Abstract
A growing body of evidence suggests that endogenous antibiotics contribute to the innate defense of mammalian mucosal surfaces. In the cow, β-defensins constitute a large family of antibiotic peptides whose members have been previously isolated from the respiratory and oral mucosa, as well as circulating phagocytic cells. A novel bovine genomic clone with sequence related to those of these α-defensins was isolated and characterized. The corresponding cDNA was isolated from a small intestinal library; its open reading frame predicts a deduced sequence of a novel β-defensin, which we designate enteric β-defensin (EBD). Northern blot analysis of a variety of bovine tissues revealed that EBD mRNA is highly expressed in the distal small intestine and colon, anatomic locations distinct from those for previously characterized β-defensins. EBD mRNA was further localized by in situ hybridization to epithelial cells of the colon and small intestinal crypts. Infection of two calves with the intestinal parasite Cryptosporidium parvum induced 5- and 10-fold increases above control levels of EBD mRNA in intestinal tissues. An anchored-PCR strategy was used to identify other β-defensin mRNAs expressed in the intestine. In addition to that of EBD, several low-abundance cDNAs which corresponded to other β-defensin mRNAs were cloned. Most of these clones encoded previously characterized β-defensins or closely related isoforms, but two encoded a previously uncharacterized prepro-β-defensin. Northern blot evidence supported that all of these other β-defensin genes are expressed at levels lower than that of the EBD gene in enteric tissue. Furthermore, some of these β-defensin mRNAs were abundant in bone marrow, suggesting that in enteric tissue their expression may be in cells of hematopoietic origin. Extracts of small intestinal mucosa obtained from healthy cows have numerous active chromatographic fractions as determined by an antibacterial assay, and one peptide was partially purified. The peptide corresponded to one of the low-abundance cDNAs. This study provides evidence of β-defensin expression in enteric tissue and that the mRNA encoding a major β-defensin of enteric tissue, EBD, is inducibly expressed in enteric epithelial cells. These findings support the proposal that β-defensins may contribute to host defense of enteric mucosa.
A striking feature of the mammalian intestinal tract is the large surface area of the mucosal epithelium. This expansive surface facilitates nutrient absorption but can, to the detriment of the host, also serve as a port of entry for invading microorganisms. Colonizing microbes in the intestinal lumen continuously pose a potential threat of infection. The relatively low occurrence of intestinally derived systemic infections, however, suggests the presence of effective host defense pathways. A more comprehensive understanding of these pathways may define therapeutic targets for enhancing host defense. Therefore, interest has focused on elucidating local defense mechanisms protecting this and other mammalian mucosal surfaces (5, 30, 35, 48, 60, 66).
Current understanding of mucosal defense suggests that the collective actions of multiple innate, nonclonal host defenses integrate with the specific clonal immune responses mediated by lymphocytes (3, 9, 22, 23, 43, 60). In the gastrointestinal tract, examples of innate defenses include physical processes, such as peristalsis and shedding of epithelial cells, and chemical barriers, including gastric acidity, mucus, bile acids, and proteins (30). Among the proteins which contribute to local defense against microbes are several antibiotic peptides recently identified in extracts from gastrointestinal mucosa (1, 2, 17, 36, 42, 45, 50, 58). In situ hybridization analysis has demonstrated that some of these peptides are synthesized by epithelial cells (32, 33, 44, 51). Other antibiotic peptides appear to be products of leukocytes which have migrated to the bowel (1, 2, 36).
The β-defensin class of antimicrobial peptides was unveiled with the discovery of tracheal antimicrobial peptide (TAP), a peptide expressed in bovine tracheal epithelial cells (13, 15). Numerous genomic sequences related to the TAP gene were identified by Southern blot analysis (13), which suggested that a large family of β-defensin genes exists in the cow. Additional β-defensin peptides have since been isolated from bovine neutrophils (59), macrophages (54), and tongue (57), supporting this notion and suggesting a wide distribution of tissue expression for this gene family. All characterized β-defensins have broad-spectrum antimicrobial activity (15, 57, 59). Inducible expression of β-defensins, by inflammatory mediators in vitro (12, 14, 53) and near sites of inflammation in vivo (57), supports a role for β-defensins in mucosal host defense. More recent investigations have identified apparent homologs of bovine β-defensins in horseshoe crabs (55), chickens (28), turkeys (20), mice (29), and humans (4, 27), indicating a evolutionary conservation of this family of host defense peptides.
We have now determined that a previously uncharacterized member of the β-defensin gene family is expressed in epithelial cells of the small intestine and colon. The gene encoding this β-defensin was isolated during our screening for the gene for TAP (13). Analysis of the gene sequence and genomic organization and its expression in a calf model of Cryptosporidium parvum infection is presented. An anchored-PCR cloning strategy revealed the expression of several other β-defensin genes in the small intestine but at much lower levels. Northern blot data supporting that many of the latter group of β-defensins are probably expressed in cells of hematopoietic origin are presented. Finally, the partial purification of a β-defensin peptide from intestinal extracts is described. The data of this study support that mammalian enteric epithelial cells express a β-defensin peptide gene and that β-defensins may contribute to local host defense of the enteric mucosa.
MATERIALS AND METHODS
General methodology.
The general methods used were as described previously (6, 13, 33). Bovine tissues were obtained from either a local abattoir or from Pel-Freeze Biologicals (Rogers, Ark.). The sequences of oligonucleotides (Keystone Labs, Menlo Park, Calif.) used for hybridization probes or PCR primers are shown in Table 1. Oligonucleotide probes were end labeled to a specific activity of ca. 107 dpm/pmol (6). A double-stranded bovine α-tubulin DNA probe was labeled to a specific activity of ca. 109 dpm/μg by using [α-32P]dCTP (800 Ci/mmol; DuPont) and T7 DNA polymerase with random oligonucleotide primers (Stratagene, La Jolla, Calif.).
TABLE 1.
Oligonucleotides used for PCR and hybridization
| Oligonucleotide | Sequence |
|---|---|
| EBD 242a | 5′-CCGCATCTCTTCCTTCTTTTACC-3′ |
| EBD 271a | 5′-CGCAGTTTCTGTCTCTGCTTAGG-3′ |
| EBD 285a | 5′-TTTCTGTCGAAGGCCGCAGTTTCTGTCTCTGCTTAGG-3′ |
| EBD 9UTa | 5′-AGAGGCTGCTCTTGCCTCTTTATAAAGGTCCCAGGTTCT-3′ |
| TAP30s | 5′-ATGAGGCTCCATCACCTGCTC-3′ |
| TAP48a | 5′-CCAAGCAGACAGGACCAGGAAGAGGAGCGCGAGGAGCAGGTGATGGAGCCTCAT-3′ |
| TAP286a | 5′-GCTCTGTCAAAGGGCGCAGTTTCTGACTGGGCATTGA-3′ |
| BNBD2/3-189a | 5′-TCTACCACGACCTGCAGCATTTTATTCGGGGCCCGAA-3′ |
| JR335.B1 | 5′-CTTTTACCACTACCTGCAGCATTTTATTTGGGGCGCT-3′ |
| JR300.C7 | 5′-GGTCCAGGGCACCTGATCAGAATACAGATGCCTCCTT-3′ |
| αTub-632a | 5′-GTGGTGTGGGTGGTGAGGATAGAGTTGTAGGGCTCAAC-3′ |
cDNA and genomic cloning.
A bovine genomic library in EMBL 3 was screened with a TAP oligonucleotide probe, TAP48a, and numerous hybridizing plaques of various intensity were identified (13). Briefly, duplicate filter lifts (Colony Plaque Screen; NEN/Dupont, Boston, Mass.) from 22 plates (3 × 104 plaques/plate) were hybridized with the probe in 25% formamide–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1× Denhardt solution–100 μg of yeast RNA per ml–1% sodium dodecyl sulfate (SDS) at 42°C overnight. The filters were washed at high stringency in 2× SSC–0.1% SDS at 65°C for 1/2 h, followed by autoradiography over 4 days at −70°C with an intensifying screen. One of the positive plaques (TAPG3) was found to encode TAP (13). Six other TAP48a-positive plaques were rescreened at a lower phage density through tertiary screens until pure. Phage DNA was isolated from liquid culture (56), and EcoRI restriction enzyme fragments of phage insert DNA were subcloned by ligation into the EcoRI site of pBluescript II SK+ plasmid DNA (Stratagene). Preliminary sequence analysis indicated that one of these clones, G11, encodes enteric β-defensin (EBD), and this clone was selected for further analysis.
A bovine small intestine cDNA library in lambda gt11 (Clontech BL1025a) was screened by using TAP48a. Duplicate filter lifts from 15 plates containing 1.5 × 104 plaques/plate were originally hybridized under the same conditions as described for the genomic screening, except that the formamide concentration in the hybridization solution was 20% and the high-stringency wash conditions were 2× SSC–0.1% SDS at 55°C for 30 min. Several dozen positive signals were identified. The filters were stripped of probe and rehybridized with TAP48a in hybridization buffer containing 50% formamide, and the high-stringency wash temperature was increased to 65°C. One strongly hybridizing plaque, BSI-13, was rescreened at progressively lower densities until pure, and recombinant phage DNA was isolated from liquid culture. The phage insert DNA was subcloned as described above. The 5′ 355-nucleotide sequence of BSI-13, which is not found in the genomic sequence of EBD, is 98% identical to that of murine 28S rRNA. Therefore, this portion of BSI-13 sequence probably is an artifact of cloning and was omitted from further analyses.
