Significance
MicroRNAs (miRNAs) are key regulators of gene expression with essential roles in postnatal brain development and function. Neuronal disruption of the miRNA-biogenesis machinery leads to neuro-inflammation and -degeneration. Here, we show that miRNAs in the adult murine forebrain determine the contribution of slow waves to the EEG (electroencephalographic), characteristic of NREM (non-rapid-eye-movement) sleep. Sleep deprivation alters forebrain miRNA expression, and particularly that of miR-709. We report that miR-709 manipulation alters EEG slow waves during sleep and regulates genes involved in glutamatergic transmission. The results causally implicate miR-709 and its targeted molecular pathways in linking neuronal activity during wakefulness to NREM sleep slow waves, which are important in synaptic plasticity and brain function.
Keywords: microRNAs, sleep deprivation, EEG delta power, glutamatergic receptor activity, synaptic transmission
Abstract
MicroRNAs (miRNAs) are key post-transcriptional regulators of gene expression that have been implicated in a plethora of neuronal processes. Nevertheless, their role in regulating brain activity in the context of sleep has so far received little attention. To test their involvement, we deleted mature miRNAs in post-mitotic neurons at two developmental ages, i.e., in early adulthood using conditional Dicer knockout (cKO) mice and in adult mice using an inducible conditional Dicer cKO (icKO) line. In both models, electroencephalographic (EEG) activity was affected and the response to sleep deprivation (SD) altered; while the rapid-eye-movement sleep (REMS) rebound was compromised in both, the increase in EEG delta (1 to 4 Hz) power during non-REMS (NREMS) was smaller in cKO mice and larger in icKO mice compared to controls. We subsequently investigated the effects of SD on the forebrain miRNA transcriptome and found that the expression of 48 miRNAs was affected, and in particular that of the activity-dependent miR-709. In vivo inhibition of miR-709 in the brain increased EEG power during NREMS in the slow-delta (0.75 to 1.75 Hz) range, particularly after periods of prolonged wakefulness. Transcriptome analysis of primary cortical neurons in vitro revealed that miR-709 regulates genes involved in glutamatergic neurotransmission. A subset of these genes was also affected in the cortices of sleep-deprived, miR-709-inhibited mice. Our data implicate miRNAs in the regulation of EEG activity and indicate that miR-709 links neuronal activity during wakefulness to brain synchrony during sleep through the regulation of glutamatergic signaling.
The molecular basis of sleep regulation has been a research focus for at least 20 y (1–3). It has become evident across species that after periods of extended wakefulness, either spontaneous or experimentally enforced (i.e., sleep deprivation or SD), brain gene expression is altered to accommodate neuronal responses in excitability and synaptic plasticity during sleep [(4, 5), reviewed in ref. 6]. Furthermore, molecular alterations, are also sufficient to modulate global brain activity during sleep and post-SD responses, as assessed by electroencephalographic (EEG) recordings (7, 8). Molecular players involved in sleep regulation include immediate early genes, adhesion and scaffolding proteins, ion channels, synapse-related genes, neurotransmitters, protein transporters, genes involved in lipid and energy metabolism (reviewed in ref. 2), and more recently, in endosomal trafficking (9) and autophagic processes (10, 11). However, the involvement of gene expression regulatory mechanisms, other than transcription factors, has yet to be fully understood. One such mechanism is the fine-tuning of gene expression at the post-transcriptional level by microRNAs (miRNAs).
MiRNAs are ~22-nucleotide short, non-coding RNAs that must undergo a multi-step maturation process requiring the enzymes DROSHA/DGCR8 and DICER. Absence of either enzyme results in a lack of mature, functional miRNAs (12). Mature miRNAs exert their function by guiding the RNA-induced silencing complex to their mRNA targets (13). Once bound to their target, miRNAs either destabilize the targeted transcript leading to repressed translation or induce degradation (14). They have been implicated in a plethora of physiological and pathological processes, such as neuronal development and neurodegeneration (15), as well as modifying neuronal excitability via the regulation of the expression of genes encoding ion-channel (16), cytoskeletal (17), and scaffolding (18), among others. Despite their known role as central transcriptional regulators in all molecular pathways studied, only a limited number of studies in zebrafish (19), mice (19, 20), rats (21–24), and flies (25–27) have demonstrated their functional involvement in the regulation of sleep. Taken together, these observations suggest that miRNAs represent an additional, evolutionarily conserved, layer of gene regulation implicated in sleep processes. Nevertheless, their roles in the regulation of brain synchrony and associated processes during physiological sleep in mice have not been assessed, and their downstream pathways in the context of sleep remain unidentified.
In the current study, we show that miRNAs regulate EEG brain activity across sleep-wake states in the adult mouse. In two conditional Dicer knockout mouse lines, each providing a different temporal blockade of miRNA maturation in forebrain excitatory neurons, the rebound in rapid-eye-movement sleep (REMS) and EEG activity was altered after SD. We identified SD-induced changes in miRNA expression in the brain and focused on the top differentially expressed miRNA, miR-709. Inhibition of miR-709 in vivo increased the number of slow waves in the slow-delta (0.75 to 1.75 Hz) band in non-REM sleep (NREMS), while its inhibition in primary cortical neuronal cultures revealed genes involved in glutamatergic receptor neurotransmission being most affected. The in vitro differentially regulated genes were also assessed in vivo in miR-709-inhibited animals, where the cortical expression of genes related to synaptic neurotransmission was again affected the most. Our observations support the involvement of miRNAs in the regulation of cortical slow-wave characteristics and implicate miR-709 in the control of pathways directly linked to neuronal synaptic plasticity (28).
Results
Constitutive Conditional Dicer Deletion Impacts EEG Spectral Power and Reduces the NREMS EEG Response to Extended Waking.
To establish a role for mature miRNAs in sleep regulation in the adult mouse, we assessed the consequences of Dicer ablation in post-mitotic excitatory neurons. First, 12-wk-old conditional Camk2a-Dicerfl/fl knockout male mice (cKO, n = 9, referred to as Dicerfl/fl) lacking forebrain Dicer from 1.5 mo of age were contrasted to their control littermates (Dicerfl/+; n = 6). cKO mice exhibited a highly aberrant low voltage EEG signal (Fig. 1A and SI Appendix, Fig. S1A), which, however, did not impede sleep-wake state identification. Both absolute and relative levels of EEG spectral power were reduced in cKO mice in all sleep-wake states and EEG frequencies >2.0 Hz compared to the littermate controls (Fig. 1D and SI Appendix, Fig. S1B). Despite these differences in EEG signal amplitude, the typical sleep-wake driven dynamics in NREMS EEG delta (1 to 4 Hz) power (29), appeared largely preserved. Nevertheless, differences in the amplitude of these relative changes were observed; at light onset, the beginning of the main rest phase in baseline, cKO mice showed significantly higher EEG delta power compared to the control group (168 vs. 155%, Fig. 1 B, Top). cKO mice slept less during the hour before and after light onset (Zeitgeber Time ZT23-1) than their control littermates and also lacked the additional resting period during the second half of the dark period (“nap” at ZT20-22; Fig. 1 B, Middle), typical in mice of this genetic background (30, 31). These differences in the baseline sleep-wake distribution might have contributed to a higher sleep homeostatic pressure and associated higher relative EEG delta levels observed in cKO mice at the beginning of light period. In the first hours of the dark period, when mice of both genotypes were spontaneously awake the most, cKO mice exhibited lower EEG delta power compared to controls (108 vs. 147% of baseline reference). After 6 h of SD, control mice exhibited the expected surge of NREMS EEG delta power (30) reaching 230% over the baseline reference (Fig. 1 B, Top). Conversely, Dicer cKO mice reached lower EEG delta power levels after SD than control mice (183 vs. 230% of reference). The levels reached after SD in cKO mice were, however, still higher than after light-onset in baseline (P = 0.002, post hoc paired t test), suggesting a slower build-up of sleep pressure in cKO mice. The lower delta power reached in cKO mice after periods of spontaneous wakefulness during the baseline dark period is consistent with this interpretation.
Fig. 1.
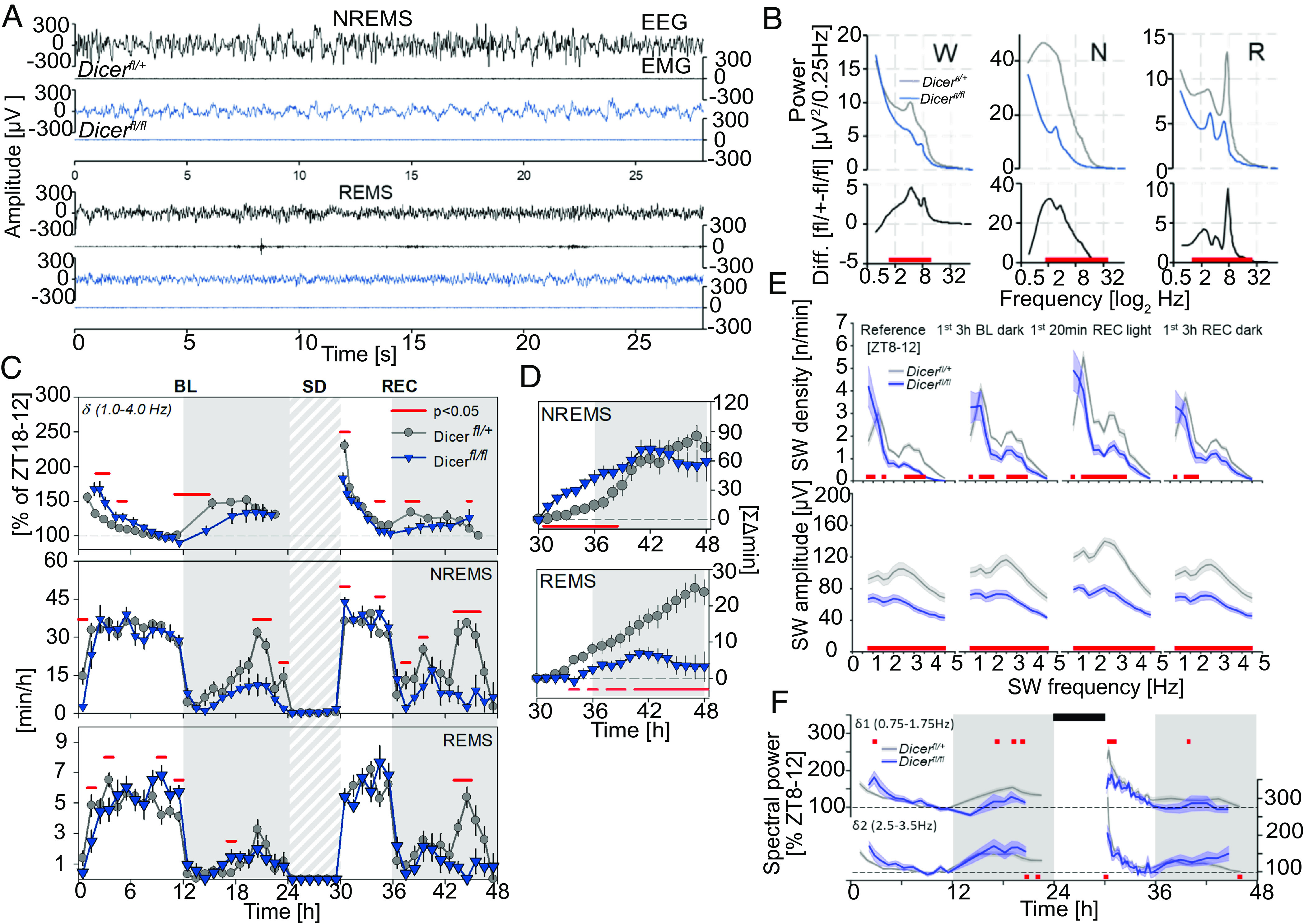
Conditional Dicer deletion from early development impacts sleep behavior and EEG spectral power. (A) Dicerfl/fl mice (dark blue) display decreased amplitude across EEG but not EMG signals compared to controls (Dicerfl/+ black), most notably during NREMS (Middle), but also REMS (Bottom; also see SI Appendix, Fig. S1A for wake). Each example represents the first consolidated (28 s of each sleep-wake state during the baseline light period), in the same two mice. A high-pass filter of 0.1 Hz is applied to decrease DC offset signal artifacts. (B) Average spectral profiles for wake (W), NREMS (N), and REMS (R), during 48 h of baseline expressed in absolute values (μV2/0.25 Hz). Differences between group means are displayed below (black). Significant genotype changes are depicted by red horizontal lines below the data (two-way rANOVA interaction factors genotype × time P < 0.001; post hoc t tests P < 0.05). (C) From Top to Bottom: Mean time course of NREMS EEG delta power [δ; expressed relative to the individual levels reached in the last 4 h of baseline light period (ZT8-12) when EEG delta power no longer declines further (29); the dashed gray line is 100%] and NREMS and REMS hourly amounts, during 24 h baseline (BL) and 18 h of recovery (REC) from a 6 h SD (gray-white pattern) starting at light onset (ZT0). Gray areas delineate the dark periods. Significant genotype changes are depicted by red horizontal lines above the data (two-way rANOVA interaction factors genotype × time P < 0.001; post hoc t tests P < 0.05). (D) NREMS (Top) and REMS (Bottom) rebound for the 18 h of recovery after SD as accumulated differences from corresponding baseline hours. Significant differences are marked by red horizontal lines underneath the data (t tests P < 0.05). (E) From Top to Bottom: SW density and amplitude at different times of the experiment. SW density is frequency corrected (n/frequency). Frequencies with significant changes in SW density or amplitude compared to controls (gray) are depicted by the red squares (two-way rANOVA interaction factors genotype × frequency; P < 0.001 for all periods; post hoc Tukey: P < 0.05). (F) Time-course of δ1 (0.75 to 1.75 Hz; Top) and δ2 (2.5 to 3.5 Hz; Bottom) spectral power during baseline and following 6 h SD (black rectangles), in Dicer knockoutmice and controls. Statistics (two-way rANOVA interaction genotype × time; P < 0.001 for both δ1 and δ2). All values represent means (solid lines) ± SEM (shaded areas). Power was expressed as % of mean δ1 and δ2 power, respectively, during the last 4 h of the light period of baseline (ZT8-12). N = Dicerfl/+: 8; Dicerfl/fl: 6.
