Abstract
During embryonic development, 2 populations of multipotent stem cells, cranial neural crest cells (NCCs) and epibranchial placode cells (PCs), are anatomically adjacent to each other. The coordinated migration of NCCs and PCs plays a major role in the morphogenesis of craniofacial skeletons and cranial nerves. It is known that ethanol-induced dysfunction of NCCs and PCs is a key contributor to the defects of craniofacial skeletons and cranial nerves implicated in fetal alcohol spectrum disorder (FASD). However, how ethanol disrupts the coordinated interaction between NCCs and PCs was not elucidated. To fill in this gap, we established a well-designed cell coculture system to investigate the reciprocal interaction between human NCCs (hNCCs) and human PCs (hPCs), and also monitored the migration behavior of NCCs and PCs in zebrafish embryos. We found that ethanol exposure resulted in a disruption of coordinated hNCCs-hPCs interaction, as well as in zebrafish embryos. Treating hNCCs-hPCs with exosomes derived from ethanol-exposed hNCCs (ExoEtOH) mimicked ethanol-induced impairment of hNCCs-hPCs interaction. We also observed that SDF1, a chemoattractant, was downregulated in ethanol-treated hPCs and zebrafish embryos. Meanwhile, miR-126 level in ExoEtOH was significantly higher than that in control exosomes (ExoCon). We further validated that ExoEtOH-encapsulated miR-126 from hNCCs can be transferred to hPCs to suppress SDF1 expression in hPCs. Knockdown of SDF1 replicated ethanol-induced abnormalities either in vitro or in zebrafish embryos. On the contrary, overexpression of SDF1 or inhibiting miR-126 strongly rescued ethanol-induced impairment of hNCCs-hPCs interaction and developmental defects.
Keywords: neural crest cells, placode cells, exosomes, SDF1, miR-126
Fetal alcohol spectrum disorders (FASD) are among the most devastating consequences of maternal ethanol exposure during pregnancy. One of the hallmarks of FASD is the birth defects of craniofacial skeletons and cranial sensory system. The neural crest cells (NCCs) are multipotent progenitor cells that can give rise to a diversity of neural and nonneural cell types, such as melanocytes, neurons, glial, and mesenchymal cells that form craniofacial skeletons and dermis (Delfino-Machin et al., 2007; Hall, 2008; Teng and Labosky, 2006). The majority of the cranial mesenchyme are made up by NCCs, which contributes significantly to the craniofacial structures (Teng and Labosky, 2006). Cranial placodes are thickened regions of ectoderm originated at the border between the neural plate and neural crest that include the olfactory, lens, otic, trigeminal, and epibranchial placodes (O’Neill et al., 2012). The epibranchial placode cells (PCs) can later differentiate into the distal ganglia of facial (VII), glossopharyngeal (IX), and vagus (X) cranial nerves (D’Amico-Martel and Noden, 1983; Harlow et al., 2011; Harlow and Barlow, 2007; Schlosser and Northcutt, 2000). NCCs and PCs are both highly migratory cell populations throughout their development, and they remain in close proximity to each other and interact in a reciprocal manner, which is crucial for the coordinated morphogenesis of head and functional sensory systems (Steventon et al., 2014). In addition, it has been demonstrated that a PCs-derived paracrine chemokine, SDF1, plays a key role in mediating the interaction between PCs and NCCs (Theveneau et al., 2013). Up to now, most studies just focused on ethanol-induced excessive apoptosis or migration impairment in NCCs (Cartwright and Smith, 1995a,b; Dunty et al., 2001; Rovasio and Battiato, 2002). How ethanol exposure disrupts the coordinated migration of NCCs and PCs in embryonic development and the detailed mechanism of ethanol-induced concurrence of the dysfunction of NCCs and PCs that leads to craniofacial and cranial nerve defects remains largely unknown.
Exosomes are approximately 40–150 nm, endosome-derived, small extracellular vesicles secreted by most cells (Jeppesen et al., 2019). Exosomes carry a variety of biologically active molecules, including proteins, lipids, and miRNAs, and can transfer these molecules from one cell to another to facilitate cell-cell communication (Lener et al., 2015; Mateescu et al., 2017; Rufino-Ramos et al., 2017). For instance, exosomes have been shown to mediate the communication between neurons and oligodendrocytes (Fruhbeis et al., 2013); During embryogenesis, they have been implicated in important classes of developmental signals such as WNT, HH, and Notch (McGough and Vincent, 2016). It has also been reported that maternal exosomes in diabetes that cross the maternal-fetal barrier can result in cardiac defects in embryos (Shi et al., 2017).
miRNAs are a class of small non-coding RNA molecules that can regulate gene expression and play a pivotal role in numerous biological events (Bartel, 2004). Among exosome cargoes, miRNAs have been considered to be crucial in the therapeutic effects of exosomes (Zhang et al., 2019). Studies have shown that exosomes can be taken up into neighboring or distant cells and release miRNAs to modulate the function of recipient cells (Bang et al., 2014; Costa-Silva et al., 2015; Fong et al., 2015; Zhang et al., 2015). A number of miRNAs have been demonstrated to be involved in the modulation of SDF1-mediated signaling (Lewellis et al., 2013; Staton et al., 2011). For instance, miR-342 and miR-137 can act as a tumor suppressor by targeting SDF1 (Dong et al., 2016; Tian et al., 2018), whereas miR-126 can directly inhibit SDF1 expression and enhance the migration of the CD34+ cells (van Solingen et al., 2011). In particular, exosome-encapsulated miR-126 transferred from endothelial cells to leukemia cells reduced SDF1 expression and cell migration which can be restored by miR-126 inhibitor (Taverna et al., 2014). These evidences indicate that exosome-derived miRNAs play an important role in exosome-mediated cell-cell communication.
Here, we aimed to uncover an exosome-mediated mechanism by which ethanol exposure destroys the coordinated hNCCs-hPCs interaction in vitro, and to elucidate whether the restoration of the SDF1-mediated chemotaxis signaling can prevent ethanol-induced teratogenesis in vivo. Using a coculture system for hNCCs-hPCs, we showed that ethanol exposure results in a significant increase of hNCCs-derived exosome cargo, miR-126, which can be horizontally transferred to hPCs to specifically inhibit SDF1 expression, leading to an impairment of coordinated migration between hNCCs and hPCs. We also demonstrated that a recovery of SDF1 expression through interfering with miR-126/SDF1 axis can reverse ethanol-induced disruption of hNCCs-hPCs interaction and developmental defects. Besides, we established an efficient way for delivering miR-126 inhibitors using the grape-derived exosome-like nanoparticles (GELNs) as a carrier, which might be a translatable strategy for the intervention for FASD.
Materials and methods
hNCCs and hPLs induction from hESCs
Human Neural crest cells (hNCCs) and human placode cells (hPCs) were both differentiated from human embryonic stem cells (hESCs) purchased from WiCell (Madison, Wisconsin). hESCs were cultured in mTeSR1 (StemCell Technologies, Inc., Vancouver, Canada) on hESC-qualified Matrigel (BD Biosciences, San Jose, California) coated plates. The differentiation procedures are as follows: For hNCCs induction, the hESCs were adapted to accutase dissociation and single-cell culture before subsequent differentiation (Lee et al., 2010). Briefly, hESCs cultures were disaggregated by accutase for 10 min. The nonadherent hESCs were washed and plated on matigel-coated dishes. hESCs at 85% confluence were ready to be passaged for hNCCs induction. The hESCs culture medium (mTeSR1) was then replaced with hNCCs-induction medium (DMEM/F-12 Medium; 14.3 M l-glutamine + β-mercapyoethanol; MEM non-essential amino acid; 10 µg/ml Fgf2; 10 µg/ml heregulin β-1; 200 µg/ml Long R3-IGF1; 10 mM CHIR 99021; 10 mM SB421542; penicillin and streptomycin) and change daily. Once reaching proper confluence (75%–85%), the differentiating cells were passaged and maintained in the hNCCs-induction medium. After 10 days, the hNCCs identity was determined by hNCCs-specific marker, HNK1 and p75, through immunocytochemistry. For hPCs induction, the initial differentiation medium includes knock out serum replacement media (KSR) supplemented with 10 μM TGF-β inhibitor SB431542 (Tocris, Minneapolis, Minnesota) and 250 ng/ml of Noggin (R&D, Minneapolis, Minnesota) (Dincer et al., 2013). Cells were grown in hPLs-induction medium for 7 days with medium changed daily and Noggin was withdrawn at Day 3 of differentiation. At Day 7, the hPC identity was examined by hPCs marker Six1 using immunocytochemistry.
Establishment of coordinated migration system in vitro
The coordinated migration of hNCCs and hPCs in vitro was evaluated by using the CytoSelect Wound Healing Assay Kit, which can be used as a coculture system for 2 types of cells (Cell Biolabs, San Diego, California). At first step, an insert was put into a cell culture plate. hNCCs and hPCs were then seeded into each side of the insert and were cultured to form a monolayer. Next, the insert was removed to generate a 0.9-mm cell-free gap between hNCCs and hPCs. Then the cocultured hNCCs and hPCs were ready for downstream experiments to evaluate the coordinated migration between hNCCs and hPCs. Finally, the cocultured hNCCs-hPCs were fixed and costained with antibodies against specific markers of hNCCs (HNK1) and hPCs (Six1), and then the images were captured under a fluorescent microscope (Olympus IMT-2, Tokyo, Japan).
Quantification of NCCs-PCs migration in vitro and in vivo
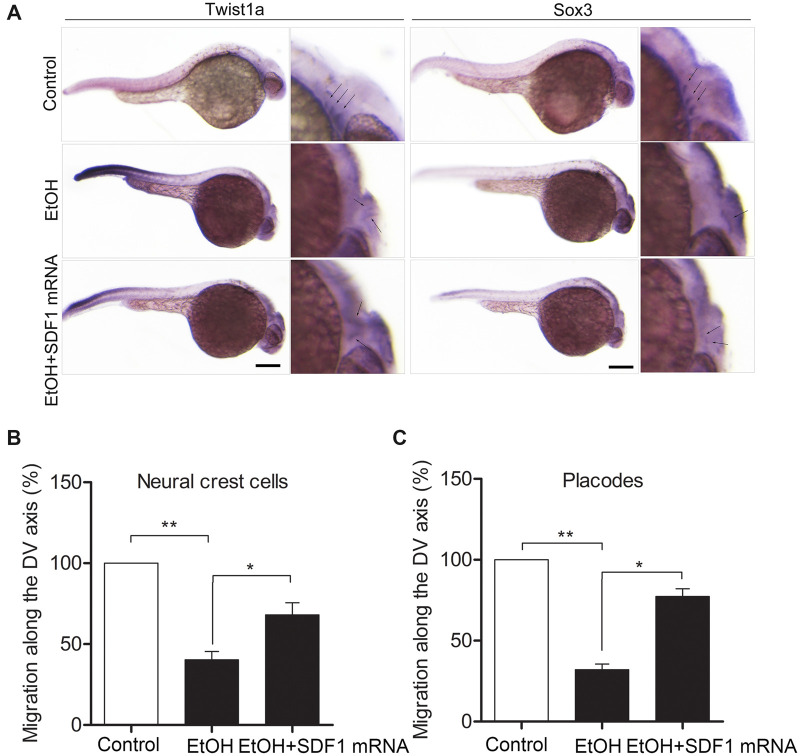
For the in vitro system (described above), the coordinated migration of hNCCs-hPCs was driven by the chemoattractant force initiated by the hPCs, therefore, the migratory ability of hNCCs reflects whether the coordinated migration between hNCCs and hPCs is disrupted or not. The migratory ability of hNCCs was measured by the migration ratio of hNCCs. The migration ratio was defined as follows: migration ratio of hNCCs = number of hNCCs inside gap/number of hNCCs outside gap (per scope). The exact number of hNCCs in each image was counted using ImageJ software. In addition, the coordinated migration of NCCs and PCs in vivo was examined by analyzing the migration patterns of NCCs and PCs at 30 hpf (hours post fertilization) by whole mount in situ hybridization (twist1a probe for NCCs and sox3 probes for PCs). All embryos in each group were photographed under a stereoscopic microscope (Olympus SZX16, Japan). The migratory distance (measured by cellSens software), being equivalent with a relative migratory length from the dorsal site of neural tube to the ventral position (migration along the DV axis [%]), was the parameter reflecting the migratory ability of NCCs and PCs in vivo.
