Abstract
Curli are thin, coiled fibers expressed on the surface of Escherichia coli that bind several matrix and plasma proteins such as fibronectin, laminin, plasminogen, tissue plasminogen activator, and H-kininogen. In this work, we examined the interactions between curli-expressing E. coli and human major histocompatibility complex class I (MHC-I) and class II (MHC-II) molecules. Curliated E. coli was found to interact with an MHC-I-expressing lymphoma cell line as shown by scanning electron microscopy, whereas the binding to a mutant variant of this cell line expressing small amounts of MHC-I molecules was significantly lower. Moreover, curli-expressing E. coli bound purified radiolabeled MHC-I but not MHC-II molecules, whereas an isogenic curli-deficient mutant strain showed no affinity for either MHC-I or MHC-II. Purified insoluble curli could also bind 125I-labeled MHC-I molecules, and in Western blot experiments the 15-kDa curlin subunit protein bound intact MHC-I molecules as well as β2-microglobulin, the light chain of MHC-I molecules. A direct interaction between monomeric MHC-I molecules and a bacterial surface protein has previously not been reported. The binding of curli to MHC-I molecules, which are present on virtually all cells in higher vertebrates, will provide curliated E. coli with ample opportunities to interact with a great variety of hosts and host cells. This should facilitate the adaptation of E. coli to different ecological niches, and in human infections the interaction between curli and MHC-I molecules could contribute to adherence and colonization.
Some Escherichia coli strains belonging to different clinical types (enterotoxigenic, enterohemorrhagic, and sepsis isolates) express fibrous surface proteins called curli (3, 23). Similar surface organelles designated thin aggregative fimbriae are also found in Salmonella enteritidis (6–8). Curli fimbriae in E. coli consist of polymers of a single 15-kDa protein encoded by the curlin subunit gene csgA (23), which is highly homologous to the AgfA subunit in thin aggregative fimbriae (6, 10). For simplicity, curli fimbriae are referred to here as curli. The production of curli in E. coli requires expression of two csg operons (15), and the polymerization of the curlin subunit to insoluble curli is dependent on the presence of a specific nucleator protein encoded by the csgB gene (16). The csgA and csgB genes are cotranscribed (1), and they show 25% sequence identity at the protein level. A prominent and noteworthy property of curli polymers is their ability to specifically interact with numerous human proteins such as the matrix proteins fibronectin and laminin (23, 24) and proteins of the fibrinolytic and contact-phase systems (3, 26). This ability should facilitate the adaptation of curli-expressing bacteria to different niches in the infected host.
Major histocompatibility complex (MHC) class I (MHC-I) molecules are highly polymorphic transmembrane glycoproteins (for references, see reference 14). They function as receptors that present foreign peptides to cytolytic T cells, resulting in the destruction of the presenting cell, i.e., a virus-infected cell. Structurally, MHC-I molecules are composed of a 40-kDa heavy chain which has three extracellular globular domains, a short transmembrane domain, and a cytoplasmic domain (5). The 12-kDa light chain, β2-microglobulin (β2m), is noncovalently associated with the three extracellular globular domains of the heavy chain. β2m and these domains all exhibit the typical immunoglobulin (Ig) fold (28), and consequently MHC-I molecules belong to the Ig superfamily of proteins. Numerous Ig-binding bacterial surface proteins have been isolated and characterized (for references, see reference 18). However, none of these or any other defined microbial surface protein has been reported to interact directly with monomeric MHC-I molecules. As these Ig-related surface proteins are found at the surface of all nucleated mammalian cells, this is somewhat surprising. Thus, it would appear a plausible microbial strategy to adhere to these abundant surface proteins during, for instance, the initial colonization of the host. This notion and the multipotent protein-binding property of curli stimulated us to analyze a possible interaction between MHC-I molecules and curli. The results demonstrate that curliated E. coli, purified curli, and the curlin subunit protein indeed have affinity for MHC-I molecules.
MATERIALS AND METHODS
Bacterial strains.
