Abstract
Cells respond to a variety of extracellular and intracellular forms of stress by down-regulating rRNA synthesis. We have investigated the mechanism underlying stress-dependent inhibition of RNA polymerase I (Pol I) transcription and show that the Pol I-specific transcription factor TIF-IA is inactivated upon stress. Inactivation is due to phosphorylation of TIF-IA by c-Jun N-terminal kinase (JNK) at a single threonine residue (Thr 200). Phosphorylation at Thr 200 impairs the interaction of TIF-IA with Pol I and the TBP-containing factor TIF-IB/SL1, thereby abrogating initiation complex formation. Moreover, TIF-IA is translocated from the nucleolus into the nucleoplasm. Substitution of Thr 200 by valine as well as knock-out of Jnk2 prevent inactivation and translocation of TIF-IA, leading to stress-resistance of Pol I transcription. Our data identify TIF-IA as a downstream target of the JNK pathway and suggest a critical role of JNK2 to protect rRNA synthesis against the harmful consequences of cellular stress.
Keywords: RNA polymerase I, TIF-IA, JNK, transcription, oxidative stress
All organisms need to sense and respond to conditions that stress their homeostatic mechanisms. The cellular response to stress represents an adaptation to environmental variables, such as nutrient supply, temperature or oxidative stress. This response to extra- or intracellular stress is accompanied by marked alterations of gene expression. Stress stimuli have been traditionally shown to activate members of mitogen-activated protein kinases (MAPKs), including BMK/ERK5, the 38-kDa protein kinases (p38 MAPKs) and the c-Jun NH2-terminal protein kinases (JNKs). JNKs belong to stress-activated protein kinases (SAPKs) that play a crucial role in the cellular response to environmental stress and balance prosurvival stimuli with opposing signals leading to apoptosis (Schreiber et al. 1995; Chen et al. 1996). The activity of JNKs is stimulated by exposure to UV-B radiation, alkylating agents, hyperosmotic shock, proinflammatory cytokines, and oxidative damage (Martindale and Holbrook 2002). Anisomycin, a widely used inducer of all SAPKs, inhibits eukaryotic peptidyl transferase and activates JNKs through an ill-defined ribosome-dependent mechanism (Iordanov et al. 1997). Upon activation, JNKs phosphorylate various target proteins, such as c-Jun, ATF-2, and Elk-1, to alter their activity and elicit biological effects. The activity of JNK is regulated through phosphorylation on threonine and tyrosine residues in the T★PY★ motif by SEK1/MKK4 (Derijard et al. 1995), which in turn is activated by upstream kinases, e.g., MEKK-1 and ASK (Barr and Bogoyevitch 2001).
All mechanisms that affect a cell's temporal need for protein biosynthesis depend and impinge on the availability of ribosomes and its constituents. Therefore, transcription of rRNA genes by RNA polymerase I (Pol I) is highly regulated to be responsive to both general metabolism and specific environmental challenges (for review, see Grummt 2003; Moss 2004). The key player in growth-dependent regulation of rDNA transcription is the transcription initiation factor TIF-IA, the mammalian homolog of yeast Rrn3p (Bodem et al. 2000; Moorefield et al. 2000) that is associated with a subpopulation of Pol I (Tower and Sollner-Webb 1987; Schnapp et al. 1993). TIF-IA interacts with the TBP-containing factor TIF-IB/SL1, thereby targeting Pol I to the rDNA promoter (Miller et al. 2001; Cavanaugh et al. 2002; Yuan et al. 2002). Importantly, the amount of Rrn3p/TIF-IA associated with Pol I, but not the overall level of Rrn3p/TIF-IA, is decreased in stationary yeast (Milkereit and Tschochner 1998) and starved or cycloheximide-treated mammalian cells (Cavanaugh et al. 2002; Yuan et al. 2002) indicating that transcriptional shut-off is caused by dissociation of TIF-IA from Pol I. These and other experiments demonstrate that most, if not all, growth-dependent control of rDNA transcription may be exerted by the formation and recruitment of TIF-IA/Pol I complexes to the rDNA promoter. In support of TIF-IA playing a central role in modulating Pol I transcription in response to external signals, TIF-IA is phosphorylated at multiple sites and both the phosphorylation pattern and the activity of TIF-IA is altered in response to environmental changes. Conditions that support growth and proliferation, such as nutrient and growth factor availability, trigger signaling pathways that activate TIF-IA by phosphorylation of serine residues 44, 633, and 649 by mTOR- and MAPK-dependent signaling cascades (Zhao et al. 2003; Mayer et al. 2004).
In this study we have investigated the response of the Pol I transcription apparatus to cellular stress. We provide evidence that activation of JNK2 inhibits rRNA synthesis in response to oxidative and ribotoxic stress. This down-regulation of Pol I transcription is due to JNK-mediated inactivation of TIF-IA by phosphorylation at one critical threonine residue (Thr 200). Upon phosphorylation at Thr 200, TIF-IA is translocated from the nucleolus into the nucleoplasm. Our data provide a mechanistic explanation for the regulation of rRNA synthesis under oxidative stress and suggest that TIF-IA exists in transcriptionally active and inactive forms depending on the environmental conditions.
Results
JNK signaling is required for stress-induced inhibition of rRNA synthesis
To study the signaling pathways that regulate rRNA synthesis upon cellular stress, HEK293T cells were treated with compounds that induce oxidative stress, such as H2O2 and BSO, or with anisomycin, a ribotoxic drug that induces both p38-MAPK and JNK cascades. As demonstrated in Figure 1A, the level of 45S pre-rRNA was decreased to different extents by the various stress-inducing agents, being lowest after treatment with H2O2 and anisomycin (lanes 2,4). If SB203580, a specific inhibitor of p38 kinases, was added prior to anisomycin treatment, the effect of anisomycin was potentiated (Fig. 1A, lane 5). In the presence of the JNK inhibitor SP600125 a partial recovery of pre-rRNA synthetic activity was observed (Fig. 1A, lane 6). Neither inhibitor alone affected pre-rRNA synthesis (data not shown). As shown in the bar diagram, there was an inverse correlation between pre-rRNA levels and JNK2 activity in response to stress-inducing agents. A >10-fold stimulation of JNK2 activity was found in cells treated with anisomycin and anisomycin plus SB203580 (Fig. 1A, lanes 4,5), suggesting that the JNK signaling pathway is responsible for inhibition of pre-rRNA synthesis. There was no change in the amount of individual components of the Pol I transcription machinery (Pol I, TIF-IA, TIF-IB/SL1, UBF) after drug treatment, indicating that the activity, but not the amount, of Pol I or basal transcription factors was altered upon short-term cellular stress (data not shown).
