Significance
Among the most common human birth defects, orofacial clefts (OFCs) are thought to result from gene–environment interactions. Our work shows that an environmentally sensitive epigenetic mechanism regulates orofacial development and that its disruption during an early embryonic window of sensitivity results in OFCs in mice. We demonstrate that DNA methylation is required for proliferation and differentiation of stem cells that form orofacial connective tissue and that the cellular mechanisms of orofacial cleft pathogenesis can be recapitulated in a tractable in vitro model. These studies establish DNA methylation as a critical regulator of orofacial morphogenesis and cleft pathogenesis and provide conceptual and experimental platforms to understand how this environmentally responsive epigenetic mechanism can be harnessed for prevention strategies.
Keywords: birth defects, epigenetics, orofacial morphogenesis, neural crest
Abstract
Orofacial clefts of the lip and palate are widely recognized to result from complex gene–environment interactions, but inadequate understanding of environmental risk factors has stymied development of prevention strategies. We interrogated the role of DNA methylation, an environmentally malleable epigenetic mechanism, in orofacial development. Expression of the key DNA methyltransferase enzyme DNMT1 was detected throughout palate morphogenesis in the epithelium and underlying cranial neural crest cell (cNCC) mesenchyme, a highly proliferative multipotent stem cell population that forms orofacial connective tissue. Genetic and pharmacologic manipulations of DNMT activity were then applied to define the tissue- and timing-dependent requirement of DNA methylation in orofacial development. cNCC-specific Dnmt1 inactivation targeting initial palate outgrowth resulted in OFCs, while later targeting during palatal shelf elevation and elongation did not. Conditional Dnmt1 deletion reduced cNCC proliferation and subsequent differentiation trajectory, resulting in attenuated outgrowth of the palatal shelves and altered development of cNCC-derived skeletal elements. Finally, we found that the cellular mechanisms of cleft pathogenesis observed in vivo can be recapitulated by pharmacologically reducing DNA methylation in multipotent cNCCs cultured in vitro. These findings demonstrate that DNA methylation is a crucial epigenetic regulator of cNCC biology, define a critical period of development in which its disruption directly causes OFCs, and provide opportunities to identify environmental influences that contribute to OFC risk.
Orofacial clefts (OFCs) of the upper lip and palate are among the most common human structural birth defects, affecting approximately 1 in 700 newborns (1). Individuals with OFCs endure feeding difficulties as infants, require multiple surgeries, dental procedures, and speech therapy during childhood and adolescence, and face higher mortality rates at all stages of life (2–5). Prevention strategies for these birth defects remain elusive because our understanding of causative factors is inadequate. Efforts to elucidate OFC etiology have largely focused on the genome, using traditional and large-scale genetic approaches to identify sequence variations in coding and noncoding regions. These efforts have identified dozens of OFC risk loci, but recognized sequence variants are rarely causative and OFCs generally do not follow Mendelian inheritance patterns (6). OFCs are now widely recognized to be etiologically complex traits that result when multiple genetic and environmental influences reach a critical threshold of insult (7–11). But while environmental influences are thought to contribute substantially to OFC etiology, specific factors, and the mechanisms by which they act, remain largely unknown (1, 10, 12).
DNA methylation is an environmentally malleable epigenetic mechanism that has been implicated in orofacial morphogenesis by several lines of evidence (13). Periconceptional intake of the dietary methyl-group donors folic acid and choline has been reported to reduce OFC risk (14–22), and DNA methylation differences have been identified among individuals with and without OFCs (23–26). In mammalian development, DNA methylation is regulated by three conserved DNA methyltransferase (DNMT) enzymes: DNMT3a and DNMT3b, which typically act as de novo methylators, and DNMT1, which is primarily responsible for maintenance methylation. Severe malformations observed in Dnmt1 and Dnmt3b knockout mice demonstrated the developmental requirement of DNMT activity, but early embryonic lethality left the role of DNMTs and DNA methylation in orofacial morphogenesis unclear (13, 27–29).
In this study, we applied genetic and pharmacologic approaches to interrogate the tissue- and timing-specific role of DNA methylation in orofacial morphogenesis. We found that disruption of DNA methylation in the multipotent cranial neural crest mesenchyme during early orofacial development results in orofacial clefts. We then investigated the cellular mechanisms by which DNA methylation regulates cranial neural crest cell (cNCC) biology during cleft pathogenesis in vivo and whether these mechanisms could be recapitulated in a tractable in vitro culture model. These studies provide conceptual insight and experimental platforms for understanding how an environmentally malleable epigenetic mechanism could be harnessed to advance OFC prevention strategies.
Results
The Key DNA Methyltransferase DNMT1 Is Expressed throughout Palate Morphogenesis.
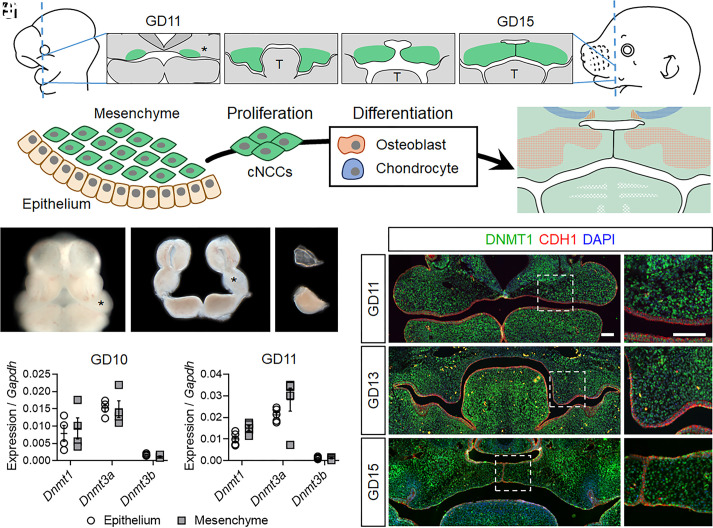
Orofacial morphogenesis requires orchestrated expansion and fusion of embryonic facial growth centers. The palatal shelves emerge as outgrowths at the oral surface of the maxillary processes, grow vertically, then elevate above the tongue, extend horizontally toward the midline, and fuse to form the secondary palate (Fig. 1A). The embryonic facial growth centers, including the maxillary processes, consist of an ectodermal epithelium surrounding dense mesenchyme that is primarily derived from cranial neural crest cells (cNCCs). cNCCs are highly proliferative multipotent stem cells that generate multiple differentiated cell types, including osteoblasts and chondrocytes that form craniofacial bone and cartilage (Fig. 1B). To assess expression of the three mammalian Dnmt genes during initial palatogenesis, maxillary processes were microdissected from wild-type mouse embryos at gestational day (GD)10 and GD11, and epithelial and mesenchymal compartments were isolated by enzymatic separation (Fig. 1 C–E). While Dnmt3b expression was minimal, Dnmt3a and Dnmt1 were readily detectable in the mesenchyme and the epithelium (Fig. 1 F and G). Dnmt1 was prioritized for further investigation because it is required for embryonic development, while Dnmt3a appears dispensable (28). Immunohistochemistry (IHC) demonstrated broad expression of DNMT1 in both the epithelium and mesenchyme throughout the key stages of palatal morphogenesis (Fig. 1 H–J and SI Appendix, Fig. S1).
Fig. 1.
DNMT1 is expressed in the epithelium and mesenchyme throughout palate morphogenesis. (A) A schematic of palatal shelf morphogenesis depicting their emergence as expansions of the maxillary processes (*), vertical growth along the sides of the tongue (T), and subsequent elevation, approximation, and fusion at the midline. (B) A cartoon of the maxillary process showing surface epithelium covering dense mesenchyme composed of multipotent cNCCs that rapidly proliferate and then differentiate into osteoblasts and chondrocytes. (C–E) Facial growth centers, including the maxillary processes (*), in an intact GD11 embryo (C), after microdissection (D), and after enzymatic separation of the epithelium and mesenchyme (E). (F and G) Expression of the three Dnmt genes in the epithelium and mesenchyme of gestational day (GD)10 (F) and GD11 (G) maxillary tissue was assessed by qRT-PCR. Data points representing pooled tissue from independent litters are shown along with mean ± SEM. (H–J’) Immunohistochemical staining for DNMT1 (green) on sections through the median aspect of the developing palate. Sections are also stained for CDH1 (red) to mark epithelium. DAPI (blue) was used to stain nuclei. (Scale bars: 100 µm.)
Dnmt1 in the Cranial Neural Crest Mesenchyme Is Required for Orofacial Morphogenesis.
