Abstract
Polygenic scores offer developmental psychologists new methods for integrating genetic information into research on how people change and develop across the life span. Indeed, polygenic scores have correlations with developmental outcomes that rival correlations with traditional developmental psychology variables, such as family income. Yet linking people’s genetics with differences between them in socially valued developmental outcomes, such as educational attainment, has historically been used to justify acts of state-sponsored violence. In this review, we emphasize that an interdisciplinary understanding of the environmental and structural determinants of social inequality, in conjunction with a transactional developmental perspective on how people interact with their environments, is critical to interpreting associations between polygenic measures and phenotypes. While there is a risk of misuse, early applications of polygenic scores to developmental psychology have already provided novel findings that identify environmental mechanisms of life course processes that can be used to diagnose inequalities in social opportunity.
Keywords: genetics, gene–environment interaction, polygenic scores, development, inequality
Too often, we pour the energy needed for recognizing and exploring difference into pretending those differences are insurmountable barriers, or that they do not exist at all. This results in a voluntary isolation or false and treacherous connections. Either way, we do not develop tools for using human difference as a springboard for creative change within our lives.
—Audre Lorde (1984, pp. 115–16)
1. INTRODUCTION
The central goal of developmental psychology is to describe, explain, and optimize how humans develop. In working to accomplish this goal, developmental psychologists cannot ignore genetic variation, because it accounts for a substantial proportion of individual differences in nearly every aspect of human development. Nevertheless, many developmental psychologists and other social scientists continue to engage in “tacit collusion” to ignore the relevance of genetics to their research (Freese 2008). In this review, we describe how recent advances in genomics offer developmental psychologists new methods for integrating genetic information into research on how people change and develop across the life span. In particular, we focus on polygenic scores. Polygenic scores can be briefly defined as genetic measures of an individual’s predisposition toward a given phenotype, but, as we describe in this review, this seemingly simple definition elides considerable uncertainty and complexity.
Polygenic scores:
indices of an individual’s genetic liability or propensity toward expressing a phenotype relative to other people, not measures of something “innate”
Phenotype:
a trait or characteristic (e.g., height, depressive symptoms) that is the result of both genetic and environmental influences
Historically, the primary tools for integrating genetic information into developmental psychology were twin and family studies. These methods generated foundational insights into the importance of the genome for personality and mental disorders (Gottesman & Shields 1972, Loehlin & Nichols 2012), and they continue to provide researchers with quasi-experimental tools to evaluate hypotheses about environmental causes of developmental phenomena (Turkheimer & Harden 2014). Yet twin studies remain contentious, in part because the studies make controversial statistical assumptions (Charney 2012), and in part because the results of twin studies have been appropriated by far-right political ideologies (e.g., Herrnstein & Murray 1996).
As with these early twin/family methods, we anticipate that developmental psychologists will also treat the introduction of polygenic scores with some degree of wariness. There are good reasons for this wariness. Linking people’s genetics with differences between them in socially valued developmental outcomes, such as cognitive skills or antisocial behavior, has historically been used to justify acts of state-sponsored violence, such as forcible sterilization and genocide (Kevles 1998). Even now, white supremacist movements appropriate genetics research to propagate narratives about the superiority or inferiority of human groups (Carlson & Harris 2020, Harmon 2018, Panofsky & Donovan 2019). Given this history, “there is an old and perhaps permanent danger that inquiries into the genetic differences among us will be appropriated to justify inequalities in the distribution of social power” (Parens 2004, p. S31). Consequently, it is imperative for developmental psychologists, as they adopt new methods and technologies, to guard against further exacerbating existing social inequalities and creating new dimensions of disparity.
Social inequalities:
unequal challenges and opportunities for different social positions, which occur at the intersection of socially constructed dimensions of race, ethnicity, skin tone, gender, wealth, education, ability, sexuality, nationality, and age, among others
In this review, we aim to highlight how scientists can incorporate polygenic measures in developmental research without compromising their commitment to social equity. To this end, we emphasize that an interdisciplinary understanding of the environmental and structural determinants of social inequality, in conjunction with a transactional and developmentally informed perspective on how people interact with their environments, is critical to interpreting associations between polygenic measures and phenotypes. This framework highlights the ethical, legal, and social concerns potentially introduced by misleading applications of polygenic measures. Yet, while there is a risk for misuse, early applications of polygenic scores to developmental psychology have shown how genetic data can be used to spotlight environmental privilege and diagnose inequalities in social opportunity.
Polygenic:
refers to phenotypes influenced by many loci dispersed across the genome, each with very small individual effects
Social equity:
the goal of fairly distributing resources, opportunities, and other social goods in ways that acknowledge historical, social, institutional, and biological differences among people
The remainder of this review is organized as follows. In Section 2, we highlight three long-standing developmental psychology questions that research integrating polygenic scores can help address. That section highlights the promise of polygenic scores, but to understand fully how polygenic scores can (and cannot) move science forward, we need to take a closer look at how these scores are generated and interpreted. Thus, in Section 3, we present a brief primer on genetic methods, with an emphasis on genome-wide association studies (GWASs) and how GWAS results are used to generate polygenic scores. In Section 4, we describe some of the greatest hits of polygenic scores, which have correlations with developmental outcomes that rival their correlations with traditional social science variables. We then describe different processes that can contribute to a correlation between a polygenic score and a developmental outcome. Our overarching goal in this section is to demonstrate that polygenic scores are not measures of something “innate” about a person. Rather, polygenic associations reflect a mix of both true causal signal and confounding by environmental contexts at different scales, and even the causal effects of genes encompass processes that depend on social and historical structures. In Section 5, we review how polygenic score research highlights the effects of environmental privilege, which informs our understanding of gene–environment interplay more generally. Finally, in Section 6, we offer some general thoughts about polygenic scores as a technology that can be used for harm but also for good.
Genome-wide association studies (GWASs):
hypothesis-free study design that associates measured genetic differences between people with differences in their phenotypes
2. THREE DEVELOPMENTAL PSYCHOLOGY QUESTIONS THAT POLYGENIC SCORES CAN HELP ADDRESS
Three long-standing areas of scientific inquiry in developmental psychology and the social sciences can leverage polygenic scores to improve their inferences. First, a central question in developmental psychology is concerned with how human behavior is transmitted across generations, for example, through parent–child interactions (Eisenberg et al. 1998). Because children typically inherit both their genes and their family environments from their parents, correlating parental characteristics with child characteristics confounds genetic and environmental pathways from parents to their children, and vice versa. For instance, a child of a more anxious parent may show stronger internalizing behaviors for a multitude of reasons: The parent’s anxiety has created a stressful environment, the child’s internalizing behaviors made the parent anxious, an external event made them both anxious, they share a genetic liability for mood disorders, or some combination of the above. Thus far, family designs that use naturally occurring variation in how strongly family members are genetically related, such as twin and adoption studies, have been used to disentangle genetic from environmental pathways of intergenerational transmission. However, such samples tend to be difficult and costly to obtain, and they make statistical assumptions that are controversial (Charney 2012).
In addition to observational studies using biological relatives, experimental studies of environmental interventions provide causal evidence of the importance of environmental influences on developmental outcomes (Duncan et al. 2017). But individuals can respond quite differently to interventions, to the degree that for some participants the intervention could exacerbate the problem they are trying to address (Freese & Shostak 2009). Therefore, the second scientific inquiry that can benefit from integrating polygenic scores is concerned with identifying who is being served by interventions, as some of the individual differences that make people react differently may be hidden, either because the relevant individual difference is difficult to measure or because it simply was not measured.
Third, developmental psychologists aim to trace the developmental precursors of adult life course outcomes, exploring the continuity and change of human behavior. However, samples that span the human life course are scarce, and can require multiple generations of researchers to maintain a costly study.
Polygenic scores have two rare and special properties that make them useful for addressing these research questions. First, a child’s genotype is a randomly assigned recombination of the parents’ genotypes. Which genetic variants people inherit from their parents (and which variants they do not inherit) is the outcome of a genetic lottery. Therefore, polygenic score data, when combined with family structure data (siblings or parent–offspring trios), allow us to root analyses in at least one variable that can be reasonably treated as exogenous. Second, one’s gene sequence, and therefore one’s polygenic score, is fixed at conception and does not change over the life span. This immutability means that polygenic scores cannot be reciprocally influenced by life experiences.
Three studies illustrate how these two special properties—that genotypes are random with respect to parental genotype and immutable over the life span—can improve psychological research. First, a study of genotyped parent–offspring trios took advantage of the fact that half of a parent’s genes are (randomly) not transmitted to their offspring and tested whether a polygenic score constructed from the parental genes that offspring did not inherit nonetheless predicted offspring educational attainment. It did. As the study design explicitly rules out genetic inheritance, this is clear evidence for the intergenerational transmission of environmental factors affecting education. Follow-up research has incorporated measured variables, such as prenatal environment and parental cognitive stimulation, to probe hypotheses about mechanisms of this effect (Armstrong-Carter et al. 2020, Wertz et al. 2019).
Second, a study of an educational policy reform in the United Kingdom found that increasing the age of compulsory schooling reduced body size, particularly in individuals who had a polygenic score associated with higher body mass index (BMI) (Barcellos et al. 2018). The policy thus mitigated genetically associated disparities in health. Notably, participants’ outcome data were collected when they were older adults, but there were no retrospective data available, to our knowledge, about participants’ body size as children or adolescents (prior to the educational reform). Polygenic score data, which are invariant across the life span, thus gave researchers a tool for time travel: They could know something about what a person’s risk for higher BMI was prior to a policy reform that was instituted in the 1970s, on the basis of a variable (DNA) that was collected in the 2000s and that was not subject to retrospective recall biases. Additionally, the integration of polygenic scores provided valuable—and otherwise invisible—information regarding whether those interventions were helping the individuals most vulnerable to adverse outcomes.
Third, several studies have used polygenic scores as an inert molecular tracer that connects data on development at different parts of the life course (Belsky et al. 2016, Belsky & Harden 2019, Nivard et al. 2017). For example, polygenic scores of Alzheimer’s disease are associated with smaller hippocampal volume throughout the life span in pediatric and adult samples (Walhovd et al. 2019). By examining the association between a single polygenic score with an array of developmental outcomes in separate samples of differing ages, researchers can highlight developmental precursors of highly heritable diseases and other phenotypes, and these precursors could become new targets of intervention.
