Abstract
We present a high-quality assembly and annotation of the periodical cicada species, Magicicada septendecula (Hemiptera: Auchenorrhyncha: Cicadidae). Periodical cicadas have a significant ecological impact, serving as a food source for many mammals, reptiles, and birds. Magicicada are well known for their massive emergences of 1 to 3 species that appear in different locations in the eastern United States nearly every year. These year classes (“broods”) emerge dependably every 13 or 17 yr in a given location. Recently, it has become clear that 4-yr early or late emergences of a sizeable portion of a population are an important part of the history of brood formation; however, the biological mechanisms by which they track the passage of time remain a mystery. Using PacBio HiFi reads in conjunction with Hi-C proximity ligation data, we have assembled and annotated the first whole genome for a periodical cicada, an important resource for future phylogenetic and comparative genomic analysis. This also represents the first quality genome assembly and annotation for the Hemipteran superfamily Cicadoidea. With a scaffold N50 of 518.9 Mb and a complete BUSCO score of 96.7%, we are confident that this assembly will serve as a vital resource toward uncovering the genomic basis of periodical cicadas’ long, synchronized life cycles and will provide a robust framework for further investigations into these insects.
Keywords: cicada, genomics, assembly, annotation, Hemiptera, HiC
Significance.
Periodical cicadas have an outsized cultural and ecological impact and a highly unusual long, synchronized, periodical life cycle that holds many yet-to-be-resolved scientific mysteries. The assembly we report here is the first whole genome Magicicada assembly, the first complete genome assembled from a species in the plant-sucking-bug family Cicadidae (∼3,000 described species), and the first whole genome assembled for the Hemipteran superfamily Cicadoidea, which dates back ∼250 Mya.
Introduction
Periodical cicadas are plant-sucking bugs in the order Hemiptera—the genus Magicicada contains 7 species, which are distributed widely across the eastern United States. They range from Nebraska to Texas at the eastern edge of the Great Plains, to the Atlantic coast from Massachusetts to Georgia (Simon 1988). The genus can be divided into 3 morphologically distinct species groups: Decim, Decula, and Cassini. These 3 species groups each contain one 17-yr species and one or two 13-yr species. During their nymphal stages, cicadas feed underground on the xylem fluid of tree roots, and after 13 or 17 yr, emerge in large year classes called “broods,” which are composed of multiple different Magicicada species (Williams and Simon 1995; Simon et al. 2022). These emergences have a major ecological impact—for example, many avian species rely heavily on adult cicadas to feed their young and even experience population growth corresponding with a brood emergence (Koenig and Leibhold 2005; Pons 2020). Periodical cicadas are one of the quintessential examples of the evolutionary strategy of predator satiation, which is likely one of the selective pressures that influenced the development of such unique life cycles (Karban 1982; Williams et al. 1993; Koenig and Leibhold 2013). Magicicada septendecula is one of three species belonging to the Great Eastern Brood, or Brood X, which last emerged in 2021 and will reappear in the next generation in 2038 (Kritsky 2021).
Several mysteries surround the evolution of these fascinating insects, including the origins of broods, the mechanisms by which they can track the passage of time, the well-documented large emergences of a portion of a population exactly 4 yr early or late and their incredibly lengthy life cycles. For most of these questions, in-depth investigations have been hindered by the lack of quality genomic resources (Berlocher 2013; White and Pirro 2021; Simon et al. 2022). Although separated into multiple different broods, the members of each species group have experienced gene flow, presumably during coemergences made more frequent by the 4-yr early and late individuals (Fujisawa et al. 2018). Despite this gene flow, life cycles have maintained their integrity. Previous studies have used mRNA sequencing to investigate gene flow and construct molecular phylogenies and have found that between 13- and 17-year species pairs, genomic divergence is minimal except in the Decim group where one of the species, Magicicada tredecim, is reproductively isolated from the other two, Magicicada septendecim and Magicicada neotredecim (Fujisawa et al 2018).
