Abstract
Plasma metabolomics profiling is an emerging methodology to identify metabolic pathways underlying cardiovascular health (CVH). The objective of this study was to define metabolomic profiles underlying CVH in a cohort of Black adults, a population that is understudied but suffers from disparate levels of CVD risk factors. The Morehouse-Emory Cardiovascular (MECA) Center for Health Equity study cohort consisted of 375 Black adults (age 53 ± 10, 39% male) without known CVD. CVH was determined by the AHA Life’s Simple 7 (LS7) score, calculated from measured blood pressure, body mass index (BMI), fasting blood glucose and total cholesterol, and self-reported physical activity, diet, and smoking. Plasma metabolites were assessed using untargeted high-resolution metabolomics profiling. A metabolome wide association study (MWAS) identified metabolites associated with LS7 score after adjusting for age and sex. Using Mummichog software, metabolic pathways that were significantly enriched in metabolites associated with LS7 score were identified. Metabolites representative of these pathways were compared across clinical domains of LS7 score and then developed into a metabolomics risk score for prediction of CVH. We identified novel metabolomic signatures and pathways associated with CVH in a cohort of Black adults without known CVD. Representative and highly prevalent metabolites from these pathways included glutamine, glutamate, urate, tyrosine and alanine, the concentrations of which varied with BMI, fasting glucose, and blood pressure levels. When assessed in conjunction, these metabolites were independent predictors of CVH. One SD increase in the novel metabolomics risk score was associated with a 0.88 higher LS7 score, which translates to a 10.4% lower incident CVD risk. We identified novel metabolomic signatures of ideal CVH in a cohort of Black Americans, showing that a core group of metabolites central to nitrogen balance, bioenergetics, gluconeogenesis, and nucleotide synthesis were associated with CVH in this population.
Subject terms: Epidemiology, Molecular medicine, Predictive markers, Metabolic pathways, Metabolomics
Introduction
Cardiovascular disease (CVD) is the leading cause of death in the United States, and there are marked disparities in CVD prevalence and outcomes between Black Americans and other racial groups1,2. High resolution metabolomic profiling is an emerging technology that has the potential to identify pathways underlying cardiovascular health (CVH). The metabolome consists of endogenous, pharmaceutical, and chemical metabolites that fall downstream of genomic, transcriptomic, and proteomic variations3. Metabolomic profiling has shown great promise in elucidating novel mechanisms underlying cardiometabolic and subclinical or clinically apparent CVD4–6. Although Black adults suffer from a greater burden of cardiometabolic risk factors1, the metabolic signatures associated with CVH remain understudied in this vulnerable population7,8.
The purpose of this study was to determine the metabolomic signatures for CVH in Black adults without known CVD and living in the greater metropolitan area of Atlanta, GA9. For this study, CVH was defined by the American Heart Association’s Life’s Simple 7 (LS7) metric (score of 0 to 14 with 14 being ideal CVH), which encompasses a set of seven CVH clinical domains (four health behaviors—smoking, weight, physical activity, and diet; three health factors—blood pressure, total cholesterol, and glucose), and has been shown to predict CVD and all-cause mortality10–13.
Methods
Study design
We studied metabolomic profiles in plasma samples from participants of the Morehouse-Emory Cardiovascular (MECA) Center for Health Equity study. Platelet-free plasma samples were obtained from 375 adults living in the greater Atlanta region, aged 30 to 70, who self-identified as Black or African American. Details on study design and recruitment strategy have been described previously9. In brief, exclusion criteria included known CVD (e.g., myocardial infarction, congestive heart failure, cerebrovascular accident, coronary artery disease, peripheral arterial disease, atrial fibrillation, and cardiomyopathies), concomitant chronic diseases (e.g., human immunodeficiency virus, lupus, or cancer), substance abuse (alcohol or illicit drug), psychiatric illness, pregnant or lactating females, and immobility such that individuals could not increase physical activity.
Enrolled participants visited either Emory University School of Medicine or Morehouse School of Medicine where they completed a physical examination, blood draw, and questionnaires. Vital signs and anthropometric measures were recorded. All blood draws were performed after > 6 h of fasting, and cholesterol and glucose levels were measured. The presence of hypertension was verified by any of the following: current use of anti-hypertensive medications, systolic blood pressure ≥ 130 mmHg, or diastolic blood pressure ≥ 80 mmHg. The presence of diabetes mellitus was determined by either current use of diabetes medications or fasting glucose ≥ 126 mg/dL. Finally, the presence of hyperlipidemia was defined by either current use of lipid-lowering medications or fasting total cholesterol ≥ 240 mg/dL. The protocol was approved by the Institutional Review Boards at Morehouse School of Medicine and Emory University and all participants provided written informed consent. The study was conducted in accordance with the relevant guidelines and regulations of these respective institutions.
Life’s Simple 7 score
Life’s Simple 7 (LS7) score10, developed by the American Heart Association, was calculated for the participants as their metric of clinical CVH. Seven clinical domains of CVH (physical activity, diet, smoking history, blood pressure, glucose, cholesterol, and BMI) were scored as 0 (poor), 1 (intermediate), or 2 (ideal), using the previously published scoring algorithm10. The summary score was computed by the summation of the 7 sub-scores with the range of 0 to 14, with higher scores indicating greater CVH (Table S1).
