Abstract
Background:
Highly expressed in various human cancers, circular RNA Protein Kinase C Iota (circPRKCI) has been reported to play an important role in cancer development and progression. Herein, we sought to reveal the prognostic and clinical value of circPRKCI expression in diverse human cancers.
Methods:
We searched the Pubmed, Web of Science, and the Cochrane Library databases from inception until May 16, 2021. The relationship between circPRKCI expression and cancer patients' survival, including overall survival (OS) and disease-free survival (DFS), was assessed by pooled hazard ratios (HR) with corresponding 95% confidence interval (CI). The correlation between circPRKCI expression and clinical outcomes was evaluated using odds ratios (OR) with corresponding 95% CI. The data were analyzed by STATA software (version 12.0) or Review Manager (RevMan 5.3).
Results:
A total of 15 studies with 1109 patients were incorporated into our meta-analysis. The results demonstrated that high circPRKCI expression was significantly related to poor OS (HR = 1.96, 95% CI: 1.61, 2.39, P <0.001) when compared with low circPRKCI expression in diverse human cancers. However, elevated circPRKCI expression was not associated with DFS (HR = 1.34, 95% CI: 0.93, 1.95, P = 0.121). Furthermore, the patient with a higher circPRKCI expression was prone to have a larger tumor size, advanced clinical stage, and lymph node metastasis, but it was not significantly correlated with age, gender, and distant metastasis.
Conclusion:
Elevated circPRKCI expression was correlated with worse OS and unfavorable clinical features, suggesting a novel prognostic and predictive role of circPRKCI in diverse human cancers.
Keywords: CircPRKCI, Cancer, Prognosis, Overall survival, Meta-analysis, Circular RNA Protein Kinase C Iota
Introduction
Circular ribonucleic acids (CircRNAs) were first identified as plant viroids in 1976. With the development of high-throughput ribonucleic acid (RNA) sequencing technology and bioinformatics, circRNAs have been established as a common feature of the human transcriptome, and are widely present in many other species.[1,2] However, the mechanism of circRNAs formation remains unclear.[3] Substantial evidence demonstrates that they have unique expression profiles and important biological functions in a variety of diseases, including tumors,[4–6] cardiovascular diseases,[7] degenerative disease,[8] and autoimmune diseases.[9] However, its mechanism has not been fully elucidated, although it is widely known to exert the biological functions by acting as a microRNA (miRNA) sponge. CircRNAs are characterized by a highly conserved and unique covalently closed loop structure without a free 5΄ cap and 3΄ tail, which differentiates them from other non-coding RNAs such as long non-coding RNA (lncRNA) and miRNA, and can avoid degradation by exonuclease. An increasing body of studies has shown the potential value of circRNAs in the diagnosis and prognosis of tumors.[10–12] Notably, circRNAs could regulate cell proliferation,[13] metastasis,[14] and drug resistance [15,16] in tumors, and thus play diverse roles in tumorigenesis and development.
Circular RNA Protein Kinase C Iota (CircPRKCI) is a transcription product of the protein kinase C Iota (PRKCI) gene mapped to chromosome 3q26.2 that has been identified as a novel oncogenic modulator in several human cancers.[17,18] Recently, CircPRKCI has been reported to be associated with disease progression in lung cancer (LC),[19] thyroid cancer (TC),[20] hepatocellular carcinomas (HCCs),[18] laryngeal cancer,[21] glioma,[22] and gastric cancer (GC).[23] Substantial evidence indicated that circPRKCI exerts oncogenic functions to facilitate cancer progression and predicts a poor prognosis for cancer patients. However, some studies that assessed circPRKCI expression in cancers could be biased by limited sample size. Accordingly, we performed this cogent meta-analysis with all related eligible studies to further investigate the feasibility of circPRKCI as a potential biomarker candidate in human cancers.
Methods
Publication search strategy
This study was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. The Pubmed, Web of Science, and the Cochrane Library databases were retrieved for potential publications from inception until May 16, 2021 by two independent authors (ZYL and XLR). The search strategies used for Pubmed, Web of Science, and the Cochrane Library were "(((((((((circPRKCI) OR (circ_PRKCI)) OR (circ-PRKCI)) OR (circRNA PRKCI)) OR (circular RNA PRKCI)) OR (circ_103510)) OR (circ_0067934)) OR (circRNA_0067934)) OR (Hsa_circRNA_0067934)) OR (Circular RNA hsa_circRNA_0067934)", "(((((((((TS = (circPRKCI)) OR TS = (circ_PRKCI)) OR TS = (circ-PRKCI)) OR TS = (circRNA PRKCI)) OR TS = (circular RNA PRKCI)) OR TS = (circ_103510)) OR TS = (circ_0067934)) OR TS = (circRNA_0067934)) OR TS = (Hsa_circRNA_0067934)) OR TS = (Circular RNA hsa_circRNA_0067934)", and "(circPRKCI) OR (circ_PRKCI) OR (circ-PRKCI) OR (circRNA PRKCI) OR (circular RNA PRKCI) OR (circ_103510) OR (circ_0067934) OR (circRNA_0067934) OR (Hsa_circRNA_0067934) OR (Circular RNA hsa_circRNA_0067934)", respectively. Additionally, references in included publications were manually searched for relevant articles.
Inclusion and exclusion criteria
All enrolled studies fulfilled the following inclusion criteria: (1) original articles on human beings; (2) definite diagnosis of cancer by histopathology; (3) studies related to the prognosis or clinical significance of circPRKCI; (4) circPRKCI were divided into high and low groups according to its relative expression levels; and (5) studies with sufficient data about the relationship between circPRKCI expression level and overall survival (OS), disease-free survival (DFS), or clinical features.
The exclusion criteria: (1) duplicate records; (2) studies irrelevant to circPRKCI or cancers; (3) reviews, meta-analysis, and comments; (4) studies that lack prognostic or clinical data; (5) studies where data could not be extracted; and (6) retracted articles.
