Abstract
Telomeres and their single stranded overhangs gradually shorten with successive cell divisions, as part of the natural aging process, but can be elongated by telomerase, a nucleoprotein complex which is activated in the majority of cancers. This prominent implication in cancer and aging has made the repetitive telomeric sequences (TTAGGG repeats) and the G-quadruplex structures that form in their overhangs the focus of intense research in the past several decades. However, until recently most in vitro efforts to understand the structure, stability, dynamics, and interactions of telomeric overhangs had been focused on short sequences that are not representative of longer sequences encountered in a physiological setting. In this review, we will provide a broad perspective about telomeres and associated factors, and introduce the agents and structural characteristics involved in organizing, maintaining, and protecting telomeric DNA. We will also present a summary of recent research performed on long telomeric sequences, nominally defined as those that can form two or more tandem G-quadruplexes, i.e., which contain eight or more TTAGGG repeats. Results of experimental studies using a broad array of experimental tools, in addition to recent computational efforts will be discussed, particularly in terms of their implications for the stability, folding topology, and compactness of the tandem G-quadruplexes that form in long telomeric overhangs.
Keywords: Telomere, Telomeric Overhang, G-quadruplex, Telomere Accessibility, FRET-PAINT
Graphical Abstract
INTRODUCTION
Eukaryotic chromosomes, which are linear and terminate with a 3’ single stranded overhang, experience end replication and end protection problems. The remedy of these problems lies in the architecture and sequence of DNA at chromosome ends in addition to proteins or nucleoprotein complexes that are specialized to function in these regions. The nucleoprotein complexes that characterize the chromosome ends, collectively called telomeres, are required to avoid genomic instability and to adapt to complex processes that eukaryotic genomes undergo [1]. The repeating non-coding sequences of telomeres provide a gene-free buffer zone that can tolerate, to a certain extent, shortening of the ends with successive cell divisions [2]. On the other hand, structural characteristics of telomeres enable distinguishing them from double strand breaks and prevent fusions with other chromosome ends through non-homologous end joining (NHEJ) [2,3]. Broadly, these sequences and structural characteristics of telomeres will be the focus of this review.
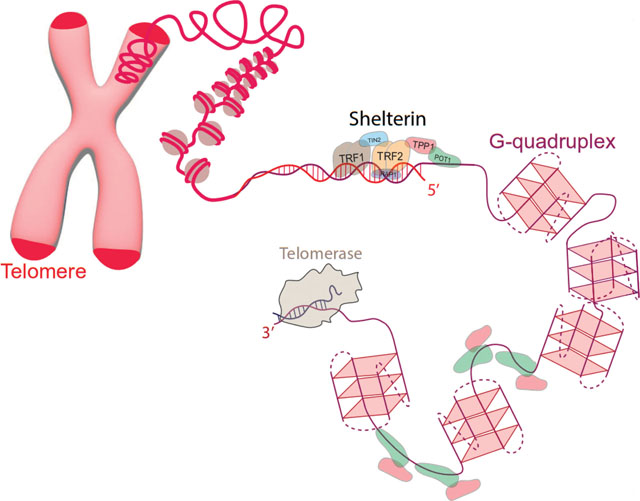
G-quadruplex (GQ or G4) structures and t-loops are important local and long-range telomeric secondary structures, respectively (Figure 1). GQs are composed of stacked G-tetrads, which contain guanines at the corners of the tetrads. The guanines in each tetrad form eight Hoogsteen hydrogen bonds with each other and their interactions are coordinated by monovalent cations that sit within or in-between the tetrad plane. The repeating telomeric sequences are widely conserved across eukaryotes and G-rich strands of almost all such sequences form GQs in vitro. By enabling the single stranded overhang to fold into a stable secondary structure, GQs are considered to cap chromosome ends. GQs have been detected in telomeres and other sites of human genome and much is now understood about their potential functions [4–7]; however, many open questions remain [8].
Figure 1.
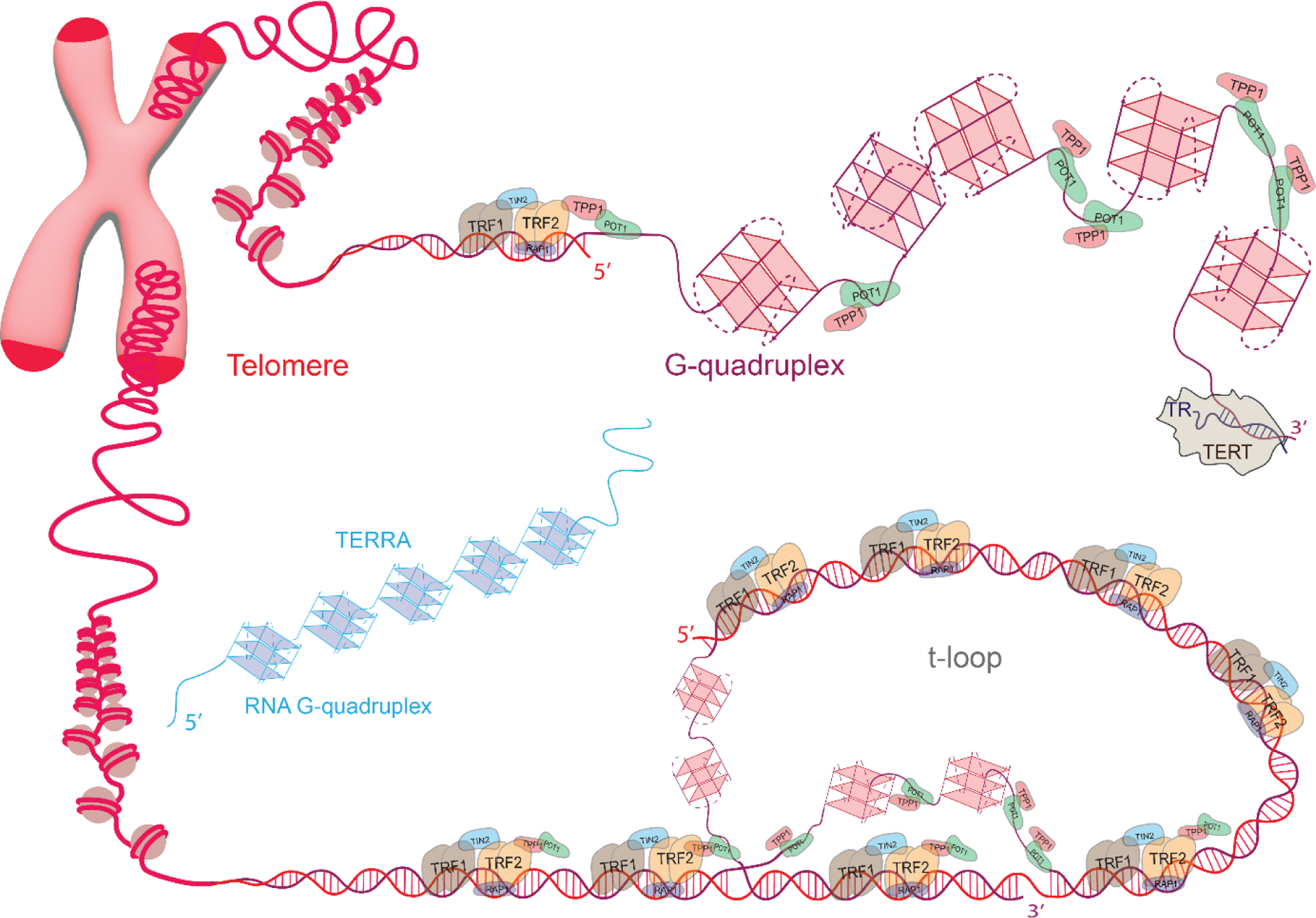
Schematic of telomeres and associated proteins. The cases for t-loop formation (bottom) and telomeric overhang decorated with G-quadruplexes (top) are illustrated for different chromosome ends. Telomeric DNA, Shelterin proteins, G-quadruplexes, telomerase reverse transcriptase (TERT), telomerase RNA (TR), and telomeric repeat containing RNA (TERRA) are shown in different segments of telomeres. POT1 binds to single stranded telomere (ssTEL) while TPP1 enhances POT1 binding and connects it to the rest of the Shelterin. TRF1 and TRF2 bind to double stranded telomere (dsTEL). TERT uses a small part of TR as a template to extend telomeric overhang at 3’ end. TERRA can fold into multiple GQs (left), which may interact with ssTEL.
The repeating telomeric sequences also facilitate t-loop formation as they provide a large number of potential target sites that can be invaded by the telomeric overhang. During t-loop formation, the G-rich single stranded telomere (ssTEL) invades the upstream double stranded telomere (dsTEL) and hybridizes with the C-rich strand; thereby, ‘hides’ the otherwise vulnerable ssTEL. In addition to remedying the end-protection problem [9–11], the t-loop could also ameliorate the end replication problem as it is structurally very similar to replication fork. This similarity could have enabled elongation of telomeres by replication enzymes before emergence of telomerase, a ribonucleoprotein complex specialized in telomere elongation [10].
GQs in ssTEL and t-loops appear to be mutually exclusive where formation of one might require resolving of the other. Therefore, they might form in different phases of the cell cycle, such as GQs forming in S-phase when t-loops need to be resolved [8]. In addition to these fundamental structural features, we observe complex mechanisms in modern eukaryotic cells to protect, maintain, and replicate telomeres. Telomerase, Shelterin complex, CST (CTC1-STN1-TEN1) complex and telomeric repeat containing RNA (TERRA) are some of the important players in this context.
Telomerase is an important agent for the remedy of end-replication problem [12,13]. Shelterin and CST complexes include proteins that bind specifically to telomeric sequences and prevent activation of DNA damage response pathways, which would otherwise recognize chromosome ends as double strand breaks [14–16]. Such a misrecognition would lead to repair, recombination, and eventually to fusion of chromosome ends. TRF2, a member of Shelterin, is proposed to facilitate t-loop formation, while POT1 specifically binds and protects the single stranded overhang. Recently, liquid-liquid phase separation and selective recruitment of shelterin proteins to telomeres have been proposed to be significant in telomere maintenance [17]. Therefore, these complexes constitute significant components of the remedy for the end-protection problem. This review will briefly highlight our current understanding about the roles of these different components, with a more detailed description of the role and architecture of GQ structures.
DISCUSSION
Significance of TTAGGG sequence:
Telomeric overhangs not only show a strong preference for guanine-rich motifs but also demonstrate a paucity of cytosines [18]. A typical telomeric repeat consists of 2–4 G’s, 1–4 T’s and an occasional A, i.e., G2–4T1–4A0–1. Example sequences include GGGTTA in vertebrates, GGGGTT in tetrahymena, and GGGTTTA in Arabidopsis thaliana. While the selection for abundance of G’s is considered to enable GQ formation, the paucity of C’s might also be significant. Interestingly, the variant GGGCTA repeat, which was identified in the proximal region of telomeres and estimated to be present in 7% of human telomeres, was highly unstable in the male germline when 11 or more homogenous repeats of it were present [19]. This length dependent instability was later proposed to be due to formation of a complex structure in which a two-tier GQ co-existed with a Watson-Crick hairpin, where the propensity of hairpin formation increased with increasing number of GGGCTA repeats [20]. The source of genomic instability in such sequences was attributed to be due to inaccessibility of hairpin structures to single stranded DNA binding proteins, such as RPA, which can effectively unfold telomeric GQs but not hairpin structures. Therefore, while readily forming and acting as roadblocks for polymerase progression, the GQs formed by GGGTTA repeat are fairly regular even at large number of repeats, preclude formation of more complex structures, and are amenable for destabilization by RPA [21–23] and helicases [24,25]. Therefore, telomeric sequences rich in G’s and poor in C’s enable formation of GQs that are stable enough to cap telomeric ends while feasible to unfold by protein activity.
TERRA:
Telomeric repeat-containing RNA is a class of noncoding RNA transcribed at telomeres and is generally considered to actively participate in regulating the telomere maintenance and chromosome end protection [26–28], such as inhibiting the telomere-telomerase interactions by hybridizing with the template sequence of telomerase in mammalian cells [29–31]. In humans, the TERRA consists of a heterogeneous population of G-rich (UUAGGG) sequences which have been shown to fold into GQs [32,33], exclusively in parallel conformation [32–36]. The long TERRA molecules consists of tens of G-repeats and can also fold into higher-order quadruplex structures [37,38]. TERRA GQs also have high affinity towards various telomere associated proteins [39–42] and the interaction between TERRA GQs and associated proteins can derive RNA aggregates promoting liquid-liquid phase separation [37,43–46]. In addition, TERRA transcripts have been shown to form hybrid R-loops at critically short telomeres and are involved in regulating the chromatin structure [47–50]. Furthermore, TERRA is involved in recruiting various epigenetic factors at telomeres and plays an essential role in establishing the heterochromatin structures [51,52]. It has been proposed that TRF2 subunit of shelterin facilitates the recruitment of TERRA at telomeres and interaction of TERRA with POT1 effectively displaces the RPA from ssTEL, hence playing an important role in maintaining the telomere integrity [53,54]. Despite being an important player in telomere maintenance and genome stability, it is still unclear how TERRA performs these functions at molecular level. A detailed discussion about emerging functions of TERRA has been reviewed in reference [55].
Maintenance of telomere length:
Telomeres in humans consist of 2–10 kb long dsTEL and 50–300 nt long 3’ ssTEL, also called G-overhang. In germline cells, the length of telomeres is actively maintained by extension of the ends by telomerase, which is abundantly present [56]. In healthy somatic cells telomerase is scarce or absent and telomere extension does not take place, which results in a progressive decrease in telomere length with every cell division [57,58]. Therefore, telomere length functions as a “molecular clock” measuring the lifespan of somatic cells [59]. When a certain threshold length is reached (Hayflick limit), cellular senescence is triggered [60]. Immortalized cells, stem cells and cancer cells generally have high telomerase activity [61]. Premature aging is observed in people with genetic disorders, such as dyskeratosis congenita (DC), where mutations in genes associated with telomere maintenance (primarily those impacting telomerase) are prevalent [62,63]. Interestingly, telomerase reactivation in an engineered telomerase-deficient mice not only arrested but reversed tissue and neuro degeneration [64], further highlighting the significance of telomere maintenance in the aging process.
Chromatin structure of telomeres:
Telomere size, shape, and compaction level were measured in intact nuclei of telomerase-positive immortalized mouse embryonic fibroblasts (MEF) and human fibrosarcoma-derived HT1080 cells, in addition to telomerase-negative (ALT cells) human bone osteosarcoma cells (U2OS) using transmission electron microscopy (TEM) and fluorescence microscopy [65]. In these studies, TRF1 or TRF2 were fused with eGFP for fluorescence signal and APEX2 for an electron-dense signal, allowing both signals to be obtained from the same cells. TEM studies showed the average telomere diameter to be ~200 nm and ~160 nm in MEF and HT1080 cells, respectively, with a round or ovoid shape. The corresponding average diameter was much larger (~350 nm) and the shape significantly more heterogenous in U2OS ALT cells. The resolution of these studies was estimated to be ~5 nm. Overall, telomeres were observed to possess a fibrous and mesh-like ultrastructure.
