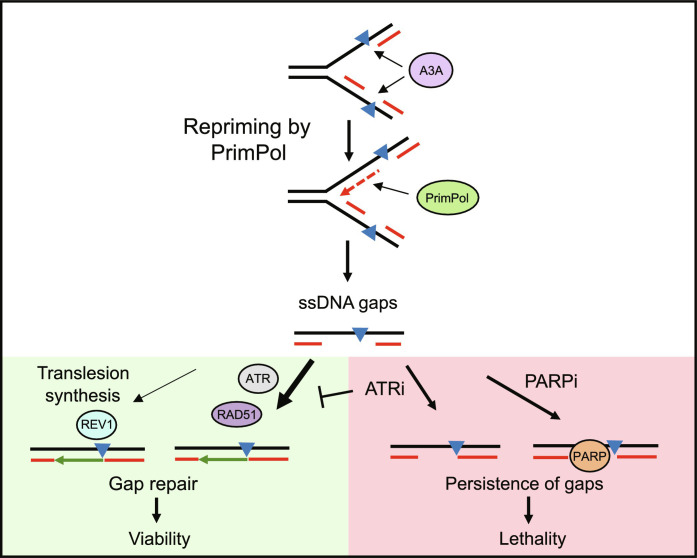
The resistance of A3A-expressing cells to ATRi and PARPi upon PrimPol loss raised a question about how these cells survive replication stress without PrimPol. Because both repriming and TLS pathways allow replication forks to continue DNA synthesis when encountering impediments, we asked whether PrimPol-depleted cells use TLS as a backup mechanism to cope with replication stress. Notably, while PrimPol depletion in A3A-expressing cells reduced their ATRi sensitivity, co-depletion of PrimPol and REV1 completely restored the ATRi sensitivity and even rendered A3A-expressing more sensitive to ATRi than before PrimPol loss (Fig. 6E). Similarly, the knockdown of Rad18, a ubiquitin ligase that promotes TLS, also restored the ATR sensitivity of PrimPol knockdown cells (Fig. 6F). Together, these results suggest that PrimPol-mediated repriming is the primary pathway to respond to A3A-induced impediments (e.g., abasic sites and uracil), but TLS can act as a backup pathway when PrimPol is lost (Fig. 7).
Fig. 7. Working model.
A3A-induced DNA lesions may temporarily stall replication forks leading to the recruitment of PrimPol. PrimPol restarts replication but leaves ssDNA gaps across the lesion. These ssDNA gaps are primarily repaired via an ATR- and RAD51-dependent mechanism with minor contribution from REV1-dependent TLS. Inhibiting ATR blocks the repair of ssDNA gaps and causes their persistence. The ssDNA gaps can also be made persistent by trapping PARP and as a result, A3A-expressing cells show hypersensitivity to combined ATRi and PARPi.
DISCUSSION
A3A/B are major drivers of mutagenesis in a broad spectrum of cancers, contributing to tumorigenesis, tumor evolution, and therapeutic resistance (5, 42). Therefore, eliminating A3A/B activities or A3A/B-expressing tumor cells is an attractive strategy to improve cancer therapy (43). In addition to the development of A3A/B-specific inhibitors, identifying cancer drugs that effectively exploit A3A/B-induced cellular vulnerabilities is another promising approach (43–45). Previous studies by us and others showed that A3A expression induces DNA replication stress, which is associated with increased levels of ssDNA (28, 29). The A3A-induced replication stress renders cells increasingly dependent on the ATR kinase for survival, providing an opportunity to selectively eliminate A3A-expressing tumor cells. Nonetheless, how ATR induces replication stress is not fully understood, why ATRi selectively kills A3A-expressing cells still needs a mechanistic explanation, and whether the efficacy of ATRi in eliminating A3A-expressing cells can be improved remains unknown. In this study, we have addressed these important questions.
Our data in this study revealed that A3A induces PrimPol-generated ssDNA gaps, explaining the elevated levels of ssDNA that we previously observed in A3A-expressing cells (28). Of note, a previous study implicated PrimPol in the suppression of APOBEC/AID-mediated mutagenesis (46), but the role of PrimPol in the response to A3A-induced replication stress has not been reported. While PrimPol-mediated repriming helps replication forks continue DNA synthesis in the presence of impediments, it leaves behind ssDNA gaps (15, 31, 47). PrimPol-generated ssDNA gaps are well tolerated in cells when replication stress is low and ssDNA gap repair is efficient, allowing PrimPol to promote stress tolerance and cell survival (25). However, when replication stress is high or gap repair is compromised, PrimPol-generated ssDNA gaps can accumulate to high levels and impose a threat to genomic integrity (17, 26, 30). We recently showed that PrimPol-generated ssDNA gaps in BRCA-deficient cells, if not repaired properly, can lead to DNA DSBs in a cell cycle–dependent manner (20). Furthermore, high levels of ssDNA can trigger a replication catastrophe, killing replicating cells (20). Thus, our data shed light on the nature of A3A-induced replication stress, explaining how modest A3A expression induces ssDNA without slowing replication forks and why high A3A expression can generate DSBs and promote cell death.
