Abstract
Acute myeloid leukemia is characterized by uncontrolled proliferation of self-renewing myeloid progenitors. PHF6 is a chromatin-binding protein mutated in myeloid leukemias, and its loss increases mouse HSC self-renewal without malignant transformation. We report here that Phf6 knockout increases the aggressiveness of Hoxa9-driven AML over serial transplantation, and increases the frequency of leukemia initiating cells. We define the in vivo hierarchy of Hoxa9-driven AML and identify a population that we term the ‘LIC-e’ (leukemia initiating cells enriched) population. We find that Phf6 loss has context-specific transcriptional effects, skewing the LIC-e transcriptome to a more stem-like state. We demonstrate that LIC-e accumulation in Phf6 knockout AML occurs not due to effects on cell cycle or apoptosis, but due to an increase in the fraction of its progeny that retain LIC-e identity. Overall, our work indicates that Phf6 loss increases AML self-renewal through context-specific effects on leukemia stem cells.
Keywords: AML, PHF6, self-renewal, differentiation, leukemia stem cell
Introduction
Acute myeloid leukemia (AML) is an aggressive bone marrow malignancy characterized by aberrant proliferation of hematopoietic stem and progenitor cells that are blocked in differentiation. Genomic profiling has revealed that combinatorial somatic mutations in gene classes spanning epigenetic regulators, signaling proteins, transcription factors, and others contribute to leukemogenesis (1). The overall prognosis of AML remains poor, with a need for improved treatments. A deeper understanding of the cellular mechanisms through which AML mutations exert their pathogenic effects is required to advance this goal.
PHF6 (Plant homeodomain-like finger protein 6) is an X-chromosome gene mutated in a variety of myeloid and lymphoid leukemias. PHF6 protein localizes to the nucleolus and nucleoplasm, and is known to interact with chromatin, but its precise molecular function is poorly understood, with reported roles ranging from cell cycle control (2–4), DNA repair (4,5), and chromatin and transcriptional regulation (6–9). Somatic PHF6 mutations were first documented in T-cell acute lymphocytic leukemia (T-ALL), where it was found to be mutated in 38% of patients (10). Subsequently, PHF6 mutations were documented in myeloid malignancies, including 3–6% of AML, myelodysplastic syndrome (MDS), and chronic myelomonocytic leukemia (CMML), as well as in 23% of mixed-phenotype acute leukemia (MPAL) and undifferentiated leukemia (1,11–16). PHF6 mutations co-occur in MDS/AML with mutations in RUNX1, ASXL1, and U2AF1 (1,12,16), and a majority of PHF6 mutations are frameshift and nonsense mutations distributed throughout the gene body (16), presumably leading to null alleles and indicating that PHF6 acts as a leukemia suppressor.
Germline Phf6 deletion in mice leads to perinatal lethality, likely due to non-hematopoietic phenotypes, while mice with hematopoietic Phf6 deletion are viable and fertile (17,18). Conditional hematopoietic knockouts using multiple Cre systems have consistently shown minimal alterations to homeostatic hematopoiesis, but striking increases in HSC self-renewal on transplantation, with the ability to engraft recipient marrow beyond five serial transplants without evidence of exhaustion, malignant transformation, or lineage skewing (17–19). Phf6 knockout HSCs from aged mice show transcriptional profiles similar to young HSCs, and deletion of Phf6 from older mice shows a shift towards a younger HSC transcriptome, suggesting that Phf6 loss may counteract HSC aging (5). Combination of Phf6 loss with overexpression of activating mutants of Notch1 (18) or Jak3 (20), or overexpression of wild-type Tlx3 (17) has been shown to cause T-ALL acceleration, while transgenic crosses of Phf6 deletion with Idh2 mutation produce mixed myeloid-lymphoid leukemias (21). Collectively, the role of PHF6 appears to be the repression of self-renewal, both in normal HSCs as well as in T-ALL (18). However, there are also reports of PHF6 being required for the growth of B-ALL (22), and more recently, for the growth of AML driven by BCR-ABL, AML1-ETO, or MLL-AF9 fusions (23). The latter publication reporting the counterintuitive finding that Phf6 loss reduces AML growth and stemness contradicts the model of PHF6 as a leukemia suppressor; however, the publication’s use of fusion protein drivers that do not co-occur with human PHF6 mutations may indicate that the chosen AML models recapitulated narrow disease subsets with particular fusion-induced phenotypes, potentially not reflective of broader AML biology. The precise role of Phf6 loss in AML therefore remains unclear.
In this study, we avoid potential confounding effects of specialized fusion proteins by using Hoxa9 retroviral transduction (without Meis1) as a model of AML (24) that is broadly relevant, given that >70% of AMLs overexpress HOXA9 (25). We examine the role of hematopoietic Phf6 deletion on AML progression and stemness, and show that Phf6 loss accelerates AML progression over serial transplantation, accompanied by an accumulation of leukemia initiating cells (LICs). We identify that LICs in the Hoxa9 transduction model are concentrated within a small Kit+ Ly6C-subpopulation that we term ‘LIC-e’ (LIC-enriched), and we show that, contrary to prior reports, Phf6 loss has no effect on cell cycle or apoptosis, but instead purely increases the fraction of LIC-e progeny that retain persistent LIC-e identity. We further show that Phf6 loss leads LIC-e cells to gain a more stem-like transcriptome, along with reduced accessibility of genomic regions bound by the transcription factors AP-1 family, GATA2, and SPI1, among others. Collectively, our data resolves a controversy in the literature by demonstrating that PHF6 suppresses AML stem cell self-renewal (and not cell cycle progression or apoptosis) in a broadly relevant AML model system, and demonstrates how the loss of a specific repressor of HSC self-renewal drives leukemia stemness.
RESULTS
PHF6 loss increases leukemia initiating cell frequency in Hoxa9-driven AML
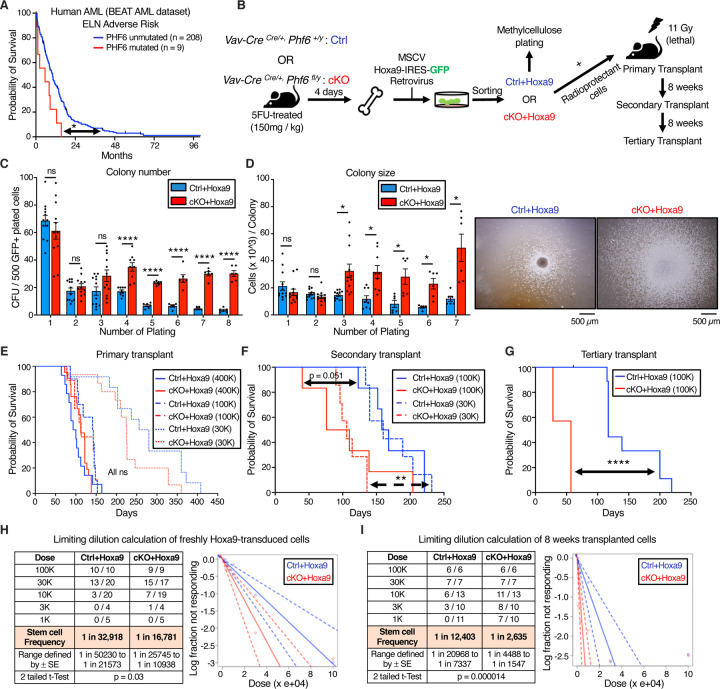
To determine the prognostic significance of PHF6 mutations in human AML, we used publicly available mutational and survival data from the BEAT AML dataset (26). Of 557 AML patients, 16 (2.87%) had PHF6 mutations. Almost all PHF6 mutations occurred in patients classified into European Leukemia Net (ELN) intermediate and adverse risk groups. PHF6 mutations were associated with reduced overall survival in adverse risk patients (median survival 5.56 months in PHF6 mutated versus 10.16 months in unmutated; p = 0.016) (Fig 1A) while survival differences did not reach statistical significance in intermediate-risk patients, potentially due to relatively small case numbers (median survival 9.21 months in PHF6 mutated versus 12.73 months in unmutated; p = 0.26) (Fig S1A). The dataset was not powered for further multivariable or co-mutation analyses, but overall indicated that PHF6 mutation is associated with worse survival.
Figure 1. Phf6 loss increases leukemia initiating cell frequency in Hoxa9-driven AML.
A, Kaplan-Meier survival curve for PHF6 mutated and unmutated adverse risk adult AML patients (ELN classification) from the BEAT AML dataset.
B, Experimental design for Hoxa9 retroviral transduction of Ctrl and cKO marrow, followed by colony forming unit assay (CFU), AML induction in mice, and serial transplantation of transformed leukemic cells.
C, Bar graph showing number of colony forming units (CFUs) obtained from 8 rounds of serial methylcellulose replating of 500 cells/plate of Ctrl+Hoxa9 and cKO+Hoxa9 transformed mouse bone marrow. (n = 6–12 biological replicates)
D, Left Bar graph showing average number of cells per colony (colony size) obtained after 7 rounds of serial methylcellulose replating of 500 cells/plate of Ctrl+Hoxa9 and cKO+Hoxa9 transformed mouse bone marrow. (n = 6–12 biological replicates) Right Representative photographs of colonies at the 3rd plating. Scale bar represents 500 μm.
