Abstract
In this work, we analyse the potential for using structural knowledge to improve the detection of the DNA-binding helix–turn–helix (HTH) motif from sequence. Starting from a set of DNA-binding protein structures that include a functional HTH motif and have no apparent sequence similarity to each other, two different libraries of hidden Markov models (HMMs) were built. One library included sequence models of whole DNA-binding domains, which incorporate the HTH motif, the second library included shorter models of ‘partial’ domains, representing only the fraction of the domain that corresponds to the functionally relevant HTH motif itself. The libraries were scanned against a dataset of protein sequences, some containing the HTH motifs, others not. HMM predictions were compared with the results obtained from a previously published structure-based method and subsequently combined with it. The combined method proved more effective than either of the single-featured approaches, showing that information carried by motif sequences and motif structures are to some extent complementary and can successfully be used together for the detection of DNA-binding HTHs in proteins of unknown function.
INTRODUCTION
DNA-binding proteins play a pivotal role in the biology of the cell, being responsible for the transfer of biological information from genes to proteins, and have been estimated to constitute ∼6–7% of all proteins expressed by eukaryotic genomes (1,2). A large number of DNA-binding proteins and protein–DNA complexes are deposited in the Protein Data Bank (PDB) (3) and in the Nucleic Acid Database (4).
With the advent of structural genomics projects (5), an increasing number of protein structures with little or no sequence similarity to current PDB entries and little function information are being solved. Consequently, the derivation of methods and tools for protein function prediction constitutes an important scientific challenge and will assume a key role in the function annotation of these structures and in the understanding of wider biological mechanisms. For instance, the numerous efforts to understand the complex control of gene expression will require as a first step the identification of all putative transcription factors, that constitute one of the major classes of DNA-binding proteins, for their subsequent experimental validation.
Many known DNA-binding proteins have been observed to bind DNA by a number of distinct structural motifs, such as the helix–turn–helix (HTH) motif, the helix–loop–helix motif, the helix–hairpin–helix motif and the zinc finger motif (6). After the determination of crystal structures of C1 and Cro repressor proteins from bacteriophage lambda (7,8), the DNA-binding HTH structural motif has become one of the most important and studied examples of the interaction between proteins and DNA. The HTH is a short motif made up of a first alpha-helix, a connecting turn and a second helix, which specifically interacts with the DNA and is known as the recognition helix. The two alpha-helices extend from the domain surface and constitute a convex unit able to fit into the major groove of DNA (9–11).
Several approaches for the detection of proteins containing the HTH motif can be found in the recent literature. Structure-based methods include scanning of 3D structural templates (12), use of the electrostatic potential to select generic DNA-binding residue patches (13,14) and a statistical model based on geometrical measures (15), such as the recognition helix/second helix hydrophobic interaction area, helix average relative solvent accessibility, etc. Apart from several consensus-based and profile-based approaches dating back to the 1990s or earlier (16) and a number of evolutionary studies (17–19), only two sequence-based methods were published recently, the first based on a pattern dictionary (16,20) and the second involving a fully connected two-layered neural network on a series of structural and sequence features for the prediction of DNA-binding proteins and residues (21). A further approach for the prediction of the nucleic-acid-binding function was based on the quantitative analysis of several structural features calculated only on positively charged electrostatic protein surfaces. In this case, the authors implemented a neural network and a generalized linear model and were able to discriminate proteins that bind double-stranded DNA among all other proteins containing positively charged electrostatic patches (22).
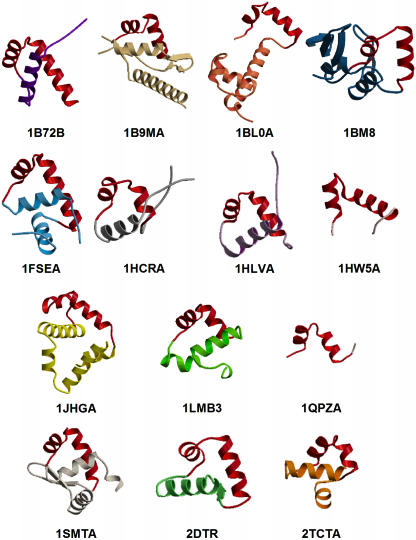
The main goal of this study was to analyse the potential for using structural knowledge to improve the detection of the DNA-binding HTH motif from sequence. In particular, we aimed to verify whether sequence information taken only from the structural motif itself is more powerful for the detection of the motif than the information derived from the corresponding whole DNA-binding domain sequence. The main difference between DNA-binding domains and their corresponding DNA-binding HTH motifs is the size. Figure 1 shows 14 DNA-binding HTH proteins from the PDB, representing as many non-homologous HTH protein families as possible. The picture clearly illustrates that HTH structural motifs have an almost constant sequence length: the average length calculated for the 14 HTHs is 25 amino acids, with a minimum of 20, a maximum of 32 and a standard deviation of 3.5 residues. In contrast, the size of whole DNA-binding domains varies quite broadly among the HTH protein families. In fact, the smallest of the shown domains, from the purine nucleotide synthesis repressor (PDB entry 1QPZ, chain A), is 25 residues long; the largest, from the transcription factor Mbp1 (PDB entry 1BM8, unique chain), is 153 residues long (average length and standard deviation within the 14 domains are 67 and 33 amino acids, respectively). Moreover, protein families with DNA-binding HTHs are known to have diverged greatly, exhibiting a near-maximal variation in both amino acid sequence and structural elements outside of the DNA-binding motif (18), and share very low similarity even in the sequence portions corresponding to the DNA-binding domains. Their evolutionary relationships are generally hidden in the ‘midnight zone’ or no longer apparent, i.e. in the area of sequence identity where an evolutionary link is only visible through their structures. Therefore, it seems likely that at least some of these domains are unrelated and may have arisen independently. In such cases, the information carried by the whole domain sequence could be unnecessary or even misleading in the development of a sequence-based approach for the detection of this motif. To explore this idea, several issues needed to be analysed: the ability of the complete domain sequence and of the structural motif sequence separately to detect the HTH motif, its comparison with the detection ability of structure-based methods, and the combination of the two approaches.
