Abstract
Nitric oxide (NO) induces deamination of guanine, yielding xanthine and oxanine (Oxa). Furthermore, Oxa reacts with polyamines and DNA binding proteins to form cross-link adducts. Thus, it is of interest how these lesions are processed by DNA repair enzymes in view of the genotoxic mechanism of NO. In the present study, we have examined the repair capacity for Oxa and Oxa–spermine cross-link adducts (Oxa–Sp) of enzymes involved in base excision repair (BER) and nucleotide excision repair (NER) to delineate the repair mechanism of nitrosative damage to guanine. Oligonucleotide substrates containing Oxa and Oxa–Sp were incubated with purified BER and NER enzymes or cell-free extracts (CFEs), and the damage-excising or DNA-incising activity was compared with that for control (physiological) substrates. The Oxa-excising activities of Escherichia coli and human DNA glycosylases and HeLa CFEs were 0.2–9% relative to control substrates, implying poor processing of Oxa by BER. In contrast, DNA containing Oxa–Sp was incised efficiently by UvrABC nuclease and SOS-induced E.coli CFEs, suggesting a role of NER in ameliorating genotoxic effects associated with nitrosative stress. Analyses of the activity of CFEs from NER-proficient and NER-deficient human cells on Oxa–Sp DNA confirmed further the involvement of NER in the repair of nitrosative DNA damage.
INTRODUCTION
The DNA molecules that carry the vital genetic information of cells continuously suffer from spontaneous decay due to exposure to water (1,2). Together with the most frequently occurring depurination (3,4), hydrolytic deamination of DNA bases takes place at slower but non-negligible rates (4,5). Deamination of DNA bases is further accelerated by nitrosative stress by nitric oxide (NO) and nitrous acid (6–9), in which nitrous anhydride is believed to be a key mediator. It has been postulated that in tissues with chronic inflammation, a high flux of NO secreted by activated macrophages can induce nitrosative damage to DNA in normal cells, thereby increasing cancer risk associated with chronic inflammation (10–13).
Nitrosative deamination of cytosine and adenine results in uracil and hypoxanthine (Hx), respectively. Uracil is repaired by the base excision repair (BER) pathway initiated by uracil-DNA glycosylase both in prokaryotic and eukaryotic cells (1,14,15), and a putative mammalian backup enzyme for uracil-DNA glycosylase has been identified (16,17). Hx is also repaired by the BER pathway initiated by 3-methyladenine-DNA glycosylase II (AlkA) and methylpurine-DNA glycosylase (MPG) in prokaryotic and eukaryotic cells, respectively (15,18,19). Unlike cytosine and adenine, nitrosative deamination of guanine gives rise to two products, xanthine (Xan) and oxanine (Oxa), via deamination of the 2-NH2 group followed by ring opening and rearrangement (20–23) [for a conflicting result, see Ref. (24)]. Furthermore, Oxa but not Xan reacts with polyamines such as spermine and spermidine and DNA binding proteins such as histone and DNA glycosylases, forming cross-link adducts (25).
In Escherichia coli, Xan is repaired either by the BER pathway involving AlkA (26,27) or by an alternative repair pathway involving endonuclease (Endo) V (15,28). Mammalian MPG, a functional homolog of AlkA, has been shown to excise Xan from DNA (27), indicating a similar BER pathway for the repair of Xan in mammalian cells. Although a human homolog of Endo V has been identified (29), it has not been tested for Xan. In contrast, very little is known about the repair of Oxa and its cross-link adducts (26,30,31).
In the present study, we examined systematically the excision capacity for Oxa and Oxa–spermine cross-link adducts (Oxa–Sp) of BER and nucleotide excision repair (NER) systems to delineate the repair mechanisms of these lesions. We report here that Oxa is a very poor substrate for E.coli and human DNA glycosylases and UvrABC nuclease. In contrast, Oxa–Sp is excised efficiently by UvrABC but not by DNA glycosylases, suggesting a role of NER in ameliorating genotoxic effects associated with nitrosative stress. The analyses of the activity of cell-free extracts (CFEs) from E.coli and human cells confirm further the involvement of NER in the repair of Oxa–Sp.
MATERIALS AND METHODS
Oligonucleotide substrates
The control oligonucleotide substrates used for the activity assay of BER enzymes (Table 1) were prepared by the phosphoramidite method (uracil, Hx and 7,8-dihydro-8-oxoguanine) or as reported previously [7-methylguanine (7mG), thymine glycol and an abasic (AP) site] (32,33). 25OXA (Figure 1A) was prepared as described previously (25). The oligonucleotide substrates used for the assay of NER activities are shown in Figure 1B and C. 60OXAe and 60OXAi carrying 5′-end and internal 32P-labels (Figure 1B), respectively, were prepared by DNA polymerase reactions using appropriate templates, primers, 2′-deoxyoxanosine 5′-triphosphate and normal dNTPs (25). Briefly, in the preparation of 60OXAe, a 5′-end 32P-labeled 30mer primer was extended on a 60mer template with 2′-deoxyoxanosine 5′-triphosphate followed by normal dNTPs. For the preparation of 60OXAi, an unlabeled 30mer primer was extended similarly with 2′-deoxyoxanosine 5′-triphosphate followed by dNTPs containing [α-32P]dCTP so that the region 3′ to Oxa was uniformly labeled by [32P]dCMP (Figure 1B). 60FL containing fluorescein (FL) was synthesized by the phosphoramidite method using the corresponding monomer unit (Glen Research). The bottom strand of 150OXA (150mer template, Figure 1C) was prepared by the enzymatic ligation of three 50mer fragments. The 80mer primer for the synthesis of the top strand of 150OXA was similarly prepared by the ligation of two 40mer fragments. 150OXAe and 150OXAi carrying 5′-end and internal 32P-labels (Figure 1C), respectively, were prepared by DNA polymerase reactions using the 150mer template and the 80mer primer as described for 60OXA substrates. 150FL was prepared by enzymatic ligation of 60FL and 40mer and 50mer fragments. 60OXA–SPe, 60OXA–SPi, 150OXA–SPe and 150OXA–SPi containing an Oxa–Sp cross-link adduct were prepared by the reaction of 60OXAe, 60OXAi, 150OXAe and 150OXA–SPi with spermine, respectively (25). Markers for the analysis of human NER products (Figure 1C) were prepared by ligation (59mer and 64mer) or DNA polymerase reactions followed by spermine modification (27mer bearing Oxa–Sp).
Table 1.
