Abstract
Although BRCA1/2 mutations are not commonly found in small cell lung cancer (SCLC), a substantial fraction of SCLC shows clinically relevant response to PARP inhibitors (PARPis). However, the underlying mechanism(s) of PARPi sensitivity in SCLC is poorly understood. We performed quantitative proteomic analyses and identified proteomic changes that signify PARPi responses in SCLC cells. We found that the vulnerability of SCLC to PARPi could be explained by the degradation of lineage-specific oncoproteins (e.g., ASCL1). PARPi-induced activation of the E3 ligase HUWE1 mediated the ubiquitin-proteasome system (UPS)–dependent ASCL1 degradation. Although PARPi induced a general DNA damage response in SCLC cells, this signal generated a cell-specific response in ASCL1 degradation, leading to the identification of HUWE1 expression as a predictive biomarker for PARPi. Combining PARPi with agents targeting these pathways markedly improved therapeutic response in SCLC. The degradation of lineage-specific oncoproteins therefore represents a previously unidentified mechanism for PARPi efficacy in SCLC.
Degradation of lineage-specific oncoproteins is a unique mechanism that drives PARPi sensitivity in small cell lung cancer.
INTRODUCTION
BReast CAncer gene 1 (BRCA1) and BReast CAncer gene 2 (BRCA2) are tumor suppressor proteins that are critical components of the homologous recombination (HR) repair pathways (1). Cancer cells with BRCA1/2 mutations, as well as other defects in the HR pathways (e.g., ATM mutations), are selectively sensitive to poly(adenosine diphosphate–ribose) polymerase (PARP) inhibitors (PARPis) via a mechanism known as synthetic lethality. The identification of PARP proteins as a therapeutic target has led to a paradigm shift for the treatment and management for BRCA1/2-mutated human malignancies (e.g., breast, ovarian, prostate, and pancreatic cancers), with the recent Food and Drug Administration approval of four PARPis (i.e., olaparib, rucaparib, niraparib, and talazoparib) in these indications (2–4).
PARP1/2 are DNA damage response (DDR) proteins that are critically involved in sensing genotoxic stimuli. Upon binding to nicked DNA, PARP1/2 become activated to synthesize many poly-ADP–ribosylated (PARylated) proteins. These protein-linked PAR polymers serve as a scaffold to recruit an array of DNA repair enzymes to resolve the DNA breaks (5–8). Besides the blockade of PAR synthesis and PAR-mediated DDR, recent studies show that PARPi also cause PARP1/2 “trapping,” which serves as a key mediator of the genotoxic, cytotoxic, and immunomodulatory roles of PARPi in BRCA1/2-mutated cancers (9–14).
Besides the existing results in BRCA1/2-mutated cancers, PARPi is being evaluated, either as a single agent or in combination with chemotherapy and radiotherapy approaches, to treat a diverse array of other solid tumors. Results from these clinical studies indicate that PARPi could represent a promising therapeutic strategy for these cancers. For example, small cell lung cancer (SCLC) is a neuroendocrine (NE) lung carcinoma (~15% of all lung cancer cases) and is one of the most aggressive human malignancies (5-year survival rate of ~6%) (15). Compared to other subtypes of lung cancer (e.g., adenocarcinoma and squamous cell carcinoma), SCLC has unique biology and genetic alterations, including the frequent deletion or inactivating mutations of the retinoblastoma (Rb) and p53 (TP53) proteins. Despite intensive research within the last several decades, the standard of care for advanced SCLC relies on chemotherapy, i.e., etoposide or irinotecan plus a platinum-based drug such as cisplatin or carboplatin (12, 16). Although this regimen often leads to initial tumor regression, recurrence is almost universal, at which point patients are left with very limited options of further systemic therapy (17, 18).
Preclinical and clinical studies have demonstrated early promise for PARPi in SCLC. Comparative proteomic studies revealed that high expression of PARP1 and PARP2 distinguishes SCLC from NSCLC, and SCLC cell lines are highly sensitive to PARPis (19–21). This discovery launched clinical trials in unselected SCLC patients with promising early-phase results. In patients with relapsed SCLC, combination of veliparib with temozolomide (TMZ) showed a marked increase in response rate (39% veliparib + TMZ versus 14% placebo + TMZ) (22). A similarly high response rate was observed with olaparib + TMZ (42%) in a single-arm study (23). However, despite these promising response rates, clear benefit was only observed in a subset of patients, PARPi sensitivity could not be predicted by tumor mutations in BRCA1/2 or other HR genes as these are not commonly found in this cancer (e.g., less than 3% of the SCLC cases contain BRCA1/2 mutations) (20, 24). Although some relapsed SCLCs are highly sensitive to PARPi combinations, BRCA1/2 mutations are not the primary determinants of this sensitivity, and the exact molecular underpinnings are poorly understood. The lack of mechanism-based, predictive biomarkers poses a substantial knowledge gap, preventing the rational clinical application of PARPi in SCLC and many other human malignancies with proficient HR pathways. Identification of these mechanisms would provide a critically important therapeutic pathway toward achieving more complete and durable responses for patients with SCLC.
Here, we assembled a large panel of molecularly annotated human SCLC cell lines and performed global, isobaric labeling-based quantitative mass spectrometry (MS) experiments to comprehensively characterize how the SCLC proteome responds to PARPi treatment. This system-wide proteomic approach led to the identification of unique proteomic signatures associated with PARPi treatment in PARPi-sensitive and PARPi-resistant SCLCs, respectively. We found that PARPi treatment led to degradation of key lineage-specific oncoproteins including ASCL1, NEUROD1, and POU2F3, only in the PARPi-sensitive, but not in the PARPi-resistant SCLC cells. Unexpectedly, we found that the degradation of these key SCLC lineage-specific oncoproteins was also induced by clinically relevant chemotherapeutic agents, suggesting that this degradation phenotype could be a key mediator of the vulnerability of SCLC to the genotoxicity induced by multiple therapeutic agents (i.e., PARPi and chemotherapy compounds). We further uncovered that the PARPi-induced degradation of the SCLC lineage-specific oncoproteins was mediated by the ubiquitin-proteasome system (UPS) via the DNA damage-responsive E3 ubiquitin ligase HUWE1 (HECT, UBA, and WWE domain-containing 1). PARPi treatment elicits a general DNA damage signal. However, individual SCLC lines derived from different patients responded differently to this DDR signal, leading to the unequal degradation of the lineage-specific oncoproteins, and hence cell death. The tumor cell line–specific oncoprotein degradation response and the associated heterogenous UPS response led to the identification of potentially predictive biomarkers (e.g., HUWE1 levels) for PARPi in SCLC. Overexpression of HUWE1 in PARPi-resistant, ASCL1high SCLC cell lines led to increased ASCL1 degradation in these cells, which then caused more cell death, under PARPi treatment conditions. We expect that the dataset will serve as a previously unknown resource to provide the foundation and biomarker for future hypothesis-driven clinical translational research that helps delineate the molecular mechanisms of the therapeutic vulnerability of PARPi beyond tumors with BRCA1/2 mutations.
RESULTS
Identification of a PARPi-induced protein degradation signature in SCLC
BRCA1/2 mutations are rarely found in SCLC, including in the SCLC lines used in this study [see table S1—where 19 of 21 SCLC tested were wild type for BRCA1/2, while potential germline alterations were found in 2: H378 (M1008I) and H446 (D1496H)]. The mechanism of selective vulnerability of some SCLCs to PARPi is not understood. Talazoparib is a highly potent PARPi that blocked the formation of PARylation in NCI-H2081 (H2081) cells (a representative SCLC cell line) (Fig. 1A and fig. S1A). Besides its catalytic inhibition activity, talazoparib treatment also led to profound PARP1/2 trapping, γH2AX formation (a marker of DNA double-strand breaks), and PARP1 cleavage (a marker of apoptotic cell death) (fig. S1, B and C).
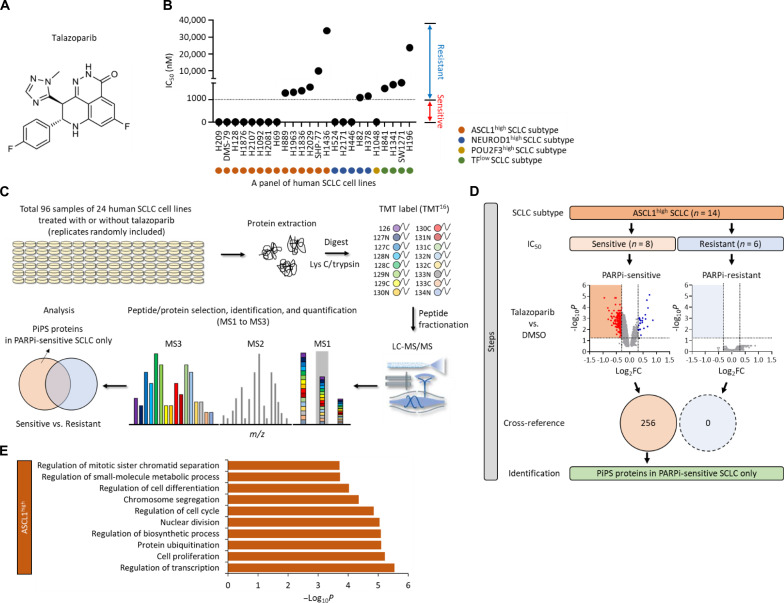
Fig. 1. Identification of a PiPS in SCLC.
(A) Structure of talazoparib. (B) Value of IC50 in a total of 24 SCLC cell lines treated with talazoparib. SCLC cell lines are indicated as ASCL1high, NEUROD1high, POU2F3high, or TFlow subtype. The sensitivity is defined as: sensitive (red), IC50 < 1 μM; resistant (blue), IC50 > 1 μM. TFlow, low expression of all three TFs. (C) Workflow of high-throughput multiplexed quantitative proteome mapping in a total of 96 proteome samples including a total of 24 SCLC lines treated with or without talazoparib (1 μM for 48 hours) in a total of six Tandem Mass Tag (TMT) experiments. (D) Steps of the identification of PARPi-induced protein degradation signature (PiPS) proteins only identified in PARPi-sensitive ASCL1high SCLC subtype. Volcano plots show differentially expressed proteins in talazoparib treatment in each PARPi-sensitive and PARPi-resistant ASCL1high SCLC subtype [log2 fold change (log2FC) < −0.3, P < 0.05]. Venn diagram shows the number of identified PiPS proteins. (E) Gene Ontology (GO) analyses of PiPS proteins identified in PARPi-sensitive ASCL1high SCLC subtype. Biological processes were analyzed using the ToppGene database (https://toppgene.cchmc.org/). LC-MS/MS, liquid chromatography tandem MS; m/z, mass-charge ratio.
SCLC can be divided into different subtypes based on the expression of certain lineage-specific transcription factors, including ASCL1, NEUROD1, and POU2F3 (25). Among them, the basic helix-loop-helix (bHLH) transcription factor (TF) ASCL1 is considered as a master regulator for a majority of these SCLC with strong NE features. In addition, two related TFs, NEUROD1 and POU2F3, characterize a smaller subset with intermediate and low NE features, respectively (26, 27). These TFs are highly expressed and are required for the establishment of the lineage of pulmonary NE cells and for the continued survival of SCLC (27–30).
To systemically characterize the proteomic response of SCLC to PARPi, we assembled a panel of 24 human SCLC cell lines that represent the major SCLC subtypes, including ASCL1high, NEUROD1high, POU2F3high, and TFlow (the TFlow subtype refers to the SCLCs that expresses low levels of ASCL1, NEUROD1, and POU2F3) (table S1). We first determined the sensitivity of these SCLC cell lines to talazoparib by measuring the IC50 (the half-maximal inhibitory concentration) required to inhibit their growth or cause cell death.
