Abstract
Climate projections predict major changes in alpine environments by the end of the 21st century. To avoid climate-induced maladaptation and extinction, many animal populations will either need to move to more suitable habitats or adapt in situ to novel conditions. Since populations of a species exhibit genetic variation related to local adaptation, it is important to incorporate this variation into predictive models to help assess the ability of the species to survive climate change. Here, we evaluate how the adaptive genetic variation of a mountain ungulate—the Northern chamois (Rupicapra rupicapra)—could be impacted by future global warming. Based on genotype-environment association analyses of 429 chamois using a ddRAD sequencing approach, we identified genetic variation associated with climatic gradients across the European Alps. We then delineated adaptive genetic units and projected the optimal distribution of these adaptive groups in the future. Our results suggest the presence of local adaptation to climate in Northern chamois with similar genetic adaptive responses in geographically distant but climatically similar populations. Furthermore, our results predict that future climatic changes will modify the Northern chamois adaptive landscape considerably, with various degrees of maladaptation risk.
Subject terms: Structural variation, Molecular ecology
Introduction
Climate models predict accelerated climate change in the coming years (Hock et al. 2019; Pörtner et al. 2019; Arneth et al. 2019), and the negative effects of this change on biodiversity have already been reported for many ecosystems (Pecl et al. 2017; IPCC 2019). Rapid climate change can significantly alter the distribution of suitable habitats, prompting populations and species to move to more suitable areas or to tolerate new environmental conditions (Bellard et al. 2012). These two responses are non-exclusive and potentially complementary. In the long term, species may also undergo genetic adaptation to novel environments in situ, which may prevent their decline and extinction (Parmesan 2006; Hoffmann and Sgrò 2011). Therefore, the response of a species to climate change will strongly depend on its dispersal ability, its degree of plasticity to adapt to the new climatic conditions and/or its capacity to evolve to track the new requirements of their local environments (Visser 2008; Hoffmann and Sgrò 2011).
Local adaptation is common in both plants and animals (e.g., Leimu and Fischer 2008; Hereford 2009). It occurs when populations of the same species are exposed to different environmental conditions, which results in the selection of locally adaptive traits in each population that maximize individual fitness (Rehfeldt et al. 2002). This implies that local populations will perform better than foreigners in their native environment and, vice versa, will show reduced aptitude when placed in a foreign environment (Blanquart et al. 2013). In the context of climate change, evolvability is likely to be facilitated when ‘standing genetic variation’ (SGV) at various adaptive traits is already present in the populations or available from adjacent populations through dispersal and gene flow (Savolainen et al. 2013). Hence, SGV is one of the essential biodiversity variables (see Hoban et al. 2022) and could become a key element of the adaptive response of populations to environmental change (Orr and Betancourt 2001; Hermisson and Pennings 2005). Therefore, identification of SGV in natural populations and its association with climate gradients are critical to informing science-based management strategies. In this regard, incorporating intraspecific genetic variation into predictive models that estimate the capacity of species to cope with future climate change is becoming increasingly popular (Waldvogel et al. 2020; Capblancq et al. 2020; Hoffmann et al. 2021; and see Lancaster et al. 2022 for a special issue).
Until very recently, the potential impact of climate change on species’ distributions has principally been predicted at the species level using species distribution models (Guisan and Thuiller 2005). These models assume that the species is homogeneous across the landscape and neglect the adaptive variation underlying local adaptation of populations (Alberto et al. 2013). In fact, the impact of climate change on populations may vary depending on the spatial distribution of adaptive alleles and the potential for them to spread in neighboring populations (Rehfeldt et al. 2002; Razgour et al. 2019; Chen et al. 2021). Therefore, more recent studies have used knowledge of adaptive genetic variation to refine classic species distribution models and provide new insights into species conservation and management, for example by identifying populations at risk of local maladaptation (when a genotype does not produce an optimal phenotype in the local environment; Capblancq et al. 2020). Firstly, these studies have used various genotype-environment association (GEA) methods to identify genetic variation that correlates with environmental parameters, then modeled the genetic ~ environment relationship to predict the changes in genetic composition that would be required for the population to track local climate change (Capblancq et al. 2020)—the so-called ‘genetic offset’ (Fitzpatrick and Keller 2015). However, contrary to species distribution models, genetic offset does not give information on spatial distribution shifts or reshuffling of adaptive variation across the changing climatic landscape. Therefore, the application of this metric for management or conservation remained mostly theoretical so far. In conservation, identifying adaptive units, corresponding to groups of individuals sharing similar adaptive genetic composition, is a promising approach in a global change context (Barbosa et al. 2018; Hohenlohe et al. 2021). Predicting how these units would be affected by future variation in climate thus appears critical for integrating that major threat into assessments of species vulnerability (Razgour et al. 2019). Here, we propose to bridge this gap using a landscape genomics approach that combines genotype-environment association methods with predictive models of adaptive potential, to determine the adaptive part of genetic variation, to identify the environmental factors that shape adaptive variation and to explore the capacity of a mountain ungulate, the Northern chamois (Rupicapra rupicapra), to adapt to climate change. Furthermore, by comparing the adaptive genetic composition of the Northern chamois under current and future conditions, our approach makes it possible to estimate a potential genetic offset (sensu Fitzpatrick and Keller 2015), that populations would have to solve to avoid local extinction.
Mountain biodiversity, from species and populations to their underlying gene pool, is particularly sensitive to ongoing climate change (Parmesan 2006). In the European Alps, changes have already been observed for abiotic parameters such as temperature, snow cover, glacier extent or avalanche risk (Gobiet et al. 2014), with consequences for alpine animal species including direct biological effects, loss of suitable habitats, and/or upslope shifts (Pauli et al. 1996; Engler et al. 2011). Animal populations inhabiting mountain habitats are particularly vulnerable to climate change because, even if suitable climates are still available at higher altitudes, these habitats are predicted to be restricted to ever smaller areas in the future (La Sorte and Jetz 2010; White et al. 2018). Therefore, species adapted to cold and extreme environment may be forced to move and/or adapt to new local conditions (Chen et al. 2011). At the species scale, the Northern chamois occupies a large diversity of habitat types, from low-elevation forested areas to high-altitude alpine meadows, or where the terrain is steep and rocky (Reiner et al. 2021; Corlatti et al. 2022). Northern chamois distribution currently covers a wide geographic gradient from the Mediterranean to the Black Sea and spans altitudes mainly ranging from 500 m to over 3000 m a.s.l. (Corlatti et al. 2011, 2022; Anderwald et al. 2021). Nevertheless, the species is considered morphologically and physiologically adapted to cold environments (Ascenzi et al. 1993).
A recent study determined that the reduction of body mass in Northern chamois, an important indicator of ungulate fitness (Gaillard et al. 2000), was due to the direct effect of climate warmings (Mason et al. 2014a, 2014b). The direct effect of rising temperatures could induce increases in the intensity of competition and decreases in time spent foraging (Mason et al. 2014a, 2014b), but indirect effects on fitness through effects on resource productivity or phenology have been documented on alpine herbivores, such as ibex, Capra ibex (Aublet et al. 2009). Behavioral response to recent environmental changes has already been observed in Northern chamois populations, where individuals select colder forest habitat to counteract the impacts of climate change (Reiner et al. 2021). Therefore, behavioral adaptation, e.g., change in habitat selection, could buffer the effects of summer warmings for alpine ungulates. In a recent study, Leugger et al. (2022) investigated how climate variation during the last millennia shaped the current distribution of the Northern chamois in Europe and influenced the species contemporary neutral genetic variation across the Alps. Yet, the potential evolutionary responses of the species to environmental change, and in particular mechanisms of local adaptation to climatic conditions, remain unexplored. This is all the more relevant because previous studies have found indications of selection for alpine environments in ungulates, for example on genes involved in immune and disease-regulating functions or respiratory health in Dall sheep (Ovis dalli dalli; Roffler et al. 2016).
In this study, we aim to: (i) investigate whether climatic variables drive genetic differentiation of Northern chamois populations along the European Alps and Dinaric Mountains; (ii) uncover genetic variation strongly associated with large climatic gradients, signaling local adaptation; (iii) use these signals of climate adaptation to delineate adaptive units; and (iv) model the maladaptation risk of adaptive units in a context of climate change. To achieve these objectives, we first used two different GEA approaches applied to a single-nucleotide polymorphism (SNP) data set obtained from double-digest restriction-site associated DNA sequencing (ddRADseq), to investigate the association between genetic variation and environmental gradient(s) across the Northern chamois distribution range. Second, using the set of putatively adaptive markers, we delineated four adaptive units along the Alps and Dinaric Mountains, distinguishing individuals genetically adapted to Mediterranean conditions from those adapted to more alpine environments. Finally, projecting the geographical distribution of adaptive units into the future, we predicted that the adaptive landscape of the northern chamois will change considerably, with varying degrees of maladaptation risk for this mountain mammal in the coming decades.
Material and methods
Genetic data
Samples of 465 Northern chamois (Rupicapra rupicapra) were obtained from the continuous range of the species in the European Alps and Dinaric Mountains (Fig. 1), thanks to a network of collaborators (e.g., hunting administrations and associations, non-governmental organizations, national parks, biobanks; see Acknowledgements). The species’ range extends beyond this geographical area, and isolated populations occupy territories at lower altitudes and on plains elsewhere e.g., in France, Germany, the Czech Republic and the Balkans (Corlatti et al. 2022). We did not take these populations into account in this study, which focused on alpine areas characterized by strong environmental gradients. A genetic dataset of 30,970 SNPs was generated for these individuals using a ddRADseq approach (following Peterson et al. 2012).
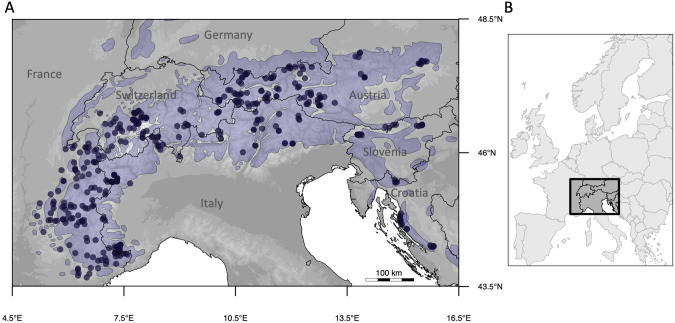
Fig. 1. Map of the study area illustrating the location of Northern chamois (Rupicapra rupicapra) sampled individuals.