High-resolution metabolomics (HRM)
Untargeted low molecular weight metabolic profiles (85–1250 daltons) were obtained from platelet free plasma (PFP) samples using the HRM platform described previously14–16. Blood was drawn into sodium citrate tubes and PFP was collected by centrifugation at 2,500 × g for 15 min followed a second centrifugation at 2,500 × g for 15 min before storage at − 80 °C. For metabolomics profiling, PFP samples were thawed and treated with acetonitrile (2:1, v/v), spiked with internal standard mix, and centrifuged at 14,000 × g for 5 min at 4 °C to remove proteins. Samples were maintained at 4 °C in an autosampler until injection and were randomized to minimize effects of instrumental drift during analysis. Three technical 10 μL aliquot replicates were analyzed on a Thermo Scientific Orbitrap Fusion Tribrid Mass Spectrometer using a Thermo Dionex Ultimate 3000 liquid chromatography system with HILIC (hydrophilic interaction liquid chromatography; ThermoFisher Scientific, Accucore, 50 × 2.1 mm, 2.6 μm) separation and electrospray ionization operated in positive mode. The flow rate of the HILIC column was maintained at 0.35 mL/min until 1.5 min, increased to 0.4 mL/min at 4 min and held for 1 min, resulting in a total analytical run time of 5 min. Mobile Phases A and B were LCMS grade water and acetonitrile, respectively. Mobile phase C was composed of 2% formic acid (v/v) in water. Mobile phase conditions consisted of 22.5% A, 75% B, 2.5% C which was held for 1.5 min, with a linear gradient to 77.5% A, 20% B, 2.5% C at 4 min, and held for 1 min. The HILIC column was then flushed for 5 min with a wash solution of 77.5% A, 20% B, 2.5% C before another injection. Mass spectral detection was completed at 120,000 resolution over a mass-to-charge ratio (m/z) range of 85–1275. A quality control pooled reference plasma sample (Q-Std3) was included at the beginning and end of each batch of 20 samples for quality control and quality assurance17. Raw data files were extracted using apLCMSv6.3.318 with xMSanalyzer v2.0.719, followed by batch correction with ComBat6. Uniquely detected ions consisting of m/z and retention time (RT) are referred to as metabolic features or respective metabolites, as appropriate. Metabolic feature annotation was performed using xMSannotator20 based on the Human Metabolome Database21, with only medium to high confidence (score 2 or 3) considered. Further, annotations were also conducted by matching to an in-house library of previously confirmed metabolites, allowing m/z difference of 10 ppm and retention time difference of 60 s, which can be considered Level 1 (highest degree of confidence in metabolite identification) per criteria of Schymanski et al.22.
Metabolome wide association study (MWAS)
Prior to data analysis, feature intensities in triplicates were median summarized based on the nonzero readings, and metabolic features that had > 20% zero readings were removed. Feature intensities were log2 transformed, mean centered and scaled by standard deviation23. Feature intensities were regressed on the LS7 score, adjusting for age and sex or age, sex, and estimated glomerular filtration rate (eGFR)24. Benjamini–Hochberg false discovery rate (FDR) method was used for correction of multiple hypothesis testing. Associations were considered significant at FDR < 0.2 threshold25.
Pathway enrichment analysis
After MWAS, metabolites associated with LS7 score at FDR < 0.2 were characterized for pathway enrichment using Mummichog v2.0.6 software26. Pathways including a minimum of 3 matched metabolites were selected and annotated using the criteria described above. Five confirmed (Level 1) metabolites (glutamine, glutamate, urate, tyrosine and alanine), selected based on significant association with LS7, enrichment in pathways identified by Mummichog, and central to the metabolite activity network (Figure S1), were converted into concentrations as previously described27.
Statistical analysis
Demographic and clinical characteristics were presented by the poor (≤ 6), intermediate (7–9) and ideal (> 10) LS7 scores for descriptive purposes only28. Continuous variables were reported as means (± standard deviation [SD]) and compared using ANOVA across the three categories. Categorical variables were reported as proportions (%) and compared using Chi-square tests. Concentrations of select metabolites were compared across the three scores (0 = poor health, 1 = intermediate health, 2 = ideal health) of each of the seven clinical domains of the LS7 using ANOVA. Then, using linear regression models adjusted for age and sex, concentrations of these select metabolites were compared across the three levels of each clinical domain based on the least squares means. Finally, multivariable models were built including these select metabolites adjusting for age and sex or age, sex, and eGFR, with the dependent variable being the LS7 score. Using the beta coefficient for each metabolite derived from the linear regression as weights, a metabolomics score was created summing over the weighted select metabolites. The score was standardized to a mean of 0, with a standard deviation of 1. It was then analyzed in linear regression of dependent variable LS7 score to explore the composite association of metabolites, adjusting for age and sex.
We performed all statistical analyses using R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria). FDR corrected q value < 0.2 was considered statistically significant for the MWAS29. For the other analyses, p-values < 0.05 were considered statistically significant.
Results
Baseline characteristics
The mean age of the 375 of participants was 53.2 ± 10.2 years with 38.9% male. Mean LS7 score was 8.0 ± 2.2 (Table 1). Lower BMI, younger age and nonsmoking status were observed in participants with higher LS7 scores. In the overall cohort, the prevalence of hypertension was 51.2%, hyperlipidemia was 30.5%, diabetes was 20.6%, and smoking was 24.3%. The mean BMI was 32.9 kg/m2.
Table 1.
Demographic and clinical characteristics of the cohort stratified by high (≥ 10), intermediate (7–9) and low (≤ 6) categories of LS7 Scores.
| Low (n = 97) | Intermediate (n = 182) | High (n = 96) | Total (n = 375) | p value | |
|---|---|---|---|---|---|
|
Demographics n(%) or mean (SD) |
|||||
| Age, years | 55.5 (8.1) | 54.4 (9.5) | 48.6 (12.0) | 53.2 (10.2) | < 0.001 |
| Male | 36 (37.1) | 65 (35.7) | 45 (46.9) | 146 (38.9) | 0.176 |
|
Objective/Clinical Measures n(%) or mean (SD) |
|||||
| Hypertension | 81 (83.5) | 95 (52.2) | 16 (16.7) | 192 (51.2) | < 0.001 |
| Hyperlipidemia | 45 (46.4) | 60 (33.0) | 9 ( 9.5) | 114 (30.5) | < 0.001 |
| Diabetes Mellitus | 46 (47.9) | 28 (15.4) | 3 ( 3.1) | 77 (20.6) | < 0.001 |
| Current Smoker | 40 (41.2) | 42 (23.1) | 9 ( 9.4) | 91 (24.3) | < 0.001 |
| Body Mass Index (BMI) | 36.4 (7.5) | 34.0 (8.3) | 27.2 (5.7) | 32.9 (8.2) | < 0.001 |
| Systolic Blood Pressure (mmHg) | 140.2 (19.5) | 131.5 (18.4) | 119.7 (15.5) | 130.7 (19.4) | < 0.001 |
| Diastolic Blood Pressure (mmHg) | 85.1 (12.2) | 81.4 (11.0) | 73.3 (9.4) | 80.3 (11.8) | < 0.001 |
| Total Cholesterol (mg/dL) | 200.9 (43.9) | 196.8 (37.9) | 172.9 (30.4) | 191.7 (39.4) | < 0.001 |
| HDL (mg/dL) | 54.1 (16.8) | 56.49 (16.6) | 61.5 (17.9) | 57.1 (17.1) | 0.008 |
| LDL (mg/dL) | 121.7 (38.1) | 119.5 (34.5) | 95.3 (25.2) | 113.8 (35.0) | < 0.001 |
| Triglycerides (mg/dL) | 129.0 (83.4) | 104.9 (46.9) | 75.5 (37.7) | 103.6 (59.9) | < 0.001 |
| Fasting Glucose (mg/dL) | 121.4 (43.7) | 98.9 (31.9) | 90.0 (23.8) | 102.4 (35.6) | < 0.001 |
| Total LS7 score | 5.2 (0.82) | 7.9 (0.82) | 10.8 (0.92) | 7.97 (2.20) | < 0.001 |
Metabolome-wide association study (MWAS)
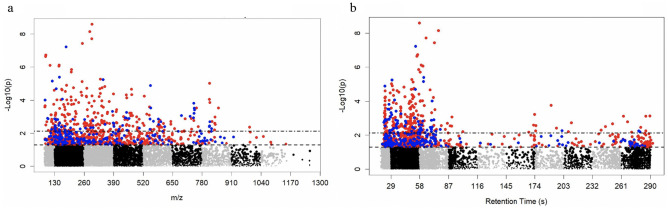
Untargeted, high resolution plasma metabolomics profiling of 375 MECA participants was performed using liquid chromatography/mass spectrometry. A total of 14,501 metabolic features were detected; metabolic features that had > 20% zero readings were removed and 8,211 underwent further analysis. After adjusting for age and sex, MWAS identified 301 metabolic features that were associated with the LS7 score at FDR cutoff < 0.2; 232 metabolic features were lower and 69 metabolic features were higher with higher LS7 (Fig. 1, Table 2, Table S2).
