Abstract
IFNγ-producing ex-Th17 cells [‘Th1/17’] were shown to play a key pathogenic role in experimental colitis and are abundant in the intestine. Here, we identified and characterised a novel, potentially colitogenic subset of Th17 cells in the intestine of patients with Crohn’s disease [CD]. Human Th17 cells expressing CCR5 [‘pTh17’] co-expressed T-bet and RORC/γt and produced very high levels of IL-17, together with IFN-γ. They had a gene signature of Th17 effector cells and were distinct from established Th1/17 cells. pTh17 cells, but not Th1/17 cells, were associated with intestinal inflammation in CD, and decreased upon successful anti-TNF therapy with infliximab. Conventional CCR5[-]Th17 cells differentiated to pTh17 cells with IL-23 in vitro. Moreover, anti-IL-23 therapy with risankizumab strongly reduced pTh17 cells in the intestine. Importantly, intestinal pTh17 cells were selectively activated by adherent-invasive Escherichia coli [AIEC], but not by a commensal/probiotic E. coli strain. AIEC induced high levels of IL-23 and RANTES from dendritic cells [DC]. Intestinal CCR5+Th1/17 cells responded instead to cytomegalovirus and were reduced in ulcerative colitis [UC], suggesting an unexpected protective role. In conclusion, we identified an IL-23–inducible subset of human intestinal Th17 cells. pTh17 cells produced high levels of pro-inflammatory cytokines, were selectively associated with intestinal inflammation in CD, and responded to CD-associated AIEC, suggesting a key colitogenic role.
1. Introduction
Inflammatory bowel diseases [IBDs] are characterised by chronic intestinal inflammation likely induced by an excessive immune response against intestinal microbes. The major forms of IBDs are Crohn’s disease [CD] and ulcerative colitis [UC], which have distinct clinical features.1 It is becoming increasingly evident that pro-inflammatory Th17 responses against intestinal bacteria play a key role in IBD pathogenesis.2,3 Th17 cells are a distinct differentiation ‘lineage’ of CD4+T cells, which is characterised by the production of IL-17, the expression of the lineage-defining transcription factor RORC/γt, the chemokine receptor CCR6, and the IL-23 receptor [IL-23R].4 Th17 cells are heterogeneous and, in addition to IL-17, can also produce pro- and/or anti-inflammatory cytokines, including IL-22, GM-CSF, IL-21, TNF-α, and IL-10, in response to polarising cytokines.4 Thus, IL-1β,IL-12, and IL-23 promote pro-inflammatory Th17 cells, and TGF-β, IL-6, and IL-21 can promote anti-inflammatory features.5–7 Th17 cells fight extracellular bacteria and fungi, and induce the release of antimicrobial peptides and of chemokines by epithelial cells. In particular, Th17 cells are enriched in the intestine, react with intestinal bacteria, and are absent in the gut of germ-free mice,8–11 demonstrating that intestinal bacteria can induce their generation. However, chronically activated Th17 cells can also exert pathogenic functions, as suggested in IBDs. Indeed, Th17 cells show a high degree of plasticity,12 and can become Th1-like cells, up-regulating T-bet and acquiring IFN-γ–producing capacities,13,14 while down-regulating IL-17 production. Notably, these ex-Th17 Th1 cells play a critical pathogenic role in mouse colitis models.15 Thus, IFN-γ produced by Th17 cells is required to drive intestinal inflammation15 and increases epithelial barrier permeability.16 Notably, IL-23 plays a central role in promoting the differentiation and proliferation of colitogenic Th17 cells in the murine gut.17 Nailing down a pathogenic T cell population in the gut of IBD patients is difficult, but cells with similar features as the highly colitogenic murine ex-Th17 cells have been identified in humans.14,18 These T cells co-express CXCR3 and CCR6, produce high levels of IFN-γ together with low levels of IL-17, and are therefore also called nonconventional Th1-, Th1*, or Th1/17 cells.14,18,19 Notably, Th1/17 cells are abundant in the inflamed mucosa of IBD patients.14
A critical feature of colitogenic T cells in mice is their reactivity to enteric dysbiotic microbiota.20,21 Thus, microbiota-specific IFN-γ–producing Th17 cells accumulate in the inflamed intestinal tissue during colitis progression.22 Moreover, recent evidence demonstrates that dysbiotic gut microbiota from IBD patients induces Th17 cells in the gut of mice upon transfer, and worsens experimental colitis.20 The detrimental changes in the gut microbiota composition have been well documented in CD.21 In particular, adherent-invasive Escherichia coli [AIEC] are significantly more prevalent in Crohn’s tissue lesions then in controls and are rarely found in UC specimens.23
Here, we characterised a previously undescribed population of intestinal, IFN-γ producing, human, Th17 cells, which were selectively associated with intestinal inflammation in CD. These Th17 cells were generated with IL-23 and activated by AIEC, suggesting a key role in CD pathogenesis.
2. Material and Methods
2.1. Human samples
Peripheral blood, lymph nodes, and intestinal specimens were isolated from patients with CD and from patients with UC. Intestinal samples isolated from the distal part of tumour from colorectal cancer [CRC] patients and peripheral blood from healthy subjects [n = 49, matched for age and sex to IBD patients] were used as non-IBD and healthy controls for intestinal and blood samples,24 respectively.
All IBD patients included in this study were categorised/classified according to clinical and endoscopic disease activity, using the Harvey–Bradshaw index [HBI] and the Simple Endoscopic Score [SES-CD] for CD patients or Mayo score for UC patients [Table 1]. For some experiments, biopsies were taken before and after treatment with biologic drugs, namely infliximab and risankizumab [Supplementary Table 1]. Infliximab was given intravenously at 5 mg/kg at Weeks 0, 2, and 6 and then every 8 weeks. Risankizumab was given intravenously at a fixed dose of 600 mg at Weeks 0, 4, and 8, and then subcutaneously at a dose of 360 mg every 8 weeks. The clinical characteristics and therapies of all IBD and non-IBD patients are listed in Table 1. The CD patients were either ileal or ileocolonic. The study was registered with Eudract ref. no. 2015-003270-32 and the institutional review board approved the study with permission no. 566_2015 quinquies, approved by the local ethics committee [Milano Area B, code 128_2018bis], and was performed in accordance with the Declaration of Helsinki protocols.
Table 1.
Baseline clinical characteristics of Crohn’s disease [CD], ulcerative colitis [UC] and non-IBD patients included in the study
| Clinical characteristics | CD | UC | Non-IBD |
|---|---|---|---|
| Number of Patients, n | 65 | 31 | 16 |
| Male/female, n | 29/36 | 15/16 | 8/8 |
|
Age at enrolment,
year [mean ± SD] |
40 ± 7 | 41 ± 13 | 69 ± 12 |
|
Disease duration,
years [mean ± SD] |
11 ± 6 | 9 ± 5 | - |
| Concomitant therapy [n] | |||
| None | 25 | 23 | |
| Mesalamine | 8 | 4 | |
| Antibiotics | 2 | 0 | |
| Thiopurines | 9 | 0 | |
| Corticosteroids | 4 | 4 | |
| Infliximab | 12 | 0 | |
| Risankizumab | 5 | 0 | |
IBD, inflammatory bowel disease; SD, standard deviation.
2.2. Cell isolation and purification
Peripheral blood mononuclear cells [PBMC] were isolated by Ficoll density gradient centrifugation [Amersham Pharmacia Biotech, Uppsala, Sweden]. Lamina propria mononuclear cells [LPMCs] were isolated as previously described.25,26 Mesenteric lymph nodes [LNs] were smashed into 70-μm nylon strainers [BD Biosciences] and erythrocytes lysed with red blood cell [RBC] lysis buffer [BD Biosciences].
For characterisation of T helper [Th] cell subsets from PBMCs, LNs, and LPMCs, T cells were stained with a combination of fluorochrome-conjugated antibodies as specified in Supplementary Table 2 and sorted [FACSAria III, BD Biosciences], according to the expression of the following specific surface markers combination: CD4+IL-7R+CD25lowCCR6-CXCR3+ [Th1 cells], CD4+IL-7R+CD25lowCCR6+CXCR3+ [Th1/17 cells] and CD4+IL-7R+CD25lowCCR6+CXCR3- [Th17 cells]. These three subsets were further subdivided according to CCR5 expression.
Human monocytes for antigen-specificity assays and for generation of monocyte-derived dendritic cells [MoDC] were purified from blood samples by Ficoll density gradient separation and by positive selection using CD14+ selection [CD14 Microbeads, Miltenyi Biotec, Bergisch Gladbach, Germany].
2.3. Flow cytometry
T cell subsets were analysed for the expression of surface markers, transcription factors, and cytokines by staining with various combinations of fluorochrome-conjugated monoclonal antibodies [Supplementary Table 2]. For intracellular cytokine detection, T cells were incubated for 4 h in the presence or absence of phorbol 12-myristate 13- acetate [PMA], ionomycin in complete RPMI [10% FBS, 1 mmol/L sodium pyruvate, 10 mmol/L nonessential amino acids, and 1% penicillin/streptomycin], and with Brefeldin A [Sigma, St Louis, MO] for an additional 2 h. After fixation with 2% paraformaldehyde [Merck, Whitehouse Station, NJ], cells were permeabilised with PBS-0.5% saponin [Sigma]. Since CD4+IL-7R+T cells produce IL-10 with delayed kinetics,27 IL-10+ cells were analysed following stimulation for 30 h with immobilised anti-CD3 antibodies, as detailed previously.28 The expression of functional IL-23 receptors [IL-23R] was assessed by IL-23–induced STAT3 phosphorylation.29 Analysis was performed with a FACSCanto II cytometer [Becton Dickinson, Franklin Lakes, NJ] and analysed using FlowJo software [BD Biosciences].
