Abstract
Sixty-one Burkholderia cepacia isolates from patients with cystic fibrosis (CF) and four plant isolates were screened for production of the siderophores salicylic acid (SA), pyochelin, cepabactin, and ornibactins and fingerprinted by a PCR-based randomly amplified polymorphic DNA (RAPD) method. Of the 24 RAPD types determined, 22 (92%) were associated with isolates that produced SA, 21 (87%) were associated with isolates that produced ornibactins, 15 (60%) were associated with isolates that produced pyochelin, and 3 (12%) were associated with isolates that produced cepabactin. Of the 24 RAPD types plus 2 phenotypic variants of types 1 and 9, 3 were associated with isolates that produced all four siderophores, 8 were associated with isolates that produced three siderophores, 12 were associated with isolates that produced two siderophores, and 3 were associated with isolates that produced only one siderophore. These results suggest that the numbers and types of siderophores produced by CF isolates of B. cepacia correlate with RAPD type and that SA and ornibactins are the most prevalent siderophores produced.
Burkholderia (formerly Pseudomonas) cepacia is an opportunistic pathogen in immunocompromised patients, particularly those with cystic fibrosis (CF) or chronic granulomatous disease (reviewed in references 8 and 9). A recent study of CF patients over a 20-year period concluded that colonization with B. cepacia significantly increased the risk of mortality at all levels of pulmonary function and that the risk was significantly higher for children (6). Differences in levels of severity of B. cepacia infections in CF patients have been noted, and these may be due to variation in the types of virulence factors expressed among strains. One potential virulence factor implicated in the pathogenesis of B. cepacia infections is the production of siderophores. B. cepacia has been reported to produce four different types of siderophores, namely, pyochelin, salicylic acid (SA), cepabactin, and ornibactins (13, 18, 19, 21, 22).
In a previous study of 43 B. cepacia isolates from CF patients from Toronto and Cleveland, 49% were found to produce pyochelin (18). The majority of pyochelin-positive strains were isolated from patients with severe pulmonary disease, with more than half of infections resulting in death. Pyochelin-negative strains were more frequently isolated from patients with moderate or mild infections (18). B. cepacia isolates that do not produce pyochelin were shown to utilize pyochelin for growth and iron transport. In a subsequent study using a rat model of chronic pulmonary infection, exogenously supplied pyochelin was shown to increase the virulence of B. cepacia pyochelin-negative strains (20). In addition to its role in iron acquisition, pyochelin has also been reported to play a role in tissue injury. Iron bound to pyochelin has been shown to be an efficient catalyst for hydroxyl radical (—OH) formation and to increase injury to pulmonary artery endothelial cells and pulmonary epithelial cells resulting from exposure to superoxide and hydrogen peroxide (4, 5).
SA, originally termed azurechelin, was previously reported to be produced by 88% of B. cepacia respiratory isolates from CF patients (19, 22). This siderophore was able to compete with transferrin for iron and promote growth of B. cepacia.
In 1989, Meyer et al. (13) identified and characterized cepabactin. Cepabactin production has been described for strains ATCC 25416, originally isolated from an onion, and ATCC 17759, a soil isolate; however, there have been no reports describing the production of cepabactin in clinical isolates of B. cepacia (13, 14).
More recently, B. cepacia strains have also been reported to produce linear hydroxamate or hydroxycarboxylate siderophores, termed ornibactins (21). Three related compounds, ornibactin-C4, -C6, and -C8, which differ only in the length of the acyl chain bound to a conserved tetrapeptide, have been identified (21).
We have shown that pyochelin and SA are commonly produced by B. cepacia respiratory isolates from CF patients (18, 19) and that they may play a role as virulence factors in these infections. Since the incidence of cepabactin and ornibactin production in clinical isolates was unknown, we determined whether these siderophores are produced in CF isolates. In light of recent epidemiological studies which suggest that CF isolates are clustered clonally and that some strains may be more transmissable (7, 10, 15, 17), we also examined the correlation of siderophore production patterns with the randomly amplified polymorphic DNA (RAPD) fingerprints of the isolates (11).