Note that the immutability of a polygenic score does not mean that phenotypes are immutable. In fact, polygenic scores can be useful for improving interventions designed to change the phenotype that a polygenic score predicts. Including polygenic scores in randomized controlled trials is likely to increase statistical power to detect average treatment effects (Rietveld et al. 2013). Moreover, as with the educational policy reform example described above, polygenic scores can be used to investigate whether interventions are indeed decreasing preexisting health differences and promoting greater health equity (Freese & Shostak 2009).
In summary, polygenic score applications can (a) improve inferences about environmental mechanisms for intergenerational transmission, (b) trace the developmental precursors of adult life course outcomes, and (c) evaluate whether interventions, policies, or treatments are mitigating versus exacerbating genetic disparities in health and well-being. But, like any method, polygenic score applications have their limitations. To fully understand how polygenic scores may advance developmental psychology research and what their limitations are, we first need to take a closer look at how they are generated.
3. A PRIMER ON GENOME-WIDE ASSOCIATION STUDIES
3.1. Beyond the Black Box of Heritability
For nearly a century, behavior genetic research in humans relied primarily on partitioning phenotypic variation among family members with known degrees of biological relatedness. Research has found that relatives who are more closely biologically related tend to more strongly resemble one another in essentially every human trait, including appearance, physical and mental health, personality, and educational attainment (Polderman et al. 2015). Consider twins as an example. If monozygotic (identical) twins sharing ~100% of their genes are more similar for a given phenotype than dizygotic (fraternal) twins sharing ~50% of their genes, then that phenotype is considered to be heritable. Heritability is a fundamental concept in genetics, referring to the proportion of variation in a particular phenotype that can be attributed to genetic factors (Visscher et al. 2008). It is a population parameter that enables comparison of the relative importance of environmental and genetic factors within that population at the time of study. Recent advances in genetics have enabled the estimation of heritability using genome-wide data from unrelated individuals, providing further evidence of the association between genotypic and phenotypic similarity. Indeed, the finding of nonzero heritability is so ubiquitous that it has been termed the first law of behavior genetics—“everything is heritable” (Turkheimer 2000).
Heritability:
the proportion of variation in a particular phenotype that can be attributed to genetic factors; it can be calculated using family designs (e.g., twin models) or using genomic methods
Heritability estimates from twin and family studies, however, are black boxes that do nothing to reveal which specific genes are relevant for individual differences. In order to identify specific genetic variants, GWASs conduct hypothesis-free tests of association between millions of measured genetic variants and a phenotype of interest (such as a psychiatric disorder). Most commonly, GWASs measure single-nucleotide polymorphisms (SNPs; pronounced snips), which are differences between people at a single DNA letter, or nucleotide. Large-scale GWASs, which include hundreds of thousands of people, are now routine, because of (a) technological changes that allow for cheap and noninvasive measurement of the human genome and (b) social changes that permit the accumulation of huge sample sizes, including the formation of international consortia (e.g., Psychiatric Genomics Consortium), national biobanks (e.g., UK Biobank), and direct-to-consumer genetic testing companies (e.g., 23andMe).
Single-nucleotide polymorphisms (SNPs):
differences between people at a single DNA letter, or nucleotide
Even with more than a million people, though, the number of SNPs measured in a GWAS typically exceeds the number of people in the study. A GWAS, then, estimates a series of linear regressions, each of which regresses the phenotype onto a SNP and a standard set of covariates, such as age, sex, and birth cohort, as well as the interactions among them. One important set of covariates is what is referred to as principal components of ancestry, which are statistical representations of patterns of genetic similarity among people due to having shared ancestors. Psychologists are likely familiar with principal components analysis, a technique that summarizes the pattern of correlations among variables in a data set by using one or more orthogonal dimensions. Analyses of principal components of ancestry work similarly, except the variables in one’s data set are people’s genetic variants rather than, say, their responses to items on a personality inventory.
Principal components of ancestry:
linear combinations of SNP genotypes, where each SNP has a loading giving its contribution to a principal component; they are used as covariates in GWASs to address the problem of population stratification
The inclusion of principal components of ancestry as covariates is important, because a GWAS aims to identify genetic associations with individual differences in a specific trait within a population that is homogeneous with regard to genetic ancestry, not to explain between-population differences. Here, we are using the word population as a geneticist might, which is different from the definitions common in psychology and the other social sciences (Waples & Gaggiotti 2006). Psychologists might use the term to refer to the group of people to whom they expect their sample-specific results to be generalizable or to a group of people living within culturally proscribed boundaries (e.g., college students in Texas). In contrast, genetic definitions of population emphasize reproductive continuity and the availability of random mating. When people are divided by physical or social barriers from mating and reproducing, their populations diverge. In the next section, we consider the genetic concept of a population and its relation to genetic ancestry more closely. Again, these ideas are important because they help us understand what GWASs and polygenic scores do and do not say about the causes of social inequalities in important developmental outcomes like education.
3.2. Within-Population Versus Between-Population Differences
Results from genetic studies have been misused by right-wing extremists and white supremacists to argue that racialized disparities in income and educational attainment are genetic in origin and thus are likely intractable to intervention or social policy (e.g., Herrnstein & Murray 1996, Murray 2020b). This conclusion is based on a serious misunderstanding of the goals and methods of GWASs and of earlier twin studies. As many psychologists’ suspicions of genetic methods stem from their misappropriation by white supremacist movements, it is necessary to clarify the errors that pseudoscientific racists make in their interpretation of genetic results (Martin et al. 2017a, Novembre & Barton 2018, Rutherford 2020, Yudell et al. 2016).
Importantly, a GWAS identifies genetic variants that are associated with phenotypic differences between individuals within a population, but, in and of itself, a GWAS is silent about the source of differences, genetic or otherwise, between populations (Coop 2019). Understanding why this is true requires, in turn, understanding how a population is defined, how populations differ in their genetics, and how GWASs deal with between-population differences in gene frequencies.
The genomics field typically refers to five superpopulations (admixed American, African, East Asian, European, and South Asian; see https://www.internationalgenome.org/category/population/). Importantly, these superpopulations are determined on the basis of analyses of patterns of genetic similarity and dissimilarity across the globe. They can be further divided into so-called ancestral subpopulations, such as the Southern Han Chinese, Chinese Dai in Xishuangbanna, Yoruba in Ibadan in Nigeria, Gujarati Indians from Houston in the USA, Americans of African ancestry in the southwestern USA, or British in England and Scotland. Notably, there are fine-grained genetic differences within ancestral subpopulations, which reflect histories of migration and mating even within an apparently homogeneous group (e.g., “White British” people in the United Kingdom; Abdellaoui et al. 2019, Sohail et al. 2019).
Populations differ genetically in terms of which variants are present, how common or rare those variants are, and how those variants are correlated with one another across the genome. These genetic differences reflect the demographic history of humans, including patterns of mating, migration, and mortality. For instance, analyses of genetic similarity and dissimilarity in people living in the USA reveal a geographical pattern of genetic similarity among Black Americans that reflects a North–South barrier to migration, with people living on either side of this geographical barrier more similar to one another than to people on the other side (Dai et al. 2020). Genotypes are records of both of these types of relatively recent migration events, but also of much deeper human history (e.g., the African diaspora). See the sidebar titled Racial/Ethnic Identity Versus Genetic Ancestry for a discussion of how genetic ancestry differs from racial/ethnic identity.
Genetic ancestry:
a description of a person’s relationship with other people in their genealogical history as reflected in their DNA; ancestry is not the same as race
In GWASs, genetic differences between populations and subpopulations can seriously confound associations of genetic variants with phenotypes (for an illustrative example, see Hamer & Sirota 2000). If groups differ, even a little bit, in how frequently people have a particular version of a gene (allele), and also differ in a phenotype for entirely cultural/environmental reasons, it can induce a spurious association. This is called the problem of population stratification. Generally, GWASs attempt to account for population stratification by restricting analyses to more or less homogeneous populations (e.g., only people who identify as being solely of recent European ancestries) and by including multiple principal components of ancestry as covariates (Price et al. 2006). It is not uncommon for analyses to include 10 or more of these components. (Consider that principal component 37, based on an analysis of genome-wide similarity among people who all identify as “White British,” has no relationship to any form of racial/ethnic identity; see the sidebar titled Racial/Ethnic Identity Versus Genetic Ancestry.) Yet even this approach might not be entirely successful in eliminating population stratification. Accordingly, researchers are increasingly turning to within-family GWAS approaches, which compare siblings to one another rather than to unrelated people (Brumpton et al. 2020).
Racial/ethnic identity:
the outcome of context-specific, dynamic social processes that connect individuals with socially constructed groups
Population stratification:
genetic differences between populations and subpopulations which could be confounded with environmental/cultural differences and could thus result in a GWAS producing false positive associations between a genotype and a phenotype
In this way, GWASs compare people who are as homogeneous as possible with regard to their genetic ancestries, even to the point of being family members. In doing so, these methods help minimize the threat of population stratification biasing very small but real effects of individual SNPs within the studied population, but they are silent about the source of between-population differences in phenotypes. If, for example, a GWAS finds genetic loci that are associated with higher BMI among Americans who have predominantly European ancestries it does not follow that differences in BMI between groups of Americans who may differ more in their genetic ancestries (e.g., White versus Black Americans) are genetic in origin (for more discussion of the logical flaws of such an argument, see Coop 2019). And, as we discuss further in the next section, the results of a GWAS conducted in one population are not portable to another population.
3.3. Genome-Wide Association Studies Are Just the Beginning: Using Summary Statistics
The millions of estimated statistical associations between analyzed genetic variants and the phenotype of interest are collectively referred to as GWAS summary statistics. These summary statistics are regression weights between phenotypes and genetic variants and are typically available in the supplementary materials of GWASs, but many have also been collected and compiled in various repositories, such as the Polygenic Score Catalog (http://www.pgscatalog.org/) and the GWAS Catalog (https://www.ebi.ac.uk/gwas/). See the sidebar titled Computing Polygenic Scores for a brief overview of how to calculate polygenic scores.