2.4. RNA isolation and sequencing
RNA was isolated from CCR6+Th cell subsets according to CXCR3 and CCR5 expression from the blood of four different healthy donors and from the intestinal lamina propria of eight CD patients, using the mirVana Isolation Kit [Ambion]. Residual contaminating genomic DNA was removed from the total RNA fraction using Turbo DNA-free [Thermo Fisher]. The RNA yields were quantified using the QuantiFluor RNA System [Promega] and RNA quality was assessed by 2100 Bioanalyser [Agilent]. Library construction was performed with Illumina TruSeq RNA Sample Preparation Kit v2 using from 30ng to 100 ng of total RNA as starting material. To obtain this amount of RNA from the rare pTh17 cells, donors were pooled and analysed in two [blood] and three [intestine] independent sequencing reactions. Final libraries were sequenced on an Illumina HiSeq with paired-end 2 × 150-bp read lengths.
2.5. RNA sequencing data analysis
Quality control of raw reads was performed with FastQC v.0.11.2. Adapter removal and read trimming was performed using Trimmomatic v.0.32. Trimmed reads were aligned to the reference genome [GRCh38] using STAR 2.5.2b and gencode annotation v.25. The raw counting of the genes obtained by STAR was then used as input for DESeq2 analysis. Downstream analyses were performed using R [3.4.4]. In particular, raw counts were normalised using DESeq2, and differentially expressed genes were selected using a false-discovery rate [FDR] lower than 5% and log2 fold-change of 1. Data transformation using ‘rlog’ function was adopted for principal component analysis [PCA] and heatmap visualisation.
2.6. Homogenization of biopsies and ELISA
Intestinal biopsies [three to four per patient] were blotted dry, weighed [range, 12–156 mg], and then frozen at -80°C until used for assay. Samples were placed in glass tubes containing 1 ml of PBS with protease inhibitor cocktail [Complete Protease Inhibitor Cocktail Tablets, Roche Diagnostics, Mannheim, Germany] and phenylmethylsulphonyl fluoride [PMSF, Sigma-Aldrich, Darmstadt, Germany] and homogenised in ice with an IKA ULTRA-TURRAX T25 homogeniser [IKA Works, Wilmington, NC]. The homogenates were then centrifuged at 13 000 rpm at 4°C in a microfuge for 15 min, and the supernatants were then transferred to fresh tubes and frozen at -80°C.
Supernatants of homogenised biopsies were analysed for IL-6 [ImmunoTools], IL-12p70 [BioLegend], IL-23p19 [R&D Systems], and IL-1β [BioLegend] by ELISA according to the manufacturer’s instructions. The ELISA plates were read on microplate reader [SAFAS MP96], and data were analysed with Prism software [version 7; GraphPad Software, La Jolla, CA].
2.7. Generation of monocyte-derived dendritic cells [MoDC] and infection assays
MoDC were obtained from CD14+ human monocytes purified from blood of healthy subjects as previously described.30 MoDCs were seeded into round-bottom, 96-well culture plates at a density of 105 cells/well in complete RPMI medium without penicillin/streptomycin and infected with 107 CFU/mL of AIEC-LF82 or E. coli Nissle 1917 strain alone [MOI 10:1 bacteria per phagocytic cell]. Following 1 h of phagocytosis, gentamicin [20 ug/mL, Sigma-Aldrich, Darmstadt, Germany] was added to kill extracellular bacteria. Subsequently, infected MoDC were washed and maintained in RPMI supplemented with gentamicin [2 ug/mL] for additional 22 h. The levels of IL-1β [BioLegend], IL-12p70 [BioLegend], IL-23p19 [R&D Systems], IL-6 [ImmunoTools], IP-10 [R&D Systems], and RANTES [R&D Systems] in supernatants of infected MoDC were analysed after 24 h by ELISA. The ELISA plates were read on a microplate reader [SAFAS MP96], and data were analysed with Prism software [version 7; GraphPad Software, La Jolla, CA].
2.8. In vitro T cell differentiation
FACS-purified cTh17 cells [CD4+IL-7R+CD25lo/-CCR6+CXCR3-CCR5[-]] from buffy-coated blood of healthy donors [HD] were stimulated in plate-bound anti-CD3 [0.02 μg/mL, BD Biosciences] and anti-CD28 antibodies [6 μg/mL, BD Biosciences] in complete RMPI medium [5% human serum, 1 mmol/L sodium pyruvate, 10 mmol/L nonessential amino acids, 1% penicillin/streptomycin] in the presence or absence of recombinant human IL-23 [10 ng/mL, R&D Systems], recombinant human IL-12 [10 ng/mL, R&D Systems], IL-1β, IL-6, TNF [10ng/ml, R&D Systems], or neutralising anti-TNF antibodies [Ebioscience]. After 4 days, cells were transferred to uncoated plates and cultured for an additional 2–6 days under the same cytokine conditions, and then analysed for CXCR3 or CCR5 surface expression.
2.9. Human intestinal T cell lines and antigen-specificity assay
Intestinal CCR5[+] and CCR5[-] Th1/17- and Th17 cells, as well as total Th1 control cells, were sorted from LPMC isolated from CD patients, and were stimulated in vitro with irradiated allogeneic feeder cells and PHA [1 μg/mL, Sigma] in complete RPMI supplemented with 200 U/mL recombinant human IL-2 [rhIL-2, Miltenyi Biotec]. Fresh, complete, RPMI medium supplemented with rhIL-2 was replaced every 6 days, and T cell lines were used in functional assays 15 to 20 days after their last stimulation. For antigen-specificity assays, T cell lines were co-cultured with autologous peripheral CD14+ monocytes isolated from blood of CD patients, as previously described,19 at a 5:1 ratio in complete RPMI in the presence or absence of viral, fungal, or bacterial antigens, for 4 h. Brefeldin A [10 ug/mL] was added for an additional 14 h of incubation, and thereafter cells were analysed for intracellular expression of IL-17, IFN-γ, and CD40L. In some experiments, neutralising anti–HLA-DR/DP/DQ antibodies [anti–MHC class II, 10 μg/mL, clone Tu39, BD Biosciences] were added together with antigens. Viral, fungal, or bacterial antigens used in the antigen-specificity assays are listed in Supplementary Table 3. AIEC DF55, PP65, and DF10 strains were collected from ileum biopsies of CD patients with active disease.31E. coli LV15, AG61, and LV40 were collected from ileum biopsies of patients with functional intestinal disorders [non-IBD] and defined as non-AIEC strains for their inability to adhere to and invade intestinal epithelial cells [HEp-2] as well as to survive within macrophages [J774], as previously described.31 For bacterial lysates preparation, S. typhimurium, S. flexneri, and all the E. coli strains were grown in LB broth [Becton Dickinson], and L. paracasei [CBA L74 strain] was grown in MRS broth [Biokar Diagostics] for 18 h at 37°C in aerobic or semi-anaerobic conditions, respectively. All bacteria cultures were then normalised at 109 CFU/ml, washed in sterile PBS, heat-inactivated at 65°C for 1 h followed by three freeze-thaw cycles, and stored at -80°C. A final concentration of 107 freeze-dried bacterial cells/ml was used.
2.10. Immunofluorescence
Immunofluorescence was performed on formalin-fixed, paraffin-embedded [FFPE] tissues. Five sections of nine human CD specimens were obtained from Ospedale Maggiore Policlinico, Milan. Sections of 3 ± 1-μm thickness were placed on positively charged, standard, microscope slides [SuperFrostRPlus; Bio-Optica]. Slices were dewaxed and rehydrated and underwent the MILAN protocol [https://doi.org/10.21203/rs.2.1646/v5]. Antigen retrieval was performed by exposing slides to a retrieval solution [10 mM EDTA in Tris-buffer pH 8; Sigma-Aldrich] and then heated with a household microwave oven for 30 min [10 min at 850W and 20 min at 300W]. Slides were transferred into a washing Tris-Buffered Saline—Tween20 sucrose [TBS-Ts] [Sigma-Aldrich] pH 7.5 1X solution, and were then permeabilised in Triton X-100 [VWR] 0.5% for 30 min. Primary antibodies were diluted to the specific final concentration with antibodies’ diluent (TBS-2% BSA, 0.05% Sodium Azide, and 100 mM Trehalose [Sigma-Aldrich]), and incubation was performed in humidity chamber racks at 4°C overnight [anti-CD3, MCA1477 BioRad 1:100; anti-CCR5, ab7346 abcam 1:50; anti-CCR6, NBP2-12131 Novus Biologicals 1:100; anti-E. coli ab20640, 1:100, Abcam, Cambridge, UK]. Secondary antibodies [Anti-Rat AlexaFluor 488, 712-545-150 Jackson Immunoresearch; Anti-Rabbit Alexa Fluor® 647, 711-605-152 Jackson Immunoresearch; Anti-mouse Cy3, 715-165-150 Jackson Immunoresearch; Anti-Rabbit Cy3, 711-165-152 Jackson Immunoresearch], were added and incubated for 30 min at room temperature. Tissues were stained with DAPI [2.5 μg/ml, Sigma-Aldrich] in the dark for 10 min. The coverslip was mounted using Fluoromount-G™ Mounting Medium [Thermofisher Scientific]. After the first acquisition at THUNDER Imager Tissue [Leica Microsystems, Wetzlar, Germany], slides underwent a stripping procedure. The stripping buffer [SDS 10% [Sigma-Aldrich], Tris HCl 0.5M pH6.8 [Sigma- Aldrich]] was preheated to 56°C in a closed, shaking water bath. Once the solution reached the temperature, β-mercaptoethanol 0.4% [Sigma-Aldrich] was added and the slides were inserted into a glass container for 30 min. Using the HALO system [Indica Labs, Albuquerque, USA], we analysed the number and spatial location of CD3 + CCR5 + CCR6 + and E. coli cells.