Southern blot analysis.
Bovine genomic DNA (10 μg) obtained from kidney tissue was digested overnight with restriction endonucleases, size separated by agarose gel (0.8%) electrophoresis, and transferred to a nylon filter (Zetabind; CUNO, Inc., Meriden, Conn.) by standard techniques (56). The filter was hybridized for 2 days in 37.5% (vol/vol) formamide–5× Denhardt solution–5× SSC–1% SDS–100 μg of yeast RNA per ml at 42°C with 32P-end-labeled EBD 285a. The filter was washed with 2× SSC–0.1% SDS three times at room temperature for 45 min and then twice at 60°C for 30 min. The moist filter was exposed to film in the presence of an intensifying screen for 4 weeks.
Northern blot analysis.
Total RNA was extracted from various tissues as described by Chirgwin et al. (8) and stored as an ethanol precipitate at −70°C. RNA was electrophoresed on a 1.2% agarose–formaldehyde gel and transferred to Zetabind (CUNO, Inc.) as described previously (33). Labeled probes were hybridized overnight to immobilized RNA under the same hybridization conditions as used for Southern blots (see above) and then washed at high stringency in 0.1× SSC–0.1% SDS at 57°C for 30 min (6). The washed blot was exposed to film with an intensifying screen at −70°C for 1 to 14 days. The filter was stripped of oligonucleotide label by incubation in 0.1× SSC–0.1% SDS at 65°C for 30 min. The filter was exposed to film to ensure removal of probe prior to hybridization with another probe. The formamide concentration was 50% in the hybridization solution with the α-tubulin cDNA probe, and the stringency of the final wash was changed to 0.1× SSC–0.1% SDS at 65°C for 30 min.
In situ hybridization.
Slides of paraffin-embedded tissue were prepared as described by Gilman (26). Tissue was hybridized with 35S-labeled riboprobes as described previously (51) and then stained and photographed as described previously (13). The DNA template for riboprobe synthesis was prepared by digesting the cDNA plasmid BSI-13 with SacI, which deleted the 5′ section, and then religating the plasmid DNA, creating G11 ΔSacI cDNA. The resulting plasmid, containing nucleotides 18 to 327 of EBD cDNA, and was then cut with either SacI or EcoRV to create templates for synthesis of antisense and sense riboprobes, respectively.
RACE-PCR.
The 5′-RACE (random amplification of cDNA ends) protocol was modified from that described previously (24) as follows. Total RNA (5 μg) from bovine distal ileum was reverse transcribed in total volume of 20 μl containing 1 μg of oligo(dT) 15-mer (Boehringer Mannheim, Indianapolis, Ind.), 40 U of RNasin (Promega, Madison, Wis.), 10 mM dithiothreitol, 0.5 mM deoxynucleoside triphosphates (dNTP), and 20 U of avian myeloblastosis virus reverse transcriptase (Promega) in buffer supplied with the enzyme at 42°C for 1 h. The excess oligo(dT) primer was removed by two sequential rounds of ultrafiltration (dilution with 10 mM Tris-HCl [pH 8.0] to a volume of 1 ml and then concentration to about 40 μl in a Centricon 100 concentrator). The resulting mixture was lyophilized to dryness and then dissolved in 35 μl of reaction buffer containing 200 mM potassium cacodylate, 25 mM Tris-HCl (pH 6.6), 1 mM dATP, 1.6 mM CoCl2, 250 μg of bovine serum albumin per ml, and 50 U of terminal transferase (Boehringer Mannheim) for 30 min at 37°C. The reaction was terminated by incubation at 65°C for 15 min. The solution was then diluted to 500 μl with 10 mM Tris-HCl–1 mM EDTA (pH 8.0). A 5-μl aliquot was amplified by PCR with 100 ng each of RACE dT17 adapter (24) and EBD 271a in 2.5 mM MgCl2–50 mM KCl–200 μM dNTP–10 mM Tris-HCl (pH 8.3) in a total volume of 50 μl. The PCR products were amplified at denaturing and extension temperatures of 94 and 72°C, respectively, by using a modification of a published ramping protocol (16). The annealing temperature was 42°C for five cycles and then was increased to 60°C for two cycles. The annealing temperature was decreased 1°C every 2 cycles until it reached 52°C; here, 10 cycles of amplification were executed and then the product ends were fully extended at 72°C for 7 min. Each segment of the first five cycles lasted 1 min, and the segments of the remaining cycles lasted 30 s. A 0.5-μl aliquot of the resulting reaction product was amplified in a second PCR with 138 ng of RACE adapter (24) and 100 ng of EBD 242a as primers. The PCR program was 25 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C. This product was extracted after gel electrophoresis with Mermaid (Bio 101, La Jolla, Calif.) and then phosphorylated and filled in with T4 polynucleotide kinase and T4 DNA polymerase (40). The resulting products were subcloned into pBluescript II SK+ at the SmaI site and subjected to dideoxynucleotide termination sequence analysis.
For 3′-RACE, a protocol based on that described by Borson (7) was employed, with reagents from Clontech Laboratories, Inc. Briefly, 1 μg of total RNA from bovine distal ileum was reverse transcribed by using the anchor primer 5′-CCTCTGAAGGTTCCAGAATCGATAGGAATTC(T)18(GCA)(GCAT)-3′ under the conditions described above. An aliquot (1/40 of the total) of the resulting cDNA product was used directly as a template in a PCR with TAP30s (0.1 μM) as a β-defensin primer and the anchor primer 5′-CTGGTTCGGCCCACCTCTGAAGGTTCCAGAATCGATAG-3′ (0.1 μM). The reaction conditions were 94°C for 30 s, 50°C for 30 s, and 68°C for 2 min for 30 cycles in 3.5 mM MgCl2–75 mM KCl–200 μM dNTP–10 mM Tris-HCl (pH 8.8). The reaction product was isolated, digested with EcoRI to cleave the DNA at the site incorporated in the anchor sequence, and then subcloned into the multiple cloning site of pBluescript II SK+ for further analysis as described previously (40).
Cryptosporidium parvum infection.
Two newborn male Holstein calves were purchased from a local dairy on the day of birth. The calves were deprived of colostrum and at 12 h of age were fed 106 C. parvum oocysts (AUCp-1 isolate) suspended in 50 ml of whole milk. This dose of C. parvum results in a nonfatal infection which usually causes diarrhea for 5 to 10 days. Fecal consistency was monitored, and calves were euthanatized by intravenous barbiturate injection 12 h after the onset of diarrhea, which occurred on the fifth day after inoculation. Two control calves were purchased from the same dairy on the day of birth, deprived of colostrum, and euthanatized at the same age as the experimental calves. The control calves did not develop diarrhea. Immediately following euthanasia, samples of ileum (10 cm proximal to ileocecal valve) and spiral colon were frozen in liquid nitrogen and then stored at −70°C until analysis. Identical regions of bowel from control and experimental animals were sampled. The presence or absence of C. parvum in control and experimental subjects was confirmed by histologic examination of formalin-fixed intestinal tissue. All experiments were performed in accordance with guidelines established by the Institutional Animal Care and Utilization Committee and the Office of Environmental Health and Safety of the University of Pennsylvania.
β-Defensin peptide isolation.
Bovine distal small intestine (200 g) was obtained at a local abattoir, immediately frozen in liquid nitrogen, and stored at −70°C. The frozen tissue was broken into small pieces with a mortar and pestle before being stirred overnight in 400 ml of 30% formic acid. The mixture was subjected to three freeze-and-thaw cycles and then filtered through nylon mesh (60 μm). The filtrate was adjusted to 1 M ammonium sulfate and stirred at 4°C for 3 h. The precipitate that formed was pelleted by centrifugation at 10,000 rpm in a GSA rotor for 30 min at 10°C. Aliquots (60 ml) of the resulting supernatant were applied to C18 SepPak cartridges (Millipore, Inc., Bedford, Mass.) which had been previously equilibrated by washing each with 4 ml of methanol and then 4 ml of 0.1% trifluoroacetic acid (TFA) in water. After the samples were loaded, the cartridges were washed with 4 ml of 0.1% TFA in water and then eluted with 4 ml of acetonitrile–0.1% TFA in water (60:40, vol/vol). The eluates were placed in polypropylene tubes and dried under vacuum at room temperature overnight. The dried samples were dissolved and pooled in a total of 1 ml of 6 M guanidinium HCl–20 mM Tris-HCl (pH 7.5) and filtered through a nylon filter (Cameo; 25-μm pore size) which had been wetted with the sample buffer. The resulting filtrate was chromatographed on a P30 (Bio-Rad, Hercules, Calif.) column (2 by 30 cm). The column had been equilibrated in 50 mM ammonium formate (pH 4.1), and the flow rate was 2.5 ml/min. Eighty fractions (2.5 ml/fraction) were collected from the column, dried under vacuum, and redissolved in 0.1 ml of 0.01% acetic acid. The antimicrobial activity of an aliquot (4 μl) of each fraction was determined in the assay described below. The active fractions (fractions 21 to 40) were pooled, dried under vacuum, and redissolved in 0.8 ml of water. The sample was applied to a sulfoethyl cation-exchange high-pressure liquid chromatography (HPLC) column (20 by 0.46 cm; Poly LC, Inc., Columbia, Md.) with acetonitrile–5 mM KH2PO4 in water (3:1, vol/vol) as a loading buffer at a flow rate of 1.0 ml/min. The column was washed with 5 ml of loading buffer and then eluted with a 45-min linear gradient to 1 M NaCl in the loading buffer. Fractions were collected at 1-min intervals, and the fractions eluting between 30 and 33 min were pooled and then dried under vacuum. Pilot experiments revealed that these fractions contained significant antibacterial activity (data not shown). The dried material was redissolved in 0.4 ml of 0.1% TFA and injected onto a C18 reverse-phase HPLC column (220 by 4.6 mm; Vydac, Hesperia, Calif.) which had been equilibrated in 0.1% TFA in water (solvent A). The column was washed with 5 ml of solvent A at 1 ml/min and then eluted with a 10-min linear gradient to 15% solvent B (0.08% TFA in acetonitrile), followed by a 49-min gradient to 30% solvent B, an isocratic elution at 30% solvent B, and finally a linear gradient over 5 min to 80% solvent B. Aliquots (50 μl) of each fraction were dried, resuspended in 5 μl of 0.01% acetic acid, and assayed for activity. Three peaks of activity were observed (eluting at 39 to 41, 52, and 59 min). The first (m/z = 5,494) and third (m/z = 6,249) active fractions had a blocked N terminus. The second active fraction of this separation, eluting at 52 min, was analyzed as follows.