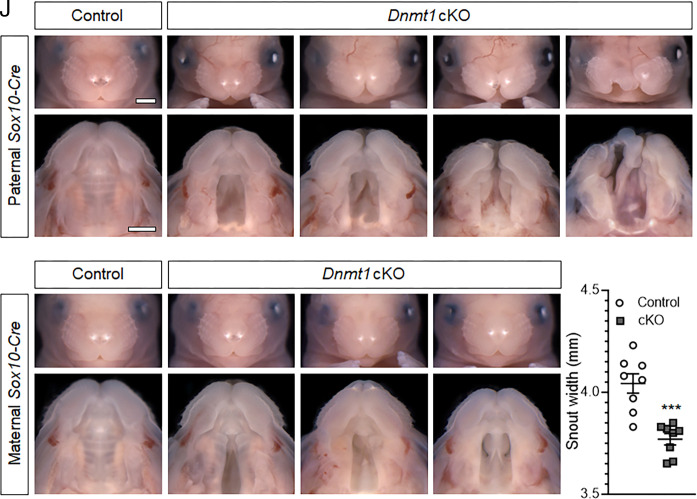
Cre/loxP approaches were then applied to examine the tissue-specific requirement of Dnmt1 during craniofacial morphogenesis. Dnmt1 inactivation was first targeted to the orofacial epithelium via Shh-Cre (30–32), but this had no apparent impact on orofacial development (SI Appendix, Fig. S2). We next targeted Dnmt1 inactivation via Sox10-Cre (33), which drives Cre recombinase activity broadly in the postmigratory cNCC mesenchyme (34–38). Sox10-Cre+;Dnmt1fl/fl(hereafter referred to as Dnmt1 cKO) fetuses exhibited overt craniofacial malformations, including wide spacing of the eyes, notching of the upper lip, and micrognathia, with varying severity (Fig. 2 A–E). Clefts of the secondary palate were observed in all Dnmt1 cKO fetuses (n = 24/24), while a subset (n = 4/24) also exhibited unilateral or bilateral clefts of the upper lip (Fig. 2 A–E and Table 1). Because Cre activity has been reported to vary depending upon parental inheritance, additional litters were generated in which the Sox10-Cre allele was maternally transmitted to complement the litters with paternally transmitted Cre transgene. This cohort of Dnmt1 cKO fetuses also exhibited mild hypertelorism and micrognathia, as well as cleft palate with near complete penetrance (n = 27 of 28) (Fig. 2 F–I and Table 1). While none exhibited overt cleft lip, a significant reduction in snout width was observed, consistent with subcleft phenotypes described in mice and humans (Fig. 2J) (39, 40).
Fig. 2.
Dnmt1 in the cranial neural crest mesenchyme is required for orofacial morphogenesis (A–E’) Representative face and palate images of targeted Dnmt1 deletion from paternally inherited Sox10-Cre of control (Sox10-Cre−;Dnmt1fl/fl) and conditional knockout (cKO, Sox10-Cre+;Dnmt1fl/fl) fetuses at GD17. (Scale bar: 1 mm.) (F–I’) Representative face and palate images of targeted Dnmt1 deletion from maternally inherited Sox10-Cre of control (Sox10-Cre−;Dnmt1fl/fl) and conditional knockout (cKO, Sox10-Cre+;Dnmt1fl/fl) fetuses at GD17. (J) Linear measurements of snout width of control (Sox10-Cre−;Dnmt1fl/fl) and cKO (Sox10-Cre+;Dnmt1fl/fl) fetuses. Data points represent individual fetuses and are shown with mean ± SEM. ***P < 0.001 by an unpaired t test.
Table 1.
Transgenic model sample size and orofacial cleft incidence
| Strain | No. of litters | Genotype | Cleft palate/total no. of fetuses | Cleft lip/total no. of fetuses |
|---|---|---|---|---|
| Paternal Sox10-Cre | 10 | Sox10-Cre − ; Dnmt1 fl/+ | 0/20 (0%) | 0/20 (0%) |
| Sox10-Cre − ; Dnmt1 fl/fl | 0/16 (0%) | 0/16 (0%) | ||
| Sox10-Cre + ; Dnmt1 fl/+ | 0/18 (0%) | 0/18 (0%) | ||
| Sox10-Cre + ; Dnmt1 fl/fl | 24/24 (100%) | 4/24 (17%) | ||
| Maternal Sox10-Cre | 12 | Sox10-Cre − ; Dnmt1 fl/+ | 0/29 (0%) | 0/29 (0%) |
| Sox10-Cre − ; Dnmt1 fl/fl | 0/10 (0%) | 0/10 (0%) | ||
| Sox10-Cre + ; Dnmt1 fl/+ | 0/19 (0%) | 0/19 (0%) | ||
| Sox10-Cre + ; Dnmt1 fl/fl | 27/28 (96%) | 0/28 (0%) | ||
| Paternal Osr2-Cre | 9 | Osr2-Cre − ; Dnmt1 fl/+ | 0/19 (0%) | 0/19 (0%) |
| Osr2-Cre + ; Dnmt1 fl/+ | 0/19 (0%) | 0/19 (0%) | ||
| Osr2-Cre − ; Dnmt1 fl/fl | 0/17 (0%) | 0/17 (0%) | ||
| Osr2-Cre + ; Dnmt1 fl/fl | 0/15 (0%) | 0/15 (0%) |
The Requirement of DNMT Activity Is Temporally Restricted to Early Orofacial Development.
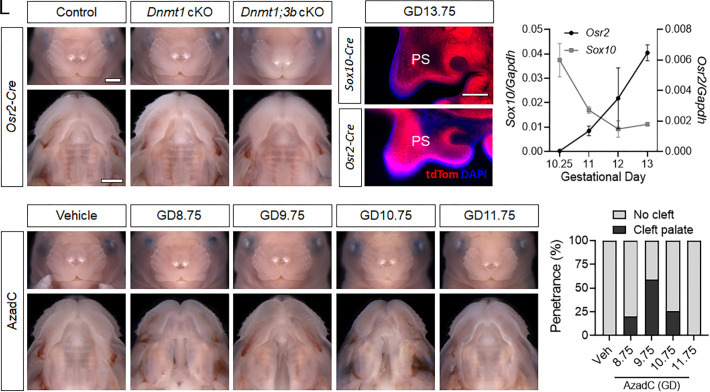
The findings from Dnmt1 cKO fetuses demonstrated that inactivation of Dnmt1 broadly targeted to the postmigratory cNCC mesenchyme causes craniofacial malformations, including cleft palate with near complete penetrance. We next examined the impact of more specific targeting of Dnmt1 using Osr2-Cre, which activates recombinase activity restricted to the palatal mesenchyme during palatal shelf outgrowth (41). In contrast to Sox10-Cre+;Dnmt1fl/flfetuses, orofacial morphogenesis was grossly normal in Osr2-Cre+;Dnmt1fl/flfetuses (Fig. 3 A–B’). This unexpected finding prompted us to assess Osr2-Cre recombinase activity. Consistent with previous reports of Osr2-Cre activity, we found that DNMT1 protein was nearly absent from the palatal mesenchyme but retained in the epithelium in Osr2-Cre+;Dnmt1fl/fltissue (SI Appendix, Fig. S3) (41, 42). Effective reduction of Dnmt1 was further confirmed by qRT-PCR, which also revealed a concurrent increase in Dnmt3b expression (SI Appendix, Fig. S3). We therefore tested potential functional compensation, as observed in other developmental contexts (43), by introducing additional Dnmt3b fl alleles (44). Remarkably, orofacial development was also grossly normal in Osr2-Cre+;Dnmt1fl/fl;Dnmt3b fl/fl fetuses (Fig. 3 C and C’ ).
Fig. 3.
The temporal requirement for Dnmt1 is restricted to initial palate morphogenesis. (A–C’) Representative face and palate images of targeted Dnmt1 deletion or Dnmt1 and Dnmt3b (Dnmt1;3b) double deletion from paternally inherited Osr2-Cre of control (Osr2-Cre−;Dnmt1fl/fl), conditional Dnmt1 knockout (cKO, Osr2-Cre+;Dnmt1fl/fl), and conditional Dnmt1;Dnmt3b double knockout (Osr2-Cre+;Dnmt1fl/fl;Dnmt3b fl/fl) fetuses at GD17. (Scale bar: 1 mm.) (D and E) Representative fluorescent microscopy images of tdTomato (tdTom, red) reporter expression from Sox10-Cre or Osr2-Cre in palatal shelves (PS) at GD13.75. Nuclear staining shown with DAPI (blue). (Scale bar: 0.2 mm.) (F) Expression of Sox10 and Osr2 in isolated maxillary/palatal shelf mesenchyme at the indicated stage of gestation as assessed by qRT-PCR. Data points represent the mean ± SEM of n = 3 samples per time point. (G–K’) Face and palate images of wild-type fetuses at GD17 exposed to vehicle (PBS) or 5-aza-2′-deoxycytidine (AzadC) at the indicated gestational day. (L) Penetrance of cleft palate in wild-type GD17 fetuses exposed to vehicle (veh) or AzadC between GD8.75 and 11.75.