Isolation, characterization, and labeling of hNCCs-Exo and GELNs
Exosomes from hNCCs (hNCCs-Exo): Differential ultracentrifugation and direct immunoaffinity capture (DIC) methods are 2 connecting steps for crude exosome isolation and further immuno-purification of the crude exosomes, respectively (Jeppesen et al., 2019). During differential ultracentrifugation process, the collected fresh conditioned media (from hNCCs) were undergone centrifugation at the speed of 300 × g, 2000 × g and 10 000 × g step by step, and the supernatants from each previous step were kept for the next step until the 10 000 × g centrifugation step was performed. Finally, the supernatants from 10 000 × g centrifugation were further ultracentrifuged at the speed of 120 000 × g for 70 min. The pellet was collected, resuspended in PBS and then ultracentrifuged for 1 h at 120 000 × g using Beckman Coulter ultracentrifuge with fixed angle rotor (Type 70 Ti Rotor, Beckman, Germany). The final volume of PBS used for resuspending the pellet was equal to 1/1000 of the initial volume of the hNCCs-conditioned media. All the centrifugation steps were performed at 4°C. The crude exosomes obtained above were proceeded to be purified by DIC method in the next step. The key idea of DIC is that exosomes can be directly immuno-captured by their specific surface marker, CD63. In this step, exosomes obtained from ultra-centrifugation were further immune-purified by CD63 Exo-Flow Capture Kit (EXOFLOW300A-1, SBI, Palo Alto, California) according to the manufacturer’s instructions. Exosomes from grapes (GELNs): Grapes skins were removed and homogenized in a high-speed blender for 1 min at 4°C. The collected juice was sequentially centrifuged at 2000 × g for 20 min and then 10 000 × g for 1 h to exclude debris. The supernatants were filtered through a 1-μm membrane filter (Millipore, Bedford, Massachusetts). Then the filtered supernatants were pelleted at 120 000 × g for 1 h, washed once with PBS and then purified and separated using sucrose gradients centrifugation (8%, 30%, 45%, and 60%, respectively). Band 1 at 8/30% and band 2 at the 30/45% interface were considered to be pure GELNs for collection. For TEM characterization, purified exosomes in PBS were fixed in 2% PFA in PBS for 2 h at room temperature. The fixed sample drop (10 μl) was placed on the surface of formvar carbon-coated copper grids (FCF200-CU, Electron Microscopy Sciences, Pennsylvania) and allowed to absorb for 5 min. Then the grids were blotted by filter paper and placed on a drop of Uranyless solution (cat. no. 22409, Electron Microscopy Sciences, Pennsylvania) for 5 min at room temperature. Finally, the grids were blotted and dried for 5 min and observed under an electron microscope (Zeiss EM 900, Germany). To quantify hNCCs-Exo or GELNs, a BCA protein assay kit (PIERCE, Rockford, Illinois) was used to measure protein content. For the cell migration assay, the work concentration of exosomes was 1 μg/105 cells, and for the microinjection experiments, each zebrafish embryo was injected with 2 ng of exosomes. To monitor the trafficking and uptake of exosomes, purified hNCCs-Exo or GELNs were labeled with PKH67 (Sigma-Aldrich, St Louis, Missouri) according to the manufacturer’s instructions.
microRNAs analysis of exosomes
DIC-purified exosomes were further proceeded for miRNA extraction using miRNeasy mini kit (Qiagen, Valencia, California) followed the manufacturer’s procedures. Extracted total miRNAs were first reverse-transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster, California) in a reaction mixture containing a miR-specific stem-loop reverse transcription primer (hsa-miR-126*: RT-000451, Thermo Fisher, Waltham, Massachusetts). Then quantitative PCR amplification was performed using TaqMan Universal PCR Master Mix kit (Applied Biosystems, Foster, California) with a sequence-specific Taqman probe (hsa-miR-126*: TM-000451, Thermo Fisher) on a Rotor-Gene 6000 Real-Time PCR system (Corbett Life Science, Sydney, Australia). Data were normalized with snoRNA202 as endogenous control, and the relative expression of miR-126 was calculated using the ΔΔCT method.
Loading of miR-126 inhibitors into exosomes, and the in vitro or in vitro treatments with exosomes
For in vitro experiments, loading of miR-126 inhibitors (MH10401, mirVana miRNA inhibitor, Thermo Fisher) into exosomes were completed by Exo-Fect exosome transfection reagent (SBI, Palo Alto, California) according to the manufacturer’s protocol. Briefly, hNCCs-Exo were initially re-suspended with sterile PBS. In a clean 1.5 ml tube, the following reagent containing 10 μl Exo-Fect solution, 20 μl miRNA inhibitors (5 μM), 70 μl sterile PBS, and 50 μl (200 μg/ml) purified exosomes were mixed together to incubate at 37°C in a shaker for 10 min, and then the tube was immediately placed on ice. After stop the reaction by adding 30 μl of the ExoQuick-TC reagent provided in the kit, the transfected exosomes were placed on ice for 30 min. Then the samples were centrifuged at 13 000 × g for 3 min. After the supernatants were removed, the pellets composed of transfected exosome were resuspended in 300 μl PBS and were ready for downstream experiments. For in vitro treatments, 1 µg of hNCCs-Exo (based on protein measurement) were added to 1 × 105 cells and cultured for 24 h. For in vivo study, zebrafish embryos were microinjected with GELNs or PHK67-labeled GELNs (2 ng/embryo) at 4 hpf.
Dual luciferase reporter assays
miR-126 targeting sites in the 3′-untranslational region (3′-UTR) of SDF1 mRNA were predicted by online database tool, Target Scan (http://www.Targetscan.org/), as described previously (Fan et al., 2019). The 3′-UTR of SDF1 containing putative miR-126 binding sites were amplified from human genomic DNA using the following primers: 5′-GTTACTGCCATCTTACTAGTCAACGCCCAGTCCCAGTGCATCCCACAGCTACAGCTT-3′ (forward primer); 5′-TAGGCTGCAGGTGATGCGTAAAGTTGGCAGTAGAACGGTGGTGAGTCACGCTGACTA-3′ (reverse primer), and were cloned into pMIR-Luciferase-Report plasmid (Applied Biosystems, Foster, California) at Mlu1/Spe1 restrictive enzyme site. Renilla luciferase pRL-TK control vector (Promega, Madison, Wisconsin) was used as a control. The pMIR-Luciferase-SDF1 (3′-UTR) constructs (200 ng of plasmid/well of 24-well plates) were co-transfected with 20 ng pRL-TK control vector and 50 nmol of miR-126a mimics or control mimics (Ambion, Austin, Texas) into hPCs by Lipofectamine 2000 (Invitrogen, Waltham, Massachusetts) according to the manufacturer’s procedures. Luciferase activity was measured 48 h after the transfection using the Dual-luciferase assay kit (Promega, Madison, Wisconsin) with a Lumat LB9507 Ultra Sensitive Tube Luminometer (Berthold Technologies, Bad Wildbad, Germany). The relative activity of luciferase of each sample was normalized to the pRL-TK driven Renilla luciferase activity.
Zebrafish maintenance and ethanol treatment
Adult AB zebrafish (Danio rerio) were obtained from the Zebrafish International Resource Center (ZIRC) at the University of Oregon, Eugene, Oregon and maintained in 14 h:10 h light: dark cycle at 28°C. Fertilized eggs were collected after natural spawning and used for this study. For ethanol treatment, the zebrafish embryos were exposed to ethanol (1% v/v) during 5–24 h post fertilization (hpf). After ethanol exposure, the embryos were rinsed 3 times and transferred to fresh system water. The embryos were collected at 30 hpf, or 2 days post fertilization (dpf) for the whole-mount in situ hybridization (WISH) to view the migration patterns of cranial neural crest and placodes, or the morphology of cranial nerves, respectively. The embryos were also collected at 5 dpf for evaluating morphology of craniofacial cartilages using Alcian Blue staining.
Microinjection of antisense morpholinos, in vitro-synthesized mRNA, or labeled exosomes
Morpholinos used for knocking down gene expression were as follows: SDF1: CTACTACGATCACTTTGAGATCCAT (Doitsidou et al., 2002). In vitro-synthesis of SDF1 mRNA was performed using mMESSAGE mMachine T7 Ultra Kit (cat. no. AM1345, Ambion, USA) following the manufacturer’s instructions. The primers used for in vitro transcription of SDF1 mRNA were as follows: SDF1: TAATACGACTCACTATAGGGATGGATCTCAAAGTGATCGTAGTA (forward), TTAGACCTGCTGCTGTTGGGCTTT (reverse). Morpholinos or synthesized mRNAs were microinjected into 1-cell stage zebrafish embryos (0.3 pmol/egg). PKH-67-labeled exosomes containing miR-126 inhibitors (2 ng/egg) were microinjected into zebrafish embryos at 4 hpf.
Whole-mount in situ hybridization
The procedure of the WISH were described as in Thisse and Thisse (2014). The PCR primers used for synthesizing digoxigenin-conjugated probes were as follows:
Twist1a: CTCAGTCTCTGAACGAGGCG (forward), TAATACGACTCACTATAGGGTCCTGCTCCCATGTCGTAGT (reverse);
Sox3: CTCGGTGCTGACTGGAAACT (forward), TAATACGACTCACTATAGGGTGCGTGTATGCTGGTGACAT (reverse);
SDF1: TGCCAAATATGCGTCCCAGT (forward), TAATACGACTCACTATAGGGGAGCGTGAAGCAACAGTGTG (reverse);
Phox2b: GAGGAGCTCGCGCTTAAGAT (forward), TAATACGACTCACTATAGGGAGAGTCCGGAATGGAGGTGA (reverse).
The dechorionated zebrafish embryos were fixed in 4% paraformaldehyde at 4°C overnight and dehydrated in methanol for 2 h. After rehydration, the embryos were permeated with proteinase K (10 μg/ml, Invitrogen, Carlsbad, California) and then pre-hybridized for 5 h at 60°C in a pre-hybridization buffer. The embryos were then incubated with digoxigenin-conjugated mRNA probes in hybridization buffer for 8 h at 60°C. Before adding the digoxigenin antibody (Roche, Basel, Switzerland), the embryos were placed in blocking buffer (containing 2 mg/ml BSA and 2% sheep serum) for 2 h. Then the embryos were incubated with digoxigenin antibody overnight at 4°C following by adding NBT and BCIP buffer to develop the hybridized gene patterns. Finally, the embryos were photographed under a stereoscopic microscope (SZX16, Olympus, Japan).
Alcian Blue staining of the zebrafish cartilages
The zebrafish embryos were collected at 5 dpf and fixed with 4% paraformaldehyde overnight at 4°C. The fixed embryos were washed and dehydrated with 50% ethanol at room temperature for 10 min, followed by staining in 0.1% Alcian Blue solution at room temperature overnight and washing in a bleach solution (Walker and Kimmel, 2007). After the washing, 1 ml solution of 20% glycerol and 0.25% KOH was added and incubated at room temperature for 30 min. The embryos were then photographed under a stereoscopic microscope (SZX16, Olympus, Japan).
Dysmorphology assessment
For dysmorphology assessment, zebrafish embryos collected from each group were examined under a stereoscopic microscope (Olympus SZX16, Japan). All embryos were photographed, and particular attention was paid to the craniofacial morphology including anomalies of craniofacial cartilages and cranial nerves. Embryos collected at 2 dpf (days post fertilization) were used for cranial nerve examination to monitor the morphology of VII, IX, and X cranial nerves that are composed of neurons (marked with Phox2b probe by in situ hybridization) generated largely from epibranchial placodes. In addition, larvae collected at 5 dpf were used for craniofacial skeleton examination by Alcian Blue staining. Because 3 main streams of NCCs coordinate with epibranchial PCs to migrate to form pharyngeal arches (PA1–PA3) at their terminal differentiation, the length and the morphology of cartilaginous pharyngeal skeletons including Meckel’s cartilage (m), palatoquadrate (pq), ceratohyal (ch), basibranchials (bb), hypobranchials (hb), and ceratobranchial (cb) were under our examination. Generally, a morphological deformity or a relatively weak staining of cranial nerves, and a significantly shorter length or a disappearance of certain craniofacial cartilages were all considered to be a developmental defect.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (GraphPad, San Diego, California). All data were expressed as means ± SEM of at least 3 independent experiments. All data from this study, except the data on the anomaly ratio of cartilages and cranial nerves, were analyzed by 1-way ANOVA. Multiple comparison post-tests between groups were conducted using Bonferroni’s comparison test. Data on the anomaly ratio of cartilages and cranial nerves were analyzed using 2-sided Fisher’s exact test. A p-value < .05 was considered statistically significant.