Control substrates and reaction buffers used in the activity assay of BER enzymes
| Substrate | Damage (X)a | Sequenceb | Enzyme testedc |
|---|---|---|---|
| 19U | U | ACAGACGCCAXCAACCAGG | Ung, hSMUG1 |
| TGTCTGCGGTGGTTGGTCC | |||
| 34MG | 7mG | GAAACACTACTXTCACCCTCCATACCCACATCCT | AlkA, hMPG |
| CTTTGTGATGACAGTGGGAGGTATGGGTGTAGGA | |||
| 25HX | Hx | GAAACACTATTCCACXCCTTCTCTC | HeLa CFE1s, hMPG |
| CTTTGTGATAAGGTGTGGAAGAGAG | |||
| 19TG | Tg | ACAGACGCCAXCAACCAGG | Endo III, Endo VIII |
| TGTCTGCGGTAGTTGGTCC | hNTH1, hNEIL1 | ||
| 25OG | 8oxoG | CATCGATAGCATCCTXCCTTCTCTC | Fpg, hOGG1 |
| GTAGCTATCGTAGGACGGAAGAGAG | |||
| 19AP | AP | ACAGACGCCAXCAACCAGG | Endo IV, hNEIL2 |
| TGTCTGCGGTGGTTGGTCC |
aU, uracil; 7mG, 7-methylguanine; Hx, hypoxanthine: Tg, thymine glycol; 8oxoG, 7,8-dihydro-8-oxoguanine; AP, abasic site.
bTop strands containing damage were 5′-end 32P-labeled.
cReaction buffer used: 20 mM Tris–HCl (pH 8.0), 1 mM EDTA and 1 mM DTT (Ung); 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.2 mM EDTA and 2 mM DTT (hSMUG1); 50 mM HEPES–KOH (pH 7.5), 1 mM EDTA and 5 mM 2-mercaptoethanol (AlkA); 50 mM HEPES–KOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, 5 mM 2-mercaptoethanol (hMPG and HeLa CFE1s); 10 mM Tris–HCl (pH 7.4), 100 mM NaCl and 1 mM EDTA (Endo III, Endo VIII, Fpg and hNEIL1); 10 mM Tris–HCl (pH 7.5), 50 mM NaCl and 1 mM EDTA (Endo IV and hNEIL2); and 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA and 1 mM DTT (hOGG1 and hNTH1).
Figure 1.
Substrates containing Oxa and Oxa–Sp. (A) Substrates for BER enzymes and human CFE1s. (B) NER substrates for UvrABC and E.coli CFEs. (C) NER substrates for human CFE2s. The abbreviations of substrates, damage (X) and the positions of 32P-labels are shown in tables under the sequences. The phosphodiester bonds incised by UvrABC and E.coli and human CFEs are indicated by vertical arrows. The sequences of oligonucleotides used as markers are indicated by horizontal arrows.
Enzymes and cells
E.coli uracil-DNA glycosylase was obtained from New England Biolabs. Other DNA glycosylases (AlkA, Endo III, Endo VIII, and formamidopyrimidine-DNA glycosylase (Fpg)) and an AP endonuclease (Endo IV) from E.coli and those from humans [single-strand-selective monofunctional uracil-DNA glycosylase (hSMUG1), an Endo III homolog (hNTH1), Endo VIII homologs (hNEIL1 and hNEIL2), 8-oxoguanine-DNA glycosylase (hOGG1)] were from laboratory stock, and purification of these enzymes has been described elsewhere (32–38). The hMPG protein lacking 1–79 residues but retaining N-glycosylase activity (39) was a gift from Dr Saparbaev. The UvrA, UvrB and UvrC proteins from Bacillus caldotenax, a thermophilic bacterium, were overexpressed in E.coli and purified as reported previously (40,41).
E.coli strains AB1157 (wild type with respect to repair capacity), BW9109 (xth) and RPC501 (xth nfo) were gifts from Dr Yonei and originally from Dr Weiss (42,43). The genotype of AB1157 is F− thr-1 ara-14 leuB6(Am) lacY1 Δ(gpt-proA2)62 tsx-33 supE44(Am) galK2 rac hisG4(Oc) rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3(Oc) thi-1, and those of BW9109 and RPC501 are the same as AB1157 but also include Δ(xthA-pncA) and Δ(xthA-pncA) nfo-1::kan, respectively. HeLa cells were from laboratory stock. NER-deficient xeroderma pigmentosum (XP) cells transformed by SV40 [XP2OS(SV) (complementation group A) and XP2YO(SV) (complementation group F)] (44,45) were obtained from the Health Science Research Resource Bank (http://www.jhsf.or.jp).
Activity assays with BER enzymes
5′-end 32P-labeled 25OXA or a control substrate (all 2 nM) was incubated with BER enzymes (10 and 100 ng) in an appropriate buffer (10 μl, Table 1) at 37°C for 30 min. The sample was mixed with gel loading buffer (0.05% xylene cyanol, 0.05% bromophenol blue, 20 mM EDTA and 98% formamide), heated and separated by 16% denaturing PAGE. Radioactivity in the gel was analyzed on a phosphorimaging analyzer Fuji BAS 2000. For monofunctional DNA glycosylases (Ung, AlkA, hSMUG1 and hMPG), the sample was mixed with 1 M NaOH (final concentration 0.1 M) after incubation with enzymes, heated briefly at 90°C to cleave AP sites, neutralized with 1 M acetic acid and subjected to PAGE analysis. For the analysis of enzymatic parameters, 5′-end 32P-labeled 25OXA and 25Hx (both 5–60 nM) were incubated with hMPG (0.6 ng) for 20 min, and products were analyzed as described above. The parameters (kcat and Km) were evaluated from Lineweaver–Burk plots.
Activity assays with UvrABC
The substrates containing Oxa (60OXAe and 60OXAi), Oxa–Sp (60OXA–SPe and 60OXA–SPi), FL (60FL) and G (60G) (all 2 nM) were incubated with UvrABC (20 nM UvrA, 60 nM UvrB and 50 nM UvrC) in UvrABC buffer [50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 50 mM KCl, 1 mM ATP and 5 mM DTT; total 20 μl] at 37 or 55°C for 1 h. The reaction was terminated by the addition of EDTA (final concentration 20 mM). The sample was heated at 95°C for 5 min and separated by 12% denaturing PAGE. Products were quantitated as described above.
Preparation of E.coli and human CFEs
E.coli cells were grown in 200 ml Luria–Bertani (LB) media (AB1157 and BW9109) or LB media supplemented with 50 μg/ml kanamycin (RPC501) until OD600 reached 0.5. Cells were harvested by centrifugation and suspended in 10 ml LB media. Half the suspended cells were irradiated with a UV lamp (40 J/m2) to induce the SOS response (46). The UV-irradiated and unirradiated cells were incubated at 37°C for 40 min with shaking. The cell suspension was mixed with an equal volume of 100 mM NaCl, 10 mM Tris–HCl (pH 8.0) and 10 mM EDTA, and cells were collected by centrifugation. The collected cells were suspended in 10 volumes of lysis buffer [10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride (PMSF) and 0.1 mg/ml lysozyme] and kept on ice for 30 min. The cells were disrupted by sonication (70 W, 15 s × 4) at an ice-cold temperature and centrifuged at 10 000 g for 15 min. The supernatant was recovered and used for activity assays. The protein concentration was determined with the BCA protein assay kit (Pierce).
