Significance
α-Synuclein aggregation is a key pathological feature of Parkinson’s disease, dementia with Lewy bodies, and multiple system atrophy, diseases for which we have no diagnostics nor disease-modifying agents. Digital seed amplification assays (SAAs) enable the quantitative measurement of α-Synuclein aggregates, which will be key to help diagnose, classify disease severity, and monitor response to pharmaceutical interventions in vivo. Applying different compartmentalization and immunocapture strategies, we developed several digital SAA platforms to detect aggregates in patient samples and characterize their growth. We demonstrate the utility of the method by quantifying the efficacy of a small-molecule inhibitor in preventing α-Synuclein aggregation, enabling these assays to be used to assess the efficacy of novel therapeutics.
Keywords: seed amplification assay (SAA), Parkinson’s disease (PD), α-synuclein, protein aggregation, digital assay
Abstract
The quantification and characterization of aggregated α-synuclein in clinical samples offer immense potential toward diagnosing, treating, and better understanding neurodegenerative synucleinopathies. Here, we developed digital seed amplification assays to detect single α-synuclein aggregates by partitioning the reaction into microcompartments. Using pre-formed α-synuclein fibrils as reaction seeds, we measured aggregate concentrations as low as 4 pg/mL. To improve our sensitivity, we captured aggregates on antibody-coated magnetic beads before running the amplification reaction. By first characterizing the pre-formed fibrils with transmission electron microscopy and size exclusion chromatography, we determined the specific aggregates targeted by each assay platform. Using brain tissue and cerebrospinal fluid samples collected from patients with Parkinson’s Disease and multiple system atrophy, we demonstrated that the assay can detect endogenous pathological α-synuclein aggregates. Furthermore, as another application for these assays, we studied the inhibition of α-synuclein aggregation in the presence of small-molecule inhibitors and used a custom image analysis pipeline to quantify changes in aggregate growth and filament morphology.
α-synuclein is an intrinsically disordered protein whose physiological functions are not entirely understood but are thought to be related to the regulation of neurotransmitter release and synaptic function (1, 2). Its accumulation in intracellular protein inclusions is a hallmark of synucleinopathies, including PD (Parkinson’s Disease), MSA (multiple system atrophy), and dementia with Lewy bodies (3). Preclinical biological processes leading to neurodegeneration are thought to evolve before symptoms occur; consequently, available treatments have limited efficacy in slowing disease progression. By the time PD is diagnosed, approximately 60% of dopaminergic neurons have died and once MSA symptoms begin, the disease advances rapidly toward severe disability and death (4, 5). There are currently no approved diagnostic tests for either synucleinopathy, and differentiating the two, especially at early stages, is challenging (6). Therefore, methods to better understand mechanisms behind α-synuclein aggregation are greatly needed. Disease severity has been strongly correlated with an increase of intraneuronal α-synuclein aggregates (7, 8), making pathological isoforms of α-synuclein an ideal biomarker candidate. Accurately measuring α-synuclein aggregates in patient biofluids could allow for earlier diagnosis, quantifiable disease staging, and clinical trial support. Recently, α-synuclein aggregates have been detected with increasing accuracy in patient biofluids using seed amplification assays (SAAs) (9–14). However, bulk SAAs suffer from irreproducibility and are not quantitative (15), whereas a digital SAA has the potential to quantify the concentration of pathological aggregates (Fig. 1A).
Fig. 1.
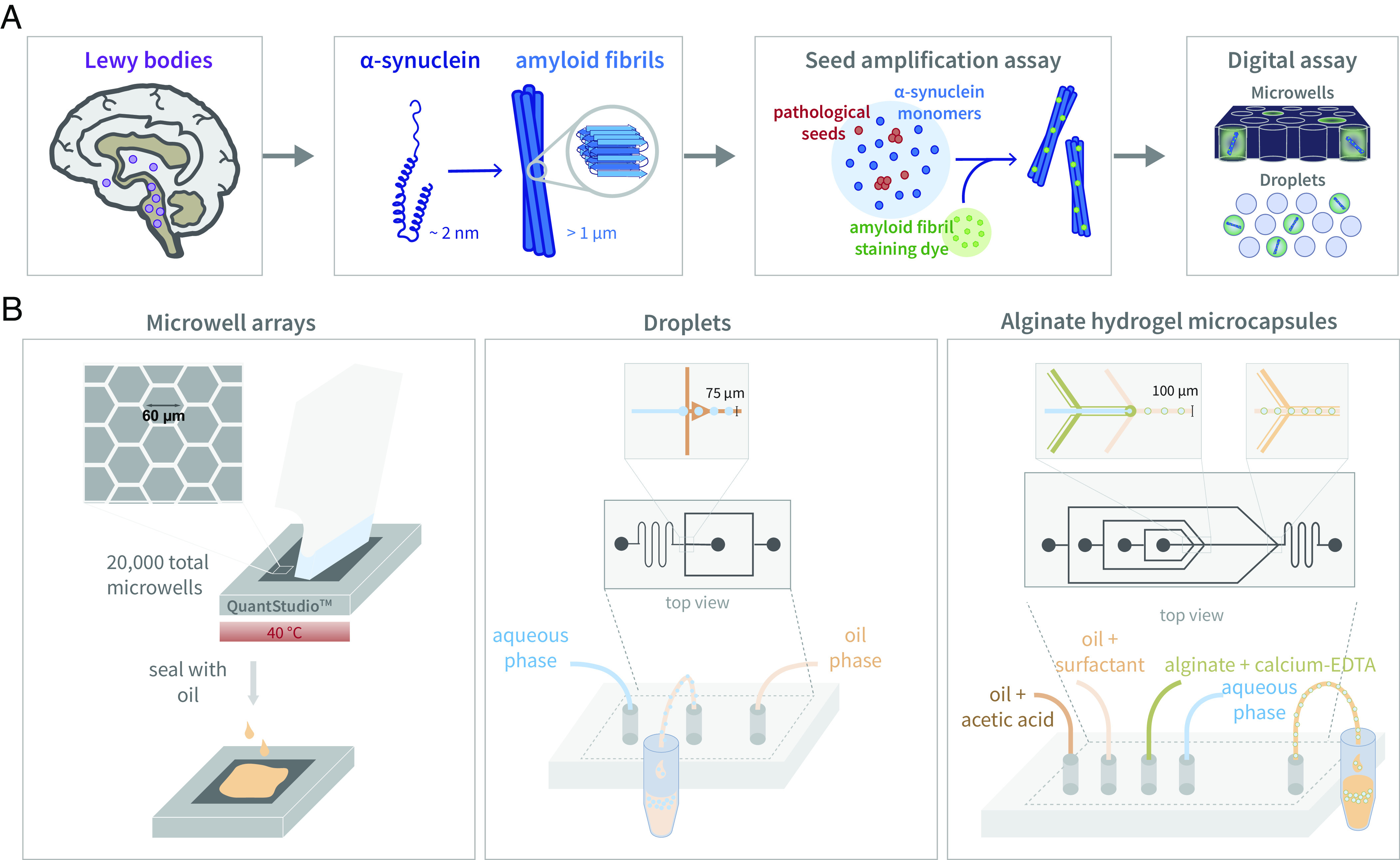
Digital SAA. (A) A pathological hallmark of PD, Lewy bodies are primarily composed of α-synuclein aggregates, which can be detected with SAAs by combining pathological seeds with wild-type monomers and an amyloid fibril staining dye. By transforming the bulk SAA into a digital format by partitioning the reaction into microcompartments, the number of single aggregate molecules can be quantified. (B) The three compartmentalization strategies used to develop digital SAA platforms, included microwell arrays, droplets, and alginate hydrogel microcapsules.
