Abstract
Campylobacter jejuni is one of the leading causes of bacterial diarrhea throughout the world. We previously found that PEB1 is a homolog of cluster 3 binding proteins of bacterial ABC transporters and that a C. jejuni adhesin, cell-binding factor 1 (CBF1), if not identical to, contains PEB1. A single protein migrating at approximately 27 to 28 kDa was recognized by anti-CBF1 and anti-PEB1. To determine the role that the operon encoding PEB1 plays in C. jejuni adherence, peb1A, the gene encoding PEB1, was disrupted in strain 81-176 by insertion of a kanamycin resistance gene through homologous recombination. Inactivation of this operon completely abolished expression of CBF1, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. In comparison to the wild-type strain, the mutant strain showed 50- to 100-fold less adherence to and 15-fold less invasion of epithelial cells in culture. Mouse challenge studies showed that the rate and duration of intestinal colonization by the mutant were significantly lower and shorter than with the wild-type strain. In summary, PEB1 is identical to a previously identified cell-binding factor, CBF1, in C. jejuni, and the peb1A locus plays an important role in epithelial cell interactions and in intestinal colonization in a mouse model.
Campylobacter jejuni is a curved, gram-negative bacterium that is the most commonly recognized cause of bacterial diarrhea in the United States and a common cause throughout the world (6, 7, 10, 37). Although the pathogenesis of Campylobacter infections is poorly understood, several significant advances recently have been made (3, 26, 32). Adherence of C. jejuni isolates to HeLa cells has been characterized and found to be associated with severity of illness. Isolates from patients with fever and bloody diarrhea are more adherent than those from patients with only diarrhea or asymptomatic infections (19). Although flagella were originally reported to be putative adhesins in C. jejuni (28), later work indicated that inactivation of flagellin genes has no effect on C. jejuni adherence to epithelial cells (22, 41). Flagella per se are not an adhesin but may be essential to the preadherence process by providing motility for the bacteria to approach the cells.
Fauchere et al. incubated glycine-extracted material from a C. jejuni strain with HeLa cells and found that at least two C. jejuni-specific bands were retained on the cells after washing (18). The major band (cell-binding factor 1 [CBF1]) has a molecular mass of ∼27 kDa, and the minor one (CBF2) is ∼29 kDa. CBF1, isolated by cutting the band from preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blocks adherence of C. jejuni to HeLa cells, but CBF2 does not (25). Antibody raised against CBF1 abolishes the adherence.
The purification of four proteins from glycine-extracted material of C. jejuni 81-176 for development of a subunit vaccine against C. jejuni infections has been described elsewhere (34). These four proteins have molecular masses of 28, 29, 30, and 31 kDa and are named PEB1, PEB2, PEB3, and PEB4, respectively. PEB1 and PEB3 are common antigens recognized by convalescent-phase sera from nearly 80% of C. jejuni-infected patients (34). By using enzyme-linked immunosorbent assay (ELISA) and Western blotting, CBF1 was found to have many characteristics identical to those of PEB1. PEB4 is at least part of, if not identical to, CBF2 (25). The predicted product of the CBF2 gene (now called peb4A) has extensive homology to gram-positive extracytoplasmic lipoproteins involved in processing exported proteins (8). Cloning and analysis of the gene encoding PEB1 (peb1A) suggests that peb1A is located within an operon homologous to those for ABC transporters in other bacteria (33).
Although physiological functions of ABC transport systems have been well identified and characterized (38), their roles in bacterial pathogenesis have not been a focus of investigation. The hypothesis of this study is that PEB1 is identical to CBF1 and that the peb1A locus enhances C. jejuni adherence to and invasion of epithelial cells and intestinal colonization in an animal model. By introduction of a mutation into peb1A, we sought to test this hypothesis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. jejuni 81-176, isolated from an outbreak of Campylobacter diarrhea and widely used in pathogenesis studies (12, 32, 34), was grown at 37°C in a microaerobic environment on brucella agar plates supplemented with 5% sheep blood as described previously (34). Escherichia coli DH5α, used for amplification of the recombinant plasmid pPB119, was grown at 37°C in LB medium supplemented with carbenicillin (50 μg/ml). Campylobacter selective medium contains polymyxin B, trimethoprim, cephalothin, amphotericin B, and vancomycin, as previously described (5).
Plasmid constructs.
Plasmid pPB119 was derived from pUC19 by insertion at the EcoRI site of a 2.6-kb chromosomal fragment from strain 81-176 (Fig. 1) as previously described (33). The 2.6-kb insert includes an opening reading frame (ORF) encoding a putative membrane receptor for PEB1 based on sequence homology, peb1A, and a partial ORF (33). A 1.4-kb kanamycin resistance gene (aphA) originally derived from C. coli (27) was digested with SmaI, creating blunt ends. pPB119 was disrupted at an NheI site within peb1A, and blunt ends were created by nucleotide filling using the Klenow fragment of DNA polymerase. After ligation of aphA into the NheI site of pPB119 by using T4 DNA ligase, transformants of E. coli DH5α cells with recombinant plasmid pPB119-km were selected on LB agar containing kanamycin (50 μg/ml) (Fig. 1).
