Abstract
Background.
The Banff system for histologic diagnosis of rejection in kidney transplant biopsies uses guidelines to assess designated features—lesions, donor-specific antibody (DSA), and C4d staining. We explored whether using regression equations to interpret the features as well as current guidelines could establish the relative importance of each feature and improve histologic interpretation.
Methods.
We developed logistic regression equations using the designated features to predict antibody-mediated rejection (AMR/mixed) and T-cell–mediated rejection (TCMR/mixed) in 1679 indication biopsies from the INTERCOMEX study (ClinicalTrials.gov NCT01299168). Equations were trained on molecular diagnoses independent of the designated features.
Results.
In regression and random forests, the important features predicting molecular rejection were as follows: for AMR, ptc and g, followed by cg; for TCMR, t > i. V-lesions were relatively unimportant. C4d and DSA were also relatively unimportant for predicting AMR: by AUC, the model excluding them (0.853) was nearly as good as the model including them (0.860). Including time posttransplant slightly but significantly improved all models. By AUC, regression predicted molecular AMR and TCMR better than Banff histologic diagnoses. More importantly, in biopsies called “no rejection” by Banff guidelines, regression equations based on histology features identified histologic and molecular rejection-related changes in some biopsies and improved survival predictions. Thus, regression can screen for missed rejection.
Conclusions.
Using lesion-based regression equations in addition to Banff histology guidelines defines the relative important of histology features for identifying rejection, allows screening for potential missed diagnoses, and permits early estimates of AMR when C4d and DSA are not available.
INTRODUCTION
The Banff system for assessing kidney transplant biopsies is central to patient management and is the standard of care for histologic assessment of antibody-mediated rejection (AMR) and T-cell–mediated rejection (TCMR).1 The Banff system uses 2 steps: step 1 assigns semiquantitative scores for selected biopsy features, including lesions, donor-specific antibody (DSA), and complement factor C4d staining; step 2 applies consensus-based rules to establish diagnoses. However, the Banff rules interpret lesions using cutoffs (eg, <2 versus ≥ 2) and therefore do not use all the information that is contained in the lesion scores, that is 0, 1, 2, 3.2–4 An alternative would be to use the actual lesion scores and features using mathematical models. Such estimates added to the existing rule-based approach in step 2 have the potential to estimate the relative importance of each feature for predicting rejection and whether some diagnoses can be estimated when certain features are not available (eg, DSA assessment).
New approaches to interpreting clinical features are increasingly used in medicine,5–7 in keeping with the principle that ensembles of independent estimates make better use of information than guidelines alone. We previously demonstrated that logistic regression could be applied to show the importance of step 1 features for assessing rejection.8,9 A recent study using a tree-based learning method (XGBoost) found that histologic diagnoses of rejection could be improved by using a mathematical approach to interpreting step 1 features.8–10 Such probabilistic estimates have the potential to be added to the existing step 2 guidelines, potentially improving the histologic diagnoses.
Probabilistic modeling of step 1 features should ideally use rejection definitions that are assigned by a system separate from those features, particularly when examining the relative importance of each feature. The emergence of molecular assessments independent of histology—the Molecular Microscope Diagnostic System (MMDx)—opens the possibility of molecular diagnoses to train step 1 feature-based regression models. Molecular rejection diagnoses can also assess the relative importance of each step 1 features because they are independent on these features. Using molecular diagnoses to train lesion-based regression equations does not depend on the assumption that molecular diagnoses are “better,” only that they are independent. (Although molecular diagnoses largely agree with histology diagnoses,11 and no test is perfect, we have presented arguments for believing that MMDx is more likely to be correct when the results are discrepant,12 eg, stronger correlations with external tests such as donor-derived cell-free DNA.13,14)
The present study explored new ways of using the step 1 biopsy features, with the goal of adding these assessments to the existing Banff step 2 algorithms, for example, for screening for rejection in biopsies diagnosed as “no rejection” by Banff guidelines. We also studied the hierarchy of importance of step 1 features, whether the inclusion of time posttransplant (TxBx) would improve the models, and whether AMR could be reliably predicted from lesions alone before C4d and DSA are available.
MATERIALS AND METHODS
Patient Population
We studied 1679 indication biopsies from the INTERCOMEX study (ClinicalTrials.gov #NCT01299168)15 performed on consenting patients under institutional review board–approved protocols as previously described.16,17 The investigators are listed in Table S1 (SDC, http://links.lww.com/TP/C864). The details of the INTERCOMEX biopsy population were previously published16,17 and are summarized in Table 1. Central histology review was not part of this study as it is not standard of care.
TABLE 1.
DSA status and %DSA-positive across histologic diagnoses and MMDx sign-outs in the kidney 1679 cohort groups (N, % of total)
| Biopsy group | All biopsies (N = 1679) | |
|---|---|---|
| Number | Number DSA-positive (% of DSA tested per row) | |
| Histologic diagnosis | ||
| Histologic rejection, N = 740 (44% of all diagnoses) | ||
| AMR-related | ||
| AMR | 333 | 219 (74%) |
| Transplant glomerulopathy | 51 | 11 (27%) |
| AMR suspected (pAMR) | 33 | 8 (30%) |
| Mixed (TCMR plus AMR) | 56 | 28 (60%) |
| TCMR-related | ||
| TCMRa | 139 | 32 (29%) |
| Borderline (pTCMR) | 128 | 32 (28%) |
| Histologic NR, N = 939 (56% of all diagnoses) | ||
| AKI | 117 | 30 (33%) |
| BK | 52 | 5 (12%) |
| Diabetic nephropathy | 24 | 7 (54%) |
| Glomerulonephritis | 108 | 27 (34%) |
| IFTA not otherwise specified | 193 | 49 (30%) |
| No major abnormalities | 371 | 113 (35%) |
| Othersb | 74 | 15 (29%) |
| All NR excluding borderline | 939 | 246 (32%) |
| MMDx sign-outs | ||
| Rejection-related | ||
| AMR-related | ||
| AMR | 509 | 271 (62%) |
| pAMR | 52 | 22 (45%) |
| Mixed | 69 | 33 (58%) |
| TCMR-related | ||
| TCMR | 123 | 25 (26%) |
| pTCMR | 21 | 7 (39%) |
| NR | 905 | 229 (31%) |
| Total | 1679 | 587 (42%) |
v-lesion frequency-v0-1492, v1-55, v2-18, v3-4, No assignment 110.
aThree biopsies had histology diagnoses of both TCMR and BK virus—we have categorized these as TCMR in this table and throughout the article.
b“Others”includes calcineurin inhibitor toxicity, C4d deposition without morphologic evidence for active rejection, donor origin vascular disease, pyelonephritis, systemic infection/diarrhea, and bacterial infection.
