Abstract
Background
A current argument in the CAM biology literature has focused on the nature of the CAM evolutionary trajectory: whether there is a smooth continuum of phenotypes between plants with C3 and CAM photosynthesis or whether there are discrete steps of phenotypic evolutionary change such as has been modelled for the evolution of C4 photosynthesis. A further implication is that a smooth continuum would increase the evolvability of CAM, whereas discrete changes would make the evolutionary transition from C3 to CAM more difficult.
Scope
In this essay, I attempt to reconcile these two viewpoints, because I think in many ways this is a false dichotomy that is constraining progress in understanding how both CAM and C4 evolved. In reality, the phenotypic space connecting C3 species and strong CAM/C4 species is both a continuum of variably expressed quantitative traits and yet also contains certain combinations of traits that we are able to identify as discrete, recognizable phenotypes. In this sense, the evolutionary mechanics of CAM origination are no different from those of C4 photosynthesis, nor from the evolution of any other complex trait assemblage.
Conclusions
To make progress, we must embrace the concept of discrete phenotypic phases of CAM evolution, because their delineation will force us to articulate what aspects of phenotypic variation we think are significant. There are some current phenotypic gaps that are limiting our ability to build a complete CAM evolutionary model: the first is how a rudimentary CAM biochemical cycle becomes established, and the second is how the ‘accessory’ CAM cycle in C3+CAM plants is recruited into a primary metabolism. The connections to the C3 phenotype we are looking for are potentially found in the behaviour of C3 plants when undergoing physiological stress – behaviour that, strangely enough, remains essentially unexplored in this context.
Keywords: Phenotype, character evolution, discrete, continuous, heuristic
INTRODUCTION
Crassulacean acid metabolism (CAM) and C4 photosynthesis are complex adaptations that alter the primary metabolism of a plant, and yet each has evolved many dozens of times across the plant tree of life (Sage et al., 2011; Gilman et al., 2023). Their extreme convergence provides both inspiration and opportunity for researchers interested in the evolutionary origins of adaptive syndromes. Many decades of research on a core set of CAM- and C4-evolving clades have revealed species exhibiting a diverse range of anatomical and physiological variables that do not fall neatly into typical C3 or C4/CAM syndromes and are often presumed to represent potential phenotypic stages that are occupied during the evolutionary transition between these major photosynthetic types. Phylogenetic analyses largely support the interpretation of these phenotypes as ‘intermediary’ between C3 and C4/CAM endpoints, and although they are all currently represented by extant species, these phenotypes have been incorporated explicitly into evolutionary models as ancestral stages to reconstruct the evolutionary changes (and their potential order) that occur along the C3-to-CAM or C3-to-C4 evolutionary trajectory (Sage, 2004; Williams et al., 2013; Edwards, 2019).
Arguably, we have been more successful in this endeavour with regard to C4 evolution, in large part because there was early adoption of the use of well-resolved phylogenies to pinpoint C4 evolutionary origins and subsequent physiological characterization of closely related C3 or intermediate-like taxa. Early comparative work focused on Flaveria, which became the first real model developed for studying C4 evolution (Cheng et al., 1989; Huber et al., 1989; Ku et al., 1991; Monson and Jaeger, 1991; McKown et al., 2005). Many specific sets of potentially intermediate photosynthetic types were defined, which corresponded largely to the physiological diversity of different Flaveria species. Once delineated in Flaveria, it was a natural progression to look for and document similar ‘types’ in other C4-evolving clades (Hattersley et al., 1982; Marshall et al., 2007; Voznesenskaya et al., 2010; Christin et al., 2011; Muhaidat et al., 2011; Fisher et al., 2015; Lauterbach et al., 2019). The discovery of recognizable, shared types of physiological diversity surrounding other C4 origins was an extremely important and productive time in C4 biology research and facilitated the development of a model of the C3-to-C4 evolutionary trajectory that has largely stood the test of time (Peisker, 1986; Sage, 2004; Edwards, 2019; Stata et al., 2019). This model has focused extensively on the evolutionary ordering of the photosynthetic types first described in Flaveria and is often presented as if the evolution of C4 proceeds via discrete jumps through phenotypic space. It is important to note that the traits used to define these discrete states are all, of course, quantitative. They include anatomical and physiological parameters such as bundle sheath-to-mesophyll (BS:M) ratio, CO2 compensation point, quantum yield, relative proportions of organelles in BS vs. M cells, relative protein abundance and activity in BS vs. M cells, and the degree of suberization of bundle sheath cell walls. In two more recent modelling approaches, both Heckmann et al. (2013) and Mallman et al. (2014) modelled C4 evolution explicitly as a continuous progression but still identified and emphasized the emergence of discrete phenotypic states.
In contrast, the development of an evolutionary model for CAM has instead focused on the idea of a continuum of CAM expression, ranging from zero expression in a full C3 plant to daily, primary expression in a strong-CAM plant, with a continuous variation of expression connecting these two purported evolutionary ‘endpoints’ (Teeri, 1982; Silvera et al., 2010; Bräutigam et al., 2017). This is understandable given an important difference between C4 and CAM plants: although C4 plants can no longer perform C3 photosynthesis, CAM is an inherently flexible system, and even typically strong-CAM species perform C3 photosynthesis in the late afternoon, with >30 % of atmospheric CO2 fixation still occurring during the day (Winter and Holtum, 2002). Additionally, the variability of CAM behaviour within individuals based on their age and physiological state further emphasizes the continuous nature of CAM expression, in contrast to the intermediate C3–C4 ‘types’ modelled on Flaveria (Dodd et al., 2002). The problem with this model is that we do not see a smooth continuum of CAM expression in the real world; based on surveys of thousands of species for 13C/12C isotopic values, it seems clear that most plants fix carbon with primarily C3 photosynthesis, or primarily CAM; relatively few species exhibit isotopic evidence that they perform an even mix of C3 and CAM primary fixation (Winter and Holtum, 2002; Winter et al., 2015; Edwards, 2019; Gilman and Edwards, 2020; even most clades studied by Messerschmid et al., 2021), and the bimodal distribution of δ13C strongly suggests discrete photosynthetic phenotypes. Furthermore, certain CAM behaviours have repeatedly been recognized and described as ‘types’; these include categories such as ‘constitutive’ vs. ‘facultative’ CAM, ‘low-level CAM’, ‘weak CAM’, ‘strong CAM’, ‘C3+CAM’, ‘CAM-cycling’ and ‘CAM-idling’ (Winter et al., 2015; Edwards, 2019; Winter, 2019). However, there have been essentially no attempts to build an evolutionary model between C3 and strong CAM by ordering these types as a trajectory, and there does not appear to be much consensus regarding the evolutionary connections between these CAM ‘types’ (e.g. Edwards, 2019; Yang et al., 2019). Rather, the CAM community tends to prefer the word ‘continuum’ (Silvera et al., 2010; Bräutigam et al., 2017; Messerschmid et al., 2021; Schiller and Bräutigam, 2021). Interestingly, these important differences in language were already established in the earliest depictions of C4 and CAM evolutionary trajectories (Teeri, 1982; Peisker, 1986; Fig. 1).