Previous work has demonstrated that it is possible to observe single-molecule protein aggregation. For example, one of the first digital methods used to characterize amyloid fibril assembly generated droplets to study the spatial and temporal evolution of growing aggregates and quantify single replicative units (16, 17). In addition, a single-molecule spectroscopy method could accurately detect different species of growing α-synuclein assemblies, from monomers to oligomers to fibrils (18). The same technique detected aggregated species in CSF (cerebrospinal fluid) samples, showing a significant difference between a small cohort of PD patients and healthy controls (19). While this method has the sensitivity to detect single aggregates, the amplification reaction occurs in bulk, where a single spontaneous aggregation event may bias the entire reaction.
Building upon this work, we developed a digital α-synuclein SAA by compartmentalizing the reaction into either microwells, droplets, or hydrogel microcapsules (Fig. 1B). Using pre-formed α-synuclein fibrils as reaction seeds, we implemented digital SAAs and observed growing aggregates. We characterized the reaction in terms of compartment surface, reaction components, temperature, and pH to determine the optimal conditions to maximize assay sensitivity. With our optimized platforms, we detected α-synuclein aggregates in brain tissue and CSF samples collected from PD and MSA patients. To improve assay sensitivity and specificity toward pathological α-synuclein aggregates, we then developed a bead-based SAA where aggregates are captured prior to compartmentalization with antibody-coated magnetic beads. Finally, we show that our digital SAAs can also be used to screen drug candidates in vitro by quantifying aggregation inhibition.
Results
Microwell Arrays.
We tested different compartmentalization strategies to partition the SAA reaction into a digital format, beginning with microwells. To run the SAA, we loaded the microwell array with reaction seeds, K23Q α-synuclein monomers, and an amyloid fibril staining dye to visualize aggregate growth. Building on previous bulk SAA work that analyzed CSF, we also included a dilution of CSF from a neurological control (NC) in our reactions to ensure that we could detect growing aggregates in relevant biomatrices. Initially, we used microwell arrays fabricated in cyclic olefin polymer (COP) (20–22), but we encountered uneven well sealing and a lack of reproducibility, which we attributed to the low nL sampling volume (SI Appendix, Fig. S1). Next, we fabricated microwell arrays in polydimethylsiloxane (PDMS) in order to screen a range of microwell dimensions, however, we had similar challenges sealing the wells with oil (SI Appendix, Fig. S1). For both the COP and PDMS arrays, we attempted to improve well loading and non-specific binding by functionalizing the surface of the microwells with polyethyleneglycol (PEG), though ultimately, neither microwell device proved to be robust. We then repurposed commercially available digital PCR chips (QuantStudio™, Thermo Fisher Scientific) for our digital SAA. These microwell array chips are fabricated from silicon and contain 20,000 uniformly sized reaction wells 60 µm in diameter.
In order to increase assay sensitivity, we explored methods to passivate the compartment surfaces and improve microwell loading. To help minimize surface adhesion that could impede aggregation, we added detergents and bovine serum albumin (BSA) to the SAA reaction before loading the silicon microwell arrays (SI Appendix, Fig. S2). We found that adding Triton-X 100 minimized non-specific binding to the microwell surface and improved loading efficiency by increasing the hydrophilicity of the solution. However, adding Triton-X 100 led to an increased background signal, which was ameliorated by adding BSA.
Apart from the effect of surface interactions, α-synuclein aggregation is particularly sensitive to pH, temperature, and salt concentration (23–26); therefore, we aimed to optimize the reaction conditions for the digital SAA. Without the fragmentation often generated by glass beads in bulk SAA reactions, secondary nucleation of α-synuclein aggregates via surface growth can be promoted at lower pH (27). For that reason, we tested different buffers ranging in pH from 4.2 to 7.4 and found that the mildly acidic piperazine-N, N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 6.5) was optimal (SI Appendix, Fig. S3). Furthermore, the sensitivity of SAAs is known to be highly influenced by the ionic environment (25), which led us to screen several Hofmeister salts, including sodium chloride (NaCl), guanidinium chloride (GdnCl), and magnesium sulfate (MgSO4) (SI Appendix, Fig. S4). While we did not observe any difference between the salts at higher initial seed concentrations, at lower seed concentrations, we observed that NaCl led to optimal aggregation in microwell arrays. Lastly, we tested a range of incubation temperatures from 30 to 55 °C and observed minimal spontaneous aggregation in the negative controls and accelerated aggregation in seeded reactions at temperatures around 40 °C.
Droplets.
Next, we generated single emulsion droplets using custom flow-focusing microfluidic devices fabricated out of PDMS with channel dimensions that produced droplets approximately 60 µm in diameter. We also fabricated PDMS “parking lot” devices in which we loaded droplets for imaging. Similar to the microwell arrays, we directly loaded droplets with the SAA reaction components, including reaction seeds, α-synuclein monomers, an amyloid fibril staining dye, and NC CSF. We screened a variety of surfactants in order to balance surface properties and droplet stability. Initially, we used a PEG krytox surfactant to create an inert inner surface with PEG (28); however, the droplets were unstable, potentially due to the high salt concentration in the SAA reaction, reducing the solubility of PEG. Adding free PEG to the aqueous solution was shown to reduce the collapse of the PEG inner surface (29), although it did not adequately improve the stability of SAA droplets when incubating at temperatures higher than 30 °C. Droplets generated with an ionic krytox surfactant were stable at temperatures up to 55 °C for weeks, although surface passivation with BSA was needed to reduce non-specific binding. Extremely stable droplets could also be generated with a “velcro” surfactant where fluorinated boronic acid random copolymers dispersed in the oil phase react with polyvinyl alcohol in the aqueous phase to form a covalently cross-linked amphiphilic interfacial film (SI Appendix, Fig. S5). Although the SAA reaction could be implemented in droplets generated with both ionic krytox and velcro surfactants with similar sensitivities, we continued with the ionic krytox due to the ease with which it could be synthesized. Optimal aggregation in droplets was seen using PIPES buffer at a pH of 6.5 with GdnCl (SI Appendix, Fig. S6) and incubating the reaction at 40 °C.
Hydrogel Microcapsules.
We hypothesized that α-synuclein aggregates would prefer to nucleate on polysaccharide surfaces. Consequently, to enhance sensitivity, we generated hydrogel microcapsules approximately 80 to 100 µm in diameter using a custom PDMS flow-focusing microfluidic device consisting of four inlets. The innermost aqueous phase contains reaction seeds and diluted NC CSF, which is co-flowed with a solution of calcium ethylenediaminetetraacetic acid (EDTA) and alginate. Droplets are then formed at a flow-focusing junction by encapsulating the aqueous phase in oil with PEG krytox surfactant. Finally, a third junction downstream introduces acetic acid, lowering the pH to release the calcium ions from EDTA and enabling the calcium and alginate to cross-link, forming the hydrogel shell. In contrast to the microwell arrays and droplets, we encapsulated only the reaction seeds and biofluid and then resuspended the microcapsules in the SAA reaction mixture, after which α-synuclein monomers and staining dye can diffuse into the microcapsules through the porous shell and attach to growing aggregates. Similar to the microwell SAA, a mildly acidic environment using PIPES buffer and added GdnCl was optimal for aggregate growth in the hydrogel microcapsules. In addition, we supplemented with additional CaCl2 to ensure the alginate hydrogel is fully cross-linked. Although we attempted to add BSA to reduce non-specific surface interactions, we found that it interfered with the hydrogel cross-linking. Furthermore, we used a slightly lower incubation temperature of 35 °C to prevent the microcapsules from adhering to each other. Due to the greater stability of the hydrogel shell, we also tried shaking the microcapsules to introduce shear stress to facilitate secondary nucleation and amplification via fibril fragmentation. However, constant and intermittent shaking led to an increase in false positives in the control reactions.
SAAs with Pre-Formed Fibrils.