Little is known about the demographic history and mechanism(s) of speciation within Magicicada. Several hypotheses have been proposed to explain multiple speciation events that have occurred in the history of these insects, most notably population fragmentation due to glacial events (Fujisawa et al 2018; Sota et al. 2013). Because periodical cicadas have been shown to time their emergence by measuring cumulative soil temperature (Heath 1968), glacial events in the last million years could provide a convenient explanation and impetus for speciation events by inducing early or late emergences that could lead to reproductively isolated populations (Cox and Carlton 1988). Scattered early and late bloomers, so to speak, have been observed appearing 1 or 2 yr before or after broods and large proportions of populations have been observed to emerge 4 yr early or late (Cooley et al. 2018; Simon et al. 2022).
While there is strong evidence that cicadas time their crawl to the surface based on accumulated soil temperature (Heath 1968), there are only unsupported hypotheses to explain how they can track the passage of years. One of the most promising hypotheses involves biological pathways that are triggered by changes in the chemical makeup of xylem fluid as the seasons change (Lloyd and Dybas 1966; Heath 1968). Annotated genome assemblies for Magicicada species will allow the discovery of genes that may be involved in these hypothesized pathways and could also allow for the comparison of genome sequences with other periodical species (Helioväära et al. 1994) or early/late bloomers to find gene variants that could explain the divergence in behavior. As this is one of nature's most sophisticated biological clocks, the results will be fascinating and provide insights into the mechanisms whereby other species track the passage of time. Recent research into the genetic control of periodicity in long-lived bamboo (Zhang et al. 2021) and temperature-based RNA expression and editing (Birk et al. 2023) could also provide fascinating hypotheses of the molecular mechanisms for timekeeping in this genus.
Further highlighting the importance of this genomic resource is the ancient estimated divergence time of the superfamilies Cercopoidea and Cicadoidea (Johnson et al. 2018). This divergence represents 250 million years of evolution, which are unrepresented (Fig. 1C) in the current genomic data. Here, we fill this gap by sequencing, assembling, and annotating a chromosome-length genome for the periodical cicada, M. septendecula, commonly known as the “little 17-year cicada.”
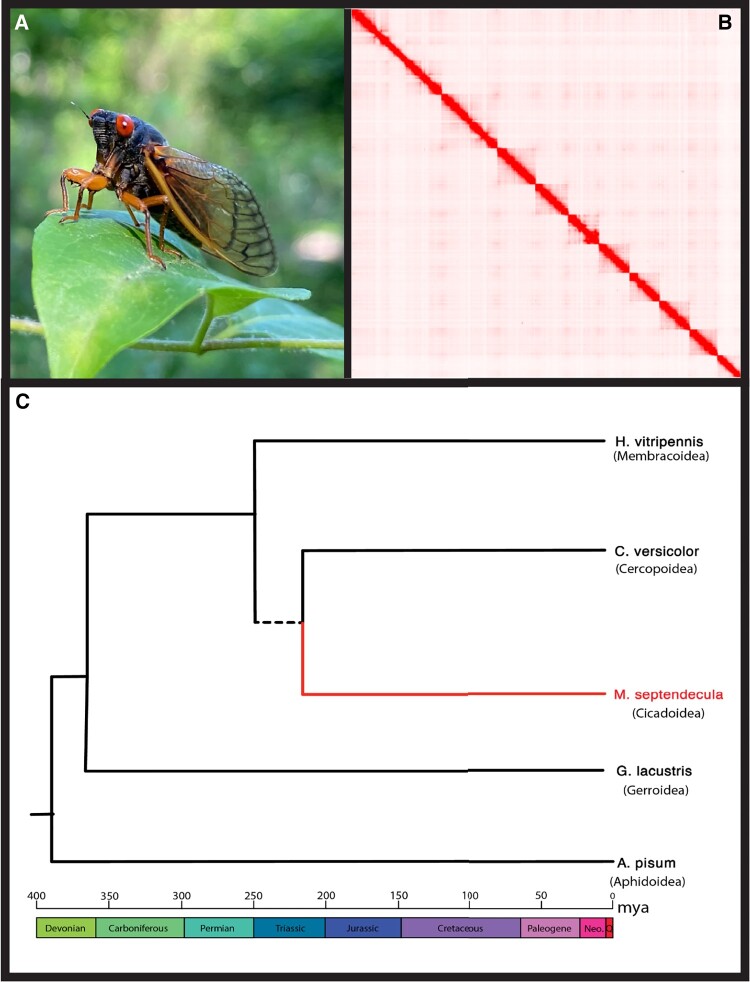
Fig. 1.