Although overall time spent in NREMS in baseline did not differ between cKO and controls, cKO mice did spend less time in NREMS at the specific time points during the dark period mentioned above (ZT20-22 and ZT23-1), while REMS was not affected (Fig. 1 C, Middle). During the dark period following SD, cKO mice again spent less time in NREMS, especially between ZT19 and −22 (Fig. 1 C, Middle). During this dark period, also REMS was reduced in cKO mice (Fig. 1 C, Bottom). After SD both genotypes accumulated more NREMS compared to corresponding baseline hours. This increase in extra time spent in NREMS was initially larger in cKO mice (ZT6-14); however, this genotype difference dissipated over the course of the subsequent dark period (Fig. 1 D, Top). In contrast, while control mice did gain extra REMS during the recovery period, SD in cKO mice failed to induce a compensatory response (+23.6 vs. +3.3 min, respectively, by the end of the recovery dark period; Fig. 1 D, Bottom).
Given the genotype differences in the dynamics of EEG delta power, we next investigated the effect of conditional Dicer deletion on EEG slow-wave (SW) features during NREMS in the time-domain, such as the number of SWs per minute of NREMS (i.e., SW density) and SW amplitude (Fig. 1E). We selected temporal windows representing different levels of sleep need, i.e., the end of the baseline rest phase [Reference (ZT8-12)], when sleep need is low, as well as after the main waking periods in the baseline and recovery dark phase (3 h BL and REC dark, respectively; ZT12-15), and the first 20 min directly following SD (20 min REC light; ZT6), when it is high. We found that regardless of differences in sleep need, both SW density and amplitude were reduced in cKO mice, except for the density of the slowest SWs (<1.25 Hz), which was increased (Fig. 1E). Consistent with our previous observations (32), two distinct SW sub-bands were evident in the distribution of SW density. We thus analyzed the delta power dynamics separately for slower (δ1: 0.5 to 1.75 Hz) and faster (δ2: 2.0 to 3.5 Hz) delta frequencies. As was the case for the entire delta frequency band (Fig. 1C), the relative power in both sub-bands was higher during the first hours of the light period, and lower after sleep loss in cKO mice, compared to mice with functional Dicer (Fig. 1F).
The aberrant EEG signal in cKO mice supports an essential role for miRNAs in brain development and function, consistent with the previous literature (33, 34). Given that previous research has reported cortical neuroinflammation starting at 2.5 mo and neurodegeneration at 6 mo of age for this particular mouse line, the deficits in cortical synchrony may be secondary to the onset of neuroinflammatory processes during early adulthood in the absence of functional Dicer. Thus, we next chose to explore miRNA regulation of sleep and EEG activity using an inducible cKO (icKO) approach.
Inducible Dicer Deletion in Excitatory Neurons of the Adult Brain Amplifies the Increase in NREMS EEG Delta Power after SD.
To determine the consequences of acute neuronal miRNA depletion in the post-mitotic neurons of adult mice, sleep and EEG features of inducible conditional Dicer knockout (icKO; Dicerfl/fl+ tamoxifen; 12 wk old, n = 7) male mice were assessed and compared to their non-induced control littermates (Dicerfl/fl + vehicle, n = 6). We previously confirmed that Dicer deletion occurred ~3 wk following tamoxifen injections in 2-mo-old mice (see ref. 35). We did not observe any major differences in sleep-wake state distribution, with the exception of a reduced REMS rebound post SD in icKO mice (Fig. 2 B, Right), reminiscent of that observed in the cKO mice (Fig. 1 D, Bottom). As in cKO mice, we observed a reduction in the absolute levels of EEG spectral power in the icKO mice in all three sleep-wake states, particularly in the range of 2.0 to 10.0 Hz for wakefulness and 2.0 to 30.0 Hz for NREMS and REMS (Fig. 2C). Nevertheless, the relative spectral profiles in icKO seemed largely unperturbed compared to those obtained in cKO mice (SI Appendix, Fig. S1C).
Fig. 2.
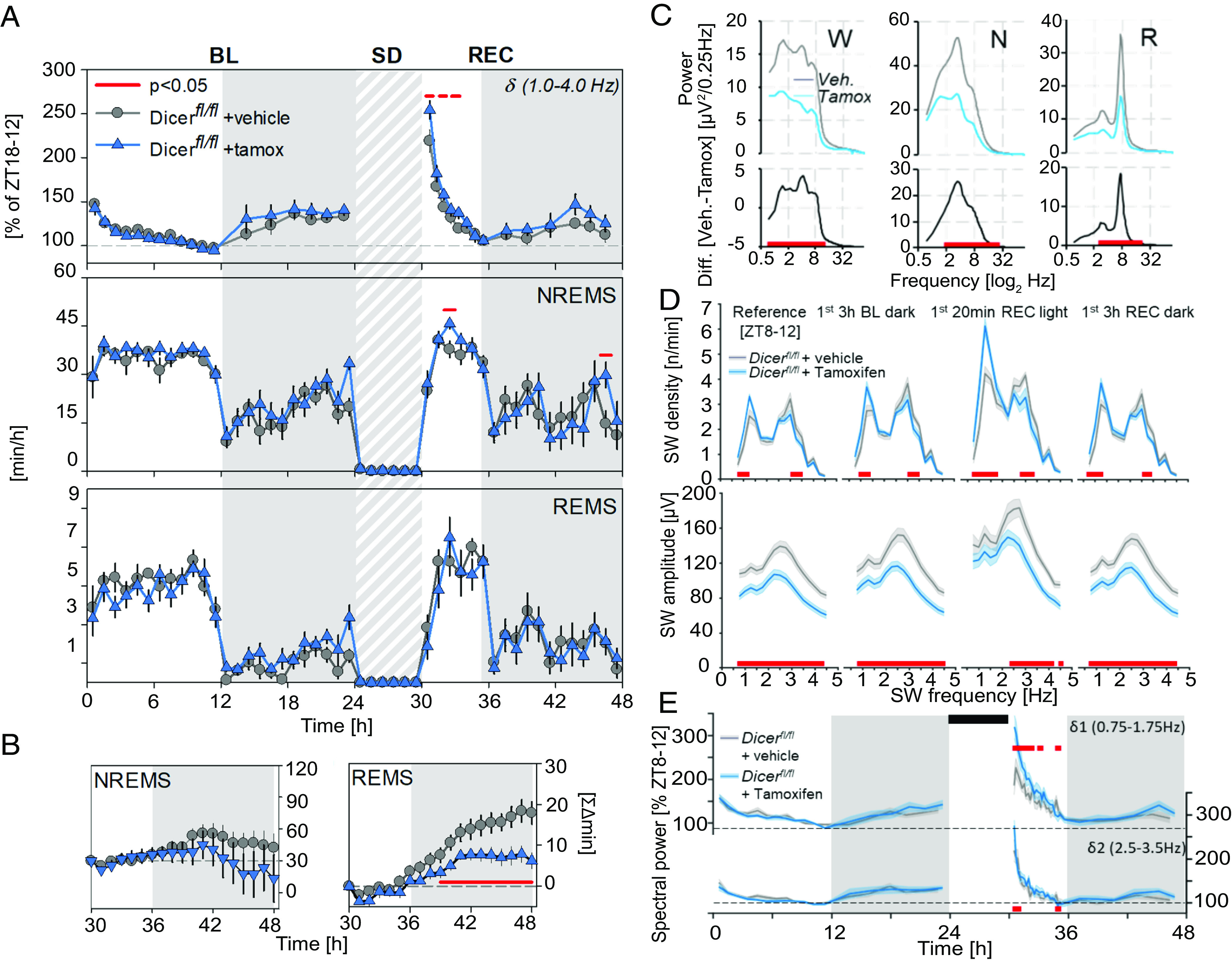
Inducible Dicer deletion modifies EEG slow-wave (SW) properties following spontaneous and enforced (SD) waking. (A) From Top to Bottom: Mean time course of NREMS EEG delta power (δ; also see Fig. 1C) and NREMS and REMS hourly amounts, during 24 h baseline (BL) and 18 h of recovery (REC) from a 6 h SD (black rectangles) starting at light onset (ZT0). Gray areas delineate the dark periods. Significant genotype changes are depicted by red horizontal lines above the data (two-way rANOVA interaction factors genotype × time P < 0.001; post hoc t tests P < 0.05). (B) NREMS (Left) and REMS (Right) rebound for the 18 h of recovery after SD as accumulated differences from corresponding baseline hours. Significant differences are marked by red horizontal lines underneath the data (t tests P < 0.05). (C) Average spectral profiles for wake (W), NREMS (N), and REMS (R), during 48 h of baseline expressed in absolute values (μV2/0.25 Hz). Differences between group means are displayed below in black. Significant genotype changes are depicted by red horizontal lines below the data (two-way rANOVA interaction factors genotype × time P < 0.001; post hoc t tests P < 0.05). (D) From Top to Bottom: SW density and amplitude at different times of the experiment during which sleep need is assumed to differ (also see description for Fig. 1E). Frequencies with significant changes in SW density or amplitude compared to controls (gray) are depicted by the red squares (two-way rANOVA interaction factors genotype × frequency; P < 0.001 for all periods; post hoc Tukey: P < 0.05). (E) Time-course of δ1 (0.75 to 1.75 Hz; Top) and δ2 (2.5 to 3.5 Hz; Bottom) spectral power during baseline and following 6 h SD (black rectangles), in icKO mice and controls. Statistics (two-way rANOVA interaction genotype × time; δ1: P < 0.001; δ2: P < 0.05). All values represent means (solid lines) ± SEM (shaded areas). Power was expressed as % of mean δ1 and δ2 power, respectively, during the last 4 h of the light period of baseline (ZT8-12). N = Dicerfl/fl + vehicle: 6; Dicerfl/fl + tamoxifen: 7.