Fifty-seven isolates were from the sputa of CF patients and were originally obtained from C. L. Prober, Hospital for Sick Children, Toronto, Ontario, Canada; J. D. Klinger, Rainbow Babies and Children’s Hospital, Cleveland, Ohio; H. R. Rabin, University of Calgary, Calgary, Alberta, Canada; J. Burns, Children’s Hospital and Medical Center, Seattle, Wash.; and D. Welch, Oklahoma University Health Science Center, Oklahoma City. Four isolates from the blood of CF patients and four isolates from plants were obtained from J. D. Klinger. Strains ATCC 17759 and ATCC 25416, previously shown to produce pyochelin, cepabactin, and ornibactins, were used as controls (14).
B. cepacia isolates were grouped into types with related genetic fingerprints by RAPD analysis. Genomic DNA was isolated from cultures which were grown overnight in 4.5 ml of Luria-Bertani broth (1) (Life-Technologies, Burlington, Ontario, Canada). Primary typing was performed with primer 270 (TGCGCGCGGG [11]), and groupings were confirmed with primer 208 (ACGGCCGACC [11]). These primers have previously been shown to discriminate among unrelated B. cepacia strains (11). Twenty-five-microliter reaction mixtures contained 40 ng of genomic B. cepacia DNA and 40 pmol of RAPD primer. PCR cycles were performed in a DNA Thermal Cycler, model 480 (Perkin-Elmer, Norfolk, Conn.), as follows: four cycles at 94°C for 5 min, 36°C for 5 min, and 72°C for 5 min; 30 cycles at 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min; and a final extension at 72°C for 10 min. PCR mixtures were electrophoresed on 1.5% agarose gels in Tris-borate-EDTA buffer. Strains were typed at least once with each primer, and the strains from each proposed group were re-run in adjacent lanes on the same gel to confirm the RAPD typing.
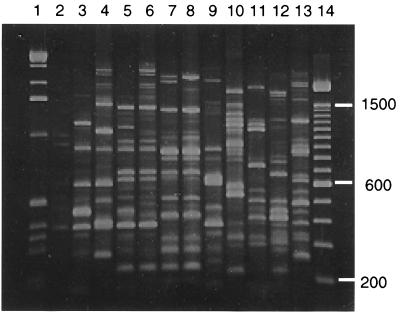
Sixty-five isolates from five different geographic locations were examined. Thirteen were found to have unique RAPD fingerprints, while the remaining 52 were grouped with isolates having similar RAPD fingerprints into one of nine types (Table 1). Figure 1 shows a representative RAPD fingerprint of each of the nine types containing multiple isolates. Five of the nine types (types 5, 6, 7, 8, and 9) were comprised of isolates from only one geographic location. Type 1 contained isolates from Toronto and Calgary, and types 2, 3, and 4 each contained isolates obtained from three different centers. Two of the isolates had fingerprints very similar to those of RAPD type 3, but fingerprints of these two isolates consistently contained several PCR products of between 1.6 and 3 kb which were not amplified from other isolates of type 3 (Fig. 1). Because of these differences, these two isolates were placed in subgroup 3A. The three type 4 isolates from Oklahoma City differed from the others of that type in that they lacked an amplified fragment of approximately 2.1 kb. In a previous study describing the use of these primers, epidemiologically related strains were categorized as having the same RAPD type as long as they differed in no more than three bands and had similarity coefficients of 0.7 (11). The three type 4 isolates from Oklahoma City, however, were subsequently also found to differ in that they do not produce pyochelin and cepabactin (Table 1). Therefore, these three isolates were placed in subgroup 4A.
TABLE 1.