A) Photograph of M. septendecula, Herndon, VA (Photo credit: Paul Frandsen). B) Contact map generated from Hi-C data showing 10 distinct blocks corresponding to chromosomes (2n = 19/20). The link for the full interactive figure can be found in the Supplementary Material (see Table S6). C) Phylogenetic tree illustrating the relationships between the included Hemipteran superfamilies (see Table 1) with relative branch lengths illustrating divergence dates estimated by Johnson et al. (2018). The dotted line subtending Cicadoidea and Cercopoidea indicates uncertainty in superfamily relationships as discussed by Skinner et al. (2020), Cao and Dietrich (2022), and Song and Zhang (2023).
Results and Discussion
HiFi Assembly and Scaffolding with Hi-C Data
We sequenced a single individual male from the periodical cicada species, M. septendecula. Upon assembly, we found that the genome length is quite large compared to most other insects as measured by flow cytometry data (Hanrahan and Johnston 2011). At 6,521,820,903 bp, this genome assembly is almost 2.5× the size of the next largest Hemipteran assembly currently on NCBI (Biello et al. 2020). Repetitive elements were found to make up a large percentage of the genome, with a repeat content of 72.78% (35.64% classified) and a GC content of 35.25% as identified by RepeatMasker (v.4.1.2) (Flynn et al. 2020). Analysis with BlobTools (v.1.1.1) (Laetsch and Blaxter 2017) revealed several sequences that were categorized into Chordata as well as viral sequences (see supplementary fig. S1, Supplementary Material online)—however, due to their considerable length and a match in GC content with the rest of the genome, we argue that it is likely that these sequences were misclassified due to a lack of database coverage, similar to findings in the genome assembly of the dragonfly Tanypteryx hageni (Tolman et al. 2023).
Hi-C sequencing revealed 9 autosomes with 1 X chromosome (Fig. 1B; supplementary table S1, Supplementary Material online) in an XX/X0 sex-determining system, consistent with prior karyotyping of the genus Magicicada (Karagyan et al. 2020). PacBio HiFi sequencing and assembly followed by Hi-C scaffolding resulted in a highly improved genome assembly, with a scaffold N50 of 518.9 Mb, an L50 of 4, and 2,030 total scaffolds. Hi-C scaffolding thus improved our initial assembly into a strong genomic resource as potential pitfalls due to high repeat content (73%) and size (6.5 Gb) are addressed by the long-read sequencing technology as well as scaffolding with Hi-C data. Using NCBI's FCS tool, we trimmed and removed any contigs flagged as contaminants, ending with 10 chromosome-length scaffolds (95.49% of the assembly length) and 2,005 unplaced scaffolds (4.51% of the assembly length). We used tidk (v.0.2.41) (Brown et al. 2023) to search for telomeric repeats in the assembly and found telomeres on the ends of each chromosome-length scaffold (see supplementary fig. S2, Supplementary Material online). The complete BUSCO score for the Hi-C-scaffolded assembly was 96.7%.
Annotation
Five Illumina, paired-end Magicicada RNA-Seq libraries were trimmed and aligned to the M. septendecula genome (supplementary table S2, Supplementary Material online). Prior to quality control, total reads ranged between 22,101,494 and 85,009,400. Following quality control, that range fell between 22,092,126 and 85,000,848. RNA alignment rates were variable, falling between 81.29% and 94.83%.
The EASEL pipeline, which leverages generalized hidden Markov models and random forests to both predict and refine gene models, produced an unfiltered and filtered structural annotation. The unfiltered prediction, derived from multiple levels of RNA and protein support, captured 140,729 genes and 303,929 transcripts. This duplication was intentionally high to maximize gene sensitivity for downstream filtering. The mono:multiexonic ratio of 2.07 was indicative of fragmentation; however, despite an inflated number of false positives, 99.7% (S: 20.8%, D: 78.9%) of Insecta single-copy orthologs were captured. Following primary and secondary feature filtering using the invertebrate training set, EASEL predicted 22,785 genes and 83,621 transcripts with a mono:multiexonic ratio of 0.200 and a BUSCO completeness score of 96.4% (S: 24.9%, D: 71.5%). The functional annotation generated by EnTAP produced 57,260 unique RefSeq similarity search alignments (68.5%). With the addition of EggNOG gene family assignment, 81,978 sequences out of 83,621 were uniquely annotated (98.0%) (supplementary table S3, Supplementary Material online). The primary gene model (longest isoform) resolved the BUSCO duplication rate for the annotation but at the expense of completeness dropping to 91.2% (S: 89.2%, D: 2.0%), the mono:multiexonic ratio increasing to 0.209, and RefSeq alignments dropping to 59.4% (supplementary table S4, Supplementary Material online). This is slightly lower than the assembly BUSCO reported previously but still within a range that indicates a high-quality annotation.