Fluorescence super resolution studies in human cells showed that the compact nature of telomeric chromatin is due to a network of interactions between shelterin proteins and telomeric DNA [66]. Removing shelterin subunits resulted in 10-fold more extended telomeres and accumulation of DNA damage response (DDR), while recompacting telomeres prevented DDR. These studies highlight the significance of having repeating telomeric sequences that could be connected to each other with homodimers of TRF1 or TRF2. The connectedness and complexity of the network could be further enhanced by RAP1 and TIN2 that interconnect TRF1, TRF2, and POT1/TPP1 with each other. This compact structure, in addition to TRF2-mediated t-loop and tandem GQs in ssTEL, appears to form a robust structure that is resilient against DDR activators.
Liquid-liquid phase separation in telomeres:
Liquid-liquid phase separation takes place when proteins and nucleic acids with locally high concentrations form condensates via weak and multivalent interactions [67,68]. These condensates phase separate from their surroundings, enable much higher reaction rates than otherwise feasible, and selectively recruit some agents while excluding others [69]. Nucleoli, Cajal bodies, P-granules, and stress granules are examples of such condensates. It was recently demonstrated that human telomeres form liquid-like condensates in vivo [17]. In vitro studies showed that TRF1 and TRF2 are prone to form condensates alone while telomeric DNA promotes the condensation [17]. The repeating sequence of telomeric DNA is proposed to serve as a super-scaffold that enables oligomerization of TRF1 and TRF2. Interestingly, TERRA and POT1 were preferentially recruited to these condensates, while RPA was not. This would help understand how POT1 can compete against the otherwise more abundant RPA in accessing ssTEL, when the two proteins have similar affinity, and prevent activation of DDR pathways [70]. As in other contexts where liquid-liquid phase separation is observed, telomeric condensates alter our understanding of telomere biology in fundamental ways, including in processes such as how telomeric ends are kept separate from each other or how telomerase is recruited to telomeres.
G-Quadruplex Topologies:
DNA GQs fold into a variety of topologies (also called ‘conformations”) (Figure 2) which depend on the solution conditions and sample preparation protocols [71,72]. The first crystal structure on a single strand of DNA that contains four G-repeats of human telomeric sequence was reported to fold in parallel topology (in the presence of K+ ions), in which all four G-repeats are oriented in parallel connected by three propeller loops [73]. In contrast, NMR studies reported hybrid structures of human telomeric sequence in the presence of K+ with three G-repeats oriented in one direction and the fourth in the opposite direction [74,75]. The lower DNA concentration in NMR studies compared to crystal structures could be a reason for observation of different conformation as crowding conditions are known to influence the GQ topology. Depending on the flanking segments, the telomeric sequence can adopt two distinct hybrid structures, i.e., hybrid-1 or hybrid-2 which coexist at equilibrium and differ in loop arrangements, strand orientations, and tetrad arrangements [74,75]. Moreover, in the presence of Na+ ions, the telomeric sequence is reported to form an antiparallel G-quadruplex structure featuring both diagonal and lateral TTA loops [76]. Form-3 is another highly stable topology formed by four telomeric repeats in K+, in which two G-tetrads are stabilized by stacking and base-pairing interactions of loop elements [77]. As illustrated by this brief review, folding topology of telomeric G-quadruplexes could be highly heterogenous under physiologically relevant ionic conditions (dominated by K+). This heterogeneity likely influences the interactions between the tandem GQs in a telomeric overhang, any potential super structure they might form, and their affinity to small molecule drugs that are designed to target them for various therapeutic purposes.
Figure 2.
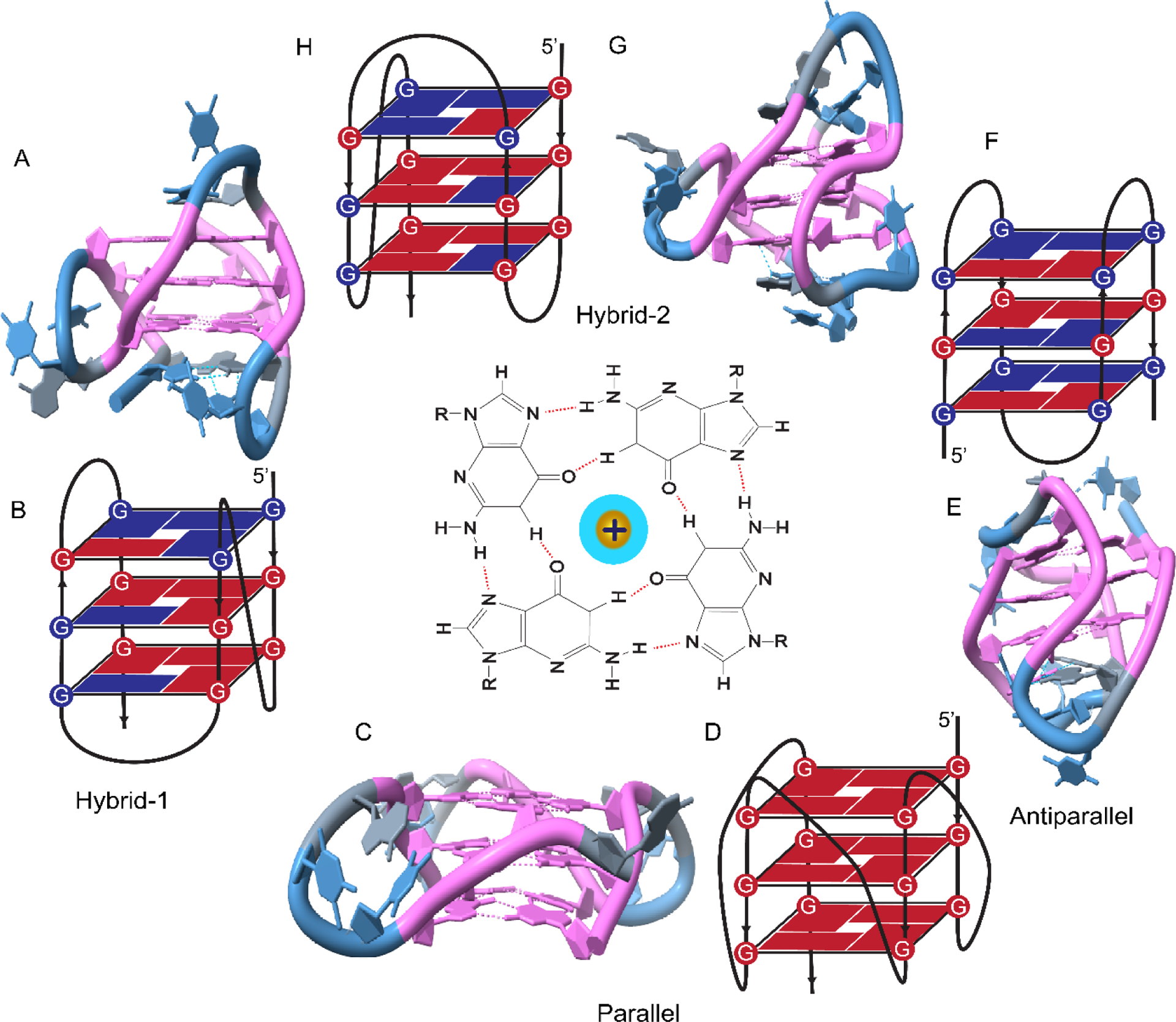
NMR structure and schematic illustration of GQ topologies. (A-B) Hybrid-1; (C-D) Parallel; (E-F) Antiparallel; (G-H) Hybrid-2 topologies. A schematic of a G-Tetrad with four guanines at the corners and a monovalent cation in the middle is shown in the center of the figure. The NMR structures in A, C, E, and F were created in Chimera software using PDB ID 2JSM, 1KF1, 2KF8 and 2JSL, respectively.
Small molecule and G-quadruplex Interactions:
Folding of telomeric overhang into quadruplex structures reduces telomerase activity while quadruplex formation in promoters of many genes, including prominent oncogenes, generally modulates transcription of associated genes [78–80]. Stabilizing telomeric GQs with small molecules might prevent their unfolding and extension by telomerase, which might eventually lead to uncapping of telomerase from the overhang [81,82], or it might sequester shelterin proteins (such as POT1) out of the telomeres [83], and thereby inhibit telomerase recruitment. Also, some GQ binding ligands, such as Braco-19, RHPS4, and telomestatin, cause cancer cells to undergo fast replication senescence. These molecules activate a DNA damage response similar to that activated after DNA double-strand breaks [84–87]. Several examples of such molecules are demonstrated in Figure 3.
Figure 3.
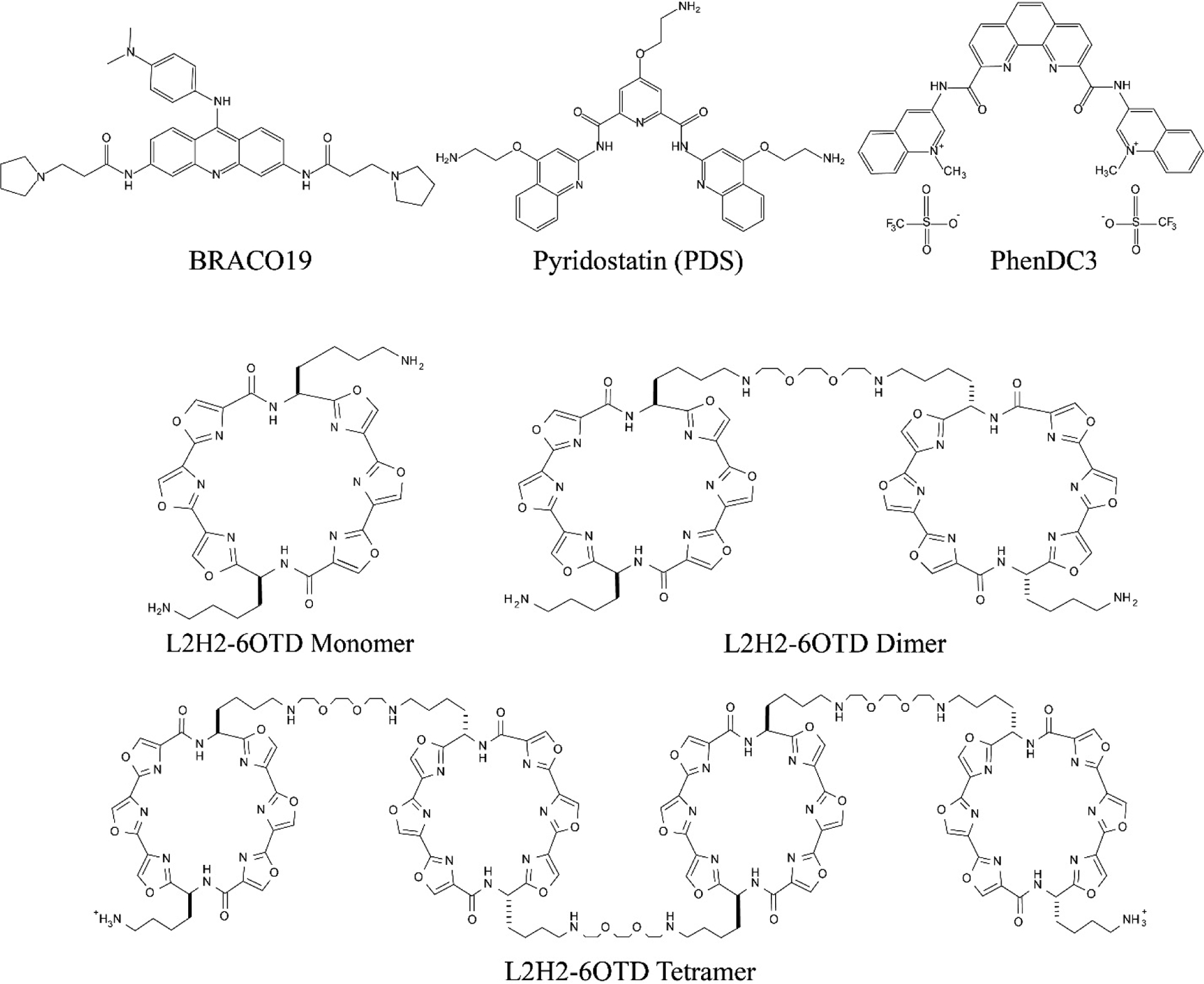
Chemical structure of example small molecules frequently used to stabilize the GQ structure. The dimer and tetramer of L2H2–6OTD represent a potential approach to increase the specificity of such small molecules to telomeric overhangs which have tandem GQs.
These considerations have motivated numerous efforts to design and synthesize small molecules (Figure 3) which would bind to and stabilize the GQ structures, and thereby serve as anti-cancer therapeutics [88]. According to G-quadruplex Ligands Database (https://www.g4ldb.com), the number of small molecules that are registered to target GQ (or i-motif) has increased from 800 in 2013 [89] to more than 3200 in 2022 (v2.2 of the database), [90] which illustrates the activity and interest in the field. However, there is currently no FDA-approved drug in the market that functions by specifically targeting GQ structures, which highlights the complications associated with the drug development efforts.
Molecules that target telomeric GQs should inhibit telomerase activity and trigger senescence in cancer cells within a relatively short period of time, while healthy cells are left relatively unaffected [82]. Also, promising drug candidates should be highly soluble in water, should be able to penetrate the nucleus, should have a high affinity for GQ and should possess high selectivity for the GQ over dsDNA. Specific targeting of a GQ structure formed by a particular sequence also requires distinguishing it from other GQ structures. While molecules with high enough affinity for GQ (dissociation constant KD~1–10 nM) have been developed, [91] selectivity for GQ over dsDNA has been limited to about two orders of magnitude with a single molecule and three orders of magnitude with dimer of a GQ binder [92]. The majority of GQ stabilizers that have been developed are based on planar organic heteroaromatic systems that interact with the terminal G-quartets through π-π stacking [93,94]. Well known and widely used examples of G-quartet binders include telomestatin [95] or oxazole telomestatin derivatives [96], porphyrins [97], PhenDC3 [98], Pyridostatin [99], and Braco-19 [84]. However, just targeting the G-quartets would naturally have limited success in distinguishing between different GQs, which essentially share the same planar quartet structure, or in selecting GQ over dsDNA. Therefore, it has become clear that other features of the GQ structures, such as the loops or grooves which have different diameters and accessibility in different GQ topologies, should also be targeted to improve selectivity. A candidate in this respect is introducing cationic arms to the aromatic structures in order to target the phosphate backbone of the loops or grooves.[100] However, such arms might also reduce the planarity of the molecule and result in lower affinity to the G-quartet. Nevertheless, deviations from a planar structure might also reduce intercalation with dsDNA (since DNA intercalators are also typically flat aromatic heterocyclic compounds) and increase the overall selectivity against dsDNA [93]. Such considerations have added to the complexity of design and synthesis of GQ binders, yet some progress has been achieved [101,102].