Our data also suggest possible mechanisms to explain how A3A activates PrimPol. The cytosine deaminase activity of A3A is required for the formation of PrimPol-generated gaps, suggesting that A3A-generated uracil is involved. Loss of UNG reduces but does not eliminate A3A-induced gaps, suggesting that the conversion of uracil to abasic sites contributes to PrimPol activation. It is possible that the stalling of DNA polymerases by abasic sites leads to uncoupling of helicase and polymerases at replication forks, generating RPA-coated ssDNA and recruiting PrimPol to reprime for DNA synthesis (47). Notably, UNG loss slows replication forks even in the absence of A3A, indicating that unprocessed uracil in the genome may also impede the progression of DNA polymerases. It is conceivable that unprocessed uracil also contributes to ssDNA formation and PrimPol activation at stressed forks, although less efficiently than abasic sites, explaining the PrimPol-dependent but UNG-independent gaps that we observed in A3A-expressing cells.
The mechanisms by which A3A-induced and PrimPol-generated gaps are repaired have emerged from this study. Although previous studies have linked HR to the suppression of APOBEC-induced mutagenesis (48–50), it was not clear whether HR is involved in the response to A3A-induced replication stress. We find that both ATR and RAD51 are critical for the repair of A3A-induced gaps. Notably, ATR and RAD51 display an epistatic relationship in the repair of A3A-induced gaps, suggesting that they act through the same pathway. Of note, the repair of A3A-induced gaps is independent of BRCA2, suggesting that the canonical HR pathway is not responsible. RAD51 is known to protect reversed forks and ssDNA gaps from nucleolytic degradation (13, 22, 51, 52), raising the possibility that ATR promotes the loading of RAD51 to PrimPol-generated gaps to protect these gaps. Furthermore, the binding of RAD51 to ssDNA gaps may promote gap repair through HR-like pathways such as TS or related mechanisms (53–55). Along these lines, yeast RAD51 paralogs bind to abasic sites and promote their bypass by facilitating RAD51-dependent error-free HR repair (56). In addition to ATRi, PARPi also blocks the repair of A3A-induced gaps, suggesting that PARP1/2 are trapped at these gaps. It is possible that PARP1/2 also contribute to gap repair. Furthermore, REV1i modestly reduces the efficiency of gap repair, suggesting that TLS is a minor contributor to the process. In the absence of PrimPol, TLS becomes more important for the survival of A3A-expressing cells in ATRi, suggesting that TLS functions as a backup pathway for PrimPol to cope with A3A-induced uracil and/or abasic sites.
The induction of ssDNA gaps by A3A provides a unique opportunity for gap-targeted therapies. ATRi blocks gap repair and renders ssDNA gaps persistent, promoting DSB formation in a cell cycle–dependent manner. ATRi also increases origin firing and the formation of ssDNA gaps at forks, exacerbating gap-associated DNA damage (57, 58). Similarly, the trapping of PARP1/2 by PARPi prevents the completion of gap repair (20), allowing A3A-induced gaps to be converted to DSBs. ATR inhibition and PARP trapping may have nonredundant effects on gap repair, which would explain the synergy between ATRi and PARPi. Notably, loss of PrimPol reduces the sensitivity of A3A-expressing cells to ATRi and PARPi, suggesting a potential mechanism of resistance. In this context, TLSi resensitizes A3A-expressing cells to ATRi, possibly by reducing the ability of replication forks to pass through unprocessed uracil and abasic sites in DNA. Loss of UNG also renders A3A-expressing cells less sensitive to ATRi by reducing ssDNA gaps, and TLSi may also overcome the resistance through the same mechanism. In future studies, it will be important to identify the oncogenic and therapeutic contexts in which A3A expression is induced (59) and investigate whether various combinations of recently developed ATRi, PARPi, and TLSi offer sufficient therapeutic windows to impede tumor progression and overcome drug resistance in preclinical models and clinical trials.
MATERIALS AND METHODS
Cell lines
U2OS-derived A3A-expressing cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 2 mM l-glutamine.
