Abstract
Progranulin is an evolutionarily conserved protein that has been implicated in human neurodevelopmental and neurodegenerative diseases. Human progranulin is comprised of multiple cysteine-rich, biologically active granulin peptides. Granulin peptides accumulate with age and stress, however their functional contributions relative to full-length progranulin remain unclear. To address this, we generated C. elegans strains that produced quantifiable levels of both full-length progranulin/PGRN-1 protein and cleaved granulin peptide. Using these strains, we demonstrated that even in the presence of intact PGRN-1, granulin peptides suppressed the activity of the lysosomal aspartyl protease activity, ASP-3/CTSD. Granulin peptides were also dominant over PGRN-1 in compromising animal fitness as measured by progress through development and stress response. Finally, the degradation of human TDP-43 was impaired when the granulin to PGRN-1 ratio was increased, representing a disease-relevant downstream impact of impaired lysosomal function. In summary, these studies suggest that not only absolute progranulin levels, but also the balance between full-length progranulin and its cleavage products, is important in regulating lysosomal biology. Given its relevance in human disease, this suggests that the processing of progranulin into granulins should be considered as part of disease pathobiology and may represent a site of therapeutic intervention.
Keywords: progranulin, granulin, TDP-43, FTD, lysosome
Introduction
Progranulin is an evolutionarily conserved glycoprotein comprised of 7.5 peptide repeats known as granulins, with established roles in growth, survival and immune regulation and emerging key functions in neuroinflammation, neurodegeneration and brain aging [1]. Encoded by the granulin (GRN, Pgrn) gene in humans, progranulin is widely expressed in multiple cell types including in the CNS and peripheral nervous system. Pgrn gene mutations resulting in progranulin haploinsufficiency are a common cause of the age-associated neurodegenerative disease frontotemporal dementia (FTD), while the protein’s complete loss results in the juvenile-onset lysosomal storage disease neuronal ceroid lipofuscinosis (NCL) [2–6]. These disorders reveal the important insight that gene dosage determines the phenotype and age of onset associated with progranulin deficiency.
Subject to complex regulation, progranulin can be proteolytically processed into biologically active granulins or multi-granulin fragments, whose functions remains an active area of investigation. While secreted protein can undergo proteolysis in the extracellular matrix, the predominant location for its processing appears to be within lysosomes [2, 7–9]. Here, progranulin can be proteolytically processed by a number of lysosomal proteases to produce granulins in a regulated manner [10, 11]. In addition, progranulin directly regulates the activity of the endo-lysosomal protease, Cathepsin D (CTSD) [12, 13], itself also a disease-associated protein. Progranulin activates CTSD activity by promoting its maturation [12, 14]. Consistent with this effect, Ctsd mutations phenocopy Pgrn mutations to a large degree, with both associated with lysosomal storage disease.
In contrast to the established role of progranulin in CTSD regulation, the role of liberated granulins on lysosomal function is less clear, particularly in the presence of the full-length protein. Some studies suggest that granulins act in a similar fashion as the full-length progranulin. For example, in vitro human recombinant enzyme activity assays suggest that both progranulin and granulin E increases CTSD activity [13]. However, other studies have demonstrated that granulins act in opposition to progranulin. For example, in the human immune system, progranulin acts in an anti-inflammatory fashion while individual granulins are pro-inflammatory [15–18]. Consistent with progranulin and granulins playing reciprocal biological roles, we have shown that C. elegans overexpressing pgrn-1 were more stress resistant, whereas those expressing individual granulin peptides were relatively less stress resistant [19].
Loss of C. elegans pgrn-1 can be rescued with human progranulin [19], suggesting conservation of function. The C. elegans progranulin protein, PGRN-1, can be cleaved into three granulins, granulins 1, 2 and 3, which are produced throughout adulthood and likely necessarily for proper embryogenesis, as efforts to create non-cleavable progranulin strains have been unsuccessful. Previously, we have shown in C. elegans that cleaved granulins increase with age and under stress, and when expressed in the absence of the full-length protein, granulins directly impair lysosomal protease activity and disrupt protein homeostasis [19]. Of the granulins, granulin 3 had the most disruptive impact, such as most impaired enzyme activity, smallest lysosome sizes, and lowest stress resistance. These studies isolated granulin peptides to better understand their functional effect. However, what remained unclear was the impact of granulin peptides when full-length protein was also present. Therefore, here we set out to understand the biological consequences of C. elegans granulin 3 peptide in the company of PGRN-1. Based on our earlier work, we hypothesized that the relative levels of granulin 3 and PGRN-1 contribute to lysosomal function and animal fitness. We found that granulin 3, even in the presence of full length PGRN-1, suppressed aspartyl protease activity and decreased animal fitness as measured by development and stress response.
