Abstract
Chemokines play critical roles in the recruitment and activation of immune cells in both homeostatic and pathologic conditions. Here, we examined chemokine ligand-receptor pairs to better understand the immunopathogenesis of cutaneous lupus erythematosus (CLE), a complex autoimmune connective tissue disorder. We used suction blister biopsies to measure cellular infiltrates with spectral flow cytometry in the interface dermatitis reaction, as well as 184 protein analytes in interstitial skin fluid using Olink targeted proteomics. Flow and Olink data concordantly demonstrated significant increases in T cells and antigen presenting cells (APCs). We also performed spatial transcriptomics and spatial proteomics of punch biopsies using digital spatial profiling (DSP) technology on CLE skin and healthy margin controls to examine discreet locations within the tissue. Spatial and Olink data confirmed elevation of interferon (IFN) and IFN-inducible CXCR3 chemokine ligands. Comparing involved versus uninvolved keratinocytes in CLE samples revealed upregulation of essential inflammatory response genes in areas near interface dermatitis, including AIM2. Our Olink data confirmed upregulation of Caspase 8, IL-18 which is the final product of AIM2 activation, and induced chemokines including CCL8 and CXCL6 in CLE lesional samples. Chemotaxis assays using PBMCs from healthy and CLE donors revealed that T cells are equally poised to respond to CXCR3 ligands, whereas CD14+CD16+ APC populations are more sensitive to CXCL6 via CXCR1 and CD14+ are more sensitive to CCL8 via CCR2. Taken together, our data map a pathway from keratinocyte injury to lymphocyte recruitment in CLE via AIM2-Casp8-IL-18-CXCL6/CXCR1 and CCL8/CCR2, and IFNG/IFNL1-CXCL9/CXCL11-CXCR3.
One Sentence Summary:
We mapped chemokine orchestrators of interface dermatitis in lupus using spatial approaches on archival tissue and confirmed with fresh tissues.
INTRODUCTION
Cutaneous lupus erythematosus (CLE) is a spectrum of clinically diverse autoimmune connective tissue disorders with shared histopathological features including interface dermatitis and lupus band reaction(1). Chronic cutaneous lupus erythematosus (CCLE, most common sub-entity discoid lupus erythematosus (DLE)), subacute cutaneous lupus (SCLE), and acute cutaneous lupus erythematosus (ACLE) include the majority of cutaneous lupus clinical subtypes, though there are other more rare entities which all have varying degrees of association with systemic lupus erythematosus (SLE). In clinical practice, CLE can be refractory to SLE treatments(2–4), but successful treatment of cutaneous disease may significantly decrease the risk of systemic involvement(5).
A common feature of CLE is the presence of interface dermatitis, which refers to inflammation and/or degenerative changes at the dermal-epidermal junction. The mechanisms by which immune cells are recruited to form interface dermatitis in CLE are unknown. Previous studies have found that interferon (IFN)-inducible chemokines are highly upregulated in CLE lesions(6, 7). One recent study characterized the associations of specific chemokine receptors and ligands in T cell subsets in cutaneous versus systemic lupus, demonstrating different profiles in SLE, DLE and SCLE(8). The cellular sources of chemokine ligands binding to cognate receptors have not been fully characterized.