AKI, acute kidney injury; AMR, antibody-mediated rejection; BK, polyoma virus; DSA, donor-specific antibody; IFTA, interstitial fibrosis and tubular atrophy; MMDx, Molecular Microscope Diagnostic System; NR, no rejection; TCMR, T-cell–mediated rejection.
Data Collection
Data were collected per standard of care at each participating local center per study protocols, then provided to the study via forms in a REDCap database.
Sample Processing
One 18-gauge biopsy core was placed immediately in RNALater (Thermo Fisher Scientific, Waltham, MA), stored overnight at 4 °C (or stored at 20 °C if longer-term storage was needed). RNA extraction followed established protocols.17 Samples were labeled and hybridized to the PrimeView 219 microarray, according to the manufacturer’s protocols. Microarrays were scanned using the Gene Array Scanner (Affymetrix) and processed with GeneChip operating software version 1.4.0 (Affymetrix). Detailed protocols for microarray processing are available in the online Affymetrix Technical Manual (www.affymetrix.com). MMDx sample processing has been described previously.11,12,15,17–19
Development of Models
All data analyses and modeling were performed using R, version 4.2.1.20 We used the R “rms” and “randomForestSRC” packages21,22 for logistic regression and random forest analyses, respectively. The molecular diagnoses used to train the regression models were based on the MMDx sign-outs, that is, diagnoses assigned by an expert observer using exclusively molecular features (ie, archetypes, classifiers, and transcript set scores). Two rejection definitions were used as the gold standard for predictions: AMR = MMDx AMR/mixed, and TCMR = MMDx TCMR/mixed. Samples called MMDx “mixed” had molecular characteristics of both TCMR and AMR. For both TCMR and AMR, 2 different models were used (generating 4 models total): model 1, including C4d/DSA/panel reactive antibody (PRA) status, and model 2, which excluded C4d/DSA/PRA status.
Ten variables were used in the regression analysis: the histologic g-, ptc-, cg-, v-, i-, and t-lesion scores (all 0/1/2/3); TxBx (entered as log-transformed days posttransplant for logistic regression); and binary (0/1) definitions of C4d, DSA, and PRA. We included HLA antibody (PRA) status because DSA-negative AMR is usually PRA-positive.23 For logistic regression, TxBx was modeled as a restricted cubic spline to handle potential nonlinearity of its relationship with rejection. Missing data were imputed using the multiple imputations by chained equation (“mice”) package.24
Statistics and Model Validation
Variable importance was assessed by ANOVA for the logistic regression models and by permutation for random forests. Importance was standardized to the variable with the highest importance for the relative importance plots. Performance statistics (sensitivities, specificities, etc) were based on the mean predicted scores (AMR and TCMR probabilities) in the out-of-bag samples over 1000 bootstrap (with replacement) iterations. Each biopsy’s predicted scores were therefore averaged over the ~368 times it was in an out-of-bag sample. Likelihood ratio tests were used to compare the final, nested models (eg, model 2 nested within model 1) using the full (nonbootstrapped) data sets.
The final logistic regression equations developed in these analyses are shown in Table S2 (SDC, http://links.lww.com/TP/C864).
Survival Analyses
For the survival curves, we selected 1 random biopsy per transplant. We only selected from biopsies with complete follow-up data (censoring/failure times, N = 1152). Time zero was defined as the time of biopsy, and all input variables, including TxBx, were measured at this time. Survival times were censored at 3 y postbiopsy if the transplant was still functioning.
Diagnostic Categories
For presentation clarity, and comparison between histologic, molecular, and regression results, we grouped the diagnoses into a small number of categories.
We used 6 classes of MMDx assignments: no rejection (NR = all samples without rejection of any type), possible TCMR (pTCMR), TCMR, mixed rejection, possible AMR (pAMR), and AMR. These classes are assigned based on an expert (PFH)’s call after assessing all the results on our MMDx report—a collection of classifiers, transcript set scores, and clustering results.
Histologic diagnoses were NR, AMR, AMR suspected (including TG), TCMR, borderline TCMR, and mixed (TCMR and AMR). For the purposes of these analyses, histologic ABMR suspected, TG, and borderline TCMR were all considered to be nonrejection.
Regression-based diagnoses for AMR and TCMR were made if the respective probabilities from the models were >0.5. When probabilities for both AMR and TCMR were >0.5, mixed rejection was assigned.
RESULTS
Population and Demographics
A total of 1679 biopsies were prospectively collected at participating centers during the INTERCOMEX study as previously described.17 The demographics and clinical features of this cohort are shown in Table 2.
TABLE 2.
Demographics and clinical features of the 1679 biopsy cohort
| Patient demographics | All patients (N = 1381) |
|---|---|
| Mean recipient age (range) | 51 (8–91) |
| Recipient gender male (%) | 761 (63%) |
| Ethnicity | |
| Caucasian | 635 |
| Black | 186 |
| Other | 152 |
| Not availablea | 408 |
| Primary disease | |
| Diabetic nephropathy | 214 |
| Hypertension/large vessel disease | 117 |
| Glomerulonephritis/vasculitis | 412 |
| Interstitial nephritis/pyelonephritis | 93 |
| Polycystic kidney disease | 131 |
| Others | 139 |
| Unknown etiology | 275 |
| Mean donor age (range) | 44 (1–85) |
| Donor gender male (%) | 433 (45%) |
| Donor type (% deceased donor transplants) | 908 (66%) |
| Latest kidney status (% of total) | |
| Functioning graft | 1001 (72%) |
| Graft failure/return to dialysis | 231 (17%) |
| Patient death with functioning graft | 20 (1%) |
| Mean (median) follow-up (functioning grafts) in days | 720 (405) |
| Biopsy data | All biopsies (N = 1679) |
| Median time of biopsy posttransplant in days (range) | 563 (1–12 371) |
| Early biopsies (<1 y) (% total) | 709 (42%)b |
| Late biopsies (≥1 y) (% total) | 966 (57%)b |
Biopsy indication: 1405 for cause biopsies as determined by standard of care by centers, 242 surveillance, and 32 no indication stated.