A Northern chamois sampling (n = 429) along the European Alps and Dinaric Mts. The bluish contours on the map show the contemporary distribution range of Northern chamois (Anderwald et al., 2021). B Location of study area in Central Europe. Black lines on the maps correspond to the country borders. The maps are projected in the world geodetic system 1984 (WGS 84) Black lines on the maps correspond to the country borders. The maps are projected in the world geodetic system 1984 (WGS 84).
The procedure regarding ddRADSeq library construction, sequencing and data processing is detailed in Leugger et al. (2022), and provided in the Supplementary Materials. Briefly, we used Stacks v2.4 (Catchen et al. 2011,2013) to demultiplex data, build a de novo SNP catalog, and call genotypes. SNPs were filtered if they were genotyped for less than 85% of the samples, when the SNP error rate was greater than 2.5% based on the analysis of replicates (~12.5% of the samples, n = 81; Figs. S2-S7), when the minor allele frequency (MAF) was lower than 1% (i.e., fewer than 8 allelic copies; Lowry et al. 2017a) and when sequencing read depth was lower than 10X or greater than 25X (equals to mean + 4 * sqrt(mean), see Supplementary Materials). The final dataset only included samples that were genotyped for > 75% of all SNPs. From the initial sample set (n = 465), we removed the isolated populations of Slovakia and Switzerland (Ticino), because they showed extreme genetic discontinuity with the rest of the sampling (see Roques 2021; Leugger et al. 2022), potentially biasing the detection of selection in the genome (Foll and Gaggiotti 2008). The final data used for subsequent analyses included 429 samples and 20,904 SNPs (only one SNP was randomly selected from each RAD-fragment, Figure S8). Missing data were imputed with the median genotype.
Overall genetic differentiation among individuals
We inferred genetic units based on overall genetic differentiation, i.e., using all 20,904 SNPs (e.g., Barbosa et al. 2018; Hohenlohe et al. 2021) using a principal component analysis (PCA) implemented in the adegenet R-package (Jombart 2008), using all 20,904 SNPs. Individual scores along the first two axes of this PCA (PC1 and PC2; Figure S9) were then used as a proxy of genetic structure to condition the search for selection in the genome (see variance partitioning section). We also investigated overall population structure using a discriminant analysis of principal components (DAPC; Jombart et al. 2010) implemented in the package adegenet. Without prior information, we used the function find.cluster to determine the optimal number of clusters; more precisely, we ran successive K-means clustering with an increasing number of clusters ranging from K = 2 to K = 6 (based on previous inspection of values of BIC versus number of clusters). We then performed a stratified cross-validation analysis to identify the optimal number of principal components (PCs) to retain in the DAPC analysis. This cross-validation procedure was carried out using the function xvalDapc, with 100 replicates for each number of PCA axes retained and using 90% of the data for training and the remaining 10% for validation. Because several runs conducted with the same number of clusters K showed different results, we performed 10 independent runs for each K and selected the run with the highest assignment success mean after cross-validation.
Environmental variables selection
To explore the environmental factors that influence chamois genetic variation, we considered the 19 bioclimatic variables available from Worldclim 2 database (Fick and Hijmans 2017). The set of variables are frequently used in similar studies (e.g., Thuiller et al. 2014; Bay et al. 2018; Rochat et al. 2021) and are already known to influence chamois distribution (Thuiller et al. 2018). We extracted the value of the bioclimatic variables within a 1500 m buffer radius around each sampling location (i.e., in area of ~5 km² in agreement with home range size and dispersal patterns observed in chamois; Loison et al. (1999), Loison et al. (2008), Nesti et al. (2010), Seigle-Ferrand et al. (2022), based on raster with a spatial resolution of 30 sec (~1 km at the equator; Fick and Hijmans 2017). To avoid collinearity among variables, we included only uncorrelated variables (based on a principal component analysis) and favored seasonal variables, because these better reflect climatic variation among sampling locations in comparison with variables measured on shorter (i.e., monthly) or longer (i.e., annual) time periods. In this way, 12 out of 19 bioclimatic variables were selected (Figure S12). Next, we wanted to select only variables that were significantly correlated with genetic variation for subsequent analyses. To identify these, we used a forward selection procedure based on Redundancy Analysis (RDA; Blanchet et al. 2008) during which the standardized variables that explained most of the model variance were added sequentially to a null model, until the addition of any remaining variable did not increase the global model R² (Capblancq and Forester 2021). This procedure was performed using the function ordiR2step from the R-package vegan (Oksanen et al. 2019). Finally, we retained the 12 variables during the forward selection because they all explained a significant and independent component of genetic variation (Table S3).
Variance partitioning
To disentangle the relative contribution of environment, geographic distance, and evolutionary history in explaining genetic variation across chamois populations, we used partial redundancy analysis (pRDA). Partitioning variance using pRDAs allows an estimation of the proportion of genetic variation explained by a particular group of variables (e.g., climate) while considering the effect of other variables (e.g., geography and/or overall population structure). If some of them are spatially correlated on a large scale, pRDAs can help disentangling their relative contribution to genetic variation. We followed Capblancq and Forester (2021) and considered 3 different sets of independent variables potentially influencing the distribution of genetic variation: 1) geography: derived from individual spatial coordinates (longitude and latitude); 2) evolutionary history: expressed as proxy of overall population structure (i.e., PC1 and PC2 from the PCA conducted above), and 3) environment: represented by the 12 bioclimatic variables retained during the forward variable selection (Table S3). We ran four models with individual genotypes as dependent variables (using the complete genetic dataset), one model for each set of variables (e.g., a pure climate model using the 12 bioclimatic variables as explanatory variables and either longitude, latitude, PC1 or PC2 as conditioning variables) and the full model using all variables. To estimate the independent contribution of each set of variables and their confounded effect with the other sets, we compared the sum of variance explained by each pRDA model. The procedure was conducted using the function rda of the R-package vegan and default parameters (Oksanen et al. 2019) and we used anova for significance testing (i.e., 999 permutations).
Genotype-environment association
To identify genetic variants that covary with environmental predictors (i.e., putatively linked to local adaptation), we used two genotype-environment association (GEA) approaches commonly used in the recent literature: a univariate method—or latent factor mixed models (LFMM; Frichot et al. 2013), and a multivariate method—or redundancy analysis (RDA; Forester et al. 2018; Capblancq et al. 2018). Both approaches use linear regressions to identify loci associated with environmental variation, with individual genotypes as response variables and environmental data as explanatory variables, while also considering the past evolutionary history of the populations. LFMM, executed using the lfmm_ridge and lfmm_test functions from the R-package lfmm (Caye et al. 2019), estimates the correlation between environmental and genetic variation considering one environmental variable at a time. Population structure is modeled during the procedure using latent factors and we used K = 4 latent factors, as suggested by the results of the genetic PCA (Fig. S13). RDA, performed using the function rda of the R-package vegan (Oksanen et al. 2019), detects adaptive variation by considering multiple environmental predictors together and identifies the environmental gradients that are the most correlated with adaptive variation. We used PC1 and PC2 from the genetic PCA conducted above to condition the RDA model and account for overall population genetic structure. We then used the first five RDA axes (based on the screenplot shown in Fig. S14) to compute Mahalanobis distances and identify outliers. For both methods, we estimated corrected p-values for multiple non-independent tests (i.e., later termed q-value) using the qvalue function implemented in the qvalue R-package (Storey et al. 2023). Outliers were then detected using a q-value threshold of 0.05, which corresponds to a false discovery rate (FDR) of 5% (Steane et al. 2014; Forester et al. 2018; González‐Serna et al. 2020). Finally, we retained the outliers identified with both LFMM and RDA for subsequent analysis.
Adaptive unit delineation
To explore the distribution of adaptive genetic variation across chamois natural range, we first used RDA to identify groups of individuals sharing a similar adaptive genetic composition (i.e., sharing similar alleles) by considering only loci that were detected as putatively under selection for climate (i.e., outliers). We then used an “adaptively enriched” RDA space (sensu Steane et al. 2014) created using these adaptive loci and the 12 environmental variables to predict the turnover of adaptive genetic composition across the species climate landscape. To do so, we followed the formula from Steane et al. (2014):
where a is the value of the standardized bioclimatic variable at the focal location, b is the RDA score of the bioclimatic variable for the concerned axis, and i corresponds to the different bioclimatic variables used in the RDA model. Adaptive indices were estimated for the two first RDA axes (based on the screenplot shown in Fig. S17) and for each raster cell of the geographic range of the species. We considered here the IUCN geographic range of the Northern chamois (Anderwald et al. 2021) at a 30-seconds spatial resolution (Fig. 1). Subsequently, the indices were not weighted by their contribution to its associated RDA axis model (i.e., eigenvalues) because each index represents a potential genetic adaptation linked to the environment and we decided to avoid favoring one adaptation over another. These indexes were then used to group the cells through a clustering procedure using the function clara of the R-package cluster (Kaufman and Rousseeuw 1990), this function is an extension to k-medoids (PAM) methods to deal with data containing a large number of objects. To select the appropriate number of clusters to be produced, the Elbow method was computed using the function fviz_nbclust from the R-package factoextra (Kassambara and Mundt 2020). These units were mapped to illustrate the species’ adaptive landscape (for an example, see Capblancq et al. 2020).
Future predictions and genetic offset
The adaptive genetic units identified above were used to investigate the potential mismatch between their current distribution and that forecasted in the coming decades under predicted climatic conditions. We considered the MIROC-ES2L global climate models (GCM) and two shared socio-economic pathways (SSP), i.e., SSP2-4.5 and SSP5-8.5), available for Worldclim 2 bioclim variables (Fick and Hijmans 2017). The moderate SSP2-4.5 scenario corresponds to a “middle of the road” world where trends broadly follow their historical patterns, and the extreme SSP5-8.5 scenario corresponds to a world of rapid and unconstrained growth in economic output and energy use (Riahi et al. 2017).
