Abstract
We have recently shown that many mutants of Legionella pneumophila exhibit similar defective phenotypes within both U937 human-derived macrophages and the protozoan host Acanthamoeba (L.-Y. Gao, O. S. Harb, and Y. Abu Kwaik, Infect. Immun. 65:4738–4746, 1997). These observations have suggested that many of the mechanisms utilized by L. pneumophila to parasitize mammalian and protozoan cells are similar, but our data have not excluded the possibility that there are unique mechanisms utilized by L. pneumophila to survive and replicate within macrophages but not protozoa. To examine this possibility, we screened a bank of 5,280 miniTn10::kan transposon insertion mutants of L. pneumophila for potential mutants that exhibited defective phenotypes of cytopathogenicity and intracellular replication within macrophage-like U937 cells but not within Acanthamoeba polyphaga. We identified 32 mutants with various degrees of defects in cytopathogenicity, intracellular survival, and replication within human macrophages, and most of the mutants exhibited wild-type phenotypes within protozoa. Six of the mutants exhibited mild defects in protozoa. The defective loci were designated mil (for macrophage-specific infectivity loci). Based on their intracellular growth defects within macrophages, the mil mutants were grouped into five phenotypic groups. Groups I to III included the mutants that were severely defective in macrophages, while members of the other two groups exhibited a modestly defective phenotype within macrophages. The growth kinetics of many mutants belonging to groups I to III were also examined, and these were shown to have a similar defective phenotype in peripheral blood monocytes and a wild-type phenotype within another protozoan host, Hartmannella vermiformis. Transmission electron microscopy of A. polyphaga infected by three of the mil mutants belonging to groups I and II showed that they were similar to the parent strain in their capacity to recruit the rough endoplasmic reticulum (RER) around the phagosome. In contrast, infection of macrophages showed that the three mutants failed to recruit the RER around the phagosome during early stages of the infection. None of the mil mutants was resistant to NaCl, and the dot or icm NaClr mutants are severely defective within mammalian and protozoan cells. Our data indicated that in addition to differences in mechanisms of uptake of L. pneumophila by macrophages and protozoa, there were also genetic loci required for L. pneumophila to parasitize mammalian but not protozoan cells. We hypothesize that L. pneumophila has evolved as a protozoan parasite in the environment but has acquired loci specific for intracellular replication within macrophages. Alternatively, ecological coevolution with protozoa has allowed L. pneumophila to possess multiple redundant mechanisms to parasitize protozoa and that some of these mechanisms do not function within macrophages.
The Legionnaires’ disease bacterium, Legionella pneumophila, is one of the common etiologic agents of pneumonia with mortality rates of up to 50% (10, 19, 31, 46). Upon transmission to humans, the bacteria invade and reside within alveolar macrophages and epithelial cells (20). The bacterium replicates within a rough endoplasmic reticulum (RER)-surrounded phagosome that is inhibited from maturation through the endosomal-lysosomal degradation pathway (2, 26, 27, 41). Within this unique intracellular niche, the bacteria undergo phenotypic modulation (3, 4, 6, 40).
Outbreaks of Legionnaires’ disease occur through droplet transmission of the bacterium from an environmental water source, predominantly air conditioning cooling towers (20). In the environment, L. pneumophila is a parasite of at least 13 species of amoebae and ciliated protozoa (20). Several investigators provided evidence that protozoa play a major factor in transmission of Legionnaires’ disease (6, 8, 9, 13, 18, 20, 38).
Uptake of L. pneumophila by macrophages and protozoa occurs by different mechanisms (5, 29). We have recently shown that attachment and uptake of L. pneumophila by the protozoan Hartmannella vermiformis are mediated by a 170-kDa Gal/GalNAc lectin receptor on the surface of protozoa (25, 44). Uptake of L. pneumophila by human macrophages has been shown to be mediated by the complement receptor, but non-complement-mediated uptake also occurs (24, 28, 32). Whether a Gal/GalNAc lectin homolog is present in mammalian macrophages is still to be determined. We have recently shown expression of multiple pili by L. pneumophila and that one of these pili is involved in attachment to mammalian and protozoan cells (39).
In contrast to differences in uptake by the two different host cells, ultrastructural characterization of the intracellular infection of protozoa by L. pneumophila has shown that it appears to be similar to that of mammalian cells (1). Within protozoa, the bacterium also multiplies within a RER-surrounded phagosome that does not fuse with lysosomes (1, 12, 23). We have recently shown that many mutants of L. pneumophila exhibit similar defective phenotypes within both human-derived macrophages and protozoa (7, 23). These observations have suggested that L. pneumophila utilizes the same mechanisms to parasitize both mammalian and protozoan cells (1, 16, 20). Moreover, it is thought that L. pneumophila has evolved as a parasite of protozoa in the natural environment and that generation of aerosols by human-made devices is the only factor that has allowed L. pneumophila to have access to humans and cause Legionnaires’ disease (1, 16, 20).
In this study, we screened a bank of transposon insertion mutants of L. pneumophila for potential mutants that exhibited defective phenotypes within macrophages but not within protozoa. We have identified 32 mutants with various degrees of defects in intracellular survival and replication within human macrophages but with a wild-type phenotype within protozoa. The data showed that some genetic loci are utilized by L. pneumophila to parasitize mammalian macrophages but not protozoa. We speculate that L. pneumophila acquired loci specific for intracellular replication within macrophages or that ecological coevolution with protozoa allowed L. pneumophila to possess multiple redundant mechanisms to parasitize protozoa and that some of these mechanisms do not function within macrophages.
MATERIALS AND METHODS
Bacterial strains and vectors.
The virulent AA100 strain of L. pneumophila has been described previously (3). AA100A is an attenuated NaClr derivative of AA100 that has been described previously (5). L. pneumophila strains were grown on buffered charcoal-yeast extract (BCYE) agar plates or in buffered yeast extract (BYE) broth. Escherichia coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was used for the majority of cloning experiments. The plasmid pUC-4K was purchased from Pharmacia (Piscataway, N.J.) and was the source of the kanamycin resistance gene used as a probe for Southern hybridization. The plasmid pCDP05 is a chloramphenicol-resistant plasmid that contains miniTn10::kan and the sacB gene from Bacillus subtilis and was a kind gift from N. Cianciotto (Northwestern University, Chicago, Ill.) (33). The sacB gene encodes levansucrase and is lethal to L. pneumophila grown in the presence of sucrose (17). The plasmid pAM10 contains a 12-kb EcoRI insert encompassing both the dot and icm loci and was kindly provided by H. A. Shuman (Columbia University, New York, N.Y.) (30).
Transposon mutagenesis and construction of a bank of mutants.