E, Kaplan-Meier survival curves of Ctrl+Hoxa9 and cKO+Hoxa9 primary transplant recipients receiving 400K, 100K or 30K GFP+ cells. (n = 7–21 mice per cohort)
F, Kaplan-Meier survival curve of Ctrl+Hoxa9 and cKO+Hoxa9 secondary transplant recipients, receiving 100K or 30K GFP+ cells harvested from bone marrow of primary recipients 8 weeks after transplantation. (n = 7–10 mice per cohort)
G, Kaplan-Meier survival curve of Ctrl+Hoxa9 and cKO+Hoxa9 tertiary transplant recipients, receiving 100K GFP+ cells harvested from bone marrow of secondary recipients 8 weeks after transplantation. (n = 7–11 mice per cohort)
H and I, Limiting dilution analysis for LIC calculation for (H) freshly Hoxa9-transduced cells, (I) 8 weeks primary transplanted leukemic cells.
All bar graphs show mean ± SEM, and statistical significance was calculated using the Student t-test. For all survival curves, statistical significance was calculated using the Log-rank (Mantel-Cox) test. *p < .05, **p < .01, ***p < .001; ****p < .0001, ns = not significant.
To assess the role of PHF6 loss in mouse AML, we generated conditional hematopoietic Phf6 knockout Vav-CreCre/+Phf6fl/y (cKO) mice and compared them to their Vav-CreCre/+Phf6+/y (Ctrl) littermates. cKO marrow was confirmed to show loss of Phf6 mRNA (Fig S1B). Published studies of hematopoietic Phf6 loss have reported no evidence of leukemic transformation either in homeostatic mice or over serial bone marrow transplantations (17–19,27). Consistent with these reports, we found no blood count abnormalities in cKO mice compared to Ctrl mice up to 9 months of age (Fig S1C–I), further indicating that Phf6 loss alone is likely insufficient to initiate overt leukemia.
We next induced AML using the Hoxa9 retroviral transduction model (Hoxa9 alone, without Meis1). We picked this model due to its ability to produce AML with a relatively longer latency (lethality in ~3–6 months) (24), allowing us to test a potential role for Phf6 loss in accelerating AML kinetics. Given that 70% of human AMLs show high HOXA9 levels (25), the Hoxa9 overexpression mouse model is broadly relevant to human leukemia biology, and avoids caveats associated with specific fusion proteins. We transduced whole bone marrow from 5-FU-treated Ctrl and cKO mice with MSCV Hoxa9-IRES-GFP retrovirus, and first investigated the effect of Phf6 loss on the ability of Hoxa9-transformed cells to form colonies in methylcellulose (Fig 1B). We observed that Ctrl+Hoxa9 cells were nearly exhausted after 4 platings, whereas cKO+Hoxa9 cells demonstrated persistent colony-forming ability up to 8 platings (Fig 1C), with larger colonies (Fig 1D). Thus, Phf6 loss gives a replating advantage to Hoxa9-transformed marrow and delays its in vitro exhaustion.
To test the role of Phf6 loss in the development of AML in vivo, we transplanted Hoxa9-transduced marrow into lethally irradiated syngeneic recipients (Fig 1B). While previous publications have reported AML development with Hoxa9-only transduction (24), detailed histological characterization of this particular model is lacking in the literature. We therefore first confirmed that Ctrl+Hoxa9 marrow produced lethality in recipient mice in ~3–5 months after transplantation (Fig 1E) due to AML characterized by accumulation of >20% blasts in the marrow (Fig S1J), peripheral leukocytosis with circulating blasts (Fig S1K), splenomegaly with effacement of splenic architecture (Fig S1L), and infiltration of leukemic cells in the liver (Fig S1M). Survival was similar between Ctrl+Hoxa9 and cKO+Hoxa9 groups in primary recipients transplanted with multiple doses (400K, 100K, or 30K cells) (Fig 1E), with similar degrees of leukemic infiltrate (Fig S2A–F) and splenomegaly at morbidity (Fig S2G). However, serial transplantation (secondary and tertiary) of bone marrow showed accelerated lethality in cKO+Hoxa9 compared to Ctrl+Hoxa9 (Fig 1F–G, S2H), with median survivals of 106 days versus 160 days in secondary recipients (p = 0.001) and 57 days versus 117 days in tertiary (p < 0.0001). Thus, loss of Phf6 accelerates Hoxa9-driven mouse AML on serial transplantation.
We next sought to determine the effect of Phf6 loss on the frequency of leukemia initiating cells (LIC), the sub-population of transformed marrow capable of initiating leukemia. We performed limiting dilution transplantation assays (LD) on freshly transduced marrow (GFP+ cells sorted 2 days after initial retroviral transduction) as well as on marrow from transplant recipients (GFP+ bone marrow cells sorted from recipients 8 weeks after primary transplantation). We picked the 8-week time point based on the initiation of lethality in this model at ~12 weeks (Fig 1E), allowing us to harvest cells from all transplanted mice at a standard time point prior to onset of lethality. LD of freshly transduced marrow showed that, at baseline, cKO+Hoxa9 cells had a 2-fold higher frequency of cells capable of leukemic transformation (1 in 16,781 versus 1 in 32,918, p = 0.03) (Fig 1H). LD on marrow extracted 8 weeks post-transplantation showed an approximately 5-fold greater frequency of leukemia initiating cells (LICs) in cKO+Hoxa9 marrow, with LIC frequency of 1 in 2,635 versus 1 in 12,403 for Ctrl+Hoxa9 marrow (p = 0.000014) (Fig 1I). Thus, Phf6 loss increases LIC frequency in Hoxa9-driven AML, with a progressive increase in LIC frequency during in vivo AML evolution.
Phf6 loss increases leukemic disease burden
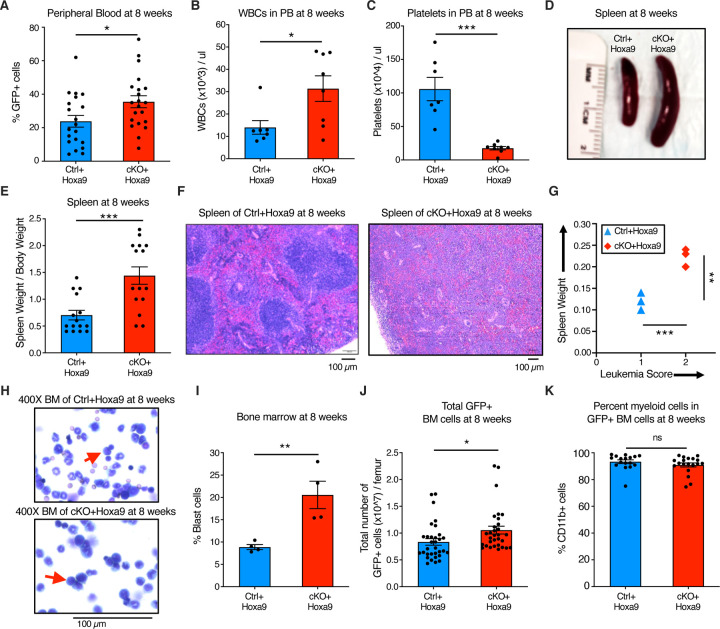
To characterize the effect of Phf6 loss further, we analyzed peripheral blood, splenic architecture, and bone marrow leukemic cell burden of primary recipient mice at 8 weeks after transplantation. Mice transplanted with cKO+Hoxa9 cells showed a higher frequency of GFP+ cells in peripheral blood at 8 weeks than mice receiving Ctrl+Hoxa9 cells (Fig 2A). The cKO+Hoxa9 group also had greater leukocytosis (Fig 2B) and more severe thrombocytopenia (Fig 2C). Mice in both groups displayed comparable levels of anemia (Fig S3A–B). The cKO+Hoxa9 group had increased spleen size and weight compared to the Ctrl+Hoxa9 group (Fig 2D–E), and histopathological analysis showed greater effacement of splenic architecture (Fig 2F). Splenic infiltration was quantified using a previously described leukemia infiltration score (28), and was found to be greater in cKO+Hoxa9 mice compared to Ctrl+Hoxa9 (Fig 2G). Giemsa stained cytospin preparations showed higher blast percentages in cKO+Hoxa9 at the 8-week timepoint (Fig 2H–I), and flow cytometry showed higher absolute and percent GFP+ cells (Fig 2J, S3C). As expected, virtually all GFP+ cells were myeloid in both groups (Fig 2K, S3D). Collectively, though mice from both groups succumbed at similar times after primary transplant (Fig 1F), analyses at matched time points before the onset of mortality revealed greater disease burden in cKO+Hoxa9 mice compared to Ctrl+Hoxa9.
Figure 2. Phf6 loss increases leukemic disease burden.
A, B, and C, Bar graphs showing peripheral blood analysis at 8 weeks after transplantation of Ctrl+Hoxa9 and cKO+Hoxa9 cells. (A) Percentage of GFP+ cells in peripheral blood. Counts of (B) WBCs and (C) platelets in peripheral blood. Normal range for WBCs: 2000–10,000/µl. Normal range for platelets: 900–1,600 x 103/µl (56).
D, Representative photograph of spleens at 8 weeks after transplantation. Ruler depicts length in centimeters.