We projected the adaptive genetic indices estimated from the adaptively enriched RDA (see above) for the 2021–2100 time-period. To allow comparison between current and future environments, the values of future bioclimatic variables were standardized using the same standardization parameters (i.e., mean and standard deviation) previously used for the current bioclimatic variables (see Breed et al. 2019 for a similar approach). Each value was also weighted by the percentage of variance explained by each RDA axis. Based on RDA1 and RDA2 adaptive genetic indexes, each pixel of the bioclimatic raster was assigned to a cluster from the adaptive units defined in the present time. If future RDA scores were outside of the current RDA score range, the adaptive unit was assigned to a new group called ‘Outsider’. In other words, the environmental conditions predicted for these locations were not experienced by Northern chamois within its current range, so we decided not to infer an optimal genetic composition for these climate conditions, because they were outside our model training space. Finally, we estimated the evolution of the spatial distribution of each of the Northern chamois adaptive units in the European Alps and Dinaric Mountains depending on different climatic scenarios, measuring changes in surface area and elevation for each adaptive unit over time.
Alongside the exploration of spatial changes for each adaptive unit, we identified the regions that would be the most severely affected by a change in climatic conditions through the estimation of genetic offset (Fitzpatrick and Keller 2015; Bay et al. 2018; Capblancq et al. 2020). Genetic offset, also known as genetic vulnerability, is defined here as a distance between the current and future optimal genetic compositions depending on the change between current and future climatic conditions. Therefore, genetic offset is a predictive measure of how much the distribution of locally adapted alleles will be perturbed by the shift between current and future environmental conditions (Fitzpatrick and Keller 2015). We estimated a rate of adaptive score change for each climatic cell included in the current range of chamois in the Alps. To do this, we calculated the Euclidean distance between each cell’s RDA score in the present and in the future. Only the first two axes (RDA1 and RDA2) of the adaptively enriched RDA were used for this estimation.
Results
Global genetic variation
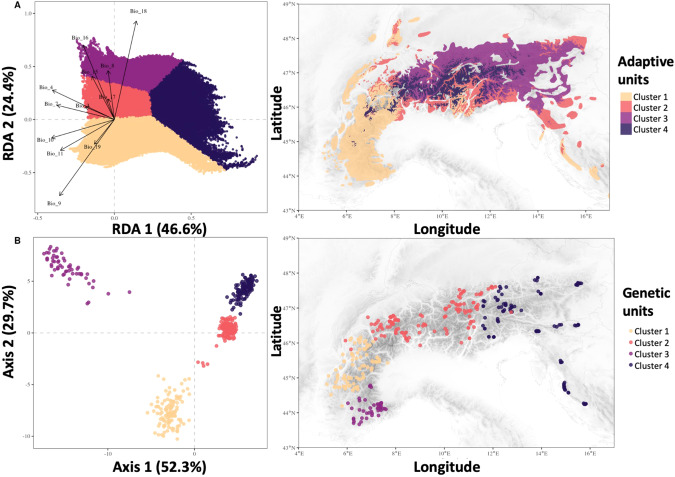
We conducted a discriminant analysis of principal components (DAPC) to characterize global genetic variation in Northern chamois (Fig. 3B). At the highest hierarchical level (i.e., K = 2), we observed genetic differentiation among individuals from the western and eastern Alps, with a transition located approximately along the France-Switzerland border (see “2 clusters” panel; Fig. S10). From K = 2 to K = 6, there were additional genetic groups identified in the western Alps (i.e., from south-eastern France to eastern Switzerland), splitting the eastern Alps in two groups along a longitudinal gradient (Fig. S10 with 6 clusters). Using the BIC criterion, we concluded that the most likely number of independent genetic clusters in the study area was four (see adaptive landscape section for details): two clusters in the western Alps, and two in the east. The genetic clusters are structured geographically, with the two most distant clusters on the first axis of the DAPC plot also the most distant across the Alps (i.e., clusters 3 and 4 in Fig. 3B).
Fig. 3. Delineation of adaptive and global genetic units.
A The four adaptive units delineated in the adaptively enriched RDA genetic space (left) and spatially mapped (right) across the species distribution range, and B the four global genetic units defined by the DAPC (left) and spatially mapped (right).
Variance partitioning
The full RDA model that included climatic variation, geography and genetic structure variables explained 12% of the total genetic variability (Table 1). The significant results from the four pRDAs showed that 37.4% of this explainable genetic variance was linked to climatic variation (p < 0.001), 6.2% was associated with geographic variation (geographic coordinates; p < 0.001), 11.0% was associated with overall genetic structure (p < 0.001), and 45.4% was not specifically attributable to any of these groups of variables. The high proportion of variance that was not attributable to specific predictors (i.e., confounded) suggested substantial covariation between environmental gradients, latitude and/or longitude and overall genetic variation across the species range. That result support the use of covariables to correct the GEA scans for the potential confounding effect of demographic history.
Table 1.
Decomposition of the influence of climate (i.e., clim.), geography (i.e., geo.) and global population genetic structure (i.e., anc.) on genetic variation using partial redundancy analyses.
| Partial RDA models | Inertia | R2 | P(>F) | Proportion of explainable variance | Proportion of total variance |
|---|---|---|---|---|---|
| Full model : F ~ clim. + anc. + geo. | 208.01 | 0.118 | 0.001 | 1.00 | 0.118 |
| Pure climate : F ~ clim. | (anc. + geo.) | 77.84 | 0.044 | 0.001 | 0.374 | 0.044 |
| Pure geography : F ~ geo. | (anc. + clim.) | 12.84 | 0.007 | 0.001 | 0.062 | 0.007 |
| Pure ancestry : F ~ anc. | (geo. + clim.) | 22.90 | 0.013 | 0.001 | 0.110 | 0.013 |
| Confounded climate/geography/ancestry | 94.43 | 0.454 | 0.054 | ||
| Total unexplained | 1553.18 | 0.882 | |||
| Total inertia | 1761.19 | 1.000 |
For each model, we calculated its statistical significance, the percentage of explained genetic variance compared to the variance explained by the full model and compared to the total variance present in the data set.
Genotype-environment association
The LFMM genome scan, using K = 5 latent factors, retained 536 loci as outliers among the 20,904 SNPs tested (Fig. S15). Those outliers were only associated with 7 of the 12 selected variables: Bio3, Bio4, Bio9, Bio15, Bio16, Bio17 and Bio18. Most of the outliers are associated with Bio9 (n = 214 outliers) and Bio18 (n = 354 outliers), which correspond to the mean temperature of the driest quarter and the precipitation of the warmest quarter, respectively (Fig. S15). The RDA-based genome scan procedure retained 2205 loci as outliers (Fig. S16). The intersection of the two outlier sets, i.e., the outliers that overlapped according the two approaches yielded 275 unique outlier loci. These 275 loci (out of 20,904 SNPs, i.e., 1.3% of all SNPs) were considered as markers of genomic regions putatively associated with environmental adaptation in Northern chamois.
Adaptive landscape
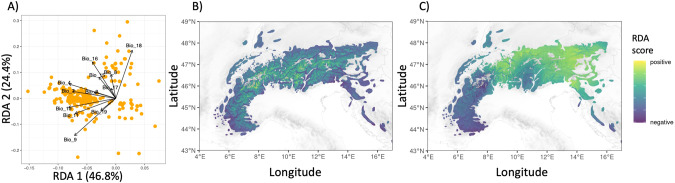
The adaptively enriched RDA was conducted using the 275 outlier SNPs (Figs. S18 and S20). Most of the adaptive genetic variance was explained by the first two RDA axes (i.e., 46.6 and 24.38%, respectively; Figs. 2A and S17). RDA1 was correlated with temperature seasonality (Bio4) and mean temperature of the warmest quarter (Bio10; Fig. 2A). RDA2 was correlated with most of the environmental variables used in the analysis but principally with mean temperature of driest quarter (i.e., Bio9), and precipitation of the warmest quarter (i.e., Bio18; Fig. 2A). We present allele and genotype frequency changes for the most significant association between outlier SNPs and climate variables in supplementary materials (Figs. S18–S21).
Fig. 2. Adaptive landscape of the Northern chamois along the European Alps and Dinaric Mts.
A Projection of bioclimatic variables (in black) and outlier SNP markers (in orange) into the adaptively enriched genetic RDA space and B, C spatial extrapolation of RDA1 and RDA2 gradients, respectively, for the current climatic conditions. See Table S3 for a description of the bioclimatic variables.
We extrapolated adaptive genetic turnover across the Alps and Dinaric Mtns by estimating RDA1 and RDA2 scores for the entire range of chamois in the study area (Fig. 2). The RDA1 index seemed to represent an altitudinal gradient (Fig. 2B). The RDA2 index mainly followed a latitudinal gradient with higher values in the south-east, south-west and western front of the Alps, and lower values in the central and north-eastern Alps (Fig. 2C). We then used those two indices to delineate different adaptive units across the species distribution range (from two to six clusters, Fig. S19). According to the clustering procedure, the most parsimonious number of clusters was four (Fig. S20). South-eastern France and Croatia were always included in the same adaptive unit as a small part in the Alps representing the Ticino region (cluster 1; Fig. 3A). Similarly, Slovenia and the center of Switzerland were grouped together (cluster 2; Fig. 3A), grouping areas located between the southern-western Alps (cluster 1; Fig. 3A) and the northern Alps (clusters 2, 3 and 4; Fig. 3A). This latter area was also characterized by higher mean temperatures and mean precipitation of the wettest quarter, low mean temperatures of the driest quarter and high mean precipitation of the warmest quarter. The difference between clusters 2, 3 and 4 was mostly linked to altitudinal gradient and temperature seasonality. Moreover, clusters 2 and 3 differed according to the mean precipitation of the warmest and wettest quarters (i.e., Bio18 and Bio16, respectively; Table S3).