E. coli strains used in this study are the curli-proficient strain YMel and the curli-deficient isogenic mutant strain YMel-1 (23). Different clinical isolates from various gastrointestinal infections (3) were kindly provided by James P. Nataro, University of Maryland School of Medicine, Baltimore, Md. To obtain maximal curli expression, the bacteria were grown on colony factor antigen-agar plates (12) for approximately 40 h at 26°C.
MHC molecules and curli.
Purified papain-solubilized human MHC-I molecules (HLA-A, -B, and -C) and detergent-solubilized MHC-II molecules (HLA-DR) were kindly provided by Lars Rask and Johan Sundelin, Department of Medical and Physiological Chemistry, Uppsala University, Uppsala, Sweden. Details concerning these preparations isolated from pooled spleens have been published elsewhere (19, 27). Human β2m was purified in this laboratory (4). Curli were purified as described elsewhere (8).
Radiolabeling of proteins.
Proteins were radiolabeled as described by Hunter and Greenwood (17); 100 μg of protein was mixed with 0.5 mCi of 125I (Amersham, Arlington Heights, Ill.) and oxidized with 20 μg of chloramine T (Sigma, St. Louis, Mo.).
Bacterial binding and inhibition assays.
Bacterial binding and inhibition analyses were performed as previously described (26). Briefly, bacteria were resuspended in phosphate-buffered saline (PBS; 0.15 M NaCl, 0.06 M phosphate [pH 7.2]) containing 0.1% Tween 20 to a concentration of 1010 cells ml−1. Dilutions of the bacteria were incubated with 125I-labeled protein (50 to 100 ng) in a total volume of 250 μl in polypropylene tubes at 20 or 37°C for 1 h. For the inhibition experiments, the radiolabeled protein was mixed with the unlabeled competitor proteins to be tested. After incubation, the bacteria were washed once in 2 ml of PBS containing 0.1% Tween 20. The radioactivity associated with the pellet was determined after a brief centrifugation. Binding was expressed as the percentage of the added radioactivity, deducting the nonspecific uptake to the propylene tubes.
Binding of MHC-I molecules to purified curli.
Purified curli preparations from YMel appearing as insoluble aggregates were incubated with 100 ng of 125I-labeled MHC-I molecules for 18 h at room temperature. After incubation, samples were centrifuged at 15,800 × g for 10 min and the supernatants were collected. The pellets were washed three times in PBS, resuspended in 20 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled, and centrifuged. The supernatants were subjected to SDS-PAGE.
SDS-PAGE and Western blot experiments.
SDS-PAGE was performed as described previously (20), and separated proteins were transferred onto polyvinylidene fluoride Immobilon-P membranes (Millipore, Bedford, Mass.). Membranes were incubated with blocking buffer (PBS containing 0.25% [wt/vol] Tween 20 and 0.25% [wt/vol] bovine serum albumin) at room temperature. The 125I-labeled proteins (≈6 × 106 cpm) were added followed by incubation at 4°C overnight. As molecular weight markers, prestained Kaleidoscope standard proteins (Bio-Rad, Hercules, Calif.) were used. The membrane was extensively washed in blocking buffer, dried, and exposed to X-ray film (Kodak, Rochester, N.Y.).
Cell culture conditions.
RMA and RMA-S cells were kindly provided by Klas Kärre, Microbiology and Tumor Biology Center, Karolinska Institute, Stockholm, Sweden. RMA and RMA-S cells were grown in Iscove’s modified Dulbecco’s medium (IMDM; Gibco BRL, Gaithersburg, Md.) supplemented with 5% fetal calf serum and antibiotics and cultured at 37°C in a 5% CO2 atmosphere. Detroit 562 (ATCC CCL 138) cells were grown in minimal essential medium (Gibco) supplemented with 0.1 mM glutamine and 10% fetal calf serum in a 5% CO2 atmosphere.
SEM.
