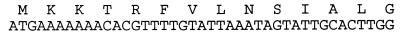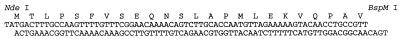Abstract
The htrA gene from two strains of nontypeable Haemophilus influenzae has been cloned and sequenced, and the encoded approximately 46-kDa HtrA proteins were found to be highly conserved. H. influenzae HtrA has approximately 55% identity with the Escherichia coli and Salmonella typhimurium HtrA stress response proteins, and expression of the H. influenzae htrA gene was inducible by high temperature. Recombinant HtrA (rHtrA) was expressed from E. coli, and the purified protein was found to have serine protease activity. rHtrA was found to be very immunogenic and partially protective in both the passive infant rat model of bacteremia and the active chinchilla model of otitis media. Immunoblot analysis indicated that HtrA is antigenically conserved in encapsulated and nontypeable H. influenzae species. Site-directed mutagenesis was performed on the htrA gene to ablate the endogenous serine protease activity of wild-type HtrA, and it was found that eight of nine recombinant mutant proteins had no measurable residual proteolytic activity. Two mutant proteins were tested in the animal protection models, and one, H91A, was found to be partially protective in both models. H91A HtrA may be a good candidate antigen for a vaccine against invasive H. influenzae type b disease and otitis media and is currently in phase I clinical trials.
Haemophilus influenzae is the cause of several serious human diseases such as meningitis, epiglottitis, septicemia, and otitis media. There are six serotypes of H. influenzae, designated a to f, which are identified by their capsular polysaccharides. H. influenzae type b (Hib) was a major cause of bacterial meningitis until the introduction of several Hib capsular polysaccharide conjugate vaccines in the 1980s (1, 30). The other serotypes of H. influenzae are associated with invasive disease at low frequencies, although there appears to be an increase in disease due to these strains as the incidence of Hib disease declines (18, 25, 34). Nonencapsulated or nontypeable H. influenzae (NTHI) is a major cause of otitis media (middle ear infection) in young children and of respiratory tract infections in adults. NTHI is the second most common bacterial cause of otitis media after Streptococcus pneumoniae and is responsible for about 25% of this disease. Otitis media affects more than 80% of children under 6 years of age, with the peak incidence in infants under the age of two. In the United States, the incidence of disease increased 2.5-fold between 1975 and 1990, and the causative bacteria are becoming increasingly antibiotic resistant (17). Although otitis media is rarely life threatening, there are serious sequelae associated with the disease, including deafness and language or learning deficits, and there is currently no vaccine.
The HtrA protein has been identified as a virulence factor in Salmonella typhimurium, Yersinia enterocolitica and Brucella abortus (8, 16, 20). The HtrA (or DegP) protein of Escherichia coli has been shown to be essential for survival of the organism at temperatures of >42°C. It is a stress response protein belonging to the ςE-dependent family of heat shock proteins (Hsps) (7). It is not related to either the ς32-regulated Hsps such as DnaK or DnaJ or the ς70-regulated Hsps such as Hsp60, Hsp70, and Hsp90 (21). The S. typhimurium HtrA protein is ∼89% identical to E. coli HtrA but is not induced by heat shock, although it is induced by oxidative stress (16).
The E. coli HtrA protein has serine protease activity (22), and two residues of the catalytic triad have been identified by site-directed mutagenesis (31). In this report, we describe the cloning and sequence analysis of the htrA genes from two strains of NTHI and the expression of recombinant HtrA (rHtrA) in E. coli. The rHtrA protein was shown to have serine protease activity, and site-directed mutagenesis was performed on the three residues of the catalytic triad to ablate the endogenous enzyme activity. The rHtrA mutant proteins were expressed at a high yield from E. coli, and the purified proteins were found to be highly immunogenic. Protection studies were performed with two animal models, the passive infant rat model of bacteremia and the active chinchilla model of otitis media.
MATERIALS AND METHODS
Recombinant DNA techniques.
Restriction endonucleases were purchased from Boehringer Mannheim, New England Biolabs, Bethesda Research Laboratories, and Pharmacia and were used according to the manufacturers’ specifications. Oligonucleotides were synthesized on an Applied Biosystems Inc. model 380B DNA synthesizer and purified by chromatography. Plasmid pUC-BgXb is a pUC8-based plasmid with additional BglII and XbaI restriction sites in its multiple-cloning site and was used for construction purposes. Other recombinant DNA methods were performed as described by Sambrook et al. (29).
Bacterial strains and media.
NTHI strains 33 and 12 are otitis media clinical isolates kindly provided by S. Barenkamp (St. Louis University, St. Louis, Mo.). NTHI strain LCDC2 was obtained from the Laboratory Centre for Disease Control (Ottawa, Ontario, Canada). Hib strain Eagan is the Pasteur Mérieux Connaught vaccine strain, and Hib strain MinnA was a kind gift from R. Munson (Ohio State University, Columbus). H. influenzae serotype a, c, d, e, and f strains were purchased from the American Type Culture Collection. H. influenzae strains were grown on Mueller-Hinton agar (BBL) supplemented with yeast extract (0.5% [wt/vol]; Sigma), hemin (15 μg ml−1; Difco), and NAD (15 μg ml−1; Difco) or in brain heart infusion (BHI) broth (Difco) as described previously (14). Chocolate agar plates were purchased from Becton Dickinson Microbiology Systems. E. coli strains were grown in NZCYM or YT medium supplemented with 50 μg of ampicillin ml−1 as required.
Cloning and sequencing of htrA genes.
The construction of an NTHI strain 33 EMBL 3 chromosomal library has been described in detail elsewhere (23). An oligonucleotide probe (41 nucleotides) was derived from sequence information presented by Weinstein et al. (36) and corresponded to the N terminus of the encoded HtrA protein. The sequence of the probe is shown in Fig. 1. The probe was radiolabelled with [γ-32P]dATP, and putative genomic clones were plaque purified three times. A 15.3-kb insert containing the NTHI strain 33 htrA gene was subcloned into pUC-BgXb, and the htrA gene was localized to a 4.7-kb BamHI fragment by restriction mapping and Southern blot analysis. Nested deletion clones were generated with an Erase-a-base kit (Promega), and sequencing was performed on the ABI model 373A DNA sequencer by using dye terminator chemistry. The first strand sequence was obtained with the universal forward primer, and second strand sequence was obtained with 17- to 25-nucleotide custom primers.
FIG. 1.
Nucleotide sequence and derived amino acid sequence for the 41-nucleotide probe used in the cloning and sequencing of htrA genes.