Materials and Methods
Specimen Collection
Samples were collected and flash frozen on dry ice by Chris Simon and Stephen Chiswell in Knox County, TN and Wilkes Co., NC in May of 2021, during the Brood X emergence. After sequencing, the samples were stored as vouchers in the Bean Life Science Museum at Brigham Young University. For the complete sample metadata, please see Table S5.
Library Prep and Sequencing
We extracted DNA from a single Wilkes Co., NC male using the Qiagen GenomicTip high molecular weight DNA extraction kit. We then sheared the DNA to 18-kb fragments using a Diagenode Megaruptor and size-selected fragments of >10 kb using a SAGE Science BluePippin. We then generated a HiFi sequencing library using the PacBio SMRTbell Express Template Prep Kito 2.0. We sequenced the library across four 30-h SMRT cells on the PacBio Sequel II instrument at the BYU DNA sequencing center. Hi-C libraries were prepared and sequenced on an Illumina NextSeq by DNAZoo at Baylor College of Medicine using methods described in earlier publications (Rao et al. 2014; Dudchenko et al. 2017; Lamb et al. 2021; Tolman et al. 2023). A Knox Co., TN male was used for Hi-C library preparation.
Assembly Generation and Contamination Screening
We used PacBio SMRTtools to generate HiFi reads from the raw PacBio subreads, which were then assembled into contigs using hifiasm (v.0.16.1) (Cheng et al. 2021). Raw PacBio reads were aligned and scaffolded to Hi-C reads using Juicer and 3D-DNA, respectively (Durand et al. 2016; Dudchenko et al. 2017). Contact maps were manually inspected using Juicebox Assembly Tools (Dudchenko et al. 2018). We used BLAST (v.2.12.0+) (Camacho et al. 2009) to create a sequence database from the initial assembly and RepeatModeler (v.2.0.1) and RepeatMasker (v.4.1.2) (Flynn et al. 2020) to identify and classify repetitive elements. We used BUSCO (v.5.2.2) (Manni et al. 2021) to evaluate gene completeness in the initial assembly and in the final annotated genome. The genome was checked for contamination using NCBI's FCS tool (v.0.4.0) (Astashyn et al. 2023), and contigs flagged as potential contaminants were removed from the final assembly. Additional sequences marked as contamination were trimmed using the faidx tool in SAMtools (v.1.15.1) (Danecek et al 2021).
Structural and Functional Annotation
Five public Illumina HiSeq RNA-Seq libraries (paired-end) of the genus Magicicada were accessed from NCBI (Table 1). Each library was trimmed with FastP (v.0.23.2) and aligned to the soft-masked M. septendecula genome with HISAT2 (v.2.2.1) (Chen et al. 2018; Kim et al. 2019). To generate the structural annotation, the soft-masked genome, RNA-Seq data and Hemiptera OrthoDB (v.11) proteins were provided to EASEL (v.1.4) (Kuznetsov et al. 2023; Webster et al. 2023). The EASEL pipeline assembled transcriptomes via StringTie2 (v.2.2.1) and PsiCLASS (v.1.0.3) and isolated complete open-reading frames with TransDecoder (v.5.7.0), culminating in the generation of a gene model (Haas 2016; Kovaka et al. 2019; Song et al. 2019). Putative transcripts and proteins were aligned to the genome with GMAP (v.2021.08.25) and miniprot (v.0.11), respectively, and converted into hints (Wu and Watanabe 2005; Li 2023). With the provided gene models and hints, AUGUSTUS (v.3.5.0) was run, resulting in an unfiltered structural annotation with alternative transcripts (Stanke et al. 2006). These transcripts were classified by primary and secondary features and filtered via a random forest algorithm using the invertebrate training set and a regressor threshold of 65. Gene-prediction accuracy of filtered and unfiltered models was summarized by the total number of genes and transcripts output by AGAT (v.1.0.0), the mono:multiexonic ratio of genes, BUSCO completeness (insecta_v10) (v.5.4.4), and a 70/70 reciprocal BLAST functional annotation rate output by EnTAP (1.0.1), referencing the complete RefSeq database (v.208) (Hart et al. 2020; Dainat et al. 2022). The final structural and functional annotations were derived from the filtered EASEL output; however, to assess BUSCO duplication rates without the added noise of alternative transcripts, the longest isoform was also extracted.