NREMS EEG delta power dynamics did not differ between the icKO and control groups under baseline conditions but, contrary to cKO mice, SD in icKO mice resulted in a significantly larger increase compared to controls (Fig. 2 A, Top). Additionally, icKO mice exhibited higher SW density in the δ1 range (0.75 to 1.75 Hz), particularly after SD (1st 20 min REC), while faster SWs (δ2; 2.5 to 3.5 Hz) were less prevalent (Fig. 2D) regardless of sleep need. Conversely, SW amplitude was reduced in icKO mice at all periods, except for that of SWs in the δ1 range after SD (Fig. 2D). The larger post-SD increase in NREMS EEG power in the delta (1 to 4 Hz) range in icKO mice (Fig. 2A) was thus largely due to the differences in the lower δ1 sub-band activity (Fig. 2E).
Together, our observations in both Dicer knockout mouse models suggest that interfering with miRNA maturation in excitatory neurons yields broad changes in brain activity during both wakefulness and sleep. Specific to NREMS SWs, their amplitude was reduced, while the incidence of slower SWs was increased and that of faster SWs decreased in both lines. The two lines also shared a reduced REMS rebound after SD, while the sleep-wake driven delta rebound showed opposite effects attributable in large part to differences in the δ1 sub-band, which we previously reported to respond less to sleep need (32). Last, although the SWs are largely a cortical phenomenon and Dicer was deleted in a neuronal subpopulation that is enriched in the cortex, other, sub-cortical, brain regions in which Dicer was deleted could have contributed to the EEG and sleep phenotypes observed in the cKO and icKO models. For example, lack of Dicer in the hippocampus, in which CamkII2a is highly expressed, might have contributed to the altered EEG activity in theta-frequency range (6 to 10 Hz).
SD Increases miR-709 Expression in the Forebrain.
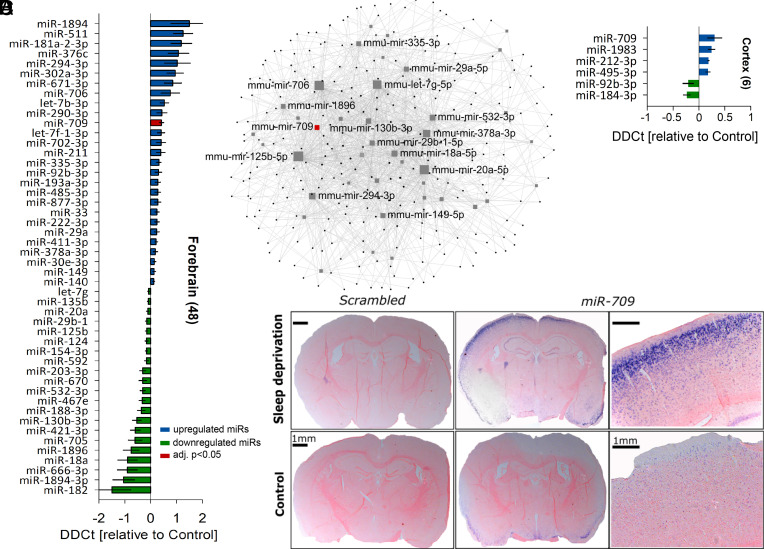
To identify specific miRNAs affected by enforced waking, we next compared brain miRNA expression after SD to that during unperturbed baseline conditions. Among the 611 miRNAs quantified, 48 miRNAs were differentially expressed in sleep-deprived forebrains (27 up-, 21 downregulated, moderate t-statistic P < 0.05), but only the increase in miR-709 levels survived correction for multiple testing (1.3-fold; FDR < 0.05; Fig. 3A).
Fig. 3.
miR-709 is the top differentially regulated miRNA after SD. Bar charts represent the relative expression levels of significantly altered miRNAs between 6 h-sleep deprived (SD, n = 5) and control (Ctrl, n = 5) animals. Error bars show the pooled SEM of SD and Ctrl samples. (A) SD significantly affected the forebrain expression of 48 (out of 611 tested miRNAs) of which 27 were up- (blue) and 21 downregulated (green; SD vs. Ctrl; moderate t-statistic, P < 0.05). Note that only miR-709 expression (red) survived multiple testing correction (FDR < 0.05). (B) Network of all differentially regulated miRNAs in the forebrain after SD and their validated targets (black dots; miRTarBase v8.0 and TarBase 8.0). Only the names of the top 15 interconnected forebrain miRNAs are shown (gray squares), including miR-709 (red square). Note that larger squares are more interconnected. Only the shortest path between two central nodes is visualized (40 out of 48 miRNA nodes; 273 gene nodes; 1,421 edges). (C) Mean differential miRNA expression in the cortex after 6 h SD, where four miRNAs increased and two decreased their expression (SD vs. Ctrl; moderate t-statistic, P < 0.05, FDR > 0.05). (D) In situ hybridization (sections approximately −2.0 mm posterior of bregma, at the somatosensory cortex) of two representative sleep-deprived (Top) and control (Bottom) mice, with either a miR-709-specific probe (four Right panels) or a scrambled miRNA probe (two Left panels). In situ hybridization confirms that SD increased miR-709 expression in the cortex. No signal was detected with the scrambled probe.
Multiple miRNAs may target the same gene(s) and signaling pathways (36). To identify these pathways, we investigated the network of shared validated targets from the 48 differentially regulated forebrain miRNAs using the miRNET resource [(37, 38), Fig. 3B]. Among the pathways that were significantly overrepresented in the set of shared target genes (SI Appendix, Table S1) were “axon guidance” (KEGG: mmu04360), “regulation of actin cytoskeleton” (KEGG: mmu04810), “regulation of intracellular transport” (GO: 0006886), “endosomal transport” (GO: 0016197), and “early endosome” (GO: 0005769). These pathways remained among the significant predictions even if the up- and downregulated miRNAs were tested separately.
Since miRNA expression and function have been found to be brain region specific (39, 40), we next investigated cortices (Fig. 3C) and hippocampi (SI Appendix, Fig. S3E) of sleep-deprived mice in a second, independent cohort. Simple t tests suggested a significant SD-driven upregulation of miR-709 also in the cortex (1.2-fold; P = 0.03). Nevertheless, miRNA expression changes in these two tissues failed to reach statistical significance after FDR correction (Fig. 3C). In situ hybridization confirmed the SD-induced increase in miR-709 cortical levels, predominantly in cortical layers 2 and 3 (Fig. 3D). Previous studies confirmed miR-709 expression in pyramidal hippocampal neurons, and its upregulation after application of L-glutamate (41) or the mGluR-agonist DHPG (42), indicating its expression in excitatory neurons. In summary, miR-709 levels were significantly increased by sleep loss in the murine forebrain. Interestingly, miR-709 was previously found to be upregulated in the mouse brain after selective agonism of group I metabotropic glutamate receptors (mGluRs) (42), as well as NMDAR-induced excitotoxicity (41). We thus further investigated the role of this miRNA in the wake-dependent changes in brain activity during sleep.
In Vivo miR-709 Inhibition Increases the Prevalence of NREMS δ1 SWs, Particularly after Periods of Prolonged Spontaneous Wakefulness.
As SD upregulated miR-709, we hypothesized that in vivo miR-709 inhibition would reveal its functional role in regulating the changes in NREMS SWs after prolonged periods of wakefulness. We therefore injected 12-wk-old male mice intracerebroventricularly (ICV) with miR-709-specific locked-nucleic-acid (LNA) inhibitors (i-miR-709, n = 7), non-specific scrambled probes (scram, n = 7), or vehicle (artificial cerebrospinal fluid; aCSF, n = 5), 48 h prior to the onset of EEG recordings (Materials and Methods). Inhibition of miR-709 in vivo resulted in a consistently higher SW density restricted to the lower delta frequencies (δ1; 0.75 to 1.75 Hz; Fig. 4 A, Upper), while SW amplitudes remained unaffected (Fig. 4 A, Lower). The higher overall levels of δ1 SW incidence observed in the miR-709-inhibited group were even more pronounced after extended periods of waking (SI Appendix, Fig. S2 B, Left) and remained significantly higher after correcting for the already higher levels at ZT8-12 in baseline (SI Appendix, Fig. S2 B, Right). Although δ1 SW incidence post SD was still higher in the miR-709-inhibited group compared to controls, this difference was not larger than the increase after spontaneous periods of wakefulness during the dark periods of baseline and second recovery day. This could be due to a ceiling effect supported by a smaller relative surge in δ1 SW incidence from the highest values in baseline to the values reached after SD in the miR-709-inhibited group (P < 0.06; SI Appendix, Fig. S2C). Subsequent analysis of NREMS EEG delta power dynamics showed that δ1 power was higher in NREMS episodes after spontaneous wakefulness during the baseline dark period but did not reach statistical significance after SD (Fig. 4B and SI Appendix, Fig. S2A). No systematic differences were observed in the time spent in NREMS and REMS between the three groups (Fig. 4 C and D). Moreover, the NREMS and REMS rebounds after SD were unaffected (Fig. 4D).
Fig. 4.