NTHI strain 12 chromosomal DNA was prepared as described previously (23), and the strain 12 htrA gene was obtained by PCR amplification with oligonucleotide primers based upon the strain 33 htrA gene sequence (Fig. 2). PCR amplification was performed with buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM potassium chloride, and 1.5 mM magnesium chloride. Each 100 μl of reaction mixture contained 5 ng of chromosomal DNA, 1 μg of each primer, 5 U of Amplitaq DNA polymerase (Perkin-Elmer Cetus), and 0.2 mM concentrations of deoxynucleoside triphosphates (Perkin-Elmer Cetus). The cycling conditions were 25 cycles of 94°C for 1.0 min, 45°C for 2.0 min, and 72°C for 1.5 min. Three independent reactions were performed, and amplified fragments were cloned into pCR II (Invitrogen) and sequenced on both strands.
FIG. 2.
Oligonucleotide primers used for PCR amplification of NTHI strain 12 DNA. The primer sequences are underlined.
Identification of active-site residues.
The NTHI HtrA sequence was initially compared with those of the E. coli and S. typhimurium HtrA proteins by using Intelligenetics Suite software. The sequence boundaries of the protease domain of NTHI HtrA were defined both manually and with XALIGN (37). To confirm the assignment of the catalytic triad residues, multiple sequence alignments and three-dimensional superimposition of mammalian and bacterial serine proteases were performed by utilizing XALIGN and the Homology module from BIOSYM Technologies, with crystal structures obtained from the Protein Data Bank (5).
Expression of recombinant HtrA and generation of mutant proteins.
The smallest Erase-a-base clone that contained all of the htrA gene was used to construct expression plasmids. By analogy with the E. coli and S. typhimurium HtrA proteins (22), a putative 26-amino-acid signal sequence was identified, and the Thr at position 27 was assumed to represent the start of the mature protein. There is a BspMI site ∼72 bp downstream of the start of the coding sequence for the mature HtrA protein and a ClaI site downstream of the end of the htrA gene. Oligonucleotides were synthesized to encode the N terminus of the mature HtrA protein up to the BspMI site (Fig. 3). Plasmid DNA was digested with BspMI and ClaI, and the 1.4-kb fragment was ligated with the NdeI-BspMI oligonucleotides and vector pT7-7, which had been digested with NdeI and ClaI, thus generating pT7-7/htrA. For site-directed mutagenesis, the T7-htrA gene fragment was cloned into M13mp18 and the Amersham oligonucleotide-directed in vitro mutagenesis system was used. The T7-htrA mutant genes were cloned into pT7-7 for expression of the recombinant proteins.
FIG. 3.
Oligonucleotides encoding the N terminus of the mature HtrA protein up to the BspM site.
E. coli BL21(DE3) cells were transformed with the T7-htrA expression plasmids by either the CaCl2 method or electroporation with a Bio-Rad Gene Pulser. The cells were grown in YT medium to an A578 of 0.3, and isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) was added to 0.4 mM for 2 h. Recombinant proteins were expressed as soluble proteins and/or inclusion bodies, depending on their mutations.
Purification of recombinant HtrA proteins.
Cells from a 500-ml culture were harvested by centrifugation at 10,000 × g for 10 min at 4°C, and the pellet was resuspended in 40 ml of 50 mM Tris-HCl (pH 8.0) and disrupted by sonication (three times for 10 min each time). The sonicate was centrifuged at 20,000 × g for 30 min at 4°C, and depending upon the location of the product, the supernatant or pellet was further purified.
For wild-type rHtrA and the H91A analog, which were expressed as soluble proteins, the supernatant was applied to a DEAE Sephacel (Pharmacia) column (1 ml of column matrix per 5 ml of extract) equilibrated in 50 mM Tris-HCl (pH 8.0). The majority of rHtrA or H91A proteins were recovered in the run-through fraction, which was loaded onto a Macro-prep ceramic hydroxylapatite column (HTP; Bio-Rad Laboratories) that had the same capacity and that was equilibrated in 10 mM sodium phosphate buffer (pH 8.0). Both rHtrA and H91A proteins bound to the HTP column. After the column was washed with 175 mM Na phosphate (pH 8.0), rHtrA or H91A was eluted from the HTP with 0.3 M Na phosphate (pH 8.0). The amount of rHtrA or H91A in the elution fractions was determined by the bicinchoninic acid protein assay (Pierce) using bovine serum albumin as a standard. The purity of rHtrA or H91A proteins was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (19).
Mutant proteins H91R, D121A, D121E, and S197A were expressed as inclusion bodies, whereas S197C and S197T were expressed partially as inclusion bodies and partially as soluble proteins. Inclusion bodies were isolated from the sonicate pellet by extraction with a mixture containing 40 ml of 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.5% Triton X-100. The extract was centrifuged at 20,000 × g for 30 min, and the pellet was solubilized in 10 ml of 50 mM Tris-HCl (pH 8.0) containing 8 M urea. To this solution was added 30 ml of 50 mM Tris-HCl (pH 8.0), and then the solubilized HtrA proteins were purified by HTP chromatography as described above.
Characterization of wild-type and mutant HtrA proteins.
The residual serine protease activities of rHtrA and its analogs were assessed with β-casein as the substrate as described by Lipinska et al. (22). To study the kinetics of the protease activity, 5 μg of β-casein was mixed with 0.5 μg of HtrA and the mixture was incubated at 37°C. Aliquots were taken at 10, 20, and 30 min and at 1, 2, 4, and 16 h, and the reaction was stopped immediately by the addition of SDS-PAGE sample buffer and heating at 100°C for 5 min. Samples were analyzed by SDS–12.5% PAGE. To determine the residual protease activities of various analogs, 5 μg of β-casein was mixed with 0.5 μg of HtrA mutant protein and the mixture was incubated at 37°C for 16 h. Samples were treated and analyzed as described above.
The effect of protease inhibitors such as Pefabloc SC (Centerchem, Inc., Stamford, Conn.), phenylmethylsulfonyl fluoride (PMSF; Sigma), and leupeptin (Sigma) on HtrA activity was determined. Various concentrations of inhibitors (2.5, 5, and 10 mM concentrations of Pefabloc SC or PMSF; 100 and 500 mM concentrations of leupeptin) were added to the β-casein–HtrA reaction mixture, and the samples were incubated at 37°C for 2 h. Samples were analyzed as described above.
Immunogenicity studies.
To study the immunogenicities of rHtrA and mutant proteins H91A, D121A, and S197A, groups of five BALB/c mice (Charles River) were injected subcutaneously (s.c.) on days 1, 29, and 43 with various doses (0.3, 1, 3, and 10 μg) of protein adsorbed to alum. Blood samples were collected on days 0, 14, 28, 42, and 54.