For scanning electron microscopy (SEM), 100 μl of lymphoma cell suspension containing approximately 5 × 105 cells in IMDM was added on top of a wet Nylaflo 0.2-μm-pore-size membrane (German Sciences, Ann Arbor, Mich.). The sample was gently drawn onto the filter by suction caused by prewetted filter paper lying underneath the Nylaflo filter. The filter was fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate–0.1 M sucrose (pH 7.2) for 1 h at 4°C and was then washed with 0.15 M cacodylate buffer (pH 7.2); 100 μl of a bacterial suspension in PBS containing approximately 107 bacteria was added on top of the fixed cells and allowed to interact for 1 h at room temperature. The filters were then washed with cacodylate buffer, fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate–0.1 M sucrose (pH 7.2) for 1 h at 4°C, and washed with cacodylate buffer. Finally, the filters were postfixed in 1% osmium tetroxide in 0.15 M sodium cacodylate (pH 7.2) for 1 h at 4°C, washed, and stored in cacodylate buffer. Fixed filter paper samples were dehydrated for 10 min at each step of an ascending ethanol series and inserted into a Balzers critical point dryer, using 100% ethanol as the intermediate solvent. The high-pressure chamber was then extensively flushed three times for 30 min with carbon dioxide to remove all traces of residual ethanol, and the samples were critical point dried, mounted on aluminum holders, palladium-gold sputtered, and examined in a Jeol JSM-T330 SEM.
For each bacterium-cell pair, five separate fields of approximately 80 to 200 cells were counted. Cells with attached bacteria were scored positive. Student’s t test was used for statistical analysis. Data are presented as mean ± standard deviation (SD).
RESULTS
E. coli bacteria expressing curli bind human MHC-I molecules.
E. coli YMel grown in vitro at 26°C expresses curli, a surface organelle composed of a 15-kDa curlin subunit. To investigate whether curli-expressing bacteria interact with human MHC molecules, purified MHC-I and MHC-II molecules were radiolabeled and tested for binding to YMel bacteria. These bacteria bound 60 to 70% of the added 125I-labeled MHC-I but less than 10% of MHC-II molecules (Fig. 1). We detected no binding of 125I-labeled MHC-I or MHC-II molecules to the isogenic mutant E. coli YMel-1 strain, which has an inactivated curlin subunit gene and lacks curli on its surface (23). When the same E. coli strains were incubated with 125I-labeled β2m, the light chain of MHC-I molecules, YMel bound 25 to 30% of the added protein (Fig. 1) whereas the curli-deficient strain YMel-1 showed no binding. The results demonstrate that curliated bacteria interact with intact MHC-I molecules and to a lesser extent also with the isolated light chain, β2m. In these experiments, the same results were obtained when the binding assays were performed at room temperature (Fig. 1) and at 37°C (data not shown). However, as mentioned above, curli expression in vitro is regulated by temperature, and curli are not expressed when YMel is grown at 37°C (2, 24). When grown at 37°C, YMel showed no binding of MHC-I molecules or β2m (data not shown), emphasizing that curli are responsible for the interaction between E. coli and MHC-I molecules. A collection of 19 E. coli isolates belonging to different clinical types previously tested for curli expression (3) was also analyzed in binding experiments with radiolabeled MHC-I molecules. The 7 curliated isolates all showed affinity for MHC-I, whereas none of the 12 nonexpressing isolates bound the added radiolabeled protein above background level (data not shown).
FIG. 1.
Curli-expressing E. coli binds MHC-I molecules and β2m. Serial dilutions of bacteria were incubated at room temperature with a constant amount of 125I-labeled purified human MHC-I (▪, □), MHC-II (▴, ▵) or β2m (•, ○). Filled symbols indicate binding to curliated YMel, while open symbols indicate binding to noncurliated YMel-1. The binding of labeled protein to the bacteria was expressed as the percent of the total amount of added radiolabeled protein. The data represent the mean ± SD of three separate experiments. Student’s t test showed a statistically significant difference in the binding of MHC-I molecules and β2m to YMel compared to the binding of MHC-II molecules (P < 0.001).