The use of miniTn10::kan in L. pneumophila has been described previously (33). Briefly, after electroporation of the plasmid pCDP05, containing miniTn10::kan (33), into the L. pneumophila wild-type strain AA100, bacteria were incubated in BYE broth for 2 h at 37°C and plated onto BCYE agar plates containing 20 μg of kanamycin per ml and 6% sucrose. For each electroporation, 200 to 300 colonies were isolated for measurements of their infectivity (23).
DNA manipulations.
Chromosomal DNA preparations, transfections, and restriction enzyme digestions were performed as described elsewhere, unless otherwise specified (37). Restriction enzymes were obtained from Bethesda Research Laboratories.
Plasmid DNA preparations were performed with the Qiagen (Chatsworth, Calif.) plasmid kit according to the manufacturer’s recommendations. Transformations were carried out by electroporation with a Bio-Rad (Hercules, Calif.) Gene Pulser as recommended by the manufacturer. Purification of DNA fragments from agarose gels for Southern hybridization was done with a QIAEX kit according to manufacturer’s recommendations (Qiagen). Transfer of DNA from agarose gels onto membranes, fluorescein labeling of DNA probes, hybridizations, and detection were performed as described before (4).
Cloning and sequencing of the chromosomal junctions of Kan insertion in the mutants.
Genomic DNA from the L. pneumophila mutants was digested with EcoRI and ligated to EcoRI-digested pBC (Stratagene Inc., La Jolla, Calif.). After ligation, the samples were concentrated by ethanol precipitation, resuspended in 2 μl of sterile water, electroporated into DH5α, and plated in the presence of kanamycin and chloramphenicol. Plasmid DNA was extracted, digested with EcoRI, and probed with the Kan cassette in Southern blots, along with EcoRI-digested genomic DNA from the mutants, to confirm the fidelity of the cloning.
For sequencing the regions flanking the Kan cassette, the flanking sequences were amplified by PCR. Two primers homologous to a region immediately upstream (5′-TGATTTTGATGACGAGCG-3′) or downstream (5′-GTGACGACTGAATCCGGT-3′) of an internal HindIII site within the Kan cassette were designed and designated UHIND and DHIND, respectively. The T7 and T3 primers of pBC were also used in the PCR. Since the orientation of the Kan cassette in these clones is not known, UHIND and DHIND were used in conjunction with T7 and T3 primers. This strategy allowed amplification of either side of the Kan cassette and the flanking chromosomal sequences, and thus the cloned fragment will contain one IS10 inverted repeat to allow sequencing. The PCR included an annealing step at 55°C for 1 min and an extension step at 72°C for 2 or 5 min. PCR products were sequenced by using a primer designated Tn10.2 (5′-TCATTAGGGGATTCATCA-3′), which is homologous to the inverted repeat of the IS10 elements of miniTn10::kan. The sequence of approximately 350 to 450 nucleotides at the junction of insertion was obtained and used in database searches.
Human U937 macrophages and peripheral blood monocytes.
Macrophage-like U937 cells were maintained as previously described (23). Prior to infection, the cells were differentiated in 96-well tissue culture plates for 48 h by using phorbol 12-myristate 13-acetate as described previously (3). For infection of monolayers, L. pneumophila grown for 20 h at 37°C in BYE broth was resuspended in RPMI 1640. The infection was carried out as described below for each experiment.
Preparation of peripheral blood monocytes was performed as described by Horwitz (27). Briefly, human mononuclear cells were isolated from heparinized blood of healthy donors with no history of Legionnaires’ disease by using Ficoll-Hypaque density gradients. Mononuclear cells were plated in 96-well tissue culture plates at 1.2 × 105 cells/well in serum-free RPMI to allow monocytes to adhere to the plastic dishes for 90 min at 37°C in 5% CO2. The cultures were washed three times with RPMI to remove nonadherent lymphocytes, and infections were performed in RPMI containing 15% heat-inactivated fetal bovine serum exactly as described above for the U937 cells.
Protozoan cultures.
Axenic Acanthamoeba polyphaga has been described as a useful model to study protozoan infections by L. pneumophila (23). Infections were performed in 96-well tissue culture plates with Acanthamoeba buffer (23) as described below for each experiment. H. vermiformis CDC-19 (ATCC 50237) has been cloned and grown in axenic culture as a model for study of the pathogenesis of L. pneumophila (1, 21). H. vermiformis was maintained in American Type Culture Collection culture medium 1034 (21).
Transmission electron microscopy.
A. polyphaga was infected with L. pneumophila at a multiplicity of infection (MOI) of 100 for 1 h, followed by washing of extracellular bacteria. Preparation of ultrathin sections was performed as described previously (23). Briefly, infected amoebae were fixed with 3.5% gluteraldehyde followed by 1% OsO4, dehydrated with ethanol, and embedded in Eponate 12 resin (Ted Pella, Redding, Calif.). Ultrathin sections were stained with uranyl acetate followed by lead citrate and examined with an H-7000/STEM electron microscope (Hitachi Inc., Tokyo, Japan) at 75 kV.
Cytopathogenicity of L. pneumophila mutants to U937 macrophages and A. polyphaga.
L. pneumophila strains were grown in BYE broth for 20 h in 48-well plates. Infection of phorbol 12-myristate 13-acetate-differentiated U937 monolayers with L. pneumophila strains was performed, in triplicate, in 96-well plates containing 105 cells/well at an MOI of 5. The infected monolayers were incubated at 37°C for 38 h. For measurements of the number of remaining viable cells in the monolayer, the monolayers were treated with 10% Alamar Blue dye (Alamar Bioscience Inc., Sacramento, Calif.), and viability of the monolayers was determined exactly as described previously (23).
Infection of A. polyphaga with L. pneumophila strains was performed, in triplicate, in 96-well tissue culture plates with 5 × 104 cells/well at an MOI of 5. After 1 h of coincubation, monolayers were washed three times with Acanthamoeba buffer (23) and incubated at 37°C for 30 h. The viability of A. polyphaga was determined by trypan blue dye exclusion as described previously (23).
Attachment of L. pneumophila mutants to U937 cells and A. polyphaga.
To examine for defects in attachment, we performed infections of U937 cell monolayers treated with cytochalasin D (1 μg/ml) and A. polyphaga treated with 20 mM methylamine to prevent uptake of the attached bacteria by both host cells, as we described previously (23). The infected monolayers (MOI = 10) were spun at 900 × g for 5 min followed by incubation for 30 min at 37°C. At the end of this infection period, monolayers were washed three times with tissue culture medium to remove nonadherent bacteria and subsequently lysed hypotonically for U937 cells or with a mild detergent (0.04% Triton X-100) for A. polyphaga. Aliquots were diluted immediately and plated on BCYE agar plates for enumeration of bacteria. To ensure that the inhibitors were effective in inhibition of uptake, control monolayers infected in presence of the inhibitor were treated with 50 μg of gentamicin per ml to kill extracellular bacteria (23). With equivalent numbers of bacteria for the mutants and the wild-type strain, the ability of the mutants to attach to U937 macrophages or to A. polyphaga was expressed as a percentage of that of the wild-type strain. It is important to note that none of the mutants were sensitive to components of the tissue culture media (data not shown).