Global versus adaptive genetic unit delineation
We observed a clear distinction between global and adaptive genetic units (Fig. 3): the spatial distribution systematically differed between global and adaptive genetic clustering, regardless of the number of clusters (Figs. S10 and S22). Adaptive genetic units clustered individuals associated with the same environmental conditions even if these were geographically distant from one another, such as south-eastern France and Croatia (Fig. 3A, Cluster 1), whereas global genetic units clearly group individuals from the same geographic area (Fig. 3B). Therefore, the most likely number of genetic units was four for adaptive genetic variation (Fig. S23), that we compare with the four global genetic units obtained with all markers using DAPC. Even if the pRDAs highlighted an effect of geography and past demographic history on chamois genetic variation, we were able to delineate multiple genetic units associated with specific environmental conditions along the European Alps and Dinaric Mountains.
Future predictions and genetic offset
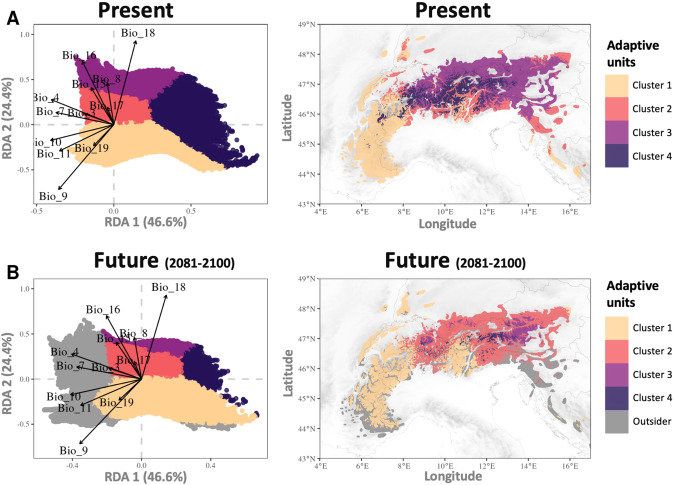
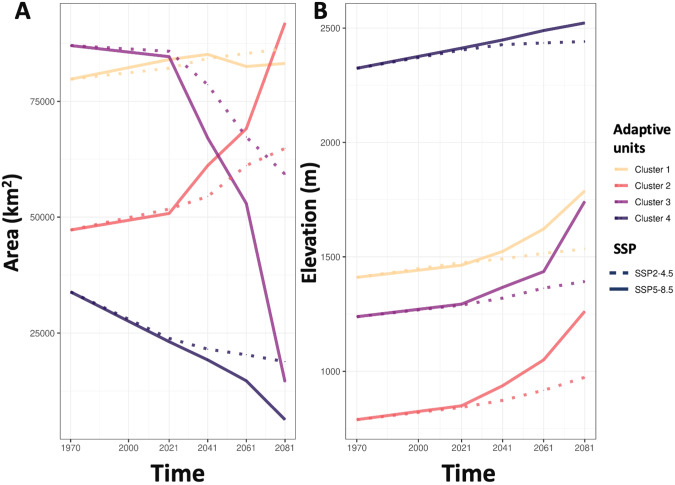
The spatial projection of the four adaptive genetic units differed substantially depending on the time-period and the SSP scenarios considered. Overall, and as expected, changes were more substantial for the SSP5-8.5 than for the SSP2-4.5 scenario. For both scenarios, we predicted an increase in mean habitat elevation for all four adaptive units by the end of the century (Fig. 4). The maximum difference between current and future elevations was 503 m, predicted for cluster 3 using SSP5-8.5, while the minimum difference was 116 m, predicted for cluster 4 for the SSP2-4.5 scenario (Table 2). We predicted an overall decrease in suitable habitat, especially for clusters 3 and 4 (Fig. 4 and Table 2). Conversely, cluster 2 will expand its distribution, up to 94% according to the SSP5-8.5 extreme scenario (Fig. 4 and Table 2). Approximately 30% of the study area will no longer be suitable according to our model based on SSP5-8.5 (Fig. 5 and Table 2). These future unsuitable areas represent climatic conditions that are either not currently experienced by the chamois we sampled and thus may represent climatic combinations that are not suitable for the Northern chamois of the study area, or for which we lack information about the species adaptive potential.
Fig. 4. Projection of the four adaptive genetic units.
A In the present and B in the future (2081–2100) for the SSP5-8.5 scenario. The left panels show the projection into the adaptively enriched RDA space and the right panels show the same on a map of the Alps and Dinaric Mts. In gray, the new group called “Outsider” depends on a bioclimatic gradient which does not currently exist meaning it is outside the current adaptive range observed for Northern chamois.
Table 2.
Changes in the area and elevation for each adaptive genetic unit from 1970–2000 to 2081–2100.
| SSP | Cluster | Area change range rate (%) | Elevation difference (m) |
|---|---|---|---|
| SSP2-4.5 | Cluster 1 | 8.28 | 124.1 |
| Cluster 2 | 37.27 | 185.18 | |
| Cluster 3 | −31.94 | 154 | |
| Cluster 4 | −44.39 | 116.95 | |
| Outsider | 326.65 | 184.63 | |
| SSP5-8.5 | Cluster 1 | 4.28 | 377.46 |
| Cluster 2 | 94.67 | 473.27 | |
| Cluster 3 | −83.38 | 503.45 | |
| Cluster 4 | −81.39 | 198.09 | |
| Outsider | 880.33 | 302.82 |
The group called “Outsider” corresponds to individuals predicted to occupy bioclimatic gradient which does not currently exist, meaning it is outside the current adaptive range observed for Northern chamois in the study area.
Fig. 5. Projection of shift in distribution of adaptive genetic units.
A Surface area change and B mean elevation shift of the four adaptive genetic clusters over time from 1970 to 2100 according to the two scenarios SSP2-4.5 and SSP5-8.5.
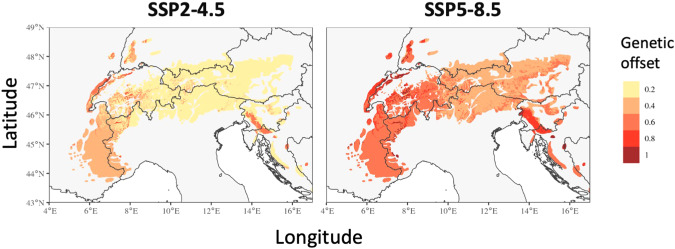
Estimates of genetic offset calculated using the differences between current and future climates in species ranges, identified areas where important genetic changes (i.e., changes in allele frequency) would need to occur for populations to track climate change. Overall, Switzerland and Slovenia were the regions with the highest required adaptive change (Fig. 6). Changes were widespread throughout the study area, but more pronounced in the Mediterranean region.
Fig. 6. Projection of genetic offset.
Representation of genetic offset between current and future climatic conditions for the two scenarios SSP2-4.5 and SSP5-8.5 for Northern chamois along the Alps and Dinaric Mtns.
Discussion
Evidence of association between genetic and environmental variation
Exploring the effects of environment, geography, and evolutionary history on Northern chamois genetic variation, we found that climatic factors played a significant role in driving population genetic differentiation. We identified putative signatures of selection linked to climate variation in the species genome, suggesting the presence of local adaptation across the global distribution of the species. According to our results, this adaptation seems to be mainly driven by temperature and precipitation during the summer months (i.e., mean temperature of the driest quarter and the precipitation of the warmest quarter). The Northern chamois exhibits physical, behavioral, and ecological adaptations to mountain life (Corlatti et al. 2022) and is known to be particularly sensitive to heat during the spring-summer period, with important effects reported on life-history traits, directly or indirectly linked to fitness (Rughetti and Festa-Bianchet 2012; Mason et al. 2014a, 2014b). Summer temperatures are known to influence the physiology and behavior of many ungulates, from northern regions (van Beest and Milner 2013) to the Mediterranean (Marchand et al. 2015) and at high elevations (Semenzato et al. 2021). Therefore, variability in chamois sensitivity to thermal stressors during summer may be due to genetic adaptation to the local environment. Similar signatures of local adaptation to cold environments have also been reported in the mountain goat Oreamnos americanus (Martchenko et al. 2020). Analyzing the relationship between genetics and environment across the European Alps, we also detected a strong collinearity between environmental gradients, geographic distances and variables reflecting the recent evolutionary history of the species. The variance partitioning analysis identified that 46.6% of the explainable genetic variation was confounded among these three factors (Table 1) and it was not possible to totally differentiate their relative influence. It confirms the difficulty of entirely untangling the impact of the environment from the influence of past demographic processes and/or isolation by distance when exploring the drivers of intraspecific genetic variation (Frichot et al. 2015; Capblancq and Forester 2021).
We mapped the outlier loci found under putative selection for the environment against the well-annotated goat reference genome (Capra hircus, ID ARS1; BioSample: SAMN03863711) in the Ensembl project (https://www.ensembl.org/). We found that some of the outliers fell in the vicinity of genes involved in skin properties such as CDH2 which was also involved in local adaptation to climate in domestic goats (Alimperti and Stelios 2015; Rochat and Joost 2019). However, we must also acknowledge that our genotyping method—ddRAD sequencing—produced a reduced representation of the Northern chamois’ large genome, which is probably ~2.5 Gbp long, similarly to close relatives (Martchenko et al. 2020). Considering this genome size, the 20,904 genotyped SNPs are, on average, 280,000 bp apart from each other. Such a low density of markers is not optimal to identify all the genes responsible for local adaptation (Lowry et al. 2017b) but likely some of them, as described above. Genome-wide diversity, estimated using a genotyping-by-sequencing approach, was also found a good indicator for adaptive variation and for predicting evolutionary responses, and better than the population history reflected by the inbreeding level (Ørsted et al. 2019). Overall, we believe our data were good enough to differentiate multiple SNPs putatively constrained (or linked to genomic regions putatively constrained) by selection for climate variations and then strongly differing from the neutral genetic pattern.