Figure 1.
Structures of DNA-binding domains of the 14 representative structures in set1. HTH structural motifs are highlighted in red. Structures are labelled by the standard four letter PDB code. The fifth letter, where found, indicates the protein chain shown. A gap in the domain structure of 1BM9A is visible and is due to the lack of coordinates for residues 69–73 in the PDB structure itself.
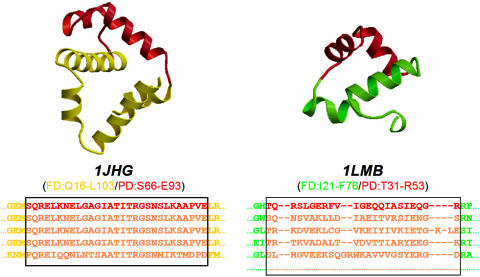
We therefore sought to derive models that are able to recognize either HTH motifs in distantly related sequences or HTH motifs that had evolved independently and are found in unrelated proteins. Hidden Markov models (HMMs), previously proved to be among the best profile-based methods (23), were chosen for the pattern detection. Two different types of HMMs were derived: one based on multiple alignments of the whole DNA-binding domain sequences (FD, full domain) and the other based on multiple alignments of the HTH structural motif sequences only (PD, partial domain), as shown in Figure 2. The figure shows the multiple alignments and the DNA-binding domains of two HTH protein families from the Pfam database (24), the family of the trp operon repressor (UniProt code TRPR_ECOLI; PDB code 1JHG, chain A) and the family of the lambda repressor/operator (UniProt code RPC1_LAMBD; PDB code 1LMB, chain 3). For both, the FD multiple alignment corresponds to the Pfam multiple alignments (shown in part below the structures owing to space constraints), while the PD multiple alignment consists only of the portion of multiple alignment delimited by the solid-lined box, that corresponds to the HTH sequence of the representative structure of the family. Therefore, two HMM libraries, corresponding, respectively, to the PD and the FD alignments, were set up. Their HTH detection ability was compared with the method of 3D structural templates described by Jones et al. (12) and evaluated using two approaches: the cross-hit detection, by jackknife tests on a set of non-homologous protein families and their representative sequences (set1), and the detection of known HTHs, by tests on a large set of structurally classified protein sequences (set2). The two methods were then combined into a sequence-plus-structure searching tool. Its ability to detect new-HTH motifs was validated by tests on a set of sequences from structures completely unrelated to any structural classification scheme (set3-unclassified) and on a set of hypothetical proteins of unknown function (set4). Results obtained from all the analyses performed showed that the HTH sequence information is complementary to the corresponding structural information and that the sequence-plus-structure method performs better than either single-feature approach.
Figure 2.
Definition of FD and PD multiple alignments: DNA-binding domains of proteins 1JHG and 1LMB from set1 are shown. Structures: the HTH motifs are highlighted in red. Multiple sequence alignments: all the shown alignments correspond to families Pfam seed alignments. The solid-lined boxes set the edges of multiple sequence alignments corresponding to the HTH sequences as found in the family templates 1JHG and 1LMB (red sequence).
MATERIALS AND METHODS
Description and definition of datasets used
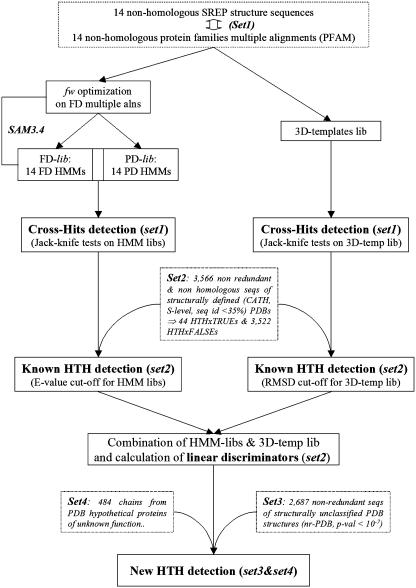
Four datasets of sequences, named set1, set2, set3 and set4, were used in the derivation of the method and in its validation. A schematic flowchart of datasets and methods is shown in Figure 3.
Figure 3.
Flowchart of methods. Dashed-lined boxes indicate descriptions of datasets; solid-lined boxes denote the steps of the methodology; fw represents the entropy value; FD and PD stands for full domain and partial domain, respectively.
Set1: A set of non-homologous protein families and structure representatives with a known DNA-binding HTH
Set1 is composed of 14 sequences of structure representatives of 14 non-homologous HTH protein families plus their multiple alignments (protein families are listed in Table 1 and their DNA-binding domains are shown in Figure 1). The structures/families were identified by CATH (25) and Pfam (24) databases and their degree of mutual similarity was checked by a PSI-BLAST search of the representative structure sequences on the PDB (number of iterations set to 20 and E-value threshold for the inclusion in the profile set to 10−10). In the CATH classification scheme, the structures included were SREPs (S-level clustering REPresentative structures), with each one representing a set of domains that clustered in the same sequence family, in which all members have sequence identities >35% to each other. Seed multiple alignments were taken from Pfam for all the representative sequences but the Hin recombinase (PDB entry 1HCR, UniProt code HIN_SALTY), for which the Pfam full alignment was taken because its sequence was not included in the seed. It is also worth highlighting that set1 does not include families with a functional ‘winged-helix’ HTH motif and families in which the HTH sequence was not fully included in the corresponding Pfam multiple alignments, so that only the strictest HTH sequences were considered.
Table 1.