The discrepant impact of Sox10-Cre- versus Osr2-Cre-mediated Dnmt1 targeting suggested either spatial domain or timing of DNMT activity in dictating normal versus abnormal palate morphogenesis. Lineage reporter assays confirmed that both Sox10-Cre and Osr2-Cre alleles produced effective recombination in the palatal mesenchyme by GD13.75 (Fig. 3 D and E), leading us to assess expression of Sox10 versus Osr2 in the maxillary/palatal mesenchyme during earlier stages of palate morphogenesis. Sox10 expression was highest at GD10 and remained relatively high throughout the time course (Fig. 3F). In contrast, Osr2 expression was minimal at GD10 and then increased with progressive developmental stage. This quantitative assessment was consistent with in situ hybridization (ISH) staining, which demonstrated clear domains of Osr2 expression in the maxillary processes by GD11.75, but not at earlier time points (SI Appendix, Fig. S4).
Findings from Cre-mediated models of Dnmt1 inactivation suggested that the requirement for DNMT activity in the cNCC mesenchyme may be stringently dependent upon developmental timing. Application of tamoxifen-inducible Cre for temporally controlled gene inactivation was not pursued to address the question because of recent evidence that tamoxifen exposure itself can cause cleft palate (45, 46). Instead, the temporal role of DNMT activity was examined by administering a single dose of the potent pharmacological DNMT enzyme inhibitor 5-aza-2′-deoxycytidine (AzadC) to timed pregnant wild-type C57BL/6J mice between GD8.75 and 11.75. Exposure to AzadC at the three earliest time points resulted in secondary palate clefts, with the highest penetrance (59%) following exposure at GD9.75 (Fig. 3 G–L and Table 2). Exposure to the same dose of AzadC at GD11.75 did not result in clefts.
Table 2.
Teratogen-exposure model sample size and orofacial cleft incidence
| Treatment (gestational day) | No. of litters | Genotype | Cleft palate/total no. of fetuses | Cleft lip/total no. of fetuses |
|---|---|---|---|---|
| Veh (9.75) | 4 | C57BL/6 J | 0/19 (0%) | 0/19 (0%) |
| Veh (11.75) | 2 | C57BL/6 J | 0/12 (0%) | 0/12 (0%) |
| AzadC (8.75) | 4 | C57BL/6 J | 8/24 (33%) | 0/24 (0%) |
| AzadC (9.75) | 12 | C57BL/6 J | 30/51 (59%) | 0/51 (0%) |
| AzadC (10.75) | 5 | C57BL/6 J | 6/31 (19%) | 0/31 (0%) |
| AzadC (11.75) | 4 | C57BL/6 J | 0/25 (0%) | 0/25 (0%) |
Dnmt1 cKO Reduces cNCC DNA Methylation during the Critical Period of OFC Sensitivity.
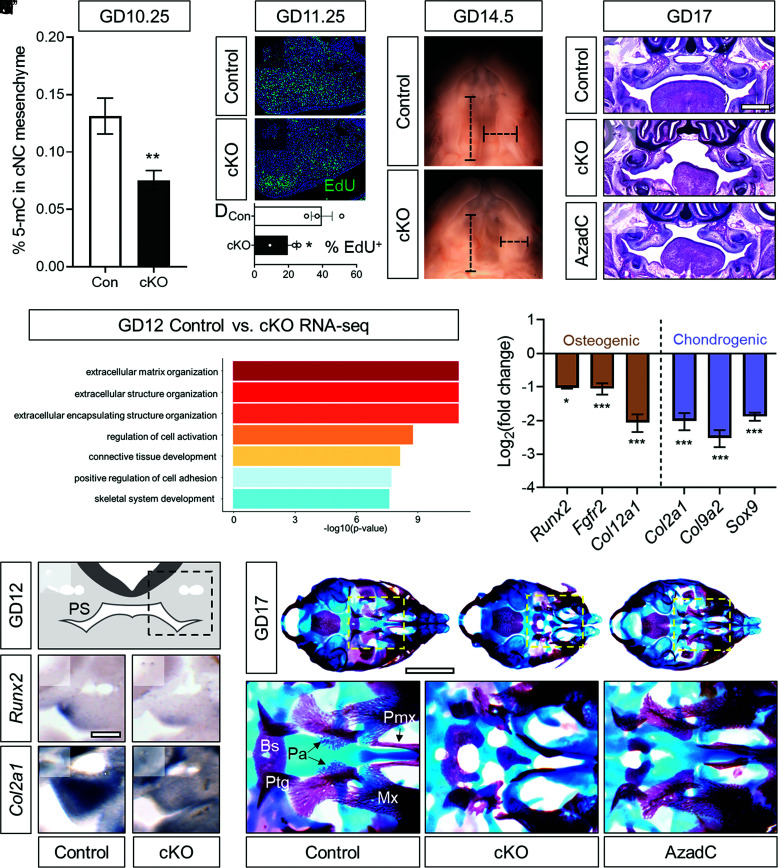
Findings from the experimental genetic and pharmacological disruption of DNMT activity suggested that DNA methylation in cNCCs is required during initial palate morphogenesis, prior to palatal shelf elevation and elongation. In this working model, genetic loss of Dnmt1 would impact DNA methylation in the cNCC on or shortly after GD9.75. We tested this premise by quantifying global 5-methylcytosine (5-mC) levels by ELISA assay. 5-mC levels were significantly reduced in maxillary mesenchymal tissue from Dnmt1 cKO embryos relative to controls at GD10.25 (Fig. 4A). These findings confirmed that conditional inactivation of Dnmt1 via Sox10-Cre effectively reduces global DNA methylation in the cNCC mesenchyme during initial palate morphogenesis.
Fig. 4.
DNA methylation regulates cNCC proliferation and differentiation. (A) Global DNA methylation as assessed by 5-mC ELISA in maxillary process mesenchyme tissue of control (Con, Sox10-Cre+;Dnmt1fl/+) and conditional knockout (cKO, Sox10-Cre+;Dnmt1fl/fl) embryos at GD10.25. N = 7 embryos were used for each genotype, and means ± SEM are shown. **P < 0.01 compared to control by unpaired t test. (B and C) Representative examples of EdU incorporation in sections through the maxillary processes of control (Sox10-Cre+;Dnmt1fl/+) and conditional knockout (cKO, Sox10-Cre+;Dnmt1fl/fl) embryos. (D) Quantification of percent EdU-positive cells. N = 3 embryos were used for each genotype, with data points representing individual embryos shown along with mean ± SEM. *P < 0.05 by the t test. (E and F) Light images of the palatal shelves of GD14.5 control (Sox10-Cre−;Dnmt1fl/+) and cKO (Sox10-Cre+;Dnmt1fl/fl) fetuses showing representative linear measurements of palatal shelf width and length. (G–I) Representative H&E-stained coronal sections showing the region of the palatal shelves in control, cKO (Sox10-Cre+;Dnmt1fl/fl), or AzadC-treated (GD9.75) fetuses. (Scale bar: 0.5 mm.) (J) GO analysis from RNA sequencing conducted on palatal shelves isolated from control (Sox10-Cre+;Dnmt1fl/+) and cKO (Sox10-Cre+;Dnmt1fl/fl) embryos at GD12. (K) Expression of established cNC osteogenic and chondrogenic markers from RNA sequencing conducted on palatal shelves isolated from control (Sox10-Cre+;Dnmt1fl/+) and cKO (Sox10-Cre+;Dnmt1fl/fl) embryos at GD12. *P < 0.05 and ***P < 0.001. (L) Schematic of a GD12 section showing the palatal shelves (PS) with the dashed box indicating field of view of (M–P). (M–P) In situ hybridization for Runx2 and Col2a1 was performed on 100 µm vibratome sections of GD12 control (Sox10-Cre−;Dnmt1fl/fl) and cKO (Sox10-Cre+;Dnmt1fl/fl) embryos. (Scale bar = 0.2 mm.) (Q–S’) Whole-mount bone and cartilage staining was performed on control (Sox10-Cre−;Dnmt1fl/+), cKO (Sox10-Cre+;Dnmt1fl/fl), or AzadC-treated (GD9.75) GD17 fetuses with Insets (dashed boxes) showing higher magnification of the palatine bone region. (Scale bar = 2 mm.) Bs, basisphenoid; Ptg, pterygoid process; Pa, palatine bones; Pmx, premaxilla; Mx, maxilla.