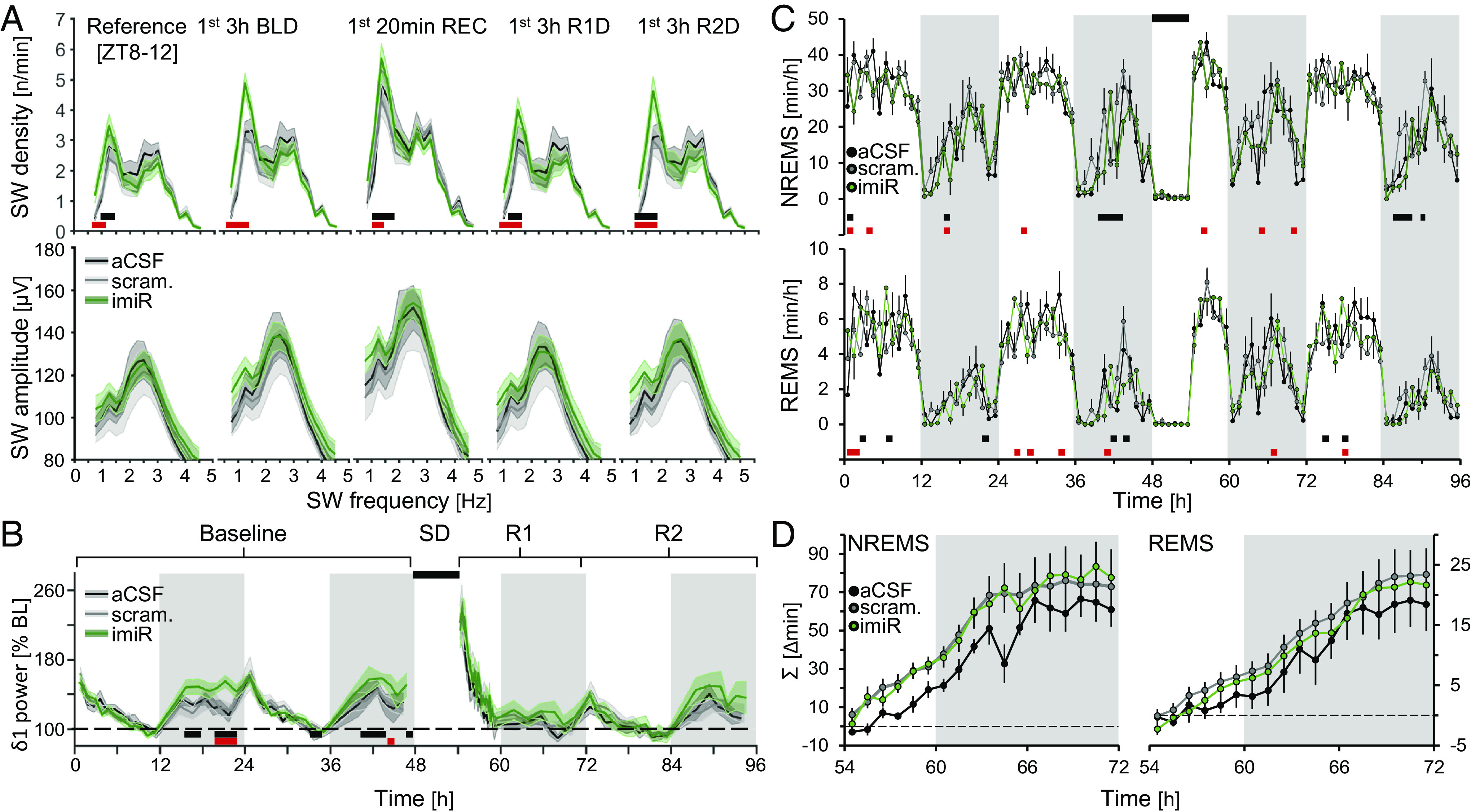
miR-709 inhibition increased SW density in the δ1 range during NREMS. (A) Mice inhibited for miR-709 (imiR, green) show increases in δ1 SW density (Top) compared to either scrambled (gray) or aCSF (black) controls during (from Left to Right): the last 4 h of light period [Reference (ZT8-12)], the first 3 h of baseline dark period NREMS (1st 3 h BLD), the first 20 min of NREMS after SD (1st 20 min REC light), and the first 3 h during the two subsequent recovery dark periods (1st 3 h R1D and R2D, respectively). Red squares represent significant changes between aCSF and imiR, and blue between scrambled and imiR, groups (two-way rANOVA interaction factors treatment × frequency; P < 0.001 for all periods; post hoc t tests P < 0.05). SW density is frequency corrected (n/frequency) to better illustrate SW bimodality. (B) Time-course of NREMS δ1 (0.75 to 1.75 Hz) power during 48 h of baseline and 42 h following a 6 h SD, i.e., recovery days R1 and R2. imiR mice had significantly higher δ1 power following spontaneous but not enforced waking (two-way rANOVA factors treatment × time; P = 0.04). Values are expressed as % of mean δ1 power during the last 4 h of the light period of baseline (ZT8-12). (C) Time-course of NREMS (Top) and REMS (Bottom) amounts per hour (two-way rANOVA interaction factors treatment × time; NREMS: P = 0.006; REMS: P = 0.03; post hoc tests < 0.05). (D) Accumulated differences in minutes between baseline and following 6 h SD, in NREMS and REMS. All values represent means (solid lines) ±1 SEM (shaded areas or bars). N = aCSF: 5; scrambled: 7; imiR: 7.
Taken together, these observations suggest a role for the mature miR-709 specifically in the regulation of NREMS EEG δ1 waves, which may also contribute to the similar δ1-density phenotype observed in the cKO and icKO mice (Fig. 2 A and C).
miR-709 Inhibition Downregulates Glutamatergic Receptor Transcripts in Primary Cortical Neurons.
To identify potential molecular pathways by which miR-709 inhibition affected the prevalence of SWs during NREMS, we next aimed at identifying its transcriptional targets. First, we quantified transcriptomic changes after inhibiting miR-709 in an in vitro assay, where inhibitors can be directly applied to the neuronal population of interest. Since miR-709 expression was previously observed in cortical neurons (43), its levels were affected by SD mainly in the cortex (Fig. 3 C and D), and a cortical origin has previously been suggested for slow SWs (32, 44), we treated immature cultured cortical neurons with either miR-709-specific LNA inhibitors (i-miR-709, n = 6), scrambled LNA inhibitors (scr, n = 6), or vehicle (ddH2O, n = 6). RNA sequencing (RNA-seq) after treatment identified a total of 13,760 expressed genes. To assess the effect of miR-709 inhibition at the individual gene level, gene expression in the inhibitor-treated mice was contrasted to those of the scrambled-treated mice. We found 111 genes to be differentially expressed (FDR < 0.05), with six genes (Akap12, s100a10, Acadm, Glipr2, Prrg3, and Hexb) being upregulated by miR-709 inhibition (Fig. 5A) and all others downregulated, which included several ion channel encoding genes. Besides determining the differential expression of individual genes, considering changes in the transcriptome as a whole, and assessing the effect of miR-709 inhibition on networks of co-expressed genes acting together in the same pathway(s), is likely to provide a more complete picture of its regulatory role. To accomplish this, we used a Weighted Gene Co-expression Network Analysis (WGCNA) on all sequenced transcripts of all 18 samples from the three experimental groups to screen for gene sets and related pathways. The analysis revealed eleven clusters, or modules, each containing a variable number of genes (from 100 to 2,000; SI Appendix, Fig. S4 A–C). Four modules were found to significantly correlate with miR-709 inhibition (SI Appendix, Fig. S4B; “green” r = −0.97; P = 9e-11; “purple” r = 0.84, P = 1e-05; “black” r = 0.52, P = 0.03; and “pink” r = 0.54, P = 0.02). The most representative genes of each of these modules, i.e., the “hub genes” identified by WGCNA (SI Appendix, Fig. S4D and Table S3), are co-expressed and often interact biologically at the protein level (45), as illustrated by the protein–protein interaction (PPI) networks (Fig. 5B). Interestingly, among the hub genes from two (black and pink) out of the three positively correlated networks, there was a strong presence of Rab GTPases and genes directly involved in endosomal trafficking (Fig. 5B; also see SI Appendix, Table S3). Conversely, the negatively correlated network (green) was heavily populated by ion-channel and synaptic-scaffold associated proteins, while it encompassed most of the 111 differentially expressed genes, with some (e.g., Camk2g, Nrg3, Tbce) emerging as hub genes. The topmost enriched molecular functions of the green hub genes (Fig. 5 C, Top) were associated with “extracellular glutamate gated ion channel activity” (GO: 0005234) and “ionotropic glutamate receptor activity” (GO: 0004970). Moreover, the main participating biological processes (Fig. 5 C, Bottom) were “regulation of vesicle docking” (GO: 0106020), “positive regulation of glutamate secretion, neurotransmission” (GO: 1903296), and “positive regulation of long-term synaptic depression” (GO: 1900454; see Fig. 5C for explanation).
Fig. 5.
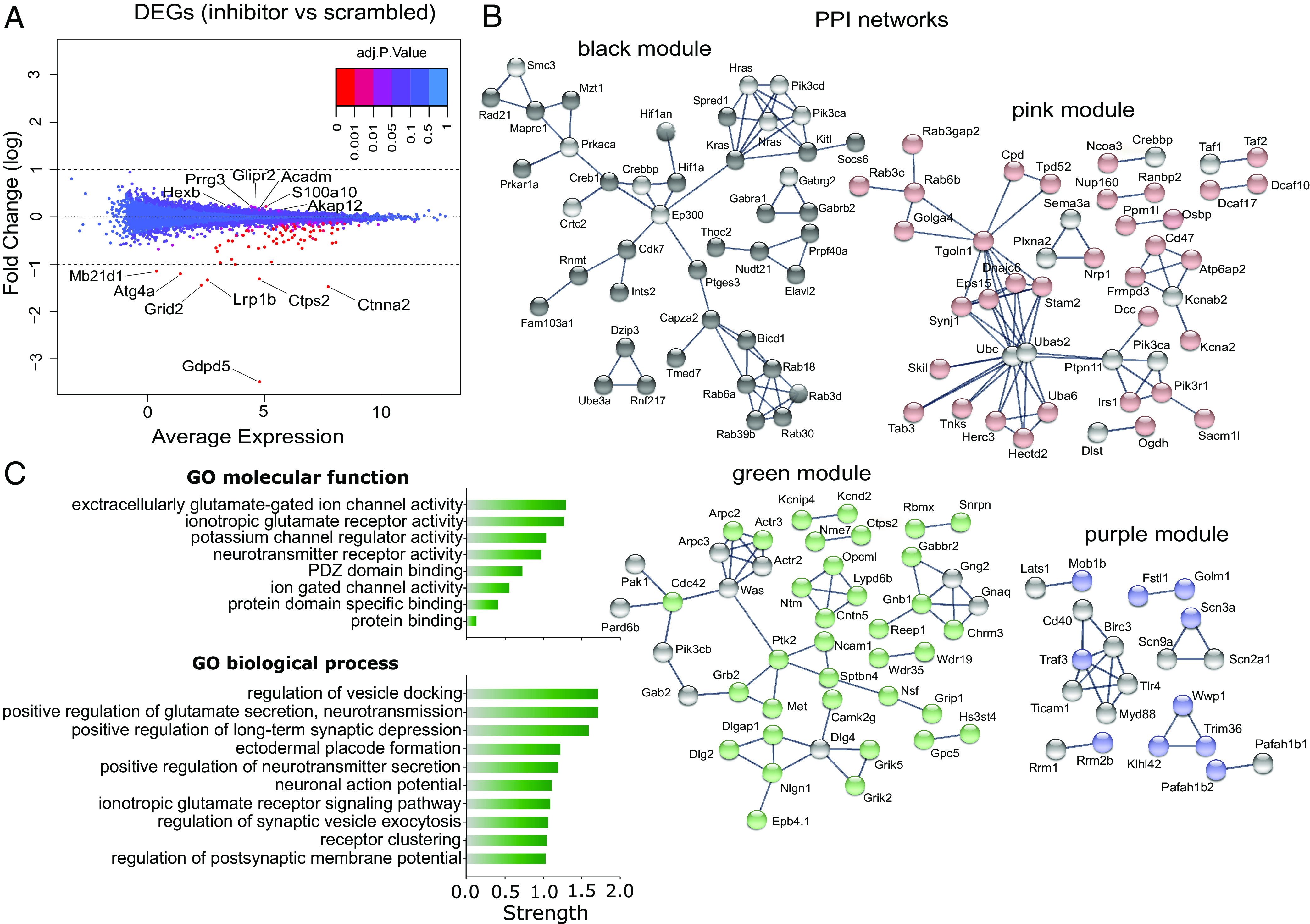
Differential expression and WGCNA of genes affected by miR-709 inhibition in primary cortical cultures. (A) Multivariate-analysis plot for the differential transcriptome analysis by RNA-seq between the inhibitor- and scramble-treated groups. Dots represent the differential expression of a gene (DEG); 111 genes were significantly altered by miR-709 inhibition (FDR < 0.05) some of which are named (for the full list, see SI Appendix, Table S2). (B and C) Weighted Gene Co-expression Network Analysis (WGCNA) for all genes identified by RNA-seq (Materials and Methods and SI Appendix, Fig. S4). (B) Visualization of the PPI networks of top interconnected (hub) genes (colored spheres) from the modules significantly correlated with in vitro miR-709 inhibition (SI Appendix, Fig. S4C) in STRING (46). Gray spheres represent genes that were directly linked to the hub genes in the modules (first shell interactors; “no more than 10” option) but were not identified in our initial dataset. (C) Top functional enrichment annotations of the most anti-correlated green module hub genes for Gene Ontology (GO) molecular function (8) and biological process (top 10). Pathway terms are hierarchically ordered, based on their “Strength” [log10 (observed/expected) number of genes in a given pathway].