Results
Ethanol exposure disrupted the coordinated migration of NCCs and PCs in vitro and in zebrafish embryos
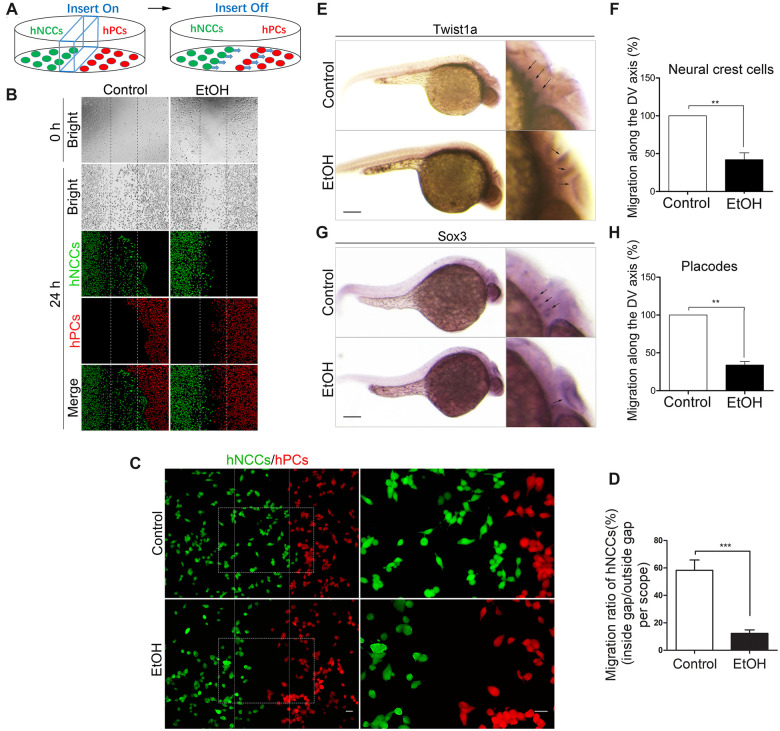
To examine whether ethanol can disrupt the coordinated migration of hNCCs and hPCs in vitro, hNCCs and hPCs were cultured and treated with 50 mM ethanol for 24 h and the migration of hNCCs and hPCs was analyzed in the CytoSelect Wound Healing Assay Kit, as described in the Materials and methods section. We designed a cell coculture system to evaluate the coordinated migration between hNCCs and hPCs (Figure 1A). Briefly, hNCCs (green) and hPCs (red) were simultaneously seeded in each semi-section of the same cell dish with an insert (blue) to physically insulate the 2 cell types. As the cells were attached to the bottom of the dish, the insert was taken away to form a central cells-free cleft on the dish bottom, and in the meantime, the hNCCs would begin to migrate towards hPCs. In control group (Figure 1B, left panel), we observed that hNCCs (stained with anti-HNK1 antibody, green) migrate towards hPCs (stained with anti-sox3 antibody, red) and hPCs move away from hNCCs, manifesting a coordinated efficient directional migration between hNCCs and hPCs in a “chase-and-run” manner as previously described by Theveneau et al. (2013). However, directional migration of hNCCs and hPCs were lost after they were exposed to ethanol for 24 h and both hNCCs and hPCs move randomly with a low directionality and poor net displacement (Figure 1B, right panel), which was also clearly shown in a higher magnified picture (Figure 1C, right panel) that there’re less cells at hNCCs-hPCs interface. In order to exactly characterize the migration difference between control and ethanol-treated group, we calculated the migration ratio as follow equation: migration ratio of hNCCs = number of hNCCs inside gap/number of hNCCs outside gap (per scope). As shown in Figure 1D, the migration ratio of hNCCs in ethanol-treated group was significant less than that in control group, indicating that ethanol can directly disrupt the coordinated migration of hNCCs and hPCs in vitro. We then investigated whether ethanol can inhibit the coordinated migration in vivo. Using WISH technique, the migration patterns of NCCs and PCs were displayed by the NCC marker twist1a and PC marker sox3 in 30 hpf zebrafish embryos, respectively. Three streams of migratory NCCs had migrated to the ventral site of neural tube in 30 hpf zebrafish embryos in control group (Figure 1E, upper panel), whereas exposure to 1% (v/v) ethanol at 5–24 hpf resulted in a significant delay of NCCs’ migration, causing 3 streams of migratory NCCs to accumulate near to the dorsal site of neural tube (Figure 1E, lower panel). The migratory distance (dorsal-to-ventral axis) of NCCs in ethanol-treated group was significantly shorter than that in control group (Figure 1F). Similarly, the migration of PCs was also strongly delayed under ethanol treatment (Figs. 1G and 1H). These data demonstrated that ethanol exposure can also cause the migration defects of NCCs and PCs in vivo.
Figure 1.
Exposure to ethanol significantly disrupted the coordinated migration in vitro and in vivo. A, A cartoon depicts hNCCs-hPCs coculture system to evaluate the interaction between hNCCs and hPCs. An insert was put into a cell culture plate, and then hNCCs and hPCs were seeded into each side of the insert and cultured to form a monolayer. The insert was then removed to generate a 0.9-mm cells-free gap between hNCCs and hPCs. hNCCs and hPCs were cultured in the medium with or without ethanol and allowed to migrate for 24 h. B, hNCCs and hPCs were treated with or without ethanol (50 mM) for 24 h and then fixed for immunofluorescence that costained with antibodies for hNCCs-specific marker HNK1 (green) and hPCs-specific marker Six1 (red). Bright-field photos of the 2 cell groups were captured at the 0 time point when the monolayer of hNCCs and hPCs was just formed (first row). The second to fourth row showed the photos of bright field and fluorescence field of control group or ethanol-treated group, in which hNCCs and hPCs had migrated for 24 h. C, Higher magnification images manifested the hNCCs-hPCs orientation after they migrated for 24 h in control and ethanol-exposed groups. Scale bar, 10 μm. D, Quantification of disruption of coordinated migration of hNCCs and hPCs. The migratory ability of hNCCs that chemoattracted by hPCs can be measured by the migration ratio of hNCCs. The migration ratio of hNCCs approximately equals to the number of hNCCs passed through the gap over the total number of hNCCs. Data represent the mean ± SEM of 3 independent experiments. ***p < .001 versus control. E, Zebrafish embryos were treated with 1% ethanol at 5–24 h post fertilization (hpf), and then the embryos were collected for WISH at 30 hpf, a time point that NCCs had already migrated to the ventral position of neural tube in developmentally normal embryos. Twist1a was used as a specific probe for detecting the NCCs in zebrafish embryos (arrows). Scale bar, 200 μm. Migration defects of NCCs were quantified in (F). The approximately migratory distance of NCCs was measured as the migrated length towards Ventral position from the Dorsal site of neural tube (labeled as “migration along the DV axis”). Data represent the mean ± SEM of 3 independent experiments. **p < .01 versus control. G, After the same treatment as in (C), Sox3 was used as a specific probe for detecting the PCs in zebrafish embryos at 30 hpf (arrows), a time point that NCCs had already migrated to the ventral position of neural tube in developmentally normal embryos. Scale bar, 200 μm. H, Quantification of migration defects of PCs. The migratory distance of PCs was also expressed as the migrated length towards ventral position from the dorsal site of neural tube (labeled as “migration along the DV axis”). Data represent the mean ± SEM of 3 independent experiments. **p < .01 versus control.
Treatment with exosomes derived from ethanol-exposed hNCCs (hNCCs-ExoEtOH) disrupted the coordinated migration of hNCCs and hPCs
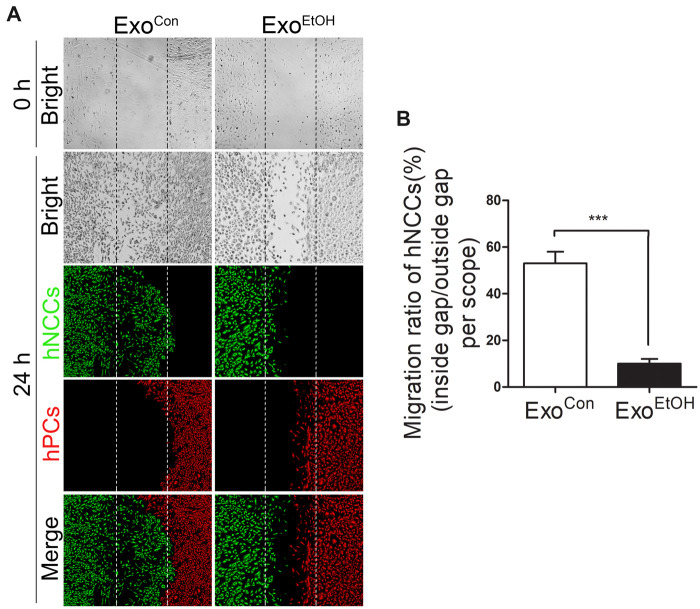
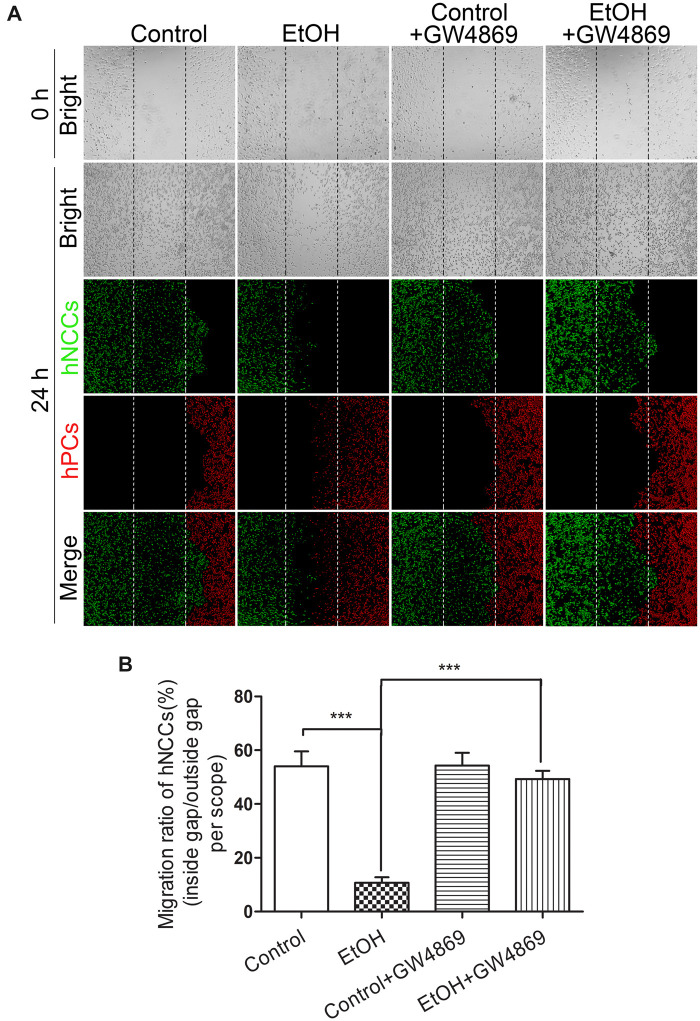
Exosomes can carry various biological molecules, such as proteins or RNAs, for intercellular communication. Regarding their emerging roles in embryonic development and neural system (McGough and Vincent, 2016; Rajendran et al., 2014; Xu et al., 2017), we hypothesized that NCCs-secreted exosomes may mediate the interaction between NCCs and PCs. To determine the role of hNCCs-ExoEtOH in ethanol-induced disruption of the coordinated migration of NCCs and PCs, control PCs were cultured with exosomes derived from control hNCCs (hNCCs-ExoCon) or hNCCs-ExoEtOH (1 μg/105 cells) for 24 h before they were cultured with control hNCCs in our cell cocultured system as described above. We found that exposure of control hPCs to hNCCs-ExoEtOH significantly disrupted the coordinated migration of control hNCCs and hPCs (Figure 2A, right panel), and the migration ratio was strongly decreased in hNCC-ExoEtOH-treated group (Figure 2B), suggesting that hNCC-ExoEtOH may mediate ethanol-induced impairment of hNCCs-hPCs interaction. To further confirm this, NCCs were pretreated with or without exosomes secretion inhibitor GW4869 (10 μM) for 24 h, to block the release of hNCCs-Exo before NCCs were cocultured with hPCs and then exposed to 50 or 0 mM ethanol for 24 h (Figure 3). As seen obviously, GW4869 restored coordinated migration of hNCCs and hPCs (Figure 3A, the fourth panel) compared with ethanol-treated group (Figure 3A, the second panel). Together, these data provide a primary foundation indicating that hNCCs-ExoEtOH may contain certain inhibitory molecules that contribute ethanol-induced disruption of coordinated migration of hNCCs and hPCs.
Figure 2.
Treatment with hNCCs-ExoEtOH significantly disrupted the coordinated migration between hNCCs and hPCs. A, At first step, hNCCs were treated with or without ethanol (50 mM) for 24 h, and then the hNCCs-cultured medium was collected for isolating hNCCs-derived exosomes. Second, hNCCs and hPCs were seeded into the cell coculture system as described in Figure 1A, and treated with control hNCCs-derived exosomes (ExoCon, 1 µg/105 cells) or ethanol-treated hNCCs-derived exosomes (ExoEtOH, 1 µg/105 cells) for 24 h. Finally, the cells were subject to immunofluorescence that costained with antibodies for with hNCCs-specific marker HNK1 (green), and hPCs-specific marker Six1 (red). Bright-field photos of ExoCon-treated group or ExoEtOH-treated group were captured at the 0 time point when the monolayer of the 2 group cells was just formed (first row). The second to fourth row displayed the photos of bright field and fluorescence field of Exocon-treated group or ExoEtOH-treated group, in which hNCCs and hPCs had migrated for 24 h. B, Quantification of migration defects of coordinated migration of hNCCs-PCs. Data represent the mean value of migration ratio ± SEM of 3 independent experiments. ***p < .001 versus control.
Figure 3.