Figure 1.
Manhattan plot (− log p vs (a) m/z or (b) retention time) of metabolites determined by MWAS to be associated with LS7 (as a continuous variable) after adjusting for age and sex. 8211 metabolic features underwent MWAS analysis. 301 metabolic features were differentially expressed at FDR < 0.2; 232 metabolic features colored red were lower with higher LS7 and 69 metabolic features colored blue were higher with higher LS7. Retention time is expressed in seconds.
Table 2.
Select metabolites from the metabolome wide association study (MWAS) that were significantly associated with ideal cardiovascular health as defined by AHA Life’s Simple 7 (LS7). For list of other metabolites, please refer to Table S2.
| Name | m/z_RT(sec) | Beta | Standard Error | p value | FDR Q value | HMDB ID |
|---|---|---|---|---|---|---|
| Lower expression with higher LS7 | ||||||
| 5-Deoxyadenosine | mz252.1078_t72.7 | − 0.131 | 0.023 | 3.68E-08 | 7.56E-05 | HMDB00101 |
| Alanine | mz90.055_t55.2 | − 0.124 | 0.024 | 2.22E-07 | 0.00026 | HMDB00056 |
| Glutamate | mz148.0605_t61.2 | − 0.117 | 0.023 | 8.51E-07 | 0.00070 | HMDB00148 |
| Urate | mz169.0359_t50.8 | − 0.087 | 0.023 | 0.00018 | 0.023 | HMDB00289 |
| N-amidino-L-aspartate | mz176.0659_t38.7 | − 0.086 | 0.024 | 0.00037 | 0.039 | HMDB03157 |
| Ferulate | mz195.0655_t59.4 | − 0.082 | 0.024 | 0.00072 | 0.061 | HMDB00954 |
| Leucine/Isoleucine | mz132.1019_t39.1 | − 0.078 | 0.023 | 0.00086 | 0.064 | HMDB00172 |
| Indole-3-acetate | mz176.0705_t24.9 | − 0.076 | 0.024 | 0.0017 | 0.097 | HMDB00197 |
| Proline | mz116.0707_t60.4 | − 0.075 | 0.024 | 0.0020 | 0.102 | HMDB00162 |
| Tyrosine | mz182.0811_t49.6 | − 0.073 | 0.024 | 0.0027 | 0.119 | HMDB00158 |
| 3-Hydroxybenzaldehyde | mz123.0441_t49.6 | − 0.073 | 0.024 | 0.0026 | 0.119 | HMDB01870 |
| Phenylpuruvate | mz165.0546_t49.3 | − 0.071 | 0.024 | 0.0036 | 0.138 | HMDB00205 |
| 1-Methyladenosine | mz282.1189_t54.1 | − 0.066 | 0.024 | 0.0060 | 0.183 | HMDB03331 |
| Higher expression with higher LS7 | ||||||
| Maleamate | mz116.0343_t74.3 | 0.067 | 0.024 | 0.0057 | 0.179 | NA |
| Oxoproline | mz130.05_t65.3 | 0.068 | 0.024 | 0.0050 | 0.165 | HMDB00267 |
| Glutamine | mz147.0766_t70.5 | 0.071 | 0.024 | 0.0033 | 0.133 | HMDB00267 |
| Homocysteine | mz136.0427_t53.5 | 0.102 | 0.024 | 2.00E-05 | 0.0064 | HMDB00742 |
| Methyladenine | mz150.0766_t62.1 | 0.110 | 0.023 | 4.08E-06 | 0.0026 | HMDB02099 |
HMDB ID- Human Metabolome Database Identification; m/z- mass-to-charge ratio; RT- Retention time, expressed in seconds; NA- Not Annotated in HMDB;
Metabolic pathways analysis
Using Mummichog, pathway enrichment analysis of the 301 metabolic features associated with LS7 identified 10 metabolic pathways (p-value < 0.05), including those involved in metabolism of glutathione, several amino acids, vitamin A, and purine (Fig. 2). Several metabolites identified by MWAS and listed in Table 2 matched to multiple pathways (Table S3). For example, alanine, glutamine, and glutamate matched to the glutathione, glutamate, alanine and aspartate, and tyrosine metabolism pathways. Visualization of the metabolite activity network, which combined analyses of identified pathways with metabolite modules showed that the amino acids glutamate, glutamine, alanine, and tyrosine were central nodes connecting different metabolic pathways (Figure S1).
Figure 2.
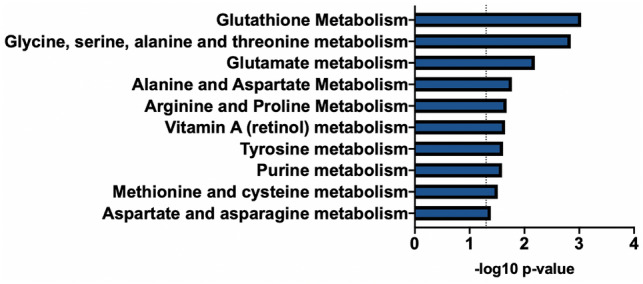
Metabolic pathways enriched for metabolites associated with LS7. Pathway analysis was performed using Mummichog software (version 2.0.6). The 301 metabolic features associated with LS7 by MWAS were entered into the analysis. Significantly enriched pathways (p < 0.05) are shown.