Identifying adaptive genetic units
Once the putative adaptive genetic component was identified, we were able to project the underlying genetic variation across the species range to characterize patterns of intraspecific adaptive divergence across the climate landscape (Steane et al. 2014; Fitzpatrick and Keller 2015). This type of approach is increasingly used to visualize spatial gradients of genetic turnover (i.e., changes in adaptive allele frequency) associated with local adaptation to climate (Bay et al. 2018; Ruegg et al. 2018). In Northern chamois, this turnover was mainly driven by variation in high temperature and low precipitation (i.e., high mean temperature of the driest quarter and low precipitation of the warmest quarter). The adaptive gradient differentiates southern from northern parts of the Alps, irrespective of longitude. We also found adaptive variation to be related to a gradient of precipitation during the wettest quarter and temperature seasonality. Rochat and Joost (2019) already found adaptive alleles associated with precipitation of the warmest quarter in domestic goats (Capra hircus) from several breeds in Morocco and Europe, for genes related to properties of the cornea and skin, possibly conferring adaptations to drought conditions such as high UV-radiation.
When looking for coherent groups along these adaptive gradients we were able to delineate four adaptive genetic units that greatly differed from inferred global genetic populations. Most prominently, two populations, one from south-eastern France and the other from Croatia were grouped into the same adaptive unit while being 700 km apart from each other and assigned to two well-differentiated global genetic clusters, suggesting different evolutionary histories. These two populations most likely clustered together because they experienced a similar climate and were probably subject to similar selective pressures. Further analyses would help to determine whether this clustering is an example of parallel evolution or more likely result from past evolutionary history of adaptation and shared ancestral polymorphism among regions. Finally, additional investigations, probably with denser genomic data from an annotated reference genome would be required to determine if the same genes are involved in local adaptation in Northern chamois and to confirm the influence of bioclimatic variables on genetic divergence in mountain ungulates.
Future of the Northern chamois in the Alps
Major environmental changes are predicted by the end of the century, especially changes in temperature and precipitation (Hoegh-Guldberg et al. 2018), with dramatic consequences on mountain ecosystems (Gobiet et al. 2014; Hoegh-Guldberg et al. 2018). Previous studies on alpine ungulates (e.g., the mountain goat; White et al. 2018) and more specifically on chamois (Thuiller et al. 2018; Lovari et al. 2020) predicted an important contraction of the future distribution ranges. Using a species distribution model approach, Mason et al. (2014a, 2014b) predicted an elevational shift of 15 to 30 meters up-slope by the year 2100 for chamois in the Gran Paradiso National Park, in Italy. However, using an integrative approach, Thuiller et al. (2018) found that the inclusion of biotic interactions in species distribution models (i.e., plant resources) produced more optimistic predictions than models based on climate only. Using the relationship between intraspecific genetic variation and climate, we went a step further than previous models and predicted a shrinking of the range for all but one adaptive unit, together with an elevation shift towards higher altitudes. By the end of the 21st century, the mean elevation inhabited by all four adaptive units of chamois is predicted to rise, on average, by 200 m. This altitudinal shift is also predicted to lead to the disappearance of the adaptive unit already living at high elevation.
By including intraspecific genetic variation in future projections, we were able to evaluate the impact of climate change on local adaptation equilibrium in Northern chamois. Overall, the integration of adaptive variation into predictive models may improve our estimation of species dynamics under scenarios of environmental changes (Waldvogel et al. 2020). Interestingly, in Northern chamois, it is not the Mediterranean region that is predicted to experience the strongest disruption of the adaptive equilibrium (i.e., genetic offset), but the central part of the species range, in Switzerland and Slovenia. Potentially because population in these areas currently have intermediate allele frequencies but will have to specialized toward the warmer end of the adaptive gradient in the future. The genetic offset metric is very complementary to the projection of shift in distribution of adaptive genetic units. Indeed, change in the distribution of adaptive units showed their future optimal location based on current genetic adaptation, while genetic offset highlighted the location where extreme adaptive genetic changes would be required to track the change in climate. In that sense, even where genetic offset is low, the future climate can be beyond the adaptive range of the current units (e.g., south-eastern France or Croatia). On the other hand, a high estimate of genetic offset for a particular unit could result in its replacement by another genetic group when such a group is already present nearby (i.e., central Switzerland).
Our results also suggest that the Northern chamois is currently genetically adapted to environments that are predicted to be no longer available in the future (by 2100), especially at higher elevation (>1000 m; see Fig. 5). Conversely, at lower elevation (<800 m), our model predicted a change in environmental conditions that would push the climate conditions beyond the adaptive space currently occupied by the species, potentially making these areas climatically unsuitable for Northern chamois in a near future. It is important to note here that this result does not necessarily mean that the Northern chamois will go extinct at low elevation, but we are currently unable to infer genome-environment association for climatic conditions that the samples we considered in this study does not currently experience. Depending on the predicted scenario, these unsuitable climatic areas represent up to approximately 50% of the chamois current distribution range across the Alps (e.g., for the SSP5-8.5 scenario). The lower elevation areas, especially Mediterranean areas such as south-eastern France and Croatia, would be the most impacted and populations would require strong genetic change (i.e., change in allele frequency) to cope with future climatic conditions. Because our projections are relatively short term (i.e., 60 to 80 years), and considering the comparatively long generation time of Northern chamois (i.e., ~6 years; Loison et al. 1999), beneficial mutations may not arise for evolutionary rescue (Bell 2013) and spread fast enough to enable the populations to adapt to the changing climates (Barrett and Schluter 2008), although adaptation may still be possible in such a short period of time, if sufficient standing genetic variation (SGV) is already present in the populations (Epstein et al. 2016; Lai et al. 2019; Bitter et al. 2019). Interestingly, Roques (2021) has shown that the areas that were predicted to experience the most marked changes here (i.e., south-eastern France) are also those with the highest global genetic diversity, giving some raw material for selection to filter and shape new patterns of adaptive variation.
Although our models can explore the current climatic niche of the species using observable adaptive variability and predict its potential future disruption, we cannot predict to what extent the species will react to these changes. In particular, the dispersal capacities of chamois could help them track suitable climates over long distances and behavioral changes could locally mitigate the impact of global warming. Our predictions also suffer from potential shortcomings. Firstly, we did not assess the strength of the relationship between adaptive genetic variability and environment, and its quantitative effects on Northern chamois fitness. Validating the negative correlation between genetic offset and fitness is critical to do in the future if one wants to use such framework to inform conservation. That would require acquiring more data on fitness related traits and potentially set up common garden experiments (Browne et al. 2019; Fitzpatrick et al. 2021), which is not easily done with wild large mammals. Secondly, the current spatial distribution of Northern chamois across the Alps is also influenced by past and current human activities, such as hunting, farming, tourism, and habitat fragmentation, which may have pushed the species towards higher elevations during the last millennia, as well as other components of the environment including topography and edaphic factors, and not only climate. This could have artificially reduced the extent of the species’ realized climatic niche that we can measure and led to missing a section of the adaptive gradient with our models. For example, the species occupied low to mid-altitude mountain areas of Western Europe, e.g., in the Jura Mountain, the Vosges Mountain, the Massif Central (Corlatti et al. 2022), and, the species appears to be continuously expanding its range into the lowlands and forested habitats, e.g., in France (Deinet et al. 2013) and also in Poland/Slovakia (Ciach and Pęksa 2019), suggesting that the species can quickly adapt to warmer habitats by moving into forests (Reiner et al. 2021). Thirdly, the Northern chamois has already evolved behavioral strategies to cope with climate fluctuations and resource availability, such as seasonal vertical migration to avoid heat stress (Papaioannou et al. 2015). In this study, we did not consider the ability of the species to react to environmental change via plastic responses, which could possibly mitigate our evolutionary predictions, especially at low elevations (Chirichella et al. 2021). Indeed, recent investigations suggest that the future of chamois will not only depend on how the species adapts to changing climatic conditions, but also on the behavioral plasticity of the species (e.g., Reiner et al. 2021), in relation to a range of factors, e.g., climate change (Lovari et al. 2020), resource availability (Thuiller et al. 2018), competition (Corlatti et al. 2019) or even disease outbreaks (Rossi et al. 2019).
Conservation and management implications
Identifying adaptive units can be directly applied to the planning and execution of conservation and management programs (e.g., Flanagan et al. 2018; Hoban et al. 2020). For example, if translocations are used to restore or reinforce populations, considering source populations belonging to the same adaptive unit would help prevent the introduction of maladapted individuals and the deleterious effect of outbreeding depression (Aitken and Whitlock 2013).
In that sense, landscape genomics is a promising framework for translating knowledge of global and adaptive genetic diversity into guidelines for conservation policy and management strategies (Hohenlohe et al. 2021). However, it is quite difficult in practical situations to use continuous proxies of adaptation such as the ones presented in Fig. 3. Often, it is more efficient to delineate discrete units of management, based on neutral genetic variation in the field of conservation (Funk et al. 2012) or seed or breeding zones in forestry (O’Neill and Aitken 2004). Here we propose what we believe is the first use of RDA to delineate such types of units and a framework to identify adaptive genetic groups within a species. Although, Fernandez-Fournier et al. (2021) recently questioned the need to “identify adaptive genetic variation when prioritizing populations for conservation” considering standing genetic variation can be used as a proxy for the adaptive genetic variation, Funk et al. (2012) and others (Hohenlohe et al. 2021) stressed the necessity to use both global and adaptive markers to make optimal management decisions (e.g., see Barbosa et al. 2018 for an empirical application). We believe that information on species’ evolutionary potential offers important new insights that can help refining conservation strategies and management policies (Borrell et al. 2020; Hohenlohe et al. 2021). It is worth noting, however that, in practice, in a rapidly changing and uncertain environment, we cannot totally rule out the importance of considering genome-wide genetic variation as an indicator of evolutionary potential in management strategies (Flanagan et al. 2018; Fernandez-Fournier et al. 2021; Hoban et al. 2022), because the foundations of adaptive variation are complex and not fully understood, it is currently impossible to make a definitive dichotomy between adaptive and neutral variation (Harrisson et al. 2014).
One management action that is gaining interest recently is the translocation of pre-adapted individuals to either genetically reinforce populations that will suffer from climate change—assisted gene flow—or colonize a previously unoccupied habitat that became suitable since the climate started to change—assisted colonization (Aitken and Whitlock 2013; Aitken and Bemmels 2016; Browne et al. 2019; Segelbacher et al. 2022). As far as we know, such actions have not yet been implemented in nature, but they could facilitate the establishment of adaptive alleles and help species with long generation times or low dispersal capacities to adapt to future climatic conditions. (Rellstab et al. 2016; Capblancq et al. 2020; Seaborn et al. 2021). It should be noted that other studies have warned of the risks and limitations of assisted migration in conservation management (Park and Talbot 2012; Montwé et al. 2018). Translocation and reintroduction programs are already common management strategies for chamois throughout its range (Soorae 2010; Apollonio et al. 2014), without any consideration of their genetic composition, and even more their adaptive genetic background. The results of the present study can help refine the selection of individual genotypes for management programs and maximize future reintroduction success.