To date, many CLE studies have focused on transcriptomics including bulk RNA sequencing and single cell RNA sequencing(9), but overlooked proteomics approaches and cellular analyses. Here, we used a blister biopsy technique(10) to sample interstitial skin fluid and cells from the interface dermatitis reaction from CLE patients and compared these to healthy donors. Blister fluid does not require enzymatic digestion for analysis, yielding high-quality liquid and cellular biopsies. We confirm elevated CXCL9/10/11 in interstitial skin fluid and the cognate receptor CXCR3 on infiltrating immune cells. We also present similarities in antiviral signatures, namely IFNL1 and IFNG, that likely drive CXCR3 ligand expression and the shared interface dermatitis tissue reaction pattern across CLE clinical subtypes. Further, we demonstrate protein-level expression of many other non-redundant chemokine ligands, cytokines, and other proteins including Caspase 8, HGF and Flt3L. Importantly, we identify CXCL6 and CCL8 as novel chemokine biomarkers that are elevated in lesional skin. The CXCL6 cognate receptor CXCR1 is predominantly expressed by CD14+CD16+ myeloid cells, which migrate preferentially towards CXCL6. In contrast, the CCL8 cognate receptor CCR2 is predominantly expressed by CD14+CD16- myeloid cells that preferentially migrate towards CCL8. We also performed matched spatial transcriptomics and proteomics of DLE and SCLE biopsies to identify the general cellular sources (keratinocytes vs immune cells) and locations of these chemokines and other immune proteins. Importantly, we were also able to identify unique gene and protein expression signatures which may underlie differences in the discoid and subacute clinical subtypes of CLE, including 475 DEGs in CD3+ ROIs, 397 DEGs in CD45+CD3- ROIs, and 735 DEGs in the epidermial/keratinocyte ROIs, in addition to unique expression of B7-H3 protein on SCLE T cells.
RESULTS
Integrating spatial and -omics approaches for characterizing the inflammatory infiltrate in cutaneous lupus interface dermatitis
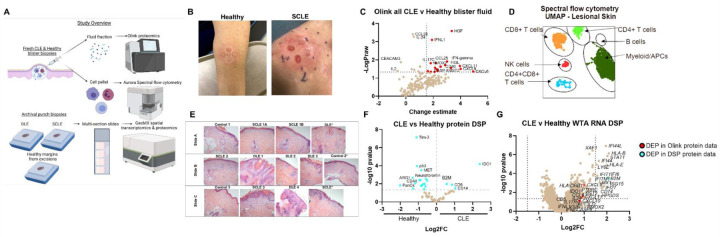
We combined spatial studies of archival tissue with analyses of fresh tissue biopsies to validate chemokine and other targets at the protein level (workflow Fig 1A). We used blister biopsies to sample interstitial skin fluid and cells from CLE patients and healthy donors (Fig 1B, Table S1). We performed Olink targeted proteomics on the interstitial skin fluid (Fig 1C), which confirmed previously reported increased levels of IFNg, CXCL9 and CXCL11 in CLE skin biopsies (6, 8). We also identified higher levels of CXCL6 and hepatocyte growth factor (HGF), which can increase CXCR3 expression on T cells (11), in CLE fluid compared to healthy controls, previously not reported in CLE skin analyses. We used a 19 color flow panel to identify cell types of interest by both UMAP and manual gating. Spectral flow cytometry analysis of blister fluid cells revealed a predominantly lymphocytic infiltrate, with increases in myeloid populations (Fig 1D).
Fig. 1. Integration of high-resolution immune techniques with spatial approaches for studying cutaneous lupus erythematosus.
(A) Study overview demonstrating workflow. We performed blister biopsies on cutaneous lupus erythematosus patients and healthy donors and analyzed the interstitial skin fluid using Olink proteomics and the cell pellets using Aurora spectral flow cytometry. We also employed biobanked FFPE skin biopsies from DLE, SCLE and healthy margins for NanoString digital spatial profiling for both protein, using 6 modules, and RNA, using the whole transcriptome assay (WTA). Created with Biorender.com. (B) Sample photographs of blister biopsies (n=3 healthy and 4 CLE patients). (C) Volcano plot of Olink data from the inflammation and neuroexploratory panels of all CLE blisters versus healthy controls (n=7 CLE lesional, 7 CLE nonlesional, and 3 healthy blisters). (D) Sample UMAP of immune infiltrates in CLE lesional skin. (E) H&E images of the slide set used for spatial transcriptomics and proteomics. (n=3 healthy margin controls, 5 SCLE biopsies from 4 patients, and 4 DLE biopsies from 4 patients). (F) Volcano plot of DSP protein data for all CLE regions of interest (ROIs) versus healthy margin ROIs. (G) Volcano plot of DSP RNA WTA data for all CLE ROIs versus healthy margin ROIs. Red dots are also differentially expressed proteins (DEPs) in the Olink protein dataset, and cyan dots are also DEPs in the DSP protein dataset. (n=41 ROIs assessed from 4 DLE, 3 SCLE, and 2 healthy margin control biopsies for WTA DSP; n=95 ROIs assessed from 4 DLE, 4 SCLE from 3 donors and 3 healthy margin controls for protein DSP. Discrepancy due to tissue not being fully seated in imaging area for WTA run, indicated by *.)