GWAS summary statistics:
the millions of estimated statistical associations (regression weights) between analyzed genetic variants and the phenotype of interest; used to calculate polygenic scores in an independent sample
GWAS summary statistics serve as the starting point for a variety of subsequent research endeavors, such as (a) fine-mapping statistical associations to identify the particular genetic variants that most likely exert a causal influence on the phenotype, (b) conducting biological annotation to map genetic discoveries with insights from biology (e.g., analyzing how genes are expressed in different tissues during different developmental epochs; Watanabe et al. 2017, 2019), and (c) calculating genetic correlations to estimate the extent of genetic sharing across phenotypes that have all been subjected to GWASs, even if those phenotypes were never measured in the same sample (Bulik-Sullivan et al. 2015a,b). The third use of GWAS results, which is called LD score regression, might be particularly surprising to psychologists. How can one calculate a genetic correlation between, say, infant head circumference and total life span if there is no group of people for whom both phenotypes have been measured? Here, the genome serves as a sort of Rosetta Stone that reveals connections between phenotypes that are otherwise never observed together. Two phenotypes, measured in two different samples (or samples of varying overlap), are regressed on the same set of SNPs, permitting inferences about the extent to which they share the same genetic underpinnings. Furthermore, once LD score regression is used to generate genetic correlation matrices, these matrices can in turn be subjected to structural equation modeling (Grotzinger et al. 2019) and other multivariate statistical techniques familiar to psychologists.
We focus on using GWASs to create polygenic scores in a new sample of people who were not included in the original GWAS but who have similar genetic ancestry as people from the original GWAS. Technically speaking, an individual’s polygenic score is calculated as the sum of the number of their alleles multiplied by the effect estimate reported in the GWAS summary statistics.
Notably, polygenic scores can be computed for any phenotype for which GWAS summary statistics are available, such as schizophrenia (Ripke et al. 2014), well-being (Okbay et al. 2016), risk tolerance (Karlsson Linnér et al. 2019), and educational attainment (Lee et al. 2018). Furthermore, polygenic scores require information only from DNA; the phenotype that was the subject of the original GWAS from which summary statistics were obtained does not have to be measured in a new data set in order for a polygenic score to be calculated in that data set. For instance, our group used summary statistics from a GWAS of schizophrenia to calculate polygenic scores for schizophrenia in a sample of typically developing university students, about whom we had no information related to schizophrenia symptoms or psychotic symptoms (Mallard et al. 2019).
Again, note that polygenic scores are based on GWAS summary statistics, so they share with GWAS results an exclusive concern with individual differences within a population. Simply comparing average polygenic scores across populations, as has been done in some pseudoscientific research published by right-wing extremists, is scientifically meaningless. Reflecting the non-Hispanic Eurocentric bias of most genetic research (Martin et al. 2019), all of the examples of polygenic score research that we describe here are specifically concerned with individual differences among people of non-Hispanic European genetic ancestries who likely identify as White in terms of their racial identity.
3.4. The Proof of the Pudding Is in the Eating
Psychologists are, by training, often suspicious of so-called fishing expeditions, and thus are often skeptical of the hypothesis-free approach of GWASs, in which every measured genetic variant is tested in relation to a phenotype. In addition, psychologists have a long tradition of valuing measurement, whereas the phenotypes examined in GWASs are typically quite crudely measured (e.g., with answers to a single survey item). As such, psychologists often embraced the hypothesis-driven candidate gene paradigm rather than GWASs (Duncan et al. 2014). In candidate gene studies, a few genetic variants, such as a polymorphism in the serotonin transporter gene, are examined in relation to a rich variety of well-measured psychological phenotypes on the basis of a priori reasoning about the gene’s functions.
But 10 years of GWAS research have definitively shown that many psychologists’ intuitions about what genetic research strategy would be successful were mistaken. Candidate gene research turned out to yield almost no replicable insights into human psychology (Border et al. 2019, Hewitt 2012), whereas very large-scale GWASs have identified thousands of replicable hits—that is, SNPs that are reliably associated with human phenotypes at very stringent levels of statistical significance (p < 5 × 10−8) and that replicate across independent samples (Visscher et al. 2017). A major reason that candidate genes failed, we now realize in the face of GWAS results, is that nearly all human traits are highly polygenic, meaning that they are influenced by many loci dispersed across the genome—each with very small individual effects (Chabris et al. 2015). Accordingly, GWASs require very large sample sizes (i.e., hundreds of thousands to millions of individuals) to accurately estimate effects. In the face of all that we do not know about the human genome, it turns out that genetic discovery has been better advanced by using brute statistical force—surveying the entire genome in as many people as possible—than by careful phenotypic measurement and a priori hypotheses.
The best validation of the GWAS approach is that polygenic scores from well-powered GWASs can now predict nonnegligible amounts of variation in outcomes in independent samples (Figure 1). Indeed, polygenic scores for some diseases and disorders have even reached a level of predictive power that has prompted the biomedical community to consider their clinical utility in medical settings (Khera et al. 2018, Lello et al. 2020). For social science outcomes, we do not expect polygenic scores to ever be so accurate at an individual level as to warrant their use for treatment or educational decisions for a specific person. At the same time, on a population level, polygenic scores now often have predictive power that is directly comparable in magnitude to commonly used variables in developmental psychology, such as family income (Harden et al. 2020, Lee et al. 2018). For instance, polygenic scores from one of the best-powered GWASs, on educational attainment, can account for ~10% of the variance in educational attainment (Lee et al. 2018) and related psychological phenotypes, such as cognitive skills (Allegrini et al. 2019) and persistence in taking mathematics classes in school (Harden et al. 2020). Similarly, current polygenic scores can capture ~8% of the variance in body size, 5% in liability to schizophrenia, and ~3% of the variance in smoking behavior and depression (Howard et al. 2019, Khera et al. 2019, Liu et al. 2019, Pardiñas et al. 2018).
Figure 1.
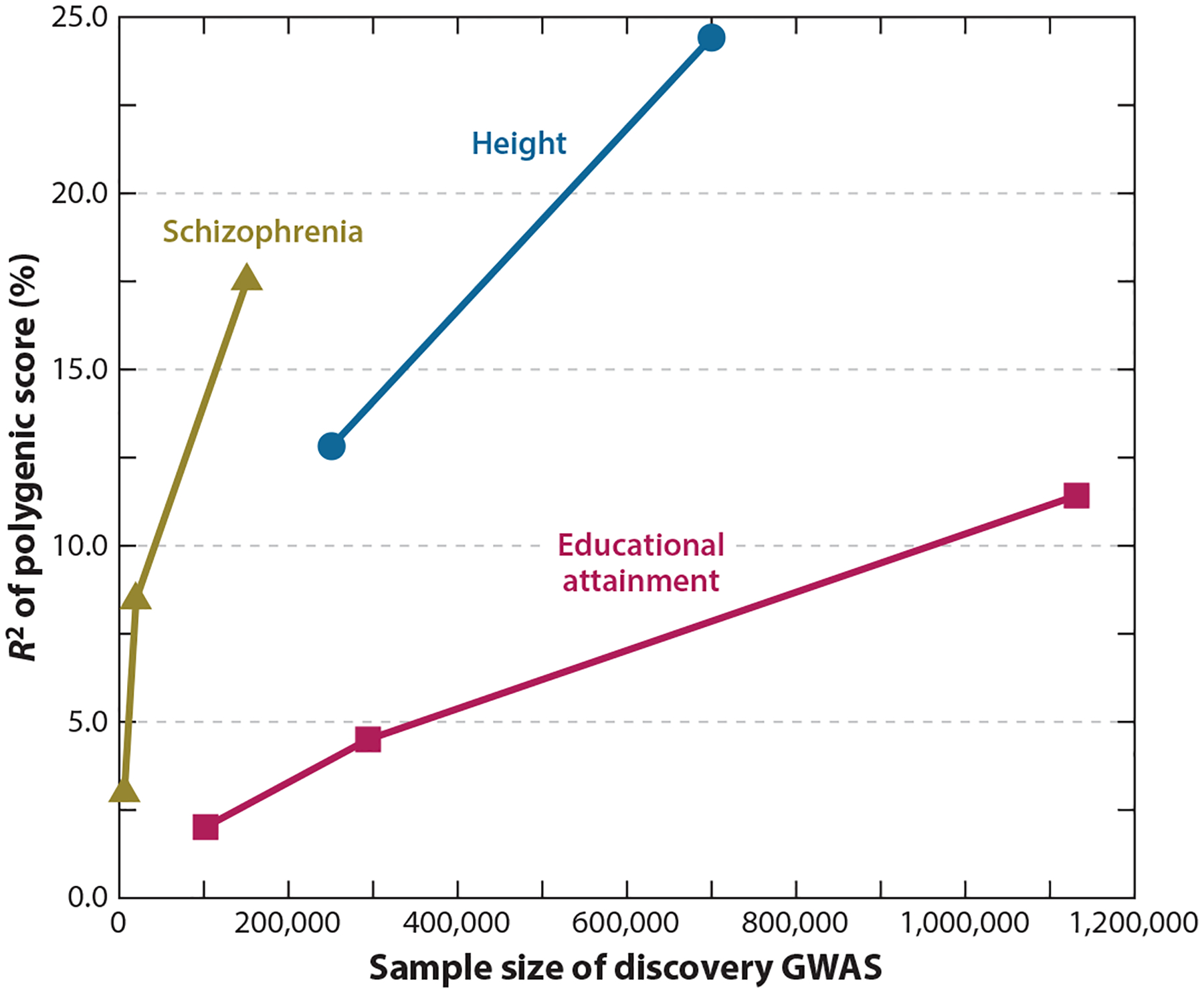
Out-of-sample variance in height, schizophrenia, and educational attainment captured by polygenic scores (R2) created by genome-wide association studies (GWASs) of progressively increasing sample size. R2 values for height were obtained from Yengo et al. (2018), for educational attainment from Harden & Koellinger (2020), and for schizophrenia from Ripke et al. (2014). Note that the R2 values are from studies that draw from different samples and are not methodologically identical (e.g., polygenic scores of schizophrenia have differing ratios of cases to controls and are calculated as Nagelkerke R2).