Conclusions
Using a combination of landscape genomic approaches, our study sheds light on the association between genetic and climate variation in a mountain ungulate, the Northern chamois, across the European Alps and the Dinaric Mts. We developed an approach that makes use of RDA to identify adaptive groups from genetic and climatic data and allow us to spatially investigate the potential response of the species to future climate changes. Our projections highlight that genetic change could be required in Northern chamois populations to be able to track climate change, especially if they cannot easily disperse, although the species may also take advantage of its behavioral plasticity to adapt to such changes. Current adaptive units are predicted to undergo a major shift in distribution, both spatially (a decrease in area and a change in location) and altitudinally, and some populations will need to adapt to new climate conditions. Considering our results, accounting for intraspecific adaptive genetic variation in management strategies appears important for future conservation efforts (Aitken and Whitlock 2013; Hoffmann et al. 2021), and our approach could be a pertinent tool for such purpose.
Supplementary information
Acknowledgements
We thank Philippe Auliac, Aurélie Barboiron, Bruno Bassano, François Biollaz, Glauco Camenisch, Marie Canut, Jérôme Cavailhès, Mathieu Garel, Veronika Grünschachner-Berger, Marie Heuret, Ludovic Imberdis, Martina Just, Christine Lettl, Laura Martinelli, Radka Poláková, Elias Pesenti, Davide Righetti, Christine Saint-Andrieux, Federico Tettamanti, Roberto Viganò, Barbora Rolečková, Barbora Zemanová, and IVB Genetic Bank (Czech Academy of Sciences) for providing help to collect samples. We acknowledge Maya Guegen, Julien Renaud, and Flurin Leugger for answering our questions on the bioclimatic variable selection. We thank Delphine Rioux, Nadine Curt Grand-Gaudin, Nathalie Tissot and Sophie Tissot for assistance with ddRADseq library preparation in the laboratory. We warmly thank the associate editor as well as three anonymous reviewers for their insightful comments on the manuscript. The research benefited from the support of AnaBM (USMB) and AEEM (UGA) laboratory facilities and we are grateful to the Roscoff Bioinformatics platform ABiMS (http://abims.sb-roscoff.fr). LP was supported by the Swiss National Science Foundation grant (N° 310030_188550), and GY received funding from the IDEX – Initiatives de Recherche Stratégiques (IRS) – Université Grenoble Alpes.
Author contributions
GY and LP coordinated the project. GY and TC designed the study and supervised AH. EB, LC, BC, HCH, and NS contributed to the sample collection. TB and GY performed the bioinformatic analysis. AH analyzed the data with TC and GY. AH, TC, and GY interpreted the results with contribution from TB, CP, and LD. AH, TC and GY wrote the manuscript with comments and editing of all authors. The final version of the manuscript has been read and approved by all authors.
Data availability
Raw reads are available under the NCBI bioproject “PRJNA813419” (Accession number for BioSamples: SAMN26501511 - SAMN26502071 and SRA: SRR18252263-SRR18252823).
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor: Paul Sunnucks.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-023-00661-2.
References
- Aitken SN, Bemmels JB. Time to get moving: assisted gene flow of forest trees. Evolut Appl. 2016;9:271–290. doi: 10.1111/eva.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken SN, Whitlock MC. Assisted Gene Flow to Facilitate Local Adaptation to Climate Change. Annu Rev Ecol Evol Syst. 2013;44:367–388. doi: 10.1146/annurev-ecolsys-110512-135747. [DOI] [Google Scholar]
- Alberto FJ, Aitken SN, Alía R, González-Martínez SC, Hänninen H, Kremer A, et al. Potential for evolutionary responses to climate change – evidence from tree populations. Glob Change Biol. 2013;19:1645–1661. doi: 10.1111/gcb.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alimperti S, Stelios TA. CDH2 and CDH11 Act as Regulators of Stem Cell Fate Decisions. Stem Cell Res. 2015;14:270–282. doi: 10.1016/j.scr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderwald P, Ambarli H, Avramov S, Ciach M, Corlatti L, Farkas A et al. (2021) Rupicapra rupicapra (amended version of 2020 assessment). The IUCN Red List of Threatened Species. Cambridge, United Kingdom
- Apollonio M, Scandura M, Šprem N, Putman R, Apollonio M. Behaviour and Management of European Ungulates. Dunbeath, Caithness: Whittles Publishing; 2014. Reintroductions as a Management tool for European Ungulates; pp. 46–77. [Google Scholar]
- Arneth A, Denton F, Agus F, Elbehri A, Erb KH, Elasha BO et al. (2019) Framing and Context. In: Shukla PR, Skea J, Calvo Buendia E, Masson-Delmotte V, Pörtner H-O, Roberts DC, Zhai P, Slade R, Connors S, van Diemen R, Ferrat M, Haughey E, Luz S, Neogi S, Pathak M, Petzold J, Portugal Pereira J, Vyas P, Huntley E, Kissick K, Belkacemi M, Malley J (eds) Climate Change and Land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. 10.1017/9781009157988.003
- Ascenzi P, Clementi ME, Condò SG, Coletta M, Petruzzelli R, Polizio F, et al. Functional, spectroscopic and structural properties of haemoglobin from chamois (Rupicapra rupicapra) and steinbock (Capra hircus ibex) Biochem J. 1993;296:361–365. doi: 10.1042/bj2960361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aublet J-F, Festa-Bianchet M, Bergero D, Bassano B. Temperature constraints on foraging behaviour of male Alpine ibex (Capra ibex) in summer. Oecologia. 2009;159:237–247. doi: 10.1007/s00442-008-1198-4. [DOI] [PubMed] [Google Scholar]
- Barbosa S, Mestre F, White TA, Paupério J, Alves PC, Searle JB. Integrative approaches to guide conservation decisions: Using genomics to define conservation units and functional corridors. Mol Ecol. 2018;27:3452–3465. doi: 10.1111/mec.14806. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Bay RA, Harrigan RJ, Underwood VL, Gibbs HL, Smith TB, Ruegg K. Genomic signals of selection predict climate-driven population declines in a migratory bird. Science. 2018;359:83–86. doi: 10.1126/science.aan4380. [DOI] [PubMed] [Google Scholar]
- Bell G. Evolutionary rescue and the limits of adaptation. Philos Trans R Soc B: Biol Sci. 2013;368:20120080. doi: 10.1098/rstb.2012.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F. Impacts of climate change on the future of biodiversity. Ecol Lett. 2012;15:365–377. doi: 10.1111/j.1461-0248.2011.01736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter MC, Kapsenberg L, Gattuso J-P, Pfister CA. Standing genetic variation fuels rapid adaptation to ocean acidification. Nat Commun. 2019;10:5821. doi: 10.1038/s41467-019-13767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet FG, Legendre P, Borcard D. Forward selection of explanatory variables. Ecology. 2008;89:2623–2632. doi: 10.1890/07-0986.1. [DOI] [PubMed] [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, Gandon S. A practical guide to measuring local adaptation. Ecol Lett. 2013;16:1195–1205. doi: 10.1111/ele.12150. [DOI] [PubMed] [Google Scholar]
- Borrell JS, Zohren J, Nichols RA, Buggs RJA. Genomic assessment of local adaptation in dwarf birch to inform assisted gene flow. Evolut Appl. 2020;13:161–175. doi: 10.1111/eva.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed MF, Harrison PA, Blyth C, Byrne M, Gaget V, Gellie NJC, et al. The potential of genomics for restoring ecosystems and biodiversity. Nat Rev Genet. 2019;20:615–628. doi: 10.1038/s41576-019-0152-0. [DOI] [PubMed] [Google Scholar]
- Browne L, Wright JW, Fitz-Gibbon S, Gugger PF, Sork VL. Adaptational lag to temperature in valley oak (Quercus lobata) can be mitigated by genome-informed assisted gene flow. Proc Natl Acad Sci USA. 2019;116:25179–25185. doi: 10.1073/pnas.1908771116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capblancq T, Fitzpatrick MC, Bay RA, Exposito-Alonso M, Keller SR. Genomic prediction of (mal)adaptation across current and future climatic landscapes. Annu Rev Ecol Evol Syst. 2020;51:245–269. doi: 10.1146/annurev-ecolsys-020720-042553. [DOI] [Google Scholar]
- Capblancq T, Forester BR. Redundancy analysis: A Swiss Army Knife for landscape genomics. Methods Ecol Evol. 2021;12:2298–2309. doi: 10.1111/2041-210X.13722. [DOI] [Google Scholar]
- Capblancq T, Luu K, Blum MGB, Bazin E. Evaluation of redundancy analysis to identify signatures of local adaptation. Mol Ecol Resour. 2018;18:1223–1233. doi: 10.1111/1755-0998.12906. [DOI] [PubMed] [Google Scholar]
- Capblancq T, Morin X, Gueguen M, Renaud J, Lobreaux S, Bazin E. Climate‐associated genetic variation in Fagus sylvatica and potential responses to climate change in the French Alps. J Evol Biol. 2020;33:783–796. doi: 10.1111/jeb.13610. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3: Genes|Genomes|Genet. 2011;1:171. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caye K, Jumentier B, Lepeule J, Fran‡ois O. LFMM 2: Fast and accurate inference of gene-environment associations in genome-wide studies. Mol Biol Evol. 2019;36:852–860. doi: 10.1093/molbev/msz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IC, Hill JK, Ohlemüller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. 2011;333:1024–1026. doi: 10.1126/science.1206432. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu Z, Régnière J, Vasseur L, Lin J, Huang S, et al. Large-scale genome-wide study reveals climate adaptive variability in a cosmopolitan pest. Nat Commun. 2021;12:7206. doi: 10.1038/s41467-021-27510-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirichella R, Stephens PA, Mason THE, Apollonio M. Contrasting effects of climate change on alpine chamois. J Wildl Manag. 2021;85:109–120. doi: 10.1002/jwmg.21962. [DOI] [Google Scholar]