GW4869, an exosome inhibitor, mitigates ethanol-induced impairment of hNCCs-hPCs interaction. A, Four groups of hNCCs and hPCs cultured in the hNCCs-hPCs coculture system were set as control, ethanol (50 mM), control plus GW4869, or ethanol (50 mM) plus GW4869 (10 µM). After the cells migrated for 24 h, hNCCs and hPCs were costained with antibodies for hNCCs-specific marker HNK1 (green) and hPCs-specific marker Six1 (red). Bright-field photos were captured at the 0 time point when the monolayer of the 4-group cells was just formed (first row). The second to fourth row showed the photos of bright field and fluorescence field for these 4 groups, in which hNCCs and hPCs had migrated for 24 h. B, Quantification of migration inhibition of coordinated migration of hNCCs-PCs. Data represent the mean value of the migration ratio ± SEM of 3 independent experiments. ***p < .001 versus control.
Ethanol exposure decreased the expression of SDF1 in hPCs and in zebrafish embryos
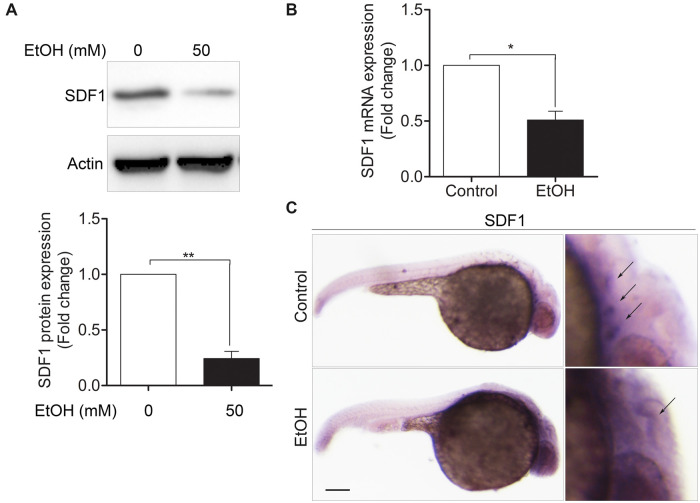
To examine the effects of ethanol on the SDF1 expression, hPCs were treated with or without 50 mM ethanol for 24 h. After that, cells were harvested for protein analysis. We found that SDF1 level was significantly decreased (Figure 4A). Next, we detected the SDF1 expression in zebrafish embryos. Zebrafish embryos were first treated with or without 1% (v/v) ethanol at 5–24 hpf, and then were collected at 30 hpf for qRT-PCR analysis and WISH. The mRNA level of zebrafish embryos in ethanol-treated group was much lower than in control group (Figure 4B). Typically, the SDF1 expression pattern in control group resembles to the pattern of Sox3 expression as they are both appeared in the position where epibranchial placodes located (Figure 4C, upper right, arrows). However, in ethanol-treated group, the normal distribution of SDF1 was abolished (Figure 4C, lower right, arrow). These results demonstrated that ethanol exposure can destroy normal expression of SDF1 in vitro and in vivo.
Figure 4.
Exposure to ethanol significantly decreased SDF1 level in hPCs and zebrafish embryos. A, hPCs were treated with or without ethanol (50 mM) for 24 h and then the cells were lysed for immunoblot analysis using anti-SDF1 or anti-actin antibodies. B, Zebrafish embryos were treated with or without ethanol (1% v/v) at 5–24 hpf. At 30 hfp, embryos were collected for detecting SDF1 expression by qRT-PCR analysis. C, Zebrafish embryos were treated with or without ethanol (1% v/v) at 5–24 hpf and then the embryos were fixed for WISH at 30 hpf, as indicated in control or ethanol-treated group, the patterns of SDF1 expression (arrows) were detected using SDF1 probe. Scale bar, 200 μm. Data are expressed as fold change over control and represent the mean ± SEM of 3 independent experiments. *p < .05, **p < .01 versus control.
Overexpression of SDF1 diminished ethanol-induced disruption of the coordinated migration of NCCs and PCs in zebrafish embryos
To investigate the role of SDF1 in mediating ethanol-induced disruption of the coordinated migration of NCCs and PCs, SDF1 was overexpressed in zebrafish embryos by the microinjection of in vitro-synthesized SDF1 mRNA. Overexpression of SDF1 significantly restored the normal migration pattern of NCCs (Figure 5A, row 3 of left panel, arrows) and PCs (Figure 5A, row 3 of right panel, arrows), as detected by their marker Twist1a and Sox3, respectively. SDF1 overexpression also strongly increased the migratory distance from dorsal site to ventral site (DV axis) of neural tube of NCCs and PCs (Figs. 5B and 5C). Together with Figure 4, these data demonstrated that SDF1, a chemoattractant cytokine, is essential for the coordinated migration of NCCs and PCs and that reduction of SDF1 in PCs may contribute to ethanol-induced disruption of the coordinated migration of NCCs and PCs.
Figure 5.
Overexpression of SDF1 diminished ethanol-induced disruption of the coordinated migration between NCCs and PCs in zebrafish embryos. A, Zebrafish embryos were divided into 3 groups. In addition to control group, embryos in the second group were treated with ethanol (1% v/v) at 5–24 hpf; in the third group, embryos were first microinjected with in vitro-synthesized SDF1 mRNA (0.6 pmol/embryo) at 1-cell stage and then treated with ethanol (1% v/v) at 5–24 hpf. At 30 hpf, the embryos were collected for WISH to visualize the migration pattern of NCCs and PCs probed with Twist1a and Sox3, respectively. Scale bar, 200 μm. B and C, As the same statistic method in Figs. 1F and 1H, the approximately migratory distance of NCCs or PCs was expressed as their migrated length toward ventral position from the dorsal site of neural tube (labeled as “migration along the DV axis”), respectively. Data are expressed as the percentage of control and represent the mean ± SEM. *p < .05, **p < .01 versus control.
Ethanol exposure resulted in a significant increase in the expression of miR-126 in hNCC-Exo
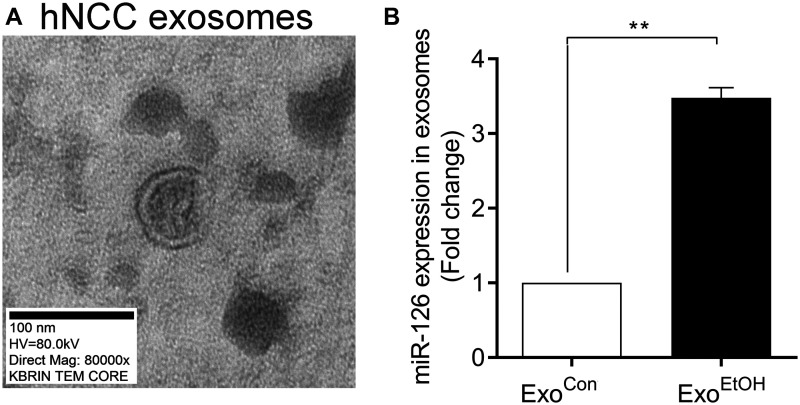
It has been reported that miR-126 wrapped in exosomes can be shuttled among different types of cells (Lee et al., 2012; Taverna et al., 2014). Particularly, miR-126 is the most relevant miRNAs in regulating migration of various tumor cells through its potential target SDF1 (Taverna et al., 2014; Zhang et al., 2013). To determine whether ethanol exposure can alter miR-126 expression, hNCCs-Exo (Figure 6A) derived from control or ethanol-treated hNCCs were isolated for qRT-PCR analysis. We found that the level of miR-126 in hNCC-ExoEtOH was significantly higher than that in hNCC-ExoCon (Figure 6B), suggesting that ethanol exposure of hNCCs can facilitates more miR-126 assembled into hNCCs-derived exosomes.
Figure 6.
Ethanol exposure resulted in a robust increase of miR-126 level in hNCCs-ExoEtOH. A, Representative of TEM image of hNCCs-Exo. Magnification, 80 000×. Scale bar, 100 nm. B, hNCCs were treated with or without ethanol (50 mM) for 24 h. Then the cultured media for hNCCs were collected for exosomes isolation. After purification, exosomes of hNCCs-ExoCon and hNCCs-ExoEtOH were lysed for detecting miR-126 expression by qRT-PCR analysis. Data are expressed as fold change over control and represent the mean ± SEM of 3 independent experiments. **p < .01 versus control.
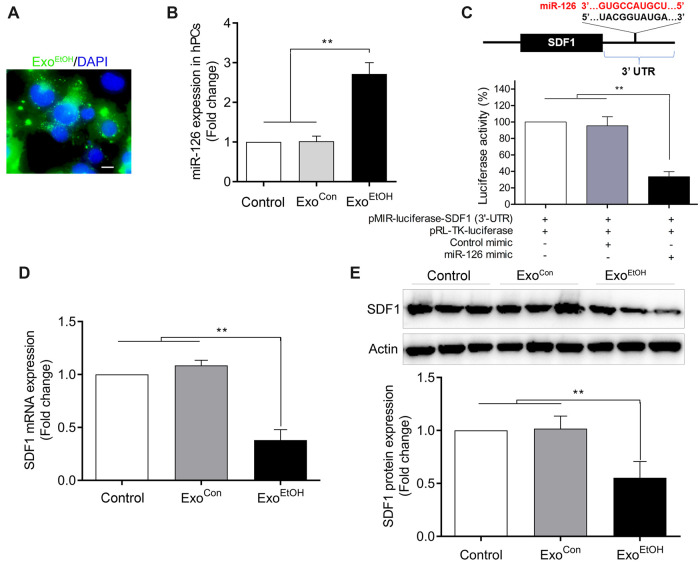
Treatment with hNCCs-ExoEtOH dramatically increased the levels of miR-126 and decreased the expression of SDF1 in hPCs
To further determine whether elevated miR-126 in hNCCs-ExoEtOH can be horizontally transferred into control hPCs to suppress the SDF1 expression, hPCs were cultured with hNCCs-ExoEtOH (1 µg/105 cells) for 24 h. We observed that hPC can efficiently uptake hNCCs-ExoEtOH (Figure 7A, labeled with green fluorescent PHK-67), and that the culture of hPCs with hNCCs-ExoEtOH significantly increased the levels of miR-126 (Figure 7B) and reduced the mRNA and protein expression of SDF1 in hPCs (Figs. 7D and 7E). Using dual luciferase reporter assay, we further validated that SDF1 can be directly targeted by miR-126 in hPCs. Taken together, these data indicated that ethanol-induced reduction of SDF1 in hPCs might be caused by the transfer of overloaded miR-126 of ExoEtOH from hNCCs to hPCs.
Figure 7.
Treatment with hNCCs-ExoEtOH significantly increased miR-126 level and decreased SDF1 expression in control hPCs. A, Representative immunofluorescence image showed the uptake of PHK67-labeled hNCCs-Exo (green dots) by hPCs, and the nuclei were stained with DAPI (blue). Scale bar, 10 µm. B, hPCs were set as blank control, or treated with ExoCon (1 µg/105 cells), or with ExoEtOH (1 µg/105 cells) for 24 h, and then the cells were harvested for detecting miR-126 expression by qRT-PCR analysis. C, As shown in the schematic figure, there is a presumable binding site at 3′-UTR of SDF1 for miR-126. 3′-UTR of SDF1-assembled pMIR-luciferase plasmid (pMIR-luciferase-SDF1 [3′-UTR]) and Renila luciferase plasmid (pRL-TK-luciferase) were cotransfected with or without control mimic or miR-126 mimic into hPCs for 48 h. Then the cells were subject to dual luciferase reporter assays. D, hPCs were treated with blank control, ExoCon (1 µg/105 cells), or ExoEtOH (1 µg/105 cells) for 24 h, and then the mRNA expression of SDF1 in hPCs were determined by qRT-PCR analysis. E, Treated as the same condition within (B), hPCs were lysed for detecting the protein level of SDF1 by immunoblot. Data are expressed as fold change over control and represent the mean ± SEM of 3 separate experiments. **p < .01 versus control.
