Abstract
Fragile-X mental retardation is caused by loss of function of a single gene encoding the Fragile-X mental retardation protein, FMRP, an RNA-binding protein that harbors two KH-type and one RGG-type RNA-binding domains. Previous studies identified intramolecular G-quartet RNAs as high-affinity targets for the RGG box, but the relationship of RNA binding to FMRP function and mental retardation remains unclear. One severely affected patient harbors a missense mutation (I304N) within the second KH domain (KH2), and some evidence suggests this domain may be involved in the proposed role of FMRP in translational regulation. We now identify the RNA target for the KH2 domain as a sequence-specific element within a complex tertiary structure termed the FMRP kissing complex. We demonstrate that the association of FMRP with brain polyribosomes is abrogated by competition with the FMRP kissing complex RNA, but not by high-affinity G-quartet RNAs. We conclude that mental retardation associated with the I304N mutation, and likely the Fragile-X syndrome more generally, may relate to a crucial role for RNAs harboring the kissing complex motif as targets for FMRP translational regulation.
Keywords: Fragile-X mental retardation, FMRP, polyribosome, loop-loop pseudoknot, kissing complex, RNA, KH domain
Fragile-X mental retardation, the most common cause of familial mental retardation, is caused by loss of function of the FMR1 gene. FMRP (Fragile-X mental retardation protein), the product of the FMR1 gene, is characterized by the presence of two tandem KH-type RNA-binding domains and a third RGG-type RNA-binding domain (RGG box). Although transcriptional silencing of FMR1 due to hypermethylation is the most frequent cause of the disorder, patients expressing mutations or deletions within FMR1 have been described (Jin et al. 2004), underscoring that loss of FMRP function leads to the Fragile-X syndrome. Given that the loss of FMRP activity leads to complex behavorial and cognitive deficits, understanding FMRP function has the potential to provide a link between molecular neurobiology and higher brain function.
Identification of the RNA ligands of FMRP will provide a key to understanding FMRP function (Kaytor and Orr 2001; Gao 2002). However, despite the finding that one severely affected patient harbors a missense mutation (I304N) in the second KH domain, no confirmed RNA targets for this domain have been identified. Interest in this mutation was heightened by the finding that this mutation reduced FMRP binding to ribohomopolymers at higher salt concentrations (Siomi et al. 1994). Moreover, X-ray crystallographic studies of an analogous protein, the KH3 domain of the neuron-specific splicing factor Nova, bound to its high-affinity RNA ligand, revealed that the FMRP I304N mutation maps to a position within the KH domain that is critical for stabilizing sequence-specific RNA-protein interactions (Lewis et al. 2000; Ramos et al. 2003). More recently, NMR structures for a number of KH-type RNA-binding proteins and nucleic acid targets have been reported, including hnRNP-K (Braddock et al. 2002a), Sf1 (Liu et al. 2001), and FUSE-binding protein (Braddock et al. 2002b), and these have supported the important role of the isoleucine 304 within the FMRP KH2 RNA-binding pocket (Ramos et al. 2003).
RNA selection with full-length FMRP has led to the identification of high-affinity FMRP RNA ligands that harbor a signature structural motif termed a G-quartet (Brown et al. 2001; Darnell et al. 2001). However, mapping experiments revealed that the G-quartet RNA was bound not by the FMRP KH domains but by the C-terminal RGG box (Darnell et al. 2001). Nevertheless, evaluation for G-quartets has been widely used to screen candidate FMRP RNA targets, identified through a variety of approaches, including bioinformatics, immunoprecipitation and DNA chip analysis, antibody-positioned RNA amplification, and biochemical approaches (Brown et al. 2001; Darnell et al. 2001; Schaeffer et al. 2001; Miyashiro et al. 2003; Todd et al. 2003).
FMRP is believed to regulate mRNA translation in the brain, although the mechanism for this action, and the RNA targets of this regulation, are unknown. Following cloning of the FMR1 gene, the FMRP protein was found to be present on polyribosome fractions of tissue culture cells (Khandjian et al. 1996; Corbin et al. 1997; Feng et al. 1997a) and brain (Feng et al. 1997b; Khandjian et al. 2004; Stefani et al. 2004). Recent data suggest that FMRP is associated with components of the RNA-induced silencing complex (RISC) complex in Drosophila and mammalian cells (Caudy et al. 2002, 2003; Ishizuka et al. 2002; Jin et al. 2004), suggesting a link between miRNA regulation and FMRP function. This observation may be consistent with localization of FMRP on polyribosomes, as miRNAs were first described to be associated with polyribosomes, where they are believed to play a role in translational regulation (Bartel 2004). FMRP has been reported to be present in neuronal dendrites, where it may be associated with polyribosomes, and where FMRP itself may be locally translated (Feng et al. 1997b; Greenough et al. 2001; Antar et al. 2004; for review, see O'Donnell and Warren 2002). Biochemical studies have suggested that FMRP may associate with specific mRNAs on polyribosomes; several mRNAs were found to have altered polyribosome distributions in lymphoblastoid cells obtained from Fragile X patient cells and to harbor G-quartet motifs (Brown et al. 2001; Darnell et al. 2001). Subsequent analysis of one of these targets, MAP1B, suggests that the Drosophila homolog of this protein (futsch) may be regulated at the translational level by the Drosophila FMRP homolog (Zhang et al. 2001).
To identify the RNA motif recognized by the FMRP KH2 domain, we undertook a new RNA selection with FMRP, using both full-length FMRP and isolated KH domains. We find that, unlike other characterized KH domains, FMRP KH2 binds to an RNA complex termed a loop-loop pseudoknot, or “kissing complex.” Detailed analysis of the FMRP KH domain-RNA interaction reveals that binding to the kissing complex is KH2 specific and is abrogated by the I304N mutation. Binding is dependent in part on the RNA structure and on several specific nucleotides, as indicated by Mg+2-dependence, chemical modification, and mutational analysis. Identification of the kissing complex as a KH2 target allowed us to re-evaluate the nature of FMRP association with polyribosomes. We find that the association of FMRP with brain polyribosomes cannot be competed off with high-affinity G-quartet RNAs, but, in contrast, is entirely competed by kissing complex RNAs. These results suggest that mental retardation associated with the I304N mutation, and likely the Fragile-X syndrome more generally, may relate to a crucial role for RNAs harboring kissing complex motifs as targets for FMRP translational regulation. Discovery of this RNA motif will facilitate identification of a new set of FMRP RNA targets that play critical roles in Fragile-X mental retardation.
Results
Identification of FMRP KH2-dependent RNA ligands
To identify RNA ligands for FMRP KH domains, we undertook in vitro RNA selection experiments (Ellington and Szostak 1990; Tuerk and Gold 1990). Oligonucleotide libraries harboring 25 or 52 random nucleotides (nt) were generated as described (Buckanovich and Darnell 1997; Jensen et al. 2000b; Darnell et al. 2001) with a fixed 5′ sequence harboring a T7 RNA polymerase promoter and a fixed 3′ sequence used for PCR amplification. RNA selection was performed with either full-length FMRP protein produced from baculovirus-infected insect cells, or with a fragment including KH1-KH2 and a short C-terminal extension (see Lewis et al. 2000). RNA selection using purified KH domains with the 25-nt random library (complexity ∼1014 25-mer RNAs; Jensen et al. 2000b) failed to produce a consensus RNA target, despite stringent control of protein and RNA integrity during each round of selection. While this negative result is not conclusive, it suggested the possibility that FMRP KH1/2 binds to a complex RNA not present within the 25-nt library due to limitations in the size or complexity of this library.
To follow up these results, we re-evaluated our experimental design. To optimize our chance of obtaining FMRP interactors, we undertook an initial six rounds of selection with full-length protein and used a longer (52-mer) random library. Since continued RNA selection with full-length FMRP results in RNAs that bind to the high-affinity RGG box (Darnell et al. 2001), we then switched to using the paired KH domains for further RNA selection. After eight rounds of RNA selection, we succeeded in purifying high-affinity KH2 ligands (Fig. 1A). Importantly, the binding of full-length FMRP to the pool of selected sequences was dependent on the integrity of the KH2 domain. FMRP harboring the I304N mutation showed greatly reduced binding to the selected RNA (Fig. 1A), while it showed binding equivalent to wild-type protein when tested against G-quartet RNA (Darnell et al. 2001; data not shown), suggesting that the KH2 domain is essential for recognizing the selected sequences.
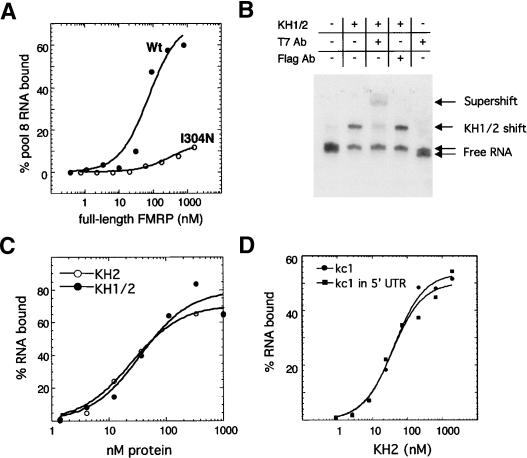
Figure 1.
Selection of FMRP KH2 RNA ligands. (A) Filter binding assays using full-length wild-type (Wt) or I304N mutant FMRP bound to the total RNA pool present after eight rounds of selection. (B) Gel-shift assays of T7-tagged FMRP KH1/2 protein with kc2 RNA, performed in the presence of indicated antibodies. Two conformations of free kc2 RNA are evident in lanes in which the RNA has not shifted, and only the faster migrating form shifts and supershifts in the presence of T7 antibody. (C) Filter binding assays comparing binding of KH1/2 and the isolated KH2 domain to kc2 RNA. (D) Filter binding assays comparing the ability of KH2 to bind isolated kc1 RNA and the kc1 RNA inserted into the middle of the 5′ UTR of luciferase mRNA.
