Abstract
This note describes the binding specificities of four lipid A monoclonal antibodies (MAbs) including Centoxin (HA-1A); these MAbs display similar binding properties. MAbs reacted with lipid A and heat-killed smooth bacteria, whereas no reactivity was observed with smooth lipopolysaccharide (LPS). Immunoblotting of bacterial extracts separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that the MAbs bound to many polypeptide bands including the molecular weight markers. Denaturation of bovine serum albumin (BSA) by boiling or dithiothreitol treatment unmasked antibody epitopes. In addition, binding both to a hydrophobic aliphatic C12 chain covalently coupled to BSA and to single-stranded DNA was observed. The polyreactivity of these clones is most likely mediated by a preferential reactivity with hydrophobic molecular patches.
Severe infections with gram-negative organisms are still an important cause of death. One of the outer membrane components of such bacteria, endotoxin (LPS) plays a pivotal role in the pathogenesis of GNB. There is substantial evidence that LPS initiates a cascade of events leading to the onset of the sepsis syndrome (13). Since lipid A is the toxic moiety of LPS, many attempts to prepare protective agents against LPS have focused on the preparation of ligands to the lipid A moiety. It has been postulated that antibodies directed against the conserved core or lipid A region of LPS may cross-react with LPS produced by phylogenetically diverse gram-negative bacteria involved in GNB (5). Centoxin (also called HA-1A) is a human monoclonal antibody raised against the rough LPS of Escherichia coli J5 (Rc chemotype) and selected on binding to lipid A. Both animal studies (16, 18) and phase III human clinical trials gave discrepant results as to the protective efficacy of Centoxin against GNB (14, 19). Since the publication of these reports, questions about the epitope specificity of this MAb have arisen. While it has been described by some authors as a lipid A-specific MAb (8, 10), others defined the antibody as polyreactive since cross-reaction was observed with i antigen present on cord erythrocytes, a ligand on human B lymphocytes, and several anionic polymers such as ssDNA, chondroitin sulfate, and cardiolipin (6, 7). In the present study we demonstrate that there might be a unifying principle to explain the cross-reactivity with several apparently different antigens. The epitope specificities of a number of anti-lipid A MAbs developed in our laboratory that showed a binding profile comparable to that of HA-1A are described. By ELISA, SDS-PAGE, and dot spot techniques it has been made plausible that these MAbs recognize hydrophobic molecular patches present in lipid A, denatured proteins and in aliphatic chains.
Abbreviations used.
BSA, bovine serum albumin; DTT, dithiothreitol; CHAPS, 3-[(3-cholamidopropyl)-dimethylammonio]propanesulfonate; ELISA, enzyme-linked immunosorbent assay; GNB, gram-negative bacteremia; LPS, lipopolysaccharide; MAb, monoclonal antibody; OD, optical density; OMPs, outer membrane proteins; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; ssDNA, single-stranded DNA; Ig, immunoglobulin.
The antibodies used in this study were murine MAbs of the IgM class (clones 28, 37, 38, 40, and 43) or of the IgG class (clone 3). Centoxin (HA-1A) is a human MAb of the IgM class and was obtained upon immunization with E. coli J5 (18). MAbs 28 and 40 were raised against alkaline- and acid-treated Salmonella minnesota R595 cells (Re chemotype), respectively (2). MAbs 37 and 38 were raised against heat-killed S. minnesota R4 cells (Rd chemotype). The epitope specificity of anti-lipid A MAb 43 has been described before (3, 11, 12); this MAb recognizes the hydrophillic part of lipid A (8a). MAb 3 was raised against heat-killed E. coli J5 bacteria (3).
ELISA was carried out as described previously (1), and the antibody titers, defined as the lowest MAb concentrations at which significant binding occurred, i.e., with an OD at 492 nm above control values by more than 0.200 without a first antibody, were determined. At 10 ng/ml MAbs 28, 37, and 40 still reacted (data not shown) with heat-killed bacterial cells of E. coli O111 and other species and with isolated lipid A; in contrast, even at 2,000 ng/ml no reaction with smooth LPS was observed. Centoxin reacted in a similar pattern, i.e., a good reaction with bacterial cells at 100 ng/ml and no reaction with smooth LPS at 32,000 ng/ml. Anti-lipid A MAb 43 reacted with lipid A but not with LPS or heat-killed bacteria.