Fig. 1.
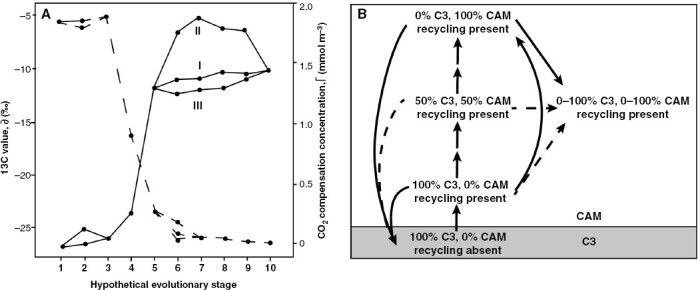
Earliest depictions of C4 and CAM evolutionary models, modified from Peisker (1986) and Teeri (1982). (A) Peisker’s model of C4 evolution. Peisker proposed nine steps to transition between C3 and C4 photosynthesis. Here, the lines represent how these nine steps influenced 13C/12C isotope ratios (continuous line) and CO2 compensation concentrations (dashed line), with the values at stage 1 being C3 values and the values at stage 10 being C4 values. Peisker modelled variation in the order in which the nine steps occurred, which would lead to different phenotypic trajectories (labelled II, I and III) that eventually evolved towards similar endpoints. Although he described these changes as discrete ‘phases’, all nine proposed changes refer to shifts in continually varying characters, such as the relative proportion of Rubisco in the bundle sheath or mesophyll, the capacity for phosphoenolpyruvate regeneration, and the increase in phosphoenolpyruvate carboxylase activity. (B) Teeri’s model of CAM evolution. Teeri proposed that the first step of CAM evolution is to evolve CAM-cycling, which is re-fixation of respired CO2 at night by the CAM cycle (labelled as ‘recycling present’ in the diagram). In the centre of the diagram, he depicts the ‘upregulation continuum’, evolving from 100 % C3 fixation to 100 % CAM fixation. The upregulation continuum has persisted as the primary CAM model in the literature, although in the text Teeri references discrete stages of CAM evolution, explaining that the double arrows in the middle indicate that there might be ‘multiple evolutionary steps’ between the phenotypes. He envisions another phenotype that is 100 % flexible between C3 and CAM pathways, modelled on Kalanchoe blossfeldiana, that he assumes must be an optimal phenotype in all conditions, because there are no evolutionary pathways away from it, only towards it. Teeri does envision reversals from the other main CAM types back to C3. The continuous arrows are hypothesized transitions supported with evidence from Crassulaceae; dashed arrows are hypothesized transitions that had not been documented at the time. Peisker and Teeri both explicitly modelled continuous characters but described the transitions as discrete evolutionary stages.
IS THE CHARACTERIZATION OF DISCRETE STATES JUSTIFIED, HELPFUL OR HARMFUL?
Biology is full of extremely useful categories and definitions that become a bit blurry when you look too closely; ‘species’ is an essential concept, yet most taxonomists would agree that individual species can be extremely difficult to delineate, and even the definition of a ‘species’ is highly debated (De Queiroz, 2007); ‘homology’ is relatively simple to understand conceptually, yet often challenging to identify in practice (Wagner, 2018). The messiness does not disqualify the concepts as exceptionally useful, however, and one could argue that the same can be said for discretely delineated phenotypes. Our description of colours such as ‘red’ and ‘blue’ belies the quantitative nature of hue, brightness and saturation, and yet these categories are essential communication tools. And there are, of course, naturally discrete phenotypic shifts, i.e. major organismal characteristics governed by single loci, such as pigmentation patterns (Hoekstra et al., 2006; Rogers et al., 2013). There is, perhaps, no better example of discrete phenotype shifting than the remarkable studies of RNA folding, which is a relatively simple phenotype of RNA molecules that is governed by biophysics and is predictable from the spatial position of single mutational changes (Stadler et al., 2001). Is the enviably simple genotype–phenotype map of RNA folding applicable to C4 and CAM evolution? Conceptually, yes, but realistically, we are still very far from this level of understanding. Both C4 and CAM are better described as adaptive syndromes, meaning that they comprise multiple shifts in different phenotypic attributes that are presumed to be genetically independent. And these elements themselves are, for the most part, quantitative characters, each likely to be influenced by multiple gene regions and the environment. The genetic architecture of C4 and CAM is still largely unknown, and reconstructing even one instantiation of a genotype–phenotype map is still out of reach.
Without much understanding of the genetic architecture of a given evolutionary trajectory, how might we best justify conceptualizing changes along the trajectory as continuous or discrete? Is the C4 model justified, or should we embrace a continuum of variation in both cases? It might seem obvious to look first at the distribution of trait values themselves and ask whether we can observe phenotypic gaps in traits that presumably have the possibility of varying continuously. Fig. 2 represents a conceptual diagram that illustrates how we create discrete phenotypic bins in the face of continuous variation of key components. In the case of a syndrome, such as C4 or CAM, there are many traits to consider, and some might have more discontinuous variation than others. In C4, for example, the CO2 compensation point appears to have significant gaps in the phenotypic possibilities between C3 and C4 values; the cluster of intermediate values represents the C2 phenotype and the instantiation of a weak carbon-concentrating mechanism (CCM) by the localization of glycine decarboxylase expression to BS cells (Fig. 2). This is similar to the 13C/12C distributions described earlier in CAM clades, which delineate real gaps in phenotypic space that can justify organizing species into discrete bins.
Fig. 2.