Figure 1.
Stress-induced activation of JNK inhibits Pol I transcription. (A) Cellular stress induces JNK activity and impairs rRNA synthesis. RNA from HEK293T cells that were mock-treated or treated with H2O2 (200 μM, 60 min), BSO (1 mM, 16 h), anisomycin (10 μM, 60 min), SB203580 (2 μM, 20 min), or SP600125 (5 μM, 30 min) followed by anisomycin (10 μM, 60 min) was analyzed on Northern blots for 45S pre-rRNA (upper panel) and JNK2 activity and levels (lower two panels). To measure JNK2 activity, HEK293T cells were transfected with pcDNA3.1HA-JNK2 and subjected to the indicated stress stimuli. HA-JNK2 was immunopurified from 2 × 106 cells and tested for kinase activity using purified GST-tagged c-Jun (amino acids 1-166). Western blots of cellular lysates were probed using antibodies against overexpressed HA-JNK2. A bar diagram shows quantification of levels of pre-rRNA (normalized to actin-mRNA signals) and phospho-c-Jun (normalized to HA-JNK western blot units). Error bars denote the standard deviation derived from three independent experiments. (B) Stress does not impair pre-rRNA synthesis in Jnk2 single and Jnk1,2 double-knockout fibroblasts. 45S pre-rRNA was monitored on Northern blots using 3 μg RNA from parental and Jnk1,2-/- MEFs (top panel) or immortalized Jnk1-/- and Jnk2-/- NIH3T3 cells (bottom panel) treated with H2O2 (200 μM, 60 min), BSO (1 mM, 16 h), or anisomycin (10 μM, 60 min). In the bottom panel, the filter was reprobed to detect actin-mRNA.
As a complementary approach to prove that JNK activation is necessary for stress-dependent shutdown of rRNA synthesis, we analyzed pre-rRNA levels in Jnk1,2-deficient MEFs (Tournier et al. 2000). While in parental MEFs a three- to fourfold reduction of pre-rRNA synthesis was observed upon stress, pre-rRNA levels in Jnk1&2 double-knockout (Jnk1,2-/-) MEFs remained unaffected (Fig. 1B, top panel). To examine which JNK subtype mediates down-regulation of Pol I transcription, we used immortalized NIH3T3 cells in which either Jnk1 or Jnk2 has been disrupted (Sabapathy et al. 1999) and monitored Pol I transcription after treatment with anisomycin and H2O2, respectively. Clearly, there were marked differences in pre-rRNA levels in Jnk1-/- compared with Jnk2-/- cells. While rRNA synthesis in Jnk1-deficient cells displayed increased sensitivity towards anisomycin and H2O2 treatment, neither treatment affected Pol I transcription in Jnk2-deficient cells (Fig. 1B, bottom panel). Thus, JNK2 plays a key role in stress-dependent regulation of rRNA synthesis.
TIF-IA is inactivated by JNK-directed phosphorylation
To analyze the molecular mechanisms that down-regulate rRNA synthesis in response to cellular stress, we assayed Pol I transcription in nuclear extracts from un-treated, anisomycin- and H2O2-treated FM3A cells in vitro. While nuclear extracts prepared from untreated control cells efficiently transcribed the rDNA template, extracts from H2O2- and anisomycin-treated cells did not support specific transcription (Fig. 2A, left panels, lane 1). Supplementation with neither exogenous UBF, nor Pol I rescued transcriptional activity (Fig. 2A, lanes 2,5), whereas addition of cellular TIF-IA relieved transcriptional repression (Fig. 2A, lane 3). Rescue of activity by exogenous TIF-IA was also observed in nuclear extracts from anisomycin- and H2O2-treated HEK293T cells (Fig. 2A, right panels). Supplementation of the extracts with TIF-IB/SL1 resulted in some, albeit much less than with TIF-IA, activation of Pol I transcription (Fig. 2A, lane 4), which may be due to contamination of the TIF-IB/SL1 fraction with trace amounts of TIF-IA.
Figure 2.
Stress-induced inhibition of Pol I transcription is due to inactivation of TIF-IA by JNK-mediated phosphorylation. (A) TIF-IA complements transcriptional activity of nuclear extracts from H2O2- and anisomycin-treated cells. Transcription reactions containing 30 μg nuclear extract protein from untreated (-), anisomycin-treated (NEaniso), or H2O2-treated (NEH2O2) FM3A (left) or HEK293T cells (right) were supplemented with fractions containing partially purified TIF-IA, TIF-IB/SL1, Pol I, or recombinant UBF, as indicated. (B) TIF-IA from H2O2- or anisomycin-treated cells is transcriptionally inactive. Forty and 80 ng of Flag-TIF-IA immunopurified from HEK293T cells that were either untreated or treated with anisomycin + SB203580, anisomycin + SP600125, or H2O2 were assayed for their capability to complement transcriptional activity of nuclear extracts from density-arrested cells. A silver-stained SDS-polyacrylamide gel with 200 ng of TIF-IA added to the transcription reactions is shown on the left. (C) In vitro phosphorylation of TIF-IA. GST-tagged TIF-IA purified from Eschericheria coli was phosphorylated with immunopurified HA-tagged JNK2 or p38. (D) Phosphorylation by JNK2 inactivates TIF-IA. (Lanes 1,2) A reconstituted TIF-IA-responsive transcription system consisting of partially purified Pol I, TIF-IB/SL1 and recombinant UBF (Schnapp and Grummt 1996) was used to assay the activity of TIF-IA. In lanes 3-5, TIF-IA was preincubated for 30 min in the presence of ATP with blocked control beads (lane 3), bead-bound HA-JNK2 (lane 4), or HA-p38 (lane 5) before adding to the transcription reactions.