E, Bar graph showing spleen weight (expressed as percentage of total body weight) of primary recipients at 8 weeks after transplantation of Ctrl+Hoxa9 and cKO+Hoxa9 cells.
F, Representative image of H&E staining of spleen from Ctrl+Hoxa9 and cKO+Hoxa9 at 8 weeks after transplantation. Scale bar is 100µm at 10X.
G, Spleen weight (y-axis) and leukemia score (x-axis) from Ctrl+Hoxa9 and cKO+Hoxa9 primary recipients at 8 weeks. X-axis represents a previously published leukemia infiltration score (28) calculated based on splenic architecture. Intact white and red pulp was scored as 0, extramedullary hematopoiesis evident by aberrant cells in disturbed white pulp was scored as 1, while infiltration with leukemic blasts with high mitotic activity was scored as 2.
H, Representative image of Wright-Giemsa staining of cytospin of Ctrl+Hoxa9 and cKO+Hoxa9 bone marrow cells at 8 weeks after transplantation. Scale bar is 100µM at 400X. Red arrows indicate representative blast cells.
I, Bar graph of percentage blast cells of total nucleated cells in bone marrow at 8 weeks after transplant.
J and K, Bar graphs showing (J) absolute number of GFP+ cells in marrow and (K) percentage of CD11b+ myeloid cells among all GFP+ cells at 8 weeks after transplantation.
All bar graphs show mean ± SEM, and statistical significance was calculated using the Student t-test. For all survival curves, statistical significance was calculated using the Log-rank (Mantel-Cox) test. *p < .05, **p < .01, ***p < .001; ****p < .0001, ns = not significant.
Phf6 loss increases the frequency of self-renewing, transplantable LICs
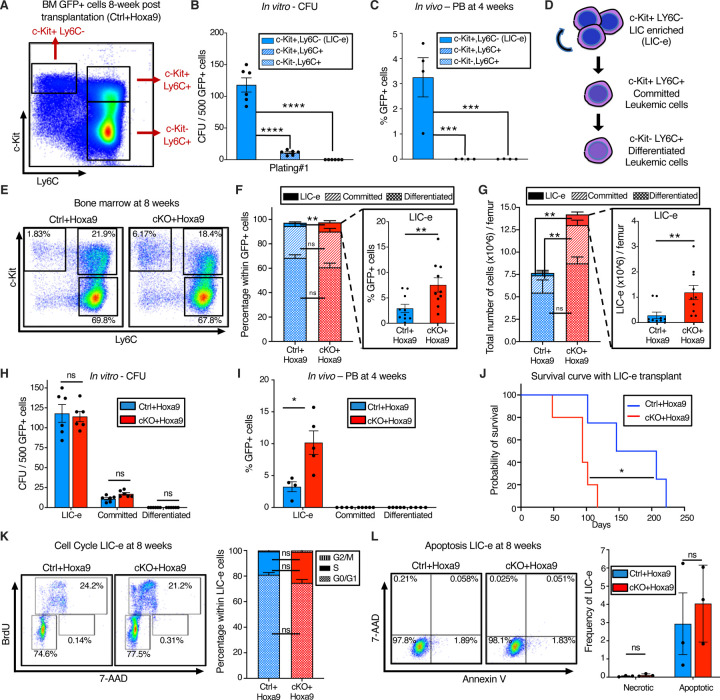
To characterize the immunophenotype of AML subpopulations (including LICs), we further analyzed the bone marrow of Ctrl+Hoxa9 recipients at 8 weeks after transplantation. As expected, GFP+ cells did not express B or T cell markers (Fig S4A). Immature AML cells are known to have high c-Kit expression (29), and leukemic stem cells (LSCs) in the MLL-AF9 retroviral mouse model aberrantly express mature myeloid lineage antigens such as Ly6C and CD11b (30). To our knowledge, corresponding AML subpopulations have not been defined in Hoxa9-only-driven AML. To identify the subpopulation containing LICs, and to characterize the differentiation hierarchy of Hoxa9-driven AML, we tested multiple gating strategies with a panel of markers, eventually settling on a simplified strategy using c-Kit and Ly6C expression to divide GFP+ bone marrow cells into three populations: i) cKit+ Ly6C-, ii) c-Kit+ Ly6C+, and iii) c-Kit-Ly6C+ (Fig 3A, S4B). We established the hierarchy of these populations based on differentiation trajectory, immunophenotype, colony-forming ability, and in vivo repopulation capacity. The population at the top of the hierarchy was the cKit+ Ly6C-population, an immature population with expression of cKit, CD34, and dim CD11b, with no expression of Ly6C, Ly6G, or Sca-1, and mixed expression of CD16/32 (Fig S4C). On sorting and culture, this population was capable of giving rise to more differentiated Ly6C+ cells within 2 days (Fig S4D), could produce colonies on methylcellulose plating (Fig 3B), and could engraft into recipient mice (Fig 3C). Based on this subpopulation’s ability to engraft, but cognizant that not all cells within it are LICs, we termed it the ‘LIC enriched’ (LIC-e) population (Fig 3D). The second population was the c-Kit+ Ly6C+ population, also expressing CD11b, CD34, and CD16/32, but not Sca-1 (Fig. S4C). On culture, this population could only give rise to Ly6C+ cells, but not to any Ly6C-cells (Fig. S4D), indicating that it is irreversibly committed to differentiation. This population could produce a small number of colonies in methylcellulose (Fig 3B), but could not engraft mice (Fig 3C). We termed it the ‘committed’ leukemic cell population (Fig 3D). The third population was the c-Kit-Ly6C+ population, which expressed Ly6C, CD11b, CD34, and CD16/32, but not Sca-1, and had mixed expression of Ly6G (Fig S4C). It could not produce any colonies (Fig 3B) nor engraft (Fig 3C). In vitro, this population only gave rise to Ly6C+ cells (Fig. S4D). We termed it the ‘differentiated’ leukemic cell population (Fig 3D). We thus established the hierarchical organization of Hoxa9-driven AML, and used it to evaluate the effects of Phf6 loss.
Figure 3. Phf6 loss increases the frequency of self-renewing, transplantable LICs.
A, Representative flow cytometry plot of bone marrow GFP+ cells at 8 weeks after transplantation with Ctrl+Hoxa9 cells, compartmentalized into three subpopulations i) LIC-e, ii) Committed, and iii) Differentiated leukemic cells using c-Kit and Ly6C expression. Note: The same flow cytometry plot has been shown in Fig S4 with detailed immunophenotypic markers.
B, Bar graph showing number of colony forming units (CFUs) obtained on methylcellulose plating of 500 cells of sorted subpopulations of Ctrl+Hoxa9 transplanted marrow. (n = 6 biological replicates)
C, Bar graph showing frequencies of GFP+ cells in peripheral blood of secondary recipient mice at 4 weeks after transplantation of sorted subpopulations from Ctrl+Hoxa9 primary recipient marrow. (n = 4–5 biological replicates)
D, Schematic of hierarchical organization of leukemic cells (LIC-e, committed, and differentiated leukemic cells) within AML produced by retroviral Hoxa9 transduction.
E, Representative flow cytometry plots depicting subpopulations of Ctrl+Hoxa9 and cKO+Hoxa9 leukemia marrow at 8 weeks after primary transplant.
F and G, Stacked bar graphs showing (F) frequencies, and (G) absolute number per femur of LIC-e, committed, and differentiated leukemic populations from Ctrl+Hoxa9 and cKO+Hoxa9 marrow at 8 weeks after transplantation. Insets show frequencies and absolute numbers of the LIC-e subpopulation. (n = 10–11)
H, Bar graph showing number of CFUs obtained on methylcellulose plating of 500 cells of sorted LIC-e, committed, and differentiated leukemic populations from Ctrl+Hoxa9 and cKO+Hoxa9 primary recipient bone marrow at 8 weeks after transplantation. (n = 6 biological replicates)
I, Bar graph showing frequencies of GFP+ cells in the peripheral blood of secondary recipient mice at 4 weeks after transplantation with sorted LIC-e, committed, and differentiated leukemic cell subpopulations from Ctrl+Hoxa9 and cKO+Hoxa9 primary recipient bone marrow at 8 weeks after transplantation. (n = 5 biological replicates). Note: The Ctrl+Hoxa9 samples in Figs 3H and 3I are the same as those depicted in 3B and 3C - the current figures are comparing Ctrl and cKO.
J, Kaplan-Meier curve of secondary transplant recipients receiving 50K sorted LIC-e cells from Ctrl+Hoxa9 and cKO+Hoxa9 primary mouse bone marrow at 8 weeks after transplantation. (n = 4–5 biological replicates)
K, Left Representative flow cytometry plots for cell cycle analysis of LIC-e cells, with BrdU (marking cells in S phase) and 7-AAD (marking DNA) 2 hours after BrdU injection into live mice at 8 weeks after transplantation. Right Stacked bar graph indicates frequencies of in vivo LIC-e cells in G0/G1, S, and G2/M phases. (n = 13 biological replicates)
L, Left Representative flow cytometry plots of apoptotic (Annexin V+, 7AAD-), and necrotic (7AAD+) LIC-e cells. Right Bar graph shows frequencies of apoptotic and necrotic LIC-e cells from Ctrl+Hoxa9 and cKO+Hoxa9 primary recipient bone marrow at 8 weeks after transplantation.