2.11. Statistics
Statistical significance was evaluated using paired or unpaired t tests for comparison of two groups [with Mann–Whitney’s or Welch’s correction to analyse variables that were not normally distributed] or by one-way ANOVA for comparison of more than two groups [Dunn’s multiple comparisons to analyse variables that were not normally distributed]. All experiments were performed at least three times. Significance was defined at p-value <0.05. Statistics were performed with Prism software [version 7; GraphPad Software, La Jolla, CA].
3. Results
3.1. CD4+T cells co-producing IL-17 and IFN-γ are increased in the inflamed intestinal mucosa of CD patients
CD4+T cells that co-produce IFN-γ and IL-17 are increased in experimental colitis in mice and in CD. We first controlled systemic and intestinal cytokine-producing capacities of T cells in the cohorts of CD and UC patients from our hospital [Table 1]. CD4+T cells from peripheral blood of healthy donors [HD] and from the uninvolved resection margin of CRC specimens were analysed as non-IBD controls.32 In peripheral blood, the production of pro-inflammatory cytokines induced by brief polyclonal stimulation of total CD4+T cells with PMA and ionomycin was not significantly different between HDs and patients with IBDs [Supplementary Figure 1A]. In marked contrast, intestinal CD4+T cells of CD patients contained significantly higher frequencies of IFN-γ, IL-17, GM-CSF, and IL-22 than intestinal CD4+T cells from CRC or UC patients [Supplementary Figure 1B]. Notably, increased pro-inflammatory cytokine production in CD was specific for CD4+T cells, since frequencies of IFN-γ and IL-17 among intestinal CD8+T cells did not differ [Supplementary Figure 1C]. Finally, in paired inflamed and non-inflamed biopsies from CD patients we detected selectively and significantly increased frequencies of CD4+T cells producing IL-17, alone or in combination with IFN-γ [Supplementary Figure 1D]. In conclusion, CD patients displayed a selective increase of Th1/17-effector cytokine production, in particular IL-17 and IFN-γ co-production, among CD4+T cells and exclusively in the inflamed intestinal mucosa.
3.2. CCR5[+]Th17 cells are a distinct pro-inflammatory T cell subset [‘pTh17’] that co-produces IL-17 and IFN-γ
Human helper T cells [phenotype CD4+IL-7R+CD25-/lo] in peripheral blood can be classified into subsets of Th1, Th17, and Th1/17 cells according to the expression of the chemokine receptors CCR6 [Th17-asociated] and CXCR3 [Th1-associated, Figure 1A].18 In addition, CCR5 is expressed on more differentiated ‘effector memory’ T cells that can home to inflamed non-lymphoid tissues, including the gut.19,33 We thus purified Th1-[CXCR3+], Th17-[CCR6+], and Th1/17 cells [CXCR3+CCR6+] according to CCR5 expression from peripheral blood [Figure 1A], and analysed their cytokine-producing capacities. All analysed T cell subsets produced high levels of the pro-inflammatory cytokine TNF-α [Supplementary Figure 2A]. However, CCR5-Th17 cells could also produce relevant amounts of the anti-inflammatory cytokine IL-10 [Supplementary Figure 2A]. The production of IL-17 and IFN-γ varied strongly between the different T cell subsets. Thus, Th1 subsets produced as expected IFN-γ, but not IL-17 or IL-22 [Figure 1B and Supplementary Figure 2A]. Reciprocally, CCR5-Th17 cells produced some IL-17 and IL-22, but not IFN-γ. Consistent with previous reports,18,19 Th1/17 cells [CXCR3+CCR6+] produced high levels of IFN-γ and low levels of IL-17, and contained some IFN-γ and IL-17 co-producing cells [Figure 1B]. Besides these well-defined T cell subsets, we identified an additional population of Th17 cells, which expressed CCR5 and contained the highest frequencies of IL-17- and IL-22-producing cells [Figure 1B and Supplementary Figure 2A]. These CCR5+Th17 cells were largely unable to produce IL-10, but contained high frequencies of GM-CSF + cells, some IFN-γ+cells, and the highest frequencies of IFN-γ/IL-17 co-producing cells [Figure 1B and Supplementary Figure 2A]. For simplicity, we will refer to these highly pro-inflammatory CCR5+Th17 cells as ‘pTh17’ cells and to CCR5-Th17 cells as conventional Th17 cells [‘cTh17’] for the rest of this report. Notably, pTh17 cells expressed only low levels of CXCR5 and CCR10, indicating that they were distinct from Tfh and Th22 cells [Supplementary Figure 2B].
Figure 1.
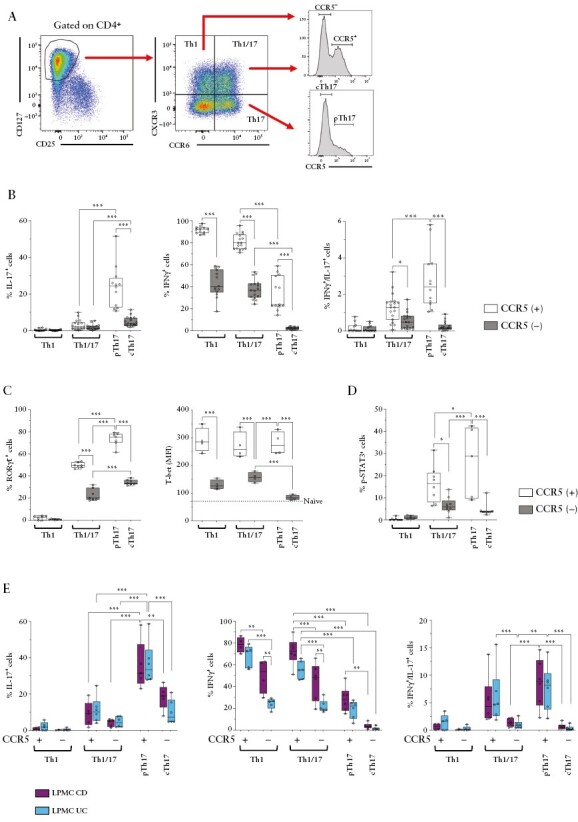
pTh17 cells express CCR5 and produce high levels of pro-inflammatory cytokines. A] Gating strategy to track pTh17 cells according to chemokine receptor expression. B] Frequencies of IL-17+, IFN-γ+, and IFN-γ/IL-17 co-producing cells among ex vivo-stimulated Th1, Th1/17, and Th17 subsets according to CCR5 expression from peripheral blood of HD [n = 20]. C] RORC/γt expression was measured ex vivo by intracellular staining of gated peripheral T cell subsets [HD, n = 6].T-bet protein expression was measured ex vivo by intracellular staining of gated peripheral T cell subsets [HD, n = 4]. MFI of T-bet in CD4+CD45RA+ naïve T cells are indicated as a dotted line as negative control. D] Percentage of T cells in sorted circulating T cell subsets that expressed a functional IL-23R, as assessed by STAT3 phosphorylation in response to brief [30 min] stimulation with IL-23. E] Frequencies of IL-17+, IFN-γ+, and IFN-γ/IL-17 co-producing cells by ex vivo-stimulated T cell subsets isolated from the lamina propria of CD [n = 6] and UC [n = 6] patients. Statistical significance was calculated using one-way ANOVA and reported as *p <0.05, **p <0.01, ***p <0.001. HD, healthy donors; CD, Crohn’s disease; MFI, Mean Fluorescence Intensity; UC, ulcerative colitis.
We next analysed the intracellular expression of the ‘lineage-defining’ transcription factors of Th1 and Th17 cells, T-bet, and RORC/γt, respectively. Th1 cells expressed as expected only T-bet, and cTh17 cells expressed exclusively RORC/γt. Conversely, pTh17 cells and Th1/17 cells expressed both T-bet and RORC/γt [Figure 1C]. Notably, pTh17 cells contained the highest frequencies of RORC/γt + cells. Next, we analysed the expression of the IL-23R. Since IL-23R staining was not convincing in our hands, we assessed the presence of functional IL-23Rs by STAT3 phosphorylation induced by brief ex vivo stimulation with IL-23, as published previously29 [Figure 1D]. pTh17 cells contained the highest frequencies of p-STAT3 + cells, indicating that they expressed the highest levels of functional IL-23Rs. Since pro-inflammatory Th1/17 cells in CD were reported to express high levels of multi-drug resistance 1 [MDR1],34 which may convey resistance to corticosteroids, we next assessed MDR1 expression by rhodamine extrusion assay. MDR1 activity was present in approximately 50% of pTh17 cells, but it was highest in Th1/17 cells [Supplementary Figure 2C].