To prove that stress-activated signaling pathways target TIF-IA, the transcriptional activity of TIF-IA from control and H2O2-treated HEK293T cells was compared using nuclear extracts from density-arrested cells. Extracts from growth-inhibited cells are defective in supporting specific initiation on rDNA templates (Fig. 2B, lane 1). Transcriptional activity was restored by TIF-IA immunopurified from mock-but not from H2O2-treated cells (Fig. 2B, lanes 2-5). Likewise, TIF-IA isolated from anisomycin/SB203580-treated cells was incapable of rescuing transcriptional activity (Fig. 2B, lanes 6,7). In contrast, TIF-IA from cells treated with anisomycin plus the JNK inhibitor SP600125 was transcriptionally active (Fig. 2B, lanes 8,9). This suggests that inhibition of Pol I transcription upon induction of oxidative stress is brought about by inactivation of TIF-IA by JNK-dependent signaling pathways.
To determine whether TIF-IA is a direct target of JNK, immunopurified JNK2 and p38 were used to phosphorylate recombinant TIF-IA in vitro. Figure 2C shows that TIF-IA was phosphorylated by JNK2 but not by p38. Most importantly, preincubation of TIF-IA with JNK2 and ATP inactivated TIF-IA, whereas preincubation with p38 did not affect TIF-IA activity (Fig. 2D, lanes 4,5). These results demonstrate that TIF-IA is targeted by JNK and phosphorylation by JNK2 abrogates TIF-IA activity.
Phosphorylation of TIF-IA at Thr 200 is required for stress-dependent inhibition of Pol I transcription
To identify the peptides that are phosphorylated by JNK, we compared the phosphorylation pattern of TIF-IA from control, H2O2- and anisomycin-treated cells expressing Flag-tagged TIF-IA. After metabolic labeling with [32P]orthophosphate, TIF-IA was immunopurified and subjected to two-dimensional tryptic phosphopeptide mapping (Fig. 3A). Consistent with previous results (Zhao et al. 2003; Mayer et al. 2004), TIF-IA is phosphorylated at multiple sites (phosphopeptides a-i). After anisomycin and H2O2 treatment, the intensity of spot a increased, while spots c, h, and i decreased. Importantly, a similar pattern was observed when a constitutively active MEKK-1, an upstream activator of JNKs (Barr and Bogoyevitch 2001), was overexpressed. Spots h and i correspond to peptides containing phospho-Ser 170 and phospho-Ser 172, target sites for casein kinase II, and spot c contains phospho-Ser 635 (J. Zhao and I. Grummt, unpubl). Peptide a harbors Ser 199, which is hyperphosphorylated after inhibition of mTOR signaling by rapamycin (Mayer et al. 2004). Ser 199 is followed by an evolutionarily conserved threonine residue (Thr 200), which is part of a p38/JNK target site (VPxTP). In support of Thr 200 being phosphorylated by JNK, phosphoamino acid analysis of peptide a revealed that in untreated cells only Ser 199 was phosphorylated, while upon stress, peptide a contained both phospho-Ser 199 and phospho-Thr 200 (Fig. 3B).
Figure 3.
The phosphorylation pattern of TIF-IA is altered upon cellular stress. (A) Two-dimensional tryptic phosphopeptide maps of TIF-IA labelled in vivo. U2OS cells stably overexpressing Flag-tagged TIF-IA were metabolically labeled for 2 h with [32P]orthophosphate in the absence or presence of H2O2 (200 μM, 60 min) or anisomycin (10 μM, 60 min), or in the presence of constitutively active (CA) MEKK-1. Immunopurified TIF-IA was subjected to two-dimensional tryptic phosphopeptide mapping. (B) Phosphoamino acid analysis of peptide a of TIF-IA from mock- and anisomycin-treated U2OS cells. A phylogenetic comparison of the amino acid sequence of the phosphopetide a is given on the left. Thr 200 is shown in bold, the conserved JNK target site TP is shaded gray. (C) Two-dimensional tryptic phosphopeptide maps of TIF-IA phosphorylated by JNK2 in vitro. GST-tagged TIF-IA (wild-type and T200V mutant) purified from E. coli was incubated with immunopurified HA-tagged JNK2 and (γ-32P)-ATP for 30 min and subjected to two-dimensional tryptic phosphopeptide mapping.
To prove that Thr 200 is phosphorylated by JNK, we generated a point mutant in which Thr 200 was replaced by valine (TIF-IAT200V) and we used the mutant as substrate for in vitro phosphorylation by JNK2. The phosphopeptide maps in Figure 3C demonstrate that wild-type TIF-IA was labeled by JNK2 at two phosphopeptides corresponding to spots a and d. Importantly, JNK2 did not phosphorylate peptide a in the T200V mutant, indicating that Thr 200 is targeted by JNK. Peptide d has been shown to be phosphorylated at Ser 649 by RSK2 in vivo (Zhao et al. 2003). The peptide contains multiple Ser/Pro sites that could serve as in vitro acceptor sites for JNK2-dependent phosphorylation.
If phosphorylation of Thr 200 by JNK2 inactivates TIF-IA, then TIF-IAT200V should be transcriptionally active, whereas replacement of Thr 200 by glutamic acid, an acidic amino acid that mimics phosphorylation, should abrogate TIF-IA activity. To test this, we overexpressed wild-type and mutant TIF-IA in HEK293T cells and monitored transcription of a cotransfected Pol I reporter plasmid on Northern blots. While TIF-IAT200V activated Pol I transcription similar to wild-type TIF-IA, the TIF-IAT200E mutant was inactive (Fig. 4A). This result is consistent with phosphorylation at Thr 200 inactivating TIF-IA. A further prediction from phosphorylation of Thr 200 being responsible for down-regulation of rRNA synthesis under oxidative stress is that TIF-IAT200V should protect cells from stress-induced transcriptional shutdown. Indeed, overexpression of TIF-IAT200V activated Pol I transcription, regardless whether or not cells were exposed to H2O2 (Fig. 4B). This result underscores the role of Thr 200 phosphorylation in regulating TIF-IA activity and demonstrates that phosphorylation of TIF-IA at Thr 200 is necessary and sufficient for inhibition of Pol I transcription upon oxidative stress.
