Significance
It is critical to determine why antibiotics frequently fail despite appropriate treatment. Unlike antibiotic resistance, persister cells are a phenomenon not routinely tested for in the clinical laboratory. The clinical significance of persisters in bacteremia is largely unknown. Our data show that mutants with higher persister formation develop during relapsed Escherichia coli bloodstream infections. We focus on a loss-of-function mutation in the ptsI gene conferring a high-persister phenotype both in vitro and in vivo. This finding suggests that recurrent infections could be more prone to treatment failure when high-persister mutants are present. Our study demonstrates the in vivo relevance of persisters in bloodstream infections and provides further evidence that targeting and killing persisters will improve treatment outcomes in patients.
Keywords: antibiotic, tolerance, bacteremia, persistence, Escherichia coli
Abstract
Gram-negative bacterial bloodstream infections (GNB-BSI) are common and frequently lethal. Despite appropriate antibiotic treatment, relapse of GNB-BSI with the same bacterial strain is common and associated with poor clinical outcomes and high healthcare costs. The role of persister cells, which are sub-populations of bacteria that survive for prolonged periods in the presence of bactericidal antibiotics, in relapse of GNB-BSI is unclear. Using a cohort of patients with relapsed GNB-BSI, we aimed to determine how the pathogen evolves within the patient between the initial and subsequent episodes of GNB-BSI and how these changes impact persistence. Using Escherichia coli clinical bloodstream isolate pairs (initial and relapse isolates) from patients with relapsed GNB-BSI, we found that 4/11 (36%) of the relapse isolates displayed a significant increase in persisters cells relative to the initial bloodstream infection isolate. In the relapsed E. coli strain with the greatest increase in persisters (100-fold relative to initial isolate), we determined that the increase was due to a loss-of-function mutation in the ptsI gene encoding Enzyme I of the phosphoenolpyruvate phosphotransferase system. The ptsI mutant was equally virulent in a murine bacteremia infection model but exhibited 10-fold increased survival to antibiotic treatment. This work addresses the controversy regarding the clinical relevance of persister formation by providing compelling data that not only do high-persister mutations arise during bloodstream infection in humans but also that these mutants display increased survival to antibiotic challenge in vivo.
Gram-negative bacterial bloodstream infections (GNB-BSI) are common and frequently lethal (1–3). It is estimated that GNB-BSI represent ~40% of nosocomial and ~50% of community-acquired BSI and are associated with high mortality of up to 24% (1, 2). Despite appropriate antimicrobial treatment, 6 to 25% of patients will experience recurrence with multiple episodes of GNB-BSI for unclear reasons (4–13). There have been numerous studies examining the clinical risk factors for recurrence, but the influence of the pathogen biology on infection relapse remains largely unknown (8–10, 13).
Antibiotic tolerance and persister cells are thought to be an underappreciated cause of persistent and recurrent bacterial infections and can be a precursor to antimicrobial resistance (AMR) (14–20). Antibiotic tolerance is generally defined as the increased capacity of an entire population to survive in the presence of bactericidal antibiotics, whereas persisters are defined as a subpopulation of tolerant bacteria within an otherwise susceptible population (21). Unlike AMR, which is routinely detected in the clinical microbiology laboratory, antibiotic tolerance and persister formation are challenging to detect, with no universally accepted testing procedure (22–24). Considering the theory that tolerant bacteria and persisters will be more difficult to eradicate with antibiotics, the connection between antibiotic treatment failure and antibiotic tolerance certainly has biological plausibility. Surprisingly, there are few animal models and even fewer clinical studies examining such an association. One seminal study in Escherichia coli revealed that high-persister, hipA7 mutants occurred in patient isolates, but the contribution of these mutations to antibiotic susceptibility in vivo remains unknown (25). There is also a single prospective clinical study that shows E. coli clinical isolates identified as “tolerant” via the tolerance disc assay are more common in patients with recurrent E. coli bacteremia (26). As of 2023, there are, to our knowledge, no reports examining if high-persister bacterial isolates exhibit decreased antibiotic susceptibility during GNB-BSI.
The mechanisms behind antibiotic tolerance and persisters in Gram-negative bacteria have been most extensively studied in E. coli (27–31). Recently, Zeng et al. reported an in vitro screen to select for pan-tolerant mutants in E. coli MG1655 (32). They identified mutations in the ptsI and cyaA genes, encoding Enzyme I and adenylate cyclase (CyaA) enzymes respectively, which conferred tolerance to all antibiotics tested. Both ptsI and cyaA play key roles in the phosphoenolpyruvate phosphotransferase system (PTS)–CRP (cAMP response protein) (phosphoenolpyruvate phosphotransferase–cyclic Adenosine monophosphate (AMP) response protein) axis, which controls the uptake of a range of carbohydrates and plays a key role in carbon catabolite repression (33). The proposed mechanism for increased antibiotic tolerance builds on work from the Collins lab, linking bactericidal antibiotic treatment with metabolic reprogramming and production of lethal quantities of reactive oxygen species (ROS) (34–38). Upon treatment with bactericidal antibiotics, E. coli is thought to increase respiratory metabolism, producing ROS which can then lead to cell death. Zeng et al. demonstrated that a functioning PTS system is required to increase the flux through the tricarboxylic acid (TCA) cycle to boost respiratory metabolism in response to bactericidal antibiotics (32). Without the ptsI-mediated metabolic reprogramming, less ROS is generated, subsequently leading to reduced cell death. While this finding is a crucial addition to the growing body of data demonstrating the metabolic dependence of bactericidal antibiotics, it was limited to in vitro experiments in broth culture with a non-pathogenic laboratory strain of E. coli. The clinical relevance of mutations in the PTS pathway remains unclear, particularly in the context of bloodstream infections. It is unknown if loss-of-function mutants in the PTS system exist outside of the basic science laboratory and therefore, the clinical significance is undetermined. Furthermore, given the metabolic flexibility required to disseminate and survive in the bloodstream, it is unknown whether disruption of the PTS system would generate an organism resilient enough to establish infection (39).
We address these issues by using a cohort of isolates from patients with relapsed GNB-BSI (>1 episode of GNB-BSI with genetically near-identical isolate) to ask two questions: First, how does the pathogen evolve within the patient between the initial and relapse episode? Second, how do these genetic changes affect the development of persister cells? We identify multiple mutations arising within patients between initial and relapsed GNB-BSI episodes in genes associated with antibiotic tolerance and persisters. Importantly, we show a loss-of-function mutation in ptsI arises during recurrent E. coli GNB-BSI in humans. We show that disruption of ptsI does not affect the ability of E. coli to establish a BSI in mice, and the high-persister phenotype seen in vitro translates to decreased antibiotic efficacy in vivo in the same model.