U2OS-A3A PRIMPOL KO cells were generated by using a pSpCas9(BB)-2A-GFP (pX458) vector containing two independent sgRNA sequences targeting exon 5 of PRIMPOL. Cells were transfected with PX458#1 or PX458#2 vectors using Lipofectamine 3000 using according to the manufacturer’s instructions. Seventy-two hours after transfection, green fluorescent protein (GFP)–positive single cells were sorted in each well of a 96-well plate and cultured until they formed colonies. Cells were scaled up and analyzed for PRIMPOL expression using Western blotting.
Non-tumorigenic human mammary epithelial cell line MCF10A was cultured in 1:1 DMEM/F12 media supplemented with 5% horse serum, epidermal growth factor (20 ng/ml), hydrocortisone (0.5 mg/ml), cholera toxin (100 ng/ml), insulin (10 μg/ml), 1% penicillin/streptomycin, and 2 mM l-glutamine. HCT15 and KM12 colon carcinoma cells were cultured in RPMI 1640 media supplemented with 10% FBS, 1% penicillin/streptomycin, and 2 mM l-glutamine. MCF10A, HCT15, and KM12 derivative cell lines expressing A3A were generated by infecting the cells with lentivirus expressing A3A under a DOX-inducible promoter (pINDUCER20) and selected with G418 (400 μg/ml). All cell lines were incubated at 37°C with 5% CO2.
NCI-H2347 and SKBR3 cells were cultured in RPMI 1640 media with 10% FBS and 1% penicillin/streptomycin solution. TOV21G and MDA-MB-453 cells were cultured in DMEM/F12 media with 10% FBS and 1% penicillin/streptomycin solution.
RNA interference
Cell transfections were performed using the reverse transfection method. A transfection mix was prepared using 250 μl of OPTI-MEM (Gibco), 4 μl of Lipofectamine RNAiMAX (Thermo Fisher Scientific), and appropriate concentration of the specific siRNA as mentioned in the figure legends. Experiments were performed 48 hours after siRNA transfections unless mentioned otherwise. siRNA sequences are listed in table S1.
DNA fiber assay
A total of 0.25 × 106 cells were seeded in six-well plates. To induce A3A expression, cells were treated with DOX (200 ng/ml) for 48 hours. After 48 hours, cells were labeled with 50 μM CldU for 20 min, washed three times with warm media, and then labeled with 250 μM IdU for 60 min. For experiments with HU treatment, HU was simultaneously added during the IdU labeling period. Cells were then washed with 1× phosphate-buffered saline (PBS) and were scraped on ice in 1 ml of 0.1% bovine serum albumin (BSA) in PBS and spun down for 5 min at 7000 rpm at 4°C. The supernatant was removed leaving ~100 μl of 0.1% BSA in PBS for resuspension of cells. Three microliters of cell suspension was spotted on a glass slide, air-dried for 2 min, and then lysed with 8 μl of fiber lysis buffer [200 mM tris (pH 7), 50 mM EDTA (pH 8), and 0.5% SDS] for 10 min. Slides were then tilted at an angle of ~15 allowing DNA to run down the length of the slide and air-dried. Slides were then fixed in 3:1 methanol:acetic acid for 10 min at room temperature (RT). Slides were then washed three times with PBS for 5 min each and were either stored at 4°C in PBS or processed immediately. For processing, DNA was denatured in 2.5 N HCl in PBS for 90 min at RT followed by washing three times with PBS. Slides were then blocked in fiber-blocking buffer (3% BSA and 0.1% Tween 20 in PBS) for 30 min at 37°C and subsequently incubated with 1:50 rat anti–5-bromo-2′-deoxyuridine (BrdU; detects CldU) and 1:100 mouse anti-BrdU (detects IdU) for 1 hour at 37°C. Slides were then washed three times with PBST (0.1% Tween 20 in PBS) for 5 min each. Slides were then incubated with 1:100 anti-rat immunoglobulin G (IgG) Alexa Flour 594 and 1:100 anti-mouse IgG Alexa Flour 488 for 1 hour at 37°C. Slides were then washed three times with PBST, air-dried, and mounted with ProLong Gold Antifade mounting media. Images were acquired using NIS-Elements software with a Nikon i90 microscope using a 60× objective and analyzed using Fiji software.
S1 nuclease DNA fiber assay
S1 nuclease fiber assays were performed as mentioned above. For experiments with siRNA treatments, reverse siRNA transfection was performed at the time of cell seeding. After IdU labeling, cells were washed with 1× PBS and then treated with CSK-100 buffer [10 mM Mops (pH 7), 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton X-100] for 10 min at RT to remove the cytoplasm and expose the nuclei. Nuclei were then washed with PBS and then once with S1 nuclease buffer [30 mM sodium acetate (pH 4.6), 10 mM zinc acetate (pH 6), 50 mM NaCl, and 5% glycerol). Nuclei were then treated with S1 nuclease (20 U/ml) for 30 min at 37°C. After 30 min, S1 nuclease was removed, and nuclei were scraped on ice in 1 ml of 0.1% BSA in PBS to precipitate nuclei and spun down for 5 min at 7000 rpm at 4°C.
