Abstract
Objectives
Early discontinuation is common among breast cancer patients taking aromatase inhibitors (AIs). Although several predictors have been identified, it is unclear how to simultaneously consider multiple risk factors for an individual. We sought to develop a tool for prediction of AI discontinuation and to explore how predictive value of risk factors changes with time.
Materials and Methods
Survival machine learning was used to predict time-to-discontinuation of AIs in 181 women who enrolled in a prospective cohort. Models were evaluated via time-dependent area under the curve (AUC), c-index, and integrated Brier score. Feature importance was analysis was conducted via Shapley Additive Explanations (SHAP) and time-dependence of their predictive value was analyzed by time-dependent AUC. Personalized survival curves were constructed for risk communication.
Results
The best-performing model incorporated genetic risk factors and changes in patient-reported outcomes, achieving mean time-dependent AUC of 0.66, and AUC of 0.72 and 0.67 at 6- and 12-month cutoffs, respectively. The most significant features included variants in ESR1 and emergent symptoms. Predictive value of genetic risk factors was highest in the first year of treatment. Decrease in physical function was the strongest independent predictor at follow-up.
Discussion and Conclusion
Incorporation of genomic and 3-month follow-up data improved the ability of the models to identify the individuals at risk of AI discontinuation. Genetic risk factors were particularly important for predicting early discontinuers. This study provides insight into the complex nature of AI discontinuation and highlights the importance of incorporating genetic risk factors and emergent symptoms into prediction models.
Keywords: survival machine learning, patient-reported outcome measures, pharmacogenomics, aromatase inhibitors, longitudinal
Introduction
Adjuvant endocrine therapy for 5-10 years has been shown to reduce recurrence and death in individuals with early-stage hormone receptor positive (HR+) breast cancer.1–6 Treatment with third-generation aromatase inhibitors (AIs), such as anastrozole, letrozole, and exemestane, has proven to be superior to tamoxifen for the treatment of HR+ breast cancer in postmenopausal women.7 However, up to 50% of patients taking AIs in the adjuvant setting discontinue their treatment before the established minimal 5-year duration and thereby reducing the beneficial effects of therapy.8–11 Consequently, recurrence and death rates have been shown to be higher among those who discontinue their medication early.12–15
Side effects are often reported as a reason for AI therapy discontinuation, with AI-induced musculoskeletal symptoms (AIMSS) being the leading reason for discontinuation.16–20 Patient-reported outcomes (PROs) indicate the development of AIMSS and are associated with early discontinuation of therapy.21–25 Additional studies have reported that age, previous taxane therapy, and depression were predictive of premature AI therapy discontinutation.26–28 Several genetic factors affecting individual’s risk for AIMSS and AIMSS-related discontinuation have been identified through candidate gene and genome-wide association studies.29–36
Despite the knowledge of risk factors affecting discontinuation of AI therapy, it is unclear how to simultaneously consider multiple risk factors for an individual, and prospective identification of patients at risk of discontinuing early remains an unmet medical need.21 Survival machine learning (ML) methods have been successful in clinical outcome prediction across many disciplines.37–44 Some recent efforts used survival ML to identify optimal treatment regimen for patients diagnosed with metastatic breast cancer, while maximizing the overall survival and time-to-discontinuation.45 Another research group developed an ML model to predict AI therapy discontinuation as a binary outcome using structured data from electronic medical records.46 This model did not incorporate genetic risk factors and PRO measures but was still able to achieve a fair performance (area under the curve [AUC] 0.65).46 The primary objective of our study was to build a survival ML model for prediction of time-to-AI discontinuation. Secondary objectives included estimating the extent to which incorporation of genetic risk factors and data collected during follow-ups improved the prediction accuracy and evaluating the predictive value of individual risk factors over time.
Methods
Data source
This work used secondary data from the Johns Hopkins Breast Cancer Program Hormone Therapy Longitudinal Database study (clinical trial id NCT01937052, registered September 3, 2013),47 a study of individuals with early-stage HR+ breast cancer taking AIs. The setting and recruitment eligibility criteria for the cohort have been described elsewhere.30,47 In short, women 18 years of age and older with stage 0-III HR+ breast cancer initiating adjuvant endocrine therapy with either selective estrogen receptor modulators (tamoxifen, raloxifene) or third-generation AIs (anastrozole, letrozole, exemestane) were eligible to participate in the study. Subjects that consented to participate could withdraw from the study at any point or become ineligible upon development of metastatic disease. This secondary data analysis was approved by the Johns Hopkins IRB (ID: NA_00051923).
Cohort selection
For this secondary data analysis, we selected the women who initiated treatment with AIs upon the enrollment in the study. Individuals with inconclusive date of therapy discontinuation were excluded from the analysis. In addition, we excluded subjects who started tamoxifen upon study enrollment and then transitioned to an AI due to our inability to determine their characteristics at the time of AI therapy initiation.
Outcome measure
The primary outcome of interest was AI therapy discontinuation due to side effects or nonadherence. Discontinuation was defined as therapy stop or switch prior to completion of 5 years of therapy. The individuals who stopped AI therapy due to the development of metastatic disease, change in menopause state, or planned pregnancy were censored at the time of therapy discontinuation. The study participants lost to follow-up were censored at the time of their last follow-up. Individuals continuing on AI therapy past 5 years were censored at the 5-year endpoint. Switching from one AI to another one was considered equivalent to discontinuing the medication. The dates of AI discontinuation and reason for discontinuation were determined by chart review.
Independent variables
Baseline and follow-up visit patient characteristics
The dataset includes baseline demographic, clinical characteristics, and endocrine therapy selected by the treating physician at enrollment. In addition, whole blood or saliva samples were collected for the participants at baseline for germline DNA isolation. Body mass index (BMI), self-reported adherence behavior via the Medication Adherence Questionnaire (MAQ), and a set of PRO measures were collected at enrollment, and at 3-, 6-, 12-, 24-, 36-, 48-, and 60-month follow-up visits.48,49 For the current study, we used the data collected at baseline and at 3-month follow-up.
Pharmacogenetics
Thirteen candidate genes previously associated with the AIMSS were selected by the investigators. The single-nucleotide polymorphisms (SNPs) considered for the analysis included CYP19A1 rs10046 and rs7176005, VDR rs11568820, TCL1A rs11849538, MIR4713HG rs16964189 and rs934635, OPG rs2073618, CYP27B1 rs4646536, CYP17A1 rs6163, RANKL rs7984870, and ESR1 rs2234693, rs9322336 and rs9340799. Table S1 summarizes the SNPs available for the analysis and their respective associations with AI-induced adverse events. Genotyping process and quality control methods are described in detail in Hertz et al.30
Patient-reported outcomes
PRO measures were collected at each study visit via the PatientViewpoint website.50–52 The measures used in this study included the Patient-Reported Outcome Measurement Information System (PROMIS) Version 1.0 sleep disturbance, physical function, pain interference, endocrine symptoms, fatigue, depression, and anxiety short forms, and a single question from the Endocrine Subscale of the Functional Assessment of Cancer Therapy—Endocrine Symptom (FACT-ES) questionnaire regarding joint pain experienced by the patients.53–56 PROMIS measures are based on the T-score metric, in which 50 represents the mean score of the US population, and higher score indicates more of the concept being measured (ie, a higher sleep disturbance on the T-scale represents more sleep disturbance, while higher T-score for the physical function indicates better physical function).54 The severity of the joint pain is assessed on the 5-point scale from 0 (“not at all”) to 4 (“very much”).30
Data pre-processing
We used survival ML to predict AI therapy discontinuation. The demographic and clinical features used to predict the outcome included age, race, prior taxane therapy, cancer stage, and BMI. Menopause state and ethnicity were excluded from the model due to the low number of premenopausal women and Hispanic or Latino individuals in the cohort. Race and prior taxane therapy were included as binary variables, while age, cancer stage, and BMI were used on a continuous scale. Cumulative adherence score was calculated from the medication adherence questionnaire (1—poor adherence, 4—good adherence).