Talazoparib displayed differential toxicity in these SCLC cell lines, and we further classified these SCLC cell lines into the talazoparib-sensitive cells (those with IC50 < 1 μM, i.e., H2081, H69, H1048, H209, H1876, DMS-79, H2107, H524, H1092, H2171, H128, and H446) and talazoparib-resistant SCLCs (those with IC50 > 1 μM, i.e., H1436, H82, H889, H378, H196, SW1271, H1836, H1341, H2029, H841, SHP-77, and H1963) (Fig. 1B; fig. S1, D and E; and table S1).
We next performed quantitative mass spectrometric analyses to systemically characterize how talazoparib treatment perturbed the proteome homeostasis in each of the aforementioned SCLC lines (Fig. 1C). We prepared a total of 96 proteome samples [the 24 different SCLC lines, each treated for 48 hours with either dimethyl sulfoxide (DMSO) or talazoparib], and these samples were divided into six Tandem Mass Tag (TMT)–16plex experiments. From these isobaric labeling-based, global quantitative proteomic experiments, we were able to quantify a total of 12,295 proteins (false discovery rate < 1%), with 5169 proteins quantified across the entire panel of the SCLC lines (fig. S1F and table S2). Correlation analyses of the resulting protein abundances indicated that an excellent reproducibility [R2 (coefficient of determination) = 0.9504] was achieved in these quantitative proteomic experiments (fig. S1G).
We then performed a series of bioinformatic analyses to identify proteomic changes that might be associated with the selective sensitivity of certain SCLC cell lines to PARPi (fig. S1H). Specifically, we first grouped the SCLC cell lines and their respective proteomic datasets into four subtypes (i.e., ASCL1high, NEUROD1high, POU2F3high, and TFlow). Because the vast majority of SCLC is characterized by the overexpression of ASCL1, we initially focused our analyses on the ASCL1high SCLC cell lines. For these cell lines, we further divided them into the ASCL1high/talazoparib-sensitive (a total of eight cell lines) and ASCL1high/talazoparib-resistant (a total of six cell lines) cell lines. We found that talazoparib was able to induce the down-regulation of 256 proteins in the ASCL1high/talazoparib-sensitive cell lines. However, we did not identify any significantly down-regulated proteins in the ASCL1high/talazoparib-resistant cell lines (Fig. 1D, fig. S1I, and table S3). We therefore termed the 256 down-regulated proteins as the PARPi-induced protein degradation signature (PiPS). Gene Ontology (GO) analyses showed that many of these PiPS proteins were involved in biological processes linked to the pathogenesis of SCLC, including regulation of transcription (P = 2.89 × 10−6), cell proliferation (P = 5.97 × 10−6), and cell cycle (P = 1.41 × 10−5) (Fig. 1E). Several SCLC lineage-specific oncoproteins, including ASCL1, were among the PiPS, and these proteins were found to be down-regulated in response to talazoparib treatment (in ASCL1high/talazoparib-sensitive cell lines).
Besides the ASCL1high SCLC subtype, we also identified 10 and 85 PiPS proteins from the NEUROD1high and POU2F3high SCLC subtype, respectively (fig. S1J and table S3). Similar to ASCL1, we found that NEUROD1 and POU2F3 were also markedly down-regulated in response to talazoparib treatment (in the respective talazoparib-sensitive cell lines) (table S3). Last, because all the TFlow SCLC cell lines were resistant to talazoparib, we did not identify any down regulated proteins in this TFlow SCLC subtype (fig. S1J and table S3).
Degradation of SCLC lineage-specific oncoproteins in response to PARPi-induced genotoxicity
We first validated our quantitative proteomic results and found that talazoparib treatment resulted in the dramatic down-regulation of ASCL1 in H2081 cells (Fig. 2A and fig. S2, A and B). Also, we established an SCLC xenograft mouse model derived from H2081 cells, which exhibited distinctive morphological characteristics associated with SCLC, including scant cytoplasm and nuclear molding, along with robust expression of SCLC marker proteins (i.e., NCAM and synaptophysin) (fig. S2C). Consistently, we observed that talazoparib also reduced the levels of ASCL1 and proliferating cell nuclear antigen (PCNA) (a widely used proliferation marker) and inhibited tumor growth in an H2081 xenograft model (Fig. 2, B and C, and fig. S2D). Quantitative real-time PCR (qRT-PCR) experiments demonstrated that talazoparib treatment did not affect the mRNA levels of this protein (Fig. 2D). These results highlight the power of quantitative proteomic analyses in the identification of these posttranslational regulation mechanisms. Besides ASCL1, we observed that other lineage-specific oncoproteins (i.e., NEUROD1 and POU2F3) were also markedly down-regulated by talazoparib treatment in talazoparib-sensitive H524 cells (NEUROD1high) and H1048 cells (POU2F3high), respectively (fig. S2, E and F).
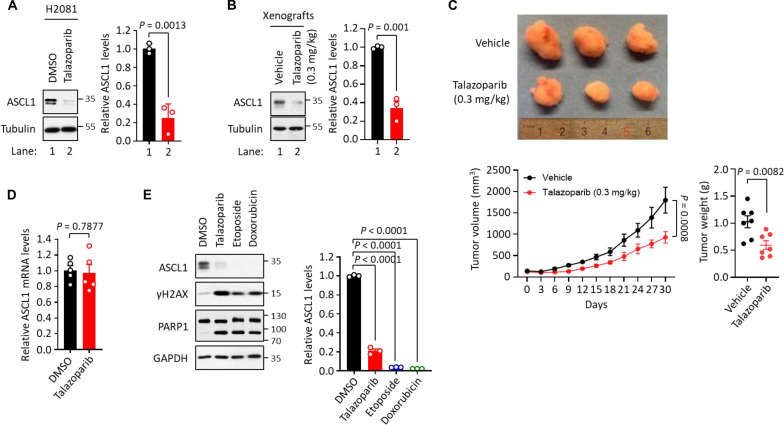
Fig. 2. Degradation of SCLC lineage-specific oncoproteins in response to PARPi-induced genotoxicity.
(A) Levels of ASCL1 in talazoparib treatment. H2081 cells were treated with or without talazoparib (1 μM for 48 hours), and the cell lysates were subjected to immunoblot analysis using the indicated antibodies. Values were presented as means ± SD (n = 3). (B) Levels of ASCL1 in vivo. H2081-implanted xenograft tumors were treated with or without talazoparib (0.3 mg/kg for 30 days), and the tumor extracts were subjected to immunoblot analysis using the indicated antibodies. Values were presented as means ± SD (n = 3). (C) Toxicity of talazoparib in vivo. Mice implanted with H2081 cells were treated with or without talazoparib (0.3 mg/kg for 30 days). Top: The image of representative tumors; bottom: tumor volume and weight. Values were presented as means ± SD (n = 6 to 10). (D) mRNA levels of ASCL1 in talazoparib treatment. H2081 cells were treated with or without talazoparib (1 μM for 48 hours), and the mRNA levels of ASCL1 were measured by qRT-PCR analysis, normalized to GAPDH mRNA levels. Values were presented as means ± SEM (n = 5). (E) The levels of ASCL1 in the treatment of DNA damaging agents. H2081 cells treated with talazoparib (1 μM), etoposide (1 μM), or doxorubicin (1 μM) for 48 hours, and the cell lysates were subjected to immunoblot analysis using the indicated antibodies. Values were presented as means ± SD (n = 3). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Although it is well recognized that SCLC is exquisitely sensitive to chemotherapy, the underlying mechanism of action is unexpectedly not well understood. In consideration of future clinical translation, we also studied the effect of clinically relevant chemotherapeutic agents used in SCLC treatment such as etoposide and doxorubicin on both DDRs and ASCL1 protein levels. We found that treatment of H2081 with etoposide and doxorubicin resulted in DDR (as shown by increased γH2AX levels) and cytotoxicity (as shown by the increase in PARP1 cleavage). These changes were accompanied by a substantial decrease of ASCL1 protein levels under these conditions (Fig. 2E). In addition, other chemotherapeutic agents, including TMZ, dinaciclib, and gemcitabine, also led to a significant increase in DDR and a decrease in ASCL1 protein levels (fig. S2G).
Collectively, our data showed that the SCLC lineage-specific oncoproteins were down-regulated in response to the genotoxicity induced by PARPi, as well as commonly used chemotherapeutic agents. Because of the critical roles of these proteins in the pathogenesis of SCLC, our data also raise the hypothesis that the induced degradation of these lineage-specific oncoproteins could be a mechanism that drives the vulnerability of SCLC to PARPi and potentially other chemotherapeutic agents.
HUWE1-mediated ASCL1 degradation in response to PARPi-induced genotoxicity
Because ASCL1 regulates a transcription program that is critical for the survival and proliferation of SCLC (31, 32), we investigated if PARPi treatment affects the expression of ASCL1 target genes in PARPi sensitive SCLCs. The gene set enrichment analysis revealed a significant presence of known ASCL1 target genes among the down-regulated proteins in ASCL1high/talazoparib-sensitive cells in our quantitative proteomic dataset (fig. S3A). The 256 PiPS proteins (the talazoparib-induced down-regulated proteins in ASCL1high/talazoparib-sensitive cell lines) contained a number of well-known ASCL1 target genes (e.g., IGFBP5, DLL3, and INSM1) (31, 32) that are associated with cell differentiation (P = 1.96 × 10−3), cell proliferation (P = 2.59 × 10−2), and transcription regulation (P = 7.12 × 10−2) (fig. S3B and table S4). qRT-PCR analyses confirmed that the mRNA levels of several representative ASCL1 transcription targets—including MYCL1, RET, SOX2, and BCL2—were significantly down-regulated in talazoparib-treated cells (Fig. 3A). To demonstrate the relevance of ASCL1 loss in mediating the cytotoxicity of PARPi, we depleted ASCL1 using two independent short hairpin RNAs (shRNAs). SCLC cells with ASCL1 knockdown (KD; shASCL1 #1 and #2) showed reduced survival and increased cell death (PARP1 cleavage) compared to control cells (fig. S3, C and D). In the Cancer Dependency Map project (DepMap; www.depmap.org/) (33), the knockdown of ASCL1 also led to decreased growth specifically in ASCL1high SCLC cells, but not in other SCLC subtypes, including NEUROD1high, POU2F3high, and TFlow SCLC cells. These results confirmed a lineage-specific function of the TF ASCL1 in the ASCL1high SCLC subtype (fig. S3E). On the contrary, overexpression of ASCL1 (ASCL1-Myc) in SCLC cells attenuated cell death induced by talazoparib treatment (fig. S3, F and G).