These effect sizes for polygenic scores are sometimes trivialized as negligible, but trivializing R2 values in the 1–10% range belies ignorance about how weakly correlated any variable—genetic or otherwise—is with complex human behavior. As a point of comparison, consider the outcome of the Fragile Families Challenge, in which 160 teams of researchers were given access to 12,942 variables collected from participants from birth to age 9 and asked to build predictive models of grade point average (GPA), grit, and other outcomes at age 15 (Salganik et al. 2020). The best-fitting models were still not very accurate: In holdout samples, the combined R2 for models (potentially including thousands of survey-based demographic variables gathered across up to six survey waves) captured only 20% of the variance in GPA and only 5% of the variance in grit. On the basis of these results, the architects of the Fragile Families Challenge suggested that “our understanding of child development and the life course is actually quite poor” (Salganik et al. 2020, p. 8402). Any evaluation of the success of genetics in predicting developmental outcomes must be made in comparison to this humbling lack of success using nongenetic variables.
4. A TRANSACTIONAL PERSPECTIVE ON GENETIC ASSOCIATIONS
4.1. Four Sources Contributing to Polygenic Score Correlations
The ability to create DNA-based measures of an individual’s likelihood of developing a particular phenotype, which might not emerge until years or even decades after conception, has been the cause of both excitement and dismay. In the excitement camp, the conservative provocateur Charles Murray opined in the Wall Street Journal that polygenic scores are “impervious to racism and other forms of prejudice.… That means polygenic scores will offer social scientists something they’ve never had before: a secure place to stand in assessing what is innate” (Murray 2020a). On the dismay side, the sociologist Catherine Bliss told the MIT Technology Review that “[t]he idea is we’ll have this [polygenic score] information everywhere you go, like an RFID tag. Everyone will know who you are, what you are about. To me that is really scary” (Regalado 2018). In our view, both the excitement and the dismay are, unfortunately, fueled by misunderstandings about what polygenic scores represent and what processes contribute to their correlation with psychological phenotypes.
Polygenic scores do offer developmental psychologists a promising new research tool, but the reasons to be excited do not include the idea that polygenic scores are an assessment of what is “innate.” They are not. With highly polygenic phenotypes, genetic effects do not reflect an innate liability or endowment or even a wholly biological process. Rather, associations between polygenic scores and developmental phenotypes correspond to four types: direct genetic effects, transactional genetic effects, indirect genetic effects, and confounded genetic effects (Figure 2). We describe these different types of effects and their developmental processes below.
Figure 2.
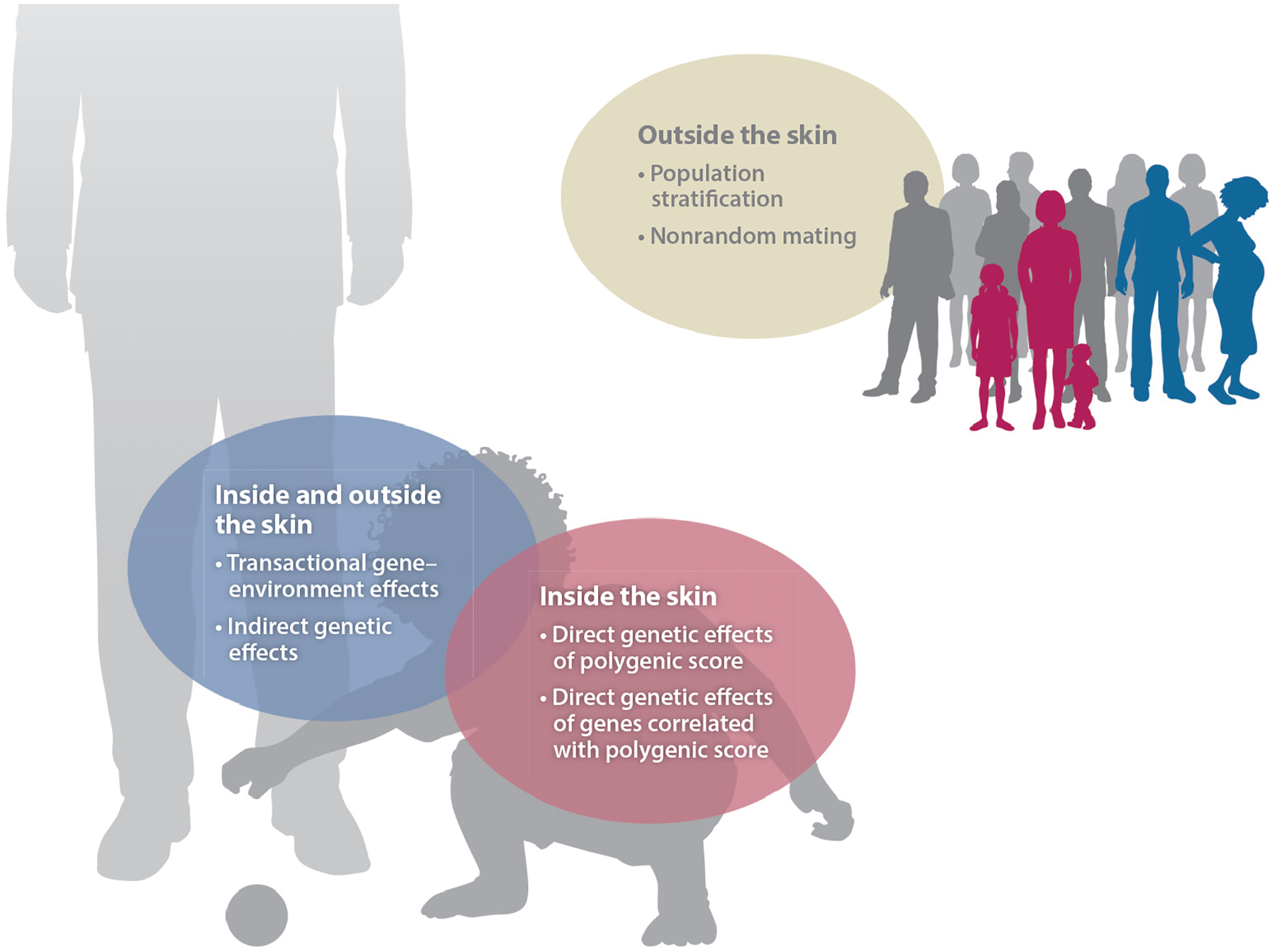
Potential contributions to a polygenic score association. First, direct genetic effects are ones that transpire largely through inside-the-skin processes that are best described in terms of the actions of molecules and cells within the body. A physical characteristic like hair texture is an example of a phenotype where inside-the-skin processes predominate. Second, transactional genetic effects originate from a person’s own genome but require interaction with the environment, thereby transpiring through processes both inside and outside the skin. Most traits of interest to developmental psychologists, such as mental health, physical health, personality, cognitive skills, and academic achievement, will depend on transactional gene–environment effects. Third, caregivers’ genes, even those that are not transmitted during reproduction, can affect child outcomes through correlated environmental processes; this process is referred to as genetic nurture or indirect genetic effects. Fourth, associations of polygenic scores with developmental outcomes may derive from confounding due to unaccounted population stratification and nonrandom mating.
Confounded genetic effects:
associations of polygenic scores with developmental outcomes that are due to confounds such as unaccounted population stratification and nonrandom mating
First, direct genetic effects transpire largely through inside-the-skin processes. By inside the skin, we mean that the chain of causal events connecting genotype to phenotype can be satisfactorily described with reference to biological components that exist within an individual’s body (e.g., molecules, cells, organs) and their interactions. For instance, genes regulate cells called melanocytes, which produce melanin within the skin, thereby affecting skin tone. Direct genetic effects do not imply that the trait is genetically simple. For instance, skin tone is a polygenic trait, especially among African populations (Martin et al. 2017b).
Direct genetic effects:
effects that transpire largely through inside-the-skin processes, where the influence of genotype on phenotype can be satisfactorily explained via biological processes that are minimally contingent upon the environment
Second, transactional genetic effects, like direct genetic effects, originate from a person’s own genome, but transpire through processes that take place both inside and outside the skin (Plomin et al. 1977, Scarr & McCartney 1983). By outside the skin, we mean that the chain of causal events connecting genotype to phenotype cannot be satisfactorily described without reference to social behaviors and the interactions among people in society. For instance, variants in the CHRNA genes affect lung cancer risk through their association with smoking quantity (Ware et al. 2011). Thus, the connection between genotype and phenotype (lung cancer) depends on the behavior of an individual (smoking), a behavior that is dependent on interactions among people in a society (e.g., laws regulating the affordability and availability of tobacco). Most traits of interest to developmental psychologists, such as mental health, physical health, personality, cognitive skills, and academic achievement, will depend on transactional gene–environment effects.
Figure 3 depicts a hypothetical example that highlights the transactional interplay among genetic propensities, environmental opportunities, and developmental idiosyncrasies. Imagine a young child who struggles to concentrate on books read by adults and prefers exploring thrilling activities. Their linguistic skills fall behind those of other children at the beginning of school, thereby decreasing their motivation to participate in school. Yet, they were born into a wealthy family and attend a well-resourced school that inhibits them from leaving before graduating with a high school diploma. Indeed, not graduating from university was never even presented as an option, even though they fathered a child in high school. Their genetic predisposition for thrilling activities is funneled into a career in risky investment banking, a field in which they find easy inclusion on the basis of other traits, such as skin tone.
Figure 3.