CFEs from HeLa, XPA and XPF cells were prepared according to the reported procedure (47). Briefly, cells were grown in a Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum at 37°C in a 5% CO2 atmosphere until they became confluent. CFEs were prepared on ice or at 4°C. Harvested cells (∼1 ml as wet cells) were washed twice with phosphate-buffered saline, suspended in 4 ml of hypotonic buffer [10 mM Tris–HCl (pH 8.0), 1 mM EDTA and 5 mM DTT], and kept standing for 20 min. To the suspension, protease inhibitors were added (0.5 mM PMSF, 0.5 μg/ml each of leupeptin, pepstatin and chymostatin), and cells were disrupted with a Dounce homogenizer. The disrupted cells were centrifuged at 100 000 g for 30 min, and part of the supernatant (designated as CFE1s) was used for BER activity assays. The rest of supernatant was mixed with 4 ml of 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2 mM DTT, 25% sucrose and 50% glycerol. Proteins were precipitated by ammonium sulfate, and those precipitated between 10 and 80% saturation were recovered, dissolved in 25 mM HEPES–KOH (pH 7.9), 0.1 M KCl, 12 mM MgCl2, 1 mM EDTA, 2 mM DTT and 17% glycerol, and dialyzed against the same buffer (4 h × 3). The sample was centrifuged at 125 000 g for 10 min, and the supernatant (designated as CFE2s) was stored as aliquots at −80°C and used for NER activity assays. The protein concentration of CFE1s and CFE2s was determined as above.
BER and NER activity assays with CFEs
In the assays for BER activity with human CFEs, 25OXA and 25HX (both 2 nM) were incubated with HeLa CFE1s (1 or 5 μg) in hMPG buffer (10 μl, Table 1) at 37°C for 1 h, and products were analyzed as described for hMPG. In the assays for lesion excision by NER with E.coli CFEs, 60OXA–SPe (2 nM) was incubated with CFEs (2–10 μg) from E.coli AB1157, BW9109 and RPC501 in UvrABC buffer (10 μl) at 37°C for 15 min. Products were separated and quantitated as described for UvrABC. The NER assays with human CFE2s were performed by following a reported procedure (47). 150OXAe, 150OXAi, 150OXA–SPe, 150OXA–SPi or 150FL (all 2 nM) was incubated with human CFE2s (100 μg) in 40 mM HEPES–KOH (pH 7.9), 70 mM KCl, 8 mM MgCl2, 0.3 mM EDTA, 1.2 mM DTT, 2 mM ATP, 10% glycerol, 20 μM each of four dNTPs and 0.1 mg/ml BSA (total 50 μl) at 30°C for 90 min. For complementation experiments, a mixture of CFE2s from XPA (50 μg) and XPF (50 μg) cells was used for NER activity assays. After incubation, protease K (Wako) was added at the final concentration of 0.2 mg/ml, and incubation was continued at 37°C for 15 min. The sample was extracted by phenol, and DNA was recovered by ethanol precipitation. Products were separated by 10% denaturing PAGE and quantitated as described for UvrABC.
RESULTS
Activity of BER enzymes for Oxa and Oxa–Sp
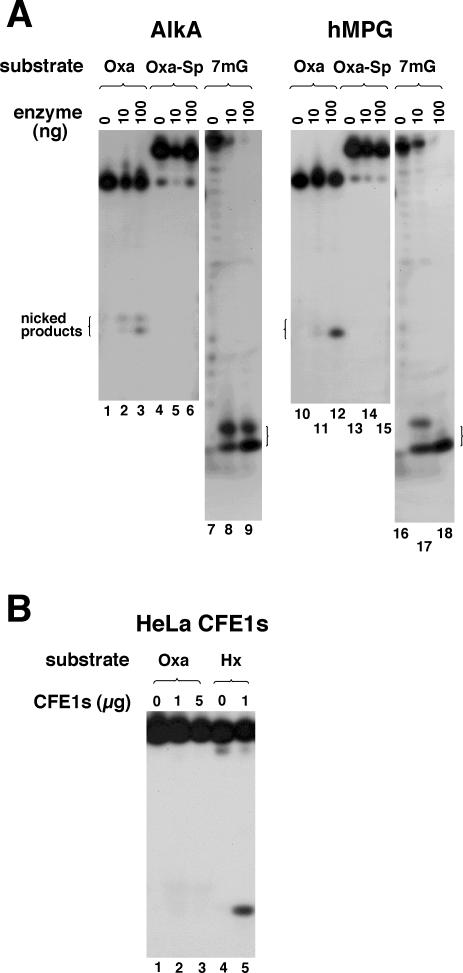
To assess the repair activity for Oxa systematically, five E.coli and six human DNA glycosylases (10 or 100 ng) were incubated with 25OXA and control substrates (all 2 nM) at 37°C for 30 min and products were analyzed by PAGE. The amounts of enzymes used were ∼15-fold (10 ng) or 150-fold (100 ng) molar excessive over the substrates so that marginal activities for Oxa could be detected. Typical gel data for AlkA and hMPG are shown in Figure 2A, where nicked products generated by enzyme and subsequent alkaline treatment are seen as a mixture of β- and β,δ-elimination products. AlkA and hMPG efficiently removed 7mG from the control substrate (lanes 8, 9, 17 and 18), but Oxa was removed poorly even with copious enzymes (lanes 3 and 12). Endo VIII, Fpg and hNEIL1 also exhibited Oxa-excising activities but they were much weaker than those of AlkA and hMPG (gel data not shown). Quantitation of the amount of damage excised per ng of enzyme indicated that the activities of the DNA glycosylases were 0.2–9% of those for their control (physiological) substrates (Table 2). hMPG excised 7mG slightly better than Hx as control substrates. Thus, the Oxa-excising activity of hMPG relative to Hx was higher than that relative to 7mG (Table 2). The enzymatic parameters of hMPG for Oxa and Hx were also evaluated for quantitative comparison of activities. The kcat and Km values for Oxa were 2.1 × 10−3 min−1 and 30 nM, respectively, and those for Hx were 1.5 × 10−2 min−1 and 15 nM, respectively, indicating that the excision efficiency (kcat/Km) of hMPG for Oxa is 14-fold lower than that for Hx. Several of the DNA glycosylases tested here react with Oxa in DNA and form cross-link adducts (25,48), which could compromise the Oxa-excising efficiency by limiting available substrates or enzymes. However, the amount of DNA glycosylases used here was excessive over the substrate (roughly by 15- and 150-fold), and the fraction of the cross-linked substrate was <5% under the present conditions (data not shown). Thus, the poor activity of DNA glycosylases for Oxa was not due to the limited availability of enzymes or substrates. We also tested Endo IV, an E.coli AP endonuclease that has been shown to recognize various oxidative and other base lesions (1,49–51), but it was not found to be active for Oxa (Table 2). Considering that BER enzymes tested here are responsible for the repair of a wide range of base damage in cells (48,52), their marginal activity for Oxa implies that this DNA lesion is poorly repaired by the BER pathway in E.coli and human cells.
Figure 2.