DNA fiber assay to measure gap repair/fill-in was performed similarly with the following modifications. After IdU labeling, cells were washed three times with media and allowed to recover in complete media in the presence of 500 μM thymidine for either 4, 8, 16, or 24 hours. Cells were then processed for S1 nuclease DNA fiber assays. For experiments with inhibitor treatments, inhibitors were added after the IdU labeling period during the recovery step. For the RAD51 inhibitor, cells were pretreated with 25 μM B02 for 6 hours, before labeling, and treatment was maintained throughout the labeling process and during the recovery process.
Western blotting
Cells were treated with the indicated treatments (siRNAs and drugs). Cells were directly lysed in six-well plates using 1× radioimmunoprecipitation assay buffer [25 mM tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1× fresh protease inhibitor cocktail], scraped, and spun down at 13,000 rpm for 5 min. Protein concentration was normalized using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) and mixed with 6× Laemmli buffer. Lysates were boiled at 95°C for 5 min, spun down, and kept on ice. Normalized lysates were loaded onto 4 to 12% Bolt Bis-Tris Plus protein gels at 100 V for 60 min. Proteins were then transferred onto polyvinylidene difluoride (PVDF) membrane in transfer buffer (20% methanol) at a constant current of 250 mA for 60 min at RT. Membranes were blocked in 5% milk in tris-buffered saline with 0.1% Tween 20 (TBST) or 5% BSA in TBST for 1 hour at RT. Membranes were then incubated with the appropriate primary antibodies overnight at 4°C shaking. Membranes were then washed three times with TBST for 10 min each and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies for 1 hour at RT with shaking. Membranes were then washed three times with TBST for 30 min each and incubated with an enhanced chemiluminescence substrate. Signals were detected using a ChemiDoc Imaging System (Bio-Rad) with ImageLab v6.0.1 software. Antibodies are listed in table S2.
Viability assay
Cell viability was measured with a CellTiter-Glo cell viability assay (Promega). Briefly, cells were seeded in triplicates in opaque 96-well flat-bottom plates at a density of 500 cells per well in 50-μl volume and incubated overnight. The next day, cells were treated with 50 μl of media containing 2× concentration of drugs such that the final concentration was 1× in 100 μl of media. For knockdown experiments or experiments using DOX-inducible A3A expression, siRNA transfection or DOX treatment was performed 48 hours before drug treatment. Plates were incubated for an additional 5 days after drug treatment. After 5 days, plates were equilibrated for 30 min to RT and 100 μl of CellTiter-Glo reagent (diluted 1:5 in PBS) was added to each well containing cells. Plates were then incubated for 20 min at RT shaking. Luminescence was measured using a PerkinElmer EnVision 2103 multilabel plate reader. Viability was calculated as the luminescence signal ratio of treated versus untreated samples.
Preparation of native cell extracts
Cells were lysed in native lysis buffer [25 mM Hepes (pH 7.9), 10% glycerol, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM MgCl2, ribonuclease A (RNase A; 0.2 μg/ml), 1 mM ZnCl2, and protease inhibitors]. Cell lysates were sonicated, incubated for 30 min at 4°C, and then centrifuged for 10 min at 13,000 rpm at 4°C. Protein concentration was determined by Bradford assay (Bio-Rad) and extracts were normalized. Twenty-microliter aliquots were prepared and flash-frozen in liquid nitrogen and stored at −80°C. A fresh vial of extract was used for each experiment.
qPCR
RNA was extracted from NCI-H2347 and SKBR3 siControl and siA3A treated cells using the RNeasy Plus Mini kit (Qiagen) according to the manufacturer’s protocol. One microgram of RNA was reverse-transcribed to cDNA using the qScript cDNA SuperMix (Quantabio) according to the manufacturer’s protocol. Gene expression was determined using SYBR Green (PowerUp SYBR Green Master Mix, Thermo Fisher Scientific) using the LightCycler 480 System (Roche Diagnostics). A3A (forward primer - GAGAAGGGACAAGCACATGG, reverse primer - TGGATCCATCAAGTGTCTGG) transcript levels were normalized to actin (forward primer - CCAACCGCGAGAAGATGA, reverse primer - CCAGAGGCGTACAGGGATAG) transcript levels.