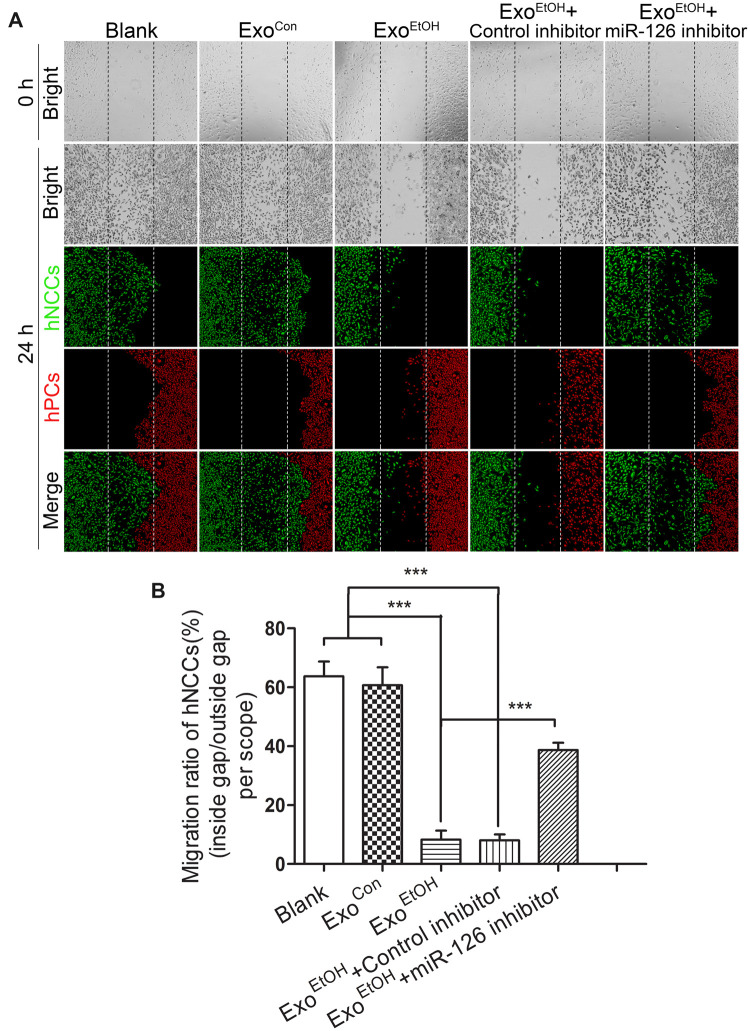
Inhibition of miR-126 significantly diminished the disruption of the coordinated migration of hNCCs and hPCs induced by hNCC-ExoEtOH
To further explore the role of exosome-derived miR-126 in mediating ethanol-induced impairment of the coordinated migration of NCCs and PCs, we examined if inhibition of miR-126 can diminish such an effect induced by hNCCs-ExoEtOH. We found that treatment with miR-126 inhibitor-loaded hNCCs-ExoEtOH (Figure 8A, the fifth panel) significantly diminished the disruption of the coordinated migration of hNCCs and hPCs induced by hNCCs-ExoEtOH (Figure 8A, the third panel), as well as the migration ratio was largely recovered in the group that treated with miR-126 inhibitor-loaded hNCCs-ExoEtOH (Figure 8B). These data provided a solid evidence that ethanol-induced elevation of miR-126 in ExoEtOH highly contributes to ethanol-induced disruption of coordinated migration between hNCCs and hPCs.
Figure 8.
Inhibition of miR-126 in hNCCs-secreted ExoEtOH significantly diminished ExoEtOH-induced disruption of coordinated migration between hNCCs and hPCs. A, hNCCs were treated with or without ethanol (50 mM) for 24 h and the hNCCs-cultured media were collected for isolating ExoCon or ExoEtOH. ExoEtOH were loaded with control inhibitor or miR-126 inhibitor. Immediately after hNCCs and hPCs were seeded in the hNCCs-hPCs coculture system, they were treated with blank control, ExoCon (1 µg/105 cells), ExoEtOH (1 µg/105 cells), ExoEtOH (1 µg/105 cells) plus control inhibitor (0.1 µM), or ExoEtOH (1 µg/105 cells) plus miR-126 inhibitor (0.1 µM). After 24 h migration, the cells were costained with antibodies for hNCCs marker HNK1 (green), and hPCs marker Six1 (red). Bright-field photos were captured at the 0 time point when the monolayers of the 5 groups of cells were just formed (first row). The second to fourth row showed the photos of bright field and fluorescence field for these 5 groups, in which hNCCs and hPCs had migrated for 24 h. B, Quantification of migration defect of coordinated migration of hNCCs-PCs. Data represent the mean value of migration ratio ± SEM of 3 independent experiments. ***p < .001 versus control.
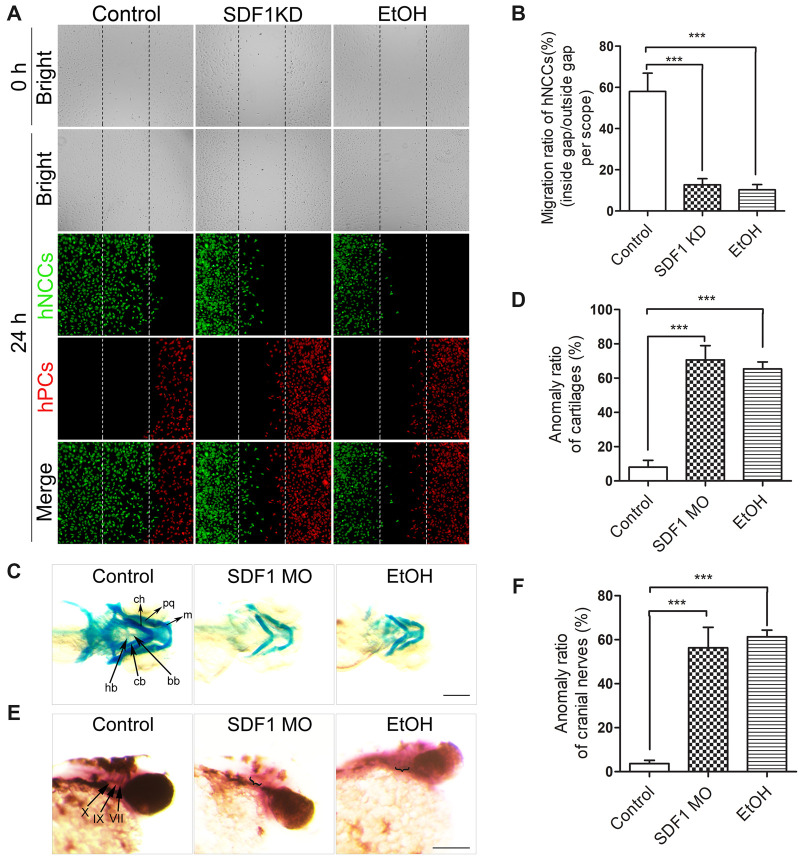
Knockdown of SDF1 recapitulated ethanol-induced disruption of coordinated migration of hNCCs and hPCs, and mimicked developmental defects induced by ethanol in zebrafish embryos
The above data have demonstrated that the ExoEtOH-miR-126-SDF1 axis is involved in mediating ethanol-induced disruption of NCCs-PCs interaction in vitro and in vivo. To further confirm the role of SDF1 in ethanol-induced teratogenesis, we silenced SDF1 expression in hPCs or in zebrafish embryos. Before seeded into the cell dishes of our cell coculture system, hPCs was transfected with SDF1 siRNAs for 24 h, and then both hNCCs and hPCs were seeded into the same cell dish to proceed the migration assay. We found that either knockdown of SDF1 (Figure 9A, middle column) or exposure to ethanol (Figure 9A, third column) can lead to a severe disruption of coordinated migration between hNCCs and hPCs, as well as a significant reduction of migration ratio of hNCCs (Figure 9B). In vertebrate embryos like zebrafish and mouse, normal development of head structures and cranial nerves needs coordinated and highly organized migration and terminal differentiation of NCCs and PCs. Seven pharyngeal arches (PA1∼PA7) in the zebrafish embryo derived largely from NCCs that form the cartilaginous skeleton, whereas epibrachial PCs, in coordinate with NCCs, would finally develop into distal part of VII, IX, and X cranial nerves (Dunty et al., 2002; Schilling et al., 1996). Therefore, the endpoints we selected to measure the developmental anomalies resulting from coordinated migration defects are: morphology of the cartilaginous pharyngeal skeleton (resulting from NCCs), and the morphology of VII, IX, and X cranial nerves (resulting from PCs). For this study, a significantly shorter length of cartilages, a morphological deformity or a relatively weak staining of cranial nerves was considered to be an endpoint anomaly. In this study, after microinjecting SDF1 morpholinos into 1-cell stage embryos, we assessed the morphological changes of craniofacial skeletons and cranial nerves using WISH and Alcian Blue staining, respectively. Knockdown of SDF1 (Figure 9C, middle column) or exposure to 1% (v/v) ethanol (Figure 9C, right column) at 5–24 hpf resulted in significant defects in craniofacial skeletons. The anomaly ratio of m, pq, ch (Meckle’s cartilage, palatoquadrate, and ceratohyal, derived from the PA1 and PA2), cb, hb, and bb (ceratobranchial, hypobranchial and basibranchial, derived from PA1–PA7) were significantly higher in SDF1 MO and EtOH groups (Figure 9D). Moreover, knockdown of SDF1 or ethanol exposure also induced cranial nerve defects in zebrafish embryos. As indicated by ganglia-specific marker Phox2b (Figure 9E, left column, arrows), the ganglia of the cranial nerves VII, IX, and X are absent, or diffused, or the ganglia volume was significantly decreased in treated embryos (Figure 9E, second and third column). The anomaly ratio of cranial nerves was also significantly higher in SDF1 MO or EtOH groups (Figure 9F). Together, these data demonstrated that downregulating SDF1 can mimic the effect of ethanol-induced impairment of hNCCs-hPCs interaction, as well as craniofacial and cranial anomalies of zebrafish embryos.
Figure 9.
The knockdown of SDF1 mimicked the effect of ethanol-induced disruption of coordinated migration between hNCCs and hPCs, as well as the effects of ethanol-induced anomalies of craniofacial skeletons and cranial nerves in zebrafish embryos. A, For the first and the third column, hNCCs-hPCs coculture system were treated with or without ethanol (50 mM) for 24 h. For the middle column, before hPCs and hNCs were seeded into the coculture system, SDF1 expression in hPCs was knocked down by SDF1 siRNA. Then, after 24 h migration of hNCCs and hPCs in the coculture system, these 2 cell types were costained with antibodies for hNCCs marker HNK1 (green), and hPCs marker Six1 (red). Bright-field photos were captured at the 0 time point when the monolayer of the cells in control, SDF1-knockdown, and ethanol-exposed groups was just formed (first row). The second to fourth row showed the photos of bright field and fluorescence field for these 3 groups, in which hNCCs and hPCs had migrated for 24 h. B, Quantification of migration defect of coordinated migration of hNCCs-PCs. Data represent the mean value of migration ratio ± SEM of 3 independent experiments. ***p < .001 versus control. C, Zebrafish embryos were microinjected with SDF1 morpholinos at 1-cell stage or exposed to ethanol (1% v/v) at 5–24 hpf, and then the craniofacial cartilages were stained with Alcian blue at 5 dpf (days post fertilization). m, Meckel’s cartilage; pq, palatoquadrate; ch, ceratohyal; bb, basibranchials; hb, hypobranchials; cb, ceratobranchial. Scale bar, 200 μm. D, Quantification of anomaly ratio. Data represent the ratio of cartilages deformities (including all the morphological anomalies observed in m, pq, ch, bb, hb, and cb) in each group (embryos’ number of anomalies/total embryos’ number in each group). ***p < .001 versus control. E, After performing the same treatment within (C), the patterns of cranial nerves at 2 dpf (a time point that the normal development of cranial nerves had completed) embryos were determined by WISH using cranial ganglia-specific probe Phox2b. VII, facial nerves; IX, glossopharyngeal nerves; X, vagal nerves. Scale bar, 200 μm. F, Quantification of the anomaly (weak or diffuse staining, or smaller volume of the ganglion) ratio of cranial nerves. Data represent mean ± SEM of 3 independent experiments. ***p < .001 versus control.
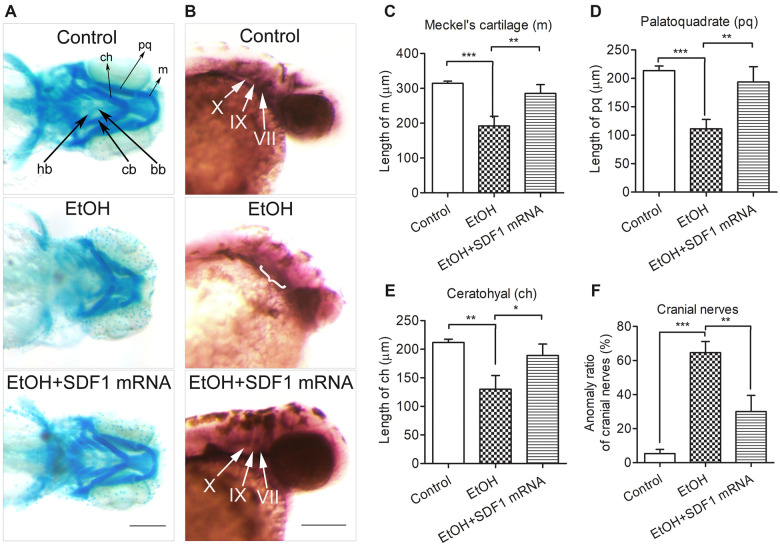
Overexpression of SDF1 diminished ethanol-induced craniofacial and cranial nerve defects in zebrafish
To determine whether exogenous supplement of SDF1 can rescue ethanol-induced malformations of craniofacial and cranial nerves, we microinjected in vitro-synthesized SDF1 mRNA into 1-cell stage zebrafish eggs. While ethanol resulted in various deformities of m, pq, ch, cb, bb, and hb (Figure 10A, second row), overexpression of SDF1 significantly diminished these anomalies (Figure 10A, third row). Also, the length of m, pq, and ch were largely recovered when supplemented with SDF1 mRNA (Figs. 10C, D, and E). Similarly, the morphological deformities of cranial nerves induced by ethanol (Figure 10B, second row) was significantly ameliorated by SDF1 overexpression (Figure 10B, third row), as well as the anomaly ratio was strongly decreased (Figure 10F). These data, in combination with Figure 9, demonstrated that lack of SDF1 expression plays a pivotal role in ethanol-induced teratogenesis.
Figure 10.