List of protein families and corresponding templates composing set1
| Protein name | PDB ID | Pfam ID | UniProt ID |
|---|---|---|---|
| Homeobox protein Hox-B1 | 1B72 | Homeobox | PBX1_HUMAN |
| Molybdate-dependent transcription regulator (Mode) | 1B9M | HTH_1 | MODE_ECOLI |
| Multiple antibiotic resistance protein (Mara) | 1BL0 | HTH_AraC | MARA_ECOLI |
| Transcription factor Mbp1 | 1BM8 | APSES | MBP1_YEAST |
| Gere regulatory protein | 1FSE | GerE | GERE_BACSU |
| Hin recombinase | 1HCR | HTH_7 | HIN_SALTY |
| Major centromere autoantigen B | 1HLV | CENP-B-N | CENB_HUMAN |
| Camp receptor protein | 1HW5 | Crp | CRP_ECOLI |
| Trp operon repressor | 1JHG | Trp_repressor | TRPR_ECOLI |
| Lambda repressor/operator complex | 1LMB | HTH_3 | RPC1_LAMBD |
| Purine nucleotide synthesis repressor | 1QPZ | lacI | PURR_ECOLI |
| Transcriptional repressor Smtb | 1SMT | HTH_5 | SMTB_SYNP7 |
| Diphtheria toxin repressor | 2DTR | Fe_dep_repress | DTXR_CORDI |
| Tetracycline repressor | 2TCT | tetR | TER4_ECOLI |
For every entry in set1, two different types of multiple alignments were considered and two subsets were defined. The first (FD-set1) was based on the FD annotated in Pfam and carries the sequence information related to the whole DNA-binding domain, the second (PD-set1) was based only on the sequence segment corresponding to the HTH structural motif. FD-set1 and PD-set1 were used to derive the two libraries of HMMs.
Set2 and set3: Two large sets of non-redundant sequences
Set2 is a large and structurally defined set of sequences derived from CATH, and consists of all the PDB entries with their CATH numbers differing at the SREP level (i.e. at the fifth digit). To reduce the danger of over-prediction, the set was made as unrelated as possible to set1 by excluding the original seed sequences used to derive the HMMs (Table 1). The final set2 includes 3566 sequences, 44 of which include a known DNA-binding HTH motif (referred to as HTHxTRUE), and 3522 non-HTH proteins (HTHxFALSE).
Set3 consists of a large number of non-redundant sequences without structural classification from the nr-PDB dataset, available at the NCBI website (http://www.ncbi.nlm.nih.gov/Structure/VAST/nrpdb.html). A P-value threshold of 10−7 was chosen and the corresponding 2687 sequences were taken.
Set4: A set of sequences of hypothetical proteins of unknown function
Set4 includes all hypothetical protein chains (477) of unknown function as found in the PDB, and includes seven chains described as putative DNA-binding proteins in the corresponding PDB file headers.
Set up of HMM libraries and jackknife tests
HMM libraries were generated from full and partial multiple alignments using the SAM package (26). For the FD-set1, the HMMs available in Pfam were not used but new models were generated, using identical parameters for all models, which then could be reliably compared with the HMMs based on PD-set1 alignments. The ability of the HMM libraries to detect cross-hits was evaluated by a series of jackknife tests, in which the sequence of the representative structure of each protein family was scored by the remaining 13 HMMs. The total number of possible predictions (Nt), also corresponding to the maximum number of detectable true hits, is Nt + N (N − 1), where N is the number of protein families. This gives 182 possible true predictions for 14 families. The number of detected hits, i.e. hits showing an E-value lower than a chosen threshold, was then used to compare the prediction effectiveness of different models and libraries.
Optimizing the entropy value for the HMM generation
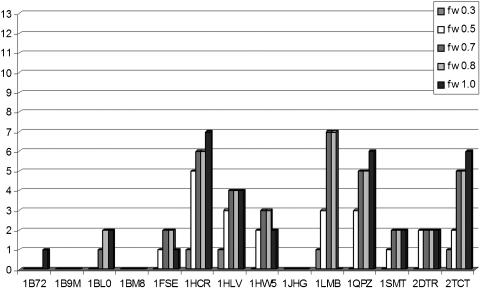
The pattern detection ability of HMMs is known to be sensitive to the length of the strings used to derive the models, meaning that longer strings generate more powerful HMMs. Therefore, owing to the very short size of PD-set1 multiple alignments, the entropy value fw for the generation of the HMM libraries was systematically optimized (fw parameter: 0.3, 0.5, 0.7, 0.8 or 1.0). The entropy value adjusts the weights assigned for insertions, deletions and replacements consistently with the data in the alignment and with the rules provided by the used regularizer (i.e. the lower the entropy value, the more specific the HMM derived). It was optimized to generate the most specific and at the same time most powerful models. Five different HMM libraries based on the 14 FD multiple alignments were built by varying the fw value and the respective predictive abilities were tested by five jackknife tests. For each HMM in the library, the number of hits with an E-value < 0.01 was counted and the total number of hits detected by all the models in the library was calculated (see Figure 4 and Table 2). As shown in Table 2, the most predictive library resulted from the FD-fw0.7, with 43 hits out of 182, followed by the FD-fw0.8, FD-fw0.5, FD-fw1.0 and FD-fw0.3, with 42, 38, 37 and 19 hits, respectively. This suggested that the method is only moderately sensitive to entropy values in the vicinity of the optimal value. Therefore, an fw value of 0.7, also corresponding to SAM default value, was chosen and two HMM libraries, FD-lib and PD-lib, corresponding to the FD and the PD multiple alignments, respectively, were built and evaluated by jackknife tests using an E-value threshold of 0.01.
Figure 4.
Number of hits detected by the 14 HMMs derived from FD multiple alignments at different fw entropy values.
Table 2.
Jackknifed prediction results at different SAM entropy values
| HMM rep structure | fw0.3a | fw0.5a | fw0.7a | fw0.8a | fw1.0a |
|---|---|---|---|---|---|
| 1B72 | 0 | 0 | 0 | 0 | 0 |
| 1B9M | 0 | 0 | 0 | 0 | 0 |
| 1BL0 | 0 | 0 | 0 | 1 | 1 |
| 1BM8 | 0 | 0 | 0 | 0 | 0 |
| 1FSE | 2 | 2 | 1 | 1 | 0 |
| 1HCR | 2 | 3 | 4 | 4 | 3 |
| 1HLV | 2 | 5 | 6 | 6 | 6 |
| 1HW5 | 0 | 2 | 3 | 3 | 2 |
| 1JHG | 0 | 0 | 0 | 0 | 0 |
| 1LMB | 2 | 8 | 10 | 8 | 8 |
| 1QPZ | 4 | 5 | 5 | 5 | 4 |
| 1SMT | 3 | 3 | 3 | 3 | 3 |
| 2DTR | 2 | 3 | 3 | 3 | 3 |
| 2TCT | 2 | 7 | 8 | 8 | 7 |
| Total hits | 19 | 38 | 43 | 42 | 37 |
afw indicates the entropy value used to derive HMMs from FD multiple alignments. The lower the entropy value, the more specific the HMM derived.