Purified peptides were analyzed by a combination of automated Edman degradation andmatrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (18, 19). Mass analysis (with 2% aliquots) was carried out with a model Voyager reverse-phase MALDI-TOF instrument (PerSeptive, Framingham, Mass.) in the linear mode and with α-cyano-4-hydroxycinnamic acid (Linear Sci., Reno, Nev.) as the matrix; a 30-kV ion acceleration voltage (grid voltage at 70%; guide wire voltage at 0.1%) and −2.0 kV multiplier voltage were used. Chemical sequencing (on 95% of the sample) was done with a model 477A instrument from Applied Biosystems (AB) (Perkin-Elmer Corp., Norwalk, Conn.). Stepwise liberated phenylthiohydantoin amino acids were identified by using an on-line 120A HPLC system (AB) equipped with a phenylthiohydantoin C18 (2.1 by 220 mm; 5-μm particle size) column (AB). Instruments and procedures had been optimized as described previously (19, 61). Average peptide isotopic masses were calculated from the predicted sequence by using ProComp version 1.2 software (P. C. Andrews, University of Michigan, Ann Arbor).
Antimicrobial assay.
The antimicrobial plate assay was based on the method described by Lehrer et al. (37). Briefly, a single colony of Escherichia coli D31 was grown overnight at 37°C in 25 ml of Trypticase soy broth (TSB) (30 g/liter). A 25-μl aliquot of the overnight growth was then grown in 25 ml of TSB for 2 h at 37°C. The bacteria were pelleted by centrifugation, and the bacterial pellet was resuspended in 10 ml of cold sterile 10 mM sodium phosphate, pH 7.4. The bacterial concentration was determined by measuring the absorbance at 620 nm, assuming that an absorbance of 1 equals 2.5 × 108 CFU/ml. Then, 2.5 × 106 CFU was mixed with 10 ml of warmed (to 55°C) underlay agarose (1% agarose [low EEO; Sigma, St. Louis, Mo.], 0.03% TSB, 0.02% Tween 20, and 10 mM sodium phosphate, pH 7.4); this mixture was poured into 100- by 100- by 15-mm square petri dishes and allowed to harden. Sample wells were made by punching holes with a 3-mm agar punch (Bio-Rad). A 4-μl sample was added to each well. In addition, 1 μg of magainin was placed in one well as a positive control. The plate was incubated at 37°C upright for 3 h in order to dry the sample and maximize the specific killing of bacteria by the applied samples before addition of 10 ml of overlay agarose (an autoclaved solution of 60 g of TSB per liter, 10 mM sodium phosphate [pH 7.4], and 1% agarose) which had been warmed to 42°C prior to pouring. The agarose was allowed to harden before incubation of the plates overnight at 37°C. Antimicrobial activity was quantitated by measuring the area of the circular clear zones on the opaque background of bacterial growth (37).
Nucleotide sequence accession numbers.
The GenBank accession number for the EBD genomic sequence is AF16539, and that for the EBD cDNA sequence is AF000362. The GenBank accession numbers for the cDNA sequences are AF016396 (BNBD-3), AF016394 (BNBD-9), and AF016395 (BBD-C7).
RESULTS
Cloning and characterization of the EBD gene and cDNA.
During the screening of a bovine genomic library for the TAP gene by using a probe common to several members of the β-defensin family (13), a related clone distinct from the TAP gene was isolated. Partial sequence analysis of this genomic clone (Fig. 1) revealed an overall identity of 88% with the TAP gene, and two potential exons similar to those of the TAP gene were identified. Pilot studies indicated that mRNA corresponding to this putative gene was expressed in enteric tissue (data not shown), and a corresponding cDNA clone was isolated from a bovine small intestine cDNA library (Fig. 2). Thus, we have designated the gene the EBD gene.
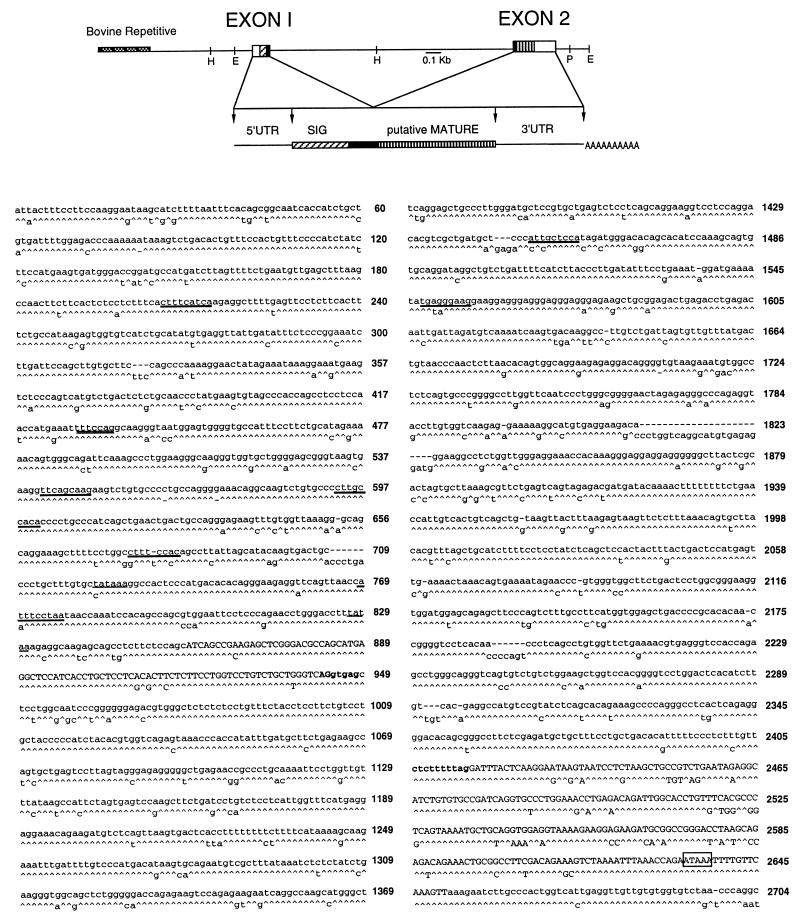
FIG. 1.
Structure and sequence of the EBD gene. (A) Restriction map of the bovine EBD gene including 0.9 kb of 5′ flanking sequence. E, EcoRI; H, HindIII; P, PstI; UTR, untranslated region; SIG, signal. Also shown is a diagram of the predicted precursor structure of EBD deduced from the gene and cDNA sequences. (B) Nucleotide sequence of the EBD gene and alignment with the TAP gene sequence (13), with carets representing nucleotide identity. Exons (capital letters) were determined by comparison with EBD cDNA sequences (Fig. 2). Consensus sequences for TATA boxes (underlined), NF–IL-6 sites (boldface underlined), and H-APF-1 (double underlined) are indicated (see text). The consensus splice junction residues are shown in boldface. The polyadenylation signal is boxed.
FIG. 2.
cDNA sequence of EBD. (A) The upstream sequences of the individual RACE clones and the relevant portion of a lambda clone, BSI-13, are aligned with the genomic sequence (SEQ); colons denote sequence identity. The major transcription start site as determined by this analysis is indicated by the thick arrow, and the first uppercase nucleotide is designated nucleotide +1. Other possible transcription sites (−2, −3, and −47) are indicated by thin arrows. A consensus TATA box sequence is double underlined, and a second putative TATA box is single underlined. The presumed translation initiation codon, ATG, is boxed. (B) The cDNA sequence and the deducedprotein sequence of EBD are aligned with protein sequence of TAP. The position of the intron in the corresponding genomic sequence is indicated by a vertical line. The brackets in the TAP sequence encompass the isolated peptide found in tracheal tissues (15).