In Vivo miR-709 Inhibition Differentially Impacts the Expression of Genes Involved in Synaptic Transmission and Regulation of Membrane Potential.
We subsequently assessed the levels of 107 out of the 111 in vitro differentially expressed transcripts in the cortex of 12-wk-old sleep-deprived male mice, ICV-injected with the same miR-709-specific (i-miR-709, n = 4) or scrambled (scr, n = 5) LNA inhibitors (Fig. 6A and SI Appendix, Fig. S6 A–D) as used previously for the EEG recordings. Inhibiting miR-709 significantly altered the expression of 23 of the 107 genes (19 up- and 4 downregulated), with two predicted targets of miR-709 (TargetScanMouse v8.0), namely Farp1 and Ctps2, being most affected (FDR < 0.05; Fig. 6A). Between the in vitro and in vivo experiments (Fig. 6B), four genes were found commonly upregulated (S100a10, Acadm, Hexb, and Glipr2), and four commonly downregulated (Grin1, Camk2g, Nrg3, and Tbce). The PCA biplot analysis of expression levels of these eight genes successfully assigned them to the correct treatment group in all in vivo samples suggesting that they were co-regulated (SI Appendix, Fig. S6D). Interestingly, Nrg3 levels were found reduced also in cortical tissue from non-sleep-deprived mice (SI Appendix, Fig. S6E).
Fig. 6.
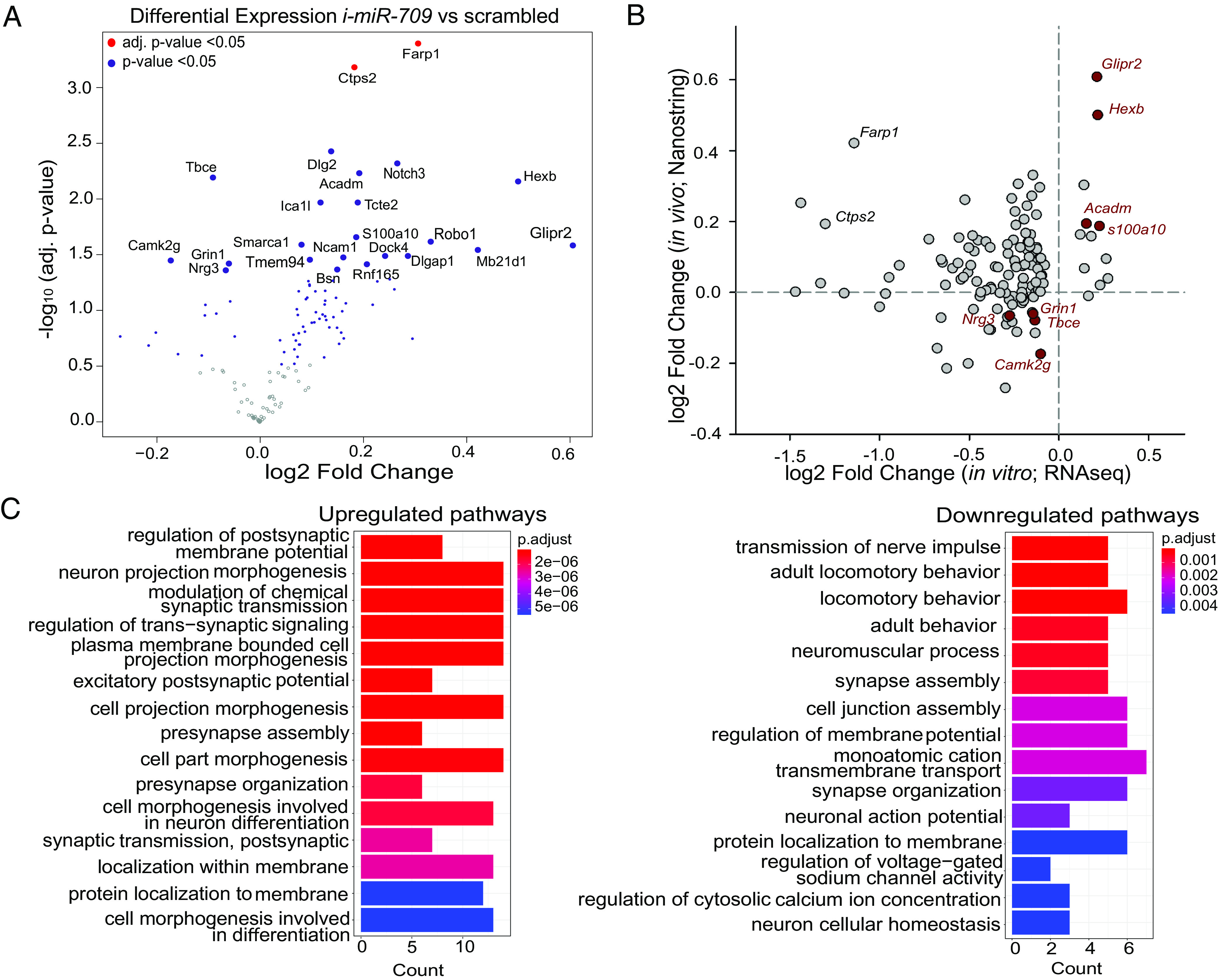
Differential gene expression after SD in the cortex of i-miR-709 treated animals. Expression levels of the 107 out of 111 differentially regulated genes from the in vitro miR-709 inhibition experiment were assessed in the cortical tissue of i-miR-709- (n = 4) or scrambled- (n = 5) injected animals after 6 h SD. (A) Volcano plot of differential gene expression between the two groups, displaying the log2 fold-change and the respective –log10 P-value for each individual gene. Genes with significantly altered expression (inhibited vs. scrambled) are depicted by the blue (t test, P-value < 0.05) and red (FDR < 0.05) dots. (B) Correlation between expression levels of the top 107 genes in the in vitro and in vivo miR-709 inhibition experiment. Note that four genes are found to be commonly up- and four downregulated in both datasets. (C) GO overrepresentation analysis (ORA) using the clusterProfiler package (Materials and Methods) for all up- (Left) and downregulated (Right) genes. Bar width of enriched terms is expressed as counts of participating genes. Bars are colored according to statistical significance (Benjamini–Hochberg multiple correction; P < 0.05; q < 0.05).
We next investigated the enriched pathways affected by the in vivo miR-709 inhibition in sleep-deprived issue. Separate enrichment analysis of all up- or downregulated genes revealed that the transcripts of genes associated with “regulation of membrane potential ” and “transmembrane cation transport” were reduced in murine cortices when miR-709 was inhibited, while genes involved in “synaptic morphogenesis” and “synaptic transmission” showed increased expression (Fig. 6C). Together with the pathways identified for the ‘green’ hub genes (Fig. 5C), these results show that “synapse assembly” and “glutamatergic neurotransmission” are regulated by miR-709 both in cortical tissue under sleep-deprived conditions, as well as in cultured cortical neurons.
Discussion
In this study, we show that ablation of mature miRNAs in two different Dicer knockout models led to alterations in brain synchrony during NREMS, as well as the time spent in REM sleep, particularly in response to sleep loss. We identified miRNAs affected by SD in the mouse forebrain and provided evidence that one among those, miR-709, regulates EEG slow waves. Using in vitro and in vivo assays, we found corroborating evidence that miR-709 exerts its effects through post-transcriptional regulation of pathways essential for activity-dependent synaptic plasticity and neuronal excitability. Our findings establish that miR-709 regulates the generation of slow waves in the mouse cortex, previously proposed to underlie synaptic plasticity among other processes (47).
Ablation of the Micro-RNA Maturation Machinery in Excitatory Neurons Dysregulates EEG Responses to Sleep Loss.
Knockout of Dicer early in development in other mouse models has been reported to be deleterious for brain development, with overt signs of neurodegeneration and severe neuroinflammation at 2.5 mo of age, resulting in early adulthood death (48, 49). In the current study, cKO mice were recorded at 2.5 to 3 mo of age, when neuroinflammatory processes set in, and thus might underlie the highly aberrant EEG signals observed in all sleep-wake states (Fig. 1 A and B and SI Appendix, Fig. S1 A and B). Indeed, inflammation has been previously reported to alter brain oscillatory function regardless of sleep-wake state (50). A neuroinflammatory phenotype may also have contributed to the lower NREMS EEG delta power levels that were reached after extended periods of spontaneous and enforced wakefulness. The profound and constitutive reduction in both the density and amplitude of EEG SWs during NREMS we observed in cKO mice seems to support an abnormally synchronized brain activity (50). Thus, the impact of disrupting the miRNA biogenesis machinery from early development on the EEG is likely not directly related to a lack of mature miRNAs but secondary to the resulting neuroinflammation and neurodegeneration. For these reasons, it is difficult to differentiate between a reduced capacity to produce slow waves and an altered underlying homeostatic process as causing the different EEG delta dynamics in the cKO mice. The use of icKO mice in which Dicer deletion occurs in adult postmitotic neurons is therefore preferable.
Although icKO mice developed normally, they are not without issues, as others have reported signs of neurodegeneration in specific brain regions several weeks after tamoxifen injection (51, 52). We previously observed a severe, but transient, metabolic disruption after tamoxifen injection in icKO mice, while brain transcriptome analysis in the cortex and hippocampus pointed to increased neuronal excitability at 4 and 8 wk after functional Dicer ablation (35). Indeed, loss of Dicer in adult forebrain neurons has been demonstrated to affect plasticity and to increase neuronal excitability, at least in CA1 pyramidal neurons (53). These authors reported that Dicer ablation increases neuronal excitation-dependent responsiveness and disrupts neuroprotective mechanisms against over-activation (53). In our hands, icKO mice exhibited a larger increase in EEG delta power in response to enforced wakefulness. Since SWs have been implicated in memory consolidation during sleep (54), this observation might provide a further link to the paradoxical transient enhancement of memory that was observed after loss of Dicer in Camk2a-expressing neurons (51, 53). In the current study, deletion of Dicer in icKO mice resulted in alteration in overall EEG power, and in the density and amplitude of NREMS slow waves. Specific inhibition of only one miRNA, miR-709, in cortical neurons exhibited a more frequency- and state-specific EEG phenotype, albeit in the same direction as icKO mice. It seems therefore plausible that miR-709 is among the miRNAs contributing to the observed phenotype in the Dicer icKO model. Together, our observations suggest that mature miRNAs in forebrain neurons modulate physiological brain synchrony expressed as δ1 density, both under physiological conditions and after sleep loss. The REMS homeostatic phenotype was shared only between the two Dicer-ablated mouse models, possibly due to additional neuronal subtypes or brain regions being affected in these models, or even as a result from a compromised cortico-hypothalamic communication (55), as compared to the miR-709-inhibited mice. Whether specific miRNAs contribute to this marked homeostatic REMS phenotype deserves further investigation.
miR-709 Inhibition Upregulates the Expression of Genes Participating in the Autophagic Machinery and Receptor Trafficking, While Genes Linked to NMDAR-Dependent Signaling Are Downregulated.