Missing Banff lesion scores: C4d-308, DSA-285, PRA-338, g-47, ptc-54, cg-50, i-142, t-41, v-110.
aSome centers preferred not to identify ethnicity.
bFour biopsies had no provided date of transplant.
Relative Importance of Banff Biopsy Features for Predicting Rejection
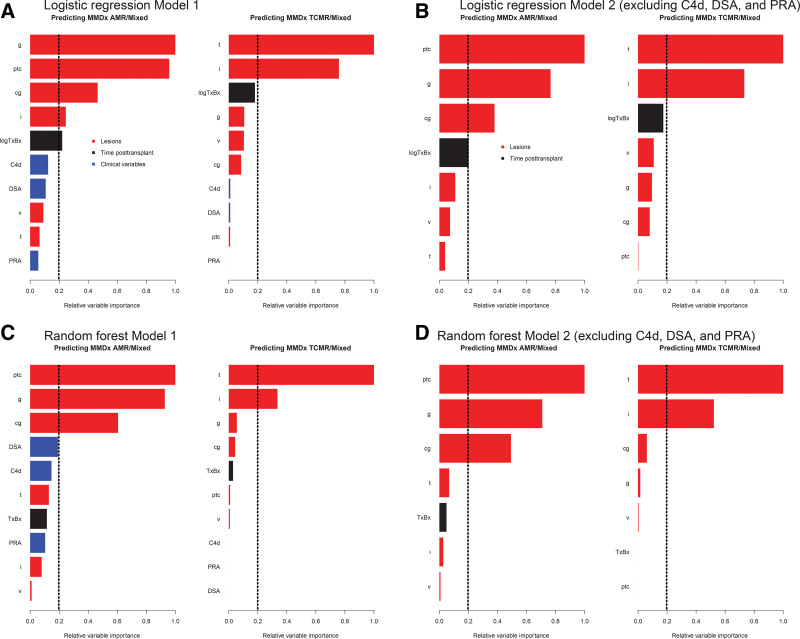
We examined the hierarchy of feature importance in models 1 and 2 using logistic regression (Figure 1A and B) and random forests (Figure 1C and D).
FIGURE 1.
Relative importance of variables in the prediction of rejection using logistic regression, (A) with and (B) without using C4d, PRA, and DSA. C and D, The equivalent random forest models using the same parameters as A and B. Vertical dashed lines designate an arbitrary cutoff for variable significance. AMR, antibody-mediated rejection; cg, transplant glomerulopathy; g, glomerulitis; i, interstitial inflammation; logTxBx, log10 of day of biopsy posttransplant; MMDx, Molecular Microscope Diagnostic System; PRA, panel reactive antibody; ptc, peritubular capillaritis; TCMR, T-cell–mediated rejection; v, arteritis.
In regression AMR model 1 (Figure 1A and C), the highest importance for predicting AMR was for g- and ptc-lesions, with moderate importance of cg-lesions. V-lesions were not important. DSA, PRA, and C4d were also relatively unimportant. For TCMR model 1 (Figure 1A and C), t-lesions were most important, followed by i-lesions and time. V-lesions were again relatively unimportant. (The frequencies of v-lesions are reported in the footnote of Table 1.)
In regression AMR model 2, excluding DSA/PRA/C4d status (Figure 1B and D), the highest importance was again for ptc- and g-lesions, with moderate importance for cg-lesions. For TCMR model 2 (Figure 1B and D), the hierarchy was similar to model 1: t-lesions followed by i-lesions.
Of interest, the error rates for predicting AMR were relatively similar whether C4d and DSA were included or excluded.
Comparing Model 1 and Model 2 Predictions of Rejection Diagnoses
By likelihood ratios, model 1 (ie, using DSA and C4d) was the better model for diagnosing AMR: P = 4.2 × 10−4. Thus, despite similar error rates, AMR diagnoses were improved when C4d and DSA were included. As expected, using DSA and C4d did not significantly improve the TCMR models (P = 0.55, Table 3).
TABLE 3.
Effect of adding DSA, C4d, and time posttransplant on the performance of regression models
| Smaller model | Larger model | Significance of added value when larger model is compared with smaller modela (P) |
|---|---|---|
| AMR model 2 (no C4d/DSA/PRA) | AMR model 1 (with C4d/DSA/PRA) | 4.2 × 10−4 |
| TCMR model 2 (no C4d/DSA/PRA) | TCMR model 1 (with C4d/DSA/PRA) | 0.55 |
| Effect of adding time of biopsy posttransplant to the models | ||
| AMR model 1 | AMR model 1 + time | 3.2 × 10−4 |
| AMR model 2 (no C4d/DSA/PRA) | AMR model 2 + time | 2.2 × 10−4 |
| TCMR model 1 | TCMR model 1 + time | 3.2 × 10−3 |
| TCMR model 2 (no C4d/DSA/PRA) | TCMR model 2 + time | 3.7 × 10−3 |
aLikelihood ratio test comparing the nested models; significant values bolded.
AMR, antibody-mediated rejection; DSA, donor-specific antibody; PRA, all HLA antibody (“panel reactive antibody”); TCMR, T-cell–mediated rejection.
Importance of Time in Predicting Rejection
Likelihood ratio tests comparing full data set models with and without TxBx showed that time significantly improved all models when included: model 1; AMR P = 3.2 × 10−4, and TCMR P = 3.2 × 10−3. For model 2; AMR P = 2.2 × 10−3 and TCMR P = 3.7 × 10−3 (Table 3).
Comparing Models Predicting Molecular Rejection by Banff Diagnoses, Regression, and Combinations
Table 4 shows a variety of estimates of the performance of Banff histology diagnoses and regression models for predicting molecular AMR and TCMR.
TABLE 4.