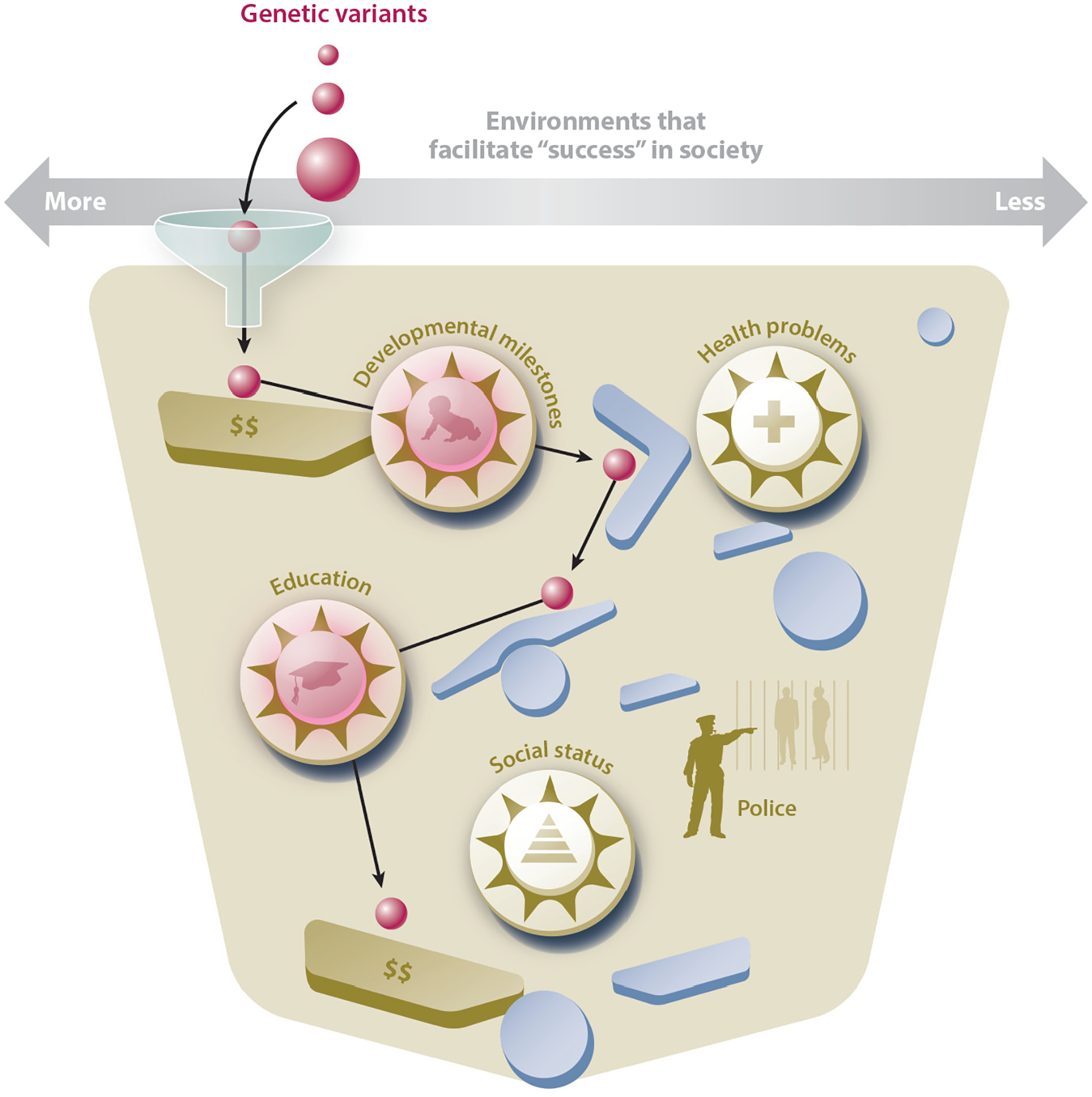
Polygenic pinball. A polygenic score is conceptualized as an inert molecular marker (a pinball) that can be used to trace how an individual moves through the environment (a gamescape). In this game, success is defined as a lucrative career and is determined by three factors. First, chance determines the inheritance of a specific combination of genetic variants, represented by the size of the pinball. A larger pinball makes for an easier game, within the context of a particular gamescape, as it is more likely to hit targets that score points, whereas a smaller pinball is more likely to fall into a hole. Second, chance determines the environmental conditions experienced at the start of the game, represented by the funnel that leads the pinball into the gamescape. Third, development itself is a source of phenotypic variability, such that the ball’s trajectory cannot be perfectly predicted given genetic and environmental information. In the example depicted, a polygenic pinball with average genetic propensity for higher educational attainment enters the game with ample environmental resources to support higher educational attainment and social status. Despite moderate genetic propensity to do well academically, the polygenic pinball’s movements are cushioned by wealth and privileges that move it away from illness and interactions with the police and toward a university degree.
Third, in addition to the child’s own genotype, caregivers’ genetic dispositions can affect child outcomes through indirect genetic effects that are not transmitted during reproduction (Koellinger & Harden 2018, Kong et al. 2018). Recall that parents have two copies of every gene, only one of which is inherited by a child. Genetic studies of trios of parents and offspring have shown that even those genetic variants that are not transmitted to offspring are nevertheless correlated with offspring outcomes. For example, mothers with higher education polygenic scores tend to have children who exhibit higher levels of academic performance and educational attainment and reach developmental milestones earlier, even after accounting for direct genetic transmission (Armstrong-Carter et al. 2020, Wertz et al. 2019). Such genetic nurture effects might be mediated by experiences during pregnancy or by cognitively stimulating and warm parenting behaviors (Armstrong-Carter et al. 2020, Wertz et al. 2019). Indirect genetic effects necessarily involve outside-the-skin processes, as the genotype–phenotype relationship transcends the boundaries of a single person.
Indirect genetic effects:
effects that are mediated through the environment, where one individual’s genotype indirectly influences another individual’s phenotype by changing the second individual’s physical or social environment
Fourth, outside-the-skin processes, including unaccounted population stratification and nonrandom mating, can confound associations of polygenic scores with child development (Selzam et al. 2019, Sohail et al. 2019). Specifically, subpopulations may differ genetically in ways that are not fully accounted for by ancestry-related principal components (see Section 2). If those subpopulations also differ in environmental or behavioral measures, then those genetic variants may be correlated with a phenotype, despite the lack of a causal genetic effect on the phenotype. In contrast, differences between genetically related individuals (e.g., full biological siblings) who live within the same family cannot arise from family-level population stratification, nonrandom mating, or indirect genetic effects. The assortment of genetic variants within families is random. Accordingly, a rigorous test of polygenic–phenotype associations is to examine whether they predict phenotypic differences between siblings, and indeed they do (Belsky et al. 2018, Morris et al. 2019, Selzam et al. 2019).
The astute reader will have noticed by now that there is no bright line separating direct, indirect, and transactional genetic effects. Rather, they exist on a spectrum. Even direct genetic effects that can be satisfactorily explained in terms of biological processes are affected by the environment. For example, skin tone can be changed through sun exposure or with highly prevalent and potentially health-damaging skin-lightening procedures (Sagoe et al. 2019). Indeed, all genetic causes have to work in the context of an environment of some kind (Dawkins 1982).
4.2. A Transactional Perspective on the Portability of Polygenic Scores
Transactional genetic effects, which involve human behavior and interactions among people in society, are expected to depend on environmental contexts that are known to differ between groups of people. Sociological (Crenshaw 1989, hooks 1995), economic (Chetty et al. 2020; see https://opportunityinsights.org/), and epidemiological (Creanga 2018, Hargrove 2018) research shows that different environmental challenges and opportunities occur at the intersection of socially constructed dimensions of race, ethnicity, skin tone, gender, wealth, education, physical ability, sexuality, nationality, and age, among others (see https://www.intersectionaljustice.org/). Genetic effects depend on these large-scale environmental contexts, rather than determining fixed phenotypes in a biodeterministic manner (Raffington et al. 2019).
Transactional genetic effects:
effects that originate from a person’s own genome but require interaction with the environment, thereby transpiring through processes both inside and outside the skin
Biodeterminism:
the belief that human behavior is determined by innate biological characteristics such as genetic makeup
A transactional perspective presumes that polygenic scores created from GWASs of largely White samples from high-income countries will have lower predictive validity in samples who live in different environments, even if disparate genomic ancestries were no longer a barrier to portability. For instance, a polygenic score of neuroticism predicts a 20-year burden of depressive symptoms for White but not Black Americans (Assari et al. 2020). Because the neuroticism genetic score does not capture an innate risk for mental health disorders, and because Black Americans are exposed to environmental challenges not experienced by White Americans (e.g., race-based interpersonal discrimination; Goosby et al. 2018), different genetic variants may be relevant to developing depression for White versus Black Americans. Accordingly, even if causal genetic variants were to be identified in a GWAS of depression, the causal pathways between genes and depression would depend on highly social mechanisms that differ across environments.
5. USING GENETICS TO SPOTLIGHT ENVIRONMENTAL PRIVILEGE
Existing GWAS research does not come close to representing the genetic and environmental diversity of the world. Indeed, ~79% of all GWAS participants (Martin et al. 2019) and ~67% of studies (Duncan et al. 2019) involve people of solely non-Hispanic European ancestries, who make up only 16% of the global population (Martin et al. 2019). GWASs also overrepresent individuals with higher socioeconomic status and better health (Fry et al. 2017, Mostafavi et al. 2020, Munafò et al. 2018). Several calls for greater diversity in genomic studies have been made (Goosby et al. 2018, Martin et al. 2019), and multiancestral GWASs are beginning to emerge (Lam et al. 2019, Mahajan et al. 2014, Peterson et al. 2019, Walters et al. 2018), but progress is slow.
Genetically informed research that is limited to White people is not representative of human development, because environmental opportunities systematically differ for White people compared with People of Color (Chetty et al. 2020). While we optimistically await genomic research on more diverse samples, developmental psychology research can use genetically informed studies to unpack the “invisible knapsack” that constitutes “White privilege” (McIntosh 1988) at the intersections of other environmental opportunities (Crenshaw 1989). In particular, early applications of polygenic scores to developmental psychology have provided novel findings that highlight the ways in which genetic effects depend on environments, which can be used to diagnose inequalities in social opportunity.
Figure 4 visualizes the theory that the expression of genetic variation is constrained by environmental opportunity. For some phenotypes a restricted environment may be beneficial (e.g., reducing the risk of developing health disorders), whereas in other cases it will limit an individual’s chances to self-actualize genetic propensities (e.g., attaining more education). For instance, a wealthy White person who attained their education in the latter half of the twentieth century in a democratic country and has a moderate to strong genetic propensity for educational attainment is more likely to attain high levels of socioeconomic status and longevity.
Figure 4.