We sequenced 96 clones following eight rounds of RNA selection and found two predominant clones (kc1 and kc2) (Table 1), together comprising 19% (18/95) of the pool. This predominance of two related RNA species suggested that the “winning” sequences had been selected. A multiple alignment analysis of the primary sequence of 19 additional clones revealed two motifs that were conserved with kc1 and kc2 (kc3-kc13; Table 1; Supplementary Fig. S1), one at the 5′ end of the random region (GGGCKAAGGARK, K is G or U, R is G or A) and one at the 3′ end (KAGCGRCUGG). Thirty-seven sequences (eight different clones) bound FMRP with lower affinity and had no obvious consensus between them. Twenty-two unique sequences did not fit any consensus, did not show detectable KH1/2 binding, and were not pursued further.
Table 1.
Consensus sequences of 13 aptamers selected with FMRP KH1/2 protein
| Clone | 5′ Consensus 18 20 22 24 26 28 GGGCK_AAGGARK | 3′ Consensus 43 45 47 49 51 KAGCGRCUGG__ | Kd (nM) | % RNA bound | Frequency |
|---|---|---|---|---|---|
| kc1 | GGACU_AAGGAGU | UAGUGGCUGG | 48.9 + 13.6 | 70 | 10 |
| kc2 | GGGCU_AAGGAAU | UAGCGGCUGG | 65.9 + 16.6 | 90 | 8 |
| kc3 | GGGCUGAAGGAAG | GUGCGACUGGGC | 80.0 + 18.5 | 50 | 5 |
| kc4 | GAGCG_AAGGGAG | UAGCAGCUGG | 54.3 + 28.9 | 50 | 4 |
| kc5 | GGGCUGAAGGAAC | GUGCGGCUGGGC | 45.3 + 26.7 | 50 | 2 |
| kc6 | GGGCG_AAGGGAG | AAGCGGCUGG | 32.7 + 22.6 | 35 | 1 |
| kc7 | GGGCG_AAGGAGU | UAGCGGCUGG | 38.0 + 12.9 | 90 | 1 |
| kc8 | GGGCU_AAGGAUU | UAGCGGCUGG | 53.8 + 20.0 | 75 | 2 |
| kc9 | GGGCU_GAGGAUG | GAGCGACUGG | 64.9 + 16.0 | 65 | 1 |
| kc10 | GGGCU_AUGGAGU | GAGCGGCUGG | 66.8 + 24.1 | 60 | 1 |
| kc11 | GGGCU_AAGGAGU | GAGCGACUGGUG | 103.3 + 24.0 | 40 | 1 |
| kc12 | GGGCA_UAGGCAC | GAGCGGCUGG | 47.3 + 26.3 | 45 | 1 |
| kc13 | GGGCG_AAGGAAG | UAGCGACUGG | 35.9 + 18.6 | 45 | 1 |
The frequency (from 37 clones in the eighth round of selection; see text) and results of filter binding assays (Kd [nM] and maximum percent RNA bound) are shown. Kds and associated errors were determined by Kaleidograph software. Maximal percent RNA bound was determined graphically and rounded to the nearest 5%. Nucleotide numbering refers to clone Δkc2; see Fig. 4B and text). IUPAC (International Union of Pure and Applied Chemistry) format is used for the consensus sequences (K = G or U, R = A or G). Clone kc13 was not identified from the eighth round of selection but rather from the seventh round of selection with full-length FMRP (Darnell et al. 2001).
Filter binding assays using the paired KH domains were performed with kc1-kc13 and Kd's ranged from 33 to 103 nM (Table 1). There was a large variation in the amount of RNA bound at saturation (percent RNA bound) ranging from ∼40%-90%. We assume that this variation was most likely due to RNAs forming competing structures (see below). Binding curves for each of the 13 clones are shown in Supplementary Fig. S2.
To further examine the interaction between FMRP and kc1 RNA, we undertook electrophoretic mobility shift analysis. The migration of radiolabeled kc1 RNA was shifted in the presence of T7-tagged FMRP KH1-KH2 domains and was supershifted by the presence of an anti-T7 antibody, but not by an irrelevant antibody (Fig. 1B). Notably, in control lanes in which no RNA is shifted, two RNA species are evident, and only the faster migrating RNA (presumably more compactly folded on this native gel) shifts with added protein, suggesting the possibility that the RNA may fold into two different conformational states and that protein binding is dependent on the RNA conformation. To confirm that the binding of kc1 required KH2 and to assess the contribution of KH1, we tested RNA binding with the isolated FMRP KH2 domain and found that it binds as well as the paired KH domains with the C-terminal extension (Fig. 1C), suggesting that KH1 is not involved in recognizing this ligand. Moreover, full-length FMRP proteins containing the I304N mutation did not bind kc2, while protein with an analogous mutation in KH1 (I241N) bound kc2 as well as wild-type FMRP (data not shown), confirming that KH2 alone is sufficient to bind this class of RNA ligands. Finally, we found that FMRP KH2 domains isolated from various sources (baculoviral, bacterial) and purification schemes (affinity chromatography, HPLC purification) bind kc RNA equally well. Taken together, these data provide strong evidence that FMRP interacts directly with kc RNA.
To assess whether kc RNA ligands can be recognized by FMRP in the context of a native RNA, we cloned kc1 into the HindIII site of the 5′UTR of the luciferase mRNA. RNA was transcribed from the cap site to 22 nt downstream of the start codon (∼280 nt) and assayed for interaction with the KH2 domain by filter binding assay. We found that the curves for KH2 binding to kc RNA in the 5′UTR and the isolated 96-mer RNA are virtually identical (Fig. 1D), suggesting that FMRP is able to recognize the kc RNA motif in the context of a natural mRNA.
Structure of FMRP RNA ligands
To characterize the nature of the FMRP-kc1 interaction, we found that binding occurred at physiologic salt concentrations and temperature (37°C) and was entirely dependent on the presence of physiologic concentrations of Mg+2 (Fig. 2A,B; data not shown). As a control for the integrity of FMRP in EDTA, the binding of FMRP to a G-quartet RNA target of the RGG box (sc1) was found to be unaffected by the absence of Mg+2 (Fig. 2A). A series of binding curves done at varying Mg+2 concentration, plotted as -log(Kd) versus log[Mg+2] concentration, demonstrated that the slope of the binding plot is ∼1.0, indicating that one Mg+2 ion is involved in the protein:RNA interaction (Fig. 2B). Since RNA typically folds into stable structures through the specific coordination of metal ions, most often Mg+2 (Draper 2004), this result provides additional evidence that the KH2:kc RNA interaction is dependent on the tertiary structure of the RNA.
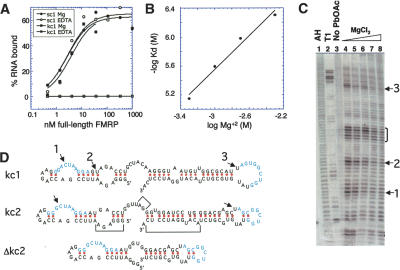
Figure 2.
Magnesium-dependent binding suggests a loop-loop interaction for the KH2 RNA ligands. (A) FMRP-kc1 binding is dependent on Mg+2. Filter binding assays were performed in the presence of Mg+2 or EDTA, as indicated, to kc1, or, as a control, to sc1 (RGG-box RNA ligand) with full-length FMRP. (B) FMRP-kc1 binding as a function of Mg+2 concentration. Analysis of kc1 binding curves to KH1/2 at indicated Mg+2 concentrations is shown; the slope is 0.94, suggesting one specifically bound Mg+2 ion participates in the RNA:protein interaction. (C) Lead-acetate cleavage of kc1 RNA in the presence of competing MgCl2. Cleavage sites (arrows labeled 1-3) that are effectively competed by increasing Mg+2 concentrations (0, 10, 25, 100, 250 mM Mg+2; lanes 4-8) indicate likely Mg+2-binding sites. Nonspecific cleavage corresponding to single-stranded “hinge” is indicated by the bracket. RNAs treated with alkaline hydrolysis (AH; lane 1) and RNase T1 (lane 2) or untreated RNA (lane 3) are shown. (D) mfold 3.1 structures of kc1, kc2, and Δkc2 RNAs, with sites of lead-acetate cleavage indicated (arrows). Δkc2 was constructed by deleting bracketed areas shown in kc2.
To evaluate where Mg+2 might be specifically bound to kc RNA, we examined the sensitivity of the folded RNA to lead acetate treatment. Lead ions (Pb(II)) are able to be chelated in place of Mg+2 and catalyze backbone cleavage at sites nearby in the 3D structure (Brown et al. 1983; Brannvall et al. 2001). Three sites of strong lead acetate cleavage were identified (arrows, Fig. 2C). The ability of lead acetate to cleave at these sites was competed by an excess of MgCl2 (Olejniczak et al. 2002), suggesting that Mg+2 and Pb(II) are competing for the same site. A fourth cleavage site was not as effectively competed and may represent nonspecifically bound Pb(II) at single-stranded regions (bracket, Fig. 2C). This experiment was repeated with kc2 (data not shown), and sites 1 and 3 were cleaved by lead acetate, but not site 2 (Fig. 2D), suggesting that sites 1 and 3 map approximate positions that are the critical ones for Mg+2 coordination, thereby supporting proper RNA folding and KH2 binding.
To assess the structure of the kc RNAs, we examined RNA folding with mfold 3.1 (Mathews et al. 1999; Zuker 2003). This revealed that the majority of the kc clones may harbor two stem-loops, with a short single-stranded region (hinge) between them (Fig. 2D, kc1 and kc2). FMRP KH2 was only able to bind to kc RNAs when both stem-loops were present within the same molecule (data not shown). Since a single Mg+2 participates in the binding of KH2 to kc RNA, and lead acetate probing demonstrated that the binding sites of this Mg+2 (sites 1 and 3, Fig. 2D) mapped to each of the loops, these results suggest that the loops might be in close proximity, facilitated by chelation of a single Mg+2 ion.
Examination of sequence covariation in ribosomal RNAs across species has provided a powerful means to determine the structure of the ribosomal subunits (Gutell et al. 2002). We aligned sequences evolved by RNA selection for their ability to bind KH2 and examined them for covariation. Inspection of the 13 high-affinity kc ligands (Table 1) revealed that two otherwise conserved nucleotides, a G20 in the 5′ conserved domain and aC45 in the 3′ conserved domain, are changed to A20 and U45 in clone kc1. Moreover, the adjacent nucleotides G19 and G46 are also changed to A19 and A46 in clone kc4. These covarying nucleotides suggest that Watson-Crick and purine-purine base pairing may occur between the two loops.