Anti-HtrA immunoglobulin G (IgG) titers in immune sera were determined by antigen-specific enzyme-linked immunosorbent assays (ELISAs) as described by Panezutti et al. (26). Briefly, microtiter wells (Nunc-MAXISORP, Nunc) were coated with 200 ng of rHtrA or mutant antigen, corresponding to the antibody being tested, for 16 h at room temperature. The plates were then blocked with 0.1% (wt/vol) bovine serum albumin (radioimmunoassay grade; Sigma) in phosphate-buffered saline. The individual sera were serially diluted, added to the wells, and incubated for 1 h at room temperature. Either affinity-purified F(ab′)2 fragments of goat anti-mouse IgG (Fc-specific) or anti-guinea pig IgG (Fc-specific) antibodies conjugated to horseradish peroxidase (HRP) (Jackson ImmunoResearch Laboratories Inc.) were used as the secondary antibodies. The reactions were developed with tetramethylbenzidine-hydrogen peroxide, and absorbancies were measured at 450 nm (with 540 nm as a reference wavelength) in a Flow Multiskan MCC microplate reader. The reactive titer of an antiserum was defined as the reciprocal of the dilution consistently showing a twofold increase in absorbance over that obtained with the prebleed serum sample.
To determine IgG subclasses, microtiter wells were coated with 200 ng of purified rHtrA or analog and pooled mouse antisera were used in ELISAs as described above. Rat anti-mouse IgG1 and IgG2a (Zymed Laboratories) and IgG2b (Serotec) antibodies, rabbit anti-mouse IgG3 (Zymed Laboratories) antibodies, and donkey anti-rabbit IgG antibodies conjugated to HRP (Jackson ImmunoResearch Laboratories Inc.) were used as reagents in the ELISAs. The reactive titers were determined as described above.
Stress response studies.
Two guinea pigs (Charles River, St. Constant, Quebec, Canada) were immunized intramuscularly (i.m.) with 10 μg of rHtrA protein emulsified in complete Freund’s adjuvant (Difco). Fourteen and 28 days later, the animals were boosted with the same amount of immunogen emulsified in incomplete Freund’s adjuvant. Antisera were collected 2 weeks after the last injection.
Hib strain Eagan and E. coli JM109 were grown under various conditions. Samples were grown to an A578 of 0.3 in BHI or YT medium at 37°C and then were aliquoted and grown in the same medium at 37, 42, or 43.5°C or in media adjusted to contain 6% ethanol, 0.2 M NaCl, or 0.3 M NaCl at 37°C. Aliquots were taken at the following times: 0, 20, 40, 60, and 90 min. They were spun at 16,000 × g for 5 min, and the cell pellets were resuspended at 40 OD ml−1. One-microliter samples were analyzed by SDS-PAGE and immunoblotting. Guinea pig antiserum which recognized both the E. coli and NTHI HtrA proteins was used to probe immunoblots. Goat anti-guinea pig IgG (Fc-specific) antibody conjugated to HRP was used as the second antibody, and the blots were visualized with the LumiGlo chemiluminescent detection system (Kirkegaard and Perry Laboratories).
Antigenic conservation.
H. influenzae serotypes a, b (Eagan), c, d, e, and f were grown in BHI medium as described above. Equivalent concentrations of whole-cell lysates were electrophoresed on an SDS–11.5% PAGE gel, electroblotted onto a nitrocellulose membrane, and probed with a 1:1,000 dilution of guinea pig anti-rHtrA antibody. Fc-specific goat anti-guinea pig IgG antibody conjugated to HRP was used as the second antibody, and the blot was visualized with the LumiGlo system.
Protection studies.
Chinchilla protection studies were performed as described by Barenkamp (2). Briefly, 1- to 2-year-old chinchillas (Moulton Chinchilla Ranch, Rochester, Minn.) were immunized i.m. with 30 μg of protein adsorbed to alum. Booster immunizations were given on days 14 and 28, and antisera were collected 2 weeks after the last injection. Negative control animals were mock immunized with alum by following the same regimen, and positive controls were convalescent animals that had recovered from previous infection. Animals were challenged with 50 to 350 CFU of live NTHI strain 12 or 200 CFU of live LCDC2 through the bulla. Otoscopy and tympanometry were performed before bacterial inoculation and repeated 4 days postchallenge. Middle ear fluid was collected and immediately mixed with 200 μl of BHI medium. Dilutions of aspirates were made, and 10 μl of undiluted aspirate and 10 μl of 1/10 and 1/100 dilutions were plated onto chocolate agar plates. Bacterial colonies were counted after 24 h.
Infant rat protection studies were performed as described by Munson and Granoff (24). Synchronized pregnant Sprague-Dawley rats were purchased from Harlan Sprague-Dawley (Indianapolis, Ind.). Five-day-old infant rats were randomized among litters and were injected s.c. in the dorsal region with 0.1 ml of the antiserum to be tested. Twenty-four hours later, animals were challenged intraperitoneally with 200 CFU of freshly grown Hib strain MinnA (0.1 ml). Blood samples were collected at 20 h postchallenge via cardiac puncture under isoflurane anesthesia and were plated on chocolate agar plates. Colonies were counted after one day, and the results were statistically analyzed by the Fisher exact test. Each study was performed at least twice, and representative results are shown in Table 3.
TABLE 3.
Protective effect of passively transferred chinchilla antiserum against Hib challenge in the infant rat model of bacteremiaa
| Group | Chinchilla serum | No. of animals bacteremic/no. challenged | Mean CFU/ 2.5 μl of bloodb | P |
|---|---|---|---|---|
| 1 | Anti-HtrA (wild type) | 4/10 | 195 | 0.0054 |
| 2 | Anti-H91A HtrA | 4/10 | 330 | 0.0054 |
| 3 | Anti-S197A HtrA | 4/10 | 450 | 0.0054 |
| 4 | Anti-MinnA | 0/10 | 0 | |
| 5 | Normal chinchilla serum | 10/10 | 500 |
Five-day-old infant rats were passively immunized s.c. with 0.1 ml of chinchilla antiserum or normal chinchilla serum. Twenty hours later, they were challenged intraperitoneally with freshly grown Hib strain MinnA (200 CFU, 0.1 ml). Blood samples were collected 20 h postchallenge via cardiac puncture and plated onto chocolate agar. Bacteria were counted after 1 day, and an animal was considered to be protected when no bacteria were recovered from its blood. The statistical significance of the number of bacteremic animals was established by the Fisher exact test.
Bacterial counts in the blood of infected animals.