To maximize the signal coming from genetic data and reduce the number of non-relevant SNPs, we developed an annotated genetic model, which included only the variants having significant associations with AI-induced side effects in Pharmacogenomics Knowledge Base (PharmGKB).57,58 We assumed an additive genetic model and encoded the number of risk alleles at a position, according to prior studies. The list of SNPs with significant associations in PharmGKB is presented in Table S1.
To assess the value of follow-up data for prediction, we conducted separate analyses with baseline data and with the data collected at 3-month follow-up. The BMI and PRO values recorded at study enrollment were used in the baseline model directly. At 3-month follow-up, we calculated the changes in BMI and PROs from the initial visit and used the obtained values for model development.
Missing values for BMI, adherence, and PROs were imputed by regressing the feature for the individual over time and extrapolating to time points at which the data were missing. Study sample means were imputed for a feature when the regression approach was unfeasible. Missing genetic data were imputed with expected genotypes based on global population allele frequency reported in the 1000 Genomes Project.59
Statistical analysis
For predictive model building, we selected 4 survival ML algorithms: Cox proportional hazards (CoxPH) model, Random Survival Forest (RSF), penalized CoxPH, and Gradient Boosted Models (GBM) with regression tree and component-wise least squares base learners. The dataset was randomly split into training and testing sets with a 70:30 ratio. Pearson coefficient was calculated to check the correlation between features. The optimal hyperparameters for each ML model were determined by an exhaustive grid search over the pre-specified parameter space with 5-fold cross-validation on the training set. The tuned models were then used to predict the outcome on the test set.
To investigate whether incorporation of genetic data improves model performance, we developed 3 genomic data integration models: (1) clinical only—the model incorporating only demographic and clinical variables; (2) clinical + selected genetics—the model that includes SNPs with significant associations in PharmGKB in addition to demographic and clinical variables; and (3) clinical + all genetics—the model that incorporates all available genetic, demographic, and clinical data.
Model evaluation
Different algorithms were compared by mean time-dependent cumulative/dynamic AUC using monthly intervals, Harrel’s concordance index (c-index), and integrated Brier score (IBS) achieved on the test set.60–63
The cumulative/dynamic AUC calculated at time t quantifies how well the model can distinguish subjects who fail by t (cumulative cases) from the individuals who fail after that time (dynamic controls).60,61 Time-dependent cumulative/dynamic AUC calculates these values for a series of pre-specified time points. Higher values of cumulative/dynamic AUC at a given time indicate a better performance of the model at that time, and higher mean time-dependent AUC indicates the better-performing model over the whole study period. C-index is a goodness-of-fit measure which evaluates the predicted ranking of failure times.62 Brier score is an extension of mean squared error for right-censored data and is used to assess model’s calibration and discrimination.63 Similarly to cumulative/dynamic AUC, Brier score can be calculated for a series of time points and integrated over the entire study period to assess overall model performance. Lower IBS indicates a better model performance.
The best-performing algorithms were chosen for each genetic data integration model at both baseline and 3-month follow-up based on mean time-dependent AUC and IBS values. Time-dependent AUC, cumulative sensitivity, dynamic specificity, and cumulative/dynamic positive predictive value (PPV) at the highest 33% of risk distribution, were assessed for each model at 6- and 12-month cutoffs to assess the ability of the models to predict early discontinuation. Different genetic data integration models were compared by their mean time-dependent AUC value and by their performance at 6 and 12 months. The robustness of the model was assessed using a bootstrapping procedure with 100 samples.
To avoid the misinterpretation of AUC values below 0.5 when assessing predictive value of individual biomarkers, we used normalized time-dependent AUC for illustration purposes. In statistical terms, AUC is defined as the probability that a randomly selected case will have a higher risk score than a randomly selected control. Thus, AUC of 0.5 corresponds to a random classifier, while values above 0.5 indicate discriminative power of the predictor when it is positively correlated with the outcome. However, according to the definition of AUC, the values below 0.5 indicate discrimination when the risk factor and the outcome are inversely related. In other words, an AUC value close to 0 suggests that a randomly selected case will have a lower risk score than a randomly selected control most of the time. We defined normalized time-dependent AUC as the improvement in discriminative ability from random class assignment, according to the following formula:
where AUC(t)N is normalized time-dependent AUC, AUC(t) is time-dependent AUC and 0.5 indicates the AUC value of a no-skill classifier. Normalized time-dependent AUC values are bound between −1 and 1 with values below 0 suggesting that the risk factor and the outcome are inversely related, and the values above 0 suggesting a positive correlation.
Feature importance
The most important features for the best-performing models at baseline and 3-month follow-up were identified with Shapley Additive Explanations (SHAP) algorithm.64 The top 5 most predictive features according to SHAP were further analyzed for time-dependence by developing a CoxPH model with only one predictor and evaluating normalized time-dependent AUC in monthly intervals.
Finally, personalized survival curves were constructed for 3 random individuals from the test set and the most predictive features locally were determined for these individuals by SHAP.
Statistical analysis was performed in Python 3.9.4. ML was performed using Scikit-learn (sklearn 1.0.2), Scikit-survival (sksurv 0.17.2) and shap (version 0.41.0) libraries.65,66
Results
Study population
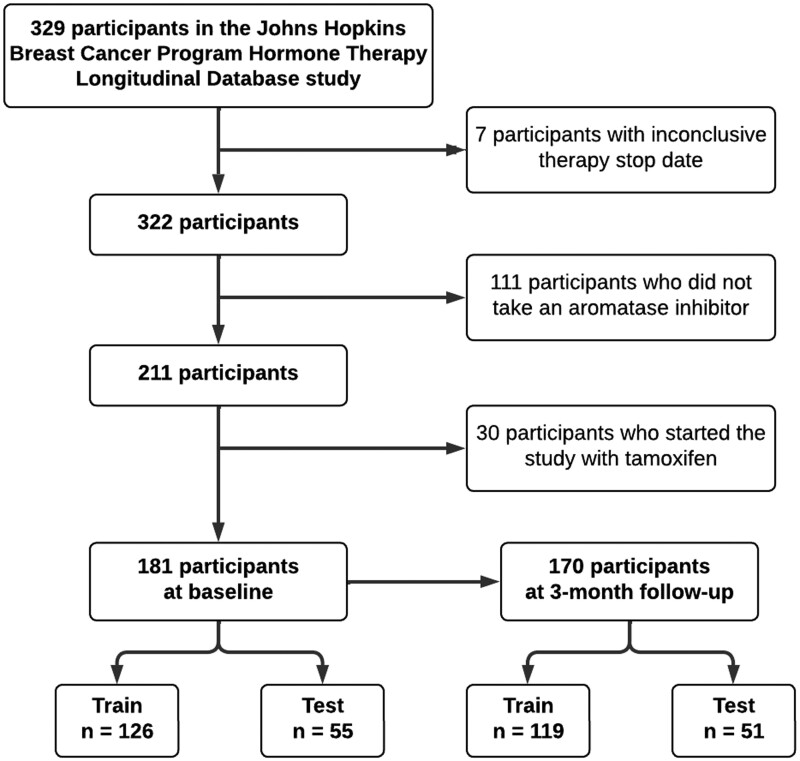
Of 329 participants enrolled in the original study, 189 subjects remained eligible for the inclusion in the study at baseline (Figure 1). The majority of the participants were White (87%), had a stage I breast cancer (62%), were prescribed anastrozole (80%), were postmenopausal (98%) and had no prior taxane chemotherapy (75%). Most participants carried risk alleles in CYP19A1 rs10046 (82%), OPG rs2073618 (79%), ESR1 rs2234693 (77%), CYP17A1 rs6163 (71%), RANKL rs7984870 (69%), and ESR1 rs9340799 (89%) and were homozygous wild-type at the other loci. On average, study participants were 63 years old upon study enrollment and had PROMIS T-scores close to 50 for all PRO measures, except endocrine symptoms, for which the sample average reached the T-score of 65. Total of 170 participants remained eligible at 3-month follow-up. The distribution of demographic and clinical characteristics at 3-month follow-up remained similar to the distribution at baseline. On average, a larger proportion of individuals who discontinued therapy at any point during the study were taking anastrozole, carried risk allele in CYP17A1 rs6163, and did not carry risk alleles in TCL1A rs11849538 and ESR1 rs9322336 loci. The distribution of other demographic and clinical characteristics was similar between those who discontinued therapy due to side effects or non-adherence and those who were censored. Descriptive statistics of the analyzed cohort are presented in Table 1. Raw genotype counts are summarized in Table S2. Means and standard deviations of continuous variables are presented in Table 2. Median time to discontinuation was 48 months.