Deamination assay
Normalized amounts of cell extracts were incubated with 0.4 μM DNA hairpin substrate TTC-HP (5′FAM-GCAAGCCTTCGGCTTGCTGA) in a 20-μl reaction containing 50 mM tris (pH 7.5), 1.5 U of uracil DNA glycosylase (New England BioLabs), RNase A (0.1 mg/ml), and 5 mM EDTA for 1 hour at 37C. NaOH (100 mM) was added to the reaction and it was incubated at 95°C for 30 min to cleave abasic sites. Twenty-two microliters (1:1) of loading buffer (0.1% Orange G in formamide) was then added to the reaction and it was further incubated at 95°C for 10 min, spun down, and kept on ice for 5 min. A 20% denaturing polyacrylamide gel [8 M urea, 1× tris-acetate-EDTA (TAE) buffer] was pre-run in 1× TAE buffer for 10 min and 10-μl samples were loaded. DNA cleavage was monitored by running the gel for 90 min at 150 V. The gel was analyzed on a ChemiDoc Imaging System (Bio-Rad) with ImageLab v6.0.1 software. DNA oligonucleotide probes were synthesized by Thermo Fisher Scientific.
Correlation between APOBEC signature and PARP inhibitor
The APOBEC signature was calculated following the previously described approach (60). Briefly, we extracted somatic single-nucleotide variants from whole-exome sequencing (WXS) data of CCLE. This resulted in a final WXS dataset comprising 1443 unique cell lines across 31 tumor types. For mutation signature analysis, we used NMF with setting K to 12, which identified mutational signatures in the cohort, matched them to known mutation processes, and quantified them in each cell line. One mutation process was APOBEC enzyme activity. We calculated the fraction of mutations assigned to the APOBEC signature, as a fraction of total mutations, for each cell line, which we termed “frac_apobec.” Cell lines with a frac_apobec value greater than or equal to 10% were considered as APOBEC+, while those below this threshold were categorized as APOBEC−. Last, we used Spearman’s method to compute the correlation between frac_apobec and the IC50 values of talazoparib, and for each cell line obtained from the Genomics of Drug Sensitivity in Cancer database.
Statistical analysis
Statistical significance was determined by one-way analysis of variance (ANOVA) using Tukey’s multiple comparisons test or using unpaired two-tailed t test. Statistical analysis was performed using GraphPad Prism (version 8). For all figures, ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, and ns = not significant where P > 0.05.
Acknowledgments
We thank members of Zou, Dyson, Lan, Mostoslavsky, Elia, and Motamedi laboratories for helpful discussions. L.Z. was the James and Patricia Poitras endowed chair in Cancer Research at Massachusetts General Hospital. P.S.P. is the recipient of Canadian Institute of Health Research Banting Postdoctoral Fellowship (FRN-188253). S.S. was supported by the James A. Harting Scientific Scholar Award from the Rivkin Center.
Funding: This work is supported by the NIH grants CA248526 and CA263934 to L.Z. and Rullo Family Innovation Award to M.S.L.
Author contributions: Conceptualization: A.S.K. and L.Z. Methodology: A.S.K. and L.Z. Investigation: A.S.K., X.R., P.S.P., and S.S. Supervision: M.S.L. and L.Z. Writing—original draft: A.S.K. and L.Z. Writing—review and editing: A.S.K., M.S.L., and L.Z.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.Swanton C., McGranahan N., Starrett G. J., Harris R. S., APOBEC enzymes: Mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov L. B., Nik-Zainal S., Wedge D. C., Aparicio S. A. J. R., Behjati S., Biankin A. V., Bignell G. R., Bolli N., Borg A., Børresen-Dale A.-L., Boyault S., Burkhardt B., Butler A. P., Caldas C., Davies H. R., Desmedt C., Eils R., Eyfjörd J. E., Foekens J. A., Greaves M., Hosoda F., Hutter B., Ilicic T., Imbeaud S., Imielinski M., Jäger N., Jones D. T. W., Jones D., Knappskog S., Kool M., Lakhani S. R., López-Otín C., Martin S., Munshi N. C., Nakamura H., Northcott P. A., Pajic M., Papaemmanuil E., Paradiso A., Pearson J. V., Puente X. S., Raine K., Ramakrishna M., Richardson A. L., Richter J., Rosenstiel P., Schlesner M., Schumacher T. N., Span P. N., Teague J. W., Totoki Y., Tutt A. N. J., Valdés-Mas R., van Buuren M. M., van ‘t Veer L., Vincent-Salomon A., Waddell N., Yates L. R., Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC Ped Brain, Zucman-Rossi J., Futreal P. A., Dermott U. M., Lichter P., Meyerson M., Grimmond S. M., Siebert R., Campo E., Shibata T., Pfister S. M., Campbell P. J., Stratton M. R., Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor B. J., Nik-Zainal S., Wu Y. L., Stebbings L. A., Raine K., Campbell P. J., Rada C., Stratton M. R., Neuberger M. S., DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2, e00534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGranahan N., Swanton C., Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 27, 15–26 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Petljak M., Alexandrov L. B., Brammeld J. S., Price S., Wedge D. C., Grossmann S., Dawson K. J., Ju Y. S., Iorio F., Tubio J. M. C., Koh C. C., Georgakopoulos-Soares I., Rodríguez-Martín B., Otlu B., O’Meara S., Butler A. P., Menzies A., Bhosle S. G., Raine K., Jones D. R., Teague J. W., Beal K., Latimer C., O’Neill L., Zamora J., Anderson E., Patel N., Maddison M., Ng B. L., Graham J., Garnett M. J., McDermott U., Nik-Zainal S., Campbell P. J., Stratton M. R., Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell 176, 1282–1294.