Summary
When evaluating the effectiveness of a treatment, policy, or intervention, the desired measure of efficacy may be expensive to collect, not routinely available, or may take a long time to occur. In these cases, it is sometimes possible to identify a surrogate outcome that can more easily, quickly, or cheaply capture the effect of interest. Theory and methods for evaluating the strength of surrogate markers have been well studied in the context of a single surrogate marker measured in the course of a randomized clinical study. However, methods are lacking for quantifying the utility of surrogate markers when the dimension of the surrogate grows. We propose a robust and efficient method for evaluating a set of surrogate markers that may be high-dimensional. Our method does not require treatment to be randomized and may be used in observational studies. Our approach draws on a connection between quantifying the utility of a surrogate marker and the most fundamental tools of causal inference—namely, methods for robust estimation of the average treatment effect. This connection facilitates the use of modern methods for estimating treatment effects, using machine learning to estimate nuisance functions and relaxing the dependence on model specification. We demonstrate that our proposed approach performs well, demonstrate connections between our approach and certain mediation effects, and illustrate it by evaluating whether gene expression can be used as a surrogate for immune activation in an Ebola study.
Keywords: Average treatment effect estimation, High-dimensional data, Surrogate marker evaluation
1. Introduction
When evaluating the effectiveness of a treatment, policy, or intervention, the desired measure of effectiveness may be expensive to collect, not routinely available, or may take a long time to occur. In these cases, it is sometimes possible to identify a surrogate outcome that can more easily, quickly, or cheaply capture the effect of interest. For example, when evaluating an intervention designed to delay dementia onset, the time required to observe enough dementia diagnoses is often very long, and surrogates that have been considered for intervention evaluation include mild cognitive impairment, adiponectin levels, neuroimages, amyloid plaques, and neurofibrillary tangles (Small, 2006; Teixeira and others 2013). Similarly, in studies evaluating treatments to prevent diabetes, surrogate measures for diabetes onset have included changes in body weight, fasting plasma glucose, and hemoglobin A1c (Caveney and Cohen, 2011; Choi and others 2011). And it is presently obvious that a surrogate marker for SARS-CoV-2 vaccine efficacy is required to develop the second generation of vaccines adapted to novel variants of concern (Karim, 2021).
Theory and methods for evaluating the strength of surrogate markers have been well studied in the context of a single surrogate marker measured in the course of one or more randomized clinical studies—see the detailed review in Joffe and Greene (2009). In particular, model-based approaches have been proposed for continuous (Alonso and others, 2004) and binary surrogates (Alonso and others, 2016) in meta-analysis and in the principal stratification framework (Gilbert and Hudgens, 2008). Robust nonparametric methods have been proposed in Parast and others (2020) and Parast and others (2016). However, fully nonparametric methods are not available or not reliable when the number of markers is more than one or two. In these cases, a parametric approach to evaluate a high-dimensional surrogate may be considered as proposed in Zhou and others (2022), which requires the correct specification of two high-dimensional linear models. Alternatively, an initial model may be used to reduce the dimensionality of the surrogate (Agniel and Parast, 2021; Parast and others 2016). However, the mis-specification of the parametric models or an inappropriate initial model may produce badly biased estimates of the utility of the surrogate.
In this work, we propose a robust and efficient method for evaluating multiple surrogate markers, with particular attention to the possibility that surrogates may be high-dimensional. Our approach revives the spirit of the estimator in Freedman and others (1992), itself based on the pioneering work of Prentice (1989) to draw on a connection between quantifying the utility of a surrogate marker and the most fundamental tools of causal inference: namely, methods for estimating the average treatment effect. This connection does not appear to have been made before in the surrogate marker literature, despite the extensive connections made between mediation and surrogacy (Taylor and others, 2005; Joffe and Greene, 2009). Making this connection facilitates the use of all of the machinery now available to robustly and efficiently estimate average treatment effects. Specifically, we take advantage of state-of-the-art methods for incorporating flexible machine learning and/or sparse high-dimensional models into the estimation of treatment effects. These methods based estimation on the efficient influence function of key quantities and use sample splitting to ensure convenient asymptotic distributions and inference while putting minimal restrictions on what types of estimators may be used. Furthermore, we show that our proposed approach has connections to certain mediation estimands when the assumptions underlying mediation analysis are satisfied.
One key benefit of our approach is that it does not require randomization of treatment, which has been required by previous nonparametric approaches to evaluating surrogate markers (Agniel and Parast, 2021; Parast and others, 2016). Assessments of surrogate markers are not solely limited to studies of randomized treatment (Obirikorang and others, 2012). In fact, using surrogate outcomes based on complex observational data is finding purchase in all corners of science (Wang and others, 2020). In Section 6, we give an example where researchers are interested in studying whether gene expression can be used as a surrogate for immune activation in an observational setting. Our approach exploits insights from observational causal inference to estimate all relevant quantities and thus is equally applicable in both observational and randomized settings.
The structure of the rest of the article is as follows. In Section 2, we lay out the notation and the setting in which we are working, and we motivate the importance of evaluating surrogate markers in light of recent advances in the surrogate marker literature. In Section 3, we detail assumptions necessary for identifying and interpreting parameters of interest. We discuss the identification, estimation, and the asymptotic behavior of our estimator in Section 4 and discuss variance estimation and inference. We evaluate the performance of the proposed approach using a simulation study in Section 5, and in Section 6, we investigate whether gene expression could be used as a surrogate for immune response to Ebola infection. We give final remarks and draw connections to other methods for evaluating surrogate markers in Section 7.
2. Evaluation of surrogate markers
2.1. Notation
Let  denote a binary treatment, and let the primary outcome of the study be
denote a binary treatment, and let the primary outcome of the study be  . Let there be a vector
. Let there be a vector 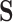 of potential surrogate information, and let
of potential surrogate information, and let 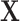 be a vector of pretreatment covariates. The primary quantity of interest is the treatment effect on the outcome
be a vector of pretreatment covariates. The primary quantity of interest is the treatment effect on the outcome 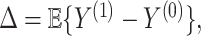 where
where 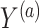 is the potential or counterfactual outcome that would have been observed if treatment were
is the potential or counterfactual outcome that would have been observed if treatment were 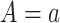 , possibly contrary to fact. Similarly, let
, possibly contrary to fact. Similarly, let 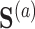 be the potential/counterfactual value the vector of surrogates would take if
be the potential/counterfactual value the vector of surrogates would take if 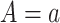 . Let the data observed in the current study be
. Let the data observed in the current study be 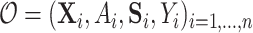 ,
, 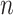 iid realizations of
iid realizations of 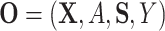 .
.
2.2. Importance of quantifying surrogate strength
Understanding the strength of a potential surrogate is important for many reasons. In practice, information about surrogate strength will inform decisions regarding whether to measure the surrogate in a future study (especially if it is costly or invasive to measure) and/or whether to use the surrogate to assess treatment effectiveness in a future study. In addition, a number of novel statistical methods for using surrogates in future studies can only be applied when the surrogate is strong. For example, Parast and others (2019) propose a robust nonparametric procedure to test for a treatment effect using surrogate marker information measured prior to the end of the study in a time-to-event outcome setting, but they rely on the assumption that the surrogate is sufficiently strong. As another example, Price and others (2018) propose constructing a function 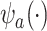 of the surrogates that leads to some optimality properties, but it can be shown that this method should only be used with a strong surrogate.
of the surrogates that leads to some optimality properties, but it can be shown that this method should only be used with a strong surrogate.