Figure 1.
Flow chart of cohort selection.
Table 1.
Descriptive statistics for categorical variables.
| All participants, N (%) | Discontinued, N (%) | Censored, N (%) | |
|---|---|---|---|
| Total | 181 | 95 | 86 |
| Race | |||
| White | 157 (87) | 83 (87) | 74 (86) |
| Black | 17 (9) | 10 (11) | 7 (8) |
| Other | 7 (4) | 2 (2) | 5 (6) |
| Therapy | |||
| Anastrozole | 144 (80) | 82 (86) | 62 (72) |
| Letrozole | 34 (19) | 12 (13) | 22 (26) |
| Exemestane | 3 (2) | 1 (1) | 2 (2) |
| Stage | |||
| 0 | 10 (6) | 8 (8) | 2 (2) |
| I | 112 (62) | 56 (59) | 56 (65) |
| II | 39 (22) | 22 (23) | 17 (20) |
| III | 20 (11) | 9 (9) | 11 (13) |
| Menopause state at diagnosis | |||
| Premenopausal | 4 (2) | 1 (1) | 3 (3) |
| Postmenopausal | 177 (98) | 99 (99) | 83 (97) |
| Prior taxane therapy | |||
| Yes | 46 (25) | 24 (25) | 22 (26) |
| No | 135 (75) | 71 (75) | 64 (74) |
| Self-reported adherence | |||
| Poor (0-1 MAQ scale) | 6 (3) | 4 (4) | 2 (2) |
| Moderate (2-3 MAQ scale) | 64 (35) | 32 (34) | 32 (37) |
| Good (4 MAQ scale) | 111 (61) | 59 (62) | 52 (60) |
| Genetic variant carriers | |||
| CYP19A1 rs10046 | 149 (82) | 78 (82) | 71 (82) |
| VDR rs11568820 | 71 (39) | 39 (41) | 32 (37) |
| TCL1A rs11849538 | 32 (18) | 13 (14) | 19 (22) |
| MIR4713HG rs16964189 | 67 (37) | 36 (38) | 31 (36) |
| OPG rs2073618 | 143 (79) | 77 (81) | 66 (77) |
| ESR1 rs2234693 | 140 (77) | 70 (74) | 70 (81) |
| CYP27B1 rs4646536 | 83 (46) | 44 (46) | 39 (45) |
| CYP17A1 rs6163 | 128 (71) | 73 (77) | 55 (64) |
| CYP19A1 rs7176005 | 46 (25) | 27 (28) | 19 (22) |
| RANKL rs7984870 | 124 (69) | 67 (71) | 57 (66) |
| ESR1 rs9322336 | 38 (21) | 15 (16) | 23 (26) |
| ESR1 rs9340799 | 161 (89) | 82 (86) | 79 (92) |
| MIR4713HG rs934635 | 41 (23) | 24 (25) | 17 (20) |
Censored group includes individuals who stopped aromatase inhibitor therapy due to reasons other than side effects or non-adherence, those who completed all 5 years of therapy, and those lost to follow-up.
Abbreviations: CYP17A1, cytochrome P450 family 17 subfamily A member 1; CYP19A1, aromatase; CYP27B1, cytochrome P450 family 27 subfamily B member 1; ESR1, estrogen receptor 1; MAQ, medication adherence questionnaire; MIR4713HG, MIR4713 host gene; OPG, osteoprotegerin; RANKL, receptor activator of nuclear factor kappa-B ligand; TCL1A, T-cell leukemia/lymphoma 1A; VDR, vitamin D receptor.
Table 2.
Descriptive statistics for continuous variables.
| Baseline, mean (σ) | 3 Months, mean (σ) | |
|---|---|---|
| Age | 63.45 (7.43) | 63.59 (7.50) |
| BMI | 28.56 (5.39) | 28.63 (5.27) |
| Joint pain | 1.14 (1.24) | 1.51 (1.40) |
| Sleep disturbancea | 48.99 (8.39) | 50.15 (8.60) |
| Pain interferencea | 48.59 (7.91) | 48.98 (8.12) |
| Physical functiona | 49.88 (7.71) | 50.03 (8.36) |
| Fatiguea | 48.47 (8.25) | 48.39 (8.05) |
| Endocrine symptomsa | 65.76 (9.11) | 64.39 (10.26) |
| Depressiona | 45.38 (8.02) | 44.73 (8.21) |
| Anxietya | 48.44 (9.10) | 47.51 (9.33) |
Means and standard deviations are presented for continuous variables. The values for joint pain indicate the FACT-ES survey responses (0—no pain, 4—severe pain).
The displayed values represent PROMIS T-scores.
Abbreviations: BMI, body mass index; σ, standard deviation.
Discontinuation prediction
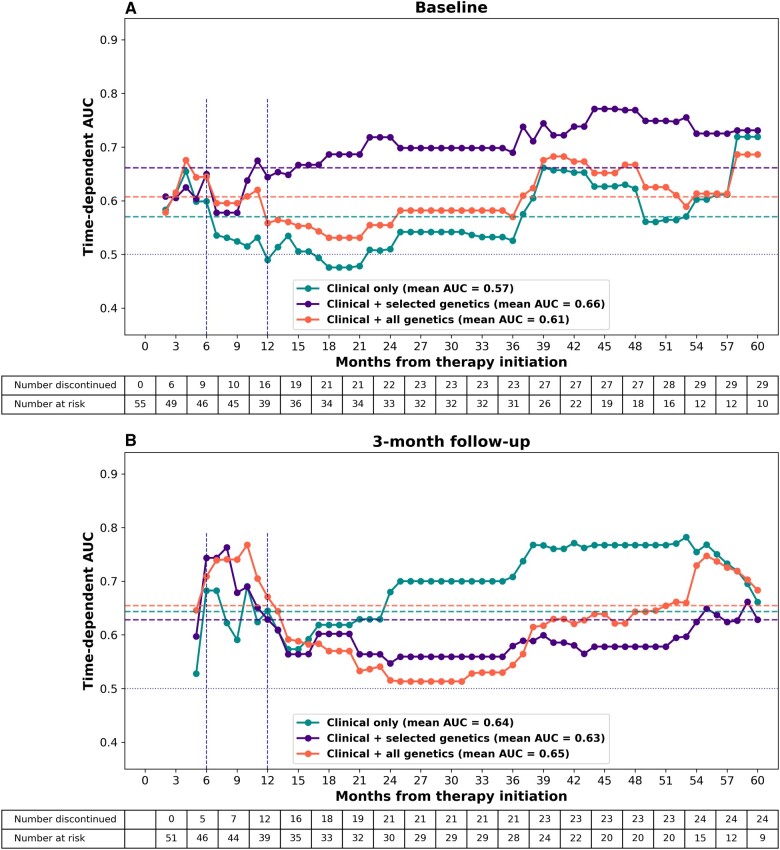
CoxPH, RSF, GBM, and penalized CoxPH models were trained on the train set consisting of randomly selected 70% of cohort at baseline (n = 126) and at 3-month follow-up (n = 119). The optimal hyperparameter values for each model are presented in Table S3. The best-performing algorithms at baseline included RSF for the clinical only model, CoxPH for the PharmGKB-annotated model, and GBM for the clinical + all genetics model. The optimal algorithms at 3-months included GBM for both the clinical only and the clinical + selected genetics models, and RSF for the clinical + all genetics model. Mean time-dependent AUC, c-index and IBS for the best-performing algorithms are reported in Table 3. The clinical + selected genetics model developed at baseline had a higher mean AUC value (0.66) and a lower IBS (0.210) than the other 2 genetic data integration models, although the c-index for this model was lower. The clinical only model had the lowest IBS (0.204) and the highest c-index (0.63), when incorporating the 3-month follow-up data. The clinical + all genetics model developed at the same time reached the highest mean AUC value (0.66). Performance metrics for the other models are summarized in Table S4.