Growth kinetics of L. pneumophila mutants in U937 macrophages, peripheral blood monocytes, A. polyphaga, and H. vermiformis.
To determine the number of intracellular bacteria within U937 macrophages, peripheral blood monocytes, and A. polyphaga at several time intervals, infections were performed exactly as described above for the attachment assays. The initial time point (0 h) represents the time at the end of the 30-min infection period after washing off of the unattached bacteria and thus represents cell-associated bacteria (attached and intracellular). Subsequently, monolayers were incubated for 1 h at 37°C in the presence of 50 μg of gentamicin per ml to kill extracellular bacteria and were further incubated for several time intervals in the absence of gentamicin. Thus, the rest of the time points represent the total number of bacteria (intracellular and released bacteria, presumably after killing of the host cell) at the corresponding time postinfection. At the end of each time interval, the supernatants were removed and monolayers were lysed as described above. The supernatants prior to lysis of the monolayers (which may contain released bacteria) were combined with the ones from the lysed monolayers, and aliquots were plated on BCYE agar plates for enumeration of intracellular bacteria.
With the exception of the mutant defective in attachment, in which case fewer bacteria enter the host cell, the decrease in the bacterial number between the first and the second time points (see Fig. 2) is due to entry by a portion of the inoculated bacteria, early partial killing within the host cell, or both.
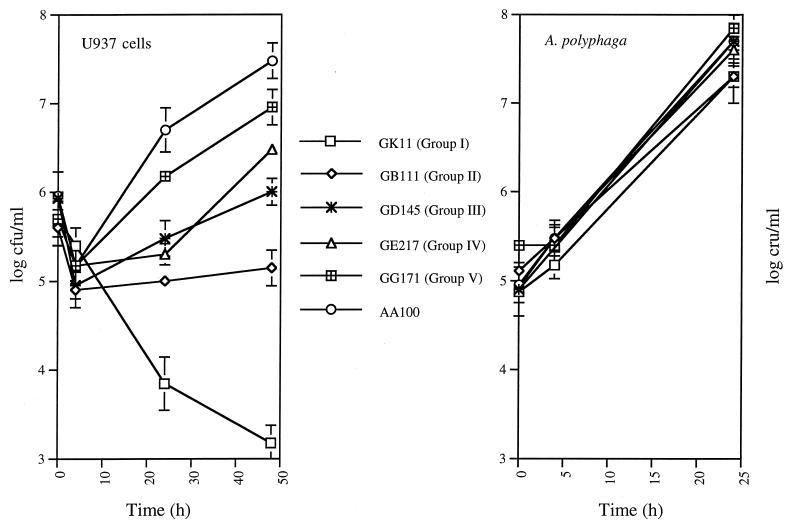
FIG. 2.
Growth kinetics of representative mil mutants belonging to each of the five phenotypic mutant groups within U937 macrophages and A. polyphaga. The initial time points (0 h) represent cell-associated bacteria (attached and intracellular) after 30 min of infection. Subsequent time points were after gentamicin treatment and represent gentamicin-protected total bacterial numbers (intracellular and released bacteria, presumably after killing of the host cell) at the corresponding times postinfection. Determinations at 48 and 72 h postinfection of A. polyphaga and at 72 h postinfection of U937 cells did not show any increase in the number of bacteria (data not shown). The data are representative of three to five independent experiments, performed in triplicate, and the error bars represent standard deviations.
To determine intracellular growth kinetics of L. pneumophila strains within the protozoan H. vermiformis, 105 amoebae/ml were infected with 103 CFU of L. pneumophila per ml in 5-ml cultures in triplicate, as described previously (7).
Other in vitro assays.
Assays of colony immunoblots, serum sensitivity, defects in iron acquisition and assimilation, and resistance of the mutants to NaCl were performed exactly as described previously (23).
RESULTS
Isolation of L. pneumophila mutants defective within human-derived macrophages but not within protozoa.
The U937 macrophages have been widely used as a reliable model to study intracellular survival and replication of L. pneumophila within macrophages (20). The ability of L. pneumophila mutants to replicate within these cells correlates with their ability to cause disease in animal models (14, 15). We and others have established the use of Acanthamoeba as a reliable protozoan model to study intracellular survival and replication of L. pneumophila within protozoa (12, 23).
We screened a bank of miniTn10::kan insertion mutants of L. pneumophila for mutants that exhibited defective phenotypes of cytopathogenicity and intracellular replication within human macrophages but had a wild-type phenotype within the protozoan A. polyphaga (see Materials and Methods). A total of 5,280 insertion mutants were isolated from 21 independent transpositions (23). Intracellular multiplication of L. pneumophila within the host cell results in eventual cytopathogenicity and killing of the host cell. To identify mutants, our method was to isolate mutants that exhibited phenotypes of cytopathogenicity to U937 macrophages different from that for A. polyphaga. In the original screening within macrophages, we isolated numerous mutants with the corresponding phenotypes. Upon subsequent testing, the phenotypes of many of the mutants were not reproducible, particularly the partially and mildly defective ones. Thirty-two mutants were isolated based on their reproducible phenotypes (three to five independent experiments) of being completely or severely defective in cytopathogenicity to macrophages but not to A. polyphaga (Fig. 1). However, approximately 6 of the 32 mutants were mildly defective in their cytopathogenicity to A. polyphaga.
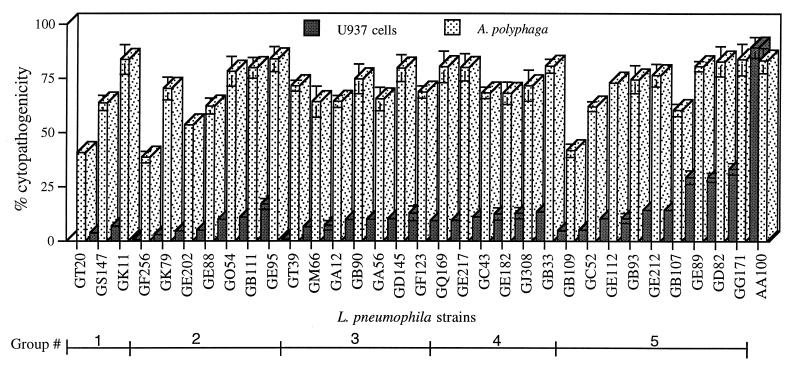
FIG. 1.