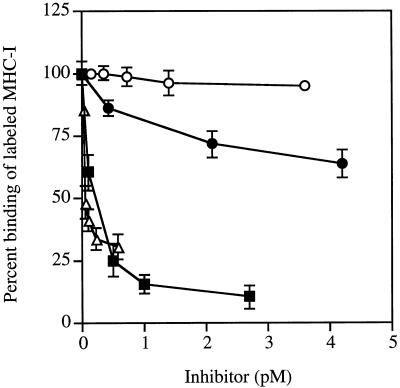
The specificity of the interaction between curli and MHC-I was investigated in experiments where the binding of radiolabeled MHC-I molecules to curliated YMel bacteria was inhibited with various proteins (Fig. 2). Human serum albumin, which shows no affinity for curli (3), did not influence the interaction, whereas unlabeled MHC-I in excess reduced the YMel–MHC-I interaction to <10% of the level observed with albumin. On the other hand, the two curli-binding proteins β2m and fibronectin partially inhibited the binding, reducing it to ≈75 and 25%, respectively, of the level of binding in the presence of albumin. These data suggest that the curli–MHC-I interaction is specific and that the binding sites for fibronectin and β2m on curli partially overlap or are located close to the binding site for intact MHC-I molecules. These results together with the data in Fig. 1, 3, and 4 (see below) suggest that MHC-I molecules interact with curli through both β2m and the heavy chain of MHC-I molecules. The data also suggest that the affinity is higher for intact MHC-I molecules than for β2m.
FIG. 2.
Inhibition of the binding of radiolabeled MHC-I molecules to curli. The binding of 125I-MHC-I molecules to curliated YMel was measured in the presence of various concentrations of unlabeled human serum albumin (○), human fibronectin (▵), β2m (•), and MHC-I molecules (▪). Binding was expressed as the percentage of the added radioactivity. The data represent the mean ± SD of two to four separate experiments. The interaction between human serum albumin and YMel was compared to the binding of MHC-I, β2m, and fibronectin to YMel. Student’s t test showed the difference to be statistically significant (P < 0.001).
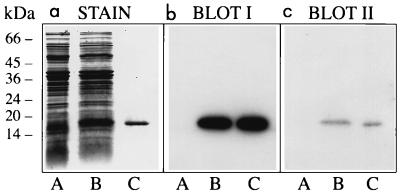
FIG. 3.
Binding of MHC-I molecules and β2m to curlin subunits. Three identical SDS-polyacrylamide gels were loaded with proteins solubilized from curli-expressing YMel bacteria by boiling in SDS-PAGE sample buffer (lane A). The same material with the addition of purified curlin subunits (approximately 1 μg) was separated in lane B. Curlin subunits alone were run in lane C. One of the gels was stained with Coomassie blue (a), whereas the proteins of the other two were transferred to Immobilon membranes and probed with 125I-MHC-I (b) or 125I-β2m (c). Molecular mass markers are indicated to the left.
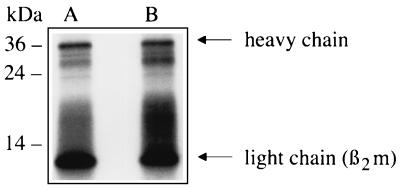
FIG. 4.
Binding of radiolabeled MHC-I molecules to insoluble curli aggregates. Curli aggregates (1 mg) were incubated with 125I-labeled MHC-I molecules (2 × 105 cpm). After incubation at room temperature, the aggregates were centrifuged and washed, and the radioactivity bound to the pellet was eluted by boiling in SDS-PAGE sample buffer. Samples were separated by SDS-PAGE (10% gel) under reducing conditions. Following electrophoresis, the gel was dried and exposed to X-ray film. Lane A, 2 × 105 cpm of 125I-labeled MHC-I molecules; lane B, radioactivity released from the curli pellet following incubation with 2 × 105 cpm of 125I-labeled MHC-I molecules. The 36- and 12-kDa bands represent the heavy and light chains, respectively, of MHC-I molecules. The migration of molecular mass markers is shown to the left.
Purified curlin subunit protein binds MHC-I molecules.
Curli are large insoluble polymers of curlin subunits. Formic acid treatment of purified intact curli organelles releases the subunit (8, 23), which migrates as a single band of 15 kDa when subjected to SDS-PAGE. The experiments summarized in Fig. 3 were set up to analyze whether purified curlin subunits have affinity for MHC-I molecules and/or β2m.