Nucleotide sequence accession numbers.
Nucleotide sequences of NTHI strain 33 htrA and strain 12 htrA have been deposited in GenBank and have accession no. AF018152 and AF018151, respectively.
RESULTS
Cloning and sequence analysis of the htrA gene.
Weinstein et al. had described the derived amino acid sequence of a 47-kDa H. influenzae protein designated Hin47 and the associated nucleotide sequence (36). An oligonucleotide probe was synthesized based upon the published sequence and the hin47 gene was cloned from a chromosomal library of NTHI strain 33. The hin47 gene was localized on the 15.3-kb chromosomal fragment and sequenced.
The structural gene encodes a protein of 49.2 kDa with a putative 26-amino-acid signal sequence, whose cleavage would result in a 46.4-kDa mature protein. When the hin47 sequence was compared with sequences in the databases, the gene was found to encode a protein with approximately 54% identity and 69% similarity to the E. coli and S. typhimurium HtrA proteins (Fig. 4). The E. coli and S. typhimurium proteins are slightly larger than the NTHI protein (49.3 kDa). (The original published sequence of the E. coli HtrA protein has been corrected at the carboxy terminus by Waller and Sauer [35].) The N-terminal ∼100 residues of the NTHI protein differ significantly from those of the E. coli and S. typhimurium HtrA proteins, which are ∼89% identical to each other (16). The htrA gene was amplified by PCR from NTHI strain 12, and its encoded mature protein sequence was found to be 99% identical to that of strain 33 HtrA. These sequences have also been compared to the published H. influenzae Rd HtrA sequence (11), and the three sequences were found to be 99% identical. In contrast to the E. coli and S. typhimurium HtrA proteins, there are no cysteine residues in the H. influenzae proteins.
FIG. 4.
Sequence alignment of HtrA proteins from NTHI strains 33 and 12, H. influenzae type d strain Rd, E. coli (Ecoli), and S. typhimurium (Styph). Stop codons are indicated by asterisks. Dots indicate residues identical to those of the NTHI strain 33 HtrA, and dashes have been used to achieve maximum alignment. The residues indicated by arrows are numbered from the putative Thr start of the mature NTHI HtrA proteins. The main numbering scheme refers to the full-length encoded protein from the start methionine of NTHI 33 HtrA. The RGD sequence is underlined in the strain 33 sequence.
There is an RGD motif found at the carboxy termini of the E. coli, S. typhimurium, NTHI strain 33, and Rd HtrA proteins, but in NTHI strain 12, the corresponding sequence is RGN. By Southern blot analysis, the htrA gene was found in H. influenzae serotype a, b (strain Eagan), c, d, e, and f strains (data not shown). A fragment encoding the RGD-containing region of HtrA was amplified by PCR from H. influenzae serotypes a, b (strain Eagan), c, d, e, and f and four additional NTHI strains. Most of the strains carried a gene encoding the RGD motif, but 5 of 13 H. influenzae strains contained the RGN motif (data not shown).
H. influenzae HtrA is a stress response protein.
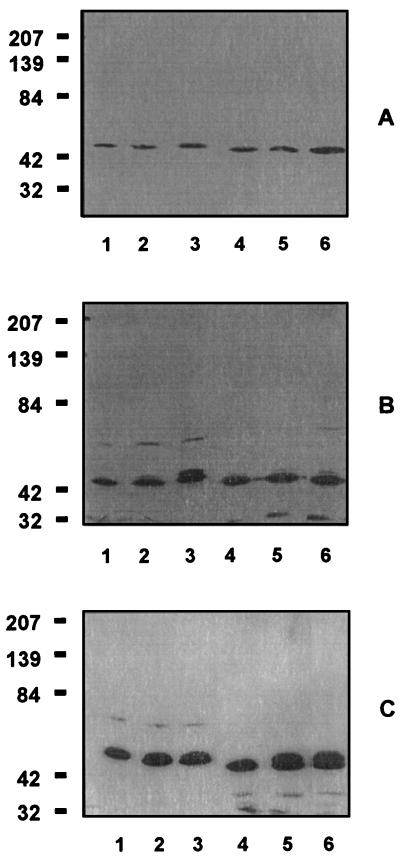
Since HtrA is known to function as a stress response protein in E. coli, experiments were performed to determine whether H. influenzae HtrA was also a stress response protein. E. coli and Hib strain Eagan were grown in parallel under standard conditions and under stress-inducing conditions such as high temperature, 6% ethanol, and high salt concentrations. A guinea pig antiserum which recognized HtrA from both organisms was used to assess expression. HtrA was found to be constitutively expressed at low levels for both organisms under all growth conditions (Fig. 5). At 43.5°C or in the presence of 6% ethanol, the expression of a second form of HtrA at a slightly higher apparent molecular weight was induced in both E. coli and H. influenzae, although the induction was significantly better in the medium containing ethanol. No effect was observed at 42°C or in the presence of 0.2 or 0.3 M NaCl (results not shown).
FIG. 5.
Stress induction of htrA expression. (A) Immunoblot of E. coli JM109 and Hib strain Eagan grown at 37°C. Lane 1, E. coli at t = 0 min; lane 2, E. coli at t = 20 min; lane 3, E. coli at t = 60 min; lane 4, Hib at t = 0 min; lane 5, Hib at t = 20 min; lane 6, Hib at t = 60 min. (B) Immunoblot of E. coli JM109 and Hib strain Eagan grown at 43.5°C. Lane 1, E. coli at t = 0 min; lane 2, E. coli at t = 20 min; lane 3, E. coli at t = 60 min; lane 4, Hib at t = 0 min; lane 5, Hib at t = 20 min; lane 6, Hib at t = 60 min. (C) Immunoblot of E. coli JM109 and Hib strain Eagan grown in the presence of 6% ethanol. Lane 1, E. coli at t = 0 min; lane 2, E. coli at t = 20 min; lane 3, E. coli at t = 60 min; lane 4, Hib at t = 0 min; lane 5, Hib at t = 20 min; lane 6, Hib at t = 60 min. In all panels, the numbers at the left represent molecular weights in thousands.
H. influenzae HtrA has serine protease activity.
The effect of protease inhibitors on rHtrA activity was determined. It was found that two serine protease inhibitors, Pefabloc SC and PMSF, completely inhibited the rHtrA activity at concentrations of 10 mM. In contrast, the thiol protease inhibitor leupeptin did not show any effect even at 500 mM, a concentration which is 250 times higher than that routinely used (27).
Identification of active-site residues.