All bar graphs show mean ± SEM, and statistical significance was calculated using the Student t-test. For all survival curves, statistical significance was calculated using the Log-rank (Mantel-Cox) test. *p < .05, **p < .01, ***p < .001; ****p < .0001, ns = non significant.
The LIC-e population, though comprising a small minority of GFP+ cells, was expanded in 8-week cKO+Hoxa9 bone marrow compared to Ctrl+Hoxa9, while the relative fractions of committed and differentiated leukemic cells were similar (Fig 3E–F). The difference in the LIC-e population was even more pronounced when absolute cell numbers were considered, showing a 5-fold increase in cKO+Hoxa9 (Fig. 3G). To determine functional differences between Ctrl+Hoxa9 and cKO+Hoxa9 subpopulations, we sorted equal numbers of cells from each subpopulation at 8 weeks after transplant, and performed methylcellulose culture and secondary transplantation into irradiated recipients. Committed and differentiated leukemic cells of either group formed few to no colonies, while LIC-e cells showed comparable colony-forming ability (Fig 3H). cKO+Hoxa9 LIC-e cells showed greater engraftment in secondary recipients at 4 weeks (Fig 3I) and led to more rapid lethality than Ctrl+Hoxa9 LIC-e cells (94 days versus 177 days, p = 0.02) (Fig 3J). We did not observe any difference in cell cycle distribution or apoptosis of LIC-e cells (Fig 3K–L). Thus, Phf6 loss led to an expanded and more transplantable LIC-enriched AML subpopulation whose enhanced leukemic potential was not explained by differences in cell cycle or apoptosis.
Phf6 loss promotes a stemness gene network
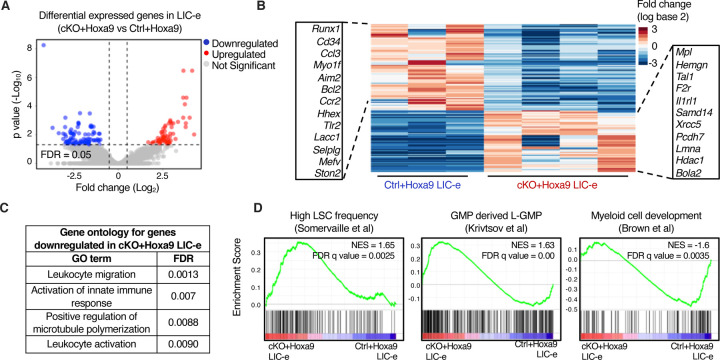
We next sought to determine the transcriptional consequences of Phf6 loss by performing bulk RNA-Seq on LIC-e and committed leukemic cells from bone marrow of transplanted recipients at 8 weeks. Committed leukemic cells showed virtually no change in gene expression as a consequence of Phf6 loss (Fig S5A–B, Table S2), while the LIC-e population showed a focused number of differentially expressed genes, with 91 downregulated and 65 upregulated genes in cKO+Hoxa9 compared to Ctrl+Hoxa9 (Fig 4A–B, Table S2). Genes downregulated in cKO+Hoxa9 LIC-e cells included known regulators of self-renewal and differentiation like Runx1 and Cd34 (31,32) and leukocyte maturation genes like Selplg (33) (Fig 4B–C). Genes upregulated in cKO+Hoxa9 LIC-e cells included the thrombopoietin receptor Mpl, known to contribute to AML pathogenesis (34,35), F2r, a G-protein coupled receptor that promotes stemness in MLL-AF9 AML (36), Hemgn, which promotes self-renewal and myeloid progenitor expansion (37), and Samd14, which promotes c-Kit signaling and hematopoietic progenitor expansion (38) (Fig 4B). Gene set enrichment analysis (GSEA) (39) showed that the cKO+Hoxa9 LIC-e transcriptome showed positive enrichment for gene sets related to high LSC potential in MLL-AF9-driven mouse AML (40) and leukemic GMPs (L-GMPs) (41), and negative enrichment for gene sets related to myeloid differentiation (42), low LSC potential (40), mature neutrophils, and mature monocytes (43) (Fig 4D, S5C). Thus, Phf6 loss has context-specific transcriptional effects, skewing the transcriptome of LIC-e cells to a more stem-like and less differentiated state, but showing no effect in more differentiated leukemic cells.
Figure 4. Phf6 loss promotes a stemness gene network.
A, Volcano plot showing differentially expressed genes in LIC-e cells from cKO+Hoxa9 compared to Ctrl+Hoxa9 bone marrow at 8 weeks after transplantation. (n = 3–4 biological replicates)
B, Heatmap of differential expression between Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells. Insets show selected downregulated (left) and upregulated (right) genes in cKO+Hoxa9 LIC-e compared with Ctrl+Hoxa9 LIC-e.
C, Top Gene Ontology terms enriched in genes downregulated in cKO+Hoxa9 LIC-e compared with Ctrl+Hoxa9 LIC-e.
D, Gene set enrichment analysis (GSEA) plots of the cKO+Hoxa9 LIC-e transcriptome compared to Ctrl+Hoxa9. Plots show positive enrichment of gene sets related to high LSC frequency (left) and leukemic GMPs (middle), and negative enrichment of a gene set related to myeloid development (right). Normalized Enrichment scores (NES) and FDR q values are shown.
Phf6 loss prevents exhaustion of LIC-e cells by maintaining their self-renewal potential
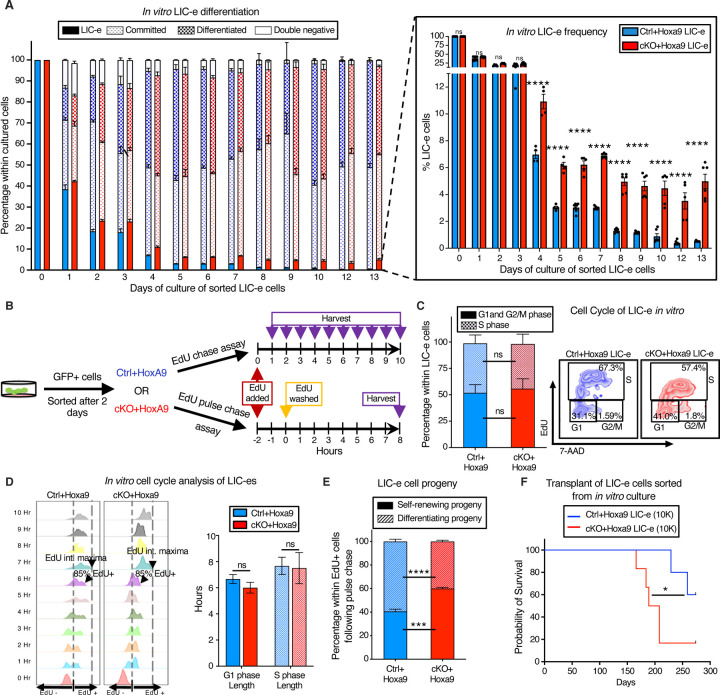
To determine the kinetics of Phf6 loss on the behavior of LIC-e cells, we cultured Hoxa9-transduced bone marrow in cytokine-supplemented media. The growth rate of bulk culture was similar for Ctrl+Hoxa9 and cKO+Hoxa9 marrow (Fig S6A). When sorted LIC-e cells were cultured, most cells lost LIC-e identity within days, and the culture was largely replaced by committed and differentiated cells by a week (Fig 5A). However, though both groups produced similar fractions of committed (c-Kit+ Ly6C+) and differentiated cells (c-Kit-Ly6C+), the Ctrl+Hoxa9 culture almost completely depleted its LIC-e population (<1%), while the cKO+Hoxa9 culture maintained this population, plateauing at 5–6% of the total culture after 5 days (Fig 5A). Thus, even though Phf6 loss does not impair the ability of cultured cells to proliferate and produce differentiated cells, it prevents exhaustion of the LIC-e population, recapitulating the in vivo LIC-e accumulation phenotype shown earlier (Fig 3E–G).
Figure 5: Phf6 loss prevents exhaustion of LIC-e cells by maintaining their self-renewal potential.
A, Bar graph showing frequencies of subpopulations resulting from in vitro culture of LIC-e cells sorted 4 days after Hoxa9 transduction of Ctrl and cKO bone marrow. Inset bar graph depicts only LIC-e frequencies in the same culture. (n = 13 biological replicates)
B, Experimental design for study of in vitro cell cycle analysis (top) and self-renewal (bottom) of LIC-e cells using Edu chase and pulse-chase assay respectively.
C, Left, Bar graph showing frequencies of G0/G1, S, and G2/M phases in Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells in culture 2 hours after addition of EdU. (n = 4–5 biological replicates) Right, Representative flow cytometry plots of same, with EdU marking cells in S phase and 7-AAD staining DNA.
D, Left, Representative flow cytometry plots showing kinetics of uptake of EdU by Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells over 10 hours of culture, performed for calculation of cell cycle length. Time to whole population (>85%) EdU uptake represents G1 phase length, and time to EdU intensity maxima represents S phase length. Right, Bar graph represents the length of cell cycle phases of LIC-e cells in culture. (n = 4–5 biological replicates)
E, Stacked bar graph showing the percentage of self-renewing and differentiating progeny produced by Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells in culture. (n = 4–5 biological replicates)
F, Kaplan-Meier curve of primary transplant recipients receiving 10K sorted Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells. (n = 5–6 biological replicates)
All bar graphs show mean ± SEM, and statistical significance was calculated using the Student t-test. For all survival curves, statistical significance was calculated using the Log-rank (Mantel-Cox) test. *p < .05, **p < .01, ***p < .001; ****p < .0001, ns = non significant.