Curliated YMel bacteria were boiled in SDS-PAGE sample buffer, and following centrifugation the proteins of the supernatant were separated by SDS-PAGE (Fig. 3a, lane A). Curlin subunits were added to this material, and the mixture was run in lane B, whereas the same amount of curlin subunits alone were separated in lane C. Three identical gels were generated; two were electroblotted onto Immobilon-P membranes and probed with radiolabeled MHC-I and β2m (Fig. 3b and c, respectively). The results demonstrate that both probes bind to curlin subunits with a high degree of specificity. None of the other proteins solubilized from curliated E. coli bacteria with SDS-PAGE sample buffer reacted with the probes. Again, the stronger signal seen with MHC-I suggests a higher affinity for intact MHC-I than for β2m.
Insoluble curli aggregates bind MHC-I molecules.
Purified curli organelles appear as insoluble aggregates (8, 23). We therefore investigated whether such purified insoluble aggregates were capable of binding MHC-I in vitro. Papain-solubilized MHC-I preparations from human spleens are typically size heterogeneous, with two dominating bands of 36 and 12 kDa corresponding to the MHC-I heavy and light chains, respectively. 125I-labeled MHC-I molecules (Fig. 4, lane A) were incubated with insoluble curli aggregates; following incubation, the aggregates were centrifuged and washed carefully. Ninety percent of the radioactivity was found to be associated with the pellet. This radioactivity was released by boiling the aggregates in SDS-PAGE sample buffer and subjecting them to SDS-PAGE followed by autoradiography. Two major bands of approximately 36 and 12 kDa were released from the curli aggregates preincubated with 125I-MHC-I molecules (Fig. 4, lane B), demonstrating that purified native curli bind MHC-I molecules.
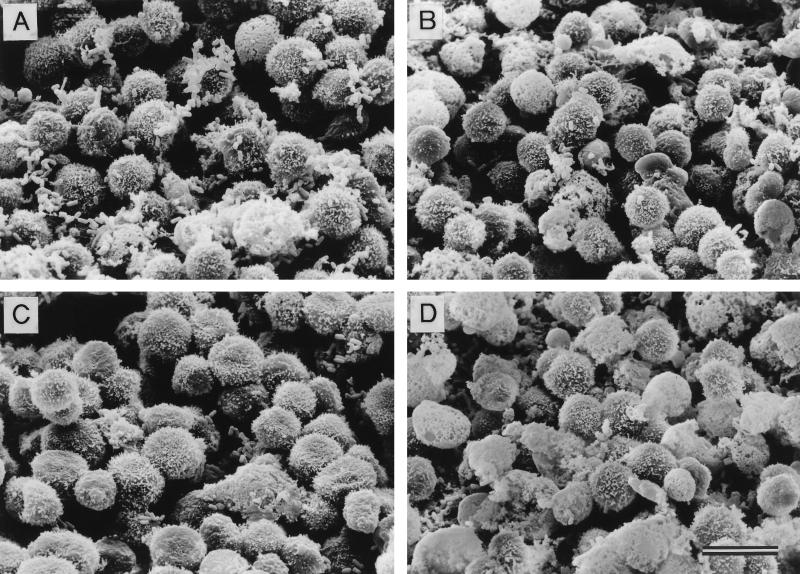
Curliated E. coli bacteria adhere to MHC-I-expressing cells.