FIG. 1.
Suicide vector and construct used for disruption of the native peb1A in C. jejuni. Restriction sites are represented by E (EcoRI), H (HindIII), and N (NheI). Three complete ORFs, B, C, and D, and two partial ORFs, A and E, are indicated below the insert. A SmaI-digested kanamycin resistance gene (aphA) from C. coli was inserted at the NheI site to disrupt peb1A. The directions of transcription for aphA and peb1A are shown by arrows. The 702-bp probe was generated by PCR amplification of the sequence encoding the mature PEB1. The disruption site is 134 bp 3′ from the start of the 702-bp probe.
Inactivation of peb1A in C. jejuni through allelic exchange.
DNA of pPB119-km was prepared by an alkaline lysis procedure, and supercoiled DNA was isolated on a cesium chloride-ethidium bromide gradient following ultracentrifugation as previously described (36). The protocol of Ferrero et al. for electrotransformation of Helicobacter pylori (20) was followed, with several modifications. In brief, a 24-h culture of C. jejuni 81-176 (consisting of 109 to 1010 CFU) was harvested from a blood agar plate and resuspended in 25 ml of electroporation buffer (15% glycerol–9% sucrose). The bacterial cells were pelleted by centrifugation at 3,500 × g for 10 min, then washed four more times in 1.5 ml of the same buffer, and finally resuspended with 50 μl of the buffer. One microliter (100 μg) of pPB119-km DNA was mixed with 50 μl of C. jejuni cells. The mixture was transferred immediately to a prechilled 0.1-cm electroporation cuvette. Pulses (Gene Pulse apparatus; Bio-Rad) were achieved with 25 F, 1.8 kV, and 200 Ω, giving a time constant ranging from 4 to 5 ms. After electric shock, 1 ml of brucella broth was immediately added to the cells, and 0.5 ml of the suspension was spread on brucella agar plates supplemented with 5% sheep blood. After incubation at 37°C for 24 h, bacteria were harvested, inoculated onto blood agar plates supplemented with kanamycin (20 mg/ml), and incubated at 37°C under microaerobic conditions for 72 h to select kanamycin-resistant transformants (27). Colony hybridization of kanamycin-resistant transformants by using two probes, aphA and pUC18 (the vector in which pPB119 is located), was performed as described previously (36).
Southern hybridization.
Chromosomal DNA was prepared from the wild-type strain and a kanamycin-resistant transformant (81-176P−) by the method of Meade et al. (29). Chromosomal DNA was digested with restriction endonucleases under conditions recommended by the manufacturer (Promega) and separated in 0.7% agarose. km and peb1A probes were prepared as described above. Prehybridization and hybridization were performed in 50% formamide at 42°C as described previously (36).
SDS-PAGE and immunoblotting.
Whole cells and glycine extracts of wild-type and mutant strains were prepared as previously described (34). SDS-PAGE was performed in a modified Laemmli gel system as described by Ames (1). Proteins were resolved by using the modified silver stain of Oakley et al. (31). Immunoblotting was performed by the method of Towbin et al. (40), with modifications (34). For two-dimensional gel electrophoresis, the Mini-Protean II 2-D system (Bio-Rad) and buffer and solution recipes from the Investigator 2-D (Millipore) system were used. First-dimension gels consisted of 9.95 M urea, 2.0% (vol/vol) Nonidet P-40, 4% acrylamide, 6% ampholytes (pH 3 to 10 [Millipore]), and 0.05% (vol/vol) 10% ammonium persulfate and were polymerized in capillary gel tubes for approximately 1 h. Gels then were prefocused sequentially at 200 V for 10 min, 300 V for 15 min, and 400 V for 15 min with 0.1 M NaCl as the cathode buffer and 0.01 M phosphoric acid as the anode buffer. Whole-cell lysates (10 μg) of C. jejuni 81-176 were solubilized in sample overlay buffer (0.5 M urea, 0.2% [vol/vol] Nonidet P-40, 0.1% [vol/vol] ampholytes, 5.0 mM dithiothreitol, 0.7 M β-mercaptoethanol) and loaded into each tube gel. Isoelectric focusing was performed at 500 V for 10 min followed by 750 V for 3.5 h. The gel was extracted from the tube and loaded atop an acrylamide gel for separation in the second dimension. The second-dimensional gel contained 4% acrylamide in the stacking gel and 15% acrylamide in the separating gel. Immunoblotting was performed as described above with 1:2,500-diluted rabbit anti-PEB1 or 1:10,000-diluted rabbit anti-CBF1 as primary antibodies.
Adherence of C. jejuni to HeLa cells.