We then analysed T-helper subsets classified according to chemokine receptor expression also in the intestine of IBD patients. Th1, Th17, and Th1/17 subsets displayed overall largely similar cytokine profiles in peripheral blood and in the intestinal lamina propria [Figure 1B, E and Supplementary Figure 2A, 2D]. Remarkably, pTh17 cells produced very high amounts of IL-17 in the intestine, and contained consequently also the highest frequencies of IL-17 and IFN-γ co-producing cells [Figure 1E]. Th1 and Th1/17 subsets in CD contained slightly higher frequencies of IFN-γ–producing cells [Figure 1E], but overall the cytokine profiles of the analysed T cell subsets were largely similar in UC and CD. Finally, we also analysed CD161 expression, which is highly expressed on pro-inflammatory Th17 cells in CD.35 Intestinal pTh17 cells expressed, indeed, very high levels of CD161, but CD161 was overall broadly expressed on intestinal Th cells [Supplementary Figure 2E].
In conclusion, pTh17 cells are a previously uncharacterised population of highly pro-inflammatory Th17 cells that infiltrate the intestinal mucosa of IBD patients.
3.3. pTh17 cells express genes characteristic for pro-inflammatory Th17 effector cells
To further characterise pTh17 cells at the molecular level, we then performed RNA sequencing of FACS-purified CCR6+Th cell subsets from peripheral blood of healthy donors and from the intestinal lamina propria of CD patients. A PCA of differentially expressed genes [DEGs] unveiled that pTh17 cells were in both cases positioned closely to CCR5+Th1/17 cells [Supplementary Figure 3A]. In the intestine, they were also in vicinity of cTh17 cells. When pTh17 cells were compared with cTh17 cells, we identified, besides the pTh17-defining gene CCR5, 498 DEGs in the blood and 462 DEGs in the gut [Figure 2A, B and Supplementary Tables 4, 7]. When pTh17 cells were compared instead with CCR5+Th1/17 cells, we identified, besides the Th1/17 marker gene CXCR3, 117 DEGs in the blood and 312 in the gut [Supplementary Tables 5, 8]. Circulating and intestinal pTh17 cells expressed in general increased levels of genes characteristic for pro-inflammatory Th17 cells, like CCL20, IL4l1,36 MSC [coding for musculin],37 IL-22, IL-17RE, and IL1R1. Among the most consistently down-regulated genes, we identified several stemness-associated genes including CCR7, SELL [coding for CD62L], CD27, and LEF1. Finally, when pTh17 cells were compared with all other CCR6+Th subsets concomitantly, we identified still 62 pTh17-specific DEGs in the blood and 366 in the intestine [Supplementary Figure 3B, Supplementary Tables 6, 9]. Among up-regulated, pTh17-specific DEGs, we confirmed several Th17-associated genes, and identified in addition PD1 in the blood and IL-23R in the gut. Among the down-regulated, pTh17-specific genes, we confirmed CD27 in the blood and several stemness-associated genes in the gut.
Figure 2.
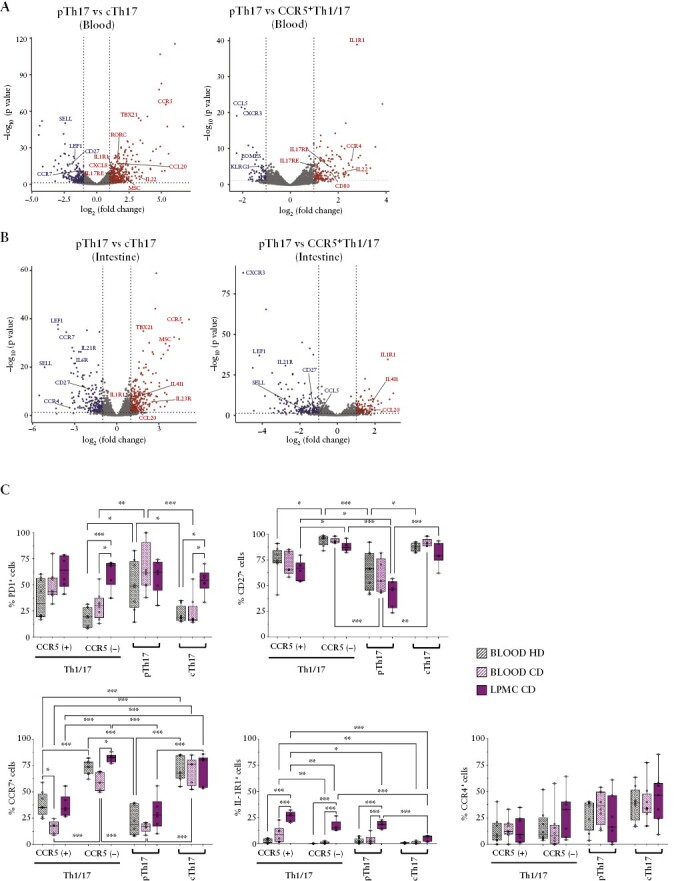
RNA sequencing of circulating and intestinal Th17 subsets indicates that pTh17 cells are highly differentiated Th17 effector cells. A/B] Volcano plots of differentially expressed genes in pTh17 cells as compared with cTh17 cells and CCR5+Th1/17 cell in peripheral blood [A] or the intestinal lamina propria [B]. Selected, up-regulated Th17-effector genes and down-regulated, stemness-associated genes are indicated. C] Surface expression of PD1, CD27, CCR7, IL-1R1, and CCR4 proteins was validated by flow cytometry in CCR6+Th subsets in unrelated healthy donors, and compared with those from peripheral blood and the intestinal lamina propria of Crohn’s disease [CD] patients.
Since differential mRNA expression does not always reflect protein expression levels, a panel of genes of interest was then validated at the protein and single cell levels by flow cytometry [Figure 2C]. CD4+T cells from peripheral blood of unrelated healthy donors were analysed and compared with CD4+T cells from peripheral blood and the intestinal lamina propria of CD patients [Figure 2C]. This analysis confirmed that pTh17 cells had a marked effector phenotype, since they expressed overall high levels of PD1 and low levels of CCR7 and CD27 in peripheral blood. CD27 expression was further reduced on intestinal pTh17 cells. PD1 was also highly expressed on intestinal pTh17 cells, but PD1 was highly expressed on all CCR6+Th cell subsets in the intestine [Figure 2C]. The Th17-associated surface marker CCR418 was confirmed to be more highly expressed on cTh17- and on pTh17 cells in peripheral blood, but was quite variable, in particular in the intestine [Figure 2C]. Finally, expression of IL-1R1, a key colitogenic cytokine receptor,38 was detectable at low levels on pTh17 cells but also on CCR5+Th1/17 cells, in peripheral blood. IL-1R1 expression was significantly increased in the intestine, but remained low on cTh17 cells [Figure 2C].
Overall, this gene expression analysis confirmed, in an unbiased manner, that pTh17 cells have features of highly differentiated Th17 effector cells, in particular in the gut.
3.4. pTh17 cells are associated with intestinal inflammation in CD patients
Given their pro-inflammatory cytokine profile, pTh17 cells were likely to contribute to the increased cytokine production in the intestinal mucosa of CD patients [Supplementary Figure 1]. We first monitored the frequencies of pTh17 in different tissues of IBD patients, ie, in peripheral blood, in the intestinal lamina propria and in intestinal lymph nodes. pTh17 cells, but also other CCR5+T cell subsets, were strongly enriched in the intestinal lamina propria, whereas they were only a minority in peripheral blood and rare in intestinal lymph nodes [Figure 3A and Supplementary Figure 4A, B].
Figure 3.