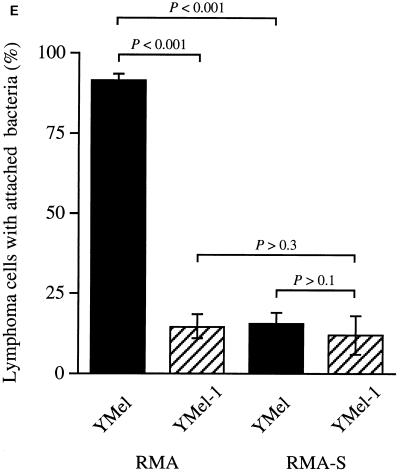
The observation that curli bound purified MHC-I molecules raised the question of whether mammalian cells could interact with curliated bacteria through MHC-I molecules. SEM revealed that curliated bacteria adhered better than noncurliated bacteria to the human epithelial cell line Detroit 562 (mean numbers of bacteria adhering/cell were 1.7 for YMel and ≤0.1 for YMel-1). However, to test our hypothesis more conclusively, the MHC-I-expressing mouse lymphoma cell line RMA (21) and its mutant variant RMA-S, which expresses less than 10% of the MHC-I level of RMA (21), were used in adhesion experiments. Curliated YMel bacteria and the isogenic mutant strain YMel-1 were incubated separately with RMA or RMA-S cells, and interactions between bacteria and eukaryotic cells were analyzed by SEM. These studies revealed that YMel bacteria adhered to the MHC-I-expressing RMA cells (Fig. 5A) whereas noncurliated YMel-1 did not (Fig. 5B). When YMel and YMel-1 bacteria were incubated with MHC-I-negative RMA-S cells, no significant binding was detected (Fig. 5C and D). The mean number of bacteria attached to RMA and RMA-S cells demonstrated that curliated YMel bacteria adhered more efficiently to RMA cells than did YMel-1 bacteria (mean values, 6.3 and 0.7 bacteria/RMA cell, respectively), whereas no significant difference was recorded when RMA-S cells were used (mean values, 0.8 and 0.3 bacterium/RMA-S cell). The expression of MHC-I is higher in lymphocytes than in epithelial cells (11, 13), which is consistent with the observation that curliated bacteria adhere better to RMA cells than to Detroit 562 cells. Furthermore, as shown in Fig. 5E, the percentage of RMA cells with adhering YMel bacteria was significantly higher than the percentage of RMA cells that bound noncurliated YMel-1 (91.2% ± 1.9%, compared to 14.3% ± 3.8%; P < 0.001). In contrast, no statistically significant difference in the binding of YMel or YMel-1 bacteria was seen with RMA-S cells (15.2% ± 3.7% and 11.8% ± 6.0%, respectively; P > 0.1). The inability of curliated YMel to interact with RMA-S cells demonstrates that autoaggregation of the bacteria is not responsible for the interaction between YMel and RMA cells. Clearly, MHC-I molecules as well as curli are required for the interactions, as demonstrated by the observation that noncurliated YMel-1 binds relatively poorly to either RMA or RMA-S cells (Fig. 5). In addition, the two cell lines bind YMel-1 to the same degree (14.3% ± 3.8%) [RMA] and 11.8% ± 6.0% [RMA-S]; P > 0.3). These results demonstrate that E. coli expressing curli can interact with host cells through MHC-I molecules.
FIG. 5.
(A to D) Adhesion between E. coli and lymphoma cells analyzed by SEM. Mouse lymphoma cells with high (RMA) and low (RMA-S) MHC-I expression were incubated with curliated (YMel) or noncurliated (YMel-1) E. coli. Shown are incubations with YMel and RMA (A), YMel-1 and RMA (B), YMel and RMA-S (C), and YMel-1 and RMA-S (D). The bar indicates 10 μm. (E) Percentage of lymphoma cells in the four different combinations with adherent bacteria. Each column represents the mean ± SD of two different experiments. The difference between sets of data were calculated by using Student’s t test, and the results were as follows: P < 0.001 for the binding of YMel and YMel-1 to RMA cells as well as for the binding of YMel to RMA and RMA-S cells, P > 0.3 for YMel-1 binding to RMA and RMA-S cells, and P > 0.1 for the binding of YMel and YMel-1 to RMA-S cells. YMel and YMel-1 bacteria are indicated with filled and hatched bars, respectively.
DISCUSSION
This report demonstrates for the first time an interaction between purified monomeric MHC-I molecules and a bacterial surface protein. Such an interaction should have several biological implications and not only influence adherence and colonization but also possibly interfere with peptide–MHC-I interactions and alter T-cell receptor recognition of peptide–MHC-I complexes. In contrast to bacterial superantigens, which bind directly to MHC-II molecules and activate T cells in an antigen-independent manner (25), curli showed no significant affinity for the detergent-solubilized MHC-II antigens used here.
