Abstract
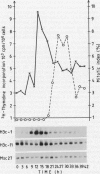
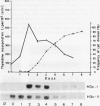
Northern analysis has revealed substantial differences in mRNA accumulation of the two histone H3 gene variants represented by pH3c-1 and pH3c-11 cDNA clones. Both in partially synchronized cell suspension cultures and in protoplast-derived cells from alfalfa, Medicago varia, the maximal level of the histone H3-1 gene transcript coincided with the peak in [3H]thymidine incorporation. Histone H3-11 mRNA was detectable in cells throughout the period of the cell cycle studied. Various stress factors such as medium replacement, enzyme digestion of the cell wall, osmotic shock, and auxin treatment considerably increased the level of the histone H3-11 transcript. In alfalfa (Medicago sativa), the presence of H3-11 mRNA in unorganized tissues of microcallus suspension and in somatic embryos induced by auxin treatment supports the idea that this H3 variant exists in a continously active state of transcription. During embryo development, the early globular stage embryos showed increased accumulation of histone H3-11 mRNA in comparison with the later stages. The highest level of the histone H3-1 transcript was detectable 1 day after treatment of callus tissues with 2,4-dichlorophenoxyacetic acid. Somatic embryos contained appreciable levels of histone H3-1 transcripts at all stages of somatic embryo development. These observations suggest that the histone H3-1 gene belòngs to the class of replication-dependent histone genes. The histone H3-11 gene showed characteristics of a constitutively expressed replacement-type histone gene, with a specific characteristic that external factors can influence the level of gene transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown D. T., Wellman S. E., Sittman D. B. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol Cell Biol. 1985 Nov;5(11):2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathala G., Savouret J. F., Mendez B., West B. L., Karin M., Martial J. A., Baxter J. D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Csordas A. On the biological role of histone acetylation. Biochem J. 1990 Jan 1;265(1):23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Wells J. R. A gene-specific promoter element is required for optimal expression of the histone H1 gene in S-phase. EMBO J. 1988 Jan;7(1):49–56. doi: 10.1002/j.1460-2075.1988.tb02782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fletcher C., Heintz N., Roeder R. G. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987 Dec 4;51(5):773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Loidl P. Towards an understanding of the biological function of histone acetylation. FEBS Lett. 1988 Jan 25;227(2):91–95. doi: 10.1016/0014-5793(88)80874-3. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Gould J., Kim S., Kane M. Y., Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986 May 23;45(4):537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- Osley M. A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Raghavan V., Olmedilla A. Spatial patterns of histone mRNA expression during grain development and germination in rice. Cell Differ Dev. 1989 Sep;27(3):183–196. doi: 10.1016/0922-3371(89)90699-0. [DOI] [PubMed] [Google Scholar]
- Waterborg J. H., Harrington R. E., Winicov I. Differential Histone Acetylation in Alfalfa (Medicago sativa) Due to Growth in NaCl : Responses in Salt Stressed and Salt Tolerant Callus Cultures. Plant Physiol. 1989 May;90(1):237–245. doi: 10.1104/pp.90.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg J. H. Sequence analysis of acetylation and methylation in two histone H3 variants of alfalfa. J Biol Chem. 1990 Oct 5;265(28):17157–17161. [PubMed] [Google Scholar]
- Waterborg J. H., Winicov I., Harrington R. E. Histone variants and acetylated species from the alfalfa plant Medicago sativa. Arch Biochem Biophys. 1987 Jul;256(1):167–178. doi: 10.1016/0003-9861(87)90435-8. [DOI] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. C., Györgyey J., Dudits D. Polyadenylated H3 histone transcripts and H3 histone variants in alfalfa. Nucleic Acids Res. 1989 Apr 25;17(8):3057–3063. doi: 10.1093/nar/17.8.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]