We also performed spatial transcriptomics and proteomics on archival tissue biopsies using NanoString Digital Spatial Profiling (DSP, Fig 1E, Table S2). Spatial proteomics captured enrichment of CD8 and CD14 as well as beta-2 microglobulin (B2M) and indoleamine 2,3-dioxygenase (IDO) (Fig 1F, dataset QC in Fig S1, QC of ROIs in Fig S2). Spatial transcriptomics analysis using the Whole Transcriptome Atlas (WTA) of aggregate ROIs recapitulated IFN signatures, and also harmonized with protein biomarkers identified in Olink and protein DSP (Fig 1G). We also validated our DSP WTA dataset using a Cancer Transcriptome Atlas (CTA) dataset (Table S3), as well as a bulk RNA microarray dataset previously generated from matched tissue blocks (Fig S3 & 4). We identified many shared DEGs, including B2m, Cxcl9, Cxcl10 and other IFN- and immune-related genes.
Characterization of the immune infiltrate in CLE interface dermatitis
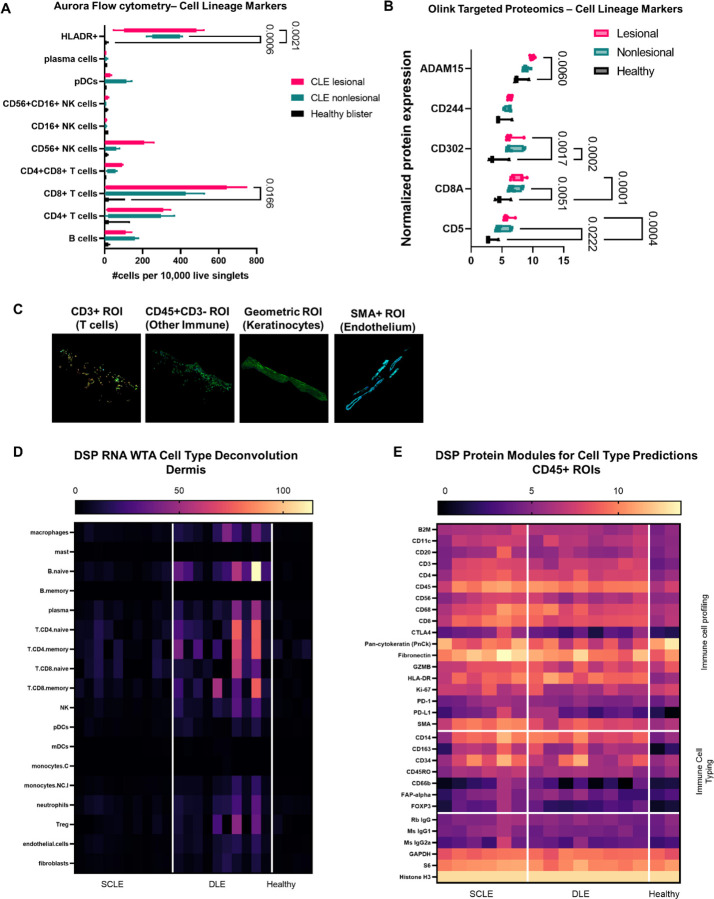
We sought to characterize which cells comprised the interface dermatitis in CLE using multimodal approaches. We performed spectral flow cytometry and Olink proteomics on blister biopsy fluid, and spatial transcriptomics and proteomics on FFPE tissues to determine the cellular composition of CLE interface dermatitis. Quantification of cellular infiltrates from blister biopsies revealed significant increases in HLA-DR+ antigen presenting cell populations (p<0.0021 lesional vs nonlesional, p<0.0006 nonlesional vs healthy) as well as CD8+ T cells (p<0.0166 lesional vs healthy, Fig 2A). We queried the Olink proteomics dataset for cell type specific proteins and found increased T cell (CD5, CD8A) and APC markers (CD302) in both lesional and nonlesional blister fluid (Fig 2B). These surface markers are likely shed due to the presence of increased matrix metalloproteinases such as ADAM15 (Fig 2B). Additional significant protein biomarkers are presented in Table S4.