The structural gene encoding the curlin subunit is present in most wild-type isolates of E. coli, but the level of expression varies considerably between different isolates and clinical types (23). For example, in the so-called EcoR collection (22), a set of natural E. coli isolates from different sources, approximately 20% of the strains expressed curli at both 26 and 37°C (23), and many enterohemorrhagic, enterotoxigenic, and sepsis isolates express curli at 26°C and retain low expression of curli when grown at 37°C in vitro (3). Enteroinvasive and enteropathogenic isolates, on the other hand, express little or no curli at either temperature (3). Thin aggregative fimbriae (8) are fibrous surface proteins in S. enteritidis closely related to curli, and some strains of Salmonella typhimurium also express these proteins at high levels at 37°C in vitro (26a). Although not yet formally demonstrated, observations of this kind support the notion that curli could indeed be expressed by E. coli growing in vivo, where the environmental conditions and selective pressures are considerably different from in vitro conditions. As mentioned above, the binding of MHC-I molecules to insoluble curli aggregates is the same at room temperature and at 37°C. Thus, once expressed, curli will have the capacity to interact with MHC-I molecules at physiological temperatures.
It has been shown that curliated E. coli as well as purified curlin subunit can bind a number of different human proteins. Interestingly, these proteins have no common structural or functional properties explaining their affinity for curli, suggesting that the curli polymer may have a unique ability to form multiple protein-binding regions with different specificities. This hypothesis is supported by inhibition experiments of this and previous investigations, demonstrating that the binding of a given protein to curli is not blocked by the simultaneous presence of other curli-interacting proteins. The remarkable ability of curli to specifically interact with a variety of host proteins should greatly facilitate the adaptation of curliated E. coli to different ecological niches. Thus, the binding to proteins like fibronectin and laminin (23, 24) may mediate interactions with cells and the cellular matrix. Furthermore, it has been demonstrated that curliated E. coli in human plasma absorbs plasminogen and tissue plasminogen activator, leading to the formation of proteolytically active plasmin (26), which may promote bacterial spreading through tissue degradation. Moreover, in the plasma environment, the proteins of the contact-phase system are assembled at the surface of curliated E. coli, resulting in the release of bradykinin (3). This potent proinflammatory peptide induces vasodilatation and increases vascular permeability and leakage of plasma, which could also contribute to the spread of the infection as well as provide growing bacteria with nutrients.
MHC-I molecules are expressed at the surface of almost all nucleated human cells, and the demonstration here that E. coli is capable of interacting with MHC-I molecules represents yet another protein-binding property of curli which could also influence the host-microbe relationship. Wild-type E. coli expresses curli at high levels in vitro when grown in poor media at temperatures below 30°C (2, 24), suggesting that E. coli infecting a new host may have a high density of curli. The interaction with MHC-I molecules should therefore facilitate initial adherence and colonization. The observation that E. coli growing on solid medium expresses more curli than when growing in liquid medium also indicates an adhesive role for curli. Depending on the type of host cells curliated E. coli is interacting with via MHC-I molecules, this could have different consequences. However, the significance of the interaction described here lies in the fact that MHC-I molecules are abundantly expressed and thus will be available for curli interactions in every possible niche of a mammalian host.
ACKNOWLEDGMENTS
This work was supported by grants from Swedish Medical Research Council (projects 7480, 11196, and 11223); the Swedish Natural Science Research Council (project 10610-301); the Royal Physiografic Society; the foundations of Crafoord, Kocks, Schybergs, Zoégas, and Österlund; the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine; ACTINOVA Ltd.; and the Medical Faculty, Lund University.
Ulla Johannesson and Ingbritt Gustafsson are acknowledged for excellent technical assistance.