Cytopathogenicities of the L. pneumophila wild-type strain AA100 and the isogenic insertion mutants to U937 macrophage-like cells 38 h postinfection and to A. polyphaga 30 h postinfection. The data are representative of three to five independent experiments, performed in triplicate, and the error bars represent standard deviations. The five phenotypic groups based on intracellular growth kinetics (shown in Fig. 2) are indicated.
Southern blot analysis of EcoRI-digested genomic DNA of each of the 32 mutants probed with the Kan cassette showed that all of them contained a single copy of the miniTn10::kan insertion (data not shown). Importantly, insertions were random and distinct, which was consistent with previous observations (23, 33).
Characteristics of intracellular survival and replication of the mutants within U937 macrophages and A. polyphaga.
We examined the intracellular survival and growth kinetics of the 32 cytopathogenicity-defective mutants within the U937 macrophages and A. polyphaga (see Materials and Methods). The mutants showed a diverse spectrum of defects in their intracellular survival and replication within macrophages but were similar to the wild-type strain in their intracellular survival and replication within A. polyphaga (Fig. 2). At least six of the mutants showed mild defects in protozoa (see below). The mutant GG171 had a prolonged lag phase in the amoebae (Fig. 2). Moreover, when the bacterial cultures in BYE broth were inoculated at a very low density at time zero, there was no detectable difference in growth of the mutants compared to that of the wild-type strain after 20 h of growth, as measured by the optical density at 550 nm (data not shown). However, we cannot exclude the possibility that there may be some mild differences in the growth rate of the mutants.
To simplify representation of these data, we grouped the 32 mutants into five phenotypic groups based on their defective intracellular survival and growth kinetics within macrophages (Fig. 1 and 2). It is important to note that there were variations among mutants within each group in the severity of the defect, indicative of their distinct defective phenotypes contributed by defects in different genetic loci. However, the general trend of the defect in intracellular growth kinetics was similar within each group. In general, the relative degree of defect in cytopathogenicity correlated with the capacity of the mutants to survive and to replicate intracellularly (Fig. 1 and 2). Since all of these mutants were defective in human-derived macrophages but exhibited wild-type or mildly defective phenotypes within protozoa, we designated the defective loci in these mutants mil (for macrophage-specific infectivity loci). The characteristics of the five mil groups are described below.
Group I included 3 mutants (GT20, GS147, and GK11) that were completely or substantially killed within macrophages but exhibited wild-type phenotypes in replication within A. polyphaga (Fig. 2). All three of these mutants were also completely defective in their cytopathogenicity to macrophages (Fig. 1). The reduced cytopathogenicity and number of intracellular bacteria of the GT20 mutant within A. polyphaga are probably due to reduced attachment and subsequent entry (see below).
Group II consisted of seven mutants that were completely defective in intracellular replication within macrophages (Fig. 2). The intracellular growth kinetics of this group of mutants showed that they were not killed and were sustained at a constant number within macrophages over a period of 72 h (Fig. 2 and data not shown).
Group III had seven mutants that exhibited a very limited replication, which did not exceed the number of the original inoculum after 48 and 72 h of infection (Fig. 2 and data not shown). Two of these mutants (GA56 and GB90) exhibited no increase in their intracellular number during the first 24 h of infection (data not shown). Subsequent intracellular growth was very limited, reaching a level that was approximately 100-fold less than that of the wild-type strain. One of these mutants (GM66) showed a slightly slower replication within A. polyphaga as evidenced by approximately sixfold-fewer bacteria recovered at 72 h postinfection compared to the wild-type strain or the other mutants (data not shown).
Intracellular growth of mutants in group IV (six mutants) was undetectable within macrophages during the first 24 h of infection. Subsequent intracellular growth was approximately 10-fold less than that of the wild type (Fig. 2).
Group V had nine members with the least defective phenotype within macrophages. These mutants were able to initiate intracellular multiplication within macrophages like the wild-type strain but at a slightly lower rate (Fig. 2).
Characteristics of attachment of the mutants to U937 macrophages and A. polyphaga.
Since there were differences in the number of bacteria derived at the first time point (attached and intracellular) for some mutants, it was essential to determine whether this difference is due to defect in attachment. To eliminate the role of intracellular killing in our assays, attachment of the mutants to macrophages and to A. polyphaga in the presence of an inhibitor of uptake was evaluated (see Materials and Methods). At an MOI of 10, two bacteria per U937 macrophage and approximately one bacterium per A. polyphaga cell attached. The data showed that GT20 was the only mutant that exhibited a dramatic defect in attachment. Attachment to macrophages was reduced to 2% of that of the parent strain AA100, and attachment to A. polyphaga was reduced to 6% of that of AA100. All of the rest of the mil mutants exhibited attachments to both host cells that were indistinguishable from those of AA100 (data not shown).
Phenotypes of the mutants in peripheral blood monocytes and in the protozoan H. vermiformis.
The U937 macrophages and A. polyphaga have been widely used as reliable models to study intracellular survival and replication of L. pneumophila within macrophages and protozoa (2, 20, 23). However, to ensure that the defective phenotype of the mil mutants within the U937 macrophages was not specific to this macrophage cell line, we examined the phenotypes of 12 mutants (all mutants belonging to groups I and II and mutants GM66 and GA56 belonging to group III) in peripheral blood monocytes and in H. vermiformis. The defective phenotypes in growth kinetics of the mutants within the U937 macrophages were highly similar to their defective phenotypes within the peripheral blood monocytes (data not shown). Similar to their wild-type phenotypes within A. polyphaga, the mutants also exhibited wild-type phenotypes in their growth kinetics within H. vermiformis (data not shown). These data showed that the defective phenotypes of the mutants within macrophages and their wild-type phenotype within A. polyphaga were reproducible in different human macrophages and protozoa, respectively.
Transmission electron microscopy of intracellular infection by mil mutants.
L. pneumophila replicates intracellularly within both mammalian macrophages and protozoa in a phagosome that is surrounded by the RER of the host cell (1, 26, 41). These organelles are recruited around the phagosome within 3 to 4 h of the intracellular infection of mammalian and protozoan cells (1, 2, 26). Mutants that are defective in recruitment of the RER are also defective in intracellular replication (11, 30). Therefore, we used transmission electron microscopy to examine three of the most defective mil mutants (GT20 and GK11, belonging to group I, and GE88, belonging to group II) for their ability to recruit the RER around the phagosome within macrophages and A. polyphaga, as well as for their intracellular replication. There was no detectable difference from the wild-type strain in the ability of the GK11 mutant to recruit the RER around the phagosome by 4 h postinfection of A. polyphaga (Fig. 3A and B).