DNA Methylation Regulates cNCC Proliferation and Differentiation.
Following colonization, postmigratory cNCCs proliferate, condense, and begin differentiating, first to a common osteochondral progenitor and then into more specific chondrocytes and osteoblasts (47). Disruption of the normal cNCC proliferation/differentiation program can result in craniofacial malformations, including OFCs (47). Apparent palatal shelf hypoplasia observed in Dnmt1 cKO fetuses prompted us to first examine cellular proliferation, which was assessed by EdU incorporation assay on GD11.25 embryos. The percentage of EdU-positive cells in the maxillary mesenchyme of Dnmt1 cKO embryos was significantly reduced relative to controls (Fig.4 B–D). Consistent with the reduction in cellular proliferation observed at GD11.25, Dnmt1 cKO embryos displayed attenuated outgrowth of the maxillary-derived palatal shelves that became clearly evident between GD12 and 13 (SI Appendix, Fig. S5). By GD14.5, when palatal shelves have normally approximated at the midline, those in Dnmt1 cKO mutant mice were significantly reduced in length and width (Fig. 4 E and F and SI Appendix, Fig. S6). Similar growth deficiency, particularly affecting palatal shelf width, was observed in wild-type mice following exposure to the pharmacologic DNMT inhibitor AzadC at GD9.75 (SI Appendix, Fig. S7). Histological assessment at GD17 illustrated gross hypoplasia of the palatal shelves in both Dnmt1 cKO and AzadC-exposed wild-type fetuses, with the former appearing more severely affected (Fig. 4 G–I).
To identify additional cellular mechanisms triggered by disruption of DNA methylation and involved in cleft pathogenesis, bulk RNA-seq was conducted on microdissected palatal shelf tissue from Dnmt1 cKO and control embryos at GD12. Applying a stringent FDR value of q < 0.05 yielded 65 differentially expressed genes (SI Appendix, Table S1), from which gene ontological (GO) analysis was conducted. GO terms centered around extracellular matrix and cartilage and bone development (Fig. 4J) led us to assess the impact of DNA methylation disruption on cNCC differentiation. Examination of RNA-seq data confirmed reduced expression levels of established cNCC osteogenic (i.e., Runx2, Fgfr2, and Col12a1) and chondrogenic (i.e., Col2a1, Col9a2, and Sox9) lineage markers in Dnmt1 cKO palatal mesenchyme tissue at GD12 (Fig. 4K). Supporting these quantitative results, reduced intensity and domain of Runx2 and Col2a1 expression was observed in the nascent palatal shelf tissue of Dnmt1 cKO embryos (Fig. 4 L–P). The impact of these changes on bone and cartilage development was then assessed in GD17 fetuses. Alizarin red and Alcian blue staining revealed defects of several cNCC-derived skeletal structures in Dnmt1 cKO mice, including the basisphenoid, maxilla, premaxilla, and palatal processes of the maxilla (Fig. 4 Q–S’). Similar, albeit less severe defects were observed in AzadC-exposed wild-type mice with cleft palate.
Regulation of cNCCs by DNA Methylation Can Be Recapitulated In Vitro.
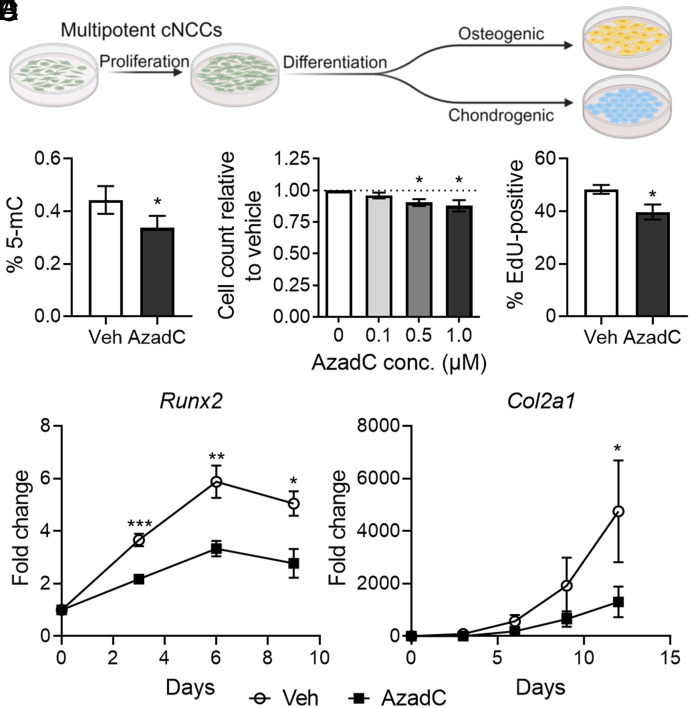
Conservation of molecular, cellular, and morphological aspects of embryogenesis makes the mouse a powerful model to investigate human development. However, lower-cost and higher-throughput in vitro platforms are better suited for efforts to identify environmental influences that alter DNA methylation and cNCC biology. We therefore tested whether the regulation of cNCC proliferation and differentiation by DNA methylation observed in mouse orofacial morphogenesis could be recapitulated in a more tractable in vitro system. To address this question, we leveraged an established murine cNCC line (O9-1) that retains the neural crest and stem cell marker signature and multipotency of in vivo cNCCs (48). These cells are highly proliferative and can be differentiated along a trajectory that parallels in vivo differentiation, first to a common osteochondral progenitor and then toward osteogenic or chondrogenic lineages (Fig. 5A) (48). In cultured cNCCs, exposure to 1 µM AzadC for 48 h significantly reduced 5-mC levels, indicating effective reduction of global DNA methylation (Fig. 5B). The same duration of AzadC exposure resulted in a concentration-dependent decrease in cell count (Fig. 5C). Parallel assays demonstrated a significant decrease in EdU-positive cNCCs after 12 h of exposure to AzadC, suggesting that decreased proliferation drives the reduced cell number observed after 48 h (Fig. 5D). To assess impact on differentiation, the same experimental approach was repeated, but after 48 h of exposure, AzadC was removed, and cNCCs were placed under osteogenic or chondrogenic differentiation conditions (SI Appendix, Fig. S8). Effective osteogenic and chondrogenic differentiation under control conditions was confirmed by observed increases in expression of the same markers used to assess in vivo differentiation (Fig. 5 E and F and SI Appendix, Fig. S8). Relative to those treated with vehicle, cNCCs exposed to AzadC prior to initiation of the differentiation protocol exhibited reduced expression of both osteogenic and chondrogenic markers. These observations suggest that the regulation of proliferation and differentiation by DNA methylation observed in vivo can be recapitulated in cNCCs cultured in vitro.
Fig. 5.
DNA methylation regulates cNCC proliferation and differentiation in cultured cNCCs. (A) Schematic depicting the proliferative and differentiation capacity of cNCCs cultured in vitro and experimental conditions to evaluate the impact of pharmacological DNMT inhibition. (B) Global DNA methylation as assessed by 5-mC ELISA in cultured cNCCs treated with PBS (vehicle, veh) or 1.0 µM AzadC for 48 h. N = 6 for each group with mean ± SEM shown. *P < 0.05 by the t test. (C) Cell counts were performed after 48-h treatments with the indicated concentrations of AzadC. N = 5 for each concentration and means ± SEM are shown. *P < 0.05 compared to the 0 µM AzadC group by ANOVA with Dunnett’s post hoc test for multiple comparisons. (D) EdU-positive cells were counted after 12 h of PBS (vehicle, veh) of 1.0 µM AzadC treatment. N = 5 biological replicates for each group and means ± SEM are shown. *P < 0.05 by the t test. (E and F) Cultured cNCCs were treated with PBS (vehicle, veh) or 1.0 µM AzadC for 48 h before undergoing differentiation conditions for osteogenic (E) or chondrogenic (F) differentiation. Expression of Runx2 or Col2a1 was assayed at the indicated days after the start of differentiation. N = 5 for each group at each time point and means ± SEM are shown. *P < 0.05, **P < 0.01, or ***P < 0.001 compared to the vehicle group at the same time point by t test.