- Ciach M, Pęksa Ł. Human-induced environmental changes influence habitat use by an ungulate over the long term. Curr Zool. 2019;65:129–137. doi: 10.1093/cz/zoy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlatti L, Bonardi A, Bragalanti N, Pedrotti L. Long-term dynamics of Alpine ungulates suggest interspecific competition. J Zool. 2019;309:241–249. doi: 10.1111/jzo.12716. [DOI] [Google Scholar]
- Corlatti L, Herrero J, Ferretti F, Anderwald P, García-González R, Hammer S et al. (2022) Northern chamois Rupicaprarupicapra (Linnaeus, 1758) and Southern chamois Rupicaprapyrenaica Bonaparte, 1845. In: L Corlatti, F Zachos (eds) Handbook of the Mammals of Europe – Terrestrial Cetartiodactyla, Springer Nature, Heidelberg, Germany, p 325–366
- Corlatti L, Lorenzini R, Lovari S. The conservation of the chamois Rupicapra spp.: Biology and conservation of the chamois. Mammal Rev. 2011;41:163–174. doi: 10.1111/j.1365-2907.2011.00187.x. [DOI] [Google Scholar]
- Deinet S, Ieronymidou C, McRae L, Burfield IJ, Foppen RP, Collen B et al. (2013) Wildlife comeback in Europe: The recovery of selected mammal and bird species. Final report to Rewilding Europe by ZSL, BirdLife International and the European Bird Census Council. London, UK: ZSL
- Engler R, Randin CR, Thuiller W, Dullinger S, Zimmermann NE, Araújo MB, et al. 21st century climate change threatens mountain flora unequally across Europe. Glob Change Biol. 2011;17:2330–2341. doi: 10.1111/j.1365-2486.2010.02393.x. [DOI] [Google Scholar]
- Epstein B, Jones M, Hamede R, Hendricks S, McCallum H, Murchison EP, et al. Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat Commun. 2016;7:12684. doi: 10.1038/ncomms12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fournier P, Lewthwaite JMM, Mooers AØ. Do we need to identify adaptive genetic variation when prioritizing populations for conservation? Conserv Genet. 2021;22:205–216. doi: 10.1007/s10592-020-01327-w. [DOI] [Google Scholar]
- Fick SE, Hijmans RJ. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37:4302–4315. doi: 10.1002/joc.5086. [DOI] [Google Scholar]
- Fitzpatrick MC, Chhatre VE, Soolanayakanahally RY, Keller SR. Experimental support for genomic prediction of climate maladaptation using the machine learning approach Gradient Forests. Mol Ecol Resour. 2021;21:2749–2765. doi: 10.1111/1755-0998.13374. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick MC, Keller SR. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol Lett. 2015;18:1–16. doi: 10.1111/ele.12376. [DOI] [PubMed] [Google Scholar]
- Flanagan SP, Forester BR, Latch EK, Aitken SN, Hoban S. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evolut Appl. 2018;11:1035–1052. doi: 10.1111/eva.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester BR, Lasky JR, Wagner HH, Urban DL. Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations. Mol Ecol. 2018;27:2215–2233. doi: 10.1111/mec.14584. [DOI] [PubMed] [Google Scholar]
- Frichot E, Schoville SD, Bouchard G, François O. Testing for Associations between Loci and Environmental Gradients Using Latent Factor Mixed Models. Mol Biol Evol. 2013;30:1687–1699. doi: 10.1093/molbev/mst063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot E, Schoville SD, de Villemereuil P, Gaggiotti OE, François O. Detecting adaptive evolution based on association with ecological gradients: Orientation matters! Heredity. 2015;115:22–28. doi: 10.1038/hdy.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WC, McKay JK, Hohenlohe PA, Allendorf FW. Harnessing genomics for delineating conservation units. Trends Ecol Evol. 2012;27:489–496. doi: 10.1016/j.tree.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J-M, Festa-Bianchet M, Delorme D, Jorgenson J. Body mass and individual fitness in female ungulates: bigger is not always better. Proc R Soc Lond Ser B: Biol Sci. 2000;267:471–477. doi: 10.1098/rspb.2000.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobiet A, Kotlarski S, Beniston M, Heinrich G, Rajczak J, Stoffel M. 21st century climate change in the European Alps-A review. Sci Total Environ. 2014;493:1138–1151. doi: 10.1016/j.scitotenv.2013.07.050. [DOI] [PubMed] [Google Scholar]
- González‐Serna MJ, Cordero PJ, Ortego J. Insights into the neutral and adaptive processes shaping the spatial distribution of genomic variation in the economically important Moroccan locust (Dociostaurus maroccanus) Ecol Evol. 2020;10:3991–4008. doi: 10.1002/ece3.6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisan A, Thuiller W. Predicting species distribution: offering more than simple habitat models. Ecol Lett. 2005;8:993–1009. doi: 10.1111/j.1461-0248.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- Harrisson KA, Pavlova A, Telonis-Scott M, Sunnucks P. Using genomics to characterize evolutionary potential for conservation of wild populations. Evolut Appl. 2014;7:1008–1025. doi: 10.1111/eva.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford J. A Quantitative Survey of Local Adaptation and Fitness Trade‐Offs. Am Nat. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Archer FI, Bertola LD, Bragg JG, Breed MF, Bruford MW, et al. Global genetic diversity status and trends: towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol Rev. 2022;97:1511–1538. doi: 10.1111/brv.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Bruford M, Jackson JD, Lopes-Fernandes M, Heuertz M, Hohenlohe PA, et al. Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biol Conserv. 2020;248:108654. doi: 10.1016/j.biocon.2020.108654. [DOI] [Google Scholar]
- Hock R, Rasul G, Adler C, Cáceres B, Gruber S, Hirabayashi Y et al. (2019) High Mountain Areas. In: IPCC Special Report on the Ocean and Cryosphere in a Changing Climate [Pörtner H-0, Roberts DC, Masson-Delmotte V, Zhai P, Tignor M, Poloczanska E, Mintenbeck K, Alegría A, Nicolai M, Okem A, Petzold J, Rama B, Weyer NM (eds). In press.
- Hoegh-Guldberg O, Jacob D, Taylor M, Bindi M, Brown S, Camilloni I et al. (2018) Impacts of 1.5°C Global Warming on Natural and Human Systems. In: Masson-Delmotte V, Zhai P, Pörtner H-O, Roberts D, Skea J, Shukla PR, Pirani A, Moufouma-Okia W, Péan C, Pidcock R, Connors S, Matthews JBR, Chen Y, Zhou X, Gomis MI, Lonnoy E, Maycock T, Tignor M, Waterfield T (eds) Global Warming of 1.5°C. An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty. In Press.
- Hoffmann AA, Miller AD, Weeks AR. Genetic mixing for population management: From genetic rescue to provenancing. Evol Appl. 2021;14:634–652. doi: 10.1111/eva.13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. doi: 10.1038/nature09670. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Funk WC, Rajora OP. Population genomics for wildlife conservation and management. Mol Ecol. 2021;30:62–82. doi: 10.1111/mec.15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2019) Climate Change and Land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. Shukla PR, Skea J, Calvo Buendia E, Masson-Delmotte V, Pörtner H-O, Roberts DC, Zhai P, Slade R, Connors S, van Diemen R, Ferrat M, Haughey E, Luz S, Neogi S, Pathak M, Petzold J, Portugal Pereira J, Vyas P, Huntley E, Kissick K, Belkacemi M, Malley J (eds). In press
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, Mundt F (2020) Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. https://CRAN.R-project.org/package=factoextra
- Kaufman L, Rousseeuw P (1990) Finding groups in data: an Introduction to cluster analysis. Wiley, New York
- La Sorte FA, Jetz W. Projected range contractions of montane biodiversity under global warming. Proc R Soc B-Biol Sci. 2010;277:3401–3410. doi: 10.1098/rspb.2010.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y-T, Yeung CKL, Omland KE, Pang E-L, Hao Y, Liao B-Y, et al. Standing genetic variation as the predominant source for adaptation of a songbird. Proc Natl Acad Sci. 2019;116:2152–2157. doi: 10.1073/pnas.1813597116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster LT, Fuller ZL, Berger D, Barbour MA, Jentoft S, Wellenreuther M. Understanding climate change response in the age of genomics. J Anim Ecol. 2022;91:1056–1063. doi: 10.1111/1365-2656.13711. [DOI] [PubMed] [Google Scholar]
- Leimu R, Fischer M. A Meta-Analysis of Local Adaptation in Plants. PLOS ONE. 2008;3:1–8. doi: 10.1371/journal.pone.0004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leugger F, Broquet T, Karger DN, Rioux D, Buzan E, Corlatti L, et al. Dispersal and habitat dynamics shape the genetic structure of the Northern chamois in the Alps. J Biogeogr. 2022;49:1848–1861. doi: 10.1111/jbi.14363. [DOI] [Google Scholar]
- Loison A, Darmon G, Cassar S, Jullien JM, Maillard D. Age- and sex-specific settlement patterns of chamois (Rupicapra rupicapra) offspring. Can J Zool-Rev Canadienne De Zoologie. 2008;86:588–593. doi: 10.1139/Z08-031. [DOI] [Google Scholar]
- Loison A, Jullien JM, Menaut P. Subpopulation structure and dispersal in two populations of chamois. J Mammal. 1999;80:620–632. doi: 10.2307/1383306. [DOI] [Google Scholar]
- Lovari S, Franceschi S, Chiatante G, Fattorini L, Fattorini N, Ferretti F. Climatic changes and the fate of mountain herbivores. Clim Change. 2020;162:2319–2337. doi: 10.1007/s10584-020-02801-7. [DOI] [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, et al. Responsible RAD: Striving for best practices in population genomic studies of adaptation. Mol Ecol Resour. 2017;17:366–369. doi: 10.1111/1755-0998.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, et al. Breaking RAD: an evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Mol Ecol Resour. 2017;17:142–152. doi: 10.1111/1755-0998.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand P, Garel M, Bourgoin G, Dubray D, Maillard D, Loison A. Sex-specific adjustments in habitat selection contribute to buffer mouflon against summer conditions. Behav Ecol. 2015;26:472–482. doi: 10.1093/beheco/aru212. [DOI] [Google Scholar]
