Abstract
Since the late 2010s, Artificial Intelligence (AI) including machine learning, boosted through deep learning, has boomed as a vital tool to leverage computer vision, natural language processing and speech recognition in revolutionizing zoological research. This review provides an overview of the primary tasks, core models, datasets, and applications of AI in zoological research, including animal classification, resource conservation, behavior, development, genetics and evolution, breeding and health, disease models, and paleontology. Additionally, we explore the challenges and future directions of integrating AI into this field. Based on numerous case studies, this review outlines various avenues for incorporating AI into zoological research and underscores its potential to enhance our understanding of the intricate relationships that exist within the animal kingdom. As we build a bridge between beast and byte realms, this review serves as a resource for envisioning novel AI applications in zoological research that have not yet been explored.
Keywords: Animal science, Data extraction, Classification model, Behavior analysis, Biomolecular sequences analysis
INTRODUCTION
Artificial intelligence (AI) stands at the forefront of modern scientific innovation (Wang et al., 2023a). Its research applications range from medical diagnostics (Moor et al., 2023) to climate tracking (Gore, 2022), with the scope of AI research expanding continuously. Of note, AI is making particularly significant strides in zoological research (Romero-Ferrero et al., 2019).
One challenge of zoological research, a discipline focused on animal classification, behavior, physiology, development, genetics and evolution, disease modeling, and paleozoology, is the management and interpretation of extensive and complex datasets. The rapid emergence of advanced AI techniques, such as machine learning (Jordan & Mitchell, 2015) and, in particular, deep learning (Hinton et al., 2006), as well as the emergence of big data, has marked the beginning of an era of intelligent data-centric zoological research.
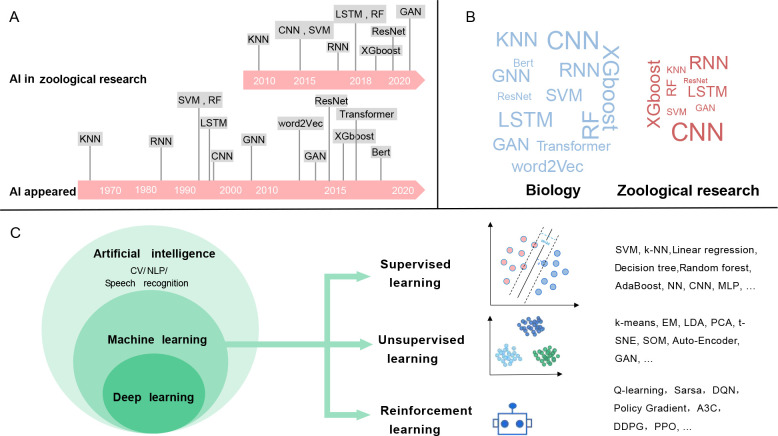
Although AI has been popular for some time, its incorporation into zoological research has not kept pace with its application in other biological fields (Figure 1A, B; Supplementary Table S1). Thus, the question arises as to why AI technologies have not been promptly adopted in animal research. A possible factor for the lack of application may be the inefficiency of computational resources and scarcity of expansive zoological datasets. Additionally, zoologists may lack the foundational knowledge required to understand and implement these approaches, creating uncertainty regarding the selection of models suitable for their objectives. Moreover, the rapid and continuous evolution of complex AI model architectures, like Bidirectional Encoder Representations from Transformers (BERT) (Devlin et al., 2019), make it challenging for zoological researchers to stay current. As access to advanced computational tools and comprehensive zoological datasets expands, it may pave the way for broader adoption of these algorithms in mainstream research. Nevertheless, unfamiliarity with these techniques persists among many zoologists, necessitating a foundational understanding of when, why, and how to employ these methods, as well as what type of data is suitable for their application.
Figure 1.
Branches and applications of AI
A: Timeline of various AI models and their applications in biological and zoological research. B: Word cloud charts displaying counts (Supplementary Table S1) of different AI models used in biological and zoological research, represented after log2 transformation. C: Branches of AI, addressed tasks, and representative models. AdaBoost: Adaptive Boosting; A3C: Asynchronous Advantage Actor-Critic; BERT: Bidirectional Encoder Representations from Transformers; CNN: Convolutional Neural Network; CV: Computer Vision; DDPG: Deep Deterministic Policy Gradient; DQN: Deep Q-Network; EM: Expectation Maximization; GAN: Generative Adversarial Network; GNN: Graph Neural Network; KNN: K-Nearest Neighbors; LDA: Latent Dirichlet Allocation; LSTM: Long Short-Term Memory Network; MLP: Multi-Layer Perception; NLP: Natural Language Processing; NN: Neural Network; PCA: Principal Component Analysis; PPO: Proximal Policy Optimization; ResNet: Residual Neural Network; RF: Random Forest; RNN: Recurrent Neural Network; SOM: Self-Organizing Map; SVM: Support Vector Machine; t-SNE: t-distributed Stochastic Neighbor Embedding; XGBoost: Extreme Gradient Boosting.
In this review, we present an introduction to AI and its primary tasks, elucidating the key models, datasets, and challenges faced. We also explore the intersection where beasts meet bytes, examining how AI applications are revolutionizing diverse areas of zoological research. By analyzing real-world case studies and predicting future directions, we offer a comprehensive overview of the role of AI in deepening our understanding of the animal kingdom and the potential fields it may unlock in the coming years.
ARTIFICIAL INTELLIGENCE
AI and its primary tasks
AI was first defined by Stanford Professor John McCarthy in 1955 as a “the science and engineering of making intelligent machines” (Shabbir & Anwer, 2018). Machine learning, a fundamental branch of AI, is characterized by the capability of systems to autonomously learn from large datasets (de Souza Filho et al., 2020). Machine learning attempts to utilize experience, usually in the form of data, to improve model performance and the process of learning (Sarker, 2021). Therefore, machine learning research primarily focuses on the development of algorithms that generate models from data, termed “learning algorithms”. In the field of zoology, machine learning is helping to shed light on tasks such as species classification, behavior identification, animal population size prediction, bird sound recognition, and nonhuman animal language learning (Layton et al., 2021; Norouzzadeh et al., 2018). These specific tasks are broadly categorized into three types of machine learning, differentiated by their respective data training approaches: supervised, unsupervised, and reinforcement learning (Dönmez, 2013) (Figure 1C).
Supervised learning, one of the most widely used machine learning methods, relies on explicit datasets labeled by experts (Jiang et al., 2020). Supervised learning algorithms build models to identify relationships within a set of feature-label pairs, utilizing the label, also known as the target, for training the system (Li, 2017c). These algorithms fall into two main categories: classification (discrete modeling) and regression (continuous modeling). Both categories are predictive modeling techniques, differing only in their target (response) variables. In classification, the target variable is discrete and takes the form of categories (class labels). For example, animal species identification (Binta Islam et al., 2023), i.e., species classification, relies on learning from diverse data types, such as images, footprints, sounds, and videos, collected from various animals. More importantly, these data are manually annotated with labels indicating the species to which each sample belongs, with the labels corresponding to a predefined list of species. In this task, a model is trained to predict the species name when presented with new input data, such as animal photos. In contrast, in regression tasks, the target variable is continuous rather than discrete. For example, predicting the weight of cultivated cattle constitutes a continuous regression analysis, as the target variable, weight, is continuous.
Unsupervised learning (Li, 2017a) analyzes and clusters unlabeled datasets. Unlike in supervised learning, the inputs in this approach are usually raw data without available labels. These algorithms uncover hidden patterns in data without requiring human intervention, thus termed “unsupervised”. Unsupervised learning models are used for three main tasks: clustering algorithms, which aim to discover unknown subgroups in unlabeled data based on their similarities or differences; dimensionality reduction techniques, which aim to minimize the dimensionality of data by discarding redundant or non-task-relevant information; and anomaly detection, which aims to identify observations that may have originated from different data generation processes. For example, application of the clustering task has enabled researchers to uncover the social structures within jackdaw populations by analyzing unlabeled data of visitation times of each individual (Valletta et al., 2017).
Reinforcement learning (Li, 2017c) involves a family of algorithms that typically operate sequentially. These algorithms are trained through interactions between agents and the (virtual) environment and applied to tasks where learning depends on executed actions and resulting consequences. A notable success is the AlphaGo computer program (Wang et al., 2016), which outperformed human players based on its integration of deep neural networks and reinforcement learning techniques. Frankenhuis et al. (2019) have advocated for the broader application of reinforcement learning methods in behavioral ecology to address the challenge of inferring the unknown reward functions of agents and to explore how biological mechanisms tackle developmental and learning problems.
In addition, AI tasks can also be classified based on the type of input data they process. The three typical subfields include computer vision (CV), natural language processing (NLP), and multi-modal learning. In the field of CV, the primary types of input data include images (e.g., grayscale, color, and binary images) and three-dimensional (3D) representations (e.g., point clouds (Liao et al., 2021)). In the field of NLP, the main data types include text data, speech data (e.g., voice and sound recordings), and time-series data (e.g., motion trajectories captured over time). In contrast, multi-modal learning (Lahat et al., 2015) integrates information from various data sources, ranging from CV to NLP and including images, text, and voice recordings. This integrative approach affords a richer data representation and more effectively captures relationships, thereby enabling a more complex and comprehensive analytical understanding.
CV seeks to automate tasks that the human visual system can perform and is concerned with the automatic extraction, analysis, and understanding of useful information from a single image or a sequence of images (e.g., videos). CV involves the development of a theoretical and algorithmic basis to achieve automatic visual understanding (Farahbakhsh et al., 2020). In this field, typical tasks (Chai et al., 2021) include image classification, retrieval, object detection, semantic segmentation, instance segmentation, object localization, action recognition, and object tracking, among other more specific tasks. Image classification (Lorente et al., 2021) is a fundamental task in CV that aims to categorize an image as a whole under a specific label. When the classification becomes highly detailed or reaches instance-level, it is often referred to as image retrieval (Chen et al., 2023b), which also involves the identification of similar images within a large database. Object detection (Zou et al., 2023) aims to detect and locate objects of interest within an image or video. Object localization focuses on locating an instance of a particular object category in an image, typically by specifying a tightly cropped bounding box centered on the instance. In the literature, “object localization” refers to locating one instance of an object category, whereas “object detection” focuses on locating all instances of a category in a given image (Chai et al., 2021). Semantic segmentation aims to categorize each pixel in an image into a class or object, producing a dense pixel-wise segmentation map where each pixel is assigned to a specific class or object. Instance segmentation involves identifying and separating individual objects within an image, including detecting the boundaries of each distinct object of interest and assigning a unique label to each one. CV is skilled at handling various scenes related to images and videos.
NLP enables a computer system to automatically process and analyze sequence data, even extracting latent information by understanding the syntax in sequences, which is natural for humans (Young et al., 2018). Notably, NLP can capture dense vector representations, known as feature embeddings, from raw sequence data (Pilehvar & Camacho-Collados, 2021). Based on these feature representations, various language models have been created for downstream NLP tasks (Young et al., 2018). Within the context of zoological research, several closely related NLP tasks are commonly applied, including tokenization (Sodhar et al., 2020) for text data processing, text classification for video recognition (Xu et al., 2016), and language modeling for text or gene sequences. With advancements in deep learning models, NLP has the potential to revolutionize research in the zoological domain.
Core models of AI
This subsection provides an overview of deep learning from various perspectives, including main concepts, architectures, and computational tools. The aim is to highlight the most important aspects of deep learning and serve as an instructive guideline for zoologists seeking to utilize this tool.
Due to its extraordinary learning capabilities, deep learning technology, which originated from artificial neural networks, has become a hot topic in the context of AI, with wide application in various areas such as CV, NLP, and speech recognition. Notably, between 2018 and 2021, the number of publications on neural network algorithms has shown a five-fold increase (Shine & Murphy, 2022).
AI models are broadly categorized into shallow or deep learning models based on the number of linear or non-linear transformations the input data undergo before yielding an output (Ahmad et al., 2018). Shallow models typically convert inputs once or twice before transmitting outputs, while deep models, derived from conventional neural networks, commonly convert inputs multiple times (Meir et al., 2023). As a result, deep models can learn more complex patterns, thereby facilitating end-to-end learning without the need for manual feature engineering and exhibit robust performance in CV and sequential data analysis tasks. The introduction of backpropagation (BP) algorithms for artificial neural networks (commonly referred to as neural networks) in the 1980s ushered in an era of machine learning dominated by statistical models (Janiesch et al., 2021), which continues to this day. In the 1990s, challenges such as overfitting and slow training speed in artificial neural networks led to the proposition of various other shallow machine learning models, including support vector machines (SVM), boosting techniques, and maximum entropy methods (e.g., logistic regression) (Xu et al., 2021). SVM and boosting are examples of hidden nodes rather than models (e.g., logistic regression) that utilize hidden nodes. Geoffrey Hinton, along with Yoshua Bengio and Yann LeCun, has been a persistent advocate for the advancement of neural networks, playing a significant role in the development of a practical and feasible deep learning framework (Wang & Duan, 2021).