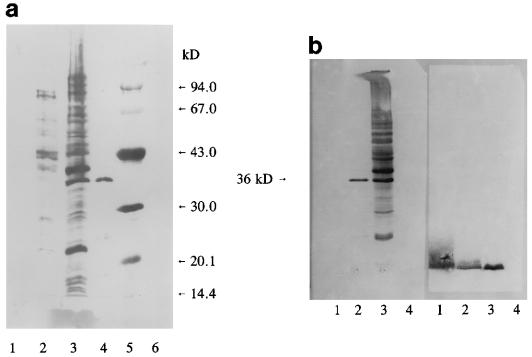
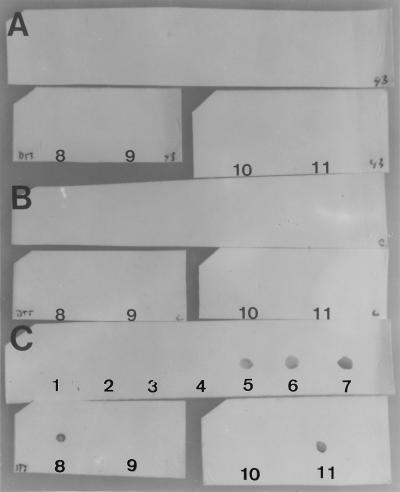
It has been proposed that HA-1A binds to bacteria by the lipid A epitope. The binding of HA-1A to smooth bacteria is enhanced by antibiotic treatment, which would unmask the lipid A epitope (17). To investigate the epitopes of MAbs 28, 37, 40, and HA-1A on bacteria, SDS-PAGE and immunoblotting were performed on heat-killed S. minnesota R5 (Rc chemotype) bacteria (OD = 3) and on an extract of the OMPs. The OMPs of S. minnesota R5 bacteria were extracted as follows. A concentrated bacterial suspension was washed three times in 50 mM Tris-HCl (pH 7.3) and incubated under agitation in the same buffer supplemented with 0.5% CHAPS for 1 h at 4°C. Proteolytic digests were prepared by incubation of a suspension of heat-killed bacteria (OD = 3) or OMPs with proteinase K (final concentration, 500 μg/ml) for 1 h at 37°C. SDS-PAGE was performed, and the gels were blotted on Immobilon-P blotting paper (Boehringer GmbH, Mannheim, Germany), blocked for 1 h with blocking buffer (Boehringer), washed in phosphate-buffered saline supplemented with 0.1% Tween 80 (Sigma), and incubated with MAb 40 (1.8 μg/ml), Centoxin (20 μg/ml), or α-Rc MAb 3 (3 μg/ml). Figure 1a shows that MAb 40 binds to a wide variety of OMPs (lane 2). No binding to proteinase K-treated OMPs was observed (lane 1). A wide range of protein bands was also recognized in heat-killed R5 bacteria (lane 3); the banding pattern was comparable to that produced by total protein staining with Congo red (data not shown). For proteinase K-treated R5 bacteria, only one predominant band of 36 kDa recognized by the antibody was left (lane 4). This band could represent either a proteinase K-resistant protein or a nonprotein component. Remarkably, all protein molecular weight markers (phosphorylase B, BSA, ovalbumin, carbonic anhydrase, soybean trypsin inhibitor, and lysozyme; Pharmacia) were also recognized by MAb 40 (lane 5). Just like MAb 40, Centoxin recognized many proteins in heat-killed S. minnesota R5 (Fig. 1b, left panel, lane 3) and one band in the proteolytic digest (lane 2). In accordance with the binding studies performed by ELISA, no binding of Centoxin to R5 LPS (lane 1), which was near the front of the gel and which was visualized with α-Rc LPS antibody, was seen (Fig. 1b, right panel, lanes 1 and 3). In contrast to what was seen with MAb 40 and Centoxin, proteinase K treatment of whole bacteria did not abolish the epitope of MAb 3 (lane 2). The binding of Centoxin to coextracted porin proteins in LPS samples has previously been described by Mascelli et al. (15). Our binding studies further emphasize the great affinity of Centoxin for proteins separated by SDS-PAGE. These proteins have undergone a series of denaturing treatments, such as breaking of cysteine bridges in DTT, dissolution in SDS, and heating. To study whether the unfolding of proteins could unmask antibody epitopes, a well-defined system was chosen for this subject of investigation. BSA (Sigma) was exposed to a number of conditions that affect its conformational characteristics: incubation in 2, 5, and 8 M urea overnight at room temperature, incubation for 1 h in 200 mM DTT at room temperature, boiling for 10 min, and a combination of these conditions. In a dot spot experiment (Fig. 2), no binding of clone 40 to native BSA (1 mg/ml; spot 10) or to BSA incubated with urea alone (spots 2, 3, and 4) was observed. Clear immunoreactivity was observed with BSA treated with 200 mM DTT (spot 8) and with boiled BSA (spot 11). By these treatments the protein is expected to unfold to expose the inner parts of the molecule to the outside. It was concluded that partial denaturation of the protein, achieved by either boiling or DTT treatment, uncovers protein patches that are hidden in native BSA. These inner parts, that are known to have a hydrophobic character, are recognized by MAb 40.