The consensus sequence surrounding the active-site serine of serine proteases is GDSGGPK (6), where the serine is the active-site nucleophile. An examination of the strain 33 HtrA sequence revealed a GNSGGAL motif around Ser197 (numbered from the proposed Thr1 residue). By comparison with other bacterial and mammalian serine proteases, the other members of the catalytic triad were predicted to be His91 and Asp121. Figure 6 shows the alignment of NTHI HtrA with the sequences of serine proteases from the Protein Data Bank.
FIG. 6.
Identification of HtrA active-site residues. Multiple sequence alignment of the serine protease domain of NTHI HtrA (residues 57 to 256) with serine proteases from the Protein Data Bank (PDB). Arrows denote the catalytic triad residues. Protease sequences obtained from PDB entries are as follows: TON, pdb1ton.ent (rat tonin); PKA, pdb2pka.ent (porcine kallikrein A); PTN, pdb2ptn.ent (bovine trypsin); CHA, pdb4cha.ent (bovine chymotrypsin); EST, pdb3est.ent (porcine elastase); RP2, pdb3rp2.ent (rat mast cell protease II); SGT, pdb1sgt.ent (Streptomyces griseus trypsin); SGB, pdb3sgb.ent (S. griseus proteinase B); SGA, pdb2sga.ent (S. griseus proteinase A); ALP, pdb2alp.ent (L. enzymogenes alpha-lytic protease).
Expression and characterization of recombinant HtrA and mutants.
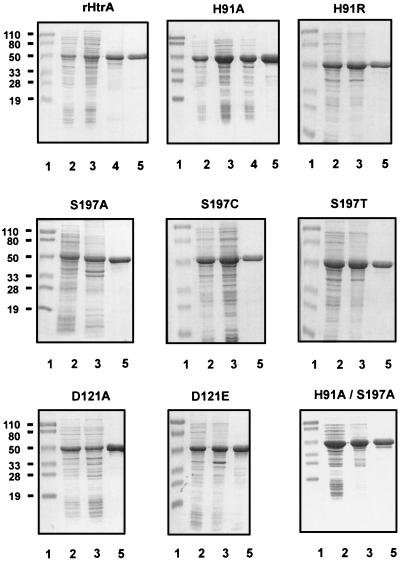
The strain 33 htrA gene was cloned behind the inducible T7 promoter in vector pT7-7 (33), and the recombinant protein was produced at high levels in E. coli BL21(DE3) cells. Nine HtrA analogs were generated at the His91, Asp121, or Ser197 residues of the catalytic triad, and all of the mutant HtrA proteins were expressed at a very high yield (∼40 to 50% of total protein) from E. coli. The recombinant proteins were expressed as soluble proteins (wild type and H91A), inclusion bodies (H91R, D121A, D121E, S197A, H91A/S197A, and H91A/D121A/S197A), or a mixture of both forms (S197C and S197T), and all were readily purified (Fig. 7).
FIG. 7.
SDS-PAGE analysis of rHtrA and analogs during purification performed on 12.5% polyacrylamide gels. Lanes 1, prestained molecular weight markers; lanes 2, total cellular proteins from E. coli BL21(DE3) expressing rHtrA or mutant proteins; lanes 3, crude extract of recombinant protein; lanes 4, flowthrough fraction from DEAE Sephacel column for soluble rHtrA and H91A; lanes 5, purified protein after the HTP column. The numbers at the left of each row represent molecular weights in thousands.
The protease activity of rHtrA was determined as described by Lipinska et al. (22), with β-casein as a substrate. As shown in Fig. 8A, digestion of casein by rHtrA was noticeable after 30 min of incubation (lane 5) and was complete after 16 h (lane 9). By the same assay, it was found that of the nine HtrA analogs, only D121E exhibited protease activity under these conditions (Fig. 8B, lane 10). Although the amount of rHtrA or analogs added to the reaction mixtures was approximately 0.5 μg, the SDS-PAGE analysis in Fig. 8B suggests that some of the analogs may actually have been present at higher concentrations and still did not show appreciable degradation of β-casein.
FIG. 8.
Protease activity of rHtrA and its analogs. (A) Wild-type rHtrA (0.5 μg) was mixed with β-casein (5 μg) and incubated at 37°C. Samples were taken at various time points and analyzed by SDS-PAGE. Lane 1, molecular weight standards; lane 2, t = 0 min; lane 3, t = 10 min; lane 4, t = 20 min; lane 5, t = 30 min; lane 6, t = 1 h; lane 7, t = 2 h; lane 8, t = 4 h; lane 9, t = 16 h; lane 10, no HtrA added at t = 16 h. (B) Wild-type rHtrA or its analogs (0.5 μg) were mixed with β-casein (5 μg) and incubated at 37°C for 16 h. Lane 1, molecular weight standards; lane 2, casein alone; lane 3, rHtrA; lane 4, S197A; lane 5, S197C; lane 6, S197T; lane 7, H91A; lane 8, H91R; lane 9, D121A; lane 10, D121E. For both panels, the numbers at the left represent molecular weights in thousands.
Immunogenicity and protection studies.
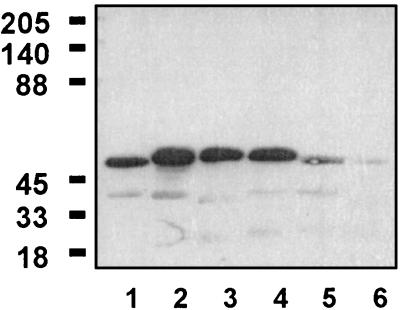
Purified rHtrA, H91A, D121A, and S197A were used to immunize BALB/c mice, and after three immunizations, all four proteins were found to be very immunogenic at doses as low as 0.3 μg when administered in alum (Table 1). The antibody isotype profiles were determined for rHtrA, H91A, and S197A and were found to be equivalent for the three proteins, with a dominant IgG1 response (Table 2). Guinea pig anti-NTHI rHtrA recognized putative HtrA from H. influenzae serotype a, b, c, d, e, and f strains, although the reactivity with serotype f was very weak (Fig. 9).
TABLE 1.