Analysis of EBD mRNA by 5′-RACE suggested that EBD gene transcription initiates at one of two sites. Three of the RACE clones started within 2 nucleotides of the 5′ end of the lambda cDNA clone (Fig. 2A), and this position was designated +1. This transcription start site was 27 nucleotides 3′ to a TATA box motif, and its position was similar to that of the transcription start site identified for the TAP gene (13). Another RACE clone, APT131.6, contained additional nucleotides of 5′ sequence not found in the other sequences. This extra 5′ sequence is contiguous with the adjacent transcribed genomic sequence, ruling out an alternative 5′ exon. The 5′ nucleotide of this clone (−47) was 32 nucleotides downstream of a second TATA sequence and thus appears to be a second site of transcription initiation. We conclude that EBD gene transcription is initiated as indicated in Fig. 2A and that EBD mRNA is derived from splicing of two exons as depicted in Fig. 1 and 2B.
The sequences of the deduced prepropeptide and predicted mature peptide of EBD are 72 and 67% identical to those of TAP, respectively (Fig. 2B). A shared feature of the deduced amino acid sequence is the cysteine array inherent to the family of β-defensins (13, 28, 59). In addition, both sequences have particularly high similarity of the 5′ cDNA sequence and the corresponding amino acids of the putative signal sequence encoded by this region.
A search of the genomic sequence for motifs recognized by transcription factors revealed several possible sites of EBD gene regulation. For example, there is an NF–interleukin-6 (NF–IL-6) consensus binding site in the 5′ flanking region of EBD positioned where a putative NF-κB site is located in the TAP gene (Fig. 1). Both the EBD and TAP genes have two additional NF–IL-6 consensus binding sites similarly located 5′ relative to the respective NF–IL-6/NF-κB site (Fig. 1). The presence of a consensus binding site for H-APF-1 (Fig. 1), a factor known to cooperate with NF–IL-6 in gene activation (39), further supports a possible functional significance of the putative NF–IL-6 sites. A database search with the entire EBD gene sequence revealed the presence of several highly repetitive elements of the bovine genome, including nucleotides −540 to −729, −370 to −540, and +1530 to +1580. The domains between these elements had similarity to the other bovine β-defensin genes previously characterized, but no other matches were notable.
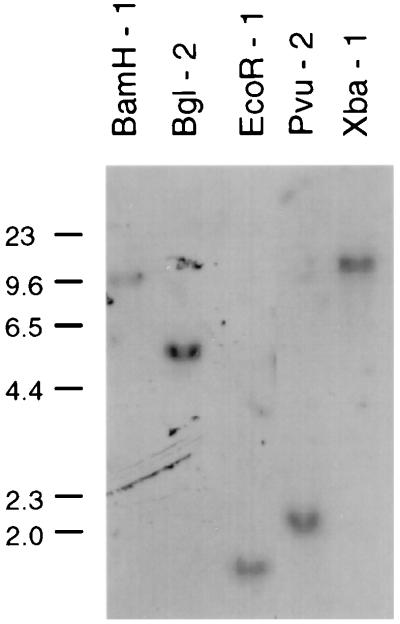
A Southern blot analysis of bovine genomic DNA revealed a single band with each of five restriction enzymes by using an EBD gene-specific probe, EBD 285a (Fig. 3). These results indicate that the EBD gene is a single-copy gene and address the specificity of hybridization conditions employed in this study. The specificity of hybridization with this probe was further demonstrated by hybridizing dot blot panels of cloned DNAs of various β-defensin genes. Only the EBD gene among a collection of nine β-defensin-encoding gene sequences was recognized by EBD 285a under the same hybridization conditions as for the Southern and Northern blots (data not shown).
FIG. 3.
Southern blot hybridization analysis of the EBD gene. Bovine genomic DNA (10 μg) was digested with restriction endonucleases, and the products were size separated by agarose gel electrophoresis. The DNA was transferred to a nylon membrane and hybridized with 32P-end-labeled oligonucleotide EBD 285a (see Materials and Methods). Hybridization conditions were 5× SSC–1% SDS–5× Denhardt solution–40 μg of RNA per ml at 42°C in the presence of 37.5% (vol/vol) formamide. The high-stringency wash of the filter was with 2× SSC–0.1% SDS at 60°C for 60 min. The autoradiographic exposure was approximately 4 weeks. Numbers on the left indicate sizes (in basepairs) of mobility standards.
Northern blot and in situ hybridization analysis of EBD mRNA.
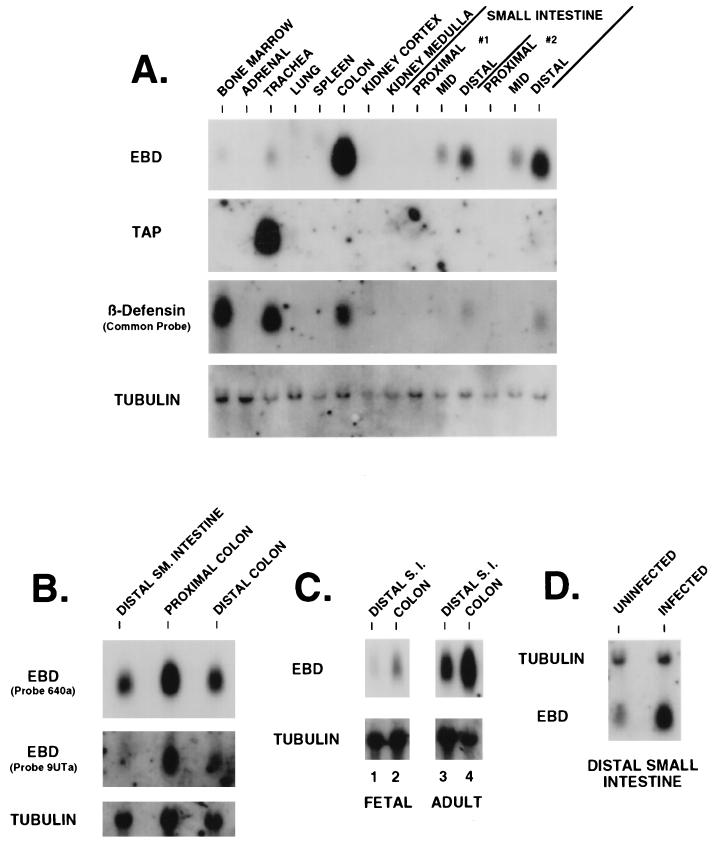
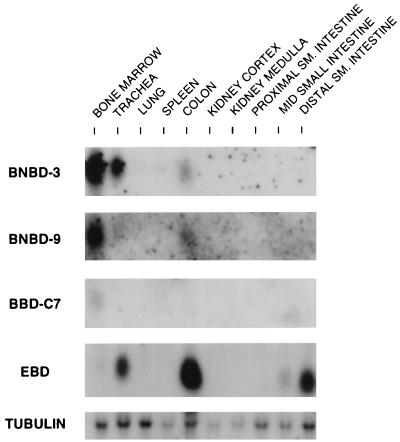
Northern blot analysis of total RNAs extracted from several bovine tissues with EBD 285a as probe revealed a message of about 600 nucleotides abundantly expressed in the colon and small intestine (Fig. 4A, EBD). The intensity of the signal detected in the distal small intestine was strong relative to the detectable mRNA in more-proximal segments of the small intestine. Comparable levels of mRNA were detected in the proximal and distal colon (Fig. 4B). Trace hybridization was detected in the trachea, and EBD mRNA was not detected in the bone marrow, adrenal gland, lung, spleen, kidney cortex, or kidney medulla (Fig. 4A). Somewhat lower levels of EBD mRNA were detected in both the distal small intestine and colon of a bovine fetus at a gestational age of 4 months as compared to in an adult cow (Fig. 4C). Hybridization of the filter with an oligonucleotide sequence specific for mRNA derived from the −47 transcription initiation site demonstrated an identical distribution of signal but uniformly at a lower relative intensity (Fig. 4B, 9UTa, and data not shown). All samples had intact RNA as evidenced by hybridization to an α-tubulin probe (Fig. 4A to C). Hybridization at low stringency with a probe from the 5′ cDNA region with sequence nearly identical in all characterized members of the bovine β-defensin gene family reveals the presence of abundant mRNA in several tissues (Fig. 4A, β-Defensin), consistent with widespread expression of various β-defensins. The relative abundances of mRNA detected in this experiment suggest that β-defensin expression is highest in the bone marrow (see below), trachea, and colon. One β-defensin mRNA expressed in bone marrow encodes BNBD-4 (65). TAP gene expression accounts for a substantial part of the β-defensin gene family expression in trachea (Fig. 4A, TAP), as previously observed (13, 53).
FIG. 4.