Chemical probing of the kc RNA structure
Prior structural studies have demonstrated that KH domains bind to single-stranded regions of RNA (Lewis et al. 2000; Liu et al. 2001; L. Malinina, M. Teplova, K. Musunuru, A. Teplov, S.K. Burley, R.B. Darnell, and D.J. Patel, unpubl.), and these may be presented in the context of stem-loop structure (Lewis et al. 2000; L. Malinina, M. Teplova, K. Musunuru, A. Teplov, S.K. Burley, R.B. Darnell, and D.J. Patel, unpubl.). To further explore the structure of the kc RNA and its relationship to KH2 binding, we analyzed positions in the RNA accessible to chemical modification, using reagents that modify unpaired nucleotides and detecting modifications by stops in primer extension. Since KH2:kc RNA binding requires the presence of Mg+2, we compared results obtained in the presence of Mg+2 versus EDTA. We used DMS, CMCT, and kethoxal (which modify unpaired N3-C and N1-A, N3-U and N1-G, and N1-G and N2-G, respectively) for chemical modification (Ehresmann et al. 1987).
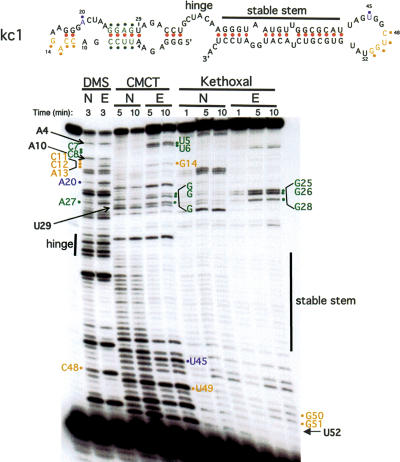
We found that kc RNA was modified in EDTA, but was protected in Mg+2 in several regions (Fig. 3). The data suggest the presence of an unanticipated 4-bp duplex forming between the two loops (a conserved 5′-C48UGG51-3′ in the 3′ loop and a 5′-C11CAG14-3′ present in the fixed sequence of the 5′ loop) (Fig. 3, labeled yellow). In addition, an A20-U45 pair previously found to covary with G20-C45 (Table 1) was also protected in the presence of Mg+2 (Fig. 3, labeled purple), suggesting formation of a Watson-Crick base pair. These experiments also provide evidence that the stem of the 5′ stem-loop predicted by mfold (5′-UUCC-3′ with 5′-GGAG-3′) (Fig. 3, labeled green) folds in a Mg+2-dependent manner. The stem of the 3′ stem-loop predicted by mfold (Fig. 3; “stable stem”) is base-paired in a Mg+2-independent manner. Our finding that there is a 4-bp interaction between the loops and that the R20 and Y45 nucleotides base pair with each other provides strong evidence that the two loops form a stable loop-loop pseudoknot, or kissing complex (Fig. 4A).
Figure 3.
Chemical modification of KH2 RNA ligand kc1. Native (“N”) or semidenatured (EDTA; “E”) RNA was treated for the indicated times with DMS (which modifies single-stranded [non-Watson-Crick-paired] A > C residues), CMCT (which modifies single-stranded U > G), or kethoxal (which modifies single-stranded G), or untreated (first lane) to control for natural stops in primer extension. Several residues protected from modification in the native structure (suggesting formation of new Watson-Crick base pairs in a Mg+2-dependent manner) are illustrated with bullets; the hinge region, which is always modified, and a stable stem, which is always protected from modification, are indicated. Several nucleotides with potential to form Watson-Crick base pairs (A4, A10, U29, and U52) are also indicated with arrows and discussed in the next section (Fig. 4). Numbers refer to positions in Δkc2, Table 1, and Figures 2 and 4B.
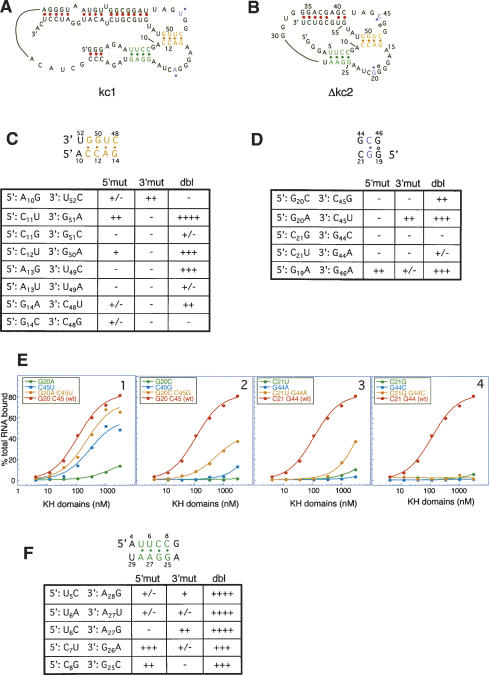
Figure 4.
Compensatory mutation analysis of Watson-Crick interactions in selected KH2 RNA ligands. (A,B) Diagram of kc1 (A) or Δkc2 (B) drawn to incorporate loop-loop interactions suggested by chemical probing. (B) Nucleotide numbering refers to Δkc2. (C) Mutations made in Δkc2 to test the presence of duplex RNA interactions (“yellow” stem) suggested by chemical probing, which would result in a loop-loop pseudoknot. Indicated mutations (A10 to G10; “A10G”, etc.) were tested for KH2 binding by filter binding assay. Binding is indicated relative to wild-type RNA (++++) to essentially non-detectable specific binding (-). For examples, see E and Supplementary Figure S3. A base pair is indicated by abrogation of binding by mutation of either the 5′ (5′ mut) or 3′ (3′ mut) nucleotide, and restoration of binding with compensatory mutations (dbl). (D) Compensatory mutation analysis to test a second potential interaction between the loops in Δkc2 (“purple” base pair), tested as in C.(E) Illustrative filter binding curves corresponding to data in D. Filter binding curves for the data in C are shown in Supplementary Figure S3. (F) Compensatory mutation analysis of a Mg+2-dependent stem (“green” stem) suggested by mfold and chemical probing in Δkc2, tested as in C.
Because this complicated structure might be formed in vitro due to the post-transcriptional heating and refolding of the RNAs but might not occur during the cotranscriptional folding of RNA in vivo, we compared the binding of KH2 to kc2 under our standard conditions (denatured and refolded) and in its newly transcribed “native” conformation. We found no difference in any aspect of binding (data not shown), suggesting that these structures can form cotranscriptionally.
Biochemical probing of the FMRP KH:kc RNA interaction
We next asked whether KH2 binding requires formation of a kissing complex. For these experiments, we generated a truncated RNA (“Δkc2”, 62 nt) (Fig. 4B) by shortening the stems and hinge region (boxed regions in kc2 in Fig. 2D) and demonstrated that KH2 bound kc2 and Δkc2 with equal affinity (data not shown). Mutations were generated in kc2 or Δkc2 and tested for KH2 binding by filter binding assay.
We first examined whether the KH2 binding is dependent on the 4-bp duplex C11-C-A-G14:C48-U-G-G51 loop-loop interaction in the proposed kissing complex (Fig. 4C, yellow). Mutations were introduced in Δkc2 to change one side or the other of each potential base pair in the duplex (Fig. 4C,D,F, “5′ and 3′ mut”) or to introduce compensatory mutations that would restore Watson-Crick interactions (Fig. 4C,D,F, “dbl”).
RNAs in which C11 was substituted with U11, permitting a U11-G51 pair, bound KH2, while substitution of G51 with A51, resulting in a C11-A51 pair, abolished binding (Fig. 4C; Supplementary Fig. S3C). A compensatory mutation generating a U11-A51 pair rescued KH2 binding (Fig. 4C; Supplementary Fig. S3C). A double compensatory mutation, G11-C51, as well as two additional double compensatory mutations in other potential base pairs (U13-A49 and C14-G48) that invert purine-pyrimidine orientation of the wild-type base pairs, fail to rescue KH2 binding (Fig. 4C,F; Supplementary Fig. S3B,F,H), suggesting the possibility that the purine-pyrimidine orientation is crucial at these positions or that introduction of one of these nucleotides changed global RNA structure.
Similarly, mutations in the adjacent C12-G50 and A13-U49 loop-loop base pairs abrogated KH2 binding and were again rescued by U12-A50 and G13-C49 compensatory mutations (Fig. 4C; Supplementary Fig. S3A,E). The final pair in the duplex is G14-C48. While a G-U pair in this position binds KH2 weakly, and an A-C pair does not bind KH2, only partial rescue occurs with the A14-U48 compensatory mutation, and no rescue was evident with C14-G48 compensatory mutation (Fig. 4C; Supplementary Fig. S3G,H). Although it is difficult to interpret these latter results definitively, it is possible that a Watson-Crick interaction is necessary for KH2 binding at the G14-C48 position, and, in addition, either the G14 or C48 are involved in further molecular interactions (with either Mg++, RNA, or KH2). Taken together, these experiments confirm that KH2 binding is dependent upon each base pair in a 4-bp duplex that is at the core of the loop-loop interaction.
A fifth possible base pair in the duplex, A10-U52, appeared to be unpaired in chemical probing experiments (Fig. 3). Indeed, mutagenesis revealed that altering the A10 to a G or U52 to a C greatly reduced KH2 binding, and the double mutation was even more deleterious (Fig. 4C; Supplementary Fig. S3D). This result indicates that base pairing at this position is not required for KH2 binding, and, together with the chemical probing experiments, suggests that an A10-U52 Watson-Crick interaction does not occur in the native structure at this position.