Figure 4.
Phosphorylation of Thr 200 inactivates TIF-IA. (A) Transcriptional activity of TIF-IA mutants in vivo. HEK293T cells were cotransfected with 8 μg of a Pol I reporter plasmid and 0.5 or 1.5 μg expression vector encoding wild-type or mutant TIF-IA (pcDNA3.1-TIF-IA or TIF-IAT200V/E). Transcripts from the reporter were analyzed on Northern blots. Quantitation of transcripts by PhosphorImager analysis is shown below.(B) Substitution of Thr 200 by valine renders TIF-IA resistant towards oxidative stress. Cells were cotransfected with 8 μg Pol I reporter plasmid and 0.5 or 1.5 μg of pcDNA3.1-TIF-IA or TIF-IAT200V. After 40 h, cells were incubated for 60 min with 200 μM H2O2 and transcripts from the reporter plasmid were analyzed on Northern blots. Quantitation of transcripts by PhosphorImager analysis is shown below.
Cellular stress impairs the assembly of initiation complexes at the rDNA promoter
To gain further insight into the molecular mechanism underlying stress-induced inhibition of Pol I transcription, promoter occupancy of individual components of the Pol I transcription apparatus was analyzed by chromatin immunoprecipitation (ChIP). Cross-linked chromatin from untreated and anisomycin-treated cells was precipitated with antibodies against Pol I (α-RPA116), TIF-IA, TIF-IB/SL1 (α-TAFI110), or UBF, and coprecipitated rDNA was quantified by real-time PCR. While the association of UBF and TIF-IB/SL1 with rDNA remained unaffected upon drug treatment, promoter occupancy of both Pol I and TIF-IA was drastically reduced (Fig. 5A, left panel). This indicates that JNK-directed phosphorylation of TIF-IA weakens the interaction of TIF-IA with TIF-IB/SL1, thereby impairing the formation of productive transcription complexes at the rDNA promoter. In support of this, the relative amount of Pol I, TIF-IA, and TIF-IB/SL1 remained unaffected or even increased after anisomycin treatment in Jnk1,2-/- MEFs (Fig. 5A, right panel), while the parental MEFs behaved similar to HEK293T cells (data not shown). This provides direct evidence that JNK-dependent phosphorylation of TIF-IA impairs the formation of transcription complexes. As a complementary approach to examine the effect of stress stimuli on the interaction of TIF-IA with TIF-IB/SL1 and Pol I, TIF-IA was overexpressed and immunoprecipitated from mock-, anisomycin-, or H2O2-treated HEK293T cells, and coprecipitated Pol I and TIF-IB/SL1 were visualized on immunoblots. Whereas in mock-treated cells Pol I and TIF-IB/SL1 coprecipitated with TIF-IA, the interaction of TIF-IA with both Pol I and TIF-IB/SL1 was greatly impaired upon anisomycin- or H2O2-treatment (Fig. 5B). This result is consistent with the ChIP data showing that stress impairs promoter occupancy of TIF-IA and Pol I and demonstrates that stress-induced inhibition of pre-rRNA synthesis is due to a failure of TIF-IA to interact with both Pol I and TIF-IB/SL1, thereby preventing transcription initiation complex formation.
Figure 5.
Cellular stress impairs the interaction of TIF-IA with TIF-IB/SL1 and Pol I. (A) Anisomycin treatment prevents the association of Pol I and TIF-IA with the rDNA promoter. Cross-linked chromatin from HEK293T cells (left) or Jnk1,2-/- MEFs (right) was immunoprecipitated with antibodies against TIF-IB/SL1 (α-TAFI110), UBF, Pol I (α-RPA116), and TIF-IA, and precipitated rDNA was monitored by real-time PCR with primers that amplify the 5′-terminal region of human rDNA. The results of three independent experiments are plotted. Light-gray and dark-gray columns represent the relative association of the analyzed proteins (calculated as bound normalized to input) from mock- and anisomycin-treated cells with the rDNA promoter, respectively. (B) Stress inhibits the interaction between TIF-IA and Pol I/TIF-IB. Lysates from mock-, H2O2-, or anisomycin-treated HEK293T cells expressing Flag-tagged TIF-IA were incubated with α-Flag antibodies, and immunoprecipitated TIF-IA, Pol I, and TIF-IB/SL1 were visualized on Western blots using α-Flag, α-RPA116, and α-TAFI68 antibodies, respectively. (C) Substitution of Thr 200 by glutamic acid interferes with the interaction of TIF-IA with Pol I and TIF-IB/SL1. Flag-tagged TIF-IA or the indicated point mutants were overexpressed in HEK293T cells and immunoprecipitated using α-Flag antibodies. Cells were either treated with H2O2 (200 μM) for 60 min or were left untreated. Coprecipitated Pol I and TIF-IB/SL1 were monitored on Western blots using α-RPA116 and α-TAFI68 antibodies, respectively.
The observation that stress signals impair the interaction of TIF-IA with Pol I and TIF-IB/SL1 suggests that phosphorylation of T200 renders TIF-IA incapable of interacting with Pol I and TIF-IB/SL1. To test this, wild-type or mutant TIF-IA was precipitated from mock- or H2O2-treated cells, and coprecipitated Pol I and TIF-IB/SL1 were monitored on Western blots. Both wild-type TIF-IA and TIF-IAT200V efficiently interacted with Pol I and TIF-IB/SL1, yet no interaction was observed with TIF-IAT200E (Fig. 5C, lanes 7,9,11). Importantly, while wild-type TIF-IA failed to interact with Pol I and TIF-IB/SL1 under oxidative stress, the interaction of TIF-IAT200V was not inhibited (Fig. 5C, lane 10). This result is consistent with the transcription data showing that transcriptional activation by TIF-IAT200V was resistant to H2O2. Moreover, these results indicate that under oxidative stress the interaction of TIF-IA with Pol I is controlled solely by the phosphorylation state of Thr 200.