Comparing performance statistics for Banff histology diagnoses, regression models, and combinations for diagnosing molecular rejection (MMDx diagnosis)
| Diagnosing molecular AMR | Diagnosing molecular TCMR | |||||||
|---|---|---|---|---|---|---|---|---|
| Banff diagnosisa | AMR model 1 | AMR model 2 (omitting C4d, DSA, and PRA) | Combined model using model 2 variables plus Banff diagnosis | Banff diagnosisa | TCMR model 1 | TCMR model 2 (omitting C4d, DSA, and PRA) | Combined model using model 2 variables plus Banff diagnosis | |
| Accuracy | 0.797 | 0.805 | 0.808 | 0.809 | 0.897 | 0.915 | 0.912 | 0.914 |
| Kappa valueb | 0.512 | 0.544 | 0.554 | 0.554 | 0.495 | 0.512 | 0.498 | 0.512 |
| Sensitivity | 0.542 | 0.611 | 0.623 | 0.614 | 0.557 | 0.464 | 0.458 | 0.474 |
| Specificity | 0.931 | 0.906 | 0.906 | 0.911 | 0.941 | 0.974 | 0.971 | 0.971 |
| AUCb | 0.736 | 0.860 | 0.853 | 0.858 | 0.741 | 0.900 | 0.901 | 0.907 |
| Positive predictive value | 0.805 | 0.774 | 0.776 | 0.779 | 0.549 | 0.695 | 0.672 | 0.730 |
| Negative predictive value | 0.795 | 0.816 | 0.821 | 0.818 | 0.943 | 0.934 | 0.933 | 0.936 |
| Balanced accuracy | 0.736 | 0.759 | 0.764 | 0.761 | 0.749 | 0.719 | 0.715 | 0.728 |
aBanff diagnosis defined as a single factor variable with 4 categories: AMR, TCMR, mixed, and no rejection.
bThese rows are bolded because they are emphasized in the text.
AMR, antibody-mediated rejection; AUC, area-under-the-curve; DSA, donor-specific antibody; MMDx, Molecular Microscope Diagnostic System; PRA, panel reactive antibody; TCMR, T-cell–mediated rejection.
For predicting AMR, the kappa values were slightly better for model 1, model 2, and the combined model than for the Banff diagnoses. Model 1 also had better AUCs: model 2 AUC = 0.853, model 1 AUC = 0.860 and Banff diagnosis AUC = 0.736.
For predicting TCMR, the combined model had the highest kappa value. The regression models had comparable AUCs (0.9–0.907), but all were higher than the Banff diagnosis (AUC = 0.741). The AMR regression models had higher sensitivities and lower specificities for predicting AMR than the histologic Banff diagnoses.
Although we acknowledge that the diagnostic accuracy, sensitivity, and specificity are only moderately different between regression and Banff diagnoses, we consider the AUC to be the most important statistic because it represents the degree of separability between other 2 classes (eg, AMR versus non-AMR) integrated over the entire range of possible cutoff values.
Comparison of Regression Diagnostic Classes With Banff Histology Diagnoses
Table 5 compares Banff histology diagnoses as reported by the centers to regression class predictions, in this case using model 2. (The tabulation for model 1 in Table S3 [SDC, http://links.lww.com/TP/C864] shows similar results). There was overall agreement between the regression output and the histologic diagnoses, with some discrepancies. For example, regression found 55 of 887 cases of Banff NR had regression-based AMR, 3 had mixed rejection, and 2 had TCMR.
TABLE 5.
Relationship between diagnoses assigned by model 2 regression equations (using the default cutoff) and Banff histologic diagnoses
| Regression diagnoses | |||||
|---|---|---|---|---|---|
| AMR | Mixed | No rejection | TCMR | Total | |
| Banff histology diagnoses | |||||
| AMR | 261 | 4 | 68 | 0 | 333 |
| Mixed | 38 | 10 | 5 | 3 | 56 |
| No rejection | 55 | 3 | 827 | 2 | 887 |
| pAMR | 50 | 0 | 34 | 0 | 84 |
| pTCMR (borderline) | 7 | 4 | 109 | 8 | 128 |
| TCMR | 13 | 18 | 43 | 65 | 139 |
| BK | 0 | 1 | 38 | 13 | 52 |
| Total | 424 | 40 | 1124 | 91 | 1679 |
Bolding denotes concordance between Banff histology diagnoses and regression diagnoses.
AMR, antibody-mediated rejection; BK, polyoma virus; pAMR, possible AMR; pTCMR, possible TCMR; TCMR, T-cell–mediated rejection.
We assessed how regression assigned the Banff “possible” AMR and Borderline classes. The regression models found that Banff possible AMR was either AMR or NR. In contrast, regression usually interpreted Banff borderline as no rejection.
Effect of Combining Banff Histology Diagnoses With Step 1 Features in a Regression Model
We generated new models that used Banff diagnoses (as a single 4-level factor: AMR, TCMR, mixed, or NR) in addition to the step 1 individual features. Table 6 shows the effect on models predicting molecular rejection of adding Banff diagnoses to the step 1 features and vice versa. The combination of Banff diagnoses with step 1 features always improved the models. However, the impact of adding step 1 input features to Banff diagnoses was far greater than that when adding the Banff diagnoses to the step 1 input features.
TABLE 6.
Effect on model performance of combining the Banff histology diagnosis with the rejection equation input variables
| Inputs for smaller model | Inputs for larger model | Significance of added predictive value when larger model is compared with smaller modela (P) |
|---|---|---|
| AMR model 1 input variables | AMR model 1 input variables + histology diagnosisb | 1.2E-04 |
| AMR model 2 input variables | AMR model 2 input variables + histology diagnosis | 1.6E-08 |
| Histology diagnosis | Histology diagnosis + AMR model 1 input variables | 1.7E-60 |
| Histology diagnosis | Histology diagnosis + AMR model 2 input variables | 2.3E-55 |
| TCMR model 1 input variables | TCMR model 1 input variables + histology diagnosis | 0.003 |
| TCMR model 2 input variables | TCMR model 2 input variables + histology diagnosis | 0.002 |
| Histology diagnosis | TCMR model 1 input variables + histology diagnosis | 1.9E-51 |
| Histology diagnosis | TCMR model 2 input variables + histology diagnosis | 3.0E-51 |
aLikelihood ratio test comparing the nested models; significant values are bolded.
bHistology diagnosis variable with 4 categories: AMR, TCMR, Mixed, and no rejection.
AMR, antibody-mediated rejection; TCMR, T cell–mediated rejection.
Using Lesion-based Regression Equations to Screen for Missed Diagnoses in Biopsies With Banff No Rejection
As an example of the potential utility of adding lesion-based regression scores in conjunction with the Banff system, we studied the impact of regression assessments on the 887 biopsies that Banff histology called NR, as outlined in Table 5. We excluded biopsies with BK because the Banff guidelines recommend that rejection should not be diagnosed in biopsies with BK. Model 2 regression equations interpreted 55 Banff NR biopsies as AMR, 2 as TCMR, and 3 as mixed (Table 7). The results were similar when model 1 was used (data not shown).