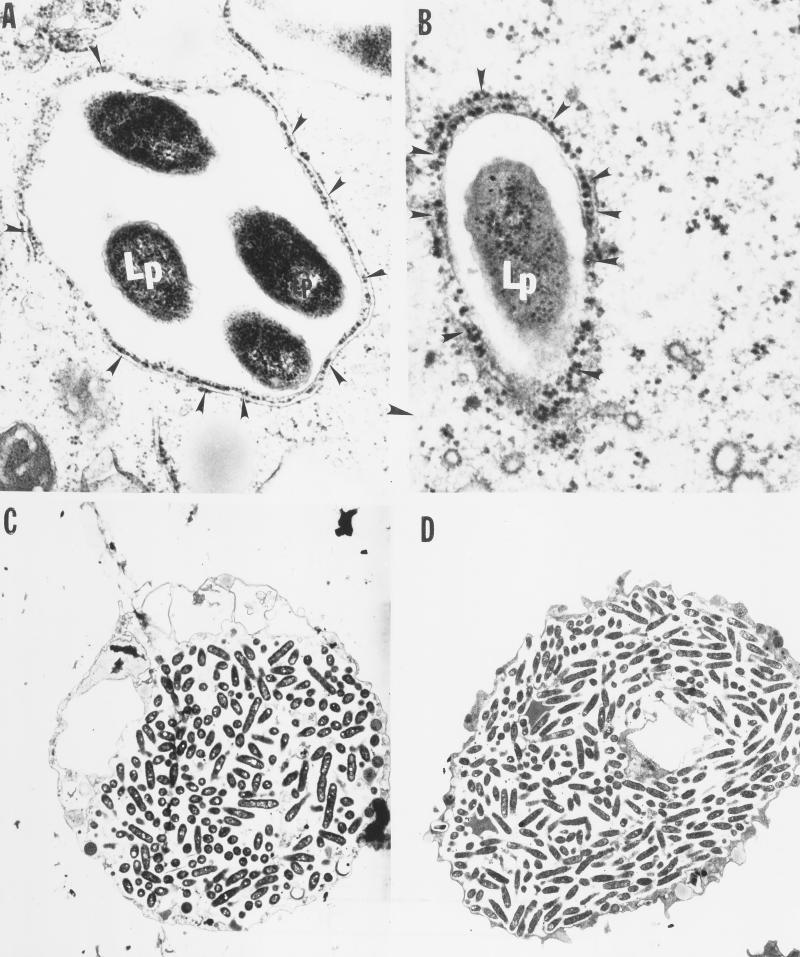
FIG. 3.
Transmission electron micrographs of A. polyphaga infected by the wild-type L. pneumophila strain AA100 (A and C) and the mil mutant GK11 (B and D) at 4 h (A and B) and 18 h (C and D) after infection. The arrowheads indicate the RER-surrounded phagosome. Lp, L. pneumophila. Similar results were obtained with the mil mutants GE88 and GT20 (data not shown). Magnifications: ×26,000 (A), ×34,000 (B), and ×3,000 (C and D).
The RER was recruited around 61 of 74 phagosomes examined (75%) containing the wild-type strain at 4 h postinfection of macrophages. In contrast, there was no detectable RER around any of the 78 examined phagosomes containing the GK11 mutant within U937 macrophages at 4 h postinfection (Fig. 4). Similar results were also obtained with the other two mutants, GT20 and GE88 (data not shown).
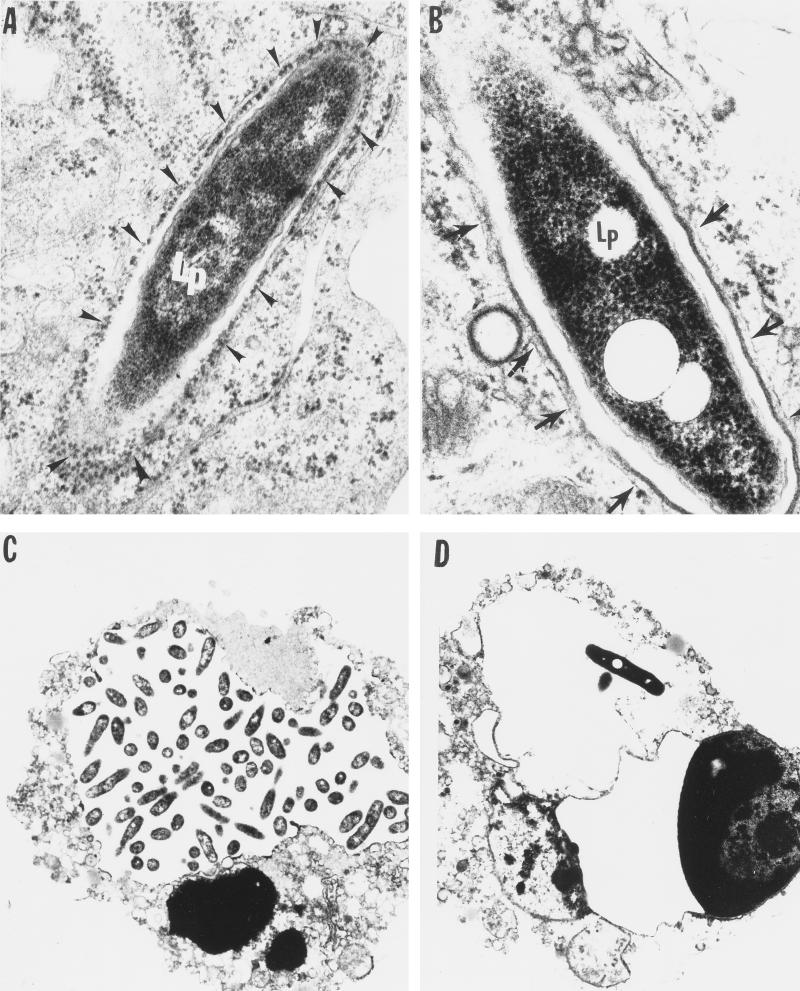
FIG. 4.
Transmission electron micrographs of U937 macrophages infected by the wild-type L. pneumophila strain AA100 (A and C) and the mil mutant GK11 (B and D) at 4 h (A and B) and 18 h (C and D) after infection. The arrowheads in panel A indicate the RER-surrounded phagosome; the arrows in panel B indicate the smooth multilayer membrane around the mutant GK11. Lp, L. pneumophila. Similar results were obtained with the mil mutants GE88 and GT20 (data not shown). Magnifications: ×36,000 (A), ×45,000 (B), and ×5,000 (C and D).
At 18 h postinfection, A. polyphaga was heavily infected by the mutant and the wild-type strain (Fig. 3C and D). In contrast, at 18 h of infection of macrophages by the GK11 mutant, a few cells contained a few visible single bacteria enclosed in RER-free phagosomes (Fig. 4C and D), but some were enclosed in RER-surrounded phagosomes (data not shown). These observations were consistent with the patterns of survival and intracellular multiplication within the two host cells. Time points between 4 and 18 h postinfection were not examined in this study.