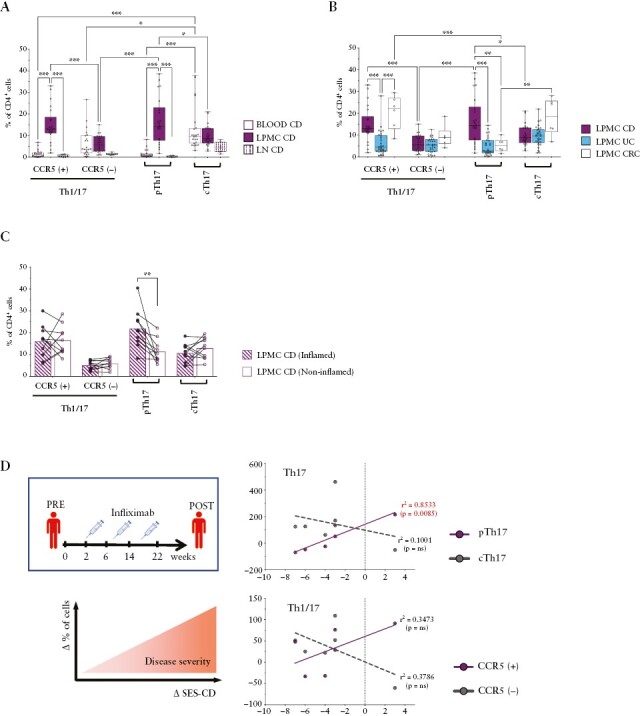
Intestinal pTh17 cells are associated with inflammation in Crohn’s disease [CD]. A] Percentages of Th1/17 and Th17 subsets gated according to CCR5 expression in blood [n = 23], lymph nodes [LN; n = 7], and lamina propria [LPMC; n = 22] of CD patients. B] Frequencies of Th1/17 and Th17 subsets gated according to CCR5 expression in the lamina propria of CD [n = 22], ulcerative colitis [UC] [n = 29] and colorectal cancer [CRC] patients [non-inflamed areas, n = 6]. A, B] Statistical significance was calculated using one-way ANOVA and reported as *p <0.05, **p <0.01, ***p <0.001. C] Percentages of Th1/17 and Th17 subsets subdivided according to CCR5 expression isolated from paired, intestinal, inflamed, and non-inflamed areas of CD patients [n = 10]. Dots and bars represent individual patients and mean values, biopsies from the same patients are connected with a line. Statistical significance was calculated using paired t test and reported as *p <0.05, **p <0.01, ***p <0.001. D] Correlation between changes in the percentage of intestinal Th1/17 and Th17 subsets [CCR5[+] violet dots, CCR5[-] grey dots], and the Simple Endoscopic Score of disease Severity [SES-CD] before and after infliximab treatment in CD patients [n = 6]. Variation of frequencies of T cell subsets was calculated setting to 100% their percentage in the lamina propria of CD patients before infliximab treatment; ΔSES-CD score were calculated as the difference between SES-CD scores at baseline and after infliximab treatment. Correlations were determined using Spearman’s correlation coefficient.
We then assessed if pTh17 cells were associated with intestinal inflammation in CD. To this end, we compared their relative abundance in: i] CD and UC patients; ii] paired inflamed and non-inflamed biopsies; and iii] pre/post therapy with infliximab [anti-TNF antibodies) that inhibits intestinal inflammation. Strikingly, in all cases there was a significant association of pTh17 cells with intestinal inflammation in CD [Figure 3B–D]. Thus, pTh17 cells were strongly and significantly increased in the intestinal mucosa of CD patients [Figure 3B]. UC patients showed no increase of pTh17 cells, but were in contrast characterised by a significant decrease of intestinal CCR5+Th1/17 cells [Figure 3B]. Th1 subsets showed a complex pattern, with CCR5-Th1 cells being increased in UC and CCR5+Th1 cells in CD [Supplementary Figure 4C]. Consistent with the cytokine data [Supplementary Figure 1], these marked differences in the intestine were not detectable in peripheral blood [Supplementary Figure 4D]. In paired intestinal biopsies from CD patients, pTh17 cells were selectively and significantly enriched in the inflamed as compared with the non-inflamed intestine [Figure 3C and Supplementary Figure 4E]. Finally, in infliximab-treated CD patients [Supplementary Tables 1], a reduction of intestinal pTh17 cells was significantly associated with the reduction of the Simple Endoscopic Score for CD [SES-CD] after infliximab therapy [Figure 3D and Supplementary Figure 4F]. Conversely, there were no significant associations of the SES-CD dynamics with Th1/17 or cTh17 cells. Notably, the production of Th1/17 effector cytokines by total intestinal CD4+T cells showed only weak associations that did not reach statistical significance [Supplementary Figure 4G].
In summary, pTh17 cells are strongly enriched in the inflamed intestinal mucosa of CD patients and are reduced upon successful infliximab treatment, suggesting that they contribute substantially to intestinal inflammation.
3.5. IL-23 promotes pTh17 cell differentiation in vitro and in vivo
IL-23 is a key colitogenic cytokine that induces the maturation of Th17 cells. We therefore wondered if IL-23 could promote the differentiation of pTh17 cells. Indeed, we and others have already documented that cTh17 cells acquire cytokine profiles that are characteristic for pTh17 cells and Th1/17 cells [Figure 1] with IL-23 and IL-12, respectively.5,39 We assessed here if IL-12 and IL-23 induced also the phenotype of these T cell subsets. Indeed, purified cTh17 cells stimulated with anti-CD3 and -CD28 antibodies up-regulated CCR5 in the presence of IL-23, whereas they up-regulated CXCR3, but not CCR5, with IL-12 [Figure 4A]. Also, other pro-inflammatory cytokines like IL-1β, IL-6, and TNF failed to induce CCR5 [Supplementary Figure 5A]. We next analysed the presence of these cytokines in intestinal biopsies of IBD patients by ELISA. Pro-inflammatory cytokines were as expected detected in all IBD patients, but IL-23 was in the main higher in CD, whereas IL-12 was more abundant in UC [Figure 4B]. To address the role of IL-23 in the generation of intestinal pTh17 cells in vivo, we analysed biopsies of CD patients at baseline and at 24 weeks following therapy with risankizumab [Supplementary Tables 1]. The latter is a therapeutic antibody that specifically neutralises IL-23 by targeting the IL-23p19 subunit,40 whereas IL-12 is not blocked. Strikingly, risankizumab induced a selective and significant decrease of pTh17 cells in the intestinal lamina propria [Figure 4C and Supplementary Figure 5B].
Figure 4.
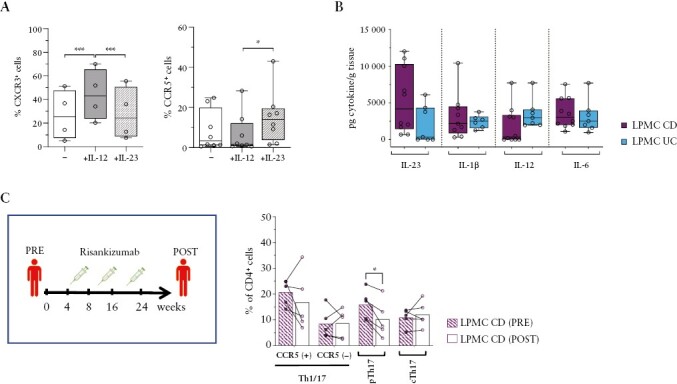
IL-23 promotes pTh17 cell generation in vitro and in vivo. A] Expression of CXCR3 [left panel] and CCR5 [right panel] on cTh17 cells [CCR6+CXCR3-CCR5-] isolated from HD [n = 4-8] after in vitro stimulation with anti-CD3/CD28 antibodies in the absence [–] or presence of IL-23 or IL-12. B] Concentrations of IL-23, IL-1β, IL-12, and IL-6 per gram of homogenised intestinal biopsies of CD [n = 10] and UC [n = 7] patients. C] Frequencies of Th1/17 and Th17 subsets among CD4+T cells in the intestinal lamina propria at baseline [before] and after 24 weeks [after] of risankizumab treatment of CD patients [n = 5]. Dots and bars represent individual patients and mean values, biopsies from the same patients are connected with a line. A–C] Statistical significance was calculated using paired t test and reported as *p <0.05, **p <0.01, ***p <0.001. HD, healthy donods; CD, Crohn’s disease; UC, ulcerative colitis.
3.6. Adherent-invasive Escherichia coli selectively activate pTh17 cells and induce IL-23 from dendritic cells
Although pTh17 cells shared several features with Th1/17 cells, only pTh17 cells were associated with intestinal inflammation in CD patients [Figure 3B]. Therefore, we asked if intestinal pTh17 cells might expand in response to specific microbial antigens. To address this question, we analysed the activation of T cell lines, expanded from FACS-purified intestinal T cell subsets of IBD patients, following stimulation with a panel of pathogen-derived antigens [Figure 5A].
Figure 5.
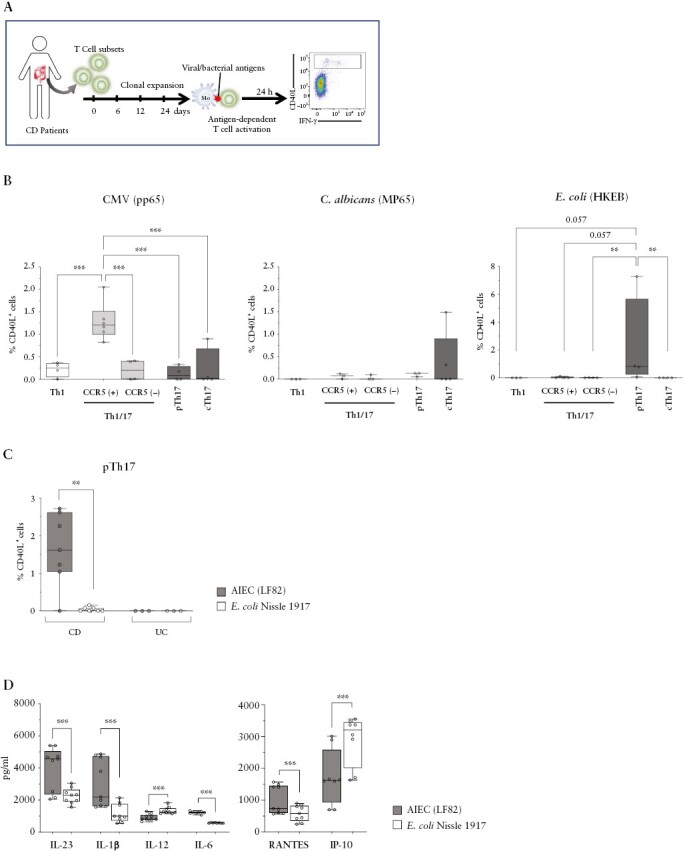
Adherent-invasive E. coli [AIEC] selectively activate pTh17 cells and induce high levels of IL-23 from DC. A] Experimental strategy to measure antigen specificities in intestinal Th subsets. B] Mean percentages of CD40L induction on the indicated intestinal T cell lines, derived from CD patients [n = 3–6], upon activation with autologous monocytes pulsed with CMV- [pp65, left], peptides derived from Candida albicans- [MP65, centre], or heat-killed E. coli [HKEB, 0111:B4 strain right]. C] CD40L induction on intestinal pTh17 cell line, derived from CD [n = 7] or UC patients [n = 3] after activation with autologous monocytes incubated with the E. coli strains AIEC-LF28 or Nissle 1917. D] Cytokine [left panel] and chemokine [right panel] production by monocyte-derived dendritic cells in response to AIEC-LF82 or E. coli Nissle 1917 strains was measured by ELISA [n = 9]. Statistical significance was calculated using Mann–Whitney’s test and reported as *p <0.05, **p <0.01, ***p <0.001. DC, dendritic cells; CD, Crohn’s disease; CMC, cytomegalovirus; UC, ulcerative colitis.