Immunogenicities of various forms of HtrAa
| Dose (μg) | Day of bleed | Log2(titer/100) for protein:
|
|||
|---|---|---|---|---|---|
| rHtrA | H91A | S197A | D121A | ||
| 0.3 | 14 | 5.00 ± 3.16 | 5.60 ± 0.55 | 7.40 ± 1.67 | 1.80 ± 0.45 |
| 28 | 6.60 ± 2.19 | 6.40 ± 2.30 | 9.40 ± 1.67 | 3.00 ± 1.63 | |
| 42 | 10.20 ± 1.79 | 10.20 ± 1.79 | 12.60 ± 0.89 | 7.40 ± 3.05 | |
| 54 | 11.00 ± 0.00 | 11.20 ± 1.30 | 13.40 ± 0.89 | 8.80 ± 1.10 | |
| 1.0 | 14 | 6.60 ± 2.61 | 6.50 ± 0.58 | 8.20 ± 1.10 | 4.60 ± 0.89 |
| 28 | 8.20 ± 1.79 | 8.00 ± 0.82 | 10.60 ± 1.67 | 5.80 ± 2.17 | |
| 42 | 11.40 ± 0.89 | 10.50 ± 0.58 | 13.80 ± 1.10 | 9.00 ± 0.71 | |
| 54 | 12.20 ± 1.10 | 11.50 ± 0.58 | 13.80 ± 1.10 | 9.40 ± 0.55 | |
| 3.0 | 14 | 8.60 ± 0.89 | 7.40 ± 0.55 | 7.80 ± 1.10 | 5.00 ± 0.71 |
| 28 | 9.00 ± 1.41 | 9.20 ± 0.84 | 10.20 ± 1.10 | 7.80 ± 1.64 | |
| 42 | 13.00 ± 0.00 | 11.80 ± 0.84 | 12.60 ± 0.89 | 10.20 ± 0.84 | |
| 54 | 13.40 ± 0.89 | 13.20 ± 0.45 | 13.00 ± 0.00 | 10.60 ± 0.89 | |
| 10.0 | 14 | 8.60 ± 1.67 | 9.40 ± 0.55 | 8.60 ± 1.67 | 6.80 ± 0.45 |
| 28 | 11.00 ± 2.00 | 9.60 ± 0.55 | 10.60 ± 1.67 | 8.80 ± 1.30 | |
| 42 | 13.00 ± 0.00 | 12.60 ± 0.89 | 13.00 ± 0.00 | 11.40 ± 0.55 | |
| 54 | 13.00 ± 0.00 | 13.80 ± 0.45 | 13.00 ± 0.00 | 12.00 ± 0.00 | |
Groups of five BALB/c mice were immunized (s.c.) on days 1, 29, and 43 with the indicated doses of rHtrA, H91A, D121A, or S197A adsorbed to alum. Blood samples were collected on days 0, 14, 28, 42, and 54. The reactive titer of an antiserum was defined as the reciprocal of the dilution consistently showing a twofold increase in absorbance over that obtained with the prebleed serum sample.
TABLE 2.
Antibody isotypes elicited by immunization with rHtrA and its analogs
| Isotype | Antibody titera for protein:
|
||
|---|---|---|---|
| rHtrA | H91A | S197A | |
| IgG (H+L) | 10 | 10 | 10 |
| IgG1 | 8 | 8 | 8 |
| IgG2a | <1 | <1 | <1 |
| IgG2b | 1.5 | <1 | 2 |
| IgG3 | <1 | <1 | <1 |
IgG isotype titers of sera collected on day 54 after three immunizations were determined by ELISA. The reactive titer was defined as the reciprocal of the dilution consistently showing a twofold increase in absorbance over that obtained with the prebleed serum sample and is expressed as log2(titer/100). H + L, heavy plus light chains.
FIG. 9.
Antigenic conservation of HtrA in H. influenzae strains. Lane 1, serotype a; lane 2, serotype b (Eagan); lane 3, serotype c; lane 4, serotype d; lane 5, serotype e; lane 6, serotype f. Guinea pig anti-NTHI rHtrA was used to probe the blots. The numbers at the left represent molecular weights in thousands.
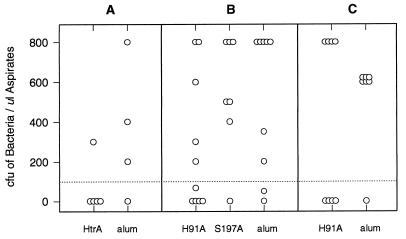
Chinchillas were immunized three times with purified strain 33-derived rHtrA, H91A, or S197A. Negative controls were immunized with alum, and positive controls were convalescent animals that had recovered from previous middle ear infection with the homologous strain. Intrabullar challenge with live NTHI strain 33 resulted in unacceptable morbidity, so animals were challenged with the heterologous NTHI strain 12 or LCDC2. All convalescent animals were protected against challenge (data not shown), whereas the majority (75 to 83%) of mock-immunized negative control animals were infected (Fig. 10). Four of five animals immunized with rHtrA and challenged with strain 12 were protected (Fig. 10A). Immunization with H91A conferred partial protection (∼50%) against challenge with either NTHI strain 12 or strain LCDC2 (Fig. 10B and C). However, no protection was observed in animals immunized with S197A (Fig. 10B).
FIG. 10.
Protection studies in the chinchilla intrabulla challenge model of otitis media. Chinchillas (1 to 2 years old) were immunized three times (i.m.) with 30 μg of antigen adsorbed to alum. Two weeks after the last injection, animals were challenged with live NTHI strain 12 (50 to 350 CFU) (A and B) or LCDC2 (200 CFU) (C) through the epitympanic bulla. Middle ear fluid was collected 4 days postchallenge. Each data point represents the bacteria recovered from a single chinchilla.
Chinchilla anti-rHtrA, anti-H91A, and anti-S197A antisera, obtained as described above, were used to passively immunize infant rats. Positive and negative control sera were antisera raised against heat-inactivated Hib strain MinnA and prebleed sera, respectively. Animals were challenged with live Hib strain MinnA. All of the HtrA-specific immune sera were partially protective in this model, with 6 of 10 animals exhibiting no or very low-level bacteremia (Table 3).
DISCUSSION
The sequences of the NTHI HtrA proteins and the Rd HtrA (HI1259) sequence (11) were found to be highly conserved, with differences at only five positions among the strains. One of these amino acid changes is in an Arg-Gly-Asp (RGD) motif found at the carboxy terminus of HtrA. The RGD motif has been identified as a cell attachment site for mammalian adhesion proteins (28). Of the 13 H. influenzae htrA genes that we studied, five encoded an RGN motif and the rest encoded RGD. E. coli expresses two homologs of HtrA (or DegP), which are called DegQ (HhoA) and DegS (HhoB) (3, 35). The DegP and DegQ proteins have 60% identical and 75% similar sequences and are serine proteases. One noticeable sequence difference is that DegP contains an RGD sequence and DegQ contains an RGN sequence (35). In a recent paper by Li et al. (20), an alignment of the HtrA proteins from Y. enterocolitica, S. typhimurium, E. coli, B. abortus, Rochalimaea hensalae, Helicobacter pylori, and Campylobacter jejuni demonstrated that only the first three of these organisms contain the carboxy-terminal RGD sequence. Since HtrA is a periplasmic protein, it is unclear what function the RGD or RGN sequence might play, but the fact that it is at least semiconserved among different species is intriguing. In addition, if it is surface expressed under stress conditions, the RGD sequence may play an attachment function.