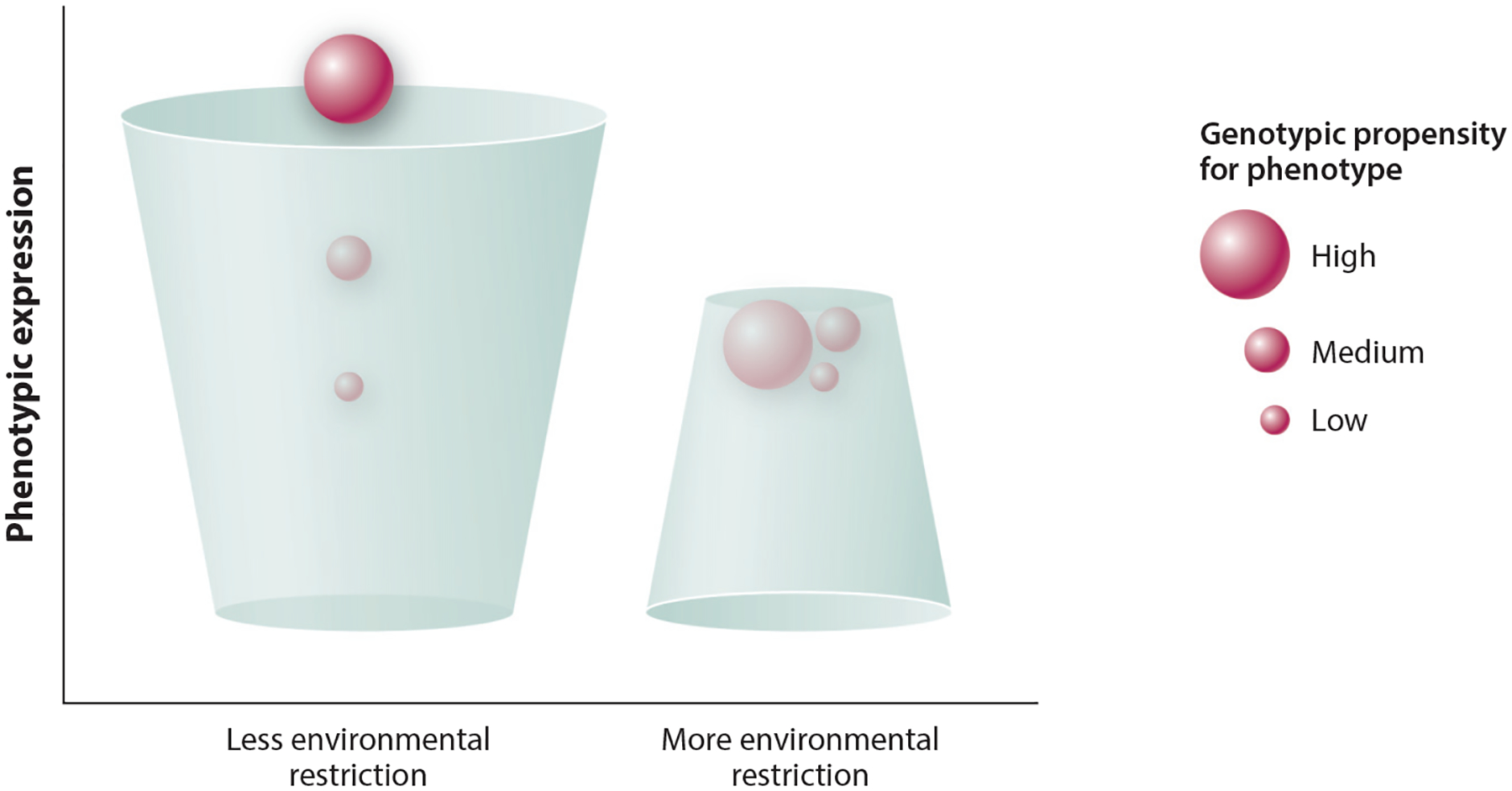
Visualization of the theory that the phenotypic expression of genetic variation is constrained and enhanced by environmental opportunity. The small upside-down cup imposes a glass ceiling effect, such that there is an attenuated association of genotypes with phenotypes imposed by restrictive environments. For some phenotypes a restricted environment may be beneficial (e.g., reducing the risk of developing health disorders), whereas in other cases it will limit an individual’s chances to self-actualize genetic propensities (e.g., attaining more education).
For example, one study comparing different birth cohorts found that education polygenic scores were better predictors of attained education in men relative to women in White American cohorts of the early twentieth century (Herd et al. 2019). As socioeconomic opportunities improved for White women, the predictive power of education polygenic scores matched and began to exceed those of White men. Similarly, studies have found that education polygenic scores became more predictive of attained education following the fall of the Soviet Union in Estonia (Rimfeld et al. 2018) and Hungary (Ujma et al. 2020). Family-based heritability studies provide corresponding evidence that genetic influences on academic outcomes and cognitive skills are maximized by environmental contexts that afford more opportunities (Heath et al. 1985, Selita & Kovas 2019, Tucker-Drob & Bates 2016).
Conversely, there is also evidence that higher socioeconomic status can buffer against genetic risk for negative health outcomes. Polygenic scores of BMI are less predictive of body size in White Germans who report higher educational attainment and income (Frank et al. 2019). These results correspond to quasi-experimental research showing that policies to increase the age of compulsory schooling preferentially benefit people with higher BMI polygenic scores (Barcellos et al. 2018).
Recent polygenic score research also helps inform how individuals with higher education polygenic scores attain their higher socioeconomic standing. Children with higher education polygenic scores achieve developmental milestones earlier and develop reading skills faster (Belsky et al. 2016). They are also more likely to enroll and persist in mathematics classes, more so when they attend a socioeconomically advantaged school (Harden et al. 2020; see also Trejo et al. 2018). At the same time, socioeconomically advantaged schools buffer students with lower education polygenic scores against dropping out of math (Harden et al. 2020). Across the life span, people born with a higher number of education-associated variants show greater upward mobility, relative to their family of origin, in occupational status, income, educational attainment, and wealth at retirement—even when they are being compared with their siblings (Belsky et al. 2016). This wealth association persists when controlling for education and income and is partially mediated by financial decision making (Barth et al. 2020). Thus, education-associated genetic variants predict socioeconomic attainment and longevity, in part because they capture individual differences in both cognitive and noncognitive skills (Demange et al. 2020). As parents, people with higher education polygenic scores provide more cognitively stimulating and warm environmental nurturance for their children, thereby transmitting their environmental privilege over generations (Armstrong-Carter et al. 2020, Wertz et al. 2019). Considered together, these genetic studies highlight the myriad intermediate phenotypes (e.g., linguistic and persistence skills) and environmental contexts (e.g., stimulating caregiving, school resources) that could be targeted to create more equitable outcomes.
6. CONCLUSION
There are clearly many risks of using polygenic scores: The limited portability of polygenic scores and a bias toward non-Hispanic European ancestries may increase existing health disparities, if polygenic scores confer a medical advantage, or may improve the development of interventions, but those benefits can be leveraged only by individuals of similar ancestries (Martin et al. 2019). As long as multiancestral polygenic scores are not technically feasible, polygenic score researchers will have to deal with the risks involved in defining and restricting their analyses within “populations” of similar genomic ancestries. Furthermore, because of the history of misuse of genetic concepts (Eigen & Larrimore 2006), researchers have a special responsibility to examine carefully the use and the language of racial and ethnic categories in their research.
Currently, biomedical research that focuses on genetic difference is garnering attention from political extremists and white nationalists, particularly when the phenotype under study is a psychological phenotype like cognition (Carlson & Harris 2020). Investigations that fail to acknowledge the full range of social mechanisms through which genetic differences can come to be correlated with phenotypes threaten to reinforce widely held stereotypes. It is not standard for either developmental psychologists or genomic researchers to be trained to handle these politically motivated misinterpretations of their work. While studying genetic propensities of individual differences in psychological phenotypes, we need to grapple with psychology’s racist past by clearly expressing that group differences in highly social outcomes (e.g., academic skills) arise from environmental disparities (e.g., racism, sexism, classism), not hypothetical genetic differences between groups.
Applying polygenic scores to individual-level decision making carries further risks of misuse. While polygenic scores for some diseases and disorders have reached a level of predictive power that has prompted the biomedical community to consider their clinical utility in medical settings (Khera et al. 2018, Lello et al. 2020), this is not true for predicting individual educational performance or personalized education (Morris et al. 2020). Therefore, discussions of the usage of polygenic scores on an individual level need to consider the potential risks and rewards of their use, which will depend on the phenotypic domain (e.g., health versus education), empirically established predictive validity for individuals, and the motivation of the organizations considering its usage. To reduce the risk of some forms of potential misuse, legislation aimed at prohibiting the discrimination of identifiable groups could include genome-based categorizations (Selita et al. 2020).
However, not using polygenic scores carries the risk of missing opportunities to understand and improve social influences on human development. For instance, when individuals receive information on their genetic risk for complex diseases alongside transactional rather than biodeterministic messaging, this can enhance positive behavioral changes (Frieser et al. 2018). In research, we finally have a tool to examine directly transmitted and environmentally mediated genetic effects of parenting on child development (Armstrong-Carter et al. 2020, Wertz et al. 2019). Moreover, we can examine the developmental precursors of highly heritable diseases (Walhovd et al. 2019) as well as the environments that effectively mitigate genetic risk for the most susceptible (Frank et al. 2019, Kuo et al. 2019). Polygenic scores therefore offer a novel opportunity to integrate genetic and environmental dimensions relevant to human development.
In summary, incorporating polygenic scores into developmental psychology has the potential to advance the identification and optimization of human development when these are cautiously interpreted in appreciation of existing social inequality and a transactional developmental perspective. The social challenge in response to genomic research lies in recognizing human genetic variation in a way that respects individual differences (Lorde 1984). If it is the “equality of opportunity” that “makes the genetic diversity among men meaningful” (Dobzhansky 1962), then only a world with equitable opportunity will allow us to make creative use of our genetic differences.
RACIAL/ETHNIC IDENTITY VERSUS GENETIC ANCESTRY.
Genetic ancestry is not the same thing as racial/ethnic identity (Rutherford 2020, Yudell et al. 2016). Racial/ethnic groups as they are defined in the USA today were inventions used as a political tool benefiting White people to justify the enslavement of Africans and genocide of indigenous Americans (Davis 2001, Kendi 2017). An adult’s racial/ethnic identity is the outcome of dynamic social processes largely occurring during childhood and adolescence (Meeus 2011). Racial/ethnic identity is context specific in terms of both how people self-identify (Pauker et al. 2018) and how they are categorized by others (Abascal 2020). In contrast, genetic ancestry describes patterns of gene frequencies that people have inherited from their genetic ancestors (see Section 2.2), and people’s genetic ancestry does not change across social contexts or over the life span.