Results
Identification of Single-Nucleotide Polymorphisms (SNPs) Arising during Relapsed GNB-BSI.
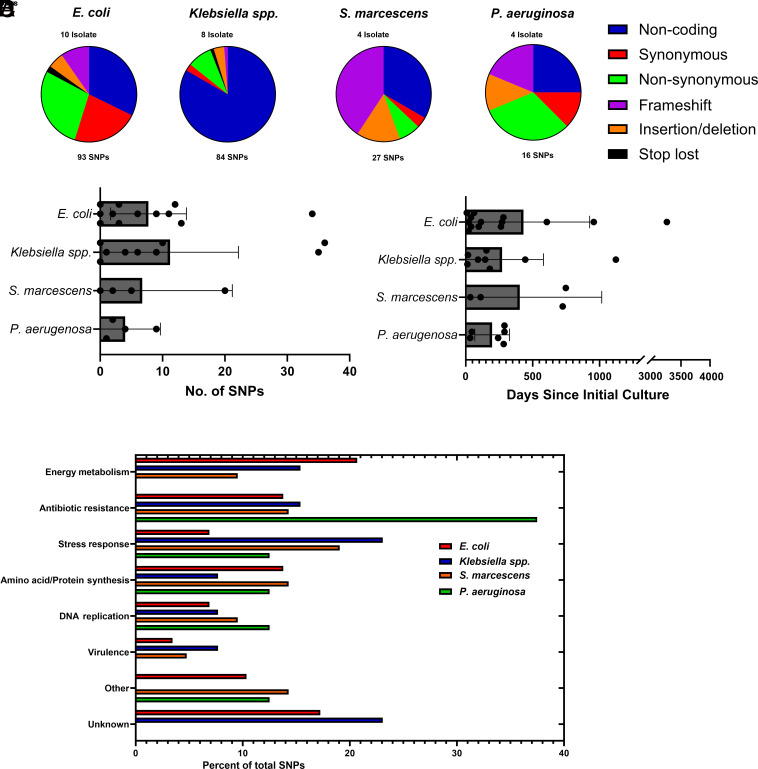
Our previous work generated a cohort of patients with recurrent GNB-BSI, defined as patients with more than one BSI episode due to a Gram-negative isolate of the same species (13). The previous study used whole-genome sequencing (WGS) as a tool to distinguish reinfection GNB-BSI (recurrent infection with genetically distinct isolate of the same species) from relapsed GNB-BSI (recurrent infection with the same isolate). For this work, we re-examined the WGS data to identify the genetic differences occurring between the initial and relapsed isolates in patients with GNB-BSI. This approach would allow us to determine the molecular changes that occur under pressure from the host and antibiotics. Our cohort consisted of 11 patients with E. coli bacteremia, 8 patients with Klebsiella species bacteremia, 4 Serratia marcescens bacteremia, and 4 patients with Pseudomonas aeruginosa bacteremia. We identified the different genetic changes between the index and relapsed isolate and classified the mutation by type (Fig. 1A). The distribution of SNPs varied by species. For example, approximately 50% of the SNPs in E. coli were in either non-coding regions or synonymous SNPs. In contrast, over 30% of the SNPs identified in S. marcescens were disruptive frameshift mutations, likely to severely impact the function of the encoded protein. The number of SNPs between the initial and relapsed isolates varied between 0 and 36 (Fig. 1B), which was surprisingly low given the length of time between initial and relapsed culture was up to 3,258 d (median; 150 d, range; 8 to 3,258 d) (Fig. 1C).
Fig. 1.
SNPs occurring between initial and recurrent GNB-BSI isolates. (A) Categorization of SNPs between species. (B) Number of SNPs occurring between initial and relapsed isolate by species, bar represents mean number of SNPs. (C) Time between initial and recurrent bacteremia episode by species. (D) Non-synonymous SNPs by gene function.
Evolution of Non-Synonymous SNPs.
We next focused on unraveling the biological relevance of the non-synonymous SNPs, as these would potentially impact protein function and affect the biology of the pathogens (SI Appendix, Tables S1–S4). The frequency of non-synonymous SNPs between the initial and relapsed isolate varied from 0 to 15 and were distributed through genes involved in a wide range of metabolic processes (Fig. 1D). There were several SNPs in genes mediating AMR, which was expected as our last study confirmed acquisition of phenotypic AMR was common in relapsed GNB-BSI (13). One notable pattern was frequent mutations in genes involved in glucose metabolism, including disruptive mutations in genes encoding critical metabolism enzymes, such as acquisition of a frameshift mutation in pfkA identified in Klebsiella spp. isolate GN02708, which resulted in a mutant unable to use glucose as a carbon source (SI Appendix, Fig. S1A). Another notable theme was multiple mutations in genes involved in the PTS system which included ptsI, gatC, and mtl in E. coli and crr in S. marcescens.
Antibiotic Resistance Unlikely to Contribute to Treatment Failure in Relapsed E. coli BSI.
From here onward, we focused on the E. coli isolates, given that it was the most frequent pathogen identified in our cohort (11 episodes of relapsed E. coli BSI). To determine whether antibiotic selection may have contributed to relapse, we compiled the antibiotic treatment data for each isolate (SI Appendix, Table S7). The classes of antibiotics used to treat these infections were exclusively cell-wall acting antibiotics (β-lactams, cephalosporins, carbapenems) and occasionally fluoroquinolones as oral step-down therapy. Despite some isolates with extensive resistance profiles (SI Appendix, Table S6), inspection of the treatment regimens demonstrated that appropriate antibiotics were selected in every case, except a single day in the treatment of GN03551. Therefore, relapse was unlikely due to inappropriate antibiotic choice or treatment duration.
Relapsed GNB-BSI Is Associated with In-Patient Development of Persister Cells.