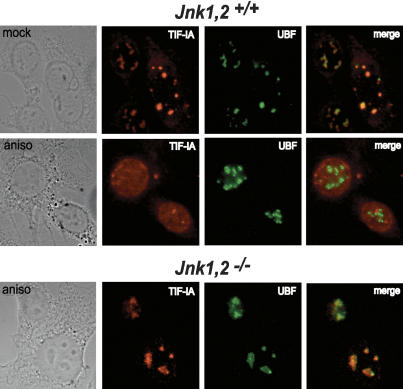
Anisomycin treatment releases TIF-IA from the nucleolus
A variety of cellular stress signals have been shown to change the subcellular distribution of transcription factors (Essers et al. 2004; Marion et al. 2004). We therefore analyzed the cellular localization of TIF-IA upon induction of JNK signaling by immunofluorescence. Both in mock-treated Jnk1,2+/+ and Jnk1,2-/- MEFs, TIF-IA colocalizes with UBF in the nucleolus (Fig. 6). After anisomycin treatment, however, there were marked differences in the localization of TIF-IA in wild-type and Jnk-deficient cells. Both in HeLa and NIH3T3 cells (data not shown) as well as wild-type MEFs, TIF-IA was released from the nucleolus to the nucleoplasm after treatment with anisomycin. This relocalization of TIF-IA was also observed after exposure to H2O2 (data not shown). In Jnk-deficient cells, on the other hand, the nucleolar localization of TIF-IA was not affected by either stress-inducing condition. Together, these results reveal that JNK2-dependent phosphorylation eliminates the interaction of TIF-IA with TIF-IB/SL1 and Pol I, which in turn leads to exclusion of TIF-IA from the nucleolus.
Figure 6.
Anisomycin treatment leads to accumulation of TIF-IA in the nucleoplasm. Cellular TIF-IA and UBF were visualized in parental or Jnk1,2-/- MEFs by indirect immunostaining. A merged image is shown on the right. Where indicated, MEFs were treated with anisomycin (10 μM) for 60 min.
Discussion
p38 kinases and JNKs constitute two important branches of the MAPK signaling cascade that function as specialized transducers of stress or injury responses. Both pathways collaborate through the usage of common upstream or downstream effectors to induce a specific program of gene expression, known as the “environmental stress response,” ESR (Gasch et al. 2000). ESR is commonly activated by a variety of environmental cues, such as osmotic and oxidative stress, inflammatory cytokines, ribotoxic agents, etc. Studies in yeast have shown that transcription of ribosomal protein genes is down-regulated in a coordinated fashion upon exposure to harmful conditions (Veinot-Drebot 1989; Lee et al. 1996; Zaragoza et al. 1998; Nierras and Warner 1999; Miyoshi et al. 2001). Moreover, rRNA synthesis was more susceptible to oxidative stress than overall mRNA synthesis (Lee et al. 1996), suggesting that yeast down-regulates the synthesis of ribosomal constituents. Yet, little is known about the molecular consequences of oxidative and ribotoxic stress on mammalian ribosome biogenesis. A systematic examination of the global transcriptional response to a variety of stresses in human cells revealed that the expression of genes encoding ribosomal proteins decreased only modestly after most stresses (Murray et al. 2004). However, in contrast to ribosomal protein genes, transcription of rRNA genes was fast and efficiently down-regulated both in human and mouse cells after oxidative stress or treatment with ribotoxic agents. Inhibition of Pol I transcription after exposure to subtoxic doses of H2O2 or anisomycin, both of which activate JNK (Iordanov et al. 1997 and references therein), was brought about by inactivation of TIF-IA, a Pol I-associated transcription initiation factor that mediates growth-dependent regulation of Pol I transcription (Tower and Sollner-Webb 1987; Schnapp et al. 1990). TIF-IA prepared from H2O2-treated cells was transcriptionally inactive, and TIF-IA from exponentially growing cells was sufficient to rescue Pol I transcription in extracts from H2O2- or anisomycin-treated cells. Moreover, in vitro phosphorylation by immunopurified JNK2 abrogated TIF-IA activity. These results reveal that TIF-IA activity is regulated by stress-induced signaling pathways and down-regulation of Pol I transcription upon stress is mediated by phosphorylation of TIF-IA. TIF-IA is phosphorylated at multiple sites that are targeted by different signaling cascades. We have previously shown that proper phosphorylation by mTOR- and ERK-dependent pathways is required for TIF-IA activity and Pol I transcription (Zhao et al. 2003; Mayer et al. 2004). We now demonstrate that stress-induced activation of JNK leads to phosphorylation of TIF-IA at Thr 200 and this phosphorylation inactivates TIF-IA. Thus, TIF-IA activity is both positively and negatively regulated by phosphorylation.