The lack of biological “reality” underlying race does not mean that race is unimportant: Racial/ethnic identity remains relevant as it pertains to people’s lived experiences, culture, community, social challenges, and opportunities. Indeed, these social constructs might affect how genetic variation is expressed. But the social identity of race will never be reducible to those patterns of genetic variation. In scientific practice, labels based on biogeographical ancestry (e.g., predominantly recent European ancestries) may be well suited for many genomic studies, socially based labels (e.g., White) may be more appropriate for health disparities and clinical research, and both types of information may be valuable for studies of gene–environment interactions (Olson et al. 2005).
COMPUTING POLYGENIC SCORES.
The common steps required to calculate polygenic scores in a target sample are as follows (for a more technical and in-depth description of these procedures, see Choi et al. 2020):
Collect biological samples from the participants that can be used for genotyping. SNP genotyping can be performed using noninvasively collected saliva samples and can be relatively inexpensive (i.e., less than $70 per sample).
Perform a series of standard quality control procedures to remove genetic variants and participants who were not successfully genotyped (for a thorough description of quality control procedures, see Anderson et al. 2010 and Turner et al. 2011). Also, estimate principal components of ancestry to account for population stratification and cryptic relatedness in your sample.
Impute missing genotypes to improve genomic coverage. One relatively easy way to impute the data is to use an imputation service such as the Michigan or Sanger Imputation Server (see https://imputationserver.sph.umich.edu/ and https://imputation.sanger.ac.uk/).
Obtain GWAS summary statistics for the polygenic score you wish to calculate from sources such as the Polygenic Score Catalog (see http://www.pgscatalog.org/). It is critical that the target sample not be included in the GWAS discovery sample.
Finally, compute the polygenic scores. PLINK can be used to calculate a polygenic score for each individual, which is a weighted sum of that individual’s genetic propensities for the designated phenotype.
ACKNOWLEDGMENTS
We thank Prof. Bridget Goosby for her comments on an earlier version of this article. L.R. is supported by the German Research Foundation. K.P.H. is a faculty research associate of the Population Research Center at the University of Texas at Austin, which is supported by a grant (5-R24-HD042849) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. K.P.H. is also supported by Jacobs Foundation research fellowships.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Abascal M 2020. Contraction as a response to group threat: demographic decline and whites’ classification of people who are ambiguously white. Am. Sociol. Rev 85(2):298–322 [Google Scholar]
- Abdellaoui A, Hugh-Jones D, Yengo L, Kemper KE, Nivard MG, et al. 2019. Genetic correlates of social stratification in Great Britain. Nat. Hum. Behav 3(12):1332–42 [DOI] [PubMed] [Google Scholar]
- Allegrini AG, Selzam S, Rimfeld K, von Stumm S, Pingault JB, Plomin R. 2019. Genomic prediction of cognitive traits in childhood and adolescence. Mol. Psychiatry 24:819–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. 2010. Data quality control in genetic case-control association studies. Nat. Protoc 5(9):1564–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong-Carter E, Trejo S, Hill LJ, Crossley KL, Mason D, Domingue BV. 2020. The earliest origins of genetic nurture: The prenatal environment mediates the association between maternal genetics and child development. Psychol. Sci 31(7):781–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assari S, Javanbakht A, Saqib M, Helmi H, Bazargan M, Smith JA. 2020. Neuroticism polygenic risk score predicts 20-year burden of depressive symptoms for Whites but not Blacks. J. Med. Res. Innov 4(1):e000183. [PMC free article] [PubMed] [Google Scholar]
- Barcellos SH, Carvalho LS, Turley P. 2018. Education can reduce health differences related to genetic risk of obesity. PNAS 115(42):E9765–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth D, Papageorge NW, Thom K. 2020. Genetic endowments and wealth inequality. J. Political Econ 128(4):1474–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Domingue BW, Wedow R, Arseneault L, Boardman JD, et al. 2018. Genetic analysis of social-class mobility in five longitudinal studies. PNAS 115(31):E7275–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Harden KP. 2019. Phenotypic annotation: using polygenic scores to translate discoveries from genome-wide association studies from the top down. Curr. Dir. Psychol. Sci 28(1):82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington HL, et al. 2016. The genetics of success: how single-nucleotide polymorphisms associated with educational attainment relate to life-course development. Psychol. Sci 27(7):957–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border R, Johnson EC, Evans LM, Smolen A, Berley N, et al. 2019. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am. J. Psychiatry 176(5):376–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumpton B, Sanderson E, Heilbron K, Hartwig FP, Harrison S, et al. 2020. Avoiding dynastic, assortative mating, and population stratification biases in Mendelian randomization through within-family analyses. Nat. Commun 11:3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, et al. 2015a. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47(11):1236–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Loh PR, Finucane HK, Ripke S, Yang J, et al. 2015b. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47(3):291–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E, Harris K. 2020. Quantifying and contextualizing the impact of bioRxiv preprints through social media audience segmentation. bioRxiv 981589. 10.1101/2020.03.06.981589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Cesarini D, Benjamin DJ, Laibson DI. 2015. The fourth law of behavior genetics. Curr. Dir. Psychol. Sci 24(4):304–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney E 2012. Behavior genetics and postgenomics. Behav. Brain Sci 35(5):331–58 [DOI] [PubMed] [Google Scholar]
- Chetty R, Friedman JN, Saez E, Turner N, Yagan D. 2020. Income segregation and intergenerational mobility across colleges in the United States. Q. J. Econ 135(3):1567–633 [Google Scholar]
- Choi SW, Mak TS, O’Reilly PF. 2020. A guide to performing polygenic risk score analyses. Nat. Protoc 15:2759–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G 2019. Reading tea leaves? Polygenic scores and differences in traits among groups. arXiv:1909.00892 [q-bio.GN] [Google Scholar]
- Creanga AA. 2018. Maternal mortality in the United States: a review of contemporary data and their limitations. Clin. Obstet. Gynecol 61(2):296–306 [DOI] [PubMed] [Google Scholar]
- Crenshaw K 1989. Demarginalizing the intersection of race and sex: a black feminist critique of antidiscrimination doctrine, feminist theory and antiracist politics. Univ. Chicago Leg. Forum 1989(1):139–67 [Google Scholar]
- Dai CL, Vazifeh MM, Yeang C-H, Tachet R, Wells RS, et al. 2020. Population histories of the United States revealed through fine-scale migration and haplotype analysis. Am. J. Hum. Genet 106(3):371–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FJ. 2001. Who Is Black? One Nation’s Definition. Philadelphia: Pa. State Univ. Press [Google Scholar]
- Dawkins R 1982. The Extended Phenotype. Oxford, UK: Oxford Univ. Press [Google Scholar]
- Demange PA, Malanchini M, Mallard TT, Biroli P, Cox SR, et al. 2020. Investigating the genetic architecture of non-cognitive skills using GWAS-by-subtraction. bioRxiv 905794. 10.1101/2020.01.14.905794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T 1962. Genetics and equality: Equality of opportunity makes the genetic diversity among men meaningful. Science 137(3524):112–15 [DOI] [PubMed] [Google Scholar]
- Duncan GJ, Magnuson K, Votruba-Drzal E. 2017. Moving beyond correlations in assessing the consequences of poverty. Annu. Rev. Psychol 68:413–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Pollastri AR, Smoller JW. 2014. Mind the gap: why many geneticists and psychological scientists have discrepant views about gene–environment interaction (G×E) research. Am. Psychol 69(3):249–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Shen H, Gelaye B, Meijsen J, Ressler K, et al. 2019. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun 10:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen S, Larrimore M, eds. 2006. The German Invention of Race. Albany: SUNY Press [Google Scholar]
- Eisenberg N, Cumberland A, Spinrad TL. 1998. Parental socialization of emotion. Psychol. Inq 9(4):241–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dragano N, Arendt M, Forstner AJ, Nöthen MM, et al. 2019. A genetic sum score of risk alleles associated with body mass index interacts with socioeconomic position in the Heinz Nixdorf Recall Study. PLOS ONE 14(8):e0221252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese J 2008. Genetics and the social science explanation of individual outcomes. Am. J. Sociol 114(Suppl. 1):1–35 [DOI] [PubMed] [Google Scholar]
- Freese J, Shostak S. 2009. Genetics and social inquiry. Annu. Rev. Sociol 35:107–28 [Google Scholar]
- Frieser MJ, Wilson S, Vrieze S. 2018. Behavioral impact of return of genetic test results for complex disease: systematic review and meta-analysis. Health Psychol. 37(12):1134–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, et al. 2017. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am. J. Epidemiol 186(9):1026–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosby BJ, Cheadle JE, Mitchell C. 2018. Stress-related biosocial mechanisms of discrimination and African American health inequities. Annu. Rev. Sociol 44:319–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Shields J. 1972. Schizophrenia and Genetics. A Twin Study Vantage Point. New York: Academic [Google Scholar]
- Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, et al. 2019. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav 3(5):513–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D, Sirota L. 2000. Beware the chopsticks gene. Mol. Psychiatry 5(1):11–13 [DOI] [PubMed] [Google Scholar]
- Harden KP, Domingue BW, Belsky DW, Boardman JD, Crosnoe R, et al. 2020. Genetic associations with mathematics tracking and persistence in secondary school. npj Sci. Learn 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Koellinger PD. 2020. Using genetics for social science. Nat. Hum. Behav 4:567–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove TW. 2018. Intersecting social inequalities and body mass index trajectories from adolescence to early adulthood. J. Health Soc. Behav 59(1):56–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon A 2018. Why white supremacists are chugging milk (and why geneticists are alarmed). New York Times, Oct. 17. https://www.nytimes.com/2018/10/17/us/white-supremacists-science-dna.html
- Heath AC, Berg K, Eaves LJ, Solaas MH, Corey LA, et al. 1985. Education policy and the heritability of educational attainment. Nature 314(6013):734–36 [DOI] [PubMed] [Google Scholar]
- Herd P, Freese J, Sicinski K, Domingue BW, Mullan Harris K, et al. 2019. Genes, gender inequality, and educational attainment. Am. Sociol. Rev 84(6):1069–98 [Google Scholar]
- Herrnstein RJ, Murray C. 1996. The Bell Curve: Intelligence and Class Structure in American Life. New York: Free [Google Scholar]
- Hewitt JK. 2012. Editorial policy on candidate gene association and candidate gene-by-environment interaction studies of complex traits. Behav. Genet 42(1):1–2 [DOI] [PubMed] [Google Scholar]
- hooks b. 1995. Killing Rage: Ending Racism. New York: Holt [Google Scholar]
- Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, et al. (23andMe Res. Team, Major Depressive Disord. Work. Group Psychiatr. Genom. Consort.). 2019. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci 22(3):343–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, et al. 2019. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat. Genet 51(2):245–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendi IX. 2017. Stamped from the Beginning: The Definitive History of Racist Ideas in America. New York: Random House [Google Scholar]
- Kevles DJ. 1998. In the Name of Eugenics: Genetics and the Uses of Human Heredity. Boston: Harvard Univ. Press. Repr. ed. [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, et al. 2018. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50(9):1219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Wade KH, Zahid S, Brancale J, et al. 2019. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell 177(3):587–96.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellinger PD, Harden KP. 2018. Using nature to understand nurture. Science 359(6374):386–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, et al. 2018. The nature of nurture: effects of parental genotypes. Science 359(6374):424–28 [DOI] [PubMed] [Google Scholar]
- Kuo SIC, Salvatore JE, Aliev F, Ha T, Dishion TJ, Dick DM. 2019. The family check-up intervention moderates polygenic influences on long-term alcohol outcomes: results from a randomized intervention trial. Prev. Sci 20(7):975–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Chen C-Y, Li Z, Martin AR, Bryois J, et al. 2019. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet 51(12):1670–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, et al. 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet 50(8):1112–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lello L, Raben TG, Hsu SDH. 2020. Within-family validation of polygenic risk scores and complex trait prediction. bioRxiv 976554. 10.1101/2020.03.04.976654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, et al. (23andMe Res. Team, HUNT All-In Psychiatry). 2019. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet 51(2):237–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin JC, Nichols RC. 2012. Heredity, Environment, and Personality: A Study of 850 Sets of Twins. Austin: Univ. Tex. Press [Google Scholar]
- Lorde A 1984. Sister Outsider: Essays and Speeches. New York: Ten Speed [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, et al. 2014. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet 46(3):234–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard TT, Harden KP, Fromme K. 2019. Genetic risk for schizophrenia is associated with substance use in emerging adulthood: an event-level polygenic prediction model. Psychol. Med 49(12):2027–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, et al. 2017a. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100(4):635–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. 2019. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51(4):584–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Lin M, Granka JM, Myrick JW, Liu X, et al. 2017b. An unexpectedly complex architecture for skin pigmentation in Africans. Cell 171(6):1340–53.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh P 1988. White privilege and male privilege: a personal account of coming to see correspondences through work in women’s studies. Work. Pap. 189, Cent. Res. Women, Wellesley Coll., Wellesley, MA [Google Scholar]
- Meeus W 2011. The study of adolescent identity formation, 2000–2010: a review of longitudinal research. J. Res. Adolesc 21(1):75–94 [Google Scholar]
- Morris TT, Davies NM, Davey Smith G. 2020. Can education be personalised using pupils’ genetic data? eLife 9:e49962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TT, Davies NM, Hemani G, Davey Smith G. 2019. Why are education, socioeconomic position and intelligence genetically correlated? bioRxiv 630426. 10.1101/630426 [DOI] [Google Scholar]
- Mostafavi H, Harpak A, Conley D, Pritchard JK, Przeworski M. 2020. Variable prediction accuracy of polygenic scores within an ancestry group. eLife 9:e48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G. 2018. Collider scope: when selection bias can substantially influence observed associations. Int. J. Epidemiol 47(1):226–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C 2020a. Genetics will revolutionize social science. Wall Street Journal, Jan. 27. https://www.wsj.com/articles/genetics-will-revolutionize-social-science-11580169106 [Google Scholar]
- Murray C 2020b. Human Diversity: The Biology of Gender, Race, and Class. New York: Twelve [Google Scholar]
- Nivard MG, Gage SH, Hottenga JJ, van Beijsterveldt CEM, Abdellaoui A, et al. 2017. Genetic overlap between schizophrenia and developmental psychopathology: longitudinal and multivariate polygenic risk prediction of common psychiatric traits during development. Schizophr. Bull 43(6):1197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Barton NH. 2018. Tread lightly interpreting polygenic tests of selection. Genetics 208(4):1351–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BML, De Neve J-E, Turley P, Nivard MG, et al. (LifeLines Cohort Study). 2016. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet 48(6):624–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S, Berg K, Bonham V, Boyer J, Brody L, et al. 2005. The use of racial, ethnic, and ancestral categories in human genetics research. Am. J. Hum. Genet 77(4):519–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panofsky A, Donovan J. 2019. Genetic ancestry testing among white nationalists: from identity repair to citizen science. Soc. Stud. Sci 49(5):653–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, et al. (GERAD1 Consort., CRESTAR Consort.). 2018. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50(3):381–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parens E 2004. Genetic differences and human identities: on why talking about behavioral genetics is important and difficult. Rep. Suppl, Hastings Cent., Garrison, NY: [PubMed] [Google Scholar]
- Pauker K, Meyers C, Sanchez DT, Gaither SE, Young DM. 2018. A review of multiracial malleability: identity, categorization, and shifting racial attitudes. Soc. Personal. Psychol. Compass 12(6):e12392 [Google Scholar]
- Peterson RE, Kuchenbaecker K, Walters RK, Chen C-Y, Popejoy AB, et al. 2019. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 179(3):589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, Loehlin JC. 1977. Genotype–environment interaction and correlation in the analysis of human behavior. Psychol. Bull 84(2):309–22 [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, et al. 2015. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet 47(7):702–9 [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet 38(8):904–9 [DOI] [PubMed] [Google Scholar]
- Raffington L, Malanchini M, Grotzinger AD, Madole JW, Engelhardt LE, et al. 2019. Changing environments reveal innovative genetic variation in children’s cortisol responses. bioRxiv 856658. 10.1101/856658 [DOI] [Google Scholar]
- Regalado A 2018. DNA tests for IQ are coming, but it might not be smart to take one. MIT Technology Review, April 2. https://getpocket.com/explore/item/dna-tests-for-iq-are-coming-but-it-might-not-be-smart-to-take-one [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, et al. 2013. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340(6139):1467–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimfeld K, Krapohl E, Trzaskowski M, Coleman JRI, Selzam S, et al. 2018. Genetic influence on social outcomes during and after the Soviet era in Estonia. Nat. Hum. Behav 2(4):269–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, et al. (Schizophr. Work. Group Psychiatr. Genom. Consort.). 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511(7510):421–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford A 2020. How to Argue with a Racist: History, Science, Race, and Reality. London: Hachette [Google Scholar]
- Sagoe D, Pallesen S, Dlova NC, Lartey M, Ezzedine K, Dadzie O. 2019. The global prevalence and correlates of skin bleaching: a meta-analysis and meta-regression analysis. Int. J. Dermatol 58(1):24–44 [DOI] [PubMed] [Google Scholar]
- Salganik MJ, Lundberg I, Kindel AT, Ahearn CE, Al-Ghoneim K, et al. 2020. Measuring the predictability of life outcomes with a scientific mass collaboration. PNAS 117(15):8398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, McCartney K. 1983. How people make their own environments: a theory of genotype–environment effects. Child Dev. 54(2):424–35 [DOI] [PubMed] [Google Scholar]
- Selita F, Kovas Y. 2019. Genes and Gini: what inequality means for heritability. J. Biosoc. Sci 51(1):18–47 [DOI] [PubMed] [Google Scholar]
- Selita F, Willers M, Kovas Y. 2020. ‘Race’ and other group discrimination in the genomic era. Leg. Issues J 8(1):16–55 [Google Scholar]
- Selzam S, Ritchie SJ, Pingault JB, Reynolds CA, O’Reilly PF, Plomin R. 2019. Comparing within- and between-family polygenic score prediction. Am. J. Hum. Genet 105(2):351–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M, Maier RM, Ganna A, Bloemendal A, Martin AR, et al. 2019. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8:e39702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo S, Belsky DW, Boardman JD, Freese J, Harris KM, et al. 2018. Schools as moderators of genetic associations with life course attainments: evidence from the WLS and Add Health. Sociol. Sci 5:513–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Bates TC. 2016. Large cross-national differences in gene × socioeconomic status interact-tion on intelligence. Psychol. Sci 27(2):138–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer E 2000. Three laws of behavior genetics and what they mean. Curr. Dir. Psychol. Sci 9(5):160–64 [Google Scholar]
- Turkheimer E, Harden KP. 2014. Behavior genetic research methods: testing quasi-causal hypotheses using multivariate twin data. In Handbook of Research Methods in Social and Personality Psychology, ed. Reis H, Judd CM, pp. 159–87. Cambridge, UK: Cambridge Univ. Press. 2nd ed. [Google Scholar]
- Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, et al. 2011. Quality control procedures for genome-wide association studies. Curr. Protoc. Hum. Genet 68(1):1.19.1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujma PP, Eszlári N, Millinghoffer A, Bruncsics B, Petschner P, et al. 2020. Genetic effects on educational attainment in Hungary. bioRxiv 905034. 10.1101/2020.01.13.905034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet 9:255–66 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, et al. 2017. 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet 101(1):5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Sørensen Ø, Mowinckel AM, Reinbold CS, et al. 2019. Genetic risk for Alzheimer’s disease predicts hippocampal volume through the lifespan. bioRxiv 711689. 10.1101/711689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, et al. 2018. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci 21(12):1656–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples RS, Gaggiotti O. 2006. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol 15(6):1419–39 [DOI] [PubMed] [Google Scholar]
- Ware JJ, Van den Bree MBM, Munafò MR. 2011. Association of the CHRNA5-A3-B4 gene cluster with heaviness of smoking: a meta-analysis. Nicotine Tob. Res 13(12):1167–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, et al. 2019. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51(9):1339–48 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D. 2017. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz J, Belsky J, Moffitt TE, Belsky DW, Harrington HL, et al. 2019. Genetics of nurture: a test of the hypothesis that parents’ genetics predict their observed caregiving. Dev. Psychol 55(7):1461–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, et al. 2018. Meta-analysis of genome-wide association studies for height and body mass index in ~700,000 individuals of European ancestry. Hum. Mol. Genet 27(20):3641–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudell M, Roberts D, DeSalle R, Tishkoff S. 2016. Taking race out of human genetics. Science 351(6273):564–65 [DOI] [PubMed] [Google Scholar]