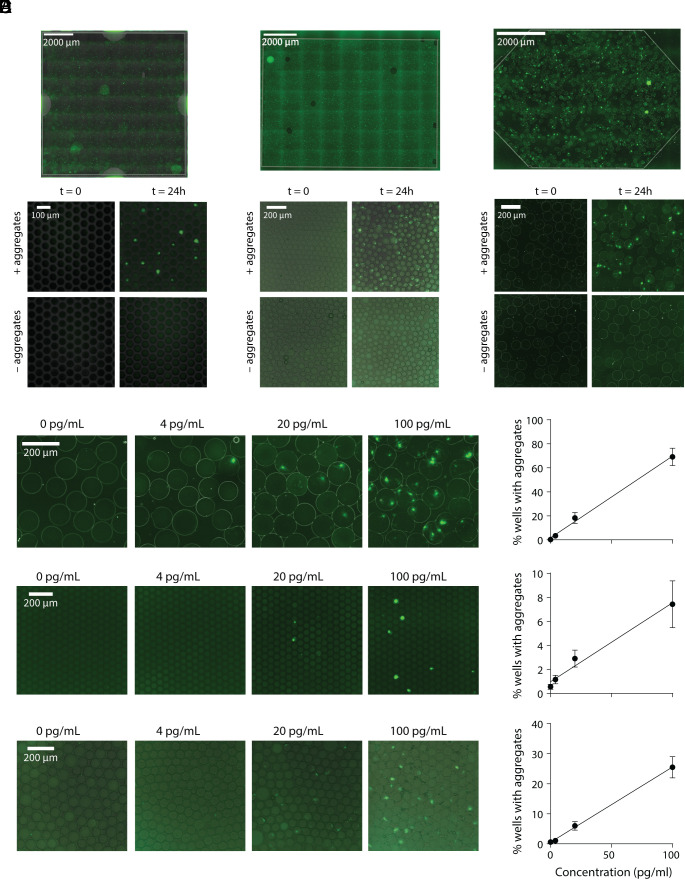
As a proof-of-concept, we used pre-formed α-synuclein fibrils as reaction seeds. Although we used the same pooled NC CSF samples to optimize our assays, we observed variable results between different preparations of pre-formed fibrils even when using the same reaction conditions. For instance, when seeding the reaction with the same concentration (0.5 ng/mL) of two different batches of pre-formed fibrils, we observe that almost every well contains a growing aggregate for the first batch vs. roughly 10% of wells for the second batch (SI Appendix, Fig. S7A), although in both cases the fibrils can be diluted as expected. To accurately quantify and compare the sensitivity of the different platforms, we then performed an in-depth characterization of a single batch of pre-formed fibrils with TEM (transmission electron microscopy), dynamic light scattering (DLS), and size exclusion chromatography (SEC) in combination with single-molecule array assays (SI Appendix, Fig. S7 B–D) and used these fibrils as the reaction seeds for all experiments presented here. Based on our analyses, the fibrils were predominantly 100 nm long, although smaller fibrils and monomers were also present. To test whether we could detect α-synuclein aggregates with the digital SAA, we spiked the characterized pre-formed fibrils into NC CSF and incubated the SAA reaction for 24 h. For all three compartmentalization strategies, we were able to visualize growing aggregates only if pre-formed filaments were added to the CSF (Fig. 2 A–C). As shown in the figure, controls without pre-formed fibrils showed no aggregates.
Fig. 2.
α-synuclein aggregate growth in microcompartments. Representative images of microwells (A), droplets (B), and hydrogel microcapsules (C) at t = 0 and t = 24 h of the SAA reaction with and without pre-formed filaments, + and – aggregates, respectively. The entire imaging area is shown on the Top for the microwell array and droplets or microcapsules loaded into parking lot devices. Displayed by decreasing sensitivity, images of alginate microcapsules (D), microwells (E), and droplets (F) for reaction seed concentrations ranging from 0 to 100 pg/mL. To the Right, the percent of wells with growing aggregates is shown vs. the concentrations of pre-formed filaments used as reaction seeds, which demonstrates the quantitative nature of all three assay formats.
For each SAA assay, using the optimized reaction conditions, we spiked in varying amounts of pre-formed fibrils, ranging from 4 to 100 pg/mL, and quantified the percent of wells or droplets containing growing aggregates (Fig. 2 D–F). We analyzed the percent of on-wells or on-droplets either with ImageJ or a custom Python image-processing pipeline (SI Appendix, Fig. S8). We analyzed approximately 13,500 wells, 5,000 droplets, and 800 alginate microcapsules to generate the calibration curves shown. Comparing the three compartmentalization methods, we observed the best sensitivity with the alginate hydrogel microcapsules. All three methods enabled us to quantify the concentration of aggregates in solution.
Detecting Pathological α-Synuclein in Patient Samples.
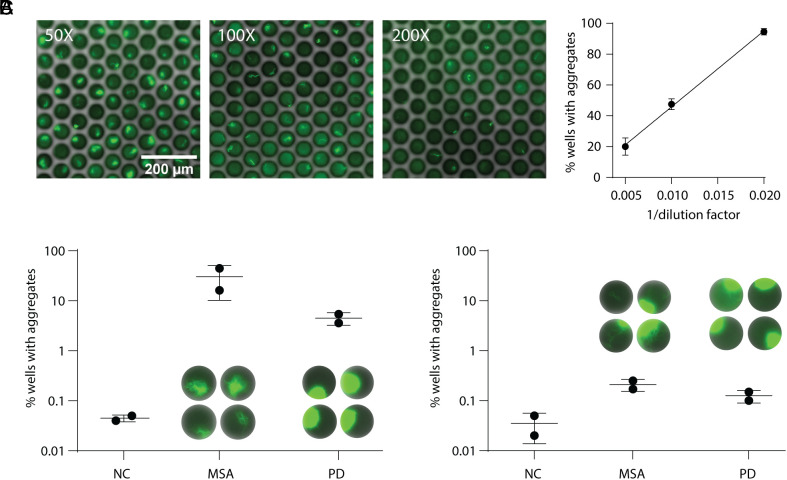
While our digital SAAs were able to detect α-synuclein aggregates formed in vitro, we wanted to ensure that endogenous forms of α-synuclein aggregates could be detected as well. To this end, we analyzed tissue lysates derived from the brain frontal cortex of individuals diagnosed with PD and MSA, in addition to NCs (SI Appendix, Table S1). We observed growing aggregates in MSA brain lysate with all platforms compared to minimal or no aggregate growth in the NC (SI Appendix, Fig. S9). We found that the optimal reaction conditions determined using pre-formed fibrils were the same for detecting aggregates in brain lysate samples in microwells and droplets, but for the hydrogel microcapsules, using NaCl as the added salt improved sensitivity and minimized false positives in the NC (SI Appendix, Fig. S10). When analyzing brain lysate, however, the microwell array platform was the most sensitive, and the only platform able to detect aggregates in both the MSA and PD brain lysates consistently. The percent of on-wells quantified in MSA brain lysate with the microwell assay decreased linearly upon sample dilution and there was a considerable difference between the percent of on-wells detected in MSA and PD samples vs. the NCs (Fig. 3 A and B). Interestingly, we observed different structures in the two synucleinopathic samples, where the growing aggregates were more fibrillar in the MSA samples and were more globular in the PD samples. Using the microwell assay, we then analyzed CSF samples collected from PD and MSA patients and NCs (Fig. 3C and SI Appendix, Table S1). Although we see an increase in the percent of on-wells in both the PD and MSA samples compared to the NCs, the difference is less marked than seen with the brain lysate samples.
Fig. 3.