Telomeric overhangs present further opportunities for selective targeting as they contain multiple tandem GQ structures that have the same TTA sequence in the loops and linkers between neighboring GQs. Extended molecules that target multiple neighboring GQs and the loops or linkers might achieve the required selectivity (Figure 2). Such molecules could also find use in targeting some promoter regions which consist of long repetitive G-rich sequences. These sequences can fold into two or three consecutive quadruplex units, such as insulin-linked polymorphic region (ILPR) located upstream of the transcriptional start site of the insulin gene [103]. Furthermore, it has been reported that the hTERT promoter consists of three contiguous quadruplex units [104,105]. It is important to note that GQs in telomeric overhangs are significantly distinct from the GQs folded in promoter regions, which are localized within a double-stranded DNA template and must compete with the complementary C-rich strand. Therefore, molecules designed for one system might not be ideal for the other.
In addition to the mentioned organic molecules, metal complexes are promising GQ ligands due to their efficient and targeted interactions with GQs, relatively regular and straightforward synthesis, and broad range of electrostatic, magnetic, and optical characteristics which facilitate their detection and tracking in cells [106]. Examples of such molecules include cisplatin derivatives, metalloporphyrins and derivatives, and metallophyhalocyanines and derivatives. These molecules are characterized by the core metal ions, which confer the entire molecule cationic characteristic that facilitates interactions with negatively charged phosphate backbone of nucleic acids and improves cell permeability. In addition, the structure can be adjusted in a controllable manner, such as made more or less planar for optimal interaction with GQs. For a detailed description of these promising molecules, we refer the reader to reviews on this topic [106,107].
Tandem G-Quadruplexes in Telomeric Overhangs:
Over the past 20 years, there has been a significant interest in studying structural properties of potential higher-order quadruplex structures due to their relevance in understanding the full length telomeric overhang [108]. Broadly, two different models (Figure 4) have been proposed for long telomeric sequences that can fold into multimeric structures consisting of tandem quadruplex units: 1) GQs are arranged independently like “beads on a string” connected by flexible TTA loops [109,110]. 2) GQs form a macrostructure where each GQ unit interacts with adjacent GQs via stacking interfaces or TTA loops [111,112]. There are also reports that propose both structural features may be present in a telomeric overhang where some GQs interact with their neighbors while others do not [113]. It should be mentioned that “beads on a string” description has been used in the literature more generally than the way we have restricted it to non-interacting GQs here [114]. Different experimental approaches and computational models have been used to explore this issue which we will briefly describe based on the experimental methods used in respective studies.
Figure 4.
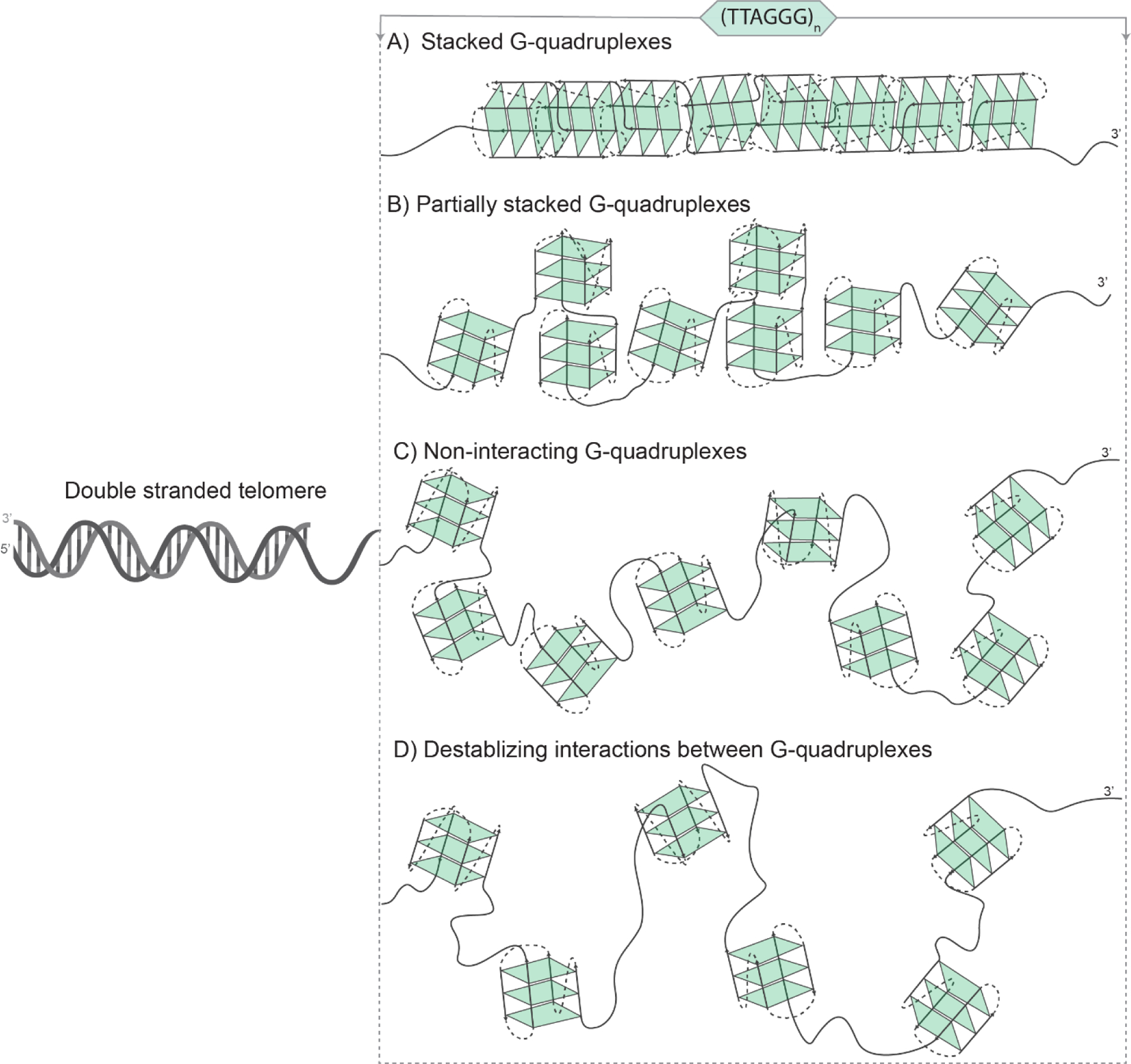
The tandem GQs in the telomeric overhang may adapt to different structures depending on how they interact with each other. Telomeric overhang formed by A) stacked GQs; B) partially stacked GQs; C) non-interacting GQs; and D) GQs with destabilizing interactions between neighboring structures. Fully stacked and destabilizing neighboring GQs are the two extremes of possible modes of interaction, with partial stacking and non-interacting models describing scenarios between these extremes. The noninteracting GQs are typically referred to as ‘beads on a string’; although, there is a certain level of loose terminology about this term in the literature. Depending on the level of folding, the GQs might adapt one or another of these models or may transition among them.
Nuclear Magnetic Resonance (NMR) spectroscopy and X-ray Crystallography
Nuclear Magnetic Resonance (NMR) spectroscopy and X-ray Crystallography have been extensively used to reveal high-resolution quadruplex structure of telomeric sequences and their interactions with small molecules [72,73,75,76,115–118]. Differently from X-ray crystallography, NMR is used in the solution state to reveal structural details in physiologically relevant solution conditions. In NMR spectroscopy, guanines in G-tetrads give rise to a characteristic chemical shift which is distinct from that in other DNA conformations. The structural features and molecular coordinates obtained from NMR or X-ray data have been used to build models to perform molecular dynamics simulations and to construct higher order structures by stacking the individual quadruplex units, with the assumption that the initial topology remains fixed [119,120].
The NMR spectra of 8- and 12-repeat telomeric sequences revealed that quadruplex units in long telomeres can adopt both antiparallel and hybrid type structures [121]. These sequences contained site specifically 15N-labelled guanines to obtain high resolution structural details. In particular, the 3’ quadruplex unit forms well defined 2-tetrad antiparallel basket and hybrid-2 structures, while the internal quadruplex units exhibited similar structural properties, but with a disordered bottom tetrad of hybrid conformation in 110 mM K+ solution. The NMR spectra did not change significantly in presence of crowding conditions, such as Xenopus oocyte extracts, which implied that longer telomeric sequences may preserve their structural properties in cellular environment. Furthermore, there are no discernible differences in the NMR spectra of GGG(TTAGGG)7 and GGG(TTAGGG)11 sequences, which can fold into two and three consecutive quadruplex units, respectively. Moreover, the inclusion of 3’-flanking nucleotides, such as TTA or TT, had no significant impact on the structure and stability of quadruplex topologies. These observations indicate that 2–3 quadruplex units are independently arranged in these telomeric sequences and support the idea of beads on a string model [121].
Despite revealing essential information about the chemical bonds in the system, these studies might have limitations when handling long telomeric sequences due to heterogeneous populations, large molecular sizes that might be generated by formation of higher order quadruplex structures, and the dynamic equilibrium between different conformations.
Circular dichroism
Circular dichroism (CD) spectroscopy is a well-established technique for elucidation of secondary structures of proteins and nucleic acids, including G-quadruplex structures. In CD spectroscopy a signal is generated based on how polarized light interacts with an optically active (chiral) molecule or a molecule with asymmetry induced by a magnetic field. Such molecules absorb left or right circularly polarized light differently, resulting in a difference in molar extinction (Δɛ), which is quantified as the CD signal (Figure 5). CD in UV region is extremely sensitive to slight changes in mutual orientation of DNA bases, measured distance between the interacting strands, and distance between bases and the axis of the structure. The relative orientation of the strands and the polarity of G-tetrads located at the core of quadruplex unit determine the spectral features of quadruplex topology observed in CD spectra [122]. Although some exceptions are noted, certain peaks (such as at 265 nm and 290 nm) and troughs (such as at 240 nm and 265 nm) are generally accepted as CD spectral features associated with particular G-quadruplex topologies [123–126]. In addition, CD spectroscopy has been used to study the kinetics and thermal melting of quadruplex structures by monitoring the temperature-dependent change in the amplitude of characteristic peaks in the CD spectra.
Figure 5.
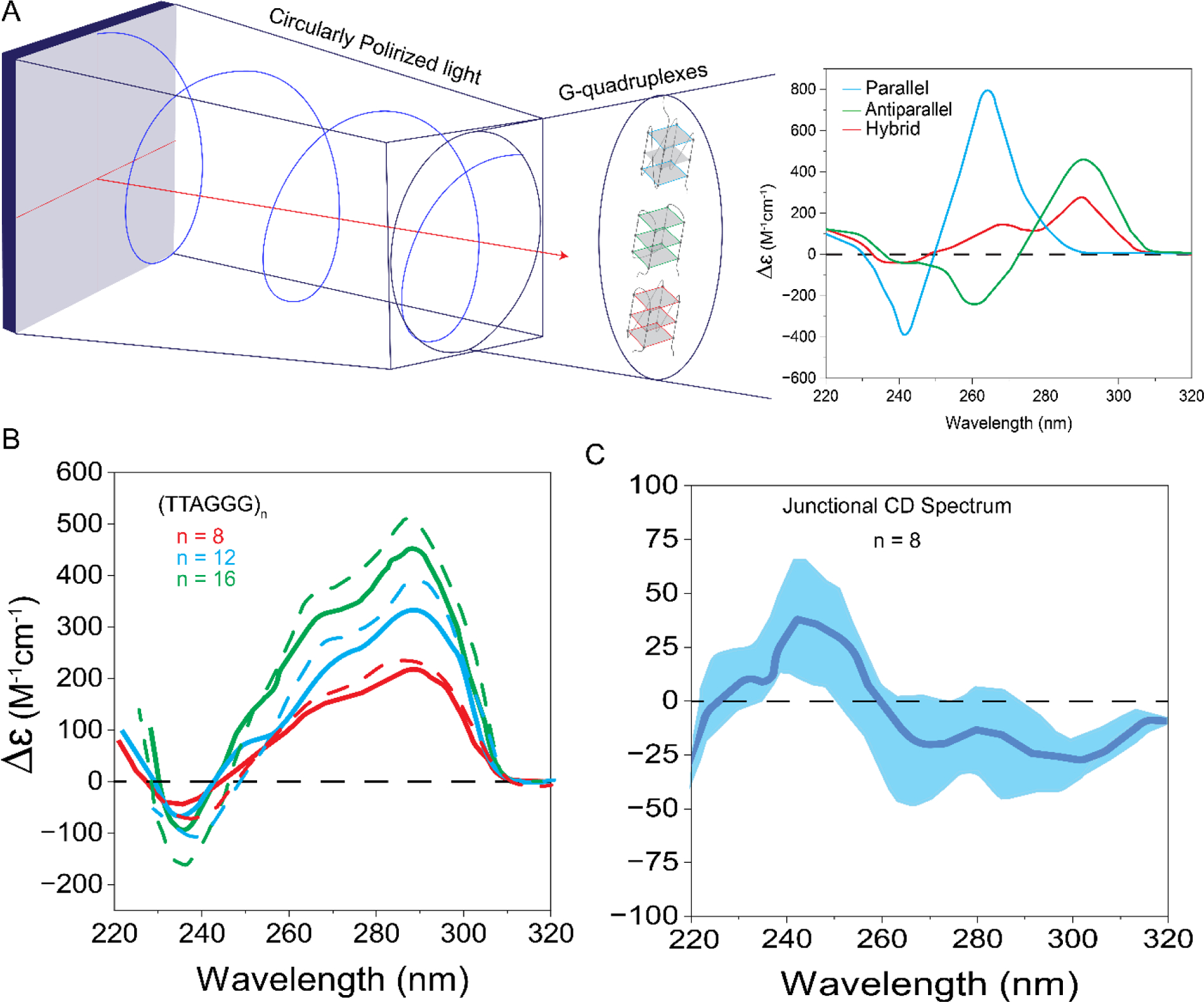
Circular dichroism assay. A) Schematic illustration of the assay and CD spectra for different GQ topologies. Graph adapted from reference [159]. B) CD spectra for sequences that have 8, 12, or 16 G-tracts, capable of forming 2–4 GQs. The signal intensity increases with the number of repeats. The solid lines show raw spectra while the dashed lines show the spectra after the GQ interaction term (junctional CD signal) is subtracted from respective the raw spectra. C) Schematic of the junctional CD spectrum which has a positive peak around 240 nm and a trough at around 260 nm, which have opposite signs to the positive peak at 260 nm and trough at 240 nm, resulting in more prominent peaks in CD spectra after junctional CD spectrum is subtracted from the raw spectra. Graph adapted from reference [129].