Results
Full-length PGRN-1 rescues granulin 3-induced abnormal lysosome morphology but not aspartyl protease activity
We have shown that in the absence of full-length progranulin, individual granulin peptides impair the function of the lysosomal aspartyl protease Cathepsin D and impair TDP-43 degradation [12, 14, 19, 20]. However, the functional impact of granulins on lysosomal biology in the presence of intact progranulin has not been carefully studied in vivo. To do so, we utilized a series of transgenic C. elegans strains with tagged versions of PGRN-1 and granulin 3 that allowed us to directly quantify and compare protein levels. These strains included an RFP-tagged PGRN-1 overexpression line (pgrn-1 OE) and a FLAG-tagged granulin 3 overexpression line (gran3 OE). These two lines were then crossed to generate a line expressing both PGRN-1::RFP and FLAG-tagged granulin 3 (pg/gr) (Fig. 1A). The pgrn-1 OE, gran3 OE, and pg/gr strains were all made on a pgrn-1 null background. Thus, the only source of progranulin and granulins were the transgenes. We focused on granulin 3 in part because we were able to follow both FLAG and liberated RFP-tagged versions of it. Of note, in the pg/gr strain could come from two sources: either proteolytic processing of PGRN-1::RFP into RFP-tagged granulin 3 or directly from FLAG-tagged granulin 3.
Figure 1.
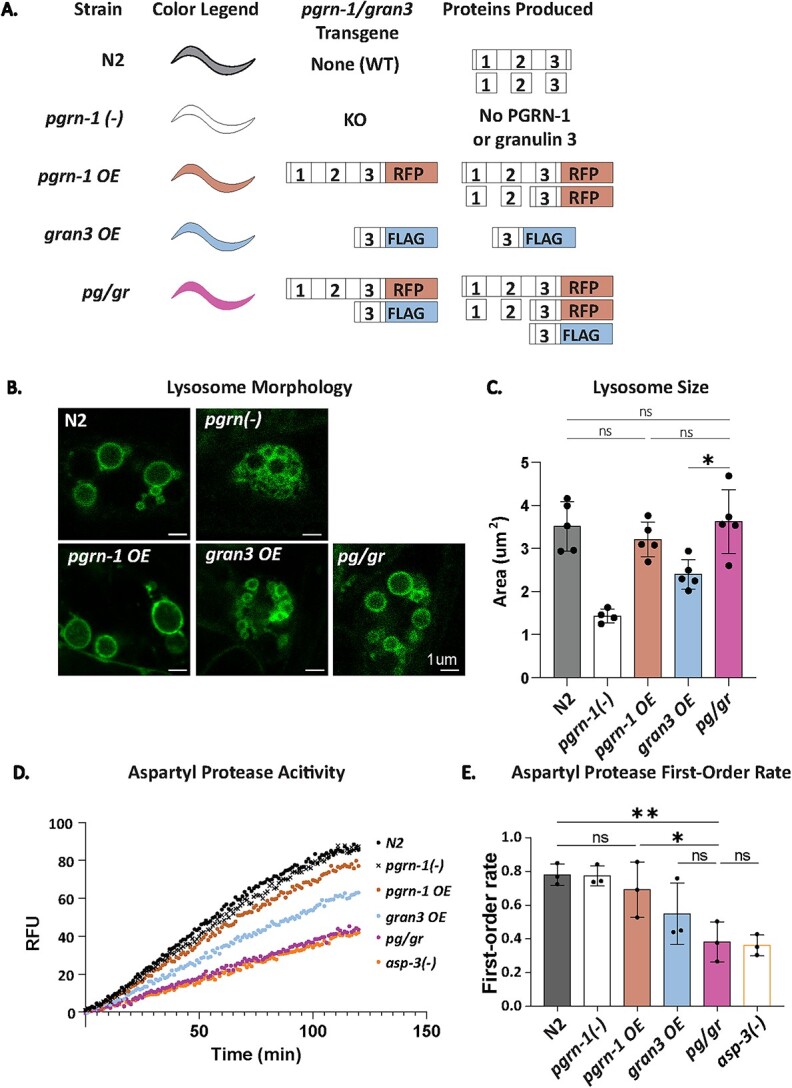
Full-length PGRN-1 rescues granulin 3-induced abnormal lysosome morphology but not aspartyl protease activity. (A) Schematic of the lines used. The color legend refers to the colors used throughout the figs. (B) Representative images of LMP-1::GFP-tagged coelomocyte lysosomes, imaged by confocal microscopy. (C) Lysosomal area was quantified for each strain by tracing the perimeter of lysosomes. n = 8 animals, 5 biological replicates. (D) Representative aspartyl protease activity traces. Data was collected by a plate reader using a fluorescence substrate mixed with worm lysate. n = 3 biological replicates of day 1 animals. (E) First-order rate of protease enzyme activity as calculated from (D). Error bars represent mean +/− SEM comparisons with one-way ANOVA with post hoc Sidak comparisons (*P < 0.05, **P < 0.005, ns = not significant).
Our group has previously shown that gran3 OE animals exhibit disrupted lysosomal morphology, with smaller size and suppressed protease activity [19]. To determine if gran3 OE still affects lysosomes in the company of full-length progranulin, we first quantified lysosomal size in our series of strains. This was done by tracing LMP-1::GFP-marked lysosomes in coelomocytes. As previously observed [19], pgrn-1(-) and gran3 OE animals caused lysosomes to become smaller, while pgrn-1 OE rescued lysosome size (Fig. 1B and C). However, pg/gr animals with both PGRN-1 and granulin 3 exhibited normal lysosomal size. This result indicated that the presence of full-length PGRN-1 restored impaired lysosome morphology produced by excess granulin 3.