The E. coli HtrA protein has been shown to be inducible by temperatures of >42°C, but the S. typhimurium HtrA protein is not temperature inducible (16). By immunoblot analysis, we demonstrated low-level constitutive expression of HtrA in both E. coli and H. influenzae under normal culture conditions, with an increased expression of a slightly larger protein under stress conditions. The amount of protein expressed was too small to allow N-terminal sequence determination, so we can only speculate as to the nature of the second protein. It is possible that it is the unprocessed precursor protein or a conformational variant of HtrA, although the latter possibility seems unlikely based upon the results of Skorko-Glonek et al. (32), who showed that E. coli HtrA was relatively thermostable up to 70°C. Since DegQ is not heat inducible, the second protein band is not likely to be DegQ (35).
The proteolytic activity of H. influenzae rHtrA was blocked by serine protease inhibitors, indicating that like the E. coli HtrA, it is a serine protease. Alignment of the NTHI, E. coli, and S. typhimurium HtrA protein sequences showed that there is a consensus sequence of GNSGGAL surrounding the putative Ser197 active-site residue. With alignments anchored on this residue, the other residues of the catalytic triad were identified from alignments with other bacterial and mammalian serine proteases of known structure. In general, bacterial trypsin-like serine proteases such as alpha-lytic protease from Lysobacter enzymogenes and proteases A and B from Streptomyces griseus are significantly smaller than mammalian ones (4). However, because the active sites and cores of these proteases can be reliably aligned to mammalian serine proteases, they provide an indication of where sequence variability, including the locations of insertions and deletions, can be expected (12, 13). Thus, the active-site histidine could be assigned to His91 of NTHI HtrA despite the presence of only a low level of local sequence homology and the absence of a conserved disulfide bond immediately following this residue. Asp121 was assigned on the basis of its local sequence alignment with a combination of identical or similar residues in other proteases. To confirm these residues, a three-dimensional model of the HtrA site was constructed by using L. enzymogenes alpha-lytic protease as a template (12) and various serine protease structures from the Protein Data Bank (5). The catalytic triad could be readily accommodated within this model in comparison to the crystal structures of mammalian and bacterial serine proteases. Using site-directed mutagenesis, Skorko-Glonek et al. (31) demonstrated that the Ser210 and His105 residues of the mature E. coli HtrA protein were part of the catalytic triad. The serine residue was identified by the homology of the sequences of the surrounding amino acids to the consensus sequence for serine proteases, and there is only one histidine residue in the molecule. Li et al. (20) and Waller and Sauer (35) predicted that the Asp136 residue of E. coli HtrA is part of the catalytic residue, a prediction which correlates very well with our finding of Asp121 in the NTHI proteins. Mutations of the His91, Asp121, and Ser197 codons were found to abolish the protease activity. The D121E analog still retained its enzymatic activity, although D121A did not, suggesting that the negative charge is important for activity.
The assignment of the catalytic triad residues provided a basis for delineating the boundaries of the protease domain, which comprises about 45% of the HtrA sequence, i.e., between residues 54 and 255. The rHtrA protein was found to contain a major degradation product that started with residue Asp54. By homology with other proteases, residues 1 to 53 may be cleaved from a zymogen form of HtrA in order for it to become active. E. coli cells overproducing wild-type recombinant E. coli HtrA were found to contain two major ∼43-kDa HtrA degradation products, beginning at Cys69 and Gln82 (31). It was speculated that these products were autocatalytic in nature since the mutant HtrA proteins did not contain them.
Antisera raised to NTHI strain 33-derived rHtrA, H91A, or S197A were all equally protective in the passive infant rat model of bacteremia (24), wherein 6 of 10 animals had no or minimal bacteremia. These data indicated that the three proteins were inducing functional antibodies. In this model, the animals are challenged with live Hib organisms, and the partial protection demonstrates that the anti-NTHI rHtrA antibodies recognize the HtrA protein in Hib. Immunoblot analysis showed that anti-NTHI rHtrA antisera recognize ∼46-kDa proteins in H. influenzae serotypes a, b, c, d, e, and f (weak), demonstrating the antigenic conservation of the HtrA proteins. In the chinchilla model of otitis media, animals were immunized with NTHI strain 33-derived rHtrA or its H91A or S197A analogs and were challenged with live NTHI strain 12 or LCDC2 organisms. The rHtrA and H91A proteins induced partial protection, but the S197A protein was not protective. The IgG subtype profiles elicited by immunization with the three immunogens were determined with mice, and it was found that all three were identical, with a predominant IgG1 response, a small IgG2b response, and no IgG2a or IgG3 response. One explanation for the observed difference in protection between H91A and S197A could be that they have different conformations after purification as soluble or inclusion body-derived proteins; however, we were unable to demonstrate any difference by circular dichroism analysis (data not shown). It is also unclear why this difference is evident only in the chinchilla model and not the infant rat model.
The exact role of the HtrA protein in the pathogenesis of H. influenzae remains to be determined; however, if HtrA serves as a target for bactericidal antibodies or opsonic activity, it must be accessible to antibodies. Since HtrA is an apparently periplasmic protein, it is not evident how such antigen-antibody interactions can occur. It is possible that during infection, some surface expression of HtrA is induced as part of a stress response mechanism, e.g., fever. Other intracellular Hsps have been shown to become surface expressed under physiological stress conditions and have been implicated as adhesion factors (9, 10, 15).
In these studies, we have cloned, sequenced, and expressed the NTHI htrA gene and have shown that the H. influenzae HtrA protein has serine protease activity. This activity can be abolished by site-directed mutagenesis of the htrA gene, and the resultant H91A and S197A HtrA analogs induce antibodies which are partially protective in a passive model of bacteremia. In the chinchilla model of otitis media, H91A was also partially protective, indicating that this antigen may be suitable for inclusion in a multicomponent otitis media vaccine. A phase I clinical trial of the H91A HtrA protein is in progress.
ACKNOWLEDGMENTS
We thank Bill Bradley for synthesis of oligonucleotides and Diane England for DNA sequencing. M. Haer, D. Persaud, and W. Xu-Li are acknowledged for their excellent technical assistance.