Validated targets of 40 out of the 48 differentially expressed miRNAs after SD in the forebrain, were found significantly enriched in genes associated with the regulation of cytoskeleton and endosomal transport. Additionally, miR-709, as well as miR-706, which was also upregulated after SD, have been found to target transcripts of protein kinase C alpha (56, 57), a suppressor of autophagy (58) and regulator of endosomal trafficking (59). miR-709-driven suppression of genes involved in cytoskeletal organization, as well as endosomal recycling and cell adhesion, was previously observed in non-neuronal tissue (40).
Since miRNAs suppress gene expression, inhibition of miR-709 levels can be expected to result in an upregulation of its direct targets. Thus, at least some of the shared upregulated genes between immature primary in vitro, and adult cortical neurons in vivo, could be direct targets of miR-709. Between the in vitro and in vivo model of miR-709 inhibition in the current study, four up- (Glipr2, S100a10, Hexb, and Acadm) and four down-regulated (Tbce, Nrg3, Camk2g, and Grin1) transcripts were shared. The levels of Glipr2, Hexb, Acadm, Camk2g, and Grin1 may also be affected by SD, as they were not significantly altered in cortical tissue of non-sleep-deprived mice after miR-709 inhibition, conversely to a consistently reduced Nrg3 expression. Glipr2 (aka Gapr-1), a negative regulator of autophagy (60), is a predicted target of miR-709 (TargetScanMouse, v8.0, 2021). S100a10 (aka p11), has been involved in autophagic pathways and controls the distribution of recycling endosomes (61), while hexosaminidase b (Hexb) is one of the main lysosomal enzymes for autophagy (62). The strong representation of Rab GTPases as biologically relevant hub genes in two out of the three upregulated gene networks after in vitro miR-709 inhibition, albeit not experimentally validated, is an additional indication that endosomal trafficking/autophagic pathways were affected. The upregulated Rab transcripts (Rab3c/d, -6a/b, -18, -30, and -39b), encode core proteins of autophagy and endosomal trafficking in neurons (63–68), in certain cases specifically involved in NMDAR trafficking (67, 69). An aberrant endosomal/autophagic process alters synaptic neurotransmission (reviewed in ref. 70), thereby modifying the electrophysiological properties of the neuron (71) and brain synchrony (72).
In agreement with these observations, the in vitro and in vivo shared downregulated genes after miR-709 inhibition were shown to be strongly involved in the electrophysiological properties of neurons, with the exception of Tbce (tubulin-specific chaperone E). More specifically, Camk2g (aka CaMKIIγ or γCaMKII) acts downstream of NMDAR signaling, to regulate experience-dependent transcription and memory (73). Camk2g is important for Ca2+/CaM shuttling to the nucleus thereby, triggering CREB phosphorylation and gene expression, a signaling pathway suggested to track and respond to neuronal depolarization rather than synaptic activity (74). NRG3 is a known mediator in the assembly of cortical excitatory-inhibitory circuits, expressed by pyramidal cells as early as E13.5 (75). It regulates glutamatergic transmission via the SNARE complex, and its dysregulation has been associated with hypofunction of the glutamatergic pathway in NRG3-related mental disorders (76). Knocking out Nrg3 increases sEPSC frequency, but not amplitude, as a result of more excitatory drive compared to controls (76, 77). As a member of the neuregulin family, it induces reduction of NMDA receptor currents, potentially by boosting the internalization of NR1, the fundamental subunit of NMDARs (reviewed in ref. 78). Last, and most importantly, the downregulated Grin1 encodes the obligatory subunit NR1, thus delineating a common pathway between three out of the four downregulated genes.
Considering its target genes and their pathways, miR-709 appears to fine-tune the balance between activity-dependent plasticity processes, such as receptor trafficking, cytoskeletal remodeling, and survival of the neuron. Lower levels of miR-709, particularly in periods of prolonged wakefulness, may contribute to an excitotoxic environment via imbalanced autophagic/endosomal trafficking pathways (79), which ultimately results in the downregulation of transcriptional levels of genes participating in glutamatergic signaling. This is also in agreement with previous observations where Grin1 transcriptional downregulation has been observed as a response to glutamate-induced excitotoxicity (80). While our results strongly suggest a role for miR-709 in glutamatergic transmission, the precise molecular mechanisms and how they relate to slow-wave generation should be investigated in follow-up work.
miR-709 Inhibition Increases NREMS Slow-Delta Density Potentially via Transcriptional Dysregulation of NMDAR-Dependent Receptor Trafficking.
The observed increase in the prevalence of δ1 SWs in the miR-709 inhibited mice was present throughout the recording (Fig. 4A). When comparing the δ1 SW density during the dark periods and after SD for the miR-709-inhibited group, it became apparent that slow waves occurred in a similar rate after either spontaneous or enforced prolonged wakefulness. This is in line with our understanding that activity in the δ1 frequency band is mostly of cortical origin and linked to aspects of extended wakefulness that do not increase proportionally with prior sleep-wake history (32). A potential mechanism for the impact of miR-709-inhibition on δ1 SW occurrence may lie with NMDAR-dependent signaling, since miR-709 was recently found to be upregulated after extra-synaptic-NMDAR-induced excitotoxicity (41). Previously, its expression was shown to increase in response to in vivo application of (S)-3,5-dihydroxyphenylglycine (DHPG), an agonist of group I mGluRs (42), as well as stress (81), and in our hands after enforced wakefulness. Indeed, cortical NMDAR downregulation or blockade was shown to increase EEG SWs during NREMS in an experience-dependent manner (82, 83). In rats, antagonism of NMDAR function suppresses spontaneous bursting activity in cortical neurons (84), and increases EEG SW activity with a peak around 1.75 Hz (85), specifically during NREMS. A causal link between cortical activity surges in the slow-wave range and NMDAR dysregulation has also been observed following the application of the NMDAR-antagonist ketamine (86), as well as in models of NMDAR hypofunction, particularly after the administration of the acute chemoconvulsant pentylenetetrazole, which enhances neuronal excitation (87). Reduced expression of NMDAR-signaling genes like Nrg3 and Grin1 also supports the notion that miR-709 inhibition alters excitation dynamics in cortical neurons, thereby boosting δ1 wave density in an activity-dependent manner (88).
To conclude, our study attributes a role for miRNAs in EEG SW regulation. Our transcriptional and electrophysiological findings underline the importance of taking developmental, temporal, and spatial aspects of miRNA expression and function into consideration for future studies. Importantly, the role of endosomal trafficking pathways and glutamatergic signaling in the regulation of plasticity and brain activity should be further elucidated in the context of sleep, particularly when their reciprocal interaction has been recently confirmed in several species (9, 89–91).
Materials and Methods
Animals and Housing Conditions.
All mice used were males, individually housed in polycarbonate cages (31 × 18 × 18 cm) in a sound-attenuated and temperature/humidity-controlled room (23 to 24 °C, 50 to 60% respectively). Mice were kept under a 12 h light/ 12 h dark cycle (light intensity 70 to 90 lx) and had access to food and water ad libitum. All experiments were approved by the Ethical Committee of the State of Vaud Veterinary Office, Switzerland (license number 2348 and 2676), and for the conditional Dicer KO mice by the Ethical Committee for Animal Experimentation of the Hôpital du Sacré-Coeur de Montréal.
Generation of Conditional Dicer Knockout Mice.
A non-inducible, forebrain-specific conditional Dicer knockout (cKO) mouse line was created by crossing homozygotes Dicerfl/fl (92) to a transgenic α-CamKII-Cre line (93, 94). The generation and characterization of the Camk2a-Cre+;Dicerfl/fl KO mice was described previously (94, 95). For all experiments, Dicerfl/+mice were used as control.
Generation of Inducible Conditional Dicer Knockout Mice.
A forebrain-specific and inducible conditional Dicer knockout (icKO) mouse line was obtained by crossing hemizygote Camk2a-CreERT2 mice (96), expressing an inducible Cre-recombinase under the control of the Camk2a promoter (Cre+; Jackson laboratory, Bar Harbor, ME, USA, stock number 012362) mice with homozygote Dicerfl/fl mice (Jackson stock #006366).
EEG/EMG (Electromyographic) Implantation.
At 10 wk of age, mice were implanted with EEG and EMG electrodes under deep anesthesia (i.p.; xylazine 10 mg/kg; ketamine 75 mg/kg) as detailed previously [(97); also see SI Appendix, Supporting Methods]. Animals were allowed to recover from surgery for approximately 5 d before they were connected to the recording cables in their home cage and an additional minimum of 6 d to habituate to the cables and the experimental room.
In Vivo miR-709 Inhibition.
Male C57BL6/J mice were implanted with a 5-mm-long cannula (PlasticsOne) reaching the right ventricle at 0.3 mm posterior from the bregma (AP), 0.84 mm lateral from the midline (ML), and 2.43 mm depth (DV). After habituation to the implants and the recording room, mice were injected ICV by means of a Hamilton 5-μL syringe, with either LNA miR-709 specific power inhibitors (i-miR-709: 5′-CTC CTG CCT CTG CCT C-3′, Exiqon; 100 mM final conc., n = 7), LNA scrambled power inhibitors (scr: 5′-ACG TCT ATA CGC CCA-3′, Exiqon; 100 mM final conc., n = 7) or vehicle (n = 6), at a speed of 0.6 μL/min. The vehicle for all injections was 4 μL of artificial cerebrospinal fluid (aCSF; composed of 150 mM Na, 3.0 mM K, 1.4 mM Ca, 0.8 mM Mg, 1.0 mM P; 155 mM Cl; pH 7.4). Post-injection, animals were left undisturbed for 48 h prior to baseline recordings, as recommended by the LNA inhibitor manufacturer (Exiqon, Vedbaek, Denmark).
Sleep Recordings and Data Acquisition.
Dicer cKO and ICV-injected mice were recorded at the age of 12 to 14 wk, while Dicer icKO mice were recorded at 16 wk. EEG and EMG signals were recorded continuously for 72 h. During the first 48 h, mice were left undisturbed and these 2 d were considered as baseline. Starting at light onset of the third day, animals were submitted to a SD by gentle handling during 6 h (ZT0–6). The remaining 18 h after the end of the SD were considered as recovery. Offline, the animal’s sleep-wake behavior was visually classified as Wakefulness, NREMS, or REMS based on the EEG and EMG signals as previously described (32, 35, 97). Four-second epochs containing EEG artifacts were marked according to the state in which they occurred.
Sleep Distribution and EEG Spectral Analysis.
Analysis of the distribution of each behavioral state was performed on 1-h values as described previously (30). Results from the two baseline days were averaged. The difference in the recovery-baseline accumulation was calculated by expressing the recovery time spent in sleep as a difference from the baseline time spent asleep for each hour. Time course analysis of EEG delta power during baseline and after SD was performed as described previously (29, 32).