TABLE 7.
Effect of considering model 2 regression scores on interpretation of biopsies with no Banff histology rejection (biopsies with BK removed)
| Recorded histology and molecular features in 887 biopsies with no rejection by Banff guidelines | Regression diagnoses | |||
|---|---|---|---|---|
| No rejection (N = 827) | TCMR (N = 2) | Mixed (N = 3) | AMR (N = 55) | |
| Histology lesion scores, plus DSA and C4d | ||||
| TCMR-related | ||||
| t (tubulitis) | 0.11 | 2.00 c | 2.67 c | 0.23 a |
| i (interstitial infiltrate) | 0.22 | 2.00 c | 3.00 c | 0.73 c |
| All rejection–related | ||||
| v (vasculitis) | 0.00 | 0.00 | 0.00 | 0.04 c |
| AMR-related | ||||
| g (glomerulitis) | 0.11 | 0.00 | 0.50 | 1.51 c |
| ptc (capillaritis) | 0.08 | 0.00 | 3.00 c | 1.04 c |
| cg (double contours) | 0.05 | 0.00 | 0.00 | 1.02 c |
| Atrophy-fibrosis-related | ||||
| ci (scarring) | 1.06 | 2.50 a | 1.67 | 1.51 c |
| ct (atrophy) | 1.00 | 2.50 | 1.67 | 1.27 |
| DSA-related | ||||
| DSA positivity | 0.35 | 0.00 | 0.00 | 0.32 |
| C4d-related | ||||
| C4d positivity | 0.06 | – | 0.50 a | 0.14 |
| Transcript set and molecular classifier scores | ||||
| TCMR-related classifiers | ||||
| TCMR classifier (TCMRProb) | 0.04 | 0.21 a | 0.22 b | 0.04 a |
| All rejection–related | ||||
| Rejection classifier (RejProb) | 0.15 | 0.33 | 0.61 a | 0.45 c |
| IFNG-inducible (GRIT3) | 0.39 | 0.87 a | 1.18 b | 0.65 c |
| AMR-related | ||||
| DSA-selective (DSAST) | 0.09 | 0.04 | 0.20 | 0.39 c |
| NK cell burden (NKB) | 0.40 | 0.60 | 0.83 a | 0.87 c |
| AMRd classifier (ABMRProb) | 0.10 | 0.06 | 0.08 | 0.32 c |
Wilcoxon test compared with the no-rejection group (significant values are bolded):
aP < 0.05;
bP < 0.01;
cP < 0.001.
dAMR is used throughout this article per journal style; however, the official classifier name is provided in this table.
AMR, antibody-mediated rejection; BK, polyoma virus; DSA, donor-specific antibody; IFNG, interferon gamma; TCMR, T-cell–mediated rejection.
The Banff NR biopsies that regression models identified as having TCMR, mixed, or AMR all not only had lesions expected for those rejection states but also had molecular abnormalities of the predicted rejection state (Table 7). Thus, Banff NR biopsies with AMR diagnoses by regression had higher mean AMR lesion scores (g-, ptc-, and cg-lesions) and were more likely to be DSA-positive than those with low AMR regression scores. Similarly, high TCMR regression scores predicted high i- or t-lesions. More importantly, relevant molecular features were also increased: Banff NR biopsies that regression called AMR had high AMR-related molecular scores (eg, ABMRProb, g classifier) and those cases regression called TCMR had higher TCMR-related molecular scores (eg, TCMRProb, t classifier). Results for model 1 were similar (data not shown).
Although Table 7 excluded biopsies with BK nephropathy diagnosed by Banff,25 the results were similar when BK was included (Table S4, SDC, http://links.lww.com/TP/C864).
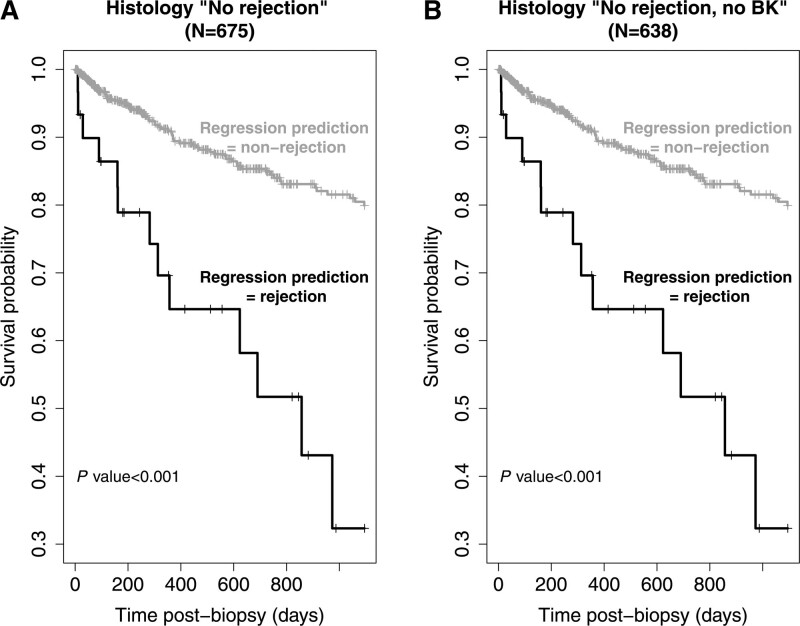
We examined how survival curves were impacted by using regression rejection diagnoses in Banff NR biopsies. The 3 y graft survival of Banff NR samples (NR + BK = 675, 1 random biopsy per patient and N = 638 for NR) split by their regression model rejection status is shown in Figure 2. There were significantly more graft losses within the Banff NR cases that the regression models identified as rejection (P < 0.001, Figure 2A). The results were similar when BK cases were excluded (P < 0.001, Figure 2B). For both analyses, survival in the regression-based nonrejection group was highly significantly better than in the rejection group whatever random sample per transplant was used, as it was when using only the most recent transplant.
FIGURE 2.
Survival curves. Survival 3 y postbiopsy in biopsies called histologic no rejection, split by whether they are called rejection by the regression models (model 2). A, All histologic no rejection samples (N = 675). B, All histologic no rejection excluding BK samples (N = 638). P are by the log-rank test. BK, polyoma virus.