Association of metabolite concentrations with clinical domains of LS7
To further examine the relationship between plasma metabolite levels and CVH, we compared concentrations of five confirmed (Level 1) metabolites (alanine, glutamine, glutamate, tyrosine, and urate) across clinical domains of LS7. All five metabolites were identified by MWAS to be associated with LS7 score when adjusted for age and sex (Table 2) or adjusted for age, sex, and eGFR (Table S5), were enriched in the metabolic pathways identified by Mummichog (Fig. 2, Table S3), and were central nodes of the metabolite activity network (Figure S1). Furthermore, the levels of these metabolites were stable across the samples as indicated by relatively low coefficients of variation (CV)—glutamine, 1.24%; glutamate—3.06%; Urate—3.39%; tyrosine—1.58%; alanine—1.89%. Metabolite concentrations were determined as described by Liu et al.27 When we compared metabolite concentrations across the three scores (0 = poor health, 1 = intermediate health, or 2 ideal health) for each clinical domain of the LS7, we found that glutamate concentrations were higher with poor blood pressure, glucose and BMI scores, while glutamine concentrations were lower with poor glucose score. Urate concentrations were higher with poor BMI score, and tyrosine concentrations were higher with poor cholesterol and BMI scores. Alanine concentrations were higher with poor cholesterol, glucose, and BMI scores and lower with ideal physical activity (Fig. 3, Table 3). Analysis of concentrations across LS7 clinical domains adjusted for age and sex demonstrated showed similar results (Table S4). These data suggest that plasma metabolite levels were influenced by individual clinical domains of LS7, indicating that the expression profile of these five metabolites can reflect individual CVH.
Figure 3.
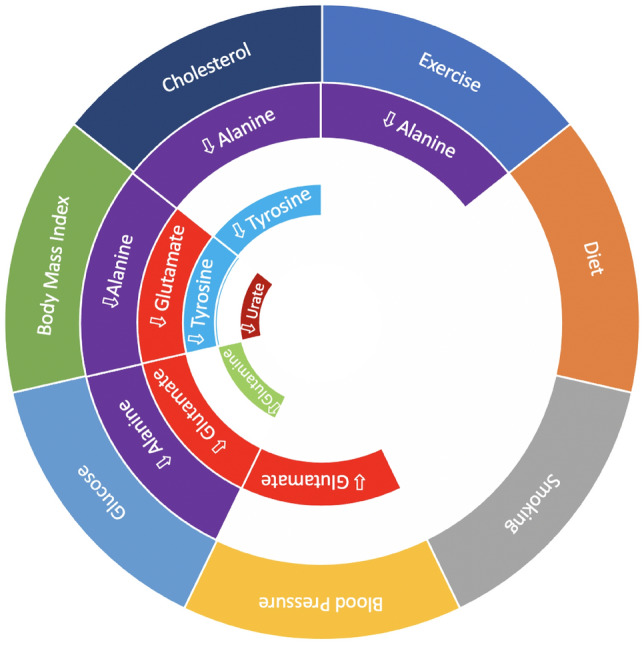
LS7 clinical domains associated with metabolite concentrations. Concentrations were compared across three scores (0, 1, 2) of the seven clinical domains of the LS7 using ANOVA. Upward arrows indicate that metabolite concentration increased with respective increase in clinical domain score, while downward arrows indicate metabolite concentration decreased with respective increase in LS7 score.
Table 3.
Concentrations in μM of select metabolites compared across LS7 clinical domains (unadjusted).
| Glutamine | Glutamate | Urate | Tyrosine | Alanine | ||
|---|---|---|---|---|---|---|
| Blood Pressure | Poor | 412 ± 121 | 35 ± 19 | 154 ± 77 | 71 ± 22 | 269 ± 81 |
| Intermediate | 406 ± 146 | 29 ± 14 | 155 ± 73 | 69 ± 23 | 261 ± 89 | |
| Ideal | 417 ± 140 | 28 ± 13 | 142 ± 60 | 70 ± 22 | 246 ± 64 | |
| p value | 0.87 | 0.001 | 0.37 | 0.77 | 0.09 | |
| Cholesterol | Poor | 431 ± 102 | 34 ± 14 | 155 ± 72 | 79 ± 21 | 279 ± 97 |
| Intermediate | 399 ± 143 | 33 ± 19 | 153 ± 82 | 68 ± 23 | 270 ± 85 | |
| Ideal | 417 ± 129 | 30 ± 15 | 149 ± 64 | 70 ± 22 | 250 ± 70 | |
| p value | 0.27 | 0.13 | 0.88 | 0.03 | 0.02 | |
| Glucose | Poor | 373 ± 137 | 37 ± 20 | 167 ± 91 | 68 ± 22 | 301 ± 110 |
| Intermediate | 417 ± 127 | 36 ± 17 | 157 ± 74 | 73 ± 23 | 277 ± 70 | |
| Ideal | 419 ±132 | 29 ± 15 | 146 ± 66 | 70 ± 23 | 247 ± 70 | |
| p value | 0.045 | < 0.001 | 0.10 | 0.39 | < 0.001 | |
| Body Mass Index (BMI) | Poor | 410 ± 131 | 33 ± 16 | 159 ± 75 | 74 ± 23 | 276 ± 85 |
| Intermediate | 415 ± 129 | 29 ± 15 | 143 ± 63 | 65 ± 19 | 248 ± 71 | |
| Ideal | 411 ± 146 | 29 ± 21 | 136 ± 72 | 65 ± 21 | 228 ± 64 | |
| p value | 0.96 | 0.05 | 0.038 | 0.001 | < 0.001 | |
| Exercise | Poor | 386 ± 136 | 36 ± 15 | 158 ± 88 | 70 ± 22 | 266 ± 57 |
| Intermediate | 415 ± 123 | 33 ± 19 | 146 ± 71 | 72 ± 22 | 279 ± 89 | |
| Ideal | 412 ± 138 | 30 ± 16 | 154 ± 71 | 70 ± 23 | 250 ± 76 | |
| p value | 0.54 | 0.068 | 0.52 | 0.75 | 0.005 | |
| Diet | Poor | 424 ± 126 | 29 ± 14 | 154 ± 71 | 72 ± 20 | 271 ± 83 |
| Intermediate | 405 ± 132 | 32 ± 18 | 149 ± 74 | 69 ± 23 | 256 ± 80 | |
| Ideal | 405 ± 168 | 36 ± 19 | 161 ± 61 | 76 ± 26 | 262 ± 71 | |
| p value | 0.45 | 0.12 | 0.68 | 0.19 | 0.26 | |
| Smoking | Poor | 409 ± 129 | 33 ± 20 | 156 ± 81 | 745 ± 24 | 255 ± 80 |
| Intermediate | 341 ± 141 | 33 ± 13 | 158 ± 89 | 71 ± 28 | 277 ± 71 | |
| Ideal | 416 ± 133 | 31 ± 16 | 150 ± 68 | 69 ± 21 | 263 ± 81 | |
| p value | 0.10 | 0.41 | 0.75 | 0.09 | 0.55 |
Metabolite risk score and cardiovascular health in black adults
A multivariable model using the concentrations of the five metabolites demonstrated that all five were associated with LS7 score, independent of each other (Tables 4, S7). After adjusting for age and sex, the model demonstrated that one standard deviation increase in the respective metabolite concentration was associated with a 0.45 unit higher LS7 score for glutamine, and 0.28 to 0.54 unit lower LS7 scores for the other metabolites (Table 4). Similar results were observed for a multivariable model that adjusted for age, sex, and eGFR (Table S7). The weighted metabolite risk score comprised of these five metabolites continued to be associated with total LS7 score, where one standard deviation increase in the score corresponded to a 0.88 unit higher LS7 score (p < 0.001, Fig. 4), which translates to a 10.4% lower incidence of lifetime CVD30. A similar relationship between metabolite risk score and LS7 score was observed when the metabolite risk score was adjusted for age, sex, and eGFR—estimate 0.88 unit higher LS7 score for one standard deviation increase in metabolite risk score (95% CI 0.69, 1.08; p = 5.69E-17). These findings emphasize the importance of non-essential amino acid and purine metabolism in CVH and demonstrate the ability of plasma concentrations of glutamate, glutamine, alanine, tyrosine, and urate to predict CVH.