Identification of dot or icm mutants among the mil mutants.
The two loci dot and icm of L. pneumophila have been shown to be involved in intracellular survival and replication of L. pneumophila in mammalian macrophages (11, 30). Mutants with mutations in dot and icm are highly resistant to NaCl (see below) (23, 36). Southern blots of EcoRI-digested genomic DNA from all the mutants probed with a 12-kb EcoRI fragment containing both dot and icm showed that none of the 32 mil mutants had an insertion in these loci (data not shown). These data are consistent with our recent observations that dot and icm insertion mutants of strain AA100 are severely defective within both U937 macrophages and A. polyphaga (23). Our data indicated that dot and icm were not mil loci.
Resistance of L. pneumophila mil mutants to NaCl.
It has been shown that NaClr strains of L. pneumophila are defective in their intracellular survival in mammalian and protozoan cell lines and are also attenuated in animal models (20). Moreover, most mutants of L. pneumophila (including dot and icm mutants) that are defective within mammalian macrophages are highly resistant to NaCl, but the relationship of the NaClr phenotype to attenuation is not known (23, 36). We examined the relationship between NaClr and the macrophage-defective phenotype of the 32 mil mutants (23). There was no increase in NaClr of any of the mil mutants (compared to that of the wild-type strain) as assessed by their plating efficiency on 0.6% NaCl-containing plates (data not shown) (23). In contrast, controls of two dot icm insertion mutants of AA100 (23) exhibited approximately a 300-fold increase in plating efficiency on 0.6% NaCl-containing plates. Interestingly, five of the mil mutants (GT20, GK79, GB111, GA12, and GE95) were dramatically more sensitive to NaCl as evident by their 600- to 1,100-fold-reduced viability following exposure to NaCl (data not shown).
Serum sensitivity.
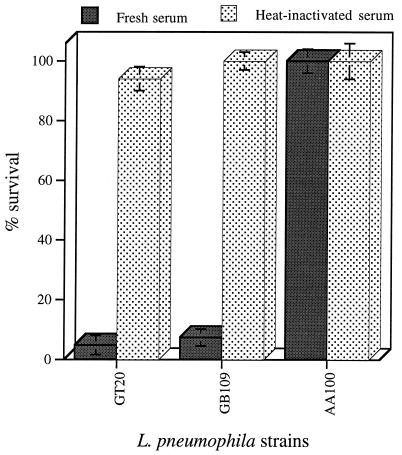
All 32 mil mutants were tested for their susceptibility to serum by using three different nonimmune human sera (23). Our data showed that two mutants (GT20 and GB109) were sensitive to all three sera tested (Fig. 5). There was at least 90% loss of viability for each of these mutants following 1 h of incubation in 50% fresh human serum (Fig. 5). The wild-type strain is completely resistant to serum, with no detectable reduction in the number of bacteria. All of the other mutants were similar to the wild-type strain in their resistance to all three sera tested (data not shown). The two serum-sensitive mil mutants were resistant to heat-inactivated serum (Fig. 5), which suggested that the serum sensitivity of these mutants was probably mediated by complement.
FIG. 5.
Serum sensitivities of mil mutants GT20 (belonging to group I) and GB109 (belonging to group V). Data are presented as percent survival in 50% normal human serum after 1 h of incubation at 37°C compared to that of the wild-type strain AA100. Values are the means of triplicate measurements, and the error bars represent standard deviations. Some of the error bars cannot be seen due to their small values.
Flagellum-defective (Fla−) mutants.
Flagellum expression has been shown to be indirectly associated with the ability of L. pneumophila to survive intracellularly in macrophages and in protozoa (35). We examined flagellar expression by all 32 mil mutants in a colony immunoblot assay with a rabbit antiserum raised against L. pneumophila flagella as a probe (23). Four mutants (GT39, GK79, GC52, and GQ54) were defective in flagellum expression (Fla−) (data not shown).
Iron acquisition and assimilation by the mil mutants.
Pope et al. have shown that some L. pneumophila mutants that are defective in iron acquisition and assimilation are also defective in intracellular replication within U937 cells and protozoa (34). We screened for sensitivity of the 32 mil mutants to the iron chelator ethylenediamine-DI(o-hydroxy-phenylacetic acid) and for resistance to the antibiotic streptonigrin, as we described previously (23). All mutants were indistinguishable from the wild-type strain in their growth in the presence of the two reagents (data not shown).
Sequence analysis of the junctions of miniTn10::kan insertions.
We cloned the Kan insert and flanking sequences from the chromosomes of four of the mil mutants (GB111, GT20, GK79, and GE88) for partial sequence analysis of the chromosomal junction of insertion (see Materials and Methods). Alignment of the predicted open reading frames to sequences in the National Center for Biotechnology Information databases by using the Blastx program showed that none of the sequences of the mutants had similarity to other known sequences. These findings suggested that many of the mil loci represent novel genes. It is important to note that the insertion may have a polar effect on the genes that were required for intracellular survival. Future characterization of these insertions will clarify this possibility.
DISCUSSION
The similarity in the intracellular infection of macrophages and protozoa by L. pneumophila is quite remarkable. Within both evolutionarily distant host cells, the bacterium multiplies within a phagosome that does not fuse with lysosomes and is surrounded by mitochondria and the RER (1, 2, 12, 26, 27, 41). Many mutants of L. pneumophila (including dot and icm insertion mutants) exhibit similar defective phenotypes in cytopathogenicity and intracellular replication within both macrophages and protozoa (7, 16, 23). These observations suggest that L. pneumophila utilizes many similar, yet unknown, mechanisms to survive and replicate within both evolutionarily distant hosts.
In this study, we have examined the possibility that L. pneumophila differentially utilizes certain loci to replicate within macrophages but not protozoa. We have identified 32 mutants that exhibit different degrees of defects in cytopathogenicity, intracellular survival, and replication within mammalian macrophages but exhibit wild-type phenotypes within protozoa. Only six of the mutants exhibited mild defects in survival and replication within A. polyphaga. Moreover, many of the mil mutants have also been tested and shown to exhibit a similar defective phenotype within peripheral blood monocytes, i.e., A/J mice peritoneal macrophages (22), and a wild-type phenotype within H. vermiformis. These data suggest that the mil loci are not cell specific in mammalian cells but rather are species specific. The dramatic differences in the pattern of survival and intracellular replication of the mil mutants within the two host cells indicate that certain genetic loci of L. pneumophila are uniquely required for survival and replication within macrophages but not protozoa. However, since the mil mutants were first isolated based on their defect within macrophages and subsequently tested in protozoa, our data did not exclude the possibility that certain L. pneumophila loci may be preferentially required for infection of protozoa.