3D-Template method
The ability of the HMM libraries was compared and subsequently combined with a previously published structure-based method for the detection of DNA-binding HTHs (12) from the 3D structure, herein referred to as the 3D-template method. The method is based on the 3D structural templates, generated from HTH-containing protein structures, that comprise the alpha carbon backbone coordinates of the residues forming the HTH motifs. Such templates are scanned against other protein structures to calculate the root mean square deviation (RMSD) of the optimal superposition of a template on a structure.
For the comparison of the 3D-template method with HMM predictions and to benchmark the combined approaches, jackknife tests were performed by scanning each structure in set1 against the HTH structural motifs of the remaining proteins in the set. All corresponding RMSDs were calculated and the lowest value was kept as the reference RMSD; the published threshold of 1.6 Å was used to discriminate hits from non-hits.
For the application of the approach to all other sequence sets and for the calculation of linear discriminators (see below), RMSD values were obtained by scanning the whole protein structures onto the seven consensus templates as published by Jones et al. (12).
Combined approaches: calculation of linear discriminators and procedures of cross-validation
A method was developed to combine 3D-templates and HMMs, i.e. a sequence-plus-structure searching. Therefore, it was necessary to define a linear discriminator from the two parameters, RMSD and log (E-value). All possible lines between true hits and false hits were generated to segregate the data, and the line optimally discriminating between true and false hits was calculated. The discrimination ability of each line was defined by calculating its corresponding error rate (KErr) and Matthew's correlation coefficient (27) (Φ), and the best line was chosen as the line showing the minimum error rate and the maximum Φ value. Error rates and Φ values were calculated as follows:
where FP is the number of false positives, FN is the number of false negatives and NTot is the total number of hits.
where TP is the number of true positives, FP is the number of false positives, FN is the number of false negatives and TN is the number of true negatives.
The best line was validated by a 10-fold cross-validation procedure (100 runs). Owing to the very strong imbalance between the number of true and false hits, the two sets were resampled independently and then randomly combined at each cross-validation run, so that at each resampling the presence of true examples was guaranteed.
RESULTS AND DISCUSSION
Evaluation of cross-hit detection ability (jackknife tests on set1)
Using HMM libraries
According to the basic premise of our work, unrelated proteins that bind DNA by an HTH motif do not share sequence similarity over the whole DNA-binding domains and, therefore, the only possible pattern relationship detectable by HMMs should be strictly at the level of the HTH motif sequence. Therefore, given a set of unrelated DNA-binding protein families, and given the corresponding HMMs for both their FD and PD domains, the FD–HMMs should not give any cross-hit within the set, i.e. should not detect any of the other protein families, while the PD–HMMs, in which the information is based on the structural motif sequence only, should ideally detect all the HTHs in the set.
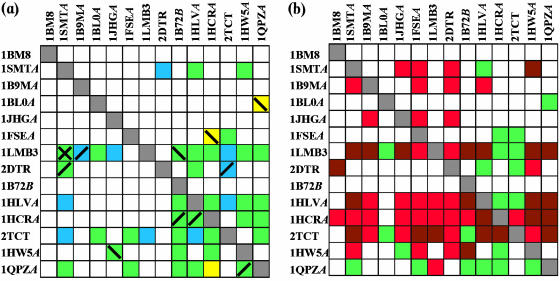
The ability of the FD and PD libraries to detect cross-hits in set1 was evaluated by a series of jackknife tests, in which the sequence of the representative structure of each protein family was scored by the remaining 13 HMMs. The theoretical maximum number of true predictions (i.e. the number of detectable true HTHs, see Materials and Methods) was 182 for the 14 protein families. The number of hits showing an E-value lower than the selected threshold (0.01) was taken to compare the prediction effectiveness of different libraries. The obtained results are shown in Figure 5a. The maximum number of cross-hits by any model in the PD-lib (yellow and green squares) was only 36 out of the possible 182 hits. The maximum number of hits detected by the FD library (blue and green squares) was 44, of which 32 were also detected by the PD library (green squares). Quite surprisingly, at this level the PD-lib was not more predictive than the FD-lib, but was slightly more restrictive. Within the FD library, the most predictive models were derived from the lambda repressor/operator complex protein family (1LMB, 10 out of 13 possible hits), from the tetracycline repressor family (2TCT, 8 hits) and from the major centromere autoantigen B family (1HLV, 6 hits). In the PD library, the highest number of hits was again detected from the lambda repressor/operator complex protein family (1LMB, 7 hits), followed by the purine nucleotide synthesis repressor family (1QPZ) and by the tetracycline receptor family (2TCT) with 6 and 5 hits, respectively. The detection of cross-hits in the FD-lib jackknife run suggested that some of the whole DNA-binding domains might indeed be related and is contrary to our original expectations. To explore this further, for each detected hit, the structural overlap between the real HTH and the predicted HTH was manually verified and the number of incorrectly aligned examples calculated (Table 3). This number was comparable for the two libraries (7 for the FD-lib and 5 for the PD-lib), but the incorrectly aligned hits were different for the FD and the PD libraries, apart from the lambda repressor/operator family in which both the FD and the PD–HMMs detected at least one incorrectly aligned example. Given the limited size of the dataset (only 14 families/structure), a robust statistical analysis could not be performed at this stage of the study. However, collected data indicated that, although the sequence information included in the two HMM libraries is different and the whole domains are not known to share evolutionary relationships, the corresponding predictive power is similar (44 and 36 detected hits over 182 possible jackknife predictions for FD and PD, respectively). Moreover, jackknife results proved that the motif information alone was not specific enough to detect HTHs, supporting the few previously published results of sequence-based methods (14,20,21) described in Introduction. In fact, 4 out of the 14 HMMs are unable to detect any hit while only 2 HMMs (corresponding to proteins 1LMB and 2TCT) detect >50% of the hits. No rationale connected to either the number of sequences in the multiple alignments or the CATH classification at the higher HREP level (percentage identity <20%) or any other observations could be found to explain these results. However, they could be interpreted in two ways: either they may reflect a hidden evolutionary relationship going back to an ancient protein family, which rapidly diverged towards several sequence-dissimilar families, or they may be due to a lack of variation in the secondary structure composition of the domains in the set, which could affect their amino acid composition. In fact, apart from the transcription factor Mbp1 family (PDB code 1BM8), all domains are mainly alpha-helical and could therefore share a local sequence similarity, which does not necessarily reflect a remote evolutionary relationship.