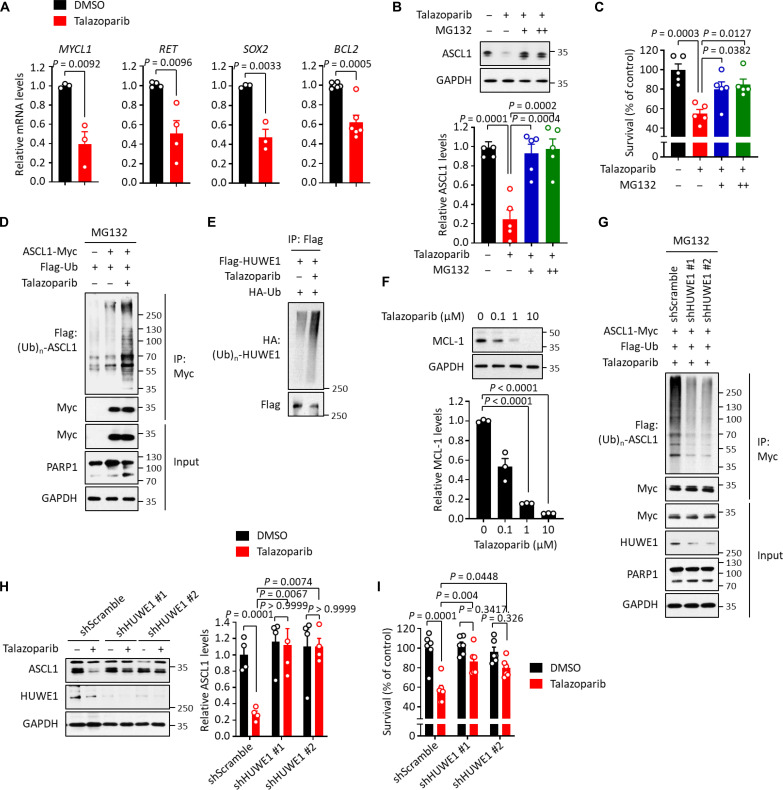
Fig. 3. HUWE1-mediated ASCL1 degradation in response to PARPi-induced genotoxicity.
(A) qRT-PCR analysis of the downstream target genes of ASCL1 in H2081 cells treating with or without talazoparib (1 μM for 48 hours). The mRNA levels were normalized to GAPDH. Values were presented as means ± SEM (n = 3 to 6). (B and C) H2081 cells were treated with talazoparib (1 μM for 48 hours) and MG132 [10 (+) and 20 (++) μM for 6 hours]. ASCL1 levels (B) and viability (C) were measured, and values were presented as means ± SD (n = 5) and ± SEM (n = 5), respectively. (D) Ubiquitination of ASCL1. H2081 cells expressing ASCL1-Myc were treated with talazoparib (1 μM for 48 hours) and MG132 (10 μM for 6 hours). Cell lysates were subjected to immunoprecipitation using anti-Myc antibody, and immunoprecipitates were subjected to immunoblot analysis. (E) Auto-ubiquitination of HUWE1. H2081 cells transfected with Flag-HUWE1 were treated with talazoparib (1 μM for 48 hours), and cell lysates were subjected to immunoprecipitation using anti-Flag M2 beads. Immunoprecipitates were subjected to immunoblot analysis. (F) Down-regulation of MCL-1. H2081 cells were treated with talazoparib for 48 hours, and cell lysates were subjected to immunoblot analysis. Values were presented as means ± SD (n = 3). (G) Inhibition of ASCL1 ubiquitination. H2081 cells expressing ASCL1-Myc were depleted with HUWE1 (shHUWE1 #1 and #2) and treated with talazoparib (1 μM for 48 hours) and MG132 (10 μM for 6 hours). Cell lysates were subjected to immunoprecipitation using anti-Myc antibody. Immunoprecipitates were subjected to immunoblot analysis. (H and I) H2081 cells depleted with HUWE1 (shHUWE1 #1 and #2) were treated with talazoparib (1 μM for 48 hours). ASCL1 levels (H) and viability (I) were measured, and values were presented as means ± SD (n = 5) and ± SEM (n = 6), respectively. IP, immunoprecipitation; HA, hemagglutinin.
We next sought to determine the molecular mechanism behind PARPi-induced ASCL1 down-regulation. A pan-caspase inhibitor, Z-VAD, did not affect talazoparib-induced ASLC1 down-regulation, suggesting that the down-regulation of ASCL1 might not be secondary to talazoparib-induced apoptotic cell death (fig. S3H). Intriguingly, we found that talazoparib-induced ASLC1 down-regulation was completely blocked by a proteasome inhibitor, MG132, suggesting that ASCL1 down-regulation could be regulated by the UPS (Fig. 3B and fig. S3I). Cell death was greatly reduced when SCLCs were treated with both talazoparib and MG132 (Fig. 3C). We used cycloheximide (CHX) to block the protein synthesis and found that talazoparib treatment led to greatly accelerated turnover of ASCL1 (fig. S3J). Consistently, we also observed that ASCL1 ubiquitination levels were markedly increased in response to talazoparib treatment compared to control (Fig. 3D and fig. S3K). Furthermore, we observed that talazoparib-induced SCLC death was attenuated by the treatment of a caspase-mediated apoptosis inhibitor Z-VAD, but not the other cell death inhibitors (i.e., an autophagy inhibitor chloroquine, a necroptosis inhibitor necrostatin-1, a ferroptosis inhibitor ferrostatin-1, and a pyroptosis inhibitor Ac-FLTD-CMK) (fig. S3L). Collectively, these data provide evidence supporting a model where PARPi leads to the UPS-dependent degradation of ASCL1 and, subsequently, apoptotic cell death of SCLC.
Because PARPi causes PARP1/2 trapping, which, in return, leads to DDR (7–10, 13, 14), we hypothesized that an E3 ubiquitin ligase, whose activity is increased during DDR, could ubiquitinate ASCL1 and mediate its degradation. Notably, an E3 ubiquitin ligase, HUWE1, was previously found to target ASCL1 for degradation in neuronal stem cells (34). HUWE1 (HECT, UBA, and WWE domain-containing 1) is a HECT domain-containing E3 ubiquitin ligase that regulates many biological processes linked to DDR (35–39). Its E3 ubiquitin ligase activity is known to increase upon sensing genotoxic stress (37–39). Intriguingly, we also identified a number of putative HUWE1 target proteins (e.g., ASCL1 and MCL-1) in the 256 PiPS proteins (the talazoparib-induced down-regulated proteins in ASCL1high/talazoparib-sensitive cell lines) (fig. S3M and table S5). These proteins are linked to biological processes related to cell survival and proliferation, including apoptotic process (P = 5.55 × 10−4), cell cycle regulation (P = 1.77 × 10−2), and regulation of transcription (P = 7.39 × 10−2) (fig. S3N). However, whether HUWE1 mediates the degradation of ASCL1 during PARPi-induced DDR in SCLC is unknown. To directly demonstrate the association between ASCL1 and HUWE1, we performed co-immunoprecipitation assays and observed that ASCL1 interacted with HUWE1 (fig. S3O). We found that: ectopic expression of HUWE1 led to the degradation of ASCL1 (fig. S3P); this effect was blocked by MG132 (fig. S3Q); the E3 ubiquitin ligase-dead (LD) mutant of HUWE1 did not degrade ASCL1 (fig. S3R); and that exogenous overexpression of HUWE1 enhanced ASCL1 ubiquitination (fig. S3S).
We next interrogated HUWE1-mediated ASCL1 degradation in the context of PARPi treatment. Immunoblotting analyses showed increased auto-ubiquitination of HUWE1 (Fig. 3E) and the degradation of known HUWE1 targets (i.e., MCL-1) (40) in talazoparib-treated cells (Fig. 3F). These results suggest that PARPi-induced genotoxicity is able to activate the E3 ubiquitin ligase activity of HUWE1. Moreover, we found that talazoparib-induced ubiquitination and degradation of ASCL1 were markedly reduced in SCLC with HUWE1 KD (shHUWE1 #1 and #2) compared to control (Fig. 3, G and H). Furthermore, HUWE1 KD also greatly decreased the levels of talazoparib-induced cell death (Fig. 3I). Collectively, these data provide evidence that PARPi-induced genotoxicity activates HUWE1, which targets the degradation of ASCL1, and promotes the cell death of ASCL1high SCLC.
The synergistic effects between PARPi and PiPS-targeting agents
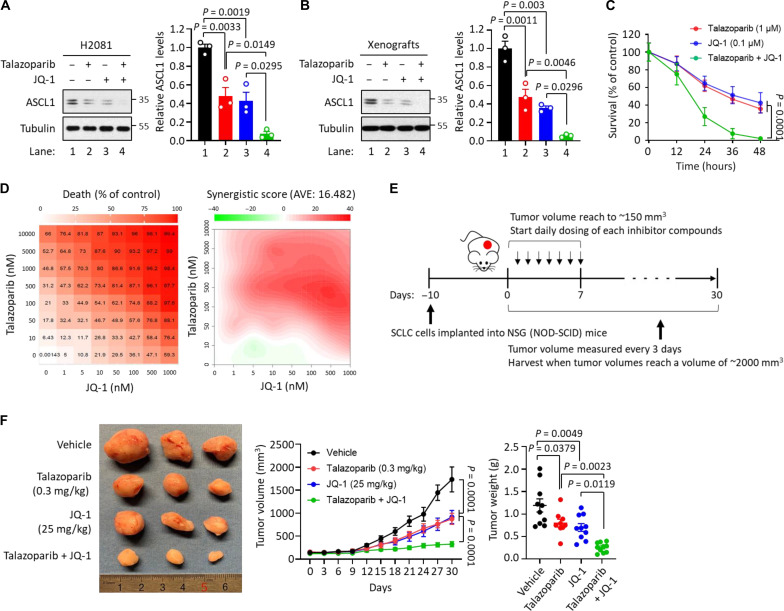
While approved chemotherapy usually results in significant antitumor responses with clinical benefit in SCLC, relapse is almost universal (17, 18). Because the down-regulation of PiPS proteins serves as one of the major therapeutic responses to PARPi in SCLC, we tested whether cotargeting these PiPS proteins and PARP proteins could offer a therapeutic strategy to achieve a more complete and durable response. Toward this, we used talazoparib to degrade ASCL1 protein and JQ-1 to reduce ASCL1 expression. Consistent with previous findings (41), ASCL1 expression was markedly reduced in ASCL1high H2081 cells and xenograft tumors treated with JQ-1 (fig. S4, A to C). JQ-1 and talazoparib reduced the survival in H2081 cells in vitro (fig. S4D) and xenograft tumor growth in vivo (Fig. 2C and fig. S4E).
We next evaluated the potential synergy of talazoparib in combination with JQ-1 in SCLC in vitro and in vivo. The combination of talazoparib and JQ-1 led a more complete suppression of ASCL1 levels in ASCL1high H2081 cells and xenograft tumors, compared to either agent alone (Fig. 4, A and B). Moreover, the combination treatment more markedly reduced the survival of SCLC cells in a time- and concentration-dependent manner (Fig. 4, C and D). Last, the growth of xenograft tumors was more markedly suppressed in mice treated with the combination of talazoparib and JQ-1 (also, as shown by a nearly complete loss of PCNA) compared to those treated with talazoparib or JQ-1 alone (Fig. 4, E and F, and fig. S4F). In addition, mice that received single or the combination treatment in xenograft experiments did not show any significant changes in body weight, providing one test of no increased toxicity (fig. S4G). These results indicate that these two compounds (i.e., talazoparib and JQ-1) lower the expression of ASCL1 via two orthogonal mechanisms (i.e., degradation and transcription, respectively), leading to profound and synergistic suppression of tumor growth in vivo. The full therapeutic potential of these combination strategies warrants further studies.
Fig. 4. The synergistic effects between talazoparib and BET inhibitor JQ-1.