The dot and icm mutants of L. pneumophila are severely defective in intracellular survival within mammalian macrophages (11, 30) and protozoa (23), and their defect is associated with failure to recruit host cell organelles, including the RER. Three of the mil mutants examined are defective in recruitment of the RER around the phagosome within macrophages during early stages of the infection but not within amoebae. However, it was evident that after 18 h of the infection, some phagosomes containing the mutants were surrounded by the RER. These observations suggest that some of the mechanisms of recruitment of the RER are different in the two host cells. Alternatively, the mutants have been trafficked and were subsequently localized in different endocytic pathways in the two host cells that did not lead to recruitment of the RER within macrophages. This further supports our hypothesis that although at the molecular and ultrastructural levels the intracellular infection of mammalian and protozoan cells is highly similar (1, 23), some of the biochemical and molecular events involved in intracellular survival within the two evolutionarily distant hosts are different.
Preliminary partial sequence analysis of the insertion junctions of many of the mil mutants showed that the insertions are within loci with no similarity to other genes in genetic databases, suggesting that many of the mil loci are novel. This may not be surprising, considering (i) that although a few other human pathogens exhibit slight multiplication within protozoa, L. pneumophila is the only documented pathogen that can invade and replicate efficiently within mammalian macrophages and protozoa and (ii) the unique replicative niche of L. pneumophila within both mammalian and protozoan cells (1, 26). We hypothesize that L. pneumophila has acquired genetic loci that are specific for survival and replication within mammalian macrophages, which has allowed the bacterium to evolve from a protozoan parasite into a Legionnaires’ disease-causing agent. Alternatively, ecological coevolution of L. pneumophila to parasitize protozoa has allowed the bacteria to possess multiple redundant mechanisms to parasitize protozoa, and some of these redundant mechanisms do not function within mammalian cells. Our recent discovery of natural competency of L. pneumophila to take up DNA supports our hypotheses (39a). It is possible that some of the mil loci are required for bacterial adaptation to different nutritional environments within the phagosomes of mammalian and protozoan cells. Once some of the mil loci are characterized, it would be interesting to examine whether they are present and functional in some of the environmental isolates of L. pneumophila strains and other Legionella species that have been shown to parasitize protozoa but have not been associated with human disease (20).
One of the mil mutants (GT20) exhibited severe defects in attachment to both macrophages and Acanthamoeba, indicating that there are similar mechanisms utilized by L. pneumophila to attach to both host cells. We have recently shown expression of multiple pili by L. pneumophila and that one of these pili is involved in attachment to mammalian and protozoan cells (39). It is highly unlikely that the GT20 mutant is defective in the respective pili, since pilin mutants that do not express the respective pili are competent for intracellular replication within mammalian and protozoan cells (39). However, there are also additional distinct mechanisms of bacterial attachment and invasion of mammalian macrophages and protozoa (5, 29). We have recently shown that one of these distinct mechanisms is present in the protozoan host H. vermiformis, in which attachment and invasion by L. pneumophila is mediated by bacterial attachment to a 170-kDa Gal/GalNAc lectin receptor (25, 44). Moreover, bacterial attachment and invasion are associated with tyrosine dephosphorylation of multiple host cell proteins, including the 170-kDa Gal/GalNAc lectin and several cytoskeletal proteins (25, 43, 44). In contrast, attachment of L. pneumophila to mammalian cells has been shown to be mediated by complement and noncomplement receptors (24, 28, 32).
Intracellular L. pneumophila undergoes phenotypic modulation during intracellular replication within macrophages (3, 40). We have recently shown that intracellular L. pneumophila manifests a stress response throughout the intracellular infection period (6). It would be interesting to contrast gene expression by L. pneumophila within macrophages to that within protozoa. Whether expression of some of the mil loci is induced during the intracellular infection is still to be determined.
Other investigators have reported the isolation of a total 87 mutants of L. pneumophila that are defective in mammalian macrophages, but these have not been tested in protozoa (36, 42, 45). Interestingly, a total of 83 of these 87 mutants are NaClr, but the relationship of NaClr to attenuation is not known. In addition, we have recently shown that many distinct mutants of L. pneumophila that are defective within both macrophages and protozoa, including dot and icm mutants, are NaClr (23). In contrast, none of the mil mutants is NaClr, suggesting that most or all of the mil mutants are different from previously identified ones. These observations indicate that the unexplained phenomenon of NaClr is unrelated to the function of the mil loci. Characterization of the role of the mil loci in the intracellular infection of mammalian cells should yield some interesting information about the pathogenic evolution of an intracellular pathogen that can efficiently parasitize both mammalian and protozoan cells.
ACKNOWLEDGMENTS
We thank N. C. Cianciotto and H. A. Shuman for their generous gifts of the pCDP05 and the pAM10 plasmids, respectively. We thank V. K. Viswanathan from the Cianciotto lab for technical suggestions. We also thank Janet Pruckler and B. S. Fields for their gifts of the antiflagellum antiserum and the A. polyphaga strain. We thank Bob Perry, Sue Straley, and members of Abu Kwaik laboratory for their comments on the manuscript.
Y.A. is supported by Public Health Service Award R29AI38410.