TIF-IA has been shown to play a central role in Pol I transcription initiation complex formation. TIF-IA interacts with both Pol I and the TBP-containing promoter selectivity factor TIF-IB/SL1, thereby targeting Pol I to the rDNA promoter (Miller et al. 2001; Yuan et al. 2002). The results of this and previous studies indicate that the interaction of TIF-IA with Pol I and/or TIF-IB/SL1 are affected by diverse regulatory pathways that link the cell's biosynthetic activities to environmental conditions. After exposure to stress, the interaction of TIF-IA with both TIF-IB/SL1 and Pol I is impaired, thus preventing initiation complex formation. In support of this, neither TIF-IA nor Pol I was found to be associated with the rDNA promoter in anisomycin-treated cells. In Jnk1,2 double-knockout MEFs, on the other hand, ribotoxic stress did not affect pre-rRNA synthesis or promoter occupancy of individual components of the Pol I transcription apparatus, demonstrating that JNK-dependent signaling events inhibit initiation complex formation, and hence rDNA transcription. To examine whether phosphorylation by JNK1, JNK2, or both inactivates TIF-IA, we monitored Pol I transcriptionin Jnk1-/- and Jnk2-/- fibroblasts after exposure to oxidative stress. We found that pre-rRNA synthesis in JNK1-deficient fibroblasts was hypersensitive toward oxidative stress, while JNK2-deficient cells were resistant, demonstrating that JNK2 is responsible for TIF-IA inactivation and Pol I repression. This finding is consistent with published data establishing antagonistic functions of different JNK isoforms in the regulation c-Jun metabolism and cell proliferation. While JNK1 has a stimulatory role on cell proliferation, JNK2 negatively regulates c-Jun stability and represses cell cycle progression (Sabapathy et al. 1999).
Various stress stimuli have been shown to induce changes of the cellular localization of nucleolar proteins. Tumor suppressor proteins like VHL and p53 are stabilized by sequestration in the nucleolus (Rubbi and Milner 2003; Mekhail et al. 2004). Likewise, nucleolin and B23 are translocated from the nucleolus into the nucleoplasm, where they play a regulatory role in inducing cell cycle arrest (Daniely et al. 2002; Kurki et al. 2004). Notably, we found that the nucleolar localization of TIF-IA was altered upon stress. Following phosphorylation by JNK2, TIF-IA was displaced from the nucleolus and accumulated in the nucleoplasm, while other components of the Pol I transcription machinery were not affected. This finding is similar to previous data showing that TIF-IA localization is controlled by mTOR-dependent signaling through hyperphosphorylation of Ser 199 (Mayer et al. 2004). Apparently, the reversible phosphorylation of two neighboring residues, Ser 199 and Thr 200, controls both the activity and nucleolar localization of TIF-IA. It is conceivable that phosphorylation at either site alters the configuration of TIF-IA such that it is incapable of interacting with both Pol I and TIF-IB/SL1 and is released into the nucleoplasm. Further studies are necessary to determine whether the mobilization of TIF-IA may serve a general function in stress response that is unrelated to rRNA synthesis. The potential connections between nucleolar localization and signaling are intriguing given the TIF-IA's link to growth control.
Materials and methods
Chemicals, antibodies, and plasmids
BSO (DL-buthionine-(S,R)-sulfoximine), anisomycin, and SB203580 were purchased from Sigma Chemical Co. SP600125 was obtain from Biomol. Polyclonal anti-TIF-IA antibodies were generated by injection of GST-tagged full-length TIF-IA into rabbits. cDNAs encoding wild-type (DDBJ/EMBL/GenBank database accession no. AJ272050) and mutant TIF-IA were tagged at the 5′-end with sequences encoding the Flag epitope peptide and cloned into pcDNA3.1 (Invitrogen). To generate mutant forms of TIF-IA, Thr 200 was replaced by valine or glutamic acid, respectively, by overlap extension PCR. pMr600, the template used for in vitro transcription in mouse extracts, contains a 622-bp PvuII fragment harboring murine rDNA sequences from -324 to +292. pHrP2 contains an 800-bp Sau3A fragment comprising human rDNA sequences from -411 to +375 and was used as template for in vitro transcription in human extracts. The Pol I reporter plasmid pHrP2-BH contains the 5′-terminal human rDNA fragment fused to a 3′-terminal rDNA fragment including two “Sal box” terminator elements.
Cells, transfections, and RNA analysis
Nuclear extracts for in vitro transcription experiments as well as Pol I and transcription factors were prepared from FM3A cells grown in RPMI1600 medium supplemented with 10% newborn calf serum (NCS) and 2 mM glutamine. All other cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 2 mM glutamine. Drug treatments were generally performed for 60 min (or 16 h for BSO) in the stated concentrations. Cellular pre-rRNA was monitored on Northern blots by hybridization to antisense RNA complementary to the first 155 nucleotides of unprocessed 45S pre-RNA (Voit et al. 1999). For reporter assays, 5 × 105 HEK293T cells were cotransfected with 8 μg of reporter plasmid pHrP2-BH and different amounts of pcDNA3.1Flag-TIF-IA. RNA was isolated after 40-48 h and transcripts from the rDNA reporter plasmid were monitored by hybridization to a riboprobe that is complementary to pUC sequences inserted between the rDNA promoter and terminator sequences of pHrP2-BH.
In vitro transcription assays
Nuclear extracts were prepared from exponentially growing, hydrogen peroxide-treated (200 μM H2O2 for 1 h), anisomycin-treated (10 μM, 1 h), or density-arrested FM3A, or HEK293T cells. For density-arrest, cells were cultured for 12-16 h at a density of 2 × 106 cells/mL. Twenty-five-microliter assays contained 50 ng of template DNA (pMr600/EcoRI or pHrP2/EcoRI); 30 μg nuclear extract proteins; 12 mM Hepes-KOH at pH 8.0; 0.1 mM EDTA; 5 mM MgCl2; 80 mM KCl; 10 mM creatine phosphate; 12% (v/v) glycerol; 0.66 mM each of ATP, GTP, and CTP; 0.01 mM UTP; and 0.5 μCi [α?32P]UTP (5000 Ci/mmol). After incubation for 60 min at 30°C, RNA was extracted and analyzed on nondenaturing 4.5% polyacrylamide gels. The reconstituted TIF-IA responsive transcription system contained 20 ng of pMR600/EcoRI, 4 μL of a Pol I-containing fraction (H-400), 3 μL of TIF-IB/SL1 (CM-400 fraction) and 5 ng of recombinant hUBF1. Four microliters TIF-IA/TIF-IC (poly-L-lysine agarose fraction) were added to complement transcription (Schnapp and Grummt 1996).