For longer telomeric DNA sequences, CD has been used to infer the presence of multiple G-quadruplex units by comparing the spectra with the known spectrum of a single quadruplex. The increase in CD signal with the number of quadruplex units is used as the indicator for multi-quadruplex formation. For example, the CD spectrum analysis of longer telomeric sequences (TTAGGG)n, where n = [4,8,12,16,20] (which can form 1–5 GQs), shows a linear increase in the peak intensity (290 nm and 265 nm) with the number of quadruplex units in 100 mM K+ or Na+ solutions [110,119,127,128]. Furthermore, the shape of the spectra for these longer sequences remains similar to that for a single quadruplex unit, the (TTAGGG)4 sequence. To illustrate, in K+ the CD spectrum of (TTAGGG)4–12 series shows two positive peaks (265 nm and 290 nm) with a dominant peak at 290 nm indicating these sequences fold into a mix of parallel/antiparallel or hybrid conformations, similar in shape to the spectrum of a single GQ. Spectra with similar shape were observed for all sequences in the n=[4–12] series in Na+, where a positive peak at 290 nm and a trough at 265 nm were observed, consistent with the antiparallel conformation [110]. Similar structural features were reported for the Oxytricha telomeric sequence (TTTGGG)4–12 in K+ and Na+ solutions. The most straightforward interpretation of these observations supports the idea that quadruplex units are independently arranged on the telomere where quadruplexes are connected to one another by TTA or TTT loops.
On the other hand, the shape of the CD spectra of GGG(TTAGGG)n, where n = [3,7,11], which differs from previously discussed sequences by the lack of a TTA overhang, shows a sequence dependent and enhanced CD signal which is not necessarily proportional to the number of quadruplex units. Even though very low K+ concentration (2.5 mM) is used in these studies, this deviation from proportionality may be due to stacking interactions between adjacent GQs that stabilize a higher order structure [111]. To directly quantify such interactions, a recent study introduced a contribution from the junction of stacking quadruplex units (called ‘junctional CD signal’) to the observed CD signal (Figure 5) [129]. This additional contribution is derived from the difference of CD signal of the dimer from its constituent monomer units. The corrected CD spectrum, obtained by subtracting the junctional CD signal from the observed spectrum, shows an intensified CD signal with a more prominent peak and trough compared to the observed CD signal. However, the increase in intensity is not linear and suggests stronger interactions at longer lengths [129].
CD has also been utilized to discern the structural features in the presence of molecular crowding agents and the effect of ion exchange on longer telomeric sequences. The longer telomeric sequences (TTAGGG)n, n = [4,8,12,16,20] were reported to fold into a mix of hybrid and parallel type conformations in the presence of 100 mM K+ ions with a dominant peak at 290 nm and shoulder around 265 nm [109,128,130]. In Na+ solution, these sequences show an antiparallel conformation with a deep trough at 265 nm, suggesting a stable topology [130]. After addition of 2 mM K+ (starting from Na+ condition) the trough at 265 nm abruptly diminishes and the stable antiparallel conformation changes to a parallel or hybrid (3+1) conformation with slow kinetics, which suggests a structural change due to re-arrangement of stacking interactions between the tetrads [130]. This structural shift is greatly facilitated by the sequence length, slow rise in temperature or incubation at 37 °C in the presence of a crowder [130]. PEG200 and ethanol induced a structural transition from known DNA conformations to parallel topology in 100 mM K+ solution whereas in 100 mM Na+, the CD spectrum remained unchanged except a deeper trough at 265 nm [128,130,131].
The normalized CD spectrum of a telomeric sequence GGG(TTAGGG)12 with and without 3’-end flanking TT bases shows slightly lower CD signal compared to similar GGG(TTAGGG)4 and GGG(TTAGGG)8 sequences. The diminished CD signal was attributed to incomplete folding of 12 repeats into three consecutive quadruplex units where the majority population (87%) consist of three quadruplex units and the remaining minor population contain only two quadruplex units [127]. Additionally, the (TTAGGG)n n=[3,7,11,15] sequences, with GGG or TTA flanking sequences added on either or both 3’-end and 5’-end showed no significant change in normalized CD spectra, which suggests that folding of the long telomeric sequences does not depend on flanking sequences [132], unlike single quadruplex units whose conformations and stability are critically impacted by flanking sequences [71]. A caveat of these observations is that CD may not be as sensitive to the effects of flanking sequences at longer lengths, or the effects may be indistinguishable due to the heterogeneity produced by multiple quadruplex subunits.
To summarize, CD spectra of long telomeric sequences provide a discernable signal of multi-quadruplex formation. For some constructs with particular overhangs, the increasing CD signal may not be linearly proportional to the number of GQ, which suggests interactions between GQs and folding frustration may impact this signal. However, the inherent polymorphism of telomeric GQs and their dynamic equilibrium between different conformations makes discerning such effects more complicated at longer telomeric sequences.
UV or CD Thermal Melting
UV or CD Thermal Melting can be used to determine the thermodynamic properties of nucleic acids, such as their melting temperature (Tm). The changes in the enthalpy (ΔH), entropy (ΔS) and free energy (ΔG) when a GQ is destabilized can be determined by analyzing its thermal melting data. In these methods, the temperature dependence of the population or amplitude of a feature, such as a peak or trough that is characteristic of the GQ structure, is used to deduce these thermodynamic properties. Various single stranded telomeric sequences with up to 20 repeats and different overhangs have been studied using these methods, as described below.
The Tm and van’t Hoff enthalpy (ΔHvH) for the (TTAGGG)n [n=8,12] sequences were similar to those of (TTAGGG)4, which suggests two and three contiguous GQ units have a similar thermal stability to that of an isolated quadruplex [110,111]. These are consistent with the long telomeric sequences folding into independent quadruplex units which are arranged as beads-on-string. However, these thermodynamic analyses are mostly performed using van’t Hoff analysis of melting profiles. This model is a good approximation for a two-state transition process whereas the long telomeric sequences have shown more complex behavior [111,127,129] questioning the validity of the two-state model.
In another study, thermodynamic analysis of (TTAGGG)n [n=4–20] showed decrease in stability with increasing number of GQ units (indicated by rise in ΔG at 37 °C) [128]. Similarly, the thermal analysis of telomeric sequences without flanking sequences, GGG(TTAGGG)n where [n=3,7,11,15], showed lower melting temperatures in 100 mM K+ as the length increased [132]. The Tm values were 68, 59, 56, and 54 °C for n=3, 7, 11, and 15, respectively. In case of symmetric TTA flanking sequences on both sides, (TTAGGG)nTTA, the Tm did not change for sequences that can form two or more GQs: Tm=60, 50, 50, and 50 °C for n=4, 8, 12, and 16, respectively. These studies suggest that the stability of telomeric overhangs with physiologically relevant lengths (~2–12 GQs) might show modest variations with telomere length. These studies also concluded the inner GQs to have similar stability to each other while the stabilities of terminal GQs are generally higher but are modulated by flanking TTA sequences, which tend to decrease their stability. Another important study in this context is that of Carrino et al. [133], which will be discussed later due to its implications about folding cooperativity and frustration.
Differential Scanning Calorimetry (DSC)
Differential Scanning Calorimetry (DSC) is an alternative model independent method to extract the thermodynamic parameters of long telomeric sequences [127]. Also, the deconvolution of DSC thermograms using statistical mechanics can characterize the multiple populations present in a sample, such as fraction of folded, unfolded and intermediate structures at any temperature. DSC thermograms of (TTAGGG)n and (TTAGGG)nTT [n=4,8,12] showed similar behavior where species corresponding to 1, 2, and 3 contiguous quadruplex units were observed for n=4, 8, and 12, respectively [127]. To quantify the interactions between adjacent quadruplex units, a coupling free-energy ΔGCoupling was introduced. The ΔGCoupling is defined as ΔGCoupling = ΔGTotal ―mΔGGQ, where ΔGTotal is the total folding free-energy change of multi-quadruplex units, ΔGGQ is the folding free energy change of single GQ unit, and m is the number of GQ units [127]. It was observed that ΔGCoupling > 0 for (TTAGGG)n, n=[8,12,16,20] which suggests destabilizing interactions between GQ units. These destabilizing interactions become stronger at longer lengths, e.g. ΔGCoupling = 13.8 kJ/mol for n=12 while ΔGCoupling ≈ 2 kJ/mol for n=8 [127,128]. The source of such interactions is not known; however, hydration analysis on (TTAGGG)n [n=8,12,16,20] sequences revealed that TTA loops between GQ units are likely more hydrated compared to the core of quadruplex unit. It is argued that hydration of TTA loops leads to a more ordered structure of these loops resulting in destabilizing interactions between quadruplex units [128]. This significant observation suggests that presence of larger unfavorable coupling free energies may limit the complete folding of the overhang, resulting in unfolded regions that could potentially aid or initiate protein binding.
Native (non-denaturing) polyacrylamide gel electrophoresis (PAGE)
Native (non-denaturing) polyacrylamide gel electrophoresis (PAGE) is extensively used to characterize biomolecules and their interactions by monitoring their migration within the electrophoretic gel. Under nondenaturing conditions, the molecules migrate with different mobilities depending on their structure, shape, and molecular size [109]. GQ forming sequences create a globular structure and migrate faster compared to unfolded single stranded nucleic acids. In addition, longer strands with greater number of telomeric repeats migrate slower than shorter strands containing smaller number of repeats [110]. Electrophoretic separation can provide information about the molecularity of GQs and the presence of multimer conformers in long telomeric sequences. For example, while the (TTTTGGGG)n, n=[4–12] series revealed two populations for each length (representing intermolecular and intramolecular GQs) with different mobilities in K+ buffer, human telomeric sequences of similar repeat number showed only a single band for each length (corresponding to intramolecular GQs), in both K+ and Na+ buffers [110].
The migration profile of (TTAGGG)n, n=[8,12,16,20] series showed similar behavior in dilute and crowding conditions suggesting that their shape and molecularity do not change in these conditions [128]. However, the strands migrated faster in Na+ compared to that in K+ suggesting more compact structure in Na+. It is possible that these different mobilities for longer sequences in Na+ vs. in K+ are due to different interactions between GQs, which impact the compactness of the overall structure. These results highlight that migration mobilities depend on the multimer structure and PAGE could be used in discerning major changes in shape and molecularity; however, determining low stability superstructures or the folding topologies are challenging due to modest resolution of the method.
A complementary technique to native PAGE, called temperature-gradient gel electrophoresis (TGGE), is used to discriminate against quadruplex polymorphism, to distinguish between conformers and to evaluate the abundant conformers in the sample. The temperature gradient allows for monitoring of the melting profiles and provide information about the thermal stability of conformers [134]. TGGE analysis of (TTAGGG)n, n=[4–12] revealed higher stability for the n=12 sequence (up to three consecutive GQs) compared to other sequences that could form one or two GQs. Similarly, sequences that could form two GQs (n=8–11) showed higher stability compared to those that can form one GQ (n=4–7). These observations imply stabilizing interactions between GQs.
Integrated Structural Biology (ISB)
Integrated Structural Biology (ISB) approach uses a set of biophysical methods integrated with computational tools to investigate the complex assembly of higher-order quadruplex structures [135]. In addition to other techniques, this collective effort combines small-angle X-ray scattering (SAXS), circular dichroism (CD), differential scanning calorimetry (DSC), analytical ultracentrifugation (AUC), size-exclusion column chromatography and molecular dynamics simulations to characterize the higher-order quadruplex units, their interactions and possible conformations adopted by long telomeric sequences.
The ISB workflow (Figure 6) involves the examination of potential sequences for non-quadruplex secondary structures and conducting biophysical and computational studies simultaneously, while using high-resolution structural information to inform and constrain the models. To illustrate, the program HYDROPRO [136] is used to compute the hydrodynamic properties of GQ structures which are then compared with experimental values obtained from AUC. In particular, the sedimentation velocity obtained from AUC is directly compared with the values predicted by the model. For example, the structural models for (TTAGGG)8 and (TTAGGG)12 (with and without TT flanking sequence at the 3’-end) sequences forming two and three contiguous quadruplex units, respectively, suggest a hybrid type conformation containing hybrid-1 and hybrid-2 structures, which are consistent with sedimentation coefficients measured experimentally [127]. Modeling these structures as GQs with parallel topology results in significant discrepancies between the models and experimental values [127]. Using the ISB approach, a library of monomeric and higher order quadruplex structures have been characterized which cover a diverse set of structures [135]. Detailed structural investigation of (TTAGGG)n revealed that multimeric quadruplex formation maximizes the use of G-repeats leaving no gaps between adjacent quadruplex subunits (other than TTA loops), with transient interactions at the interfaces between individual quadruplex units [129].
Figure 6.
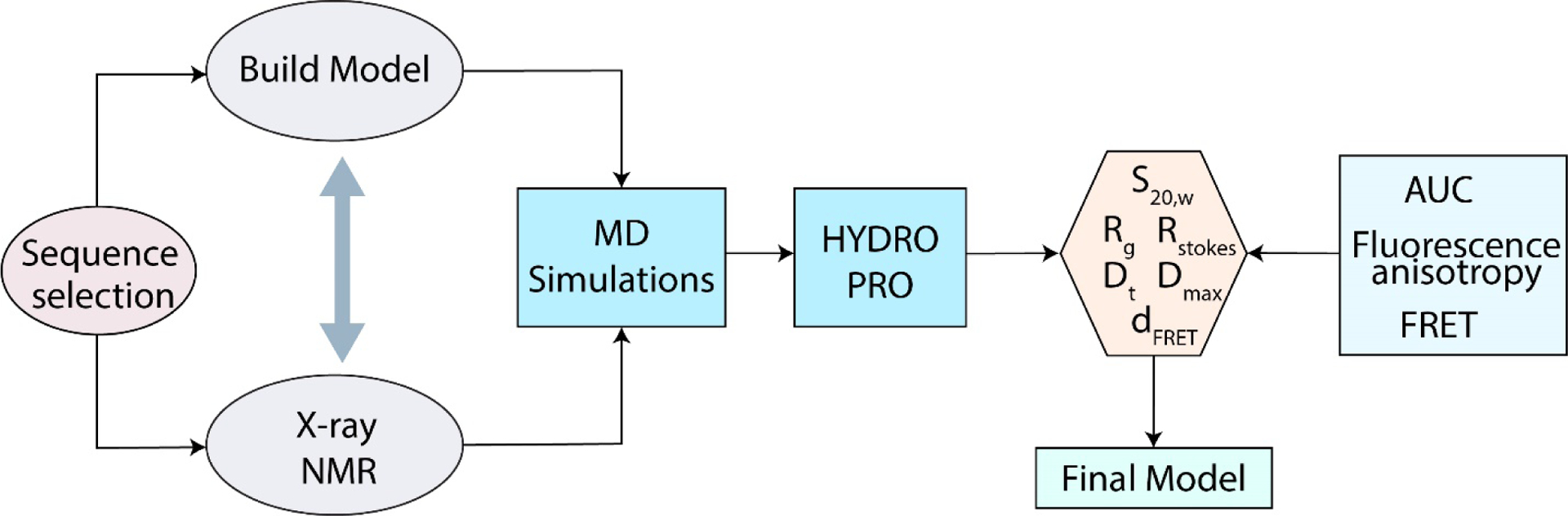
Workflow of Integrated Structural Biology approach where data from multiple experimental approaches are used to refine the computational models GQ structures.
Single molecule methods
Single molecule methods provide a set of potent tools to detect minority populations, dynamics, and weak interactions, which are often inaccessible to the ensemble methods described above. These techniques have made significant contributions to our understanding of biological processes at the nanoscale level, and long telomeric overhangs have recently become accessible for these techniques. Below, we will summarize findings of several prominent single molecule methods, before discussing modeling approaches.