The effect of mutagenesis of peb1A on C. jejuni adherence was tested in a system previously used to identify CBF1 (18, 19). In brief, 24-h cultures of the wild-type and 81-176P− mutant strains were harvested from plates, washed once, and resuspended in Eagle’s minimum essential medium (MEM). HeLa cells were cultured for 48 h in a humidified atmosphere containing 5% CO2 in 24-well plates in MEM supplemented with 10% fetal bovine serum, streptomycin (50 μg/ml), penicillin (200 U/ml), and amphotericin B (2.5 μg/ml). For the adherence assay, the cell monolayers were washed three times with serum- and antibiotic-free MEM, and 1 ml of bacterial suspension was added to each well. After incubation at 37°C in 5% CO2 for 1 h, each well was washed five times with 1 ml of MEM, and then cells were lysed in 1 ml of ice cold water for 1 h. The bacteria released from HeLa cells were quantified by viable cell culture on blood agar plates.
Invasion of INT407 cells.
C. jejuni was grown in biphasic Mueller-Hinton (M-H) medium at 37°C as described previously (32). To ensure that invasion differences did not reflect changes in motility, the cells were stabbed in 0.4% M-H motility agar, and after incubation for 24 h at 37°C, the leading edge of growth at the periphery of the motility zone was picked with a sterile needle for use as the inoculum for invasion assays. After overnight growth to mid-log phase (optical density at 600 nm of 0.6) in M-H biphasic flasks, 50 μl of the culture supernatant was used as an inoculum for each invasion assay, representing a starting multiplicity of infection of approximately 20 to 40. INT407 cells (human embryonic intestine 407 cells; American Type Culture Collection, Rockville, Md.) were cultured in MEM with 10% fetal calf serum and 2 mM l-glutamine under 5% CO2 in 75-cm2 tissue culture flasks maintained at 37°C as described previously (15, 32). INT407 monolayers were trypsinized, washed, and split 1:4 into fresh culture medium, reaching confluence within 3 days, at which point they were used in invasion assays. For invasion assays, 6 × 105 of the split INT407 cells in 1 ml of culture medium were grown in each well of 24-well tissue culture plates (Sarstedt, Inc.). Invasion assays were performed essentially as described previously (32) except that the bacterial inoculum was not centrifuged to initiate contact with epithelial cells. Mid-log-phase bacteria in 50 μl of medium were added to each monolayer for a 2-h invasion period, washed twice in MEM, and then incubated for 2 h in medium containing gentamicin (100 μg/ml) to kill extracellular bacteria. After treatment with 0.1% Triton X-100 for 10 min and serial dilution in phosphate-buffered saline (pH 7.4), released intracellular bacteria were enumerated by plate count on M-H agar. Bacterial invasion efficiency was calculated as (number of internalized bacteria at the end of the assay/starting inoculum) × 100. All assays were conducted in triplicate and repeated independently six times.
C. jejuni colonization in mice.
BALB/c mice, 6 to 8 weeks old, were purchased from Jackson Laboratory (Bar Harbor, Maine) and housed in laminar-flow cages for a minimum of 8 days before being used for experiments. The experiments reported herein were conducted according to the principles set forth in reference 24a. Frozen stocks of 81-176 and 81-176P− were grown for 18 h on Trypticase soy blood agar (TSBA; Remel, Lenexa, Kans.) and then inoculated into a biphasic culture of brain heart infusion (BHI) supplemented with 1% yeast extract (BHI-YE) and incubated for 18 to 20 h at 42°C in an atmosphere of 10% CO2, 5% O2, and 85% N2. The broth phase of the culture was used to initiate infection in mice (2, 3). Initially, the number of Campylobacter cells in the suspension was estimated spectrophotometrically, and then the challenge dose was more exactly determined by plating serial dilutions of the inoculum on TSBA.
Inoculations.
The procedure for oral feeding of mice has been reported in detail elsewhere (2, 4). In brief, gastric acidity was neutralized by orally feeding 0.5 ml of sodium bicarbonate solution followed by 0.5 ml of BHI-YE containing the desired number of CFU of C. jejuni. Control mice received 0.5 ml of BHI-YE alone. The procedure for intranasal inoculation and illness index calculations after challenge also have been described previously (3). After light anesthesia, 30 μl of BHI-YE alone (control) or containing selected bacteria was delivered to the external nares. Following challenge, mice were observed for 6 consecutive days and assigned a daily numerical score (0 = apparently healthy; 1 = ill, lethargic, and with ruffled fur; 2 = dead). The means of these daily indices are presented as the illness indices of various experimental immunization-challenge groups.
Fecal excretion.
Fecal excretion of C. jejuni was monitored by culturing fecal homogenate (approximately 5% suspension in phosphate-buffered saline, using four to five fresh pellets) on TSAB supplemented with cefoperazone, vancomycin, and amphotericin B (Remel). After 48 h of incubation as described above, selected Campylobacter colonies were confirmed by morphology and oxidase reactions. A mouse was considered negative if no C. jejuni colonies were detected in fecal homogenates on 3 consecutive days.
RESULTS
Characterization of kanamycin-resistant transformants.