Responses to autologous monocytes in the absence of exogenous antigens were low in all T cell subsets, indicating that they were poorly auto-reactive [Supplementary Figure 6A]. Notably, the majority of tested bacterial [Salmonella tyhphimurium FB62, Shigella flexneri serotype 5a M90T, Lactobacillus paracasei CBA L74] and viral [Epstein–Barr virus, John Cunningham virus, varicella zoster virus] antigens failed to induce a detectable activation of the analysed intestinal T cell subsets in CD patients [data not shown]. In contrast, we detected robust antigen-specific responses, as assessed by up-regulation of CD40L and cytokines, to E. coli [HKEB] and to cytomegalovirus [CMV, pp65], and more variable responses to Candida albicans [C. albicans, MP65] [Figure 5B and Supplementary Figure 6A]. Pathogen-induced activation of intestinal T cells was prevented by neutralising anti-MHC class-II antibodies [Supplementary Figure 6B], indicating that activation was TCR-dependent. Importantly, intestinal T cell subsets had strongly biased antigen specificities. Thus, Th1 cells responded only weakly to CMV, whereas cTh17 cells reacted with C. albicans. Most important, pTh17 cells were the only subset that reacted with E. coli [Figure 5B and Supplementary Figure 6A]. Intriguingly, CCR5+Th1/17 cells responded instead strongly and selectively to CMV. Given the strong and selective association of AIEC with CD,23 we next compared responses of pTh17 cells to LF82, the reference strain for AIEC pathovar, and to the probiotic strain E. coli Nissle 1917, commonly used for the treatment of gastrointestinal disorders.41 Strikingly, pTh17 cells from CD patients reacted very poorly with E. coli Nissle 1917 strain, but strongly with AIEC-LF82 [Figure 5C, supplementary Figures 6B and 7]. Notably, monocytes expressed comparable levels of MHC class II, CD80, and CD86 under these conditions [Supplementary Figure 6C], suggesting that they had similar T cell-stimulatory capacities, but presented different antigens. Consistently, a comparison of genes expressed by AIEC-LF82 and by E. coli Nissle 1917 unveiled that 415 genes were expressed exclusively by LF82 [Supplementary Figure 8], and could thus code for antigens that selectively activate pTh17 cells. In a fraction of IBD patients, we also tested six additional AIEC and non-AIEC strains, and found that AIEC strains induced in general stronger responses [Supplementary Figure 7]. Indeed, the responses of pTh17 cells from CD patients to two different non-AIEC strains were significantly weaker as compared with AIEC-LF82. Importantly, pTh17 cells from three UC patients did in contrast not respond to any of the tested E.coli strains [Figure 5C and Supplementary Figure 7]. Finally, we tested the capacity of E. coli strains to induce polarising cytokines and chemokines from monocyte-derived dendritic cells [DC]. AIEC-LF82 induced higher levels of IL-23, IL-1β,IL-6, and the CCR5 ligand RANTES, whereas E. coli strain Nissle 1917 induced higher amounts of IL-12 and IP-10 [Figure 5D].
In summary, whereas intestinal Th1/17 cells are strongly activated by CMV, pTh17 cells react selectively and strongly with AIEC, indicating that they fight different classes of pathogens. Moreover, the CD-associated colitogenic bacteria [AIEC] activated DC to secrete the soluble mediators that promote the generation and migration of pTh17 cells.
3.7. T cells co-expressing CCR6 and CCR5 are frequent in areas colonized by Escherichia coli
Finally, in order to assess the localisations of pTh17 cells and E.coli in the intestine, we performed immunofluorescence analysis of intestinal lamina propria sections of CD patients [Figure 6A and Supplementary Figure 9A]. Areas where T cells co-expressing CCR6 and CCR5, the defining chemokine receptors of pTh17 cells, were frequent [Supplementary Figure 9B] contained significantly higher frequencies of E. coli cells [Figure 6B]. These findings suggest that T cells co-expressing CCR6 and CCR5, including pTh17 cells, may be preferentially recruited to sites of intestinal E.coli colonisation in CD.
Figure 6.
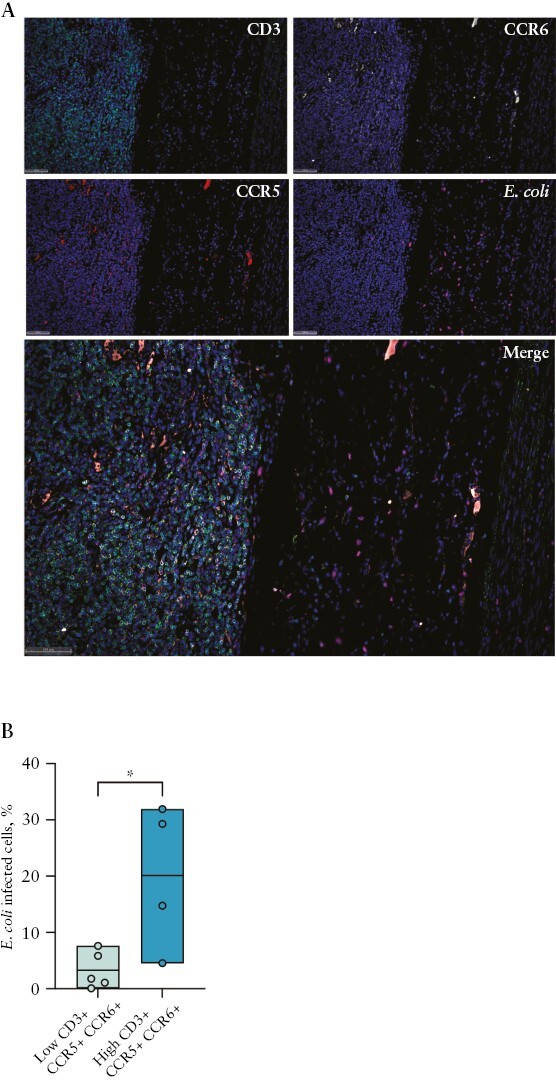
T cells co-expressing CCR6 and CCR5 are enriched in E.coli-colonised areas. A] Representative image of CCR6 + CCR5 + T cells and E. coli localisation within CD-intestinal tissue. Sections were stained with DAPI [blue], CD3 [green], CCR6 [grey], CCR5 [red], and E. coli [orange]. B] E. coli cell abundance among CD patients subdivided into high and low groups of CCR6 + CCR5 + T cells. Statistical significance was calculated using unpaired Student’s t test for comparison within two groups.
4. Discussion
IBDs are associated with pro-inflammatory CD4+T cells, but the identity of colitogenic T cells is still elusive. Here, we identified and characterised a highly pro-inflammatory population of human intestinal Th17 cells, called pTh17 cells here, and showed that they are associated with intestinal inflammation in CD and respond to AIEC, ie, to CD-associated bacteria.
Human IL-17/IFN-γ co-producing CD4+ T cells were reported to co-express CCR6 and CXCR3, are termed here Th1/17 cells, and were extensively characterised.14,18,34,39 They can be induced with IL-12 from Th17 cells, but also with Th17-promoting cytokines from Th1 cells in the gut.42 They produce very high levels of IFN-γ, but only low levels of IL-17, play a pathogenic role in autoimmune diseases like multiple sclerosis,19 and are abundant in the intestine. They were thus likely candidates to drive also pathogenic inflammation in IBDs.34,35 In healthy individuals, Th1/17 cells from peripheral blood react with both intracellular and extracellular pathogens, including E. coli19,39 Little is however known about the antigen specificities of human T cells in the intestine,10 which are most relevant in IBDs. We showed here that intestinal Th1/17 cells in CD patients reacted strongly and selectively with CMV, but not with a panel of enterobacteria-derived antigens. Notably, CMV infections occur frequently in IBD patients, with high incidence of CMV reactivations particularly in steroid-refractory UC patients, and are associated with worse clinical outcomes in IBDs.43,44 Intriguingly, we found that intestinal CCR5+Th1/17 cells, the subset that reacted strongly with CMV, were selectively reduced in UC. These findings suggest that intestinal Th1/17 cells contain CMV infections, and may thus play an unexpected protective role in IBDs.