REFERENCES
- 1.Arnqvist A, Olsén A, Normark S. ςs-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by ς70 in the absence of nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 2.Arnqvist A, Olsén A, Pfeifer J, Russell D G, Normark S. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol Microbiol. 1992;6:2443–2452. doi: 10.1111/j.1365-2958.1992.tb01420.x. [DOI] [PubMed] [Google Scholar]
- 3.Ben Nasr A, Olsén A, Sjöbring U, Müller-Esterl W, Björck L. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol Microbiol. 1996;20:927–935. doi: 10.1111/j.1365-2958.1996.tb02534.x. [DOI] [PubMed] [Google Scholar]
- 4.Berggård I, Bearn A G. Isolation and properties of a low molecular weight β2-globulin occurring in human biological fluids. J Biol Chem. 1968;243:4095–4103. [PubMed] [Google Scholar]
- 5.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 6.Collinson S K, Clouthier S C, Doran J L, Banser P A, Kay W W. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collinson S K, Doig P C, Doran J L, Clouthier S, Trust T J, Kay W W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collinson S K, Emödy L, Müller K-H, Trust T J, Kay W W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson S K, Emödy L, Trust T J, Kay W W. Thin aggregative fimbriae from diarrheagenic Escherichia coli. J Bacteriol. 1992;174:4490–4495. doi: 10.1128/jb.174.13.4490-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doran J L, Collinson S K, Burian J, Sarlós G, Todd E C D, Munro C K, Kay C M, Banser P A, Peterkin P I, Kay W W. DNA-based diagnostic tests for Salmonella species targeting agfA, the structural gene for thin aggregative fimbriae. J Clin Microbiol. 1993;31:2263–2273. doi: 10.1128/jcm.31.9.2263-2273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorval G, Welsh K I, Nilsson K, Wigzell H. Quantitation of β2-microglobulin and HLA on the surface of human cells. I. T and B lymphocytes and lymphoblasts. Scand J Immunol. 1977;6:255–263. doi: 10.1111/j.1365-3083.1977.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 12.Evans D G, Evans D J, Tjoa W. Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect Immun. 1977;18:330–337. doi: 10.1128/iai.18.2.330-337.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evrin P E, Pertoft H. β2-Microglobulin in human blood cells. J Immunol. 1973;111:1147–1154. [PubMed] [Google Scholar]
- 14.Germain R N, Margulies D H. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 15.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 16.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter W M, Greenwood F C. Preparation of iodine-131 labelled human growth hormone of high specific activity. Nature. 1962;194:495–496. doi: 10.1038/194495a0. [DOI] [PubMed] [Google Scholar]
- 18.Kehoe M A. Cell-wall-associated proteins in Gram-positive bacteria. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier; 1994. pp. 217–261. [Google Scholar]
- 19.Klareskog L, Trägårdh L, Rask L, Peterson P A. Isolation and characterization of detergent-solubilized human HLA-DR transplantation antigens. Biochemistry. 1979;18:1481–1489. doi: 10.1021/bi00575a015. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Ljunggren H G, Stam N J, Öhlén C, Neefjes J J, Höglund P, Heemels M-T, Bastin J, Schumacher T N M, Townsend A, Kärre K, Ploegh H L. Empty MHC class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 22.Ochman H, Selander R K. Standard reference strains from Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsén A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin binding curli in Escherichia coli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 24.Olsén A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 25.Scherer M T, Ignatowicz L, Winslow G M, Kappler J W, Marrack P. Superantigens: bacterial and viral proteins that manipulate the immune system. Annu Rev Cell Biol. 1993;9:101–128. doi: 10.1146/annurev.cb.09.110193.000533. [DOI] [PubMed] [Google Scholar]
- 26.Sjöbring U, Pohl G, Olsén A. Plasminogen, absorbed by Escherichia coli expressing curli or by Salmonella enteritidis expressing thin aggregative fimbriae, can be activated by simultaneously captured tissue-type plasminogen activator (t-PA) Mol Microbiol. 1994;14:443–452. doi: 10.1111/j.1365-2958.1994.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 26a.Sukupolvi S, Edelstein A, Rhen M, Normark S J, Pfeifer J D. Development of a murine model of chronic Salmonella infection. Infect Immun. 1997;65:838–842. doi: 10.1128/iai.65.2.838-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trägårdh L, Curman B, Wiman K, Rask L, Peterson P A. Chemical, physical-chemical, and immunological properties of papain-solubilized human transplantation antigens. Biochemistry. 1979;18:2218–2226. doi: 10.1021/bi00578a013. [DOI] [PubMed] [Google Scholar]
- 28.Williams A F, Barclay A N. The immunoglobulin superfamily—domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]