FIG. 1.
(a) Western blot analysis of S. minnesota R5 bacteria, R5 OMPs, and proteolytic digests with MAb 40. Blots were incubated for 2 h at 37°C with MAb 40 (1.8 μg/ml) in phosphate-buffered saline supplemented with 0.1% Tween 80. Lane 1, S. minnesota R5 OMPs (7.5 μl) incubated with proteinase K (500 μg/ml); lane 2, S. minnesota R5 OMPs (7.5 μl); lane 3, S. minnesota R5 bacteria (OD = 3; 7.5 μl); lane 4, S. minnesota R5 bacteria (OD = 3; 7.5 μl) treated with proteinase K (500 μg/ml); lane 5, molecular weight markers; lane 6, proteinase K (12.5 μg). kD, kilodaltons. (b) Western blot analysis of S. minnesota R5 bacteria, R5 LPS, and proteolytic digests with anti-lipid A and anti-Rc MAb. Left panel: blot incubated with MAb HA-1A (20 μg/ml); right panel: blot incubated with α-Rc MAb 3 (3 μg/ml). Lanes 1, R5 LPS (7.5 μg); lanes 2, S. minnesota R5 bacteria (OD = 3; 7.5 μl) treated with proteinase K (500 μg/ml); lanes 3, S. minnesota R5 bacteria (OD = 3; 7.5 μl); lanes 4, proteinase K (12.5 μg).
FIG. 2.
Dot spot analysis of BSA (1 mg/ml) preincubated at various denaturing conditions (DTT, urea, boiling) with anti-lipid A MAb 43 (1.8 μg/ml) (A), without a first antibody (B), and with MAb 40 (1.8 μg/ml) (C). Spot 1, 8 M urea plus 200 mM DTT; spots 2, 3, and 4, BSA incubated for 24 h in 2, 5, and 8 M urea, respectively; spots 5, 6, and 7, BSA incubated in 2, 5, and 8 M urea, respectively, plus 200 mM DTT; spot 8, BSA incubated in 200 mM DTT; spot 9, 200 mM DTT; spot 10, native BSA; spot 11, boiled BSA.
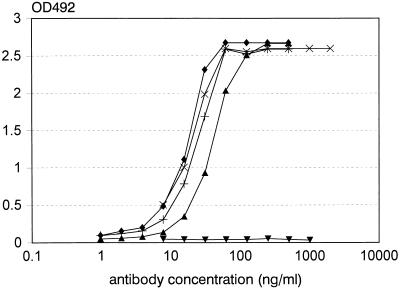
The binding of MAbs 40 and HA-1A to all molecular weight markers and to bacterial proteins in SDS-PAGE and to denatured BSA but not to native BSA in a dot spot assay suggests that hydrophobic protein patches are recognized. In additional ELISA experiments it was observed that MAbs 28, 37, 40, and HA-1A also have great affinity for non-protein-derived hydrophobic ligands, such as a polymeric aliphatic chain (C12, dodecyl) covalently coupled to BSA (9) (Fig. 3). No binding to native BSA was observed (data not shown). In addition to observing the binding to dodecyl-BSA, we confirmed the finding of Bhat et al. (6) that Centoxin binds to ssDNA (origin: calf thymus type I; Sigma) and found that MAbs 28, 37, and 40 also bind to ssDNA (data not shown); DNase treatment of the DNA sample completely abolished the binding (data not shown). The binding of MAb 40 and Centoxin to solid-phase bound ssDNA could be inhibited by fluid-phase dodecyl-BSA and heat-killed bacteria (data not shown); this suggests that the same antigen-binding site is involved in the interaction with the various ligands. Bhat et al. (6) provided convincing evidence for a highly specific binding of Centoxin to i antigen, a carbohydrate structure present on cord erythrocytes. In agreement with this was the observation that the MAb was coded for by VH4.21, a gene often utilized by anti-i antibodies (7). They explained the cross-reactivity with lipid A by assuming that an acyl-substituted disaccharide structure, present both in i antigen and lipid A, was the common epitope (4). Alternatively, the data provided by us suggest that the hydrophobicity of the antigen-binding site is responsible for its polyreactivity. The specificity of MAb 28 differs from that of Centoxin, i.e., MAb 28 reacts specifically and strongly with LPS of S. minnesota R595 (2) and does not bind to LPS of other chemotypes. Yet MAb 28 displays a polyreactivity similar to that of Centoxin; in Centoxin and MAb 28 hydrophobicity is a common property.