Next, we investigated if lysosomal protease activity was restored in animals with both full-length PGRN-1 and granulin 3. In contrast to lysosomal size, the presence of PGRN-1 with granulin 3 in pg/gr animals did not rescue lysosomal aspartyl protease activity (Fig. 1D and E, Supplementary Figure 1A and B). In fact, pg/gr animals had the lowest aspartyl protease activity among all lines, at a level that was similar to that of the aspartyl protease knockout mutant asp-3. The asp-3 gene encodes for the C. elegans CTSD ortholog, ASP-3, which we have previously used as a negative control [19]. It is one of several C. elegans aspartyl proteases. These data indicate that while full-length PGRN-1 rescues the lysosome morphology deficits of granulin 3, the lysosomal enzyme activity is not restored. This suggested when progranulin and granulins are both present, their impact on specific lysosomal metrics was not always straightforward, and that their relative levels may be important in determining which exerted a dominant effect.
Quantification of PGRN-1 and granulin 3 levels
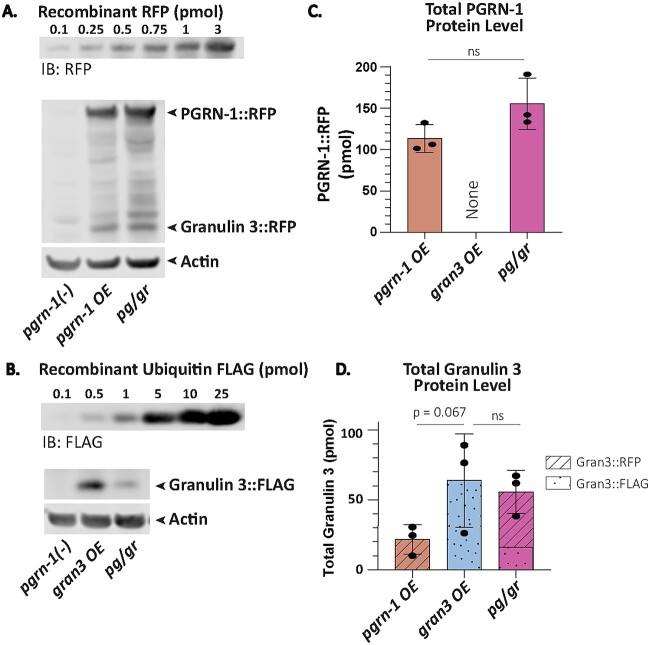
To decipher the mechanism underlying the disparate effects of progranulin and granulin on lysosomal biology, their relative levels required quantification. To do so, we took advantage of the C-terminal RFP and FLAG tags marking PGRN-1 and granulin 3, respectively. These tags allowed us to directly compare their levels to that of standard curves generated with recombinant RFP and FLAG (Fig. 2A and B, Supplementary Fig. S2A and B). As a result, we could quantify, with picomolar sensitivity, the amounts of full-length PGRN-1 protein and granulin 3 peptides produced, regardless of whether the latter is released from PGRN-1::RFP or generated directly as flag-tagged granulin 3 (See Fig. 1A). To ensure accurate measurements, dilution curves were generated to identify the linear range of detection, from which 30 ug of lysate was determined to be appropriate (Supplementary Fig. S2D–F).
Figure 2.
Quantification of PGRN-1 and granulin 3 levels. Day 1 animals were collected for Western blot. (A) A representative blot of the recombinant RFP standard curve, the RFP tagged PGRN-1 protein and RFP-tagged granulin 3 peptide from pgrn-1 OE and pg/gr animals’ whole worm lysate immunoblotted with an anti-RFP antibody, and the corresponding actin immunoblotted with an anti-actin antibody. (B) A representative blot of the standard curve of recombinant ubiquitin tagged FLAG and the FLAG-tagged granulin 3 peptide levels in gran3 OE and pg/gr animals’ whole worm lysate immunoblotted with an anti-FLAG antibody, and the corresponding actin immunoblotted with an anti-actin antibody. (C) Total PGRN-1::RFP as calculated using the standard curve in (A), normalized to actin. n = 3 biological replicates. Statistics done by student t-test. (D) Total granulin 3 amounts as calculated using the standard curves in (A) and (B) and normalized to actin. n = 3 biological replicates. Statistics done by one-way ANOVA with post-hoc Sidak comparisons as indicated. (*P < 0.05, ns = not significant).
Having established a method for quantifying PGRN-1 and granulin 3, we then measured their levels in our transgenic strains. The pgrn-1 OE strain produced about 113 pmol of PGRN-1::RFP and 21 pmol of RFP-tagged granulin 3 per 30 ug of total protein (Fig. 2C and D and Supplementary Fig. S2C and G). The pg/gr strain produced 155 pmol of PGRN-1 and 42 pmol of RFP-tagged granulin 3 per 30ug mg of total protein, in both cases slightly more than the pgrn-1 OE strain (Fig. 2C and D and Supplementary Fig. S2C and G).
The gran3 OE strain produced about 66 pmol of FLAG-tagged granulin 3 per 30 ug of total protein (Fig. 2D and Supplementary Fig. S2H). Despite containing the same transgene, the pg/gr strain produced significantly less FLAG-tagged granulin 3, 13 pmol. Nonetheless, when the FLAG-tagged granulin 3 and the RFP-tagged granulin 3 were summed, there was a total of 55 pmol. Thus, the total granulin 3 protein in gran3 OE and pg/gr animals is more than twice that of pgrn-1 OE animals (Fig. 2D). Protein levels were also normalized to total protein (as quantified by LiCOR’s REVERT total protein staining) to confirm the findings (Supplementary Fig. S2I–L). When granulin 3 and PGRN-1 levels were compared in each of the strains, it became apparent that the granulin 3 levels were highest in gran3 OE and pg/gr animals. Since aspartyl protease activity was lowest in these two strains, this would be consistent with the granulins being responsible for inhibiting aspartyl proteases, regardless of the presence of progranulin.