Two pairs of nucleotides within the loop sequences covaried (G20-C45 and G19-G46; Table 1), suggesting molecular interactions that could be relevant to KH2 binding. Mutations in these nucleotides were tested for KH2 binding (Fig. 4D,E, purple). Substitution of G20-C45 with A20-U45 (Fig. 4E, panel 1; “G20AC45U” double mutation, yellow curve) restored binding compared with either single mutation alone, including A20-C45 (Fig. 4E, panel 1; “G20A” mutation, green curve), which didn't support KH2 binding, and G20-U45 (Fig. 4E, panel 1; “C45U”, blue curve), which was an intermediate ligand for KH2. Reversing nucleotides G20-C45 to C20-G45 bound KH2 poorly (Fig. 4E, panel 2, yellow curve), suggesting either that the purine-pyrimidine orientation of this base pair is important or that introduction of either the C20 or G45 may generate a structure competing with the kissing complex. Taken together, these results strongly suggest that a canonical Watson-Crick base pair between the loops at this position N20-N′45 is necessary to support KH2 binding.
Immediately adjacent to this critical base pair are G19 and G46, which were invariant in the 13 kc clones except in kc4, where both G's were changed to A's. This covariation suggests that an A19-A46 purine pair may isosterically substitute for G19-G46. Substitution of G19 with A19 lowered the Kd approximately eightfold (from 58 to 453 nM), and G46 to A46 ∼28-fold (from 58 to 1.7 μM) compared with wild-type kc2 (Fig. 4D). The double substitution of A19-A46 substantially rescued the G46-A46 mutation (Kd of 289 nM), though not to wild-type levels (Fig. 4D). Together with the covariation at this position, these results suggest but do not prove that R19-R′46 may form a purine-purine loop-loop interaction.
Examination of the sequences of the selected 13 clones also revealed that the adjacent loop nucleotides C21 and G44 are 100% invariant in the selected KH2 ligands, suggesting that they too might base-pair. Mutation of C21-G44 to G21-C44 failed to bind KH2 (Fig. 4E, panel 4, yellow curve) and mutation to U21-A44 bound weakly (Fig. 4E, panel 3, yellow curve), and we are therefore unable to make a conclusion regarding the role of the potential base pair C21-G44 in KH2 binding.
We also examined whether KH2 binding to Δkc2 RNA was dependent on the stems evident in the RNA by mfold. Mutagenesis of the predicted 5′ stem (U5UCC8-G25GAA28) in Δkc2 confirmed that formation of each base pair was necessary for KH2 binding (Fig. 4F, green). mfold predicted a fifth base pair in the duplex, A4-U29 (Fig. 2D), which did not appear base-paired by chemical modification (Fig. 3) and KH2 binding was not rescued by compensatory mutations (data not shown), confirming that the length of this stem required for KH2 binding is limited to 4 bp.
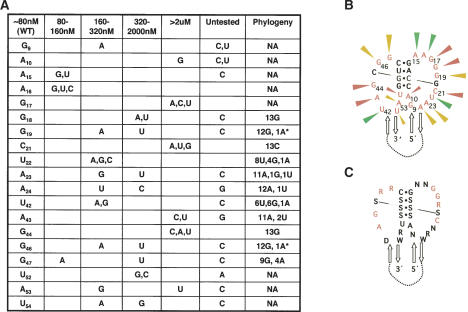
FMRP KH2 interaction with single-stranded loop nucleotides
The results of chemical probing and mutagenesis experiments allowed us to reach a tentative model about what nucleotides are likely to be single stranded or double stranded in the loop-loop region of the kissing complex RNA ligands (Fig. 5B). To fully understand the single-stranded selected regions in the kissing loops, we re-evaluated their conservation from RNA selection and undertook exhaustive mutagenesis of these nucleotides. We first used mfold to identify and discard mutations that would globally alter the structure of the Δkc2 RNAs and then generated all other mutations and tested these for KH2 binding by filter binding assay. The results (Fig. 5A) largely confirm the sequence specificity evident by analysis of the phylogeny of evolved ligands (kc1-kc13). Notably, A10, G17, G18, C21, and to a lesser extent A23 and A24 in loop 1, and A43, G44, and U52 in loop 2 are invariant by phylogeny and/or mutagenesis. In addition, G19 and G46, as mentioned, are invariant except for an A19-A46 double substitution. It is likely that requirement for a particular nucleotide implies a role in Mg+2 chelation, tertiary interactions within the RNA, or direct binding by KH2. These results suggest a preliminary model for the sequence specificity and structure of the kc RNA ligands (Fig. 5C).
Figure 5.
Mutational analysis of the non-Watson-Crick-paired regions of Δkc2. (A) Indicated nucleotides and substituted mutations for each were assessed for binding to KH2 by filter binding assay. Results are summarized as a comparison to wild-type Δkc2 Kd (80 nM) and grouped by descending Kd. Untested mutations were those in which substitution of the indicated nucleotide at that position resulted in global misfolding of Δkc2 as predicted by mfold 3.1. Phylogeny refers to the number of occurrences of a particular nucleotide out of 13 different sequences (kc1-kc13). NA (not applicable) means those nucleotides are from the fixed regions of the selected clones and are invariant. Asterisks denote the G19-G46 pair that covaries with one A19-A46 base pair. (B) Δkc2 RNA ligand redrawn to focus on loop sequences. Probable non-Watson-Crick-paired nucleotides (tested in A) are shown in red, Watson-Crick interacting nucleotides are shown in black, and 5′ and 3′ stems are shown as anti-parallel arrows. Nucleotides in which mutational analysis showed essentially no tolerance for mutation and phylogenetic variation are indicated by red arrowheads, those with some tolerance for mutation and phylogenetic variation are indicated by yellow arrowheads, and those that are most permissive for mutation and phylogenetic variation are indicated by green arrowheads. (C) Consensus of Δkc2 RNA KH2 ligand based upon experimental data. Red nucleotides are those most likely to be involved in KH2 binding. K is a G or U, R is an A or G, W is an A or U, N can be any nucleotide, and S refers to a Watson-Crick base pair.
Kissing complex RNAs disrupt FMRP-polyribosome association
Some of the strongest data regarding FMRP function have been reports that it is associated with polyribosomes (Khandjian et al. 1996; Corbin et al. 1997; Feng et al. 1997a). Whether these findings are relevant in the brain has been questioned (Zalfa et al. 2003), prompting us to re-evaluate the association of FMRP with brain polyribosomes. We developed a method of preparing intact brain polyribosomes (Stefani et al. 2004), which clearly demonstrates that FMRP cosediments on sucrose gradients with brain polyribosomes (Fig. 6; Stefani et al. 2004), coincident with or slightly larger than the ribosomal S6 (Figs. 7, 8) protein. FMRP sedimentation was shifted to light fractions when polyribosomes were prepared in the presence of EDTA, consistent with recently published data demonstrating the presence of FMRP on polyribosomes in the mouse brain (Khandjian et al. 2004; Stefani et al. 2004).
Figure 6.
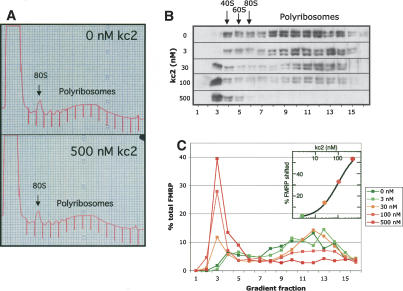
kc2 RNA competes FMRP off of mouse brain polyribosomes in a dose-dependent manner. (A) P8 mouse cerebral cortical extracts were separated by sucrose density gradient (20%-50%) centrifugation; positions of the 80S monosome and polyribosomes are indicated. Indicated amounts of kc2 RNA were added to brain lysates and incubated for 15 min at room temperature prior to centrifugation. A254 profile of sucrose density gradients for the lowest and highest kc2 RNA concentrations are shown; A254 profiles for all kc2 concentrations were indistinguishable (data not shown). (B) Western blots of indicated fractions from sucrose density gradients incubated with the indicated kc2 concentration were probed with the FMRP monoclonal antibody 1C3. Fractions corresponding to 40S, 60S, and 80S ribosomal peaks are indicated. Ribosomal protein S6 was unshifted at all kc2 concentrations (data not shown). (C) Quantitation by chemiluminescence of the Western blot data in B (Versadoc imaging); the percentage of FMRP present in each fraction as a function of the total FMRP signal in that gradient is indicated. Inset shows a plot of the percent of FMRP shifted off the polyribosomes plotted against kc2 concentration (nanomolar). The half-maximal concentration of kc2 able to compete FMRP off of polysome fractions is ∼100 nM kc2 RNA.
Figure 7.
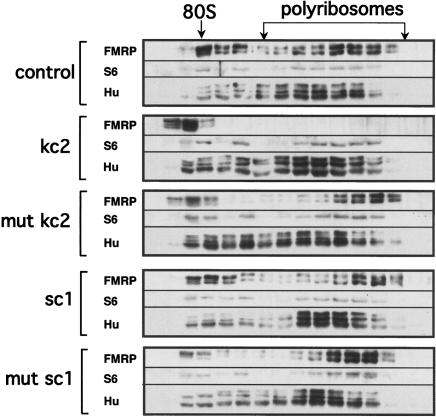
kc2 RNA competes FMRP off polyribosomes. Western blot as in Figure 6 of sucrose density gradients in which indicated RNAs were used to compete FMRP polyribosomal association; mut kc2 harbors a single point mutation in kc2 RNA (G20 to C20; see Fig. 4E for binding curve), sc1 is a high-affinity G-quartet ligand for FMRP RGG box, and mut sc1 harbors a single point mutation that destroys G-quartet formation and FMRP binding. Western blots were probed with antibodies to the indicated proteins, including FMRP, ribosomal protein S6, and the Hu family of RNA-binding proteins.
Figure 8.
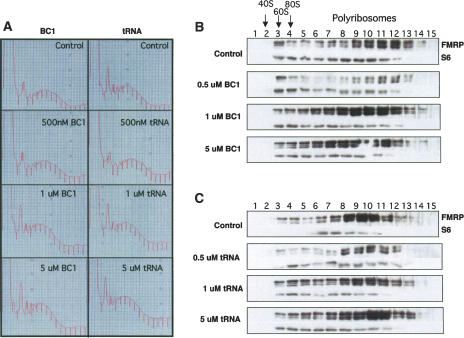
Specificity of FMRP interaction with polyribosomal targets. A254 traces (A) and Western blot analysis for FMRP or ribosomal S6 protein (B), as in Figure 6. No RNA was added to the control lysates (control). BC1 RNA (produced by in vitro transcription and gel purified) and yeast tRNA at the indicated concentrations fail to compete with FMRP binding to polyribosomes.