Discussion
Understanding how development is influenced through malleable epigenetic mechanisms could directly inform strategies to prevent birth defects like OFCs. Inherent environmental sensitivity makes DNA methylation a particularly attractive focus for these efforts, but the role of this epigenetic mechanism in orofacial development was unclear. In this study, we present direct evidence that DNA methylation regulates orofacial development and demonstrate that its disruption results in craniofacial malformations, including OFCs. Genetic and pharmacological interrogation revealed a specific requirement in the postmigratory cNCC mesenchyme during a narrow, sensitive developmental window. Leveraging in vivo OFC models, we found that DNA methylation regulates cNCC proliferation and subsequent differentiation and that these cellular mechanisms of OFC pathogenesis can be recapitulated in multipotent cNCCs cultured in vitro.
While the requirement of DNMT activity for early embryogenesis is well established, initial evidence suggested that it may be dispensable for cNCC biology and orofacial development. While global deletion of Dnmt3b results in early embryonic lethality, mice with conditional Dnmt3b deletion targeted to the neural crest via Wnt1-Cre or Sox10-Cre exhibited normal craniofacial development (36). On the other hand, we found that Sox10-Cre-driven conditional deletion of Dnmt1 overtly disrupts craniofacial development and results in OFCs. These discrepant results may be explained by DNMT-specific compensatory capacity or a unique requirement of DNMT1-regulated maintenance methylation in postmigratory cNCCs during early orofacial morphogenesis. The phenotype observed in Dnmt1 cKO mice is also consistent with the observation here and in previous studies that systemic in utero AzadC exposure can cause cleft palate in rodent models (49–52). While AzadC is known to have off-target effects (53, 54), the degree of phenotypic overlap in Dnmt1 cKO mice to AzadC-exposed wild-type mice observed in this study suggests that the teratogenic effect of AzadC is mediated through its well documented mechanism of DNMT enzyme inhibition.
The genetic and pharmacologic interrogations of DNA methylation employed in this study defined a narrow developmental period in which disruption of DNMT activity in cNCCs causes OFCs. Early targeting of Dnmt1 via Sox10-Cre resulted in OFCs with near complete penetrance, while later targeting via Osr2-Cre did not. Accordingly, we found that AzadC administration at GD9.75 led to peak cleft incidence, while no clefts resulted from exposure just 48 h later. Collectively, these findings place the most sensitive window for disruption of DNA methylation around GD10. Narrowing of the critical period for DNA methylation in orofacial development provides valuable context for future efforts to define the specific molecular mechanisms by which DNA methylation regulates cNCC biology. This developmental contextualization also has important implications for understanding environmental influences in human OFC risk, as GD10 in the mouse corresponds approximately to the beginning of the fifth week of human development, when many pregnancies may not yet be recognized.
After demonstrating the requirement of Dnmt1 in the cNCC mesenchyme, focus was placed on DNA methylation-regulated cellular mechanisms of OFC pathogenesis. The neural crest originates as neuroectodermal cells at the dorsal margins of the anterior neural folds that undergo epithelial-to-mesenchymal transition and migrate into the field of facial morphogenesis. Once colonization is complete around GD9.5, cNCCs rapidly proliferate and condense, forming the mesenchyme of the facial growth centers. By GD11.5, postmigratory cNCCs are undergoing fate determination and begin differentiating, first to a common osteochondral progenitor and then into more specific chondrocytes and osteoblasts (47). By narrowing the critical period to around GD10, our studies suggest that DNA methylation is particularly critical when postmigration cNCCs are rapidly proliferating and at the onset of differentiation. Indeed, we found that disruption of DNA methylation was immediately followed by reduced cNCC proliferation, leading to attenuated outgrowth of the palatal shelves. These observations from Dnmt1 cKO mice are consistent with a previous investigation showing that AzadC exposure reduces cell proliferation in the epithelium and mesenchyme of the facial growth centers that form the palate (55). Here, we further reveal that reduced cNCC proliferation is followed by attenuated differentiation capacity. The present study provides instructive context and tools for future investigation of how DNA methylation regulates the balance of cNC stem cell proliferation and differentiation during orofacial morphogenesis.
Consistent with a critical role of Dnmt1 during initial orofacial morphogenesis, we found that global DNA methylation was significantly reduced in Dnmt1 cKO mice at GD10.25. However, neither assessment of global 5-mC levels at GD10.25 or RNA-seq analyses at GD12 were directed toward assessing specific DNA methylation–regulated transcriptional events that drive cNCC biology and cleft pathogenesis. For example, whether the expression of identified osteogenic and chondrogenic genes is directly regulated by DNMT1-mediated DNA methylation or by DNA methylation-regulated upstream mechanisms is not known. Addressing these knowledge gaps will require future investigation that should benefit from the conceptual and experimental advances presented here. A growing body of literature identifies DNA methylation differences between human cohorts with and without OFCs (14–22). While suggesting that DNA methylation status influences human orofacial cleft risk, these studies rely on surrogate tissue collected long after orofacial morphogenesis is complete and cannot discern the impact of DNA methylation changes on gene expression during orofacial development. Identification of DNA methylation events that regulate transcription and directly influence cNCC biology and orofacial morphogenesis will require tractable models like those presented in the present study. For example, applying DNA methylation and RNA sequencing in tandem to the Dnmt1 cKO model could define correlated methylome and transcriptome changes involved in OFC pathogenesis. Modified CRISPR/Cas9-based approaches for epigenome editing could also be leveraged to functionally assess the impact of specific OFC-associated methylation events on cNC biology.
The in vivo and in vitro models presented here also provide opportunities to define environmental modulators of DNA methylation that influence OFC risk. DNA methylation is inherently malleable and influenced by diverse environmental factors, including maternal stress, diet, and exposure to drugs, toxins, and environmental pollutants (56–61). Several specific environmental DNA methylation modifiers are already linked to OFC risk. For example, supplementation of the dietary methyl donors folic acid and choline appears to reduce the risk for OFCs, while first-trimester exposure to folic acid antagonists appears to increase OFC risk (15, 16, 20, 21, 62, 63). Another example supporting this relationship is the common environmental contaminant cadmium, which has been shown to reduce DNA methylation, possibly by inhibiting DNMT activity (59, 64, 65). Prenatal cadmium exposure can cause OFCs in rodent models, while human studies have found that umbilical cord and placental tissue cadmium concentrations are associated with increased OFC risk (66–69). Comparison of molecular drivers of OFC pathogenesis in Dnmt1 cKO mice against those elicited by agents like folic acid, choline, and cadmium should help delineate specific methylome and transcriptome responses that modulate OFC susceptibility. This knowledge could also be harnessed to identify additional environmental influences that may alter OFC risk by modulating DNA methylation. Indeed, demonstration that cultured cNCCs recapitulate the cellular mechanisms of OFC pathogenesis supports the use of this tractable model as a high throughput screening platform to identify additional environmental factors that influence cNCC biology via DNA methylation. Collectively, the experimental and conceptual advances gained from the present study provide a foundation to functionally define environmental- and dietary-induced methylome-transcriptome responses that alter cNC biology and modulate OFC susceptibility.
Materials and Methods
Mouse Strains.
The following strains of mice were used: C57BL/6J (wild type, The Jackson Laboratory strain number 000664), Sox10-Cre (B6;CBA-Tg(Sox10-cre)1Wdr/J, The Jackson Laboratory strain number 025807), Osr2-Cre (B6;129S1-Osr2tm2(cre)Jian/J, The Jackson Laboratory strain number 009388, https://www.ncbi.nlm.nih.gov/pubmed/17941042), Dnmt1fl (B6.129S4-Dnmt1tm2Jae/Mmucd, Mutant Mouse Resource & Research Centers strain number 014114-UCD, https://www.ncbi.nlm.nih.gov/pubmed/11137995), Dnmt3bfl (B6;129S4-Dnmt3btm1Jae/Mmnc, MMRRC strain number 31043), and R26tdTomato (Ai9, B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, The Jackson Laboratory strain number 007909). Mice were genotyped using the Phire Tissue Direct PCR Master Mix (ThermoFisher Scientific) according to manufacturer recommendations using the primer sequences listed in SI Appendix, Table S2.
Animal Husbandry.
This study was conducted in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was approved by the University of Wisconsin School of Veterinary Medicine Institutional Animal Care and Use Committee (protocol number V005396). Mice (Mus musculus) were housed in specific pathogen-free conditions in disposable, ventilated cages (Innovive). Rooms were maintained at 22 ± 2 °C and 30 to 70% humidity on a 12-h light, 12-h dark cycle. Mice were fed Irradiated Soy Protein-Free Extruded Rodent Diet (Catalog No. 2920x; Envigo Teklad Global) until day of plug. Pregnant mice were then were fed Irradiated Teklad Global 19% Protein Extruded Rodent Diet (Catalog No. 2919; Envigo Teklad Global).