After transformation of 109 to 1010 CFU of strain 81-176 with 100 μg of DNA of pPB119-km, ∼1,000 colonies grew on kanamycin-selective agar plates, yielding a transformation rate of approximately 10−6 to 10−7. Two types of colonies were observed: small round colonies ∼1.0 mm in diameter and large flat colonies 0.5 to 1.0 cm in diameter. The small colonies were composed of nonmotile organisms, while the large ones consisted of motile cells, a variation reflecting the phenomenon of flagellar phase shift (9). Although flagella are not directly involved in C. jejuni adherence (22, 41), they are essential for this process by providing motility for bacteria to approach cells and are essential for colonization in vivo (9). Therefore, the large colonies consisting of motile organisms were chosen for further characterization. All 19 kanamycin-resistant strains tested were found by colony blotting to contain aphA but not vector sequence, indicating that the insertion of aphA was mediated by double crossovers in these strains. Southern hybridizations using HindIII or BamHI-PstI digestion indicated that in the mutant strain, aphA had been correctly inserted into peb1A in the bacterial chromosome (data not shown). To test whether the mutant strains continued to express PEB1 protein, protein profiles of whole cells from the wild-type and mutant strains were analyzed by SDS-PAGE and silver staining and found to be indistinguishable. However, by immunoblotting using antibody to PEB1, as expected, PEB1 antigen was present in the parental strain but absent in the mutant, which we now called 81-176P− (data not shown).
Relatedness of PEB1 to CBF1.
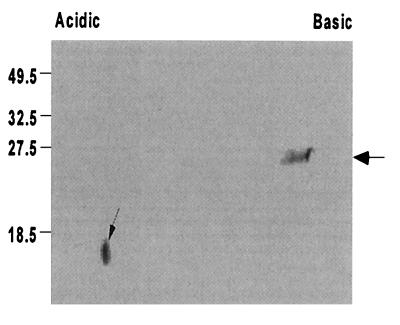
With the construction of the peb1A mutant that does not express PEB1, it now was possible to further clarify the relationship of PEB1 to CBF1. As expected, in immunoblotting with whole bacterial cells as antigens, rabbit antibody to CBF1 recognized a single band at 28 kDa in the wild-type strain but none in 81-176P− (data not shown). To confirm that CBF1 is encoded by peb1A, E. coli XL-1 blue was transformed with pPB203 harboring peb1A (33). In immunoblotting of cells of the parental strain XL-1 blue, normal rabbit serum, anti-PEB1, or anti-CBF1 did not recognize any antigens. In contrast, anti-CBF1 recognized a single band at ∼28 kDa in the transformant that was identical to the recognition by anti-PEB1; as expected, there was no recognition by normal rabbit serum (data not shown). In two-dimensional immunoblotting of whole C. jejuni cell lysates, anti-CBF1 recognized a 28-kDa band with a pI of approximately 8 to 9 and a lower-molecular-mass spot of less than 18 kDa in a more acidic region of approximately pH 5 (Fig. 2). Anti-PEB1 yielded a pattern nearly identical to that yielded by anti-CBF1 except that the recognition of the lower-molecular-mass spot was much weaker (data not shown).
FIG. 2.
Relatedness of PEB1 and CBF1 as detected by two-dimensional immunoblotting. Whole-cell antigens of wild-type C. jejuni 81-176 were separated first by isoelectric focusing (pH 3 to 10) and then by molecular weight in 15% acrylamide. After being blotted to nitrocellulose paper, C. jejuni proteins were immunostained with rabbit antibody to CBF1 and a secondary alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G. Anti-CBF1 recognized a band of approximately the same molecular weight as PEB1 (large arrow) and a lower-molecular-weight spot with a pI of approximately 5 (small arrow); results for immunoblotting with anti-PEB1 were nearly identical (not shown). Positions of molecular mass markers in kilodaltons are shown at the left.
Adherence to HeLa cells.
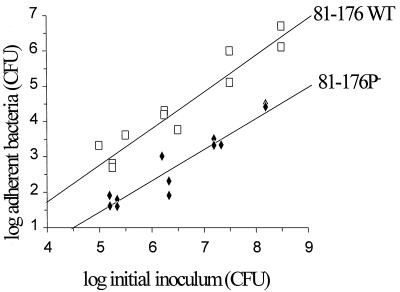
To test the effect of inactivation of the peb1A locus on adherence of C. jejuni to eukaryotic cells, cells of the wild-type and the mutant strains from 24-h cultures on blood agar plates were incubated with HeLa cells. In independent experiments, adherence of the mutant (81-176P−), although not completely abolished, was consistently ∼50- to 100-fold lower than for the wild-type strain across a broad range of bacterial inocula (Fig. 3).
FIG. 3.
Effect of mutation of the peb1A locus on adherence of C. jejuni to HeLa cells. Cells of the wild-type (81-176 WT) or the peb1A mutant (81-176P−) strain were incubated with HeLa cell monolayers for 1 h. Nonadherent bacteria were removed by washing, and adherent bacteria were released from HeLa cells after lysis of the cells and counted by viable cell culture. The relationship between log bacterial inoculum and log bacterial adherence was determined by linear regression based on data of two independent experiments. Mutant strain 81-176P− was 50- to 100-fold lower in adherence than the wild-type strain.