The effectiveness of machine learning algorithms is highly dependent on the integrity of the input data representation. Therefore, feature engineering has long been an important research area in machine learning, aiming to extract features from raw data with considerable human investment. In contrast, deep learning algorithms automate the feature extraction process, thus reducing reliance on extensive human labor and domain expertise to extract salient features. These algorithms possess a multi-layer data representation architecture, with the first layer extracting low-level features and the last layer extracting high-level features (Wang & Duan, 2021). Due to its considerable success, deep learning has emerged as a prominent research trend. In this context, convolutional neural networks (CNNs), recurrent neural networks (RNNs), and pretrained foundation models (PFMs) have been increasingly employed in zoological research.
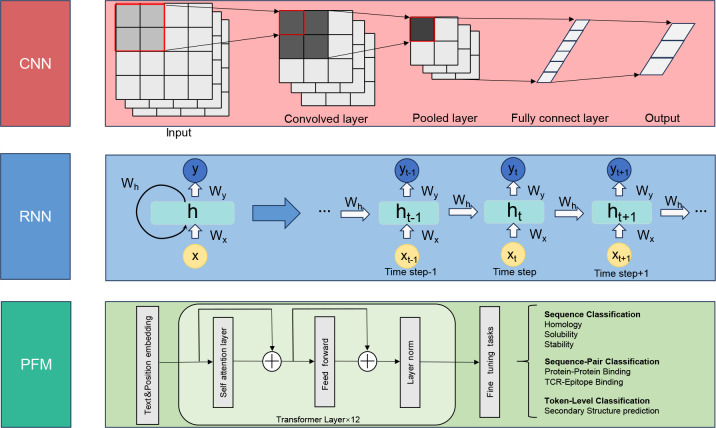
Within the field of deep learning, CNNs are the most commonly used algorithms, with extensive application in image recognition, speech recognition, and NLP. The three principal advantages of CNN are equivariant representation, sparse interaction, and parameter sharing. Unlike traditional fully connected networks, CNNs leverage shared weights and localized connections to fully exploit two-dimensional (2D) input data structures, such as those found in image data. This operation employs a very small number of parameters, which simplifies the training process and accelerates network speed. This concept mirrors the functioning of cells in the visual cortex, as elucidated by Alzubaidi et al. ( 2021), where each unit is responsive to only a subset of the visual field, thereby capturing spatial localities within the input similar to the application of localized filters. A common type of CNN, akin to a multi-layer perceptron, features numerous convolutional layers preceding the subsampling (pooling) layer and concluding with a fully connected layer (Figure 2).
Figure 2.
Concise architectures of three deep learning models
Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), and Pretrained Foundation Models (PFM).
The high performance achieved by CNN architectures in challenging benchmark task competitions indicates that innovative architectural concepts and parameter optimization can improve CNN performance in various visually related tasks (Khan et al., 2020). The exploration of grid topology data (image and time-series data) by LeCun et al. (1989) marked the initial recognition of the capabilities of CNNs. Since 2012, different innovations have been proposed regarding CNN architecture. The performance improvements in CNNs can primarily be attributed to the reconstruction of processing units and design of new modules. With the introduction of AlexNet (Krizhevsky et al., 2017) and its exceptional performance on ImageNet datasets, the application of CNNs has become increasingly popular. The development of the Inception module by the Google team, characterized by its split-transform-merge strategy, marked a substantial advancement in CNN architectures. Its novel introduction of intra-layer branching facilitated feature extraction across varying spatial dimensions. In 2015, ResNet revolutionized CNN training by introducing residual connections (He et al., 2015), a concept incorporated in many subsequent networks, including Inception ResNet, wide residual networks, and ResNext. Similarly, certain architectures, including wide residual networks, Pyramid Nets, and Xception, have introduced multi-layer transformations, implemented through additional cardinality and increased width.
Typical deep CNN models, in addition to a fully connected layer (sometimes not included because of Global Average Pooling), also include convolutional and pooling layers, used to extract meaningful features from locally associated data sources and reduce the number of parameters, respectively (Khan et al., 2020) (Figure 2). Compared to other methods, the minimal preprocessing requirements of CNNs have solidified their status as the de-facto standard computation framework in CV. The achievements of CNNs have garnered widespread attention, both inside and outside academia, leading to the proliferation of diverse CNN models. For instance, the partialS/HIC model (Xue et al., 2021), which is based on a CNN for image processing, addresses the increasing demand for scanning tools capable of tracking in-progress evolutionary dynamics. Similarly, the DeepBehavior toolbox (Arac et al., 2019), which integrates many different CNNs, can automatically analyze animal behavior from both video and 3D image data. The landscape of CNN-based frameworks includes ResNet, MobileNet, DenseNet, ShuffleNet, EfficientNet, R-CNN, and YOLO (Zhu & Zhang, 2018). Among them, YOLO is an advanced algorithm for object detection, and includes different versions such as YOLOv2, YOLOv3, YOLOv4 (Bochkovskiy et al., 2020), and YOLOR (Wang et al., 2021). Each algorithm designed for object detection has the capability to identify objects both in real-time and with high accuracy.
RNNs, designed to handle sequential data (e.g., text, speech, and time series data), are commonly employed in the field of deep learning (Figure 2), including speech processing and NLP (Lipton et al., 2015). RNNs learn the features of time series data by memorizing previous inputs in the internal state of NNs. They can also predict future information based on past and present data but struggle to learn long sequences structures due to the vanishing or exploding gradient issue. Long short-term memory (LSTM) (Staudemeyer & Rothstein Morris, 2019) networks and their variants, such as the gated recurrent unit (GRU) networks (Chung et al., 2014), have resolved the gradient issue using various gates that control how information flows. These algorithms can be used in fields requiring the analysis of sequential data and the prediction of future events based on present data. Research in NLP frequently addresses time series data, analogous to that encountered in zoology, such as text, sound recordings, and sequence data. To capture the positional dependencies inherent in sequential data and retain abundance information from raw data, several deep learning-based models have been developed. RNN-based models (like RNN (Rumelhart et al., 1986), LSTM (Hochreiter & Schmidhuber, 1997)) and transformer-based models (Vaswani et al., 2017) (like BERT) have proven effective in automatically modeling sequential data and conducting downstream task prediction and analysis.
Transformer-based models are fundamentally structured around the attention mechanism and its derived framework, multi-head attention (Choi & Lee, 2023). Leveraging this core architecture, transformer-based models can overcome the limitations of RNN models, which cannot parallelize input processing across all time steps, while still effectively capturing the positional dependencies inherent in sequential data in tasks such as language translation, text summarization, image captioning, and speech recognition. The transformer includes encoder and decoder structures for processing sequence inputs and generating corresponding outputs, respectively (Choi & Lee, 2023). The architectural variant BERT, which only incorporates the encoder structure, employs random sequence masking during its pre-training tasks, leading to superior outcomes in protein 3D structure prediction (Lin et al., 2022b) and single-cell annotation (Yang et al., 2022).
Pretrained foundation models (Zhou et al., 2023) are essential and significant components of AI in the era of big data. These models demonstrate enhanced proficiency in multi-task learning with large-scale datasets and exhibit increased efficiency during fine-tuning for targeted, smaller-scale tasks, resulting in rapid data handling capabilities (Bommasani et al., 2021) (Figure 2). The most famous application among them is ChatGPT, a conversational model derived from the generative pre-trained transformer architecture developed by OpenAI (Gozalo-Brizuela & Garrido-Merchan, 2023). ChatGPT applies reinforcement learning from human feedback (RLHF) (Bai et al., 2022), a promising approach for aligning large language models with human intent, i.e., pretrained language models (Wulff et al., 2023). Many open-source pretrained language models are currently available, operable on individual computing systems and trainable on private datasets, including Llama, Alpaca, Vicuna, and Falcon models (Zhang et al., 2023). Given their success (Wang et al., 2023b) in various general-domain NLP tasks, these open-source large language models exhibit significant potential for application when fine-tuned using knowledge-based instruction data.
Inspired by the success of pretrained language models in NLP, pre-trained visual models in the field of CV have also achieved great success. These models are pre-trained on massive image datasets and can analyze image content and extract rich semantic information. Furthermore, multi-modal visual models like CLIP (Radford et al., 2021) and ALIGN (Cohen, 1997; Lahat et al., 2015) use contrastive learning to align textual and visual information. This alignment allows the pre-trained models to apply learned semantic information to the visual domain, thereby facilitating efficient generalization in downstream tasks, including zoological applications.
Generative models and contrastive learning are two other important types of models. Generative models gained popularity after the introduction of generative adversarial networks (GANs) in 2014, which formed the foundation for many subsequent architectures, including CycleGAN (Zhu et al., 2017), StyleGAN (Karras et al., 2019), and DiscoGAN (Kim et al., 2017). Unlike generative models, contrastive learning is a discriminative approach that aims to group similar samples closer together and diverse samples farther apart (Jaiswal et al., 2021). To achieve this, a similarity metric is used to measure the closeness of two embeddings. Self-supervised learning, a type of unsupervised learning (Wang, 2022), integrates both generative and contrastive approaches. Notably, it utilizes unlabeled data to learn underlying representations, thereby avoiding the labor-intensive task of data labeling. Thus, this approach offers the potential for better utilization of unlabeled data in zoological research.
Given that deep learning has proven more effective in data extraction, feature representation, and prediction than non-deep learning models, our emphasis is on discussing deep learning models used in zoological research and explaining why they are more effective compared to non-deep learning models, especially when dealing with unstructured data such as images, text, videos, and sequence data. Furthermore, while supervised task is frequently discussed in this review due to their relevance to species classification and identification (fundamental concerns in Zoology), we also address unsupervised and reinforcement learning.
Datasets in zoological research
A variety of datasets are available for research in zoology, each tailored to specific tasks, as summarized in Supplementary Table S2. These datasets primarily encompass text and image data. Text data can be processed using a range of models, such as RNN, transformer, and pretrained foundation models, depending on the specific task requirements. Image data, including videos, are compatible with models developed for CV tasks. Among these datasets, the Paleobiology Database, maintained by an international consortium of non-governmental paleontologists, is a publicly accessible repository of paleontological data (Alroy et al., 2008). The AP-10K dataset (Yu et al., 2021) represents the first comprehensive resource for general animal pose estimation, featuring 10 015 images from 23 animal families and 54 species, with high-quality keypoint annotations. This highly versatile dataset is suitable for supervised, self-supervised, semi-supervised, and cross-domain transfer learning, as well as intra- and inter-family domain analyses, with annotation files provided in Common Objects in Context (COCO) format. The KaoKore dataset, established by the ROIS-DS Center for Open Data in the Humanities, comprises a curated collection of facial expressions and has been publicly accessible since 2018 (Tian et al., 2020).
Complex unstructured data in zoological research, including images, videos, sounds, and text, present considerable challenges for AI application. Addressing these challenges often requires expert selection of appropriate models and significant data preprocessing efforts. The following sections provide detailed discussion on AI models tailored for different data types and tasks (Supplementary Table S3), as well as their specific processing procedures. This information should help researchers in selecting models best suited for their specific data and research goals.
AI IN ANIMAL CLASSIFICATION AND RESOURCE PROTECTION
The rapid identification and classification of wild animals is critical for the categorization and protection of biological resources. However, such tasks typically require extensive labor and time investment from experts to process data (Vélez et al., 2022) and manually extract information from field-captured photographs (Kulkarni et al., 2020). To handle these challenges, AI algorithms, especially deep learning models effective in graphic identification studies, have been increasingly applied for the automatic identification of wildlife (Figure 3A). Previous research using deep neural networks (DNNs) to automatically identify, count, and describe wild animals in images captured by motion-sensor cameras not only reduced human labor costs but achieved a high identification accuracy of 96.6%, exceeding that of expert identification (Norouzzadeh et al., 2018). Moreover, such innovative approaches demonstrate that deep learning techniques, in contrast with traditional methodologies for single-species classification, have marked advancements in the classification of multiple species, enhancing the versatility available to researchers in the field of animal resource studies. CNN algorithms have also been used to identify endangered animals in images captured by unmanned aerial vehicles, enabling non-invasive surveillance and protection (Kellenberger et al., 2018). Similar studies have likewise demonstrated success (Fergus et al., 2023; Tabak et al., 2019), highlighting the potential for AI to accelerate advancements in wildlife classification, re-identification, and biodiversity monitoring (Thalor et al., 2023).