Relating Rejection Predictions by Regression to Risk of Graft Loss in All Biopsies
Given the above evidence that regression scores stratified Banff NR samples into useful subgroups, we looked in all biopsies to see if regression models that used step 1 features added to Banff histology diagnoses would give better predictions of 3 y survival. Models that added step 1 inputs to histologic diagnoses gave significantly improved predictions compared with the histologic diagnoses alone (P = 2.7 × 10−5 for model 1 and 2.0 × 10−5 for model 2). In contrast, adding Banff histologic diagnoses to step 1 lesions did not improve the models (P = 0.85 for regression model 1 and 0.91 for regression model 2). Thus, the regression models reveal information in histology features that can impact clinical risk assessment compared with using the Banff rejection diagnoses alone.
A random forest assessing which features of TCMR and ABMR are more important in a prediction of 3 y graft survival is shown in Figure S1 (SDC, http://links.lww.com/TP/C864). We found that ABMR features (eg, cg, PRA) were the most important for this prediction.
DISCUSSION
Following the belief that machine learning approaches can add value to existing diagnostic systems, we explored whether logistic regression estimates using step 1 features could add value to the current Step 2 guidelines in diagnosing rejection. Having independent molecular assessments for 1679 biopsies allowed us to use the step 1 features (histology lesion scores, C4d, and DSA) in logistic regression and random forest models predicting rejection and defined the hierarchy of relative importance for each step 1 feature. This highlighted the importance of the canonical histologic lesions for TCMR (t-scores and i-scores) and AMR (ptc-, g-, and cg-scores) but also indicated that including TxBx improved the model. Of interest, DSA and C4d status had relatively little value compared with the histologic lesions and TxBx in regression or random forest estimates using step 1 features to predict AMR, suggesting that AMR could be estimated by regression even when C4d and DSA is not available. Thus, logistic regression scores using 10 step 1 features—6 ordinal lesion scores plus TxBx (adding 3 binary predictors for DSA, PRA, and C4d for the model 1s)—provide a richer way of using the step 1 information, particularly when added to the Banff diagnoses.
We believe that the addition of models using step 1 features to the usual Banff-guided assessments offers opportunities to get additional insights from a biopsy by making better use of the histology lesion scores. The logistic regression estimates were in general agreement with the Banff diagnoses and predicted molecular rejection as well as Banff diagnoses in terms of kappa values—better for AMR. However, in Banff NR biopsies, regression scores found some biopsies with subtle rejection-like changes—both molecular and histologic—that had been missed, and improved the prediction of risk of failure. Signing out such cases by their Banff histology diagnosis alone is concerning because it may miss opportunities for increased clinical monitoring and intervention. The NR biopsies that regression indicated had an increased probability of AMR also have an increased frequency of DSA positivity, consistent with the recent finding of subtle AMR-related states in biopsies considered to have NR.16,26 The model 2 regression predictions of AMR also offer the ability to estimate AMR when DSA and C4d are not available, a situation that often arises in the clinic at the time of first biopsy readings.
These results agree with Labriffe et al,10 with similarities in the importance of ptc and g-lesions in their AMR model and similar t- and i-lesion in their TCMR model. The results also expand our earlier regression analyses of step 1 features in a smaller population.8,9
TCMR regression and random forest modeling strongly confirmed tubulitis lesions as the most important variable for the prediction of molecular TCMR.8,27 Tubulitis as a feature has always been strongly associated with interstitial inflammation even in native kidneys, as described by Ooi et al in 1975,28 a correlation that may explain why i-scores are less important than t-scores in modeling.
AMR regression and random forest models confirmed the importance of the Banff ptc-, g-, and cg-lesions but indicated surprisingly low relative importance of C4d and DSA, consistent with the recent recognition that a large proportion of AMR cases can be both DSA-negative and C4d-negative.23,29–34 We previously found a high frequency of DSA-negative molecular AMR in the INTERCOMEX23 and Trifecta studies35 and showed that DSA-negative AMR releases just as donor-derived cell-free DNA as its DSA-positive counterpart. However, such population-wide analyses used DSA as a binary variable—positive versus negative—as specified in the Banff guidelines, which may not use the potential of these technologies optimally. Enhanced granularity for defining DSA for individual cases is available in many centers, including de novo DSA, MFI, titer, specificity, complement binding, IgG subclass, non-HLA antibodies, etc. Such methods need to be standardized and their utility established in multicenter trials.
These results suggest that it is opportune to critically re-examine the interpretation of v-lesions. In our models, v-lesions were always of relatively low importance once the other lesions were considered. In our previous analyses, we explored the complexity of v-lesions, which can occur in TCMR, AMR, or early posttransplant injury and can be particularly difficult to interpret when they occur in isolation.36 The ambiguity of v-lesions creates the potential for serious errors affecting treatment. The addition of regression models to the Banff interpretations may help prevent these errors because they consider the impact of a v-lesion in the context of the other measurements.37
Time (TxBx) added weakly but significantly to the performance of all regression models and should be considered for inclusion in the new iterations of the Banff guidelines. TxBx was of higher relative importance than C4d, PRA, and DSA in the AMR model 1, all features of which are already part of the Banff guidelines for AMR. Although some lesions themselves are associated with time posttransplant, TxBx remained important as a separate variable in our analyses, indicating that some element of this measurement is not captured using time-dependent lesions alone. Additionally, TxBx may be influenced by some clinical aspects of the case, most notably medication nonadherence, which increases over time.38–41
Some limitations to this study include the binary nature of most DSA status reporting by the local center (excluding class or further details), a limited amount of missing data including follow-up for some biopsies, and the size of the TCMR subset in relation to the ABMR subset.
We propose that interested groups such as ourselves and the Labriffe collaborators10 develop simple software apps to make various model-based predictions available to pathologists and clinicians—permitting availability of these assessments within minutes during biopsy interpretation. Consensus about how to add them to the current Banff guidelines should also be sought. This could enhance Banff biopsy interpretation at the local center while adding no cost to the biopsy assessment, and the Banff process could indicate how the output should be used in conjunction with other elements of the patient case, that is, histology, cell-free DNA, or clinical symptoms. Use of regression models through an app could aid the interpretation of clinical problems, even when C4d and DSA are pending, and could be used to decide when to seek molecular assessments. It may also be useful to more closely investigate and monitor biopsies that have been designated NR by the Banff rules but have positive regression scores, given the increased risk for graft failure in this subpopulation. Overall, adding regression models to the current Banff guidelines for interpreting step 1 lesions may improve our clinical management of these patients by noting cases with increased risk, allowing for earlier interpretation when other binary elements (eg, DSA status) are missing, and flagging concerning cases for the clinician and the pathologist.