Figure 5.
(a) Jackknife results for the PD-lib and FD-lib libraries: yellow squares indicate hits detected from the PD-lib library, blue squares indicate hits detected from the FD-lib library and green squares indicate hits detected from both the libraries. Incorrectly aligned hits are highlighted with slashes and crosses: forward slashes (/) indicate incorrectly aligned hits detected from the FD-lib library, backward slashes (\) indicate incorrectly aligned hits detected from the PD-lib library and crosses (X) indicate incorrectly aligned hits detected from both the libraries. (b) Jackknife results for the combination of PD-lib and FD-lib libraries and the method of 3D-templates. Green squares indicate hits detected from either the FD-lib library or the PD-lib library or both; red squares indicate hits detected from the method of 3D-templates; and brown squares indicate hits detected from all the three methods.
Table 3.
HTH cross-hits
| HMM rep | Len | FD-liba | FPFD-liba | PD-liba | FPPD-liba | NSeqsa | FDmaxa | PDmaxa | Δmax |
|---|---|---|---|---|---|---|---|---|---|
| 1BM8 | 833 | 0 | 0 | 0 | 0 | 6 | 161 | 27 | 134 |
| 1SMT | 122 | 3 | 0 | 2 | 0 | 42 | 108 | 28 | 80 |
| 1B9M | 262 | 0 | 0 | 0 | 0 | 14 | 104 | 25 | 79 |
| 1BL0 | 129 | 0 | 0 | 1 | 1 | 45 | 90 | 26 | 64 |
| 1JHG | 107 | 0 | 0 | 0 | 0 | 5 | 88 | 28 | 60 |
| 1FSE | 74 | 1 | 0 | 2 | 1 | 30 | 73 | 32 | 41 |
| 1LMB | 236 | 10 | 2 | 7 | 2 | 194 | 66 | 27 | 39 |
| 2DTR | 226 | 3 | 2 | 2 | 0 | 10 | 62 | 29 | 33 |
| 1B72 | 430 | 0 | 0 | 0 | 0 | 182 | 60 | 33 | 27 |
| 1HLV | 599 | 6 | 0 | 4 | 0 | 8 | 55 | 21 | 34 |
| 1HCR | 190 | 4 | 2 | 4 | 0 | 14 | 54 | 22 | 32 |
| 2TCT | 217 | 8 | 0 | 5 | 0 | 117 | 49 | 23 | 26 |
| 1HW5 | 210 | 3 | 0 | 3 | 1 | 12 | 34 | 29 | 5 |
| 1QPZ | 340 | 5 | 1 | 6 | 0 | 27 | 34 | 24 | 10 |
| Total | 43 | 7 | 36 | 5 |
aLen, protein sequence length; FD-lib, number of its detected from the corresponding FD–HMM; PD-lib, number of hits detected from the corresponding PD–HMM; FPFD-lib, number of FP detected from the corresponding FD–HMM; FPPD-lib, number of FP detected from the corresponding PD–HMM; Nseq, number of sequences in the Pfam multiple alignment; FDmax, maximum sequence length in the FD multiple alignment; and PDmax, maximum sequence length in the PD multiple alignment.
Using 3D-templates
The sequence-based approach was then compared with a previously published structure-based approach (3D-template method) (12). The 3D-template method was applied to the 14 representative structures in set1 and the jackknife test was performed: each structure in the set was scanned against the HTH structural motifs of the remaining 13 proteins, corresponding RMSD values were calculated and the best RMSD was kept as the reference value. Results reported in Figure 5b (red squares indicate 3D-template hits, green squares PD-lib or FD-lib hits and brown square hits detected by both 3D-templates and HMM libraries) showed that the 3D-templates were better than the HMM approach, but their prediction was still in some way restricted: only 56 hits over the possible 182 were detected and again 4 structural templates detected no hit. Interestingly, only 2 of these 4 3D-templates were found to represent protein families in which the associated HMMs were also unable to detect any hit. Such evidence suggested that the motif sequence information could to some extent be complementary to the motif structure information. In fact, a simple addition of results from the two methods improved the cross-hit detection of the single-feature approaches, with an increase in the number of detected hits to 76 for the FD-lib/3D and to 67 for the PD-lib/3D.
Detection of known HTHs: scan against a structurally defined sequence set (set2)
The detection of known DNA-binding HTHs was analysed to test if the libraries can identify this motif in sequences unrelated to the protein families used to derive the HMMs. Both the FD and the PD libraries were scanned against sequences in set2. A single E-value corresponding to the best hit was calculated for each sequence in set2 by each library (i.e. the lowest E-value among the 14 values generated by the 14 HMMs constituting the library). Within each library, both the total number of hits and the fractions of HTHxTRUE andHTHxFALSE hits were calculated with different cut-offs corresponding to log(E-value) intervals of 0.2. The fraction distributions showed a strong overlapping of HTHxTRUEs and HTHxFALSEs at very high E-values, so that the definition of any E-value threshold would cause either a low sensitivity (i.e. high number of FNs) or a low specificity (i.e. a high number of FPs). Moreover, the results were again comparable for both the libraries, the only difference being the spread range of E-values. E-values resulting from the PD library were found to be between 10−6 and 105, while E-values from the FD-lib spread between 10−34 and 105. Such a difference was expected and is ascribable to the difference in lengths between the PD and the FD multiple alignments. In fact, the shortness of PD–HMMs was expected to affect the E-value itself, making it unlikely to assume very low values. However, neither library was able to discriminate satisfactorily between HTHxTRUEs and HTHxFALSEs at high E-values.