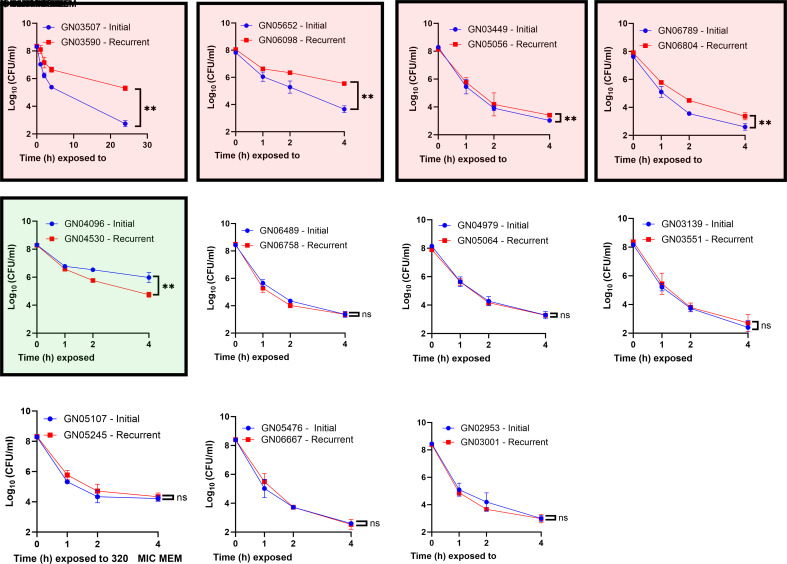
We hypothesized that recurrent GNB-BSI is associated with increased persister formation. We defined increased persister formation as a statistically significant decrease in antibiotic killing following antibiotic challenge. To test this hypothesis, we focused on the 11 pairs of initial/relapsed E. coli isolates in our cohort. We performed a time-kill curve screen with meropenem on each pair of isolates to quantify persisters. Meropenem was selected for the screen for two reasons: first, it was one of the few antibiotics with activity against every E. coli isolate in our cohort (SI Appendix, Table S6). Second, the mechanism of action is clinically relevant, given that every patient in our cohort received a treatment regimen including a cell-wall active antibiotic (SI Appendix, Table S7). Our data show four of eleven (36%) relapsed isolates displayed increased persister formation relative to the initial isolate (Fig. 2). To ensure the persister phenotypes were not meropenem specific, we expanded the screen to include ceftriaxone, a second clinically relevant cell wall targeting antibiotic (SI Appendix, Fig. S4). Three of 9 isolate pairs tested (two isolate pairs were excluded due to resistance to ceftriaxone) showed increased ceftriaxone persisters. There was one discordant result with the GN04979 and GN05064 pair showing no change in meropenem persisters but a small, but statistically significant increase in ceftriaxone persisters. The relapsed isolates with the high-persister phenotype differed from the initial isolate by just 2 to 7 SNPs when compared to the initial strain (Table 1), providing clues regarding the mechanism. The GN03590 isolate demonstrated over 100-fold increased persisters over a 24-h period (Fig. 2) without a change in the minimum inhibitory concentration (MIC) for meropenem (SI Appendix, Table S5). The relapsed isolate GN03590 differs from the initial isolate by just two non-synonymous SNPs. These SNPs resulted in two amino acid substitutions: one in PtsI (V488F) and one in ArgD (P85L).
Fig. 2.
Increase in persister cell formation in relapsed GNB-BSI isolates. Screen performed with 80 to 320× MIC meropenem (MEM) added to mid-exponential E. coli grown in LB broth. At a specified time point, aliquot removed, washed, and enumerated. Isolate pairs highlighted in red show increased persisters, isolate pair in green with reduced persisters. Data are summary of six biological replicates. Error bars represent SD. Significance determined by Mann–Whitney test of the 4-h or 22-h time point (significance level: P = <0.05).
Table 1.
Nonsynonymous SNPs occurring between the initial and relapsed isolates in E. coli pairs demonstrating increased persister formation
| Initial isolate | Recurrent isolate | Species | Mutation type | Gene | Product | Amino acid substitution | Function |
|---|---|---|---|---|---|---|---|
| GN06789 | GN06804 | E. coli | Missense | pfkB | ATP-dependent 6-phosphofructokinase isozyme 2 | M225L | CHO metabolism |
| Missense | ompR | Transcriptional regulatory protein OmpR | K88Q | Drug resistance | |||
| Frameshift | pal | Peptidoglycan-associated lipoprotein | L9fs | Virulence | |||
| GN05652 | GN06098 | E. coli | Missense | gpmI | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | G410V | CHO metabolism |
| Missense | mtl | PTS system mannitol-specific EIICB component | A122V | CHO metabolism | |||
| Frameshift | mnmC | tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein MnmC | S30fs | Amino acid/Protein synthesis | |||
| Missense | yedI | Inner membrane protein YedI | A122V | Unknown | |||
| Missense | rimI | [Ribosomal protein S18]-alanine N-acetyltransferase | Stop130Y | Amino acid/Protein synthesis | |||
| Missense | oxyR | Hydrogen peroxide-inducible genes activator | A100T | Stress response | |||
| Conservative deletion | traD | Coupling protein TraD | Q638-P640 | Conjugation | |||
| GN03507 | GN03590 | E. coli | Missense | argD | Acetylornithine/succinyldiaminopimelate aminotransferase | P85L | Amino acid/Protein synthesis |
| Missense | ptsI | Phosphoenolpyruvate-protein phosphotransferase | V488F | CHO metabolism | |||
| GN03449 | GN05056 | E. coli |
Disruptive Deletion |
rplW | 50S ribosomal protein L23 | E56-V57 del | Protein Synthesis |
| Missense | ygaV | putative HTH-type transcriptional regulator YgaV | D73G | Unknown | |||
| Missense | fliT | Flagellar protein | V93I | Flagella |
PtsI V488F Isoform Is a Loss-of-Function Substitution Conferring a High-Persister Phenotype.