Analysis of clinical samples with digital SAA. (A) Representative images acquired after analyzing different dilutions of MSA brain lysate using the microwell SAA. On the Right, the percent of wells with detectable aggregates vs. one over the lysate dilution factor is shown. Below, the percentage of wells with growing aggregates is shown for NC, MSA, and PD brain lysate (B) and CSF (C) samples. Exemplary images of the aggregate structures are shown for the MSA and PD samples.
Bead-Based SAA.
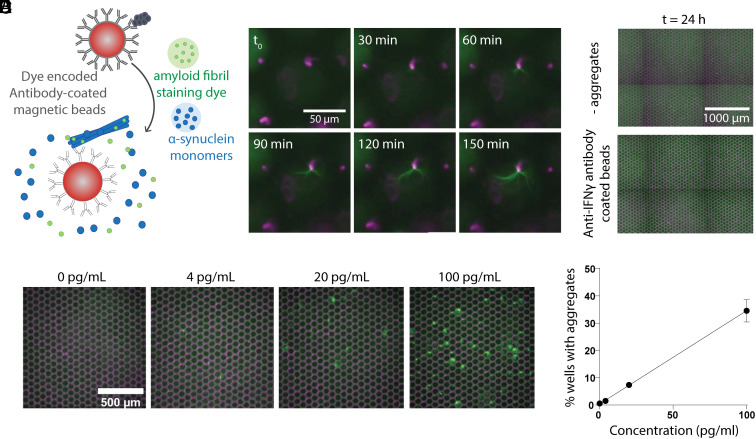
Given that we achieved the best sensitivity analyzing clinical samples with the microwell-based assay, we attempted to further improve assay sensitivity by first capturing aggregates on antibody-coated magnetic beads (Fig. 4A). This allowed us to increase the sample volume used and, therefore, increase the number of seeds analyzed. We tested two previously validated antibodies, including one antibody targeting fibrillar α-synuclein (Syn-F1, Creative Biolabs) (30) and another that was generated to target pathological pre-formed α-synuclein fibrils but can detect aggregates in Lewy bodies as well (Syn7015) (31). These antibodies were chosen based on previous work that demonstrated their specificity toward α-synuclein oligomers and fibrils with minimal cross-reactivity to monomers (32–34). Each antibody was conjugated to fluorescently dyed magnetic beads and incubated with pre-formed α-synuclein fibrils spiked into NC CSF. The beads were then resuspended in the SAA reaction mixture, and the solution was loaded into the microwell array. Over 3 h, we could visualize single aggregates growing on the surface of the beads (Fig. 4B), whereas when beads coated with a non-specific antibody were used, we did not see any non-specific aggregate binding or spontaneous aggregation reactions (Fig. 4C). Incubating α-synuclein aggregates with beads before running the digital SAA in microwell arrays led to ~fivefold increase in sensitivity, which can be seen by the increase in percent of on-wells for varying concentrations of pre-formed α-synuclein fibrils (Fig. 4 D and E) compared with the percent of on-wells without bead pre-capture (Fig. 2E).
Fig. 4.
Bead-based digital SAA. (A) Schematic depicting the assay where aggregates are first captured on antibody-coated magnetic beads and then combined with SAA reaction components before microwell loading. (B) Images showing filament growth from a single bead over the course of 150 min. (C) Representative images after 24 h of running the bead-based assay when the beads were incubated without pre-formed filaments (− aggregates) and when pre-formed filaments were incubated with non-specific antibody-coated beads (anti-IFNγ). (D) Microwell images 24 h after incubating the bead-based SAA at varying concentrations of pre-captured pre-formed filaments. (E) The percent of wells with growing aggregates vs. the concentration of pre-formed fibrils incubated with antibody-coated beads.
One of the limitations of using an antibody against α-synuclein aggregates is that the bead-based SAA could be biased toward larger aggregates. To better characterize the size specificity, we separated the pre-formed fibrils using SEC and visualized the size differences in each fraction with TEM (SI Appendix, Fig. S11). We subsequently used fibrils obtained from the various fractions as reaction seeds in either the microwell SAA or the bead-based microwell SAA. Quantification of the on-wells revealed a strong bias toward the larger aggregates obtained in the SEC 9 to 10 fractions for the bead-based assay, whereas aggregates could be detected using all SEC fractions for the standard microwell SAA with a slight bias toward larger aggregates. Nonetheless, we were able to detect α-synuclein aggregates in both MSA and PD brain lysate samples, but not in NC brain lysates (SI Appendix, Fig. S12), indicating that the antibodies used were specific to pathological strains of α-synuclein.
Additionally, incorporating beads is advantageous because it allows for multiplexed assays. We conjugated each antibody, Syn-F1 and Syn7015, to two different colored beads and captured pre-formed filaments before running the amplification reaction. We observed single aggregates growing from both labeled beads (SI Appendix, Fig. S13), showing that it is possible to target different pathological epitopes on α-synuclein aggregates and monitor their amplification in parallel. Targeting different pathological epitopes could enhance the assay’s specificity, allow for the differentiation of synucleinopathies, and possibly inform the stage of the disease.
Quantifying Aggregate Growth and Morphology.
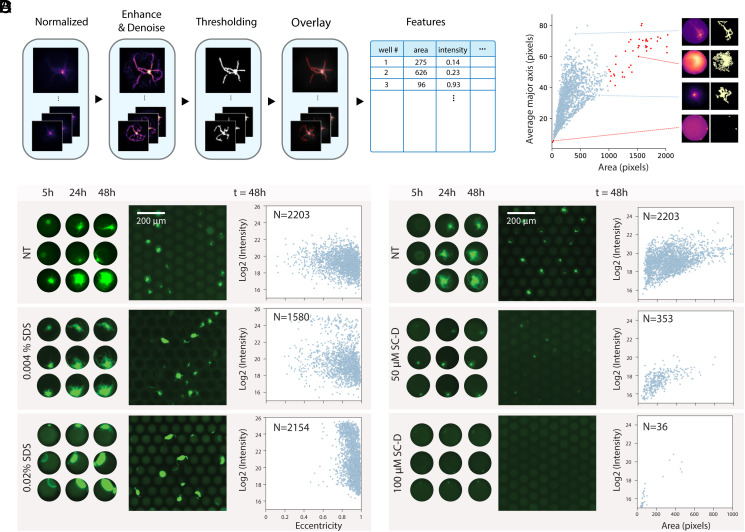
As a first demonstration of the utility of the digital SAA, we screened potential anti-aggregation therapeutic candidates. To understand how specific drug molecules may affect the aggregation of α-synuclein, we expanded our image processing pipeline to analyze the structures of growing aggregates by converting aggregate features into skeleton lines that could be quantified by structural parameters, including area, intensity, and eccentricity (Fig. 5A). While aggregate branches could be detected accurately in many cases, false positives did occur and could be identified as outliers when plotting the average major axis vs. the aggregate area (Fig. 5B). As a proof-of-concept, we ran SAA reactions with increasing concentrations of sodium dodecyl sulfate (SDS), a compound we observed to significantly alter aggregate morphology at higher concentrations. Plotting the eccentricity values vs. the intensity for the detected aggregates reveals a shift toward higher eccentricity values regardless of the intensity as the SDS concentration increases (Fig. 5C). Interestingly, we also see a second subpopulation emerging at higher intensities with increasing SDS concentration, reflecting a shift from branched filament structures toward more dense globular structures. We performed the same experiment with a known inhibitor, SynuClean-D (SC-D), designed to specifically block α-synuclein aggregation (35). In the presence of increasing concentrations of SC-D, we observed that aggregate growth was inhibited, which can be visualized quantitatively by a reduction in both area and intensity (Fig. 4D).
Fig. 5.