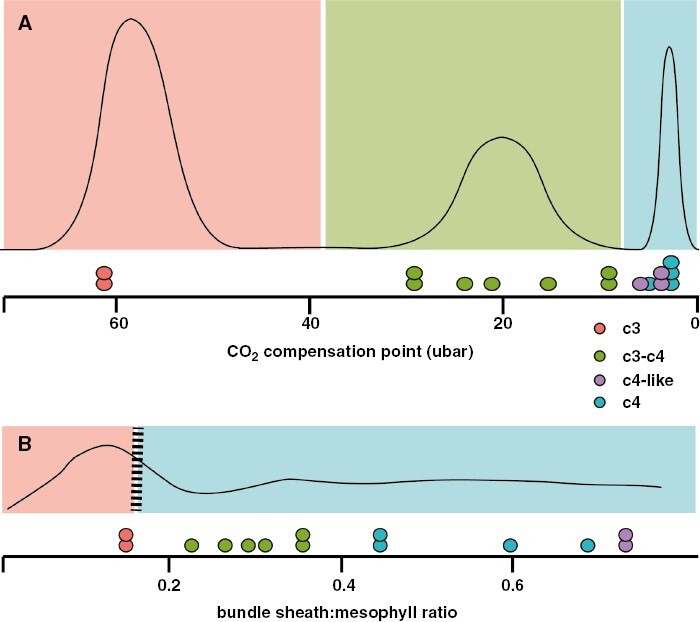
Discretizing quantitative characters to delineate stages of an evolutionary trajectory. (A) Hypothetical distribution of trait values for a character that is naturally multi-modal. In this case, it is straightforward to bin the standing variation into ‘low’, ‘medium’ and ‘high’ values. Dots below the hypothetical distribution curve are real data from Flaveria for the CO2 compensation point, demonstrating a clear phenotypic gap between C3 and C3–C4 species, less so between C3–C4 and C4. (B) Discretization can also be justified when the distribution is not multi-modal, perhaps with other analyses that support threshold values where a particular value of one trait influences the evolvability of another. In this case, the bundle sheath-to-mesophyll (BS:M) ratio of leaves might show a relatively flat distribution, but phylogenetic modelling studies demonstrate that a BS:M ratio of 0.15 is required for C4 evolution (although the BS:M ratio of many C4 plants is much higher than that number). Dots below the hypothetical distribution curve are real data from Flaveria for the BS:M ratio, demonstrating no clear phenotypic gaps, but the C3 species lie right at the value of the modelled threshold, with all other C3–C4 and C4 species on the other ‘C4 side’ of the threshold. We may use thresholds to discretize traits into ‘low’ and ‘high’ bins that are biologically relevant. Flaveria data are from Lyu et al. (2021).
However, many of the traits that vary along the C3-to-CAM/C3-to-C4 spectrum do not show such clumped distribution of values, but we still utilize them to create phenotypic boundaries. A more nuanced approach to binning continuously varying characters is the identification of threshold values, above or below which a new element of the syndrome can evolve. This requires us to think non-linearly about how different elements of the phenotype might be interacting and influencing one another. Again, we have learned of these dynamics in C4 evolution precisely because so much attention has been given to quantifying potentially relevant variables. The BS:M ratio is a classic example. This is a quantitative character and is influenced both by the vein density of leaf tissue and by the size of BS cells. A high BS:M ratio is crucial to efficient C4 function, because the Calvin–Benson–Bassham cycle occurs only in BS cells and therefore they must occupy a certain proportion of the leaf photosynthetic tissue. We often refer to this anatomical requirement as a discrete phenotype, e.g. plants with a ‘high BS:M’ vs. a ‘low BS:M’, but the BS:M ratio varies continuously across species. In a study of grasses, Christin et al. (2013) identified a threshold value of BS:M of 15 %, above which C4 evolved repeatedly and below which C4 appeared to be evolutionarily inaccessible. The existence of thresholds in a quantitative character can essentially create two distinct phenotypic spaces that we might then interpret as discrete evolutionary ‘steps’ in a trajectory. In this case, it is logical to assign BS:M ratios of >15 % as ‘high’ and those <15 % as ‘low’, despite the lack of a natural break in BS:M ratio distributions, because this threshold places the organism in a new qualitative state where new evolutionary trajectories are now possible (Fig. 2).
To summarize, when looked at individually, the various characters that are measured to delineate the ‘stepwise’ evolution of C4 photosynthesis are, for the most part, continuously varying, and it might be more rational to think of the C3-to-C4 trajectory as a continuum. Yet the creation of discrete phenotypes has great heuristic value, in that it is an essential communication tool and allows us to discuss roughly the evolutionary phases of trait origination, even if the discrete states are not entirely real (Wimsatt, 1987). If binning variation is nothing more than a mental tool that humans invoke to make sense of the world, we should take advantage of it, because it is helpful with orientation and framing of a problem. And spending our energy on improving our bins can lead us to discover the very real existence of thresholds in quantitative characters, which is a phenomenon I would argue is central to both C4 and CAM evolution and probably a host of other major evolutionary transitions.
WHY ARE OUR CURRENT CAM CATEGORICAL BINS NOT INFORMING US ABOUT CAM EVOLUTION?
As mentioned earlier, CAM photosynthesis has, at the outset, as many discrete types currently described as the C4 trajectory contains, yet this has not appeared to influence the way in which researchers describe the dynamics of CAM evolution. This is very curious and might stem, in part, from the explicit identity of an ‘evolutionary origin’ (in both C4 and CAM communities) as the emergence of the basic biochemical pathway of the photosynthetic system, rather than all of the other organismal attributes that the biochemistry requires in order to function optimally. In the case of C4, many of these essential structural changes had to occur before even a weak C4 cycle could become established, which meant that these elements were noticed, studied and incorporated into the evolutionary model. In the case of CAM, a weak CAM cycle can be expressed without much additional organismal change or, at least, none that has been identified definitively. All previously described CAM phenotypes essentially refer to different variations of weakly expressed CAM, whereby CAM plays a secondary role in plant carbon fixation (lumped into a single ‘C3+CAM’ category by Edwards, 2019). The C3+CAM plants tend to be fleshy, but the anatomical characteristics of their photosynthetic tissue are woefully understudied, nor do we know much about possible variation in their stomatal behaviour, carbohydrate regulation or any other phenotypic character that might be relevant to CAM expression. Given that the CAM biochemical cycle is apparently expressed at a low level in the context of a relatively unspecialized phenotype, the rhetoric describing CAM evolution quickly becomes one of ‘upregulation’, i.e. a simple continuum between weak and strong expression of an established biochemical pathway. This bias is growing stronger as genomics is quickly becoming the preferred approach to study CAM evolution (Ming et al., 2015; Yang et al., 2017; Heyduk et al., 2018; Schiller and Bräutigam, 2021). There are certainly many important expression and regulatory changes that occur during the emergence of a CAM biochemistry, in addition to the transition to CAM as a primary metabolism, but what are we missing by focusing solely on the biochemistry? What other organismal changes might be evolving that would enable the upregulation of CAM? The correlation of strong CAM and succulence has long been acknowledged, but there are scant few studies that quantify anatomical differences associated with succulence in relationship to CAM expression across species (Griffiths et al., 2008; Nelson and Sage, 2008; Barrera Zambrano et al., 2014; Males, 2018; Luján et al., 2021; Leverett et al., 2023), and especially neglected is the anatomical spectrum occupied by C3 and C3+CAM phenotypes. Are there ‘hidden’ yet crucially important intermediate types within C3+CAM that we have not yet identified, because we have not yet done the focused phylogenetic comparative biology that the C4 community has? Ironically, it might be that we need to spend far more time quantifying relevant phenotypic characters that are potentially associated with CAM expression before we can conceive of qualitatively delineated intermediate stages along the CAM evolutionary trajectory.