We had already determined that in vivo LIC-e cells showed no difference in cell cycle distribution (Fig 3K), but to confirm that LIC-e accumulation wasn’t due to subtle cycling differences, we performed a 10-hour cell cycle analysis in culture by adding EdU to sorted Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells and harvesting them at serial time points (Fig 5B). We observed that the distribution of Ctrl+Hoxa9 or cKO+Hoxa9 LIC-e cells in G1 and S phases showed no difference at the start of culture, and showed no difference in the kinetics or magnitude of EdU incorporation (Fig 5C). Based on previously published rationale (44) that cells that had just begun G1 would be the last to enter S phase and incorporate EdU, and that the maximum EdU fluorescence intensity reached by the population as a whole would represent cells that had incorporated EdU into their entire genomes, we defined the time required for 85% of each sample to become EdU+ as the ‘G1 phase length’, and the time required for each sample to reach a maximal EdU fluorescent intensity as the ‘S phase length’. We did not observe any difference in G1 and S phase lengths between Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells (Fig 5D), further demonstrating that the phenotype of LIC-e accumulation (Fig 5A) is not explained by increased cycling of that subpopulation. We therefore hypothesized that cKO+Hoxa9 LIC-e cells, while capable of producing differentiated progeny, may have a greater tendency to produce progeny with persistent LIC-e identity. To test this hypothesis, we performed an EdU pulse-chase experiment by incubating sorted LIC-e cells with EdU for 2 hours (pulse), followed by washing off the EdU and further culturing cells for 8 more hours (chase, for the length of S phase) (Fig 5B). The goal was to allow all cells that were in S phase during the initial EdU pulse to complete mitosis, so that all EdU+ cells at the end of the 8 hour EdU-free chase would be daughter cells/progeny of the original EdU-uptaking cells. We determined the percentage of self-renewing progeny by calculating the percentage of total progeny (total EdU+ cells) that had LIC-e markers (Ly6C-EdU+), while the rest (Ly6C+ EdU+) were differentiating progeny. We observed that while 40.6% of the progeny of Ctrl+Hoxa9 LIC-e cells were also LIC-e cells, this fraction was increased to 60.0% in cKO+Hoxa9 (Fig 5E). To confirm that in vitro LIC-e cells are functionally equivalent to their in vivo counterparts, we performed primary transplantation with 10K LIC-e cells sorted after 2 days of culture. Recipients of cKO+Hoxa9 LIC-e cells succumbed faster than Ctrl+Hoxa9 LIC-e (p = 0.047) (Fig 5F). Collectively, Phf6 loss prevents the exhaustion of LIC-e cells by increasing the fraction of their progeny that retain persistent LIC-e identity.
Effects of Phf6 loss on chromatin accessibility in LIC-e cells
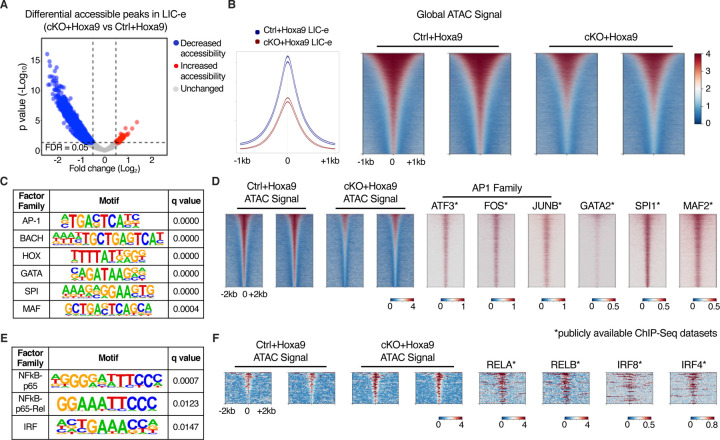
To profile the effects of Phf6 loss on the accessibility landscape of LIC-e cells, we performed ATAC-Seq on sorted LIC-e cells from freshly transduced cKO+Hoxa9 and Ctrl+Hoxa9 marrow. We observed that cKO+Hoxa9 LIC-e cells showed a surprising global reduction in chromatin accessibility compared to Ctrl+Hoxa9, with only a few regions showing increased accessibility (Fig 6A–B). Regions that lost accessibility in cKO+Hoxa9 LIC-e cells showed enrichment for AP-1, HOX, SPI, and GATA family motifs, among others (Fig 6C). Public ChIP-Seq tracks from leukemic or myeloid cells showed co-occupancy of these factors at ATAC peaks with reduced accessibility (Fig 6D, Table S3). Promoters of multiple genes like Runx1, Selplg, and Aim2, which are downregulated in cKO+Hoxa9 LIC-e cells (Fig 4B), showed occupancy by these factors and showed reduced chromatin accessibility in cKO+Hoxa9 LIC-e cells (Fig S6B). Conversely, the small number of regions that gained accessibility in cKO+Hoxa9 LIC-e cells showed enrichment for NF-kB, and IRF family motifs (Fig 6E). Public ChIP-Seq tracks showed co-occupancy of NF-kB (RELA, RELB) and IRF8, IRF4 at regions of increased accessibility (Fig 6F, Table S3). Overall, Phf6 loss, likely via a combination of direct and indirect effects, led to altered accessibility at sites bound by key hematopoietic transcription factors.
Figure 6: Effects of Phf6 loss on chromatin accessibility in LIC-e cells.
A, Volcano plot showing differentially accessible regions in LIC-e cells from cKO+Hoxa9 compared Ctrl+Hoxa9. (n = 3 biological replicates)
B, Representative signal profile (left) and metagene plots (right) showing genome-wide intensity of ATAC signal in Ctrl+Hoxa9 and cKO+Hoxa9 LIC-e cells.
C, HOMER analysis for regions of decreased chromatin accessibility in cKO+Hoxa9 LIC-e cells, showing enrichment of AP-1, HOX, GATA, SPI1, and MAF motifs.
D, Representative metagene plots at regions of decreased chromatin accessibility in cKO+Hoxa9 LIC-e, show ChIP-Seq signal for select proteins whose motifs are seen to be enriched through HOMER in (C).
E, HOMER analysis for regions of increased chromatin accessibility in cKO+Hoxa9 LIC-e cells showing enrichment of NF-kB and IRF motifs.
F, Representative metagene plots at regions of increased chromatin accessibility in cKO+Hoxa9 LIC-e, show ChIP-Seq signal for select proteins whose motifs are seen to be enriched through HOMER in (E).
Publicly available ChIP-Seq datasets in leukemia or myeloid cells were used in metagene heatmaps (Table S3). All plots were centered around ATAC-Seq peaks. SeqPlots was used to draw all metagene plots.
DISCUSSION
A majority of PHF6 mutations in myeloid and lymphoid leukemias are frameshift and nonsense mutations scattered throughout the gene body, presumed to cause loss of gene function and indicating that PHF6 acts as a leukemia suppressor. In concordance with this genomic observation, multiple mouse studies have reported increased HSC self-renewal with Phf6 knockout (17–19,27), and have reported enhanced T-ALL progression when Phf6 knockout is combined with activating mutations in Notch1 or Jak3, or overexpression of Tlx3 (17,18,20). In contrast, a recent publication (Hou et al, (23)) reported that AML induced by BCR-ABL, AML1-ETO, and MLL-AF9 fusions is impaired by Phf6 loss, reaching the counterintuitive conclusion that Phf6 is required for myeloid leukemogenesis. Our paper addresses this controversy, and we demonstrate that Phf6 loss accelerates AML in a broadly-relevant model, and that it does so by increasing the frequency and persistence of leukemic stem cells, a finding that is harmonious with the known role of Phf6 as a repressor of HSC self-renewal.
We first interrogated the BEAT AML dataset, which showed that PHF6 mutations are associated with worsened survival in human AML. Then, by using the mouse Hoxa9 transduction AML model, we demonstrated that Phf6 knockout led to increased colony replating, increased disease burden in vivo, progressively worsened survival on serial transplantation, and increased LIC frequency. To our knowledge, AML hierarchy and LIC immunophenotypes had not been previously defined in the Hoxa9 model, and we determined that a simplified gating scheme could identify an LIC-enriched population that we termed ‘LIC-e’, which was the only population capable of robust colony plating and engraftment. We found that Phf6 loss led to an expansion of LIC-e cells, and that this expansion could be recapitulated in vitro, with Phf6 knockout LIC-e cells showing the ability to persist in culture while wild-type LIC-e cells were depleted. Contrary to reports that Phf6 loss alters cell cycle or apoptosis, we found evidence of neither, and instead found that Phf6 loss leads LIC-e cells to produce more progeny with persistent LIC-e identity, thus indicating that Phf6 specifically controls the balance between LIC self-renewal and differentiation. RNA-Seq and ATAC-Seq showed that Phf6 loss skews the LIC-e transcriptome towards a more stem-like state, and leads to reduced accessibility at loci bound by AP-1, GATA2, and SPI1.