Overexpression of SDF1 diminished ethanol-induced defects of craniofacial skeletons and cranial nerves in zebrafish embryos. A, The first row of control group showed a normal morphology of craniofacial cartilages (black arrows). In the second row, zebrafish embryos were treated with ethanol (1% v/v) at 5–24 hpf. For the third row, the embryos were microinjected with in vitro-synthesized SDF1 mRNA (0.6 pmol/embryo) at 1-cell stage and then treated with ethanol (1% v/v) at 5–24 hpf. Zebrafish embryos in these 3 groups were stained with Alcian Blue at 5 dpf for visualizing the morphology of craniofacial cartilages. m, Meckel’s cartilage; pq, palatoquadrate; ch, ceratohyal; bb, basibranchials; hb, hypobranchials; cb, ceratobranchial. Scale bar, 200 μm. B, After the same treatment within (A), zebrafish embryos were subject to WISH at 2 dpf. The patterns of cranial nerves were indicated by ganglia-specific probe Phox2b (white arrows in first and third row). VII, facial nerves; IX, glossopharyngeal nerves; X, vagal nerves. Scale bar, 200 μm. Statistics analyses of cartilaginous length were shown in (C), (D), and (E). F, Quantification of the anomaly (weak or diffuse staining, or smaller volume of the ganglion) ratio of cranial nerves. Data represent mean ± SEM of 3 independent experiments. * p < .05 versus control, **p < .01 versus control, ***p < .001 versus control.
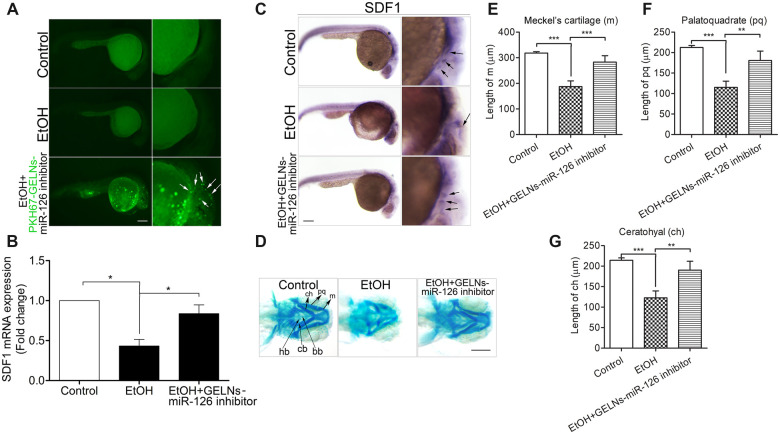
Uptake of miR-126 inhibitor-loaded GELNs significantly rescued SDF1 expression, and diminished ethanol-caused craniofacial anomalies in zebrafish embryos
It has been reported that edible plant-derived exosome-like nanoparticles (EPDENs) can be used for therapeutic or delivery purposes (Ju et al., 2013; Wang et al., 2014; Zhuang et al., 2015). To figure out whether delivery of miR-126 inhibitors by GELNs can prevent ethanol-induced reduction of SDF1, and malformations of craniofacial skeletons in zebrafish embryos, GELNs-loaded miR-126 inhibitor (GELNs-miR-126 inhibitor) was used for delivering to zebrafish embryos. Using PKH67 as a green fluorescence tag for GELNs, miR-126 inhibitor-loaded GELNs were microinjected into 1-cell stage zebrafish eggs. We found that the assembly of GELNs-miR-126 inhibitor (AGmiR126I) can be efficiently taken up by zebrafish embryos and distributed to the position where NCCs and PCs are located (Figure 11A, third row, arrows). Interestingly, we observed that in AGmiR126I-injected group, the level of SDF1 (Figure 11B) and the migration pattern of SDF1 were strikingly restored (Figure 11C, third row, arrows) compared with ethanol-treated group. Furthermore, the ethanol-induced severe defects of craniofacial skeletons of zebrafish embryos were significantly diminished by exogenous supplement with AGmiR126I (Figure 11D, third column), as well as an obvious improvement of the length of m, pq, and ch (Figs. 11E–G). Collectively, these results demonstrated that GELNs can be used as a carrier for miR-126 inhibitor to efficiently attenuate ethanol-caused downregulation of SDF1 and subsequently to prevent ethanol-induced teratogenesis.
Figure 11.
Uptake of miR-126 inhibitor-loaded GELNs diminished ethanol-induced repression of SDF1 expression, and rescued the defects of craniofacial skeletons induced by ethanol in zebrafish embryos. A, For the first and second rows, zebrafish embryos were treated without or with ethanol (1% v/v) at 5–24 hpf. In the third row, the embryos were microinjected with miR-126 inhibitor-loaded GELNs that labeled with PHK67 (PHK67-GELNs-miR-126 inhibitor) at 1-cell stage and then were treated with ethanol (1% v/v) at 5–24 hpf. The embryos were dechorionized at 30 hpf and photographed under a fluorescence microscope. The PHK67-GELNs-miR-126 inhibitors were distributed throughout the craniofacial position of the embryos as indicated by the white arrows. Scale bar, 200 μm. (B) For the first and second groups, zebrafish embryos were treated without or with ethanol (1% v/v) at 5–24 hpf. For the third group, the embryos were microinjected with miR-126 inhibitor-loaded GELNs (GELNs-miR-126 inhibitor) at 1-cell stage and then were treated with ethanol (1% v/v) at 5–24 hpf. The embryos of these 3 groups were collected at 30 hpf and subject to qRT-PCR analysis for detecting SDF1 expression. C and D, As treated with the same methods in (B), zebrafish embryos were subject to WISH for visualizing the expression pattern of SDF1 (indicated by black arrows in the 3 rows) detected by SDF1 probes at 30 hpf, or were stained by Alcian Blue to view the morphology of craniofacial cartilages at 5 dpf, respectively. m, Meckel’s cartilage; pq, palatoquadrate; ch, ceratohyal; bb, basibranchials; hb, hypobranchials; cb, ceratobranchial. Scale bars, 200 μm. Statistics analyses of cartilaginous length of (D) were shown in (E), (F), and (G). Data represent mean ± SEM of 3 independent experiments. *p < .05 versus control, **p < .01 versus control, ***p < .001 versus control.
Discussion
While craniofacial anomaly is considered as a key diagnostic feature of FASD, the defects of the cranial nerves might be associated with functional deficits that have been reported in up to 95% of children with FAS, such as voice dysfunction, and impaired tongue, mouth and larynx movements (Becker et al., 1990; Church and Gerkin, 1988; Church and Kaltenbach, 1997; Lipson et al., 1989). Over the past decades, by using different animal models for FASD, investigators recapitulated craniofacial defects or the dysfunctions of sensory system that were remarkably similar to those of affected humans (Moore et al., 2001, 2007; Muralidharan et al., 2013; Sulik, 2005; Sulik et al., 1981; 1986). Mechanism study for these malformations and malfunctions have revealed that exposure to ethanol results in excessive apoptosis in NCCs and PCs (Cartwright and Smith, 1995a,b; Chen et al., 2013a,b, 2015; Dunty et al., 2001; Kotch and Sulik, 1992b; Rovasio and Battiato, 2002; Sun et al., 2014; Yuan et al., 2017, 2018) and that ethanol-induced apoptosis in NCCs contributes heavily to the subsequent craniofacial defects (Cartwright and Smith, 1995a; Kotch and Sulik, 1992a; Sulik et al., 1981). In addition, ethanol-induced cell death in NCCs and PCs resulted in anomalies of cranial nerves, including the fusion of the trigeminal (V) and the facial vestibulocochlear (VII–VIII) ganglia complex, a loss of the dorsal root of the IX nerve and partial fusion between the roots of the IX and X cranial nerves, and the disorganization of the dorsal roots of the X nerve (Dunty et al., 2002). Moreover, ethanol exposure has been demonstrated to strongly suppress NCC migration, resulting in short travel distance and less directional movements of NCCs (Czarnobaj et al., 2014; Rovasio and Battiato, 2002). Studies have also shown that fewer NCCs emigrated from the forebrain, midbrain and hindbrain in ethanol-exposed embryos (Cartwright and Smith, 1995b; Rovasio and Battiato, 1995). Real-time mapping of the abnormal NCC migration in ethanol-exposed zebrafish embryos demonstrated that the migration of NCCs loses left-right symmetry and that NCCs travel a shorter distance (Boric et al., 2013). Based on above evidences, 3 points can be generated: (1) dysfunctions of NCCs and PCs contribute to the craniofacial abnormalities and cranial nerves, respectively; (2) NCCs migration was intensively studied whereas PCs migration was seldom paid attention, although these 2 tightly neighbored cell populations are simultaneously affected by ethanol exposure; (3) how ethanol impact the NCCs-PCs interaction remains poorly understood. In our study, we acquired solid evidences demonstrating that ethanol exposure can directly disrupt NCCs-PCs interaction and directional migration of NCCs and PCs in vitro or in vivo.
Reported by Ammann decades ago, FASD and DiGeorge Syndrome share high similarities of clinical and laboratory features in human (Ammann et al., 1982). Through acute maternal alcohol administration, Sulik et al. reproduced the typical FASD craniofacial phenotype that is similar to that noted in DiGeorge anomaly (Sulik et al., 1986). Further genetic studies revealed that disruption of a chemokine SDF1, and its receptor CXCR4-mediated signaling plays a key role in the pathogenesis of DiGeorge Syndrome (Toritsuka et al., 2013). Specifically, downregulation of SDF1/CXCR4 signaling resulted in misrouting of pharyngeal NCCs migration and remarkable morphological defects in the craniofacial skeletons and cranial sensory ganglia (Duband et al., 2016; Escot et al., 2016). Repression of SDF1/CXCR4 signaling has also been shown to impair NCCs migration (George et al., 2007; Olesnicky Killian et al., 2009; Rezzoug et al., 2011) or to cause major defects in many organisms (Braun et al., 2002; Escot et al., 2013; George et al., 2007; Kasemeier-Kulesa et al., 2010; Rezzoug et al., 2011). In particular, an outstanding study by Theveneau and colleagues have demonstrated that SDF1-postive PCs can attract CXCR4-positive NCCs to move toward PCs, which result in a physical contact between these 2 cell populations and the subsequent contact inhibition of locomotion (CIL), leading to the moving away of PCs from the NCCs, a process termed “chase and run” (Theveneau et al., 2013). This coordinated and efficient directional migration of cranial NCCs and epibranchial PCs toward lateral and ventral regions of embryo is pivotal for craniofacial development. In addition, it has been shown that alcohol consumption can reduce blood SDF1 levels in patients (Xiao et al., 2008). Based on these existed findings, we come up with the hypothesis that dysregulation of SDF1/CXCR4 signaling induced by ethanol is highly correlated with the defects of craniofacial structures and cranial sensory ganglia observed in FASD. As expected, we found that ethanol exposure can decrease the SDF1 expression in hPCs and in zebrafish embryos. Furthermore, the silence of SDF1 expression either in vitro or in vivo mimicked ethanol-induced interruption of hNCC-hPC interaction or FASD-related morphological defects in zebrafish, respectively. Conversely, restoration of SDF1 expression by delivering exogenous SDF1 mRNA into zebrafish embryos protected against ethanol-induced craniofacial anomalies.
PCs-secreted SDF1 is considered to be a motor to initiate the PCs-guided NCCs migration. As demonstrated in our experiments, NCCs and PCs synergistically migrate in a chemotaxis-mediated mechanism, whereas ethanol exposure can disrupt such a process. However, through what exact mechanism that ethanol impairs the NCCs-PCs interaction is unclear. During NCCs-PCs crosstalk, PCs can emit a chemoattractant signaling molecule, SDF1, to NCCs, so it is of particular interest to quest whether NCCs can also send feedback signal to PCs through the similar way as the PCs utilize or through other unknown mechanisms. With versatile biological activities, exosomes have recently emerged as an important mode of intercellular communication especially in brain development and nervous system (Budnik et al., 2016; Kramer-Albers and Hill, 2016; Morton and Feliciano, 2016; Rajendran et al., 2014; Sharma et al., 2013), which carry a defined but mixed cargo of bioactive molecules to modulate the molecular configuration and behavior of target cells. For instance, during neurons communication, cells can deploy a membrane protein, EphB2 receptor into exosomes to activate ephrinB signaling resulting in growth cone collapse (Gong et al., 2016; Pasquale, 2016). During embryogenesis of zebrafish, neurons can secrete miR-132-containing exosomes to regulate endothelial integrity of brain (Xu et al., 2017). Regarding these existed knowledge, we came up with the idea that it is of high possibility that NCCs-derived exosomes may transmit negative or positive signal to PCs to regulate NCCs-PCs interaction. To address this issue, we have demonstrated that ethanol-treated hNCCs-derived exosomes (ExoEtOH) can strongly suppress the coordinated migration of hNCCs and hPCs in our coculture system, indicating that certain inhibitory signaling might be conducted through ExoEtOH. Then, we proceeded to pretreat hNCCs with exosome inhibitor, GW4869, to block exosome releasing. We observed that ethanol-induced disruption on hNCCs-hPCs interaction was dramatically blocked, indicating that hNCCs-derived exosomes play a key role in mediating ethanol’s detrimental effect.