WHAT WILL BE THE FLAVERIA OF THE CAM WORLD?
Some might argue that the characterization of variation in Flaveria has had an oversized influence on our view of C4 evolution or even that it has positively misled us (e.g. Kadereit et al., 2017). Not surprisingly, I would argue the opposite, because the establishment of Flaveria as the original ‘model clade’ of C4 evolution motivated a period of exceptional discovery, during which researchers built up general predictions from finding similar types of variation in other C4-evolving clades from across angiosperms (Hattersley et al., 1982; Marshall et al., 2007; Voznesenskaya et al., 2010; Christin et al., 2011; Muhaidat et al., 2011; Fisher et al., 2015; Lauterbach et al., 2019).
Does the CAM community have our own Flaveria yet? It seems that we don’t; instead, we have a set of clades that have each received some attention, but all of them lack the detailed understanding of physiology and anatomy in every species that makes the Flaveria story so complete and informative. Part of the problem is that CAM-evolving clades tend to be hyperdiverse, such as orchids, cacti, bromeliads, ice plants and relatives, and euphorbias, making complete species-level sampling for phylogenetics and physiological surveys extremely challenging. In some cases, efforts on phylogenetic vs. phenotypic sampling have been mismatched historically, but they are now catching up to each other (e.g. Clusia; Luján et al., 2023). Given that individual CAM research programmes have developed naturally around different clades, we are now potentially poised to develop multiple Flaverias simultaneously.
I am hopeful that the coming decade will harbour the emergence of multiple model clades in parallel, potentially spanning all vascular plants and different CAM life forms (e.g. leaf succulents, stem succulents, epiphytes and aquatic CAM). I use ‘model clade’ in the aspirational sense of Donoghue and Edwards (2019), with multiple research groups using these systems to understand all dimensions of organismal biology and evolution, including biogeography, reproductive biology, plant–animal interactions, development and life history. At first glance, these subjects do not appear to relate directly to CAM evolution, but when integrated into a whole-organism perspective, they might reveal important new factors that we have not yet considered. The challenges will be in delineating the ‘right-sized’ sub-clades in these systems that allow for complete taxonomic sampling and also contain pure C3 species (that are not evolutionary reversals) and a variety of CAM phenotypes. The requirement for a closely related C3 species might be the most difficult element in some cases; are there pure C3 species in all of Crassulaceae? In all of Portulacineae? It is certainly true that some clades will be more appropriate for investigating certain evolutionary transitions than others. The Portulacineae, for example, harbour multiple transitions from C3+CAM to strong CAM, in addition to a diversity of C3+CAM phenotypes (Hancock et al., 2019), but it might not be especially helpful in understanding how a nascent CAM cycle becomes established. For this fundamental question, we might need to develop clades that we currently think include only a handful of C3+CAM species, nested within an otherwise C3 lineage; clades such as Pelargonium (Jones et al., 2003) and Pilea (Winter et al., 2021) come to mind.
DEVELOPING A CAM EVOLUTIONARY MODEL: IS THERE A PHENOTYPIC GAP BETWEEN C3 AND C3+CAM PLANTS?
A recent conversation in the literature illustrates a renewed interest in generating a CAM evolutionary model and was the primary inspiration for this essay. Bräutigam and colleagues (Bräutigam et al., 2017; Schiller and Bräutigam, 2021) have argued that, unlike C4 photosynthesis, there is a clear ‘continuum’ between C3 and CAM plants, because some C3 plants have been shown to store malate and citrate in their vacuoles overnight and to use these stores for amino acid synthesis during the day. They cite 13C labelling studies that demonstrate a time lag in the incorporation of 13C-labelled CO2 into malate, citrate and derived amino acids, suggesting that these molecules are not generated during the day directly from photosynthate, but are instead generated and accumulated during the night. They argue further that this, essentially, is a CAM biochemical module and that all plants express it routinely. This fits their definition of an evolutionary continuum, in that no metabolic rewiring would be required to initiate a CAM metabolism, because it exists already and simply has to be upregulated.
In response, Winter and Smith (2022) strongly assert the opposite, i.e. that C3 plants typically accumulate malate and citrate in their vacuoles over the course of the day, not the night, and that this accumulation is a key element of balancing cytosolic pH and charge during daytime nitrogen assimilation. They argue that the results of the labelling study might be anomalous owing to the growing conditions of the plants, and regardless, the fluxes are minimal in comparison to the more standard and opposite fluxes associated with the nitrogen metabolism of a plant. They conclude by asserting that there is no simple continuum between C3 and CAM plants, but that CAM behaviour is complex and distinct from what typical C3 plants do and, indeed, requires significant metabolic rewiring. They provide new data for a good sampling of C3 plants to demonstrate their lack of nocturnal acid accumulation, supporting the argument that most C3 plants do not store detectable quantities of malic acid at night.
So, which is it? Both positions are perhaps exaggerated on purpose, and the truth is likely to lie in the middle. In the same sense that phenotypic continua and recognizably discrete phenotypes are compatible concepts, it seems worth stating (the obvious) that the expression of elements of CAM in some C3 plants provides the only logical path to the evolution of CAM and, furthermore, that those elements will undoubtedly become altered during the evolution of CAM. The biochemical modules recruited for both C4 and CAM are common elements of the daily physiology of a C3 plant, itself a partial explanation of why they have evolved so many times (Heyduk et al., 2019a). No element of these modules had to be derived de novo; instead, a new coordination and degree of expression had to evolve. It seems straightforward to view this sort of trajectory as a continuum such that, once a new configuration of reactions becomes expressed predictably, a qualitatively distinct new phenotype emerges.
LOOKING FOR NASCENT CAM IN DROUGHT-STRESSED C3 PLANTS
If typical C3 plants behave according to Winter and Smith (2022), i.e. they accumulate malate as malic acid in their vacuoles throughout the day, rather than the night, where should we look for elements of an inverted behaviour (a nascent CAM cycle) that Bräutigam was rightfully highlighting as a crucial element in understanding CAM evolution? Winter and Smith (2022) drop a hint when they dismiss the 13C labelling study in part because the plants might have been suffering from light limitation. The implication is that in an altered physiological state, typical fluxes and the timing and strength of biochemical reactions will differ, either predictably or not, and this means it is not a good example of what a ‘normal’ C3 plant would do. But an interesting question is to ask explicitly, what do ‘not-normal’ C3 plants do? A key area that has hardly been explored regarding C4 and CAM evolution is how these biochemical modules are potentially altered in C3 plants as they are exposed to particular stressors that we suspect drive the evolution of C4 and CAM (but see Gonzalez et al., 2003; Doubnerová and Ryšlavá, 2011).