Northern blot analysis of EBD gene expression in bovine tissues. (A) Tissue distribution of β-defensins. Total RNA (20 μg) extracted from 14 different tissues was resolved by denaturing gel electrophoresis, capillary blotted to a nylon filter, and probed with either EBD 285a as an EBD probe, TAP286a as a TAP-specific probe, TAP48a as a common probe for β-defensins, or an α-tubulin probe. The hybridization and wash conditions were as described in Materials and Methods. Small intestine samples designated #1 and #2 represent RNAs extracted from the tissues of two healthy cows. (B) Expression of EBD in colonic tissue and usage of the putative upstream transcription start site. Total RNAs from the distal small (SM.) intestine, proximal colon (10 cm from the ileocecal junction), and distal colon (10 cm from the rectum) were analyzed as for panel A. The Northern blot was hybridized with EBD 285a to assess distribution of expression. The same blot was stripped of probe and rehybridized with EBD 9UTa, a probe from the unique sequence found in the 5′-extended RACE clone (Fig. 2, clone APT131.5) (see text). (C) Comparison of fetal and adult tissue expression of EBD. Total RNAs were isolated from the distal small intestine (S.I.) and colon of a bovine fetus at 4 months gestational age and from corresponding tissues of an adult cow and then analyzed as for panel A. (D) Northern blot analysis of EBD mRNA in enteric tissue from a calf infected with C. parvum. Total RNA was isolated from the distal 20 cm of small intestine from a C. parvum-infected calf and from a control uninfected calf. Analysis was as for panel A.
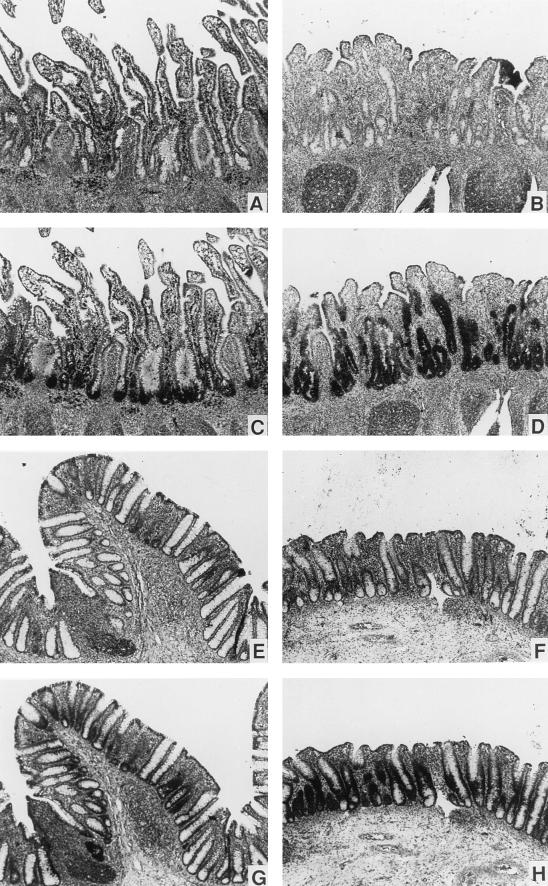
The cellular localization of the EBD mRNA was determined by in situ hybridization. Tissue sections of intestinal mucosa were probed with sense and antisense 35S-labeled riboprobes. Signal was observed with the antisense riboprobe in epithelial cells lining the small intestinal crypts in the ileum and colon of healthy cows (Fig. 5C and G, respectively). No signal in epithelial cells was observed in parallel sections when the sense riboprobe was used (Fig. 5A and E) or when the sections were treated with RNase prior to hybridization with the antisense probe (data not shown). Leukocytes present in the lamina propria of numerous small intestinal sections also appeared to be positive with the antisense riboprobe used in these experiments (Fig. 5C); however, the signal was not attenuated by pretreatment with RNase, and the sense riboprobe appeared equally positive (data not shown). The simplest explanation from these control experiments is that the EBD signal in these leukocytes may be artifactual, as previously described (46), and not from specific hybridization. However, Northern blot and anchored-PCR data suggest that cells of hematopoietic origin present in enteric tissue may express other β-defensin mRNAs (see below).
FIG. 5.
Detection of EBD mRNA by in situ hybridization. Paraffin-embedded sections of bovine ileum (A to D) and colon (E to H) were hybridized with EBD riboprobes labeled with [35S]UTP, washed under high-stringency conditions, and processed as described in Materials and Methods. (A, C, E, and G) Normal, uninfected intestinal tissue; (B, D, F, and H) intestinal tissue infected by C. parvum. The parasites are not visible at this magnification, but the blunting of the small intestinal villi and the inflammation of the lamina propria induced by the infection are easily seen in panels B and D. In normal ileum and colon, EBD mRNA is localized within epithelial cells at the base of the crypts (C and G). In C. parvum-infected ileum and colon, abundant EBD mRNA is present in epithelial cells throughout the elongated crypt (D and H). Magnification, ×80.
Expression of EBD in C. parvum infection.
We sought to determine if expression of EBD was modulated in the presence of an inflammatory gastrointestinal infection. The calf model of C. parvum infection is characterized by invasion of intestinal epithelial cells by the parasite, blunting of villi in the small intestine, crypt epithelial cell hyperplasia, and an inflammatory infiltrate in the lamina propria (64). As described in Materials and Methods, tissue samples of distal small intestine and colon were obtained from an infected calf, as well as a healthy control, in each of two experiments. Total RNA was isolated from the distal small intestine for each specimen and analyzed by Northern blot analysis (Fig. 4D). The signal for EBD mRNA was increased 10-fold in the infected calf intestine compared to the control for one experiment (Fig. 4D) and was increased 5-fold in the second experiment. Samples of small intestine and colon were also analyzed by in situ hybridization. Although less quantitative, the signal for EBD mRNA was dramatically elevated in epithelial cells in the sections from infected calves, for both ileum (Fig. 5D versus C) and cecum (Fig. 5H versus G). Hybridization appeared to be more intense within crypt cells of the small intestine and colon than in cells covering the small intestinal villi (absorptive cells) or on the surface of the colonic mucosa. For individual epithelial cells, the localization of EBD mRNA did not correlate with localization of C. parvum infection, since the heavily infected villus tip cells produced little EBD mRNA while the minimally infected crypt cells produced abundant EBD mRNA. Therefore, C. parvum does not appear to directly induce EBD mRNA production in individual infected cells but rather causes the hyperplasia of uninfected crypt cells which produce EBD mRNA.
Identification of β-defensin isoforms expressed in intestinal tissue.
To investigate the possibility that additional β-defensin mRNA isoforms are expressed in the bovine small intestine, an anchored-PCR strategy was employed. Invariant nucleotide sequences have been found in the 5′ portions of all bovine β-defensin cDNAs cloned to date (13, 53, 57, 65). A PCR primer, BTAP-27s, was selected from this region of high nucleotide similarity. RNA from the distal ileum was reverse transcribed by using a modified oligo(dT) primer which contained a flanking anchor sequence. The resulting cDNA template was then used in a PCR amplification with the β-defensin primer and an anchor sequence primer. A product of approximately 300 bp was obtained, and Southern blot hybridization with an internal probe also from the common sequence of β-defensin cDNA (TAP48a) demonstrated strong hybridization (data not shown). The PCR products were subcloned, and 118 individual colonies that appeared to encode members of the β-defensin family were characterized. High-stringency dot blot hybridization analysis of these clones with oligonucleotide probes which corresponded to several different β-defensins allowed us to segregate the clones into several groups. Plasmid DNAs from one (or more) clones from each group were sequenced in entirety from both strands. Ninety-six of the 118 β-defensin-related clones (81%) (Table 2) hybridized with each of two EBD oligonucleotide probes under high-stringency conditions, and the nucleotide sequence obtained from five randomly selected clones from this group corresponded exactly to that of the EBD gene (Table 2). Several clones excluded from this group encoded other known β-defensins whose cDNAs have been cloned previously, including TAP (n = 4) (15), TAP(S20N) (n = 2) (54), BNBD-4(n = 6) (65), and lingual antibiotic peptide (LAP) (n = 3) (53, 57). Other clones were found to encode previously identified β-defensins, BNBD-3 (n = 3) and BNBD-9 (n = 1), whose cDNAs had not been cloned. Two clones encoded a previously uncharacterized β-defensin, designated β-defensin c7 (BBD-C7). Northern blot analysis revealed that BNBD-3, BNBD-9, and BBD-C7 are expressed in bone marrow (Fig. 6). Previous studies have shown that BNBD-4 mRNA is found chiefly in bone marrow, but significant signal was also present in the distal small intestine, lung, trachea, and spleen (65). Other reports have localized TAP mRNA expression to the trachea and adjoining portions of the conducting airway (13), LAP to the tongue (57) and trachea (53), and TAP(S20N) to alveolar macrophages (54) and tracheal tissue (52).
TABLE 2.
3′-RACE clone analysis
| cDNA clone | No. of clonesa (% of total) |
|---|---|
| EBD | 96 (81) |
| BNBD-4 | 6 (5) |
| TAP | 4 (3) |
| LAP | 3 (3) |
| BNBD-3 | 3 (3) |
| BNBD-9 | 2 (2) |
| TAP(S20N) | 2 (2) |
| BBD-C7 | 2 (2) |
A total of 118 clones were analyzed.
FIG. 6.