Invasion of INT407 cells.
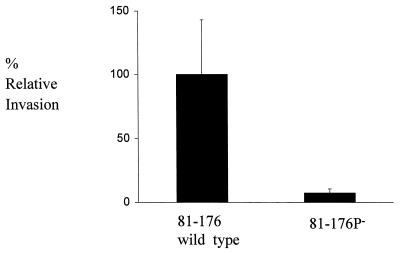
In previous studies, C. jejuni 81-176 has been shown to be able to invade intestinal epithelial cells (19, 26, 32). To test the effect of the insertion in peb1A, we compared the wild-type and mutant strains for the ability to invade INT407 cells. The wild-type strain showed an invasion efficiency similar to that reported previously (2), but the 81-176P− mutant was approximately 15-fold less invasive (Fig. 4). These results are consistent with the lowered adherence of the mutant strain and indicate that mutation in the peb1A locus affects the interaction of C. jejuni with epithelial cells.
FIG. 4.
Effect of mutation of the peb1A locus on C. jejuni invasion of INT407 cells. Results are expressed as percent relative invasion, with the parent strain set at 100%. Levels of invasion (mean ± standard deviation) were 0.345 ± 0.151 for wild-type strain 81-176 and 0.0255 ± 0.019 for 81-176P− (peb1A mutant), averaged over six independent assays.
Intestinal colonization of the mutant strain in mice and protection studies.
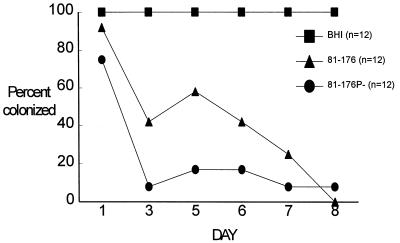
After oral challenge, the wild-type strain colonized all 16 BALB/c mice studied for at least 8 days. At day 38, 12 (75%) of the 16 mice remained colonized. Similarly, the mutant (81-176P−) was recovered from all 16 mice challenged on day 1. However, at day 8, only 3 (19%) of 16 mice were colonized, and at day 38, none were colonized (Fig. 5).
FIG. 5.
Effect of mutation of the peb1A locus on intestinal colonization of mice. Separate groups of 16 mice each were immunized orally with 2.3 × 109 CFU of mutant strain 81-176P−. At each indicated interval, fresh fecal pellets were collected from individual mice and cultured for the presence of Campylobacter. By day 2 after infection, the mutant strain showed a marked reduction in colonization frequency.
Oral immunization by infection with sublethal doses of either the wild-type or mutant strain protected mice from intestinal colonization following intranasal challenge compared to immunization with culture medium (BHI) alone (Fig. 6). By 8 days after challenge, all broth-immunized mice remained colonized, whereas, 0 or 1 of 12 mice in the 81-176 or 81-176P− group, respectively, was excreting C. jejuni in the feces. Comparative mean illness indices (mean ± standard deviation) for the control group and the groups immunized with 81-176 and 81-176P− were 0.92 ± 0.13, 0.42 ± 0.39, and 0.33 ± 0.43, respectively. Compared to those of controls, illness indices were significantly lower (BHI versus 81-176, P = 0.025; BHI versus 81-176P−, P = 0.018; 81-176 versus 81-176P−, P = 0.73) in mice immunized with 81-176P− or 81-176.
FIG. 6.
Effect of inactivation of the peb1a locus on protection of mice after intranasal challenge with virulent C. jejuni. Separate groups of 12 mice each were immunized orally with BHI-YE or with 5 × 107 CFU of wild-type strain 81-176 or mutant strain 81-176P−. Thirty days after immunization, all mice were assayed for fecal excretion of C. jejuni and found to be negative. At 32 days after immunization, mice were challenged intranasally with 4.3 × 109 CFU of wild-type strain 81-176 and observed for the development of colonization.
DISCUSSION
Relatedness of CBF1 to PEB1.
We previously found by ELISA and one-dimensional immunoblotting that CBF1 is antigenically related and potentially identical to PEB1 (25). We now provide evidence demonstrating that CBF1 is identical to PEB1, since an E. coli strain transformed with pPB203 harboring peb1A produced a molecule recognized by anti-CBF1, and inactivation of peb1A by the aphA insertion abolished expression of CBF1 by C. jejuni. Both experiments indicate that CBF1 is encoded by the PEB1 operon, but whether CBF1 is encoded by peb1A cannot be deduced from these experiments due to the possible polar effects caused by the insertion. The nonsense (translation termination) mutation introduced by aphA not only blocks translation of the remainder of peb1A (ORF D) but also can decrease the transcription of downstream genes, such as ORF E. Insertion of aphA into peb1A (ORF D) could block expression of ORF E and any other unidentified downstream genes, and the possibility that CBF1 contains other proteins of 27 to 28 kDa encoded by the downstream genes in addition to PEB1 cannot be ruled out. In other ABC transport systems, members of the same system are encoded by a single operon, which usually contains three to five genes (38). It is highly unlikely that any of the products of these downstream genes have both the same molecular weight and the same isoelectric point as does PEB1. Therefore, we tentatively conclude that CBF1 contains only one component, PEB1, which is encoded by peb1A.