In a recent series of papers, Wagner and colleagues (Erkenbrack et al., 2018; Wagner et al., 2019; Love and Wagner, 2022) present a new model for the origin of evolutionary novelties, which they call ‘stress-induced evolutionary innovation (SIEI)’. Their work focuses on the origin of the decidual stromal cell (DSC), a specialized cell type of eutherian mammals that plays a key role in implantation and placental development during pregnancy. Evidence suggests that DSCs are derived from endometrial stromal fibroblast (ESF) cells, which are more widespread amongst amniotes. When they exposed non-eutherian ESF cells to the hormones that initiate DSC development, they discovered the expression of a shared gene regulatory network between ESF cells and DSCs, but in the ESF cells their expression initiated a general cell stress response, rather than the differentiation of a new cell type. They conclude that DSCs evolved by co-opting a gene regulatory network that originally responded to cellular oxidative stress. SIEI might be considered a specialized case of phenotypic plasticity and eventual genetic assimilation as a mechanism of organismal evolution (Waddington, 1953; West-Eberhard, 2003); their particular case also demonstrates how pre-existing regulatory networks might be recruited wholesale to control expression of an entirely new module of genes. I would argue that both phenomena are relevant to the emergence of a nascent CAM cycle from a C3 ancestor.
How might we invoke SIEI in CAM evolution? It seems that we need to study carefully the stress biology of both closely related and distantly related C3 species, especially to understand what they do at night. Nocturnal physiology of both C3 and C4 plants is woefully understudied relative to CAM plants, perhaps understandably. A recent study of C3, C3+CAM and strong-CAM Yucca species revealed that CAM-like patterns of gene expression exist in C3Yucca, even if they do not result in metabolic fluxes large enough to be detectable with our standard protocols (Heyduk et al., 2019b). Is C3Yucca a typical C3 species, or is this a distinct phenotype that we might consider ‘CAM enabled’? What gene regulatory networks are upregulated at night under stress in C3 plants, and are any of these associated with regulating CAM expression in C3+CAM or strong-CAM species? What conditions might facilitate nocturnal malate accumulation as a byproduct of the physiological stress response of a plant, and what other organismal attributes might contribute to this malate accumulation being beneficial?
As our knowledge of the phylogenetic distribution of C3+CAM phenotypes grows (Gilman et al., 2023), it appears that even a weak CAM cycle seems to be found consistently in plants with at least slightly fleshy photosynthetic tissue. We must be careful to consider the possibility that our decisions about which species to test for CAM are highly biased, in that we examine CAM activity only in plants that are fleshy, thus creating a self-fulfilling (and potentially false!) prophesy. We need more reports of negative results (examples of phenotypes where a CAM cycle is not found), such as the recent and important table in the paper by Winter and Smith (2022). That said, it is tempting to hypothesize that mildly succulent C3 species present a particular nighttime stress response environment that differs from other C3 plants and might elevate any benefits to what might initiate as a relatively stochastic and unregulated nocturnal malate accumulation. Perhaps the low cuticular and mesophyll conductance typical of mildly succulent plants works to increase internally respired CO2 concentrations, providing more substrate for phosphoenolpyruvate carboxylase to interact with. Perhaps the generally higher cellular water potentials of mildly succulent plants facilitate greater accumulation of malate without negatively impacting the osmotic environment. These are the types of questions that I hope we might all start to address in a diversity of lineages, as our emergent ‘CAM clades’ grow to maturity in the coming years.
A STRESS-FREE ALTERNATIVE
The ubiquity of stress-induced facultative-CAM species and their phylogenetic proximity to strong-CAM species provides an initial motivation to invoke SIEI as a possible mechanism for CAM evolution. SIEI predicts a particular evolutionary order of events (Fig. 3), in which a facultative-CAM phenotype evolves first, and CAM is later recruited into the primary metabolism of the plant. This order certainly appears to be true for the evolution of strong CAM in cacti, with drought-induced facultative CAM being present in all the close relatives of core cacti, including the leafy cactus Pereskia (Edwards and Donoghue, 2006; Brilhaus et al., 2016; Gilman et al., 2022; Moreno-Villena et al., 2022).
Fig. 3.
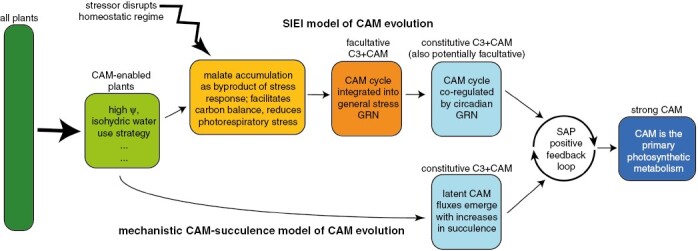
Model of CAM evolution showing two potential trajectories from C3 to strong CAM. Understanding which characteristics of C3 plants facilitate the establishment of a nascent CAM cycle (‘CAM-enabled’ box) remains an open research question; likely candidates include species that maintain high tissue water potentials and operate with a conservative water-use strategy. The upper trajectory follows the SIEI model, hypothesizing that stress induction results in variation in malate accumulation, which is selected for and eventually genetically assimilated, with a full CAM cycle becoming regulated by stress-induced gene regulatory networks (GRNs). Eventually, a CAM cycle also becomes constitutively expressed, potentially via co-regulation with circadian GRNs and/or increases in succulence (see lower trajectory). The lower trajectory follows a ‘mechanistic CAM–succulence model’, whereby all plants have a latent CAM cycle such that increased fluxes may be selected for when expressed in a facilitating anatomical context. Both trajectories result in a plant with a constitutive but weakly expressed CAM cycle with moderate succulence. The evolutionary transition to strong CAM is driven by a ‘synergistic anatomical pleiotropy’, whereby further anatomical changes towards increased succulence positively influence both CAM and water-storage functions, driving convergent evolution of strong-CAM succulent life forms across the tree of life (sensuEdwards, 2019). Abbreviations: Ψ, photosynthetic tissue water potential; GRN, gene regulatory network; SAP, synergistic anatomical pleiotropy.