Northern blot analysis of several low-abundance β-defensin cDNAs cloned from small intestine by anchored PCR. Total RNAs (20 μg) extracted from 10 different tissues were resolved by denaturing gel electrophoresis, capillary blotted to a nylon filter, and probed with either JR335.B1 as a BNBD-9 probe, BNBD2/3-189a as an BNBD-3 probe, JR300.C7 as a BBD-C7 probe, EBD 285a as an EBD probe, or an α-tubulin probe. The hybridization and wash conditions were as for Fig. 4.
Peptide isolation.
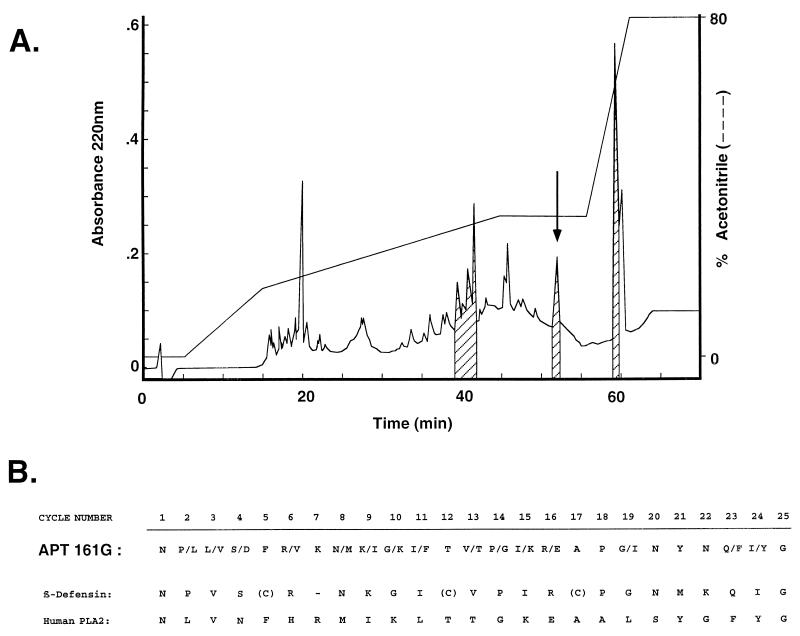
If the putative prepropeptides predicted from the characterized cDNA sequences were processed similarly to the mature TAP (15), peptides of 38 to 42 amino acids with masses of approximately 4 kDa and net charges of +8 under acidic conditions should be detected in intestinal extracts. These properties are similar to those of TAP, which has 38 amino acids, a molecular mass of 4,085 Da, and a net charge of +10. We employed an antibacterial assay with the gram-negative bacterium E. coli D31 to isolate active fractions with chromatographic properties similar to those observed for TAP. Numerous active fractions from the distal small intestine were detected, and several were partially characterized. As described in Materials and Methods, a combination of gel filtration, ion-exchange chromatography, and reverse-phase chromatography was used to partially purify an active fraction relevant to this report, APT161G (Fig. 7A). This fraction was composed of one component which had a molecular mass-to-charge ratio (m/z) of 4,113 ± 1.6 and minor species with m/z = 7,220 and 14,440. The two minor species are almost certainly related, with either the 7,220 species representing the doubly charged form of a 14,440-Da component or the 14,440 species being the dimer of a 7,220-Da component. APT161G was subjected to amino-terminal sequence analysis (Fig. 7B). Two recognizable motifs were identified in the mixed sequence; one corresponded to a β-defensin, and the other was similar (68% identity) to human group II phospholipase A2 (41). Of the various β-defensin cDNAs identified in this study and elsewhere, the presumed β-defensin component of the isolated material had a primary sequence most consistent with TAP(S20N) (54), with the sequence identical at 19 of 22 positions. Assuming that the six cysteines in the predicted primary sequence of TAP(S20N) all participated in intracellular disulfide bonds and that the amino-terminal residue was asparagine, the calculated mass of the peptide would be 4,112 Da, consistent with that obtained for the isolated material (Fig. 7).
FIG. 7.
Isolation of β-defensin peptide from distal small intestine. An extract of bovine distal small intestine (200 g) was fractionated by using a combination of gel filtration and ion-exchange chromatography, and individual fractions were tested for antibacterial activity by an antibacterial plate assay (see Materials and Methods). (A) An active fraction from ion-exchange chromatography, eluting under conditions for previously characterized β-defensins, was applied to a C18 reverse-phase HPLC column and eluted with a gradient of acetonitrile (dashed line). The eluate was monitored by UV light absorbance (solid line), and each fraction (1 ml) was assayed for antibacterial activity (hash marks indicate active fractions). Fraction APT161G (arrow) was subjected to mass spectral analyses (see text) and N-terminal sequence analysis. (B) N-terminal sequence analysis. Two amino acids were identified at several positions, consistent with peptide heterogeneity within this fraction (see text). The mixed sequence is aligned with the sequences of TAP(S20N) (β-defensin) (54) and human group II phospholipase A2 (PLA2) (41).
DISCUSSION
Previous work in defining the structure of the TAP gene had demonstrated the presence of several TAP gene-related sequences in the bovine genome (13). We have now determined the structure of one of these related genes, the EBD gene (Fig. 1), which has led us to discover a new anatomical site of β-defensin expression. The EBD gene is expressed in the distal small intestine and colon (Fig. 4), and specifically in epithelial cells in these two sites (Fig. 5). Moreover, EBD mRNA levels are dramatically elevated in association with C. parvum infection (Fig. 5 and 7). These data support the hypothesis that EBD may contribute to host defense of the enteric mucosa.
EBD mRNA, like other β-defensin-encoding mRNAs, has a selective tissue distribution. Abundant mRNA is detected in epithelial cells throughout the length of the colon and the distal small intestine (Fig. 4 and 5). This alludes to a possible functional link for cells at these two anatomical locations that was not previously recognized. The location and morphology of the small intestinal epithelial cells expressing this mRNA may be analogous to those of the so-called undifferentiated crypt cells, previously described for human and rodent small intestinal crypts. This cell type has ultrastructural characteristics of a secretory cell, including membrane-enclosed secretory-type granules (63). Paneth cells, another secretory cell type of the small intestinal crypt of many mammals, were shown previously to be the site of expression of α-defensins in mice (44) and humans (32, 33, 49) but are not prominent in the cow (34). Paneth cells are most abundant in the ileum and are much less common in the colon. The pattern of EBD expression reported here contrasts with this distribution pattern of Paneth cells, because colonic tissue is a major site of EBD expression and at levels comparable to those of the distal small intestine. Several questions that emerge from these findings are as follows. (i) Do humans, mice, and other species with known enteric α-defensin expression in Paneth cells also express β-defensins in enteric epithelial cells? (ii) Conversely, do cows also express α-defensins in enteric tissue, and if so, where are they expressed? (iii) What is the common and distinguishing physiological role(s) of enteric α-defensins and β-defensins?
C. parvum, a protozoan parasite, has emerged as an important mammalian pathogen whose anatomic site of infection usually is the alimentary tract (10) but can also include other mucosa (62). This pathogen has emerged as a significant cause of devastating diarrheal disease in immunocompromised individuals (47). The levels of EBD mRNA are elevated in specimens from calves infected with C. parvum, suggesting that this gene may be part of a dynamic host response to inflammation and/or infection of the enteric mucosa. While our data show a correlation of EBD mRNA levels and parasitic infection, it is unclear whether induction is a direct consequence of tissue infection or the result of a general inflammatory response. The histologic changes produced by C. parvum in the intestinal mucosa (villus blunting, crypt cell hyperplasia, and inflammatory infiltration of the lamina propria) are not unique to cryptosporidiosis and may be seen in other infections, such as viral diarrhea and giardiasis (11, 21). Crypt cell hyperplasia is widely believed to represent an attempt to regenerate the villus epithelium damaged by invading microbes (31). Our studies suggest that these hyperplastic crypt cells may also play a role in immunity by producing antimicrobial peptides to defend against the insulting agent. It is likely that other intestinal infections which cause epithelial damage and crypt cell hyperplasia also induce EBD expression.
Analysis of EBD genomic and cDNA sequences (Fig. 1 and 2) indicates a two-exon gene structure, similar to that of the TAP gene. Its nucleotide identity with the TAP gene is 84% across the gene. This finding is most consistent with the two genes arising from relatively recent gene duplication and/or conversion events. Yount et al. have recently determined a similar two-exon structure for the gene encoding the hematopoietic β-defensin BNBD-4 (65). The nucleotide identity between the EBD and BNBD-4 genes is also quite high (88%) in the 969 nucleotides of available BNBD-4 gene sequence. All three of these genes have been localized to bovine chromosome 27 (25), further supporting an evolutionary history of divergence from a common ancestral gene. A striking feature of this comparison is the high similarity of nucleotide sequence throughout these three genes, given the dramatic difference in tissue expression (Fig. 4) (65). Thus, an important question that remains to be addressed is the identity of sequences within these genes which mediate their distinct patterns of expression. A second feature is that although the sequences of these three β-defensin genes are highly similar to one another, the genes have no detectable sequence similarity to those of mammalian α-defensins (data not shown). Recent data indicate that α- and β-defensin genes are located in the same chromosomal cluster (38), suggesting that they are part of a single gene family which arose from a common ancestral gene. The lack of significant sequence similarity suggests that these two subfamilies diverged long ago and/or that sequence divergence in this gene family has occurred at an accelerated pace. Nevertheless, the expression of β-defensin genes in tissues which come in frequent contact with bacteria and other microbes supports the idea that this gene subfamily, like the α-defensin subfamily, is part of a first-line host defense system.