PAGE analysis of BER activities of AlkA, hMPG and HeLa CFE1s for Oxa and Oxa–Sp. (A) 25OXA, 25OXA–SP and 34MG (all 2 nM, Figure 1A and Table 1) were incubated with the indicated amounts of AlkA and hMPG at 37°C for 30 min. After incubation, the reaction mixture was treated with 0.1 M NaOH to cleave AP sites, and products were separated by 16% denaturing PAGE. The nicked products due to β-elimination (upper bands) and β,δ-elimination (lower bands) are indicated by open brackets. (B) 25OXA and 25HX (both 2 nM, Figure 1A and Table 1) were incubated in hMPG buffer with the indicated amounts of HeLa CFE1s at 37°C for 1 h. Products were analyzed as described above.
Table 2.
Activity of BER enzymes for Oxa and Oxa–Sp
| Enzyme | Control substratea | Activity Oxab,c | Oxa–Spb |
|---|---|---|---|
| E.coli | |||
| Ung | U | n.d. | n.d. |
| AlkA | 7mG | Weak (1%) | n.d. |
| Endo III | Tg | n.d. | n.d. |
| Endo VIII | Tg | Weak (0.2%) | n.d. |
| Fpg | 8oxoG | Weak (0.5%) | n.d. |
| Endo IV | AP | n.d. | n.d. |
| Human | |||
| hSMUG1 | U | n.d. | n.d. |
| hMPG | 7mG | Weak (4%) | n.d. |
| hMPG | Hx | Weak (9%) | n.d. |
| hNTH1 | Tg | n.d. | n.d. |
| hNEIL1 | Tg | Weak (0.2%) | n.d. |
| hNEIL2 | AP | n.d. | n.d. |
| hOGG1 | 8oxoG | n.d. | n.d. |
aU, uracil; 7mG, 7-methylguanine; Tg, thymine glycol; 8oxoG, 7,8-dihydro-8-oxoguanine; AP, abasic site; Hx, hypoxanthine. The sequences of substrates are given in Table 1.
bn.d., activity was not detectable.
cThe percentage in parentheses indicates activity relative to that for the control substrate.
The activity of BER enzymes for Oxa–Sp was similarly tested using 25OXA–SP as a substrate, but none of the enzymes showed detectable activity (Figure 2A, lanes 4–6 and 13–15, and Table 2). It is likely that BER enzymes (DNA glycosylases) can not accommodate the bulky Oxa–Sp lesion in the active site pocket since they generally recognize relatively minor base modifications induced by oxidation, alkylation and deamination.
Activity of UvrABC for Oxa and Oxa–Sp
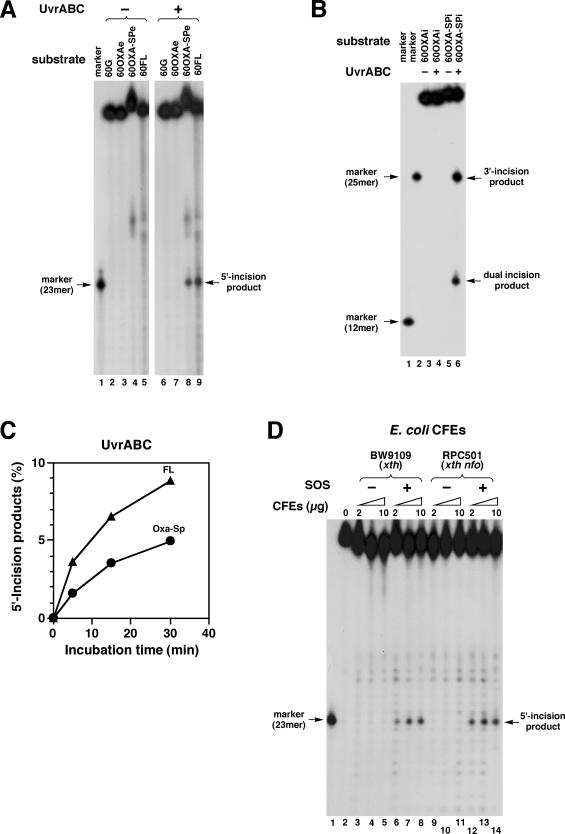
The thermophilic UvrABC from B.caldotenax was used for activity assays because of its stable and robust in vitro NER activity (40,41). To assess the activity of UvrABC for Oxa and Oxa–Sp, UvrABC was incubated with 60OXAe and 60OXA–SPe (both were 5′-end labeled and 2 nM) at 37°C for 1 h. Experiments with control substrates containing G (60G) and FL (60FL) were also performed in parallel. It has been shown that UvrABC recognizes FL, an artificial substrate, and efficiently makes incisions on DNA containing FL (41). No incision was observed for 60OXAe, indicating that Oxa is not a substrate for UvrABC (Figure 3A, lane 7). In contrast, the PAGE analysis of products for 60OXA–SPe and 60FL revealed incision products (lanes 8 and 9), whose gel mobilities were comparable to that of a 23mer marker (lane 1, see also Figure 1B for the sequence). Thus, UvrABC made an incision at the 8th phosphodiester bond 5′ to Oxa–Sp and FL. Considering that the UvrABC proteins are from a thermophilic bacterium, UvrABC was also incubated with substrates at 55°C. However, the increase in temperature led to no significant increase in the activity. To clarify whether UvrABC introduced an additional 3′ incision, 60OXAi and 60OXA–SPi carrying internal labels (Figure 1B) were similarly incubated with UvrABC, and products were analyzed by PAGE. Again no incision was observed for 60OXAi (Figure 3B, lane 4). For 60OXA–SPi, two labeled fragments were generated (lane 6). The gel mobility of a large fragment was comparable to that of a 25mer marker (lane 1, see also Figure 1B for the sequence), indicating an incision at the 5th phosphodiester bond 3′ to Oxa–Sp. The small fragment was a dual incision product, a 12mer bearing an Oxa–Sp adduct (Figure 1B), and it migrated slower than a 12mer marker without damage (Figure 3B, lane 1). It is noted that the parental oligonucleotide containing Oxa–Sp also exhibited slower gel mobility than those containing G or Oxa (Figure 3A, lanes 2–4). All components of UvrABC were essential for the incision of substrates containing Oxa–Sp so that no incision occurred in the absence of either UvrA, UvrB or UvrC (data not shown).
Figure 3.
Analysis of NER activities of UvrABC and E.coli CFEs for Oxa and Oxa–Sp. (A) 60G, 60OXAe, 60OXA–SPe and 60FL (all 2 nM and 5′-end labeled, Figure 1B) were incubated with UvrABC (20 nM UvrA, 60 nM UvrB and 50 nM UvrC) at 37°C for 1 h. Products were analyzed by 12% denaturing PAGE. The sequence of the 23mer marker is given in Figure 1B. (B) 60OXAi and 60OXA–SPi (all 2 nM and internally labeled, Figure 1B) were incubated with UvrABC, and products were analyzed as in panel A. The sequences of the 12mer and 25mer markers are given in Figure 1B. (C) 60OXA–SPe and 60 FL were incubated with UvrABC as described in panel A for up to 30 min. The amounts of 5′ incision products are plotted against incubation time. Symbols: circles, Oxa–Sp; triangles, FL. (D) 60OXA–SPe (2 nM) was incubated at 37°C for 15 min with CFEs (2, 5 and 10 μg) from E.coli BW9109 (xth) and RPC501 (xth nfo) that were pre-irradiated without (−SOS) or with (+SOS) UV. Products were analyzed as in panel A.