Table 3.
Summary of performance metrics for the best models.
| Mean AUC | C-index | Integrated Brier score | ||
|---|---|---|---|---|
| Baseline | Clinical only | 0.57 | 0.58 | 0.226 |
| Clinical + selected genetics | 0.66 | 0.55 | 0.210 | |
| Clinical + all genetics | 0.61 | 0.59 | 0.225 | |
| 3-month follow-up | Clinical only | 0.64 | 0.54 | 0.204 |
| Clinical + selected genetics | 0.63 | 0.56 | 0.233 | |
| Clinical + all genetics | 0.65 | 0.56 | 0.228 |
The integrated Brier scores produced by the random prediction model were 0.256 at baseline and 0.258 at 3-month follow-up.
Abbreviations: AUC, cumulative/dynamic area under the curve; c-index, concordance index.
Time-dependent AUC curves for the best-performing models are presented in Figure 2. The PharmGKB-annotated model had the best overall performance at the baseline, reaching mean time-dependent AUC of 0.66 over the entire study period (Figure 2A). At 3-month follow-up, the clinical + all genetics model displayed the highest overall performance (mean AUC 0.65, Figure 2B). When considering the performance at 12 months, the best discriminative ability was displayed by the clinical + all genetics models at both baseline and 3-month follow-up. The peak AUC value in the early discontinuation period was achieved by the clinical + all genetics model at 4 months when using the baseline data for longitudinal markers (AUC = 0.68), and by the clinical + selected genetics model at 8 months when incorporating the 3-month follow-up data (AUC = 0.76). The clinical + selected genetics model developed at baseline retained high performance throughout the entire follow-up period, achieving the highest discriminative ability at 44 months (AUC = 0.77). On the contrary, the best-performing 3-month follow-up model had a decline in performance after the initial peak at 10 months, while the clinical only model improved with time, reaching the highest AUC of 0.78 at 53 months. The discriminative ability of the models was low for the prediction of discontinuation between 1 and 3 years of treatment. The bootstrapping procedure demonstrated significant overlap between the models. The time-dependent AUC curves obtained in each bootstrapping run are presented in Figure S1.
Figure 2.
Time-dependent AUC for different genetic data integration models. Cumulative/dynamic AUC evaluated on the test set is plotted in monthly intervals for 5 years. All models included demographics, clinical features, and patient-reported outcomes. Labels are intended to emphasize the amount of genetic data included in the model. (A) The baseline values for longitudinal features were used for model development. (B) Changes in values for longitudinal features from baseline to 3-month follow-up were used for model development. Horizontal dashed lines represent the mean AUC values over the whole study period. Horizontal blue dotted line represents performance of a model making predictions based on random guess. Vertical dashed lines indicate the 3-, 6-, and 12-month cutoffs for prediction of early discontinuation. Abbreviation: AUC, area under the curve.
To evaluate the ability of our models to predict early discontinuation, we assessed time-dependent AUC, sensitivity, specificity, and PPV at the highest 33% of risk distribution at 6 and 12 months (Table 4). Among the models developed at baseline, the clinical + selected genetics model displayed the best predictive ability at 12 months, achieving AUC of 0.64, sensitivity of 0.44, specificity of 0.72, and PPV of 0.39. When incorporating 3-month follow-up data, the clinical + all genetics model outperformed the other 2 models at 12 months, reaching AUC of 0.67, sensitivity of 0.58, specificity of 0.74, and PPV of 0.41. At 6 months, the 3 different genetic data integration models performed similarly. Overall, the models that incorporated 3-month follow-up data performed better at the 6- and 12-month cutoffs than the models developed at baseline.
Table 4.
Time-dependent performance metrics for the best models at 6 and 12 months.
| 6 months |
12 months |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | Sensitivity | Specificity | PPV | AUC | Sensitivity | Specificity | PPV | ||
| Baseline | Clinical only | 0.60 | 0.44 | 0.70 | 0.22 | 0.49 | 0.25 | 0.64 | 0.22 |
| Clinical + selected genetics | 0.65 | 0.44 | 0.70 | 0.22 | 0.64 | 0.44 | 0.72 | 0.39 | |
| Clinical + all genetics | 0.64 | 0.44 | 0.70 | 0.22 | 0.56 | 0.31 | 0.67 | 0.28 | |
| 3-Month follow-up | Clinical only | 0.68 | 0.60 | 0.70 | 0.18 | 0.64 | 0.50 | 0.72 | 0.35 |
| Clinical + selected genetics | 0.74 | 0.60 | 0.72 | 0.19 | 0.63 | 0.50 | 0.74 | 0.38 | |
| Clinical + all genetics | 0.71 | 0.60 | 0.70 | 0.18 | 0.67 | 0.58 | 0.74 | 0.41 | |
Abbreviations: AUC, cumulative/dynamic area under the receiver operating characteristic curve; PharmGKB, Pharmacogenomics Knowledge Base; PPV, positive predictive value.
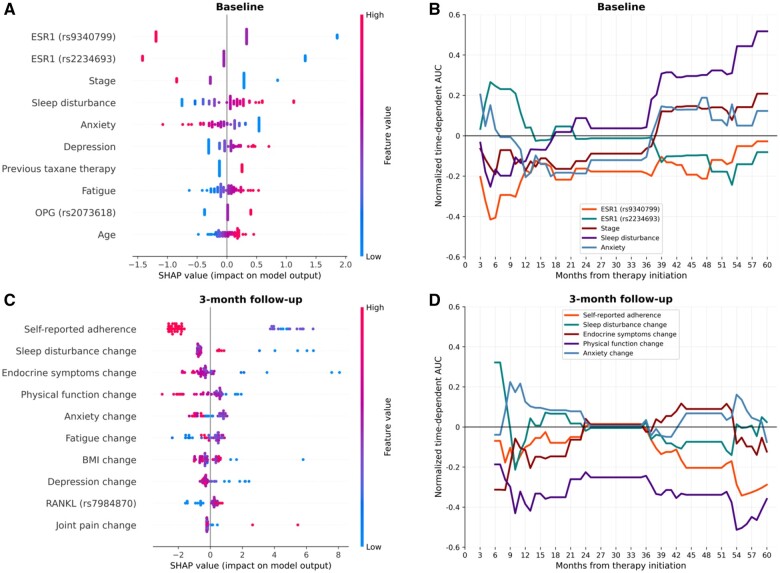
We then selected the clinical + selected genetics model at baseline and the clinical + all genetics model at 3-month follow-up for feature importance assessment. SHAP summary plots for the 2 models are presented in Figure 3A and C. Each dot along the x-axis represents an individual, and the color of the dot indicates the value of the feature for the given subject. The position of the dot along the x-axis indicates the SHAP value, representing the impact of the given feature on the prediction. Positive SHAP values suggest a higher predicted risk of discontinuation, while negative SHAP values suggest a lower predicted risk. In the example of the ESR1 rs9340799 in Figure 3A, being homozygous for the risk allele (red dots) resulted in lower predicted risk of discontinuation. According to SHAP analysis, the 2 most predictive features in the baseline clinical + selected genetics model were variants in ESR1—rs9340799 and rs2234693 (Figure 3A). Interestingly, carrying risk allele at these positions was considered protective by the model. Higher cancer stage and baseline anxiety were associated with lower predicted risk of discontinuation, while prior taxane therapy and greater baseline sleep disturbance, depression, fatigue, and age at therapy initiation were suggestive of higher risk. Carrying both OPG rs2073618 risk alleles resulted in an increased predicted risk. For the model incorporating 3-month follow-up data, the most predictive feature was self-reported adherence, with higher adherence suggesting lower risk of discontinuation (Figure 3C). Changes in PROs comprised the majority of the most important features. However, the direction of effect for several PRO measures contradicted univariate associations, suggesting the complex interplay between features. Specifically, the lowest values for change in sleep disturbance, endocrine symptoms, and depression were associated with higher predicted risk of discontinuation. Increases in endocrine symptoms, physical function, and BMI were generally associated with decreased risk of discontinuation, while higher changes in fatigue and joint pain suggested increased risk. Interestingly, high changes in anxiety levels were associated with lower predicted risk, whereas moderate changes had an opposite effect on the model’s prediction. Finally, RANKL rs7984870 risk allele carriers were predicted to have a higher risk of discontinuation.