Animal Study Design.
For timed matings, one or two nulliparous female mice were placed with a single male mouse for 1 to 2 h and then examined for copulation plugs. The beginning of the mating period was designated as gestational day (GD)0, and pregnancy was confirmed by assessing weight gain between GD7 and GD10, as previously described (70). Dams were killed by carbon dioxide inhalation followed by cervical dislocation between GD10-14 ± 1 h for embryo collection or GD14.5-19 ± 2 h for fetal collection.
Separation of mesenchyme and surface ectoderm of GD10, GD10.25, and GD11 maxillary processes was performed as previously described (71, 72). Tissues from entire litters were pooled for GD10 and GD11 wild-type samples, and N = 4 litters were used for subsequent RNA expression analyses. Paired maxillary process mesenchyme tissues were combined from individual GD10.25 Dnmt1 cKO and control embryos, and N = 7 embryos of each genotype were used for subsequent DNA analyses. Timed-pregnant wild-type dams were dosed intraperitoneally with 0.5 mg/kg body weight 5-aza-2′-deoxycytidine (AzadC, MP Biomedicals, catalog number IC15480305) dissolved in PBS, or PBS alone (vehicle), on GD8.75, 9.75, 10.75, or 11.75 ± 20 min.
Morphometric Assessment.
Snout width morphometric assessment was conducted on GD17 fetuses fixed in 10% formalin. Frontal images of heads were taken using a Micropublisher 5.0 camera (QImaging) mounted on an Olympus SZX-10 stereomicroscope. Snout width was measured using ImageJ by a single investigator blinded to fetus genotype. Palatal shelf morphometric assessment was conducted on GD14.5 fetuses fixed in 10% formalin. Cuts between the upper and lower jaw were made with a scalpel to expose the roof of the oral cavity, which was then imaged with a Micropublisher 5.0 camera (QImaging) mounted on an Olympus SZX-10 stereomicroscope. Images were analyzed for palatal shelf elevation, growth, and fusion to assess for clefting pathogenesis. Linear measurements of palatal shelf length and width were performed using ImageJ. Fetuses were excluded from this analysis if connective tissue obstructed clear ascertainment of palatal borders or if the initial cut removed palatal tissue from either shelf. Palatal shelf measurements were performed by a single investigator blinded to fetus genotype.
Immunohistochemistry.
Mouse embryos and fetuses at indicated time points were fixed in 4% paraformaldehyde overnight and dehydrated through a graded series into 100% methanol for storage at −20 °C. Embryos and fetuses were rehydrated, and immunohistochemistry was performed as previously described (73). The following primary and secondary antibodies were used: 1:50 rabbit anti-DNMT1 (#5032, Cell Signaling Technologies), 1:200 mouse anti-CDH1 (BD 610181), 1:250 DyLight 488-conjugated goat anti-mouse IgG, and 1:250 DyLight 594-conjugated goat anti-rabbit IgG (#35502 and #35560, Jackson ImmunoResearch). Sections were imaged using a Keyence BZ-X700 (Keyence) fluorescence microscope.
Endogenous Reporter Imaging.
Embryos were fixed in 4% PFA for 1 h at room temperature and embedded in 4% agarose. Sections (100 μm) were produced using a vibrating microtome, incubated with DAPI, and mounted on microscope slides with Vectashield. Images were taken on a Keyence BZ-X700 (Keyence) fluorescence microscope.
EdU Proliferation on Tissue Sections.
Pregnant dams were dosed via IP injection with 5 mg/kg EdU in PBS 1 h prior to harvest at GD11.25. Embryos were fixed in 4% paraformaldehyde in PBS overnight prior to graded dehydration into methanol and storage at −80 °C. Embryos were subsequently rehydrated into PBS with 0.1% Triton X-100, paraffin processed, and embedded. Immunostaining was performed on 5-μm sections using the Alexa Fluor 488 Click-iT EdU Imaging Kit (C10337; ThermoFisher Scientific) according to manufacturer recommendations. Sections were imaged at 20× magnification on a Keyence BZ-X700 series microscope. The BZ-X analyzer program was used to reduce background and optimize signal accuracy. A rectangular section spanning from the nasal pit to maxillary process ectoderm was selected for analysis. EdU-positive cells/Hoechst-positive cells in each image were determined using the macro cell count batch analysis tool.
In Situ Hybridization.
Riboprobes were synthesized with gene-specific primers (listed in SI Appendix, Table S3), and in situ hybridization was performed on 100-μm vibratome sections of GD12 embryos as previously described (74, 75). Images were captured using a MicroPublisher 5.0 camera (QImaging) mounted on an Olympus SZX-10 stereomicroscope.
Hematoxylin and Eosin Staining.
Fetuses were collected at GD17 and fixed in Bouin’s Solution. After paraffin embedding, 10-μm sections were produced and stained with hematoxylin and eosin using standard protocols. Slides were imaged with a MicroPublisher 5.0 camera (QImaging) mounted on an Olympus SZX-10 stereomicroscope.
Bone and Cartilage Staining.
GD17 fetuses were fixed in 100% ethanol, and bone and cartilage staining was performed as previously described (46). Images were captured using a MicroPublisher 5.0 camera (QImaging) mounted on an Olympus SZX-10 stereomicroscope.
Tissue Microdissection and RNA Isolation for RNA Sequencing.
Palatal shelves from GD12 embryos were microdissected under a stereomicroscope, snap-frozen in liquid nitrogen, and then stored at −80 °C. Embryos were genotyped, and palatal shelves from two embryos of the same genotype were pooled for RNA extraction. Three RNA samples (from six embryos) for each group were used for subsequent sequencing. No more than three embryos of the same genotype were used from a single litter, and embryos from at least three litters were included for each genotype. RNA was isolated using the Illustra RNAspin kit (GE) using the manufacturer recommendations with on-column DNase I digestion. RNA was eluted in 30 μL of water, and RNA concentration was quantified using a Qubit fluorometer (ThermoFisher Scientific).
RNA Library Preparation and Sequencing.
Approximately 100 ng of total RNA from three pooled RNA samples (from six embryos) for each group was used for sequence library construction following instructions of the NuGen mRNA sample prep kit (cat# 0348). In brief, total RNA was copied into first strand cDNA using reverse transcriptase and random primers. This was followed by second strand cDNA synthesis using DNA Polymerase I and RNaseH. These cDNA fragments went through an end repair process, the addition of a single “A” base, and then ligation of the adapters. These products were gel purified and enriched with PCR to create the final cDNA libraries. The library constructs were run on the bioanalyzer to verify the size and concentration before sequencing on the Illumina HiSeq2500 machine where 100-cycle single-end sequencing was performed by the University of Wisconsin Biotechnology Center.
RNAseq Processing and Analysis.
An average of 31.5 million paired-end reads were sequenced per sample. An initial check of quality was assessed for each pair-mate using FastQC. After reads were assessed for quality, paired-end reads were trimmed for adapter contamination using trim_galore run with default parameters. Trimmed and filtered reads were aligned to the Mus musculus genome (mm10) using RSEM v1.3.1 (76), which utilized STAR v2.7.0 (77). An average of 27.3 million reads passed quality filtering through STAR, and an average of 23.4 million reads (85.6%) were successfully aligned for each sample. Gene expression was calculated using RSEM which employed a forward probability of 0.0, followed by differential expression analysis using DESeq2 (78). To account for multiple comparisons and to reduce false positives, a false discovery rate (FDR) of 0.05 was employed in the determination of differentially expressed genes. Gene ontological analyses were performed using R package clusterProfiler (79), using an FDR threshold of 0.05 to determine significant enrichment of biological processes.
Global DNA Methylation Assay.
DNA was isolated from paired maxillary process mesenchyme tissue from GD10.25 embryos (N = 7 samples) using a QIAamp DNA Micro Kit (Qiagen) or from cells (N = 6 biological replicates) using Wizard SV DNA Isolation Kit (Promega) according to manufacturer recommendations and quantified with NanoDrop (ThermoFisher). Global 5-mC levels were assayed using a MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit (EpigenTek) following manufacturer recommendations.
Gene Expression Analysis.