Data extraction and quality assessment
Two researchers (ZYL and XLR) independently retrieved the eligible studies. The following information was extracted from the enrolled studies: (1) the baseline information of the studies, including first author's name, published year, cancer type, number of samples, cut-off value, follow-up interval, survival analysis method, outcome measure method and the Newcastle–Ottawa Scale (NOS) score, and so on; (2) the related clinical characteristics, including age, gender, tumor size, clinical stage, distant metastasis (DM), and lymph node metastasis (LNM); and (3) hazard ratios (HR) with corresponding 95% confidence interval (CI) of circPRKCI for OS and DFS. The software Engauge Digitizer (version 4.1, http://markummitchell.github.io/engauge-digitizer/) was used to estimate the HR with corresponding 95% CI based on Kaplan–Meier curve (KM curve) if these data were not directly reported in the studies. The NOS score was adopted to measure the quality of the eligible literature.
Data synthesis and statistical analysis
HR or odds ratios (OR) with their corresponding 95% CI were used to evaluate the correlation between circPRKCI expression and OS or clinical features by STATA software (version 12.0, StataCorp, College Station, Texas, USA) or Review Manager (RevMan 5.3, Cochrane Collaboration, Oxford, UK), respectively. The heterogeneity among the enrolled studies was assessed by the chi-squared test and I2 statistics. A random-effect model was adopted when the heterogeneity among these studies was significant (P <0.05, I2 >50%). Otherwise, a fixed-effect model was applied. Sensitivity analysis was conducted by omitting one study at a time to verify the consistency of the result. Begg's and Egger's tests were used to detect the potential publication bias and the corresponding results were presented in funnel plot.
Results
Selection and description of enrolled studies
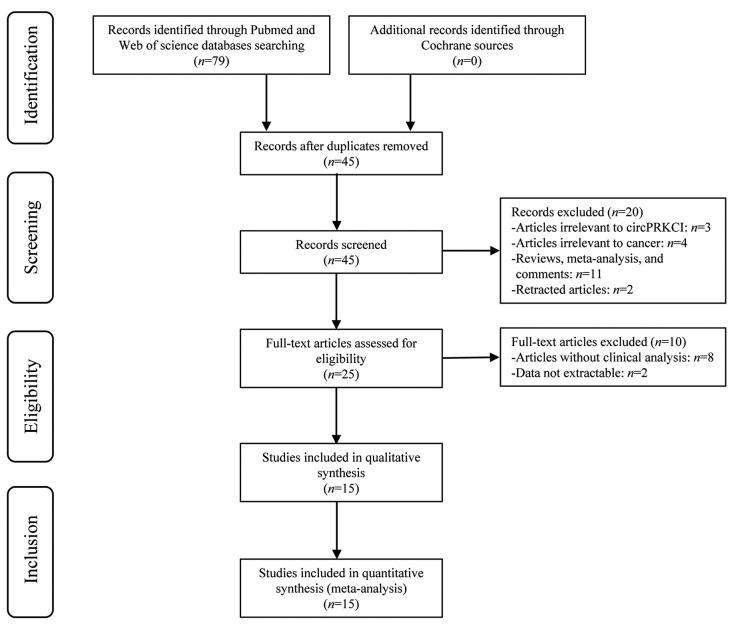
Overall, 79 published articles were initially enrolled in this meta-analysis, and 45 remained after duplicates were removed. Then, titles and abstracts were carefully screened; twenty publications were excluded as reviews, meta-analyses, comments (n = 11), studies on irrelevant genes (n = 3) or diseases (n = 4), and retracted articles (n = 2). Subsequently, ten out of the remaining full-text publications (n = 25) were removed due to the lack of clinical data (n = 8) and data that could not be extracted (n = 2). Finally, 15 articles involving 1109 patients were ultimately enrolled in this study. The flowchart of the selection process is shown in Figure 1.
Figure 1.
Flowchart of the study selection process.
As shown in Table 1, all studies were published from 2018 to 2021, with a follow-up interval ranging from 56 to 84 months. There were eight kinds of cancers among the included 15 studies, including five about LC (two of lung adenocarcinoma [LUAC] and two of non-small cell lung cancer [NSCLC]), three about TC, two about HCC, one about bladder cancer (BC), one about cervical cancer (CC), one about glioblastoma (GBM), one about GC, and one about laryngeal squamous cell cancer (LSCC). The circPRKCI expression was detected by quantitative real-time polymerase chain reaction (qRT-PCR) with the sample size ranging from 40 to 159, 8 of which adopted the median expression of circPRKCI as the cut-off value while the cut-off value of the other 7 were not available (NA). A total of 14 out of the 15 publications evaluated the correlation between circPRKCI expression and prognosis (fourteen and two studies provided the OS and DFS, respectively), and eight explored the relationship between circPRKCI expression and clinical characteristics such as tumor size, clinical stage, and metastasis by univariate or multivariate analysis. Among these 14 prognostic studies, the raw data of three studies were directly presented while the related data from the other 11 studies were extracted from KM curves in the absence of raw data. NOS scores of the enrolled studies were not inferior to 6, indicating the high quality of these studies [Supplementary Table 1, http://links.lww.com/CM9/B713].
Table 1.