Figure 3.
Most important predictors of discontinuation. Top 10 most important features for prediction were determined by the SHAP algorithm for the baseline clinical + selected genetics model (A), and the 3-month follow-up clinical + all genetics model (C). Top 5 features as determined by SHAP were further assessed for time-dependent importance (panels B and D). Dashed lines in panels B and D represent mean AUC over the whole follow-up period. Abbreviations: AUC, area under the curve; ESR1, estrogen receptor 1; OPG, osteoprotegerin; PharmGKB, Pharmacogenomics Knowledge Base; RANKL, receptor activator of nuclear factor kappa-B ligand; SHAP, Shapley additive explanations; SNP, single-nucleotide polymorphism.
We then analyzed the time-dependence of the most important features determined by SHAP. The variants in the ESR1 gene were highly predictive of the outcome in the first year of treatment, becoming less important in the later periods (Figure 3B). Time-dependent analysis suggests that ESR1 rs2234693 variant increased the risk of early discontinuation but had an opposite effect after 4 years of therapy. The direction of effect of ESR1 rs9340799 stayed consistent over time. Higher baseline sleep disturbance was associated with slightly decreased risk of discontinuing between 3 and 12 months but was highly predictive of discontinuation after 3 years of therapy. A similar trend was observed for cancer stage. Lower baseline anxiety was predictive of discontinuation within the first 3 years, while having the opposite effect in the later periods. Regarding the model incorporating the 3-month follow-up data, decrease in physical function was highly predictive of discontinuation when used as an independent predictor (Figure 3D). Increases in sleep disturbance and anxiety levels and decreases in endocrine symptoms were predictive of early discontinuation, while lower self-reported adherence had an effect past 36 months of therapy. In general, the independent effect of individual risk factors was the most pronounced in the first 6-12 months and after 36 months of treatment, remaining low between 1.5 and 3 years.
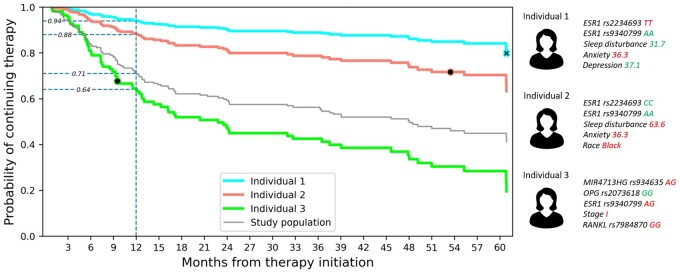
Finally, we used the best baseline model to construct personalized survival curves for 3 randomly selected individuals from the test set (Figure 4). The 5 most influential features for these individuals are presented next to the survival curves. Red color of the feature value indicates increased predicted risk, while green color suggests otherwise.
Figure 4.
Personalized survival curve. The PharmGKB-annotated model developed at baseline was used to predict survival curves for 3 random individuals from the test set. The markers along the curve indicate the actual times when the individual discontinued the medication (black dot) or was censored (black cross). Gray line represents the survival curve of the study sample. Vertical dashed blue line indicates the 12-month cutoff, and the values along the line display the probability that the individual continues the medication at 12 months. Top 5 features affecting the prediction for the selected individuals were determined by SHAP and are presented on a side panel. Abbreviations: ESR1, estrogen receptor 1; MIR4713HG, MIR4713 host gene; OPG, osteoprotegerin; PharmGKB, Pharmacogenomics Knowledge Base; RANKL, receptor activator of nuclear factor kappa-B ligand; SHAP, Shapley additive explanations.
Discussion
In this retrospective analysis of women with HR+ breast cancer initiating AI therapy who were enrolled in a prospective cohort study, we utilized survival ML approaches to predict the time of therapy discontinuation. We demonstrated the time-dependence of the signal coming from the data and showed that incorporation of genetic risk factors improved the ability of the model to identify early discontinuers. In addition, we determined the most influential features for the prediction and explored their predictive value as a function of time. Finally, we presented an example of a personalized survival curve that may be constructed for an individual initiating AI therapy and presented to a clinician for personalized risk assessment. To our knowledge, this is the first report of using survival ML to predict AI therapy discontinuation. Furthermore, there have been no prior studies demonstrating how the predictive value of risk factors for discontinuation changes with time.
For each data model, we tested several survival ML algorithms and focused on the mean cumulative/dynamic AUC and the IBS metrics for evaluation. We chose those metrics over c-index, since the latter only assesses the order of the event times, without taking into consideration how close the predicted time is to the true value.67 Time-dependent AUC and IBS avoid this problem, and as a result were deemed more appropriate evaluation metrics, given our goal of predicting time of discontinuation. We were specifically interested in evaluating the ability of our models to identify the individuals at risk of discontinuing early, as such patients are difficult to determine for a physician. Therefore, the best-performing models were also assessed by their time-dependent AUC, sensitivity, specificity, and PPV calculated at 6 months and 12 months. None of the models were able to achieve a high PPV, since the fraction of study participants discontinuing by that time remained low (29% at 12 months). Among the baseline models, the PharmGKB-annotated model showed the best performance, according to all evaluation metrics. Notably, the other 2 baseline models had a moderate discriminative power within the first year and after the third year of treatment, while not being able to distinguish the patients discontinuing between 1 and 3 years. A similar pattern was observed for both 3-month follow-up models that incorporated genetics. The clinical + all genetics and the clinical only models developed at 3-month follow-up had equally good overall performance, with the former performing better within the first year of treatment. The addition of genetic predictors had lower contribution to the overall model performance at the 3-month follow-up compared to baseline. This may partially be explained by the fact that we selected the SNPs that were associated with AI-induced toxicity. Thus, at baseline, genetic factors predicted which patients would experience toxicity, while at the 3-month follow-up, this toxicity was accounted for by the incorporation of emergent symptoms, thereby reducing the contribution of genetics to the overall model performance. Given our focus on early prediction, we selected the model incorporating genetics as the best model developed at 3-month follow-up, which achieved the AUC of 0.71 at 6 months, outperforming the previously published model.46 In general, the observed patterns suggest that genetic risk factors are important determinants of early discontinuation and their inclusion in predictive models is of value for the identification of patients at-risk of discontinuing within the first year of treatment.
It is important to note that, due to the small sample size, there was a large overlap between the models, and the best-performing models at both baseline and follow-up achieved higher time-dependent AUC values than an average bootstrap run (Figure S1B and C). This can be partially explained by the fact that the algorithm selection and hyperparameter tuning were conducted on the original sample of training data, which may not have been optimal for the samples used during bootstrapping. In general, the bootstrapping procedure demonstrates how well our model learns from new data, but it does not estimate the confidence in the predictions made by the trained model. Future validation studies on independent datasets are necessary to assess this aspect.