Atomic Force Microscopy (AFM)
Atomic Force Microscopy (AFM) has emerged as a powerful single molecule tool to investigate the longer telomeric sequences and their interactions with proteins. The first AFM experiment on long telomeric sequences used (TTAGGG)16 sequence as a substrate to demonstrate the formation of multiple quadruplex structures in 100 mM K+. In these studies, it was possible to visualize four distinct blobs (each of 2–3 nm height, consistent with 2.4 nm diameter of G-quartets) corresponding to four consecutive quadruplex structures connected with TTA loops in this 16-repeat telomeric sequence [137].
AFM images and analysis on (TTAGGG)16 sequence, which was flanked by a dsDNA at the 5’-side, did not achieve maximum folding in 150 mM K+ [138]. The majority of (TTAGGG)16 sequences only had two distinct peaks (bright blobs of 1.32±0.22 nm height) that could be attributed to two GQs, about 1% of the population showed three distinct peaks while none showed four distinct peaks. The flanking dsDNA (32-bp), which is not present in any of the bulk measurements described above, might have prevented achieving maximal folding as flanking dsDNA is known to destabilize the GQ [139,140]. Unlike telomeric sequence which showed distinct peaks, the AFM images of G-wires (intermolecular GQs connected with each other through overlapping short strands, which by design form a long array of stacked GQs) showed a smooth surface without distinct peaks [138]. This suggested the telomeric overhang resembles beads-on-string with the GQs separated from each other by unfolded regions, and that GQs do not form stable stacking interactions. In addition, the impact of POT1 on folding of GQ was studied for the same construct with the (TTAGGG)16 overhang, where POT1 and GQs coexisting on the same DNA construct could be distinguished based on their peak height and occupied volume. The fraction of molecules that showed a folded GQ structure drastically dropped from 100% to 24% when the telomeric DNA was mixed with 200 nM POT1 [138]. This was interpreted as POT1 disrupting the GQ structures by sterically impairing the adjacent telomeric repeats from folding into GQ structures. Whether POT1 is as effective in unfolding GQ structures in a physiological setting where it is about an order of magnitude less concentrated is not clear. Finally, despite their impressive resolution, the AFM images could not reveal the specific conformations of individual quadruplex units.
Optical Tweezers (OT)
Optical Tweezers (OT) is another single molecule assay (Figure 7) which was employed to investigate the unfolding characteristics of GQ units folded in long telomeric sequences in 100 mM K+ [113,141]. Force-extension analysis revealed 1, 2 and 3 consecutive GQ units folded in 4, 8 and 12 repeat telomere sequences, respectively, in 100 mM K+. Further analysis of a longer construct containing 24 repeats of TTAGG sequence showed that only 5% of the folded GQs are interacting via stabilizing stacking interactions, while 95% of GQs are non-interacting, which might be consistent with the partial stacking scenario illustrated in Figure 4 [113]. Shorter sequences 4 and 8 repeats showed no evidence for stacking interactions (consistent with beads on a string), while the 12-repeat sequence showed less than 5% of folded GQs interacted. A subsequent study from the same group reported GQs randomly fold along the long telomeric sequences [(TTAGGG)4–48] and may have single stranded gaps between them. In addition, free energy analysis revealed that the folding is governed by kinetic pathways, rather than thermodynamics which require the maximal folding of G-repeats [141].
Figure 7.
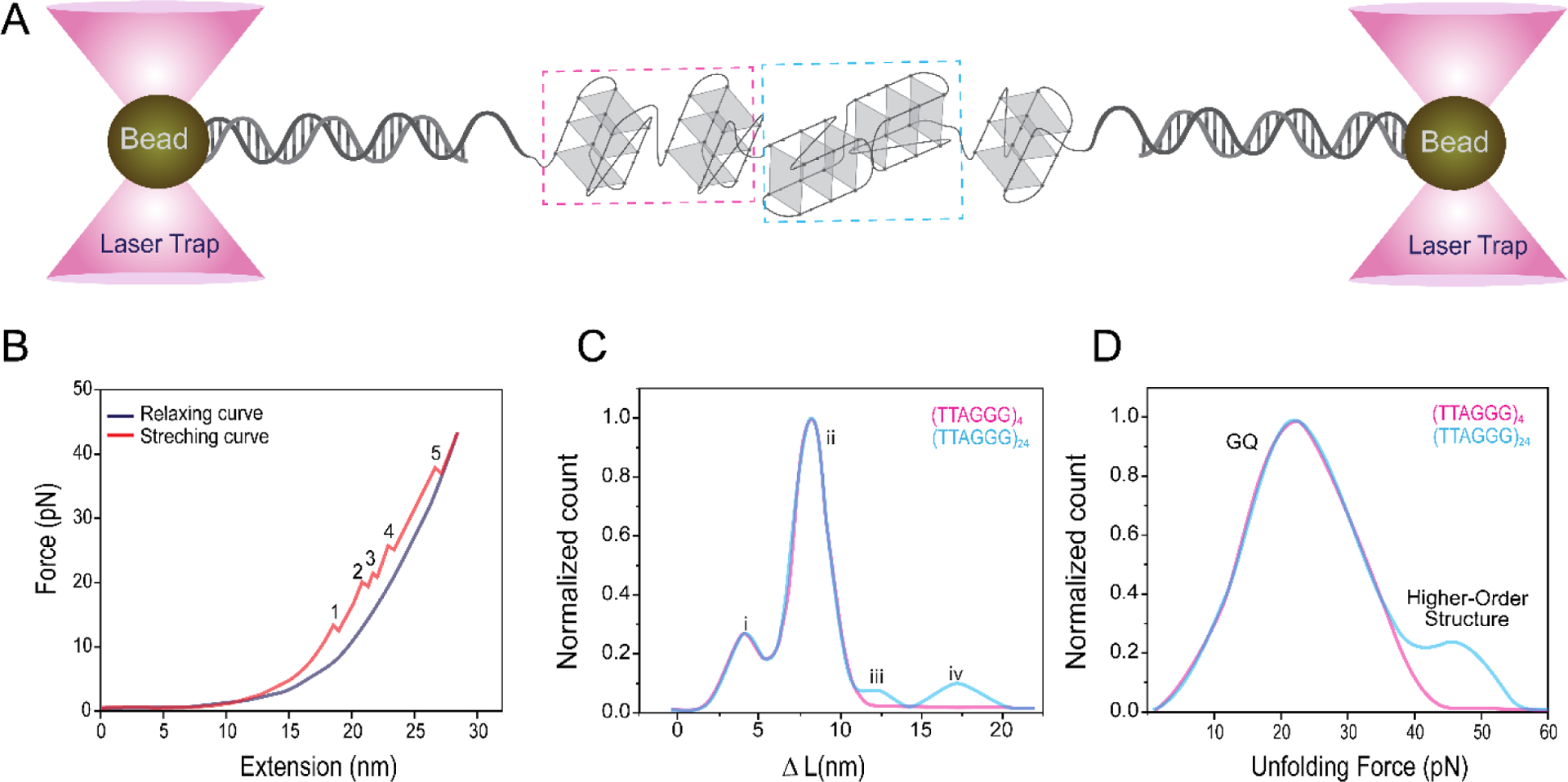
Schematic illustration of optical tweezers assay. (A) Glass beads of typically micrometer diameter are attached to dsDNA handles which are connected to the DNA construct of interest, in this case an ssDNA that can form five GQs. (B) Force extension assay. Increasing the separation between the beads (extension) via the trapping laser results in an increased force that is applied on the GQs, which unfold at large enough forces. Each of the regions marked by numbers 1–5, where an abrupt drop in force is observed, represent unfolding of a GQ. (C-D) For longer telomeric overhangs (such as that containing 24 repeats), it is possible to identify what fraction of GQ molecules interact with each other and achieve a higher order structure by comparing the change in contour length (ΔL) and unfolding forces with those of a similar data on a single GQ. The four marked populations in (C) represent G-triplex, single GQ, GQ with long loops, and higher order structures, respectively. Figure adapted from reference [141].
Electron Microscopy (EM)
Electron Microscopy (EM) is another prominent single molecule approach that provides sub nanometer resolution, which is required to study structural characteristics of telomeric overhangs. EM was used to demonstrate the presence of stacking interactions, including multiple quadruplex structure formation, and unfolded regions between GQs in 100 mM K+ on a 20 kb long single stranded telomeric sequence, much longer than physiological telomeric overhangs [112]. EM data revealed two different bead sizes composed of tetramer or octamer GQ structures in 100 mM K+. The tetramers (octamers) were formed by 20 (40) telomeric repeats where four (eight) repeats were unfolded within the unit. Combined with single-molecule magnetic tweezer data, it was further proposed that unfolded single stranded regions orchestrate the interactions between quadruplex units resulting in higher-order structures that compact the telomeric DNA by 12-fold [112]. The EM images clearly show that stabilizing stacking interactions are possible between neighboring GQs under certain conditions; however, the telomeric overhangs are considerably shorter than the strands used in this study, and it is not clear whether such features are reflective of physiological telomeres or require the presence of large number of tandem GQs. Another exciting development in this field is the recently published cryo-EM structure (with 7.4 Å resolution) on a non-telomeric GQ that was embedded within a double stranded DNA [142]. Whether such studies could be successfully extended to GQs in telomeric overhangs remains to be seen given the challenges associated with the significant flexibility in the structure (which stem from TTA linkers between consecutive GQs or unfolded regions within the overhang) and the potential folding heterogeneity when telomeric sequences are long enough to accommodate several GQs.
Single-molecule Förster resonance energy transfer (smFRET)
Single-molecule Förster resonance energy transfer (smFRET) has been extensively used to study GQs formed by short telomeric sequences (eight telomeric repeats or less), their conformations [143–145], their folding and unfolding kinetics, and their interactions with small molecules and proteins [146–151]. In addition, single-molecule fluorescence-force assay has shown that human telomeric GQs exhibit six distinct conformations in K+ and Na+ solutions [152]. However, it has been challenging to extend such methods to study telomeric sequences of physiologically relevant lengths due to multiplicity of possible structures and the different topologies they adapt [153].
Until recently, the cost and feasibility of synthesizing long telomeric sequences with fluorescent markers had made it impractical to study such sequences using these established single molecule assays. Our lab has recently adopted FRET-PAINT [154], which combines smFRET with the super-resolution technique DNA-PAINT (Point Accumulation for Imaging in Nanoscale Topography), to overcome some of these challenges [155]. We have primary used this technique to investigate the accessibility of telomeric overhangs in presence of physiologically relevant factors and to deduce insights about the impact of such factors on accessibility, stability, and folding patterns of the overhang. A major distinction of our approach compared to earlier studies is that we directly probe accessibility, rather than study the stability, kinetics, or folding topologies and project the implications of these studies on telomere accessibility. In addition, this approach has shown significant promise to be extended to more complex settings which include shelterin proteins, nucleases, DNA binding proteins, TERRA, telomerase, and small molecules.
FRET-PAINT
FRET-PAINT takes advantage of transient bindings of a fluorescently labelled small probe which is complementary to the telomeric sequence, such as 6–9 nt long DNA, peptide nucleic acid (PNA) or locked nucleic acid (LNA) strand (Figure 8A–B). An ideal probe should be as short as possible for maximum resolution to distinguish between different G-tracts, bind to ssTEL for a long enough time (a few seconds) to be distinguished from non-specific binding events (shorter than 1 s) but not too long to overlap with other binding events, should cause minimal disturbance to the folded GQ structures, and have minimal interaction with proteins under investigation. We identified a Cy5-labeled PNA strand which is complementary to the TTAGGGT sequence based on its binding characteristics and negligible probe induced unfolding of GQ structure, although a systematic effort to identify the optimal probe has not been performed yet. The validity and sensitivity of the assay to investigate longer telomeric overhangs is established by studying the accessibility of telomeric sequences as long as 28 repeats, which can form up to seven GQ units [140,156,157]. Significantly, DNA constructs that consist of dsDNA on the 5’side and a free ssTEL at the 3’ side were used in these studies, which better mimic the physiological telomere.
Figure 8.
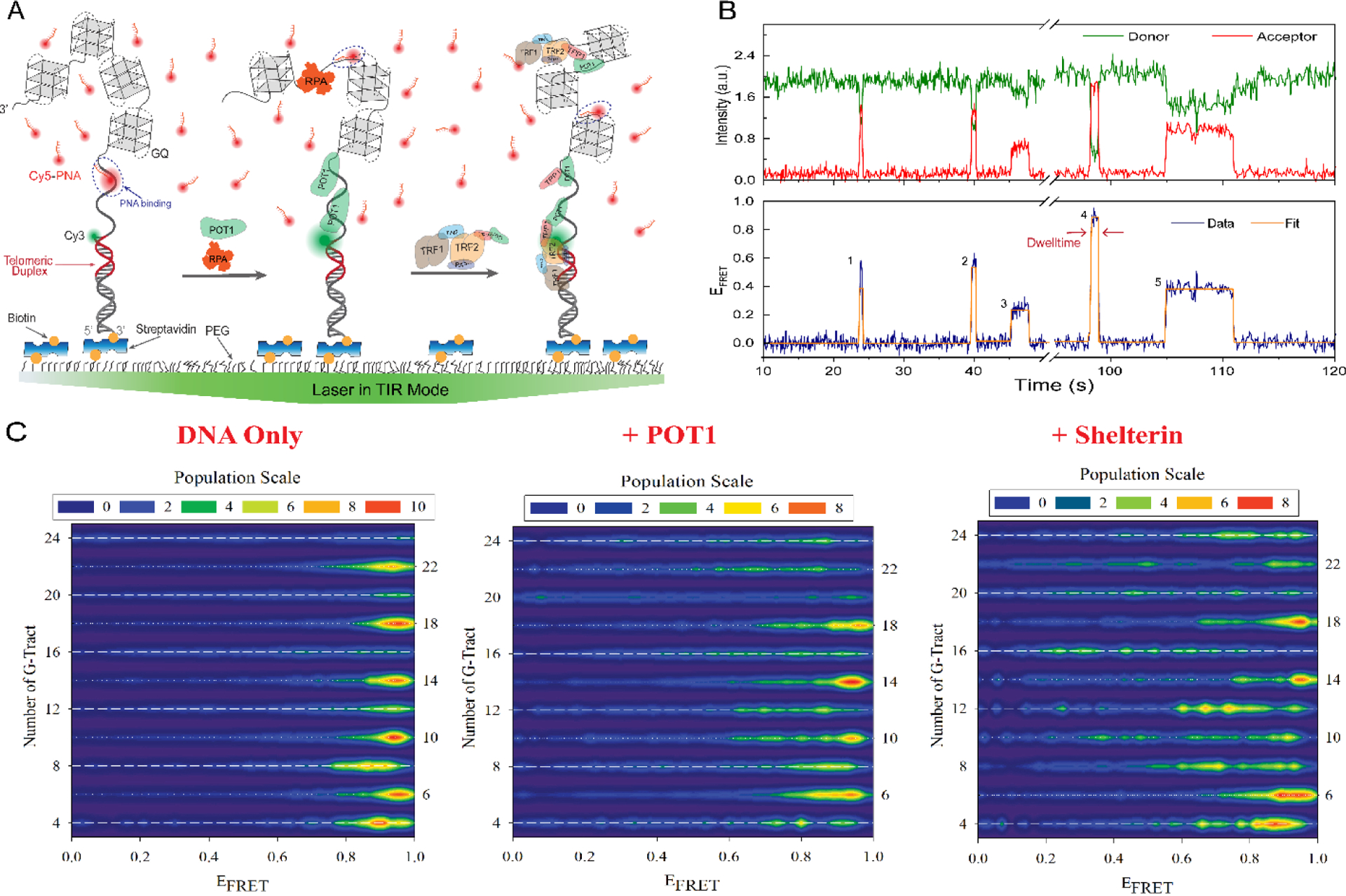
Schematic illustration of FRET-PAINT assay. A) Partial duplex DNA constructs containing dsTEL (red duplex) and ssTEL (overhang that forms multiple GQs) are immobilized on the surface. DNA-only case (in the absence of proteins, left), DNA with POT1 (middle) or with shelterin (right) are shown. The shelterin is a four-component construct with POT1-TPP1-PIN2-TRF1. B) A single molecule trace showing five binding events marked with numbers 1–5. C) Accessibility patterns for telomeric DNA constructs containing 4–24 GGGTTA repeats (as in A), obtained using FRET-PAINT, in the absence of proteins (‘DNA Only’, left) or in the presence of POT1 (middle) or shelterin (right). The bright red regions at high FRET indicate higher accessibility of the vicinity of ssTEL/dsTEL junction. In the absence of proteins, alternating narrow (dominated by a bright red population at high FRET) and broad (yellow-green regions throughout the FRET range) accessibility patterns are observed for 4n+2 and 4n constructs, respectively. POT1 and shelterin generally convert the narrow red distributions to broader green-yellow distributions, suggesting better protection of the junction and more uniform accessibility. Figure adapted from reference [157].