60OXA–SPe and 60FL were incubated with UvrABC for up to 30 min, and activities for Oxa–Sp and FL were quantitatively compared by following the time course of incision. The amount of incision products for Oxa–Sp was approximately two-thirds of that for FL throughout the reaction (Figure 3C). Considering that FL serves as a good substrate for UvrABC (41), the present result indicates that Oxa–Sp is a fairly good substrate for UvrABC.
Activity of human CFE1s for Oxa
25OXA and 25HX (2 nM) were incubated with HeLa CFE1s (1 and 5 μg) in hMPG buffer at 37°C for 1 h, and reaction products were subjected to alkaline treatment to cleave AP sites. PAGE analysis of products revealed that Hx was efficiently removed by HeLa CFE1s (Figure 2B, lane 5), whereas Oxa was removed very poorly (lanes 2 and 3). The amount of Oxa excised from DNA (per μg of CFE1s) was at most a few percentage points of that of Hx, in keeping with the result with purified hMPG (Table 2).
Activity of E.coli CFEs for Oxa–Sp
60OXA–SPe was incubated with E.coli CFEs, and products were analyzed by PAGE. When 60OXA–SPe (2 nM) was incubated with CFEs (2–10 μg) from E.coli AB1157 (xth wild type) in UvrABC buffer containing Mg2+, the substrate was degraded exonucleolytically (data not shown). Thus, it was not clear whether damage-specific incision occurred on the substrate. However, when 60OXA–SPe was incubated with CFEs from SOS-induced BW9109 or RPC501 (both xth mutants), specific incision products were observed (Figure 3D, lanes 6–8 and 12–14). These incision products were 23mers (lane 1), indicating that the incision site with the CFEs was the same as that with UvrABC (Figure 3A). The incision was dependent on the SOS response (i.e. UV-induction of UvrA and UvrB proteins) so that no obvious incision occurred with CFEs without SOS induction (Figure 3D, lanes 3–5 and 9–11). These results indicate that DNA containing Oxa–Sp is specifically incised by cellular UvrABC and probably repaired by the NER pathway.
NER activity of human CFEs for Oxa–Sp
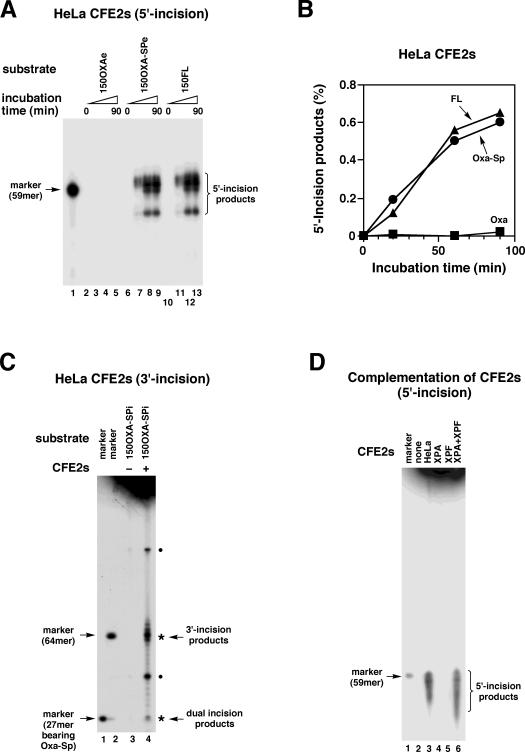
The NER activity of human cells for Oxa–Sp was initially examined using 150OXA–SPe together with 150OXAe and 150FL as control substrates (Figure 1C). The substrates (2 nM) were incubated with HeLa CFE2s (100 μg) at 30°C for up to 90 min, and products were analyzed by PAGE. Although no incision was observed with 150OXAe (Figure 4A, lanes 2–5), 150OXA–SPe and 150FL were incised by HeLa CFE2s (lanes 6–13). The cleavage pattern of DNA was similar for 150OXA–SPe and 150FL. The incision products were tentatively assigned as 54mer and 58–60mers by comparison with the gel mobility of a 59mer marker (lane 1), suggesting that the 21st–23rd and 27th phosphodiester bonds 5′ to the lesions were cleaved (Figure 1C). Figure 4B shows the relationship between incubation time and the amount of 5′ incision products for 150OXA–SPe and 150FL, which reveals that HeLa CFE2s exhibit comparable activities for Oxa–Sp and FL. To analyze additional 3′ incisions, 150OXA–SPi carrying internal labels (Figure 1C) was similarly incubated with HeLa CFE2s. Product analysis by PAGE revealed labeled fragments indicative of damage-specific 3′ and dual incisions by NER (indicated by asterisks in Figure 4C) together with other fragments (indicated by dots). Considering the different specific radioactivities of damage-specific incision products (Figure 1C), the yields of 3′ and dual incision products were virtually comparable, indicating coupled incisions on both sides of Oxa–Sp. These products exhibited gel mobilities comparable to those of a 64mer marker and a 27mer bearing Oxa–Sp (lanes 1 and 2), respectively. More detailed size analyses of damage-specific incision products by long electrophoresis showed that incisions occurred at the 4th–7th phosphodiester bonds 3′ to Oxa–Sp (Figure 1C) and that dual incision products were 26mer and 27mer in size. The origins of two other fragments (indicated with dots in Figure 4C) were unknown, but the estimated size of large one (∼92mer) implied that it was generated by a rarely occurring uncoupled 5′ incision. The human NER complex generally incises the 16th–26th phosphodiester bonds on the 5′ side and the 4th–10th bonds on the 3′ side of a lesion, resulting in 24–32mer fragments bearing a lesion (1,53–55). Thus, coupled incisions on the 5′ and 3′ sides of Oxa–Sp observed here are hallmarks of the human NER system. To confirm the involvement of NER further, activity complementation experiments were performed with CFE2s from NER-deficient XPA and XPF cells. 150OXA–SPe was incubated with CFE2s and products were analyzed by denaturing PAGE (Figure 4D). Damage-specific 5′ incisions indicative of NER were not observed with CFE2s from XPA and XPF cells (lanes 4 and 5). However, incision activity for Oxa–Sp was restored with the mixture of CFE2s from XPA and XPF cells (lane 6), clearly demonstrating the involvement of the NER system in the incision of Oxa–Sp DNA.
Figure 4.