Comparison of abilities of isolates of B. cepacia RAPD types to produce siderophores
| RAPD type | Strain | Sourcea | Ability to produceb:
|
|||
|---|---|---|---|---|---|---|
| SA | Pyochelin | Cepabactin | Ornibactins | |||
| 1 | K56-2 | Toronto | + | ± | − | + |
| K55-1 | Toronto | + | ± | − | + | |
| K61-3 | Toronto | + | ± | − | ||
| K53-2 | Toronto | + | ± | − | ||
| R5231 | Toronto | + | − | − | ||
| R2196 | Toronto | + | − | − | ||
| K37-3 | Toronto | + | − | − | ||
| K67-6 | Toronto | + | − | − | ||
| 12106-2 | Calgary | + | − | − | ||
| 7466ty | Calgary | + | − | − | ||
| 2 | K63-1 | Toronto | + | + | − | |
| K63-2 | Toronto | + | + | − | + | |
| K63-3 | Toronto | + | + | − | ||
| 10201 | Calgary | + | + | − | ||
| 10285 | Calgary | + | + | − | + | |
| 13858 | Calgary | + | + | − | ||
| Pc715j | Cleveland | + | + | − | ||
| 3 | H1724-1 | Toronto | + | ± | − | + |
| Pc67b | Cleveland | + | ± | − | ||
| Pc707g | Cleveland | + | + | − | ||
| Pc90ee | Cleveland | + | + | − | ||
| Pc706k | Cleveland | + | + | − | + | |
| Pc275c | Cleveland | + | + | − | ||
| Pc535a | Cleveland | + | + | − | + | |
| cc-7 | Oklahoma City | + | + | − | + | |
| cc-18 | Oklahoma City | + | + | − | ||
| 3A | P120 | Seattle | + | + | − | + |
| P107 | Seattle | + | + | − | ||
| 4 | 5530pk | Calgary | + | + | + | |
| 34930 | Calgary | + | + | + | + | |
| 34192 | Calgary | + | + | + | ||
| 7459 | Calgary | + | + | + | ||
| 10243 | Calgary | ± | + | + | ||
| P109 | Seattle | + | + | + | + | |
| 4A | cc-2 | Oklahoma City | + | − | − | + |
| cc-8 | Oklahoma City | + | − | − | ||
| cc-23 | Oklahoma City | + | − | − | ||
| 5 | cc-9 | Oklahoma City | + | + | − | + |
| cc-19 | Oklahoma City | + | + | − | ||
| cc-21 | Oklahoma City | + | + | − | ||
| 6 | cc-17 | Oklahoma City | + | − | − | + |
| cc-22 | Oklahoma City | + | − | − | ||
| 7 | 394g | Cleveland | + | − | − | |
| 554d | Cleveland | + | − | − | + | |
| 43-3 | Cleveland (blood) | + | − | − | + | |
| 48-36 | Cleveland (blood) | + | − | − | ||
| 56-31 | Cleveland (blood) | + | − | − | ||
| 69-14 | Cleveland (blood) | + | − | − | ||
| 8 | Pc22-20 | Cleveland (plant) | ± | + | + | + |
| Pc22-22 | Cleveland (plant) | ± | + | + | + | |
| 9 | P118 | Seattle | + | + | − | |
| Pc104 | Seattle | − | − | − | + | |
| 10 | P114 | Seattle | + | + | + | + |
| 11 | Pc701j | Cleveland | + | + | − | + |
| 12 | Pc102 | Seattle | + | + | − | − |
| 13 | 174 | Seattle | − | + | − | − |
| 14 | P108 | Seattle | + | − | − | + |
| 15 | cc-26 | Oklahoma City | + | − | − | + |
| 16 | 12544 | Calgary | + | − | − | + |
| 17 | Pc105 | Seattle | + | − | − | + |
| 18 | Pc224c | Cleveland | + | + | − | + |
| 19 | Pc22-12 | Cleveland (plant) | + | − | − | − |
| 20 | Pc22-18 | Cleveland (plant) | + | + | − | + |
| 21 | cc-14 | Oklahoma City | + | − | − | + |
| 22 | P121 | Seattle | − | + | − | + |
Respiratory isolates from CF patients unless otherwise indicated.
+, easily detectable within 24 h of growth; ±, detectable at very low concentrations after 40 h of growth; −, not detectable.
FIG. 1.
RAPD fingerprints of DNAs from B. cepacia isolates amplified with primer 208. Lane 1, molecular size markers (1-kb ladder); lane 2, control reaction without DNA; lane 3, type 1; lane 4, type 2; lane 5, type 3; lane 6, type 3A; lane 7, type 4; lane 8, type 4A; lane 9, type 5; lane 10, type 6; lane 11, type 7; lane 12, type 8; lane 13, type 9; lane 14, molecular size markers (100-bp ladder; sizes are shown on the right). A photograph of the gel was scanned with a Hewlett-Packard Scan Jet 4C scanner with Hewlett-Packard Deskscan II software.