An anchored-reverse-transcription-PCR strategy has revealed that several additional β-defensins [TAP, TAP(S20N), LAP, BNBD-3, BNBD-4, BNBD-9, and BBD-C7] are also expressed in the distal small intestine but at much lower relative abundances. The cellular sources of these low-abundance β-defensin mRNAs in enteric tissue remain to be rigorously established, but Northern blot evidence (Fig. 6) showing principal expression in bone marrow supports that BNBD-3, BNBD-9, and BBD-C7 are probably expressed in cells of hematopoietic origin and that these cells, present in the lamina propria, contribute β-defensin mRNA to the pool of enteric RNA analyzed here. Also, Ryan et al. (54) have shown that lung macrophages, obtained by bronchoalveolar lavage, express several β-defensins at the mRNA level, including BNBD-4, BNBD-5, and TAP(S20N). We have previously shown that tracheal epithelial cells express at least two different β-defensin genes, the TAP gene at high levels and the LAP gene at much reduced levels. It is also possible that enteric epithelial cells, which express EBD at high levels (Fig. 5), also express other β-defensins at lower levels. Because of the very high sequence similarity between these cDNAs, it will be technically challenging to unambiguously determine by in situ hybridization the cellular origins of these low-abundance β-defensins. Monoclonal antibodies, which are not currently available, may help identify the β-defensin-containing cells.
Many chromatographic fractions of distal small intestinal tissue extracts had antimicrobial activity in a screening assay with the gram-negative bacterium E. coli, suggesting the presence of numerous molecules that may have physiologically relevant antimicrobial function. Characterization of each of these fractions is outside the scope of this study. The isolation of a partially purified β-defensin isoform, TAP(S20N), from biologically active fractions of the intestinal extracts demonstrates the expression of β-defensin peptide in this tissue. Expression of TAP(S20N)-encoding mRNA has been observed in alveolar macrophages (54) and tracheal extracts, and an isoform containing a 2-amino-acid N-terminal extension has been isolated and characterized (8a). Identification of a β-defensin peptide corresponding to a cDNA present in a very low proportion was unanticipated, as we had sought here to isolate and characterize the EBD peptide. It is possible that the more abundant class of cDNA, that of EBD, is subject to translational regulation, which would account for abundant mRNA and low peptide levels. However, another possibility is that our isolation strategy, which focused on chromatographic fractions likely to contain TAP-like peptides, may have introduced a strong bias on β-defensin isoforms that would be identified. It is possible that EBD is processed differently from TAP (15) and some other characterized β-defensins (57, 59). Furthermore, a focus on colon tissue, where EBD mRNA is even more abundant than in the small intestine, may be helpful. Additional studies will be required to more fully characterize the structure and activity of the EBD peptide of bovine enteric tissue.
Conclusion.
The intestinal lumen of mammals is in contact with the external environment and is continuously colonized with microorganisms. As the intestinal mucosa may serve as a port of entry for invading microorganisms, effective host defense mechanisms are required. Our data reveal that a new member of the β-defensin family of antibiotic peptides is inducibly expressed in enteric epithelial cells of the colon and distal small intestine, suggesting the dynamic participation of enteric epithelial cells in local host defense and/or regulation of enteric flora. These studies provide a groundwork for future investigations into host defense responses of these mucosal surfaces. Further studies will be necessary to elucidate the specific role of the β-defensin EBD in host defense of this tissue.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI32738 and AI32234), the National Science Foundation (DBI-9420123), and the Irma T. Hirschl Trust (to P.T.); the Sloan-Kettering sequencing lab is supported by a National Cancer Institute Core Grant (5 P30 CA08748).
ADDENDUM
Stolzenberg et al. (60a) have recently also reported in situ hybridization evidence of inducible β-defensin expression in the bovine intestinal tract.
REFERENCES
- 1.Agerberth B, Boman A, Andersson M, Jörnvall H, Mutt V, Boma H G. Isolation of three antibacterial peptides from pigintestine: gastric inhibitory polypeptide(7-42), diazepam-binding inhibitor(32-86) and a novel factor, peptide 3910. Eur J Biochem. 1993;216:623–629. doi: 10.1111/j.1432-1033.1993.tb18182.x. [DOI] [PubMed] [Google Scholar]
- 2.Agerberth B, Lee J-Y, Bergman T, Carlquist M, Boman H G, Mutt V, Jörnvall H. Amino acid sequence of PR-39, isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur J Biochem. 1991;202:849–854. doi: 10.1111/j.1432-1033.1991.tb16442.x. [DOI] [PubMed] [Google Scholar]
- 3.Beagley K W, Elson C O. Cells and cytokines in mucosal immunity and inflammation. Gastroenterol Clin N Am. 1992;21:347–366. [PubMed] [Google Scholar]
- 4.Bensch K W, Raida M, Mägert H-J, Schulz-Knappe P, Forssmann W-G. hBD-1: a novel β-defensin from human plasma. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 5.Bevins C L. Antimicrobial peptides as agents of mucosal immunity. Ciba Found Symp. 1994;186:250–269. doi: 10.1002/9780470514658.ch15. [DOI] [PubMed] [Google Scholar]
- 6.Bevins C L, Diamond G. Molecular biological strategies in the analysis of antibiotic peptide gene families: the use of oligonucleotides as hybridization probes. Methods Mol Biol. 1997;78:151–166. doi: 10.1385/0-89603-408-9:151. [DOI] [PubMed] [Google Scholar]
- 7.Borson N D. A lock-docking oligo(dT) primer for 5′ and 3′ RACE PCR. PCR Methods Appl. 1992;2:144–148. doi: 10.1101/gr.2.2.144. [DOI] [PubMed] [Google Scholar]
- 8.Chirgwin J M, Pryzybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 8a.Cohen, K., and C. L. Bevins. Unpublished observations.
- 9.Cohen M S, Weber R D, Mårdh P-A. Genitourinary mucosal defenses. In: Holmes K K, Mårdh P-A, Sparling P F, Wiesner P J, editors. Sexually transmitted diseases. 2nd ed. New York, N.Y: McGraw-Hill, Inc.; 1990. pp. 117–127. [Google Scholar]
- 10.Current W, Garcia L. Cryptosporidiosis. Clin Microbiol Rev. 1991;4:325–358. doi: 10.1128/cmr.4.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davidson G. Viral diarrhoea. Clin Gastroenterol. 1986;15:39–53. [PubMed] [Google Scholar]
- 12.Diamond, G., and C. L. Bevins. 1994. Endotoxin up-regulates expression of an antimicrobial peptide gene in mammalian airway epithelial cells. Chest 105(Suppl.):51–52S. [DOI] [PubMed]
- 13.Diamond G, Jones D E, Bevins C L. Airway epithelial cells are the site of expression of a mammalian antimicrobial peptide gene. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond G, Russell J P, Bevins C L. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamond G, Zasloff M, Eck H, Brasseur M, Maloy W L, Bevins C L. Tracheal antimicrobial peptide, a novel cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenhauer P B, Harwig S S S L, Lehrer R I. Cryptdins: antimicrobial defensins of the murine small intestine. Infect Immun. 1992;60:3556–3565. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elicone C, Lui M, Geromanos S, Erdjument-Bromage H, Tempst P. Microbore reversed-phase high-performance liquid chromatographicpurification of peptides for combined chemical sequencing—laser-desorption mass spectrometric analysis. J Chromatogr. 1994;676:121–137. doi: 10.1016/0021-9673(94)00089-1. [DOI] [PubMed] [Google Scholar]
- 19.Erdjument-Bromage H, Lui M, Sabatini D M, Snyder S H, Tempst P. High-sensitivity sequencing of large proteins: partial structure of the rapamycin-FKBP12 target. Protein Sci. 1994;3:2435–2446. doi: 10.1002/pro.5560031227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans E W, Beach G G, Wunderlich J, Harmon B G. Isolation of antimicrobial peptides from avian heterophils. J Leukocyte Biol. 1994;56:661–665. doi: 10.1002/jlb.56.5.661. [DOI] [PubMed] [Google Scholar]
- 21.Farthing, M. 1993. Diarrhoeal disease: current concepts and future challenges. Pathogenesis of giardiasis. Trans. Royal Soc. Trop. Med. Hyg. 87(Suppl. 3):17–21. [DOI] [PubMed]
- 22.Fearon D T, Locksley R M. The instructive role of innate immunity in the acquired immune response. Science. 1996;272:50–54. doi: 10.1126/science.272.5258.50. [DOI] [PubMed] [Google Scholar]
- 23.Fiocchi C. Cytokines and intestinal inflammation. Transplant Proc. 1996;28:2442–2443. [PubMed] [Google Scholar]