Of note, C. elegans PGRN-1 is cleaved into three granulins and our previous work suggests that all three C. elegans granulins impair animal fitness [20]. The granulin quantification utilized here only allowed the measurement of granulin 3. Thus, this method likely underestimated the total granulins levels since it did not account for granulin 1 and 2. This may explain why despite having slightly less granulin 3, the pg/gr animals exhibited lower aspartyl protease than the gran3 OE. Nonetheless, this method is notable as it is the first to quantitatively measure full-length progranulin and granulin and correlate them to behavioral outputs in vivo.
Granulin 3 impairs animal fitness, even in the presence of PGRN-1
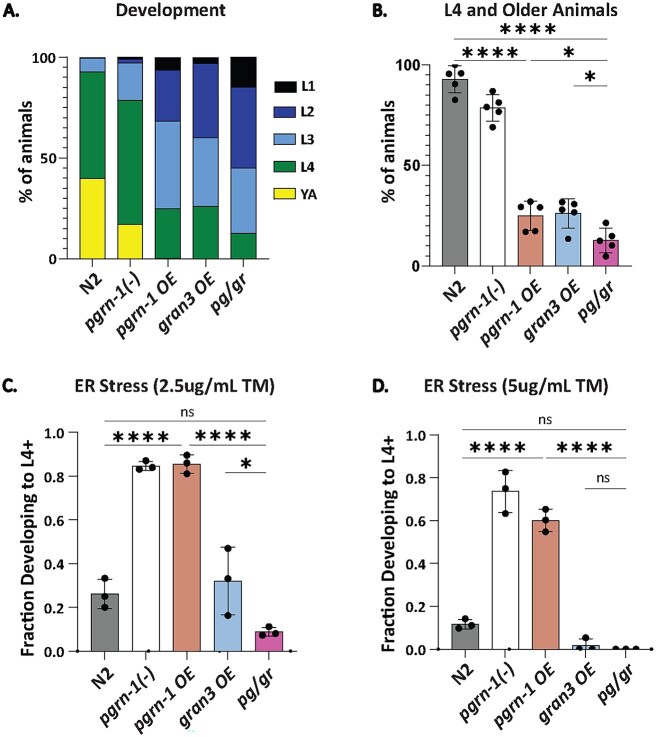
We previously showed that granulin 3 impairs animal fitness [20]. To assess whether this impaired fitness could be rescued by progranulin, we measured the development rate of each of our transgenic lines. We found that gran3 OE animals developed slower than wildtype, consistent with previously published results [20] (Fig. 3A and B). Progranulin overexpression, in both the pgrn-1 OE and pg/gr animals did not rescue the developmental delay (Fig. 3A and B). The slowest development of all lines was observed in pg/gr animals (Fig. 3B). Development was also delayed in pgrn-1 OE animals, which is consistent with the effect of increased in granulin 3 as demonstrated in the gran OE and pg/gr animals (Fig. 3A and B). These results affirmed the negative effect of granulins on animal development, regardless of the presence of PGRN-1.
Figure 3.
Granulin 3 impairs animal fitness, even in the presence of PGRN-1. (A) Forty-eight hours after egg staging, development stage distribution of animals was scored. n = 5 biological replicates, 50 animals each. (B) L4 and older animals are represented from (A). (C) ER stress resistance as measured by percent of animals successfully developed from egg to L4 and older after 72 h on plates supplemented with 2.5ug/ml of Tunicamycin. n = 3 biological replicates, 150 animals each. (D) ER stress resistance repeated at 5ug/ml of Tunicamycin. n = 3 biological replicates, 150 animals each. Error bars represent mean +/− SEM, statistics was done by one-way ANOVA with post hoc Sidak comparisons indicated (*P < 0.05, ****P < 0.00005. ns = not significant).
One of the most important functional features of lysosomes is to dynamically respond to proteotoxic stressors and maintain protein homeostasis, such as during endoplasmic reticulum (ER) stress [21–23]. To test if increased granulin 3 levels decreased resistance to proteotoxic stress even in the presence of PGRN-1, we induced ER stress in animals by growing them in media containing tunicamycin and determining the fraction of animals able to develop to L4 and beyond [19, 20]. We found that pgrn-1 OE were more stress resistant relative to wildtype and confirmed our earlier finding [20] that gran3 OE animals were stress sensitive relative to N2 and pgrn-1(-) animals (Fig. 3C and D). Notably, pg/gr animals were more stress sensitive than pgrn-1 OE (Fig. 3C and D). These results again supported the ability of granulin 3 to impair stress resistance, even in the presence of PGRN-1.