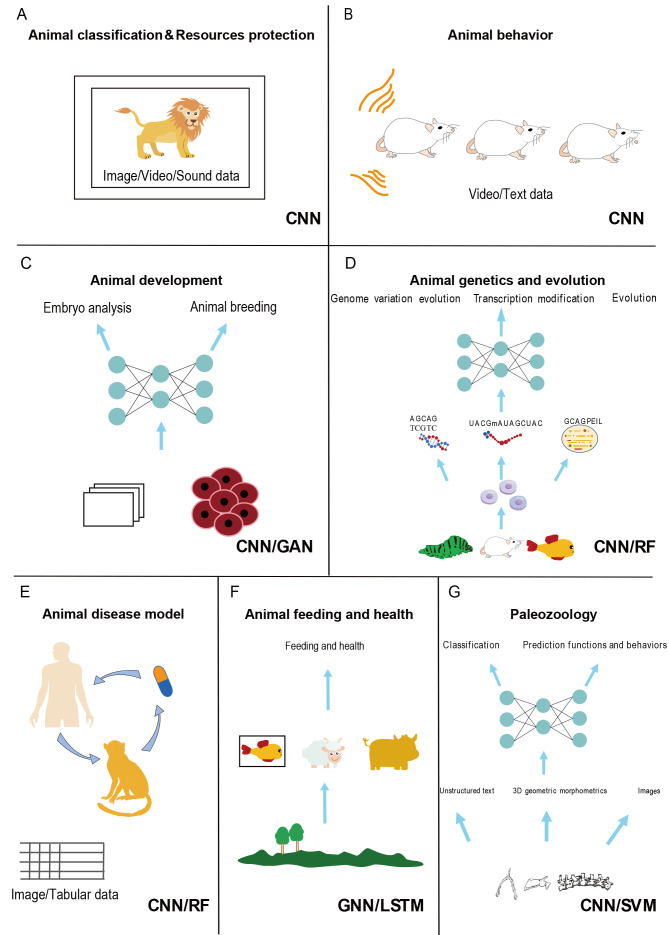
Figure 3.
Application of AI across different fields
Each panel highlights examples of input data type, model type, and applied species of AI. A: Animal classification and resource protection, B: Behavior and neuroscience, C: Development, D: Disease models, E: Breeding and feeding, F: Evolution and genetics, and G: Paleozoology. CNN: Convolutional Neural Network; GAN: Generative Adversarial Network: GNN: Graph Neural Network; LSTM: Long Short-Term Memory Network; RF: Random Forest; SVM: Support Vector Machine.
The conflict between human activity and wildlife is becoming ever more pronounced, manifesting in the extinction of species and degradation of natural habitats. To alleviate these issues, there is an urgent need for effective management and monitoring of wildlife ecological preserves to reduce negative human-animal encounters. The development of DeepVerge, a CNN-based approach, represents a significant advance is this regard, enabling judicious planning of road construction and environmental management by governments. Notably, DeepVerge has shown a mean accuracy of 88% in the identification of positive indicator species in grassland ecosystems, a traditionally challenging task in wildlife resource surveys (Perrett et al., 2022). Nandutu et al. (2022) also employed CNN technology to analyze images captured from unmanned aerial vehicles for the detection of wildlife fences along roadsides, thereby reducing the frequency of human-animal encounters and enhancing the protection of wildlife from traffic-related hazards.
Furthermore, AI models have the ability to collect wildlife images from social media platforms to expand biodiversity preservation efforts, thereby compensating for the scarcity of corresponding data from traditional wildlife monitoring techniques (Foglio et al., 2019). In addition, automatic AI-assisted analysis of aerial images can facilitate accurate counting of various species, such as elephants and dolphins (Singh et al., 2023). These models generally outperform conventional analytical methods, primarily due to their superior capacity for extracting features from images, as well as their regularization and denoising capabilities. These strengths facilitate their application across distinct categories of images, including both wildlife and aerial photographs, irrespective of apparent disparities in image type.
Insect taxa are numerous and characterized by fine-grained distribution patterns. (Høye et al., 2021). Traditional insect detection methods face challenges in performance, particularly when identifying small pests, due to a lack of adequate learning samples and models. To address this, Xiang et al. (2023) developed Yolo-Pest, a CNN-based model optimized for the effective detection of small target pests, which showed significant improvement in performance compared to existing methods when tested on the Teddy Cup pest dataset. Tsetse flies serve as important vectors of human disease in Africa. Geldenhuys et al. (2023) developed a CNN-based method for classifying tsetse fly images and precisely predicting wing landmarks to facilitate vector identification and control. Additionally, Lee et al. (2023) developed a deep learning-based automated object detection technique to identify mosquito species from image data, thereby reducing manual labor in the field.
However, it is important to note that the application of AI in the classification and protection of animal resource is still in its early stages. Although advanced DNN and CNN models have yielded good results in analyzing wildlife pictures, the adaptability and robustness of AI algorithms in dealing with variations in image quality and lighting conditions still require improvement. Classification accuracy typically diminishes when species are recorded live within their natural environments (Van Horn et al., 2017; Wu et al., 2019). Consequently, future work should focus on these issues and refine AI models for application in diverse natural environments. Moreover, wildlife data covers not only images but also an abundance of text and voiceprint data, which are inherently sequential and more amenable for analysis using transformer-based frameworks, such as BERT (Khan et al., 2022). Although the use of CNNs for classifying bird species based on raw sound waveforms has achieved suboptimal accuracy (Bravo Sanchez et al., 2021), the application of transformer-based approaches, though presently limited (Supplementary Table S3), holds considerable promise for future research.
AI IN ANIMAL BEHAVIOR
Animal behavior and neuroscience
Animal behavior is critically related to neural activity, neurological function, and cognitive states. Research has uncovered the mechanisms that govern a variety of complex animal behaviors, including those related to feeding (Shafiullah et al., 2019), anxiety, and mating (Moulin et al., 2021), as well as the analysis of daily behaviors (Liu et al., 2022) and recognition of behaviors based on facial expressions (Liu et al., 2023). However, the core problem lies in the extraction of meaningful information from the wealth of data on animal behavior within the context of neuroscience, propelled by advancements in experimental techniques (Bath et al., 2014; Scaplen et al., 2019; Svensson et al., 2018). Such data are often presented as high-dimensional time series (Scaplen et al., 2019), video recordings (Liu et al., 2022), still images, and 3D image data. These data types are complex, high-dimensional, and sometimes unstructured, rendering them unsuitable for analysis using traditional methods (Figure 3B).
In comparison, deep learning-based models are effective in extracting features and representations from unstructured data. For instance, DeepLabCut, using deep learning transfer techniques, presents a novel approach for estimating the gestures of unlabeled objects, and has produced outstanding results with minimal training data (Mathis et al., 2018). The CNN-based DeepBehavior toolbox enables the automatic analysis of animal behavior data, including both video and 3D image data and has proven effective in neuroscientific investigations of five distinct mouse behaviors (Arac et al., 2019). Coffey et al. (2019) recently developed DeepSqueak, an automatic vocalization analytical framework trained and validated using a comprehensive dataset of mouse and rat vocalizations, which revealed that the grammatical structure of these vocalizations was related to social behaviors, especially mating behaviors. Similarly, Graving et al. (2019) developed DeepPoseKit, a deep learning model adept at the rapid and accurate analysis of animal poses, a critical component for behavioral estimation with important implications in neuroscience research. Sainburg and Gentner (2021) developed CNN and RNN-based computational neurobehavioral models to elucidate the physiological characteristics of animals, especially in the context of acoustic communication. In addition to deep learning techniques, Klibaite et al. (2017) also established a machine learning-based Gaussian mixture model to automatically extract information from animal behavior video recordings, supporting studies in the field of neuroscience
The study of animal behavior, informed by neural activity and complex interactions, can guide the design and construction of deep network models (Richards et al., 2019). This research draws connections between animal neurology and behavior and the foundational principles inherent to reinforcement learning, highlighting the parallels in strategic optimization, task resolution, and reward allocation within these domains.
Animal feeding behavior monitoring
Feeding behavior is a key concern in animal behavior and a crucial factor for health and survival. Achour et al. (2020) used four CNNs to analyze the feeding behavior of cows. Their analytical framework was designed to recognize individual cows and distinguish whether they were eating or standing in a feedlot area, as well as whether there were different food categories in the feedlot. In addition, Chen et al. (2020) developed a video-based deep learning method to identify piglet feeding behavior and calculate feeding durations based on multiple one-second feeding and non-feeding events obtained from video footage. The CNN architecture was used to extract spatial features, which were then fed into a LSTM framework to extract temporal features. These features were then propagated through a fully connected layer, and a Softmax classifier was applied to categorize the one-second segments into feeding or non-feeding events, demonstrating excellent sensitivity (98.8%), specificity (98.3%), and accuracy (95.9%) in identifying feeding behavior.
Data generated in animal husbandry, including feeding data, physical characteristics, and health conditions, can be used to predict other animal characteristics, such as weight and growth trajectories. He et al. (2021) used LASSO regression and two machine learning algorithms, random forest and LSTM network, to predict the weight of pigs aged 159 to 166 days. The predictions were made in four scenarios: individual-specific prediction, individual and population-specific prediction, breed-specific individual and population-specific prediction, and population-specific prediction. For each scenario, four models were developed, and their predictive performance was evaluated using the Pearson correlation coefficient, root mean square error, and binary diagnostic tests. Notably, they successfully predicted pig weights based on feeding conditions and yielding favorable outcomes, confirming the feasibility of using predictive methods to determine later-stage characteristics of animals. Taylor et al. (2023) used machine learning to estimate the growth trajectories of individual pigs based on their weight, subsequently validating trajectory predictions using root-mean-square deviation scores. The findings revealed that, on average, the random forest model exhibited the most accurate predictions, achieving the best score of 2.00 kg per pig and the worst score of 2.45 kg per pig, outperforming traditional approaches and showing considerable prediction potential.
Using AI models, it is also possible to predict feed supply stages and initiate adjustments to avoid feed shortages, as well as to predict delivery behavior to reduce the mortality rate. Shafiullah et al. (Shafiullah et al., 2019) assessed various machine learning algorithms, including SVM, random forest, XGBoost, and neural networks, to predict shortages in spring forage by analyzing changes in cow feeding behavior indicative of potential feed insufficiency. Turner et al. (2023) developed deep learning models to identify the behavior of ewes during childbirth and postpartum licking, using data obtained from accelerometer sensors located on the neck of the sheep. Behavior labels derived from video recordings and an LSTM model were then used to classify different behaviors. The model achieved an accuracy of 84.8% and a weighted F1 score of 0.85 in identifying birthing and licking actions, effectively differentiating between parturition behaviors. This provides a critical framework for prompt intervention during challenging births, potentially decreasing neonatal lamb mortality.
Although AI models have been successfully utilized in feeding behavior monitoring, capable of processing different forms of data, it is important to note that CNNs and their variants play dominant roles in this realm (Supplementary Table S3). Given their sequential nature, time series and video recording data may be better processed using transformer-based architectures. The pre-trained large language models underpinning BERT (homologous transformer architecture) have already set new benchmarks in sequence-dependent bioresearch (Khan et al., 2022; Lin et al., 2022). Hence, we propose that future studies, particularly those involving time-series analyses of continuous data from animal vocalizations, locomotion positions, and text data, may benefit from the utilization of language models, rather than solely relying on CNNs.
AI IN ANIMAL DEVELOPMENT
Advancements in AI within developmental biology predominantly center on the application of CV and machine learning algorithms for the automated processing of images, particularly embryonic images (Figure 3C). Assessment of embryo quality is crucial for improving pregnancy rates following in vitro transfer. Although morphological analysis remains the standard for evaluating embryo quality, it is subject to variation based on the expertise of the evaluator (Rocha et al., 2017). In contrast, AI methods enable systematic and automatic embryo quality evaluations, demonstrating enhanced robustness compared to conventional assessments. Researchers initially developed a computational model using dynamic Bayesian networks and machine learning to understand the regulatory networks governing cell differentiation processes in multicellular organisms, with a focus on Caenorhabditis elegans, and reported significant enhancements in inferential accuracy through the integration of interaction data from various species (Sun & Hong, 2009). Following this, the automated DevStaR system was developed, employing CV and SVM to provide rapid and accurate measurements of embryonic viability and high-throughput quantification of developmental stages in C. elegans, a principal model organism in developmental and behavioral studies (White et al., 2013).