REFERENCES
- 1.Barbour M L, Mayon-White R T, Coles C, Crook D W M, Moxon E R. The impact of conjugate vaccine on carriage of Haemophilus influenzae type b. J Infect Dis. 1995;171:93–98. doi: 10.1093/infdis/171.1.93. [DOI] [PubMed] [Google Scholar]
- 2.Barenkamp S. Protection by serum antibodies in experimental nontypable [sic] Haemophilus influenzae otitis media. Infect Immun. 1986;52:572–578. doi: 10.1128/iai.52.2.572-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass S, Gu Q, Christen A. Multicopy suppressors of Prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DskA, and a truncated RlpA. J Bacteriol. 1996;178:1154–1161. doi: 10.1128/jb.178.4.1154-1161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazan J F, Fletterick R J. Structural and catalytic models of trypsin-like viral proteases. Semin Virol. 1990;1:311–322. [Google Scholar]
- 5.Bernstein F C, Koetzle T F, Williams G J B, Meyer E F, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988;334:528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- 7.Chuang S-E, Blattner F R. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elzer P H, Phillips R W, Robertson G T, Roop R M., II The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect Immun. 1996;64:4838–4841. doi: 10.1128/iai.64.11.4838-4841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensgraber M, Loos M. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect Immun. 1992;60:3072–3078. doi: 10.1128/iai.60.8.3072-3078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans D J, Jr, Evans D G, Engstrand L, Graham D Y. Urease-associated heat shock protein of Helicobacter pylori. Infect Immun. 1992;60:2125–2127. doi: 10.1128/iai.60.5.2125-2127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Fujinaga M, Delbaere L T J, Brayer G D, James M N G. Refined crystal structure of α-lytic protease at 1.7 Å resolution. J Mol Biol. 1985;183:479–502. doi: 10.1016/0022-2836(85)90296-7. [DOI] [PubMed] [Google Scholar]
- 13.Greer J. Comparative molecular modeling methods: application to the family of the mammalian serine proteases. Proteins. 1990;7:317–334. doi: 10.1002/prot.340070404. [DOI] [PubMed] [Google Scholar]
- 14.Harkness R E, Chong P, Klein M H. Identification of two iron-repressed periplasmic proteins in Haemophilus influenzae. J Bacteriol. 1992;174:2425–2430. doi: 10.1128/jb.174.8.2425-2430.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartmann E, Lingwood C. Brief heat shock treatment induces a long-lasting alteration in the glycolipid receptor binding specificity and growth rate of Haemophilus influenzae. Infect Immun. 1997;65:1729–1733. doi: 10.1128/iai.65.5.1729-1733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson K, Charles I, Dougan G, Pickard D, O’Gaora P, Costa G, Ali T, Miller I, Hormaeche C. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 17.Klein J O. Lessons from recent studies on the epidemiology of otitis media. Pediatr Infect Dis J. 1994;13:1031–1034. doi: 10.1097/00006454-199411000-00031. [DOI] [PubMed] [Google Scholar]
- 18.Kroll J S, Moxon R, Loynds B M. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J Infect Dis. 1994;169:676–679. doi: 10.1093/infdis/169.3.676. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Li S-R, Dorrell N, Everest P H, Dougan G, Wren B W. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect Immun. 1996;64:2088–2094. doi: 10.1128/iai.64.6.2088-2094.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipinska B, Fayet O, Baird L, Georgopoulos C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J Bacteriol. 1989;171:1574–1584. doi: 10.1128/jb.171.3.1574-1584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loosmore S M, Yang Y-P, Coleman D C, Shortreed J M, England D M, Harkness R E, Chong P S-C, Klein M H. Cloning and expression of the Haemophilus influenzae transferrin receptor genes. Mol Microbiol. 1996;19:575–586. doi: 10.1046/j.1365-2958.1996.406943.x. [DOI] [PubMed] [Google Scholar]
- 24.Munson R S, Jr, Granoff D M. Purification and partial characterization of outer membrane proteins P5 and P6 from Haemophilus influenzae type b. Infect Immun. 1985;49:544–549. doi: 10.1128/iai.49.3.544-549.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitta D M, Jackson M A, Burry V F, Olson L C. Invasive Haemophilus influenzae type f disease. Pediatr Infect Dis J. 1995;14:157–160. [PubMed] [Google Scholar]
- 26.Panezutti H, James O, Hanson E J, Choi Y, Harkness R E, Klein M H, Chong P. Identification of surface-exposed B-cell epitopes recognized by Haemophilus influenzae type b P1-specific monoclonal antibodies. Infect Immun. 1993;61:1867–1872. doi: 10.1128/iai.61.5.1867-1872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohl T. Concentration of proteins and removal of solutes. Methods Enzymol. 1990;182:68–89. doi: 10.1016/0076-6879(90)82009-q. [DOI] [PubMed] [Google Scholar]
- 28.Ruoslahti E, Pierschbacher M D. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Schoendorf K C, Adams W G, Kiely J L, Wenger J D. National trends in Haemophilus influenzae meningitis mortality and hospitalization among children, 1980–1991. Pediatrics. 1994;93:663–668. [PubMed] [Google Scholar]
- 31.Skorko-Glonek J, Wawrzynow A, Krzewski K, Kurpierz K, Lipinska B. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene. 1995;163:47–52. doi: 10.1016/0378-1119(95)00406-v. [DOI] [PubMed] [Google Scholar]
- 32.Skorko-Glonek J, Krzewski K, Lipinska B, Bertoli E, Tanfani F. Comparison of the structure of wild-type HtrA heat shock protease and mutant HtrA proteins. J Biol Chem. 1995;270:11140–11146. doi: 10.1074/jbc.270.19.11140. [DOI] [PubMed] [Google Scholar]
- 33.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waggoner-Fountain L A, Hendley J O, Cody E J, Perriello V A, Donowitz L G. The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin Infect Dis. 1995;21:1322–1324. doi: 10.1093/clinids/21.5.1322. [DOI] [PubMed] [Google Scholar]
- 35.Waller P R H, Sauer R T. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J Bacteriol. 1996;178:1146–1153. doi: 10.1128/jb.178.4.1146-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstein D L, Turkovski S M, Kerry C F, Krivan H C, Samuel J E. Abstracts of the 92nd General Meeting of the American Society for Microbiology 1992. Washington, D.C: American Society for Microbiology; 1992. Cloning and characterization of an [sic] Haemophilus influenzae type b adhesin, abstr. D-243; p. 136. [Google Scholar]
- 37.Wishart D S, Boyko R F, Sykes B D. Constrained multiple sequence alignment using XALIGN. Comput Appl Biosci. 1994;10:687–688. doi: 10.1093/bioinformatics/10.6.687. [DOI] [PubMed] [Google Scholar]