As mentioned above, exosomes have a biological effect that can transfer their contents between cells as natural carriers, most important of which are miRNAs. Among numerous miRNAs, miR-126 was highly related with SDF1/CXCR4 signaling in modulating cell migration and cancer metastasis (Li et al., 2013; Liu et al., 2014; Qian et al., 2016; Yuan et al., 2016). Also, studies have shown that SDF1 is directly repressed by miR-126 in breast cancer cells and endothelial cells (van Solingen et al., 2011; Zhang et al., 2013). More interestingly, exosomal shuttling of miR-126 in endothelial cells can modulates adhesive and migratory abilities of leukemia cells through targeting SDF1 (Taverna et al., 2014). Taken together, these studies provided us a clue that miR-126 might act as a negative regulator encapsulated in NCCs-secreted exosomes during coordinated migration of NCCs and PCs. To test this hypothesis, we examined the miR-126 level in hNCC-derived exosomes and found that miR-126 level in ExoEtOH was significantly higher than that in control exosomes (ExoCon). Furthermore, consistent with the other studies, we confirmed that miR-126 can specifically inhibit SDF1 expression in hPCs. Then we loaded ethanol-treated hNCCs-derived exosomes with miR-126 inhibitors and demonstrated that inhibiting exosome-shuttled miR-126 can significantly diminish ethanol-induced impairment of hNCCs-hPCs interaction. Also in zebrafish experiments, we observed that fruit-derived exosomal delivery of miR-126 inhibitors into zebrafish embryos can strongly rescue ethanol-induced repression of SDF1 and the subsequent craniofacial defects.
Exosome therapy is recently emerging as a promising treatment for many diseases. The advantages of exosome therapy include that they are natural carriers of proteins and RNAs with a favorable size and are well-tolerated in vivo (Shtam et al., 2013; Valadi et al., 2007; Zhang et al., 2017). However, the difficulties in large scale production of exosomes from mammalian cells limit their clinical application. A promising solution for this problem is to produce exosome-like nanoparticles from edible plants. Recently, EPDENs that share similar properties as mammalian exosomes have been reported in grapes, grapefruit, ginger, and carrots (Ju et al., 2013; Mu et al., 2014; Wang et al., 2014; Zhuang et al., 2015). Several studies have demonstrated that EPDENs can be used for therapeutic or delivery purposes. For instance, it has been demonstrated that systemic deliver siRNA by using ginger exosome-like nanoparticles as a siRNA delivery vesicle could suppress tumor growth (Li et al., 2018). In our experiments, we used GELNs as a carrier for miR-126 inhibitors, showing that GELNs-encapsulated miR-126 inhibitors can be easily absorbed by zebrafish embryos and exert a strongly protective effect against ethanol-induced craniofacial anomalies.
In summary, we uncovered a novel mechanism by which exosomal delivery of miR-126 act as a negative regulator for SDF1-mediated chemotaxis process involved in ethanol-induced disruption of the coordinated NCCs-PCs interaction and developmental defects. Moreover, efficient delivery of miR-126 inhibitors by GELNs could potentially be a new strategy of intervention for FASD.
Contributor Information
Yihong Li, Ningbo No.2 Hospital, Ningbo 315099, China; Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo 315000, China; Lab of Nanopharmacology Research for Neurodegeneration, Department of Research and Development of Science and Technology, Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo, Zhejiang Province 315000, China; Department of Pharmacology and Toxicology, University of Louisville Health Sciences Center, Louisville, Kentucky 40292, USA.
Ting Cai, Ningbo No.2 Hospital, Ningbo 315099, China; Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo 315000, China.
Huina Liu, Ningbo No.2 Hospital, Ningbo 315099, China; Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo 315000, China.
Jie Liu, Department of Pharmacology and Toxicology, University of Louisville Health Sciences Center, Louisville, Kentucky 40292, USA.
Shao-Yu Chen, Department of Pharmacology and Toxicology, University of Louisville Health Sciences Center, Louisville, Kentucky 40292, USA.
Huadong Fan, Ningbo No.2 Hospital, Ningbo 315099, China; Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo 315000, China; Department of Pharmacology and Toxicology, University of Louisville Health Sciences Center, Louisville, Kentucky 40292, USA; Lab of Dementia and Neurorehabilitation Research, Department of Research and Development of Science and Technology, Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences, Ningbo, Zhejiang Province 315000, China.
Author contributions
H.F. and Y.H.L. designed research, performed the experiments, and participated in data analysis and manuscript preparation. H.F., Y.L., T.C., H.L., J.L., and S.C. participated in data interpretation and discussion. All authors reviewed the manuscript.
Declaration of conflicting interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
Young Innovative Talent Project of YongJiang Talent Introduction Programme (2021A-012-G to H.F.); Ningbo Natural Science Foundation (2021J321 to H.F., 2021J328 to Y.L.); Ningbo Public Service Technology Foundation (2022S030 to H.F.); Special Funding for Microfluidic Chip of Biomedicine of Ningbo Institute of Life and Health Industry, University of Chinese Academy of Sciences (2021YJY1006 to H.F., 2021YJY1005 to Y.L.); Medical Scientific Research Foundation of Zhejiang Province (2023KY299 to Y.L.); National Institute of Health Grants (R01AA028435, R01AA021434. to S.C.).
References
- Ammann A. J., Wara D. W., Cowan M. J., Barrett D. J., Stiehm E. R. (1982). The DiGeorge syndrome and the fetal alcohol syndrome. Am. J. Dis. Child. 136, 906–908. [DOI] [PubMed] [Google Scholar]
- Bang C., Batkai S., Dangwal S., Gupta S. K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A., et al. (2014). Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 124, 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Becker M., Warr-Leeper G. A., Leeper H. A. (1990). Fetal alcohol syndrome: A description of oral motor, articulatory, short-term-memory, grammatical, and semantic abilities. J. Commun. Disord. 23, 97–124. [DOI] [PubMed] [Google Scholar]
- Boric K., Orio P., Vieville T., Whitlock K. (2013). Quantitative analysis of cell migration using optical flow. PLoS One. 8, e69574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M., Wunderlin M., Spieth K., Knochel W., Gierschik P., Moepps B. (2002). Xenopus laevis stromal cell-derived factor 1: Conservation of structure and function during vertebrate development. J. Immunol. 168, 2340–2347. [DOI] [PubMed] [Google Scholar]
- Budnik V., Ruiz-Canada C., Wendler F. (2016). Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 17, 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright M. M., Smith S. M. (1995a). Increased cell death and reduced neural crest cell numbers in ethanol-exposed embryos: Partial basis for the fetal alcohol syndrome phenotype. Alcohol. Clin. Exp. Res. 19, 378–386. [DOI] [PubMed] [Google Scholar]
- Cartwright M. M., Smith S. M. (1995b). Stage-dependent effects of ethanol on cranial neural crest cell development: Partial basis for the phenotypic variations observed in fetal alcohol syndrome. Alcohol. Clin. Exp. Res. 19, 1454–1462. [DOI] [PubMed] [Google Scholar]
- Chen X., Liu J., Chen S. Y. (2013a). Over-expression of Nrf2 diminishes ethanol-induced oxidative stress and apoptosis in neural crest cells by inducing an antioxidant response. Reprod. Toxicol. 42, 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu J., Chen S. Y. (2013b). Sulforaphane protects against ethanol-induced oxidative stress and apoptosis in neural crest cells by the induction of Nrf2-mediated antioxidant response. Br. J. Pharmacol. 169, 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu J., Feng W. K., Wu X., Chen S. Y. (2015). Mir-125b protects against ethanol-induced apoptosis in neural crest cells and mouse embryos by targeting Bak 1 and PUMA. Exp. Neurol. 271, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church M. W., Gerkin K. P. (1988). Hearing disorders in children with fetal alcohol syndrome: Findings from case reports. Pediatrics 82, 147–154. [PubMed] [Google Scholar]
- Church M. W., Kaltenbach J. A. (1997). Hearing, speech, language, and vestibular disorders in the fetal alcohol syndrome: A literature review. Alcohol. Clin. Exp. Res. 21, 495–512. [DOI] [PubMed] [Google Scholar]
- Costa-Silva B., Aiello N. M., Ocean A. J., Singh S., Zhang H., Thakur B. K., Becker A., Hoshino A., Mark M. T., Molina H., et al. (2015). Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnobaj J., Bagnall K. M., Bamforth J. S., Milos N. C. (2014). The different effects on cranial and trunk neural crest cell behaviour following exposure to a low concentration of alcohol in vitro. Arch. Oral Biol. 59, 500–512. [DOI] [PubMed] [Google Scholar]
- D’Amico-Martel A., Noden D. M. (1983). Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. Am. J. Anat. 166, 445–468. [DOI] [PubMed] [Google Scholar]
- Delfino-Machin M., Chipperfield T. R., Rodrigues F. S., Kelsh R. N. (2007). The proliferating field of neural crest stem cells. Dev. Dyn. 236, 3242–3254. [DOI] [PubMed] [Google Scholar]
- Dincer Z., Piao J., Niu L., Ganat Y., Kriks S., Zimmer B., Shi S. H., Tabar V., Studer L. (2013). Specification of functional cranial placode derivatives from human pluripotent stem cells. Cell Rep. 5, 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M., Reichman-Fried M., Stebler J., Koprunner M., Dorries J., Meyer D., Esguerra C. V., Leung T., Raz E. (2002). Guidance of primordial germ cell migration by the chemokine sdf-1. Cell 111, 647–659. [DOI] [PubMed] [Google Scholar]
- Dong S., Jin M., Li Y., Ren P., Liu J. (2016). Mir-137 acts as a tumor suppressor in papillary thyroid carcinoma by targeting CXCL12. Oncol. Rep. 35, 2151–2158. [DOI] [PubMed] [Google Scholar]
- Duband J. L., Escot S., Fournier-Thibault C. (2016). SDF1-CXCR4 signaling: A new player involved in DiGeorge/22q11-deletion syndrome. Rare Dis. 4, e1195050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunty W. C. Jr., Chen S. Y., Zucker R. M., Dehart D. B., Sulik K. K. (2001). Selective vulnerability of embryonic cell populations to ethanol-induced apoptosis: Implications for alcohol-related birth defects and neurodevelopmental disorder. Alcohol. Clin. Exp. Res. 25, 1523–1535. [PubMed] [Google Scholar]
- Dunty W. C. Jr., Zucker R. M., Sulik K. K. (2002). Hindbrain and cranial nerve dysmorphogenesis result from acute maternal ethanol administration. Dev. Neurosci. 24, 328–342. [DOI] [PubMed] [Google Scholar]
- Escot S., Blavet C., Faure E., Zaffran S., Duband J. L., Fournier-Thibault C. (2016). Disruption of CXCR4 signaling in pharyngeal neural crest cells causes DiGeorge syndrome-like malformations. Development 143, 582–588. [DOI] [PubMed] [Google Scholar]
- Escot S., Blavet C., Hartle S., Duband J. L., Fournier-Thibault C. (2013). Misregulation of SDF1-CXCR4 signaling impairs early cardiac neural crest cell migration leading to conotruncal defects. Circ. Res. 113, 505–516. [DOI] [PubMed] [Google Scholar]
- Fan H., Yuan F., Yun Y., Wu T., Lu L., Liu J., Feng W., Chen S. Y. (2019). MicroRNA-34a mediates ethanol-induced impairment of neural differentiation of neural crest cells by targeting autophagy-related gene 9a. Exp. Neurol. 320, 112981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong M. Y., Zhou W., Liu L., Alontaga A. Y., Chandra M., Ashby J., Chow A., O’Connor S. T., Li S., Chin A. R., et al. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruhbeis C., Frohlich D., Kuo W. P., Kramer-Albers E. M. (2013). Extracellular vesicles as mediators of neuron-glia communication. Front. Cell. Neurosci. 7, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L., Chaverra M., Todd V., Lansford R., Lefcort F. (2007). Nociceptive sensory neurons derive from contralaterally migrating, fate-restricted neural crest cells. Nat. Neurosci. 10, 1287–1293. [DOI] [PubMed] [Google Scholar]
- Gong J., Korner R., Gaitanos L., Klein R. (2016). Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance. J. Cell Biol. 214, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. K. (2008). The neural crest and neural crest cells: Discovery and significance for theories of embryonic organization. J. Biosci. 33, 781–793. [DOI] [PubMed] [Google Scholar]