Importantly, the FMRP KH2 domain is critical for the interaction of FMRP with polyribosomes, as it is disrupted by the I304N mutation, and there are several proposed mechanisms to explain this finding (Feng et al. 1997a; Lewis et al. 2000; Laggerbauer et al. 2001; Ramos et al. 2003; see Discussion). Given the relationship between the KH2 domain and polyribosome association, we evaluated whether kissing complex RNA ligands may interfere with the FMRP:polyribosome complex. When brain polyribosomes were incubated with increasing concentrations of kc2 (3-500 nM; or kc1; data not shown), we observed a dramatic dose-dependent shift in FMRP sedimentation from the polysomal fractions to light fractions corresponding to a size smaller than the 40S ribosomal subunit (Fig. 6B,C). The global polyribosomal profile was unchanged (Fig. 6A) and no change was seen in the sedimentation of ribosomal protein S6 (Fig. 7; data not shown). Quantitation revealed that the half-maximal FMRP shift off polyribosomes occurred at an RNA concentration of ∼100 nM (Fig. 6C), similar to the Kd for FMRP binding kc2 in vitro. The ability of kissing complex RNA ligands to compete endogenous FMRP off polyribosomes suggests that this ligand mimics the site FMRP uses to regulate translation in neurons.
To assess the specificity of these results, the experiment was repeated with a G-quartet ligand for the RGG box (sc1; Darnell et al. 2001), a point mutant in kc2 that destroys a critical base pair in the loop-loop interaction (G20-C20, Fig. 4D,E, mut kc2) or a point mutant in sc1 that destroys the G-quartet (Darnell et al. 2001). In each case, the association of FMRP with polyribosomes was unaffected (Fig. 7). To control for the integrity of RNA, the same preparations of the four RNAs (kc2, mut kc2, sc1, and mut sc1) were used in filter binding assays with KH2 (kc2 and mut kc2) and the RGG box of FMRP (sc1 and mut sc1) and these preparations of RNA were found to bind with the expected affinity (data not shown). As an additional control, we evaluated sedimentation of the neuronal Hu RNA-binding proteins (Okano and Darnell 1997), since the nonneuronal Hu isoform is known to be polyribosome associated (Gallouzi et al. 2000), and found that the neuronal Hu proteins are polyribosome associated in mouse brain, but that kc2 did not compete these proteins off polyribosomes (Fig. 7). These results demonstrate that kc RNA specifically disrupts FMRP polyribosome association. Taken together, these experiments suggest that the binding of KH2 to kissing complex RNA ligands may mediate polyribosomal association in the brain, while association of the RGG box with G-quartets is not required for stability of the complex.
It has recently been reported that FMRP binding to BC1, a neuron-specific, dendritically localized nontranslatable RNA (Brosius and Tiedge 2001), may play a role in modulating mRNA translation (Zalfa et al. 2003). It has also been reported that FMRP binds to highly structured RNAs such as tRNA (Gabus et al. 2004). We tested whether an excess of BC1 RNA or tRNA was able to compete FMRP off polyribosomes (Fig. 8). Addition of up to 5 μM BC1 or tRNA to cortical lysates fails to compete with the in vivo binding site of FMRP on polyribosomes and displace it (Fig. 8B,C). These results demonstrate that FMRP is specifically associated with brain polyribosomes, and that this interaction is effectively competed by kissing complex RNAs.
Discussion
Fragile-X mental retardation results from mutations that lead to loss of FMRP function. One mutation of particular interest is the I304N point mutation within the second KH domain, which leads to a severe mental retardation phenotype (DeBoulle et al. 1993). Crystallographic and NMR structures of KH RNA-binding domains have lent support to the idea that the I304N mutation is likely to disrupt sequence-specific RNA recognition (Lewis et al. 2000; Liu et al. 2001; Ramos et al. 2003). Together with our current results, these observations suggest that the disease in the patient with the I304N mutation, and more generally the loss of FMRP function in the Fragile-X syndrome, may result from a loss of KH2 recognition of kissing complex RNA targets.
Such a complex structure has not previously been described as a target for a KH-type RNA-binding domain. We have previously found RNA targets for KH domains of Nova-1 (Buckanovich and Darnell 1997; Jensen et al. 2000b), Nova-2 (Yang et al. 1998), and zipcode-binding protein 1 (Farina et al. 2003), which are stem-loop RNAs harboring 4-7-nt loop motifs, and several of these interactions have been confirmed by X-ray crystallography (Lewis et al. 2000; L. Malinina, M. Teplova, K. Musunuru, A. Teplov, S.K. Burley, R.B. Darnell, and D.J. Patel, unpubl.) or biologically (Kislauskis et al. 1993; Ross et al. 1997; Jensen et al. 2000a; Dredge and Darnell 2003; Ule et al. 2003). Other examples include 7-12-nt RNA motifs identified as ligands for hnRNP K and E (Ostareck et al. 1997), and the Star-KH domain proteins SF1 (Liu et al. 2001) and GLD-1 (Ryder et al. 2004). Our analysis of the kissing complex suggests that relatively few nucleotides may be required for FMRP KH2 binding [in one loop, AG(bp)RR, and in the second loop, GGR(bp)C] (Fig. 5C), which is in the range of the size of previous KH targets. What differs here appears to be the 3D presentation of this RNA to FMRP KH2.
We have related the kissing complex ligand to the known biology of FMRP. The laboratories of Warren and Khandjian first provided evidence that FMRP associates with polyribosomes (Khandjian et al. 1996; Corbin et al. 1997; Feng et al. 1997a). The I304N mutant FMRP has been found to be associated with abnormally small mRNPs rather than polyribosomes (Feng et al. 1997a), although it is unknown whether this is due to a loss of protein-protein interactions (Feng et al. 1997a; Laggerbauer et al. 2001), protein-RNA interactions (Lewis et al. 2000; Ramos et al. 2003), or other phenomena such as protein unfolding (Musco et al. 1996). We find that the association of FMRP with brain polyribosomes is specifically competed by the KH2 kissing complex RNA, but not by G-quartet RNAs that bind to the FMRP RGG domain. Therefore, the kissing complex RNA provides a new link between the neurologic disease (via the I304N mutation) and the presumed role FMRP plays in translational regulation, as such RNA elements in vivo are likely to mediate the association of FMRP with brain polyribosomes.
Kissing complex RNAs as FMRP targets
Kissing loop-loop interactions were first described in tRNA (Kim et al. 1974; Moras et al. 1980). Some biologically important kissing complexes are intramolecular, for example, those found in tRNA and the Neurospora VS ribozyme (Rastogi et al. 1996), while others are intermolecular, for example, the bicoid 3′ UTR mRNA (Ferrandon et al. 1997) and HIV DIS element (Paillart et al. 1996a,b), which dimerize when stem-loops on two different RNA molecules interact. Although we cannot rule out the possibility that FMRP could bind to two stem-loops on different RNA molecules present in high enough local concentration, our data indicate that an essential element of KH2 binding is the interaction of two intramolecular stem-loops to form a kissing complex consisting of a 4-bp duplex and at least a single nucleotide loop-loop Watson-Crick interaction.
Several functions have been described for kissing complexes that may provide insight into the FMRP:kissing complex RNA interaction. For example, in prokaryotes, two instances have been reported in which small non-translated RNAs (OxyS and CopA) complementary to 5′ noncoding sequences form intermolecular kissing complexes that subsequently form intermolecular RNA duplexes that suppress translation (Argaman and Altuvia 2000; Kolb et al. 2000a,b). Such interactions might be relevant to the presence of FMRP in the RISC complex, where, by analogy, FMRP interactions with kissing complex RNAs could contribute to stabilization of miRNA: target RNA duplexes. In fact, FMRP promotes nucleic acid strand annealing in vitro (Gabus et al. 2004). Additionally, intermolecular loop-loop interactions of bicoid 3′ UTR elements, which again initiate RNA duplex formation, are essential for forming localized mRNA particles containing Staufen. These observations could relate to reports that FMRP is present in Staufen containing granules (Ohashi et al. 2002; Villace et al. 2004) and to the proposed role for FMRP in regulating mRNAs localized to neuronal dendrites (Greenough et al. 2001; Jin and Warren 2003; Zalfa et al. 2003; Antar et al. 2004).
FMRP KH2:kissing complex RNA interactions
Deletional and mutational analysis indicates that the FMRP KH2 domain is necessary and sufficient for interaction with kissing complex RNA; the isolated KH2 domain interacts with this RNA with no loss of affinity, and RNA binding is abrogated by the I304N mutation within KH2, but not by the I241N mutation in KH1. Interestingly, RNA selection experiments using libraries of smaller RNAs failed to produce a consensus RNA ligand for FMRP KH2, and deletional analysis failed to identify kissing complex RNAs smaller than 60 nt able to bind FMRP, suggesting that FMRP KH2 may only be able to recognize a complex RNA target. This is in contrast to other KH domain:RNA interactions; KH domains present in Nova (Buckanovich and Darnell 1997; Jensen et al. 2000b; Lewis et al. 2000), SF1 (Liu et al. 2001), hnRNP-K/hnRNP E1/αCP2 (Ostareck et al. 1997; Thisted et al. 2001), ZBP1 (Farina et al. 2003), and PSI (Amarasinghe et al. 2001) recognize RNAs with small 4-7-nt consensus sequences. One potential difference between these proteins and FMRP is the extended variable loop present within the KH2 domain (Lewis et al. 1999); however, we found no evidence that this domain affects the KH2:kissing complex interaction (J.C. Darnell, C.E. Fraser, O. Mostovetsky, and R.B. Darnell, unpubl.). Detailed understanding of the features of FMRP KH2 that mediate kissing complex RNA interactions will require structural studies.