Purification of cellular and recombinant TIF-IA
Cellular TIF-IA was purified from nuclear extracts of FM3A cells by chromatography on DEAE-Sepharose and Heparin-Ul-trogel (Schnapp and Grummt 1996) followed by fractionation on polylysine-agarose (PL-650 fraction). Recombinant TIF-IA was immunopurified either from baculovirus-infected Sf9 cells or from HEK293T cells overexpressing Flag-tagged TIF-IA as previously described (Mayer et al. 2004).
Tryptic phosphopeptide analysis of TIF-IA and in vitro kinase assays
For metabolic labeling of cells, 3-5 × 106 U2OS cells overexpressing Flag-tagged TIF-IA were cultured in DMEM/10% FCS and transferred to phosphate-free DMEM with 10% dialyzed FCS for 45 min before labeling with 0.5 mCi/mL [32P]orthophosphate for 2-4 h. Cells were lysed in RIPA buffer (20 mM Tris-HCl at pH 7.4, 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100, 0.1% SDS, 10 mM β-glycerophosphate, 10 mM KH2PO4, 1 mM Na3VO4, 0.5 mM PMSF, 1 mg/mL aprotinin, 5 mg/mL leupeptin, and 1 mg/mL pepstatin A). TIF-IA was immunoprecipitated, separated by SDS-PAGE, transferred to nitrocellulose and visualized by autoradiography. TIF-IA was digested overnight at 37°C with trypsin (Promega, sequencing grade) in 50 mM ammonium bicarbonate. After lyophilization, peptides were resolved on cellulose thin-layer plates as described (Mayer et al. 2004). Phosphoamino acid analysis was carried out as described (Boyle et al. 1991).
HA-tagged protein kinases were purified from HEK293T cells overexpressing HA-tagged JNK or p38, respectively. Cells (5 &rimes; 106 were lysed in 0.8 mL IP buffer (20 mM Tris-HCl at pH 7.4, 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100), centrifuged at 10,000 × g for 20 min, and the supernatants were incubated for 4 h at 4°C with 15 μg anti-HA antibodies bound to 20 μL protein G-agarose. Beads were washed three times with AM-400 and AM-100 and twice in 1.5× kinase buffer (37 mM Tris-HCl at pH 7.8, 10 mM MgCl2, 1.5 mM DTT, 1.5 mM β-glycerophosphate, 0.1 mM Na3VO4). To phosphorylate TIF-IA, 0.5 μg GST/TIF-IA was incubated in 15 μL kinase buffer containing 10 μCi (γ-32P(ATP (5000 Ci/mmol) and 20-50 fmol kinase.
Coimmunoprecipitation and immunoblotting
To analyze the interaction of TIF-IA with Pol I and TIF-IB/SL1, cells overexpressing Flag-tagged TIF-IA were lysed in IP buffer, cleared by centrifugation at 10,000 × g for 30 min, and the supernatants were incubated for 4 h at 4°C with anti-Flag M2-agarose (Sigma). The immunoprecipitates were washed with IP buffer (for associated TIF-IB/SL1) or with IP buffer containing 600 mM NaCl (for Pol I). TIF-IB/SL1 was visualized on immunoblots with α-TAFI110 antibodies and Pol I with antibodies against the second largest subunit of Pol I (RPA116).
ChIP assays
ChIP assays were performed according to Boyd et al. (1998). Briefly, chromatin from 5 × 107 cross-linked (1% formaldehyde, 15 min at 37°C) cells was sonicated to yield average fragments of ∼500 bp, was pre-cleared with protein A/G agarose (blocked with 1% BSA, 0.5 mg/mL sheared salmon sperm DNA) and an equivalent of 1 × 107 cells were incubated with 5-10 μL of polyclonal antisera against TIF-IA, Pol I (RPA116), TIF-IB/SL1 (TAFI110) and UBF overnight. Blocked protein A/G-agarose beads were then added and incubation was continued for 60 min. After extensive washing, cross-linked DNA was eluted from beads with 1% SDS/0.1 M NaHCO3 and incubated for 5 h at 65°C to reverse cross-linking, followed by ethanol precipitation. Recovered chromatin was digested with proteinase K and purified on Qiaquick PCR purification columns. Quantitative real-time PCR was performed with primers amplifying the mouse rDNA promoter from -165 to +130 or human rDNA promotor from -150 to +130 using a LightCycler (Roche) and the SYBR Green detection system.
Immunofluorescence
Cells grown on cover-slips were fixed with 2% paraformaldehyde/PBS for 10 min. After three washes in PBS, cells were permeabilized with methanol for 90 sec at -20°C and washed again three times in PBS. Endogenous TIF-IA was visualized using purified rabbit α-TIF-IA antibodies (dilution 1:150) in combination with Cy3-conjugated α-rabbit antibodies (dilution 1:150). UBF was detected using human α-UBF auto-immune serum (dilution 1:1200) followed by FITC-conjugated α-human antibodies (dilution 1:50). Immunofluorescence was monitored by microscopy (Zeiss Axiophot using air/oil-objective lenses 10×/63×) and images were digitally recorded using a Nikon DXM1200 camera. Image aquisition was done using the Nikon ACT-1 software, Adobe Photoshop 5.0.2 was used for image processing.
Acknowledgments
We thank Bettina Doerr for expert technical assistance. We are grateful to P. Angel for expression vectors encoding HA-tagged p38, JNK2, and MEKK-1, and E.F. Wagner for Jnk1-/-Jnk2-/- MEFs as well as 3T3Jnk1-/- and 3T3Jnk2-/- cells. We are grateful to all members of the lab for sharing reagents and advice. This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.333205.