(A) Effect of talazoparib with JQ-1 in ASCL1 degradation. H2081 cells were treated with talazoparib (1 μM for 48 hours), JQ-1 (0.1 μM for 48 hours), or talazoparib + JQ-1 as indicated, and the cell lysates were subjected to immunoblot analysis using the indicated antibodies. Values were presented as means ± SD (n = 3). (B) Effect of talazoparib with JQ-1 in ASCL1 degradation in vivo. Mice implanted with H2081 cells were treated with talazoparib (0.3 mg/kg), JQ-1 (25 mg/kg), or talazoparib + JQ-1 as indicated, and the tumor extracts were subjected to immunoblot analysis using the indicated antibodies. Values were presented as means ± SD (n = 3). (C) Effect of talazoparib with JQ-1 in a time-dependent manner. H2081 cells were treated with talazoparib (1 μM), JQ-1 (0.1 μM), or talazoparib + JQ-1 in a time-dependent manner. Viability was measured using a CellTiter-Glo assay, and all values were normalized by the DMSO control (100%) in each time point and presented as means ± SEM (n = 3). (D) Antitumor effect of talazoparib with JQ-1 in a concentration-dependent manner demonstrated synergy. H2081 cells were treated with talazoparib, JQ-1, or talazoparib + JQ-1 in a concentration-dependent manner. Viability was measured using a CellTiter-Glo assay. Left: Cell death ratio; right: synergistic score. AVE, average. (E) Overview of H2081-implanted xenograft tumor models. (F) Synergistic effects of talazoparib with JQ-1 in vivo. Mice implanted with H2081 cells were treated with talazoparib (0.3 mg/kg), JQ-1 (25 mg/kg), or talazoparib + JQ-1 as indicated. Left: The image of representative tumors; right: tumor volume and weight. Values were presented as means ± SD (n = 6 to 10).
Predictive biomarkers for PARPi-mediated cytotoxicity in SCLC
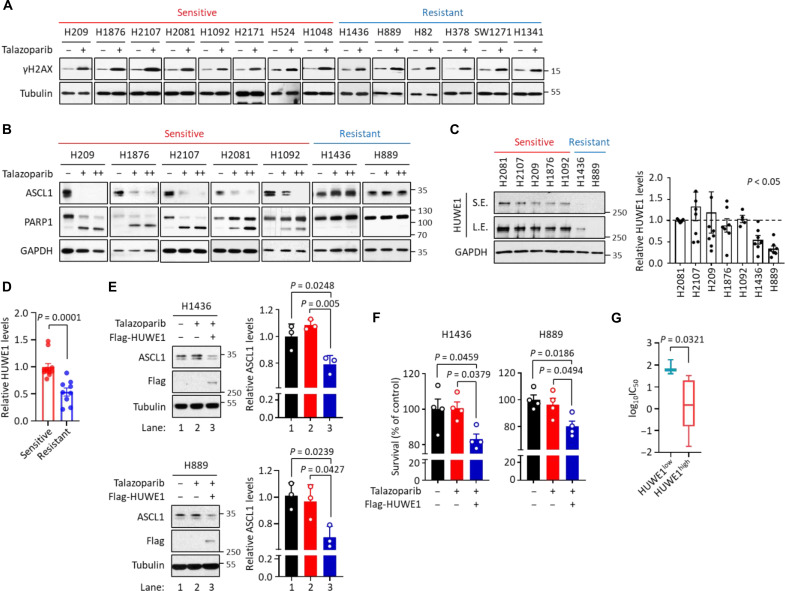
PARP proteins have been proposed as a promising therapeutic target for SCLC (42, 43). However, BRCA1/2 mutations are rarely found in SCLC, and the mechanism for the selective PARPi toxicity in certain SCLC cell lines is poorly understood. Despite the heterogeneous response of these SCLC cell lines to PARPi (Fig. 1B), we found that talazoparib treatment was able to elicit a general DDR in these cells, as shown by the universal increase in γH2AX levels (Fig. 5A and fig. S5A). These results suggest that the selective toxicity of PARPi in certain SCLC cells could result from a cell context-specific response to PARPi-induced DDR stimuli, rather than an unequal DDR signal itself.
Fig. 5. Predictive biomarkers for PARPi-mediated cytotoxicity in SCLC.
(A) Extent of DNA damage in a panel of SCLC cells treated with talazoparib. Cells were treated with talazoparib (1 μM for 48 hours), and cell lysates were subjected to immunoblot analysis. Red: Sensitive SCLC cells; blue: resistant SCLC cells. (B) Levels of ASCL1 and cleaved PARP1 in ASCL1high SCLC cells treated with talazoparib. ASCL1high SCLC cells were treated with talazoparib [1 (+) and 10 (++) μM for 48 hours]. Cell lysates were subjected to immunoblot analysis. Red: Sensitive ASCL1high SCLC cells; blue: resistant ASCL1high SCLC cells. (C) Levels of HUWE1 in ASCL1high SCLC cells. Values were presented as means ± SD from (n = 3 to 7). Red: Sensitive ASCL1high SCLC cells; blue: resistant ASCL1high SCLC cells. S.E., short exposure; L.E., long exposure. (D) Relative levels of HUWE1 in ASCL1high SCLC cells from TMT-based proteomic datasets. The values of HUWE1 from the DMSO-treated samples in PARPi-sensitive and PARPi-resistant ASCL1high SCLC cells used in this study were collected, normalized, and presented as means ± SD. (E) HUWE1-mediated ASCL1 degradation in PARPi-resistant ASCL1high SCLC cells. H1436 and H889 cells expressing Flag-HUWE1 were treated with talazoparib (1 μM for 48 hours), and cell lysates were subjected to immunoblot analysis. Values were presented as means ± SD (n = 3). (F) HUWE1-mediated cell death in PARPi-resistant ASCL1high SCLC cells. H1436 and H889 cells expressing Flag-HUWE1 were treated with talazoparib (1 μM for 48 hours). Viability was measured and values were presented as means ± SEM (n = 4). (G) Boxplots showing the sensitivity of a panel of ASCL1high SCLC cells to PARPi. ASCL1high SCLC cells were grouped by the relative levels of HUWE1 expression, and the sensitivity was determined by the analysis of the GDSC database measuring IC50 against talazoparib treatment. Values were presented as means ± SD.
Using PARP1 cleavage as a marker for apoptosis, we found that the SCLC cells demonstrated varying degrees of talazoparib sensitivity (fig. S5, B to D). We then studied the level of ASCL1 degradation induced by talazoparib treatment in ASCL1high talazoparib-sensitive and talazoparib-resistant SCLC cells, (i.e., H209, H1876, H2107, H2081, H1092, H1436, and H889). We observed that the degree of ASCL1 degradation was highly correlated with the level of talazoparib toxicity (Fig. 5B). Specifically, talazoparib treatment led to increased ASCL1 degradation in ASCL1high/talazoparib-sensitive cells, but not in ASCL1high/talazoparib-resistant cells (Fig. 5B and fig. S5F). Similarly, we observed the degradation of NEUROD1 in NEUROD1high/talazoparib-sensitive cells, but not in NEUROD1high/talazoparib-resistant cells (fig. S5, E and F). We were only able to study one POU2F3high SCLC and observed robust degradation of POU2F3 in H1048 cells, which is a POU2F3high/talazoparib-sensitive cell line (fig. S5, E and F). Furthermore, we examined two TFlow SCLC lines (SW1271 and H1341) and found both to be resistant to talazoparib (fig. S5, E and F). Collectively, these results are all consistent with the hypothesis that talazoparib induced degradation of lineage-specific TFs correlates with PARPi sensitivity in SCLC cell lines.
Because HUWE1 plays a critical role in regulating the PARPi-induced degradation of ASCL1 (Fig. 3), we hypothesized that expression levels of HUWE1 could be a predictive biomarker for SCLC sensitivity to PARPi. Toward this, we first measured the abundance of HUWE1 in a representative panel of ASCL1high SCLC cell lines. We observed that the PARPi-sensitive ASCL1high SCLC cell lines expressed significantly higher levels of HUWE1, compared to PARPi-resistant ASCL1high SCLC cell lines (Fig. 5C). From our TMT-based quantitative mass spectrometric analyses (Fig. 1C and table S2), we also observed that the ASCL1high/talazoparib-sensitive cells had higher HUWE1 levels, compared to the ASCL1high/talazoparib-resistant cells (Fig. 5D). However, we did not observe that talazoparib treatment regulates HUWE1 expression in SCLC cells (fig. S5G and table S2).
We selected two ASCL1high SCLC cells (i.e., H1436 and H889) that displayed relatively low levels of HUWE1 expression and demonstrated resistance to talazoparib treatment. We examined how HUWE1 overexpression influenced the following two aspects in response to talazoparib treatment: (i) ASCL1 degradation and (ii) cell survival. HUWE1 overexpression resulted in a significantly increased level of talazoparib-induced ASCL1 degradation and cell death in both cell lines (Fig. 5, E and F). These data again provided compelling evidence pointing to the critical role of HUWE1 in the regulation of talazoparib sensitivity in ASCL1high SCLC. To further investigate the potential impact of HUWE1 expression on the sensitivity of ASCL1high SCLC cells to PARPi, we conducted an analysis using the Genomics of Drug Sensitivity in Cancer dataset (GDSC; www.cancerrxgene.org/) (44, 45). Our analysis revealed that ASCL1high SCLC cell lines with higher levels of HUWE1 exhibited increased sensitivity to talazoparib (Fig. 5G). A similar trend was observed with olaparib treatment (fig. S5H).
In addition, we explored the impact of HUWE1 expression on the survival of patients with SCLC by analyzing a survival dataset and matching genotype and expression datasets from publicly available sources (20). Our findings from this SCLC database indicated that SCLC patients with high HUWE1 expression had a significantly better survival when treated with chemotherapy alone or in combination with radiotherapy compared to those with low HUWE1 expression (fig. S5, I and J). Collectively, these results suggest that HUWE1 abundance could serve, at least in part, as a predictive biomarker for the therapeutic response to genotoxic agents (i.e., PARPi and other chemotherapeutic agents) for ASCL1high SCLC.
DISCUSSION
The critical roles of PARP1/2 in mediating DDR provide the rationale for the usage of PARPi to treat human malignancy. In particular, it is well known that cancers with HR deficiencies (HRDs; as a result, for example, BRCA1/2 mutations) are particularly sensitive to PARPi, as a result of the synthetic lethality mechanism (4). In addition to BRCA1/2-mutated breast and ovarian cancers, recent studies have indicated that a number of other human malignancies could potentially benefit from PARPi. For example, it is known that at least a subset of SCLC responds favorably to PAPRi, and based on these results, multiple clinical trials have been initiated for the evaluation of PARPi in SCLC (46). However, the underlying mechanism(s) of PARPi sensitivity in SCLC is poorly understood. Mutations of BRCA1/2 are not commonly found in this tumor type (e.g., less than 3% of the SCLC cases contain BRCA1/2 mutations) (20, 24). Using a series of PDX (patient-derived xenograft) models, a recent study evaluated whether HRD scores could predict PARPi sensitivity in SCLC (47). In this study, no association was found between the in vivo sensitivity to talazoparib and the HRD score or the mutation status of common DDR genes. These results point to a disease-specific context for the HRD to predict PARPi sensitivity. It was also concluded from this study that standard predictive biomarkers for PARPi in other cancer types (HRD scores, mutational burden, and DDR gene mutations) are unlikely to be applicable in SCLC.
Although gene expression profiling experiments have been widely used to characterize tumor adaptive response to chemical perturbations, mRNA levels alone do not fully recapitulate these adaptive changes. Toward this, we used isobaric labeling-based, quantitative MS experiments to evaluate how the SCLC proteome is remodeled upon the treatment of PARPi. To do so, the global quantitative proteomic experiments were performed in a panel of 24 SCLC cell lines that were treated with talazoparib. These cells signify the common subtypes of SCLC, i.e., ASCL1high, NEUROD1high, POU2F3high, and low expression of all three TFs (TFlow).