Specifically, they construct an optimal transformation of the surrogates 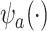 in terms of minimizing the following mean squared error
in terms of minimizing the following mean squared error
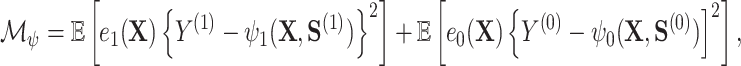 |
(2.1) |
with 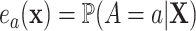 , under the constraint that it satisfies the so-called Prentice definition:
, under the constraint that it satisfies the so-called Prentice definition:
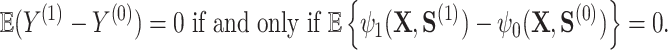 |
The optimal transformations in this case are shown to be 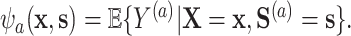 In addition to this appealing optimality property, this proposal also resolves the so-called “surrogate paradox.” The paradox states that the treatment could have a positive causal effect on a univariate surrogate
In addition to this appealing optimality property, this proposal also resolves the so-called “surrogate paradox.” The paradox states that the treatment could have a positive causal effect on a univariate surrogate 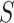 which could have a positive correlation with the outcome, but the treatment could yet have a negative effect on the outcome. The Price surrogate resolves the surrogate paradox by definition. The treatment effect on the transformed surrogate is by definition equivalent to the treatment effect on the outcome:
which could have a positive correlation with the outcome, but the treatment could yet have a negative effect on the outcome. The Price surrogate resolves the surrogate paradox by definition. The treatment effect on the transformed surrogate is by definition equivalent to the treatment effect on the outcome: 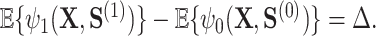
However, the Price surrogate’s resolution of the surrogate paradox is in some sense too good. The surrogate paradox is thus resolved for every potential set of surrogates, good and bad. Even if the surrogates 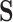 are completely unrelated to the outcome
are completely unrelated to the outcome 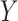 ,
, 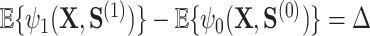 . In fact, the power to detect a treatment effect in a future study using
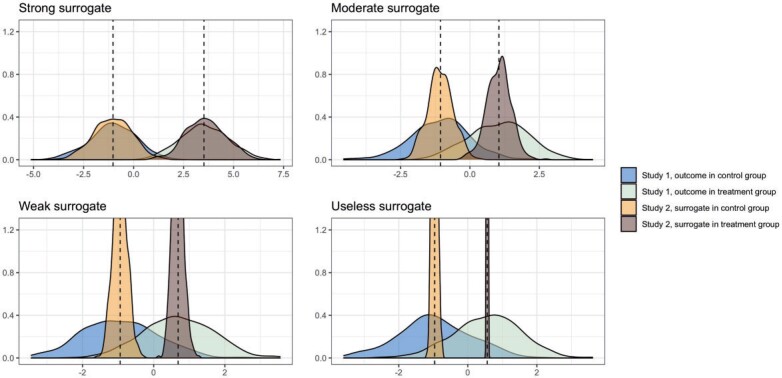
. In fact, the power to detect a treatment effect in a future study using 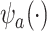 actually increases as the surrogate becomes weaker (explaining less of the treatment effect), if the Prentice definition does not hold. Consider the toy example depicted in Figure 1 (see Appendix A of the Supplementary material available at Biostatistics online for details of the data generation). As the strength of the surrogate increases,
actually increases as the surrogate becomes weaker (explaining less of the treatment effect), if the Prentice definition does not hold. Consider the toy example depicted in Figure 1 (see Appendix A of the Supplementary material available at Biostatistics online for details of the data generation). As the strength of the surrogate increases, 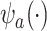 becomes more like the true outcome
becomes more like the true outcome 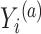 . As the surrogate becomes weaker,
. As the surrogate becomes weaker, 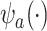 becomes more like
becomes more like 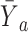 the mean outcome in treatment group
the mean outcome in treatment group 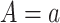 from the first study. This means that a weak surrogate ensures that the treatment effect in the second study will be identical to the treatment effect in the first study because the distribution of
from the first study. This means that a weak surrogate ensures that the treatment effect in the second study will be identical to the treatment effect in the first study because the distribution of 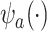 will cluster closely around the estimate of
will cluster closely around the estimate of 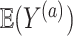 from the first study (see specifically, the fourth panel of Figure 1). In this scenario, the second study is providing no new information about the treatment. Thus, the Price surrogate should only be used with a strong
from the first study (see specifically, the fourth panel of Figure 1). In this scenario, the second study is providing no new information about the treatment. Thus, the Price surrogate should only be used with a strong 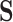 , and its use with a weak or possibly even moderately strong set of surrogates may not be advisable.
, and its use with a weak or possibly even moderately strong set of surrogates may not be advisable.
Fig. 1.
Distribution of outcomes 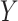 in an initial study (Study 1) and surrogate functions (approximating the outcome)
in an initial study (Study 1) and surrogate functions (approximating the outcome) 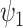 (surrogate in the treatment group) and
(surrogate in the treatment group) and 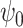 (surrogate in the control group) in a future study (Study 2) based on simulated data, arranged according to the strength of the surrogate marker—i.e., the capacity of the surrogate to explain the treatment effect. The wider distribution on the left corresponds to the control group in Study 1, the wider distribution on the right to the treated group in Study 1, the narrower distribution on the left to
(surrogate in the control group) in a future study (Study 2) based on simulated data, arranged according to the strength of the surrogate marker—i.e., the capacity of the surrogate to explain the treatment effect. The wider distribution on the left corresponds to the control group in Study 1, the wider distribution on the right to the treated group in Study 1, the narrower distribution on the left to 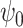 in Study 2, and the narrower distribution on the right to
in Study 2, and the narrower distribution on the right to 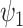 in Study 2. Vertical dotted lines correspond to the means of the treatment groups in the first study. See Appendix A of the Supplementary material available at Biostatistics online for simulation details. Surrogate functions
in Study 2. Vertical dotted lines correspond to the means of the treatment groups in the first study. See Appendix A of the Supplementary material available at Biostatistics online for simulation details. Surrogate functions 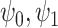 were estimated via a correctly specified linear regression in the first study and then applied to the second study population. When the surrogate was strong, the distribution of the surrogate in the second study was quite close to the distribution of the outcome in the first study. However, when the surrogate was weak, the distribution of the surrogate in the second study clustered around the group mean from the first study.
were estimated via a correctly specified linear regression in the first study and then applied to the second study population. When the surrogate was strong, the distribution of the surrogate in the second study was quite close to the distribution of the outcome in the first study. However, when the surrogate was weak, the distribution of the surrogate in the second study clustered around the group mean from the first study.
Therefore, for both decision-making purposes and to use statistical methods that take advantage of strong surrogates, methods are needed to rigorously quantify the strength of the proposed set of surrogates before using (functions of) the surrogates in practice. In the following section, we propose an efficient method for estimating the strength of a possibly high-dimensional set of surrogates in observational studies. Our aim is for this proposed method to complement existing work, like that of Parast and others (2019) and Price and others (2018), that rely on the availability of a strong surrogate but currently lack tools to assess surrogate strength, particularly in a high-dimensional surrogate setting.
2.3. Quantity to evaluate surrogate strength
In this section, we present a general method for evaluating the usefulness of a set of surrogates. This can be used to evaluate the suitability of any set of surrogates or surrogate transformations and is appropriate for 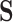 of any dimension. To evaluate the surrogates’ usefulness, we use the proportion of treatment effect explained by
of any dimension. To evaluate the surrogates’ usefulness, we use the proportion of treatment effect explained by 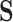 (PTE) defined as
(PTE) defined as 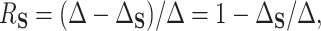 with
with 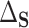 the residual treatment effect, or the treatment effect that remains after controlling for the surrogate information. This measure has been used frequently in the evaluation of surrogate markers, as far back as the pioneering works of Prentice (1989) and Freedman and others (1992) and more recently in Parast and others (2020; 2016) and Agniel and Parast (2021). If
the residual treatment effect, or the treatment effect that remains after controlling for the surrogate information. This measure has been used frequently in the evaluation of surrogate markers, as far back as the pioneering works of Prentice (1989) and Freedman and others (1992) and more recently in Parast and others (2020; 2016) and Agniel and Parast (2021). If 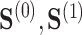 are independent of
are independent of 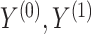 conditional on
conditional on  , then
, then  and
and 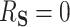 . In contrast, if all of the treatment effects can be attributed to
. In contrast, if all of the treatment effects can be attributed to 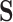 , then
, then 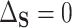 and
and 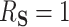 .
.