Main characteristics of the included studies on circPRKCI as a potential biomarker candidate in human cancers.
| Author | Year | Country | Cancer type | Clinical stage | Sample size | Cut-off value | Follow-up interval (months) | Detection method | Adjuvant therapy | Survival analysis | Outcome measure | Source of HR/CI | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chu et al[21] | 2020 | China | LSCC | – | 40 | Median | 80 | qRT–PCR | None | Univariate | OS and CP | KM curve | 8 |
| Hu et al[24] | 2019 | China | CC | I–II | 61 | Median | 60 | qRT–PCR | None | Univariate | OS | KM curve | 7 |
| Liu et al[25] | 2020 | China | BC | – | 54 | Median | 56 | qRT–PCR | None | Univariate | OS, DFS, and CP | KM curve | 6 |
| Liu et al[20] | 2021 | China | PTC | I–III | 48 | Median | – | qRT–PCR | None | Univariate | CP | – | 6 |
| Meng et al[19] | 2020 | China | LC | – | 60 | N/A | 60 | qRT–PCR | None | Univariate | OS | KM curve | 8 |
| Qiu et al[17] | 2018 | China | LUAC | I–III | 89 | N/A | 84 | qRT–PCR | N/A | Multivariate | OS and CP | KM curve | 9 |
| Sui et al[26] | 2021 | China | LUAC | I–IV | 50 | N/A | 60 | qRT–PCR | None | Univariate | OS | KM curve | 8 |
| Wang et al[27] | 2019 | China | TC | I–IV | 57 | Median | 72 | qRT–PCR | N/A | Univariate | OS and CP | Raw data | 8 |
| Wang et al[28] | 2018 | China | NSCLC | I–IV | 159 | Median | 60 | qRT–PCR | None | Multivariate | OS and CP | Raw data | 9 |
| Wu et al[29] | 2019 | China | GC | I–IV | 50 | N/A | 60 | qRT–PCR | N/A | Univariate | OS | KM curve | 8 |
| Xin et al[30] | 2019 | China | GBM | – | 157 | N/A | 60 | qRT–PCR | None | Univariate | OS and DFS | KM curve | 8 |
| Zhang et al[31] | 2019 | China | TC | I–IV | 50 | Median | 60 | qRT–PCR | None | Univariate | OS | KM curve | 8 |
| Zhou et al[18] | 2020 | China | HCC | I–IV | 106 | N/A | 60 | qRT–PCR | None | Univariate | OS and CP | KM curve | 8 |
| Zhu et al[32] | 2018 | China | HCC | I–III | 49 | N/A | 60 | qRT–PCR | None | Univariate | OS | KM curve | 8 |
| Zou et al[33] | 2018 | China | NSCLC | I–IV | 79 | Median | 80 | qRT–PCR | None | Multivariate | OS and CP | Raw data | 9 |
BC: Bladder cancer; CC: Cervical cancer; CP: Clinical parameters; DFS: Disease-free survival; GBM: Glioblastoma; GC: Gastric cancer; HCC: Hepatocellular carcinoma; HR/CI: Hazard ratios with corresponding 95% confidence interval; KM curve: Kaplan–Meier curve; LC: Lung cancer; LSCC: Laryngeal squamous cell cancer; LUAC: Lung adenocarcinoma; N/A: Not available; NOS: Newcastle–Ottawa Scale; NSCLC: Non-small cell lung cancer; OS: Overall survival; PTC: Papillary thyroid cancer; qRT-PCR: Quantitative real-time polymerase chain reaction; TC: Thyroid cancer; –: Not available.
Association between circPRKCI expression and prognosis
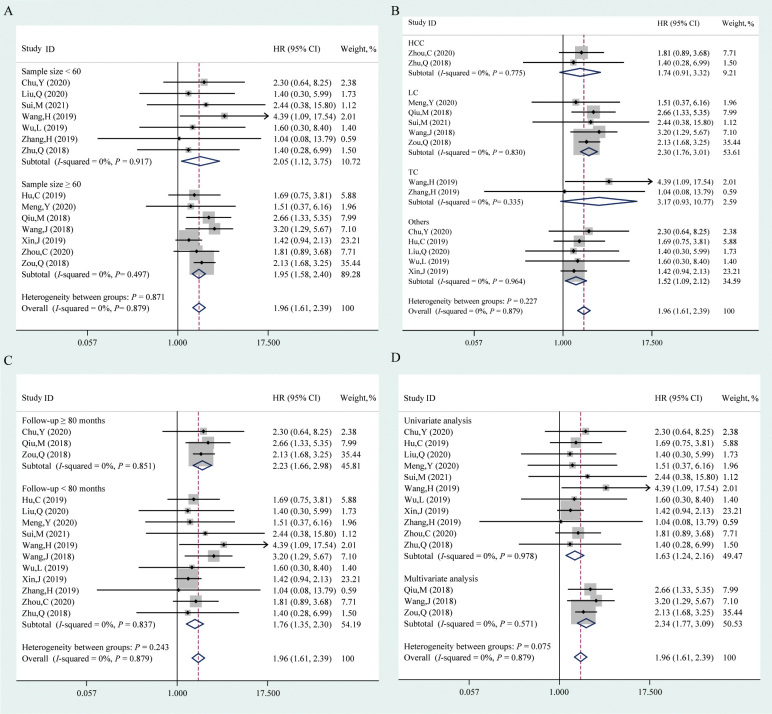
In this study, the pooled OS was analyzed from 14 studies comprising 1061 patients and the association between circPRKCI expression and DFS was assessed in two studies, including 211 patients. As shown in Figure 2, the fixed-effect model was adopted to analyze the HR and corresponding 95% CI given the absence of heterogeneity (I2 = 0, P = 0.762). High circPRKCI expression correlated with a poor OS (HR = 1.96, 95% CI: 1.61, 2.39, P <0.001); however, the association between elevated circPRKCI expression and DFS was not robust (HR = 1.34, 95% CI: 0.93, 1.95, P = 0.121).
Figure 2.