Fish embryo models are commonly used for assessing the efficacy and toxicity of chemicals. Genest et al. (2019) automated this process by classifying fish embryos based on the presence or absence of spinal malformations using 2D images, feature extraction based on mathematical morphology operators, decision trees, and random forest classifiers. Quantitative analysis of cerebral vasculature is crucial for understanding vascular development. Chen et al. (2023a) developed a deep learning method to analyze cerebral vasculature in transgenic zebrafish embryos. Using 3D light-sheet imaging and FE-Unet to enhance 3D structures, they transformed incomplete vascular structures into continuous forms, allowing for the accurate extraction of eight key topological parameters. Polarization of mammalian embryos at the correct time is crucial for development. Shen et al. (2022a) developed an automated stain-free detection method for detecting embryo polarization using a deep CNN binary classification model, achieving an accuracy of 85% and significantly outperforming human volunteers trained on the same data (61% accuracy). In addition, Qiu et al. (2021) introduced a deep learning pipeline for automating the segmentation and classification of high-frequency ultrasound images of mouse embryo brain ventricles and bodies based on a volumetric CNN classifier, thus providing a powerful tool for developmental biologists.
In the industrial sector, integrating AI and animal development can facilitate the generation of offspring with ideal characteristics or improved performance to meet human needs. A critical component of this process involves the identification of parents exhibiting superior phenotypes for reproduction. Robson et al. (2021) implemented an automated system combining generative adversarial networks with a CV pipeline to extract phenotypic data from ovine medical images, achieving an accuracy of 98% and faster speed (0.11 s vs. 30 min), thereby greatly reducing processing time. The phenotypes identified using this approach can provide valuable insights to guide genetic and genomic breeding programs and benefit animal breeding. Furthermore, Rocha et al. (2017) developed a novel method for embryo analysis by combining a genetic algorithm and artificial neural network, which was applied to test features extracted from 482 bovine embryo images, achieving an accuracy of 76.4% compared with evaluations conducted by embryologists.
AI IN ANIMAL GENETICS AND EVOLUTION
Recent advances in third-generation high-throughput sequencing and assembly technologies (Mao et al., 2021) have streamlined the acquisition of genomic, transcriptomic, and expression profiles in animals (Zhang et al., 2018). However, the challenge of processing huge multiomics data has created an urgent need for the application of AI technologies. This has significantly enhanced the efficiency of data analysis and elucidated previously difficult to observe biological phenomena (Badia et al., 2023; Gomes & Ashley, 2023). Notably, various machine learning techniques, such as random forest and gradient boosting, as well as deep learning approaches, such as CNN, DNN, and RNN, have been widely employed (Figure 3D) to study genetic variation, transcriptional modification, and evolutionary issues, primarily based on genomic and proteomic data.
Genetic variation
Traditional genetic variation analyses (Durward-Akhurst et al., 2021; Mashayekhi & Sedaghat, 2023) often struggle to handle large-scale genomic data. In contrast, AI can rapidly and effectively process these vast datasets (Kaushik et al., 2021), offering greater precision in identifying meaningful mutation sites and expediting the inference of biological functions.
Next-generation sequencing has revolutionized genetic testing but has also produced considerable amounts of noisy data, necessitating extensive bioinformatics analysis for meaningful interpretation. The Genome Analysis Toolkit (GATK) has stood as the gold standard for reliable genotype calling and variant detection (Brouard et al., 2019; Lin et al., 2022a). However, the development of cutting-edge deep neural networks, such as DeepVariant, has shown exceptional accuracy in identifying genomic variations, from single nucleotide polymorphisms (SNPs) to indels (Yun et al., 2020). In comparison to GATK, DeepVariant showed a 2% reduction in data errors, highlighting its potential in the ever-evolving field of genomics (Lin et al., 2022). In addition, Babayan et al. (2018) employed a gradient boosting machine algorithm to extract sequence data from over 500 single-stranded RNA viruses and analyzed the evolutionary signals within them, resulting in the rapid identification of pathogens and their potential hosts and vectors, thus providing new avenues for disease control and prevention. Furthermore, Layton et al. (2021) integrated genomic diversity with environmental relevance and a random forest model to identify vulnerable populations of Arctic migratory fish (migratory charr) and reconstruct population sizes and climate-related declines throughout the 20th century. This innovative approach revealed the impacts of climate change on species survival, providing important scientific support for the conservation and genetic breeding of Arctic ecosystems.
Deep learning models, including CNNs and RNNs, have been recently applied in animal omics analyses. Researchers have addressed challenges such as the detection of selective sweeps within mosquito populations (Xue et al., 2021) and identification of short viral sequences within metagenomes (Lee et al., 2021). Xue et al. (2021) devised partialS/HIC, a deep learning approach based on CNNs, to analyze the African mosquito genome, enabling detection of different stages and regions of selective sweeps and insecticide resistance, thus identifying adaptive loci critical for disease control. Additionally, Kaplow et al. (2023) explored the use of CNNs for elucidating genetic variations in enhancers and identifying associations between enhancers and phenotypes across various mammals. Furthermore, Jiang et al. (2023) developed RNN-VirSeeker, a deep learning method tailored to detect short viral sequences within pig metagenomes. Trained on 500 bp samples of known viral host RefSeq genomes, RNN-VirSeeker surpassed three established methods in testing, achieving superior performance in receiver operating characteristic curve (AUROC), recall, and precision, particularly for the CAMI gut metagenome dataset. These studies integrating AI with genomics not only lay the groundwork for understanding genetic variation within animal populations but also forge new pathways for linking genomic research with phenotypes.
Transcriptional modification
Transcriptional modification (Li et al., 2017b), a complex process central to the regulation of gene expression, presents a challenge to traditional analysis due to its dynamic and complex nature. Deep learning models, which excel at handling large datasets and identifying modifications (Salekin et al., 2020), offer a powerful and efficient solution for transcriptional modification analysis in the era of big data. Luo et al. (2022) developed im6APred, an advanced model designed to predict N6-methyladenosine (m6A) sites—a pivotal RNA modification implicated in many biological processes (Zhu et al., 2020)—across a spectrum of mammalian tissues. The architecture of im6APred was grounded in the comprehensive assessment of seven distinct classification methods, including four traditional algorithms and three deep learning techniques, along with their integration. This ensemble approach enabled im6APred to ascertain the general methylation patterns on RNA bases and extend this knowledge to the entire m6A transcriptome across different tissues and biological environments, with increased prediction accuracy and robustness.
Evolutionary issues
The integration of AI with omics data has proven efficacious in extracting vital features from such datasets, which are subsequently employed in the construction of phylogenetic trees for species classification and in the adaptive analysis of gene flow. Barrow et al. (2021) highlighted the potential of machine learning algorithms, particularly the random forest model, in analyzing intraspecific diversity among Nearctic amphibians, integrating over 42 000 gene sequences across 299 species. Similarly, Derkarabetian et al. (2019) applied genomics to study species classification within arachnid taxa, such as Metanonychus, known for a high degree of population genetic structuring. Using three unified machine learning methods, namely, random forest, variational autoencoders, and t-distributed Stochastic Neighbor Embedding (t-SNE), they constructed a phylogenetic tree conducive to species delimitation, demonstrating effectiveness across diverse natural systems and taxa with different biological characteristics.
Deep learning techniques have also been applied in the study of animal evolution. Gower et al. (2021) introduced an innovative CNN-based approach that analyzes the genome to explore adaptive introgression in human evolution, particularly the process of adaptive gene flow. This approach, which aims to distinguish between genomic regions influenced by adaptive gene flow from those affected by neutral evolution or selective sweeps, has been successfully applied to the human genome dataset, achieving 95% accuracy in identifying candidate regions implicated in adaptive gene flow, thus offering insights into the evolutionary history of humans.
Deep learning approaches have also been successfully integrated with protein sequences. Li et al. (2018) introduced DNN-PPI, a deep neural network framework for analysis of protein interaction datasets from humans, mice, and other species, which showed excellent prediction accuracy and significant generalization ability. Its innovation lies in its ability to make predictions entirely from features automatically learned from protein sequences, thus avoiding the need to rely on complex biochemical information. These examples emphasize the advantages of AI in evolutionary studies, showcasing its proficiency in handling large-scale datasets, revealing hidden evolutionary patterns, and enhancing analytical efficiency.
AI IN ANIMAL DISEASE MODELS
Animal disease models are essential for advancing our understanding of human pathologies and drug development. In the face of challenges associated with the automatic and effective processing of diverse data, various AI models have been adopted to process complex datasets related to animal disease (Figure 3E). Tree-based methodologies, including random forest and Extreme Gradient Boosting (XGBoost) models, are more adept at handling structured tabular data than popular deep learning models (Grinsztajn et al., 2022). Conversely, non-deep learning algorithms, such as logistic regression and SVM, are often employed for the modeling and analysis of structured data due to their simplicity in training. For example, Leung et al. (2019) analyzed amyloidosis in Indian-origin rhesus macaques by applying logistic regression to structured tabular data with 62 variables. After analyzing the model output, they identified significant factors associated with diagnoses of colitis, gastric adenocarcinoma, and endometriosis, along with clinical issues including trauma and pregnancies. Gardiner et al. (2020) utilized several machine learning models, including linear regression, XGBoost, random forest, k-Nearest Neighbors, and light gradient boosting, to extract information from gene expression and chemical structure data, predicting potential renal dysfunction induced by drugs in rats. These studies highlight the potential of non-deep learning algorithms to effectively handle structured tabular data and benefit research on animal disease models.
As discussed above, deep learning techniques are more suitable for unstructured data, as demonstrated across various studies. Bouteldja et al. (2021) leveraged CNN models to analyze whole-slide images from mouse models of disease, including unilateral ureteral obstruction, adenine-induced kidney disease, renal ischemia-reperfusion injury, and nephrotoxic serum nephritis. Based on computational comparative pathology, Abduljabbar et al. (2023) utilized a CNN model trained on human samples to accurately assess immune responses in 18 vertebrate species. Li et al. (2023) used a ResNet-based toolkit to investigate active and depressive behaviors in mutant monkeys, serving as a disease model for Rett syndrome. Zhang et al. (2022) developed an enhanced YOLOv5 model algorithm to significantly increase the accuracy and efficiency of counting retinal ganglion cells in glaucoma research.
Although non-deep learning and deep learning algorithms are employed for various types of data, it is important to note that structured data often originate from raw unstructured data, such as chemical structures. This conversion, known as feature engineering (Verdonck et al., 2021), can lead to the loss of original information, potentially impairing the predictive accuracy of models. Additionally, gene expression and key variable datasets may be transformed or regarded as pseudo-sequential (Yang et al., 2022) or graph-structured data (Cao & Gao, 2022). Therefore, deep learning algorithms, with their inherent ability to fit unstructured data, may dominate in this research area. These algorithms offer enhanced precision, end-to-end manipulation of unprocessed data for the extraction of predictive outcomes, and more effective exploitation of the intrinsic relationships embedded within the data.
AI IN ANIMAL BREEDING AND HEALTH
Traditional methods of recording animal behavior, size, weight, and feeding data to predict growth patterns and living habits require substantial manpower and material resources. Moreover, intensive efforts to optimize efficiency and output in animal husbandry can result in substantial social deprivation for livestock, leading to adverse effects on behavior and welfare (Neethirajan, 2021b). AI has the potential to enhance farm management and breeding practices by advancing the automation of anomaly detection, body condition evaluation, and disease diagnosis (Figure 3F). Such advancements can substantially reduce the burden of manual data analysis while increasing overall accuracy.
Abnormal detection in breeding
Although conventional methods based on sensors are well established, they often fail to detect anomalies promptly due to the extensive coverage of farming facilities. In contrast, machine learning technology offers rapid, efficient, and contactless automatic recognition of individual animals for breeding farm management. Shen et al. (2020) developed a recognition method using the YOLO model to detect side view images of cows and a fine-tuning CNN for classification, achieving a recognition accuracy rate of 96.65%. Similarly, Li et al. (2017a) extracted shape feature descriptors from captured cow tail root images, employing four classifiers, namely linear discriminant analysis, quadratic discriminant analysis, artificial neural network, and SVM, with the quadratic discriminant analysis and SVM classifiers demonstrating superior performance, obtaining an F1 score of 0.995 and the highest accuracy and precision, respectively. Additionally, Bakoev et al. (2020) used nine machine learning algorithms, including random forest and k-Nearest Neighbors, to assess pig limb conditions based on growth and meat characteristics, thus addressing the issue of leg weakness and lameness in pig farming.