Characterizing aggregate growth in the presence of small inhibitor molecules. (A) Schematic outlining the image analysis pipeline used to extract aggregate features. (B) Average major axis plotted vs. the area for each aggregate detected, highlighting the skeleton lines extracted from four individual wells. Correctly identified aggregates are shown in blue, and outliers that arise due to background noise are shown in red. (C) Representative images of aggregates growing in the absence (NT) and in the presence of two concentrations of SDS (0.004% and 0.02%). On the Left, the growth of three single aggregates over time is shown and on the Right, log2 of the intensity vs. the eccentricity of all detected aggregates is displayed for each condition. (D) Example images of aggregates amplified in the absence (NT) and presence of SC-D at two concentrations (50 µM and 100 µM). Single aggregates growing over time are shown on the Left and log2 of the intensity is plotted vs. area on the Right.
Discussion
The molecular mechanisms underlying the abnormal aggregation of α-synuclein remain largely unknown. SAAs offer an in vitro method to study protein aggregation while probing biochemical and biophysical reaction parameters that promote or inhibit aggregation. From a therapeutic standpoint, this system provides a platform to screen drugs and better understand how they constrain aggregate growth. In addition, it can potentially serve as a diagnostic tool to detect pathological forms of α-synuclein in patient biofluids.
Here, we developed digital α-synuclein SAAs by partitioning the reaction into microwells, droplets, and hydrogel microcapsules. To begin, we tested COP and PDMS microwell arrays with microwell dimensions ranging from 3 to 10 µm. Although suboptimal surface interactions were observed with both materials, we determined that our sampling volume was too small to robustly detect low concentrations of aggregates. We then partitioned the reaction into larger microwell and droplet compartments between 60 and 100 µm in diameter, allowing us to sample 5 to 10 µL of CSF per assay. The microwell arrays used were commercially available and easy to use, compared to the droplet and hydrogel microcapsules, which were more technically challenging to implement. Basic microfluidic fabrication knowledge is needed to fabricate the droplet and microcapsule-generating devices; however, once the system was in place, it was more adaptable, enabling us to explore different surfactants and types of surface functionalization.
To develop digital SAAs and study aggregation under different conditions, we used α-synuclein filaments formed in vitro as reaction seeds. We acknowledge that pre-formed filaments may not precisely reflect the pathological forms of α-synuclein that we expect to detect in patient biofluids, yet when carefully characterized, they provide the best available model system. Due to the batch-to-batch variability we observed with pre-formed fibrils, we characterized our working solution with TEM, DLS, and SEC to ensure that we used comparable reaction seeds and could accurately report the size distribution of our reaction seeds.
While it was easier to develop and optimize digital SAAs using pre-formed filaments spiked into biological matrices, we also analyzed a small cohort of brain tissue and CSF samples. We were able to detect α-synuclein aggregates in MSA brain lysates with all of the SAA platforms, however only with the microwell array assay were we able to detect aggregates in PD brain lysates, as well as in CSF samples collected from PD and MSA patients. Furthermore, we observed that the growing aggregates detected in PD and MSA samples were structurally different. Previous work found that the aggregates in PD vs. MSA CSF samples represent distinct conformational strains and amplify differently using bulk SAAs (14). In particular, the maximum fluorescence values recorded were substantially lower for MSA samples, a trend that could be explained by our observation of more diffuse branched structures in MSA samples vs. densely bright globular aggregates in PD samples. Although we analyzed a small number of clinical samples, we demonstrate that our assay is capable of detecting endogenous α-synuclein aggregates in addition to providing structural and quantitative information that is not possible to readily obtain with bulk assays. Further work is necessary, however, to improve assay sensitivity to better distinguish between CSF samples collected from patients with neurodegenerative synucleinopathies and NCs.
One of the limitations of the bulk SAA is that they are not quantitative. The calibration curves for all three platforms described here show that the digital SAA is quantitative and robust. We consistently detected single-growing aggregates at around 4 pg/mL using pre-formed α-synuclein fibrils as reaction seeds. By approximating the hydrodynamic radius based on the average size of our pre-formed filaments (36, 37), we can estimate their molecular weight and expect to observe around one filament per ten wells or droplets at that concentration. Although we observed the best sensitivity with the hydrogel microcapsule SAA when using pre-formed filaments, we still detected fewer growing aggregates than expected. This result may be due to suboptimal assay conditions, including adhesion of aggregates to the loading or compartment surfaces. However, we may also be overestimating the concentration of pre-formed fibrils, given that we also detected monomers in our fibril solution after separation by SEC (SI Appendix, Fig. S1). In addition, we rely on secondary nucleation by monomer attachment on the fibril surface rather than by fragmentation, which is often used in bulk SAAs. For that reason, we explored hydrogel microcapsules because they can be incubated with shaking due to their stability, but even intermittent shaking led to an increased rate of false positives. The pore size of alginate hydrogels is around 16 nm; thus, small aggregates can diffuse out of the microcapsules, turning the outer solution into a bulk reactor where shaking only seemed to accelerate any spontaneous aggregation. Furthermore, using brain tissue lysates we observed better sensitivity with the microwell assay, suggesting that smaller pathological aggregates may diffuse out of the hydrogel microcapsules or other endogenous components might interfere with seed amplification.
Apart from the type of compartment, we screened reaction parameters, including the ionic environment, pH, and temperature. Depending on the compartmentalization strategy used, the optimal reaction conditions varied. For all digital SAAs, the reaction was buffered to a pH of 6.5. NaCl proved to be ideal for the microwell array and hydrogel microcapsules, whereas GdnCl was optimal for the droplets. Furthermore, the microwell chips and droplets were incubated at a temperature of 40 °C compared to 35 °C for the hydrogel microcapsules. A lower incubation temperature for the hydrogel microcapsules reduced the adherence between microcapsules and likely reduced the probability that spontaneous aggregation occurred in the external solution. Additional components, such as BSA and Triton-X 100 were added to the reaction to reduce non-specific binding to the compartment surface in both the microwell arrays and droplets. Similar to previous findings, we found that aggregation of α-synuclein was sensitive to reaction conditions, and converting the SAA reaction to a digital format required different compartment surfaces and reaction parameters to be varied independently. While we were able to detect α-synuclein aggregates in MSA brain lysate samples using the optimized reaction conditions for each platform, the sensitivity of the platforms changed. We detected more growing aggregates in clinical samples using the microwell-based assay, suggesting that the reaction conditions and parameters should be further tuned to promote seed amplification of endogenous aggregates.
After optimizing the SAA reaction for each different type of compartment, we aimed to further improve the microwell assay sensitivity by pre-capturing aggregates onto beads. By first incubating CSF containing α-synuclein fibrils with antibody-coated magnetic beads, we could sample 50 to 70 times the volume of CSF compared to the standard assay, increasing our sensitivity. Although we found that the bead-based assay was biased toward larger aggregates, its utility still proved to be relevant given that we could detect α-synuclein aggregates in PD and MSA brain lysate samples. Although we tested only two aggregate-specific antibodies, screening additional antibodies that target distinct pathological conformations could potentially enable the classification of different synucleinopathies. Alternatively, the assay could be modified to target other protein aggregates associated with neurodegenerative diseases, such as tau and amyloid-ß. Since we also showed that it is possible to multiplex the bead-based assay, different aggregate conformations could then be detected simultaneously. When working with precious samples, such as CSF, the ability to run multiplexed assays with less sample volume is advantageous. Lastly, by coupling this assay with SEC, it is possible to not only obtain isotype information but also to characterize the size distribution of captured aggregates and measure their concentrations.
Finally, we used our digital SAA platform to characterize aggregate inhibition with SDS and an anti-aggregation molecule, SC-D. With custom image analysis, we extracted values for a range of variables, enabling us to quantify aggregate growth and structural changes in the presence or absence of SDS and SC-D. When the concentration of either molecule increased, distinct subpopulations emerged with different aggregate areas, intensities, and eccentricities. While we focused on analyzing the inhibition of fibril growth, the platform has the potential not only to characterize potential aggregation inhibitor candidates but also better understand the biochemistry of molecules that enhance aggregation. As we observe distinct aggregate structures in a small number of PD and MSA brain tissue and CSF samples, it would be interesting to further characterize the structural parameters of detected aggregates in a larger number of patient samples as well.