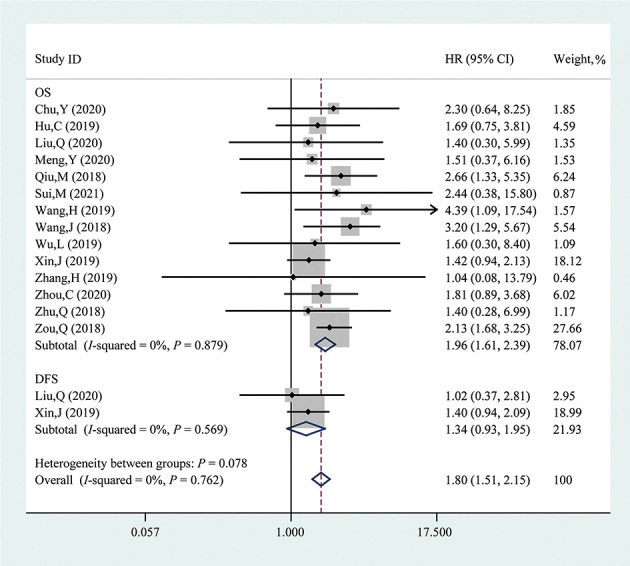
Forest plot evaluating the association between circPRKCI expression and OS or DFS. The dotted line in the figure means the hazard ratio of the overall pooled result while the rhombuses in the figure mean the hazard ratios and the 95% CIs of the pooled results (overall and subtotal). circPRKCI: Circular RNA Protein Kinase C Iota; CI: Confidence interval; DFS: Disease-free survival; HR: Hazard ratio; ID: Identity document; OS: Overall survival.
Subgroup analyses were further performed based on sample size (<60 and ≥60), cancer type (HCC, LC, TC, and others), follow-up interval (≥80 and <80 months), and survival analysis method (univariate and multivariate) [Figure 3]. CircPRKCI was found to be a poor prognostic factor in studies with a sample size less than 60 (HR = 2.05, 95% CI: 1.12, 3.75, P = 0.019) or no less than 60 (HR = 1.95, 95% CI: 1.58, 2.40, P <0.001). After stratifying based on cancer type, higher circPRKCI expression indicated a worse OS in LC (HR = 2.30, 95% CI: 1.76, 3.01, P <0.001) and other cancers (HR = 1.52, 95% CI: 1.09, 2.12, P = 0.015), but not in HCC (HR = 1.74, 95% CI: 0.91, 3.32, P = 0.096) or TC (HR = 3.17, 95% CI: 0.93, 10.77, P = 0.065). Of note, the results indicated a significant positive relationship between the elevated circPRKCI expression and poor OS in studies with a follow-up interval of more than or equal to (HR = 2.23, 95% CI: 1.66, 2.98, P <0.001) or less than (HR = 1.76, 95% CI: 1.35, 2.30, P <0.001) 80 months. Moreover, the high circPRKCI expression predicted an unfavorable OS with univariate analysis (HR = 1.63, 95% CI: 1.24, 2.16, P = 0.001) and multivariate analysis (HR = 2.34, 95% CI: 1.77, 3.09, P <0.001). The detailed information is provided in Supplementary Table 2, http://links.lww.com/CM9/B713.
Figure 3.
Forest plots of the subgroups analysis evaluating the correlation between circPRKCI expression and OS, including sample size (A), cancer type (B), follow-up interval (C), and survival analysis method (D). The dotted lines in the figure mean the hazard ratios of the overall pooled results, while the rhombuses in the figure mean the hazard ratios and the 95% CIs of the pooled results (overall and subtotal). circPRKCI: Circular RNA Protein Kinase C Iota; CI: Confidence interval; HCC: Hepatocellular carcinoma; HR: Hazard ratio; ID: Identity document; LC: Lung cancer; OS: Overall survival; TC: Thyroid cancer.
Association between circPRKCI expression and clinical characteristics
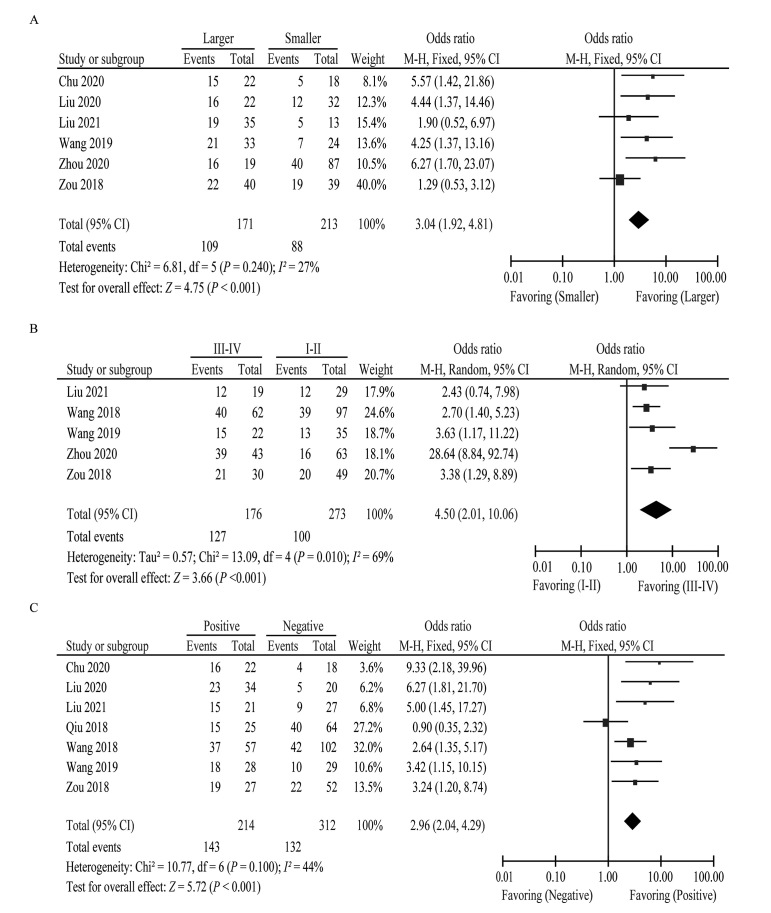
We further assessed the correlation between high circPRKCI expression and clinical characteristics involving age, gender, tumor size, clinical stage, DM, and LNM. Six studies consisting of 384 patients explored the association between circPRKCI expression and tumor size. The pooled estimates indicated that elevated circPRKCI expression correlated with larger tumor size (OR = 3.04, 95% CI: 1.92, 4.81, P <0.001) [Figure 4]. Similarly, upregulated circPRKCI expression was positively correlated with an advanced clinical stage (OR = 4.50, 95% CI: 2.01, 10.06, P <0.001) and higher risk of LNM (OR = 2.96, 95% CI: 2.04, 4.29, P <0.001). However, more evidence is needed to assess the association between abnormal expression of circPRKCI and age (OR = 1.29, 95% CI: 0.89, 1.86, P = 0.180), gender (OR = 1.16, 95% CI: 0.82, 1.62, P = 0.410), and DM (OR = 2.22, 95% CI: 0.62, 7.92, P = 0.220) [Supplementary Figure 1, http://links.lww.com/CM9/B713]. The relevant information is presented in Supplementary Table 3, http://links.lww.com/CM9/B713.