- Harlow D. E., Barlow L. A. (2007). Embryonic origin of gustatory cranial sensory neurons. Dev. Biol. 310, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow D. E., Yang H., Williams T., Barlow L. A. (2011). Epibranchial placode-derived neurons produce BDNF required for early sensory neuron development. Dev. Dyn. 240, 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen D. K., Fenix A. M., Franklin J. L., Higginbotham J. N., Zhang Q., Zimmerman L. J., Liebler D. C., Ping J., Liu Q., Evans R., et al. (2019). Reassessment of exosome composition. Cell 177, 428–445.e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju S., Mu J., Dokland T., Zhuang X., Wang Q., Jiang H., Xiang X., Deng Z. B., Wang B., Zhang L., et al. (2013). Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol. Ther. 21, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasemeier-Kulesa J. C., McLennan R., Romine M. H., Kulesa P. M., Lefcort F. (2010). CXCR4 controls ventral migration of sympathetic precursor cells. J. Neurosci. 30, 13078–13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotch L. E., Sulik K. K. (1992a). Experimental fetal alcohol syndrome: Proposed pathogenic basis for a variety of associated facial and brain anomalies. Am. J. Med. Genet. 44, 168–176. [DOI] [PubMed] [Google Scholar]
- Kotch L. E., Sulik K. K. (1992b). Patterns of ethanol-induced cell death in the developing nervous system of mice; neural fold states through the time of anterior neural tube closure. Int. J. Dev. Neurosci. 10, 273–279. [DOI] [PubMed] [Google Scholar]
- Kramer-Albers E. M., Hill A. F. (2016). Extracellular vesicles: Interneural shuttles of complex messages. Curr. Opin. Neurobiol. 39, 101–107. [DOI] [PubMed] [Google Scholar]
- Lee G., Chambers S. M., Tomishima M. J., Studer L. (2010). Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 5, 688–701. [DOI] [PubMed] [Google Scholar]
- Lee Y., El Andaloussi S., Wood M. J. (2012). Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 21, R125–134. [DOI] [PubMed] [Google Scholar]
- Lener T., Gimona M., Aigner L., Borger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F. A., Del Portillo H. A., et al. (2015). Applying extracellular vesicles based therapeutics in clinical trials: an ISEV position paper. J. Extracell. Vesicles. 4, 30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellis S. W., Nagelberg D., Subedi A., Staton A., LeBlanc M., Giraldez A., Knaut H. (2013). Precise SDF1-mediated cell guidance is achieved through ligand clearance and microRNA-mediated decay. J. Cell Biol. 200, 337–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li N., Wu M., Li X., Luo Z., Wang X. (2013). Expression of miR-126 suppresses migration and invasion of colon cancer cells by targeting CXCR4. Mol. Cell. Biochem. 381, 233–242. [DOI] [PubMed] [Google Scholar]
- Li Z., Wang H., Yin H., Bennett C., Zhang H. G., Guo P. (2018). Arrowtail RNA for ligand display on ginger exosome-like nanovesicles to systemic deliver siRNA for cancer suppression. Sci. Rep. 8, 14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson A. H., Webster W. S., Brown-Woodman P. D., Osborn R. A. (1989). Moebius syndrome: Animal model–human correlations and evidence for a brainstem vascular etiology. Teratology 40, 339–350. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhou Y., Feng X., An P., Quan X., Wang H., Ye S., Yu C., He Y., Luo H. (2014). MicroRNA-126 functions as a tumor suppressor in colorectal cancer cells by targeting CXCR4 via the Akt and Erk1/2 signaling pathways. Int. J. Oncol. 44, 203–210. [DOI] [PubMed] [Google Scholar]
- Mateescu B., Kowal E. J., van Balkom B. W., Bartel S., Bhattacharyya S. N., Buzas E. I., Buck A. H., de Candia P., Chow F. W., Das S., et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA: an ISEV position paper. J. Extracell. Vesicles. 6, 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I. J., Vincent J. P. (2016). Exosomes in developmental signalling. Development 143, 2482–2493. [DOI] [PubMed] [Google Scholar]
- Moore E. S., Ward R. E., Jamison P. L., Morris C. A., Bader P. I., Hall B. D. (2001). The subtle facial signs of prenatal exposure to alcohol: An anthropometric approach. J. Pediatr. 139, 215–219. [DOI] [PubMed] [Google Scholar]
- Moore E. S., Ward R. E., Wetherill L. F., Rogers J. L., Autti-Ramo I., Fagerlund A., Jacobson S. W., Robinson L. K., Hoyme H. E., Mattson S. N., et al. ; CIFASD (2007). Unique facial features distinguish fetal alcohol syndrome patients and controls in diverse ethnic populations. Alcohol. Clin. Exp. Res. 31, 1707–1713. [DOI] [PubMed] [Google Scholar]
- Morton M. C., Feliciano D. M. (2016). Neurovesicles in brain development. Cell. Mol. Neurobiol. 36, 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Zhuang X., Wang Q., Jiang H., Deng Z. B., Wang B., Zhang L., Kakar S., Jun Y., Miller D., et al. (2014). Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 58, 1561–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan P., Sarmah S., Zhou F. C., Marrs J. A. (2013). Fetal alcohol spectrum disorder (FASD) associated neural defects: Complex mechanisms and potential therapeutic targets. Brain Sci. 3, 964–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill P., Mak S. S., Fritzsch B., Ladher R. K., Baker C. V. (2012). The amniote paratympanic organ develops from a previously undiscovered sensory placode. Nat. Commun. 3, 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky Killian E. C., Birkholz D. A., Artinger K. B. (2009). A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev. Biol. 333, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale E. B. (2016). Exosomes expand the sphere of influence of Eph receptors and ephrins. J. Cell Biol. 214, 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Wang X., Lv Z., Guo C., Yang Y., Zhang J., Wang X. (2016). MicroRNA126 is downregulated in thyroid cancer cells, and regulates proliferation, migration and invasion by targeting CXCR4. Mol. Med. Rep. 14, 453–459. [DOI] [PubMed] [Google Scholar]
- Rajendran L., Bali J., Barr M. M., Court F. A., Krämer-Albers E.-M., Picou F., Raposo G., van der Vos K. E., van Niel G., Wang J., et al. (2014). Emerging roles of extracellular vesicles in the nervous system. J. Neurosci. 34, 15482–15489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzoug F., Seelan R. S., Bhattacherjee V., Greene R. M., Pisano M. M. (2011). Chemokine-mediated migration of mesencephalic neural crest cells. Cytokine 56, 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovasio R. A., Battiato N. L. (1995). Role of early migratory neural crest cells in developmental anomalies induced by ethanol. Int. J. Dev. Biol. 39, 421–422. [PubMed] [Google Scholar]
- Rovasio R. A., Battiato N. L. (2002). Ethanol induces morphological and dynamic changes on in vivo and in vitro neural crest cells. Alcohol. Clin. Exp. Res. 26, 1286–1298. [DOI] [PubMed] [Google Scholar]
- Rufino-Ramos D., Albuquerque P. R., Carmona V., Perfeito R., Nobre R. J., Pereira de Almeida L. (2017). Extracellular vesicles: Novel promising delivery systems for therapy of brain diseases. J. Control. Release 262, 247–258. [DOI] [PubMed] [Google Scholar]
- Schilling T. F., Piotrowski T., Grandel H., Brand M., Heisenberg C. P., Jiang Y. J., Beuchle D., Hammerschmidt M., Kane D. A., Mullins M. C., et al. (1996). Jaw and branchial arch mutants in zebrafish I: Branchial arches. Development 123, 329–344. [DOI] [PubMed] [Google Scholar]
- Schlosser G., Northcutt R. G. (2000). Development of neurogenic placodes in Xenopus laevis. J. Comp. Neurol. 418, 121–146. [PubMed] [Google Scholar]
- Sharma P., Schiapparelli L., Cline H. T. (2013). Exosomes function in cell-cell communication during brain circuit development. Curr. Opin. Neurobiol. 23, 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R. J., Zhao L. B., Cai W. B., Wei M. Y., Zhou X. Y., Yang G. D., Yuan L. J. (2017). Maternal exosomes in diabetes contribute to the cardiac development deficiency. Biochem. Biophys. Res. Commun. 483, 602–608. [DOI] [PubMed] [Google Scholar]
- Shtam T. A., Kovalev R. A., Varfolomeeva E. Y., Makarov E. M., Kil Y. V., Filatov M. V. (2013). Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 11, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton A. A., Knaut H., Giraldez A. J. (2011). Mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 43, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B., Mayor R., Streit A. (2014). Neural crest and placode interaction during the development of the cranial sensory system. Dev. Biol. 389, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik K. K. (2005). Genesis of alcohol-induced craniofacial dysmorphism. Exp. Biol. Med. (Maywood) 230, 366–375. [DOI] [PubMed] [Google Scholar]
- Sulik K. K., Johnston M. C., Daft P. A., Russell W. E., Dehart D. B. (1986). Fetal alcohol syndrome and DiGeorge anomaly: Critical ethanol exposure periods for craniofacial malformations as illustrated in an animal model. Am. J. Med. Genet. Suppl. 2, 97–112. [DOI] [PubMed] [Google Scholar]
- Sulik K. K., Johnston M. C., Webb M. A. (1981). Fetal alcohol syndrome: Embryogenesis in a mouse model. Science 214, 936–938. [DOI] [PubMed] [Google Scholar]
- Sun H., Chen X., Yuan F., Liu J., Zhao Y., Chen S. Y. (2014). Involvement of seven in absentia homolog-1 in ethanol-induced apoptosis in neural crest cells. Neurotoxicol. Teratol. 46, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S., Amodeo V., Saieva L., Russo A., Giallombardo M., De Leo G., Alessandro R. (2014). Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol. Cancer. 13, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L., Labosky P. A. (2006). Neural crest stem cells. Adv. Exp. Med. Biol. 589, 206–212. [DOI] [PubMed] [Google Scholar]
- Theveneau E., Steventon B., Scarpa E., Garcia S., Trepat X., Streit A., Mayor R. (2013). Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 15, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Thisse C. (2014). In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol. Biol. 1211, 53–67. [DOI] [PubMed] [Google Scholar]
- Tian Y., Matsui S., Touma M., Wu Q., Sugimoto K. (2018). MicroRNA-342 inhibits tumor growth via targeting chemokine CXCL12 involved in macrophages recruitment/activation. Genes Cells 23, 1009–1022. [DOI] [PubMed] [Google Scholar]
- Toritsuka M., Kimoto S., Muraki K., Landek-Salgado M. A., Yoshida A., Yamamoto N., Horiuchi Y., Hiyama H., Tajinda K., Keni N., et al. (2013). Deficits in microRNA-mediated CXCR4/CXCL12 signaling in neurodevelopmental deficits in a 22q11 deletion syndrome mouse model. Proc. Natl. Acad. Sci. U.S.A. 110, 17552–17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. [DOI] [PubMed] [Google Scholar]
- van Solingen C., de Boer H. C., Bijkerk R., Monge M., van Oeveren-Rietdijk A. M., Seghers L., de Vries M. R., van der Veer E. P., Quax P. H., Rabelink T. J., et al. (2011). MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1(+)/Lin(-) progenitor cells in ischaemia. Cardiovasc. Res. 92, 449–455. [DOI] [PubMed] [Google Scholar]
- Walker M. B., Kimmel C. B. (2007). A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28. [DOI] [PubMed] [Google Scholar]
- Wang B., Zhuang X., Deng Z. B., Jiang H., Mu J., Wang Q., Xiang X., Guo H., Zhang L., Dryden G., et al. (2014). Targeted drug delivery to intestinal macrophages by bioactive nanovesicles released from grapefruit. Mol. Ther. 22, 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Ye S., Oberhollenzer F., Mayr A., Jahangiri M., Willeit J., Kiechl S., Xu Q. (2008). SDF1 gene variation is associated with circulating SDF1alpha level and endothelial progenitor cell number: The Bruneck study. PLoS One. 3, e4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Zhang Y., Du X. F., Li J., Zi H. X., Bu J. W., Yan Y., Han H., Du J. L. (2017). Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 27, 882–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Chen X., Liu J., Feng W., Cai L., Wu X., Chen S. Y. (2018). Sulforaphane restores acetyl-histone H3 binding to Bcl-2 promoter and prevents apoptosis in ethanol-exposed neural crest cells and mouse embryos. Exp. Neurol. 300, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Chen X., Liu J., Feng W., Wu X., Chen S. Y. (2017). Up-regulation of Siah1 by ethanol triggers apoptosis in neural crest cells through p38 MAPK-mediated activation of p53 signaling pathway. Arch. Toxicol. 91, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Guo Y. Q., Li X. Y., Deng M. Z., Shen Z. H., Bo C. B., Dai Y. F., Huang M. Y., Yang Z. Y., Quan Y. S., et al. (2016). MicroRNA-126 inhibits colon cancer cell proliferation and invasion by targeting the chemokine (C-X-C motif) receptor 4 and Ras homolog gene family, member A, signaling pathway. Oncotarget 7, 60230–60244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Lee H., Zhu Z., Minhas J. K., Jin Y. (2017). Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 312, L110–L121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Zhang S., Yao J., Lowery F. J., Zhang Q., Huang W. C., Li P., Li M., Wang X., Zhang C., et al. (2015). Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang P., Sun T., Li D., Xu X., Rui Y., Li C., Chong M., Ibrahim T., Mercatali L., et al. (2013). miR-126 and miR-126 repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 15, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. G., Buller B., Chopp M. (2019). Exosomes-beyond stem cells for restorative therapy in stroke and neurological injury. Nat. Rev. Neurol. 15, 193–203. [DOI] [PubMed] [Google Scholar]
- Zhuang X., Deng Z. B., Mu J., Zhang L., Yan J., Miller D., Feng W., McClain C. J., Zhang H. G. (2015). Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles. 4, 28713. [DOI] [PMC free article] [PubMed] [Google Scholar]