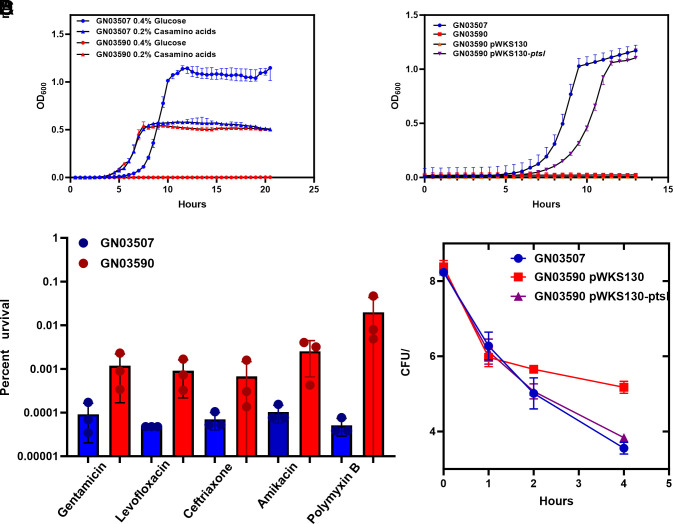
PTS is the main system mediating the transport and phosphorylation of carbohydrates, including glucose, in E. coli (33, 40). The PTS system also governs carbon catabolite repression, which leads to the preferential consumption of glucose by transcriptional downregulation of the machinery involved in the uptake of other carbon sources. With a non-functioning PTS, the cell is unable to utilize glucose as a sole carbon source. Disruption of ptsI has previously been shown to induce multidrug tolerance in E. coli and has been associated with resistance to certain cell-wall active antibiotics, including fosfomycin (32, 41, 42). To determine the effect of the V488F substitution on the function of PtsI, we performed a growth curve comparing the initial (from here onward referred to as GN03507-I) with the relapsed isolate (from here onward referred to as GN03590-R) containing the mutation in ptsI. There was an increased lag phase noted in the ptsI mutant when grown in rich media (SI Appendix, Fig. S1B). To determine whether the high persister formation of the ptsI mutant was associated with a sub-population of bacteria remaining in a prolonged lag phase, we grew GN03507-I and GN03590-R to mid-exponential phase and then used this exponential culture to inoculate a new culture and grew that to mid-exponential phase, thereby drastically reducing any bacteria remaining in a lag state before challenging with meropenem. This sequential sub-culture did not have any significant impact on persister numbers and the ptsI mutant still displayed a 100-fold increase in persisters relative to the wild-type (SI Appendix, Fig. S1C). When grown in M9 minimal media using 0.2% casamino acids as a carbon source, there was no difference between the growth GN03507-I and GN03590-R (Fig. 3A). When glucose was used as a sole carbon source, GN03590-R was unable to grow, indicating a defect in the ability to utilize glucose as a carbon source consistent with complete loss-of-function of ptsI (43). When GN03590-R was complemented with a plasmid expressing wild-type ptsI, its ability to use glucose was restored (Fig. 3B). Similarly, the GN03590-R isolate exhibited at least fivefold increased persister cells compared to the GN03507-I isolate when challenged with multiple different classes of antibiotic (Fig. 3C). The relapsed isolate GN03590-R differed from the initial isolate by just two missense mutations in argD and ptsI. The argD gene encodes the N-acetylornithine aminotransferase enzyme involved in arginine biosynthesis, which is not essential for arginine biosynthesis. As disruption of the ptsI gene is known to result in a pan-tolerant E. coli (32) and argD is redundant in arginine biosynthesis (44), we suspected that the high-persister phenotype was exclusively due to the mutation in ptsI. This was confirmed by complementing the defective ptsI with the wild-type ptsI gene on a plasmid in the GN03590-R isolate, which restored the level of persisters to the level of the GN03507-I (Fig. 3D). These data are consistent with the findings of Zeng et al, who determined disruption of ptsI alone in E. coli MG1655 is sufficient to increase persister formation (32).
Fig. 3.
PtsI [V488F) substitution causes loss-of-function of PtsI and multidrug antibiotic tolerance. (A) Growth of initial (GN03507) and recurrent (GN03590-PtsI(V488F)) isolate in M9 minimal media supplemented with 0.4% glucose or 0.2% casamino acids. (B) Growth of initial and recurrent isolate in M9 minimal media supplemented with 1% glucose, complemented with empty vector (pWKS130) or vector expressing ptsI (pWKS130-ptsI). (C) Antibiotic killing with different antibiotics of initial isolate (GN03507) compared to recurrent isolate (GN03590). Antibiotic conditions: Gentamicin 2× MIC for 1 h, levofloxacin 2× MIC for 2 h, ceftriaxone 160× MIC for 4 h, ertapenem 210× MIC for 4 h, amikacin 2× MIC for 1 h, polymyxin B 10× MIC for 1 h. The dashed line represents the lower limit of detection. Data are averages of six biological replicates. Statistical significance was determined by Mann–Whitney test. Results considered significant if P = <0.05. Complete time–kill curves are provided in SI Appendix, Fig. S3. (D) Time–kill curve of GN03507, GN03590 pWKS530 (empty vector), and GN03590 pWKS530-ptsI complemented with wild-type ptsI gene treated with 160× MIC meropenem. Data are average of six biological replicates. Error bars represent SD.
Klebsiella pneumoniae ΔptsI Displays High Persister Phenotype.
The effect of ptsI deletion on persister formation in species other than E. coli remains unknown. To confirm that the persister phenotype can be extrapolated to other species, we tested K. pneumoniae KPPR1 ptsI transposon multidrug tolerance relative to wild type (SI Appendix, Fig. S2). Our data show that the ptsI mutant showed an increase in persister formation to all antibiotics tested, with the exception of gentamicin. It is unclear why disruption of ptsI would result in decreased gentamicin-induced persister formation in K. pneumoniae but not E. coli. Regardless, loss of Enzyme I activity in K. pneumoniae, and potentially other species of Gram-negative bacteria, results in decreased antibiotic killing and an increase in persisters.
Functioning PTS System Is Not Required to Establish BSI.
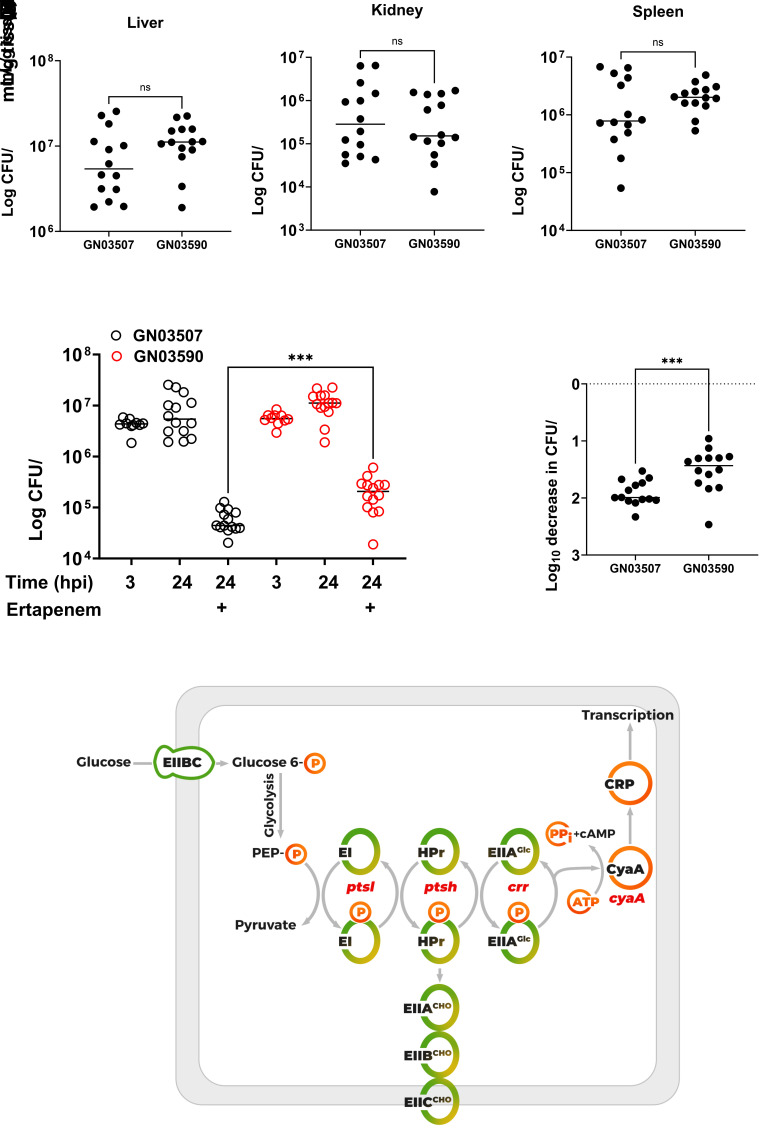