The results of our feature importance analysis provide further evidence for early impact of genetics on the outcome. Interestingly, the direction of effect for the 2 most important genetic markers at baseline, ESR1 rs9340799 and ESR1 rs2234693, was the opposite from what has been reported in the previous studies, as was confirmed by univariate analysis via Kaplan-Meier estimator (results not shown). Possible reasons for such discrepancy include the different outcome definition used in our study, or low sample size of our cohort leading to a false signal. One possible explanation for the negative correlation of cancer stage and baseline anxiety with predicted risk of discontinuation is the fact that patients with higher anxiety and more advanced cancer stage may be more concerned about their outcome, leading to higher adherence, despite the development of side effects. The low predictive value of baseline sleep disturbance in early periods followed by an increase after third year of treatment may indicate that the patients may tolerate short periods of sleep disturbance but cannot endure chronic deprivation of sleep. When 3-month follow-up data were incorporated into the model, self-reported adherence was the most important predictor, followed by a set of features indicating changes in PROs and BMI. Given better performance of this model at 6 and 12 months than that of the best model developed at baseline, this finding highlights the utility of follow-up data for prediction.
Although the best model developed at 3-month follow-up was better than the best baseline model at identifying the individuals at risk of discontinuing within the first 6 and 12 months, the baseline model can make its prediction and communicate the risk to the clinician earlier than the model developed at a follow-up, allowing for early implementation of appropriate interventions. The individual’s risk at baseline may be communicated with a physician via a personalized survival curve similar to the one presented in Figure 4, along with the survival curve of general population. The individual’s probability of continuing the medication past a time point of interest or the probability of discontinuing the medication by that time may be presented to a clinician, who will use this information to intervene, if deemed necessary. Furthermore, patient’s values for the most important predictors along with their effect on risk estimate can be displayed and may help choose the appropriate intervention. Some possible interventions may include prescribing tamoxifen instead of an AI, closer monitoring of the symptoms, sending regular reminders, physical activity, integrative medicine approaches, or pharmacotherapy to alleviate the symptoms. In addition, this information may be communicated with the patient when discussing potential risks and benefits of the therapy. Importantly, our model is not meant to be prescriptive but is rather intended to assist clinicians in estimating and communicating overall patient’s risk given their complex multifactorial profile.
It is important to acknowledge some limitations of our study. First, the sample size was small for an ML problem, which may have led to overfitting of the model on the training set. Future studies on broader populations are needed to increase the power of the ML approach. Second, the population used in this study included mostly White Non-Hispanic individuals and did not accurately represent general US population. Further studies should include more diverse populations to assess generalizability of the model. Third, our assessment of genetic determinants was limited to the 13 SNPs available for the analysis. In the future, it would be interesting to explore the extent of genetic contribution to the phenotype using genome-wide data. In addition, this was a retrospective study demonstrating the proof-of-concept on a case study involving AIs. Future prospective studies are needed to validate the ability of the model to predict discontinuation, to identify risk thresholds for guidance regarding potential interventions and to extrapolate to other medication classes. Finally, PRO and genetic data are not commonly integrated within the electronic health record (EHR), making external validation of our model challenging. However, our study shows the importance of integrating such data into medical records. As healthcare institutions incorporate these measures into their EHR, it will become possible to implement the model in the clinical setting.
Conclusion
In this study, we developed a survival ML model to predict AI discontinuation in women diagnosed with HR+ breast cancer. The model incorporating all genetic markers and follow-up data performed best, achieving mean time-dependent AUC of 0.65, and AUC values of 0.71 and 0.67 at 6 and 12 months, respectively. We demonstrated that incorporation of genomic data improves model’s performance and that genetic risk factors are particularly important for prediction of early discontinuers. Our model suggests that changes in PROs are better predictors of discontinuation than the PROs at baseline and that there is a complex interplay between emergent symptoms driving the prediction. Finally, we presented an example of a personalized survival that may be used to communicate the risk of discontinuation to a physician.
Supplementary Material
Contributor Information
Ilia Rattsev, Institute for Computational Medicine, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, 21218, United States; Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, 21218, United States.
Vered Stearns, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, 21287, United States.
Amanda L Blackford, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, 21287, United States.
Daniel L Hertz, Department of Clinical Pharmacy, University of Michigan College of Pharmacy, Ann Arbor, MI, 48109, United States.
Karen L Smith, Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, 21287, United States.
James M Rae, Department of Internal Medicine, University of Michigan Medical School, Ann Arbor, MI, 48109, United States; Department of Pharmacology, University of Michigan Medical School, Ann Arbor, MI, 48109, United States.
Casey Overby Taylor, Institute for Computational Medicine, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, 21218, United States; Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, 21218, United States; Department of General Internal Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, 21205, United States.
Author contributions
I.R., V.S., A.L.B., D.L.H., and C.O.T. contributed to conceptualization and design of the study. Data collection was performed by V.S., A.L.B., D.L.H., and K.L.S. Data analysis was performed by I.R. I.R. wrote the original draft, which was reviewed and edited by all co-authors. All authors contributed to writing and revising the article and approved the submitted version.
Funding
This work was supported by funding from the Susan G. Komen Foundation and the National Institutes of Health [P30 CA006973]. This work was also supported in part by an NIH NHGRI Genomic Innovator award [R35 HG010714 to C.O.T.] and The Breast Cancer Research Foundation [BCRF; N003173 to J.M.R].
Conflicts of interest
The authors declare no competing interests.
Data availability
The data underlying this article cannot be shared publicly due to patient privacy concerns. The researchers interested in data access and collaboration are encouraged to contact the corresponding author.