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petljak M., Dananberg A., Chu K., Bergstrom E. N., Striepen J., von Morgen P., Chen Y., Shah H., Sale J. E., Alexandrov L. B., Stratton M. R., Maciejowski J., Mechanisms of APOBEC3 mutagenesis in human cancer cells. Nature 607, 799–807 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts S. A., Lawrence M. S., Klimczak L. J., Grimm S. A., Fargo D., Stojanov P., Kiezun A., Kryukov G. V., Carter S. L., Saksena G., Harris S., Shah R. R., Resnick M. A., Getz G., Gordenin D. A., An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wörmann S. M., Zhang A., Thege F. I., Cowan R. W., Rupani D. N., Wang R., Manning S. L., Gates C., Wu W., Levin-Klein R., Rajapakshe K. I., Yu M., Multani A. S., Kang Y., Taniguchi C. M., Schlacher K., Bellin M. D., Katz M. H. G., Kim M. P., Fleming J. B., Gallinger S., Maddipati R., Harris R. S., Notta F., Ross S. R., Maitra A., Rhim A. D., APOBEC3A drives deaminase domain-independent chromosomal instability to promote pancreatic cancer metastasis. Nat. Cancer 2, 1338–1356 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Zhao K., Zhang Q., Flanagan S. A., Lang X., Jiang L., Parsels L. A., Parsels J. D., Zou W., Lawrence T. S., Buisson R., Green M. D., Morgan M. A., Cytidine deaminase APOBEC3A regulates PD-L1 expression in cancer cells in a JNK/c-JUN-dependent manner. Mol. Cancer Res. 19, 1571–1582 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinet A., Tirman S., Cybulla E., Meroni A., Vindigni A., To skip or not to skip: Choosing repriming to tolerate DNA damage. Mol. Cell 81, 649–658 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena S., Zou L., Hallmarks of DNA replication stress. Mol. Cell 82, 2298–2314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berti M., Cortez D., Lopes M., The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 21, 633–651 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Bhat K. P., Cortez D., RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 25, 446–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinet A., Lemaçon D., Vindigni A., Replication fork reversal: Players and guardians. Mol. Cell 68, 830–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mourón S., Rodriguez-Acebes S., Martínez-Jiménez M. I., García-Gómez S., Chocrón S., Blanco L., Méndez J., Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Bai G., Kermi C., Stoy H., Schiltz C. J., Bacal J., Zaino A. M., Hadden M. K., Eichman B. F., Lopes M., Cimprich K. A., HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol. Cell 78, 1237–1251.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirman S., Quinet A., Wood M., Meroni A., Cybulla E., Jackson J., Pegoraro S., Simoneau A., Zou L., Vindigni A., Temporally distinct post-replicative repair mechanisms fill PRIMPOL-dependent ssDNA gaps in human cells. Mol. Cell 81, 4026–4040.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., Marino F., Lucic B., Biasin V., Gstaiger M., Aebersold R., Sidorova J. M., Monnat R. J. J., Lopes M., Vindigni A., Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20, 347–354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genois M.-M., Gagné J.-P., Yasuhara T., Jackson J., Saxena S., Langelier M.-F., Ahel I., Bedford M. T., Pascal J. M., Vindigni A., Poirier G. G., Zou L., CARM1 regulates replication fork speed and stress response by stimulating PARP1. Mol. Cell 81, 784–800.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simoneau A., Xiong R., Zou L., The trans cell cycle effects of PARP inhibitors underlie their selectivity toward BRCA1/2-deficient cells. Genes Dev. 35, 1271–1289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nayak S., Calvo J. A., Cong K., Peng M., Berthiaume E., Jackson J., Zaino A. M., Vindigni A., Hadden M. K., Cantor S. B., Inhibition of the translesion synthesis polymerase REV1 exploits replication gaps as a cancer vulnerability. Sci. Adv. 6, eaaz7808 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V., Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17, 1305–1311 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su X., Bernal J. A., Venkitaraman A. R., Cell-cycle coordination between DNA replication and recombination revealed by a vertebrate N-end rule degron-Rad51. Nat. Struct. Mol. Biol. 15, 1049–1058 (2008). [DOI] [PubMed] [Google Scholar]