Our findings agree with the study by Zeng et al, showing inactivation of ptsI results in a pan-tolerant mutant of E. coli. Until now, antibiotic-killing assays in our work and theirs have been limited to in vitro experiments in broth culture using rich medium and artificial conditions. We sought to investigate how disruption of PTS may negatively affect pathogenesis during BSI in a murine infection model. As E. coli transitions from its natural reservoir (gastrointestinal tract or urinary tract) to the bloodstream, it needs to rapidly adapt to stark alterations in nutrient availability and varying carbon sources (39, 45). The role of the PTS system in dissemination is unknown, but given its role in carbon catabolite repression, one can hypothesize that it would play a key role in metabolic adaptation. To test this, we developed a murine bacteremia model by infecting BALB/c mice with 5 × 107 colony-forming units (CFU) via the lateral tail vein (LTV) with either GN03507-I or GN03590-R. This dose was selected as 5 × 108 CFU resulted in mortality in 70% of mice infected with GN03507-I in less than 24 h and 5 × 106 CFU did not establish a robust infection. At 24 hours post infection (hpi), the mice were killed and hematogenous dissemination quantified by recovering and enumerating viable bacteria from the liver, spleen, and kidney. There was no significant difference in murine mortality or the quantity of bacteria in any of the organs examined, indicating that ptsI is not required for adaptation and survival inside the bloodstream of the host (Fig. 4 A–C). To ensure the c1462g mutation in the ptsI gene of GN03590 did not revert to wild-type inside the host, we screened 60 colonies recovered from each mouse (20 from each organ) by patching onto M9 minimal media plates supplemented with either 0.2% casamino acids of 0.2% glucose. Then, 100% of the colonies screened maintained disrupted PTS function (growth on casamino acids but not glucose).
Fig. 4.
Virulence and antibiotic susceptibility of ptsI mutant in vivo. (A–C) Eight-week-old BALB/c mice infected via LTV with 5 × 107 CFU. After 24 h, mice were killed and organs harvested for enumeration of CFU. (D and E) Eight-week-old BALB/c mice infected via LTV with 5 × 107 CFU. Mice treated with 50 mg/kg subcutaneous ertapenem for two doses at 3 hpi and 9 hpi. After 24 h, mice killed and organs harvested for enumeration of CFU. The data show infection burden at 24 hpi after ertapenem treatment at 3 hpi and 9 hpi. Significance determined by Mann–Whitney test (significance level” P = <0.05). (F) Schematic illustrating role of ptsI in the PTS phosphorelay. The phosphate is transferred from EI to the carrier protein HPr and subsequently to the EIIA domain of the sugar transporter. The glucose EIIA is unique as it can transfer phosphate to the EIIBC subunit to facilitate transport and phosphorylation of glucose, or to CyaA to stimulate cyclic adenosine monophosphate (cAMP) formation and activation of CRP inducing the expression of the CRP regulon. The EIIABC complex for other carbohydrates is abbreviated as EIIABCCHO.
Loss-of-Function Mutation in ptsI Increases Survival to Antibiotic Treatment In Vivo.
In contrast to ideal conditions provided in the laboratory, the bloodstream provides a hostile environment for bacteria due to the constant barrage from the innate immune system and relatively low nutrient availability (39, 45). Given the environmental dependence of many persister phenotypes, it is often unknown whether the high persister phenotype observed in the ptsI mutant would affect survival to antibiotic treatment in vivo. We addressed this question using mice infected via LTV with GN03507-I and GN03590-R and treated with subcutaneous ertapenem. Ertapenem was selected for three reasons. First, this was the antibiotic the patient was treated with during their relapsed infection. Second, there are studies indicating that subcutaneous ertapenem dosing every 6 h emulates pharmacokinetics seen in humans with favorable time above the MIC (46). Third, ertapenem treatment also resulted in a high-persister phenotype, comparable to meropenem (Fig. 3C). Our data show significantly less antibiotic killing of GN03590-R in the liver compared to GN03507-I (Fig. 4 D and E). This important finding demonstrates that loss of ptsI function results in high-persister phenotype, even in the environment of the host. This finding provides further evidence that persister cells and the in-patient evolution of high-persister forming strains may contribute to antibiotic treatment failure during human BSI.
Discussion
In this work, we performed a molecular and phenotypic assessment of clinical isolates obtained from patients with relapsed GNB-BSI. By comparing the genetic changes in the relapsed isolate compared to the initial isolate, we were able to establish how the bacteria evolves inside the host between episodes. It was surprising how few SNPs occurred (range 0 to 36) despite one of the episodes being almost 10 y apart. This could be explained by the inhospitable environment in the host for any mutations resulting in decreased fitness. There were several frameshift mutations in important genes such as pfkA, encoding phosphofructokinase A, a key component of glycolysis. Studies in S. marcescens (47) and Citrobacter freundii (48) have shown deletion of genes involved in glycolysis (including pfkA) results in a fitness defect during mouse BSI. As mutants from this collection were isolated from the bloodstream of a patient, it suggests that experiments showing significant fitness defects in vitro do not necessarily mean thar the mutant is unable to establish a life-threatening BSI in vivo.
Importantly, we found that the in-patient evolution of E. coli led to increases in persister cell formation in some isolates, as measured in vitro using meropenem. We previously showed acquisition of AMR is common in this cohort of isolates, which is unsurprising given the exposure to antibiotics in the initial episode (13). The acquisition of antibiotic tolerance and persister formation in recurrent or persistent GNB-BSI has not previously been reported.