In the more recent works using this PTE, 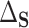 is defined in terms of a particular reference distribution, e.g., taking
is defined in terms of a particular reference distribution, e.g., taking 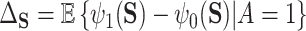 to be the residual treatment effect among the treated group. This choice of reference distribution is often arbitrary and was not required by the older (model-based) approaches. To obviate the need for this choice of reference distribution and to take advantage of the extensive development in average treatment effect estimation (and the associated intuition and machinery that have been built up around it), we define
to be the residual treatment effect among the treated group. This choice of reference distribution is often arbitrary and was not required by the older (model-based) approaches. To obviate the need for this choice of reference distribution and to take advantage of the extensive development in average treatment effect estimation (and the associated intuition and machinery that have been built up around it), we define 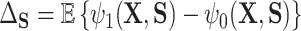 , the average treatment effect conditional on the distribution of
, the average treatment effect conditional on the distribution of 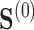 and
and 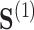 both being equal to the distribution of
both being equal to the distribution of 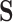 (all conditional on
(all conditional on 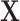 ). Other choices of reference distribution could be used when there are substantive reasons to prefer them. When there are no interactions with treatment –
). Other choices of reference distribution could be used when there are substantive reasons to prefer them. When there are no interactions with treatment – 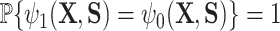 – choice of reference distribution does not change
– choice of reference distribution does not change 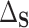 . See Section 7 for further connections in this vein to Agniel and Parast (2021) and similar recent methods.
. See Section 7 for further connections in this vein to Agniel and Parast (2021) and similar recent methods.
This formulation of PTE also has deep connections to quantities important to mediation analysis without requiring some of the restrictive assumptions common in the mediation literature. In particular, we show in Appendix B of the Supplementary material available at Biostatistics online that 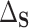 is a function of conditional natural direct effects and that
is a function of conditional natural direct effects and that 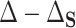 is a function of conditional natural indirect effects (Joffe and Greene, 2009; VanderWeele 2013), when the assumptions for identifying those effects are met. While many previous reviews of surrogate methods identify the causal effects framework for surrogate evaluation solely with mediation (Joffe and Greene, 2009; Conlon and others, 2017), we use causal tools without requiring all of the assumptions necessary for mediation. Importantly, we do not require that all confounders of the surrogate–outcome relationship are measured and included in the study because the aims of mediation and surrogate marker evaluation are different (VanderWeele, 2013). The aim of identifying a mediator is determining whether the effect of treatment operates through the mediator itself, e.g., through some biological pathway. Often, a good surrogate marker is similarly conceptualized as a variable through which the treatment operates, but this is not necessarily required; a variable can be a good surrogate if it captures the treatment effect on the outcome, even if the treatment effect does not operate through the variable itself (sometimes called a nonmechanistic correlate of protection; Plotkin and Gilbert, 2012).
is a function of conditional natural indirect effects (Joffe and Greene, 2009; VanderWeele 2013), when the assumptions for identifying those effects are met. While many previous reviews of surrogate methods identify the causal effects framework for surrogate evaluation solely with mediation (Joffe and Greene, 2009; Conlon and others, 2017), we use causal tools without requiring all of the assumptions necessary for mediation. Importantly, we do not require that all confounders of the surrogate–outcome relationship are measured and included in the study because the aims of mediation and surrogate marker evaluation are different (VanderWeele, 2013). The aim of identifying a mediator is determining whether the effect of treatment operates through the mediator itself, e.g., through some biological pathway. Often, a good surrogate marker is similarly conceptualized as a variable through which the treatment operates, but this is not necessarily required; a variable can be a good surrogate if it captures the treatment effect on the outcome, even if the treatment effect does not operate through the variable itself (sometimes called a nonmechanistic correlate of protection; Plotkin and Gilbert, 2012).
3. Assumptions
3.1. Identifying assumptions
We first require 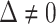 , without which requirement the goals of identifying surrogate markers are practically and theoretically not meaningful. We further make the three typical assumptions of treatment effect estimation: consistency, positivity, and no unmeasured confounding. Specifically, we assume that the observed values of
, without which requirement the goals of identifying surrogate markers are practically and theoretically not meaningful. We further make the three typical assumptions of treatment effect estimation: consistency, positivity, and no unmeasured confounding. Specifically, we assume that the observed values of 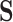 and
and 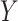 when
when 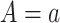 are identical to the counterfactuals
are identical to the counterfactuals 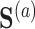 and
and 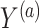 such that
such that 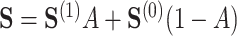 and
and 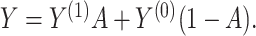 We furthermore assume that
We furthermore assume that 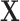 contains all confounders of the effects of
contains all confounders of the effects of 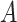 on the surrogates and the outcome, such that the treatment
on the surrogates and the outcome, such that the treatment 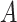 is as good as randomized conditional on the covariates
is as good as randomized conditional on the covariates 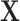 : (A.1)
: (A.1) 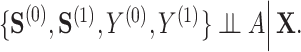 In addition, we assume two forms of positivity, which ensures that individuals in the two study arms are not too different from one another. First, the usual positive probability of receiving either treatment for some
In addition, we assume two forms of positivity, which ensures that individuals in the two study arms are not too different from one another. First, the usual positive probability of receiving either treatment for some 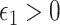 ,
, 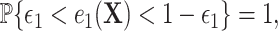 and a related assumption that further conditions on the surrogates,
and a related assumption that further conditions on the surrogates, 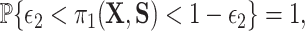 for some
for some 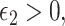 where
where 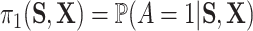 . Notably, letting
. Notably, letting 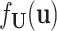 be the density of the random variable
be the density of the random variable 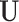 evaluated at
evaluated at 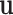 , because
, because 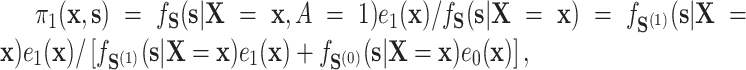 these two positivity conditions ensure that the conditional distribution of the counterfactual surrogates under treatment and control cannot be too different from one another, i.e., ensuring overlap. When the treatment has a large effect on
these two positivity conditions ensure that the conditional distribution of the counterfactual surrogates under treatment and control cannot be too different from one another, i.e., ensuring overlap. When the treatment has a large effect on 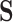 , this additional overlap requirement may be suspect—e.g., there may be some values of
, this additional overlap requirement may be suspect—e.g., there may be some values of 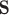 such that
such that 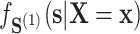 approaches 0 and thus
approaches 0 and thus 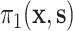 also approaches 0. We discuss a generalization of our approach that removes this overlap condition in Appendix C of the Supplementary material available at Biostatistics online.
also approaches 0. We discuss a generalization of our approach that removes this overlap condition in Appendix C of the Supplementary material available at Biostatistics online.
3.2. Required assumptions to ensure interpretation of 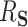 as a proportion
as a proportion
The interpretation of 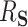 as the PTE depends on it actually being a proportion, lying between 0 and 1. We adapt conditions in Wang and Taylor (2002) and Agniel and Parast (2021) which ensure that
as the PTE depends on it actually being a proportion, lying between 0 and 1. We adapt conditions in Wang and Taylor (2002) and Agniel and Parast (2021) which ensure that 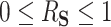 . These conditions are as follows (assuming without loss of generality that
. These conditions are as follows (assuming without loss of generality that 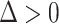 ): (A.2)
): (A.2)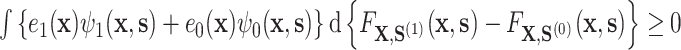 ; and (A.3)
; and (A.3) 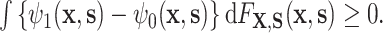 The condition (A.3) ensures that
The condition (A.3) ensures that 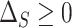 , i.e., that the residual treatment effect is in the same direction as the overall treatment effect
, i.e., that the residual treatment effect is in the same direction as the overall treatment effect 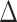 . Condition (A.2), which ensures that
. Condition (A.2), which ensures that 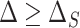 , requires that, roughly speaking, a propensity-weighted mixture of the two conditional mean functions
, requires that, roughly speaking, a propensity-weighted mixture of the two conditional mean functions 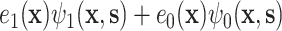 is larger when
is larger when 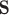 takes values from the distribution of the counterfactual surrogates under treatment than if it took values from the distribution under control. These two conditions are analogs of conditions (A6) and (A7) in Agniel and Parast (2021). See Appendix D of the Supplementary material available at Biostatistics online for an alternative approach that may be considered if these conditions are not or unlikely to be met, though the alternative approach lacks the interpretability and connections to previous literature of the proposed approach.
takes values from the distribution of the counterfactual surrogates under treatment than if it took values from the distribution under control. These two conditions are analogs of conditions (A6) and (A7) in Agniel and Parast (2021). See Appendix D of the Supplementary material available at Biostatistics online for an alternative approach that may be considered if these conditions are not or unlikely to be met, though the alternative approach lacks the interpretability and connections to previous literature of the proposed approach.