That both anti-CBF1 and anti-PEB1 also recognized a low-molecular-mass (18-kDa) antigen with an acidic pI was unexpected. Recognition of this antigen by anti-PEB1 may be explained if even a small amount of this antigen was present in the fast protein liquid chromatography fraction used to immunize rabbits for production of antiserum to PEB1. Since the CBF1 used to immunize the rabbits was purified by cutting a 27- to 28-kDa CBF1 protein band, there also could have been minor contamination with an 18-kDa antigen (25). However, since neither anti-CBF1 nor anti-PEB1 reacts with this antigen in one-dimensional immunoblots (25, 34), it also is possible that the 18-kDa antigen recognized in two-dimensional immunoblotting was due to degradation of PEB1 in the electrophoresis buffer system.
That the band recognized by anti-CBF1 completely disappeared in the peb1A mutant indicates that CBF1, a band of ∼28 kDa cut from preparative SDS-PAGE, is a homogeneous molecule. Although there are at least 14 proteins migrating at 28 to 31 kDa in C. jejuni, most are present only in whole bacterial cells and not in glycine-extracted material which is enriched for PEB1 (13, 14, 34). In glycine extracts from which CBF1 is isolated, PEB1 is the major protein, migrating at ∼28 kDa (34). Coisolation of proteins other than PEB1 as part of CBF1 may occur but has not been detectable by the methods used thus far.
Role of PEB1 in C. jejuni interactions with epithelial cells.
Since previous studies have demonstrated that the isolated CBF1 (PEB1) is adherent to HeLa cells and adherence of C. jejuni can be blocked either by isolated CBF1 or by anti-CBF1 (11, 25), and in the present study CBF1 was found to be a homogeneous molecule identical to PEB1, it is not surprising that inactivation of the peb1A locus significantly reduced C. jejuni adherence to HeLa cells. However, it is novel that PEB1, a putative binding component of an ABC transporter (33), may play a role in C. jejuni adherence to epithelial cells. This possibility cannot be ruled out unless genes downstream of the aphA insertion site are demonstrated to encode an adhesin.
Investigations of bacterial ABC transport systems have focused mainly on their physiological roles in nutrient transport or chemotaxis (38). Although PEB1 has not been directly shown to be a member of the ABC transport system, indirect evidence suggests that PEB1 is the binding component since it is ∼24 to 30% identical to the binding components of well-characterized ABC transport systems, such as GlnH, HisJ, and LAO (23, 24, 30, 42), all belonging to cluster 3 of the binding protein superfamily (38). Two signature sequences and eight of nine position-specific amino acid residues unique for cluster 3 are found in PEB1 (33). In the partial PEB1 operon, ORF C is more than 50% identical to the membrane receptors of the binding components of ABC transport systems in other bacterial species, such as GlnQ and HisP (33).
In gram-negative bacteria, the binding components of ABC transport systems are freely located in the periplasmic space, a location not permitting them to directly participate in interactions with epithelial cells. In contrast, in gram-positive bacteria, due to the absence of an outer membrane, the binding component is a surface-exposed lipoprotein and is linked to the bacterial cell wall through a fatty acid chain (21). Although C. jejuni is a gram-negative bacterium, its tinctorial characteristics are atypical since it is not counterstained with safranin. Although homologous to the binding components of ABC transport systems in gram-negative bacteria (33), PEB1 also exhibits characteristics of a binding component of gram-positive ABC transport systems; it is more closely related to GlnH of gram-positive Bacillus stearothermophilus (∼37% identity) than to GlnH of gram-negative E. coli (30.5% identity) (33). It contains a typical lipoprotein signal peptidase cleavage site at the amino terminus (33) and is surface exposed on C. jejuni (25). Similarly, amino acid sequences of the tetracycline resistance determinant from C. jejuni, TetO, and the chloramphenicol resistance determinant, chloramphenicol acetyltransferase, are significantly more homologous to respective gram-positive tetracycline resistance proteins and to chloramphenicol acetyltransferase than to those of gram-negative organisms (39). The surface location of PEB1 distinguishes it from binding components of other gram-negative ABC transport systems and is consistent with the present observations suggesting that PEB1 plays a direct role in C. jejuni adherence.
That inactivation of the peb1A locus significantly reduced but did not completely abolish C. jejuni adherence suggests that PEB1 is an important but not the only adhesin in C. jejuni. The presence of multiple adhesins is a common finding in other pathogenic bacteria (16, 17, 35). Similarly, C. jejuni may use several other components in addition to PEB1 in adherence to epithelial cells, such as lipopolysaccharide, pili, and antigens at ∼29 to 32, 36, and 42 kDa, as suggested previously (11, 12, 18, 28).