Table 4.
Multivariable linear regression demonstrated that in a model adjusting for age and sex, all five of the metabolites were independently associated with LS7.
| Name | m/z_RT(sec) | Beta* | Standard Error | p value |
|---|---|---|---|---|
| Glutamine | mz147.0766_t70.5 | 0.45 | 0.12 | < 0.001 |
| Glutamate | mz148.0605_t61.2 | − 0.32 | 0.11 | < 0.001 |
| Urate | mz169.0359_t50.8 | − 0.38 | 0.11 | < 0.001 |
| Tyrosine | mz182.0811_t49.6 | − 0.28 | 0.12 | 0.02 |
| Alanine | mz90.055_t55.2 | − 0.54 | 0.12 | < 0.001 |
*Beta: increase in LS7 per one standard deviation increase in respective metabolite concentration.
Figure 4.
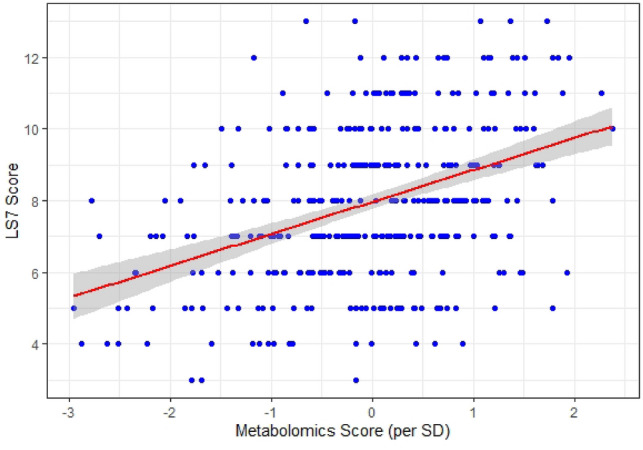
Relationship between metabolite risk score and LS7. One standard deviation increase in the score corresponded to 0.88 higher LS7 score.
Discussion
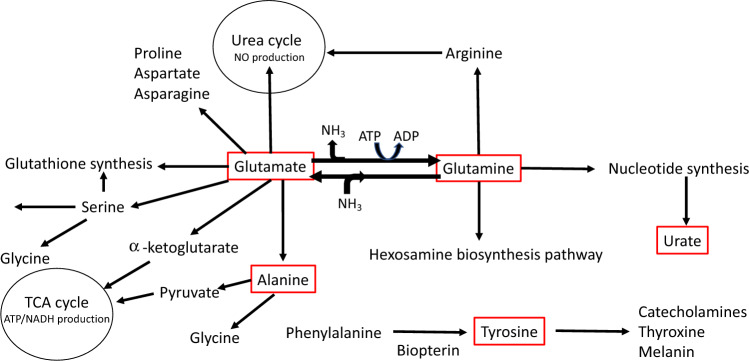
Herein, we used high resolution metabolomics profiling, advanced data extraction algorithms, and pathways analysis to examine metabolites and pathways underlying CVH in a cohort of Black participants without known CVD and living in the greater metro area of Atlanta, GA. CVH was assessed by AHA LS7 score, which incorporates key clinical risk factors for CVD and has been previously demonstrated to be a surrogate for CVH30. We identified novel metabolomic signatures that included five key metabolites (glutamine, glutamate, urate, tyrosine, and alanine) linked to energy production, nitrogen balance, gluconeogenesis, and the metabolic syndrome (Fig. 5)5,31–40. Subsequently, we created a novel metabolomics risk score to predict CVH in Black adults.
Figure 5.
Metabolites predictive of CVH in MECA are central to essential metabolic pathways. Five metabolites (red rectangles) that were associated with CVH and were the focus of current study are central components of key metabolic processes.
We found that the concentrations of these five metabolites in the plasma were specifically associated with poor, intermediate, or ideal scores in the cardiometabolic domains of the LS7 score. Prior studies have demonstrated similar association between levels of non-essential amino acids (e.g. glutamate, glutamine, alanine, etc.) and cardiometabolic risk or disease, such as diabetes, in multiethnic populations not enriched for Black adults8,31–34. Cheng et al. corroborated a metabolomic signature involving non-essential amino acids, such as alanine, with metabolic syndrome within a mainly white cohort enrolled in the Framingham Heart Study31. They also demonstrated an association of high glutamine to glutamate ratio with lower risk of incident diabetes. Similar associations between non-essential amino acid levels and cardiometabolic risk were noted in Asian32,33 and Mediterranean34 cohorts. To our knowledge, only two other studies, both conducted within the Jackson Heart Study cohort, have assessed metabolomic signatures in Black adults but in the context of incident coronary heart disease7 and heart failure8. Thus, the current study is the first to demonstrate that CVH (or CV risk factors) in Black Americans is associated with metabolomic signatures similar to those reported for other racial/ethnic cohorts. In particular, the current study demonstrated the importance of glutamate and glutamine metabolism in this cohort. In addition, other novel metabolic pathways associated with CVH were identified. Whether these other pathways are only relevant to CVH in Black adults require further study in multi-ethnic cohorts.