Granulin 3 impairs TDP-43 degradation
TDP-43 is an RNA binding protein that mislocalizes to the cytoplasm and accumulates in frontotemporal lobar degeneration (FTLD) due to progranulin mutations [1, 24]. When expressed in C. elegans neurons, human TDP-43 (huTDP-43) slows animal development [20]. We previously showed that granulins 2 and 3, but not 1, further slow the development of animals expressing huTDP-43 when in a pgrn-1(-) background [20]. To determine if full-length progranulin ameliorated this, we assessed development in our series of strains in the presence of huTDP-43. We found that the high granulin 3 levels in the gran3 OE and pg/gr lines enhanced the toxicity of huTDP-43 (Fig. 4A). Likewise, in these two strains, the steady-state levels of huTDP-43 levels were highest (Fig. 4B and C). This was consistent with our earlier observation that granulins impaired the degradation of huTDP-43, but not tau [20]. Interestingly, in the gran3 OE line, huTDP-43 mRNA levels were quite elevated, whereas they were unchanged compared to controls in the pg/gr line (Fig. 4D).
Figure 4.
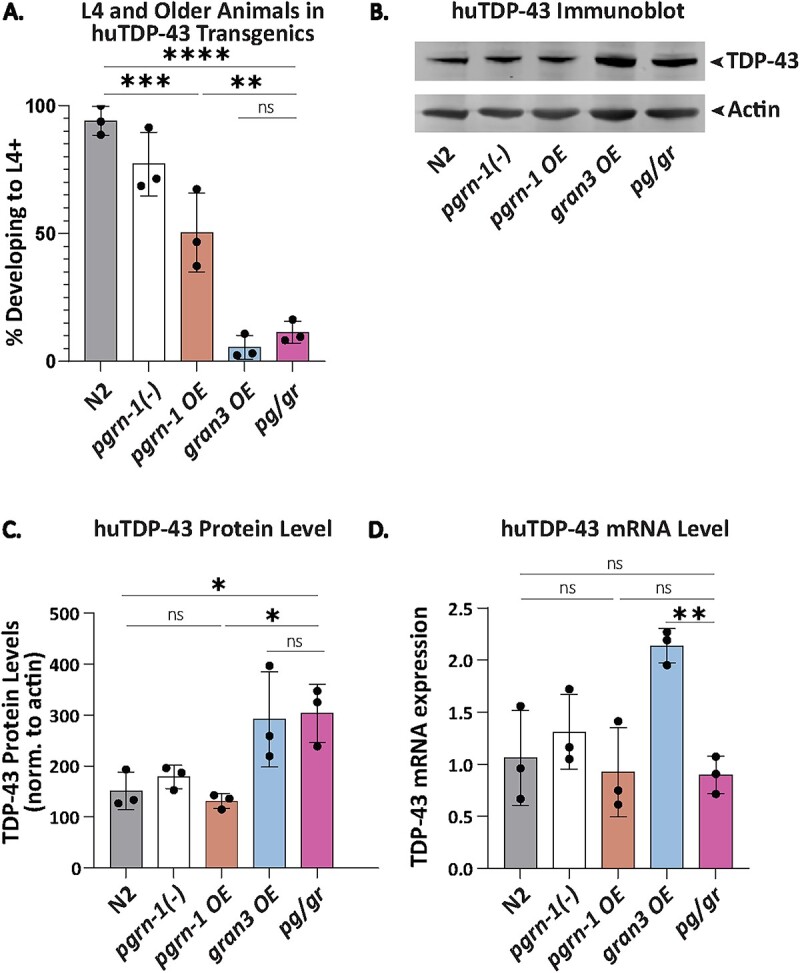
Granulin 3 impairs huTDP-43 degradation. (A) L4 and older animals were scored 48 h after egg staging. n = 5 biological replicates, 50 animals each. (B) huTDP-43 immunoblotted with an anti-huTDP-43 antibody on a Western blot across with the corresponding anti-actin antibody staining as a loading control. n = 3 biological replicates of day 1 adult animals. (C) Quantification of huTDP-43 protein levels as shown in (B), normalized to actin. (D) TDP-43 mRNA levels as measured by qPCR. n = 3 biological replicates of day 1 adult animals. Error bars represent mean +/− SEM, statistics was done by one-way ANOVA with post hoc Sidak comparisons indicated (*P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.00005 ns = not significant).
Discussion
This work addressed a gap in the field regarding whether granulins will continue to exert a negative effect on animal fitness and lysosomal function when in the presence of full-length progranulin. We found that reintroduction of PGRN-1 to granulin 3 overexpressing strains rescued lysosomal morphology but did not ameliorate the effects of granulin on aspartyl protease activity, development, stress resistance or TDP-43 toxicity. Progranulin has an established role in regulating the lysosomal enzyme CTSD, an aspartyl protease. Thus, it is likely that granulin 3 exerted its effects through suppression of CTSD activity, which would be consistent with our data.
While our experimental design focused on quantifying granulin 3 and PGRN-1 levels, granulin 1 and 2 are also cleavage products from PGRN-1. Thus, both pgrn-1 OE and pg/gr animals contained more total granulins than we measured, potentially 3 times. In the case of the pg/gr animals, this could explain the more severe phenotypes relative to gran3 OE animals, such as in development, ER stress resistance, and aspartyl protease activity. In addition, multi-granulin fragments have also been shown to promote CSTD maturation and activity [12]. This complexity was also not explicitly captured here.