Figure 4.
Forest plots of the association between circPRKCI expression and clinical characteristics, including tumor size (A), clinical stage (B), and LNM (C). circPRKCI: Circular RNA Protein Kinase C Iota; CI: Confidence interval; LNM: Lymph node metastasis; M-H: Mantel–Haenszel (a statistical method).
Sensitivity analysis and publication bias
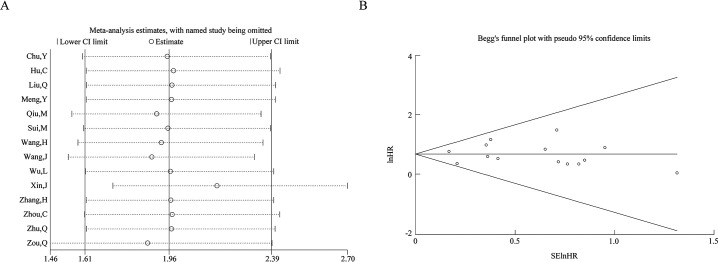
Sensitivity analysis was conducted by calculating the HR with corresponding 95% CI via omitting each study, and the results suggested the robustness of the correlation between circPRKCI expression and OS [Figure 5A].
Figure 5.
Sensitivity analysis (A) and funnel plot for publication bias (B) for circPRKCI on OS. circPRKCI: Circular RNA Protein Kinase C Iota; CI: Confidence interval; lnHR: ln (hazard ratio); SElnHR: Standard error of ln (hazard ratio).
Begg's funnel plot and Egger's tests were applied to explore the publication bias, and the symmetrically distributed funnel plot suggested the absence of potential publication bias [Figure 5B]. Moreover, the Begg's and Egger's tests (P = 0.274 and P = 0.888, respectively) exhibited no publication bias.
Regulation mechanism of circPRKCI in cancers
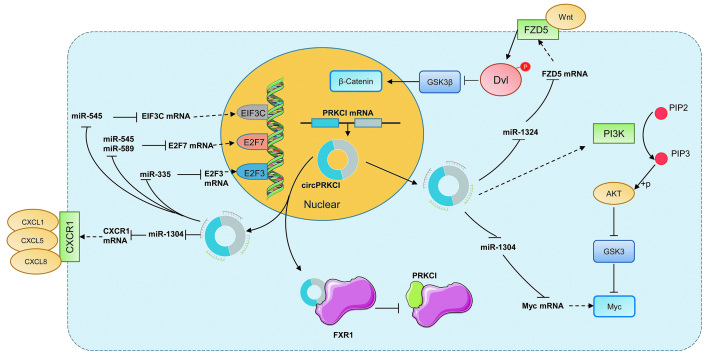
Recently, multiple studies have reported the potential mechanism of circPRKCI in cell proliferation, apoptosis, migration, invasion, and metabolism in a wide spectrum of cancers. The competing endogenous RNA (ceRNA) mechanism has been shown to regulate cancer development [Table 2]. It was reported that circPRKCI was remarkably elevated in BC and played a tumor-promoting role. CircPRKCI was reported to promote BC cell proliferation, migration, and invasion via the miR-1304/Myc axis.[25] Moreover, the circPRKCI level was significantly increased in LC tissues and exhibited various important mechanisms in cancer prognosis. CircPRKCI has been shown to modulate the invasiveness and proliferation of LC through binding to miR-1324 via promoting MECP2 expression.[19] In NSCLC, it was shown that circPRKCI promoted proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT) and inhibited apoptosis by sponging miR-1182 to increase KLF8 expression to activate the Wnt/β-catenin pathway.[34] CircPRKCI has been demonstrated to promote the proliferation and colony formation of LUAC by adsorbing both miR-545 and miR-589 in a sponge-like manner to abolish their suppressive effect on the protumorigenic transcription factor E2F7.[17] Meanwhile, circPRKCI regulated proliferation, migration, and cell cycle progression of LUAC cells by targeting miR-219a-5p regulated CAMK1D.[26] In addition, circPRKCI could bind to the cellular protein directly and interfere with its function. It has been reported that circPRKCI decoyed the RNA-binding protein FXR1 to interrupt the formation of the FXR1/PRCKI complex to facilitate the tumorigenic capacity in LC and decrease cell invasion and drug resistance [Figure 6].[35] Furthermore, circPRKCI has been documented as an oncogene that can promote CC progression through the miR-545/EIF3C and miR-765/HMGA1 axis.[24,36] Importantly, circPRKCI promoted proliferation, colony formation, and cell cycle progression and decreased radiosensitivity by sponging miR-186-5p to upregulate PARP9 expression.[37] Moreover, the circPRKCI/miR-1290/FOXC1 axis,[38] circPRKCI/miR-545/AKT3 axis,[39] circPRKCI/miR-1324/FZD5 axis,[32] and circPRKCI/miR-1294, miR-186-5p/FOXK1 axis[40] promoted proliferation, migration, invasion, and glycolysis and inhibited apoptosis in HCC. Besides, circPRKCI was found to be upregulated in papillary thyroid cancer tissues and promoted proliferation, metastasis, and glycolysis via the miR-335/E2F3 axis,[20] and increased proliferation, migration, invasion and inhibited apoptosis via miR-1304/CXCR1 in thyroid cancer.[31]
Table 2.