Adherent bacteria most likely have improved chances for subsequent triggering of effective invasion-specific ligand-receptor interactions. That inactivation of the peb1A locus also has a substantial effect on C. jejuni invasion of INT407 intestinal epithelial cells may particularly reflect the importance of a primary adherence event in the process of invasion. It is possible that the invasion-specific ligand-receptor complex is different from the adherence ligand-receptor complex, or alternatively, PEB1 may serve as one of several invasion-specific ligands, and its absence would be expected to reduce but not eliminate invasion ability.
Mouse studies.
Bacterial colonization of the gut is in part a function of the ability of the bacteria to adhere to intestinal epithelial cells and to survive in that environment. In the present study, inactivation of the peb1A locus significantly reduced the rate and duration of mouse intestinal colonization compared with the wild-type strain. Although the possibility that this reduction is due to a decrease in the mutant’s fitness after inactivation of a gene putatively related to nutrient transport cannot be ruled out, this is unlikely since growth rates of the wild-type and mutant strains in nutrient media or synthetic basal media were not significantly different (unpublished data). The marked reduction of its in vitro adherence to and invasion of HeLa cells and of its in vivo colonization consistently indicate that PEB1/CBF1 plays an important role, probably a direct role in bacterium-epithelial cell surface interactions. Previous findings that purified PEB1/CBF1 adheres to HeLa cells and that adherence of C. jejuni can be inhibited by anti-PEB1 and anti-CBF1 (25) also support this conclusion. Finally, the evidence that mutant strain 81-176P− provides protection from disease as well as intestinal colonization in mice suggests that it could have a future role as a live attenuated vaccine candidate.
ACKNOWLEDGMENTS
This work was supported in part by an interagency agreement between the Department of Defense and the Department of Veterans Affairs, by the Medicine Research Service of the Department of Veterans Affairs, and by the Naval Medical Research and Development Command work unit no. 63002AM00101.810.HOX1294.
REFERENCES
- 1.Ames G F L. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. J Biol Chem. 1974;249:634–644. [PubMed] [Google Scholar]
- 2.Baqar S, Applebee L A, Bourgeois A L. Immunogenicity and protective efficacy of a prototype Campylobacter killed whole-cell vaccine in mice. Infect Immun. 1995;63:3731–3735. doi: 10.1128/iai.63.9.3731-3735.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baqar S, Bourgeois A L, Applebee L A, Mourad A S, Kleinosky M T, Mohran A, Murphy J R. Murine intranasal challenge model for the study of Campylobacter pathogenesis and immunity. Infect Immun. 1996;64:4933–4939. doi: 10.1128/iai.64.12.4933-4939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baqar S, Pacheco N D, Rollwagen F M. Modulation of mucosal immunity against Campylobacter jejuni by orally administered cytokines. Antimicrob Agents Chemother. 1993;37:2688–2692. doi: 10.1128/aac.37.12.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser M J, Berkowitz I D, LaForce F M, Cravens J, Reller L B, Wang W-L L. Campylobacter enteritis: clinical and epidemiological features. Ann Intern Med. 1979;91:179–185. doi: 10.7326/0003-4819-91-2-179. [DOI] [PubMed] [Google Scholar]
- 6.Blaser M J, Black R E, Duncan D J, Amer J. Campylobacter jejuni-specific serum antibodies are elevated in healthy Bangladeshi children. J Clin Microbiol. 1985;21:164–167. doi: 10.1128/jcm.21.2.164-167.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser M J, Wells J G, Feldman R A, Pollard R A, Allen J G. Campylobacter enteritis in the United States. A multicenter study. Ann Intern Med. 1983;98:360–365. doi: 10.7326/0003-4819-98-3-360. [DOI] [PubMed] [Google Scholar]
- 8.Burucoa C, Fremauc C, Pei Z, Tummuru M, Blaser M J, Centiempo Y, Fauchere J L. Nucleotide sequence and characterization of peb4A encoding an antigenic protein in Campylobacter jejuni. Res Microbiol. 1995;146:467–476. doi: 10.1016/0923-2508(96)80292-0. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell M B, Guerry P, Lee E C, Burans J P, Walker R I. Reversible expression of flagella in Campylobacter jejuni. Infect Immun. 1985;50:941–943. doi: 10.1128/iai.50.3.941-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury M N H. Campylobacter jejuni enteritis: a review. Trop Geogr Med. 1984;36:215–222. [PubMed] [Google Scholar]
- 11.de Melo M A, Pechere J-C. Identification of Campylobacter jejuni surface proteins that bind to eukaryotic cells in vitro. Infect Immun. 1990;58:1749–1756. doi: 10.1128/iai.58.6.1749-1756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doig P, Yao R, Burr D H, Guerry P, Trust T J. An environmentally regulated pilus-like appendage involved in Campylobacter pathogenesis. Mol Microbiol. 1996;20:885–894. doi: 10.1111/j.1365-2958.1996.tb02526.x. [DOI] [PubMed] [Google Scholar]