Progranulin and granulins also exist extracellularly and exert effects on the immune system and in lipid metabolism [5, 8, 9, 25–30]. It’s possible that the phenotypes we measured were in part affected by these other biological processes. Nonetheless, this experimental approach provided a framework for studying progranulin and granulins in a quantitative, biologically relevant, in vivo paradigm. These findings confirmed the negative effects of granulins on animal fitness. Therefore, in addition to the progranulin repletion therapeutics approach [31–35], they suggest that granulin lowering strategies should also be considered as a site of therapeutic intervention.
Materials and Methods
Elegans husbandry and strains
Elegans strains were grown at 20C by standard procedures. Strains were provided by the Caenorhabditis Genetics Center (CGC) at the University of Minnesota. The N2E control strain was used as the wildtype strain. PHX7061 [Ppgrn-1a::rfp] was created by SunyBiotech and outcrossed 4x with the wildtype strain to make AWK600. The following strains were used:
CF3050 pgrn-1(tm985) I
CF3778 pgrn-1(tm985) I; muIs213[Ppgrn-1::pgrn-1::RFP]
AWK107 pgrn-1(tm985) I; rocIs5[Ppgrn-1+SignalSequence::granulin3::FLAG::polycistronic mCherry+Pmyo-2::GFP]
AWK485 pgrn-1(tm985) I; muIs213[Ppgrn-1::pgrn-1::RFP]; rocIs5[Ppgrn-1+SignalSequence::granulin3::FLAG::polycistronic mCherry+Pmyo-2::GFP]
RT258 unc-119(ed3) III; pwIs50 [lmp-1::GFP+Cbr-unc-119(+)]
AWK188 pgrn-1(tm985)I; unc-119(ed3) III; pwIs50 [lmp-1::GFP+Cbr-unc-119(+)]
AWK361 pgrn-1(tm985); muIs213[Ppgrn-1::pgrn-1::RFP]; unc-119(ed3) III; pwIs50 [lmp-1::GFP+Cbr-unc-119(+)]
AWK239 pgrn-1(tm985)I; unc-119(ed3) III; pwIs50 [lmp-1::GFP+Cbr-unc-119(+)]; rocIs5 [Ppgrn-1+SS::granulin3::FLAG::polycistronic mCherry+Pmyo-2::GFP]
AWK592 pgrn-1(tm985); muIs213[Ppgrn-1::pgrn-1::RFP]; unc-119(ed3) III; pwIs50 [lmp-1::GFP+Cbr-unc-119(+)]; rocIs5 [Ppgrn-1+SS::granulin3::FLAG::polycistronic mCherry+Pmyo-2::GFP]
AWK579 muIs206[Pegl-3::TDP-43::GFP]
CF3588 pgrn-1(tm985)I;muIs206[Pegl-3::TDP-43::GFP]
AWK576 pgrn-1(tm985)I;muIs206[Pegl-3::TDP-43::GFP]; muIs213[Ppgrn-1::pgrn-1::RFP]
AWK137 pgrn-1(tm985)I; muIs206[Pegl-3::TDP-43::GFP]; rocEx7[Ppgrn-1+SS::granulin3::FLAG::polycistronic mCherry+Pmyo-2::GFP]
AWK578 pgrn-1(tm985)I;muIs206[Pegl-3::TDP-43::GFP]; muIs213[Ppgrn-1::pgrn-1::RFP];rocEx7[Ppgrn-1+SS::granulin3::FLAG::polycistronic mCherry+Pmyo-2::GFP]
AWK153 asp-3(tm4450) X
Behavioral assays
ER stress. Eggs were transferred to 2.5 cm NGM plates with 5ug/ml tunicamycin or 0.01% DMSO. After 72 h, the fraction of L4 animals and the total number of animals were recorded. To adjust for development differences between strains, each strain’s fraction of L4 animals treated with tunicamycin was normalized to its DMSO condition. 3 replicates were recorded, each comprising of three 2.5 cm plates with 50 eggs for tunicamycin and DMSO treatment for each strain.
Development Assay. 50 eggs per replicate were placed on 2.5 cm NGM plates seeded with OP50. After 48 h, animals were scored by development stage.
Worm length. L4s animals were transferred to 2.5 cm NGM plates. Day 1 adult animals were imaged the following day. Day 5 animals were transferred every day for the next 5 days before imaging. Animals were imaged on a Zeiss AxioImager microscope. Worm length was measured in Fiji ImageJ by drawing a segmented line from the tail to head through the midline of the animal. 3 replicates were recorded with 10 animals for each strain.
Animal synchronization for western blot and enzyme activity assay
Forty to ninety adult animals were used to lay eggs for 6 h on a 10 cm NGM plate seeded with OP50. After 6 h, the adult animals were removed. The progeny were grown at 20C and visually checked after 3 and 4 days for development stage. When the progeny reached adulthood, the animals were collected and washed with M9, then frozen in 1x RIPA buffer (Alfa Aesar, Cat J62524) supplemented with Halt protease inhibitor cocktail (Thermo Scientific, Cat #PI87786) for Western Blot or frozen in 1% NP-40 (Thermo scientific, Cat #85124) for enzyme activity assays.