ACKNOWLEDGMENTS
The authors thank their valued clinicians in the INTERCOMEX study group who partnered with them for this study by contributing biopsies and feedback (Carmen Lefaucheur and Alexandre Loupy).
The INTERCOMEX Investigators are Roslyn Mannon, Daniel Serón, Joanna Sellares, Enver Akalin, Declan de Freitas, Michael Picton, Jonathan Bromberg, Matt Weir, Klemens Budde, Timm Heinbokel, Gunilla Einecke, Harold Yang, Seth Narins, Milagros Samaniego-Picota, Marek Myslak, Agnieszka Perkowska-Ptasinska, Adam Bingaman, Daniel Brennan, Andrew Malone, Bertram Kasiske, Arthur Matas, Arjang Djamali, Georg Böhmig, Farsad Eskandary, and Gaurav Gupta.
Supplementary Material
Footnotes
INTERCOMEX Study Clinical Trial Notation: ClinicalTrials.gov ID NCT01299168.
The list of the INTERCOMEX Investigators is given in the Acknowledgments.
M.L.N.S. contributed to manuscript preparation, discussion of results, and interpretation. J.R. and K.S.M-T. contributed to manuscript writing/reviewing, data analysis, and interpretation. P.F.H. was the principal investigator and contributed to manuscript writing/reviewing, data interpretation, and study design. The INTERCOMEX Investigators contributed to biopsy collection and manuscript reviewing.
P.F.H. holds shares in Transcriptome Sciences Inc (TSI), a University of Alberta research company dedicated to developing molecular diagnostics, supported in part by a licensing agreement between TSI and Thermo Fisher Scientific, and by a research grant from Natera Inc. P.F.H. is a consultant to Natera Inc. The other authors declare no conflicts of interest.
The Microarray biopsy assessment project is supported in part by a licensing agreement with One Lambda/Thermo Fisher Scientific. This research has been principally supported by grants from Genome Canada, Canada Foundation for Innovation, the University of Alberta Hospital Foundation, the Alberta Ministry of Advanced Education and Technology, the Mendez National Institute of Transplantation Foundation, and Industrial Research Assistance Program. Partial support was also provided by funding from a licensing agreement with the One Lambda division of Thermo Fisher Scientific. P.F.H. held a Canada Research Chair in Transplant Immunology until 2008 and currently holds the Muttart Chair in Clinical Immunology.
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
CEL files will be available on the Gene Expression Omnibus website (GSE124203).
Contributor Information
Collaborators: Roslyn Mannon, Daniel Serón, Joanna Sellares, Enver Akalin, Declan de Freitas, Michael Picton, Jonathan Bromberg, Matt Weir, Klemens Budde, Timm Heinbokel, Gunilla Einecke, Harold Yang, Seth Narins, Milagros Samaniego-Picota, Marek Myslak, Agnieszka Perkowska-Ptasinska, Adam Bingaman, Daniel Brennan, Andrew Malone, Bertram Kasiske, Arthur Matas, Arjang Djamali, Georg Böhmig, Farsad Eskandary, and Gaurav Gupta
REFERENCES
- 1.Mengel M, Loupy A, Haas M, et al. Banff 2019 Meeting Report: molecular diagnostics in solid organ transplantation-consensus for the Banff Human Organ Transplant (B-HOT) gene panel and open source multicenter validation. Am J Transplant. 2020;20:2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nankivell BJ, Agrawal N, Sharma A, et al. The clinical and pathological significance of borderline T cell-mediated rejection. Am J Transplant. 2019;19:1452–1463. [DOI] [PubMed] [Google Scholar]
- 3.Becker JU, Chang A, Nickeleit V, et al. Banff borderline changes suspicious for acute t cell-mediated rejection: where do we stand? Am J Transplant. 2016;16:2654–2660. [DOI] [PubMed] [Google Scholar]
- 4.Feinstein AR. The inadequacy of binary models for the clinical reality of three-zone diagnostic decisions. J Clinl Epidemiol. 1990;43:109–113. [DOI] [PubMed] [Google Scholar]
- 5.Decruyenaere A, Decruyenaere P, Peeters P, et al. Prediction of delayed graft function after kidney transplantation: comparison between logistic regression and machine learning methods. BMC Med Inform Decis Mak. 2015;15:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lasserre J, Arnold S, Vingron M, et al. Predicting the outcome of renal transplantation. J Am Med Inform Assn. 2011;19:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krikov S, Khan A, Baird BC, et al. Predicting kidney transplant survival using tree-based modeling. ASAIO J. 2007;53:592–600. [DOI] [PubMed] [Google Scholar]
- 8.Reeve J, Chang J, Salazar ID, et al. Using molecular phenotyping to guide improvements in the histologic diagnosis of t cell-mediated rejection. Am J Transplant. 2016;16:1183–1192. [DOI] [PubMed] [Google Scholar]
- 9.Halloran PF, Famulski KS, Chang J. A Probabilistic approach to histologic diagnosis of antibody-mediated rejection in kidney transplant biopsies. Am J Transplant. 2017;17:129–139. [DOI] [PubMed] [Google Scholar]
- 10.Labriffe M, Woillard JB, Gwinner W, et al. Machine learning-supported interpretation of kidney graft elementary lesions in combination with clinical data. Am J Transplant. 2022;22:2821–2833. [DOI] [PubMed] [Google Scholar]
- 11.Madill-Thomsen K, Perkowska-Ptasinska A, Bohmig GA, et al. ; MMDx-Kidney Study Group. Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant. 2020;20:1341–1350. [DOI] [PubMed] [Google Scholar]
- 12.Reeve J, Bohmig GA, Eskandary F, et al. ; INTERCOMEX MMDx-Kidney Study Group. Generating automated kidney transplant biopsy reports combining molecular measurements with ensembles of machine learning classifiers. Am J Transplant. 2019;19:2719–2731. [DOI] [PubMed] [Google Scholar]
- 13.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; Trifecta Investigators. The trifecta study: comparing plasma levels of donor-derived cell-free DNA with the molecular phenotype of kidney transplant biopsies. J Am Soc Nephrol. 2022;33:387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor-derived cell-free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation. 2022;106:1061–1070. [DOI] [PubMed] [Google Scholar]
- 15.Reeve J, Bohmig GA, Eskandary F, et al. ; MMDx-Kidney study group. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight. 2017;2:e94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madill-Thomsen KS, Bohmig GA, Bromberg J, et al. ; INTERCOMEX Investigators. Donor-specific antibody is associated with increased expression of rejection transcripts in renal transplant biopsies classified as no rejection. J Am Soc Nephrol. 2021;32:2743–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: the INTERCOMEX study. Am J Transplant. 2017;17:2851–2862. [DOI] [PubMed] [Google Scholar]
- 18.Halloran P, Reeve J, Grp IS. Real time assessment of kidney transplant indication biopsies by microarrays: first results of the INTERCOMEX study. Am J Transplant. 2016;16:796–796. [DOI] [PubMed] [Google Scholar]
- 19.Madill-Thomsen KS, Wiggins RC, Eskandary F, et al. The effect of cortex/medulla proportions on molecular diagnoses in kidney transplant biopsies: rejection and injury can be assessed in medulla. Am J Transplant. 2017;17:2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (2019) RCT. R: A language and environment for statistical computing. Available at http://www.r-project.org/. Accessed June 9, 2023. [Google Scholar]
- 21.rms: Regression Modeling Strategies. R package version 6.0-0. Available at https://CRAN.R-project.org/package=rms. Accessed June 9, 2023.