An alternative model is inspired by Bräutigam’s arguments and consideration of a unique study that has, for the most part, hardly been considered by the CAM research community. Yuan et al. (2012) reported on a remarkable phenomenon in Camellia oleifera, the tea-oil camelia cultivated and grown extensively as a crop in China. A common pathogen of C. oleifera is the fungus Exobasidium vexans, and C. oleifera leaf tissue that is infected by E. vexans becomes visibly succulent. Yuan et al. (2012) monitored tissue acidity, stomatal conductance and RNA expression and protein abundances in succulent and non-succulent C. oleifera leaves, and all measured variables were consistent with the induction of a CAM cycle in the succulent tissue and absence of a CAM cycle in non-succulent tissue. If substantiated, this would provide the strongest evidence yet for a mechanistic link between a succulent anatomy and CAM expression. It also supports Bräutigam’s model that a ‘latent CAM cycle’ is present in all plants and simply needs to be upregulated or, as a slight addendum to that argument, simply needs to be expressed in the correct internal anatomical environment that can accommodate CAM metabolic fluxes. What is perhaps most remarkable about this phenomenon is that C. oleifera is nested well within the Ericales, a diverse clade that diverged from other angiosperms >100 Mya and, aside from this report, does not include a single other known CAM plant. Owing to this substantial evolutionary isolation from any other known CAM occurrence, if C. oleifera can express a CAM cycle when a fungal infection produces succulent tissue, we might as well assume for now that any other plant could do the same. A tight mechanistic CAM–succulence relationship, coupled with a latent CAM biochemistry present in all plants, suggests an alternative CAM evolutionary model to SIEI (Fig. 3), whereby a constitutive CAM cycle is established once a certain threshold of succulent anatomy is present. In this model, unlike the SIEI model, there is no need to invoke the origin of variation of the trait in question (e.g. a nascent CAM cycle in stressed C3 plants), because the assumption is that the CAM cycle is already essentially present and can be selected to be expressed more fully once it is operating in a permissive anatomical context.
CONCLUDING THOUGHTS
I present the CAM evolutionary models in Fig. 3, not necessarily because I think they are true, but because they are relatively reasonable hypotheses based on the comparative data that have been assembled to date and because they present ideas that can be tested, rejected, expanded and improved. The stepwise model of C4 evolution has been an exceptionally useful heuristic tool to conceptualize complex physiological transitions, and I have argued here that we need to build a similar model (or models) for CAM that embraces the concept of discrete phases of evolution. Somewhat ironically, this might require the accumulation of more quantitative datasets of other organismal attributes that we think could be important for CAM expression (rather than only the strength/timing of the CAM cycle itself). Given that the CAM community has focused research programmes on multiple, diverse CAM-evolving groups for the past decades, we are potentially poised to develop multiple model systems in parallel in the coming years, if we focus on finding the ‘right-sized’ sub-clades in these groups that facilitate complete taxon sampling and contain the full spectrum of C3 to strong CAM phenotypes.
ACKNOWLEDGEMENTS
The many works of Dr Klaus Winter have been a constant source of understanding and inspiration throughout my entire career, beginning with my introduction to CAM biology as a graduate student studying the evolution of Pereskia and continuing to this day. It is impossible to imagine where the CAM research community would be without his many foundational contributions, in addition to his leadership and care in maintaining a collaborative global community of CAM biologists. I thank all members of the Edwards laboratory, past and present, for many discussions over the years, all of which have influenced me in many ways. I’m also grateful for many insightful discussions about these issues with Andrea Bräutigam, Michael Donoghue, Ian Gilman, Howard Griffiths, Karolina Heyduk, Joe Holtum, Mark Olson, Rowan Sage, Andrew Smith, Gunter Wagner and Klaus Winter.
FUNDING
This work was supported, in part, by National Science Foundation grant NSF-IOS 1754662.
LITERATURE CITED
- Barrera Zambrano VA, Lawson T, Olmos E, Fernández-García N, Borland AM.. 2014. Leaf anatomical traits which accommodate the facultative engagement of crassulacean acid metabolism in tropical trees of the genus Clusia. Journal of Experimental Botany 65: 3513–3523. doi: 10.1093/jxb/eru022. [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Schlüter U, Eisenhut M, Gowik U.. 2017. On the evolutionary origin of CAM photosynthesis. Plant Physiology 174: 473–477. doi: 10.1104/pp.17.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhaus D, Bräutigam A, Mettler-Altman T, Winter K, Weber APM.. 2016. Reversible burst of transcriptional changes during induction of Crassulacean acid metabolism in Talinum triangulare. Plant Physiology 170: 102–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-H, Moore B, Wu J, Edwards GE, Ku MSB.. 1989. Photosynthetic plasticity in Flaveria brownii. Plant Physiology 89: 1129–1135. doi: 10.1104/pp.89.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P, Sage TL, Edwards EJ, Ogburn RM, Khoshravesh R, Sage RF.. 2011. Complex evolutionary transitions and the significance of C3–C4 intermediate forms of photosynthesis in Molluginaceae. Evolution; International Journal of Organic Evolution 65: 643–660. doi: 10.1111/j.1558-5646.2010.01168.x. [DOI] [PubMed] [Google Scholar]
- Christin PA, Osborne CP, Chatelet DS, et al. 2013. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proceedings of the National Academy of Sciences USA 110: 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Queiroz K. 2007. Species concepts and species delimitation. Systematic Biology 56: 879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Borland AM, Haslam RP, Griffiths H, Maxwell K.. 2002. Crassulacean acid metabolism: plastic, fantastic. Journal of Experimental Botany 53: 569–580. doi: 10.1093/jexbot/53.369.569. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, Edwards EJ.. 2019. Model clades are vital for comparative biology, and ascertainment bias is not a problem in practice: a response to Beaulieu and O’Meara. American Journal of Botany 106: 327–330. doi: 10.1002/ajb2.1255. [DOI] [PubMed] [Google Scholar]