Kissing complex RNA, mental retardation, and FMRP polyribosome association
Abrogation of the FMRP:kissing complex interaction by the KH2 I304N mutation links the identification of this RNA target to the Fragile X mental retardation disorder. This relationship in turn provides a further connection to the proposed role of FMRP in translational regulation. FMRP was first suggested to be involved in translational regulation based on findings that it associated with polyribosomes in tissue culture cell lines and that this association was abrogated by the I304N mutation. However, there have been various interpretations of the latter observation, including suggestions that the I304N mutation interferes with protein homodimerization (Laggerbauer et al. 2001) or heterodimerization with FXR2 (Feng et al. 1997a), causes protein instability (Musco et al. 1996), or abrogates sequence-specific RNA binding (Lewis et al. 2000; Ramos et al. 2003). Moreover, it has been suggested that FMRP associates with polyribosomes in tissue culture cells, but not in brain (Zalfa et al. 2003).
We have found that FMRP is associated with brain polyribosomes (Figs. 6, 7, 8; Stefani et al. 2004) and that kissing complex RNA is able to displace FMRP from polyribosomes at ∼100-nM concentration. This disruption is very specific for the kc RNA ligand: FMRP polyribosome association is not affected by kc RNA harboring a single point mutation, nor by G-quartet RNA or 50-fold higher concentrations of tRNA or BC1 RNA. These observations clearly indicate that FMRP associates with brain polyribosomes via a specific interaction between the KH2 domain and RNA.
The mechanism by which FMRP might interact with polyribosomes has not been well understood. FMRP has been reported to interact with a number of RNA-binding proteins, including FXR1P/FXR2P (Schenck et al. 2001), NUFIP (Bardoni et al. 1999), YB-1 (Ceman et al. 2000), staufen (Ohashi et al. 2002; Villace et al. 2004), and nucleolin (Ceman et al. 1999), consistent with data suggesting that FMRP may be part of large mRNPs (Mazroui et al. 2002; O'Donnell and Warren 2002; Ohashi et al. 2002; Villace et al. 2004), some of which may be associated with polyribosomes. In addition, FMRP copurifies with the RISC complex in Drosophila (Caudy et al. 2002; Ishizuka et al. 2002) and coimmunoprecipitates with the dAgo-1 homolog, a protein component of the RISC complex in mammalian cells in culture (Jin et al. 2004). These observations have suggested that FMRP might bind to polyribosomes indirectly, via protein-RNA networks, and/or RISC complex-targeting of miRNAs to mRNAs. For example, it has been suggested that FMRP might “scan” mRNAs for G-quartet elements that then become associated with RISC complex proteins in a translationally regulated polyribosome complex (Jin et al. 2004). Our current data indicate that this model needs revision. G-quartet RNAs may be involved in RNA binding, translational control, and even polyribosome association in the brain. However, G-quartets do not compete FMRP off polyribosomes (Fig. 7), and deletion of the RGG-box does not affect FMRP polyribosome association (Mazroui et al. 2003; Darnell et al. 2005), suggesting that G-quartets are not required for polyribosome association. Instead, our data suggest that direct association of FMRP with RNAs harboring kissing complex motifs fitting the consensus described in this study mediate FMRP association with brain polyribosomes.
The current study predicts that the in vivo RNA target(s) of FMRP KH domains harbor kissing complex motifs but does not identify those targets. Such studies will be aided by software capable of folding and aligning RNAs to sequence databases for RNAs with structural and sequence features as complex as the kissing complex (Dowell and Eddy 2004). Clearly, structural analysis of the KH2:kissing complex RNA interaction would facilitate such a screen.
Kissing complex and G-quartet RNA targets
Despite the primary role of kissing complex RNA in mediating FMRP polyribosome binding, G-quartet RNA binding is still likely to be relevant to FMRP biology. Previous data indicate that the majority of transcripts that both coimmunoprecipitate with FMRP and are altered in their polyribosome association in FMRP null cells contain G-quartets (Brown et al. 2001; Darnell et al. 2001). While this may appear contradictory with the current findings, the immunoprecipitations in Brown et al. were done under conditions (30 mM EDTA) that our current data demonstrate would dissociate FMRP from kissing complex RNAs (Supplementary Fig. S4). Therefore, RNAs bound solely by the KH2 domain are likely to have been inadvertently lost in those experiments. While G-quartet binding may have an unidentified role in mediating translational control by FMRP, it is neither necessary nor sufficient for polyribosomal association and may have additional roles in FMRP biology, including RNA export, localization, or interaction with components of the miRNA pathway.
FMRP may independently bind RNAs harboring G-quartets or kissing complex motifs, or may interact with both motifs at the same time. Such interactions could be in trans, such that FMRP binds both motifs in independent RNA molecules, facilitating interaction between different RNA-protein complexes. Alternatively, FMRP may bind single transcripts harboring both G-quartet and kissing complex RNA targets. Either model could accommodate an association with RISC complex proteins and further RNA selectivity via miRNA binding.
Conclusion
The association of FMRP with brain polyribosomes is specifically disrupted by competition with kissing complex RNAs fitting the consensus described here. Furthermore, the FMRP:kissing complex interaction is mediated by KH2 and is sensitive to the I304N mutation. Taken together, these findings indicate that these kissing complex RNAs may provide a crucial link between the association of FMRP in brain polyribosomes, its proposed role in regulating mRNA translation, and neurologic dysfunction in the Fragile-X syndrome. Redirecting the ongoing search for FMRP RNA targets to those RNAs harboring kissing complex motifs may be of importance in identifying RNA ligands central to the Fragile-X syndrome.
Materials and methods
Expression and purification of FMRP fusion proteins
Individual KH domains of FMRP were cloned by PCR amplification into pET21b (Novagen) to produce N-terminally T7-tagged and C-terminally histidine-tagged fusion proteins. The KH1/2 domain, including a C-terminal extension, spans amino acids SRQLA (nucleotide 758; GenBank accession no. XM_010288; human FMR1 mRNA, lacking exon 12) to GMGR (nucleotide 1469), and the KH2 domain spans amino acids AEDVI (nucleotide 968) to HLNYL (nucleotide 1327). The iso7 forms (lacking exon 12) of full-length FMRP and I304N FMRP were purified from a pMelBacB expression system in insect cells as described (Darnell et al. 2001).
In vitro RNA selection
RNA selection, including RNA transcription and gel purification, were carried out as described (Buckanovich and Darnell 1997; Darnell et al. 2001) with the following exceptions. Following the sixth round of selection with full-length FMRP in selection binding buffer (SBB, 200 mM KOAc, 50 mM TrisOAc at pH 7.7, 5 mM MgOAc), KH1/2 was used for two additional rounds of selection. RNA coeluting with protein was reverse transcribed with Superscript (GIBCO-BRL, Life Technologies) and PCR amplified with selection primers (5′primer is 5′-AGTAATACGACTCACTATAGGGAGAATTCCGACCAGAAG3′; 3′primer is 5′-TGAGGATCCATGTAGACGCACATA-3′) (Buckanovich and Darnell 1997). Ninety-six clones from round eight were sequenced following TA cloning (Invitrogen) of the PCR reaction.
In vitro transcription
In vitro transcription of RNA for binding curves was performed with 13 μL of PCR-generated DNA template, 0.4 μM NTPs, 1 μL RNAsin (Promega), 40 μCi α-32P-UTP, 1× transcription buffer (Stratagene), and 1 μL T7 RNA polymerase (Stratagene). RNA was treated with 3 units of RQ1 DNase for 45 min at 37°C and followed by gel purification on 8% denaturing polyacrylamide gels.
Nitrocellulose filter binding assays
Ten-thousand counts per minute (1-5 fmol) of internally labeled RNA (preheated to 75°C and cooled at room temperature for 5 min) was incubated with the indicated concentrations of protein in a total volume of 50 μL in SBB, 10 min at room temperature. Binding solutions were passed through MF-membrane filters (0.45 HA, Millipore) and washed with 4 mL SBB. Filters were air dried and counted in 5-mL ReadiSafe scintillant. Data were plotted as percentage of total RNA bound versus log of the protein concentration and Kd's determined using Kaleidograph software (Synergy Software).
Production of a 5′ UTR containing the in vitro selected sequence kc1
The sequence encoding kc1 was inserted into pGEMTEZ (Promega) and then subcloned into the HindIII site of the pGL3-promoter plasmid (Promega). The 5′ UTR was transcribed in vitro after PCR amplification with a 5′ primer harboring a T7 RNA polymerase promoter such that the first nucleotide transcribed is the natural cap site of the RNA, and a 3′ primer (GL2; Promega) hybridizing just downstream of the translation initiation site, to generate a 284-nt long RNA.
Electrophoretic mobility shift assay
Native gel electrophoresis was performed at 4°C in 0.5× TBM (45 mM Tris at pH 8.3, 45 mM borate, 2.5 mM MgCl2)ingelsof 8% polyacrylamide (acrylamide:bis ratio 39:1). RNA was diluted to 5000 cpm/μL in RNA dilution buffer (100 mM KOAc, 50 mM TrisOAc at pH 7.7, 5 mM MgOAc), heated for 5 min to 60°C and an equal volume of 15% Ficoll added. Five microliters RNA was added to 1 μM KH1-KH2 in a total volume of 20 μL in gel shift buffer (50 mM KOAc, 50 mM TrisOAc at pH 7.7, 10 mM DTT, 5 mM Mg(OAc)2, 30 μg/mL tRNA), and samples incubated for 20 min at 4°C. Gels (Bio-Rad minigels) were prerun at 100 V for 20 min and voltage increased to 200 V at the time of sample loading, run for 20 min, and exposed to autoradiographic film (Kodak MR). Where indicated, monoclonal antibodies, 1 μL (anti-T7 from Novagen, anti-Flag (M2) from Sigma), were mixed with the proteins before the addition of RNA.
Lead acetate cleavage of RNA
RNA (40 pmol) produced by in vitro RNA transcription was 5′ end labeled with T4 polynucleotide kinase (New England Biolabs) and γ-32P-ATP following dephosphorylation with calf intestinal alkaline phosphatase. Following gel purification, RNA was digested with RNase T1 (GIBCO-BRL, 1180 U/mL) by pre-heating 2 μL RNA with 6 μL 1.25× T1 buffer (8.75 M urea, 625 mM Na-Citrate at pH 5.0, 1.25 mM EDTA) for 2 min at 50°C. Two microliters T1 was added for 4 min at 50°C and stopped by addition of 6 μL loading buffer (USB). RNAs were subject to mild alkaline hydrolysis by incubating 10 μL RNA in 50 mM Na carbonate at pH 9.0, for 3 min at 95°C and ethanol precipitated.