Approximately 10 CF isolates were randomly selected from each geographic center for analysis. The isolates from patients in Toronto belonged to RAPD types 1 and 2, with the exception of one type 3 isolate. The isolates from Calgary belonged to either type 2 or type 4, with the exception of one isolate with a unique RAPD type, designated type 16. The Cleveland isolates were predominantly grouped as type 3 and type 7, and the Oklahoma City isolates were split between types 3, 4A, 5, and 6. There was no predominant RAPD type in the group of strains from Seattle. The fingerprints of the plant isolates were not related to those of any of the CF patient isolates.
The 61 CF isolates and 4 plant isolates were grown in succinate medium (12) and analyzed for the production of pyochelin, SA, and cepabactin. For some isolates, we also examined SA and pyochelin production in cultures grown in 0.5% Casamino Acids (Difco, Detroit, Mich.)–0.2 mM MgCl2 (CAA medium), deferrated as previously described (20). Chloroform extracts of acidified supernatants were dried and resuspended in 250 μl of methanol and chromatographed on a thin layer (250 μM) of Silica Gel G (Mandel Scientific), with chloroform-acetic acid-ethanol (90:5:2.5) as the development solvent. The limits of detection for SA and pyochelin by fluorescence on thin-layer-chromatography plates were 0.5 and 20 μg, respectively.
Pyochelin was detected in chloroform extracts of culture supernatants in 38 (62%) of the 61 strains (Table 1). Pyochelin was easily detected in the supernatants of 32 isolates, whereas it was detected only at very low concentrations after 40 h of growth in the supernatants of 6 isolates (Table 1). In general, the isolates of each RAPD type had similar patterns of pyochelin production. The pyochelin-negative isolates were clustered among RAPD types 1, 4A, 6, and 7. The variation in levels of pyochelin produced by isolates of RAPD type 1 may be a result of the very low levels of pyochelin produced by these strains. It is possible that the six strains of this group which were reported as negative may produce pyochelin at a concentration of less than 0.1 μg/ml in the culture medium after 40 h, which is the limit of detection in this assay. Only one of the two RAPD type 9 strains produced pyochelin. Strain P118 produced significant amounts of pyochelin, so the differences between these two isolates do not appear to be due to the sensitivity of the assay. Three of the four plant isolates produced pyochelin.
SA was detectable in culture supernatants from 57 (93%) of the 61 CF isolates (Table 1). The two RAPD type 9 isolates also differed in terms of SA production. P118 produced SA and pyochelin, whereas Pc104 did not produce either siderophore. Two of the plant isolates tested produced significant amounts of SA; the other two produced SA, which was detectable only at very low levels after 40 h of growth.
Cepabactin was detected in the supernatants of only nine of the isolates tested: two plant isolates (RAPD type 8) and seven (11%) CF isolates. Of the CF isolates that produced cepabactin, six were of RAPD type 4 and one was a unique RAPD type. The two American Type Culture Collection (ATCC) strains used as positive controls were soil and onion isolates. Therefore, four of six of the environmental isolates examined were cepabactin positive, which suggests that cepabactin may be more common in environmental strains than in clinical isolates; however, a larger number of environmental isolates need to be examined to determine the frequencies of cepabactin production in these strains.