The FRET-PAINT data showed onsetting of periodic accessibility patterns for ssTEL with 10 or more TTAGGG repeats (Figure 8C). These patterns, which are influenced by the underlying GQ folding characteristics and interactions between the quadruplex units, revealed a frustrated folding landscape and varying folding stabilities in different regions of the overhang. Broader distributions of accessible sites along the ssTEL for overhangs with integral multiple of four repeats (4n) were interpreted as evidence for higher folding frustration. Sequences with extra repeats (particularly the 4n+2 constructs with two additional repeats) displayed narrower distributions where the accessible sites were concentrated at the vicinity of ssTEL/dsTEL junction and were interpreted to illustrate lower folding frustration [140,156].
These observations were supported by complementary computational studies that revealed stabilizing interactions between adjacent quadruplex units (positive cooperativity) and lower probability of GQ formation at the ssTEL/dsTEL junction [140]. An earlier study that combined systematic UV thermal melting studies in 110 mM K+ with Monte-Carlo simulations on long single stranded telomeres (without a dsTEL on one side) reported similar patterns in folding probability and proposed the folding frustration to be length dependent. These patterns were explained in terms of combinatorial factors, which influence the probability of a particular G-repeat to be part of a GQ, folding stabilities in different regions of the overhang, and end-effects. This study concluded that interactions between quadruplex units are destabilizing (negative cooperativity) [133]. The difference in DNA constructs might be a reason for the difference in the conclusions of this study from the FRET-PAINT study.
The FRET-PAINT approach was recently extended to include shelterin proteins, which significantly reduced ssTEL accessibility (2.5-fold by POT1 and 5-fold by shelterin) [157]. Furthermore, the asymmetry in the accessibility of different regions of ssTEL was diminished in the presence of shelterin, particularly for the junction region (Figure 8C). These observations suggest shelterin restructures the junction region between ssTEL and dsTEL, which reduces its accessibility and helps create a robust protective cap at the chromosome ends. This approach has exciting potential to understand how telomeric structures and associated protein complexes regulate access (including that of telomerase) to these critical genomic regions and protect it against DNA processing enzymes and nucleases.
Modeling Approaches:
The basic building blocks of modeling approaches to investigate higher order structures have been the NMR or crystal structures of the four-repeat human telomeric sequence. An early model for a multiple quadruplex structure was based on the parallel topology, which was observed in X-ray crystallography, and individual quadruplexes were connected to each other by TTA loops [120]. Such an arrangement created a highly stable higher order extended GQ stack where the terminal G-tetrad of each quadruplex subunit was stacked with the initial G-tetrad of subsequent quadruplex [120,127,158]. The stacking interactions and structural stability increased with the number of GQ subunits (between 1–4 GQs), whereas the loops remained the most flexible parts of the model without affecting the overall structure. Such extensive stacking interactions forming higher-order structures might be plausible for parallel GQs; however, there is extensive experimental evidence that human telomeric GQ attains a hybrid topology (hydrid-1 and hybrid-2) at physiological K+ concentration (although not at physiologically relevant crowding conditions), which should be considered for its potential to form higher order structures [119,129,135,158].
The HYDROPRO program was used to model higher-order quadruplex structures based on hybrid topologies, which resulted in a range of stacking interaction between GQ subunits [119,158]. For example, four different models were created for the (TTAGGG)8TT sequence, which can fold into two contiguous GQ units, each of which could be in hybrid-1 or hybrid-2 topology. These models resulted in different stacking and stability of the higher-order structure [158]. The combination of hybrid-1 and hybrid-2 (hybrid-12) model was proposed as a plausible model for the (TTAGGG)8TT sequence, where the interface of hybrid-1 and hybrid-2 GQs is stabilized by specific interactions and the overall compactness of the structure matched with sedimentation coefficient observed in K+ solution. This was also supported by the analysis of solvent accessibility surface area (SASA) of adenine bases localized at the junction between quadruplex units in hybrid-12 topology. The adenine bases localized in specific loops have different surroundings depending on the folding topology of quadruplex subunits. These adenine bases were systematically replaced with 2-aminopurine bases (2-AP) because of their enhanced sensitivity to local environment [158]. The SASA is experimentally measured by monitoring the photophysical properties of 2-AP bases. However, the interactions between quadruplex subunits mediated by loops are reported to be small and are lost before the higher order structure melts [119], which might limit the applicability of SASA to longer sequences.
Using a similar approach, both hybrid-121 model (hybrid-1 topology for terminal GQs and hybrid-2 for the middle GQ) [127] and hybrid-212 (hybrid-2 topology for terminal GQs and hybrid-1 for the middle GQ) [129] were proposed for the (TTAGGG)12 sequence. For longer sequences consisting of four consecutive quadruplex subunits, molecular dynamics (MD) simulations reported many hybrid structures but hybrid-2122 was found to be a representative model based on light scattering data and radius of gyration calculations. These studies reported very similar free energy barriers between different hybrid-hybrid junctions; however, the hybrid-2 topology at 5’-end was preferred to maximize folding of G-repeats into GQs [129,135]. Despite variations in some details, these studies suggest hybrid-type topology with stacking interactions between neighboring GQs and close to maximal folding are robust features of long telomeric sequences.
Perspectives:
As this review aims to illustrate, many questions regarding the structure and interactions of telomeric overhangs of physiological length remain open, with different studies supporting one picture or another. Variations in the DNA constructs, environmental conditions, sample preparation protocols, resolution of the methods and whether they are sensitive to minority populations, transient or weak interactions, and inherent heterogeneities in the system are among the likely sources of contrasting results in different studies. However, based on our current understanding, the following evaluation can be made with moderate confidence. In the absence of associated proteins, telomeric GQs are stable under physiological ionic conditions and the telomeric overhangs attain near maximal folding with occasional unfolded repeats separating the GQs. Beads on a string type model is supported by many studies; however, whether these beads or a subset of them stably or transiently interact with each other via stabilizing, neutral, or destabilizing interactions has not been established yet. The somewhat loose terminology in the literature about what level of interaction makes the model other than “beads on a string” contributes to the controversy. Furthermore, GQs with different topologies likely interact differently with their neighbors. GQs with parallel topology (partially based on prominent demonstrations in non-telomeric GQs with parallel topology) are generally accepted to have higher propensity to form stabilizing interactions with each other compared to GQs with anti-parallel topology. In the case of hybrid topologies, the nature of their interactions might be impacted with the details of how the strand is arranged, such as whether it enters and exits the GQ in the same or opposite directions. The contrasting results about the nature of interactions in different studies could be due to the GQs attaining different topologies in these studies, which could be a result of different ionic conditions, different levels of molecular crowding or DNA concentrations, or different protocols to prepare the samples, and whether the samples reach a thermodynamic equilibrium or are trapped in a kinetically favorable state.
Even though the impact of overhangs on a single GQ is well-established, their effects on longer sequences that can form several GQs is not clear, which might indicate a weaker effect. However, flanking double stranded DNA is generally accepted to destabilize the neighboring GQ. The impact of TTA flanking sequences, compared to blunt ends, on GQ stability might be more modest. However, such variations in design might be a cause for different conclusions about whether the GQs at the 3′-end (free end in the cell), 5′-end (neighbor of dsTEL), or in the middle are more stable compared to others. Nevertheless, there is strong evidence for the GQs away from the edges to have similar stability to each other. Most studies demonstrate GQs at the 3′ end to be more stable than those in the middle, although the probability of the G-repeats in this end to be protected in a GQ is diminished due to combinatorial effects, i.e., there are a smaller number of GQs in which such repeats could be part of compared to the internal repeats. Similar combinatorial disadvantages are valid for the telomeric repeats at the 5′-end, which results in significantly less stability when combined with the destabilizing effects of neighboring dsTEL. This less stable region is the junction between ssTEL and dsTEL which is a likely location for a protein-bridge between POT1/TPP1 and TRF1(or TRF2)/TIN2. Therefore, shelterin might provide a physiological cover for this otherwise exposed region.
Small molecules targeting telomeric overhangs have been actively investigated as potential anti-cancer therapeutics and will continue to be an active research field in the near future. However, it is clear that molecules that only target the planar surface of the G-tetrads do not provide adequate specificity and more elaborate designs that add elements to target the loops, grooves, or the backbone are required. In the particular case of telomeric overhangs, the availability of multiple GQs in close proximity provides opportunities for multimeric small molecules which are promising candidates for enhanced specificity.
In the near future, the in vitro investigations on long telomeric overhangs, which provide the required control to attain mechanistic information, will continue to increase in sophistication and will better mimic the cellular complexity. There is already some work where shelterin proteins, small molecules, or crowding agents are introduced in the assays; and it is feasible to extend these to include telomerase, TERRA, DDR activators, and nucleases in the near future. The significance of telomeres in cancer and aging will continue to drive advances in terms of techniques, particularly approaches which combine multiple methods, high-resolution structural investigations, new single molecule methodologies that are adapted for the study of long telomeric sequences, and more sophisticated computational model; all of which will make the next decade very exciting for this important field.
Research Highlights.
Review of structural features of telomeric overhangs
Review of impact of telomeric sequence, liquid-liquid phase separation, and chromatin structure on telomere structure and function
Review of small molecules that stabilize G-quadruplexes
Review of ensemble and single molecule studies on telomeric overhangs of physiologically relevant lengths to probe the structure of tandem G-quadruplexes and potential interactions between them.
Acknowledgements:
This work was funded by NIH [1R15GM123443, 1R15GM146180 to H.B].