References
- 1. Davies C, Godwin J, Gray R, et al. ; Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378(9793):771-784. 10.1016/S0140-6736(11)60993-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davies C, Pan H, Godwin J, et al. ; Adjuvant Tamoxifen: Longer Against Shorter (ATLAS) Collaborative Group. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet. 2013;381(9869):805-816. 10.1016/S0140-6736(12)61963-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goss PE, Ingle JN, Martino S, et al. A randomized trial of letrozole in postmenopausal women after five years of tamoxifen therapy for early-stage breast cancer. N Engl J Med. 2003;349(19):1793-1802. 10.1056/NEJMoa032312 [DOI] [PubMed] [Google Scholar]
- 4. Goss PE, Ingle JN, Pritchard KI, et al. Extending aromatase-inhibitor adjuvant therapy to 10 years. N Engl J Med. 2016;375(3):209-219. 10.1056/NEJMoa1604700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blok EJ, Kroep JR, Meershoek-Klein Kranenbarg E, et al. ; IDEAL Study Group. Optimal duration of extended adjuvant endocrine therapy for early breast cancer; results of the IDEAL trial (BOOG 2006-05). J Natl Cancer Inst. 2018;110(1):40-48. 10.1093/jnci/djx134 [DOI] [PubMed] [Google Scholar]
- 6. Tjan-Heijnen VCG, van Hellemond IEG, Peer PGM, et al. ; Dutch Breast Cancer Research Group (BOOG) for the DATA Investigators. Extended adjuvant aromatase inhibition after sequential endocrine therapy (DATA): a randomised, phase 3 trial. Lancet Oncol. 2017;18(11):1502-1511. 10.1016/S1470-2045(17)30600-9 [DOI] [PubMed] [Google Scholar]
- 7. Francis PA, Pagani O, Fleming GF, et al. ; SOFT and TEXT Investigators and the International Breast Cancer Study Group. Tailoring adjuvant endocrine therapy for premenopausal breast cancer. N Engl J Med. 2018;379(2):122-137. 10.1056/NEJMoa1803164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Partridge AH, LaFountain A, Mayer E, Taylor BS, Winer E, Asnis-Alibozek A.. Adherence to initial adjuvant anastrozole therapy among women with early-stage breast cancer. J Clin Oncol. 2008;26(4):556-562. 10.1200/JCO.2007.11.5451 [DOI] [PubMed] [Google Scholar]
- 9. Hershman DL, Kushi LH, Shao T, et al. Early discontinuation and nonadherence to adjuvant hormonal therapy in a cohort of 8,769 early-stage breast cancer patients. J Clin Oncol. 2010;28(27):4120-4128. 10.1200/JCO.2009.25.9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murphy CC, Bartholomew LK, Carpentier MY, Bluethmann SM, Vernon SW.. Adherence to adjuvant hormonal therapy among breast cancer survivors in clinical practice: a systematic review. Breast Cancer Res Treat. 2012;134(2):459-478. 10.1007/s10549-012-2114-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lambert-Côté L, Bouhnik A, Bendiane M, et al. Adherence trajectories of adjuvant endocrine therapy in the five years after its initiation among women with non-metastatic breast cancer: a cohort study using administrative databases. Breast Cancer Res Treat. 2020;180(3):777-790. 10.1007/s10549-020-05549-x [DOI] [PubMed] [Google Scholar]
- 12. Kuba S, Ishida M, Nakamura Y, Taguchi K, Ohno S.. Persistence and discontinuation of adjuvant endocrine therapy in women with breast cancer. Breast Cancer. 2016;23(1):128-133. 10.1007/s12282-014-0540-4 [DOI] [PubMed] [Google Scholar]
- 13. Hershman DL, Shao T, Kushi LH, et al. Early discontinuation and non-adherence to adjuvant hormonal therapy are associated with increased mortality in women with breast cancer. Breast Cancer Res Treat. 2011;126(2):529-537. 10.1007/s10549-010-1132-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makubate B, Donnan PT, Dewar JA, Thompson AM, McCowan C.. Cohort study of adherence to adjuvant endocrine therapy, breast cancer recurrence and mortality. Br J Cancer. 2013;108(7):1515-1524. 10.1038/bjc.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chirgwin JH, Giobbie-Hurder A, Coates AS, et al. Treatment adherence and its impact on disease-free survival in the breast international group 1-98 trial of tamoxifen and letrozole, alone and in sequence. J Clin Oncol. 2016;34(21):2452-2459. 10.1200/JCO.2015.63.8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crew KD, Greenlee H, Capodice J, et al. Prevalence of joint symptoms in postmenopausal women taking aromatase inhibitors for early-stage breast cancer. J Clin Oncol. 2007;25(25):3877-3883. 10.1200/JCO.2007.10.7573 [DOI] [PubMed] [Google Scholar]
- 17. Moscetti L, Agnese Fabbri M, Sperduti I, et al. Adjuvant aromatase inhibitor therapy in early breast cancer: What factors lead patients to discontinue treatment? Tumori. 2015;101(5):469-473. 10.5301/tj.5000376 [DOI] [PubMed] [Google Scholar]
- 18. Nabieva N, Fehm T, Häberle L, et al. Influence of side-effects on early therapy persistence with letrozole in post-menopausal patients with early breast cancer: results of the prospective EvAluate-TM study. Eur J Cancer. 2018;96:82-90. 10.1016/j.ejca.2018.03.020 [DOI] [PubMed] [Google Scholar]
- 19. Aiello Bowles EJ, Boudreau DM, Chubak J, et al. Patient-reported discontinuation of endocrine therapy and related adverse effects among women with early-stage breast cancer. J Oncol Pract. 2012;8(6):e149-e157. 10.1200/JOP.2012.000543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brett J, Fenlon D, Boulton M, et al. Factors associated with intentional and unintentional non-adherence to adjuvant endocrine therapy following breast cancer. Eur J Cancer Care (Engl). 2018;27(1):e12601. 10.1111/ecc.12601 [DOI] [PubMed] [Google Scholar]
- 21. Smith KL, Verma N, Blackford AL, et al. Association of treatment-emergent symptoms identified by patient-reported outcomes with adjuvant endocrine therapy discontinuation. NPJ Breast Cancer. 2022;8(1):53. 10.1038/s41523-022-00414-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kadakia KC, Snyder CF, Kidwell KM, et al. Patient-reported outcomes and early discontinuation in aromatase inhibitor-treated postmenopausal women with early stage breast cancer. Oncologist. 2016;21(5):539-546. 10.1634/theoncologist.2015-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sestak I, Smith SG, Howell A, Forbes JF, Cuzick J.. Early participant-reported symptoms as predictors of adherence to anastrozole in the international breast cancer intervention studies II. Ann Oncol. 2018;29(2):504-509. 10.1093/annonc/mdx713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chim K, Xie SX, Stricker CT, et al. Joint pain severity predicts premature discontinuation of aromatase inhibitors in breast cancer survivors. BMC Cancer. 2013;13:401. 10.1186/1471-2407-13-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wagner LI, Zhao F, Goss PE, et al. Patient-reported predictors of early treatment discontinuation: treatment-related symptoms and health-related quality of life among postmenopausal women with primary breast cancer randomized to anastrozole or exemestane on NCIC clinical trials group (CCTG) MA.27 (E1Z03). Breast Cancer Res Treat. 2018;169(3):537-548. 10.1007/s10549-018-4713-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henry NL, Azzouz F, Desta Z, et al. Predictors of aromatase inhibitor discontinuation as a result of treatment-emergent symptoms in early-stage breast cancer. J Clin Oncol. 2012;30(9):936-942. 10.1200/JCO.2011.38.0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ribi K, Luo W, Walley BA, et al. Treatment-induced symptoms, depression and age as predictors of sexual problems in premenopausal women with early breast cancer receiving adjuvant endocrine therapy. Breast Cancer Res Treat. 2020;181(2):347-359. 10.1007/s10549-020-05622-5 [DOI] [PubMed] [Google Scholar]
- 28. Mausbach BT, Schwab RB, Irwin SA.. Depression as a predictor of adherence to adjuvant endocrine therapy (AET) in women with breast cancer: a systematic review and meta-analysis. Breast Cancer Res Treat. 2015;152(2):239-246. 10.1007/s10549-015-3471-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry NL, Skaar TC, Dantzer J, et al. Genetic associations with toxicity-related discontinuation of aromatase inhibitor therapy for breast cancer. Breast Cancer Res Treat. 2013;138(3):807-816. 10.1007/s10549-013-2504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hertz DL, Smith KL, Zong Y, et al. Further evidence that OPG rs2073618 is associated with increased risk of musculoskeletal symptoms in patients receiving aromatase inhibitors for early breast cancer. Front Genet. 2021;12:662734. 10.3389/fgene.2021.662734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hertz DL, Henry NL, Rae JM.. Germline genetic predictors of aromatase inhibitor concentrations, estrogen suppression and drug efficacy and toxicity in breast cancer patients. Pharmacogenomics. 2017;18(5):481-499. 10.2217/pgs-2016-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johansson H, Gray KP, Pagani O, et al. ; the TEXT principal investigators. Impact of CYP19A1 and ESR1 variants on early-onset side effects during combined endocrine therapy in the TEXT trial. Breast Cancer Res. 2016;18(1):110-118. 10.1186/s13058-016-0771-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Giralt N, Rodríguez-Sanz M, Prieto-Alhambra D, et al. Genetic determinants of aromatase inhibitor-related arthralgia: the B-ABLE cohort study. Breast Cancer Res Treat. 2013;140(2):385-395. 10.1007/s10549-013-2638-3 [DOI] [PubMed] [Google Scholar]