Ornibactins were isolated as described by Meyer et al. (14) with minor modifications from cultures grown in succinate medium supplemented with 10 mM ornithine. The cells were pelleted from 150 ml of succinate medium by centrifugation at 7,000 × g, and the supernatants were dried by lyophilization. The residues were extracted three times with 15 ml of methanol. The pooled methanol extracts were treated with anhydrous sodium sulfate, filtered, and concentrated to dryness by rotary evaporation. The residues were resuspended in 1 ml of methanol and applied to a Sephadex LH-20 column (35 by 1.5 cm), with methanol as the eluting solvent. Column fractions were screened for iron-binding compounds by the chrome azurol S assay (16). Depending on the isolate, up to three peaks with iron-binding activities were eluted from the column. The first peak with chrome azurol S activity contained ornibactins and eluted between 36 and 48 ml. The second and third peaks were identified as cepabactin and pyochelin, respectively. The fractions from the first peak were dried, and 150 nmol was spotted on Silica Gel G and developed in butanol-acetic acid-water (3:1:1). Ornibactins were visible as three brown bands (Rf 0.25, Rf 0.18, and Rf 0.09) when the plate was sprayed with iron (0.1 M FeCl3 in 0.1 N HCl). Previous studies had used reversed-phase high-pressure liquid chromatography to separate ornibactins (14, 21). Therefore, to confirm that these three iron-binding compounds were indeed ornibactins, portions of the three bands from strain ATCC 17759 were eluted from the thin-layer chromatography gel and the molecular masses of the iron-free forms were determined by electrospray mass spectrometry. All mass spectrometric experiments were conducted with an API 300 triple-quadrupole mass spectrometer (Perkin-Elmer/SCIEX, Concord, Ontario, Canada.) Nanoelectrospray mass spectra were obtained in the positive-ion mode with a modified MicroIonspray interface (Perkin-Elmer/SCIEX) comprising a small tee insert used to establish electrical contact between a fused-silica transfer line (inside diameter, 80 cm by 50 μm) and the emitter tip essentially as previously described (2). The molecular masses of the three compounds were 680, 708, and 736 Da (data not shown), which correspond to the deferrated forms of ornibactin-C4 (ornibactin B), ornibactin-C6 (ornibactin D), and ornibactin-C8 (ornibactin F), respectively (21).
Since production of pyochelin, SA, and cepabactin generally correlated with RAPD type, one or two representative strains from each RAPD type were screened for the production of ornibactins. Twenty-nine of 32 (91%) isolates were found to produce ornibactins. Two of the unique CF isolates from Seattle and one of the plant isolates were found to be ornibactin negative.
Patterns of siderophore production of members of the same RAPD type were generally identical, with the exception of those of types 1 and 9. The differences in detection of pyochelin produced by RAPD type 1 strains, however, may be due to the low levels produced by these strains. Of the 24 RAPD types, 22 (92%) were associated with isolates that produced SA, 21 (87%) were associated with isolates that produced ornibactins, 15 (60%) were associated with isolates that produced pyochelin, and 3 (12%) were associated with isolates that produced cepabactin. Only isolates of 3 of the 26 types (24 RAPD types plus 2 variants of types 1 and 9), RAPD types 4, 8, and 10, produced all four siderophores. All of the strains which produced cepabactin also produced SA, pyochelin, and ornibactins. Isolates of eight types produced three types of siderophores: SA, pyochelin, and ornibactins. Isolates of 12 types produced two siderophores; isolates of 9 of these types produced SA and ornibactins, isolates of 2 produced SA and pyochelin, and isolates of 1 produced pyochelin and ornibactins. Isolates of each of three RAPD types produced only one type of siderophore, SA, pyochelin, or ornibactins. Four of the isolates of RAPD type 7 were bacteremic isolates from CF patients in Cleveland. These isolates produced two types of siderophores: SA and ornibactins.
Bevivino et al. (3), in a study of four strains, previously suggested that clinical isolates of B. cepacia could be differentiated from rhizosphere isolates by their production of pyochelin and SA. In our study, three of four additional plant isolates were also shown to produce pyochelin and all of the plant isolates produced SA. Meyer et al. examined pyochelin, cepabactin, and ornibactin production in B. vietnamiensis as well as two clinical isolates of B. cepacia and the two ATCC strains (13, 14). All of the B. vietnamiensis strains and the two B. cepacia clinical isolates produced only ornibactins, whereas the ATCC strains produced all three siderophores. Meyer et al. suggested that the B. cepacia isolates which produce only ornibactins are more related to B. vietnamiensis than to the two B. cepacia ATCC strains and that production of siderophores may be used to discriminate between these two species (14). We have determined that there are a variety of siderophore production patterns among B. cepacia CF isolates and that it is therefore not possible to speciate strains based on siderophore production. The ATCC strains, however, may not be considered typical of CF isolates, as the majority of CF isolates do not produce cepabactin. Our study indicates that ornibactins and SA are the predominant siderophores produced by CF isolates and that pyochelin may also be an important siderophore for B. cepacia. Analysis of siderophores or other potential virulence factors in this organism should be performed in conjunction with a molecular typing method such as RAPD analysis with representative groups of strains prior to forming conclusions regarding their significance in particular patient populations.