With the increasing need to boost fishery productivity, AI models have been applied to aquaculture management to monitor fish growth and improve aquatic product quality. Chang et al. (2021) developed an intelligent cage farming management system, known as AIoT, to collect extensive data on fish and feed within the cage environment. They trained two deep learning-based object detectors, Faster-RCNN and YOLOv3, to specifically detect fish bodies and monitor growth and feeding conditions, thereby reducing feed waste, enhancing feed conversion rates, and increasing fish survival rates. The YOLO model is capable of tracking multiple objects in real-time within images, making it ideal for automated systems designed to monitor large populations in both aquaculture and fisheries. Fish classification is essential for quality control and population statistics. However, the task becomes challenging due to changes that occur when fish leave the water. Abinaya et al. (2021) implemented a deep learning model augmented with a naive Bayes classifier for fish classification, achieving better accuracy and robustness compared to original neural networks. Additionally, Liao et al. (2021) developed 3DPhenoFish to provide a more efficient and objective approach for semantic segmentation and extraction of 3D fish morphological phenotypes from point cloud data, which has implications in artificial breeding, functional gene mapping, and population-based studies in aquaculture and ecology.
Animal health
AI technologies have also been applied to streamline animal healthcare by improving body condition assessment and disease diagnosis (Ezanno et al., 2021). Accurate body condition scoring is crucial for evaluating health and energy balance in cows, guiding dietary and reproductive management (Cockburn, 2020; Rodríguez Alvarez et al., 2019). Rodríguez Alvarez et al. (2019) developed a CNN-based model combined with transfer learning and ensemble modeling to assess body condition in cows from depth image features, achieving an accuracy of 82% within 0.25 units and 97% within 0.50 units of the real value. In addition, bluetongue disease represents a common infectious disease in ruminants. Gouda et al. (2022) utilized several algorithms, including logistic regression, decision tree, random forest, and feedforward artificial neural network, to predict the risk of bluetongue disease using seroepidemiological data, with the centralized machine learning model outperforming traditional methods. Wagner et al. (2020) used machine learning techniques, such as K-nearest neighbor regression, decision tree regression, Multi-Layer Perceptron, and LSTM, to detect subtle behavioral changes in dairy cows based on data from the previous day. Furthermore, AI algorithms have been applied to automatically monitor the health and emotional state of animals (Neethirajan, 2021a). Neethirajan (2021b) developed an AI model based on generative adversarial network architecture capable of analyzing different modes of animal communication behavior and generating virtual conspecific companion videos to improve animal well-being. Such AI-driven approaches can successfully detect abnormal behavior and stress in animals, even before clinical symptoms appear.
AI IN PALEOZOOLOGY
The field of paleozoology, dedicated to the study of prehistoric life through fossil analysis, has traditionally demanded significant labor and time. In recent years, however, advancements in AI algorithms capable of learning, prediction, and automation have revolutionized this field by enhancing fossil analysis, species identification, and the extrapolation of functions and behaviors from fossil evidence (Figure 3G).
AI has been effectively applied for the classification of animal fossils based on both text and image data. For instance, in their study on marine invertebrates, Kopperud et al. (2019) used an automated pipeline based on supervised machine learning to extract observations of fossils and their inferred ages from unstructured text, achieving high accuracy compared to human annotators and greatly reducing the time required. In a broader study, Hoyal Cuthill et al. (2020) used a machine learning spatial embedding method to measure distance among 171 231 species and analyze the impacts of speciation and extinction based on an evolutionary decay clock. In regard to primates, AI has been utilized to infer behaviors from fossils based on species classification (Marcé-Nogué et al., 2020; Püschel et al., 2018). Püschel et al. (2018) applied machine learning classification methods to differentiate between various locomotor categories in primates, using both biomechanical and morphometric data. Similarly, Marcé-Nogué et al. (2020) used an SVM algorithm to classify primate fossils into broad ingesta-related hardness categories.
The application of deep learning algorithms for image data analysis has become increasingly prevalent in resolving classification issues related to animal fossils. CNNs have demonstrated considerable accuracy in the classification of modern and ancient bone surface modifications within multiple mammals (Domínguez-Rodrigo et al., 2020). Using both unsupervised and supervised AI algorithms, studies have achieved high accuracy (88%–98%) in classifying carnivore species based on 3D modeling and geometric morphometrics of tooth pits (Courtenay et al., 2021). Furthermore, deep CNNs have distinguished between theropod and ornithischian dinosaur tracks based on outline silhouettes, consistently demonstrating superior performance compared to human experts (Lallensack et al., 2022).
CHALLENGES FOR USING AI IN ZOOLOGICAL RESEARCH
As discussed in the introduction, there is a lag in the adoption of AI in zoological research (Figure 1A, B). One reason for this delay is the unfamiliarity of zoologists with various models, coupled with the challenges arising from data format complexity, data insufficiency, and reliance on small sample learning tasks. Comprehensive zoological research includes complex unstructured data, spanning images, videos, sound recordings, text sequences, and protein structures, presenting significant challenges for AI model applications, requiring experts to select appropriate models for specific data types and, in some instances, expend additional effort in data processing. Small sample learning is another common challenge, especially when studying specific species or ecosystems, with the extreme rarity of certain animal species posing considerable challenges in gathering adequate data. Furthermore, while identifying animals based on their sounds can provide valuable insights, gathering high-quality sound data can be time and resource-intensive, leading to limitations in sample sizes (Bravo Sanchez et al., 2021). In addition, observing animal behavior typically requires time and effort, often resulting in limited data and constraints on in-depth studies of specific behaviors (Arac et al., 2019). These circumstances give rise to imbalanced datasets and fewer samples from rare species for training neural networks (Høye et al., 2021).
Although the application of AI models in zoology has been slow compared to the broader biological sciences, the introduction of new technologies in specialized subfields often comes with an inherent time lag. To address these challenges, it is recommended to enhance data collection initiatives, focusing on long-term accumulation and integration with existing databases to mitigate data insufficiency. In situations of limited sample sizes, employing pre-training and transfer learning methods would be beneficial ((Høye et al., 2021; Vélez et al., 2022). For data concerning rare species or of low quality, applying data augmentation methods such as image manipulation and geometric transformations can expand training datasets (Klasen et al., 2022). Model-agnostic meta-learning (Shui et al., 2023) and multiset feature learning (Jing et al., 2021) could offer innovative solutions to these issues (Shui et al., 2023; Jing et al., 2021). When encountering species not present in existing databases, the challenge becomes even more significant. Addressing this situation necessitates the application of multiple class anomaly/novelty detection or open set/world recognition (Perera & Patel, 2019; Turkoz et al., 2020). Moreover, simpler models such as logistic regression, K-Nearest Neighbor Regression, and SVM may be more suitable in these cases. To address generalization issues, emphasis should be placed on data integrity through meticulous cleaning and quality control processes. Leveraging pre-trained models and fine-tuning them on specific tasks can also aid in adapting to varied species data (Lin et al., 2023). Lastly, the broader dissemination and implementation of these AI technologies should be encouraged, alongside fostering collaborative efforts between AI experts and domain specialists.
Supervised learning plays a pivotal role in zoological research due to its close association with species classification, behavior identification, and regression tasks in feeding behaviors. Supervised learning requires high-quality annotated data to train models and produce highly accurate predictions. As discussed in the “Animal Classification and Resource Protection” section, certain species classification models have shown suboptimal accuracy (Bravo Sanchez et al., 2021), which may be partly attributed to a lack of labeled data for supervised learning. To address this issue, techniques like semi-supervised, which combines supervised and unsupervised learning, and weakly supervised learning have been applied in the field of AI, offering potential methodologies for zoological research.
PROSPECTS FOR FUTURE WORK
Deep learning models, particularly CNNs, are favored by researchers as predictive models due to their local receptive fields and convolutional architecture, which make them effective at processing image data. However, their efficacy in handling sequential data, such as text and video recordings, is less satisfactory compared to linguistic models such as LSTM and BERT. The main limitation of CNNs is their inability to effectively capture and preserve the positional dependencies inherent in sequential data. Thus, zoological researchers may adopt different trajectories, which is not necessarily problematic but warrants further exploration. Supervised learning is also prevalent in zoological studies, commonly chosen for fitting models and performing classification tasks. In contrast, unsupervised learning is less frequently used, primarily for generative and clustering tasks, due to the absence of supervised labels. Reinforcement learning also draws inspiration from zoological studies exploring animal decision-making behavior derived from neuronal activation. Such integration is expected, as the field of zoological research emphasizes using AI models to address specific, domain-related challenges spanning supervised, unsupervised, and reinforcement learning.
The integration of AI into zoological research promises a paradigm shift, enhancing efficiency, accuracy, and scope (Wang et al., 2023a). AI will enhance data collection and interpretation by increasing efficiency, reducing time expenditure, and improving accuracy, especially in handling complex data like intricate sounds and high-resolution images (Shen et al., 2022b). AI will enable real-time, non-invasive monitoring of wildlife, in contrast to traditional methods that are often invasive and limited. AI technologies also promote objectivity in behavioral analysis by automating processes and providing quantitative metrics. The focus of zoological research is shifting from mere descriptive to predictive and explanatory models. This technology will also improve the verifiability and reproducibility of research, influencing the broader acceptance of theories. The interdisciplinary nature of AI also allows for the integration of computer science, statistics, and biology, facilitating the development of new theoretical frameworks. AI further democratizes zoological research by enabling public participation in data collection, such as through social media-based wildlife monitoring (Foglio et al., 2019). Paradigm shifts are not usually completed in the short term but are gradual, evolving transformations. However, as the application of AI in zoology becomes increasingly widespread, we anticipate a series of profound changes in this field, including in the evolution of theoretical frameworks. These changes will advance our understanding of the complexity of the animal world in a more comprehensive and in-depth manner.
In the future, beyond the establishment and use of cameras in nature reserves, AI will also be able to utilize satellite images, climate data, and drones to detect changes in animal populations or potential threats like poaching, helping to predict areas where endangered species can reproduce and thrive. The integration of AI with biological recording devices will also revolutionize the study of animal migration, behavior, and physiology under natural conditions. The potential development of a global platform will enable zoologists worldwide to collaborate, share data, and utilize AI tools collectively for analysis.
Zoological research can also directly influence model design, drawing insights from animal neuroscience and behavioral studies (Yu, 2016). Furthermore, the zoological domain provides a vast and diverse dataset, supporting comparative analysis of models across different architectures and theoretical paradigms. As a result, it serves as an ideal experimental platform for AI models requiring extensive data resources. Therefore, the relationship is not just about applying AI to zoology but is mutually beneficial for both disciplines.