Because of the profound heterogeneity in the SCLC proteome and the limited number of the talazoparib-sensitive and talazoparib-resistant cell lines for each SCLC subtype (i.e., ASCL1high, NEUROD1high, POU2F3high, and TFlow), the aggregation of the ratios (talazoparib versus DMSO) across the cell lines and biological replicates appears to have caused the low average log2 fold change (log2FC). The comparison of the talazoparib-sensitive or talazoparib-resistant cell lines based on the log2FC was also less ideal to capture the proteomic changes associated with each subgroup. We hereby used a workflow where we first divided each subtype (e.g., ASCL1high) into two categories based on their sensitivities to talazoparib treatment (i.e., talazoparib-sensitive and talazoparib-resistant cell lines) (fig. S1H). The protein abundances were directly extracted from the DMSO and talazoparib treatment samples for each cell line. In this step, we also calculated the P value and adjusted P value for every protein and identified a list of proteins that were significantly down-regulated in either the talazoparib-sensitive or the talazoparib-resistant subgroup. Next, we performed cross-reference analyses to extract the proteins that were down-regulated (in response to talazoparib treatment) only in the talazoparib-sensitive groups. Using this workflow, we found that ASCL1 was down-regulated only in the talazoparib-sensitive ASCL1high groups. Using similar analyses, we also identified NEUROD1 and POU2F3 as talazoparib-induced down-regulated proteins from the NEUROD1high and POU2F3high subtypes, respectively.
These analyses led to the identification of key proteomic changes that might explain talazoparib sensitivity in SCLC. As an example, ASCL1 is a TF that is highly expressed in ~70% of the SCLC cases. ASCL1 is required for the establishment of pulmonary NE lineage and therefore is a lineage-specific oncoprotein for SCLC (29, 31). Furthermore, it is required for the continued survival of SCLC in vitro, and for tumor formation in a genetically engineered mouse model of SCLC (25, 48). We confirmed that talazoparib treatment down-regulates TFs (e.g., ASCL1). ASCL1 was also down-regulated upon the treatment of other clinically relevant chemotherapeutic agents. These results suggest that the degradation of these oncoproteins in SCLC could be a critical mediator of the cytotoxicity induced by PARPi and chemotherapy.
How does talazoparib regulate the abundance of these PiPS proteins? We found that talazoparib treatment did not alter the mRNA abundances of these proteins (e.g., ASCL1), suggesting that the PARPi-mediated regulation occurred at the protein level. These results also highlighted the power of quantitative proteomics in identifying translational or posttranslational regulation events. Ubiquitination is known to play a crucial role in regulating various forms of DDR. We performed biochemical characterization and showed that ASCL1 was degraded by E3 ligase HUWE1. HUWE1 is known to be induced in DDR to regulate genomic stability by promoting the repair of DSB through ubiquitin signaling (38, 39, 49). HUWE1 has been previously proposed as a tumor suppressor (36, 50–52). Specifically, HUWE1 KO results in increased oncogenesis in a mouse model of skin cancer. Consistent with its role as a tumor suppressor, besides ASCL1, several additional oncoproteins are also known substrates of HUWE1, including MYC and MCL-1. Here, we found that HUWE1 could function as a “sensor” to detect PARPi-induced DDR. The activation of HUWE1 E3 ligase subsequently causes the degradation of ASCL1, leading to the death of the corresponding SCLC cells. However, the E3 ubiquitin ligases that are involved in degrading the other lineage-specific oncoproteins (e.g., NEUROD1 and POU2F3) are currently unknown.
Although SCLC usually responds to initial chemo- and/or radiotherapy approaches, resistance often rapidly develops (12, 17, 53, 54). While we have not explored the development of talazoparib resistance in SCLCs that are initially talazoparib sensitive, it would be important to find a companion treatment that would enhance the anti-SCLC effects of talazoparib potentially leading to more complete and durable control of the disease. Toward this and given our finding that talazoparib led to dramatic decreases in expression of ASCL1, we explored the combination of talazoparib with other agents that could also potentially lower ASCL1 expression. This led us to discover that cotreatment of talazoparib and the BET inhibitor, JQ-1 (targeting ASCL1) markedly increased cell death both in vitro and in vivo (Fig. 4). In this case, ASCL1 protein levels were greatly reduced by two different mechanisms, i.e., JQ-1–mediated ASCL1 transcription reduction and PARPi-mediated ASCL1 degradation. Therefore, the combination of PARPi with agents targeting the lineage-specific oncoprotein (e.g., ASCL1) could offer a strategy to achieve a more complete and durable control of the disease.
Previous studies and our current study show that there is considerable heterogeneity among SCLC in their response to PARPi (47). However, we found that PARPi was able to induce robust DDR (as shown by γH2AX) in all SCLC cell lines tested, regardless of their sensitivity to PARPi (Fig. 5A and fig. S5A). These observations raise the hypothesis that the selective toxicity of PARPi in certain SCLC does not originate from a cell-specific generation of the DNA damage signal, but rather how each SCLC cell line senses and responds to this genotoxic stimulus. We found that the selective toxicity of PARPi could be explained, at least in part, by the cell-specific regulatory mechanisms of the PiPS proteins. Specifically, among the ASCL1high SCLC cells, ASCL1 was degraded by PARPi in PARPi-sensitive cells, but not in those cells that were resistant to PARPi. PARPi-sensitive cells appeared to express higher levels of HUWE1 compared to PARPi-resistant cells. These results suggest that HUWE1 might serve as a potential predictive biomarker for PARPi treatment in SCLC. However, it is important to note that serial biopsy samples are not generally available for patients with SCLC treated with PARPi or other therapeutic agents. The potential connection between ASCL1 and HUWE1 in the context of PARPi treatment for patients with SCLC needs to be addressed in future studies. Nevertheless, the Lok et al. (19) study showed that SCLC cell lines with high expression of SLFN11 are sensitive to PARPi. SLFN11 has been shown to interact with RPA1, which leads to the destabilization of RPA1-ssDNA complexes. This results in a “BRCA-like” state that renders these cells sensitive to PARPi treatment. In summary, these results suggest that a combination of all these factors might predict the therapeutic response of SCLC to PARPi. Future clinical investigation is warranted to define the full biomarker potential of these protein factors.
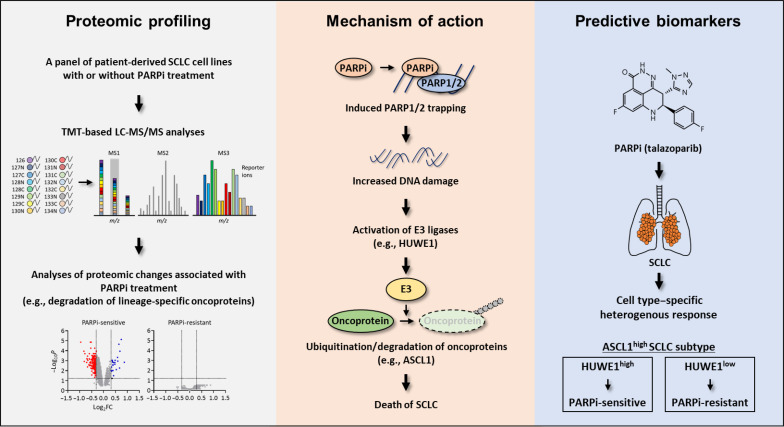
In conclusion, using quantitative MS, we performed proteomic characterization of the adaptive response of SCLC to PARPi (Fig. 6). Through these unbiased studies, we identified a set of PARPi-responsive proteins that contained key lineage-specific oncoproteins for SCLC (e.g., ASCL1). Combinatorial treatment of PARPi with the agents targeting the ASCL1 showed synergistic effects. We found that PARPi induces the degradation of these proteins via a set of DDR-responsive E3 ligases (e.g., HUWE1). Last, we found that the context-specific sensitivity of SCLC to PARPi originates from the heterogeneous expression of the relevant E3 ligases (e.g., HUWE1). Together, our efforts highlight PARP proteins as a relevant therapeutic target for SCLC. The identification of the molecular underpinnings of these processes could provide previously unknown patient stratification strategies to accelerate the clinical utility of PARPi in SCLC. Last, in addition to SCLC, a number of other human malignancies are driven by certain TFs that function as lineage-specific oncogenes. Whether these proteins will be degraded as a result of genotoxicity (e.g., as induced by chemotherapeutic agents), and whether this phenomenon underlies the therapeutic vulnerability warrant future studies. Together, our data will serve as an invaluable resource, providing the foundation for future hypothesis-driven research that helps delineate the molecular mechanisms that underlie the therapeutic potential of PARPi in SCLC and, more broadly, BRCA1/2-proficient cancers.
Fig. 6. A Schematic of the workflow in this study.
Proteomic changes were identified that signify PARPi responses in a large panel of molecularly annotated human SCLC cells. The therapeutic vulnerability of SCLC to PARPi in SCLC could be explained, at least in part, by the PARPi-induced degradation of key lineage-specific oncoproteins including ASCL1. Although PARPi resulted in a general DDR in all SCLCs, this signal is sensed differently by individual SCLC cells to generate a cell-specific response. PARPi-induced activation of the E3 ubiquitin ligase HUWE1 mediated the UPS-dependent ASCL1 degradation and eventually, led to SCLC cell death. In addition, PARPi-sensitive ASCL1high SCLC cells expressed significantly higher levels of HUWE1 compared to PARPi-resistant ASCL1high SCLC cells. Therefore, these observations suggested HUWE1 as a potentially predictive biomarker for PARPi.
MATERIALS AND METHODS
Cell lines and culture procedures
Cells used in this study—human SCLC cell lines NCI-H209, NCI-H1876, NCI-H2107, NCI-H2081, NCI-H128, NCI-H69, NCI-H1092, DMS-79, NCI-H2171, NCI-H524, NCI-H446, NCI-H1048, NCI-H1436, NCI-H889, NCI-H1836, NCI-H2029, SHP-77, NCI-H196, NCI-H1963, NCI-H378, NCI-H82, SW1271, NCI-H1341, and NCI-H84—were either obtained from the Hamon Center for Therapeutic Oncology Research at UT Southwestern Medical Center or purchased from American Type Culture Collection. Cells were maintained in RPMI 1640 (MilliporeSigma) supplemented with 10% FBS (MilliporeSigma) at 37°C in 5% CO2. Human embryonic kidney (HEK) 293T (HEK293T), HEK293TD, and HeLa cell lines were maintained in high glucose Dulbecco’s modified Eagle’s medium (MilliporeSigma) supplemented with 10% FBS (MilliporeSigma) at 37°C in 5% CO2. All cells were found to be mycoplasma-free using the e-Myco kit (Boca Scientific). Cell lines were authenticated by DNA fingerprints for cell line individualization using Promega Stem Elite ID system, a short tandem repeat (STR)–based assay at UT Southwestern Medical Center Genomics Core.
Mice
All animal experiments were performed in compliance with the Institutional Animal Care and Use Committee (IACUC) at the UT Southwestern Medical Center. Female NOD Scid gamma (NSG; NOD-SCID) mice (the Jackson Laboratory) at 6 to 8 weeks of age were used.
Antibodies and reagents
Antibodies against the following proteins were used: Cell Signaling Technology: PARP1 (#9542), γH2AX (#9718), Caspase 3 (#9665), α-tubulin (#3873), Histone H3 (#4499), NEUROD1 (#4373), Myc (#2276), hemagglutinin (#3724), HUWE1 (#5695), and MCL-1 (#94296); Santa Cruz Biotechnology: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (#sc-32233), ASCL1 (#sc-374550), POU2F3 (#sc-293402), PCNA (#sc-56), Myc (#sc-40), and ubiquitin (#sc-8017); Trevin: PAR (#4335-MC-100); MilliporeSigma: Flag (#F7425); and ActiveMotif: PARP2 (#39743). The following reagents were used (1 μM for 48 hours, if not indicated): Talazoparib (Pfizer Inc.), MG132 (10 μM), and Z-VAD-FMK (Z-VAD, 50 μM) were all purchased from Selleck; Necrostatin-1 (20 μM), Ferrostatin-1 (10 μM), Ac-FLTD-CMK (10 μM), DMSO, and Lipofectamine 2000 were all purchased from Thermo Fisher Scientific; JQ-1 was purchased from AddoQ Bioscience; chloroquine diphosphate salt (chloroquine; 50 μM), CHX (10 μg/ml), polybrene (8 μg/ml), puromycin (2 μg/ml), NEM (25 mM), sodium orthovanadate (2 mM), and sodium fluoride (20 mM) were purchased from MilliporeSigma. See also table S7.