- Doubnerová V, Ryšlavá H.. 2011. What can enzymes of C4 photosynthesis do for C3 plants under stress? Plant Science 180: 575–583. doi: 10.1016/j.plantsci.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Edwards EJ. 2019. Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytologist 223: 1742–1755. doi: 10.1111/nph.15851. [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Donoghue MJ.. 2006. Pereskia and the origin of the cactus life form. The American Naturalist 167: 777–793. doi: 10.1086/504605. [DOI] [PubMed] [Google Scholar]
- Erkenbrack EM, Maziarz JD, Griffith OW, et al. 2018. The mammalian decidual cell evolved from a cellular stress response. PLoS Biology 16: e2005594. doi: 10.1371/journal.pbio.2005594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AE, McDade LA, Kiel CA, et al. 2015. Evolutionary history of Blepharis (Acanthaceae) and the origin of C4 photosynthesis in section Acanthodium. International Journal of Plant Sciences 176: 770–790. doi: 10.1086/683011. [DOI] [Google Scholar]
- Gilman IS, Edwards EJ.. 2020. Crassulacean acid metabolism. Current Biology: CB 30: R57–R62. doi: 10.1016/j.cub.2019.11.073. [DOI] [PubMed] [Google Scholar]
- Gilman IS, Moreno-Villena JJ, Lewis ZR, Goolsby EW, Edwards EJ.. 2022. Gene co-expression reveals the modularity and integration of C4 and CAM in Portulaca. Plant Physiology 189: 735–753. doi: 10.1093/plphys/kiac116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman IS, Smith WAC, Holtum J, et al. 2023. The CAM lineages of planet Earth. Annals of Botany, this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MC, Sanchez R, Cejudo FJ.. 2003. Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 216: 985–992. doi: 10.1007/s00425-002-0951-x. [DOI] [PubMed] [Google Scholar]
- Griffiths H, Robe WE, Girnus J, Maxwell K.. 2008. Leaf succulence determines the interplay between carboxylase systems and light use during Crassulacean acid metabolism in Kalanchöe species. Journal of Experimental Botany 59: 1851–1861. doi: 10.1093/jxb/ern085. [DOI] [PubMed] [Google Scholar]
- Hancock LP, Holtum JAM, Edwards EJ.. 2019. The evolution of CAM photosynthesis in Australian Calandrinia reveals lability in C3+CAM phenotypes and a possible constraint to the evolution of strong CAM. Integrative and Comparative Biology 59: 517–534. doi: 10.1093/icb/icz089. [DOI] [PubMed] [Google Scholar]
- Hattersley PW, Watson L, Johnston CR.. 1982. Remarkable leaf anatomical variations in Neurachne and its allies (Poaceae) in relation to C3 and C4 photosynthesis. Botanical Journal of the Linnean Society 84: 265–272. doi: 10.1111/j.1095-8339.1982.tb00364.x. [DOI] [Google Scholar]
- Heckmann D, Schulze S, Denton A, et al. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153: 1579–1588. doi: 10.1016/j.cell.2013.04.058. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Ray JN, Ayyampalayam S, Leebens-Mack J.. 2018. Shifts in gene expression profiles are associated with weak and strong Crassulacean acid metabolism. American Journal of Botany 105: 587–601. doi: 10.1002/ajb2.1017. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Moreno-Villena JJ, Gilman IS, Christin P-A, Edwards EJ.. 2019a. The genetics of convergent evolution: insights from plant photosynthesis. Nature Reviews. Genetics 20: 485–493. doi: 10.1038/s41576-019-0107-5. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Ray JN, Ayyampalayam S, et al. 2019b. Shared expression of crassulacean acid metabolism (CAM) genes pre-dates the origin of CAM in the genus Yucca. Journal of Experimental Botany 70: 6597–6609. doi: 10.1093/jxb/erz105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP.. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313: 101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Huber WE, Brown RH, Bouton JH, Sternberg LO.. 1989. CO2 exchange, cytogenetics, and leaf anatomy of hybrids between photosynthetically distinct Flaveria species. Plant Physiology 89: 839–844. doi: 10.1104/pp.89.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CS, Cardon ZG, Czaja AD.. 2003. A phylogenetic view of low-level CAM in Pelargonium (Geraniaceae). American Journal of Botany 90: 135–142. doi: 10.3732/ajb.90.1.135. [DOI] [PubMed] [Google Scholar]
- Kadereit G, Bohley K, Lauterbach M, Tefarikis DT, Kadereit JW.. 2017. C3–C4 intermediates may be of hybrid origin – a reminder. The New Phytologist 215: 70–76. doi: 10.1111/nph.14567. [DOI] [PubMed] [Google Scholar]
- Ku MS, Wu J, Dai Z, Scott RA, Chu C, Edwards GE.. 1991. Photosynthetic and photorespiratory characteristics of Flaveria species. Plant Physiology 96: 518–528. doi: 10.1104/pp.96.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach M, Zimmer R, Alexa AC, et al. 2019. Variation in leaf anatomical traits relates to the evolution of C4 photosynthesis in Tribuloideae (Zygophyllaceae). Perspectives in Plant Ecology, Evolution and Systematics 39: 125463. doi: 10.1016/j.ppees.2019.125463. [DOI] [Google Scholar]
- Leverett A, Hartzell S, Winter K, et al. 2023. Dissecting succulence: Crassulacean acid metabolism and hydraulic capacitance are independent adaptations in Clusia leaves. Plant, Cell & Environment 46: 1472–1488. doi: 10.1111/pce.14539. [DOI] [PubMed] [Google Scholar]
- Love AC, Wagner GP.. 2022. Co-option of stress mechanisms in the origin of evolutionary novelties. Evolution; International Journal of Organic Evolution 76: 394–413. doi: 10.1111/evo.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luján M, Oleas NH, Winter K.. 2021. Evolutionary history of CAM photosynthesis in Neotropical Clusia: insights from genomics, anatomy, physiology and climate. Botanical Journal of the Linnean Society 199: 538–556. doi: 10.1093/botlinnean/boab075. [DOI] [Google Scholar]
- Luján M, Leverrett A, Winter K.. 2023. Forty years of research into crassulacean acid metabolism in the genus Clusia: anatomy, ecophysiology and evolution. Annals of Botany, this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu MA, Gowik U, Kelly S, et al. 2021. The coordination of major events in C4 photosynthesis evolution in the genus Flaveria. Scientific Reports 11: 15618. doi: 10.1038/s41598-021-93381-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Males J. 2018. Concerted anatomical change associated with crassulacean acid metabolism in the Bromeliaceae. Functional Plant Biology: FPB 45: 681–695. doi: 10.1071/FP17071. [DOI] [PubMed] [Google Scholar]
- Mallmann J, Heckmann D, Bräutigam A, et al. 2014. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 3: e02478. doi: 10.7554/eLife.02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall DM, Muhaidat R, Brown NJ, et al. 2007. Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesis. The Plant Journal 51: 886–896. doi: 10.1111/j.1365-313x.2007.03188.x. [DOI] [PubMed] [Google Scholar]