4. Identification, estimation, and inference
4.1. Identifiability
In this section, we show that the effects of interest are identifiable, propose a robust and efficient estimation procedure, and describe the asymptotic properties of our proposed estimators. The effects of interest may be identified as average treatment effects identifiable from the data, one of which conditions on the surrogates (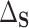 ) and one which does not (
) and one which does not (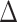 ). In particular, the residual treatment effect may be identified as
). In particular, the residual treatment effect may be identified as
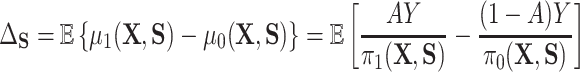 |
(4.2) |
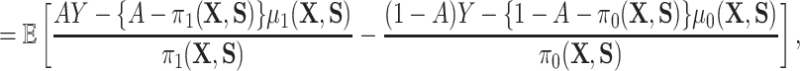 |
(4.3) |
where 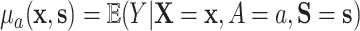 . The result in the first equality of (4.2) follows from the definition of
. The result in the first equality of (4.2) follows from the definition of 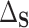 and the fact that
and the fact that 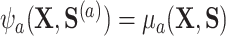 because of (A.1), and the other results follow shortly thereafter, following familiar paths as arguments for the identification of the average treatment effect in other contexts. These results show that the residual treatment effect may be identified without knowledge of the mean function for the outcome via the second equality in (4.2) and gives an augmented inverse probability weighting (Bang and Robins, 2005) version of the estimand (4.3).
because of (A.1), and the other results follow shortly thereafter, following familiar paths as arguments for the identification of the average treatment effect in other contexts. These results show that the residual treatment effect may be identified without knowledge of the mean function for the outcome via the second equality in (4.2) and gives an augmented inverse probability weighting (Bang and Robins, 2005) version of the estimand (4.3). 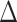 may also be identified, following similar standard arguments and using similar functionals with
may also be identified, following similar standard arguments and using similar functionals with 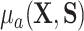 replaced by
replaced by 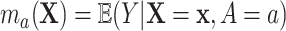 and
and 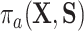 replaced by
replaced by 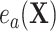 .
.
4.2. Proposed estimation
This identification of 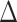 and
and 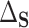 in terms of average treatment effects allows us to take advantage of the rich literature on robust estimation of these quantities. We propose to estimate
in terms of average treatment effects allows us to take advantage of the rich literature on robust estimation of these quantities. We propose to estimate 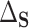 and
and 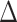 as
as
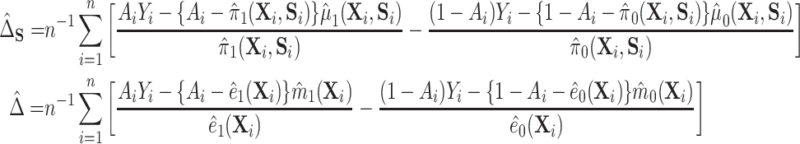 |
and thus, estimate  as
as  , where we leave estimation of the components:
, where we leave estimation of the components:  ,
, 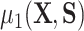 ,
, 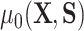 ,
, 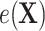 ,
, 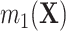 ,
, 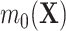 to be quite general. We propose and evaluate two different versions of this proposed estimator: one where we estimate these components using the Super Learner (Van der Laan and others, 2007), which finds an optimal combination of a set of candidate models or learners (denoted “DR-SL”) and another that uses the relaxed lasso (Meinshausen, 2007) (denoted “DR-lasso”). In addition, our proposed estimators use a sample-splitting scheme (Chernozhukov and others, 2017), which we describe in detail in Appendix C of the Supplementary material available at Biostatistics online, to avoid placing restrictive conditions on the estimation of the nuisance functions.
to be quite general. We propose and evaluate two different versions of this proposed estimator: one where we estimate these components using the Super Learner (Van der Laan and others, 2007), which finds an optimal combination of a set of candidate models or learners (denoted “DR-SL”) and another that uses the relaxed lasso (Meinshausen, 2007) (denoted “DR-lasso”). In addition, our proposed estimators use a sample-splitting scheme (Chernozhukov and others, 2017), which we describe in detail in Appendix C of the Supplementary material available at Biostatistics online, to avoid placing restrictive conditions on the estimation of the nuisance functions.
Remark 1.
While
depends on the counterfactual quantities
and is thus not the same target treatment effect estimated by Chernozhukov and others (2017), because it is identified by the same observed data quantities in (4.3), we can use the same machinery used in Chernozhukov and others (2017), though requiring the stronger assumption (A.1).
4.3. Inference
Under very general conditions, including the standard causal assumptions identified in Section 3.1, 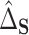 will converge at the parametric
will converge at the parametric 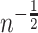 rate and be asymptotically normal (Chernozhukov and others, 2017; Farrell, 2015) so long as
rate and be asymptotically normal (Chernozhukov and others, 2017; Farrell, 2015) so long as
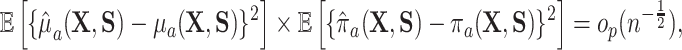 |
(4.4) |
with similar results holding for 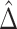 if
if
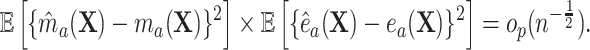 |
(4.5) |
As these estimators are built on the efficient influence functions, they are also semiparametrically efficient. Furthermore, as we show in Appendix C of the Supplementary material available at Biostatistics online, as long as the sample splits are the same for 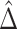 and
and 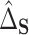 , we will have
, we will have 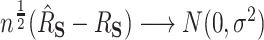 if both (4.4) and (4.5) hold. Specifically, we have that
if both (4.4) and (4.5) hold. Specifically, we have that
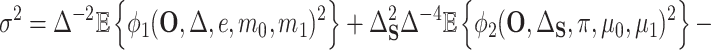 |
(4.6) |
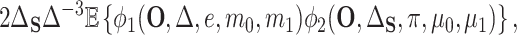 |
(4.7) |
where
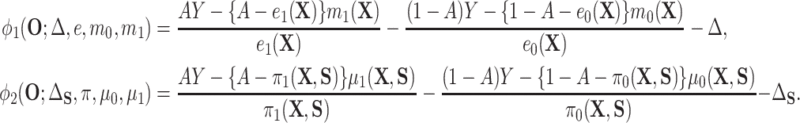 |
Remark 2.
When the dimension of
is not high, (4.4) and (4.5) are not in general restrictive, as many parametric and nonparametric methods are able to achieve the slow rates required of the estimators under certain conditions, including the lasso (Tibshirani, 1996), random forests (Wager and Walther, 2015), and deep neural networks (Farrell and others, 2021), when used with a sample-splitting scheme. Ensemble methods such as the Super Learner (Van der Laan and others, 2007) may also be used to combine these and other methods to obtain good performance if any of the methods achieves the required rates of consistency. As described in the prior section, our proposed approach combines both ensemble estimation of nuisance functions and sample splitting; we demonstrate the good performance of this approach in finite samples in Section 5.
In higher dimensions, the convergence in (4.4) and (4.5) is more difficult to ensure without further restrictions. If only
is high-dimensional, then a typical approach is to assume that
may be specified as a sparse linear model
and
may be specified as a sparse logistic regression model
for
, and with
sparse enough to ensure (4.4). Because of the fact that
is related to
, restricting
to this class of models is unproblematic so long as
is low-dimensional and
may be estimated nonparametrically (i.e., without being restricted to the class of sparse linear models). However, if
is also high-dimensional, sparse logistic models for
and
may not in general be compatible with one another because of the well-known noncollapsibility of logistic regression (Guo and Geng, 1995), unless
(which would imply
) or
(which would imply that
does not confound the relationship between
and
). In these cases, the approach of Tan (2020) may be used to estimate the nuisance functions, in which case only one of
and
are required to be a correctly specified logistic regression. However, this approach did not appear to perform well in our simulations in Section 5. Despite these theoretical considerations, our simulations suggest that an ensemble approach using the Super Learner may outperform approaches based on sparse linear models, as in Tan (2020) or in the version of our estimator that uses only lasso regression. More theoretical development in this area may be warranted.