Table 1.
Hemipteran genome assembly statistics
| Order | Superfamily | Species | Source | GenBank Accession | Assembly Length (Mb) | N50 (Mb) | Assembly BUSCO Results (OrthoDB: Insecta) | Genes Annotated |
|---|---|---|---|---|---|---|---|---|
| Hemiptera | Cicadoidea | Magicicada septendecula, little 17-yr cicada | Current study | … | 6,521 | 518.9 | C: 96.8% (S: 93.4%, D: 3.4%), F: 2.4%, M: 0.8% | 22,785 |
| Hemiptera | Membracoidea | Homalodisca vitripennis, glassy-winged sharpshooter | Li et al. (2022) | GCA_021130785.2 | 2,305 | 168.8 | C: 92.7% (S: 82.0%, D: 10.8%), F: 1.0%, M: 6.2% | 22,591 |
| Hemiptera | Cercopoidea | Callitettix versicolor, rice spittlebug | Chen et al. (2022) | GCA_022606455.1 | 975 | 5.6 | C: 94.7% (S: 92.3%, D: 2.3%), F: 0.5%, M: 4.7% | 21,937 |
| Hemiptera | Membracoidea | Nephotettix cincticeps, rice green leafhopper | Yan et al. (2021) | GCA_023375725.1 | 753 | 85.4 | C: 97.0% (S: 95.2%, D: 1.8%), F: 2.0%, M: 1.0% | 14,337 |
| Hemiptera | Aphidoidea | Acyrthosiphon pisum, pea aphid | Li et al. (2019) | GCA_005508785.2 | 542 | 132.5 | C: 93.3% (S: 89.7%, D: 3.6%), F: 1.9%, M: 4.8% | 20,307 |
| Hemiptera | Gerroidea | Gerris lacustris, common water strider | Wellcome Sanger Institute (2023) | GCA_951217055.1 | 938 | 76.4 | … | … |
Assembly BUSCO results = C, complete; S, single copy; D, duplicates; F, fragmented; M, missing.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
E.L.A. was supported by the Welch Foundation (Q-1866), a McNair Medical Institute Scholar Award, an NIH Encyclopedia of DNA Elements Mapping Center Award (UM1HG009375), a US-Israel Binational Science Foundation Award (2019276), the Behavioral Plasticity Research Institute (NSF DBI-2021795), National Science Foundation Physics Frontiers Center Award (NSF PHY-2019745) and by the National Human Genome Research Institute of the National Institutes of Health (RM1HG011016-01A1). DNA Zoo also acknowledges support from Illumina, IBM, and the Pawsey Supercomputing Centre. Additionally, C.W. was supported by NSF DBI 194337, and C.S. acknowledges support from NSF DEB 16-55891. Finally, this work was also supported by a BYU Life Sciences College Undergraduate Research Award.
Contributor Information
Jonas Bush, Huck Life Sciences Institute, The Pennsylvania State University, State College, PA, USA; Department of Plant and Wildlife Sciences, Brigham Young University, Provo, UT, USA.
Cynthia Webster, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT, USA.
Jill Wegrzyn, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT, USA.
Chris Simon, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT, USA.
Edward Wilcox, Department of Plant and Wildlife Sciences, Brigham Young University, Provo, UT, USA.
Ruqayya Khan, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
David Weisz, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
Olga Dudchenko, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA; The Center for Theoretical Biological Physics, Rice University, Houston, TX, USA.