EEG spectral analysis was performed for different time periods, as described previously (29, 32). In the in vivo miR-709-inhibited mice, interindividual differences in overall EEG signal power were normalized by expressing EEG spectral density in each frequency bin as a percentage of a baseline reference calculated as the mean total EEG power over all frequencies and sleep-wake states over the 48 h of baseline. The relative contribution of each sleep-wake state to this individual reference was weighted as to avoid that, e.g., individuals that spent more time in NREMS (during which overall EEG power is higher compared to wake and REMS) obtain a higher reference power as a result. For slow-wave parameter analysis, see SI Appendix, Supporting Methods.
miRNA Microarrays.
Sixteen and 8 C57BL/6J mice (12 wk old) were used in experiments 1 (forebrain analysis) and 2 (cortex and hippocampus analysis), respectively. In each experiment, half of the mice were submitted to a 6-h SD performed by the so-called “gentle handling protocol,” starting at light onset (ZT0). The other half was left undisturbed and used as controls. At the end of the SD (ZT6), animals from both groups were killed and the forebrain (experiment 1) or cortex and hippocampus (experiment 2) were dissected and collected. In the first experiment, a total of 16 arrays (i.e., 8 SD and 8 control animals) were used. For the second experiment, samples were pooled per group of 2 animals for each condition (SD/control) and for each brain region (cortex/hippocampus), resulting in a total of 8 arrays. Statistical analysis was performed with the R Bioconductor package limma by fitting a linear model and computing moderated t tests, comparing miRNA expression levels in the SD vs. the control group. For multiple testing correction within each experiment, the Benjamini–Hochberg method was applied (98).
In Situ Hybridization.
A separate group of SD (n = 5) and control (n = 5) mice was used to perform in situ hybridization on brain samples after 6 h of SD or undisturbed sleep. Paraformaldehyde-fixed mouse brains were cryosected into 20-μm sections and hybridized with a miRCURY LNA™ miRNA Detection kit according to the manufacturer’s instructions [Optimization Kit 4 (miR-124), Exiqon, Vedbaek, Denmark].
Transfection of Primary Cortical Cultures.
Dissociated cortical primary cultures were prepared from C57BL/6J mouse brains at embryonic day 17 to 18 (E17-E18; of either sex) as detailed previously (99, 100). After 3 days in vitro (DIV3), half of the medium was removed and kept at 37 °C. Subsequently, primary cortical cells were treated for 12 h with 2 μL of Lipofectamine® 2000 (Invitrogen) transfection reagent, and 2 μL of either specific miR-709 inhibitors (i-miR-709, 40 nM final conc., n = 6), scrambled (scr, 40 nM final conc., n = 6) or ddH2O (ctrl, n = 6; for the exact constructs see Materials and Methods, In Vivo miR-709 Inhibition), diluted in a total of 100 μL Gibco™ Opti-MEM™ Reduced Serum Media (ThermoFisher Scientific), according to the manufacturer’s protocol. Forty-eight hours post-transfection (DIV5), cells were lysed for RNA extraction. Based on previous studies (101–103), DIV5 was within the optimal window (DIV5-7) for immature cortical neurons to start forming functional networks and synapses, but without any apoptosis signaling which would have altered miRNA and gene expression, and without astrocytes which would have added “noise” in our subsequent transcriptomic assay.
RNA-seq.
Following treatment, RNA from primary cortical cells (DIV5) was extracted and purified using the RNeasy Mini Kit 50 (Qiagen), according to the manufacturer instructions for cells grown in a monolayer. Differential expression was computed with the limma-trend approach (104) by fitting all sample results into one linear model and adding the experimental batches in the design matrix. Moderated t tests were used for comparing i-miR-709 injected to the scrambled group. As for the miRNA arrays, the adjusted P-values were computed by the Benjamini–Hochberg method (98), controlling for FDR.
Weighted Gene Co-Expression Network Analysis (WGCNA).
To investigate pathways impacted by miR-709 inhibition, the freely available statistical analysis software [WGCNA R package, (45)] was used. All 13,760 genes were clustered into modules, taking into consideration their expression in all samples for all treatments (SI Appendix, Supporting Methods). Visualization of the interactions between the top representative genes for each of the four significant modules with respect to the treatment, was performed with the help of STRING v11.0 resource (46) (Fig. 5 and SI Appendix, Fig. S5). For the network creation, only the interaction with “highest confidence (0.9)” and “no more than 10” first shell interactors were used. Finally, the functional annotation for the green module was performed with the build-in Functional Enrichment tool of the STRING resource (46, 105), using the entire set of 13,760 genes identified by RNA-seq as background.
Nanostring Assays and Analysis of Gene Expression.
Cortical tissues dissected from i-miR-709 and scrambled-ICV-injected SD mice (n = 5/group) were collected at ZT6, snap frozen, and stored at −80 °C. After RNA extraction (SI Appendix, Supporting Methods), the expression analysis was performed using a custom CodeSet designed to investigate the 107 out of the 111 transcripts that were found differentially regulated in the in vitro miR-709 inhibition (SI Appendix, Table S2), due to lack of trustworthy probes for the four remaining genes. The pathway enrichment barplots were created using the enrichGO function of the clusterProfiler 4.8.1 R package (106) and using separately all up- (75) and downregulated (33) genes (also see SI Appendix, Supporting Methods for details).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This study was supported by the Swiss National Science Foundation (nos. 130825 and 136201 to P.F. supporting S.J. and C.N.H.), the state of Vaud supporting K.K., G.M.M., J.H., Y.E., and P.F., the Canada Research Chair in Sleep Molecular Physiology and a grant from the Canadian Institutes of Health Research to V.M. The in vitro experiments were supported by a grant from the Sophie Afenduli Foundation to K.K. We acknowledge the Lausanne Genomics Technologies Facility for their invaluable contribution to all transcriptomic assays and Prof. Antoine Adamantidis (University of Bern) for his critical support in the completion of this study.
Author contributions
K.K., G.M.M., C.N.H., V.M., and P.F. designed research; K.K., G.M.M., S.J., Y.E., and V.M. performed research; K.K., S.S.H., V.M., and P.F. contributed new reagents/analytic tools; K.K., G.M.M., J.H., C.P., L.W., and P.F. analyzed data; and K.K., G.M.M., J.H., C.N.H., L.W., S.S.H., V.M., and P.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Konstantinos Kompotis, Email: konstantinos.kompotis@pharma.uzh.ch.
Paul Franken, Email: paul.franken@unil.ch.
Data, Materials, and Software Availability
Raw read files (.fastq), nanostring files (.rcc) andgene counts (.txt) are publicly available in the Gene Expression Omnibus repository under accession no. GSE189353 (107).
Supporting Information
References
- 1.Maret S., et al. , Homer1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. U.S.A. 104, 20090–20095 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cirelli C., The genetic and molecular regulation of sleep: From fruit flies to humans. Nat. Rev. Neurosci. 10, 549–560 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allada R., Cirelli C., Sehgal A., Molecular mechanisms of sleep homeostasis in flies and mammals. Cold Spring Harb. Perspect. Biol. 9, a027730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Helou J., et al. , Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 9974–9979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diering G. H., et al. , Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science 355, 511–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirelli C., A molecular window on sleep: Changes in gene expression between sleep and wakefulness. Neuroscientist 11, 63–74 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Ahnaou A., Raeymaekers L., Steckler T., Drinkenbrug W. H. I. M., Relevance of the metabotropic glutamate receptor (mGluR5) in the regulation of NREM-REM sleep cycle and homeostasis: Evidence from mGluR5 (-/-) mice. Behav. Brain Res. 282, 218–226 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Holst S. C., et al. , Cerebral mGluR5 availability contributes to elevated sleep need and behavioral adjustment after sleep deprivation. Elife 6, e28751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Y., et al. , Chronic sleep fragmentation shares similar pathogenesis with neurodegenerative diseases: Endosome-autophagosome-lysosome pathway dysfunction and microglia-mediated neuroinflammation. CNS Neurosci. Ther. 26, 215–227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chauhan A. K., Mallick B. N., Association between autophagy and rapid eye movement sleep loss-associated neurodegenerative and patho-physio-behavioral changes. Sleep Med. 63, 29–37 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Li Y., et al. , Autophagy triggered by oxidative stress appears to be mediated by the AKT/mTOR signaling pathway in the liver of sleep-deprived rats. Oxid. Med. Cell Longev. 2020, 6181630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J., Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Martin H. C., et al. , Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol. 15, R51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketting R. F., MicroRNA biogenesis and function: An overview. Adv. Exp. Med. Biol. 700, 1–14 (2010). [PubMed] [Google Scholar]
- 15.Saba R., Schratt G. M., MicroRNAs in neuronal development, function and dysfunction. Brain Res. 1338, 3–13 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Wang Z., miRNA in the regulation of ion channel/transporter expression. Compr. Physiol. 3, 599–653 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Hawley Z. C. E., Campos-Melo D., Droppelmann C. A., Strong M. J., MotomiRS: MiRNAs in motor neuron function and disease. Front. Mol. Neurosci. 10, 127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand C. M., et al. , Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm A., et al. , The evolutionarily conserved miRNA-137 targets the neuropeptide hypocretin/orexin and modulates the wake to sleep ratio. Proc. Natl. Acad. Sci. U.S.A. 119, e2112225119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surbhi, et al. , miR-155 deletion modulates lipopolysaccharide-induced sleep in female mice. Chronobiol. Int. 36, 188–202 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Davis C. J., et al. , MicroRNA 132 alters sleep and varies with time in brain. J. Appl. Physiol. 111, 665–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis C. J., Clinton J. M., Krueger J. M., MicroRNA 138, let-7b, and 125a inhibitors differentially alter sleep and EEG delta-wave activity in rats. J. Appl. Physiol. 113, 1756–1762 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis C. J., Bohnet S. G., Meyerson J. M., Krueger J. M., Sleep loss changes microRNA levels in the brain: A possible mechanism for state-dependent translational regulation. Neurosci. Lett. 422, 68–73 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balakathiresan N. S., et al. , Micrornas in basolateral amygdala associated with stress and fear memories regulate rapid eye movement sleep in rats. Brain Sci. 11, 489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Rosbash M., MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat. Commun. 8, 14707 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodwin P. R., et al. , MicroRNAs regulate sleep and sleep homeostasis in Drosophila. Cell Rep. 23, 3776–3786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobin M., et al. , The Drosophila microRNA bantam regulates excitability in adult mushroom body output neurons to promote early night sleep. iScience, 25, 104847 (2022). 10.2139/ssrn.3942131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes J. R., et al. , The relationship between glutamate dynamics and activity-dependent synaptic plasticity. J. Neurosci. 40, 2793–2807 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franken P., Chollet D., Tafti M., The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 21, 2610–2621 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franken P., Malafosse A., Tafti M., Genetic determinants of sleep regulation in inbred mice. Sleep 22, 155–169 (1999). [PubMed] [Google Scholar]