5. Simulations
5.1. Simulation overview
Our proposed estimator based on the Super Learner (“DR-SL”) is implemented using the 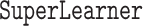 package (Polley and others, 2021) in
package (Polley and others, 2021) in  . The candidate learners we included were the lasso, ridge regression, ordinary least squares, support vector machines, and random forests for
. The candidate learners we included were the lasso, ridge regression, ordinary least squares, support vector machines, and random forests for 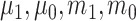 and the lasso, logistic regression, linear discriminant analysis, quadratic discriminant analysis, support vector machines, and random forests for
and the lasso, logistic regression, linear discriminant analysis, quadratic discriminant analysis, support vector machines, and random forests for 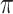 and
and 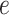 . Our second estimator using the relaxed lasso (“DR-lasso”) is implemented using the
. Our second estimator using the relaxed lasso (“DR-lasso”) is implemented using the 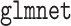 package (Friedman and others, 2010), to estimate all needed functions. For all simulations, we truncated estimates of the propensity and surrogate scores so that
package (Friedman and others, 2010), to estimate all needed functions. For all simulations, we truncated estimates of the propensity and surrogate scores so that 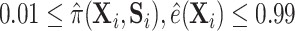 for all
for all 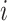 . We used the default cross-validation procedure to select the tuning parameters for the Super Learner. For the DR-lasso estimator, we used the default crossvalidation to select the tuning parameters. It is possible that performance could be improved by tweaking the tuning parameters for the candidate learners.
. We used the default cross-validation procedure to select the tuning parameters for the Super Learner. For the DR-lasso estimator, we used the default crossvalidation to select the tuning parameters. It is possible that performance could be improved by tweaking the tuning parameters for the candidate learners.
It is our understanding that there are no currently available methods to robustly estimate the PTE of a high-dimensional surrogate. However, since there are available methods to measure the strength of high-dimensional mediators, we compare our proposed approach to these available methods. While the goals of mediation analysis and surrogate markers analysis are different and the necessary assumptions differ, this allows us to offer some reasonable comparison to methods that are currently available, rather than no comparison at all. Thus, to fairly compare, we set up the majority of our simulations such that 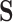 is a mediator in that it lies on the causal pathway between
is a mediator in that it lies on the causal pathway between 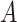 and
and 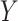 . While mediation methods are attempting to estimate a distinct quantity from
. While mediation methods are attempting to estimate a distinct quantity from 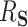 , in three of the four following simulation settings, the estimation of the “proportion mediated” (which is used in mediation) and
, in three of the four following simulation settings, the estimation of the “proportion mediated” (which is used in mediation) and  are the same.
are the same.
In our simulations, we compare our proposed approach to: high-dimensional mediation analysis (HIMA, Zhang and others, 2016) as implemented in the  package (Zheng and others, 2018); Bayesian mediation analysis (BAMA, Song and others, 2020) as implemented in the
package (Zheng and others, 2018); Bayesian mediation analysis (BAMA, Song and others, 2020) as implemented in the  package (Rix and Song, 2021); and high-dimensional linear mediation analysis (Zhou and others, 2020) as implemented in the
package (Rix and Song, 2021); and high-dimensional linear mediation analysis (Zhou and others, 2020) as implemented in the  package. All three of these methods propose
package. All three of these methods propose 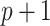 linear models:
linear models: 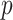 models for the surrogates/mediators—
models for the surrogates/mediators— 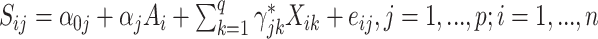 — and one outcome model—
— and one outcome model— 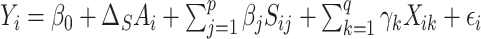 — though the implementation of
— though the implementation of  does not allow for the inclusion of covariates
does not allow for the inclusion of covariates 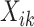 . Using these models, the overall treatment effect can be identified as
. Using these models, the overall treatment effect can be identified as 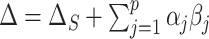 , and as above the PTE (or proportion mediated) may be identified as
, and as above the PTE (or proportion mediated) may be identified as 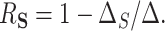
We computed 95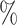 confidence intervals (CIs) for
confidence intervals (CIs) for 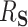 as
as 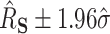 , where
, where 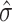 is an estimate of (4.6). We computed 95
is an estimate of (4.6). We computed 95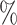 credible intervals for the BAMA estimator from the 2.5
credible intervals for the BAMA estimator from the 2.5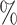 and 97.5
and 97.5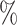 quantiles of the posterior distribution of
quantiles of the posterior distribution of 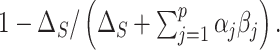 CIs were not computed for the HIMA and freebird implementations because they are not available.
CIs were not computed for the HIMA and freebird implementations because they are not available.
5.2. Simulation setup
We constructed four sets of simulations for a total of 18 simulation settings to assess the performance of our proposed approach in both low- and high-dimensional settings.
For the first set of simulations, the data-generating mechanism for 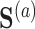 was linear in
was linear in 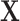 , the data-generating mechanism for
, the data-generating mechanism for 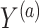 was linear in
was linear in 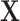 and
and 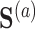 , and the propensity score was linear on the log-odds scale. Given this data-generating mechanism, we would expect that all methods (proposed and comparisons) should perform reasonably well. We set the dimension of
, and the propensity score was linear on the log-odds scale. Given this data-generating mechanism, we would expect that all methods (proposed and comparisons) should perform reasonably well. We set the dimension of 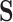 (
(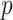 ) and of
) and of 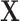 (
(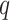 ) to be 100. We let
) to be 100. We let 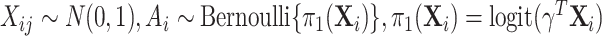 , where
, where 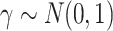 . The surrogates were generated as
. The surrogates were generated as 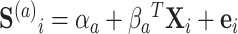 , where
, where 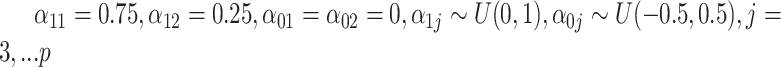 . And only five of the covariates were important for determining the surrogate:
. And only five of the covariates were important for determining the surrogate: 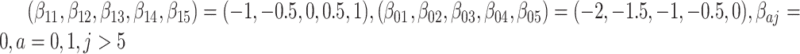 . The outcome counterfactuals were given by
. The outcome counterfactuals were given by 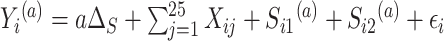 , suggesting that only the first two surrogates and 25 of the covariates were important for determining the outcome. The errors
, suggesting that only the first two surrogates and 25 of the covariates were important for determining the outcome. The errors 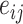 and
and 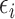 were
were 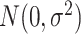 . We let the sample size vary between
. We let the sample size vary between 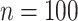 and
and 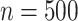 and the level of noise between
and the level of noise between 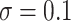 and
and 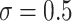 —for a total of four settings—and we set
—for a total of four settings—and we set 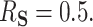
In the second set of simulations (six more settings), the data-generating mechanism was less linear, including interactions between covariates in both the propensity score and the model for 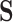 , though the outcome model was still a simple linear combination of
, though the outcome model was still a simple linear combination of 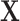 and
and 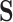 . In the third set of simulations (two more settings), the dimension of the surrogates (
. In the third set of simulations (two more settings), the dimension of the surrogates (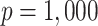 ) was much larger than the sample size (
) was much larger than the sample size (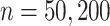 ), and correct specification of the outcome functions also included nonlinear terms. These simulations should mimic what may happen in practice since all models are typically subject to some amount of mis-specification. We expected the nonlinearity to induce bias in the competing methods (which require linear models to hold). In the fourth set of simulations (six more settings), we specified
), and correct specification of the outcome functions also included nonlinear terms. These simulations should mimic what may happen in practice since all models are typically subject to some amount of mis-specification. We expected the nonlinearity to induce bias in the competing methods (which require linear models to hold). In the fourth set of simulations (six more settings), we specified 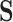 so that it was not a mediator but was instead downstream of a true mediator. We performed 1000 replications for each simulation setting. We give full details and results for these simulations in Appendix E of the Supplementary material available at Biostatistics online.
so that it was not a mediator but was instead downstream of a true mediator. We performed 1000 replications for each simulation setting. We give full details and results for these simulations in Appendix E of the Supplementary material available at Biostatistics online.