We found that concentrations of both glutamine and glutamate varied with glucose levels, while glutamate concentrations also correlated with blood pressure and BMI. Glutamine is one of the most abundant amino acids in the body, classified as a conditionally essential amino acid, and has a critical role in nitrogen balance, providing intermediates to the tricarboxylic acid (TCA) cycle (anapleurosis), immunity, and pH homestasis5,35–40. Glutamine is mainly synthesized from glutamate and ammonia by the enzyme glutamine synthetase (Fig. 5)7. The pathophysiology of cardiometabolic disease is complex and disturbances in glutamine/glutamate metabolism have been implicated in the development of metabolic risk in multiethnic communities5,31–34. The role of glutamine/glutamate in CVH is likely mediated through multiple mechanisms. First, glutamine has potent antioxidant, anti-inflammatory, and anti-apoptotic effects by stimulating glutathione, heat shock proteins, and heme oxygenase-15,41. Glutamine also stimulates nitric oxide bioavailability by increasing arginine synthesis that may maintain normal blood pressure42. We note here that in the current study glutamate levels were higher with higher blood pressure.
Glutamine can improve glucose homeostasis by stimulating release of glucagon-like peptide-1, externalization of glucose transporters, insulin release by pancreatic β-cells, transcription of insulin-dependent genes, and increased insulin disposition43. Glutamine also increases the transamination of pyruvate to alanine, which is directly involved with the TCA cycle and is involved in modulating obesity by being a strong promoter of gluconeogenesis44. Dysregulation in glutamate/glutamine metabolism can lead to higher levels of alanine45, which has been shown to be associated with increased Type 2 diabetes risk46. In our study, we noted that lower levels of alanine were associated with ideal glucose, BMI, cholesterol scores, as well as the modifiable component of physical activity. Based on these data, we hypothesize that the well-recognized beneficial effects of physical activity on CVH are mediated through changes in the metabolome, including changes in alanine metabolism, but future studies are required to determine whether health interventions that involve increased physical activity can modify alanine levels.
Tyrosine was another one of the select metabolites that was associated with BMI, as well as the cholesterol subcomponent of LS7 score. Previously, elevated levels of tyrosine have shown positive associations with insulin resistance and Type 2 diabetes in multiple non-Black cohorts31,47–50. Catecholamines (dopamine, norepinephrine, epinephrine) are neurotransmitters synthesized from tyrosine51,52. We speculate that changes in tyrosine can likely lead to changes in physiological and behavioral functions that can impact CVH in Black adults but future studies are required.
Our final metabolite of interest was urate or uric acid, the concentration of which was associated with BMI. Urate has long been established as associated with cardiovascular disease53. Urate is a major product of purine metabolism and has been found to impair nitric oxide synthesis and promote endothelial dysfunction, which is pro-thrombotic, pro-inflammatory, pro-vasoconstrictive, and increases risk of incident CVD54,55. While we did not find an association between urate levels and risk factors directly linked to endothelial dysfunction such as hypertension, we did note, in accordance with prior studies, that high urate levels were associated with obesity and as such, remain an important marker for CVH56.
As shown in Fig. 5, the five metabolites (glutamine, glutamate, alanine, tyrosine, and urate) that were the focus of this study are known to be central elements to key metabolic processes. Each of them has been previously shown to be associated with CVH. While Pearson correlation analysis revealed associations between expression of each metabolite and expression of other metabolites (Table S6), we uniquely demonstrate that when assessed together, these metabolites were independent predictors of CVH in Black adults, and the novel metabolomics risk score derived from the concentrations of these metabolites was similarly predictive. Prior risk scores have been developed from metabolomic signatures to predict incident coronary heart disease29,57,58, but, to our knowledge, we are the first to incorporate the concentration of these metabolites into a risk prediction tool for CVH.
While major limitations of this study are that it is single center, and it examined a relatively small cohort, we are among the first to demonstrate the molecular basis of CVH within Black Americans, a population underrepresented in scientific literature but suffers from disparate levels of CVD. Other limitations of the study are the need for replication in a separate cohort, and, given the cross-sectional design, the inability to address causality, which would require longer term follow-up. This study used the LS7 score as a surrogate for CVH, so longer-term follow-up of participants’ outcomes would also be needed to determine whether the five highlighted metabolites can predict CVH better than clinical risk factors. Furthermore, while we explored several metabolic pathways that were associated with CVH, we did not further explore others that were identified in our analysis, including pathways responsible for retinol and arginine metabolism, which require consideration in future studies. Lastly, while the concentrations of the metabolites examined in this study varied with cardiometabolic risk factors, none of them varied with diet or smoking, factors known to be important determinants of CVH. Future study involving a more detailed assessment of diet will be needed to determine its influence on the metabolomic signature associated with CVH in Black adults. Future studies will also need to examine the interaction of social and environmental factors with metabolomic signatures. Despite these limitations, we report several novel findings and present a new metabolomics risk score, which demonstrated that CVH was associated with concentrations of five metabolites, pointing to the importance of non-essential amino acid and purine metabolism in CVH and the potential of the five highlighted metabolites as therapeutic targets. Additional studies are required to validate this score in multiethnic populations and prospectively follow participants for development of cardiovascular disease.
Supplementary Information
Abbreviations
- BMI
Body Mass Index
- CI
Confidence interval
- CV
Cardiovascular
- CVD
Cardiovascular disease
- CVH
Cardiovascular health
- MECA
Morehouse-Emory Cardiovascular Center for Health Equity
- LS7
Life’s Simple 7
- SD
Standard deviation
Author contributions
C.D.S, S.J.I., Y.V.S., S.B.D., T.T.L, H.A.T., D.P.J, and A.A.Q. designed and conceived the study. S.J.I., C.L., A.A.Q., D.P.J. and C.D.S. wrote the main manuscript text. K.R. and A.N.M. processed blood samples. S.J.I. and C.L. performed analyses. S.J.I., C.L., and C.D.S. prepared the figures and tables. All authors reviewed data and the manuscript.