FIG. 3.
Binding of anti-lipid A MAbs to immobilized dodecyl-BSA by ELISA. Ninety-six-well microtiter plates were coated with dodecyl-BSA (1 μg/ml). Data for twofold dilution series for MAbs 28 (+), 37 (▴), 40 (⧫), and HA-1A (×) and control MAb 38 (▾) are shown. The OD was measured at 492 nm and expressed as a function of antibody concentration. Data are representative for two independent experiments.
Our study emphasizes the preferential reactivity of Centoxin and MAbs 28, 37, and 40 with hydrophobic structures. These are present in lipid A (acyl chains), in unfolded proteins (hydrophobic amino acid side chains), and in dodecyl-BSA (polymeric aliphatic chain). The binding of Centoxin to ssDNA can be explained by the hydrophobic nature of the DNA bases, which are only exposed in the single-stranded conformation. Indeed Centoxin does not bind to double-stranded DNA (7, 8). Moreover it was found that MAbs 37 and Centoxin had a greater affinity for monophosphoryl lipid A than for diphosphoryl lipid A (data not shown), which is less hydrophobic than monophosphoryl lipid A.
In summary, this binding study shows that a number of MAbs raised against rough mutants of bacteria involved in GNB and selected on binding to lipid A are polyreactive. The multispecific binding pattern can be explained by a strong interaction with hydrophobic molecular patches, and in addition, these Mabs might react more specifically with carbohydrate epitopes such as those present in the i antigen or in R595 LPS (2). As it has now been demonstrated that Centoxin has a great affinity for denatured proteins and likely also for hydrophic patches present in native proteins such as bacterial porins, contamination of LPS samples with coextracted proteins may have led to misinterpretations in studies of Centoxin binding to LPS performed in the past.
Acknowledgments
We thank H. Brade (Borstel, Germany) for providing lipid A, LPS, and bacterial strains.
REFERENCES
- 1.Appelmelk B J, Verweij-van Vught A M J J, MacLaren D M, Thijs L G. An enzyme-linked immunosorbent assay (ELISA) for measurement of antibodies to different parts of the Gram-negative lipopolysaccharide core region. J Immunol Methods. 1985;82:199–207. doi: 10.1016/0022-1759(85)90351-5. [DOI] [PubMed] [Google Scholar]
- 2.Appelmelk B J, Verweij-van Vught A M J J, Maaskant J J, Schouten W F, Thijs L G, MacLaren D M. Monoclonal antibodies detecting novel structures in the core region of Salmonella minnesota lipopolysaccharide. FEMS Microbiol Lett. 1987;40:71–74. [Google Scholar]
- 3.Appelmelk B J, Verweij-van Vught A M J J, Maaskant J J, Schouten W F, de Jonge A J R, Thijs L G, MacLaren D M. Production and characterization of mouse monoclonal antibodies reacting with the lipopolysaccharide core region of Gram-negative bacilli. J Med Microbiol. 1988;26:107–114. doi: 10.1099/00222615-26-2-107. [DOI] [PubMed] [Google Scholar]
- 4.Appelmelk B J, Qing A Y, Helmerhorst E J, Maaskant J J, Abraham P R, Thijs L G, MacLaren D M. Diversity in lipid A binding ligands: comparison of lipid A monoclonal antibodies with BPI. Prog Clin Biol Res. 1995;392:453–463. [PubMed] [Google Scholar]
- 5.Baumgartner J D, Glauser M P. Immunotherapy of endotoxemia and septicemia. Immunobiology. 1993;187:464–477. doi: 10.1016/S0171-2985(11)80357-8. [DOI] [PubMed] [Google Scholar]
- 6.Bhat N M, Bieber M M, Chapman C J, Stevenson F K, Teng N N H. Human antilipid A monoclonal antibodies bind to human B cells and the i antigen on cord red blood cells. J Immunol. 1993;151:5011–5021. [PubMed] [Google Scholar]
- 7.Bieber M M, Bhat N M, Teng N N H. Anti-endotoxin human monoclonal antibody A6H4C5 (HA-1A) utilizes the VH4.21 gene. Clin Infect Dis. 1995;21:S186–S189. doi: 10.1093/clinids/21.supplement_2.s186. [DOI] [PubMed] [Google Scholar]