- McKown AD, Moncalvo J-M, Dengler NG.. 2005. Phylogeny of Flaveria (Asteraceae) and inference of C4 photosynthesis evolution. American Journal of Botany 92: 1911–1928. doi: 10.3732/ajb.92.11.1911. [DOI] [PubMed] [Google Scholar]
- Messerschmid TFE, Wehling J, Bobon N, et al. 2021. Carbon isotope composition of plant photosynthetic tissues reflects a Crassulacean Acid Metabolism (CAM) continuum in the majority of CAM lineages. Perspectives in Plant Ecology, Evolution and Systematics 51: 125619. [Google Scholar]
- Ming R, VanBuren R, Wai CM, et al. 2015. The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics 47: 1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson RK, Jaeger CH.. 1991. Photosynthetic characteristics of C3-C4 intermediate Flaveria floridana (Asteraceae) in natural habitats: evidence of advantages to C3-C4 photosynthesis at high leaf temperatures. American Journal of Botany 78: 795–800. doi: 10.1002/j.1537-2197.1991.tb14481.x. [DOI] [Google Scholar]
- Moreno-Villena JJ, Zhou H, Gilman IS, Tausta SL, Cheung CYM, Edwards EJ.. 2022. Spatial resolution of an integrated C4+CAM metabolism. Science Advances 8: eabn2349. doi: 10.1126/sciadv.abn2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhaidat R, Sage TL, Frohlich MW, Dengler NG, Sage RF.. 2011. Characterization of C3–C4 intermediate species in the genus Heliotropium L. (Boraginaceae): anatomy, ultrastructure and enzyme activity. Plant, Cell & Environment 34: 1723–1736. doi: 10.1111/j.1365-3040.2011.02367.x. [DOI] [PubMed] [Google Scholar]
- Nelson EA, Sage RF.. 2008. Functional constraints of CAM leaf anatomy: tight cell packing is associated with increased CAM function across a gradient of CAM expression. Journal of Experimental Botany 59: 1841–1850. doi: 10.1093/jxb/erm346. [DOI] [PubMed] [Google Scholar]
- Peisker M. 1986. Models of carbon metabolism in C3-C4 intermediate plants as applied to the evolution of C4 photosynthesis. Plant, Cell and Environment 9: 627–635. doi: 10.1111/j.1365-3040.1986.tb01620.x. [DOI] [Google Scholar]
- Rogers WA, Salomone JR, Tacy DJ, et al. 2013. Recurrent modification of a conserved cis-regulatory element underlies fruit fly pigmentation diversity. PLoS Genetics 9: e1003740. doi: 10.1371/journal.pgen.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis. The New Phytologist 161: 341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ.. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62: 3155–3169. doi: 10.1093/jxb/err048. [DOI] [PubMed] [Google Scholar]
- Schiller K, Bräutigam A.. 2021. Engineering of crassulacean acid metabolism. Annual Review of Plant Biology 72: 77–103. doi: 10.1146/annurev-arplant-071720-104814. [DOI] [PubMed] [Google Scholar]
- Silvera K, Neubig KM, Whitten WM, Williams NH, Winter K, Cushman JC.. 2010. Evolution along the crassulacean acid metabolism continuum. Functional Plant Biology 37: 995–1010. [Google Scholar]
- Stadler BM, Stadler PF, Wagner GP, Fontana W.. 2001. The topology of the possible: formal spaces underlying patterns of evolutionary change. Journal of Theoretical Biology 213: 241–274. doi: 10.1006/jtbi.2001.2423. [DOI] [PubMed] [Google Scholar]
- Stata M, Sage TL, Sage RF.. 2019. Mind the gap: the evolutionary engagement of the C4 metabolic cycle in support of net carbon assimilation. Current Opinion in Plant Biology 49: 27–34. doi: 10.1016/j.pbi.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Teeri J. 1982. Photosynthetic variation in the Crassulaceae. In: Ting IP, Gibbs M, eds. Crassulacean acid metabolism: proceedings of the Fifth Annual Symposium in Botany (January 14–16, 1982). Rockville, MD: American Society of Plant Physiologists, 244–259. [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Edwards GE, Ocampo G.. 2010. Revealing diversity in structural and biochemical forms of C4 photosynthesis and a C3–C4 intermediate in genus Portulaca L. (Portulacaceae). Journal of Experimental Botany 61: 3647–3662. doi: 10.1093/jxb/erq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 1953. Genetic assimilation of an acquired character. Evolution; International Journal of Organic Evolution 7: 118–126. doi: 10.2307/2405747. [DOI] [Google Scholar]
- Wagner GP. 2018. Homology, genes, and evolutionary innovation. Princeton, NJ: Princeton University Press. [Google Scholar]
- Wagner GP, Erkenbrack EM, Love AC.. 2019. Stress-induced evolutionary innovation: a mechanism for the origin of cell types. BioEssays 41: 1800188. doi: 10.1002/bies.201800188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. 2003. Developmental plasticity and evolution. New York, NY: Oxford University Press. [Google Scholar]
- Williams BP, Johnston IG, Covshoff S, Hibberd JM.. 2013. Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2: e00961. doi: 10.7554/eLife.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimsatt WC. 1987. False models as means to truer theories. In: Nitecki M, Hoffman A, eds. Neutral models in biology. London: Oxford University Press, 23–55. [Google Scholar]
- Winter K. 2019. Ecophysiology of constitutive and facultative CAM photosynthesis. Journal of Experimental Botany 70: 6495–6508. doi: 10.1093/jxb/erz002. [DOI] [PubMed] [Google Scholar]
- Winter K, Holtum JAM.. 2002. How closely do the δ13C values of Crassulacean acid metabolism plants reflect the proportion of CO2 fixed during day or night? Plant Physiology 129: 1843–1851. doi: 10.1104/pp.002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Smith JAC.. 2022. CAM photosynthesis: the acid test. New Phytologist 233: 599–609. doi: 10.1111/nph.17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Holtum JAM, Smith JAC.. 2015. Crassulacean acid metabolism: a continuous or discrete trait? The New Phytologist 208: 73–78. doi: 10.1111/nph.13446. [DOI] [PubMed] [Google Scholar]
- Winter K, Garcia M, Virgo A, Smith JAC.. 2021. Low-level CAM photosynthesis in a succulent-leaved member of the Urticaceae, Pilea peperomioides. Functional Plant Biology 48: 683–690. doi: 10.1071/FP20151. [DOI] [PubMed] [Google Scholar]
- Yang X, Hu R, Yin H, et al. 2017. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nature Communications 8: 1899. doi: 10.1038/s41467-017-01491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Liu D, Tschaplinski TJ, Tuskan GA.. 2019. Comparative genomics can provide new insights into the evolutionary mechanisms and gene function in CAM plants. Journal of Experimental Botany 70: 6539–6547. doi: 10.1093/jxb/erz408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Xu F, Wang S-D, et al. 2012. A single leaf of Camellia oleifera has two types of carbon assimilation pathway, C3 and crassulacean acid metabolism. Tree Physiology 32: 188–199. [DOI] [PubMed] [Google Scholar]