In one relapse isolate, displaying a 100-fold increase in persister formation, we identified a loss-of-function mutation in the ptsI gene, encoding Enzyme I of the PTS pathway (Fig. 4F). Loss of ptsI function was previously found to be associated with E. coli multidrug tolerance in vitro (32). We provide evidence that this mutation arises in humans during recurrent BSI. As discussed above, it is unclear whether this mutation was acquired de novo during antibiotic treatment of the initial BSI episode or whether the ptsI polymorphism was present as a subpopulation of the gastrointestinal or genitourinary flora in the patient and this ptsI-negative population survived the initial antibiotic treatment course due to the decreased antibiotic-mediated cell death. The prevalence of loss-of-function ptsI mutations in nature remains unknown, as do the particular niches where a functioning ptsI is advantageous. A search using Basic Local Alignment Search Tool shows one instance of E. coli with a V488F substitution in a cohort of E. coli urinary isolates (49), although there is no clinical or biological context. It is highly likely that there are several other mutations in ptsI that would result in a defective Enzyme I and a multidrug tolerant strain. While apparently rare, there are multiple studies reporting the existence of mutations in ptsI and cyaA in E. coli clinical isolates associated with fosfomycin resistance (50–52). The mechanism of ptsI-mediated fosfomycin resistance is thought to result from decreased transporter expression due to reduced PTS-mediated intracellular cAMP (42). Given the mechanism results from dysfunctional PTS-CRP axis, we suspect these mutations would also negatively affect ptsI function. Mutations in ptsI have also been identified in other fosfomycin-resistant Enterobacterales, including K. pneumoniae (53). Another study identified multiple ptsI nonsense mutations in fosfomycin-resistant K. pneumoniae clinical isolates, which would almost certainly lead to an Enzyme I deficient, high-persister strain based on our data (54). These findings suggest that mutations in ptsI are likely more widespread in nature than previously thought with potential implications for antibiotic treatment failure and increased morbidity and mortality.
Importantly, inactivation of ptsI reduces antibiotic efficacy in vivo. Using a murine bacteremia model we show that not only is the ptsI mutant as virulent as the wild type, but it is significantly more tolerant to antibiotics when the mice are treated with ertapenem. This highlights the impact persister cells could have on decreased bacterial clearance during BSI in humans and identified ptsI as a clinically relevant persister gene.
This work represents a significant contribution to the understanding of how persister formation, antibiotic tolerance, and treatment failure may be intertwined. Unlike AMR, which has a clear association with antibiotic failure, determining the causality of persisters with treatment failure has long been a challenge in the field of antibiotic tolerance and persistence (18, 19, 55). The increased abundance of antibiotic-tolerant persisters is widely accepted in certain chronic infections, such as cystic fibrosis chronic lung infections (56), and chronic biofilm-associated infections (57). However, it is difficult to pinpoint whether chronic infections enrich for persisters or if persisters increase the frequency of chronic infections. The role of persisters in refractory or recurrent acute infections, such as bacteremia and urinary tract infections (UTIs) is less well understood. While there have been several genes associated with persister formation in E. coli, almost all have been identified on the bench and therefore have uncertain clinical and biological relevance (55). The exception is the hipA gene encoding a serine-protein kinase in E. coli. The hipA7 locus containing two missense mutations (resulting in G22S and D291A substitutions) has been shown to increase persister formation 1000-fold and was identified in 23/477 isolates patients with an E. coli UTI (25). While we do not know whether the hipA7 locus is associated with lower rates of clinical success in patients with UTI, the identification and prevalence of this gene underlined the potential clinical importance in human disease.
We propose that ptsI should be considered a clinically important persister gene in patients with E. coli and Klebsiella spp. bacteremia. This suggestion is based on the isolation of this mutant from the bloodstream of a patient who had experienced antibiotic treatment failure (infection relapse) combined with the in vitro phenotype of 100-fold increase in persister formation and increased survival to antibiotic treatment observed in a murine bacteremia model. It is critical that we identify bacterial genes associated with persistence and potentially treatment failure in BSI. There is a systematic push to reduce the duration of antibiotic therapy for GNB-BSI from 14 d to 7 d or less in an effort to blunt the development of AMR (58). The identification of bacterial genotypes associated with persistence could help distinguish patients who could benefit from a longer course of antibiotics from those who can be treated with a shorter course. Additionally, the increase in persister formation associated with relapsed GNB-BSI also suggests a shorter antibiotic course may not be suitable for recurrent BSIs. Further work is needed to investigate the prevalence of PTS-disrupting mutations in clinical bloodstream isolates and the biological relevance of the mutations of other PTS components identified in this study (crr, gatC, mtl).
These data warrant that we consider the broad applicability of antibiotic tolerance and persister cells in other species of pathogenic bacteria. Our data show deletion of ptsI in K. pneumoniae, generates a phenotype similar to E. coli with tolerance to multiple different classes of antibiotics. It is likely that loss of ptsI function affects persister formation in other species, especially Gram-negative pathogens possessing an cAMP-CRP axis. Loss of function of Enzyme I has been shown to induce penicillin tolerance in Streptococcus gordonii (59), which is likely due to a mechanism distinct from E. coli due to lack of CRP global regulator. Similarly, a mutation in ptsI was also identified in a multidrug tolerant isolate of vancomycin-resistant Enterococcus faecium isolated from a patient with persistent E. faecium bacteremia (60), but the contribution of ptsI to the tolerance phenotype was not specifically determined. Our study underlines the importance of further studies examining the role of antibiotic tolerance and persisters in treatment failure in BSI.
Materials and Methods
Bacteria, Reagents, and Growth Conditions.
GNB-BSI clinical isolates were obtained from the Duke Bloodstream Infection Biorepository as described previously (13). K. pneumoniae KPPR1 and ptsI transposon mutant were obtained from Laura Mike (61). E. coli cultures were grown aerobically in Luria Broth (LB) at 200 rpm. Colony formation was on LB agar at 37 °C. Meropenem and ertapenem were obtained from MedChemExpress, Ceftriaxone, levofloxacin, polymyxin B, amikacin from Cayman Medical Company, gentamicin from Fisher. LB broth (Lennox) was obtained from Fisher.
WGS.
Described previously (13). Qiagen Bacterial DNA Isolation Kits—20G tips—were used to extract genomic DNA. Genomic DNA was prepared in parallel for Illumina short-read sequencing and Oxford Nanopore long-read sequencing. Illumina DNA Prep kit with unique dual indexes (Illumina, CA, USA) was used to prepare libraries for sequencing on Illumina NextSeq 2000. An Oxford Nanopore GridION X5 was used to sequence the prepared libraries using the Rapid Barcoding Kit and the FLO-MIN106D chemistry (Oxford Nanopore, Oxford, UK).
Bioinformatic Analysis.
A bespoke pipeline was used to generate hybrid genome assemblies (Shropshire, W GitHub: https://github.com/wshropshire/flye_hybrid_assembly_pipeline). Briefly, long-read assemblies were created with Flye-v2.9-b1768 (62) and polished with the long-reads using Racon-v1.4.21 (63) and Medaka-v1.4.4 (nanoporetech GitHub: https://github.com/nanoporetech/medaka). Each high-quality long-read-based assembly was polished using the high-accuracy Illumina short-read sequencing data with Racon. Errors in regions with multiple copies or low complexity were corrected using a pathogendb script (powerpak GitHub: https://github.com/powerpak/pathogendb-pipeline/tree/master/scripts). Data uploaded under BioProject PRJNA1007270.