For lead acetate treatment 5′-labeled RNA was heated for 10 min to 75°C in 1× HEPES-SBB (25 mM HEPES at pH 7.5, 200 mM potassium acetate, 5 mM Mg-acetate) and the indicated concentration of MgCl2 and cooled at room temperature for 5 min. Lead acetate was added to 10 mM for 5 min at 37°C. The reaction was stopped by addition of 50 mM EDTA, 1/10th volume of 3 M sodium acetate at pH 5.2, and two volumes of ethanol:isopropanol (1:1) and precipitated at -80°C. Washed pellets were run on 8% urea PAGE gels run in 1× TBE, visualized by autoradiography, and quantitated by phosphorimaging (Bio-Rad Molecular Imager FX).
Chemical modifications of RNA
All reactions were performed with 0.1 pmol of unlabeled kc1 transcript added to 100 μL native buffer (N) or semidenaturing buffer (E; see below), heated for 10 min at 75°C and cooled at room temperature for 5 min followed by addition of the indicated amounts of chemicals for the indicated times. Reactions were stopped by addition of 15 μg tRNA, 12 μL sodium acetate, and 250 μL ethanol:isopropanol (1:1). Following precipitation and washing, the pellets were dissolved in 7 μL water, and 1 × 106 cpm of 5′ labeled primer was added, denatured for 5 min at 55°C, and 2 μL RT buffer (BRL) added. Following annealing for 5 min at room temperature, 2 μL 5× buffer, 2 μL 0.1 M DTT, 2 μL 10 mM dNTP, 2 μL water, 0.5 μL RNAsin, and 0.5 μL Superscript II RT (BRL) were added. Reactions were incubated for 30 min at 37°C and 30 min at 48°C, and 5 μL 3 M KOH was added to remove the RNA template. After incubation for 3 min at 95°C, 25 μL of 50 mM Tris-HCl at pH 8.0, 7.5 mM EDTA, and 0.5% SDS was added. RT products were precipitated with 6 μL 3 M acetic acid, 6 μL 3 M Na-acetate at pH 5.2, and 200 μL isopropanol:ethanol (1:1) and pellets were loaded on 8% urea PAGE gels and phosphorimaged.
Buffers for dimethyl sulfate (DMS, Sigma) modification were DMS-N (50 mM sodium cacodylate at pH 7.5, 50 mM KCl, 5 mM MgCl2) or DMS-E (50 mM sodium cacodylate at pH 7.5, 1 mM EDTA). Three microliters of DMS was added for 5 min at room temperature. Buffers for 1-cyclohexyl-3-(2-morpholino-ethyl)carbodiimide metho-p-toluenesulfonate (CMCT, Sigma) were CMCT-N (50 mM sodium borate at pH 8.0, 50 mM KCl, 5 mM MgCl2) and CMCT-E (50 mM sodium borate, 1 mM EDTA). CMCT was added to 14 mg/mL final from a fresh 42 mg/mL stock and the reaction incubated for 10 or 30 min at 30°. Buffers for 2-keto-3-ethoxybutyraldehyde (kethoxal, ICN) were K-N (50 mM sodium cacodylate at pH 7.0, 50 mM KCl, 5 mM MgCl2) and K-E (50 mM sodium cacodylate at pH 7.0, 1 mM EDTA). Kethoxal was added to 2 mg/mL from a 10× stock in ethanol for 1, 5, or 10 min.
Point mutations in kc2 and Δkc2
Mutations were in kc2 or Δkc2 were generated by PCR amplification with one mutant primer. The PCR products were passed over a G-25 column (Amersham Pharmacia Biotech) and used as templates for in vitro transcription. Mutations were confirmed by sequencing the PCR products on both strands.
Mouse cerebral cortical polyribosome analysis
One-week-old to 2-wk-old CD1 mice were killed by decapitation. The brain was removed and placed in ice-cold dissection buffer (10 mM HEPES-KOH at pH 7.4, 150 mM KCl, 5 mM MgCl2, 100 μg/mL cycloheximide). The cerebral cortex was dissected free of underlying brain and white matter, homogenized in 1 mL per cortex of lysis buffer (10 mM HEPES-KOH at pH 7.4, 150 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 100 μg/mL cycloheximide, 1× Complete EDTA-free protease inhibitor cock-tail [Roche], 40 U/mL rRNasin [Promega]) with 12 strokes at 900 rpm in a motor-driven glass-Teflon homogenizer. Homogenate was spun at 2000 × g for 10 min at 4°C. In vitro transcribed RNAs were prepared as described above, incubated in 100 μL of 1× SBB for 10 min at 75°C, then incubated for 15 min at room temperature. The supernatant (S1) from the homogenized material was collected and adjusted to 1% NP-40, v/v. Two microliters of rRNasin, and 100 μL of in vitro transcribed RNA (or yeast tRNA [Roche]) in 1× SBB were added to 1.1 mL of S1 and mixed by inversion. After incubation for 15 min at room temperature, S1 lysate was spun at 20,000 × g for 10 min at 4δCand the supernatant (S2) was loaded onto a 20%-50% w/w linear density gradient of sucrose in 10 mM HEPES-KOH at pH 7.4, 150 mM KCl, and 5 mM MgCl2. Material obtained from one cortex was loaded onto each gradient. Gradients were centrifuged at 40,000 × g for 2 h at 4°C in a Beckman SW 41 rotor. Fractions of 0.5 or 0.75 mL volume were collected with continuous monitoring at 254 nm using an ISCO UA-6 UV detector.
Western blotting
The proteins contained in each fraction of the sucrose gradients were TCA-precipitated and analyzed by Western blot using anti-FMRP monoclonal antibody diluted 1:2000 (1C3, Chemicon International mAb 2160); anti-S6 ribosomal protein (Cell Signalling); and anti-Hu B, C, and D (human Hu syndrome patient sera) and the appropriate anti-HRP secondary antibody (Jackson Immunochemicals). Chemiluminescence was quantitated with a Versadoc Imaging System (Bio-Rad).
Acknowledgments
We thank Dinshaw Patel, Anh Tuan Phan, Alexander Serganov, Kirk Jensen, Kate Dredge, and members of the lab for helpful discussions and critical review of the manuscript. This work was supported by the National Institute of Health (R01 NS34389 and NS40955 [R.B.D.], R01 HD40647 [J.C.D.], and R01 HG01363 [S.R.E.]) and the Tri-Institutional MD-PhD Program of The Rockefeller University, Memorial Sloan-Kettering Cancer Center and Weill Cornell Medical College (MSTP-GM07739 [C.E.F.]). R.B.D. and Sean Eddy are Investigators of the Howard Hughes Medical Institute.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1276805.
References
- Amarasinghe A.K., MacDiarmid, R., Adams, M.D., and Rio, D.C. 2001. An in vitro-selected RNA-binding site for the KH domain protein PSI acts as a splicing inhibitor element. RNA 7: 1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar L.N., Afroz, R., Dictenberg, J.B., Carroll, R.C., and Bassell, G.J. 2004. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J. Neurosci. 24: 2648-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L. and Altuvia, S. 2000. fhlA repression by OxyS RNA: Kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300: 1101-1112. [DOI] [PubMed] [Google Scholar]
- Bardoni B., Schenck, A., and Mandel, J.L. 1999. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum. Mol. Genet. 8: 2557-2566. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. [DOI] [PubMed] [Google Scholar]
- Braddock D.T., Baber, J.L., Levens, D., and Clore, G.M. 2002a. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 21: 3476-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock D.T., Louis, J.M., Baber, J.L., Levens, D., and Clore, G.M. 2002b. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature 415: 1051-1056. [DOI] [PubMed] [Google Scholar]
- Brannvall M., Mikkelsen, N.E., and Kirsebom, L.A. 2001. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various divalent metal ions. Nucleic Acids Res. 29: 1426-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. and Tiedge, H. 2001. Neuronal BC1 RNA: Intracellular transport and activity-dependent modulation. Results Probl. Cell Differ. 34: 129-138. [DOI] [PubMed] [Google Scholar]
- Brown R.S., Hingerty, B.E., Dewan, J.C., and Klug, A. 1983. Pb(II)-catalysed cleavage of the sugar-phosphate backbone of yeast tRNAPhe—Implications for lead toxicity and self-splicing RNA. Nature 303: 543-546. [DOI] [PubMed] [Google Scholar]
- Brown V., Jin, P., Ceman, S., Darnell, J.C., O'Donnell, W.T., Tenenbaum, S.A., Jin, X., Feng, Y., Wilkinson, K.D., Keene, J.D., et al. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X Syndrome. Cell 107: 477-487. [DOI] [PubMed] [Google Scholar]
- Buckanovich R.J. and Darnell, R.B. 1997. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell. Biol. 17: 3194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Myers, M., Hannon, G.J., and Hammond, S.M. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes & Dev. 16: 2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Ketting, R.F., Hammond, S.M., Denli, A.M., Bat-hoorn, A.M., Tops, B.B., Silva, J.M., Myers, M.M., Hannon, G.J., and Plasterk, R.H. 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411-414. [DOI] [PubMed] [Google Scholar]
- Ceman S., Brown, V., and Warren, S.T. 1999. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol. Cell. Biol. 19: 7925-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S., Nelson, R., and Warren, S.T. 2000. Identification of mouse YB1/p50 as a component of the FMRP-associated mRNP particle. Biochem. Biophys. Res. Commun. 279: 904-908. [DOI] [PubMed] [Google Scholar]
- Corbin F., Bouillon, M., Fortin, A., Morin, S., Rousseau, F., and Khandjian, E.W. 1997. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 6: 1465-1472. [DOI] [PubMed] [Google Scholar]
- Darnell J.C., Jensen, K.B., Jin, P., Brown, V., Warren, S.T., and Darnell, R.B. 2001. Fragile X mental retardation protein targets G Quartet mRNAs important for neuronal function. Cell 107: 489-499. [DOI] [PubMed] [Google Scholar]
- Darnell J.C., Mostovetsky, O. and Darnell, R.B. 2005. FMRP RNA targets: Identification and validation. Genes, Brain and Behavior (in press). [DOI] [PubMed]
- DeBoulle K., Verkerk, A., Reyniers, E., Vits, L., Hendrickx, J., Van Roy, B., Van Den Bos, F., de Graaff, E., Oostra, B., and Willems, P. 1993. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 3: 31-35. [DOI] [PubMed] [Google Scholar]
- Dowell R.D. and Eddy, S.R. 2004. Evaluation of several light-weight stochastic context-free grammars for RNA secondary structure prediction. BMC Bioinformatics 5: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. 2004. A guide to ions and RNA structure. RNA 10: 335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge B.K. and Darnell, R.B. 2003. Nova regulates GABA(A) receptor γ2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol. Cell. Biol. 23: 4687-4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin, F., Mougel, M., Romby, P., Ebel, J.P., and Ehresmann, B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15: 9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. and Szostak, J. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818-822. [DOI] [PubMed] [Google Scholar]