There could be multiple reasons for why our results stand in contrast to those of Hou et al. First, as stated earlier, we specifically picked Hoxa9 transduction as a driver that broadly recapitulates AML biology, while Hou et al used fusions that are not known to co-occur with PHF6 in patients. It is therefore unclear if their models reflected the in vivo context in which PHF6 mutations gain a clonal advantage in humans. Second, though our group and theirs both used Vav-Cre to knock out Phf6, Hou et al used Flox-only mice as negative controls, while we used Vav-Cre-only mice. Cre toxicity has been reported with other Cre models (45,46), and may be a contributory factor to these discordant phenotypes; our approach eliminates Cre toxicity as a potential confounder. Third, it is highly likely that Phf6 loss produces different downstream transcriptional effects in different epigenetic contexts; we ourselves observed that Phf6 loss produces stemness-skewing changes only in LIC-e cells but not in committed cells, and it is therefore conceivable that Phf6 loss produces divergent effects in certain AML contexts compared to others. Careful work in human genomics and mouse modeling will be required to define the co-mutational contexts in which Phf6 loss is cooperative, neutral, or actively deleterious to AML stemness and growth.
We observed a striking reduction in global chromatin accessibility in LIC-e cells on Phf6 loss, with a third of all peaks showing significantly reduced ATAC signal. Collectively, this led to reduced accessibility at sites with AP-1 family, GATA2, and SPI1 occupancy, and increased accessibility at a small number of sites with NF-kB and IRF family occupancy. This observation does not allow us to draw any immediate conclusion about the direct molecular action of PHF6 protein on chromatin, and we believe it to be a consequence of indirect effects that may cumulatively lead to reduced overall accessibility. We also acknowledge that the LIC-e population is not homogenous, and likely contains cells with higher or lower global chromatin accessibility. Future work using bulk and single-cell ATAC-Seq studies will be required to map out the precise chromatin effects of Phf6 loss in the Hoxa9 model, as well as in other models such as co-mutation with RUNX1 or ASXL1.
In summary, our work defines an LIC-enriched population in Hoxa9-driven AML, and the hierarchy through which it differentiates and expands to produce the bulk of the AML population. We show that Phf6 loss increases the number of these LIC-e cells, not through increased cycling, but by producing progeny with persistent LIC-e identity. Taken together with the relatively pure phenotype of increased HSC self-renewal observed when Phf6 is knocked out in homeostatic marrow, this presents a useful system to demonstrate how the normal process of self-renewal is co-opted in AML to drive the self-renewal of leukemic stem cells.
MATERIALS AND METHODS
Analysis of BEAT AML data
Mutational and survival information for 557 human adults with AML was obtained from the BEAT AML (26) dataset (through the cBioPortal website https://www.cbioportal.org/, RRID:SCR_014555). Patients were stratified based on ELN 2017 classification provided on cBioPortal. Kaplan-Meier survival curves were plotted separately for adverse and intermediate ELN risk groups (PHF6 mutations were rare in favorable risk AML), and statistical significance was assessed using the Log-rank (Mantel-Cox) test.
Mice
Cryopreserved sperm from Phf6fl/Y mice (serial# 4621-2 / G4621) was purchased from the Mouse Clinical Institute at GIE-CERBM (GIE-Centre Européen de Recherche en Biologie et Médecine, France), and pups were generated by the Children’s Hospital of Philadelphia Transgenic Core by IVF using C57BL/6J oocytes. Vav-Cre mice were originally generated by Thomas Graf (47) and were provided as a generous gift by Warren Pear (Department of Pathology, University of Pennsylvania Perelman School of Medicine). Both alleles were backcrossed with pure C57BL/6J for over 10 generations. All animals were maintained and experiments were carried out according to the University of Pennsylvania’s Animal Resources Center and IACUC protocols.
Hoxa9 overexpression construct and retrovirus preparation
pMSCV Hoxa9-IRES-GFP retroviral construct plasmid was a generous gift from Kathrin Bernt (Division of Oncology, Children's Hospital of Philadelphia). Retroviruses were generated by transient co-transfection of 293T cells with retroviral vector and ecotropic packaging plasmids. Supernatant containing retroviruses was collected after 48 and 72 hrs of culture, filtered through a 0.45 μm filter, and used to transduce mouse bone marrow cells.
Hoxa9 transduction of mouse bone marrow
Age-matched Vav-CreCre/+ Phf6+/Y (Ctrl) or Vav-CreCre/+ Phf6fl/Y (cKO) littermate mice were intraperitoneally injected with 150 mg/kg 5-fluorouracil (5-FU, Fresenius Kabi, 101720), and bone marrow cells were isolated from femurs after 4 days. Red blood cells were lysed with ACK buffer (Lonza, #10-548E), and cells were cultured for 2 days in IMDM (Thermo Fisher Scientific, #12440053) with 15% fetal bovine serum (Gemini Bio, #100-100), 20 ng/ml mSCF (Peprotech, #250-03), 20 ng/ml mTPO (Peprotech, #315-14), 20 ng/ml mFLT3 ligand (Peprotech, #250-31L), 10 ng/ml mIL3 (Peprotech, #213-13), and 10 ng/ml mIL6 (Peprotech, #216-16). Cultured cells were then transduced with spinofection (2400 RPM, 90 min) with pMSCV Hoxa9-IRES-GFP retroviral supernatant and 4 ug/ml polybrene (Millipore Sigma, #TR-1003), and transferred to fresh media 18 hours after spin. GFP+ cells were purified after 2 days by fluorescence-activated cell sorting (BD FACSAria II).
mRNA isolation and qPCR
To confirm that Phf6 cKO does not express Phf6 mRNA, we isolated mRNA from bone marrow of 3 Ctrl and 3 cKO animals using TRizol (Thermo Scientific, #15596026) and prepared cDNA using 500 ng of total RNA using high capacity cDNA reverse transcriptase (Thermo Scientific, #4368813). qPCR for Phf6 was carried out using primers TGTTTTCGTCTGCTTTGGTG (forward) and ATCATGCAATGCACAGTGGT (reverse), and for Gapdh (endogenous control) using primers CCCAGCTTAGGTTCATCAGG (forward) and GAATTTGCCGTGAGTGGAGT (reverse).
Immunophenotypic analysis and cell sorting
Total bone marrow was isolated by flushing and crushing bones (femur, tibia, humerus, ilium and column vertebralis) in PBS supplemented with 2% FCS. Red blood cell lysis was performed for 5 min at room temperature in ACK lysing buffer (Lonza, #10-548E). For flow cytometry, a single-cell suspension of bone marrow cells was stained with the requisite panel of fluorochrome-conjugated antibodies on ice for 30 minutes to 1 hour. A complete list of antibodies is provided in Table S1. Cells were analyzed using an LSR Fortessa flow cytometer (BD Biosciences), and data were analyzed using FlowJo (FlowJo, LLC). When purification of cell populations was desired, cells were sorted using a BD Aria II sorter (BD Biosciences) into chilled PBS with 10% BSA for further processing.
Colony Forming assay
For myeloid clonogenic progenitor assays, Hoxa9-transduced cells were plated on 35 mm Petri dishes (Corning) in a 1.1 mL culture mixture containing Methocult (STEMCELL Technologies, #M3234) with 20 ng/ul mIL3 (Peprotech, #213-13), 20 ng/ul mIL6 (Peprotech, #216-16), and 20 ng/ul mSCF (Peprotech, #250-03). Colonies propagated in culture were scored and replated (500 cells/plate) every 7 days. Average colony size was determined by counting the number of cells obtained per plate and dividing by the number of colonies. A total of 8 replating rounds were performed.
In vitro culture of Hoxa9-transduced cells
Hoxa9-transduced bone marrow cells from Ctrl or cKO mice were sorted for GFP positivity, and cultured in IMDM (Thermo Scientific, #12440053) with 15% fetal bovine serum (Gemini Bio), 20 ng/ml mSCF (Peprotech, #250-03), 20 ng/ml mTPO (Peprotech, #315-14), 20 ng/ml mFLT3 ligand (Peprotech, #250-31L), 10 ng/ml mIL3 (Peprotech, #213-13), and 10 ng/ml mIL6 (Peprotech, #216-16). Cells were diluted to 100K/ml every other day.
AML mouse model and serial bone marrow transplantation
For all transplantation experiments, recipient mice were wild-type C57BL/6J mice aged 8–20 weeks, lethally irradiated with 1100cGy administered as two split doses of 550cGy given 4 hours apart. Hoxa9-transduced bone marrow cells from Ctrl or cKO mice were injected retro-orbitally into primary recipient mice along with 400K radioprotectant cells of healthy age-matched bone marrow. Mice were monitored for activity and body conditioning score. For survival curve cohorts, mice were euthanized when humane endpoints for morbidity were met. For serial transplantation, bone marrow from primary recipient mice was harvested at 8 weeks after transplantation and lysed with ACK buffer. Sorted GFP+ cells along with 400K radioprotectant cells were retro-orbitally injected into lethally irradiated secondary recipients. Subsequent tertiary transplants were similarly performed using sorted GFP+ cells from marrow harvested from secondary recipients at 8 weeks after transplantation. Each cohort contained 8–20 week old age and sex-matched recipient mice.