- Martchenko D, Chikhi R, Shafer ABA. Genome Assembly and Analysis of the North American Mountain Goat (Oreamnos americanus) Reveals Species-Level Responses to Extreme Environments. G3 Genes|Genomes|Genet. 2020;10:437–442. doi: 10.1534/g3.119.400747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason THE, Apollonio M, Chirichella R, Willis SG, Stephens PA. Environmental change and long-term body mass declines in an alpine mammal. Front Zool. 2014;11:69. doi: 10.1186/s12983-014-0069-6. [DOI] [Google Scholar]
- Mason T, Stephens PA, Apollonio M, Willis SG. Predicting potential responses to future climate in an alpine ungulate: interspecific interactions exceed climate effects. Glob Change Biol. 2014;20:3872–3882. doi: 10.1111/gcb.12641. [DOI] [PubMed] [Google Scholar]
- Montwé D, Isaac-Renton M, Hamann A, Spiecker H. Cold adaptation recorded in tree rings highlights risks associated with climate change and assisted migration. Nat Commun. 2018;9:1574. doi: 10.1038/s41467-018-04039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesti I, Posillico M, Lovari S. Ranging behaviour and habitat selection of Alpine chamois. Ethol Ecol Evol. 2010;22:215–231. doi: 10.1080/03949370.2010.502316. [DOI] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2019) vegan: Community Ecology Package. Software http://CRAN.R-project.org/package=vegan
- O’Neill GA, Aitken SN. Area-based breeding zones to minimize maladaptation. Can J Res. 2004;34:695–704. doi: 10.1139/x03-227. [DOI] [Google Scholar]
- Orr HA, Betancourt AJ. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørsted M, Hoffmann AA, Sverrisdóttir E, Nielsen KL, Kristensen TN. Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLOS Genet. 2019;15:1–18. doi: 10.1371/journal.pgen.1008205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou H, Sgardelis S, Chondropoulos B, Vassilakis D, Kati V, Dimopoulos P. Demographic characteristics, seasonal range and habitat topography of Balkan chamois population in its southernmost limit of its distribution (Giona mountain, Greece) J Nat Hist. 2015;49:327–345. doi: 10.1080/00222933.2013.869365. [DOI] [Google Scholar]
- Park A, Talbot C. Assisted migration: uncertainty, risk and opportunity. For Chron. 2012;88:412–419. doi: 10.5558/tfc2012-077. [DOI] [Google Scholar]
- Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst. 2006;37:637–669. doi: 10.1146/annurev.ecolsys.37.091305.110100. [DOI] [Google Scholar]
- Pauli H, Gottfried M, Grabherr G. Effects of climate change on mountain ecosystems - Upward shifting of alpine plants. World Resour Rev. 1996;8:382–390. [Google Scholar]
- Pecl GT, Araújo MB, Bell JD, Blanchard J, Bonebrake TC, Chen I-C, et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science. 2017;355:eaai9214. doi: 10.1126/science.aai9214. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. Plos One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pörtner H-O, Roberts DC, Masson-Delmotte V, Zhai P, Tignor M, Poloczanska E et al. (2019) IPCC, 2019: Summary for Policymakers. IPCC Special Report on the Ocean and Cryosphere in a Changing Climate. Cambridge University Press, Cambridge, UK and New York, NY, USA, p 3–35
- Razgour O, Forester B, Taggart JB, Bekaert M, Juste J, Ibáñez C, et al. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc Natl Acad Sci USA. 2019;116:10418–10423. doi: 10.1073/pnas.1820663116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt GE, Tchebakova NM, Parfenova YI, Wykoff WR, Kuzmina NA, Milyutin LI. Intraspecific responses to climate in Pinus sylvestris: Responses to climate in Pinus Sylvestris. Glob Change Biol. 2002;8:912–929. doi: 10.1046/j.1365-2486.2002.00516.x. [DOI] [Google Scholar]
- Reiner R, Zedrosser A, Zeiler H, Hackländer K, Corlatti L (2021) Forests buffer the climate‐induced decline of body mass in a mountain herbivore. Glob Change Biol. 10.1111/gcb.15711 [DOI] [PMC free article] [PubMed]
- Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR, Graf R, et al. Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Mol Ecol. 2016;25:5907–5924. doi: 10.1111/mec.13889. [DOI] [PubMed] [Google Scholar]
- Riahi K, van Vuuren DP, Kriegler E, Edmonds J, O’Neill BC, Fujimori S, et al. The Shared Socioeconomic Pathways and their energy, land use, and greenhouse gas emissions implications: An overview. Glob Environ Change. 2017;42:153–168. doi: 10.1016/j.gloenvcha.2016.05.009. [DOI] [Google Scholar]
- Rochat E, Joost S (2019) Spatial Areas of Genotype Probability (SPAG): predicting the spatial distribution of adaptive genetic variants under future climatic conditions. bioRxiv: 2019.12.20.884114
- Rochat E, Selmoni O, Joost S. Spatial areas of genotype probability: Predicting the spatial distribution of adaptive genetic variants under future climatic conditions. Divers Distrib. 2021;27:1076–1090. doi: 10.1111/ddi.13256. [DOI] [Google Scholar]
- Roffler GH, Amish SJ, Smith S, Cosart T, Kardos M, Schwartz MK, et al. SNP discovery in candidate adaptive genes using exon capture in a free‐ranging alpine ungulate. Mol Ecol Resour. 2016;16:1147–1164. doi: 10.1111/1755-0998.12560. [DOI] [PubMed] [Google Scholar]
- Roques J (2021) Disentangling the effects of landscape on the functional connectivity of the Northern chamois as assessed by a population genomics approach. MSc Thesis. Université Savoie Mont Blanc, Chambéry, France
- Rossi L, Tizzani P, Rambozzi L, Moroni B, Meneguz PG. Sanitary Emergencies at the Wild/Domestic Caprines Interface in Europe. Animals. 2019;9:922. doi: 10.3390/ani9110922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegg K, Bay RA, Anderson EC, Saracco JF, Harrigan RJ, Whitfield M, et al. Ecological genomics predicts climate vulnerability in an endangered southwestern songbird. Ecol Lett. 2018;21:1085–1096. doi: 10.1111/ele.12977. [DOI] [PubMed] [Google Scholar]
- Rughetti M, Festa-Bianchet M. Effects of spring-summer temperature on body mass of chamois. J Mammal. 2012;93:1301–1307. doi: 10.1644/11-MAMM-A-402.1. [DOI] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nat Rev Genet. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- Seaborn T, Griffith D, Kliskey A, Caudill CC. Building a bridge between adaptive capacity and adaptive potential to understand responses to environmental change. Glob Change Biol. 2021;27:2656–2668. doi: 10.1111/gcb.15579. [DOI] [PubMed] [Google Scholar]
- Segelbacher G, Bosse M, Burger P, Galbusera P, Godoy JA, Helsen P, et al. New developments in the field of genomic technologies and their relevance to conservation management. Conserv Genet. 2022;23:217–242. doi: 10.1007/s10592-021-01415-5. [DOI] [Google Scholar]
- Seigle-Ferrand J, Marchand P, Morellet N, Gaillard J-M, Hewison AJM, Saïd S, et al. On this side of the fence: Functional responses to linear landscape features shape the home range of large herbivores. J Anim Ecol. 2022;91:443–457. doi: 10.1111/1365-2656.13633. [DOI] [PubMed] [Google Scholar]
- Semenzato P, Cagnacci F, Ossi F, Eccel E, Morellet N, Hewison AJM, et al. Behavioural heat-stress compensation in a cold-adapted ungulate: Forage-mediated responses to warming Alpine summers. Ecol Lett. 2021;24:1556–1568. doi: 10.1111/ele.13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soorae PS (ed.) (2010) Global re-introduction perspectives: Additional case-studies from around the globe. IUCN/ SSC Re-introduction Specialist Group, Abu Dhabi, UAE, p xii + 352
- Steane DA, Potts BM, McLean E, Prober SM, Stock WD, Vaillancourt RE, et al. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol Ecol. 2014;23:2500–2513. doi: 10.1111/mec.12751. [DOI] [PubMed] [Google Scholar]
- Storey JD, Bass AJ, Dabney A, Robinson D (2023) qvalue: Q-value estimation for false discovery rate control. R package version 2.34.0, 10.18129/B9.bioc.qvalue
- Thuiller W, Gueguen M, Bison M, Duparc A, Garel M, Loison A, et al. Combining point-process and landscape vegetation models to predict large herbivore distributions in space and time-A case study of Rupicapra rupicapra. Divers Distrib. 2018;24:352–362. doi: 10.1111/ddi.12684. [DOI] [Google Scholar]
- Thuiller W, Guéguen M, Georges D, Bonet R, Chalmandrier L, Garraud L, et al. Are different facets of plant diversity well protected against climate and land cover changes? A test study in the French Alps. Ecography. 2014;37:1254–1266. doi: 10.1111/ecog.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beest FM, Milner JM (2013) Behavioural responses to thermal conditions affect seasonal mass change in a heat-sensitive northern ungulate. PLOS ONE 8:1–10 [DOI] [PMC free article] [PubMed]
- Visser ME. Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc R Soc B. 2008;275:649–659. doi: 10.1098/rspb.2007.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldvogel A-M, Feldmeyer B, Rolshausen G, Exposito-Alonso M, Rellstab C, Kofler R, et al. Evolutionary genomics can improve prediction of species’ responses to climate change. Evol Lett. 2020;4:4–18. doi: 10.1002/evl3.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KS, Gregovich DP, Levi T. Projecting the future of an alpine ungulate under climate change scenarios. Glob Change Biol. 2018;24:1136–1149. doi: 10.1111/gcb.13919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads are available under the NCBI bioproject “PRJNA813419” (Accession number for BioSamples: SAMN26501511 - SAMN26502071 and SRA: SRR18252263-SRR18252823).