- Farina K.L., Huttelmaier, S., Musunuru, K., Darnell, R.B., and Singer, R.H. 2003. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160: 77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Absher, D., Eberhart, D.E., Brown, V., Malter, H.E., and Warren, S.T. 1997a. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1: 109-118. [DOI] [PubMed] [Google Scholar]
- Feng Y., Gutekunst, C.A., Eberhart, D.E., Yi, H., Warren, S.T., and Hersch, S.M. 1997b. Fragile X mental retardation protein-nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J. Neurosci. 17: 1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon D., Koch, I., Westhof, E., and Nusslein-Volhard, C. 1997. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16: 1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C., Mazroui, R., Tremblay, S., Khandjian, E.W., and Darlix, J.L. 2004. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 32: 2129-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi I.E., Brennan, C.M., Stenberg, M.G., Swanson, M.S., Eversole, A., Maizels, N., and Steitz, J.A. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. 97: 3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F.B. 2002. Understanding fragile X syndrome: Insights from retarded flies. Neuron 34: 859-862. [DOI] [PubMed] [Google Scholar]
- Greenough W.T., Klintsova, A.Y., Irwin, S.A., Galvez, R., Bates, K.E., and Weiler, I.J. 2001. Synaptic regulation of protein synthesis and the fragile X protein. Proc. Natl. Acad. Sci. 98: 7101-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R.R., Lee, J.C., and Cannone, J.J. 2002. The accuracy of ribosomal RNA comparative structure models. Curr. Opin. Struct. Biol. 12: 301-310. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi, M.C., and Siomi, H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes & Dev. 16: 2497-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., and Darnell, R.B. 2000a. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25: 359-371. [DOI] [PubMed] [Google Scholar]
- Jensen K.B., Musunuru, K., Lewis, H.A., Burley, S.K., and Darnell, R.B. 2000b. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova KH3 domain. Proc. Natl. Acad. Sci. 97: 5740-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P. and Warren, S.T. 2003. New insights into fragile X syndrome: From molecules to neurobehaviors. Trends Biochem. Sci. 28: 152-158. [DOI] [PubMed] [Google Scholar]
- Jin P., Zarnescu, D.C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T.A., Nelson, D.L., Moses, K., and Warren, S.T. 2004. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7: 113-117. [DOI] [PubMed] [Google Scholar]
- Kaytor M.D. and Orr, H.T. 2001. RNA targets of the fragile x protein. Cell 107: 555-557. [DOI] [PubMed] [Google Scholar]
- Khandjian E.W., Corbin, F., Woerly, S., and Rousseau, F. 1996. The fragile X mental retardation protein is associated with ribosomes. Nat. Genet. 12: 91-93. [DOI] [PubMed] [Google Scholar]
- Khandjian E.W., Huot, M.E., Tremblay, S., Davidovic, L., Mazroui, R., and Bardoni, B. 2004. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. 101: 13357-13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Suddath, F.L., Quigley, G.J., McPherson, A., Sussman, J.L., Wang, A.H., Seeman, N.C., and Rich, A. 1974. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435-440. [DOI] [PubMed] [Google Scholar]
- Kislauskis E.H., Li, Z., Singer, R.H., and Taneja, K.L. 1993. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 123: 165-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb F.A., Engdahl, H.M., Slagter-Jager, J.G., Ehresmann, B., Ehresmann, C., Westhof, E., Wagner, E.G., and Romby, P. 2000a. Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 19: 5905-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb F.A., Malmgren, C., Westhof, E., Ehresmann, C., Ehresmann, B., Wagner, E.G., and Romby, P. 2000b. An unusual structure formed by antisense-target RNA binding involves an extended kissing complex with a four-way junction and a side-by-side helical alignment. RNA 6: 311-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Ostareck, D., Keidel, E.M., Ostareck-Lederer, A., and Fischer, U. 2001. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10: 329-338. [DOI] [PubMed] [Google Scholar]
- Lewis H.A., Chen, H., Edo, C., Buckanovich, R.J., Yang, Y.Y., Musunuru, K., Zhong, R., Darnell, R.B., and Burley, S.K. 1999. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure 7: 191-203. [DOI] [PubMed] [Google Scholar]
- Lewis H.A., Musunuru, K., Jensen, K.B., Edo, C., Chen, H., Darnell, R.B., and Burley, S.K. 2000. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323-332. [DOI] [PubMed] [Google Scholar]
- Liu Z., Luyten, I., Bottomley, M.J., Messias, A.C., Houngninou-Molango, S., Sprangers, R., Zanier, K., Kramer, A., and Sattler, M. 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098-1102. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911-940. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Huot, M.E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E.W. 2002. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11: 3007-3017. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Huot, M.E., Tremblay, S., Boilard, N., Labelle, Y., and Khandjian, E.W. 2003. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 12: 3087-3096. [DOI] [PubMed] [Google Scholar]
- Miyashiro K.Y., Beckel-Mitchener, A., Purk, T.P., Becker, K.G., Barret, T., Liu, L., Carbonetto, S., Weiler, I.J., Greenough, W.T., and Eberwine, J. 2003. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron 37: 417-431. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond, M.B., Fischer, J., Weiss, R., Thierry, J.C., Ebel, J.P., and Giege, R. 1980. Crystal structure of yeast tRNAAsp. Nature 288: 669-674. [DOI] [PubMed] [Google Scholar]
- Musco G., Stier, G., Joseph, C., Morelli, M.A.C., Nilges, M., Gibson, T., and Pastore, A. 1996. Three-dimensional structure and stability of the KH domain: Molecular insights into the Fragile X syndrome. Cell 85: 237-245. [DOI] [PubMed] [Google Scholar]
- O'Donnell W.T. and Warren, S.T. 2002. A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci. 25: 315-338. [DOI] [PubMed] [Google Scholar]
- Ohashi S., Koike, K., Omori, A., Ichinose, S., Ohara, S., Kobayashi, S., Sato, T.A., and Anzai, K. 2002. Identification of mRNA/protein (mRNP) complexes containing Purα, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 277: 37804-37810. [DOI] [PubMed] [Google Scholar]
- Okano H.J. and Darnell, R.B. 1997. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 17: 3024-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M., Gdaniec, Z., Fischer, A., Grabarkiewicz, T., Bielecki, L., and Adamiak, R.W. 2002. The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucleic Acids Res. 30: 4241-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D.H., Ostarecklederer, A., Wilm, M., Thiele, B.J., Mann, M., and Hentze, M.W. 1997. mRNA silencing in erythroid differentiation—hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89: 597-606. [DOI] [PubMed] [Google Scholar]
- Paillart J.C., Marquet, R., Skripkin, E., Ehresmann, C., and Ehresmann, B. 1996a. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie 78: 639-653. [DOI] [PubMed] [Google Scholar]
- Paillart J.C., Skripkin, E., Ehresmann, B., Ehresmann, C., and Marquet, R. 1996b. A loop-loop `kissing' complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. 93: 5572-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A., Hollingworth, D., and Pastore, A. 2003. The role of a clinically important mutation in the fold and RNA-binding properties of KH motifs. RNA 9: 293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T., Beattie, T.L., Olive, J.E., and Collins, R.A. 1996. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 15: 2820-2825. [PMC free article] [PubMed] [Google Scholar]
- Ross A.F., Oleynikov, Y., Kislauskis, E.H., Taneja, K.L., and Singer, R.H. 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17: 2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder S.P., Frater, L.A., Abramovitz, D.L., Goodwin, E.B., and Williamson, J.R. 2004. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 11: 20-28. [DOI] [PubMed] [Google Scholar]
- Schaeffer C., Bardoni, B., Mandel, J.L., Ehresmann, B., Ehresmann, C., and Moine, H. 2001. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 20: 4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A., Bardoni, B., Moro, A., Bagni, C., and Mandel, J.L. 2001. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. 98: 8844-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Choi, M., Siomi, M., Nussbaum, R., and Dreyfuss, G. 1994. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 77: 33-39. [DOI] [PubMed] [Google Scholar]
- Stefani G., Fraser, C.E., Darnell, J.C., and Darnell, R.B. 2004. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24: 9272-9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T., Lyakhov, D.L., and Liebhaber, S.A. 2001. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and α CP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem. 276: 17484-17496. [DOI] [PubMed] [Google Scholar]
- Todd P.K., Mack, K.J., and Malter, J.S. 2003. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl. Acad. Sci. 100: 14374-14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C. and Gold, L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505-510. [DOI] [PubMed] [Google Scholar]
- Ule J., Jensen, K.B., Ruggiu, M., Mele, A., Ule, A., and Darnell, R.B. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212-1215. [DOI] [PubMed] [Google Scholar]
- Villace P., Marion, R.M., and Ortin, J. 2004. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32: 2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.Y.L., Yin, G.L., and Darnell, R.B. 1998. The neuronal RNA binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl. Acad. Sci. 95: 13254-13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B., and Bagni, C. 2003. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112: 317-327. [DOI] [PubMed] [Google Scholar]
- Zhang Y.Q., Bailey, A.M., Matthies, H.J., Renden, R.B., Smith, M.A., Speese, S.D., Rubin, G.M., and Broadie, K. 2001. Drosophila fragile X-related gene regulates the MAP1B homolog futsch to control synaptic structure and function. Cell 107: 591-603. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]