RNA was isolated using the Illustra RNAspin (GE) or RNeasy Mini kit (Qiagen) with on-column DNase I digestion according to the manufacturer recommendations. cDNA was synthesized from 100 to 500 ng of total RNA using the GoScript reverse transcription reaction kits (Promega). Singleplex quantitative real-time PCR (qRT-PCR) was performed using SSoFast EvaGreen Supermix (Bio-Rad) on a Bio-Rad CFX96 real-time PCR detection system (Bio-Rad Laboratories). qRT-PCR primers were designed using PrimerQuest (IDT), and sequences are listed in SI Appendix, Table S4. Target gene specificity was confirmed using the National Center for Biotechnology Information Primer Basic Local Alignment Search Tool (NCBI Primer-BLAST). Gapdh was used as the housekeeping gene, and analyses were conducted with the 2−ΔΔCt method.
Cell Culture.
O9-1 cells, a mouse cNCC line, were cultured as described previously (48). For AzadC treatment, O9-1 cells were plated at 1.25 × 105 cells/mL (0.4 mL per well in a 24-well plate) and allowed to attach for 24 h before media were replaced with complete medium ± AzadC (0, 0.1, 0.5, or 1 μM in PBS).
After 48 h of AzadC treatment, cell counts were performed with a CyQUANT Cell Proliferation Assay (ThermoFisher) according to manufacturer recommendations. For EdU proliferation assays, cells were treated with 10 µM EdU after 8 h of AzadC treatment and fixed in 4% PFA for 15 min after 12 h of AzadC treatment. EdU detection was performed using a Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 488 dye (ThermoFisher) according to manufacturer recommendations. A Keyence BZ-X700 microscope was used for fluorescent imaging, and the Keyence BZ-X Image Analyzer software was used to count EdU-positive nuclei. Nuclei were counted in two random fields in technical duplicates and averaged for each sample. N = 5 biological replicates were performed in each treatment group for each assay.
cNCC Osteogenic and Chondrogenic Differentiation.
After 48 h of AzadC or vehicle (PBS) treatment, O9-1 cells were differentiated as follows. For osteogenic differentiation, media were changed to osteogenic differentiation media (α-MEM, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 0.1 μM dexamethasone, 10 mM β-glycerophosphate, 50 μg/mL ascorbic acid, and 100 ng/mL BMP2). For chondrogenic differentiation, cells were initially treated with osteogenic differentiation media for 3 d. Then, cells were trypsinized and cultured as pellets by centrifuging 500,000 cells in 1.5-mL microcentrifuge tubes at 2,000 g for 5 min. Holes were made in the tube caps with a 21-G needle to allow for gas exchange. Media were changed to chondrogenic differentiation media (α-MEM, 5% FBS, 1% ITS, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 ng/mL TGF-β3, 50 μg/mL ascorbic acid, 10 ng/mL BMP2, 0.1 μM dexamethasone, and 1 mM sodium pyruvate) without disturbing the cell pellet. Osteogenic or chondrogenic differentiation media were changed every other day during differentiation.
Statistics.
All statistical analyses were performed using GraphPad Prism 9. Unpaired, two-tailed t tests were used to compare snout widths, palatal shelf widths, palatal shelf lengths, and % 5-mC in maxillary process tissue. Paired, two-tailed t tests were used to compare RNA expression in tissues and cultured cells, % 5-mC in cultured cells, and EdU-positive cells. One-way ANOVA with Dunnett’s post hoc test for multiple comparisons was used to analyze cell counts in cNCCs after AzadC treatments. An alpha value of 0.05 was maintained for determination of significance.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Research reported in this publication was supported by the NIH under award numbers R03DE027162, R56DE030917, R01DE032710, U01 DK110807, R01DK099328, and T32ES007015. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Support was also provided by the University of Wisconsin Hilldale Undergraduate Research Award. We thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility and the BRMS Mouse Breeding Core and Research Services for their support and expertise and Dr. Rulang Jiang for generously providing Osr2-Cre mice. We also thank Tyler Beames and Jacob Kracke-Bock for careful review of the manuscript.
Author contributions
C.M.U., M.R.S., C.M.V., R.S.A., and R.J.L. designed research; C.M.U., M.R.S., C.R.J., A.C.S., K.S.R.-G., J.C., A.A.M., M.B., L.W., and D.B.J. performed research; C.M.U., M.R.S., C.R.J., A.C.S., K.S.R.-G., J.C., A.M., L.A.P., and R.S.A. analyzed data; and R.J.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
RNA-seq data have been deposited in Gene Expression Omnibus and are available under accession number GSE250370 (80).
Supporting Information
References
- 1.Rahimov F., Jugessur A., Murray J. C., Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 49, 73–91 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulet S. L., Grosse S. D., Honein M. A., Correa-Villaseñor A., Children with orofacial clefts: Health-care use and costs among a privately insured population. Public Health Rep. 124, 447–453 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semb G., Brattström V., Mølsted K., Prahl-Andersen B., Shaw W. C., The Eurocleft study: Intercenter study of treatment outcome in patients with complete cleft lip and palate. Part 1: Introduction and treatment experience. Cleft Palate Craniofac J. 42, 64–68 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Wehby G. L., et al. , Academic achievement of children and adolescents with oral clefts. Pediatrics 133, 785–792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wehby G. L., Cassell C. H., The impact of orofacial clefts on quality of life and healthcare use and costs. Oral Dis. 16, 3–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leslie E. J., Marazita M. L., Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C Semin. Med. Genet. 163C, 246–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon M. J., Marazita M. L., Beaty T. H., Murray J. C., Cleft lip and palate: Understanding genetic and environmental influences. Nat. Rev. Genet. 12, 167–178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krauss R. S., Hong M., Gene-environment interactions and the etiology of birth defects. Curr. Top. Dev. Biol. 116, 569–580 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Lovely C., Rampersad M., Fernandes Y., Eberhart J., Gene-environment interactions in development and disease. Wiley Interdiscip Rev. Dev. Biol. 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray J. C., Gene/environment causes of cleft lip and/or palate. Clin. Genet. 61, 248–256 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Marazita M. L., Gene×environment associations in orofacial clefting. Curr. Top. Dev. Biol. 152, 169–192 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Juriloff D. M., Harris M. J., Mouse genetic models of cleft lip with or without cleft palate. Birth Defects Res. A Clin. Mo l. Teratol. 82, 63–77 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Seelan R. S., Pisano M., Greene R. M., Nucleic acid methylation and orofacial morphogenesis. Birth Defects Res. 111, 1593–1610 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Mendonca V. J., Maternal folic acid intake and risk of nonsyndromic orofacial clefts: A hospital-based case-control study in Bangalore, India. Cleft Palate Craniofac. J. 57, 678–686 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Shaw G. M., Carmichael S. L., Laurent C., Rasmussen S. A., Maternal nutrient intakes and risk of orofacial clefts. Epidemiology 17, 285–291 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Wallenstein M. B., Shaw G. M., Yang W., Carmichael S. L., Periconceptional nutrient intakes and risks of orofacial clefts in California. Pediatr. Res. 74, 457–465 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Jahanbin A., Shadkam E., Miri H. H., Shirazi A. S., Abtahi M., Maternal folic acid supplementation and the risk of oral clefts in offspring. J. Craniofac. Surg. 29, e534–e541 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Butali A., et al. , Folic acid supplementation use and the MTHFR C677T polymorphism in orofacial clefts etiology: An individual participant data pooled-analysis. Birth Defects Res. A Clin. Mol. Teratol. 97, 509–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilcox A. J., et al. , Folic acid supplements and risk of facial clefts: National population based case-control study. BMJ 334, 464 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw G. M., Lammer E. J., Wasserman C. R., O’Malley C. D., Tolarova M. M., Risks of orofacial clefts in children born to women using multivitamins containing folic acid periconceptionally. Lancet 346, 393–396 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., et al. , Folate intake, markers of folate status and oral clefts: An updated set of systematic reviews and meta-analyses. Birth Defects Res. 112, 1699–1719 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Kelly D., O’Dowd T., Reulbach U., Use of folic acid supplements and risk of cleft lip and palate in infants: A population-based cohort study. Br. J. Gen. Pract. 62, e466–e472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonseth S., et al. , Epigenomic profiling of newborns with isolated orofacial clefts reveals widespread DNA methylation changes and implicates metastable epiallele regions in disease risk. Epigenetics 14, 198–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe L. J., et al. , Evidence for DNA methylation mediating genetic liability to non-syndromic cleft lip/palate. Epigenomics 11, 133–145 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z., Lie R. T., Wilcox A. J., Saugstad O. D., Taylor J. A., A comparison of DNA methylation in newborn blood samples from infants with and without orofacial clefts. Clin. Epigenet. 11, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young J. I., Slifer S., Hecht J. T., Blanton S. H., DNA methylation variation is identified in monozygotic twins discordant for non-syndromic cleft lip and palate. Front. Cell Dev. Biol. 9, 656865 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dan J., Chen T., Genetic studies on mammalian DNA methyltransferases. Adv. Exp. Med. Biol. 945, 123–150 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Li E., Bestor T. H., Jaenisch R., Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992). [DOI] [PubMed] [Google Scholar]
- 30.Lan Y., Jiang R., Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development 136, 1387–1396 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harfe B. D., et al. , Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Jackson-Grusby L., et al. , Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27, 31–39 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Matsuoka T., et al. , Neural crest origins of the neck and shoulder. Nature 436, 347–355 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schock E. N., Brugmann S. A., Neural crest cells utilize primary cilia to regulate ventral forebrain morphogenesis via Hedgehog-dependent regulation of oriented cell division. Dev. Biol. 431, 168–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He F., Soriano P., Sox10ER(T2) CreER(T2) mice enable tracing of distinct neural crest cell populations. Dev. Dyn. 244, 1394–1403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacques-Fricke B. T., Roffers-Agarwal J., Gammill L. S., DNA methyltransferase 3b is dispensable for mouse neural crest development. PLoS One 7, e47794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji Y., et al. , Single cell transcriptomics and developmental trajectories of murine cranial neural crest cell fate determination and cell cycle progression. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.05.10.443503v1 (Accessed 10 May 2021).