REFERENCES
- 1.Abu Kwaik Y. The phagosome containing Legionella pneumophila within the protozoan Hartmanella vermiformis is surrounded by the rough endoplasmic reticulum. Appl Environ Microbiol. 1996;62:2022–2028. doi: 10.1128/aem.62.6.2022-2028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik Y. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect Immun. 1998;66:203–212. doi: 10.1128/iai.66.1.203-212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik Y, Eisenstein B I, Engleberg N C. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect Immun. 1993;61:1320–1329. doi: 10.1128/iai.61.4.1320-1329.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu Kwaik Y, Engleberg N C. Cloning and molecular characterization of a Legionella pneumophila gene induced by intracellular infection and by various in vitro stress stimuli. Mol Microbiol. 1994;13:243–251. doi: 10.1111/j.1365-2958.1994.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 5.Abu Kwaik Y, Fields B S, Engleberg N C. Protein expression by the protozoan Hartmannella vermiformis upon contact with its bacterial parasite Legionella pneumophila. Infect Immun. 1994;62:1860–1866. doi: 10.1128/iai.62.5.1860-1866.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu Kwaik Y, Gao L-Y, Harb O S, Stone B J. Transcriptional regulation of the macrophage-induced gene (gspA) of Legionella pneumophila and phenotypic characterization of a null mutant. Mol Microbiol. 1997;24:629–642. doi: 10.1046/j.1365-2958.1997.3661739.x. [DOI] [PubMed] [Google Scholar]
- 7.Abu Kwaik Y, Pederson L L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 8.Barker J, Brown M R W, Collier P J, Farrell I, Gilbert P. Relationships between Legionella pneumophila and Acanthamoebae polyphaga: physiological status and susceptibility to chemical inactivation. Appl Environ Microbiol. 1992;58:1420–2425. doi: 10.1128/aem.58.8.2420-2425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barker J, Scaife H, Brown M R W. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob Agents Chemother. 1995;39:2684–2688. doi: 10.1128/aac.39.12.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates J H, Capmpbell G D, Baron A L. Microbial etiology of acute pneumonia in hospitalized patients. Chest. 1992;101:1005–1012. doi: 10.1378/chest.101.4.1005. [DOI] [PubMed] [Google Scholar]
- 11.Berger K H, Isberg R R. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 12.Bozue J A, Johnson W. Interaction of Legionella pneumophila with Acanthamoeba catellanii: uptake by coiling phagocytosis and inhibition of phagosome-lysosome fusion. Infect Immun. 1996;64:668–673. doi: 10.1128/iai.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brieland J K, Fantone J C, Remick D G, LeGendre M, McClain M, Engleberg N C. The role of Legionella pneumophila-infected Hartmanella vermiformis as an infectious particle in a murine model of Legionnaires’ disease. Infect Immun. 1997;65:5330–5333. doi: 10.1128/iai.65.12.5330-5333.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cianciotto N P, Eisenstein B I, Mody C H, Engleberg N C. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis. 1990;162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 15.Cianciotto N P, Eisenstein B I, Mody C H, Toews G B, Engleberg N C. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cianciotto N P, Fields B S. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cianciotto N P, Long R, Eisenstein B I, Engleberg N C. Site-specific mutagenesis in Legionella pneumophila by allelic exchange using counterselectable ColE1 vectors. FEMS Microbiol Lett. 1988;56:203–208. [Google Scholar]
- 18.Cirillo J D, Tompkins L S, Falkow S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang G D, Fine M, Orloff J. New and emerging etiologies of community-acquired pneumonia with implications for therapy, a prospective multicenter study of 359 cases. Medicine (Baltimore) 1990;69:307–316. doi: 10.1097/00005792-199009000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 21.Fields B S, Nerad T A, Sawyer T K, King C H, Barbaree J M, Martin W T, Morrill W E, Sanden G N. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J Protozool. 1990;37:581–583. doi: 10.1111/j.1550-7408.1990.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 22.Gao, L.-Y., B. J. Stone, J. K. Brieland, M. Gutzman, and Y. Abu Kwaik. Unpublished data.
- 23.Gao L-Y, Harb O S, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect Immun. 1997;65:4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson F C, III, Tzianabos O A, Rodgers F G. Adherence of Legionella pneumophila to U-937 cells, guinea-pig alveolar macrophages, and MRC-5 cells by a novel, complement-independent binding mechanism. Can J Microbiol. 1993;39:718–722. doi: 10.1139/m94-137. [DOI] [PubMed] [Google Scholar]
- 25.Harb O S, Venkataraman C, Haack B J, Gao L-Y, Abu Kwaik Y. Heterogeniety in the attachment and uptake mechanisms of the Legionnaires’ disease bacterium, Legionella pneumophila, by protozoan hosts. Appl Environ Microbiol. 1998;64:126–132. doi: 10.1128/aem.64.1.126-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitz M A. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwitz M A. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Husmann L K, Johnson W. Adherence of Legionella pneumophila to guinea pig peritoneal macrophages, J774 mouse macrophages, and undifferentiated U937 human monocytes: role of Fc and complement receptors. Infect Immun. 1992;60:5212–5218. doi: 10.1128/iai.60.12.5212-5218.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King C H, Fields B S, Shotts E B, Jr, White E H. Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect Immun. 1991;59:758–763. doi: 10.1128/iai.59.3.758-763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marra A, Blander S J, Horwitz M A, Shuman H A. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci USA. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marston B J. Epidemiology of community-acquired pneumonia. Infect Dis Clin Prac. 1995;4:S232–S239. [Google Scholar]
- 32.Payne N R, Horwitz M A. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J Exp Med. 1987;166:1377–1389. doi: 10.1084/jem.166.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pope C D, Dhand L, Cianciotto N P. Random mutagenesis of Legionella pneumophila with mini-Tn10. FEMS Microbiol Lett. 1994;124:107–112. doi: 10.1111/j.1574-6968.1994.tb07269.x. [DOI] [PubMed] [Google Scholar]
- 34.Pope C D, O’Connell W A, Cianciotto N P. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect Immun. 1996;64:629–636. doi: 10.1128/iai.64.2.629-636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruckler J M, Benson R F, Moyenuddin M, Martin W T, Fields B S. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect Immun. 1995;63:4928–4932. doi: 10.1128/iai.63.12.4928-4932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadosky A B, Wiater L A, Shuman H A. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning; a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Steinert M, Emody L, Amann R, Hacker J. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl Environ Microbiol. 1997;63:2047–2053. doi: 10.1128/aem.63.5.2047-2053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone, B. J., and Y. Abu Kwaik. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to and intracellular replication within mammalian and protozoan cells, in press. [DOI] [PMC free article] [PubMed]
- 39a.Stone, B. J., and Y. Abu Kwaik. Submitted for publication.
- 40.Susa M, Hacker J, Marre R. De novo synthesis of Legionella pneumophila antigens during intracellular growth in phagocytic cells. Infect Immun. 1996;64:1679–1684. doi: 10.1128/iai.64.5.1679-1684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson M S, Isberg R R. Formation of the Legionella pneumophila replicative phagosome. Infect Agents Dis. 1995;2:269–271. [PubMed] [Google Scholar]
- 42.Swanson M S, Isberg R R. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venkataraman, C., L.-Y. Gao, S. Bondada, and Y. Abu Kwaik. Submitted for publication.
- 44.Venkataraman C, Haack B J, Bondada S, Abu Kwaik Y. Identification of a Gal/GalNAc lectin in the protozoan Hartmanella vermiformis as a potential receptor for attachment and invasion by the Legionnaires’ disease bacterium, Legionella pneumophila. J Exp Med. 1997;186:537–547. doi: 10.1084/jem.186.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel J P, Roy C, Isberg R R. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann NY Acad Sci. 1996;797:271–272. doi: 10.1111/j.1749-6632.1996.tb52975.x. [DOI] [PubMed] [Google Scholar]
- 46.Woodhead M A, Macfarlane J T, McCracken J S, Rose D H, Finch R G. Prospective study of the aetiology and outcome of pneumonia in the community. Lancet. 1987;i:671–674. doi: 10.1016/s0140-6736(87)90430-2. [DOI] [PubMed] [Google Scholar]