Funding
The American Heart Association 15SFCRN23910003, Abraham J. & Phyllis Katz Foundation, Byron Williams Jr, MD Fellowship Fund and the National Institutes of Health T32 HL130025 & T32 HL007745-26A1. A.A.Q. has been supported by NIH grants 4R61HL138657-04, U54AG062334-01, 1P30DK111024-03S1, 15SFCRN23910003, 5P01HL086773-09, 1R01HL141205-01, 5P01HL101398-05, 1P20HL113451-04, 3RF1AG051633-01S2.
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available due to sensitive nature of personalized healthcare data but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-51920-z.
References
- 1.Carnethon MR, Pu J, Howard G, et al. Cardiovascular health in African Americans: A scientific statement from the American Heart Association. Circulation. 2017;136(21):e393–e423. doi: 10.1161/CIR.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 2.Van Dyke, M., Greer, S. & Odom, E, et al. Heart disease death rates among blacks and whites aged ≥35 years—United States, 1968–2015. Morbidity and mortality weekly report Surveillance summaries (Washington, DC : 2002). 67(5):1–11 (2018). 10.15585/mmwr.ss6705a1 [DOI] [PMC free article] [PubMed]
- 3.Sharon D, Chen R, Snyder M. Systems biology approaches to disease marker discovery. Dis. Mark. 2010;28(4):209–224. doi: 10.3233/DMA-2010-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: Form and function. Circulation. 2012;126(9):1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durante W. The emerging role of l-glutamine in cardiovascular health and disease. Nutrients. 2019;11:2092. doi: 10.3390/nu11092092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 7.Cruz DE, Tahir UA, Hu J, et al. Metabolomic analysis of coronary heart disease in an African American cohort from the Jackson Heart Study. JAMA Cardiol. 2022;7(2):184–194. doi: 10.1001/jamacardio.2021.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahir UA, Katz DH, Zhao T, et al. Metabolomic profiles and heart failure risk in black adults: Insights from the Jackson Heart Study. Circ. Heart Fail. 2021;14(1):e007275. doi: 10.1161/CIRCHEARTFAILURE.120.007275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Islam S, Kim J, Topel M, et al. Cardiovascular risk and resilience among blacks: Rationale and design of the MECA study. J. Am. Heart Assoc. 2020;9:e015247. doi: 10.1161/JAHA.119.015247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd-Jones Donald M, Hong Y, Labarthe D, et al. Defining and setting national goals for cardiovascular health promotion and disease reduction. Circulation. 2010;121(4):586–613. doi: 10.1161/CIRCULATIONAHA.109.192703. [DOI] [PubMed] [Google Scholar]
- 11.Effoe VS, Carnethon MR, Echouffo-Tcheugui JB, et al. The American Heart association ideal cardiovascular health and incident type 2 diabetes mellitus among blacks: The Jackson Heart Study. J. Am. Heart Assoc. 2017;6(6):e005008. doi: 10.1161/JAHA.116.005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph JJ, Echouffo-Tcheugui JB, Carnethon MR, et al. The association of ideal cardiovascular health with incident type 2 diabetes mellitus: The multi-ethnic study of atherosclerosis. Diabetologia. 2016;59(9):1893–1903. doi: 10.1007/s00125-016-4003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, N., Jiang, M. & Fan, Y. Ideal cardiovascular health metrics and risk of cardiovascular disease or mortality: A meta-analysis (1874–1754 (Electronic)). [DOI] [PubMed]
- 14.Li S, Park Y, Duraisingham S, et al. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roede JR, Uppal K, Park Y, et al. Serum metabolomics of slow vs. rapid motor progression Parkinson's disease: A pilot study. PLoS ONE. 2013;8(10):e77629. doi: 10.1371/journal.pone.0077629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman JM, Soltow QA, Li S, Sidik A, Jones DP, Promislow DE. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, Drosophila melanogaster. Aging Cell. 2014;13(4):596–604. doi: 10.1111/acel.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Go Y-M, Walker DI, Liang Y, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol. Sci. 2015;148(2):531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alves G, Yu Y-K. Improving peptide identification sensitivity in shotgun proteomics by stratification of search space. J. Proteome Res. 2013;12(6):2571–2581. doi: 10.1021/pr301139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uppal K, Ma C, Go YM, Jones DP, Wren J. xMWAS: A data-driven integration and differential network analysis tool. Bioinformatics. 2018;34(4):701–702. doi: 10.1093/bioinformatics/btx656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uppal K, Walker DI, Jones DP. xMSannotator: An R Package for network-based annotation of high-resolution metabolomics data. Anal. Chem. 2017;89(2):1063–1067. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS, Feunang YD, Marcu A, et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–d617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schymanski, E. L., Jeon, J. & Gulde, R., et al. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. ACS Publications; (2014). [DOI] [PubMed]
- 23.Patel RM, Roback JD, Uppal K, Yu T, Jones DP, Josephson CD. Metabolomics profile comparisons of irradiated and nonirradiated stored donor red blood cells. Transfusion. 2015;55(3):544–552. doi: 10.1111/trf.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inker LA, Eneanya ND, Coresh J, et al. New creatinine- and cystatin C-based equations to estimate GFR without Race. N. Engl. J. Med. 2021;385(19):1737–1749. doi: 10.1056/NEJMoa2102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.). 1995;57(1):289–300. [Google Scholar]
- 26.Uppal K, Walker DI, Liu K, Li S, Go Y-M, Jones DP. Computational metabolomics: A framework for the million metabolome. Chem. Res. Toxicol. 2016;29(12):1956–1975. doi: 10.1021/acs.chemrestox.6b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu KH, Nellis M, Uppal K, et al. Reference standardization for quantification and harmonization of large-scale metabolomics. Anal. Chem. 2020;92(13):8836–8844. doi: 10.1021/acs.analchem.0c00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thacker EL, Gillett SR, Wadley VG, et al. The American Heart Association Life's Simple 7 and Incident Cognitive Impairment: The REasons for Geographic And Racial Differences in Stroke (REGARDS) Study. J. Am. Heart Assoc.3(3), e000635. 10.1161/JAHA.113.000635 [DOI] [PMC free article] [PubMed]
- 29.Mehta A, Liu C, Nayak A, et al. Untargeted high-resolution plasma metabolomic profiling predicts outcomes in patients with coronary artery disease. PLoS ONE. 2020;15(8):e0237579. doi: 10.1371/journal.pone.0237579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xanthakis V, Enserro Danielle M, Murabito Joanne M, et al. Ideal cardiovascular health. Circulation. 2014;130(19):1676–1683. doi: 10.1161/CIRCULATIONAHA.114.009273. [DOI] [PubMed] [Google Scholar]