5.3. Results
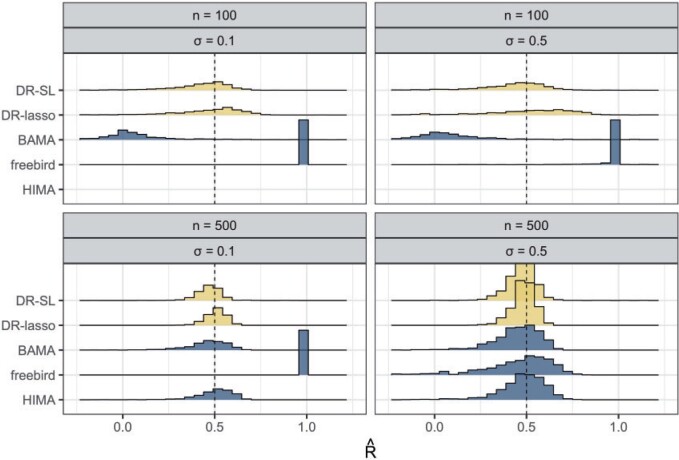
As expected, the first data-generating mechanism was well approximated using linear models. When the sample size was large (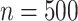 ) and
) and 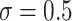 , all methods performed well, with a relatively small bias (see Figure 2). The median
, all methods performed well, with a relatively small bias (see Figure 2). The median 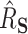 estimates were 0.46 (BAMA), 0.48 (freebird), 0.49 (HIMA), 0.48 (DR-SL), and 0.49 (DR-lasso), all of which compare favorably to the true
estimates were 0.46 (BAMA), 0.48 (freebird), 0.49 (HIMA), 0.48 (DR-SL), and 0.49 (DR-lasso), all of which compare favorably to the true 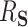 value of 0.5. The distribution of estimates for the proposed approaches was a bit tighter than for the competing methods. The empirical 2.5
value of 0.5. The distribution of estimates for the proposed approaches was a bit tighter than for the competing methods. The empirical 2.5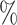 and 97.5
and 97.5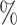 quantiles were 0.32 and 0.60 for DR-SL, 0.35 and 0.59 for DR-lasso, 0.15 and 0.67 for HIMA, 0.11 and 0.63 for BAMA, and
quantiles were 0.32 and 0.60 for DR-SL, 0.35 and 0.59 for DR-lasso, 0.15 and 0.67 for HIMA, 0.11 and 0.63 for BAMA, and  0.85 and 0.71 for freebird. Results were similar for the sample size with less noise (
0.85 and 0.71 for freebird. Results were similar for the sample size with less noise (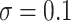 ), except for the freebird approach which began to estimate the PTE to be exactly 1 in almost all simulations: the empirical 2.5
), except for the freebird approach which began to estimate the PTE to be exactly 1 in almost all simulations: the empirical 2.5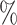 and 97.5
and 97.5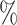 quantiles of the
quantiles of the 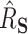 distribution for freebird were exactly 1. At the lower sample size (
distribution for freebird were exactly 1. At the lower sample size (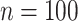 ), HIMA did not run, while BAMA estimates tended to be near 0 and freebird estimates were again clustered tightly around 1. Our proposed approaches had median
), HIMA did not run, while BAMA estimates tended to be near 0 and freebird estimates were again clustered tightly around 1. Our proposed approaches had median 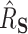 values of 0.45 (DR-SL) and 0.56 (DR-lasso), though with more variability than when
values of 0.45 (DR-SL) and 0.56 (DR-lasso), though with more variability than when 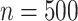 . The median absolute deviation (
. The median absolute deviation (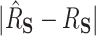 ) was smaller for both DR-SL and DR-lasso than any of the competing approaches for all settings.
) was smaller for both DR-SL and DR-lasso than any of the competing approaches for all settings.
Fig. 2.
Distribution of estimates of 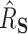 in the first data-generating mechanism. Lighter shaded regions represent the distribution of the proposed estimators (“DR-SL” and “DR-lasso”); darker shaded regions represent the distribution of the comparison estimators (“BAMA,” “freebird,” and “HIMA”). The true value of
in the first data-generating mechanism. Lighter shaded regions represent the distribution of the proposed estimators (“DR-SL” and “DR-lasso”); darker shaded regions represent the distribution of the comparison estimators (“BAMA,” “freebird,” and “HIMA”). The true value of 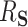 is given as a vertical dotted line at 0.5. At the lower sample size (
is given as a vertical dotted line at 0.5. At the lower sample size (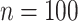 ), HIMA did not run and so is not shown.
), HIMA did not run and so is not shown.
CI coverage for the proposed estimators tended to be quite good for DR-SL in all settings: coverage was 95 (
(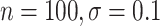 ), 96
), 96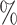 (
(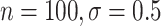 ), 94
), 94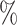 (
(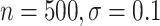 ), and 97
), and 97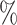 (
(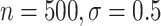 ). Coverage for the DR-lasso estimator was worse in general: coverage was 88
). Coverage for the DR-lasso estimator was worse in general: coverage was 88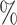 (
(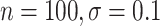 ), 82
), 82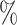 (
(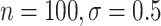 ), 89
), 89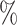 (
(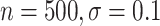 ), and 96
), and 96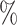 (
(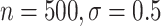 )
)
However, BAMA CIs also did not uniformly obtain nominal coverage: 96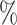 (
(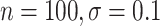 ), 99
), 99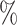 (
(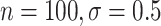 ), 92
), 92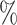 (
(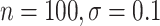 ), and 85
), and 85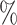 (
(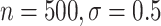 ). Even when BAMA CIs had nominal coverage, they were more than four times larger than the CIs for the proposed estimators: for example, BAMA CI half-lengths were 1.40 (
). Even when BAMA CIs had nominal coverage, they were more than four times larger than the CIs for the proposed estimators: for example, BAMA CI half-lengths were 1.40 (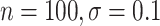 ) and 1.61 (
) and 1.61 (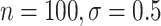 ), while the corresponding half-lengths were 0.25 and 0.30 for DR-SL and 0.28 and 0.44 for DR-lasso.
), while the corresponding half-lengths were 0.25 and 0.30 for DR-SL and 0.28 and 0.44 for DR-lasso.
The proposed estimators also outperformed the competing methods in the additional simulation settings where models were subject to mis-specification and the dimensionality of the surrogates was much larger than the sample size. See Appendix E of the Supplementary material available at Biostatistics online for complete results.
6. Ebola immune response application
The concentration of binding antibodies is often used as the primary outcome of interest in studies of Ebola vaccine efficacy, being itself a surrogate of vaccine efficacy as measured by the effect on the incidence of infections (Roozendaal and others, 2020). Because gene expression is the means by which DNA is turned into RNA and eventually proteins, it is associated with cellular function. Thus, the establishment of the humoral immune response may be captured by changes in gene expression as suggested by early works on systems vaccinology (Li and others, 2014). Furthermore, gene expression changes may occur days or even weeks before traditional measures of immune function (Rechtien and others, 2017). If gene expression can act as a surrogate for immune response, then it could possibly be used to shorten vaccine trials or to quickly measure the effect of vaccination in a population. Finally, genome-wide expression data offer the opportunity at looking at very different ways of influencing the response to intervention which constitutes potential surrogate markers. In this study, we sought to use observational data on long-term Ebola survivors and healthy controls to shed light on the possibility of gene expression’s use as a surrogate for antibody response to Ebola virus. This aim is inspired by the study of potential surrogates of protection among Ebola disease survivors (Sullivan and others, 2009).