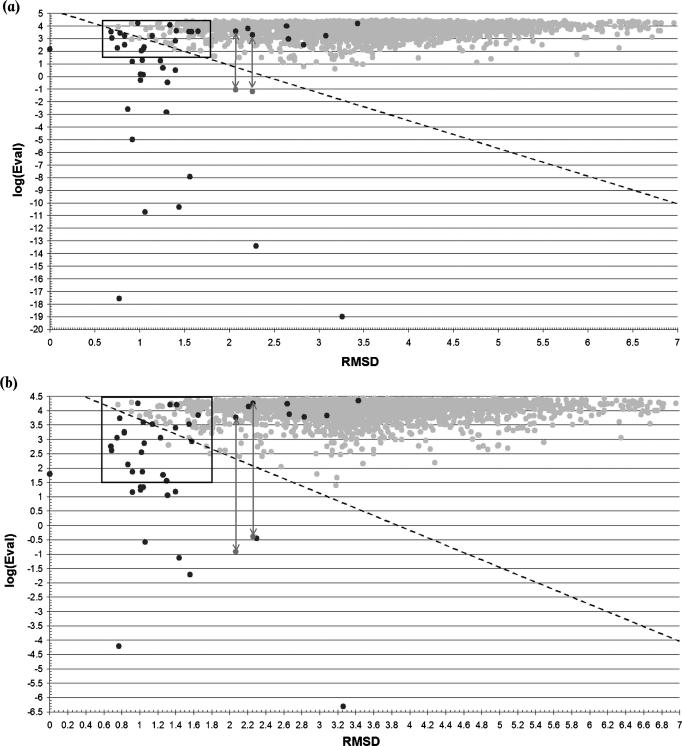
The combination of the HMM libraries and the 3D-template approach (Figure 6a and b) led to an improvement in discrimination, as results of the cross-hit detection already suggested. The linear discriminators were defined as follows (see Materials and Methods):
generate all possible lines joining each HTHxTRUE with each HTHxFALSE within a ‘critical box’, defined as the area where the maximum overlapping between HTHxTRUEs and HTHxFALSEs was observed;
calculate the corresponding error rates and Matthew's correlation coefficient (Φ) values;
identify the threshold line that optimally separates HTHxTRUEs from HTHxFALSEs (i.e. showing the best error rate and Φ);
validate using a 10-fold cross-validation procedure.
Figure 6.
RMSD/log (E-value) plots for the FD-lib/3D (a) and PD-lib/3D (b) methods. Dark grey dots correspond to HTHxFALSEs and light grey dots to HTHxTRUEs. The solid line limits the critical box used to derive the best threshold line (dashed diagonal line) as described in Materials and Methods. The two pairs of points connected by an arrow correspond to the PDB structures 1LNW (chain A) and 1IXC (chain A), detected as FNs. However, sequences of both entries show MSE instead of the canonical amino acid. The replacement of all MSE with normal Met makes the proteins to be detected as TP (points below arrows).
The same procedure was performed on the single-feature methods, i.e. FD library, PD library and 3D-templates, to allow for a statistical measure and comparison of their effectiveness. Results are reported in Table 4. It should be noted that the statistical data corresponding to best lines were calculated before the cross-validation and are liable to a certain amount of over-prediction. Therefore, cross-validated parameters were chosen as reference parameters for a better measure of the performance of each method.
Table 4.
Statistics of the 3D/log(E-value) predictions
| Method | NHBTa | TP | FP | FN | Φ | Φ (cv)a | Err% | Err%(cv)a |
|---|---|---|---|---|---|---|---|---|
| 3D | 42 | 29 | 14 | 16 | 0.610 | 0.538 | 0.81 | 0.81 |
| PD-lib | 20 | 18 | 3 | 27 | 0.570 | 0.524 | 0.84 | 0.87 |
| FD-lib | 16 | 16 | 1 | 29 | 0.562 | 0.483 | 0.84 | 0.89 |
| 3D/PD-lib | 33 | 31 | 3 | 14 | 0.785 | 0.758 | 0.47 | 0.47 |
| 3D/FD-lib | 29 | 29 | 1 | 16 | 0.782 | 0.747 | 0.48 | 0.48 |
aNHBT, number of hits below the thresholds; (cv), parameter calculated after 100 runs of 10-fold cross-validation.
Cross-validated parameters obtained for the three single-featured methods were comparable, although the FD library, in contrast to results from jackknife tests, performed slightly worse than both the PD-lib and the 3D method. Moreover, an interesting difference between the structure-based and the sequence-based methods is noticeable: HMM libraries detect a smaller number of FPs than the 3D method and therefore the result is more specific, while the 3D method detects fewer FNs with a subsequent increase in sensitivity.
It is worth highlighting that chains A of entries 1LNW and 1IXC, both known to include an HTH motif, were not detected at first by the combined approaches (PD-lib/3D and FD-lib/3D) because all methionines in their sequences are replaced by selenomethionines (MSE). Conversion of all MSEs to classical methionines shifted the two E-values to put the two hits back among the TPs.
The combined approaches improves over the single performances, keeping both the highest specificity and the highest sensitivity found in the single-featured predictions, and, although the obtained cross-validated parameters were again comparable, the PD-lib/3D method gave a better statistics overall, showing that the addition of the 3D structural information to sequence information might make the PD–HMMs to some extent more discriminating than the FD–HMMs for DNA-binding HTH motif assignments.