- 22.Ishwaran H, Kogalur UB. Fast Unified Random Forests for Survival, Regression, and Classification (RF-SRC). R package version 3.2.2. Available at https://cran.r-project.org/package=randomForestSRC. Accessed June 9, 2023.
- 23.Halloran PF, Madill-Thomsen KS, Pon S, et al. ; INTERCOMEX Investigators. Molecular diagnosis of ABMR with or without donor-specific antibody in kidney transplant biopsies: Differences in timing and intensity but similar mechanisms and outcomes. Am J Transplant. 2022;22:1976–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Buuren S, Groothuis-Oudshoorn K. mice: multivariate imputation by chained equations in R. J Statistical Software. 2011;45:1–67. [Google Scholar]
- 25.Halloran PF, Madill-Thomsen KS, Bohmig GA, et al. ; INTERCOMEX Investigators. A 2-fold approach to polyoma virus (BK) nephropathy in kidney transplants: distinguishing direct virus effects from cognate T cell-mediated Inflammation. Transplantation. 2021;105:2374–2384. [DOI] [PubMed] [Google Scholar]
- 26.Rosales IA, Mahowald GK, Tomaszewski K, et al. Banff human organ transplant transcripts correlate with renal allograft pathology and outcome: importance of capillaritis and subpathologic rejection. J Am Soc Nephrol. 2022;33:2306–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solez K, Axelsen RA, Benediktsson H, et al. International standardization of criteria for the histologic diagnosis of renal allograft rejection: the Banff working classification of kidney transplant pathology. Kidney Int. 1993;44:411–422. [DOI] [PubMed] [Google Scholar]
- 28.Ooi BS, Jao W, First MR, et al. Acute interstitial nephritis. A clinical and pathologic study based on renal biopsies. Am J Med. 1975;59:614–628. [DOI] [PubMed] [Google Scholar]
- 29.Koenig A, Mezaache S, Callemeyn J, et al. Missing self-induced activation of NK cells combines with non-complement-fixing donor-specific antibodies to accelerate kidney transplant loss in chronic antibody-mediated rejection. J Am Soc Nephrol. 2021;32:479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callemeyn J, Lamarthée B, Koenig A, et al. Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 2022;101:692–710. [DOI] [PubMed] [Google Scholar]
- 31.Callemeyn J, Lerut E, de Loor H, et al. Transcriptional changes in kidney allografts with histology of antibody-mediated rejection without anti-HLA donor-specific antibodies. J Am Soc Nephrol. 2020;31:2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senev A, Coemans M, Lerut E, et al. Histological picture of antibody-mediated rejection without donor-specific anti-HLA antibodies: clinical presentation and implications for outcome. Am J Transplant. 2019;19:763–780. [DOI] [PubMed] [Google Scholar]
- 33.Sablik KA, Clahsen-van Groningen MC, Looman CWN, et al. Chronic-active antibody-mediated rejection with or without donor-specific antibodies has similar histomorphology and clinical outcome - a retrospective study. Transplant Int. 2018;31:900–908. [DOI] [PubMed] [Google Scholar]
- 34.Delville M, Lamarthee B, Pagie S, et al. Early acute microvascular kidney transplant rejection in the absence of anti-HLA antibodies is associated with preformed IgG antibodies against diverse glomerular endothelial cell antigens. J Am Soc Nephrol. 2019;30:692–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halloran PF, Reeve J, Madill-Thomsen KS, et al. ; the Trifecta Investigators. Antibody-mediated rejection without detectable donor-specific antibody releases donor-derived cell-free DNA: results from the Trifecta Study [published correction appears in Transplantation. 2023 Jan 1;107(1):e43]. Transplantation. 2023;107:709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar IDR, Lopez MM, Chang J, et al. Reassessing the significance of v-lesions in kidney transplant biopsies. J Am Soc Nephrol. 2015;26:3190–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar ID, Merino Lopez M, Chang J, et al. Reassessing the significance of intimal arteritis in kidney transplant biopsy specimens. J Am Soc Nephrol. 2015;26:3190–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couzi L, Moulin B, Morin MP, et al. Factors predictive of medication nonadherence after renal transplantation: a French observational study. Transplantation. 2013;95:326–332. [DOI] [PubMed] [Google Scholar]
- 39.De Geest S, Burkhalter H, Bogert L, et al. Describing the evolution of medication nonadherence from pretransplant until 3 years post-transplant and determining pretransplant medication nonadherence as risk factor for post-transplant nonadherence to immunosuppressives: The Swiss Transplant Cohort Study. Transplant Int. 2014;27:657–666. [DOI] [PubMed] [Google Scholar]
- 40.Tsapepas D, Langone A, Chan L, et al. A longitudinal assessment of adherence with immunosuppressive therapy following kidney transplantation from the Mycophenolic Acid Observational REnal Transplant (MORE) study. Ann Transplant. 2014;19:174–181. [DOI] [PubMed] [Google Scholar]
- 41.Nevins TE, Robiner WN, Thomas W. Predictive patterns of early medication adherence in renal transplantation. Transplantation. 2014;98:878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.