In total, 26 Ebola survivors of the 2013–2016 Ebola outbreak in West Africa were recruited from the Postebogui cohort (Etard and others, 2017) as well as 33 healthy donors as described in Wiedemann and others (2020), each of whom had an expression for 29 624 genes quantified from whole blood RNA-seq (freely available from the Gene Expression Omnibus repository with accession code GSE143549) as well as the concentration of specific Ebola binding antibodies measured. Clearly, this is a setting where the number of potential surrogate markers (the genes) is substantially larger than the sample size. Given the superior performance of the DR-SL in our simulation study, and the high-dimensional setting, we focus only on using the DR-SL estimator to quantify how much of the overall difference in humoral immune response between survivors and healthy donors could be captured by the measured gene expression data. We used the same candidate learners as specified in Section 5. Propensity and surrogate scores (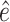 and
and 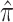 ) were truncated at 0.05 and 0.95 to prevent instability due to extreme weights, and
) were truncated at 0.05 and 0.95 to prevent instability due to extreme weights, and 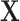 included age and sex.
included age and sex.
Ebola survivors were estimated to have a much higher abundance of Ebola-specific antibodies (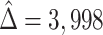 , SE
, SE  851.5). Using DR-SL, the residual treatment effect,
851.5). Using DR-SL, the residual treatment effect, 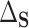 , was estimated as
, was estimated as 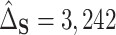 with SE
with SE  727.8, and the proportion of the difference explained by gene expression was estimated as
727.8, and the proportion of the difference explained by gene expression was estimated as 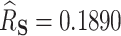 , with a SE of 0.07923. Thus, a large part of the humoral immune response cannot be explained by the differences in gene expression. Of note, we assumed no unmeasured confounding factors although it cannot be guaranteed in such a real-life context where survivors and healthy volunteers are two selected populations. If unmeasured confounding inflated both
, with a SE of 0.07923. Thus, a large part of the humoral immune response cannot be explained by the differences in gene expression. Of note, we assumed no unmeasured confounding factors although it cannot be guaranteed in such a real-life context where survivors and healthy volunteers are two selected populations. If unmeasured confounding inflated both 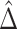 and
and 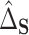 roughly equally, this could have the effect of artificially deflating
roughly equally, this could have the effect of artificially deflating 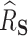 . Another explanation for the low estimated
. Another explanation for the low estimated 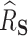 could be that, while gene expression measured shortly after infection may potentially be a good surrogate, measuring it long after infection (as in this study), does not capture the treatment effect as well. Measurement error in
could be that, while gene expression measured shortly after infection may potentially be a good surrogate, measuring it long after infection (as in this study), does not capture the treatment effect as well. Measurement error in 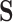 could also deflate the PTE. Importantly, one other potential violation of our assumptions is that about one-third of the observations have truncated surrogate scores—
could also deflate the PTE. Importantly, one other potential violation of our assumptions is that about one-third of the observations have truncated surrogate scores—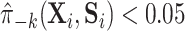 or
or 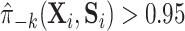 . This suggests that positivity might be (nearly) violated. We discuss this further in Appendix C of the Supplementary material available at Biostatistics online along with a potential solution.
. This suggests that positivity might be (nearly) violated. We discuss this further in Appendix C of the Supplementary material available at Biostatistics online along with a potential solution.
7. Discussion
We have proposed a very general approach to evaluating surrogate markers which can be applied in randomized experiments or in observational studies and can be used regardless of the dimensionality of the surrogates. Our approach is robust in that we have defined the PTE of the surrogates without reference to any models, and we have shown how machine learning approaches like Super Learner may be used to very flexibly estimate nuisance functions. Our simulation results suggest that our Super Learner estimator (DR-SL) outperforms competing methods even when the underlying data-generating mechanism is linear and still gives reasonable results even when high-dimensional linear models are mis-specified.
Our approach here is intimately tied to previous approaches for evaluating surrogate markers. The approaches in Agniel and Parast (2021) and in Parast and others (2016) can be seen to be similar to a version of our approach where the average treatment effect among controls is estimated under further assumptions. In Agniel and Parast (2021), they further require strict randomization of treatment, and they assume that 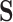 is a realization of a smooth continuous function. Parast and others (2016) make similar assumptions but take
is a realization of a smooth continuous function. Parast and others (2016) make similar assumptions but take 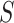 to be a scalar surrogate. They estimate a version of (4.2) among controls:
to be a scalar surrogate. They estimate a version of (4.2) among controls: 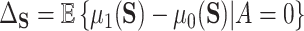 using kernel smoothing and taking advantage of the fact that treatment is randomized. Their estimates have the form
using kernel smoothing and taking advantage of the fact that treatment is randomized. Their estimates have the form 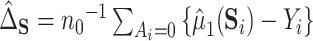 , where
, where 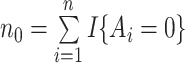 and
and 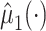 is estimated via kernel smoothing (possibly after dimension reduction). Our approach here could easily be adapted to estimate a similar quantity by using methods for doubly robust estimation of the average treatment effect on the treated (Shu and Tan, 2018; Moodie and others, 2018; Chernozhukov and others, 2017) by, for example,
is estimated via kernel smoothing (possibly after dimension reduction). Our approach here could easily be adapted to estimate a similar quantity by using methods for doubly robust estimation of the average treatment effect on the treated (Shu and Tan, 2018; Moodie and others, 2018; Chernozhukov and others, 2017) by, for example, 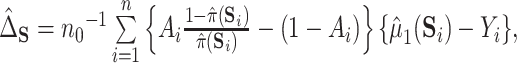 where
where 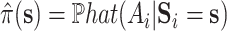 may be estimated using a model similar to the one used for
may be estimated using a model similar to the one used for 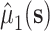 . Furthermore, our approach could be used to facilitate the use of machine learning methods in the estimation of
. Furthermore, our approach could be used to facilitate the use of machine learning methods in the estimation of 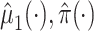 , to include covariates to control for confounding, or to simplify or strengthen asymptotic results. For example, results for the kernel estimator used in Agniel and Parast (2021) obtain rates of convergence of
, to include covariates to control for confounding, or to simplify or strengthen asymptotic results. For example, results for the kernel estimator used in Agniel and Parast (2021) obtain rates of convergence of 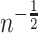 only under very limited technical conditions, but using sample-splitting parametric rates of convergence could be obtained under very general conditions.
only under very limited technical conditions, but using sample-splitting parametric rates of convergence could be obtained under very general conditions.
Supplementary Material
Acknowledgments
We thank Yves Lévy and all of the investigators at the Vaccine Research Institute for providing the Ebola survivor data. We thank the Postebogui team for their daily work and the survivors. This work was supported by grant R01 DK118354 from the National Institute of Diabetes and Digestive and Kidney Diseases as well as by the Investissements d’Avenir program managed by the ANR under reference ANR-10-LABX-77-01.
Conflict of Interest: None declared.
Contributor Information
Denis Agniel, RAND Corporation, 1776 Main St. Santa Monica, CA, 90401, USA.
Boris P Hejblum, Univ. Bordeaux, INSERM, INRIA, BPH, U1219, SISTM, F-33000 Bordeaux, France and Vaccine Research Institute, F-94000 Créteil, France.
Rodolphe Thiébaut, Univ. Bordeaux, INSERM, INRIA, BPH, U1219, SISTM, F-33000 Bordeaux, France, CHU de Bordeaux, Service d’Information médicale, F-33000 Bordeaux, France and Vaccine Research Institute, F-94000 Créteil, France.
Layla Parast, University of Texas at Austin, Department of Statistics and Data Sciences, 3925 West Braker Lane, Austin, TX 78759, USA.
8. Software
We include software to implement our proposed methods in the R package  available at github.com/denisagniel/crossurr.
available at github.com/denisagniel/crossurr.
Supplementary material
Supplementary material is available online at http://biostatistics.oxfordjournals.org.