CONCLUSIONS
This review comprehensively examines the integration of AI within zoological research, outlining the primary tasks, core models, datasets, and the challenges encountered in this field. AI algorithms have been instrumental in advancing various subfields, managing complex data forms, and achieving leading-edge predictive accuracy. The combination of AI and zoological research harnesses the computational power of “bytes” to successfully decipher the inner secrets of “beasts”. Given the increasing demand for analyzing data generated from laboratories, natural habitats, and agricultural settings—including but not limited to images, text, tables, videos, gene sequences, and molecular structures—the strategic selection and application of AI models tailored to specific scientific inquiries is expected to gain momentum.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Y.J.Z. and Z.C. conceived the review. Y.J.Z., Y.S., J.L., Z.L, and Z.C. prepared the manuscript, figures, and tables. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31871274), Natural Science Foundation of Chongqing, China (CSTB2022NSCQ-MSX0650), Science and Technology Research Program of Chongqing Municipal Education Commission (KJQN202100508), Team Project of Innovation Leading Talent in Chongqing (CQYC20210309536), and “Contract System” Project of Chongqing Talent Plan (cstc2022ycjh-bgzxm0147)
References
- Abduljabbar K, Castillo SP, Hughes K, et al Bridging clinic and wildlife care with AI-powered pan-species computational pathology. Nature Communications. 2023;14(1):2408. doi: 10.1038/s41467-023-37879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abinaya NS, Susan D, Kumar SR Naive Bayesian fusion based deep learning networks for multisegmented classification of fishes in aquaculture industries. Ecological Informatics. 2021;61:101248. doi: 10.1016/j.ecoinf.2021.101248. [DOI] [Google Scholar]
- Achour B, Belkadi M, Filali I, et al Image analysis for individual identification and feeding behaviour monitoring of dairy cows based on Convolutional Neural Networks (CNN) Biosystems Engineering. 2020;198:31–49. doi: 10.1016/j.biosystemseng.2020.07.019. [DOI] [Google Scholar]
- Ahmad A, Asif M, Ali SR Review paper on shallow learning and deep learning methods for network security. International Journal of Scientific Research in Computer Science and Engineering. 2018;6(5):45–54. doi: 10.26438/ijsrcse/v6i5.4554. [DOI] [Google Scholar]
- Alroy J, Aberhan M, Bottjer DJ, et al Phanerozoic trends in the global diversity of marine invertebrates. Science. 2008;321(5885):97–100. doi: 10.1126/science.1156963. [DOI] [PubMed] [Google Scholar]
- Alzubaidi L, Zhang JL, Humaidi AJ, et al Review of deep learning: concepts, CNN architectures, challenges, applications, future directions. Journal of Big Data. 2021;8(1):53. doi: 10.1186/s40537-021-00444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac A, Zhao PP, Dobkin BH, et al DeepBehavior: a deep learning toolbox for automated analysis of animal and human behavior imaging data. Frontiers in Systems Neuroscience. 2019;13:20. doi: 10.3389/fnsys.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babayan SA, Orton RJ, Streicker DG Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science. 2018;362(6414):577–580. doi: 10.1126/science.aap9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badia-I-Mompel P, Wessels L, Müller-Dott S, et al Gene regulatory network inference in the era of single-cell multi-omics. Nature Reviews Genetics. 2023;24(11):739–754. doi: 10.1038/s41576-023-00618-5. [DOI] [PubMed] [Google Scholar]
- Bai YT, Jones A, Ndousse K, et al. 2022. Training a helpful and harmless assistant with reinforcement learning from human feedback. arXiv, doi:https://doi.org/10.48550/arXiv.2204.05862.
- Bakoev S, Getmantseva L, Kolosova M, et al PigLeg: prediction of swine phenotype using machine learning. PeerJ. 2020;8:e8764. doi: 10.7717/peerj.8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow LN, da Fonseca EM, Thompson CEP, et al Predicting amphibian intraspecific diversity with machine learning: challenges and prospects for integrating traits, geography, and genetic data. Molecular Ecology Resources. 2021;21(8):2818–2831. doi: 10.1111/1755-0998.13303. [DOI] [PubMed] [Google Scholar]
- Bath DE, Stowers JR, Hörmann D, et al. 2014. FlyMAD: rapid thermogenetic control of neuronal activity in freely walking Drosophila. Nature Methods, 11(7): 756–762.
- Binta Islam S, Valles D, Hibbitts TJ, et al Animal species recognition with deep convolutional neural networks from ecological camera trap images. Animals. 2023;13(9):1526. doi: 10.3390/ani13091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkovskiy A, Wang CY, Liao HYM. 2020. YOLOv4: optimal speed and accuracy of object detection. arXiv, doi:https://doi.org/10.48550/arXiv.2004.10934.
- Bommasani R, Hudson DA, Adeli E, et al. 2021. On the opportunities and risks of foundation models. arXiv, doi:https://doi.org/10.48550/arXiv.2108.07258.
- Bouteldja N, Klinkhammer BM, Bülow RD, et al Deep learning–based segmentation and quantification in experimental kidney histopathology. Journal of the American Society of Nephrology. 2021;32(1):52–68. doi: 10.1681/ASN.2020050597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Sanchez FJ, Hossain R, English NB, et al Bioacoustic classification of avian calls from raw sound waveforms with an open-source deep learning architecture. Scientific Reports. 2021;11(1):15733. doi: 10.1038/s41598-021-95076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouard JS, Schenkel F, Marete A, et al The GATK joint genotyping workflow is appropriate for calling variants in RNA-seq experiments. Journal of Animal Science and Biotechnology. 2019;10(1):44. doi: 10.1186/s40104-019-0359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao ZJ, Gao G Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nature Biotechnology. 2022;40(10):1458–1466. doi: 10.1038/s41587-022-01284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JY, Zeng H, Li AM, et al Deep learning in computer vision: a critical review of emerging techniques and application scenarios. Machine Learning with Applications. 2021;6:100134. doi: 10.1016/j.mlwa.2021.100134. [DOI] [Google Scholar]
- Chang CC, Wang JH, Wu JL, et al Applying artificial intelligence (AI) techniques to implement a practical smart cage aquaculture management system. Journal of Medical and Biological Engineering. 2021;41(5):652–658. [Google Scholar]
- Chen C, Tang YJ, Tan Y, et al Three-dimensional cerebral vasculature topological parameter extraction of transgenic zebrafish embryos with a filling-enhancement deep learning network. Biomedical Optics Express. 2023a;14(2):971–984. doi: 10.1364/BOE.484351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhu WX, Steibel J, et al Recognition of feeding behaviour of pigs and determination of feeding time of each pig by a video-based deep learning method. Computers and Electronics in Agriculture. 2020;176:105642. doi: 10.1016/j.compag.2020.105642. [DOI] [Google Scholar]
- Chen W, Liu Y, Wang WP, et al Deep learning for instance retrieval: a survey. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2023b;45(6):7270–7292. doi: 10.1109/TPAMI.2022.3218591. [DOI] [PubMed] [Google Scholar]
- Choi SR, Lee M Transformer architecture and attention mechanisms in genome data analysis: a comprehensive review. Biology. 2023;12(7):1033. doi: 10.3390/biology12071033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Gulcehre C, Cho K, et al. 2014. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv, doi:https://doi.org/10.48550/arXiv.1412.3555.
- Cockburn M Review: application and prospective discussion of machine learning for the management of dairy farms. Animals. 2020;10(9):1690. doi: 10.3390/ani10091690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey KR, Marx RE, Neumaier JF DeepSqueak: a deep learning-based system for detection and analysis of ultrasonic vocalizations. Neuropsychopharmacology. 2019;44(5):859–868. doi: 10.1038/s41386-018-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GH ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. Journal of Applied Crystallography. 1997;30(6):1160–1161. doi: 10.1107/S0021889897006729. [DOI] [Google Scholar]
- Courtenay LA, Herranz-Rodrigo D, González-Aguilera D, et al Developments in data science solutions for carnivore tooth pit classification. Scientific Reports. 2021;11(1):10209. doi: 10.1038/s41598-021-89518-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Filho EM, de Amorim Fernandes F, de Abreu Soares C, et al Inteligência artificial em cardiologia: conceitos, ferramentas e desafios – “quem corre é o cavalo, você precisa ser o jóquei”. Arquivos Brasileiros de Cardiologia. 2020;114(4):718–725. doi: 10.36660/abc.20180431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkarabetian S, Castillo S, Koo PK, et al A demonstration of unsupervised machine learning in species delimitation. Molecular Phylogenetics and Evolution. 2019;139:106562. doi: 10.1016/j.ympev.2019.106562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J, Chang MW, Lee K, et al. 2019. BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Minneapolis: ACL, 4171–4186.
- Domínguez-Rodrigo M, Cifuentes-Alcobendas G, Jiménez-García B, et al Artificial intelligence provides greater accuracy in the classification of modern and ancient bone surface modifications. Scientific Reports. 2020;10(1):18862. doi: 10.1038/s41598-020-75994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dönmez P Introduction to Machine Learning, 2nd ed., by Ethem Alpaydın. Cambridge, MA: The MIT Press2010. ISBN: 978-0-262-01243-0. $54/£ 39.95 + 584 pages. Natural Language Engineering. 2013;19(2):285–288. doi: 10.1017/S1351324912000290. [DOI] [Google Scholar]
- Durward-Akhurst SA, Schaefer RJ, Grantham B, et al Genetic variation and the distribution of variant types in the horse. Frontiers in Genetics. 2021;12:758366. doi: 10.3389/fgene.2021.758366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezanno P, Picault S, Beaunée G, et al Research perspectives on animal health in the era of artificial intelligence. Veterinary Research. 2021;52(1):40. doi: 10.1186/s13567-021-00902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahbakhsh E, Chandra R, Olierook HKH, et al Computer vision-based framework for extracting tectonic lineaments from optical remote sensing data. International Journal of Remote Sensing. 2020;41(5):1760–1787. doi: 10.1080/01431161.2019.1674462. [DOI] [Google Scholar]
- Fergus P, Chalmers C, Longmore S, et al Empowering wildlife guardians: an equitable digital stewardship and reward system for biodiversity conservation using deep learning and 3/4G camera traps. Remote Sensing. 2023;15(11):2730. doi: 10.3390/rs15112730. [DOI] [Google Scholar]
- Foglio M, Semeria L, Muscioni G, et al. 2019. Animal wildlife population estimation using social media images collections. arXiv, doi:https://doi.org/10.48550/arXiv.1908.01875.
- Frankenhuis WE, Panchanathan K, Barto AG Enriching behavioral ecology with reinforcement learning methods. Behavioural Processes. 2019;161:94–100. doi: 10.1016/j.beproc.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Gardiner LJ, Carrieri AP, Wilshaw J, et al Using human in vitro transcriptome analysis to build trustworthy machine learning models for prediction of animal drug toxicity. Scientific Reports. 2020;10(1):9522. doi: 10.1038/s41598-020-66481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldenhuys DS, Josias S, Brink W, et al Deep learning approaches to landmark detection in tsetse wing images. PLoS Computational Biology. 2023;19(6):e1011194. doi: 10.1371/journal.pcbi.1011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest D, Puybareau E, Léonard M, et al High throughput automated detection of axial malformations in Medaka embryo. Computers in Biology and Medicine. 2019;105:157–168. doi: 10.1016/j.compbiomed.2018.12.016. [DOI] [PubMed] [Google Scholar]
- Gomes B, Ashley EA Artificial intelligence in molecular medicine. The New England Journal of Medicine. 2023;388(26):2456–2465. doi: 10.1056/NEJMra2204787. [DOI] [PubMed] [Google Scholar]
- Gore AL Measure emissions to manage emissions. Science. 2022;378(6619):455. doi: 10.1126/science.adf5788. [DOI] [PubMed] [Google Scholar]
- Gouda HF, Hassan FAM, El-Araby EE, et al Comparison of machine learning models for bluetongue risk prediction: a seroprevalence study on small ruminants. BMC Veterinary Research. 2022;18(1):394. doi: 10.1186/s12917-022-03486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower G, Picazo PI, Fumagalli M, et al Detecting adaptive introgression in human evolution using convolutional neural networks. eLife. 2021;10:e64669. doi: 10.7554/eLife.64669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalo-Brizuela R, Garrido-Merchan EC. 2023. ChatGPT is not all you need. A state of the art review of large generative AI models. arXiv, doi:https://doi.org/10.48550/arXiv.2301.04655.
- Graving JM, Chae D, Naik H, et al DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. eLife. 2019;8:e47994. doi: 10.7554/eLife.47994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinsztajn L, Oyallon E, Varoquaux G. 2022. Why do tree-based models still outperform deep learning on typical tabular data?. In: Proceedings of the 36th Conference on Neural Information Processing Systems. New Orleans.
- He KM, Zhang XY, Ren SQ, et al. 2015. Deep residual learning for image recognition. In: Proceedings of 2016 IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas: IEEE.