- 24.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallagher D S, Ryan A M, Diamond G, Bevins C L, Womack J E. Somatic cell mapping of β-defensin genes to cattle syntenic group U25 and fluorescent in situ hybridization localization to chromosome 27. Mamm Genome. 1995;6:554–556. doi: 10.1007/BF00356177. [DOI] [PubMed] [Google Scholar]
- 26.Gilman M. In situ hybridization, sections 14.1–8 and 14.3–14. In: Ausubel R M, Brent R, Kinston R E, Moore D L, Seidman J G, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 27.Harder J, Bartels J, Christophers E, Schroder J M. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 28.Harwig S S L, Swiderek K M, Kokryakov V N, Tan L, Lee T D, Panyutich E A, Aleshina G M, Shamova O V, Lehrer R I. Gallinacins: cysteine-rich antimicrobial peptides of chicken leukocytes. FEBS Lett. 1994;342:281–285. doi: 10.1016/0014-5793(94)80517-2. [DOI] [PubMed] [Google Scholar]
- 29.Huttner K M, Kozak C A, Bevins C L. The mouse genome encodes a single homolog of the antimicrobial peptide human b-defensin 1. FEBS Lett. 1997;413:45–49. doi: 10.1016/s0014-5793(97)00875-2. [DOI] [PubMed] [Google Scholar]
- 30.Israel E J, Walker W A. Host defense development in gut and related disorders. Pediatr Clin N Am. 1988;35:1–15. doi: 10.1016/s0031-3955(16)36396-9. [DOI] [PubMed] [Google Scholar]
- 31.Johnson L, McCormack S. Regulation of gastrointestinal mucosal growth. In: Johnson L, editor. Physiology of the gastrointestinal tract. 3rd ed. New York, N.Y: Raven Press; 1994. pp. 611–641. [Google Scholar]
- 32.Jones D E, Bevins C L. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 33.Jones D E, Bevins C L. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 34.Jubb K V F, Kennedy P C, Palmer N. Pathology of domestic animals. 4th ed. Vol. 2. San Diego, Calif: Academic Press; 1993. [Google Scholar]
- 35.Kaliner M A. Human nasal respiratory secretions and host defense. Am Rev Respir Dis. 1991;144:S52–S56. doi: 10.1164/ajrccm/144.3_pt_2.S52. [DOI] [PubMed] [Google Scholar]
- 36.Lee J-Y, Boman A, Chuanxin S, Anderson M, Jornvall H, Mutt V, Boman H G. Antibacterial peptides from the pig intestine. Proc Natl Acad Sci USA. 1989;86:9159–9162. doi: 10.1073/pnas.86.23.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehrer R I, Rosenman M, Harwig S S, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 38.Liu L, Zhao C, Heng H H Q, Ganz T. The human β-defensin-1 and α-defensins are encoded by adjacent genes: two peptide families with differing disulphide topology share a common ancestry. FEBS Lett. 1997;43:316–320. doi: 10.1006/geno.1997.4801. [DOI] [PubMed] [Google Scholar]
- 39.Majello B, Arcone R, Toniatti C, Ciliberto G. Constitutive and IL-6-induced nuclear factors that interact with the human x C-reactive protein promoter. EMBO J. 1990;9:457–465. doi: 10.1002/j.1460-2075.1990.tb08131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallow E B, Harris A, Salzman N, Russell J P, DeBerardinis J R, Ruchelli E, Bevins C L. Human enteric defensins: gene structure and developmental expression. J Biol Chem. 1996;271:4038–4045. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
- 41.Minami T, Tojo H, Shinomura Y, Matsuzawa Y, Okamoto M. Purification and characterization of a phospholipase A2 from human ileal mucosa. Biochim Biophys Acta. 1993;1170:125–130. [PubMed] [Google Scholar]
- 42.Moore K S, Bevins C L, Brasseur M M, Tomasini N, Turner K, Eck H, Zasloff M. Antimicrobial peptides in the stomach of Xenopus laevis. J Biol Chem. 1991;266:19851–19857. [PubMed] [Google Scholar]
- 43.Newhouse M T, Bienenstock J. Respiratory tract defense mechanisms. In: Baum G L, Wolinsky E, editors. Textbook of pulmonary disease. Boston, Mass: Little, Brown & Co.; 1989. pp. 21–47. [Google Scholar]
- 44.Ouellette A J, Greco R M, James M, Frederick D, Naftilan J, Fallon J T. Developmental regulation of cryptdin, a corticostatin/defensin precursor mRNA in mouse small intestinal crypt epithelium. J Cell Biol. 1989;108:1687–1695. doi: 10.1083/jcb.108.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouellette A J, Miller S I, Henschen A H, Selsted M E. Purification and primary structure of murine cryptdin-1, a Paneth cell defensin. FEBS Lett. 1992;304:146–148. doi: 10.1016/0014-5793(92)80606-h. [DOI] [PubMed] [Google Scholar]
- 46.Patterson S, Gross J, Webster A B D. DNA probes bind nonspecifically to eosinophils during in situ hybridization: carbol chromotrope blocks binding to eosinophils but does not inhibit hybridization to specific nucleotide sequences. J Virol Methods. 1989;23:105–109. doi: 10.1016/0166-0934(89)90124-9. [DOI] [PubMed] [Google Scholar]
- 47.Petersen C. Cryptosporidiosis in patients infected with the humanimmunodeficiency virus. Clin Infect Dis. 1992;15:903–909. doi: 10.1093/clind/15.6.903. [DOI] [PubMed] [Google Scholar]
- 48.Pleyer U, Baatz H. Antibacterial protection of the ocular surface. Ophthalmologica. 1997;1:2–8. doi: 10.1159/000310878. [DOI] [PubMed] [Google Scholar]
- 49.Porter E, Liu L, Oren A, Anton P, Ganz T. Localization of human intestinal defensin 5 in Paneth cell granules. Infect Immun. 1997;65:2389–2395. doi: 10.1128/iai.65.6.2389-2395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porter E, van Dam E, Valore E, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65:2396–2401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reilly D S, Tomassini N, Bevins C L, Zasloff M. A Paneth cell analogue in Xenopus small intestine expresses antimicrobial peptide genes: conservation of an intestinal host-defense system. J Histochem Cytochem. 1994;42:697–704. doi: 10.1177/42.6.8189032. [DOI] [PubMed] [Google Scholar]
- 52.Russell, J. P., and C. L. Bevins. Unpublished observations.
- 53.Russell J P, Diamond G, Tarver A, Bevins C L. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor alpha. Infect Immunity. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan L K, Rhodes J, Bhat M, Diamond G. Expression of β-defensin genes in bovine alveolar macrophages. Infect Immun. 1998;66:878–881. doi: 10.1128/iai.66.2.878-881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito T, Kawabata S, Shigenaga T, Takayenoki Y, Cho J, Nakajima H, Hirata M, Iwanaga S. A novel big defensin identified in horseshoe crab hemocytes: isolation, amino acid sequence, and antibacterial activity. J Biochem. 1995;117:1131–1137. doi: 10.1093/oxfordjournals.jbchem.a124818. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 57.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 58.Selsted M E, Miller S I, Henschen A H, Ouellette A J. Enteric defensins: antibiotic peptide components of intestine host defense. J Cell Biol. 1992;118:929–936. doi: 10.1083/jcb.118.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selsted M E, Tang Y-Q, Morris W L, McGuire P A, Novotny M J, Smith W, Henschen A H, Cullor J S. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J Biol Chem. 1993;268:6641–6648. [PubMed] [Google Scholar]
- 60.Sherman P M, Forstner J F, Forstner G G. Mucosal barrier and its defense during the perinatal period. In: Lebenthal E, editor. Human gastrointestinal development. New York, N.Y: Raven Press; 1989. pp. 697–698. [Google Scholar]
- 60a.Stolzenberg E D, Anderson G M, Ackermann M R, Whitlock R H, Zasloff M. Epithelial antibiotic induced in states of disease. Proc Natl Acad Sci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tempst P, Geromanos S, Elicone C, Erdjument-Bromage H. Improvements in microsequencer performance for low picomole sequence analysis. Methods Companion Methods Enzymol. 1994;6:248–261. [Google Scholar]
- 62.Travis W, Schmidt K, MacLowry J, Masur H, Condron K, Fojo A. Respiratory cryptosporidiosis in a patient with malignant lymphoma. Report of a case and review of the literature. Arch Pathol Lab Med. 1990;114:519–522. [PubMed] [Google Scholar]
- 63.Trier J S. Studies on small intestinal crypt epithelium. I. The fine structure of the crypt epithelium of the proximal small intestine of fasting humans. J Cell Biol. 1963;18:599–620. doi: 10.1083/jcb.18.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tzipori S. Cryptosporidiosis in animals and humans. Microbiol Rev. 1983;47:84–96. doi: 10.1128/mr.47.1.84-96.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yount, N. Y., J. Yuan, A. P. Tarver, G. Diamond, J. N. Levy, P. A. McGuire, C. McCullogh, J. S. Cullor, C. L. Bevins, and M. E. Selsted. Molecular cloning and expression of bovine neutrophil β-defensin 4. Characterization of cDNA and genomic sequences, pattern of tissue expression, andlocalization of the fully-processed peptide to neutrophil dense granules. Submitted for publication.
- 66.Zasloff M. Antibiotics peptides as mediators of innate immunity. Curr Opin Immunol. 1992;4:3–7. doi: 10.1016/0952-7915(92)90115-u. [DOI] [PubMed] [Google Scholar]