Accurately detecting pathological forms of α-synuclein in patient biofluids is an ongoing challenge, and platforms to study α-synuclein aggregation quantitatively are limited. The developed platforms offer assays to characterize the aggregation of α-synuclein in the presence of therapeutic compounds and the ability to detect multiple pathological isoforms in parallel. While they presently do not meet the stringent criteria of robust diagnostic assays, they provide a promising advance toward measuring discrete concentrations of α-synuclein aggregates in clinical samples. Future work is needed both to optimize the assays for CSF samples and to analyze larger sample cohorts to determine their specificity and sensitivity.
Methods
SAA Reaction Components.
Pre-formed α-synuclein fibrils, used as our reaction seeds, were prepared according to previously described protocols by our collaborators at the University of Pennsylvania (38). Using an ultrasonic bath, we sonicated a solution of pre-formed fibrils at a concentration of 1 mg/mL for 5 min and then diluted the fibrils to the desired concentration in UltraPure™ water (Invitrogen) or phosphate-buffered saline (PBS, Life Technologies). We tested four buffers, including potassium phosphate buffer (pH 4.2 Sigma-Aldrich), sodium acetate (NaOAc, pH 5.2, Sigma-Aldrich), PIPES (100 mM, pH 6.5, Thermo Fisher Scientific), and PBS (pH 7.4, Life Technologies). In addition, we screened the following salts, NaI, NaCl, CaCl2, MgSO4, and GdnCl. NaCl was optimal for the microwell SAA, and GdnCl was ideal for the droplets and alginate hydrogel microcapsule SAAs (Sigma-Aldrich and Thermo Fisher Scientific). We tested two amyloid fibril staining dyes: thioflavin T (ThT, Thermo Fisher Scientific) and X-34 (Sigma-Aldrich). ThT was dissolved in water before each experiment. X-34 was dissolved in DMSO to a stock concentration of 6 mM, aliquoted, and stored at −80 °C. Before each experiment, a fresh aliquot was thawed and diluted in UltraPure™ water (Invitrogen). All prepared reagent solutions were filtered with 0.2-µm syringe filters before use. We tested recombinant wild-type (Sigma-Aldrich) and K23Q (Impact Biologicals) α-synuclein monomers. Monomers were filtered using 30 kD spin filters (Amicon™) and the flow through was retained to remove any possible larger molecular weight aggregates.
Negative Staining and TEM Imaging.
Carbon-coated grids (CF-400CU, Electron Microscopy Sciences) were glow discharged and 5 μL of the sample was absorbed onto the grid for 1 min. Excess sample was blotted with a Whatmann paper and the grid was stained with 5 μL 1% Uranyl Acetate for 15 s and excess staining solution was blotted away. Samples were imaged on a JEOL 1200EX – 80 kV transmission electron microscope with an AMT 2k CCD camera.
SEC.
Econo-Pac Chromatography columns (Bio-Rad) were packed with 10 mL of Sepharose CL-6B resin as previously described (39). The columns were washed with 10 mL PBS and the sample was loaded once the PBS finished dripping. PBS was then added and 0.5-mL fractions were collected.
Microwell Array SAA.
QuantStudio™ 3D Digital PCR 20K chips (Thermo Fisher Scientific) were loaded according to the manufacturer’s instructions. Briefly, the chips were placed on a digital heating block at 40 °C, and the SAA reaction mixture was loaded into the chips using the provided sample loading blade. After allowing the excess liquid on top of the wells to evaporate for approximately 10 to 20 s, the immersion oil was applied dropwise to the top of the array until all wells were sealed. The chip lid was cleaned with nitrogen and sealed on top of the array. Immersion oil was inserted into the lid inlet until the chip cavity was filled, and only a small air bubble remained. Lastly, the lid inlet was sealed, and the chips were incubated at 40 °C with the lid facing down on the heating block, which was tilted at a slight angle to ensure that the remaining air bubble floated toward the top part of the chip above the microwell array. The optimal SAA reaction components for the microwell arrays included 100 mM PIPES at pH 6.5, 100 mM NaCl, 0.1 mg/mL K23Q α-synuclein monomers, 0.02% Triton-X 100, 0.1% BSA, 1 µM X-34, and CSF diluted fourfold. The chips were incubated on a heating block at 40 °C and were mounted onto a 3D printed chip-holder for imaging.
Bead-Based SAA.
Antibody-coated beads were prepared using dye-encoded carboxylated paramagnetic beads and EDC conjugation as previously described (34). The bead-based SAA consisted of three main steps. In the first step, 100 µL solution containing PBS pH 7, 0.05% Triton-X 100, 0.1% BSA, and CSF diluted fourfold was mixed with 100,000 antibody-coated beads. The beads were agitated on a rotator for 30 min at room temperature and then washed three times with 100 µL of PBS on a magnet. In the second step, the beads were resuspended in the working solution containing 100 mM NaCl, 100 mM PIPES pH 6.5, 6% OptiPrep, 0.02% Triton-X 100, 0.1% BSA, 1 µM X-34, and 0.1 mg/mL K23Q α-synuclein monomers. To increase the assay’s sensitivity, the beads were incubated at 37 °C for 3 h and shook every 30 min for 1 min in a microplate reader (Tecan). In the third step, the beads were loaded into the QuantStudio™ 3D Digital PCR 20K chips and incubated at 40 °C on a hot plate until imaging.
Microfluidic Device Fabrication.
Microfluidic devices for generating and imaging droplets and hydrogel microcapsules were fabricated using soft lithography (40). Standard photolithography techniques were used to make structures with negative photoresist (SU8 2050, Kayaku) on a 3-inch silicon wafer. These structures served as master molds for PDMS casting. Droplet generators and parking lot devices for droplet imaging were fabricated with a 50 µm channel height. Droplet generators for alginate hydrogel microcapsule preparation were fabricated with a 100 µm channel height. To facilitate removal of PDMS from the silicon wafer, each mold was treated with 1% 1H,1H,2H,2H-perfluorooctyltrichlorosilane (Thermofisher) in HFE 7500 oil (3M) following fabrication. Sylgard 184 PDMS curing agent and elastomer base (Dow) were mixed at a 1:10 ratio, poured on top of the silicon master mold, degassed in a vacuum chamber, and baked at 60 °C for at least 4 h. The polymerized PDMS was then removed from the silicon master and inlet and outlet holes were punched using a 1-mm Integra Militex biopsy punch (Thermo Fisher Scientific). PDMS devices were bonded to glass slides using oxygen or air plasma (Plasma Etch, Inc.). Next, the device surface was rendered hydrophobic by flushing the device with 3% trichloroperfluorooctylsilane in HFE 7500 oil. The silane was left to incubate in the device for 30 min and then washed three times with isopropanol, dried with air, and baked at 60 °C for 1 h. Parking lot devices were not silane treated.
Surfactant Preparation.
HFE 7500 soluble Krytox surfactant was prepared by converting Krytox FSH 157 (Miller-Stephenson) into an ammonium carboxylate salt. Krytox FSH 157 was dissolved into methanol (20% w/w) and placed on a stir plate in a fume hood. Ammonium hydroxide (Thermo Fisher Scientific) was added dropwise until the solution turned clear. The solution was then left to evaporate overnight under gentle stirring. Once the solvent had completely evaporated and the Dupont FSH 157 was crystallized, HFE 7500 was added to achieve a final concentration of 20% (w/w). Any residual methanol then formed an emulsion in the HFE 7500 oil. The solution was then spun down in a centrifuge at 4,000 × g for 10 min, and methanol in the HFE emulsion was removed from the top of the centrifuge tube and discarded. The pH of the solution was then adjusted to 7 by the dropwise addition of glacial acetic acid (Thermo Fisher Scientific).