Fig. 2. Characterizing immune cell subsets in CLE interface dermatitis reveals significant increases in HLA-DR+ cells and CD8+ T cells.
(A) Aurora flow cytometry for cell lineage markers in CLE lesional and nonlesional skin compared to healthy blisters. Enumeration of cellular infiltrates were normalized to 10,000 live singlet cells. One-way ANOVA with Tukey’s post hoc tests revealed significant increases in HLA-DR+ cells and CD8+ T cells compared to healthy control blisters, with trends towards increased B cells, CD4+ T cells and NK cells. pDCs were trending higher in nonlesional blister fluid. (B) Olink targeted proteomics for cell lineage markers in blister fluid. T cell markers (CD5, CD8A) and antigen presenting cell markers (CD302) were also significantly elevated in CLE blister fluid compared to healthy controls. NK marker CD244 was not significantly elevated, matching flow cytometry findings. Note that shedding of surface CD molecules is likely due to the presence of proteases such as ADAM15. (C) Examples of Regions of interest (ROIs) chosen for spatial analysis of FFPE CLE biopsies. We used cell masking approaches for CD3+ T cells, CD45+ immune cells, geometric autofluorescent keratinocytes/epidermis and SMA+ endothelium. (D) Cell type deconvolution in the CD45+ dermal ROIs from the WTA dataset was calculated using NanoString’s prediction module. We noted increases in T cells and B cells, as well as neutrophils which were not assessed by flow cytometry due to neutrophil death upon extraction from skin. (E) Spatial proteomics was assessed for the immune cell profiling and immune cell typing modules to predict the presence of immune cells. Housekeeping proteins are depicted in the bottom module.
Next, we examined our cell type regions of interest (ROIs) in the spatial datasets. We used CD3+CD45+ masks to enrich for T cells, CD45+CD3- masks to enrich for other immune cells, geometric autofluorescent ROIs to enrich for keratinocytes and SMA+ ROIs to enrich for endothelium (Fig 2C). We were not able to capture many ROIs from endothelium in cross section, therefore we focused our analyses on the T cell, immune cell and keratinocyte ROIs. NanoString GeoMX software deconvolution of the immune cell ROIs in the WTA dataset revealed increases in many immune cell types in the dermis including B cells, T cells, dendritic cells and monocytes/macrophages as assessed by cell-type specific genes (Fig 2D). Examination of protein-level expression of CD surface markers using the immune cell profiling and immune cell typing protein modules confirmed CD20+ B cell, CD14+ and CD68+ monocyte/macrophage expression (Fig 2E). Taken together, the predominant cell types comprising CLE interface dermatitis are HLADR+ antigen presenting cells and CD8+ T cells.