The ceRNA regulation of circPRKCI in various cancers.
| Cancer type | miRNA | mRNA | Function |
|---|---|---|---|
| BC | miR-1304 | Myc | Proliferation, migration, and invasion[25] |
| CC | miR-545 | EIF3C | Proliferation, colony formation, migration, invasion, and EMT[24] |
| miR-765 | HMGA1 | Proliferation, migration, and invasion[36] | |
| EC | miR-186-5p | PARP9 | Proliferation, colony formation, cell cycle, and radiosensitivity[37] |
| miR-3680-3p | AKT3 | Proliferation, migration, and invasion[41] | |
| GC | miR-545 | – | Proliferation, invasion, and apoptosis[29] |
| HCC | miR-1290 | FOXC1 | Proliferation and apoptosis[38] |
| miR-545 | AKT3 | Proliferation, migration, invasion, and apoptosis[39] | |
| miR-1324 | FZD5/Wnt/beta-catenin | Proliferation, migration, and invasion[32] | |
| miR-1294, miR-186-5p | FOXK1 | Proliferation, invasion, migration, and glycolysis[40] | |
| LC | miR-1324 | MECP2 | Proliferation and invasion[19] |
| miR-1182 | KLF8 | Proliferation, migration, invasion, apoptosis, and EMT[34] | |
| LUAC | miR-545, miR-589 | E2F7 | Proliferation and colony formation[17] |
| miR-219a-5p | CAMK1D | Proliferation, migration, and cell cycle[26] | |
| LSCC | miR-1324 | – | Proliferation and migration[21] |
| PTC | miR-335 | E2F3 | Proliferation, metastasis, and glycolysis[20] |
| TC | miR-1304 | CXCR1 | Proliferation, migration, invasion, and apoptosis[31] |
BC: Bladder cancer; CC: Cervical cancer; ceRNA: Competing endogenous RNA; EC: Esophageal cancer; EMT: Epithelial–mesenchymal transition; GC: Gastric cancer; HCC: Hepatocellular carcinoma; LC: Lung cancer; LSCC: Laryngeal squamous cell cancer; LUAC: Lung adenocarcinoma; miRNA: MicroRNA; mRNA: Messenger RNA; PTC: Papillary thyroid cancer; TC: Thyroid cancer; –: Not available.
Figure 6.
Major mechanism pattern of circPRKCI in the regulation of cancer development. AKT: Protein kinase B; circPRKCI: Circular RNA Protein Kinase C Iota; CXCL: C-X-C motif chemokine ligand; CXCR1: C-X-C motif chemokine receptor 1; Dvl: Dishvelled; E2F: E2F transcription factor 3; EIF3C: Eukaryotic initiation factor subunit c; FXR1: Fragile-X mental retardation autosomal 1; FZD5: Frizzled 5; GSK Glycogen synthase kinase-3; miR: miRNA; mRNA: Messenger RNA. PI3K: Phosphatidylinositol-3 kinase; PIP2: Phosphatidylinositol 4,5-biphosphate; PIP3: Phosphatidylinositol 3,4,5-trisphosphate; PRKCI: Protein Kinase C Iota.
Discussion
CircRNAs have been reported as clinical diagnostic biomarkers and treatment targets with high stability, specificity, and conservation.[42] Nowadays, the isolation and detection of circRNAs from peripheral blood or tissue are feasible. Chinnaiyan et al[43] evaluated more than 2000 tumor samples and cell lines by exome capture RNA sequencing, involving more than ten kinds of tumors such as lung, breast, liver, and pancreatic cancer. This study contributed to the development of circRNAs as diagnostic or therapeutic targets across cancer types.[44,45] The Cancer Genome Atlas (TCGA) analysis of PRKCI genes demonstrated amplification in more than 20% of clinical squamous cell carcinoma samples, correlated with decreased OS. CircPRKCI is a circular RNA derived from PRKCI exons via back-splicing. There is a rich literature suggesting that circPRKCI is overexpressed in various cancers, including LC, TC, HCC, laryngeal cancer, glioma, and GC.[20–23,35,40] Besides, circPRKCI has been proved to correlate with poor prognosis and progression in various cancers.
Here, we first estimated the correlation between circPRKCI expression and the prognosis of cancer patients. A total of 1061 patients from 14 studies were included for pooled OS analysis. The pooled HR demonstrated high circPRKCI expression correlated with a poor OS without significant heterogeneity. Moreover, only two studies composed of 211 patients assessed the association between circPRKCI and DFS. Similarly, the association between elevated circPRKCI expression and DFS also exhibited the same trend, although not statistically significant. Interestingly, higher circPRKCI expression indicated a worse OS in LC. Then, we evaluated the association between circPRKCI and the major clinical characteristics, including age, gender, tumor size, clinical stage, DM, and LNM. Higher circPRKCI expression was associated with larger tumor size, advanced clinical stage, and higher possibility of LNM. No significant correlation was found between circPRKCI expression and age, gender, and DM. Indeed, more studies are warranted to analyze the heterogeneity of pooled HR for the clinical stage and DM.