Sample preparation for MS
Following procedures previously described (11), all SCLC cell lines were treated with either DMSO or talazoparib for 48 hours. Cells were lysed with 1% SDS lysis buffer containing 10 mM Hepes (pH 7.0), 2 mM MgCl2, universal nucleases (20 U/ml). Protein concentrations were determined with the BCA assay (Thermo Fisher Scientific). Samples were reduced with 3 mM dithiothreitol for 20 min and alkylated with 25 mM iodoacetamide for 30 min at room temperature (RT) in dark. The detergents were removed by methanol/chloroform precipitation. The proteins were resolubilized in 8 M urea and digested by Lys-C at a 1:100 (w/w) enzyme/protein ratio for 2 hours, followed by trypsin digestion at 1:100 (w/w) enzyme/protein ratio overnight at RT in 2 M urea. The peptides were desalted using Oasis HLB solid-phase extraction cartridges (Waters), and approximately 100 μg of peptides for each sample was resuspended in 200 mM Hepes (pH 8.5). The peptides were then labeled with either the amine-based TMT 16-plex reagents (Thermo Fisher Scientific) for 1 hour at RT. Hydroxylamine solution was added to quench the reaction, and the labeled peptide samples were combined. Next, the TMT samples were lyophilized and were reconstituted in buffer A (10 mM ammonium formate, pH 10.0). It was then centrifuged at 10,000g for 3 min using Corning Costar Spin-X Plastic Centrifuge Tube Filters (MilliporeSigma) before loading onto a ZORBAX 300 Extend-C18 high-performance liquid chromatography (HPLC) column (Agilent; narrow bore RR, 2.1 mm by 100 mm, 3.5 μm in particle size, 300 Ǻ in pore size). Peptides were fractioned by bRPLC (basic pH reversed-phase HPLC) at a flow rate of 0.2 ml/min using a gradient from 0 to 70% buffer B (1% ammonium formate, pH 10.0 and 90% acetonitrile). The collected 17 fractions were lyophilized, desalted, and analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) as described previously (55). Briefly, peptides were separated on a PicoFrit microcapillary column (75 μm × 15 cm; New Objective). A 180-min linear gradient was developed ranging from 7 to 32% acetonitrile in 0.1% formic acid at 300 nl/min to elute the peptides (Thermo EASY-nLC system).
Quantitative proteomic analysis by LC-MS/MS
Following procedures previously described (11), the TMT sample was analyzed by LC-MS/MS on an Orbitrap Eclipse Tribrid Mass Spectrometer (Thermo Fisher Scientific) using a multi-notch [synchronous precursor selection (SPS)]–MS3 approach (56–58). Briefly, MS1 spectra within 375 to 1500 mass-charge ratio (m/z) were acquired at 120,000 resolving power with a maximum of 50-ms ion injection in the Orbitrap. MS2 spectra were acquired by selection of the most abundant features within 3 s via collisional-induced dissociation in the ion trap using an automatic gain control (AGC) setting of 10,000, quadrupole isolation width of 0.5 m/z and a maximum ion accumulation time of 100 ms. Then, an SPS-MS3 scan was performed using up to 10 b- and y-type fragment ions as precursors with an AGC of 200 K for a maximum of 86 ms, with a normalized collision energy setting of 45. MS spectra were searched against a composite database of human protein sequences (Uniprot) and their reversed complement using the Sequest algorithm (Ver28) embedded in an in-house–developed software suite (59) or Proteome Discoverer Software (Thermo Fisher Scientific). MS1 mass tolerance was set to be 50 ppm. Search parameters allowed for full tryptic peptides with a static modification of 57.02146 Da on cystine (Carbamidomethyl), a variable modification of 15.994915 Da on methionine (oxidation), and a static modification of TMT labels (295.1896 Da, TMT 16-plex) on peptide N terminus and lysine. Search results were filtered to include <1% matches (both peptide and protein level filtering) to the reverse database by the linear discriminator function using parameters including Xcorr, dCN, missed cleavage, charge state (exclude 1 + peptides), mass accuracy, peptide length, and fraction of ions matched to MS/MS spectra. Peptide quantification was performed by using the CoreQuant algorithm implemented in an in-house–developed software suite (60) or Proteome Discoverer Software (Thermo Fisher Scientific). The labeling scheme for the TMT experiments is listed in table S2. For TMT quantification, a 0.03-Th window was scanned around the theoretical m/z of each reporter ion to detect the presence of these ions. The maximum intensity of each ion was extracted, and the signal-to-noise value of each protein was calculated by summing the reporter ion counts across all identified peptides. Because the same amount of peptides was used for each TMT channel, the total reporter ion intensity of each channel was summed across all quantified proteins and was then normalized and reported. Data were exported to Excel for further analysis.
Immunoprecipitation and immunoblot analysis
Cellular lysates were prepared using a Triton X-100 lysis buffer, consisting of 50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM N-ethylmaleimide, 2 mM Na3VO4, 20 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail and 0.5% (v/v) Triton X-100. Cellular lysates were clarified by centrifugation at 14,000g at 4°C for 15 min. The resulting supernatants were subjected to immunoprecipitation and immunoblot analysis with the corresponding antibodies. For immunoprecipitation, proteins (1 mg) were pre-incubated with Protein G Sepharose beads (MilliporeSigma) for preclearing and further incubated with 1 to 2 μg of the corresponding antibodies or anti-Flag M2 affinity gel beads (MilliporeSigma) overnight at 4°C. The immunocomplexes were collected with Protein G Sepharose beads followed by centrifugation at 3000g at 4°C for 2 min. Proteins were eluted from the beads by addition of 2× protein sample buffer, denatured by boiling, separated on SDS–polyacrylamide gel electrophoresis, and subjected to immunoblot analysis. Indicated antibodies were used. Enhanced chemiluminescence was used to detect specific bands using standard methods. The relative band intensity was measured using the ImageJ imaging software.
Cellular fractionation
Following procedures previously described (11), cells were fractioned using a subcellular protein fractionation kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, cells were harvested with trypsin-EDTA, centrifuged at 500g for 5 min, and washed with ice-cold phosphate-buffered saline (PBS). After adding cytoplasmic extraction buffer (CEB) buffer to the cell pellet, the tube was incubated at 4°C for 10 min with gentle mixing. Following centrifugation at 500g for 5 min, the supernatant (cytoplasmic extract) was transferred to a clean prechilled tube on ice. Next, the MEB buffer was added to the pellet. The tube was briefly vortexed and was incubated at 4°C for 10 min with gentle mixing. The tube was then centrifuged at 3000g for 5 min, and the supernatant (membrane extract) was transferred to a clean prechilled tube on ice. An ice-cold nuclear extraction buffer (NEB) buffer was added to the pellet, and the tube was vortexed at the highest setting for 15 s. Following incubation at 4°C for 30 min with gentle mixing, the tube was centrifuged at 5000g for 5 min and the supernatant (soluble nuclear extract) was transferred to a clean prechilled tube on ice. Last, RT NEB buffer containing micrococcal nuclease and CaCl2 was added to the pellet. The tube was vortexed for 15 s and was incubated at RT for 15 min. After incubation, the tube was centrifuged at 16,000g for 5 min and the supernatant (chromatin-bound nuclear extract) was transferred to a clean prechilled tube on ice.
In vivo drug treatment experiments
All animal experiments were performed in compliance with the IACUC at the UT Southwestern Medical Center. Female NSG (NOD-SCID) mice (the Jackson Laboratory) at 6 to 8 weeks of age were used. Compounds were dissolved in 2% DMSO + 30% polyethylene glycol, molecular weight 300 + 5% Tween 80 + ddH2O (for JQ-1) and in 10% Dimethylacetamide (DMAc) + 6% solutol + 84% PBS (for talazoparib), respectively. Tumors were engrafted in NSG mice by subcutaneous injection of 1 × 106 cells of H2081 in RPMI 1640 medium supplemented with 50% Matrigel (BD Biosciences). Ten days after the injection, mice carrying 100- to 150-mm3 subcutaneous tumors were assigned randomly to control and various treatment groups (n = 6 to 10 for each group). Tumor-bearing mice were orally (for talazoparib) or intraperitoneally injected (for JQ-1) with vehicle, talazoparib (0.3 mg/kg), or JQ-1 (25 mg/kg) daily for 30 days. The weight of the mice was monitored every 3 days, and the tumor volume was also measured with calipers every 3 days. Tumor volumes were calculated using a modified ellipsoid formula: tumor volume = ½(length × width2). Mice were euthanized at 31 days after injection. Fresh tumor samples were harvested, and the weight was measured with an electronic scale, followed by extraction for further experiments.
Histological analysis
For tissue preparation, freshly dissected xenograft tumor samples were fixed overnight at 4°C in 10% neutral-buffered formalin. Tissue sections were cut at a thickness of 4 μm from formalin-fixed paraffin-embedded (FFPE) tissue blocks using a microtome. Slides containing FFPE sections were warmed for 30 min in a 60°C oven before deparaffinization. Tissue sections were deparaffinized with xylene and then rehydrated through a series of decreasing ethanol concentration. After a 10 min wash in deionized water, tissue sections were stained with hematoxylin and eosin. For immunohistochemistry, post-rehydration and antigen retrieval were performed in SignalStain Citrate Unmasking Solution (Cell Signaling Technology, #14746) at 98°C for 10 min. After antigen retrieval and inhibition of endogenous peroxidase activity with 3% H2O2, the slides were incubated with 10% normal goat serum for 1 hour at RT. Tissue sections were then incubated overnight at 4°C in a humid chamber with primary antibodies, neural cell adhesion molecule (Cell Signaling Technology, #99746) and synaptophysin (Cell Signaling Technology, #36406), and diluted in SignalStain Antibody Diluent (Cell Signaling Technology, #8112). After washing with Tris Buffered Saline with Tween 20 (TBST), tissue sections were incubated with horseradish peroxidase–conjugated secondary antibody (Vector Laboratories) for 1 hour. The signal was developed using the chromogenic substrate DAB (SignalStain DAB Substrate Kit, Cell Signaling Technology, #8059). Slides were scanned, and images were captured using a NanoZoomer (Hamamatsu).
Plasmids
Human ASCL1 complementary cDNA was inserted into a pCI/3’Myc (modified pCI vector containing C-terminal Myc tag) by subcloning. Flag-Ub and HA-Ub (all in pcDNA3.1) and Flag-HUWE1 WT (wild type) and LD were gifts from Y.J. Oh (Yonsei University, South Korea), and Q. Zhong (UT Southwestern Medical Center, USA) (40), respectively. Lentiviral plasmids (VSVG and ∆8.9) were gifts from A. Kung (Dana Farber Cancer Institute, USA) and D. Baltimore (California Institute of Technology, USA). All plasmids were subjected to DNA sequencing for verification.
RNA interference and ectopic overexpression using mammalian lentiviral system and transfection
To produce the lentiviruses, shRNA plasmids were cotransfected into HEK293TD cells along with packaging (Δ8.9) and envelope (VSVG) expression plasmids using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. After 2 days, viral supernatants were collected and were filtered using a 0.45-μm filter. Recipient cells were infected in the presence of a serum-containing medium supplemented with polybrene (8 μg/ml). Two days after infection, cells were used for the indicated experiments. Lipofectamine 2000 reagents were also used to transiently knock down or overexpress the target genes, according to the manufacturer’s instructions. The knockdown or overexpression of target genes was validated by immunoblot assays. The shRNA constructs and overexpression plasmids were listed in tables S6 and S7.