Western blot
Samples were lysed by sonication, then centrifuged at 21000rcf for 10 min at 4C. The supernatant was collected and the protein concentration measured by BCA (Pierce, Cat #PI23225). The linear range of detection for each protein was found to be around 10–30ug of lysate, verified by a serial dilution blot. Lysate was prepared with reducing agent (NuPAGE Sample Reducing Agent, Cat NP0009) and LDS buffer (NuPAGE LDS Sample Buffer, Cat NP0007), then boiled at 80C for 10 min. The samples were loaded onto a 4%–12% Bis Tris gel (NuPAGE) with MES buffer (NuPAGE), ran at 100 V until the dye front reached the end of the first stack, at which point voltage was increased to 130 V. Protein was transferred to a Nitrocellulose membrane at 300 mA for 60 min. Where applicable, membranes were stained for total protein using LiCOR’s REVERT kit (LiCOR LIC-929-11016). Briefly, membranes were dried at 37C for 10 min then rehydrated using TBS. Revert Total Protein Staining was applied for 5 min before two 30 s washes with the Wash buffer. Membranes were immediately imaged using LiCOR Odyssey. After imaging, membranes were stripped using the Destaining buffer for 2–3 min. Membranes were then blocked for 1 h using LiCOR buffer. Primary antibodies were applied overnight. Secondary antibodies were applied for one hour. Membranes were imaged on the LiCOR Odyssey machine and analyzed using ImageStudio Lite.
Recombinant standard curves
Recombinant RFP and FLAG curves were created by serial dilution. Dilution stocks were prepared with reducing agent (NuPAGE Sample Reducing Agent, Cat NP0009) and LDS buffer (NuPAGE LDS Sample Buffer, Cat NP0007), boiled, and stored at −80C to guarantee each experiment used the same dilution. The FLAG standard curve was created with recombinant FLAG Ubiquitin (Fischer, U12001M). The RFP standard curve was created with recombinant RFP (Novus, NBP1-99583).
RT-PCR
Forty to sixty adults were placed onto 10 cm NGM plates and removed after 5 h. The remaining eggs were allowed to grow to adulthood, around 72 to 85 h after the egg lay, depending on the line’s development rate. Animals were collected with M9 supplemented with 0.5% Polyethylene Glycol 8000 and washed twice. M9 supernatant was removed and 700ul of Qiazol (Qiagen Cat #79306) added. Worms were flash frozen and stored at −80C. RNA extraction was done with Qiagen miRNeasy kit’s tissue RNA extraction protocol. 1ug of RNA was reverse transcribed using SuperScript III Reverse Transcriptase (Life Technologies, Cat #18080044). RT-PCR was conducted with 1ul of cDNA and custom primers using Sybr Green (Life Technologies, Cat #4309155) on Applied Biosystem’s 7900 T. Ct values were calculated on the software using auto-thresholding and normalized to three housekeeping genes.
ASP-3 protease activity
ASP-3 activity was measured using the BioVision kit for human CTSD (Biovision, #K143-100). Samples were lysed by sonication, centrifuged at 21000rcf for 15 min at 4C. The supernatant was collected and the protein concentration measured by BCA (Pierce, Cat #PI23225). 0.25ug of total protein for each sample was used. Each biological triplicate was measured with a technical triplicate. Measurements were taken across 2 h at 25C on an Infinite M Plex plate reader (Tecan, 30190085). Enzyme activity rate was calculated using the slope of the linear region for each condition.
Antibodies
The following antibodies were used: M2 Flag antibody (Sigma, F3165) at 1:1000 dilution, RFP6g6 antibody (Chromotek, 6G6-150) at 1:1000 dilution, anti-TDP-43 antibody (Abcam, ab57105) at 1:1000 dilution, anti-actin C4 antibody (Millipore, MAB1501) at 1:1000 dilution, and anti-granulin 3b antibody (Biomatik, rb2587) at 1:200 dilution.
qPCR primers
To measure gran3::flag mRNA, the forward primer was 5′-ccatctgctgtgagaacact-3′ and the reverse primer was 5′-CCTaccggtacatCAACTTTGTA-3′. To measure tdp-43 mRNA, the forward primer was 5′-gtgctgctctccacggttac-3′ and the reverse primer was 5′-ttctaccagccggacacctc-3′. Three housekeeping genes were used as previously described20. For cdc-42, the forward primer was 5′-ctgctggacaggaagattacg-3′ and the reverse primer was 5′-ctcggacattctcgaatgaag-3′. For pmp-3, the forward primer was 5′-gttcccgtgttcatcactcat-3′ and the reverse primer was 5′-acaccgtcgagaagctgtaga-3′. For y510d0.4, the forward primer was 5′-gtcgcttcaaatcagttcagc-3′ and the reverse primer was 5′-gttcttgtcaagtgatccgaca-3′.
Supplementary Material
Contributor Information
Austin L Wang, Memory and Aging Center, Weill Institute for Neuroscience, Department of Neurology, University of California, San Francisco, San Francisco, CA, United States.
Edwina A Mambou, Memory and Aging Center, Weill Institute for Neuroscience, Department of Neurology, University of California, San Francisco, San Francisco, CA, United States.
Aimee W Kao, Memory and Aging Center, Weill Institute for Neuroscience, Department of Neurology, University of California, San Francisco, San Francisco, CA, United States; Bakar Aging Research Institute, University of California, San Francisco, San Francisco, CA, United States.
Conflict of interest statement: Unrelated to this work, A.W.K. serves on the Scientific Advisory Board for Nine Square Therapeutics and has received a research award from Eli Lilly.