Erez Lieberman Aiden, The Center for Genome Architecture, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA; The Center for Theoretical Biological Physics, Rice University, Houston, TX, USA; Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Paul Frandsen, Department of Plant and Wildlife Sciences, Brigham Young University, Provo, UT, USA; Data Science Lab, Office of the Chief Information Officer, Smithsonian Institution, Washington, DC, USA.
Data Availability
The final assembly (filtered and scaffolded), all corresponding annotation files, and the plot of telomeric regions can be found in a Figshare repository (doi:10.6084/m9.figshare.24488050). The final assembly is also currently being processed on NCBI under BioProject PRJNA966940. The raw reads, original unscaffolded assembly, Hi-C data and unfiltered assembly are available publicly on DNAZoo's website (https://www.dnazoo.org/assemblies/magicicada_septendecula).
Literature Cited
- Astashyn A, Tvedte ES, Sweeney D, Sapojnikov V, Bouk N, Joukov V, Mozes E, Strope PK, Sylla PM, Wagner L, et al. Rapid and sensitive detection of genome contamination at scale with FCS-GX. bioRxiv. 2023. 10.1101/2023.06.02.543519., 6 June 2023, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Berlocher SH. Regularities and irregularities in periodical cicada evolution. Proc Natl Acad Sci U S A. 2013:110(17):6620–6621. 10.1073/pnas.1304228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biello R, Mathers TC, Mugford ST, Liu Q, Rodrigues ASB, Neto AC, Rebelo MT, Paulo OS, Seabra SG, Hogenhout SA. Draft genome assembly version 1 of the meadow spittlebug Philaenus spumarius (Linnaeus, 1758) (Hemiptera, Aphrophoridae). Zenodo. 2020. 10.5281/ZENODO.3368385. [DOI] [Google Scholar]
- Birk MA, Liscovitch-Brauer N, Dominguez MJ, McNeme S, Yue Y, Hoff JD, Twersky I, Verhey KJ, Sutton RB, Eisenberg E, et al. Temperature-dependent RNA editing in octopus extensively recodes the neural proteome. Cell. 2023:186(12):2544–2555.e13. 10.1016/j.cell.2023.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M, González De la Rosa PM, Mark B. A Telomere Identification Toolkit. Zenodo. 2023. 10.5281/zenodo.10091385. [DOI]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009:10(1):421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Dietrich CH. Phylogenomics of flavobacterial insect nutritional endosymbionts with implications for Auchenorrhyncha phylogeny. Cladistics. 2022:38(1):38–58. 10.1111/cla.12474. [DOI] [PubMed] [Google Scholar]
- Chen H, Qiao G, Liang A. Chromosome-level genome assembly of Callitettix versicolor (rice spittlebug). Genome Biol Evol. 2022:14(9):evac130. 10.1093/gbe/evac130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021:18(2):170–175. 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley JR, Arguedas N, Bonaros E, Bunker G, Chiswell SM, DeGiovine A, Edwards M, Hassanieh D, Haji D, Knox J, et al. The periodical cicada four-year acceleration hypothesis revisited and the polyphyletic nature of Brood V, including an updated crowd-source enhanced map (Hemiptera: Cicadidae: Magicicada). PeerJ. 2018:6:e5282. 10.7717/peerj.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Carlton CE. Paleoclimatic influences in the evolution of periodical cicadas (Insecta: Homoptera: Cicadidae: Magicicada spp.). Am Midland Nat. 1988:120(1):183–193. 10.2307/2425898. [DOI] [Google Scholar]
- Dainat J, Hereñú D, Pucholt P. NBISweden/AGAT: AGAT-v1.0.0. Zenodo. 2022. 10.5281/ZENODO.7255559. [DOI]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021:10(2):giab008. 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017:356(6333):92–95. 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Shamim MS, Batra SS, Durand NC, Musial NT, Mostofa R, Pham M, Glenn St Hilaire B, Yao W, Stamenova E, et al. The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. bioRxiv. 2018. 10.1101/254797, 28 January 2018, preprint: not peer reviewed. [DOI]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016:3(1):95–98. 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020:117(17):9451–9457. 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Koyama T, Kakishima S, Cooley JR, Simon C, Yoshimura J, Sota T. Triplicate parallel life cycle divergence despite gene flow in periodical cicadas. Commun Biol. 2018:1(1):1–14. 10.1038/s42003-018-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ. TransDecoder. 2016. https://github.com/TransDecoder/TransDecoder.