Measurement of IC50
Cells were plated into 96-well plates at densities of 1000 to 2000 cells per well. The next day, cells were treated with the indicated concentration of talazoparib for 4 days. The value of IC50 was measured using the CellTiter-Glo assay (Promega) according to the manufacturer’s instructions. Briefly, after incubation, RT CellTiter-Glo reagent was added 1:1 to each well, and the plates were incubated at RT for 2 min. Luminescence was measured with the Synergy HT Multi-Detection Microplate Reader and was normalized against control cells treated with DMSO.
Cell viability measurement
Cells were plated into 96-well plates at densities of 1000 to 2000 cells per well. Two days later, cells were treated with indicated compounds. Cell viability was measured using the CellTiter-Glo assay (Promega) according to the manufacturer’s instructions. For synergistic effect measurement, cells were treated with talazoparib or JQ-1, alone or in combination as indicated for 1 to 4 days. Briefly, after incubation, RT CellTiter-Glo reagent was added 1:1 to each well, and the plates were incubated at RT for 2 min. Luminescence was measured with the Synergy HT Multi-Detection Microplate Reader and was normalized against control cells treated with vehicle.
Quantitative real-time polymerase chain reaction
The mRNA extraction was performed using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Subsequently, total RNAs were converted into cDNA using the SuperScript III Reverse Transcriptase (Thermo Fisher Scientific) following the manual for first-strand cDNA synthesis. qRT-PCR reactions were performed on a CFX384 Touch Real-Time PCR Detection System using 2× Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). For each condition, technical triplicates were prepared and the quantitation cycle (Cq) was calculated. For normalization, GAPDH levels were used as an internal reference and relative expression levels were presented.
Ubiquitination assays
A cell-based in vivo ubiquitination and auto-ubiquitination assays were performed as previously described (61). Briefly, for cell-based in vivo ubiquitination assays, cells were incubated with MG132 (10 μM for 6 hours) and were lysed in the abovementioned Triton X-100 lysis buffer. The cell lysates were subjected to immunoprecipitation using the indicated antibodies and beads followed by immunoblot analysis with the corresponding antibodies. For auto-ubiquitination assays, the denaturing immunoprecipitation method was performed. To disrupt noncovalent protein-protein interactions, the cell lysates were heated at 98°C in a lysis buffer containing 1% SDS and then were diluted with Triton X-100 lysis buffer (1:10 ratio).
DepMap analysis
To determine whether the identified proteins play a role in regulating SCLC cell proliferation and survival, we examined their dependency scores using the data available from the Cancer Dependency Map portal (https://depmap.org/portal/download/) (33). Gene dependency scores were derived using the DEMETER2 algorithm applied to combined RNAi screen data (62). A negative gene dependency score corresponds to greater gene essentiality, such that the median gene dependency score of pan-essential genes is normalized to −1 and that of negative control genes is set to 0. We analyzed the distribution of dependency scores from SCLC cell lines (n = 25). Genes were organized in ascending order by median gene dependency scores.
Drug response data analysis
To quantify the association between PARPi sensitivity and the expression level of the HUWE1 gene, we focused our analysis on ASCL1high SCLC cell lines, which were previously defined as having significantly higher expression of the ASCL1 gene (45). Drug response data across cell lines of SCLC lineage was downloaded from the GDSC (61 human SCLC cell lines; www.cancerrxgene.org/) (44). The GDSC cohorts utilized the half maximal inhibitory concentration (IC50) as a measure of a small-molecule compound’s potency in inhibiting the growth of a specific cell line. Briefly, we initially stratified the cell lines into two equally populated groups based on the log-normalized expression level of the HUWE1 gene (value = 3.89). We then conducted a nonparametric Mann-Whitney U test on the IC50 values to examine whether PARPi exhibited significantly different sensitivity between the two groups (HUWE1high and HUWE1low).
Survival association analysis
Gene expression and clinical data was downloaded from (20). As we observed bimodal distribution for both ASCL1 and HUWE1 gene expression, model-based clustering was used to determine the cutoff to dichotomize the low and high groups. For each gene, the Mclust R package was used to classify subgroups. The package automatically estimates models with different numbers of clusters and covariance structures, the best model was then selected by the Bayesian Information Criterion. For both ASCL1 and HUWE1, two clusters with variable variance stood out as the best model and the groups were classified accordingly. In the Kaplan Meier plot, P value was computed by log-rank test. As for the table, Cox proportional hazards regression models were used to assess the association between the HUWE1 expression level; sex; T, N, and M stages; and overall survival. Both univariate and multivariate Cox proportional hazards regression analyses were performed. In the univariate analysis, each selected feature was individually examined in separate models, without adjusting for other variables. The hazard ratio (HR) and corresponding P value were calculated for each feature to determine their individual association with overall survival. For the multivariate analysis, a comprehensive model was constructed that included all selected features simultaneously. This model allowed for the assessment of each feature’s association with overall survival while controlling for the effects of other variables. The HR and P value were estimated for each feature in the multivariate model.
Quantification and statistical analysis
All the other statistical analyses including unpaired Student’s t tests, one- and two-way analysis of variance (ANOVA) were performed using the GraphPad Prism software (v9.2.0). Data were calculated as means ± SEM or SD. For the proteomic data analyses, we first grouped the SCLC cell lines into four subtypes (ASCL1high, NEUROD1high, POU2F3high, and TFlow). For the ASCL1high and NEUROD1high subtypes, we then divided the cells into two categories based on their sensitivities to talazoparib treatment (i.e., talazoparib-sensitive and talazoparib-resistant). The changes of the protein abundances between DMSO and talazoparib were analyzed in each subgroup using the Limma package (3.50.0) in R (4.1.1). The POU2F3high and TFlow subtypes were not further divided because they only contained either talazoparib-sensitive cell lines (POU2F3high) or talazoparib-resistant cell lines (TFlow). The paired t test analyses were used when comparing PARPi versus DMSO in each group. The significance was calculated, and the resulting P values were corrected for multiple testing using the Benjamini-Hochberg method in R. (Synergy statistical tests used).
Acknowledgments
We thank S. Niessen for providing talazoparib, R. Hu for the initiation of the quantitative MS experiments, and Y.J. Oh for providing the Flag- and HA-Ub plasmids. We thank S. Niessen, S. Bagrodia, X. Ding, and M. White (all current or previous employees of Pfizer) for helpful discussions and for funding this work.
Funding: This work was supported, in part, by grants from NIH (R21CA261018 and R35GM134883 to Y.Y. and CA213338, CA213274, and CA070907 to J.D.M.), Welch foundation (I-1800 to Y.Y.), and a sponsored research agreement from Pfizer.
Author contributions: Conceptualization: C.K., X.-D.W., and Y.Y. Methodology: C.K., X.-D.W., and Y.Y. Investigation: C.K. and Y.Y. Visualization: C.K. and X.-D.W. Writing (original draft): C.K. Writing (review and editing): C.K., J.D.M., and Y.Y. Formal analysis: C.K., X.-D.W., P.L., Z.Z., L.C., J.K., Y.L., H.L., S.J., S.P.C., and B.J.D. Resources: C.K., Z.L., J.H., S.W., Q.D., S.d.R., and K.E.H. Supervision: Y.Y. Funding acquisition: J.D.M. and Y.Y.
Competing interests: Y.Y. is a cofounder and shareholder of ProteoValent Therapeutics. Y.Y. receives or has received research funding from Pfizer and Biosplice Inc. J.D.M. receives royalties from the NIH and UT Southwestern for distribution of human lung cancer cell lines. The authors declare that they have no other competing interests.
Data and material availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository with the dataset identifier nos. PXD029968 and PXD029972.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Legends for tables S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S7
REFERENCES AND NOTES
- 1.Welcsh P. L., King M. C., BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10, 705–713 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Farmer H., McCabe N., Lord C. J., Tutt A. N. J., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M. B., Jackson S. P., Smith G. C. M., Ashworth A., Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T., Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., Poirier G. G., PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satoh M. S., Lindahl T., Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Schreiber V., Dantzer F., Ame J. C., de Murcia G., Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Yélamos J., Schreiber V., Dantzer F., Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol. Med. 14, 169–178 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Hopkins T. A., Ainsworth W. B., Ellis P. A., Donawho C. K., DiGiammarino E. L., Panchal S. C., Abraham V. C., Algire M. A., Shi Y., Olson A. M., Johnson E. F., Wilsbacher J. L., Maag D., PARP1 Trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol Cancer Res. 17, 409–419 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Murai J., Huang S. Y. N., Renaud A., Zhang Y., Ji J., Takeda S., Morris J., Teicher B., Doroshow J. H., Pommier Y., Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 13, 433–443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim C., Wang X. D., Yu Y., PARP1 inhibitors trigger innate immunity via PARP1 trapping-induced DNA damage response. eLife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara P. N. Jr., Natale R., Crowley J., Lenz H. J., Redman M. W., Carleton J. E., Jett J., Langer C. J., Kuebler J. P., Dakhil S. R., Chansky K., Gandara D. R., Phase III trial of irinotecan/cisplatin compared with etoposide/cisplatin in extensive-stage small-cell lung cancer: Clinical and pharmacogenomic results from SWOG S0124. J. Clin. Oncol. 27, 2530–2535 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Y., Rehman F. L., Feng Y., Boshuizen J., Bajrami I., Elliott R., Wang B., Lord C. J., Post L. E., Ashworth A., BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 19, 5003–5015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murai J., Huang S. Y. N., das B. B., Renaud A., Zhang Y., Doroshow J. H., Ji J., Takeda S., Pommier Y., Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 72, 5588–5599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gazdar A. F., Bunn P. A., Minna J. D., Small-cell lung cancer: What we know, what we need to know and the path forward. Nat. Rev. Cancer 17, 725–737 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Chan B. A., Coward J. I., Chemotherapy advances in small-cell lung cancer. J. Thorac. Dis. 5, S565–S578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asai N., Ohkuni Y., Kaneko N., Yamaguchi E., Kubo A., Relapsed small cell lung cancer: Treatment options and latest developments. Ther Adv Med Oncol. 6, 69–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong J., Salgia R., Managing Patients With Relapsed Small-Cell Lung Cancer. J. Oncol. Pract. 