The pTh17 cells here characterised lacked CXCR3 expression, which is expressed on human Th1/17 cells by definition,18 indicating that they are a distinct population. Indeed, unlike Th1/17 cells, they expressed very high amounts of IL-17 and IL-22, but only intermediate levels of IFN-γ. Nevertheless, they shared several features of potentially colitogenic T cells with Th1/17 cells, including co-expression of RORC/γt and T-bet, co-production of IL-17 and IFN-γ, and responsiveness to IL-1 and IL-23. Importantly, only pTh17 cells were consistently associated with intestinal inflammation in CD patients, as demonstrated in both paired inflamed and non-inflamed intestinal biopsies, and by their significant decrease after successful anti-TNF therapy. The most relevant difference between pTh17- and Th1/17 cells was their antigen specificities. This is a key finding, because antigenic stimulation is the principal trigger for T cells to produce cytokines, which in turn drive intestinal inflammation. Most importantly, pTh17 cells reacted selectively with E. coli, in particular with AIEC, which are associated with ileal CD.23,45 Intriguingly, they largely failed to respond to the probiotic E. coli Nissle 1917 strain.
This is to our knowledge the first report that identifies AIEC-specific T cells in the human intestine. AIEC are able to persist in infected macrophages,45 and pTh17 cells that produce IFN-γ to activate macrophages appear to be the appropriate type of effector cells to contain these colitogenic bacteria. Indeed AIEC, and potentially also other bacterial species,11,46 are likely to induce their differentiation in the gut. Notably, pTh17 cells produce high amounts of IL-17 and IL-22, which have crucial functions in promoting mucosal barrier functions, induction of antimicrobial peptides, and recruitment of phagocytic cells. AIEC strains are however highly virulent, having a high resistance to antimicrobial peptides, a strong ability to persist within macrophages, and also triggering a higher IL-23 release by DCs from CD compared with UC patients.30,45 Consequently, the activity of pTh17 cells may not be sufficient to control these pathogenic bacteria in CD, leading to chronic and excessive inflammation.
pTh17 cells could differentiate from conventional cTh17 cells in vitro upon stimulation with IL-23, a key colitogenic cytokine17 that was efficiently induced from DC by AIEC. Importantly, selectively blocking IL-23 with risankizumab, a drug currently under investigation for active CD patients,40 resulted in a selective reduction of intestinal pTh17 cells. Infliximab inhibits the production of several pro-inflammatory cytokines in the intestine, including IL-23.47 It seems therefore plausible that also the infliximab-induced reduction of pTh17 cells is a consequence of impaired IL-23 signalling. Consistently, TNF failed to induce CCR5 expression on conventional Th17 cells in vitro, but we can of course not exclude that other pro-inflammatory cytokines besides IL-23 contribute to the induction of pTh17 cells in the gut. Mucosal CCR5 expression was previously shown to be associated with IBDs,48 and CCR5 antagonists were able to reduce mucosal inflammation in a mouse colitis model.49 Thus, CCR5 is not only a surface marker that identifies pTh17 cells, but is also likely to be important for their functions, by mediating their recruitment to infected DC. Notably, CCR5 is also expressed on intestinal, anti-inflammatory Tr1 cells, but the latter lack IL-7R and CCR6 expression and display an anti-inflammatory cytokine profile.25
Collectively, our data suggests that pTh17 cells play a key, pro-inflammatory role in CD. They are generated and activated by DC that present antigens derived from colitogenic bacteria like AIEC and secrete IL-23. pTh17 cells might be exploited as immunological targets for the future development of new therapeutic strategies in CD.
Supplementary Material
Contributor Information
Moira Paroni, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy; Department of Biosciences, Università degli Studi di Milano, Milan, Italy.
Gabriella Leccese, Department of Biosciences, Università degli Studi di Milano, Milan, Italy.
Valeria Ranzani, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Giorgia Moschetti, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Matteo Chiara, Department of Biosciences, Università degli Studi di Milano, Milan, Italy.
Federica Perillo, Department of Experimental Oncology, European Institute of Oncology, Milan, Italy.
Sara Ferri, Department of Biotechnology and Bioscience, University of Milano-Bicocca, Milan, Italy.
Francesca Clemente, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Daniele Noviello, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy; Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy.
Francesco Simone Conforti, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy.
Stefano Ferrero, Pathology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy; Department of Biomedical, Surgical, and Dental Sciences, Università degli Studi di Milano, Milan, Italy.
Bhavna Karnani, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Roberto Bosotti, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Chiara Vasco, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Serena Curti, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Maria Cristina Crosti, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Paola Gruarin, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy.
Grazisa Rossetti, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy; Molecular Oncology and Immunology, FIRC Institute of Molecular Oncology [IFOM], Milan, Italy.
Maria Pia Conte, Department of Public Health and Infectious Diseases, ‘Sapienza’ University of Rome, Rome, Italy.
Maurizio Vecchi, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy; Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy.
Massimiliano Pagani, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy; Molecular Oncology and Immunology, FIRC Institute of Molecular Oncology [IFOM], Milan, Italy; Department of Medical Biotechnology and Translational Medicine, University of Milan, Milan, Italy.
Paolo Landini, Department of Biosciences, Università degli Studi di Milano, Milan, Italy.
Federica Facciotti, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy; Department of Experimental Oncology, European Institute of Oncology, Milan, Italy; Department of Biotechnology and Bioscience, University of Milano-Bicocca, Milan, Italy.
Sergio Abrignani, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy; DISCCO, Department of Clinical Science and Community Health, University of Milan, Milan, Italy.
Flavio Caprioli, Gastroenterology and Endoscopy Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy; Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy.
Jens Geginat, INGM-National Institute of Molecular Genetics ‘Romeo ed Enrica Invernizzi’, Milan, Italy; DISCCO, Department of Clinical Science and Community Health, University of Milan, Milan, Italy.
Funding
This study was financially supported by the Italy’s Ministry of Health [Young Researchers ‘Giovani Ricercatori’ Program Ref. No. GR-2011-02352001] to FC and MP, by Italian Ministry of Health [GR-2016-02361741] to FC, and by Cariplo Foundation-Biomedical Research Conducted by Young Researchers [GR-2017–0816] to MP and FC. J G is supported by Associazione Italiana per la Ricerca sul Cancro [AIRC, IG 2019 23581].
Conflict of Interest
MV served as a consultant to Abbvie, MSD, Takeda, Janssen-Cilag, and Celgene; and received lecturer fees from Abbvie, Ferring, Takeda, MSD, Janssen-Cilag, and Zambon. FC has served as consultant to Mundipharma, AbbVie, MSD, Takeda, Janssen, Roche, and Celgene; received lecture fees from AbbVie, Ferring, Takeda, Allergy Therapeutics, and Janssen; and received unrestricted research grants from Giuliani, SOFAR, MS&D, Takeda, Pfizer and AbbVie. All other authors have no conflicts of interest to declare.
Author Contributions
DN, SC, and FC enrolled patients; MP, GL, VR, GM FC, BK, RB, CV, SC, FP, SaF, StF, MCC, PG and GR performed experiments; MPC provided E coli strains and gave important intellectual contributions; MP, VR, MC, and CV analysed and interpreted data; MP, FC, and JG initiated, designed, and supervised the study and wrote the manuscript; MP, FC, and JG obtained funding; MV, MP, PL, FF, and SA contributed to interpretation of the data and study supervision, and gave important intellectual contributions. All authors have read and agreed to the published version of the manuscript. JG and FC share the senior authorship.
Data Availability
The RNA sequencing data of intestinal and circulating CCR6 + T cell subsets have been deposited in the ArrayExpress [https://www.ebi.ac.uk/biostudies/arrayexpress] (accession no. E-MTAB-13178).