- 31.Cheng S, Rhee EP, Larson MG, et al. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation. 2012;125(18):2222–2231. doi: 10.1161/CIRCULATIONAHA.111.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang SM, Yang RY, Wang M, et al. Identification of serum metabolites associated with obesity and traditional risk factors for metabolic disease in Chinese adults. Nutr. Metab. Cardiovasc. Dis. 2018;28(2):112–118. doi: 10.1016/j.numecd.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Yu X, Zhang W, et al. Association of serum metabolites with impaired fasting glucose/diabetes and traditional risk factors for metabolic disease in Chinese adults. Clin. Chim. Acta. 2018;487:60–65. doi: 10.1016/j.cca.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 34.Ntzouvani A, Nomikos T, Panagiotakos D, et al. Amino acid profile and metabolic syndrome in a male Mediterranean population: A cross-sectional study. Nutr. Metab. Cardiovasc. Dis. 2017;27(11):1021–1030. doi: 10.1016/j.numecd.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Bruce M, Constantin-Teodosiu D, Greenhaff PL, Boobis LH, Williams C, Bowtell JL. Glutamine supplementation promotes anaplerosis but not oxidative energy delivery in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2001;280(4):E669–E675. doi: 10.1152/ajpendo.2001.280.4.E669. [DOI] [PubMed] [Google Scholar]
- 36.Roth E. Nonnutritive effects of glutamine. J. Nutr. 2008;138(10):2025s–2031s. doi: 10.1093/jn/138.10.2025S. [DOI] [PubMed] [Google Scholar]
- 37.Young VR, Ajami AM. Glutamate: An amino acid of particular distinction. J. Nutr. 2000;130(4S Suppl):892s–900s. doi: 10.1093/jn/130.4.892S. [DOI] [PubMed] [Google Scholar]
- 38.Xi P, Jiang Z, Zheng C, Lin Y, Wu G. Regulation of protein metabolism by glutamine: Implications for nutrition and health. Front. Biosci. (Landmark Ed). 2011;16:578–597. doi: 10.2741/3707. [DOI] [PubMed] [Google Scholar]
- 39.DeBerardinis RJ, Cheng T. Q's next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruzat V, Macedo Rogero M, Noel Keane K, Curi R, Newsholme P. Glutamine: Metabolism and immune function, supplementation and clinical translation. Nutrients. 2018 doi: 10.3390/nu10111564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todorova V, Vanderpool D, Blossom S, et al. Oral glutamine protects against cyclophosphamide-induced cardiotoxicity in experimental rats through increase of cardiac glutathione. Nutrition. 2009;25(7–8):812–817. doi: 10.1016/j.nut.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Boelens PG, van Leeuwen PA, Dejong CH, Deutz NE. Intestinal renal metabolism of L-citrulline and L-arginine following enteral or parenteral infusion of L-alanyl-L-[2,15N]glutamine or L-[2,15N]glutamine in mice. Am. J. Physiol. Gastrointestinal Liver Physio. 2005;289(4):G679–G685. doi: 10.1152/ajpgi.00026.2005. [DOI] [PubMed] [Google Scholar]
- 43.Greenfield JR, Farooqi IS, Keogh JM, et al. Oral glutamine increases circulating glucagon-like peptide 1, glucagon, and insulin concentrations in lean, obese, and type 2 diabetic subjects. Am. J. Clin. Nutr. 2009;89(1):106–113. doi: 10.3945/ajcn.2008.26362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stumvoll M, Perriello G, Nurjhan N, et al. Glutamine and alanine metabolism in NIDDM. Diabetes. 1996;45(7):863–868. doi: 10.2337/diab.45.7.863. [DOI] [PubMed] [Google Scholar]
- 46.Yang SJ, Kwak S-Y, Jo G, Song T-J, Shin M-J. Serum metabolite profile associated with incident type 2 diabetes in Koreans: findings from the Korean Genome and Epidemiology Study. Sci. Rep. 2018;8(1):8207. doi: 10.1038/s41598-018-26320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Floegel A, Stefan N, Yu Z, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stančáková A, Civelek M, Saleem NK, et al. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes. 2012;61(7):1895–1902. doi: 10.2337/db11-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tillin T, Hughes AD, Wang Q, et al. Diabetes risk and amino acid profiles: Cross-sectional and prospective analyses of ethnicity, amino acids and diabetes in a South Asian and European cohort from the SABRE (Southall And Brent REvisited) Study. Diabetologia. 2015;58(5):968–979. doi: 10.1007/s00125-015-3517-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Würtz P, Soininen P, Kangas AJ, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36(3):648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagatsu T. The catecholamine system in health and disease—Relation to tyrosine 3-monooxygenase and other catecholamine-synthesizing enzymes. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2007;82(10):388–415. doi: 10.2183/pjab.82.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernstrom JD, Fernstrom MH. Tyrosine, phenylalanine, and catecholamine synthesis and function in the brain. J. Nutr. 2007;137(6):1539S–1547S. doi: 10.1093/jn/137.6.1539S. [DOI] [PubMed] [Google Scholar]
- 53.Muiesan ML, Agabiti-Rosei C, Paini A, Salvetti M. Uric acid and cardiovascular disease: An update. Eur. Cardiol. 2016;11(1):54–59. doi: 10.15420/ecr.2016:4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi YJ, Yoon Y, Lee KY, et al. Uric acid induces endothelial dysfunction by vascular insulin resistance associated with the impairment of nitric oxide synthesis. FASEB J. 2014;28(7):3197–3204. doi: 10.1096/fj.13-247148. [DOI] [PubMed] [Google Scholar]
- 55.Chang C-C, Wu C-H, Liu L-K, et al. Association between serum uric acid and cardiovascular risk in nonhypertensive and nondiabetic individuals: The Taiwan I-Lan Longitudinal Aging Study. Sci. Rep. 2018;8(1):5234–5234. doi: 10.1038/s41598-018-22997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masuo K, Kawaguchi H, Mikami H, Ogihara T, Tuck ML. Serum uric acid and plasma norepinephrine concentrations predict subsequent weight gain and blood pressure elevation. Hypertension. 2003;42(4):474–480. doi: 10.1161/01.HYP.0000091371.53502.D3. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Zhu C, Nambi V, et al. Metabolomic pattern predicts incident coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2019;39(7):1475–1482. doi: 10.1161/ATVBAHA.118.312236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaarhorst AA, Verhoeven A, Weller CM, et al. A metabolomic profile is associated with the risk of incident coronary heart disease. Am. Heart J. 2014;168(1):45–52.e7. doi: 10.1016/j.ahj.2014.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available due to sensitive nature of personalized healthcare data but are available from the corresponding author on reasonable request.