Examining T cell phenotypes in discoid and subacute cutaneous lupus erythematosus reveals unique immune checkpoint expression
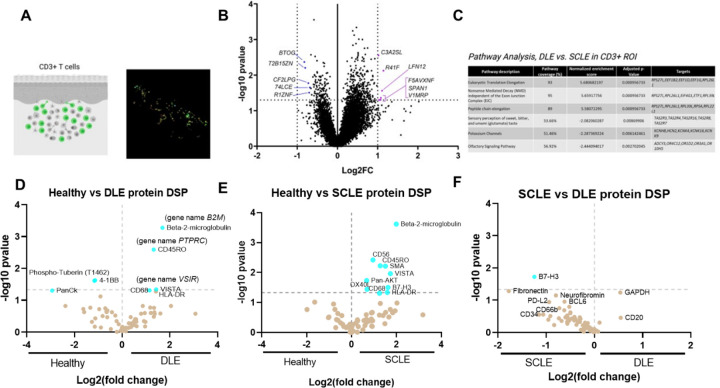
One of the remaining questions in the field of cutaneous lupus is what immunologic differences underlie the pathogenesis of discoid lupus erythematosus (DLE) versus subacute cutaneous lupus erythematosus (SCLE). To this end, we separated out the CLE subtypes and performed both transcriptomic and proteomic analyses comparing the 2 entities. Spatial transcriptomic analysis of DLE vs. SCLE skin detected 475 DEGs in CD3+ ROIs, 397 DEGs in CD45+CD3- ROIs, and 735 DEGs in the epidermis segment (Fig S5). We noted most of the differences existed in the T cell ROI (Fig 3A & B). Pathway analysis in the CD3+ ROI revealed terms including eukaryotic translation elongation, nonsense mediated decay and peptide chain elongation in DLE, versus sensory perception, potassium channels and olfactory signaling pathway in SCLE, which may be of interest given neuroimmune connections that have been identified in other chronic skin disorders (Fig 3C) (12). Querying the T cell ROIs in the protein DSP dataset revealed similar expression patterns in B2M, the memory marker CD45RO, and the immune checkpoint molecule VISTA (Fig 3D & E). Most interestingly, the immune inhibitory family member B7-H3 was uniquely upregulated in SCLE (Fig 3F). Taken together, these data suggest that checkpoint agonism might be a novel treatment approach for CLE, and that these checkpoints may differ between DLE and SCLE.
Fig. 3. Examining differential expressed genes in DLE versus SCLE T cells reveals biological differences that may underlie pathogenesis and/or susceptibility to systemic disease.
(A) Example CD3+ ROI and GeoMX image. Created with Biorender.com. (B) Volcano plot of the CD3+ ROI in DLE vs SCLE biopsies. (C) Pathway analysis of the CD3+ ROI in DLE vs SCLE biopsies as performed in GeoMX software. (D) Protein DSP analysis of the CD3+ ROI in healthy vs DLE, (E) healthy vs SCLE and (F) SCLE vs DLE. There are several shared DEGs and DEPs including B2M, CD45RO and VISTA. However, the immune checkpoint B7-H3 is unique to SCLE and may serve as a novel treatment target.
Keratinocytes at the inflammatory and lesional sites shape chemotactic signals for leukocyte recruitment
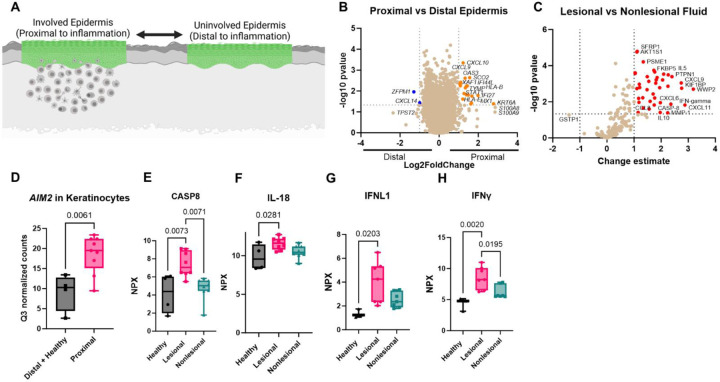
We next asked whether we could identify immune signaling pathways in keratinocytes that could lead to the initiation of inflammation. Comparing keratinocytes that were above interface dermatitis (proximal) versus distal to an infiltrate (Fig 4A) revealed upregulation of key chemokines including Cxcl9 and Cxcl10 in keratinocytes proximal to the immune infiltrate (Fig 4B). We also compared Olink proteomic data in lesional and nonlesional blister fluid, and found that CXCL9 and CXCL11 are significantly upregulated in lesional blister fluid (Fig 4C). Additionally, we identified increased levels of chemokine ligands CCL8 and CXCL6 in lesional versus nonlesional blister fluid (Fig 4C). We re-queried the WTA dataset to analyze potential innate immune pathways that could lead to chemokine expression and found increased AIM2 expression (Fig 4D). We therefore queried specific members of this pathway in our Olink dataset and found significant increases in caspase 8 (13) and the IL1 family member IL18 (14) in lesional skin relative to both healthy and nonlesional samples (Fig 4E & F). We also examined interferons upstream of CXCL9 and CXCL11 and found significant increases in IFNL1 and IFNG protein levels in lesional CLE skin (Fig 4G & H). Of note, IL18 induces CXCL6 expression in murine lung (15), and CCL8 is optimally induced by combinations of IL1, IFNG and dsRNA (16). Taken together, these data suggest that keratinocyte injury can induce AIM2-caspase 8-IL18 pathways and IFN pathways (17) to turn on chemokines including CCL8, CXCL6, CXCL9 and CXCL11 for recruitment of immune cells in CLE.