- 8.Bogard W C, Siegel S A, Leone A O, Damiano E, Shealy D J, Ely T M, Frederick B, Mascelli M A, Siegel R C, Machielse B, Naveh D, Kaplan P M, Daddona P E. Human monoclonal antibody HA-1A binds to endotoxin via an epitope in the lipid A domain of lipopolysaccharide. J Immunol. 1993;150:4438–4449. [PubMed] [Google Scholar]
- 8a.Brade H, Holst O, Brade H. An artificial glycoconjugate containing the biphosphorylated glucosamine disaccharide backbone of lipid A binds lipid A monoclonal antibodies. Infect Immun. 1993;61:4114–4117. doi: 10.1128/iai.61.10.4514-4517.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claassen E, van Rooijen N. A comparative study on the effectiveness of various procedures for attachment of two proteins (l-asparagine and horseradish peroxidase) to the surface of liposomes. Prep Biochem. 1983;13:167–174. doi: 10.1080/00327488308068746. [DOI] [PubMed] [Google Scholar]
- 10.Fujihara Y, Lei M, Morrison D C. Characterization of specific binding of a human immunoglobulin M monoclonal antibody to lipopolysaccharide and its lipid A domain. Infect Immun. 1993;61:910–918. doi: 10.1128/iai.61.3.910-918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn H-M, Brade L, Appelmelk B J, Kusumoto S, Rietschel E T, Brade H. Characterization of the epitope specificity of murine monoclonal antibodies directed against lipid A. Infect Immun. 1992;60:2201–2210. doi: 10.1128/iai.60.6.2201-2210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn H M, Brade L, Appelmelk B J, Kusomoto S, Rietschel E T, Brade H. The antibody reactivity of monoclonal lipid A antibodies is influenced by the acylation pattern of lipid A and the assay system employed. Immunobiology. 1993;189:457–471. doi: 10.1016/S0171-2985(11)80417-1. [DOI] [PubMed] [Google Scholar]
- 13.Lagrange P H, Blanchard H S, Felten A. Bacterial endotoxin and the human monoclonal antibody HA-1A: specificity, potential mechanisms of action, and limits to its effectiveness. J Endotoxin Res. 1995;2:371–386. [Google Scholar]
- 14.MacCloskey R V, Straube R C, Sanders C, Smith S M, Smith C R. Treatment of septic shock with human monoclonal antibody HA-1A. Ann Intern Med. 1994;121:1–5. doi: 10.7326/0003-4819-121-1-199407010-00001. [DOI] [PubMed] [Google Scholar]
- 15.Mascelli M A, Frederick B, Ely T, Neblock D S, Shealy D J, Pak K Y, Daddona P E. Reactivity of the human antiendotoxin immunoglobulin M monoclonal antibody HA-1A with lipopolysaccharides from rough and smooth gram-negative organisms. Infect Immun. 1993;61:1756–1763. doi: 10.1128/iai.61.5.1756-1763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quezado Z M N, Natanson C, Alling D W, Banks S M, Koev C A, Elin R J, Hosseini J M, Bacher J D, Danner R L, Hoffman W D. A controlled trial of HA-1A in a canine model of Gram-negative septic shock. JAMA. 1993;269:2221–2227. [PubMed] [Google Scholar]
- 17.Siegel S A, Evans M E, Pollack M, Leone A O, Kinney C S, Tam S H, Daddona P E. Antibiotics enhance binding by human lipid A-reactive monoclonal antibody HA-1A to smooth gram-negative bacteria. Infect Immun. 1993;61:512–519. doi: 10.1128/iai.61.2.512-519.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng N N H, Kaplan H S, Hebert J M, Moore C, Douglas H, Wunderlich A, Braude A I. Protection against Gram-negative bacteremia and endotoxemia with human monoclonal IgM antibodies. Proc Natl Acad Sci USA. 1985;82:1790–1794. doi: 10.1073/pnas.82.6.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler E J, Fisher C J, Sprung C L, Straube R C, Sadoff J C, Foulke G E, Wortel C H, Fink M P, Dellinger P, Teng N N H, Allen E, Berger H J, Knatterud G L, LoBuglio A F, Smith C R. Treatment of Gram-negative bacteremia and septic shock with HA-1A human monoclonal antibody against endotoxin. N Engl J Med. 1991;324:429–436. doi: 10.1056/NEJM199102143240701. [DOI] [PubMed] [Google Scholar]