- 24.González-Prieto R., Muñoz-Cabello A. M., Cabello-Lobato M. J., Prado F., Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J. 32, 1307–1321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piberger A. L., Bowry A., Kelly R. D. W., Walker A. K., González-Acosta D., Bailey L. J., Doherty A. J., Méndez J., Morris J. R., Bryant H. E., Petermann E., PrimPol-dependent single-stranded gap formation mediates homologous recombination at bulky DNA adducts. Nat. Commun. 11, 5863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taglialatela A., Leuzzi G., Sannino V., Cuella-Martin R., Huang J.-W., Wu-Baer F., Baer R., Costanzo V., Ciccia A., REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol. Cell 81, 4008–4025.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belan O., Sebald M., Adamowicz M., Anand R., Vancevska A., Neves J., Grinkevich V., Hewitt G., Segura-Bayona S., Bellelli R., Robinson H. M. R., Higgins G. S., Smith G. C. M., West S. C., Rueda D. S., Boulton S. J., POLQ seals post-replicative ssDNA gaps to maintain genome stability in BRCA-deficient cancer cells. Mol. Cell 82, 4664–4680.e9 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Buisson R., Lawrence M. S., Benes C. H., Zou L., APOBEC3A and APOBEC3B activities render cancer cells susceptible to ATR inhibition. Cancer Res. 77, 4567–4578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green A. M., Landry S., Budagyan K., Avgousti D. C., Shalhout S., Bhagwat A. S., Weitzman M. D., APOBEC3A damages the cellular genome during DNA replication. Cell Cycle 15, 998–1008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinet A., Tirman S., Jackson J., Šviković S., Lemaçon D., Carvajal-Maldonado D., González-Acosta D., Vessoni A. T., Cybulla E., Wood M., Tavis S., Batista L. F. Z., Méndez J., Sale J. E., Vindigni A., PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells. Mol. Cell 77, 461–474.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinet A., Carvajal-Maldonado D., Lemacon D., Vindigni A., DNA fiber analysis: Mind the gap! Methods Enzymol. 591, 55–82 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Biayna J., Garcia-Cao I., Álvarez M. M., Salvadores M., Espinosa-Carrasco J., McCullough M., Supek F., Stracker T. H., Loss of the abasic site sensor HMCES is synthetic lethal with the activity of the APOBEC3A cytosine deaminase in cancer cells. PLOS Biol. 19, e3001176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehta K. P. M., Lovejoy C. A., Zhao R., Heintzman D. R., Cortez D., HMCES maintains replication fork progression and prevents double-strand breaks in response to APOBEC deamination and abasic site formation. Cell Rep. 31, 107705 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schormann N., Ricciardi R., Chattopadhyay D., Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 23, 1667–1685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand J., Chiou L., Sciandra C., Zhang X., Hong J., Wu D., Zhou P., Vaziri C., Roles of trans-lesion synthesis (TLS) DNA polymerases in tumorigenesis and cancer therapy. NAR Cancer 5, zcad005 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mutreja K., Krietsch J., Hess J., Ursich S., Berti M., Roessler F. K., Zellweger R., Patra M., Gasser G., Lopes M., ATR-mediated global fork slowing and reversal assist fork traverse and prevent chromosomal breakage at DNA interstrand cross-links. Cell Rep. 24, 2629–2642.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanjiv K., Hagenkort A., Calderón-Montaño J. M., Koolmeister T., Reaper P. M., Mortusewicz O., Jacques S. A., Kuiper R. V., Schultz N., Scobie M., Charlton P. A., Pollard J. R., Berglund U. W., Altun M., Helleday T., Cancer-specific synthetic lethality between ATR and CHK1 kinase activities. Cell Rep. 14, 298–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J., Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 (2000). [PMC free article] [PubMed] [Google Scholar]
- 39.Buisson R., Niraj J., Rodrigue A., Ho C. K., Kreuzer J., Foo T. K., Hardy E. J.-L., Dellaire G., Haas W., Xia B., Masson J.-Y., Zou L., Coupling of homologous recombination and the checkpoint by ATR. Mol. Cell 65, 336–346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yazinski S. A., Comaills V., Buisson R., Genois M.-M., Nguyen H. D., Ho C. K., Todorova Kwan T., Morris R., Lauffer S., Nussenzweig A., Ramaswamy S., Benes C. H., Haber D. A., Maheswaran S., Birrer M. J., Zou L., ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ianevski A., Giri A. K., Aittokallio T., SynergyFinder 3.0: An interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 50, W739–W743 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Robinson P. S., Coorens T. H. H., Moore L., Lee-Six H., Noorani A., Sanders M. A., Jung H., Katainen R., Heuschkel R., Brunton-Sim R., Weston R., Read D., Nobbs B., Fitzgerald R. C., Saeb-Parsy K., Martincorena I., Campbell P. J., Rushbrook S., Zilbauer M., Buczacki S. J. A., Stratton M. R., APOBEC mutagenesis is a common process in normal human small intestine. Nat. Genet. 