- 31.Collins B., et al. , Circadian VIPergic neurons of the suprachiasmatic nuclei sculpt the sleep-wake cycle. Neuron 108, 486–499 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbard J., et al. , Rapid fast-delta decay following prolonged wakefulness marks a phase of wake-inertia in NREM sleep. Nat. Commun. 11, 3130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Follert P., Cremer H., Béclin C., MicroRNAs in brain development and function: A matter of flexibility and stability. Front. Mol. Neurosci. 7, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajman M., Schratt G., MicroRNAs in neural development: From master regulators to fine-tuners. Development 144, 2310–2322 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Mang G. M., et al. , A neuron-specific deletion of the microRNA-processing enzyme dicer induces severe but transient obesity in mice. PLoS One 10, e0116760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimoto Y., Akiyama Y., Yuasa Y., Multiple-to-multiple relationships between micrornas and target genes in gastric cancer. PLoS One 8, e62589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang L., Zhou G., Soufan O., Xia J., miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48, W244–W251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malkani S., et al. , Circulating miRNA spaceflight signature reveals targets for countermeasure development. Cell Rep. 33, 108448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isakova A., Fehlmann T., Keller A., Quake S. R., A mouse tissue atlas of small noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 117, 25634–25645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surendran S., et al. , Gene targets of mouse miR-709: Regulation of distinct pools. Sci. Rep. 6, 18958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bas-Orth C., Koch M., Lau D., Buchthal B., Bading H., A microRNA signature of toxic extrasynaptic N-methyl-D-aspartate (NMDA) receptor signaling. Mol. Brain 13, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lusardi T. A., et al. , Effect of (S)-3,5-DHPG on microRNA expression in mouse brain. Exp. Neurol. 235, 497–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki Y., Gross C., Xing L., Goshima Y., Bassell G. J., Identification of axon-enriched MicroRNAs localized to growth cones of cortical neurons. Dev. Neurobiol. 74, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steriade M., Nunez A., Amzica F., A novel slow (<1 Hz) oscillation of neocortical neurons in vivo: Depolarizing and hyperpolarizing components. J. Neurosci. 13, 3252–3265 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyamoto D., Hirai D., Murayama M., The roles of cortical slow waves in synaptic plasticity and memory consolidation. Front. Neural Circuits 11, 92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis T. H., et al. , Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hébert S. S., et al. , Genetic ablation of dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 19, 3959–3969 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Mamad O., Islam M. N., Cunningham C., Tsanov M., Differential response of hippocampal and prefrontal oscillations to systemic LPS application. Brain Res. 1681, 64–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konopka W., et al. , MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinnikov I. A., et al. , Hypothalamic miR-103 protects from hyperphagic obesity in mice. J. Neurosci. 34, 10659–10674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fiorenza A., et al. , Blocking miRNA biogenesis in adult forebrain neurons enhances seizure susceptibility, fear memory, and food intake by increasing neuronal responsiveness. Cereb. Cortex 26, 1619–1633 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Diekelmann S., Born J., Slow-wave sleep takes the leading role in memory reorganization. Nat. Rev. Neurosci. 11, 218 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Krone L. B., et al. , A role for the cortex in sleep–wake regulation. Nat. Neurosci. 24, 1210–1215 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin R., Guo D., Zhang S., Zhang X., MiR-706 inhibits the oxidative stress-induced activation of PKCα/TAOK1 in liver fibrogenesis. Sci. Rep. 6, 37509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng Y., et al. , miR-709 inhibits GHRP6 induced GH synthesis by targeting PRKCA in pituitary. Mol. Cell Endocrinol. 506, 110763 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Wang F., et al. , Protein kinase C-alpha suppresses autophagy and induces neural tube defects via miR-129-2 in diabetic pregnancy. Nat. Commun. 8, 15182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alvi F., Idkowiak-Baldys J., Baldys A., Raymond J. R., Hannun Y. A., Regulation of membrane trafficking and endocytosis by protein kinase C: Emerging role of the pericentrion, a novel protein kinase C-dependent subset of recycling endosomes. Cell. Mol. Life Sci. 64, 263–270 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y., et al. , GLIPR2 is a negative regulator of autophagy and the BECN1-ATG14-containing phosphatidylinositol 3-kinase complex. Autophagy 17, 2891–2904 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zobiack N., Rescher U., Ludwig C., Zeuschner D., Gerke V., The annexin 2/S100A10 complex controls the distribution of transferrin receptor-containing recycling endosomes. Mol. Biol. Cell 14, 4896–4908 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brekk O. R., et al. , Upregulating β-hexosaminidase activity in rodents prevents α-synuclein lipid associations and protects dopaminergic neurons from α-synuclein-mediated neurotoxicity. Acta Neuropathol. Commun. 8, 127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlüter O. M., Schmitz F., Jahn R., Rosenmund C., Südhof T. C., A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 24, 6629–6637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mallard F., et al. , Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nian F. S., et al. , Rab18 collaborates with Rab7 to modulate lysosomal and autophagy activities in the nervous system: An overlapping mechanism for Warburg micro syndrome and charcot-marie-tooth neuropathy type 2B. Mol. Neurobiol. 56, 6095–6105 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Zulkefli K. L., et al. , A role for Rab30 in retrograde trafficking and maintenance of endosome-TGN organization. Exp. Cell Res. 399, 112442 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Niu M., et al. , RAB39B deficiency impairs learning and memory partially through compromising autophagy. Front. Cell Dev. Biol. 8, 598–622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Passemard S., et al. , Golgi trafficking defects in postnatal microcephaly: The evidence for “Golgipathies”. Prog. Neurobiol. 153, 46–63 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Wang J., et al. , Postsynaptic RIM1 modulates synaptic function by facilitating membrane delivery of recycling NMDARs in hippocampal neurons. Nat. Commun. 9, 2267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng Z., Zhou X., Lu J. H., Yue Z., Autophagy deficiency in neurodevelopmental disorders. Cell Biosci. 11, 214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuijpers M., et al. , Neuronal autophagy regulates presynaptic neurotransmission by controlling the axonal endoplasmic reticulum. Neuron 109, 299–313.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Linda K., et al. , Imbalanced autophagy causes synaptic deficits in a human model for neurodevelopmental disorders. Autophagy 18, 423–442 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cohen S. M., et al. , Calmodulin shuttling mediates cytonuclear signaling to trigger experience-dependent transcription and memory. Nat. Commun. 9, 2451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma H., et al. , γcaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell 159, 281–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bartolini G., et al. , Neuregulin 3 mediates cortical plate invasion and laminar allocation of GABAergic interneurons. Cell Rep. 18, 1157–1170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y. N., et al. , Controlling of glutamate release by neuregulin3 via inhibiting the assembly of the SNARE complex. Proc. Natl. Acad. Sci. U.S.A. 115, 2508–2513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Müller T., et al. , Neuregulin 3 promotes excitatory synapse formation on hippocampal interneurons. EMBO J. 37, e98858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu Z., Jiang Q., Fu A. K. Y., Ip N. Y., Yan Z., Regulation of NMDA receptors by neuregulin signaling in prefrontal cortex. J. Neurosci. 25, 4974–4984 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sadasivan S., et al. , Acute NMDA toxicity in cultured rat cerebellar granule neurons is accompanied by autophagy induction and late onset autophagic cell death phenotype. BMC Neurosci. 11, 21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gascón S., et al. , Transcription of the NR1 subunit of the N-methyl-D-aspartate receptor is down-regulated by excitotoxic stimulation and cerebral ischemia. J. Biol. Chem. 280, 35018–35027 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Babenko O., Golubov A., Ilnytskyy Y., Kovalchuk I., Metz G. A., Genomic and epigenomic responses to chronic stress involve miRNA-mediated programming. PLoS One 7, e29441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell I. G., Feinberg I., NREM delta stimulation following MK-801 is a response of sleep systems. J. Neurophysiol. 76, 3714–3720 (1996). [DOI] [PubMed] [Google Scholar]
- 83.Miyamoto H., Katagiri H., Hensch T., Experience-dependent slow-wave sleep development. Nat. Neurosci. 6, 553–564 (2003). [DOI] [PubMed] [Google Scholar]
- 84.Armstrong-James M., Fox K., Evidence for a specific role for cortical NMDA receptors in slow-wave sleep. Brain Res. 451, 189–196 (1988). [DOI] [PubMed] [Google Scholar]
- 85.Duncan W. C., et al. , Concomitant BDNF and sleep slow wave changes indicate ketamine-induced plasticity in major depressive disorder. Int. J. Neuropsychopharmacol. 16, 301–311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chauvette S., Crochet S., Volgushev M., Timofeev I., Properties of slow oscillation during slow-wave sleep and anesthesia in cats. J. Neurosci. 31, 14998–15008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosch R. E., et al. , NMDA-receptor antibodies alter cortical microcircuit dynamics. Proc. Natl. Acad. Sci. U.S.A. 115, E9916–E9925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pafundo D. E., Pretell Annan C. A., Fulginiti N. M., Belforte J. E., Early NMDA receptor ablation in interneurons causes an activity-dependent E/I imbalance in vivo in prefrontal cortex pyramidal neurons of a mouse model useful for the study of schizophrenia. Schizophr. Bull. 47, 1300–1309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Vivo L., et al. , Loss of sleep affects the ultrastructure of pyramidal neurons in the adolescent mouse frontal cortex. Sleep 39, 861–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng Y., Kim W. K., Wellman L. L., Sanford L. D., Guo M. L., Short-term sleep fragmentation dysregulates autophagy in a brain region-specific manner. Life 11, 1098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bedont J. L., et al. , Short and long sleeping mutants reveal links between sleep and macroautophagy. Elife 10, e64140. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harfe B. D., McManus M. T., Mansfield J. H., Hornstein E., Tabin C. J., The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. U.S.A. 102, 10898–10903 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu H., et al. , APP processing and synaptic plasticity in presenilin-1 conditional knockout mice. Neuron 31, 713–726 (2001). [DOI] [PubMed] [Google Scholar]
- 94.Cheng S., et al. , Age-dependent neuron loss is associated with impaired adult neurogenesis in forebrain neuron-specific Dicer conditional knockout mice. Int. J. Biochem. Cell Biol. 57, 186–196 (2014). [DOI] [PubMed] [Google Scholar]
- 95.Fukaya M., Kato A., Lovett C., Tonegawa S., Watanabe M., Retention of NMDA receptor NR2 subunits in the lumen of endoplasmic reticulum in targeted NR1 knockout mice. Proc. Natl. Acad. Sci. U.S.A. 100, 4855–4860 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mang G. M., Franken P., Sleep and EEG phenotyping in mice. Curr. Protoc. Mouse Biol. 2, 55–74 (2012). 10.1002/9780470942390.mo110126. [DOI] [PubMed] [Google Scholar]
- 98.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc.: Series B (Methodol.) 57, 289–300 (1995). [Google Scholar]
- 99.Mikhail C., Vaucher A., Jimenez S., Tafti M., ERK signaling pathway regulates sleep duration through activity-induced gene expression during wakefulness. Sci. Signal. 10, eaai9219 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Hinard V., et al. , Key electrophysiological, molecular, and metabolic signatures of sleep and wakefulness revealed in primary cortical cultures. J. Neurosci. 32, 12506–12517 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cotterill E., et al. , Characterization of early cortical neural network development in multiwell microelectrode array plates. J. Biomol. Screen 21, 510–519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Costantini C., et al. , Astrocytes regulate the expression of insulin-like growth factor 1 receptor (IGF1-R) in primary cortical neurons during in vitro senescence. J. Mol. Neurosci. 40, 342–352 (2010). [DOI] [PubMed] [Google Scholar]
- 103.Janzen D., et al. , Cortical neurons form a functional neuronal network in a 3D printed reinforced matrix. Adv. Healthc. Mater. 9, e1901630 (2020). [DOI] [PubMed] [Google Scholar]
- 104.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Szklarczyk D., et al. , The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu T., et al. , clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kompotis K., et al. , Superseries of microarray, RNA seq and Nanostring data. NCBI Gene Expression Omnibus. GSE189353. Deposited 22 November 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Raw read files (.fastq), nanostring files (.rcc) andgene counts (.txt) are publicly available in the Gene Expression Omnibus repository under accession no. GSE189353 (107).