Droplet SAA.
Water in oil droplets were generated using conventional microfluidic flow-focusing geometries (41). The SAA solution containing 100 mM PIPES at pH 6.5, 100 mM GdnCl, 0.2 mg/mL K23Q α-synuclein monomers, 0.025% Triton-X 100, 0.5% BSA, 5 µM X-34, and CSF diluted fourfold was loaded into the device with polyethylene tubing (Scientific Commodities, Inc.) as the inner phase flowed at rates ranging from 300 to 500 µL/h with syringe pumps (New Era Pump Systems, Inc.). For the outer phase, 5% ionic krytox in HFE 7500 (w/w) was flowed at a rate typically around three times greater than that of aqueous phase, 900 to 1,500 µL/h. The above flow rates generated monodisperse droplets with an average diameter of 70 µm. The droplets were collected in a 1.5-mL tube (Eppendorf) and 200 µL of mineral oil (Sigma) was added on top of the droplets to avoid evaporation. The tubes were incubated at 40 °C in a digital block heater (Thermo Fisher Scientific). Droplets were loaded into either a custom PDMS parking lot devices or cell counters (Invitrogen) for imaging.
Alginate Hydrogel Microcapsule SAA.
Four solutions were prepared to generate hydrogel microcapsules. The innermost solution consisted of CSF diluted fourfold in UltraPure™ water. The next solution consisted of 1% alginate in 100 mM CaCl2 and 100 mM EDTA at a pH of approximately 7. The third outermost solution contained 1% 008-fluorosurfactant (RAN Biotechnologies) in HFE 7500 oil and the outermost solution contained 0.4% acetic acid in HFE 7500 oil. Each solution was drawn into PTFE tubing and loaded into the device at the following flow rates: 10 µL/min, 4 µL/min, 70 µL/min, and 35 µL/min, listed from innermost to outermost phases. Hydrogel microcapsules were collected in 1.5-mL tubes (Eppendorf) only after flow rates had stabilized since the formation of the hydrogel capsule depends on the uniform mixture with acetic acid. All oil was removed from the tube and the drops were briefly resuspended in HFE-7500 oil with 0.4% acetic acid to ensure the gelation of the microcapsules and to remove excess surfactant. The drops were then resuspened twice in HFE-7500 oil only. Excess oil was removed and 200 µL of 2 mM CaCl2 with 0.05% Triton-X was added on top of the microcapsule layer. To transfer the microcapsules into the aqueous phase, 50 µL (or greater than 20 v/v%) of 1H,1H,2H,2H-perfluorooctanol (Alfa Aesar) was added to the oil phase and the tube was gently inverted until the microcapsules could no longer be seen in the oil phase. Any remaining oil was removed using gel loading pipette tips and the microcapsules were centrifuged at 200 × g for 2 min. If any residual oil remained, it was again removed and the microcapsules were washed with fresh 2 mM CaCl2 solution three times. After the final wash, the microcapsules were resuspended in the SAA reaction mixture, containing 100 mM PIPES at pH 6.5, 100 mM NaCl, 0.1 mg/mL K23Q α-synuclein monomers, 5 µM X-34, and 2 mM CaCl2. The microcapsules were incubated at 35 °C and loaded into PDMS parking lot devices for imaging.
Clinical Sample Preparation.
Postmortem human brain was collected as previously described. The brains are part of the Mass General Brigham SCiN (Stem Cells in Neurodegeneration) study. Then, 5% w/v brain homogenates were prepared in 1% Triton in PBS supplemented with proteinase inhibitors (cOmplete, Roche). After mechanical homogenization in TissueLyser LT with one 7-mm steel bead at 50 Hz for 1 min, lysates were centrifuged at 2,000 × g for 5 min at 4 °C. The supernatant was aliquoted and frozen in liquid nitrogen. CSF was collected using an atraumatic technique according to the Institutional Review Board (IRB) protocol 2003P000541 into polypropylene falcon tubes. After a 10-min centrifugation at 400 × g, the CSF was aliquoted for storage at −80 °C.
Image Acquisition and Analysis.
Images of the microwell array chips or droplets were acquired using a fluorescent microscope (Olympus IX83) equipped with an LED light source (CoolLED pE-300) and a 10× objective. The imaging area was scanned, and images were taken on each field of view with both a reflected brightfield channel (to identify the position of each well/droplet) and the CFP fluorescence channel (433/475 nm). Image processing and data analysis were performed using an automated custom Python (3.10) code and the scikit-image (0.19), numpy (1.23), and pandas (1.4) packages. All image analysis code is available on Github.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Alice Chen-Plotkin for providing materials for this study and we thank Aviad Levin and Tuomas Knowles for helpful discussions. The research described in this manuscript was supported by a grant from the Michael J. Fox Foundation (Grant number 2021A011886). T.G. is an awardee of the Weizmann Institute of Science Women’s Postdoctoral Career Development Award.
Author contributions
T.G., Z.S., M.N., and D.R.W. designed research; T.G., Z.S., R.T., K.H.O., and E.A.F. performed research; T.G., Z.S., R.T., B.T.D., A.C., E.B., A.K., A.N., V.K., and D.A.W. contributed new reagents/analytic tools; T.G., Z.S., R.A.G., and K.H.O. analyzed data; and T.G., Z.S., and D.R.W. wrote the paper.
Competing interests
D.R.W. is a founder and equity holder in Quanterix. His interests were reviewed and are managed by BWH and Partners HealthCare in accordance with their conflict of interest policies.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Python code data have been deposited in Github (https://github.com/Wyss/chip-extract) (42).