Funding
This work is supported by funding from the National Institutes of Health R01NS095257, RF1NS127414 and the Bakar Aging Research Institute grants (to A.W.K.) and 3RF1NS127414-01S1 (to E.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
Manuscript data will be made available upon reasonable request to the Corresponding Author.
References
- 1. Kao AW, McKay A, Singh PP. et al. Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci 2017;18:325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith KR, Damiano J, Franceschetti S. et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 2012;90:1102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gass J, Cannon A, Mackenzie IR. et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet 2006;15:2988–3001. [DOI] [PubMed] [Google Scholar]
- 4. Baker M, Mackenzie IR, Pickering-Brown SM. et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006;442:916–9. [DOI] [PubMed] [Google Scholar]
- 5. Bateman A, Bennett HPJ. The granulin gene family: from cancer to dementia. BioEssays 2009;31:1245–54. [DOI] [PubMed] [Google Scholar]
- 6. Cruts M, Gijselinck I, van der Zee J. et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006;442:920–4. [DOI] [PubMed] [Google Scholar]
- 7. Cenik B, Sephton CF, Kutluk Cenik B. et al. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem 2012;287:32298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu F, Padukkavidana T, Vægter CB. et al. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 2010;68:654–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou X, Sun L, Bastos de Oliveira F. et al. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol 2015;210:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohan S, Sampognaro PJ, Argouarch AR. et al. Processing of progranulin into granulins involves multiple lysosomal proteases and is affected in frontotemporal lobar degeneration. Mol Neurodegener 2021;16:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holler CJ, Taylor G, Deng Q. et al. Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro 2017;4:ENEURO.0100-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butler VJ, Cortopassi WA, Argouarch AR. et al. Progranulin stimulates the in vitro maturation of pro-cathepsin D at acidic PH. J Mol Biol 2019;431:1038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Valdez C, Wong YC, Schwake M. et al. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet 2017;26:4861–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Butler VJ, Cortopassi WA, Pierce OM. et al. Multi-granulin domain peptides bind to pro-cathepsin D and stimulate its enzymatic activity in vitro. Biochemistry 2019;58:2670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kessenbrock K, Fröhlich L, Sixt M. et al. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J Clin Invest 2008;118:2438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med 2003;81:600–12. [DOI] [PubMed] [Google Scholar]
- 17. Tolkatchev D, Malik S, Vinogradova A. et al. Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci 2008;17:711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu J, Nathan C, Jin W. et al. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 2002;111:867–78. [DOI] [PubMed] [Google Scholar]
- 19. Butler VJ, Gao F, Corrales CI. et al. Age- and stress-associated C. elegans granulins impair lysosomal function and induce a compensatory HLH-30/TFEB transcriptional response. PLoS Genet 2019;15:e1008295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salazar DA, Butler VJ, Argouarch AR. et al. The progranulin cleavage products, granulins, exacerbate TDP-43 toxicity and increase TDP-43 levels. J Neurosci 2015;35:9315–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calfon M, Zeng H, Urano F. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 MRNA. Nature 2002;415:92–6. [DOI] [PubMed] [Google Scholar]
- 22. Lin T, Lee JE, Kang JW. et al. Endoplasmic reticulum (ER) stress and unfolded protein response (UPR) in mammalian oocyte maturation and preimplantation embryo development. Int J Mol Sci 2019;20:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee W-S, Yoo W-H, Chae H-J. ER stress and autophagy. Curr Mol Med 2015;15:735–45. [DOI] [PubMed] [Google Scholar]
- 24. Zhang J, Velmeshev D, Hashimoto K. et al. Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency. Nature 2020;588:459–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serrero G. Progranulin/GP88, a complex and multifaceted player of tumor growth by direct action and via the tumor microenvironment. Adv Exp Med Biol 2021;1329:475–98. [DOI] [PubMed] [Google Scholar]
- 26. Boland S, Swarup S, Ambaw YA. et al. Deficiency of the frontotemporal dementia gene GRN results in gangliosidosis. Nat Commun 2022;13:5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Houser MC, Uriarte Huarte O, Wallings RL. et al. Progranulin loss results in sex-dependent dysregulation of the peripheral and central immune system. Front Immunol 2022;13:105647. 10.3389/fimmu.2022.1056417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Devireddy S, Ferguson SM. Efficient progranulin exit from the ER requires its interaction with prosaposin, a Surf4 cargo. J Cell Biol 2022;221:e202104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marschallinger J, Iram T, Zardeneta M. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat Neurosci 2020;23:194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evers BM, Rodriguez-Navas C, Tesla RJ. et al. Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep 2017;20:2565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davis SE, Roth JR, Aljabi Q. et al. Delivering progranulin to neuronal lysosomes protects against excitotoxicity. J Biol Chem 2021;297:100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Logan T, Simon MJ, Rana A. et al. Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell 2021;184:4651–4668.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arrant AE, Filiano AJ, Unger DE. et al. Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain 2017;140:1447–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Minami SS, Min S-W, Krabbe G. et al. Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat Med 2014;20:1157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith DM, Niehoff ML, Ling K. et al. Targeting nonsense-mediated RNA decay does not increase progranulin levels in the Grn R493X mouse model of frontotemporal dementia. PLoS One 2023;18:e0282822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Manuscript data will be made available upon reasonable request to the Corresponding Author.