Minimum Inhibitory Concentration Determination.
MIC was determined by the broth microdilution method. Cultures were grown overnight in LB, diluted 1:2,000 in a 96-well plate containing different antibiotic concentrations, and incubated for 24 h. The MIC was defined as the concentration of antibiotic required to inhibit growth by >90%.
Antibiotic Tolerance Assay.
Time-kill curves were performed using the following protocol. Overnight cultures were grown in LB and diluted 1:100 to 1:500 in fresh LB media. Cultures were grown at 37 °C with shaking at 200 rpm to exponential phase (OD600 = 0.4 or approximately 2 × 108 CFU/mL), except the K. pneumoniae culture used to test levofloxacin tolerance, which was grown to late exponential phase (OD600 = 1.3, approximately 1 × 109 CFU) due to excessive killing. At that time, antibiotics were added at a fixed multiple of the MIC. At designated time points, 100 mL was removed, washed twice with equal volume of phosphate buffered saline (PBS) and 10 mL serially diluted. Surviving cells were enumerated by plating onto LB agar. Each experiment was performed using six biological replicates.
Plasmid Construction and Complementation.
Complementation of the ptsI disruption in GN03590 was accomplished using pWSK130 (64). Primers 5′-GCGGTCGACAACTCGAGTAATTTC and 5′-GCGGGATCCGAGATTTCAGTTTATC were used to amplify the ptsI gene and ligated into the BamHI and SalI sites of pWSK130. The empty vector and plasmid containing ptsI gene were transformed into GN03590 using electroporation.
Murine Bacteremia Infection.
Eight-week-old BALB/cJ mice were obtained from The Jackson Laboratory. Overnight cultures of GN03507 and GN03590 were grown in an LB medium. The following day, 100 mL of culture was centrifuged at 12, 000 × g for 2 min. The supernatant was removed, and the culture resuspended in 1 mL phosphate-buffered saline, which equates to approximately 5 × 108 CFU/mL. The mice were infected via LTV with 5 × 107 CFU of either clinical isolate. Where indicated, 50 mg/kg ertapenem was administered via subcutaneous injection at 3 and 9 hpi. Mice were euthanized and kidney, liver, and spleen harvested at 24 hpi.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by NIH Grant R56AI158511. C.A.A. is supported by NIH/NIAID grants K24AI121296, R01AI134637, R01AI148342, R01AI173138, and P01AI152999 to C.A.A. Thanks to Laura Mike for providing the K. pneumoniae ptsI mutant. Appreciate Kim Walker for discussion and assisting with acquiring plasmids and bacterial strains.
Author contributions
J.B.P., B.M.H., F.R., S.E.R., C.A.A., V.G.F., J.T.T., and B.P.C. designed research; J.B.P., A.E.S., A.Z.V., B.M.H., and M.A.-S. performed research; J.B.P. and B.M.H. analyzed data; and J.B.P., C.A.A., V.G.F., J.T.T., and B.P.C. wrote the paper.
Competing interests
V.G.F. reports personal fees from Novartis, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., MedImmune, Bayer, Basilea, Affinergy, Janssen, Contrafect, Regeneron, Destiny, Amphliphi Biosciences, Integrated Biotherapeutics; C3J, Armata, Valanbio; Akagera, Aridis, Roche, grants from NIH, MedImmune, Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Merck; Medical Biosurfaces; Locus; Affinergy; Contrafect; Karius; Genentech, Regeneron, Deep Blue, Basilea, Janssen; Royalties from UpToDate, stock options from Valanbio and ArcBio, Honoraria from Infectious Diseases of America for his service as Associate Editor of Clinical Infectious Diseases, and a patent sepsis diagnostics pending. CA.A. reports honoraria from UptoDate and salary support from the American Society for Microbiology as Editor in Chief of Antimicrobial Agents and Chemotherapy.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Joshua B. Parsons, Email: joshua.parsons@duke.edu.
Brian P. Conlon, Email: brian_conlon@med.unc.edu.
Data, Materials, and Software Availability
Whole genome sequencing data have been deposited in NCBI (BioProject PRJNA1007270) (65).
Supporting Information
References
- 1.Diekema D. J., et al. , The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 63, e00355-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diekema D. J., et al. , Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J. Clin. Microbiol. 41, 3655–3660 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verway M., et al. , Prevalence and mortality associated with bloodstream organisms: A population-wide retrospective cohort study. J. Clin. Microbiol. 60, e0242921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samet A., et al. , Leukemia and risk of recurrent Escherichia coli bacteremia: Genotyping implicates E. coli translocation from the colon to the bloodstream. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1393–1400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanz-Garcia M., et al. , Recurrent Escherichia coli bloodstream infections: Epidemiology and risk factors. Medicine (Baltimore) 88, 77–82 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Maslow J. N., Mulligan M. E., Arbeit R. D., Recurrent Escherichia coli bacteremia. J. Clin. Microbiol. 32, 710–714 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giannella M., et al. , Risk factors for recurrent carbapenem resistant Klebsiella pneumoniae bloodstream infection: A prospective cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1965–1970 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi T., et al. , Clinical and microbiological characteristics of recurrent Escherichia coli bacteremia. Microbiol. Spectr. 9, e0139921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marschall J., Doherty J., Warren D. K., The epidemiology of recurrent Gram-negative bacteremia in a tertiary-care hospital. Diagn. Microbiol. Infect. Dis. 66, 456–459 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Kim Y. C., et al. , Risk factors and microbiological features of recurrent Escherichia coli bloodstream infections. PLoS One 18, e0280196 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Hasan M. N., Eckel-Passow J. E., Baddour L. M., Recurrent gram-negative bloodstream infection: A 10-year population-based cohort study. J. Infect. 61, 28–33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mylotte J. M., McDermott C., Recurrent gram-negative bacteremia. Am. J. Med. 85, 159–163 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Bock A., et al. , Clinical and molecular analyses of recurrent gram-negative bloodstream infections. Clin. Infect. Dis. 76, e1285–e1293 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin-Reisman I., et al. , Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Windels E. M., Michiels J. E., Van den Bergh B., Fauvart M., Michiels J., Antibiotics: Combatting tolerance to stop resistance. mBio 10, e02095-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin-Reisman I., Brauner A., Ronin I., Balaban N. Q., Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. U.S.A. 116, 14734–14739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehl R., Morata L., Meylan S., Mensa J., Soriano A., When antibiotics fail: A clinical and microbiological perspective on antibiotic tolerance and persistence of Staphylococcus aureus. J. Antimicrob. Chemother. 75, 1071–1086 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Westblade L. F., Errington J., Dorr T., Antibiotic tolerance. PLoS Pathog. 16, e1008892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huemer M., Mairpady Shambat S., Brugger S. D., Zinkernagel A. S., Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 21, e51034 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang M., et al. , Ampicillin-controlled glucose metabolism manipulates the transition from tolerance to resistance in bacteria. Sci. Adv. 9, eade8582 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balaban N. Q., et al. , Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gefen O., Chekol B., Strahilevitz J., Balaban N. Q., TDtest: Easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci. Rep. 7, 41284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herren S. C., et al. , Assessing antibiotic tolerance of Staphylococcus aureus derived directly from patients by the replica plating tolerance isolation system (REPTIS). Antimicrob. Agents Chemother. 