Supporting Information
References
- 1.Bendor J. T., Logan T. P., Edwards R. H., The function of α-synuclein. Neuron 79, 1044–1066 (2013), 10.1016/j.neuron.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lashuel H. A., Overk C. R., Oueslati A., Masliah E., The many faces of α-synuclein: From structure and toxicity to therapeutic target. Nat. Rev. Neurosci. 14, 38–48 (2013), 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poewe W., et al. , Parkinson disease. Nat. Rev. Dis. Primer. 3, 1–21 (2017), 10.1038/nrdp.2017.13. [DOI] [PubMed] [Google Scholar]
- 4.Dauer W., Przedborski S., Parkinson’s disease: Mechanisms and models. Neuron 39, 889–909 (2003), 10.1016/S0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 5.Poewe W., et al. , Multiple system atrophy. Nat. Rev. Dis. Primer. 8, 1–21 (2022), 10.1038/s41572-022-00382-6. [DOI] [PubMed] [Google Scholar]
- 6.Wenning G. K., et al. , What clinical features are most useful to distinguish definite multiple system atrophy from Parkinson’s disease? J. Neurol. Neurosurg. Psychiatry 68, 434–440 (2000), 10.1136/jnnp.68.4.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler C. H., et al. , Unified staging system for lewy body disorders: Clinicopathologic correlations and comparison to braak staging. J. Neuropathol. Exp. Neurol. 78, 891–899 (2019), 10.1093/jnen/nlz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozawa T., et al. , The spectrum of pathological involvement of the striatonigral and olivopontocerebellar systems in multiple system atrophy: Clinicopathological correlations. Brain 127, 2657–2671 (2004), 10.1093/brain/awh303. [DOI] [PubMed] [Google Scholar]
- 9.Rossi M., et al. , Ultrasensitive RT-QuIC assay with high sensitivity and specificity for Lewy body-associated synucleinopathies. Acta Neuropathol. (Berl.) 140, 49–62 (2020), 10.1007/s00401-020-02160-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo M. J., et al. , High diagnostic performance of independent alpha-synuclein seed amplification assays for detection of early Parkinson’s disease. Acta Neuropathol. Commun. 9, 179 (2021), 10.1186/s40478-021-01282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iranzo A., et al. , Detection of α-synuclein in CSF by RT-QuIC in patients with isolated rapid-eye-movement sleep behaviour disorder: A longitudinal observational study. Lancet Neurol. 20, 203–212 (2021), 10.1016/S1474-4422(20)30449-X. [DOI] [PubMed] [Google Scholar]
- 12.L. Concha-Marambio, S. Pritzkow, M. Shahnawaz, C. M. Farris, C. Soto, Seed amplification assay for the detection of pathologic alpha-synuclein aggregates in cerebrospinal fluid. Nat. Protoc. 18, 1179–1196 (2023), 10.1038/s41596-022-00787-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahnawaz M., et al. , Development of a biochemical diagnosis of Parkinson disease by detection of α-synuclein misfolded aggregates in cerebrospinal fluid. JAMA Neurol. 74, 163–172 (2017), 10.1001/jamaneurol.2016.4547. [DOI] [PubMed] [Google Scholar]
- 14.Shahnawaz M., et al. , Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 578, 273–277 (2020), 10.1038/s41586-020-1984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paciotti S., Bellomo G., Gatticchi L., Parnetti L., Are we ready for detecting α-synuclein prone to aggregation in patients? The case of “protein-misfolding cyclic amplification” and “real-time quaking-induced conversion” as diagnostic tools. Front. Neurol. 9, 415 (2018), 10.3389/fneur.2018.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles T. P. J., et al. , Observation of spatial propagation of amyloid assembly from single nuclei. Proc. Natl. Acad. Sci. U.S.A. 108, 14746–14751 (2011), 10.1073/pnas.1105555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfammatter M., et al. , Absolute quantification of amyloid propagons by digital microfluidics. Anal. Chem. 89, 12306–12313 (2017), 10.1021/acs.analchem.7b03279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau D., et al. , Single molecule fingerprinting reveals different amplification properties of α-synuclein oligomers and preformed fibrils in seeding assay. ACS Chem. Neurosci. 13, 883–896 (2022), 10.1021/acschemneuro.1c00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhumkar A., et al. , Single molecule counting coupled to rapid amplification enables detection of αSynuclein aggregates in cerebrospinal fluid of PD patients. Angew. Chem. Int. Ed. 60, 11874–11883 (2021), 10.1002/anie.202014898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rissin D. M., et al. , Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010), 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kan C. W., et al. , Digital enzyme-linked immunosorbent assays with sub-attomolar detection limits based on low numbers of capture beads combined with high efficiency bead analysis. Lab Chip. 20, 2122–2135 (2020), 10.1039/D0LC00267D. [DOI] [PubMed] [Google Scholar]
- 22.Duan B. K., Cavanagh P. E., Li X., Walt D. R., Ultrasensitive single-molecule enzyme detection and analysis using a polymer microarray. Anal. Chem. 90, 3091–3098 (2018), 10.1021/acs.analchem.7b03980. [DOI] [PubMed] [Google Scholar]
- 23.Munishkina L. A., Henriques J., Uversky V. N., Fink A. L., Role of protein−water interactions and electrostatics in α-synuclein fibril formation. Biochemistry 43, 3289–3300 (2004), 10.1021/bi034938r. [DOI] [PubMed] [Google Scholar]
- 24.Buell A. K., et al. , Solution conditions determine the relative importance of nucleation and growth processes in α-synuclein aggregation. Proc. Natl. Acad. Sci. U.S.A. 111, 7671–7676 (2014), 10.1073/pnas.1315346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metrick M. A., et al. , Million-fold sensitivity enhancement in proteopathic seed amplification assays for biospecimens by Hofmeister ion comparisons. Proc. Natl. Acad. Sci. U.S.A. 116, 23029–23039 (2019), 10.1073/pnas.1909322116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pálmadóttir T., Malmendal A., Leiding T., Lund M., Linse S., Charge regulation during amyloid formation of α-synuclein. J. Am. Chem. Soc. 143, 7777–7791 (2021), 10.1021/jacs.1c01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaspar R., et al. , Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Q. Rev. Biophys. 50, e6 (2017), 10.1017/S0033583516000172. [DOI] [PubMed] [Google Scholar]
- 28.Holtze C., et al. , Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip. 8, 1632–1639 (2008), 10.1039/B806706F. [DOI] [PubMed] [Google Scholar]
- 29.Etienne G., Kessler M., Amstad E., Influence of fluorinated surfactant composition on the stability of emulsion drops. Macromol. Chem. Phys. 218, 1600365 (2017), 10.1002/macp.201600365. [DOI] [Google Scholar]
- 30.Vaikath N. N., et al. , Generation and characterization of novel conformation-specific monoclonal antibodies for α-synuclein pathology. Neurobiol. Dis. 79, 81–99 (2015), 10.1016/j.nbd.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Covell D. J., et al. , Novel conformation-selective alpha-synuclein antibodies raised against different in vitro fibril forms show distinct patterns of Lewy pathology in Parkinson’s disease. Neuropathol. Appl. Neurobiol. 43, 604–620 (2017), 10.1111/nan.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaikath N. N., et al. , Antibodies against alpha-synuclein: Tools and therapies. J. Neurochem. 150, 612–625 (2019), 10.1111/jnc.14713. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S. T., et al. , How specific are the conformation-specific α-synuclein antibodies? Characterization and validation of 16 α-synuclein conformation-specific antibodies using well-characterized preparations of α-synuclein monomers, fibrils and oligomers with distinct structures and morphology. Neurobiol. Dis. 146, 105086 (2020), 10.1016/j.nbd.2020.105086. [DOI] [PubMed] [Google Scholar]
- 34.Norman M., Gilboa T., Walt D. R., High-sensitivity single molecule array assays for pathological isoforms in Parkinson’s disease. Clin Chem. 68, 431–440 (2022), 10.1093/clinchem/hvab251. [DOI] [PubMed] [Google Scholar]
- 35.Pujols J., et al. , Small molecule inhibits α-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 115, 10481–10486 (2018), 10.1073/pnas.1804198115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nath S., Meuvis J., Hendrix J., Carl S. A., Engelborghs Y., Early aggregation steps in α-synuclein as measured by FCS and FRET: Evidence for a contagious conformational change. Biophys. J. 98, 1302–1311 (2010), 10.1016/j.bpj.2009.12.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krouglova T., Vercammen J., Engelborghs Y., Correct diffusion coefficients of proteins in fluorescence correlation spectroscopy. Application to tubulin oligomers induced by Mg2+ and paclitaxel. Biophys. J. 87, 2635–2646 (2004), 10.1529/biophysj.104.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luk K. C., et al. , Exogenous α-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 106, 20051–20056 (2009), 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ter-Ovanesyan D., et al. , Framework for rapid comparison of extracellular vesicle isolation methods. eLife 10, e70725 (2021), 10.7554/eLife.70725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia Y., Whitesides G. M., Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184 (1998), 10.1146/annurev.matsci.28.1.153. [DOI] [Google Scholar]
- 41.Tan Y. C., Cristini V., Lee A. P., Monodispersed microfluidic droplet generation by shear focusing microfluidic device. Sens. Actuators B Chem. 114, 350–356 (2006), 10.1016/j.snb.2005.06.008. [DOI] [Google Scholar]
- 42.Gould R. A., Gilboa T., Swank Z., Digital seed amplification assay image processing pipeline. Chip extract. https://github.com/Wyss/chip-extract. Accessed 24 December 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Python code data have been deposited in Github (https://github.com/Wyss/chip-extract) (42).