- He YQ, Tiezzi F, Howard J, et al Predicting body weight in growing pigs from feeding behavior data using machine learning algorithms. Computers and Electronics in Agriculture. 2021;184:106085. doi: 10.1016/j.compag.2021.106085. [DOI] [Google Scholar]
- Hinton GE, Osindero S, Teh YW A fast learning algorithm for deep belief nets. Neural Computation. 2006;18(7):1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- Hochreiter S, Schmidhuber J Long short-term memory. Neural Computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Hoyal Cuthill JF, Guttenberg N, Budd GE Impacts of speciation and extinction measured by an evolutionary decay clock. Nature. 2020;588(7839):636–641. doi: 10.1038/s41586-020-3003-4. [DOI] [PubMed] [Google Scholar]
- Høye TT, Ärje J, Bjerge K, et al Deep learning and computer vision will transform entomology. Proceedings of the National Academy of Sciences of the United States of America. 2021;118(2):e2002545117. doi: 10.1073/pnas.2002545117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal A, Babu AR, Zadeh MZ, et al A survey on contrastive self-supervised learning. Technologies. 2021;9(1):2. [Google Scholar]
- Janiesch C, Zschech P, Heinrich K Machine learning and deep learning. Electronic Markets. 2021;31(3):685–695. doi: 10.1007/s12525-021-00475-2. [DOI] [Google Scholar]
- Jiang SY, Zhang S, Kang XP, et al Risk assessment of the possible intermediate host role of pigs for coronaviruses with a deep learning predictor. Viruses. 2023;15(7):1556. doi: 10.3390/v15071556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Gradus JL, Rosellini AJ Supervised machine learning: a brief primer. Behavior Therapy. 2020;51(5):675–687. doi: 10.1016/j.beth.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing XY, Zhang XY, Zhu XK, et al Multiset feature learning for highly imbalanced data classification. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2021;43(1):139–156. doi: 10.1109/TPAMI.2019.2929166. [DOI] [PubMed] [Google Scholar]
- Jordan MI, Mitchell TM Machine learning: trends, perspectives, and prospects. Science. 2015;349(6245):255–260. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- Kaplow IM, Lawler AJ, Schäffer DE, et al Relating enhancer genetic variation across mammals to complex phenotypes using machine learning. Science. 2023;380(6643):eabm7993. doi: 10.1126/science.abm7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras T, Laine S, Aila T. 2019. A style-based generator architecture for generative adversarial networks. In: Proceedings of 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Long Beach: IEEE.
- Kaushik AC, Mehmood A, Peng SL, et al A-CaMP: a tool for anti-cancer and antimicrobial peptide generation. Journal of Biomolecular Structure and Dynamics. 2021;39(1):285–293. doi: 10.1080/07391102.2019.1708796. [DOI] [PubMed] [Google Scholar]
- Kellenberger B, Marcos D, Tuia D Detecting mammals in UAV images: best practices to address a substantially imbalanced dataset with deep learning. Remote Sensing of Environment. 2018;216:139–153. doi: 10.1016/j.rse.2018.06.028. [DOI] [Google Scholar]
- Khan A, Sohail A, Zahoora U, et al A survey of the recent architectures of deep convolutional neural networks. Artificial Intelligence Review. 2020;53(8):5455–5516. doi: 10.1007/s10462-020-09825-6. [DOI] [Google Scholar]
- Khan YF, Kaushik B, Rahmani MKI, et al HSI-LFS-BERT: novel hybrid swarm intelligence based linguistics feature selection and computational intelligent model for Alzheimer’s prediction using audio transcript. IEEE Access. 2022;10:126990–127004. doi: 10.1109/ACCESS.2022.3223681. [DOI] [Google Scholar]
- Kim T, Cha M, Kim H, et al. 2017. Learning to discover cross-domain relations with generative adversarial networks. In: Proceedings of the 34th International Conference on Machine Learning. Sydney: PMLR, 1857–1865.
- Klasen M, Ahrens D, Eberle J, et al Image-based automated species identification: can virtual data augmentation overcome problems of insufficient sampling? Systematic Biology. 2022;71(2):320–333. doi: 10.1093/sysbio/syab048. [DOI] [PubMed] [Google Scholar]
- Klibaite U, Berman GJ, Cande J, et al An unsupervised method for quantifying the behavior of paired animals. Physical Biology. 2017;14(1):015006. doi: 10.1088/1478-3975/aa5c50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopperud BT, Lidgard S, Liow LH Text-mined fossil biodiversity dynamics using machine learning. Proceedings of the Royal Society B:Biological Sciences. 2019;286(1901):20190022. doi: 10.1098/rspb.2019.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE ImageNet classification with deep convolutional neural networks. Communications of the ACM. 2017;60(6):84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- Kulkarni S, Gadot T, Luo C, et al. 2020. Unifying data for fine-grained visual species classification. arXiv, doi:https://doi.org/10.48550/arXiv.2009.11433.
- Lahat D, Adali T, Jutten C Multimodal data fusion: an overview of methods, challenges, and prospects. Proceedings of the IEEE. 2015;103(9):1449–1477. doi: 10.1109/JPROC.2015.2460697. [DOI] [Google Scholar]
- Lallensack JN, Romilio A, Falkingham PL A machine learning approach for the discrimination of theropod and ornithischian dinosaur tracks. Journal of the Royal Society Interface. 2022;19(196):20220588. doi: 10.1098/rsif.2022.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton KKS, Snelgrove PVR, Dempson JB, et al Genomic evidence of past and future climate-linked loss in a migratory Arctic fish. Nature Climate Change. 2021;11(2):158–165. doi: 10.1038/s41558-020-00959-7. [DOI] [Google Scholar]
- LeCun Y, Boser B, Denker JS, et al Backpropagation applied to handwritten zip code recognition. Neural Computation. 1989;1(4):541–551. doi: 10.1162/neco.1989.1.4.541. [DOI] [Google Scholar]
- Lee K, Ray J, Safta C The predictive skill of convolutional neural networks models for disease forecasting. PLoS One. 2021;16(7):e0254319. doi: 10.1371/journal.pone.0254319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim H, Cho BK Deep learning-based image classification for major mosquito species inhabiting korea. Insects. 2023;14(6):526. doi: 10.3390/insects14060526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung ET, Raboin MJ, McKelvey J, et al Modelling disease risk for amyloid A (AA) amyloidosis in non-human primates using machine learning. Amyloid. 2019;26(3):139–147. doi: 10.1080/13506129.2019.1625038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Xiao ZF, Li YR, et al Deep learning-based activity recognition and fine motor identification using 2D skeletons of cynomolgus monkeys. Zoological Research. 2023;44(5):967–980. doi: 10.24272/j.issn.2095-8137.2022.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gong XJ, Yu H, et al Deep neural network based predictions of protein interactions using primary sequences. Molecules. 2018;23(8):1923. doi: 10.3390/molecules23081923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WY, Ji ZT, Wang L, et al Automatic individual identification of Holstein dairy cows using tailhead images. Computers and Electronics in Agriculture. 2017a;142:622–631. doi: 10.1016/j.compag.2017.10.029. [DOI] [Google Scholar]
- Li XY, Xiong XS, Yi CQ Epitranscriptome sequencing technologies: decoding RNA modifications. Nature Methods. 2017b;14(1):23–31. doi: 10.1038/nmeth.4110. [DOI] [PubMed] [Google Scholar]
- Li YX. 2017c. Deep reinforcement learning: an overview. arXiv, doi:https://doi.org/10.48550/arXiv.1701.07274.
- Liao YH, Zhou CW, Liu WZ, et al 3DPhenoFish: application for two- and three-dimensional fish morphological phenotype extraction from point cloud analysis. Zoological Research. 2021;42(4):492–501. doi: 10.24272/j.issn.2095-8137.2021.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YL, Chang PC, Hsu C, et al Comparison of GATK and DeepVariant by trio sequencing. Scientific Reports. 2022a;12(1):1809. doi: 10.1038/s41598-022-05833-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZM, Akin H, Rao RS, et al. 2022b. Evolutionary-scale prediction of atomic level protein structure with a language model. BioRxiv, doi:https://doi.org/10.1101/2022.07.20.500902.
- Lin ZM, Akin H, Rao RS, et al Evolutionary-scale prediction of atomic-level protein structure with a language model. Science. 2023;379(6637):1123–1130. doi: 10.1126/science.ade2574. [DOI] [PubMed] [Google Scholar]
- Lipton ZC, Berkowitz J, Elkan C. 2015. A critical review of recurrent neural networks for sequence learning. arXiv, doi:https://doi.org/10.48550/arXiv.1506.00019.
- Liu MS, Gao JQ, Hu GY, et al MonkeyTrail: a scalable video-based method for tracking macaque movement trajectory in daily living cages. Zoological Research. 2022;43(3):343–351. doi: 10.24272/j.issn.2095-8137.2021.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Gan L, Zhang ZT, et al Probing the processing of facial expressions in monkeys via time perception and eye tracking. Zoological Research. 2023;44(5):882–893. doi: 10.24272/j.issn.2095-8137.2023.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente Ò, Riera I, Rana A. 2021. Image classification with classic and deep learning techniques. arXiv, doi:https://doi.org/10.48550/arXiv.2105.04895.
- Luo ZT, Lou LL, Qiu WR, et al Predicting N6-methyladenosine sites in multiple tissues of mammals through ensemble deep learning. International Journal of Molecular Sciences. 2022;23(24):15490. doi: 10.3390/ijms232415490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YF, Yuan XG, Cun YP A novel machine learning approach (svmSomatic) to distinguish somatic and germline mutations using next-generation sequencing data. Zoological Research. 2021;42(2):246–249. doi: 10.24272/j.issn.2095-8137.2021.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcé-Nogué J, Püschel TA, Daasch A, et al Broad-scale morpho-functional traits of the mandible suggest no hard food adaptation in the hominin lineage. Scientific Reports. 2020;10(1):6793. doi: 10.1038/s41598-020-63739-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi S, Sedaghat S Study the genetic variation using Eta functions. Computational and Applied Mathematics. 2023;42(2):95. doi: 10.1007/s40314-023-02242-9. [DOI] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, et al DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nature Neuroscience. 2018;21(9):1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- Meir Y, Tevet O, Tzach Y, et al Efficient shallow learning as an alternative to deep learning. Scientific Reports. 2023;13(1):5423. doi: 10.1038/s41598-023-32559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor M, Banerjee O, Abad ZSH, et al Foundation models for generalist medical artificial intelligence. Nature. 2023;616(7956):259–265. doi: 10.1038/s41586-023-05881-4. [DOI] [PubMed] [Google Scholar]
- Moulin TC, Covill LE, Itskov PM, et al Rodent and fly models in behavioral neuroscience: an evaluation of methodological advances, comparative research, and future perspectives. Neuroscience & Biobehavioral Reviews. 2021;120:1–12. doi: 10.1016/j.neubiorev.2020.11.014. [DOI] [PubMed] [Google Scholar]
- Nandutu I, Atemkeng M, Okouma P, et al. 2022. Creating awareness about security and safety on highways to mitigate wildlife-vehicle collisions by detecting and recognizing wildlife fences using deep learning and drone technology. arXiv, doi:https://doi.org/10.48550/arXiv.2301.07174.
- Neethirajan S. 2021a. Happy cow or thinking pig? WUR wolf—facial coding platform for measuring emotions in farm animals. AI, 2(3): 342–354.
- Neethirajan S Is seeing still believing? Leveraging deepfake technology for livestock farming. Frontiers in Veterinary Science. 2021b;8:740253. doi: 10.3389/fvets.2021.740253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzzadeh MS, Nguyen A, Kosmala M, et al Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(25):E5716–E5725. doi: 10.1073/pnas.1719367115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera P, Patel VM. 2019. Deep transfer learning for multiple class novelty detection. In: Proceedings of 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Long Beach: IEEE, 11544–11552.
- Perrett A, Pollard H, Barnes C, et al DeepVerge: classification of roadside verge biodiversity and conservation potential. Computers, Environment and Urban Systems. 2022;102:101968. [Google Scholar]
- Pilehvar MT, Camacho-Collados J Embeddings in natural language processing: theory and advances in vector representations of meaning. Computational Linguistics. 2021;47(3):699–701. doi: 10.1162/coli_r_00410. [DOI] [Google Scholar]
- Püschel TA, Marcé-Nogué J, Gladman JT, et al Inferring locomotor behaviours in Miocene New World monkeys using finite element analysis, geometric morphometrics and machine-learning classification techniques applied to talar morphology. Journal of the Royal Society Interface. 2018;15(146):20180520. doi: 10.1098/rsif.2018.0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu ZM, Xu TD, Langerman J, et al A deep learning approach for segmentation, classification, and visualization of 3-D high-frequency ultrasound images of mouse embryos. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. 2021;68(7):2460–2471. doi: 10.1109/TUFFC.2021.3068156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford A, Kim JW, Hallacy C, et al. 2021. Learning transferable visual models from natural language supervision. In: Proceedings of the 38th International Conference on Machine Learning. PMLR, 8748–8763.