References
- Agniel, D. and Parast, L. (2021). Evaluation of longitudinal surrogate markers. Biometrics 77, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, A., Molenberghs, G.. and others. (2004). Prentice’s approach and the meta-analytic paradigm: a reflection on the role of statistics in the evaluation of surrogate endpoints. Biometrics 60, 724–728. [DOI] [PubMed] [Google Scholar]
- Alonso, A., Van der Elst, W., Molenberghs, G., Buyse, M. and Burzykowski, T. (2016). An information-theoretic approach for the evaluation of surrogate endpoints based on causal inference. Biometrics 72, 669–677. [DOI] [PubMed] [Google Scholar]
- Bang, H. and Robins, J. M. (2005). Doubly robust estimation in missing data and causal inference models. Biometrics 61, 962–973. [DOI] [PubMed] [Google Scholar]
- Caveney, E. J. and Cohen, O. J. (2011). Diabetes and biomarkers. Journal of Diabetes Science and Technology 5, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernozhukov, V., Chetverikov, D., Demirer, M., Duflo, E., Hansen, C. and Newey, W. (2017). Double/debiased/Neyman machine learning of treatment effects. American Economic Review 107, 261–265. [Google Scholar]
- Choi, S. H., Kim, T. H., Lim, S., Park, K. S., Jang, H. C. and Cho, N. H. (2011). Hemoglobin A1c as a diagnostic tool for diabetes screening and new-onset diabetes prediction: a 6-year community-based prospective study. Diabetes Care 34, 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon, A., Taylor, J., Li, Y., Diaz-Ordaz, K. and Elliott, M. (2017). Links between causal effects and causal association for surrogacy evaluation in a Gaussian setting. Statistics in Medicine 36, 4243–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etard, J.-F., Sow, M. S., Leroy, S., Touré, A., Taverne, B., Keita, A. K., Msellati, P., Baize, S., Raoul, H., Izard, S. and Kpamou, C. (2017). Multidisciplinary assessment of post-Ebola sequelae in Guinea (Postebogui): an observational cohort study. The Lancet Infectious Diseases 17, 545–552. [DOI] [PubMed] [Google Scholar]
- Farrell, M. H. (2015). Robust inference on average treatment effects with possibly more covariates than observations. Journal of Econometrics 189, 1–23. [Google Scholar]
- Farrell, M. H., Liang, T. and Misra, S. (2021). Deep neural networks for estimation and inference. Econometrica 89, 181–213. [Google Scholar]
- Freedman, L. S., Graubard, B. I. and Schatzkin, A. (1992). Statistical validation of intermediate endpoints for chronic diseases. Statistics in Medicine 11, 167–178. [DOI] [PubMed] [Google Scholar]
- Friedman, J., Hastie, T. and Tibshirani, R. (2010). Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Gilbert, P. B. and Hudgens, M. G. (2008). Evaluating candidate principal surrogate endpoints. Biometrics 64, 1146–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J. and Geng, Z. (1995). Collapsibility of logistic regression coefficients. Journal of the Royal Statistical Society. Series B (Methodological) 57, 263–267. [Google Scholar]
- Joffe, M. M. and Greene, T. (2009). Related causal frameworks for surrogate outcomes. Biometrics 65, 530–538. [DOI] [PubMed] [Google Scholar]
- Karim, S. S. A. (2021). Vaccines and SARS-CoV-2 variants: the urgent need for a correlate of protection. The Lancet 397, 1263–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Rouphael, N.. and others. (2014). Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nature Immunology 15, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinshausen, N. (2007). Relaxed lasso. Computational Statistics & Data Analysis 52, 374–393. [Google Scholar]
- Moodie, E. E. M., Saarela, O. and Stephens, D. A. (2018). A doubly robust weighting estimator of the average treatment effect on the treated. Stat 7, e205. [Google Scholar]
- Obirikorang, C., Quaye, L. and Acheampong, I. (2012). Total lymphocyte count as a surrogate marker for CD4 count in resource-limited settings. BMC Infectious Diseases 12, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parast, L., Cai, T. and Tian, L. (2019). Using a surrogate marker for early testing of a treatment effect. Biometrics 75, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parast, L., McDermott, M. M. and Tian, L. (2016). Robust estimation of the proportion of treatment effect explained by surrogate marker information. Statistics in Medicine 35, 1637–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parast, L., Tian, L. and Cai, T. (2020). Assessing the value of a censored surrogate outcome. Lifetime Data Analysis 26, 245–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin, S. A. and Gilbert, P. B. (2012). Nomenclature for immune correlates of protection after vaccination. Clinical Infectious Diseases 54, 1615–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley, E., LeDell, E., Kennedy, C., Lendle, S. and van der Laan, M. (2021). SuperLearner: Super Learner Prediction. R package version 2.0-28. https://CRAN.R-project.org/package=SuperLearner. [Google Scholar]
- Prentice, R. L. (1989). Surrogate endpoints in clinical trials: definition and operational criteria. Statistics in Medicine 8, 431–440. [DOI] [PubMed] [Google Scholar]
- Price, B. L., Gilbert, P. B. and van der Laan, M. J. (2018). Estimation of the optimal surrogate based on a randomized trial. Biometrics 74, 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtien, A., Richert, L.. and others. (2017). Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the Ebola vaccine rVSV-ZEBOV. Cell Reports 20, 2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rix, A., Kleinsasser, M. and Song, Y. (2021). BAMA: High Dimensional Bayesian Mediation Analysis. R package version 1.2. https://CRAN.R-project.org/package=bama [Google Scholar]
- Roozendaal, R., Hendriks, J.. and others. (2020). Nonhuman primate to human immunobridging to infer the protective effect of an Ebola virus vaccine candidate. NPJ Vaccines 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, H. and Tan, Z. (2018). Improved estimation of average treatment effects on the treated: local efficiency, double robustness, and beyond. arXiv preprint arXiv:1808.01408. [Google Scholar]
- Small, G. W. (2006). Diagnostic issues in dementia: neuroimaging as a surrogate marker of disease. Journal of Geriatric Psychiatry and Neurology 19, 180–185. [DOI] [PubMed] [Google Scholar]
- Song, Y., Zhou, X., Zhang, M., Zhao, W., Liu, Y., Kardia, S. L., Roux, A.V.D., Needham, B.L., Smith, J. A. and Mukherjee, B. (2020). Bayesian shrinkage estimation of high dimensional causal mediation effects in omics studies. Biometrics 76, 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, N. J., Martin, J. E., Graham, B. S. and Nabel, G. J. (2009). Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nature Reviews Microbiology 7, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Z. (2020). Model-assisted inference for treatment effects using regularized calibrated estimation with high-dimensional data. The Annals of Statistics 48, 811–837. [Google Scholar]
- Taylor, J. M. G., Wang, Y. and Thiébaut, R. (2005). Counterfactual links to the proportion of treatment effect explained by a surrogate marker. Biometrics 61, 1102–1111. [DOI] [PubMed] [Google Scholar]
- Teixeira, A. L., Diniz, B. S., Campos, A. C., Miranda, A. S., Rocha, N. P., Talib, L. L., Gattaz, W. F. and Forlenza, O. V. (2013). Decreased levels of circulating adiponectin in mild cognitive impairment and Alzheimer’s disease. Neuromolecular medicine 15, 115–121. [DOI] [PubMed] [Google Scholar]
- Tibshirani, R. (1996). Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological) 58, 267–288. [Google Scholar]
- van der Laan, M. J., Polley, E. C., and Hubbard, A. E. (2007). Super Learner. Statistical Applications in Genetics and Molecular Biology 6, 1–23. [DOI] [PubMed] [Google Scholar]
- VanderWeele, T. J. (2013). Surrogate measures and consistent surrogates. Biometrics 69, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager, S. and Walther, G. (2015). Adaptive concentration of regression trees, with application to random forests. arXiv preprint arXiv:1503.06388. [Google Scholar]
- Wang, S., McCormick, T. H., and Leek, J. T. (2020). Methods for correcting inference based on outcomes predicted by machine learning. Proceedings of the National Academy of Sciences 117, 30266–30275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Taylor, J. M. G. (2002). A measure of the proportion of treatment effect explained by a surrogate marker. Biometrics 58, 803–812. [DOI] [PubMed] [Google Scholar]
- Wiedemann, A., Foucat, E.. and others. (2020). Long-lasting severe immune dysfunction in Ebola virus disease survivors. Nature Communications 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Zheng, Y.. and others. (2016). Estimating and testing high-dimensional mediation effects in epigenetic studies. Bioinformatics 32, 3150–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Zhang, H., Hou, L. and Liu, L. (2018). HIMA: High-Dimensional Mediation Analysis. R package version 1.0.7. [Google Scholar]
- Zhou, R. R., Wang, L. and Zhao, S. D. (2020). Estimation and inference for the indirect effect in high-dimensional linear mediation models. Biometrika 107, 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, R. R., Zhao, S. D. and Parast, L. (2022). Estimation of the proportion of treatment effect explained by a high-dimensional surrogate. Statistics in Medicine 41, 2227–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.