References
- Barr R.K. and Bogoyevitch, M.A. 2001. The c-jun N-terminal kinase family of mitogen-activated protein kinases (JNK MAPKs). Int. J. Biochem. Cell Biol. 33: 1047-1063. [DOI] [PubMed] [Google Scholar]
- Bodem J., Dobreva, G., Hoffmann-Rohrer, U., Iben, S., Zentgraf, H., Delius, H., Vingron, M., and Grummt, I. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1: 171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd K.E., Wells, J., Gutman, J., Bartley, S.M., and Farnham, P.J. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. 95: 13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle W.J., van der Geer, P., and Hunter, T. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Meth. Enzymol. 201: 110-149. [DOI] [PubMed] [Google Scholar]
- Cavanaugh A.H., Hirschler-Laszkiewicz, I., Hu, Q., Dundr, M., Smink, T., Misteli, T., and Rothblum, L.I. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277: 27423-27432. [DOI] [PubMed] [Google Scholar]
- Chen Y.-R., Wang, X., Templeton, D., Davis, R.J., and Tan, T.-H. 1996. The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and γ-radiation. J. Biol. Chem. 271: 31929-31936. [DOI] [PubMed] [Google Scholar]
- Daniely Y., Dimitrova, D.D., and Borowiec, J.A. 2002. Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol. Cell. Biol. 22: 6014-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B., Raingeaud, J., Barrett, T., Wu, I.-H., Han, J., Ulevitch, R.J., and Davis, R.J. 1995. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science 267: 682-684. [DOI] [PubMed] [Google Scholar]
- Essers M.A.G, Weijzen, S., de Vries-Smits, A.M.M., Saarloos, I., de Ruijter, N.D., Bos, J.L., and Burgering, B.M.T. 2004. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 23: 4802-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., and Brown, P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11: 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. 2003. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17: 1691-1702. [DOI] [PubMed] [Google Scholar]
- Iordanov M.S., Pribnow, D., Magun, J.L., Dinh, T.-H., Pearson, J.A., Chen, S.L.-Y., and Magun, B.E. 1997. Ribotoxic stress response: Activation of the stress-activated protein kinae JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the α-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol. 17: 3373-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurki S., Peltonen, K., Latonen, L., Kiviharju, T.M., Ojala, P.M., Meek, D., and Laiho M. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5: 465-475. [DOI] [PubMed] [Google Scholar]
- Lee J., Romeo, A., and Kosman, D.J. 1996. Transcriptional remodeling and G1 arrest in dioxygen stress in Saccharomyces cerevisiae. J. Biol. Chem. 271: 24885-24893. [DOI] [PubMed] [Google Scholar]
- Marion R.M., Regev, A., Segal, E., Barash, Y., Koller, D., Friedman, N., and O'Shea, E.K. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal gene expression. Proc. Natl. Acad. Sci. 101: 14315-14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale J.L. and Holbrook, N.J. 2002. Cellular response to oxidative stress: Signaling for suicide and survival. J. Cell. Physiol. 192: 1-15. [DOI] [PubMed] [Google Scholar]
- Mayer C., Zhao, J., Yuan, X., and Grummt, I. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18: 423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K., Gunaratnam, L., Bonicalzi, M.E., and Lee, S. 2004. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell. Biol. 6: 642-647. [DOI] [PubMed] [Google Scholar]
- Milkereit P. and Tschochner, H. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J. 17: 3692-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Panov, K.I., Friedrich, J.K., Trinkle-Mulcahy, L., Lamond, A.I., and Zomerdijk, J.C. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20: 1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K., Miyakawa, T., and Mizuta, K. 2001. Repression of rRNA synthesis due to a secretory defect requires the C-terminal silencing domain of Rap1p in Saccharomyces cerevisiae. Nucleic Acids Res. 29: 3297-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorefield B., Greene, E.A., and Reeder, R.H. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. 97: 4724-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. 2004. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 14: 210-217. [DOI] [PubMed] [Google Scholar]
- Murray J.I., Whitfield, M.L., Trinklein, N.D., Myers, R.M., Brown, P.O., and Botstein, D. 2004. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15: 2361-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierras C.R. and Warner, J.R. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274: 13235-13241. [DOI] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner, J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22: 6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabapathy K., Jochum, W., Hochedlinger, K., Chang, L., Karin, M., and Wagner, E.F. 1999. Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89: 115-124. [DOI] [PubMed] [Google Scholar]
- Schnapp A. and Grummt, I. 1996. Purification, assay, and properties of RNA polymerase I and class I-specific transcription factors in mouse. Meth. Enzymol. 273: 233-248. [DOI] [PubMed] [Google Scholar]
- Schnapp A., Pfleiderer, C., Rosenbauer H., and Grummt, I. 1990. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 9: 2857-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A., Schnapp, G., Erny, B., and Grummt, I. 1993. Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell. Biol. 13: 6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M., Baumann, B., Cotten, M., Angel, P., and Wagner, E.R. 1995. Fos is an essential component of the mammalian UV response. EMBO J. 14: 5338-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier C., Hess, P., Yang, D.D., Xu, J., Turner, T.K., Nimnual, A., Bar-Sagi, D., Jones, S.N., Flavell, R.A., and Davis, R.J. 2000. Requirement of JNK for stress-induced activation to the cytochrome c-mediated death pathway. Science 288: 870-874. [DOI] [PubMed] [Google Scholar]
- Tower J. and Sollner-Webb, B. 1987. Transcription of mouse rDNA is regulated by an activated subform of RNA polymerase I. Cell 50: 873-883. [DOI] [PubMed] [Google Scholar]
- Veinot-Drebot L.M., Singer, R.A., and Johnston, G.C. 1989. Heat shock causes transient inhibition of yeast rRNA gene transcription. J. Biol. Chem. 264: 19473-19474. [PubMed] [Google Scholar]
- Voit R., Hoffman, M., and Grummt, I. 1999. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 18: 1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Zhao, J., Zentgraf, H., Hofmann-Rohrer, U., and Grummt, I. 2002. Multiple interactions between RNA polymerase I, TIF-IA, and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3: 1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza D., Ghavidel, A., Heitman, J., and Schultz, M.C. 1998. Rapamycin induces the Go program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell Biol. 18: 4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan, Y., Frödin, M. and Grummt, I. 2003. ERK-dependent phosphorylation of the transcription factor TIF-IA is required for RNA polymerase I transcription. Mol. Cell 11: 405-413. [DOI] [PubMed] [Google Scholar]