Analysis of NER activities of human CFE2s for Oxa and Oxa–Sp. (A) 150OXAe, 150OXA–SPe and 150FL (all 2 nM and 5′-end labeled, Figure 1C) were incubated with HeLa CFE2s (100 μg) at 30°C for 0, 20, 60 and 90 min, and products were analyzed by 10% denaturing PAGE. Note that the only gel region containing incised products is shown. The sequence of the 59mer marker is given in Figure 1C. (B) The amount of 5′ incision products shown in panel A was quantitated for Oxa–Sp (circles), FL (triangles) and Oxa (squares), and are plotted against incubation time. (C) 150OXA–SPi (2 nM and internally labeled, Figure 1C) was incubated with HeLa CFE2s (100 μg), and products were analyzed as in panel A. The sequences of a 64mer marker and a 27mer bearing Oxa–Sp are given in Figure 1C. The fragments derived from damage-specific 3′ and dual incisions are indicated by asterisks. The fragments from unknown origin are indicated by dots (see also text). (D) 150OXA–SPe (2 nM) was incubated with CFE2s (100 μg) from HeLa, XPA and XPF cells, or a mixture of CFE2s from XPA (50 μg) and XPF (50 μg) cells at 30°C for 90 min. Products were analyzed as in panel A. The CFE2s used are indicated on the gel.
DISCUSSION
In the present study, we have assessed systematically the Oxa- and Oxa–Sp-excising activities of BER and NER enzymes and CFEs to delineate the potential repair mechanisms of guanine lesions generated by nitrosative stress. Several DNA glycosylases from E.coli (AlkA, Endo VIII and Fpg) and humans (hMPG and hNEIL1) exhibited Oxa-excising activity but the activity was weak at only 0.2–9% of those for their physiological substrates (Table 2). The results of AlkA and Endo VIII are consistent with our previous data (26). Towards the end of this study, it was reported that mammalian MPG (also called AAG or ANPG) acts as Oxa-DNA glycosylase (30), which agrees qualitatively, but not quantitatively, with the present result for hMPG. According to the reported data (30), hMPG excises Oxa with compromised (∼30%) or comparable efficiencies relative to a control substrate (Hx). In addition, Oxa-excising activity is found in CFEs from the spleen of wild-type but not MPG-knockout mice. Conversely, we found that the Oxa-excising activity of hMPG was much lower than that for control substrates (4 and 9% relative to 7mG and Hx, respectively, Table 2). The low activity for Oxa was not due to DNA–protein cross-link formation, since hMPG did not form cross-links with Oxa (data not shown). Consistent with the weak activity of hMPG for Oxa, the Oxa-excising activity of HeLa CFE1s was very weak (a few percent of the Hx-excising activity) under the present conditions (Figure 2B). Accordingly, we tentatively assume that Oxa is a poor substrate for hMPG and therefore is not processed efficiently by the BER pathway in human cells. However, further studies are necessary to clarify whether the Oxa-excising activity of mammalian MPG is a physiologically relevant function.
Oxa was also a poor substrate for E.coli DNA glycosylases (Table 2), suggesting that Oxa is poorly repaired by the BER pathway in E.coli. This is in contrast to Xan, another major guanine lesion generated by nitrosative stress, being efficiently removed from DNA by E.coli and human DNA glycosylases that initiate the BER process (26,27). It has been shown very recently that prokaryotic Endo V incises Oxa-containing DNA at the second phosphodiester bond on the 3′ side of the lesion (31). The cleavage efficiency is 6-fold lower than that for Hx-containing DNA, a control substrate. In addition to Oxa, Endo V recognizes a wide range of aberrant DNA structures including deaminated bases (uracil, Hx and Xan), base mismatches and flap and pseudo Y structures (15). The genetic analysis of E.coli strains deficient in Endo V suggests that Endo V plays a role in ameliorating genotoxic effects conferred by nitrous acid (56–58), and hence provides an alternative repair pathway for deaminated bases. Combining the present and previous biochemical and genetic data, it is likely that in E.coli Xan is repaired by BER and/or an alternative repair pathway, whereas Oxa is apparently not repaired by BER and thus probably processed by an alternative repair pathway (15), albeit slowly.
Oxa produced by nitrosative stress reacts further with polyamines and DNA binding proteins in cells to form cross-link adducts (25). Therefore, we have examined whether DNA glycosylases can remove cross-link adducts from DNA using Oxa–Sp-containing DNA as a representative adduct. However, none of the DNA glycosylases tested were able to remove Oxa–Sp from DNA (Table 2). We reason that DNA glycosylases that recognize relatively minor base modifications cannot accommodate the bulky Oxa–Sp adduct in the active site pocket. Accordingly, we examined the excision capacity of NER systems for Oxa–Sp that can recognize bulky adducts, such as UV-induced pyrimidine dimers and polycyclic aromatic hydrocarbon adducts (1,54,55,59,60). In keeping with the damage specificity, UvrABC, the prokaryotic NER system, made dual incisions on DNA containing Oxa–Sp but not Oxa (Figure 3A and B), resulting in a 12mer fragment bearing Oxa–Sp. The incision of Oxa–Sp DNA by UvrABC was fairly efficient (Figure 3C). The specific incision by UvrABC was further substantiated by CFEs from SOS-induced E.coli where the transcription of the uvrA and uvrB genes are activated (Figure 3D). Previous genetic analyses of E.coli strains defective in various DNA repair pathways suggest that NER and recombination repair constitute a defense mechanism against the genotoxic effects conferred by NO and nitrous acid (11,61–64). Nitrosative stress also induce the SOS response in Salmonella enterica, though the response seems to be attributable to the inhibition of DNA replication due to zinc mobilization rather than DNA damage (65). Assuming a similar response to NO in E.coli, it is possible that UvrABC contributes to the defense against nitrosative stress by removing genotoxic cross-link adducts.
We have also found that DNA containing Oxa–Sp is specifically incised by CFE2s from NER-proficient HeLa cells (Figure 4D), and that the incision efficiency for Oxa–Sp is comparable to that for FL (a control substrate) (Figure 4B). The coupled dual incisions and their sites observed for Oxa–Sp (Figure 4C) are characteristic of the human NER system (1,53–55). Together with the activity complementation experiments with XPA and XPF CFE2s (Figure 4D), the present data demonstrate that Oxa–Sp is processed by the NER system in human cells when the lesion is generated by NO. Conversely, in keeping with its non-bulky nature, Oxa-containing DNA was not cleaved by human CFE2s. In this study, we have not tested Oxa-protein cross-link adducts for NER enzymes or CFEs, which are more bulky than Oxa–Sp. It has been recently shown that UvrABC nuclease makes dual incisions on a substrate containing a cross-link adduct between DNA and phage T4 Endo V, which was prepared by the borohydride trapping of a reaction intermediate (66,67). This suggests that Oxa–protein cross-links adducts generated by nitrosative stress could be processed similarly by the NER pathway.