14, 359–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lok B. H., Gardner E. E., Schneeberger V. E., Ni A., Desmeules P., Rekhtman N., de Stanchina E., Teicher B. A., Riaz N., Powell S. N., Poirier J. T., Rudin C. M., PARP Inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin. Cancer Res. 23, 523–535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George J., Lim J. S., Jang S. J., Cun Y., Ozretić L., Kong G., Leenders F., Lu X., Fernández-Cuesta L., Bosco G., Müller C., Dahmen I., Jahchan N. S., Park K. S., Yang D., Karnezis A. N., Vaka D., Torres A., Wang M. S., Korbel J. O., Menon R., Chun S. M., Kim D., Wilkerson M., Hayes N., Engelmann D., Pützer B., Bos M., Michels S., Vlasic I., Seidel D., Pinther B., Schaub P., Becker C., Altmüller J., Yokota J., Kohno T., Iwakawa R., Tsuta K., Noguchi M., Muley T., Hoffmann H., Schnabel P. A., Petersen I., Chen Y., Soltermann A., Tischler V., Choi C. M., Kim Y. H., Massion P. P., Zou Y., Jovanovic D., Kontic M., Wright G. M., Russell P. A., Solomon B., Koch I., Lindner M., Muscarella L. A., la Torre A., Field J. K., Jakopovic M., Knezevic J., Castaños-Vélez E., Roz L., Pastorino U., Brustugun O. T., Lund-Iversen M., Thunnissen E., Köhler J., Schuler M., Botling J., Sandelin M., Sanchez-Cespedes M., Salvesen H. B., Achter V., Lang U., Bogus M., Schneider P. M., Zander T., Ansén S., Hallek M., Wolf J., Vingron M., Yatabe Y., Travis W. D., Nürnberg P., Reinhardt C., Perner S., Heukamp L., Büttner R., Haas S. A., Brambilla E., Peifer M., Sage J., Thomas R. K., Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byers L. A., Wang J., Nilsson M. B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y. H., Diao L., Masrorpour F., Shen L., Liu W., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B. S., Kalhor N., Wistuba I. I., Girard L., Lippman S. M., Mills G. B., Coombes K. R., Weinstein J. N., Minna J. D., Heymach J. V., Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2, 798–811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietanza M. C., Waqar S. N., Krug L. M., Dowlati A., Hann C. L., Chiappori A., Owonikoko T. K., Woo K. M., Cardnell R. J., Fujimoto J., Long L., Diao L., Wang J., Bensman Y., Hurtado B., de Groot P., Sulman E. P., Wistuba I. I., Chen A., Fleisher M., Heymach J. V., Kris M. G., Rudin C. M., Byers L. A., Randomized, Double-Blind, Phase II Study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386–2394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farago A. F., Yeap B. Y., Stanzione M., Hung Y. P., Heist R. S., Marcoux J. P., Zhong J., Rangachari D., Barbie D. A., Phat S., Myers D. T., Morris R., Kem M., Dubash T. D., Kennedy E. A., Digumarthy S. R., Sequist L. V., Hata A. N., Maheswaran S., Haber D. A., Lawrence M. S., Shaw A. T., Mino-Kenudson M., Dyson N. J., Drapkin B. J., Combination olaparib and temozolomide in relapsed small cell lung cancer. Cancer Discov. 9, 1372–1387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudin C. M., Durinck S., Stawiski E. W., Poirier J. T., Modrusan Z., Shames D. S., Bergbower E. A., Guan Y., Shin J., Guillory J., Rivers C. S., Foo C. K., Bhatt D., Stinson J., Gnad F., Haverty P. M., Gentleman R., Chaudhuri S., Janakiraman V., Jaiswal B. S., Parikh C., Yuan W., Zhang Z., Koeppen H., Wu T. D., Stern H. M., Yauch R. L., Huffman K. E., Paskulin D. D., Illei P. B., Varella-Garcia M., Gazdar A. F., de Sauvage F. J., Bourgon R., Minna J. D., Brock M. V., Seshagiri S., Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudin C. M., Poirier J. T., Byers L. A., Dive C., Dowlati A., George J., Heymach J. V., Johnson J. E., Lehman J. M., MacPherson D., Massion P. P., Minna J. D., Oliver T. G., Quaranta V., Sage J., Thomas R. K., Vakoc C. R., Gazdar A. F., Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pozo K., Minna J. D., Johnson J. E., Identifying a missing lineage driver in a subset of lung neuroendocrine tumors. Genes Dev. 32, 865–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y. H., Klingbeil O., He X. Y., Wu X. S., Arun G., Lu B., Somerville T. D. D., Milazzo J. P., Wilkinson J. E., Demerdash O. E., Spector D. L., Egeblad M., Shi J., Vakoc C. R., POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 32, 915–928 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borges M., Linnoila R. I., van de Velde H. J. K., Chen H., Nelkin B. D., Mabry M., Baylin S. B., Ball D. W., An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386, 852–855 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Jiang T., Collins B. J., Jin N., Watkins D. N., Brock M. V., Matsui W., Nelkin B. D., Ball D. W., Achaete-scute complex homologue 1 regulates tumor-initiating capacity in human small cell lung cancer. Cancer Res. 69, 845–854 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborne J. K., Larsen J. E., Shields M. D., Gonzales J. X., Shames D. S., Sato M., Kulkarni A., Wistuba I. I., Girard L., Minna J. D., Cobb M. H., NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc. Natl. Acad. Sci. U.S.A. 110, 6524–6529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augustyn A., Borromeo M., Wang T., Fujimoto J., Shao C., Dospoy P. D., Lee V., Tan C., Sullivan J. P., Larsen J. E., Girard L., Behrens C., Wistuba I. I., Xie Y., Cobb M. H., Gazdar A. F., Johnson J. E., Minna J. D., ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 14788–14793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borromeo M. D., Savage T. K., Kollipara R. K., He M., Augustyn A., Osborne J. K., Girard L., Minna J. D., Gazdar A. F., Cobb M. H., Johnson J. E., ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 16, 1259–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McFarland J. M., Ho Z. V., Kugener G., Dempster J. M., Montgomery P. G., Bryan J. G., Krill-Burger J. M., Green T. M., Vazquez F., Boehm J. S., Golub T. R., Hahn W. C., Root D. E., Tsherniak A., Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 9, 4610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbán N., Urbán N., van den Berg D. L. C., Forget A., Andersen J., Demmers J. A. A., Hunt C., Ayrault O., Guillemot F., Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science 353, 292–295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D., Kon N., Li M., Zhang W., Qin J., Gu W., ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121, 1071–1083 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Myant K. B., Cammareri P., Hodder M. C., Wills J., Kriegsheim A. V., Győrffy B., Rashid M., Polo S., Maspero E., Vaughan L., Gurung B., Barry E., Malliri A., Camargo F., Adams D. J., Iavarone A., Lasorella A., Sansom O. J., HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling. EMBO Mol Med. 9, 181–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao S. H., Wu H. T., Wu K. J., Ubiquitination by HUWE1 in tumorigenesis and beyond. J. Biomed. Sci. 25, 67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall J. R., Kow E., Nevis K. R., Lu C. K., Luce K. S., Zhong Q., Cook J. G., Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell 18, 3340–3350 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandemaker I. K., van Cuijk L., Janssens R. C., Lans H., DNA damage-induced histone H1 ubiquitylation is mediated by HUWE1 and stimulates the RNF8-RNF168 pathway. Sci. Rep. 7, 15353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Q., Gao W., Du F., Wang X., Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121, 1085–1095 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Lenhart R., Kirov S., Desilva H., Cao J., Lei M., Johnston K., Peterson R., Schweizer L., Purandare A., Ross-Macdonald P., Fairchild C., Wong T., Wee S., Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ascl1 gene expression. Mol. Cancer Ther. 14, 2167–2174 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Sen T., Gay C. M., Byers L. A., Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl. Lung Cancer Res. 7, 50–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Den Borg R., Leonetti A., Tiseo M., Giovannetti E., Peters G. J., Novel targeted strategies to overcome resistance in small-cell lung cancer: Focus on PARP inhibitors and rovalpituzumab tesirine. Expert Rev. Anticancer Ther. 19, 461–471 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Yang W., Soares J., Greninger P., Edelman E. J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J. A., Thompson I. R., Ramaswamy S., Futreal P. A., Haber D. A., Stratton M. R., Benes C., McDermott U., Garnett M. J., Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–D961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gay C. M., Stewart C. A., Park E. M., Diao L., Groves S. M., Heeke S., Nabet B. Y., Fujimoto J., Solis L. M., Lu W., Xi Y., Cardnell R. J., Wang Q., Fabbri G., Cargill K. R., Vokes N. I., Ramkumar K., Zhang B., Della Corte C. M., Robson P., Swisher S. G., Roth J. A., Glisson B. S., Shames D. S., Wistuba I. I., Wang J., Quaranta V., Minna J., Heymach J. V., Byers L. A., Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 39, 346–360.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barayan R., Ran X., Lok B. H., PARP inhibitors for small cell lung cancer and their potential for integration into current treatment approaches. J. Thorac. Dis. 12, 6240–6252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allison Stewart C., Tong P., Cardnell R. J., Sen T., Li L., Gay C. M., Masrorpour F., Fan Y., Bara R. O., Feng Y., Ru Y., Fujimoto J., Kundu S. T., Post L. E., Yu K., Shen Y., Glisson B. S., Wistuba I., Heymach J. V., Gibbons D. L., Wang J., Byers L. A., Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8, 28575–28587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shue Y. T., Lim J. S., Sage J., Tumor heterogeneity in small cell lung cancer defined and investigated in pre-clinical mouse models. Transl Lung Cancer Res. 7, 21–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett E. J., Harper J. W., DNA damage: Ubiquitin marks the spot. Nat. Struct. Mol. Biol. 15, 20–22 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Inoue S., Hao Z., Elia A. J., Cescon D., Zhou L., Silvester J., Snow B., Harris I. S., Sasaki M., Li W. Y., Itsumi M., Yamamoto K., Ueda T., Dominguez-Brauer C., Gorrini C., Chio I. I. C., Haight J., You-ten A., McCracken S., Wakeham A., Ghazarian D., Penn L. J. Z., Melino G., Mak T. W., Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 27, 1101–1114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X., Heng J. I. T., Guardavaccaro D., Jiang R., Pagano M., Guillemot F., Iavarone A., Lasorella A., The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat. Cell Biol. 10, 643–653 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clements K. E., Schleicher E. M., Thakar T., Hale A., Dhoonmoon A., Tolman N. J., Sharma A., Liang X., Imamura Kawasawa Y., Nicolae C. M., Wang H. G., de S., Moldovan G. L., Identification of regulators of poly-ADP-ribose polymerase inhibitor response through complementary CRISPR knockout and activation screens. Nat. Commun. 11, 6118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noda K., Nishiwaki Y., Kawahara M., Negoro S., Sugiura T., Yokoyama A., Fukuoka M., Mori K., Watanabe K., Tamura T., Yamamoto S., Saijo N., Irinotecan plus cisplatin compared with etoposide plus cisplatin for extensive small-cell lung cancer. N. Engl. J. Med. 346, 85–91 (2002). [DOI] [PubMed] [Google Scholar]
- 54.William W. N. Jr., Glisson B. S., Novel strategies for the treatment of small-cell lung carcinoma. Nat. Rev. Clin. Oncol. 8, 611–619 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Hu R., Huffman K. E., Chu M., Zhang Y., Minna J. D., Yu Y., Quantitative secretomic analysis identifies extracellular protein factors that modulate the metastatic phenotype of non-small cell lung cancer. J. Proteome Res. 15, 477–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ting L., Rad R., Gygi S. P., Haas W., MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McAlister G. C., Huttlin E. L., Haas W., Ting L., Jedrychowski M. P., Rogers J. C., Kuhn K., Pike I., Grothe R. A., Blethrow J. D., Gygi S. P., Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wühr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., Gygi S. P., MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P., A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erickson B. K., Rose C. M., Braun C. R., Erickson A. R., Knott J., McAlister G. C., Wühr M., Paulo J. A., Everley R. A., Gygi S. P., A Strategy to combine sample multiplexing with targeted proteomics assays for high-throughput protein signature characterization. Mol. Cell 65, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim C., Yun N., Lee J., Youdim M. B. H., Ju C., Kim W. K., Han P. L., Oh Y. J., Phosphorylation of CHIP at Ser20 by Cdk5 promotes tAIF-mediated neuronal death. Cell Death Differ. 23, 333–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marcotte R., Sayad A., Brown K. R., Sanchez-Garcia F., Reimand J., Haider M., Virtanen C., Bradner J. E., Bader G. D., Mills G. B., Pe’er D., Moffat J., Neel B. G., Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell 164, 293–309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for tables S1 to S7
Tables S1 to S7