At present, circRNAs exert their biological functions mainly through the following four patterns: acting as miRNA molecular sponge through the mechanism of ceRNA, interacting with RBPs as the protein sponge or scaffold, translation into proteins or small peptides, and regulating mRNA selective splicing, chromatin looping, or transcription regulation of genes. However, the function and mechanism of circPRKCI in tumorigenesis and progression warrant further exploration. We found that circPRKCI could regulate progression in various cancers via ceRNA mechanism.[31,38] Only a few studies reported that circPRKCI modulates cancer progression via interacting with RBPs. For example, circPRKCI can bind to FXR1 protein competitively and block the formation of FXR1/PRKCI complex, thereby reducing cell invasion and drug resistance of NSCLC.[35] Nevertheless, the regulatory mechanism of circPRKCI in cancers remains largely unknown and warrants further extensive studies.
However, this study also has several limitations. First, all included patients were from China, which decreased the applicability of our results across different geographic regions. Coincidentally, with the inclusion and exclusion criteria mentioned in this study, the enrolled patients were all Chinese. However, there have been no reported studies about other races from other countries so far. Admittedly, this may introduce potential bias to this study. We hope there will be more studies about this topic in the future to further consolidate the conclusions. Second, since several studies did not report HR with 95% CI, we had to extract the data and calculate them based on the KM curve, which may lead to potential bias. Third, only 14 studies were included in the prognosis and few studies for pooled analysis of the clinical characteristics, which may result in a significant heterogeneity and decrease the robustness of the pooled results. Accordingly, studies with a greater sample size are still needed to analyze the correlation between circPRKCI expression and age, gender, and DM. Further studies investigating the circPRKCI expression and function in cancers may lead to further insights into this remarkable observation.
To sum up, this study revealed that aberrant circPRKCI expression was significantly associated with unfavorable OS, and clinical characteristics comprising tumor size, clinical stage, and LNM. Therefore, circPRKCI could be served as a potential prognostic predictor of OS and clinical characteristics in human cancers.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81902745, 82172500, 82272664, and 82103228), Hunan Provincial Innovation Foundation for Post-graduate (No. CX20190160), and China Postdoctoral Science Foundation (No. 2021M693557).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Liu ZY, Ren XL, Yang ZM, Mei L, Li WY, Tu C, Li ZH. Prognostic and clinical value of circPRKCI expression in diverse human cancers. Chin Med J 2024;137:152–161. doi: 10.1097/CM9.0000000000002844
References
- 1.Capel B Swain A Nicolis S Hacker A Walter M Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993;73: 1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 2.Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993;7: 155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12: 381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med 2018;7: 3101–3109. doi: 10.1002/cam4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang W Yang Y Wu J Niu Y Yao Y Zhang J, et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ 2020;27: 1709–1727. doi: 10.1038/s41418-019-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and Exosomes: a mysterious frontier for human cancer. Mol Ther Nucleic Acids 2020;19: 384–392. doi: 10.1016/j.omtn.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y Li C Zhao R Qiu Z Shen C Wang Z, et al. CircUbe3a from M2 macrophage-derived small extracellular vesicles mediates myocardial fibrosis after acute myocardial infarction. Theranostics 2021;11: 6315–6333. doi: 10.7150/thno.52843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W Qi L Chen R He J Liu Z Wang W, et al. Circular RNAs in osteoarthritis: indispensable regulators and novel strategies in clinical implications. Arthritis Res Ther 2021;23: 23. doi: 10.1186/s13075-021-02420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Zou Y, Chen C, Tang Y, Guo J. Current understanding of circular RNAs in systemic lupus erythematosus. Front Immunol 2021;12: 628872. doi: 10.3389/fimmu.2021.628872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao F, Wang S, Zhang C, Han D, Ma Z, Chen G. Circular RNA circSMARCA5 is a prognostic biomarker in patients with malignant tumor: a meta-analysis. BMC Cancer 2021;21: 600. doi: 10.1186/s12885-021-08316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aishanjiang K Wei XD Fu Y Lin X Ma Y Le J, et al. Circular RNAs and hepatocellular carcinoma: new epigenetic players with diagnostic and prognostic roles. Front Oncol 2021;11: 653717. doi: 10.3389/fonc.2021.653717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C He J Qi L Wan L Wang W Tu C, et al. Diagnostic and prognostic significance of dysregulated expression of circular RNAs in osteosarcoma. Expert Rev Mol Diagn 2021;21: 235–244. doi: 10.1080/14737159.2021.1874922. [DOI] [PubMed] [Google Scholar]
- 13.Meng S Zhou H Feng Z Xu Z Tang Y Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 2017;16: 94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei S Zheng Y Jiang Y Li X Geng J Shen Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine 2019;44: 182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu K, Ma X, Zhang L, Zhang C, Hu J, Zhan T. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics 2018;10: 1327–1346. doi: 10.2217/epi-2018-0023. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids 2019;18: 24–33. doi: 10.1016/j.omtn.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Qiu M Xia W Chen R Wang S Xu Y Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res 2018;78: 2839–2851. doi: 10.1158/0008-5472.can-17-2808. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Li R, Mi W. circ_0067934: a potential biomarker and therapeutic target for hepatocellular carcinoma. Ann Clin Lab Sci 2020;50: 734–738. [PubMed] [Google Scholar]