In summary, we have analyzed the excision activity of E.coli and human BER and NER enzymes for Oxa and Oxa–Sp. Oxa is poorly excised from DNA by BER and NER enzymes, whereas DNA containing Oxa–Sp is efficiently incised by NER but not BER enzymes. Combining the present and published data on the repair of guanine lesions generated by nitrosative stress (Xan, Oxa and Oxa–Sp), Xan is repaired by the BER pathway (or an alternative repair pathway in E.coli), and the Oxa–Sp cross-link adduct is repaired by the NER pathway. The repair mechanism of Oxa remains to be established and further studies are necessary.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to H.I.) and for Cancer Research (16-7) from the Ministry of Health, Labor and Welfare of Japan (to T.S.). T.N. is supported by the JSPS fellowship for young scientists. We thank Drs Murat Saparbaev (Centre National de la Recherche Scientifique) and Shuji Yonei (Kyoto University) for the generous gifts of the hMPG protein and E.coli strains, respectively. We are also grateful to Drs Yoshihiko Ohyama (Hiroshima University) and Leroy Worth (NIEHS) for valuable discussions and critical reading of this manuscript. Funding to pay the Open Access publication charges for this article was provided by the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg E.C., Walker G.C., Siede W. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology; 1995. [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro R. Damage to DNA caused by hydrolysis. In: Seeberg E., Kleppe K., editors. Chromosome Damage and Repair. New York: Plenum; 1982. pp. 3–18. [Google Scholar]
- 5.Karran P., Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry. 1980;19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 6.Wink D.A., Kasprzak K.S., Maragos C.M., Elespuru R.K., Misra M., Dunams T.M., Cebula T.A., Koch W.H., Andrews A.W., Allen J.S., et al. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 7.Hartman Z., Henrikson E.N., Hartman P.E., Cebula T.A. Molecular models that may account for nitrous acid mutagenesis in organisms containing double-stranded DNA. Environ. Mol. Mutagen. 1994;24:168–175. doi: 10.1002/em.2850240305. [DOI] [PubMed] [Google Scholar]
- 8.Caulfield J.L., Wishnok J.S., Tannenbaum S.R. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem. 1998;273:12689–12695. doi: 10.1074/jbc.273.21.12689. [DOI] [PubMed] [Google Scholar]
- 9.Spencer J.P., Whiteman M., Jenner A., Halliwell B. Nitrite-induced deamination and hypochlorite-induced oxidation of DNA in intact human respiratory tract epithelial cells. Free Radic. Biol. Med. 2000;28:1039–1050. doi: 10.1016/s0891-5849(00)00190-8. [DOI] [PubMed] [Google Scholar]
- 10.Ohshima H., Bartsch H. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat. Res. 1994;305:253–264. doi: 10.1016/0027-5107(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 11.Tamir S., Burney S., Tannenbaum S.R. DNA damage by nitric oxide. Chem. Res. Toxicol. 1996;9:821–827. doi: 10.1021/tx9600311. [DOI] [PubMed] [Google Scholar]
- 12.Wink D.A., Vodovotz Y., Laval J., Laval F., Dewhirst M.W., Mitchell J.B. The multifaceted roles of nitric oxide in cancer. Carcinogenesis. 1998;19:711–721. doi: 10.1093/carcin/19.5.711. [DOI] [PubMed] [Google Scholar]
- 13.Dedon P.C., Tannenbaum S.R. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 14.Krokan H.E., Drablos F., Slupphaug G. Uracil in DNA—occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 15.Kow Y.W. Repair of deaminated bases in DNA. Free Radic. Biol. Med. 2002;33:886–893. doi: 10.1016/s0891-5849(02)00902-4. [DOI] [PubMed] [Google Scholar]
- 16.Haushalter K.A., Todd Stukenberg P., Kirschner M.W., Verdine G.L. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 17.Nilsen H., Haushalter K.A., Robins P., Barnes D.E., Verdine G.L., Lindahl T. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saparbaev M., Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl Acad. Sci. USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saparbaev M., Mani J.C., Laval J. Interactions of the human, rat, Saccharomyces cerevisiae and Escherichia coli 3-methyladenine-DNA glycosylases with DNA containing dIMP residues. Nucleic Acids Res. 2000;28:1332–1339. doi: 10.1093/nar/28.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki T., Yamaoka T., Nishi M., Ide H., Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligonucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc. 1996;118:2515–2516. [Google Scholar]
- 21.Suzuki T., Ide H., Yamada M., Endo N., Kanaori K., Tajima K., Morii T., Makino K. Formation of 2′-deoxyoxanosine from 2′-deoxyguanosine and nitrous acid: mechanism and intermediates. Nucleic Acids Res. 2000;28:544–551. doi: 10.1093/nar/28.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayat S., Majumdar P., Tipton P., Glaser R. 5-Cyanoimino-4-oxomethylene-4,5-dihydroimidazole and 5-cyanoamino-4-imidazolecarboxylic acid intermediates in nitrosative guanosine deamination: evidence from 18O-labeling experiments. J. Am. Chem. Soc. 2004;126:9960–9969. doi: 10.1021/ja049835q. [DOI] [PubMed] [Google Scholar]
- 23.Rayat S., Wu Z., Glaser R. Nitrosative guanine deamination: ab initio study of deglycation of N-protected 5-cyanoimino-4-oxomethylene-4,5-dihydroimidazoles. Chem. Res. Toxicol. 2004;17:1157–1169. doi: 10.1021/tx0499416. [DOI] [PubMed] [Google Scholar]
- 24.Dong M., Wang C., Deen W.M., Dedon P.C. Absence of 2′-deoxyoxanosine and presence of abasic sites in DNA exposed to nitric oxide at controlled physiological concentrations. Chem. Res. Toxicol. 2003;16:1044–1055. doi: 10.1021/tx034046s. [DOI] [PubMed] [Google Scholar]
- 25.Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., Ide H. DNA–protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 26.Terato H., Masaoka A., Asagoshi K., Honsho A., Ohyama Y., Suzuki T., Yamada M., Makino K., Yamamoto K., Ide H. Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid. Nucleic Acids Res. 2002;30:4975–4984. doi: 10.1093/nar/gkf630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wuenschell G.E., O'Connor T.R., Termini J. Stability, miscoding potential, and repair of 2′-deoxyxanthosine in DNA: implications for nitric oxide-induced mutagenesis. Biochemistry. 2003;42:3608–3616. doi: 10.1021/bi0205597. [DOI] [PubMed] [Google Scholar]
- 28.He B., Qing H., Kow Y.W. Deoxyxanthosine in DNA is repaired by Escherichia coli endonuclease V. Mutat. Res. 2000;459:109–114. doi: 10.1016/s0921-8777(99)00063-4. [DOI] [PubMed] [Google Scholar]
- 29.Moe A., Ringvoll J., Nordstrand L.M., Eide L., Bjoras M., Seeberg E., Rognes T., Klungland A. Incision at hypoxanthine residues in DNA by a mammalian homologue of the Escherichia coli antimutator enzyme endonuclease V. Nucleic Acids Res. 2003;31:3893–3900. doi: 10.1093/nar/gkg472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hitchcock T.M., Dong L., Connor E.E., Meira L.B., Samson L.D., Wyatt M.D., Cao W. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 31.Hitchcock T.M., Gao H., Cao W. Cleavage of deoxyoxanosine-containing oligodeoxyribonucleotides by bacterial endonuclease V. Nucleic Acids Res. 2004;32:4071–4080. doi: 10.1093/nar/gkh747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asagoshi K., Yamada T., Terato H., Ohyama Y., Monden Y., Arai T., Nishimura S., Aburatani H., Lindahl T., Ide H. Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J. Biol. Chem. 2000;275:4956–4964. doi: 10.1074/jbc.275.7.4956. [DOI] [PubMed] [Google Scholar]