- Hanrahan SJ, Johnston JS. New genome size estimates of 134 species of arthropods. Chromosome Res. 2011:19(6):809–823. 10.1007/s10577-011-9231-6. [DOI] [PubMed] [Google Scholar]
- Hart AJ, Ginzburg S, Xu MS, Fisher CR, Rahmatpour N, Mitton JB, Paul R, Wegrzyn JL. EnTAP: bringing faster and smarter functional annotation to non-model eukaryotic transcriptomes. Mol Ecol Resour. 2020:20(2):591–604. 10.1111/1755-0998.13106. [DOI] [PubMed] [Google Scholar]
- Heath JE. Thermal synchronization of emergence in periodical ‘17-year’ cicadas (Homoptera, Cicadidae, Magicicada). Am Midland Nat. 1968:80(2):440–448. 10.2307/2423537. [DOI] [Google Scholar]
- Heliövaara K, Väisänen R, Simon C. Evolutionary ecology of periodical insects. Trends Ecol Evol. 1994:9(12):475–480. 10.1016/0169-5347(94)90312-3. [DOI] [PubMed] [Google Scholar]
- Johnson KP, Dietrich CH, Friedrich F, Beutel RG, Wipfler B, Peters RS, Allen JM, Petersen M, Donath A, Walden KKO, et al. Phylogenomics and the evolution of hemipteroid insects. Proc Natl Acad Sci U S A. 2018:115(50):12775–12780. 10.1073/pnas.1815820115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagyan G, Golub N, Sota T. Cytogenetic characterization of periodical cicadas (Hemiptera: Cicadidae: Magicicada). Eur J Entomol. 2020:117:474–480. 10.14411/eje.2020.050. [DOI] [Google Scholar]
- Karban R. Increased reproductive success at high densities and predator satiation for periodical cicadas. Ecology. 1982:63(2):321–328. 10.2307/1938949. [DOI] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig WD, Liebhold AM. Effects of periodical cicada emergences on abundance and synchrony of avian populations. Ecology. 2005:86(7):1873–1882. 10.1890/04-1175. [DOI] [Google Scholar]
- Koenig WD, Liebhold AM. Avian predation pressure as a potential driver of periodical cicada cycle length. Am Nat. 2013:181(1):145–149. 10.1086/668596. [DOI] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019:20(1):278. 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsky G. One for the books: the 2021 emergence of the periodical cicada brood X. Am Entomol. 2021:67(4):40–46. 10.1093/ae/tmab059. [DOI] [Google Scholar]
- Kuznetsov D, Tegenfeldt F, Manni M, Seppey M, Berkeley M, Kriventseva EV, Zdobnov EM. OrthoDB v11: annotation of orthologs in the widest sampling of organismal diversity. Nucleic Acids Res. 2023:51(D1):D445–D451. 10.1093/nar/gkac998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML. BlobTools: interrogation of genome assemblies. F1000Res. 2017:6:1287. 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- Lamb S, Taylor AM, Hughes TA, McMillan BR, Larsen RT, Khan R, Weisz D, Dudchenko O, Aiden EL, Edelman NB, et al. De novo chromosome-length assembly of the mule deer (Odocoileus hemionus) genome. GigaByte. 2021:2021:gigabyte34. 10.46471/gigabyte.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Protein-to-genome alignment with miniprot. Bioinformatics. 2023:39(1):btad014. 10.1093/bioinformatics/btad014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li Y, Xue AZ, Dang V, Holmes VR, Johnston JS, Barrick JE, Moran NA. The genomic basis of evolutionary novelties in a leafhopper. Mol Biol Evol. 2022:39(9):msac184. 10.1093/molbev/msac184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Park H, Smith TE, Moran NA. Gene family evolution in the pea aphid based on chromosome-level genome assembly. Mol Biol Evol. 2019:36(10):2143–2156. 10.1093/molbev/msz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd M, Dybas HS. The periodical cicada problem. II. Evolution. Evolution. 1966:20(4):466–505. 10.2307/2406585. [DOI] [PubMed] [Google Scholar]
- Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol. 2021:38(10):4647–4654. 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons P. True cicadas (Cicadidae) as prey for the birds of the Western Palearctic: a review. Avian Res. 2020:11(1):14. 10.1186/s40657-020-00200-1. [DOI] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014:159(7):1665–1680. 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C. Evolution of 13-and 17-year periodical cicadas (Homoptera: Cicadidae: Magicicada). Bull Entomol Soc Am. 1988:34(4):163–176. 10.1093/besa/34.4.163. [DOI] [Google Scholar]