Limiting dilution transplantation assay
To calculate LIC (Leukemia initiating cell) frequency in freshly transduced marrow or in 8-week transplanted marrow, cohorts of lethally irradiated recipient mice were transplanted with 400K, 100K, 30K, 3K or 1K GFP+ cells from the respective source. Mice were euthanized when humane endpoints for morbidity were met or when >375 days had passed (experiment termination), and analyzed for the presence of GFP+ cells in bone marrow. All mice that achieved humane endpoints for morbidity had >80% GFP+ cells in marrow, and were counted as responders to the corresponding dose. At experiment termination (>375 days), a small number of mice with >1% GFP were also counted as responders (most had nearly 80% GFP positivity), while mice with <1% GFP+ cells in marrow and peripheral blood were counted as non-responders. LIC frequency was calculated using ELDA software (48).
Histopathology
Mouse femur, spleen, liver, and lungs were harvested and fixed in formaldehyde. Femurs were decalcified and tissue paraffin blocks and slides were prepared and stained by the University of Pennsylvania Molecular Pathology and Imaging Core. Bone marrow was cytospun and peripheral blood was smeared onto glass slides, following which the slides were air-dried and stained using HEMA 3 stain (Thermo Fisher Scientific, #22-122911). Slides were imaged using a Leica microscope.
In vivo cell cycle analysis
The BD Pharmingen™ APC BrDU Kit (BD Biosciences, #552598) was used as per the manufacturer’s protocol. 1 mg/mouse BrdU (BD bioscience, #552598) was administered intraperitoneally at 8 weeks after primary transplant, and bone marrow was harvested after 2 hours. Following ACK lysis, cells were stained for surface antigens, fixed and treated with DNase to expose BrdU antigen, and incubated with Pacific Blue conjugated anti-BrdU antibody as per the instruction manual. Flow cytometry was performed as described above.
In vitro cell cycle analysis
For cell cycle analysis and to calculate the length of the cell cycle, the Click-iT™ EdU Pacific Blue™ Flow Cytometry Assay kit (Invitrogen™, #C10418) was used as per the manufacturer’s protocol. Briefly, 50K sorted LIC-e cells were incubated for 10 hours in media containing 10 µM EdU. Cells were harvested every hour, washed with 1% BSA in PBS, and stained for surface antigens. Cells were then fixed using the Click-iT fixative for 15 minutes at room temperature, and subsequent washes were performed using the 1X Click-iT saponin-based permeabilization and wash reagent. Cells were incubated for 30 minutes at room temperature in the dark with the Click-iT reaction cocktail composed of CuSO4, Pacific Blue dye, reaction buffer additive and PBS, following the manufacturer’s instructions. Flow cytometry was performed as described above.
For EdU pulse-chase experiments, 50K purified LIC-e cells were incubated for 2 hours in media containing 10 µM EdU (pulse). Cells were washed 3 times with fresh media to remove EdU, and were cultured for an additional 8 hours (chase) before being harvested and stained as described above.
Apoptosis
Apoptotic and necrotic cell analysis was performed using Pacific Blue™ Annexin V Apoptosis Detection Kit with 7-AAD (BioLegend, #640926) as per manufacturer’s instructions. Cells were distinguished as live (AnnexinV-, 7AAD-), apoptotic (AnnexinV+, 7AAD-), necrotic (7AAD+).
RNA sequencing
50K LIC-e and 150K committed leukemic cells were sorted from bone marrow of primary recipients of Ctrl+Hoxa9 and cKO+Hoxa9 cells at 8 weeks after transplantation. Total RNA was extracted using TRIzol LS reagent (Thermo Fisher Scientific, #10296010) and RNeasy Mini Kit (Qiagen, #74106). Total RNA concentration and RNA integrity number was determined using a Bioanalyzer (Agilent Technologies) and high sensitivity RNA Screentape (Agilent, #5067-5579). Samples with RNA integrity number 8 or higher were used for RNA library preparation. For LIC-e cells, RNA libraries were prepared using Smarter Stranded Total RNA-Seq Kit - Pico Input Mammalian (Takara Bio, #635005). For committed cells, RNA libraries were prepared using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, #E7530L). All libraries were prepared using manufacturer's instructions. Libraries were quantified using high sensitivity D1000 Screentape (Agilent Technologies, #5067-5584) and KAPA Library Quantification Kit (Thermo Fisher Scientific, #KK4824). Equimolar libraries from each sample were pooled and subjected to 75-bp paired-end sequencing on a NextSeq 550 machine (Illumina). Raw reads were demultiplexed and FASTQ files were generated using Bcl2fastq v2.20.0.422 (Illumina). The reads were trimmed for low quality/adapter sequence using Trimmomatic (RRID:SCR_011848) (49), followed by aligning to the mouse genome (mm10) with STAR v2.7.10b aligner (50). Gene-level read counts were generated using the featureCounts tool from Rsubread v2.0.3) (51). Read count normalization and differential gene expression testing was performed using DESeq2 v1.30.1 (52). GSEA analysis was performed using DESeq2 normalized read counts using GSEAv4.1.0. Mac application (SeqGSEA, RRID:SCR_005724, Broad institute). Gene signatures were downloaded from the MSigDB (Broad) or from the GSE datasets directly in the GSEA application. Gene set enrichment data was represented using default parameters and an FDR cutoff of 0.05.
ATAC sequencing
After 4 days of Hoxa9 transduction, 50K LIC-e cells were sorted from Ctrl+Hoxa9 and cKO+Hoxa9 bone marrow cultures. Cells were lysed and their nuclei were isolated, followed by tagmentation performed using Nextera Tn5 transposase (Illumina Nextera ATAC seq kit, 20034197). Libraries were prepared using dual-end indexing using NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, #E7645L) using the manufacturer's protocol. The size and concentration of each sample’s library were determined using Agilent 2200 Tapestation and KAPA library quantification kit (Roche, #KK4824) respectively. Libraries from all samples were pooled in equimolar concentrations and denatured along with 1% of PhiX for paired-end sequencing. The pooled library was quantified using KAPA library quantification kit (Roche KK4824) and equimolar libraries were pooled and subjected to 75-bp paired-end sequencing on a NextSeq 550 machine (Illumina). Raw reads were demultiplexed and FASTQ files were generated. Reads were trimmed of contaminating adapter sequences using Trimmomatic (49) and aligned to the mouse genome (mm10) using Bowtie2 (RRID:SCR_016368) (53). Peaks of transposase-accessible chromatin were called and quantified using MACS2 v2.2.7.1 (54). Differential and consensus peak analyses were performed using the DiffBind v3.0.15 package in R software (RRID:SCR_012918). Motif analysis was also performed for all regions with significant differential accessibility after filtering out low-confidence regions using HOMER (55). Public ChIP-Seq datasets (Table S3) for ATF3, FOS, JUN, GATA2, SPI1, MAF2, IRF4, IRF8, RELA and RELB were mapped to mm10 genome and used for generating metagene plots using SeqPlot software.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism v9.5.1 (GraphPad Software). We assumed equal distribution of variance between groups and performed either 1-way Anova with Sidak’s multiple comparison testing or Student’s t-test. Kaplan-Meier curves were used to represent survival, where significance was calculated with the log-rank (Mantel-Cox) test. We considered results p < 0.05 as statistically significant and represented using *p < 0.05, **p < 0.01, ***p < 0.001; ****p < 0.0001. We analyzed serial limited dilution leukemia-initiating cell data using the ELDA tool(48).
Blinding
All histopathological analyses were performed in a blinded fashion.
Replication
All transplant experiments were performed in multiple cohorts of recipients, and data were compiled for final analyses.
Power Analysis
Power analyses for mouse experiments were at 90% confidence, significance level of 0.05.
Inclusion and Exclusion Criteria
N/A, no clinical trial data in this study
Attrition
N/A, no clinical trial data in this study
Cell Line Authentication
N/A, no cell lines used in this study
Supplementary Material
Statement of Significance.
Leukemia stem cell self-renewal is critical to the pathophysiology of AML. Phf6 deletion accelerates mouse AML by increasing LSC self-renewal, specifically by increasing the fraction of LSC progeny that retain LSC identity. Our work shows how a repressor of HSC self-renewal is inactivated in AML to drive LSC stemness.
ACKNOWLEDGEMENTS
We thank Nancy Speck, Ivan Maillard, Wei Tong, and Kathrin Bernt for helpful discussions. V.R.P. is supported by National Institute of Health (NIH) grant R01-HL155144 (NHLBI), American Cancer Society (ACS) grant 129784-IRG-16-188-38-IRG, an American Society of Hematology (ASH) Faculty Scholar Award, and a University of Pennsylvania Covid-19 Research Disruption Mitigation Fund. S.S.J. is supported by an ASH Restart Award. C.A is supported by a Scholar Award from the American Society of Hematology (ASH) and Co-Operative Center for Excellence in Hematology (CCEH) grant by the National Institute of Diabetes and Digestive and Kidney diseases (NIDDK). Data for this manuscript were generated in the Penn Cytomics and Cell Sorting Shared Resource Laboratory at the University of Pennsylvania and is partially supported by the Abramson Cancer Center NCI Grant (P30 016520). The research identifier number is RRid: SCR_022376.