- 33.Katafuchi A., Nakano T., Masaoka A., Terato H., Iwai S., Hanaoka F., Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 34.Masaoka A., Terato H., Kobayashi M., Honsho A., Ohyama Y., Ide H. Enzymatic repair of 5-formyluracil. I. Excision of 5-formyluracil site-specifically incorporated into oligonucleotide substrates by alka protein (Escherichia coli 3-methyladenine DNA glycosylase II) J. Biol. Chem. 1999;274:25136–25143. doi: 10.1074/jbc.274.35.25136. [DOI] [PubMed] [Google Scholar]
- 35.Asagoshi K., Yamada T., Okada Y., Terato H., Ohyama Y., Seki S., Ide H. Recognition of formamidopyrimidine by Escherichia coli and mammalian thymine glycol glycosylases. Distinctive paired base effects and biological and mechanistic implications. J. Biol. Chem. 2000;275:24781–24786. doi: 10.1074/jbc.M000576200. [DOI] [PubMed] [Google Scholar]
- 36.Asagoshi K., Odawara H., Nakano H., Miyano T., Terato H., Ohyama Y., Seki S., Ide H. Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry. 2000;39:11389–11398. doi: 10.1021/bi000422l. [DOI] [PubMed] [Google Scholar]
- 37.Masaoka A., Matsubara M., Hasegawa R., Tanaka T., Kurisu S., Terato H., Ohyama Y., Karino N., Matsuda A., Ide H. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 38.Matsubara M., Tanaka T., Terato H., Ohmae E., Izumi S., Katayanagi K., Ide H. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res. 2004;32:5291–5302. doi: 10.1093/nar/gkh859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau A.Y., Scharer O.D., Samson L., Verdine G.L., Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 40.Theis K., Chen P.J., Skorvaga M., Van Houten B., Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorvaga M., Theis K., Mandavilli B.S., Kisker C., Van Houten B. The beta-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J. Biol. Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
- 42.White B.J., Hochhauser S.J., Cintron N.M., Weiss B. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J. Bacteriol. 1976;126:1082–1088. doi: 10.1128/jb.126.3.1082-1088.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunningham R.P., Saporito S.M., Spitzer S.G., Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takebe H., Nii S., Ishii M.I., Utsumi H. Comparative studies of host-cell reactivation, colony forming ability and excision repair after UV irradiation of xeroderma pigmentosum, normal human and some other mammalian cells. Mutat. Res. 1974;25:383–390. doi: 10.1016/0027-5107(74)90067-0. [DOI] [PubMed] [Google Scholar]
- 45.Yagi T., Takebe H. Establishment by SV40 transformation and characteristics of a cell line of xeroderma pigmentosum belonging complementation group F. Mutat. Res. 1983;112:59–66. doi: 10.1016/0167-8817(83)90024-x. [DOI] [PubMed] [Google Scholar]
- 46.Crowley D.J., Hanawalt P.C. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6–4 photoproducts, in UV-irradiated Escherichia coli. J. Bacteriol. 1998;180:3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J.C., Sancar A. Determination of minimum substrate size for human excinuclease. J. Biol. Chem. 1994;269:19034–19040. [PubMed] [Google Scholar]
- 48.Ide H., Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol. Pharm. Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- 49.Ishchenko A.A., Sanz G., Privezentzev C.V., Maksimenko A.V., Saparbaev M. Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res. 2003;31:6344–6353. doi: 10.1093/nar/gkg812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ide H., Tedzuka K., Shimzu H., Kimura Y., Purmal A.A., Wallace S.S., Kow Y.W. Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV. Biochemistry. 1994;33:7842–7847. doi: 10.1021/bi00191a011. [DOI] [PubMed] [Google Scholar]
- 51.Ishchenko A.A., Ide H., Ramotar D., Nevinsky G., Saparbaev M. Alpha-anomeric deoxynucleotides, anoxic products of ionizing radiation, are substrates for the endonuclease IV-type AP endonucleases. Biochemistry. 2004;43:15210–15216. doi: 10.1021/bi049214+. [DOI] [PubMed] [Google Scholar]
- 52.Wallace S.S. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 53.Huang J.C., Svoboda D.L., Reardon J.T., Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 55.Wood R.D. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 56.Guo G., Weiss B. Endonuclease V (nfi) mutant of Escherichia coli K-12. J. Bacteriol. 1998;180:46–51. doi: 10.1128/jb.180.1.46-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schouten K.A., Weiss B. Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res. 1999;435:245–254. doi: 10.1016/s0921-8777(99)00049-x. [DOI] [PubMed] [Google Scholar]
- 58.Weiss B. Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res. 2001;461:301–309. doi: 10.1016/s0921-8777(00)00062-8. [DOI] [PubMed] [Google Scholar]
- 59.Van Houten B. Nucleotide excision repair in Escherichia coli. Microbiol. Rev. 1990;54:18–51. doi: 10.1128/mr.54.1.18-51.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Friedberg E.C. How nucleotide excision repair protects against cancer. Nature Rev. Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 61.Routledge M.N. Mutations induced by reactive nitrogen oxide species in the SupF forward mutation assay. Mutat. Res. 2000;450:95–105. doi: 10.1016/s0027-5107(00)00018-x. [DOI] [PubMed] [Google Scholar]
- 62.Spek E.J., Wright T.L., Stitt M.S., Taghizadeh N.R., Tannenbaum S.R., Marinus M.G., Engelward B.P. Recombinational repair is critical for survival of Escherichia coli exposed to nitric oxide. J. Bacteriol. 2001;183:131–138. doi: 10.1128/JB.183.1.131-138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frankel A.D., Duncan B.K., Hartman P.E. Nitrous acid damage to duplex deoxyribonucleic acid: distinction between deamination of cytosine residues and a novel mutational lesion. J. Bacteriol. 1980;142:335–338. doi: 10.1128/jb.142.1.335-338.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sidorkina O., Saparbaev M., Laval J. Effects of nitrous acid treatment on the survival and mutagenesis of Escherichia coli cells lacking base excision repair (hypoxanthine-DNA glycosylase-ALK A protein) and/or nucleotide excision repair. Mutagenesis. 1997;12:23–28. doi: 10.1093/mutage/12.1.23. [DOI] [PubMed] [Google Scholar]
- 65.Schapiro J.M., Libby S.J., Fang F.C. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc. Natl Acad. Sci. USA. 2003;100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minko I.G., Zou Y., Lloyd R.S. Incision of DNA–protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl Acad. Sci. USA. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou Y., Ma H., Minko I.G., Shell S.M., Yang Z., Qu Y., Xu Y., Geacintov N.E., Lloyd R.S. DNA damage recognition of mutated forms of UvrB proteins in nucleotide excision repair. Biochemistry. 2004;43:4196–4205. doi: 10.1021/bi035992a. [DOI] [PMC free article] [PubMed] [Google Scholar]