- Richards BA, Lillicrap TP, Beaudoin P, et al A deep learning framework for neuroscience. Nature Neuroscience. 2019;22(11):1761–1770. doi: 10.1038/s41593-019-0520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson JF, Denholm SJ, Coffey M Automated processing and phenotype extraction of ovine medical images using a combined generative adversarial network and computer vision pipeline. Sensors. 2021;21(21):7268. doi: 10.3390/s21217268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha JC, Passalia FJ, Matos FD, et al A method based on artificial intelligence to fully automatize the evaluation of bovine blastocyst images. Scientific Reports. 2017;7(1):7659. doi: 10.1038/s41598-017-08104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Alvarez J, Arroqui M, Mangudo P, et al Estimating body condition score in dairy cows from depth images using convolutional neural networks, transfer learning and model ensembling techniques. Agronomy. 2019;9(2):90. doi: 10.3390/agronomy9020090. [DOI] [Google Scholar]
- Romero-Ferrero F, Bergomi MG, Hinz RC, et al idtracker. ai: tracking all individuals in small or large collectives of unmarked animals. Nature Methods. 2019;16(2):179–182. doi: 10.1038/s41592-018-0295-5. [DOI] [PubMed] [Google Scholar]
- Rumelhart DE, Hinton GE, Williams RJ Learning representations by back-propagating errors. Nature. 1986;323(6088):533–536. doi: 10.1038/323533a0. [DOI] [Google Scholar]
- Sainburg T, Gentner TQ Toward a computational neuroethology of vocal communication: from bioacoustics to neurophysiology, emerging tools and future directions. Frontiers in Behavioral Neuroscience. 2021;15:811737. doi: 10.3389/fnbeh.2021.811737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salekin S, Mostavi M, Chiu YC, et al Predicting sites of epitranscriptome modifications using unsupervised representation learning based on generative adversarial networks. Frontiers in Physics. 2020;8:196. doi: 10.3389/fphy.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker IH Machine learning: algorithms, real-world applications and research directions. SN Computer Science. 2021;2(3):160. doi: 10.1007/s42979-021-00592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaplen KM, Mei NJ, Bounds HA, et al. 2019. Automated real-time quantification of group locomotor activity in Drosophila melanogaster. Scientific Reports, 9(1): 4427.
- Shabbir J, Anwer T. 2018. Artificial intelligence and its role in near future. arXiv, doi:https://doi.org/10.48550/arXiv.1804.01396
- Shafiullah AZ, Werner J, Kennedy E, et al Machine learning based prediction of insufficient herbage allowance with automated feeding behaviour and activity data. Sensors. 2019;19(20):4479. doi: 10.3390/s19204479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Lamba A, Zhu M, et al Stain-free detection of embryo polarization using deep learning. Scientific Reports. 2022a;12(1):2404. doi: 10.1038/s41598-022-05990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WZ, Hu HQ, Dai BS, et al Individual identification of dairy cows based on convolutional neural networks. Multimedia Tools and Applications. 2020;79(21-22):14711–14724. doi: 10.1007/s11042-019-7344-7. [DOI] [Google Scholar]
- Shen Y, Ding LF, Yang CY, et al Mapping big brains at subcellular resolution in the era of big data in zoology. Zoological Research. 2022b;43(4):597–599. doi: 10.24272/j.issn.2095-8137.2022.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine P, Murphy MD Over 20 years of machine learning applications on dairy farms: a comprehensive mapping study. Sensors. 2022;22(1):52. doi: 10.3390/s22010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui K, Wang CW, Zhang XD, et al Small-sample learning reveals propionylation in determining global protein homeostasis. Nature Communications. 2023;14(1):2813. doi: 10.1038/s41467-023-38414-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T, Gangloff H, Pham MT. 2023. Object counting from aerial remote sensing images: application to wildlife and marine mammals. In: Proceedings of 2023 IEEE International Geoscience and Remote Sensing Symposium. Pasadena: IEEE.
- Sodhar IH, Jalbani AH, Buller AH. 2020. Tokenization of sindhi text on information retrieval tool. doi: 10.13140/RG.2.2.10555.95529.
- Staudemeyer RC, Rothstein Morris E. 2019. Understanding LSTM -- a tutorial into long short-term memory recurrent neural networks. arXiv, doi:https://doi.org/10.48550/arXiv.1909.09586.
- Sun XY, Hong PY Automatic inference of multicellular regulatory networks using informative priors. International Journal of Computational Biology and Drug Design. 2009;2(2):115–133. doi: 10.1504/IJCBDD.2009.028820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson E, Williams MJ, Schioth HB Neural cotransmission in spinal circuits governing locomotion. Trends in Neurosciences. 2018;41(8):540–550. doi: 10.1016/j.tins.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Tabak MA, Norouzzadeh MS, Wolfson DW, et al Machine learning to classify animal species in camera trap images: applications in ecology. Methods in Ecology and Evolution. 2019;10(4):585–590. doi: 10.1111/2041-210X.13120. [DOI] [Google Scholar]
- Taylor C, Guy J, Bacardit J Estimating individual-level pig growth trajectories from group-level weight time series using machine learning. Computers and Electronics in Agriculture. 2023;208:107790. doi: 10.1016/j.compag.2023.107790. [DOI] [Google Scholar]
- Thalor MA, Nagabhyrava R, Rajkumar K, et al. 2023. Deep learning insights and methods for classifying wildlife. In: Proceedings of the 3rd International Conference on Advance Computing and Innovative Technologies in Engineering. Greater Noida: IEEE, 403–407.
- Tian YT, Suzuki C, Clanuwat T, et al. 2020. Kaokore: a pre-modern japanese art facial expression dataset. arXiv, doi:https://doi.org/10.48550/arXiv.2002.08595.
- Turkoz M, Kim S, Son Y, et al Generalized support vector data description for anomaly detection. Pattern Recognition. 2020;100:107119. doi: 10.1016/j.patcog.2019.107119. [DOI] [Google Scholar]
- Turner KE, Sohel F, Harris I, et al Lambing event detection using deep learning from accelerometer data. Computers and Electronics in Agriculture. 2023;208:107787. doi: 10.1016/j.compag.2023.107787. [DOI] [Google Scholar]
- Valletta JJ, Torney C, Kings M, et al Applications of machine learning in animal behaviour studies. Animal Behaviour. 2017;124:203–220. doi: 10.1016/j.anbehav.2016.12.005. [DOI] [Google Scholar]
- Van Horn G, Mac Aodha O, Song Y, et al. 2017. The iNaturalist species classification and detection dataset. arXiv, doi: https://doi.org/10.48550/arXiv.1707.06642.
- Vaswani A, Shazeer N, Parmar N, et al. 2017. Attention is all you need. arXiv, doi:https://doi.org/10.48550/arXiv.1706.03762.
- Vélez J, Castiblanco-Camacho PJ, Tabak MA, et al. 2022. Choosing an appropriate platform and workflow for processing camera trap data using artificial intelligence. arXiv, doi:https://doi.org/10.48550/arXiv.2202.02283.
- Verdonck T, Baesens B, Óskarsdóttir M, et al. 2021. Special issue on feature engineering editorial. Machine Learning,doi: 10.1007/s10994-021-06042-2.
- Wagner N, Antoine V, Mialon MM, et al Machine learning to detect behavioural anomalies in dairy cows under subacute ruminal acidosis. Computers and Electronics in Agriculture. 2020;170:105233. doi: 10.1016/j.compag.2020.105233. [DOI] [Google Scholar]
- Wang CY, Yeh IH, Liao HYM. 2021. You only learn one representation: unified network for multiple tasks. arXiv, doi:https://doi.org/10.48550/arXiv.2105.04206.
- Wang FY, Zhang JJ, Zheng XH, et al Where does AlphaGo go: from church-turing thesis to AlphaGo thesis and beyond. IEEE/CAA Journal of Automatica Sinica. 2016;3(2):113–120. doi: 10.1109/JAS.2016.7471613. [DOI] [Google Scholar]
- Wang HC, Fu TF, Du YQ, et al Scientific discovery in the age of artificial intelligence. Nature. 2023a;620(7972):47–60. doi: 10.1038/s41586-023-06221-2. [DOI] [PubMed] [Google Scholar]
- Wang HC, Liu C, Xi NW, et al. 2023b. HuaTuo: tuning LLaMA model with chinese medical knowledge. arXiv, doi:https://doi.org/10.48550/arXiv.2304.06975.
- Wang Y, Duan XW. 2021. Research on face recognition algorithm based on deep learning. In: Proceedings of 2021 IEEE 21st International Conference on Communication Technology. Tianjin: IEEE, 1139–1142.
- Wang ZB. 2022. Self-supervised learning in computer vision: a review. In: Liu Q, Liu XD, Cheng JR, et al. Proceedings of the 12th International Conference on Computer Engineering and Networks. Singapore: Springer, 1112–1121.
- White AG, Lees B, Kao HL, et al DevStaR: high-throughput quantification of C. elegans developmental stages. IEEE Transactions on Medical Imaging. 2013;32(10):1791–1803. doi: 10.1109/TMI.2013.2265092. [DOI] [PubMed] [Google Scholar]
- Wu XP, Zhan C, Lai YK, et al. 2019. IP102: a large-scale benchmark dataset for insect pest recognition. In: Proceedings of 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition. Long Beach: IEEE, 8779–8788.
- Wulff P, Mientus L, Nowak A, et al. 2023. Correction to: utilizing a pretrained language model (BERT) to classify preservice physics teachers’ written refections. International Journal of Artificial Intelligence in Education,doi: 10.1007/s40593-023-00330-9.
- Xiang QC, Huang XN, Huang ZX, et al Yolo-pest: an insect pest object detection algorithm via CAC3 module. Sensors. 2023;23(6):3221. doi: 10.3390/s23063221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JM, Shivakumara P, Lu T, et al A new method for multi-oriented graphics-scene-3D text classification in video. Pattern Recognition. 2016;49:19–42. doi: 10.1016/j.patcog.2015.07.002. [DOI] [Google Scholar]
- Xu YY, Zhou Y, Sekula P, et al Machine learning in construction: From shallow to deep learning. Developments in the Built Environment. 2021;6:100045. doi: 10.1016/j.dibe.2021.100045. [DOI] [Google Scholar]
- Xue AT, Schrider DR, Kern AD, et al Discovery of ongoing selective sweeps within anopheles mosquito populations using deep learning. Molecular Biology and Evolution. 2021;38(3):1168–1183. doi: 10.1093/molbev/msaa259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wang WC, Wang F, et al scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nature Machine Intelligence. 2022;4(10):852–866. doi: 10.1038/s42256-022-00534-z. [DOI] [Google Scholar]
- Young T, Hazarika D, Poria S, et al Recent trends in deep learning based natural language processing [review article] IEEE Computational Intelligence Magazine. 2018;13(3):55–75. doi: 10.1109/MCI.2018.2840738. [DOI] [Google Scholar]
- Yu H, Xu YF, Zhang J, et al. 2021. AP-10K: a benchmark for animal pose estimation in the wild. arXiv, doi:https://doi.org/10.48550/arXiv.2108.12617.
- Yu S New challenge for bionics-brain-inspired computing. Zoological Research. 2016;37(5):261–262. doi: 10.13918/j.issn.2095-8137.2016.5.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T, Li H, Chang PC, et al Accurate, scalable cohort variant calls using DeepVariant and GLnexus. Bioinformatics. 2020;36(24):5582–5589. doi: 10.1093/bioinformatics/btaa1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huo YB, Yang JL, et al Automatic counting of retinal ganglion cells in the entire mouse retina based on improved YOLOv5. Zoological Research. 2022;43(5):738–749. doi: 10.24272/j.issn.2095-8137.2022.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SY, Dong LF, Li XY, et al. 2023. Instruction tuning for large language models: a survey. arXiv, doi:https://doi.org/10.48550/arXiv.2308.10792.
- Zhang YJ, Zhu CX, Ding YR, et al Subcellular stoichiogenomics reveal cell evolution and electrostatic interaction mechanisms in cytoskeleton. BMC Genomics. 2018;19(1):469. doi: 10.1186/s12864-018-4845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Li Q, Li C, et al. 2023. A comprehensive survey on pretrained foundation models: a history from BERT to ChatGPT. arXiv, doi:https://doi.org/10.48550/arXiv.2302.09419.
- Zhu JY, Park T, Isola P, et al. 2017. Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of 2017 IEEE International Conference on Computer Vision. Venice: IEEE.
- Zhu QY, Zhang RX. 2018. HENet: a highly efficient convolutional neural networks optimized for accuracy, speed and storage. arXiv, doi:https://doi.org/10.48550/arXiv.1803.02742.
- Zhu S, Wang JZ, Chen D, et al An oncopeptide regulates m6A recognition by the m6A reader IGF2BP1 and tumorigenesis. Nature Communications. 2020;11(1):1685. doi: 10.1038/s41467-020-15403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou ZX, Chen KY, Shi ZW, et al Object detection in 20 years: a survey. Proceedings of the IEEE. 2023;111(3):257–276. doi: 10.1109/JPROC.2023.3238524. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.