References
- 1. Uhlig HH, Powrie F.. Translating immunology into therapeutic concepts for inflammatory bowel disease. Annu Rev Immunol 2018;36:755–81. [DOI] [PubMed] [Google Scholar]
- 2. Biancheri P, Di Sabatino A, Ammoscato F, et al. Absence of a role for interleukin-13 in inflammatory bowel disease. Eur J Immunol 2014;44:370–85. [DOI] [PubMed] [Google Scholar]
- 3. Brand S. Crohn’s disease: Th1, th17 or both? The change of a paradigm: New immunological and genetic insights implicate th17 cells in the pathogenesis of crohn’s disease. Gut 2009;58:1152–67. [DOI] [PubMed] [Google Scholar]
- 4. Peters A, Lee Y, Kuchroo VK.. The many faces of th17 cells. Curr Opin Immunol 2011;23:702–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kastirr I, Maglie S, Paroni M, et al. Il-21 is a central memory t cell-associated cytokine that inhibits the generation of pathogenic th1/17 effector cells. J Immunol 2014;193:3322–31. [DOI] [PubMed] [Google Scholar]
- 6. McGeachy MJ, Bak-Jensen KS, Chen Y, et al. Tgf-beta and il-6 drive the production of il-17 and il-10 by t cells and restrain t[h]-17 cell-mediated pathology. Nat Immunol 2007;8:1390–7. [DOI] [PubMed] [Google Scholar]
- 7. Gagliani N, Vesely MC, Iseppon A, et al. Th17 cells transdifferentiate into regulatory t cells during resolution of inflammation. Nature 2015;523:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huber S, Gagliani N, Flavell RA.. Life, death, and miracles: Th17 cells in the intestine. Eur J Immunol 2012;42:2238–45. [DOI] [PubMed] [Google Scholar]
- 9. Yang Y, Torchinsky MB, Gobert M, et al. Focused specificity of intestinal th17 cells towards commensal bacterial antigens. Nature 2014;510:152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hegazy AN, West NR, Stubbington MJT, et al.; Oxford IBD Cohort Investigators. Circulating and tissue-resident cd4[+] t cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 2017;153:1320–37.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geginat J, Paroni M, Maglie S, et al. Plasticity of human cd4 t cell subsets. Front Immunol 2014;5:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lexberg MH, Taubner A, Albrecht I, et al. Ifn-gamma and il-12 synergize to convert in vivo generated th17 into th1/th17 cells. Eur J Immunol 2010;40:3017–27. [DOI] [PubMed] [Google Scholar]
- 14. Maggi L, Santarlasci V, Capone M, et al. Distinctive features of classic and nonclassic [th17 derived] human th1 cells. Eur J Immunol 2012;42:3180–8. [DOI] [PubMed] [Google Scholar]
- 15. Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT.. Th17 cells give rise to th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci U S A 2015;112:7061–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nizzoli G, Burrello C, Cribiu FM, et al. Pathogenicity of in vivo generated intestinal th17 lymphocytes is ifngamma dependent. J Crohns Colitis 2018;12:981–92. [DOI] [PubMed] [Google Scholar]
- 17. Ahern PP, Schiering C, Buonocore S, et al. Interleukin-23 drives intestinal inflammation through direct activity on t cells. Immunity 2010;33:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Acosta-Rodriguez EV, Rivino L, Geginat J, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing t helper memory cells. Nat Immunol 2007;8:639–46. [DOI] [PubMed] [Google Scholar]
- 19. Paroni M, Maltese V, De Simone M, et al. Recognition of viral and self-antigens by th1 and th1/th17 central memory cells in patients with multiple sclerosis reveals distinct roles in immune surveillance and relapses. J Allergy Clin Immunol 2017;140:797–808. [DOI] [PubMed] [Google Scholar]
- 20. Britton GJ, Contijoch EJ, Mogno I, et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut th17 and rorgammat[+] regulatory t cells and exacerbate colitis in mice. Immunity 2019;50:212–224.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaur N, Chen CC, Luther J, Kao JY.. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes 2011;2:211–6. [DOI] [PubMed] [Google Scholar]
- 22. Feng T, Qin H, Wang L, Benveniste EN, Elson CO, Cong Y.. Th17 cells induce colitis and promote th1 cell responses through il-17 induction of innate il-12 and il-23 production. J Immunol 2011;186:6313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive escherichia coli associated with ileal mucosa in crohn’s disease. Gastroenterology 2004;127:412–21. [DOI] [PubMed] [Google Scholar]
- 24. Monteleone G, Biancone L, Marasco R, et al. Interleukin 12 is expressed and actively released by crohn’s disease intestinal lamina propria mononuclear cells. Gastroenterology 1997;112:1169–78. [DOI] [PubMed] [Google Scholar]
- 25. Alfen JS, Larghi P, Facciotti F, et al. Intestinal ifn-gamma-producing tr1 cells co-express ccr5 and pd-1, and down-regulate il-10 in the inflamed guts of ibd patients. J Allergy Clin Immunol 2018;142:1537–1547.e8. [DOI] [PubMed] [Google Scholar]
- 26. Paroni M, Magarotto A, Tartari S, et al. Uncontrolled il-17 production by intraepithelial lymphocytes in a case of non-ipex autoimmune enteropathy. Clin Transl Gastroenterol 2016;7:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rivino L, Gruarin P, Haringer B, et al. Ccr6 is expressed on an il-10-producing, autoreactive memory t cell population with context-dependent regulatory function. J Exp Med 2010;207:565–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cossarizza A, Chang HD, Radbruch A, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies [third edition]. Eur J Immunol 2021;51:2708–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kastirr I, Crosti M, Maglie S, et al. Signal strength and metabolic requirements control cytokine-induced th17 differentiation of uncommitted human t cells. J Immunol 2015;195:3617–27. [DOI] [PubMed] [Google Scholar]
- 30. Leccese G, Bibi A, Mazza S, et al. Probiotic lactobacillus and bifidobacterium strains counteract adherent-invasive escherichia coli [aiec] virulence and hamper il-23/th17 axis in ulcerative colitis, but not in crohn’s disease. Cells 2020;9:1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conte MP, Longhi C, Marazzato M, et al. Adherent-invasive escherichia coli [aiec] in pediatric crohn’s disease patients: phenotypic and genetic pathogenic features. BMC Res Notes 2014;7:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Monteleone G, Monteleone I, Fina D, et al. Interleukin-21 enhances t-helper cell type i signaling and interferon-gamma production in crohn’s disease. Gastroenterology 2005;128:687–94. [DOI] [PubMed] [Google Scholar]
- 33. Geginat J, Paroni M, Facciotti F, et al. The cd4-centered universe of human t cell subsets. Semin Immunol 2013;25:252–62. [DOI] [PubMed] [Google Scholar]
- 34. Ramesh R, Kozhaya L, McKevitt K, et al. Pro-inflammatory human th17 cells selectively express p-glycoprotein and are refractory to glucocorticoids. J Exp Med 2014;211:89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cosmi L, De Palma R, Santarlasci V, et al. Human interleukin 17-producing cells originate from a cd161+cd4+ t cell precursor. J Exp Med 2008;205:1903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santarlasci V, Maggi L, Mazzoni A, et al. Il-4-induced gene 1 maintains high tob1 expression that contributes to tcr unresponsiveness in human t helper 17 cells. Eur J Immunol 2014;44:654–61. [DOI] [PubMed] [Google Scholar]
- 37. Santarlasci V, Mazzoni A, Capone M, et al. Musculin inhibits human t-helper 17 cell response to interleukin 2 by controlling stat5b activity. Eur J Immunol 2017;47:1427–42. [DOI] [PubMed] [Google Scholar]
- 38. Coccia M, Harrison OJ, Schiering C, et al. Il-1beta mediates chronic intestinal inflammation by promoting the accumulation of il-17a secreting innate lymphoid cells and cd4[+] th17 cells. J Exp Med 2012;209:1595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duhen T, Campbell DJ.. Il-1beta promotes the differentiation of polyfunctional human ccr6+cxcr3+ th1/17 cells that are specific for pathogenic and commensal microbes. J Immunol 2014;193:120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kashani A, Schwartz DA.. The expanding role of anti-il-12 and/or anti-il-23 antibodies in the treatment of inflammatory bowel disease. Gastroenterol Hepatol [N Y] 2019;15:255–65. [PMC free article] [PubMed] [Google Scholar]
- 41. Schultz M. Clinical use of e. Coli nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis 2008;14:1012–8. [DOI] [PubMed] [Google Scholar]
- 42. Geginat J, Paroni M, Kastirr I, Larghi P, Pagani M, Abrignani S.. Reverse plasticity: Tgf-beta and il-6 induce th1-to-th17 cell transdifferentiation in the gut. Eur J Immunol 2016;46:2306–10. [DOI] [PubMed] [Google Scholar]
- 43. Kalappurayil NB, Thomas J, Mankuni B, Thomas V.. Assessment of disease severity and role of cytomegalovirus infection in patients with ulcerative colitis. J Clin Diagn Res 2017;11:EC07–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Siegmund B. Cytomegalovirus infection associated with inflammatory bowel disease. Lancet Gastroenterol Hepatol 2017;2:369–76. [DOI] [PubMed] [Google Scholar]
- 45. Chervy M, Barnich N, Denizot J.. Adherent-invasive e. coli: update on the lifestyle of a troublemaker in crohn’s disease. Int J Mol Sci 2020;21:3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015;163:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caprioli F, Bose F, Rossi RL, et al. Reduction of cd68+ macrophages and decreased il-17 expression in intestinal mucosa of patients with inflammatory bowel disease strongly correlate with endoscopic response and mucosal healing following infliximab therapy. Inflamm Bowel Dis 2013;19:729–39. [DOI] [PubMed] [Google Scholar]
- 48. Ye X, Liu S, Hu M, Song Y, Huang H, Zhong Y.. Ccr5 expression in inflammatory bowel disease and its correlation with inflammatory cells and beta-arrestin2 expression. Scand J Gastroenterol 2017;52:551–7. [DOI] [PubMed] [Google Scholar]
- 49. Mencarelli A, Cipriani S, Francisci D, et al. Highly specific blockade of ccr5 inhibits leukocyte trafficking and reduces mucosal inflammation in murine colitis. Sci Rep 2016;6:30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data of intestinal and circulating CCR6 + T cell subsets have been deposited in the ArrayExpress [https://www.ebi.ac.uk/biostudies/arrayexpress] (accession no. E-MTAB-13178).