55, 246–254 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petljak M., Green A. M., Maciejowski J., Weitzman M. D., Addressing the benefits of inhibiting APOBEC3-dependent mutagenesis in cancer. Nat. Genet. 54, 1599–1608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurup H. M., Kvach M. V., Harjes S., Jameson G. B., Harjes E., Filichev V. V., Seven-membered ring nucleobases as inhibitors of human cytidine deaminase and APOBEC3A. Org. Biomol. Chem. 21, 5117–5128 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano J. C., von Trentini D., Berríos K. N., Barka A., Dmochowski I. J., Kohli R. M., Structure-guided design of a potent and specific inhibitor against the genomic mutator APOBEC3A. ACS Chem. Biol. 17, 3379–3388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilzecker B., Buoninfante O. A., Pritchard C., Blomberg O. S., Huijbers I. J., van den Berk P. C. M., Jacobs H., PrimPol prevents APOBEC/AID family mediated DNA mutagenesis. Nucleic Acids Res. 44, 4734–4744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tirman S., Cybulla E., Quinet A., Meroni A., Vindigni A., PRIMPOL ready, set, reprime! Crit. Rev. Biochem. Mol. Biol. 56, 17–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cano-Linares M. I., Yáñez-Vilches A., García-Rodríguez N., Barrientos-Moreno M., González-Prieto R., San-Segundo P., Ulrich H. D., Prado F., Non-recombinogenic roles for Rad52 in translesion synthesis during DNA damage tolerance. EMBO Rep. 22, e50410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giannattasio M., Zwicky K., Follonier C., Foiani M., Lopes M., Branzei D., Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol. 21, 884–892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenbaum J. C., Bonilla B., Hengel S. R., Mertz T. M., Herken B. W., Kazemier H. G., Pressimone C. A., Ratterman T. C., MacNary E., De Magis A., Kwon Y., Godin S. K., Van Houten B., Normolle D. P., Sung P., Das S. R., Paeschke K., Roberts S. A., VanDemark A. P., Bernstein K. A., The Rad51 paralogs facilitate a novel DNA strand specific damage tolerance pathway. Nat. Commun. 10, 3515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saini N., Roberts S. A., Sterling J. F., Malc E. P., Mieczkowski P. A., Gordenin D. A., APOBEC3B cytidine deaminase targets the non-transcribed strand of tRNA genes in yeast. DNA Repair 53, 4–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoopes J. I., Hughes A. L., Hobson L. A., Cortez L. M., Brown A. J., Roberts S. A., Avoidance of APOBEC3B-induced mutation by error-free lesion bypass. Nucleic Acids Res. 45, 5243–5254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mertz T. M., Rice-Reynolds E., Nguyen L., Wood A., Cordero C., Bray N., Harcy V., Vyas R. K., Mitchell D., Lobachev K., Roberts S. A., Genetic inhibitors of APOBEC3B-induced mutagenesis. Genome Res. 33, 1568–1581 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., Lopes M., Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 8, 859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemaçon D., Jackson J., Quinet A., Brickner J. R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., Vindigni A., MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8, 860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Branzei D., Szakal B., Building up and breaking down: Mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol. 52, 381–394 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Couch F. B., Bansbach C. E., Driscoll R., Luzwick J. W., Glick G. G., Bétous R., Carroll C. M., Jung S. Y., Qin J., Cimprich K. A., Cortez D., ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moiseeva T., Hood B., Schamus S., O’Connor M. J., Conrads T. P., Bakkenist C. J., ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat. Commun. 8, 1392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oh S., Bournique E., Bowen D., Jalili P., Sanchez A., Ward I., Dananberg A., Manjunath L., Tran G. P., Semler B. L., Maciejowski J., Seldin M., Buisson R., Genotoxic stress and viral infection induce transient expression of APOBEC3A and pro-inflammatory genes through two distinct pathways. Nat. Commun. 12, 4917 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langenbucher A., Bowen D., Sakhtemani R., Bournique E., Wise J. F., Zou L., Bhagwat A. S., Buisson R., Lawrence M. S., An extended APOBEC3A mutation signature in cancer. Nat. Commun. 12, 1602 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2