- 13.Dubreuil J D, Kostrzynska M, Logan S M, Harris L A, Austin J W, Trust T J. Purification, characterization, and localization of a protein antigen shared by thermophilic campylobacters. J Clin Microbiol. 1990;28:1321–1328. doi: 10.1128/jcm.28.6.1321-1328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn B E, Blaser M J, Snyder E L. Two-dimensional gel electrophoresis and immunoblotting of Campylobacter outer membrane proteins. Infect Immun. 1987;55:1564–1572. doi: 10.21236/ada265461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsinghorst E A, Baron L S, Kopecko D J. Penetration of human intestinal epithelial cells by Salmonella: molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:5173–5177. doi: 10.1073/pnas.86.13.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans D G, Evans D J. New surface-associated heat-labile colonization factor antigen (CFA/II) produced by enterotoxigenic Escherichia coli for serogroups O6 and O8. Infect Immun. 1978;21:638–647. doi: 10.1128/iai.21.2.638-647.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans D G, Silver R P, Evans D J, Chase D G, Gorbach S L. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fauchere J-L, Kervella M, Rosenau A, Mohanna K, Veron M. Adhesion to HeLa cells of Campylobacter jejuni and C. coli outer membrane components. Res Microbiol. 1989;140:379–392. doi: 10.1016/0923-2508(89)90014-4. [DOI] [PubMed] [Google Scholar]
- 19.Fauchere J L, Rosenau A, Veron M, Moyen E N, Richard S, Pfister A. Association with HeLa cells of Campylobacter jejuni and C. coli from human feces. Infect Immun. 1986;54:283–287. doi: 10.1128/iai.54.2.283-287.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilson E, Alloing G, Schmidt T, Claverys J-P, Dudler R, Hofnung M. Evidence for high affinity binding protein dependent transport systems in Gram-positive bacteria and in mycoplasma. EMBO J. 1988;7:3997–4004. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant C C R, Konkel M E, Cieplak J W, Tompkins L S. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins C F, Ames G F L. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequence. Proc Natl Acad Sci USA. 1981;78:6038–6042. doi: 10.1073/pnas.78.10.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogg R W. The amino acid sequence of the histidine binding protein of Salmonella typhimurium. J Biol Chem. 1981;256:1935–1939. [PubMed] [Google Scholar]
- 24a.Institute of Laboratory Animal Resources. Guide for the care and use of laboratory animals. Washington, D.C: Institute of Laboratory Animal Resources, National Research Council, National Academy Press; 1996. [Google Scholar]
- 25.Kervella M, Pages J-M, Pei Z, Grollier G, Blaser M, Fauchere J-L. Isolation and characterization of two Campylobacter glycine-extracted proteins that bind to HeLa cell membranes. Infect Immun. 1993;61:3440–3448. doi: 10.1128/iai.61.8.3440-3448.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konkel M, Cieplak W. Altered synthetic response of Campylobacter jejuni to cocultivation with human epithelial cells is associated with enhanced internalization. Infect Immun. 1992;92:4945–4949. doi: 10.1128/iai.60.11.4945-4949.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McSweegan E, Walker R I. Identification and characterization of two Campylobacter adhesins for cellular and mucous substrates. Infect Immun. 1986;53:141–148. doi: 10.1128/iai.53.1.141-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meade H M, Long S R, Ruvkun C B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nohno T, Saito T, Hong J S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ) Mol Gen Genet. 1986;205:260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
- 31.Oakley B R, Kirsch D R, Morris N R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- 32.Oelschlaeger T A, Guerry P, Kopecko D J. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc Natl Acad Sci USA. 1993;90:6884–6888. doi: 10.1073/pnas.90.14.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei Z, Blaser M J. PEB1, the major cell-binding factor of Campylobacter jejuni, is a homolog of the binding component in Gram-negative nutrient transport systems. J Biol Chem. 1993;268:18717–18725. [PubMed] [Google Scholar]
- 34.Pei Z, Ellison I R T, Blaser M J. Identification, purification, and characterization of major antigenic proteins of Campylobacter jejuni. J Biol Chem. 1991;266:16363–16368. [PubMed] [Google Scholar]
- 35.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Skirrow M B. Campylobacter enteritis: a “new” disease. Br Med J. 1977;2:9–11. doi: 10.1136/bmj.2.6078.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam R, Saier M H. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor D E. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli to tetracycline, chloramphenicol, and erythromycin. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C: American Society for Microbiology; 1992. pp. 74–86. [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon T. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar T M, Bleumink-Pluym N M C, van den Zeijst B A M. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 1991;10:2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Welker N E. Cloning and characterization of a glutamine transport operon of Bacillus stearothermophilus NUB36: effect of temperature on regulation of transcription. J Bacteriol. 1992;173:4877–4888. doi: 10.1128/jb.173.15.4877-4888.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]