Using the combined method to detect new HTHs in a non-redundant PDB sequence set (set3) and in a set of sequences from the PDB proteins of unknown function (set4)
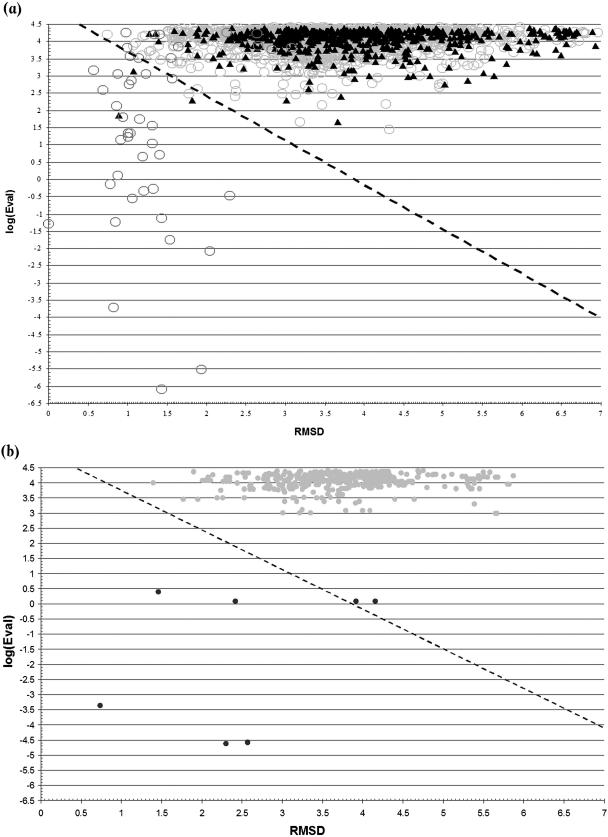
The PD-lib/3D method was applied on set3 to verify its ability to detect DNA-binding HTHs in proteins with no structural classification, and the results are shown in Figure 7a. It is worth noting that the set includes a large number of sequences corresponding to structures found in CATH and classifiable as HTHxTRUEs or HTHxFALSEs (2116 proteins: 12 with CATH SREP numbers found in set1, 34 with CATH SREP numbers found in set2-HTHxTRUE, and 2070 with CATH SREP numbers found in the set2-HTHxFALSE). Therefore, only 571 entries are structurally unclassified and represent really ‘new’ HTHs relevant for the test (set3-unclassified subset). Among the unclassified entries, three hits were detected as positive, namely PDBs 1PP8, 1Q1H and 1OKR corresponding to Ibp39 initiator binding protein, transcription factor E/Iie A and the methicillin resistance regulator protein MECI, respectively. According to their reference literature (28–30), the three hits are winged-helix proteins that bind DNA and, therefore, include an HTH motif in their DNA-binding domains. For the sake of knowledge, the subset of CATH-related entries was also analysed: 11 FNs (over HTHxTRUEs) and only 1 FP were detected, with a Φ of 0.857 and a percentage error rate of 0.57%.
Figure 7.
(a) PD-lib/3D results for the nr-PDB dataset. Light grey circles correspond to the entries related to the HTHxFALSE set; dark grey circles correspond to the entries related to the HTHxTRUE set and to the 14 family templates in the HMM set; and closed triangles correspond to the PDB entries unrelated to CATH structural classification. (b) PD-lib/3D results for hypothetical proteins in the PDB. Light grey dots correspond to the hypothetical but not DNA-binding proteins; and dark grey dots correspond to the hypothetical DNA-binding proteins.
Finally, to verify its applicability to function prediction for proteins of unknown function, the method was applied to all hypothetical proteins found in the PDB (set4). The set included 477 chains from putative not DNA-binding proteins and 7 chains from putative DNA-binding proteins; results are shown in Figure 7b. All the hypothetical falses were predicted as not-HTH chains. Five putative DNA-binding proteins were correctly assigned, while the remaining two predictions were wrong. However, the two hypothetical chains predicted as falses (1NFJ and 1NFH, chains A) were found to belong to a unique protein, the chromatin protein Alba, which is supposed to bind DNA by a flexible beta-hairpin motif and not by an HTH motif (31) and should therefore be included in the hypothetical falses. The other five trues (i.e. 1S7O, 1R7J, 1TBX, 1KU9 and 1SGM, chains A) were all found to have a putative DNA-binding winged-helix or HTH motif in their structures.
Likewise, the FD-lib/3D method was applied to set3-unclassified and set4 to verify if any significant difference between the two approaches was detectable at this level. As expected, the obtained results were comparable. Three hits from the set3-unclassified were detected as positive, two of them, 1PP8 and 1Q1H, were already been detected by the PD-lib/3D while the remaining, 1XCB (32), corresponds to a Rex family transcriptional repressor and include a winged-helix DNA-binding motif in its N-terminal domain. Predictions on set4 were identical to predictions seen for the PD-lib/3D.
CONCLUSIONS
Protein families that bind DNA by an HTH motif are known to vary widely in sequence over the whole DNA-binding domains and their relationships can often only be based on structural similarity. One might therefore assume that pattern recognition will work better if based on HTH-only motif sequences than on those from whole DNA-binding domains. The main aim of this study was to develop more powerful models to identify such motifs and to verify this hypothesis. In particular, we have explored whether using the limited HTH sequence proved more powerful than using the whole domain sequence.
Given a set of unrelated DNA-binding protein families, we were expecting that the HMMs from the HTH-only PDs would have a better detection ability than those from the full DNA-binding domain. Two separate HMM libraries were built from the two different types of multiple alignment, and used to detect DNA-binding HTH motifs in ‘unknown’ sequences. Their detection ability was analysed at three different levels, cross-hit detection, known-HTH detection and new-HTH detection, compared with the ability of a previously published structure-based method based on 3D structural templates and combined with it. Generally speaking, sequence information alone was insufficient to detect HTHs, but the addition of 3D structural information resulted in significant improvements and satisfactory performances. The results also highlighted some interesting and controversial issues. In the cross-hit detection, contrary to expectations, HMMs based on FDs were slightly more effective than models based on the HTH-only sequence, either when used separately or when combined with 3D information. This might reflect hidden evolutionary relationships but more probably can be explained by the very high percentage of alpha-helices in these domains that could influence the recognition of the HTH pattern. In contrast to that, in the known-HTH detection, in which a much larger set of proteins was considered and therefore a statistical analysis was performable, the FD–HMMs were slightly less powerful than the partial HTH-only HMMs. The best performance was obtained for the HTH-only sequence-plus-structure tool, with a significant statistical improvement over the single-feature methods (error rate ∼ halved and Matthew's correlation coefficient increased by 0.2).