Abbreviations:
- ssTEL
Single Stranded Telomere
- dsTEL
Double Stranded Telomere
- GQ
G-quadruplex
- TERRA
Telomeric repeat containing RNA
- ISB
Integrated Structural Biology
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Blackburn EH, B. E.H., Structure and function of telomeres, Nature. 350 (1991) 569–573. 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- [2].De Lange T, How telomeres solve the end-protection problem, Science. 326 (2009) 948–952. 10.1126/SCIENCE.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Smith EM, Pendlebury DF, Nandakumar J, Structural biology of telomeres and telomerase, Cell. Mol. Life Sci 77 (2020) 61–79. 10.1007/s00018-019-03369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hänsel-Hertsch R, Di Antonio M, Balasubramanian S, DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential, Nat. Rev. Mol. Cell Biol 18 (2017) 279–284. 10.1038/NRM.2017.3. [DOI] [PubMed] [Google Scholar]
- [5].Biffi G, Tannahill D, McCafferty J, Balasubramanian S, Quantitative visualization of DNA G-quadruplex structures in human cells, Nat. Chem 5 (2013) 182–186. 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Di Antonio M, Ponjavic A, Radzevičius A, Ranasinghe RT, Catalano M, Zhang X, Shen J, Needham LM, Lee SF, Klenerman D, Balasubramanian S, Single-molecule visualization of DNA G-quadruplex formation in live cells, Nat. Chem 12 (2020) 832–837. 10.1038/s41557-020-0506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee WTC, Yin Y, Morten MJ, Tonzi P, Gwo PP, Odermatt DC, Modesti M, Cantor SB, Gari K, Huang TT, Rothenberg E, Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling, Nat. Commun 12 (2021). 10.1038/S41467-021-22830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bryan TM, G-Quadruplexes at Telomeres: Friend or Foe?, Molecules. 25 (2020). 10.3390/MOLECULES25163686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, De Lange T, Mammalian telomeres end in a large duplex loop, Cell. 97 (1999) 503–514. 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- [10].De Lange T, A loopy view of telomere evolution, Front. Genet 6 (2015) 321. 10.3389/FGENE.2015.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tomaska L, Nosek J, Kar A, Willcox S, Griffith JD, A New View of the T-Loop Junction: Implications for Self-Primed Telomere Extension, Expansion of Disease-Related Nucleotide Repeat Blocks, and Telomere Evolution, Front. Genet 10 (2019) 792. https://doi.org/ 10.3389/fgene.2019.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Greider CW, Blackburn EH, Identification of a specific telomere terminal transferase activity in tetrahymena extracts, Cell. 43 (1985) 405–413. 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- [13].Blackburn EH, Greider CW, Henderson E, Lee MS, Shampay J, Shippen-Lentz D, Recognition and elongation of telomeres by telomerase, in: Genome, 1989: pp. 553–560. 10.1139/g89-104. [DOI] [PubMed] [Google Scholar]
- [14].De Lange T, Shelterin-mediated telomere protection, Annual Reviews Inc, 2018. 10.1146/annurev-genet-032918-021921. [DOI] [PubMed] [Google Scholar]
- [15].Lyu X, Sang PB, Chai W, CST in maintaining genome stability: Beyond telomeres, DNA Repair (Amst). 102 (2021). 10.1016/j.dnarep.2021.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lim CJ, Cech TR, Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization, Nature Reviews Molecular Cell Biology, 2021. 10.1038/S41580-021-00328-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jack A, Kim Y, Strom AR, Lee DSW, Williams B, Schaub JM, Kellogg EH, Finkelstein IJ, Ferro LS, Yildiz A, Brangwynne CP, Compartmentalization of telomeres through DNA-scaffolded phase separation, Dev. Cell 57 (2022) 277–290.e9. 10.1016/J.DEVCEL.2021.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fulnečková J, Ševčíková T, Fajkus J, Lukešová A, Lukeš M, Vlček Č, Lang BF, Kim E, Eliáš M, Sýkorová E, A Broad Phylogenetic Survey Unveils the Diversity and Evolution of Telomeres in Eukaryotes, Genome Biol. Evol 5 (2013) 468–483. 10.1093/GBE/EVT019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mendez-bermudez A, Hills M, Pickett HA, Phan AT, Mergny JL, Riou JF, Royle NJ, Human telomeres that contain (CTAGGG)n repeats show replication dependent instability in somatic cells and the male germline, Nucleic Acids Res. 37 (2009) 6225–6238. 10.1093/nar/gkp629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chatain J, Blond A, Phan AT, Saintomé C, Alberti P, GGGCTA repeats can fold into hairpins poorly unfolded by replication protein A: a possible origin of the length-dependent instability of GGGCTA variant repeats in human telomeres, Nucleic Acids Res. 49 (2021) 7588–7601. 10.1093/NAR/GKAB518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lancrey A, Safa L, Chatain J, Delagoutte E, Riou JF, Alberti P, Saintomé C, The binding efficiency of RPA to telomeric G-strands folded into contiguous G-quadruplexes is independent of the number of G4 units, Biochimie. 146 (2018) 68–72. 10.1016/j.biochi.2017.11.017. [DOI] [PubMed] [Google Scholar]
- [22].Qureshi MH, Ray S, Sewell AL, Basu S, Balci H, Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency, J. Phys. Chem. B 116 (2012) 5588–5594. 10.1021/jp300546u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Audry J, Maestroni L, Delagoutte E, Gauthier T, Nakamura TM, Gachet Y, Saintomé C, Géli V, Coulon S, RPA prevents G-rich structure formation at lagging‐strand telomeres to allow maintenance of chromosome ends , EMBO J. 34 (2015) 1942–1958. 10.15252/embj.201490773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chatterjee S, Zagelbaum J, Savitsky P, Sturzenegger A, Huttner D, Janscak P, Hickson ID, Gileadi O, Rothenberg E, Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures, Nat. Commun 2014 51. 5 (2014) 1–12. 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Budhathoki JB, Stafford EJ, Yodh JG, Balci H, ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity, Nucleic Acids Res. 43 (2015) 5961–5970. 10.1093/nar/gkv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Azzalin CM, Lingner J, Telomere functions grounding on TERRA firma, Trends Cell Biol. 25 (2015) 29–36. 10.1016/j.tcb.2014.08.007. [DOI] [PubMed] [Google Scholar]
- [27].De Lange T, T-loops and the origin of telomeres, Nat. Rev. Mol. Cell Biol 5 (2004) 323–329. 10.1038/NRM1359. [DOI] [PubMed] [Google Scholar]
- [28].Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J, Telomeric repeat-containing RNA and RNA surveillance factors at mammalian chromosome ends, Science. 318 (2007) 798–801. 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- [29].Schoeftner S, Blasco MA, Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II, Nat. Cell Biol 10 (2008) 228–236. 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- [30].Redon S, Reichenbach P, Lingner J, The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase, Nucleic Acids Res. 38 (2010) 5797–5806. 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lalonde M, Chartrand P, TERRA, a Multifaceted Regulator of Telomerase Activity at Telomeres, J. Mol. Biol 432 (2020) 4232–4243. 10.1016/j.jmb.2020.02.004. [DOI] [PubMed] [Google Scholar]
- [32].Collie GW, Haider SM, Neidle S, Parkinson GN, A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex, Nucleic Acids Res. 38 (2010) 5569–5580. 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xu Y, Suzuki Y, Ito K, Komiyama M, Telomeric repeat-containing RNA structure in living cells, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 14579–14584. 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Phan AT, Human telomeric G-quadruplex: structures of DNA and RNA sequences, FEBS J. 277 (2010) 1107–1117. 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- [35].Arora A, Maiti S, Differential biophysical behavior of human telomeric rna and dna quadruplex, J. Phys. Chem. B 113 (2009). 10.1021/jp810638n. [DOI] [PubMed] [Google Scholar]
- [36].Martadinata H, Phan AT, Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution, J. Am. Chem. Soc 131 (2009). 10.1021/ja806592z. [DOI] [PubMed] [Google Scholar]
- [37].Randall A, Griffith JD, Structure of long telomeric RNA transcripts: The G-rich RNA forms a compact repeating structure containing G-quartets, J. Biol. Chem 284 (2009) 13980–13986. 10.1074/jbc.M900631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Martadinata H, Heddi B, Lim KW, Phan AT, Structure of long human telomeric RNA (TERRA): G-quadruplexes formed by four and eight UUAGGG repeats are stable building blocks, Biochemistry. 50 (2011). 10.1021/bi200569f. [DOI] [PubMed] [Google Scholar]
- [39].Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, Youmans DT, Cech TR, Davidovich C, Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines, Mol. Cell 65 (2017) 1056–1067.e5. 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- [40].Takahama K, Takada A, Tada S, Shimizu M, Sayama K, Kurokawa R, Oyoshi T, Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS, Chem. Biol 20 (2013) 341–350. 10.1016/j.chembiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- [41].Biffi G, Tannahill D, Balasubramanian S, An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2, J. Am. Chem. Soc 134 (2012) 11974–11976. 10.1021/JA305734X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kondo K, Mashima T, Oyoshi T, Yagi R, Kurokawa R, Kobayashi N, Nagata T, Katahira M, Plastic roles of phenylalanine and tyrosine residues of TLS/FUS in complex formation with the G-quadruplexes of telomeric DNA and TERRA, Sci. Rep 8 (2018) 1–11. 10.1038/s41598-018-21142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V, Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures, J Am Chem Soc. 132 (2010) 9328–9334. 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- [44].Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T, Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation, Mol. Cell 70 (2018) 1038–1053.e7. 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- [45].Phan AT, Mergny JL, Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix, Nucleic Acids Res. 30 (2002) 4618–4625. 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].De Silanes IL, D’Alcontres MS, Blasco MA, TERRA transcripts are bound by a complex array of RNA-binding proteins, Nat. Commun 1 (2010) 1–10. 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- [47].Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT, Luke B, Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle, Cell. 170 (2017) 72–85.e14. 10.1016/j.cell.2017.06.006. [DOI] [PubMed] [Google Scholar]
- [48].Grunseich C, Wang IX, Watts JA, Burdick JT, Guber RD, Zhu Z, Bruzel A, Lanman T, Chen K, Schindler AB, Edwards N, Ray-Chaudhury A, Yao J, Lehky T, Piszczek G, Crain B, Fischbeck KH, Cheung VG, Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters, Mol. Cell 69 (2018) 426–437.e7. 10.1016/j.molcel.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Castellano-Pozo M, Santos-Pereira J, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, García-Muse T, Aguilera A, R loops are linked to histone H3 S10 phosphorylation and chromatin condensation, Mol. Cell 52 (2013) 583–590. 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- [50].Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ, R-loops induce repressive chromatin marks over mammalian gene terminators, Nature. 516 (2014) 436–439. 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Arnoult N, Van Beneden A, Decottignies A, Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α, Nat. Struct. Mol. Biol 19 (2012) 948–956. 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- [52].Balk B, Dees M, Bender K, Luke B, The differential processing of telomeres in response to increased telomeric transcription and RNA-DNA hybrid accumulation, RNA Biol. 11 (2014) 95–100. 10.4161/rna.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mei Y, Deng Z, Vladimirova O, Gulve N, Johnson FB, Drosopoulos WC, Schildkraut CL, Lieberman PM, TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity, Sci. Reports 2021 111. 11 (2021) 1–14. 10.1038/s41598-021-82406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Flynn R, Centore R, O’Sullivan R, Rai R, Tse A, TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA., Nature. 471 (2011) 532–536. https://doi.org/ 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bettin N, Oss Pegorar C, Cusanelli E, The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability., Cells. 8 (2019) 246. 10.3390/cells8030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW, Telomerase activity in human germline and embryonic tissues and cells., Dev. Genet 18 (1996) 173–179. . [DOI] [PubMed] [Google Scholar]
- [57].Stewart SA, Ben-Porath I, Carey VJ, O’Connor BF, Hahn WC, Weinberg RA, Erosion of the telomeric single-strand overhang at replicative senescence, Nat. Genet 33 (2003) 492–496. 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- [58].Lansdorp P, Telomere Length Regulation., Front. Oncol 12 (2022) 943622. 10.3389/fonc.2022.943622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB, Telomere length predicts replicative capacity of human fibroblasts, Proc. Natl. Acad. Sci. U. S. A 89 (1992) 10114–10118. 10.1073/PNAS.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Herbig U, Jobling WA, Chen BPC, Chen DJ, Sedivy JM, Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a), Mol. Cell 14 (2004) 501–513. 10.1016/S1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- [61].Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW, Specific association of human telomerase activity with immortal cells and cancer, Science. 266 (1994) 2011–2015. 10.1126/SCIENCE.7605428. [DOI] [PubMed] [Google Scholar]
- [62].Armanios M, Blackburn EH, The telomere syndromes., Nat. Rev. Genet 13 (2012) 693–704. 10.1038/NRG3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wong JMY, Collins K, Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita, Genes Dev. 20 (2006) 2848–2858. 10.1101/GAD.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadiñanos J, Horner JW, Maratos-Flier E, Depinho RA, Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice, Nature. 469 (2011) 102–106. 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hübner B, von Otter E, Ahsan B, Wee ML, Henriksson S, Ludwig A, Sandin S, Ultrastructure and nuclear architecture of telomeric chromatin revealed by correlative light and electron microscopy, Nucleic Acids Res. 50 (2022) 5047–5063. 10.1093/NAR/GKAC309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bandaria JN, Qin P, Berk V, Chu S, Yildiz A, Shelterin protects chromosome ends by compacting telomeric chromatin, Cell. 164 (2016) 735–746. 10.1016/j.cell.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev. Mol. Cell Biol 2017 185. 18 (2017) 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hyman AA, Weber CA, Jülicher F, Liquid–liquid phase separation in biology, Annu. Rev. Cell Dev. Biol 30 (2014) 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- [69].Söding J, Zwicker D, Sohrabi-Jahromi S, Boehning M, Kirschbaum J, Mechanisms for Active Regulation of Biomolecular Condensates, Trends Cell Biol. 30 (2020) 4–14. 10.1016/J.TCB.2019.10.006. [DOI] [PubMed] [Google Scholar]
- [70].Palm W, de Lange T, How shelterin protects mammalian telomeres., Annu. Rev. Genet 42 (2008) 301–334. 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- [71].Dai J, Carver M, Yang D, Polymorphism of human telomeric quadruplex structures, Biochimie. 90 (2008). 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S, Quadruplex DNA: sequence, topology and structure, Nucleic Acids Res. 34 (2006) 5402–5415. 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Parkinson GN, Lee MPH, Neidle S, Crystal structure of parallel quadruplexes from human telomeric DNA, Nature. 417 (2002) 876–880. 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- [74].Dai J, Carver M, Punchihewa C, Jones RA, Yang D, Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence, Nucleic Acids Res. 35 (2007). 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D, Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution, Nucleic Acids Res. 34 (2006) 2723–2735. 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wang Y, Patel DJ, Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex, Structure. 1 (1993). 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- [77].Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT, Structure of the human telomere in K + solution: A stable basket-type G-quadruplex with only two G-tetrad layers, J. Am. Chem. Soc 131 (2009). 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou J-F, Mergny J-L, Targeting telomeres and telomerase., Biochimie. 90 (2008) 131–55. 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- [79].Koirala D, Dhakal S, Ashbridge B, Sannohe Y, Rodriguez R, Sugiyama H, Balasubramanian S, Mao H, A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands, Nat Chem. 3 (2011) 782–787. 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH, Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription, Proc Natl Acad Sci U S A. 99 (2002) 11593–11598. 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Neidle S, Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer, FEBS J, 2010. 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- [82].Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH, Inhibition of human telomerase by a G-quadruplex-interactive compound, J Med Chem. 40 (1997) 2113–2116. 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- [83].Rodriguez R, Muller S, Yeoman JA, Trentesaux C, Riou JF, Balasubramanian S, A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres, J Am Chem Soc. 130 (2008) 15758–15759. 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S, The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function, Cancer Res. 65 (2005) 1489–1496. 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- [85].Phatak P, Cookson JC, Dai F, Smith V, Gartenhaus RB, Stevens MFG, Burger AM, Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism, Br. J. Cancer 96 (2007) 1223–1233. 10.1038/sj.bjc.6603691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K, Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways., Oncogene. 22 (2003) 5338–5347. 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- [87].Zell J, Sperti FR, Britton S, Monchaud D, DNA folds threaten genetic stability and can be leveraged for chemotherapy, RSC Chem. Biol 2 (2021) 47–76. 10.1039/d0cb00151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Balasubramanian S, Neidle S, G-quadruplex nucleic acids as therapeutic targets, Curr Opin Chem Biol. 13 (2009) 345–353. 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li Q, Xiang JF, Yang QF, Sun HX, Guan AJ, Tang YL, G4LDB: a database for discovering and studying G-quadruplex ligands, Nucleic Acids Res. 41 (2013) D1115–D1123. 10.1093/NAR/GKS1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wang YH, Yang QF, Lin X, Chen D, Wang ZY, Chen B, Han HY, Di Chen H, Cai KC, Li Q, Yang S, Tang YL, Li F, G4LDB 2.2: a database for discovering and studying G-quadruplex and i-Motif ligands, Nucleic Acids Res. 50 (2022) D150–D160. 10.1093/NAR/GKAB952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Iida K, Nagasawa K, Macrocyclic polyoxazoles as G-quadruplex ligands, Chem Rec. 13 (2013) 539–548. 10.1002/tcr.201300015. [DOI] [PubMed] [Google Scholar]