Footnotes
DECLARATION OF INTERESTS
None
DATA AVAILABILITY
All generated datasets have been deposited to GEO: GSE249108 for RNA-Seq on LIC-e cells, GSE249109 for RNA-Seq on committed leukemic cells, and GSE249110 for ATAC-Seq datasets on LIC-e cells.
REFERENCES
- 1.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang C. The X-linked Intellectual Disability Protein PHF6 Associates with the PAF1 Complex and Regulates Neuronal Migration in the Mammalian Brain. Neuron. 2013;78:986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Leung JW-C, Gong Z, Feng L, Shi X, Chen J. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J Biol Chem. 2013;288:3174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warmerdam DO, Alonso-de Vega I, Wiegant WW, van den Broek B, Rother MB, Wolthuis RM, et al. PHF6 promotes non-homologous end joining and G2 checkpoint recovery. EMBO Rep. 2020;21:e48460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wendorff AA, Aidan Quinn S, Alvarez S, Brown JA, Biswas M, Gunning T, et al. Epigenetic reversal of hematopoietic stem cell aging in Phf6-knockout mice. Nat Aging. 2022;2:1008–23. [DOI] [PubMed] [Google Scholar]
- 6.Soto-Feliciano YM, Bartlebaugh JME, Liu Y, Sánchez-Rivera FJ, Bhutkar A, Weintraub AS, et al. PHF6 regulates phenotypic plasticity through chromatin organization within lineage-specific genes. Genes Dev. 2017;31:973–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todd MAM, Picketts DJ. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res. 2012;11:4326–37. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez S, da Silva Almeida AC, Albero R, Biswas M, Barreto-Galvez A, Gunning TS, et al. Functional mapping of PHF6 complexes in chromatin remodeling, replication dynamics, and DNA repair. Blood. 2022;139:3418–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh S, Boo K, Kim J, Baek SA, Jeon Y, You J, et al. The chromatin-binding protein PHF6 functions as an E3 ubiquitin ligase of H2BK120 via H2BK12Ac recognition for activation of trophectodermal genes. Nucleic Acids Res. 2020;48:9037–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 2011;25:130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W, Bharadwaj M, Levine M, Farnhoud N, Pastore F, Getta BM, et al. and mutations are enriched in distinct subgroups of mixed phenotype acute leukemia with T-lineage differentiation. Blood Adv. 2018;2:3526–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bataller A, Chien KS, Sasaki K, Montalban-Bravo G, Kanagal-Shamanna R, Urrutia S, et al. PHF6 mutations in myelodysplastic syndromes, chronic myelomonocytic leukemia and acute myeloid leukemia. Leuk Res. 2023;127:107044. [DOI] [PubMed] [Google Scholar]
- 15.Huang K, Wang L, Zheng Y, Yue C, Xu X, Chen H, et al. PHF6 mutation is associated with poor outcome in acute myeloid leukaemia. Cancer Med. 2023;12:2795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McRae HM, Garnham AL, Hu Y, Witkowski MT, Corbett MA, Dixon MP, et al. PHF6 regulates hematopoietic stem and progenitor cells and its loss synergizes with expression of TLX3 to cause leukemia. Blood. 2019;133:1729–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendorff AA, Quinn SA, Rashkovan M, Madubata CJ, Ambesi-Impiombato A, Litzow MR, et al. Loss Enhances HSC Self-Renewal Driving Tumor Initiation and Leukemia Stem Cell Activity in T-ALL. Cancer Discov. 2019;9:436–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu Y-C, Chen T-C, Lin C-C, Yuan C-T, Hsu C-L, Hou H-A, et al. Phf6-null hematopoietic stem cells have enhanced self-renewal capacity and oncogenic potentials. Blood Adv. 2019;3:2355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan S, Wang X, Hou S, Guo T, Lan Y, Yang S, et al. PHF6 and JAK3 mutations cooperate to drive T-cell acute lymphoblastic leukemia progression. Leukemia. 2022;36:370–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T-C, Yao C-Y, Chen Y-R, Yuan C-T, Lin C-C, Hsu Y-C, et al. Oncogenesis induced by combined Phf6 and Idh2 mutations through increased oncometabolites and impaired DNA repair. Oncogene. 2022;41:1576–88. [DOI] [PubMed] [Google Scholar]
- 22.Meacham CE, Lawton LN, Soto-Feliciano YM, Pritchard JR, Joughin BA, Ehrenberger T, et al. A genome-scale in vivo loss-of-function screen identifies Phf6 as a lineage-specific regulator of leukemia cell growth. Genes Dev. 2015;29:483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou S, Wang X, Guo T, Lan Y, Yuan S, Yang S, et al. PHF6 maintains acute myeloid leukemia via regulating NF-κB signaling pathway. Leukemia. 2023;37:1626–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spencer DH, Young MA, Lamprecht TL, Helton NM, Fulton R, O’Laughlin M, et al. Epigenomic analysis of the HOX gene loci reveals mechanisms that may control canonical expression patterns in AML and normal hematopoietic cells. Leukemia. 2015;29:1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyagi S, Sroczynska P, Kato Y, Nakajima-Takagi Y, Oshima M, Rizq O, et al. The chromatin-binding protein Phf6 restricts the self-renewal of hematopoietic stem cells. Blood. 2019;133:2495–506. [DOI] [PubMed] [Google Scholar]
- 28.Spring J, Khan AA, Lara S, O’Grady K, Wilks J, Gurbuxani S, et al. Gut commensal bacteria enhance pathogenesis of a tumorigenic murine retrovirus. Cell Rep. 2022;40:111341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda H, Kanakura Y, Tamaki T, Kuriu A, Kitayama H, Ishikawa J, et al. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 1991;78:2962–8. [PubMed] [Google Scholar]
- 30.Somervaille TCP, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–68. [DOI] [PubMed] [Google Scholar]
- 31.North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MFTR. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22:158–68. [DOI] [PubMed] [Google Scholar]
- 32.Bellissimo DC, Speck NA. Mutations in Inherited and Sporadic Leukemia. Front Cell Dev Biol. 2017;5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Y-Q, Plow EF, Geng J-G. P-selectin binding to P-selectin glycoprotein ligand-1 induces an intermediate state of alphaMbeta2 activation and acts cooperatively with extracellular stimuli to support maximal adhesion of human neutrophils. Blood. 2004;104:2549–56. [DOI] [PubMed] [Google Scholar]
- 34.Pulikkan JA, Madera D, Xue L, Bradley P, Landrette SF, Kuo Y-H, et al. Thrombopoietin/MPL participates in initiating and maintaining RUNX1-ETO acute myeloid leukemia via PI3K/AKT signaling. Blood. 2012;120:868–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauch PJ, Ellegast JM, Widmer CC, Fritsch K, Goede JS, Valk PJM, et al. MPL expression on AML blasts predicts peripheral blood neutropenia and thrombocytopenia. Blood. 2016;128:2253–7. [DOI] [PubMed] [Google Scholar]
- 36.Goyama S, Shrestha M, Schibler J, Rosenfeldt L, Miller W, O’Brien E, et al. Protease-activated receptor-1 inhibits proliferation but enhances leukemia stem cell activity in acute myeloid leukemia. Oncogene. 2017;36:2589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Yu H, Shou Y, Neale G, Zhou S, Lu T, et al. Hemgn is a direct transcriptional target of HOXB4 and induces expansion of murine myeloid progenitor cells. Blood. 2010;116:711–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hewitt KJ, Kim DH, Devadas P, Prathibha R, Zuo C, Sanalkumar R, et al. Hematopoietic Signaling Mechanism Revealed from a Stem/Progenitor Cell Cistrome. Mol Cell. 2015;59:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3. [DOI] [PubMed] [Google Scholar]
- 40.Somervaille TCP, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL–AF9. Nature. 2006;442:818–22. [DOI] [PubMed] [Google Scholar]
- 42.Brown AL, Wilkinson CR, Waterman SR, Kok CH, Salerno DG, Diakiw SM, et al. Genetic regulators of myelopoiesis and leukemic signaling identified by gene profiling and linear modeling. J Leukoc Biol. 2006;80:433–47. [DOI] [PubMed] [Google Scholar]
- 43.Franzén O, Gan L-M, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database. 2019;2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira PD, Serra-Caetano A, Cabrita M, Bekman E, Braga J, Rino J, et al. Quantification of cell cycle kinetics by EdU (5-ethynyl-2’-deoxyuridine)-coupled-fluorescence-intensity analysis. Oncotarget. 2017;8:40514–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higashi AY, Ikawa T, Muramatsu M, Economides AN, Niwa A, Okuda T, et al. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J Immunol. 2009;182:5633–40. [DOI] [PubMed] [Google Scholar]
- 46.Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–75. [DOI] [PubMed] [Google Scholar]
- 47.Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development. 2005;132:203–13. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ginhoux F, Guilliams M, Naik S. Dendritic Cell and Macrophage Nomenclature and Classification. Frontiers Media SA; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connell KE, Mikkola AM, Stepanek AM, Vernet A, Hall CD, Sun CC, et al. Practical murine hematopathology: a comparative review and implications for research. Comp Med. 2015;65:96–113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All generated datasets have been deposited to GEO: GSE249108 for RNA-Seq on LIC-e cells, GSE249109 for RNA-Seq on committed leukemic cells, and GSE249110 for ATAC-Seq datasets on LIC-e cells.