- 19.Meng Y, Sun WL, Wang M, Sun J. Circular RNA circ-PRKCI promotes lung cancer progression by binding to microRNA-1324 to regulate MECP2 expression. Eur Rev Med Pharmacol Sci 2020;24: 10557–10565. doi: 10.26355/eurrev_202010_23410. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Chen G, Wang B, Wu H, Zhang Y, Ye H. Silencing circRNA protein kinase C iota (circ-PRKCI) suppresses cell progression and glycolysis of human papillary thyroid cancer through circ-PRKCI/miR-335/E2F3 ceRNA axis. Endocrine journal 2021;68: 713–727. doi: 10.1507/endocrj.EJ20-0726. [DOI] [PubMed] [Google Scholar]
- 21.Chu Y. Circ_0067934 correlates with poor prognosis and promotes laryngeal squamous cell cancer progression by sponging miR-1324. Eur Rev Med Pharmacol Sci 2020;24: 4320–4327. doi: 10.26355/eurrev_202004_21013. [DOI] [PubMed] [Google Scholar]
- 22.Cui X, Wang X, Lin S, Miao C, Wu M, Wei J. Circular RNA circ_0067934 functions as an oncogene in glioma by targeting CSF1. Eur Rev Med Pharmacol Sci 2020;24: 7558. doi: 10.26355/eurrev_202007_22210. [DOI] [PubMed] [Google Scholar]
- 23.He S, Guan S, Wu M, Li W, Xu M, Tao M. Down-regulated hsa_circ_0067934 facilitated the progression of gastric cancer by sponging hsa-mir-4705 to downgrade the expression of BMPR1B. Transl Cancer Res 2019;8: 2691–2703. doi: 10.21037/tcr.2019.10.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu C, Wang Y, Li A, Zhang J, Xue F, Zhu L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol 2019;234: 9225–9232. doi: 10.1002/jcp.27601. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Zhou Q, Zhong P. circ_0067934 increases bladder cancer cell proliferation, migration and invasion through suppressing miR-1304 expression and increasing Myc expression levels. Exp Ther Med 2020;19: 3751–3759. doi: 10.3892/etm.2020.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sui M, Zhang W, Geng D, Sun D. CircPRKCI regulates proliferation, migration and cycle of lung adenocarcinoma cells by targeting miR-219a-5p-regulated CAMK1D. Eur Rev Med Pharmacol Sci 2021;25: 1899–1909. doi: 10.26355/eurrev_202102_25085. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Yan X, Zhang H, Zhan X. CircRNA circ _0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med Sci Monit 2019;25: 1342–1349. doi: 10.12659/msm.913463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci 2018;22: 3053–3060. doi: 10.26355/eurrev_201805_15063. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Li Y, Xu XM, Zhu X. Circular RNA circ-PRKCI promotes cell proliferation and invasion by binding to microRNA-545 in gastric cancer. Eur Rev Med Pharmacol Sci 2019;23: 9418–9426. doi: 10.26355/eurrev_201911_19435. [DOI] [PubMed] [Google Scholar]
- 30.Xin J, Zhang XY, Sun DK, Tian LQ, Xu P. Up-regulated circular RNA hsa_circ_0067934 contributes to glioblastoma progression through activating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci 2019;23: 3447–3454. doi: 10.26355/eurrev_201904_17709. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Ma X, Li X, Deng F. Circular RNA circ_0067934 exhaustion expedites cell apoptosis and represses cell proliferation, migration and invasion in thyroid cancer via sponging miR-1304 and regulating CXCR1 expression. Eur Rev Med Pharmacol Sci 2019;23: 10851–10866. doi: 10.26355/eurrev_201912_19789. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Q Lu G Luo Z Gui F Wu J Zhang D, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun 2018;497: 626–632. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 33.Zou Q Wang T Li B Li G Zhang L Wang B, et al. Overexpression of circ-0067934 is associated with increased cellular proliferation and the prognosis of non-small cell lung cancer. Oncol Lett 2018;16: 5551–5556. doi: 10.3892/ol.2018.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao M, Ma W, Ma C. Circ_0067934 promotes non-small cell lung cancer development by regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway. Biomed Pharmacother 2020; 129: 110461. doi: 10.1016/j.biopha.2020.110461. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Zhang M, Zhang Y. Circ_0000079 decoys the RNA-binding protein FXR1 to interrupt formation of the FXR1/PRCKI complex and decline their mediated cell invasion and drug resistance in NSCLC. Cell Transplantation 2020;29: 963689720961070. doi: 10.1177/0963689720961070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y, Li Y. Circ-protein kinase C iota type promotes cervical carcinoma cell invasion and migration by sponging miR-765. Nanosci Nanotech let 2019;11: 576–583. doi: 10.1166/nnl.2019.2905. [Google Scholar]
- 37.Ma Y Zhang D Wu H Li P Zhao W Yang X, et al. Circular RNA PRKCI silencing represses esophageal cancer progression and elevates cell radiosensitivity through regulating the miR-186-5p/PARP9 axis. Life Sci 2020;259: 118168. doi: 10.1016/j.lfs.2020.118168. [DOI] [PubMed] [Google Scholar]
- 38.Yu Y, Han S, Li M, Song Y, Qi F. Circ_0004913 sponges miR-1290 and regulates FOXC1 to inhibit the proliferation of hepatocellular carcinoma. Cancer Cell Int 2020;20: 431. doi: 10.1186/s12935-020-01521-3. [Google Scholar]
- 39.Qi S, Sun H, Liu H, Yu J, Jiang Z, Yan P. Role and mechanism of circ-PRKCI in hepatocellular carcinoma. World J Gastroenterol 2019;25: 1964–1974. doi: 10.3748/wjg.v25.i16.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W, Li Y, Zhong J, Wen G. circ-PRKCI targets miR-1294 and miR-186-5p by downregulating FOXK1 expression to suppress glycolysis in hepatocellular carcinoma. Mol Med Rep 2021; 23: 464. doi: 10.3892/mmr.2021.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi N, Shan B, Gu B, Song Y, Chu H, Qian L. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem 2019;120: 10021–10030. doi: 10.1002/jcb.28285. [DOI] [PubMed] [Google Scholar]
- 42.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol 2019;13: 669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slack FJ, Chinnaiyan AM. The Role of Non-coding RNAs in Oncology. Cell 2019;179: 1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vo JN Cieslik M Zhang Y Shukla S Xiao L Zhang Y, et al. The landscape of circular RNA in cancer. Cell 2019;176: 869–881.e813. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu C He J Qi L Ren X Zhang C Duan Z, et al. Emerging landscape of circular RNAs as biomarkers and pivotal regulators in osteosarcoma. J Cell Physiol 2020;235: 9037–9058. doi: 10.1002/jcp.29754. [DOI] [PubMed] [Google Scholar]