66, e0096721 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose W. E., Fallon M., Moran J. J., Vanderloo J. P., Vancomycin tolerance in methicillin-resistant Staphylococcus aureus: Influence of vancomycin, daptomycin, and telavancin on differential resistance gene expression. Antimicrob. Agents Chemother. 56, 4422–4427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher M. A., et al. , HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524, 59–64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarovits G., et al. , Prevalence of antibiotic tolerance and risk for reinfection among Escherichia coli bloodstream isolates: A prospective cohort study. Clin. Infect. Dis. 75, 1706–1713 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keren I., Shah D., Spoering A., Kaldalu N., Lewis K., Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y., Vulic M., Keren I., Lewis K., Role of oxidative stress in persister tolerance. Antimicrob. Agents Chemother. 56, 4922–4926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keren I., Kaldalu N., Spoering A., Wang Y., Lewis K., Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Shan Y., Lazinski D., Rowe S., Camilli A., Lewis K., Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6, e00078-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulaiman J. E., Lam H., Evolution of bacterial tolerance under antibiotic treatment and its implications on the development of resistance. Front. Microbiol. 12, 617412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng J., et al. , A broadly applicable, stress-mediated bacterial death pathway regulated by the phosphotransferase system (PTS) and the cAMP-Crp cascade. Proc. Natl. Acad. Sci. U.S.A. 119, e2118566119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deutscher J., et al. , The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong Y., Zeng J., Wang X., Drlica K., Zhao X., Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 116, 10064–10071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobritz M. A., et al. , Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112, 8173–8180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohanski M. A., Dwyer D. J., Wierzbowski J., Cottarel G., Collins J. J., Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dwyer D. J., et al. , Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, E2100–E2109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belenky P., et al. , Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 13, 968–980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmes C. L., Anderson M. T., Mobley H. L. T., Bachman M. A., Pathogenesis of gram-negative bacteremia. Clin. Microbiol. Rev. 34, e00234-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escalante A., Salinas Cervantes A., Gosset G., Bolivar F., Current knowledge of the Escherichia coli phosphoenolpyruvate-carbohydrate phosphotransferase system: Peculiarities of regulation and impact on growth and product formation. Appl. Microbiol. Biotechnol. 94, 1483–1494 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Sloan R., Surber J., Roy E. J., Hartig E., Morgenstein R. M., Enzyme 1 of the phosphoenolpyruvate:sugar phosphotransferase system is involved in resistance to MreB disruption in wild-type and ∆envC cells. Mol. Microbiol. 118, 588–600 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silver L. L., Fosfomycin: Mechanism and resistance. Cold Spring Harb. Perspect. Med. 7, a025262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long C. P., Au J., Sandoval N. R., Gebreselassie N. A., Antoniewicz M. R., Enzyme I facilitates reverse flux from pyruvate to phosphoenolpyruvate in Escherichia coli. Nat. Commun. 8, 14316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal P. B., Schneider B. L., Vu K., Reitzer L., The redundant aminotransferases in lysine and arginine synthesis and the extent of aminotransferase redundancy in Escherichia coli. Mol. Microbiol. 94, 843–856 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Alteri C. J., Mobley H. L., Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 15, 3–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xuan D., et al. , Pharmacodynamic assessment of ertapenem (MK-0826) against Streptococcus pneumoniae in a murine neutropenic thigh infection model. Antimicrob. Agents Chemother. 46, 2990–2995 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson M. T., Mitchell L. A., Zhao L., Mobley H. L. T., Capsule production and glucose metabolism dictate fitness during serratia marcescens bacteremia. mBio 8, e00740-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson M. T., Mitchell L. A., Zhao L., Mobley H. L. T., Citrobacter freundii fitness during bloodstream infection. Sci. Rep. 8, 11792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garretto A., et al. , Genomic survey of e. coli from the bladders of women with and without lower urinary tract symptoms. Front. Microbiol. 11, 2094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ballestero-Tellez M., et al. , Molecular insights into fosfomycin resistance in Escherichia coli. J. Antimicrob. Chemother. 72, 1303–1309 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Takahata S., et al. , Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 35, 333–337 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Ohkoshi Y., et al. , Mechanism of reduced susceptibility to fosfomycin in Escherichia coli clinical isolates. Biomed. Res. Int. 2017, 5470241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abbott I. J., et al. , Oral fosfomycin activity against Klebsiella pneumoniae in a dynamic bladder infection in vitro model. J. Antimicrob. Chemother. 77, 1324–1333 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P., et al. , Mechanisms of fosfomycin resistance in clinical isolates of carbapenem-resistant Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 22, 238–243 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Van den Bergh B., Fauvart M., Michiels J., Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 41, 219–251 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Mulcahy L. R., Burns J. L., Lory S., Lewis K., Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192, 6191–6199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dengler Haunreiter V., et al. , In-host evolution of Staphylococcus epidermidis in a pacemaker-associated endocarditis resulting in increased antibiotic tolerance. Nat. Commun. 10, 1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yahav D., et al. , Seven versus 14 days of antibiotic therapy for uncomplicated gram-negative bacteremia: A noninferiority randomized controlled trial. Clin. Infect. Dis. 69, 1091–1098 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Bizzini A., et al. , A single mutation in enzyme I of the sugar phosphotransferase system confers penicillin tolerance to Streptococcus gordonii. Antimicrob. Agents Chemother. 54, 259–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Honsa E. S., et al. , RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. mBio 8, e02124-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mike L. A., et al. , A systematic analysis of hypermucoviscosity and capsule reveals distinct and overlapping genes that impact Klebsiella pneumoniae fitness. PLoS Pathog. 17, e1009376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kolmogorov M., Yuan J., Lin Y., Pevzner P. A., Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Vaser R., Sović I., Nagarajan N., Šikić M., Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27, 737–746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang R. F., Kushner S. R., Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199 (1991). [PubMed] [Google Scholar]
- 65.Hanson B., whole Recurrent Gram-negative bloodstream infection genome sequences. National Center for Biotechnology Information (NCBI). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1007270. Deposited 19 August 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Whole genome sequencing data have been deposited in NCBI (BioProject PRJNA1007270) (65).