- [92].Iida K, Tera M, Hirokawa T, Shin-ya K, Nagasawa K, G-quadruplex recognition by macrocyclic hexaoxazole (6OTD) dimer: greater selectivity than monomer, Chem Commun. (2009) 6481–6483. 10.1039/b910242f. [DOI] [PubMed] [Google Scholar]
- [93].Monchaud D, Teulade-Fichou MP, A hitchhiker’s guide to G-quadruplex ligands, Org. Biomol. Chem 6 (2008) 627–636. 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- [94].Ou TM, Lu YJ, Tan JH, Huang ZS, Wong KY, Gu LQ, G-quadruplexes: Targets in anticancer drug design, ChemMedChem. 3 (2008) 690–713. 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- [95].Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH, Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex, J Am Chem Soc. 124 (2002) 2098–2099. [DOI] [PubMed] [Google Scholar]
- [96].Iida K, Tera M, Shin-Ya K, Nagasawa K, G-quadruplex recognition by macrocyclic hexaoxazole (6OTD) dimer, Nucleic Acids Symp Ser. (2009) 233–234. 10.1093/nass/nrp117. [DOI] [PubMed] [Google Scholar]
- [97].Han FX, Wheelhouse RT, Hurley LH, Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition, J. Am. Chem. Soc 121 (1999) 3561–3570. https://doi.org/ 10.1021/ja984153m. [DOI] [Google Scholar]
- [98].De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou MP, Shin-Ya K, Lacroix L, Lingner J, Mergny JL, Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action, Proc Natl Acad Sci U S A. 104 (2007) 17347–17352. 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S, High-throughput sequencing of DNA G-quadruplex structures in the human genome, Nat. Biotechnol 33 (2015) 877–881. 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- [100].Amato J, Morigi R, Pagano B, Pagano A, Ohnmacht S, De Magis A, Tiang YP, Capranico G, Locatelli A, Graziadio A, Leoni A, Rambaldi M, Novellino E, Neidle S, Randazzo A, Toward the Development of Specific G-Quadruplex Binders: Synthesis, Biophysical, and Biological Studies of New Hydrazone Derivatives, J Med Chem. 59 (2016) 5706–5720. 10.1021/acs.jmedchem.6b00129. [DOI] [PubMed] [Google Scholar]
- [101].Gai W, Yang Q, Xiang J, Jiang W, Li Q, Sun H, Guan A, Shang Q, Zhang H, Tang Y, A dual-site simultaneous binding mode in the interaction between parallel-stranded G-quadruplex [d(TGGGGT)]4 and cyanine dye 2,2′-diethyl-9-methyl-selenacarbocyanine bromide, Nucleic Acids Res. 41 (2013) 2709. 10.1093/NAR/GKS1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cosconati S, Marinelli L, Trotta R, Virno A, De Tito S, Romagnoli R, Pagano B, Limongelli V, Giancola C, Baraldi PG, Mayol L, Novellino E, Randazzo A, Structural and conformational requisites in DNA quadruplex groove binding: Another piece to the puzzle, J. Am. Chem. Soc 132 (2010) 6425–6433. 10.1021/ja1003872. [DOI] [PubMed] [Google Scholar]
- [103].Schonhoft JD, Bajracharya R, Dhakal S, Yu Z, Mao H, Basu S, Direct experimental evidence for quadruplex-quadruplex interaction within the human ILPR, Nucleic Acids Res. 37 (2009). 10.1093/nar/gkp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO, The hTERT core promoter forms three parallel G-quadruplexes, Nucleic Acids Res. 48 (2020) 5720–5734. 10.1093/NAR/GKAA107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Palumbo SML, Ebbinghaus SW, Hurley LH, Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands, J. Am. Chem. Soc 131 (2009). 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Georgiades SN, Abd Karim NH, Suntharalingam K, Vilar R, Interaction of metal complexes with G-quadruplex DNA, Angew. Chemie - Int. Ed 49 (2010) 4020–4034. 10.1002/anie.200906363. [DOI] [PubMed] [Google Scholar]
- [107].Cao Q, Li Y, Freisinger E, Qin PZ, Sigel RKO, Mao Z-W, G-quadruplex DNA targeted metal complexes acting as potential anticancer drugs, Inorg. Chem. Front 4 (2017) 10. 10.1039/c6qi00300a. [DOI] [Google Scholar]
- [108].Tran PLT, De Cian A, Gros J, Moriyama R, Mergny J-L, Tetramolecular Quadruplex Stability and Assembly, 2012. 10.1007/128_2012_334. [DOI] [PubMed] [Google Scholar]
- [109].Vorlíčková M, Chládková J, Kejnovská I, Fialová M, Kypr J, Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats, Nucleic Acids Res. 33 (2005) 5851–5860. 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yu HQ, Miyoshi D, Sugimoto N, Characterization of structure and stability of long telomeric DNA G-quadruplexes, 128 (2006) 15461–15468. 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- [111].Bauer L, Tlučková K, Tóthová P, Viglaský V, G-quadruplex motifs arranged in tandem occurring in telomeric repeats and the insulin-linked polymorphic region, Biochemistry. 50 (2011) 7484–7492. 10.1021/bi2003235. [DOI] [PubMed] [Google Scholar]
- [112].Kar A, Jones N, Ozlem Arat N, Fishel R, Griffith JD, Long repeating (TTAGGG)n single-stranded DNA self-condenses into compact beaded filaments stabilized by G-quadruplex formation, J. Biol. Chem 293 (2018) 9473–9485. 10.1074/jbc.RA118.002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Abraham Punnoose J, Cui Y, Koirala D, Yangyuoru PM, Ghimire C, Shrestha P, Mao H, Interaction of G-quadruplexes in the full-length 3’ human telomeric overhang, J Am Chem Soc. 136 (2014) 18062–18069. 10.1021/ja510079u. [DOI] [PubMed] [Google Scholar]
- [114].Stefan L, Denat F, Monchaud D, Deciphering the DNAzyme activity of multimeric quadruplexes: Insights into their actual role in the telomerase activity evaluation assay, J. Am. Chem. Soc 133 (2011) 20405–20415. https://doi.org/ 10.1021/ja208145d. [DOI] [PubMed] [Google Scholar]
- [115].Sen D, Gilbert W, A sodium-potassium switch in the formation of four-stranded G4-DNA, Nature. 344 (1990). 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- [116].Maizels N, Dynamic roles for G4 DNA in the biology of eukaryotic cells, Nat. Struct. Mol. Biol 13 (2006). 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- [117].Balagurumoorthy P, Brahmachari SK, Structure and stability of human telomeric sequence, J. Biol. Chem 269 (1994). 10.1016/s0021-9258(17)31882-3. [DOI] [PubMed] [Google Scholar]
- [118].Smith FW, Feigon J, Quadruplex structure of Oxytricha telomeric DNA oligonucleotides, Nature. 356 (1992). 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- [119].Petraccone L, Trent JO, Chaires JB, The tail of the telomere, J. Am. Chem. Soc 130 (2008) 16530–16532. 10.1021/ja8075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Haider S, Parkinson GN, Neidle S, Molecular dynamics and principal components analysis of human telomeric quadruplex multimers, Biophys J. 95 (2008) 296–311. 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Hänsel R, Löhr F, Trantirek L, Dötsch V, High-resolution insight into G-overhang architecture, J. Am. Chem. Soc 135 (2013). 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- [122].Kejnovská I, Renčiuk D, Palacký J, Vorlíčková M, CD Study of the G-Quadruplex Conformation, in: Methods Mol. Biol, 2019. 10.1007/978-1-4939-9666-7_2. [DOI] [PubMed] [Google Scholar]
- [123].Randazzo A, Spada GP, Da Silva MW, Circular dichroism of quadruplex structures, Top. Curr. Chem 330 (2013). 10.1007/128-2012-331. [DOI] [PubMed] [Google Scholar]
- [124].Vorlíčková M, Kejnovská I, Sagi J, Renčiuk D, Bednářová K, Motlová J, Kypr J, Circular dichroism and guanine quadruplexes, Methods. 57 (2012). 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- [125].Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP, A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures, Org. Biomol. Chem 8 (2010). 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- [126].Paramasivan S, Rujan I, Bolton PH, Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding, Methods. 43 (2007). 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- [127].Petraccone L, Spink C, Trent JO, Garbett NC, Mekmaysy CS, Giancola C, Chaires JB, Structure and stability of higher-order human telomeric quadruplexes, J. Am. Chem. Soc 133 (2011) 20951–20961. 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yu H, Gu X, Nakano SI, Miyoshi D, Sugimoto N, Beads-on-a-string structure of long telomeric dnas under molecular crowding conditions, J. Am. Chem. Soc 134 (2012) 20060–20069. https://doi.org/ 10.1021/ja305384c. [DOI] [PubMed] [Google Scholar]
- [129].Monsen RC, Chakravarthy S, Dean WL, Chaires JB, Trent JO, The solution structures of higher-order human telomere G-quadruplex multimers, Nucleic Acids Res. 49 (2021) 1749–1768. 10.1093/NAR/GKAA1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Renčiuk D, Kejnovská I, Školáková P, Bednářová K, Motlová J, Vorlíčková M, Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions, Nucleic Acids Res. 37 (2009) 6625–6634. 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Petraccone L, Malafronte A, Amato J, Giancola C, G-quadruplexes from human telomeric DNA: How many conformations in PEG containing solutions?, J. Phys. Chem. B 116 (2012). 10.1021/jp209170v. [DOI] [PubMed] [Google Scholar]
- [132].Bugaut A, Alberti P, Understanding the stability of DNA G-quadruplex units in long human telomeric strands, Biochimie. 113 (2015) 125–133. 10.1016/j.biochi.2015.04.003. [DOI] [PubMed] [Google Scholar]
- [133].Carrino S, Hennecker CD, Murrieta AC, Mittermaier A, Frustrated folding of guanine quadruplexes in telomeric DNA, Nucleic Acids Res. 49 (2021) 3063–3076. 10.1093/nar/gkab140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Víglaský V, Bauer L, Tlučková K, Structural features of intra- and intermolecular G-quadruplexes derived from telomeric repeats, Biochemistry. 49 (2010). 10.1021/bi902099u. [DOI] [PubMed] [Google Scholar]
- [135].Monsen RC, Trent JO, Chaires JB, G-quadruplex DNA: A Longer Story, Acc. Chem. Res 2022 (2022) 54. 10.1021/acs.accounts.2c00519. [DOI] [PubMed] [Google Scholar]
- [136].García De La Torre J, Huertas ML, Carrasco B, Calculation of hydrodynamic properties of globular proteins from their atomic-level structure, Biophys. J 78 (2000). 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Xu Y, Ishizuka T, Kurabayashi K, Komiyama M, Consecutive formation of G-Quadruplexes in human telomerieoverhang DNA: A protective capping structure for telomere ends, Angew. Chemie - Int. Ed 48 (2009) 7833–7836. 10.1002/anie.200903858. [DOI] [PubMed] [Google Scholar]
- [138].Wang H, Nora GJ, Ghodke H, Opresko PL, Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding, J. Biol. Chem 286 (2011) 7479–7489. 10.1074/jbc.M110.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Vesco G, Lamperti M, Salerno D, Marrano CA, Cassina V, Rigo R, Buglione E, Bondani M, Nicoletto G, Mantegazza F, Sissi C, Nardo L, Double-stranded flanking ends affect the folding kinetics and conformational equilibrium of G-quadruplexes forming sequences within the promoter of KIT oncogene, Nucleic Acids Res. 49 (2021) 9724–9737. 10.1093/nar/gkab674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Shiekh S, Mustafa G, Kodikara SG, Hoque ME, Yokie E, Portman JJ, Balci H, Emerging accessibility patterns in long telomeric overhangs, Proc. Natl. Acad. Sci. U. S. A 119 (2022). 10.1073/pnas.2202317119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Abraham Punnoose J, Ma Y, Hoque ME, Cui Y, Sasaki S, Guo AH, Nagasawa K, Mao H, Random Formation of G-Quadruplexes in the Full-Length Human Telomere Overhangs Leads to a Kinetic Folding Pattern with Targetable Vacant G-Tracts, Biochemistry. 57 (2018) 6946–6955. 10.1021/acs.biochem.8b00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Monsen RC, Chua EYD, Hopkins JB, Chaires JB, Trent JO, Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 Å resolution with cryo-EM, Nucleic Acids Res. 51 (2023) 1943–1959. 10.1093/NAR/GKAD014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Maleki P, Budhathoki JB, Roy WA, Balci H, A practical guide to studying G-quadruplex structures using single-molecule FRET, Mol. Genet. Genomics 292 (2017) 483–498. 10.1007/s00438-017-1288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Okumus B, Ha T, Real-time observation of G-quadruplex dynamics using single-molecule FRET microscopy, Methods Mol Biol. 608 (2010) 81–96. 10.1007/978-1-59745-363-9_6. [DOI] [PubMed] [Google Scholar]
- [145].Hwang H, Kreig A, Calvert J, Lormand J, Kwon Y, Daley JM, Sung P, Opresko PL, Myong S, Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins, Structure. 22 (2014) 842–853. 10.1016/j.str.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H, G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding., Proc. Natl. Acad. Sci. U. S. A 111 (2014) 2990–5. 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Maleki P, Mustafa G, Gyawali P, Budhathoki JB, Ma Y, Nagasawa K, Balci H, Quantifying the impact of small molecule ligands on G-quadruplex stability against Bloom helicase, Nucleic Acids Res. 47 (2019) 10744–10753. 10.1093/nar/gkz803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Noer SL, Preus S, Gudnason D, Aznauryan M, Mergny JL, Birkedal V, Folding dynamics and conformational heterogeneity of human telomeric G-quadruplex structures in Na+ solutions by single molecule FRET microscopy, Nucleic Acids Res. 44 (2016) 464–471. 10.1093/nar/gkv1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Tippana R, Xiao W, Myong S, G-quadruplex conformation and dynamics are determined by loop length and sequence, Nucleic Acids Res. 42 (2014) 8106–8114. 10.1093/nar/gku464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Chen MC, Tippana R, Demeshkina NA, Murat P, Balasubramanian S, Myong S, Ferre-D’Amare AR, Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36, Nature. 558 (2018) 465–469. 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].V Jena P, Shirude PS, Okumus B, Laxmi-Reddy K, Godde F, Huc I, Balasubramanian S, Ha T, G-quadruplex DNA bound by a synthetic ligand is highly dynamic, J Am Chem Soc. 131 (2009) 12522–12523. 10.1021/ja903408r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Mitra J, Makurath MA, Ngo TTM, Troitskaia A, Chemla YR, Ha T, Extreme mechanical diversity of human telomeric DNA revealed by fluorescence-force spectroscopy, Proc Natl Acad Sci U S A. 116 (2019) 8350–8359. 10.1073/pnas.1815162116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Kristoffersen EL, Coletta A, Lund LM, Schiøtt B, Birkedal V, Inhibited complete folding of consecutive human telomeric G-quadruplexes., Nucleic Acids Res. 51 (2023) 1571–1582. 10.1093/nar/gkad004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Deußner-Helfmann NS, Auer A, Strauss MT, Malkusch S, Dietz MS, Barth HD, Jungmann R, Heilemann M, Correlative Single-Molecule FRET and DNA-PAINT Imaging, Nano Lett. 18 (2018) 4626–4630. 10.1021/acs.nanolett.8b02185. [DOI] [PubMed] [Google Scholar]
- [155].Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC, Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami, Nano Lett. 10 (2010) 4756–4761. 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- [156].Mustafa G, Shiekh S, Gc K, Abeysirigunawardena S, Balci H, Interrogating accessibility of telomeric sequences with FRET-PAINT: Evidence for length-dependent telomere compaction, Nucleic Acids Res. 49 (2021) 3371–3380. 10.1093/nar/gkab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Shiekh S, Jack A, Saurabh A, Mustafa G, Kodikara SG, Gyawali P, Hoque ME, Pressé S, Yildiz A, Balci H, Shelterin reduces the accessibility of telomeric overhangs, Nucleic Acids Res. 50 (2022) 12885–12895. 10.1093/NAR/GKAC1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Petraccone L, Garbett NC, Chaires JB, Trent JO, An integrated molecular dynamics (MD) and experimental study of higher order human telomeric quadruplexes, Biopolymers. 93 (2010) 533–548. 10.1002/bip.21392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ray S, Qureshi MH, Malcolm DW, Budhathoki JB, Çelik U, Balci H, RPA-mediated unfolding of systematically varying G-quadruplex structures, Biophys. J 104 (2013) 2235–2245. 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