The obtained results did not meet our initial expectations regarding the detection ability of PD sequences towards FD sequences. However, single-feature methods were less powerful than those combined in all tests, suggesting some non-obvious complementarity between sequence and structure information that can be successfully used for the assignments of the DNA-binding function to proteins of unknown function.
Acknowledgments
We would like to thank Dr Hannes Ponstingl for useful discussion and advices in the statistical validation, Dr Richard J. Morris for critical reading, and Professor Helen Berman and the Department of Energy of USA for funding (grant DE-FG02-96ER62166). Funding to pay the Open Access publication charges for this article was provided by the EMBL-EBI (UK).
Conflict of interest statement. None declared.
REFERENCES
- 1.Luscombe N.M., Austin S.E., Berman H.M., Thornton J.M. An overview of the structures of protein–DNA complexes. Genome Biol. 2001;1:1–37. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S., Thornton J.M. Protein–DNA interactions: the story so far and a new method for prediction. Comp. Funct. Genomics. 2003;4:428–431. doi: 10.1002/cfg.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman H.M., Olson W.K., Beveridge D.L., Westbrook J., Gelbin A., Demeny T., Hsieh S.-H., Srinivasan A.R., Schneider B. The Nucleic Acid Database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sali A. 100,000 protein structures for the biologist. Nature Struct. Biol. 1998;5:1029–1032. doi: 10.1038/4136. [DOI] [PubMed] [Google Scholar]
- 6.Harrison S.C. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- 7.Pabo C.O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982;298:443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- 8.Anderson W.F., Ohlendorf D.H., Takeda Y., Matthews B.W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981;290:754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- 9.Pabo C.O., Sauer R.T. Transcription factors: structural families and principle of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 10.Steitz T.A., Ohlendorf D.H., McKay D.B., Anderson W.F., Matthews B.W. Structural similarity in the DNA-binding domains of catabolite gene activator and cro repressor protein. Proc. Natl Acad. Sci. USA. 1982;79:3097–3100. doi: 10.1073/pnas.79.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan R.G. DNA recognition by the helix–turn–helix motif. Curr. Opin. Struct. Biol. 1992;2:100–108. [Google Scholar]
- 12.Jones S., Barker J.A., Nobeli I., Thornton J.M. Using structural motif templates to identify proteins with DNA binding function. Nucleic Acids Res. 2003;31:2811–2823. doi: 10.1093/nar/gkg386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S., Shanahan H.P., Berman H.M., Thornton J.M. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–7198. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shanahan H.P., Garcia M.A., Jones S., Thornton J.M. Identifying DNA-binding proteins using structural motifs and the electrostatic potential. Nucleic Acids Res. 2004;32:4732–4741. doi: 10.1093/nar/gkh803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin W.A., Berman H.M. Statistical models for discerning protein structures containing the DNA-binding helix–turn–helix motif. J. Mol. Biol. 2003;330:43–55. doi: 10.1016/s0022-2836(03)00532-1. [DOI] [PubMed] [Google Scholar]
- 16.Mathee K., Narasimhan G. Detection of DNA-binding helix–turn–helix motifs in proteins using the pattern dictionary method. Methods Enzymol. 2003;370:250–264. doi: 10.1016/S0076-6879(03)70022-6. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L., Koonin E.V. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosinski J.A., Atchley W.R. Molecular evolution of helix–turn–helix proteins. J. Mol. Evol. 1999;49:301–309. doi: 10.1007/pl00006552. [DOI] [PubMed] [Google Scholar]
- 19.Roy S., Sahu A., Adhya S. Evolution of DNA binding motifs and operators. Gene. 2002;285:169–173. doi: 10.1016/s0378-1119(02)00413-4. [DOI] [PubMed] [Google Scholar]
- 20.Narasihman G., Bu C., Gao Y., Wang X., Xu N., Mathee K. Mining protein sequences for motifs. J. Comp. Biol. 2002;9:707–720. doi: 10.1089/106652702761034145. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad S., Gromiha M.M., Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- 22.Stawiski E.W., Gregoret L.M., Mandel-Gutfreund Y. Annotating nucleic acid-binding function based on protein structure. J. Mol. Biol. 2003;326:1065–1079. doi: 10.1016/s0022-2836(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 23.Park J., Karplus K., Barrett C., Hughey R., Haussler D., Hubbard T., Chothia C. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J. Mol. Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- 24.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S.R., Griffiths-Jones S., Howe K.L., Marshall M., Sonnhammer E.L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 26.Hughey R., Krogh A. Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput. Appl. Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- 27.Matthews B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochem. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 28.Schumacher M.A., Lau A.O.T., Johnson P.J. Structural basis of core promoter recognition in a primitive eukaryote. Cell. 2003;115:413–424. doi: 10.1016/s0092-8674(03)00887-0. [DOI] [PubMed] [Google Scholar]
- 29.Meinhart A., Blobel J., Cramer P. An extended winged helix domain in general transcription factor E/IIE alpha. J. Biol. Chem. 2003;278:48267–48274. doi: 10.1074/jbc.M307874200. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Castellanos R., Marrero A., Mallorqui-Fernandez G., Potempa J., Coll M., Gomis-Ruth F.X. Three-dimensional structure of MecI. Molecular basis for transcriptional regulation of staphylococcal methicillin resistance. J. Biol. Chem. 2003;278:39897–39905. doi: 10.1074/jbc.M307199200. [DOI] [PubMed] [Google Scholar]
- 31.Zhao K., Chai X., Marmorstein R. Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding. J. Biol. Chem. 2003;278:26071–26077. doi: 10.1074/jbc.M303666200. [DOI] [PubMed] [Google Scholar]
- 32.Sickmier E.A., Brekasis D., Paranawithana S., Bonanno J.B., Paget M.S., Burley S.K., Kielkopf C.L. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]