- Simon C, Cooley JR, Karban R, Sota T. Advances in the evolution and ecology of 13- and 17-year periodical cicadas. Annu Rev Entomol. 2022:67(1):457–482. 10.1146/annurev-ento-072121-061108. [DOI] [PubMed] [Google Scholar]
- Skinner RK, Dietrich CH, Walden KKO, Gordon E, Sweet AD, Podsiadlowski L, Petersen M, Simon C, Takiya DM, Johnson KP. Phylogenomics of Auchenorrhyncha (Insecta: Hemiptera) using transcriptomes: examining controversial relationships via degeneracy coding and interrogation of gene conflict. Syst Entomol. 2020:45(1):85–113. 10.1111/syen.12381. [DOI] [Google Scholar]
- Song L, Sabunciyan S, Yang G, Florea L. A multi-sample approach increases the accuracy of transcript assembly. Nat Commun. 2019:10(1):5000. 10.1038/s41467-019-12990-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Zhang H. A comprehensive analysis of higher-level phylogenetic relationships of Hemiptera based on transcriptome data. J Syt Evol. 2023:61(4):572–586. 10.1111/jse.12855. [DOI] [Google Scholar]
- Sota T, Yamamoto S, Cooley JR, Hill KBR, Simon C, Yoshimura J. Independent divergence of 13- and 17-y life cycles among three periodical cicada lineages. Proc Natl Acad Sci U S A. 2013:110(17):6919–6924. 10.1073/pnas.1220060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006:34(Web Server):W435–W439. 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman ER, Beatty CD, Bush J, Kohli M, Moreno CM, Ware JL, Weber KS, Khan R, Maheshwari C, Weisz D, et al. A chromosome-length assembly of the black petaltail (Tanypteryx hageni) dragonfly. Genome Biol Evol. 2023:15(3):evad024. 10.1093/gbe/evad024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C, Fetter K, Zaman S, Vuruputoor V, Bhattarai A, Chinta V, Wegrzyn J. EASEL. GitLab [retrieved 2023 August 22]. 2023. https://gitlab.com/PlantGenomicsLab/easel.
- White HB, Pirro S. The complete genome sequences of two species of seventeen-year cicadas: Magicicada septendecim and Magicicada septendecula. F1000Res. 2021:10:215. 10.12688/f1000research.27309.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KS, Simon C. The ecology, behavior, and evolution of periodical cicadas. Annu Rev Entomol. 1995:40(1):269–295. 10.1146/annurev.en.40.010195.001413. [DOI] [Google Scholar]
- Williams KS, Smith KG, Stephen FM. Emergence of 13-yr periodical cicadas (Cicadidae: Magicicada): phenology, mortality, and predators satiation. Ecology. 1993:74(4):1143–1152. 10.2307/1940484. [DOI] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005:21(9):1859–1875. 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Yan B, Yu X, Dai R, Li Z, Yang M. Chromosome-level genome assembly of Nephotettix cincticeps (Uhler, 1896) (Hemiptera: Cicadellidae: Deltocephalinae). Genome Biol Evol. 2021:13(11):evab236. 10.1093/gbe/evab236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang H, Wang Y, Xi F, Wang H, Kohnen MV, Gao P, Wei W, Chen K, Liu X, et al. Whole-genome characterization of chronological age-associated changes in methylome and circular RNAs in moso bamboo (Phyllostachys edulis) from vegetative to floral growth. Plant J. 2021:106(2):435–453. 10.1111/tpj.15174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final assembly (filtered and scaffolded), all corresponding annotation files, and the plot of telomeric regions can be found in a Figshare repository (doi:10.6084/m9.figshare.24488050). The final assembly is also currently being processed on NCBI under BioProject PRJNA966940. The raw reads, original unscaffolded assembly, Hi-C data and unfiltered assembly are available publicly on DNAZoo's website (https://www.dnazoo.org/assemblies/magicicada_septendecula).