- 38.Hari L., et al. , Temporal control of neural crest lineage generation by Wnt/β-catenin signaling. Development 139, 2107–2117 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Lipinski R. J., et al. , Characterization of subtle brain abnormalities in a mouse model of Hedgehog pathway antagonist-induced cleft lip and palate. PLoS One 9, e102603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinberg S. M., et al. , Face shape of unaffected parents with cleft affected offspring: Combining three-dimensional surface imaging and geometric morphometrics. Orthod. Craniofac Res. 12, 271–281 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan Y., Wang Q., Ovitt C. E., Jiang R., A unique mouse strain expressing cre recombinase for tissue-specific analysis of gene function in palate and kidney development. Genesis 45, 618–624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan Y., Qin C., Jiang R., Requirement of hyaluronan synthase-2 in craniofacial and palate development. J. Dent. Res. 98, 1367–1375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elliott E. N., Sheaffer K. L., Kaestner K. H., The “de novo” DNA methyltransferase Dnmt3b compensates the Dnmt1-deficient intestinal epithelium. Elife 5, e12975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin H., et al. , Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol. Cell Biol. 26, 2976–2983 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J., et al. , Tamoxifen exposure induces cleft palate in mice. Br. J. Oral Maxillofac Surg. 59, 52–57 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Sun M. R., Steward A. C., Sweet E. A., Martin A. A., Lipinski R. J., Developmental malformations resulting from high-dose maternal tamoxifen exposure in the mouse. PLoS One 16, e0256299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dash S., Trainor P. A., The development, patterning and evolution of neural crest cell differentiation into cartilage and bone. Bone 137, 115409 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Ishii M., et al. , A stable cranial neural crest cell line from mouse. Stem. Cells Dev. 21, 3069–3080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Branch S., Chernoff N., Brownie C., Francis B. M., 5-AZA-2’-deoxycytidine-induced dysmorphogenesis in the rat. Teratog. Carcinog. Mutagen. 19, 329–338 (1999). [PubMed] [Google Scholar]
- 50.Branch S., Francis B. M., Brownie C. F., Chernoff N., Teratogenic effects of the demethylating agent 5-aza-2’-deoxycytidine in the Swiss Webster mouse. Toxicology 112, 37–43 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Bulut H. E., Ozdemir O., Başimoglu-Koca Y., Korkmaz M., Atalay A., Effects of a DNA demethylating agent–5-azacytidine–on testicular morphology during mouse embryo development. Okajimas Folia Anat. Jpn. 76, 47–53 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Rogers J. M., et al. , Cell death and cell cycle perturbation in the developmental toxicity of the demethylating agent, 5-aza-2’-deoxycytidine. Teratology 50, 332–339 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Kondo Y., et al. , Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 40, 741–750 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Wozniak R. J., Klimecki W. T., Lau S. S., Feinstein Y., Futscher B. W., 5-Aza-2’-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 26, 77–90 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Mukhopadhyay P., et al. , Determinants of orofacial clefting I: Effects of 5-Aza-2’-deoxycytidine on cellular processes and gene expression during development of the first branchial arch. Reprod. Toxicol. 67, 85–99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ladd-Acosta C., et al. , Epigenetic marks of prenatal air pollution exposure found in multiple tissues relevant for child health. Environ. Int. 126, 363–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee H.-S., Impact of maternal diet on the epigenome during in utero life and the developmental programming of diseases in childhood and adulthood. Nutrients 7, 9492–9507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li S., Chen M., Li Y., Tollefsbol T. O., Prenatal epigenetics diets play protective roles against environmental pollution. Clin. Epigenet. 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin E. M., Fry R. C., Environmental influences on the epigenome: Exposure-associated DNA methylation in human populations. Ann. Rev. Public Health 39, 309–333 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Oni-Orisan O., et al. , DNA methylation in children with prenatal methamphetamine exposure and environmental adversity. Pediatric Res. 89, 1152–1156 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breton-Larrivee M., Elder E., McGraw S., DNA methylation, environmental exposures and early embryo development. Animal Reproduction 16, 465–474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Millacura N., Pardo R., Cifuentes L., Suazo J., Effects of folic acid fortification on orofacial clefts prevalence: A meta-analysis. Public Health Nutr. 20, 2260–2268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anthony B., Zhou F. C., Ogawa T., Goodlett C. R., Ruiz J., Alcohol exposure alters cell cycle and apoptotic events during early neurulation. Alcohol. Alcoholism 43, 261–273 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takiguchi M., Achanzar W. E., Qu W., Li G., Waalkes M. P., Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp. Cell Res. 286, 355–365 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Vilahur N., Vahter M., Broberg K., The epigenetic effects of prenatal cadmium exposure. Curr. Environ. Health Rep. 2, 195–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hovland D. N., Machado A. F., Scott W. J., Collins M. D., Differential sensitivity of the SWV and C57BL/6 mouse strains to the teratogenic action of single administrations of cadmium given throughout the period of anterior neuropore closure. Teratology 60, 13–21 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Salvatori F., Talassi C. B., Salzgeber S. A., Spinosa H. S., Bernardi M. M., Embryotoxic and long-term effects of cadmium exposure during embryogenesis in rats. Neurotoxicol. Teratol. 26, 673–680 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Ni W., et al. , Umbilical cord concentrations of selected heavy metals and risk for orofacial clefts. Environ. Sci. Technol. 52, 10787–10795 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Suhl J., et al. , Maternal occupational cadmium exposure and nonsyndromic orofacial clefts. Birth Defects Res. 110, 603–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heyne G. W., et al. , A simple and reliable method for early pregnancy detection in inbred mice. J. Am. Assoc. Lab Anim. Sci. 54, 368–371 (2015). [PMC free article] [PubMed] [Google Scholar]
- 71.Li H., Williams T., Separation of mouse embryonic facial ectoderm and mesenchyme. J. Vis. Exp. 74, 50248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Everson J. L., Fink D. M., Chung H. M., Sun M. R., Lipinski R. J., Identification of sonic hedgehog-regulated genes and biological processes in the cranial neural crest mesenchyme by comparative transcriptomics. BMC Genomics 19, 497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Everson J. L., et al. , Sonic hedgehog regulation of Foxf2 promotes cranial neural crest mesenchyme proliferation and is disrupted in cleft lip morphogenesis. Development 144, 2082–2091 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Everson J. L., et al. , Developmental toxicity assessment of piperonyl butoxide exposure targeting sonic hedgehog signaling and forebrain and face morphogenesis in the mouse: An in vitro and in vivo study. Environ. Health Perspect. 127, 107006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abler L. L., et al. , A high throughput in situ hybridization method to characterize mRNA expression patterns in the fetal mouse lower urogenital tract. J. Vis. Exp. 54, 2912 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li B., Dewey C. N., RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobin A., et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ulschmid C. M., et al. , Disruption of DNA methylation-mediated cranial neural crest proliferation and differentiation causes orofacial clefts in mice. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE250370. Deposited 20 December 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA-seq data have been deposited in Gene Expression Omnibus and are available under accession number GSE250370 (80).