Acknowledgments
This study was supported by a grant from the Canadian Cystic Fibrosis Foundation. M.C. was the recipient of an Alberta Heritage Foundation for Medical Research summer studentship award.
The CF isolates used in this study were provided by D. E. Woods. We also thank E. Mahenthiralingam for helpful advice and assistance with the RAPD typing methodology and P. Thibault and D. Krajcarski for assistance with the mass spectroscopy.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1 1989. , chapter 2. John Wiley & Sons, New York, N.Y. [Google Scholar]
- 2.Bateman K P, White R L, Thibault P. Disposable emitters for on-line capillary zone electrophoresis/nanoelectrospray mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:307–315. [Google Scholar]
- 3.Bevivino A, Tabacchioni S, Chiarini L, Carusi M V, Del Gallo M, Visca P. Phenotypic comparison between rhizosphere and clinical isolates of Burkholderia cepacia. Microbiology. 1994;140:1069–1077. doi: 10.1099/13500872-140-5-1069. [DOI] [PubMed] [Google Scholar]
- 4.Britigan B E, Rasmussen G T, Cox C D. Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect Immun. 1997;65:1071–1076. doi: 10.1128/iai.65.3.1071-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffman T J, Cox C D, Edeker B L, Britigan B E. Possible role of bacterial siderophores in inflammation. Iron bound to the Pseudomonas siderophore pyochelin can function as a hydroxyl radical catalyst. J Clin Invest. 1990;86:1030–1037. doi: 10.1172/JCI114805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corey M, Farewell V. Determinants of mortality from cystic fibrosis in Canada, 1970–1989. Am J Epidemiol. 1996;143:1007–1017. doi: 10.1093/oxfordjournals.aje.a008664. [DOI] [PubMed] [Google Scholar]
- 7.Govan J R W, Brown P H, Maddison J, Doherty C J, Nelson J W, Dodd M, Greening A P, Webb A K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 8.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 10.Johnson W M, Tyler S D, Rozee K R. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J Clin Microbiol. 1994;32:924–930. doi: 10.1128/jcm.32.4.924-930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahenthiralingam E, Campbell M E, Henry D A, Speert D P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J Clin Microbiol. 1996;34:2914–2920. doi: 10.1128/jcm.34.12.2914-2920.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer J-M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physico-chemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 13.Meyer J-M, Hohnadel D, Halle F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J Gen Microbiol. 1989;135:1479–1487. doi: 10.1099/00221287-135-6-1479. [DOI] [PubMed] [Google Scholar]
- 14.Meyer J-M, Tran Van V, Stintzi A, Berge O, Winkelmann G. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia) Biometals. 1995;8:309–317. doi: 10.1007/BF00141604. [DOI] [PubMed] [Google Scholar]
- 15.Pegues D A, Carson L A, Tablan O C, FitzSimmons S C, Roman S B, Miller J M, Jarvis W R the Summer Camp Study Group. Acquisition of Pseudomonas cepacia at summer camps for patients with cystic fibrosis. J Pediatr. 1994;124:694–702. doi: 10.1016/s0022-3476(05)81357-5. [DOI] [PubMed] [Google Scholar]
- 16.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 17.Smith D L, Gumery L B, Smith E G, Stableforth D E, Kaufmann M E, Pitt T L. Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: evidence of person-to-person transmission. J Clin Microbiol. 1993;31:3017–3022. doi: 10.1128/jcm.31.11.3017-3022.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol P A. Production and untilization of pyochelin by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1986;23:560–562. doi: 10.1128/jcm.23.3.560-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 20.Sokol P A, Woods D E. Effect of pyochelin on Pseudomonas cepacia respiratory infections. Microb Pathog. 1988;5:197–205. doi: 10.1016/0882-4010(88)90022-8. [DOI] [PubMed] [Google Scholar]
- 21.Stephan H, Freund S, Beck W, Jung G, Meyer J-M, Winkelmann G. Ornibactins—a new family of siderophores from Pseudomonas. Biometals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- 22.Visca P, Ciervo A, Sanfilippo V, Orsi N. Iron-regulated salicylate synthesis by Pseudomonas spp. J Gen Microbiol. 1993;139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]