- 34. Fontein DBY, Houtsma D, Nortier JWR, et al. Germline variants in the CYP19A1 gene are related to specific adverse events in aromatase inhibitor users: a substudy of Dutch patients in the TEAM trial. Breast Cancer Res Treat. 2014;144(3):599-606. 10.1007/s10549-014-2873-2 [DOI] [PubMed] [Google Scholar]
- 35. Wang J, Lu K, Song Y, et al. RANKL and OPG polymorphisms are associated with aromatase inhibitor-related musculoskeletal adverse events in Chinese han breast cancer patients. PLoS One. 2015;10(7):e0133964. 10.1371/journal.pone.0133964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J, Lu K, Song Y, et al. Indications of clinical and genetic predictors for aromatase inhibitors related musculoskeletal adverse events in chinese han women with breast cancer. PLoS One. 2013;8(7):e68798. 10.1371/journal.pone.0068798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaparro M, Baston-Rey I, Fernández Salgado E, et al. Using interpretable machine learning to identify baseline predictive factors of remission and drug durability in Crohn's disease patients on ustekinumab. J Clin Med. 2022;11(15):4518. 10.3390/jcm11154518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hamidi O, Poorolajal J, Farhadian M, Tapak L.. Identifying important risk factors for survival in kidney graft failure patients using random survival forests. Iran J Public Health. 2016;45(1):27-33. [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar N, Qi S, Kuan L, Sun W, Zhang J, Greiner R.. Learning accurate personalized survival models for predicting hospital discharge and mortality of COVID-19 patients. Sci Rep. 2022;12(1):4472-4476. 10.1038/s41598-022-08601-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang L, Huang T, Xu F, et al. Prediction of prognosis in elderly patients with sepsis based on machine learning (random survival Forest). BMC Emerg Med. 2022;22(1):26. 10.1186/s12873-022-00582-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen H, Li C, Zheng L, Lu W, Li Y, Wei Q.. A machine learning-based survival prediction model of high grade glioma by integration of clinical and dose-volume histogram parameters. Cancer Med. 2021;10(8):2774-2786. 10.1002/cam4.3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mosquera Orgueira A, Díaz Arias JÁ, Cid López M, et al. Improved personalized survival prediction of patients with diffuse large B-cell lymphoma using gene expression profiling. BMC Cancer. 2020;20(1):1017. 10.1186/s12885-020-07492-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Z, Huang L, Li J, Wang P.. Bioinformatics analysis reveals immune prognostic markers for overall survival of colorectal cancer patients: a novel machine learning survival predictive system. BMC Bioinformatics. 2022;23(1):124-123. 10.1186/s12859-022-04657-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wan G, Nguyen N, Liu F, et al. Prediction of early-stage melanoma recurrence using clinical and histopathologic features. NPJ Precis Oncol. 2022;6(1):79-74. 10.1038/s41698-022-00321-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cui ZL, Kadziola Z, Lipkovich I, Faries DE, Sheffield KM, Carter GC.. Predicting optimal treatment regimens for patients with HR+/HER2- breast cancer using machine learning based on electronic health records. J Comp Eff Res. 2021;10(9):777-795. 10.2217/cer-2020-0230 [DOI] [PubMed] [Google Scholar]
- 46. Ni C, Warner JL, Malin BA, Yin Z.. Predicting hormonal therapy medication discontinuation for breast cancer patients using structured data in electronic medical records. AMIA Annu Symp Proc. 2022;2022:359-368. [PMC free article] [PubMed] [Google Scholar]
- 47. Clinical trial. Primary investigator and study chair: Vered Stearns, MD. Accessed January 13, 2024. https://clinicaltrials.gov/ct2/show/NCT01937052.
- 48. Morisky DE, Ang A, Krousel-Wood M, Ward HJ.. Predictive validity of a medication adherence measure in an outpatient setting. J Clin Hypertens (Greenwich). 2008;10(5):348-354. 10.1111/j.1751-7176.2008.07572.x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49. Garcia SF, Cella D, Clauser SB, et al. Standardizing patient-reported outcomes assessment in cancer clinical trials: a patient-reported outcomes measurement information system initiative. J Clin Oncol. 2007;25(32):5106-5112. 10.1200/JCO.2007.12.2341 [DOI] [PubMed] [Google Scholar]
- 50. Snyder CF, Jensen R, Courtin SO, Wu AW; Website for Outpatient QOL Assessment Research Network. PatientViewpoint: a website for patient-reported outcomes assessment. Qual Life Res. 2009;18(7):793-800. 10.1007/s11136-009-9497-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Snyder CF, Blackford AL, Wolff AC, Carducci MA, Herman JM, Wu AW; PatientViewpoint Scientific Advisory Board. Feasibility and value of PatientViewpoint: a web system for patient-reported outcomes assessment in clinical practice. Psychooncology. 2013;22(4):895-901. 10.1002/pon.3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu AW, White SM, Blackford AL, et al. ; PatientViewpoint Scientific Advisory Board. Improving an electronic system for measuring PROs in routine oncology practice. J Cancer Surviv. 2016;10(3):573-582. 10.1007/s11764-015-0503-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fallowfield LJ, Leaity SK, Howell A, Benson S, Cella D.. Assessment of quality of life in women undergoing hormonal therapy for breast cancer: Validation of an endocrine symptom subscale for the FACT-B. Breast Cancer Res Treat. 1999;55(2):189-199. 10.1023/a:1006263818115 [DOI] [PubMed] [Google Scholar]
- 54. Jensen RE, Potosky AL, Moinpour CM, et al. United States population-based estimates of patient-reported outcomes measurement information system symptom and functional status reference values for individuals with cancer. J Clin Oncol. 2017;35(17):1913-1920. 10.1200/JCO.2016.71.4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schalet BD, Pilkonis PA, Yu L, et al. Clinical validity of PROMIS depression, anxiety, and anger across diverse clinical samples. J Clin Epidemiol. 2016;73:119-127. 10.1016/j.jclinepi.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Teresi JA, Ocepek-Welikson K, Kleinman M, Ramirez M, Kim G.. Measurement equivalence of the patient reported outcomes measurement information system® (PROMIS®) anxiety short forms in ethnically diverse groups. Psychol Test Assess Model. 2016;58(1):183-219. [PMC free article] [PubMed] [Google Scholar]
- 57. Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414-417. 10.1038/clpt.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barbarino JM, Whirl-Carrillo M, Altman RB, Klein TE.. PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med. 2018;10(4):e1417. 10.1002/wsbm.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Heagerty PJ, Lumley T, Pepe MS.. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337-344. 10.1111/j.0006-341x.2000.00337.x [DOI] [PubMed] [Google Scholar]
- 61. Lambert J, Chevret S.. Summary measure of discrimination in survival models based on cumulative/dynamic time-dependent ROC curves. Stat Methods Med Res. 2016;25(5):2088-2102. 10.1177/0962280213515571 [DOI] [PubMed] [Google Scholar]
- 62. Harrell FEJ, Lee KL, Mark DB.. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Statist Med. 1996;15(4):361-387. 10.1002/(SICI)1097-0258(19960229)15:4 [DOI] [PubMed] [Google Scholar]
- 63. Graf E, Schmoor C, Sauerbrei W, Schumacher M.. Assessment and comparison of prognostic classification schemes for survival data. Statist Med. 1999;18(17-18):2529-2545. 10.1002/(sici)1097-0258(19990915/30)18:17/18 [DOI] [PubMed] [Google Scholar]
- 64. Lundberg S, Lee S. A unified approach to interpreting model predictions. In: Proceedings in Advances in Neural Information Processing Systems 30 (NIPS 2017). Long Beach, CA: Neural Information Processing Systems Foundation, Inc. (NeurIPS); 2017.
- 65. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in python. JMLR. 2011;12(85):2825-2830. [Google Scholar]
- 66. Pölsterl S. Scikit-survival: a library for time-to-event analysis built on top of scikit-learn. J Mach Learn Res. 2020;21(212):1-6.34305477 [Google Scholar]
- 67. Blanche P, Kattan MW, Gerds TA.. The c-index is not proper for the evaluation of $t$-year predicted risks. Biostatistics. 2019;20(2):347-357. 10.1093/biostatistics/kxy006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article cannot be shared publicly due to patient privacy concerns. The researchers interested in data access and collaboration are encouraged to contact the corresponding author.