Fig 4. Examining proximal versus distal keratinocytes, and lesional versus nonlesional blister fluid, reveals pathways that may be required for initiation of chemokine cascades for leukocyte recruitment to form interface dermatitis.
(A) Schematic of proximal versus distal analysis. Created with Biorender.com. (B) Volcano plot of DSP WTA data for proximal versus distal epidermis. (C) Examination of lesional versus nonlesional blister fluid in Olink as performed with NPX software. (D) Box plot of AIM2 expression in keratinocytes from distal plus healthy margins versus proximal keratinocytes from Q3 normalized counts in the WTA dataset. (E) Caspase 8, (F) IL-18, (G) IFNL1 and (H) IFNG were elevated in CLE blisters compared to healthy controls in the Olink dataset (Normalized Protein eXpression, NPX).
Combining proteomic analysis of blister fluid with spatial transcriptomics of skin tissue reveals cell-specific expression patterns of chemokine ligands in CLE skin
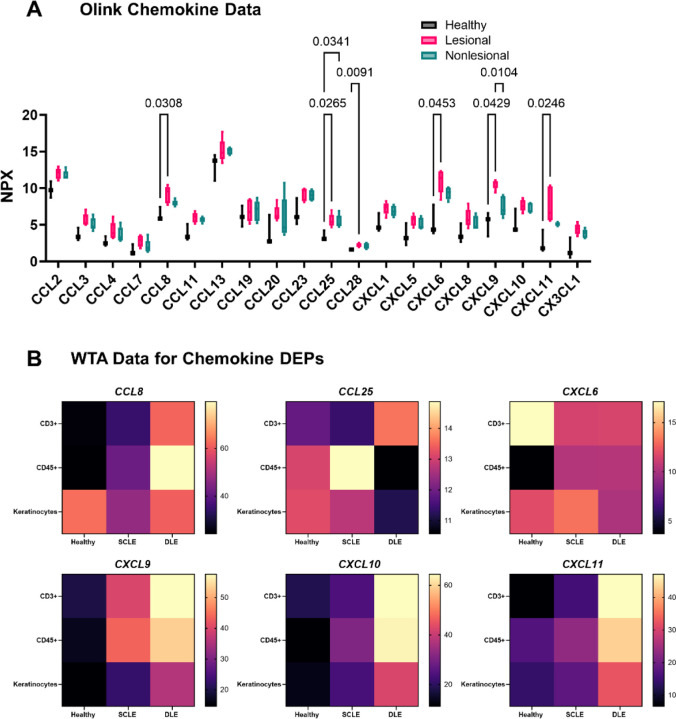
We (18–20) and others (9, 21, 22) have demonstrated upregulation of chemokine ligands in CLE by RNA sequencing and microarray. Using the Olink proteomic platform, we found significant increases in many chemokine ligands, including CXCR3 family, CCL8, CCL25 and CXCL6 chemokines (Fig 5A). Chemokines were fairly stable over time (Fig S6). Querying the WTA dataset revealed cell-specific context expression of these ligands which differed by both ROI type and disease state (Fig 5B). Specifically, Ccl8 was expressed highly in the CD45+ ROI from DLE biopsies, whereas Ccl25 was expressed in SCLE biopsies, indicating a different cellular context of these signals. In contrast, Cxcl6 was expressed by both keratinocytes and immune cells. Cxcl9/10/11 were expressed mainly by the immune system, with the highest upregulation in DLE, which fits with prior literature describing an IFNG “node” in DLE (9).
Fig. 5. Combining proteomics and spatial transcriptomics using a cell masking approach allows assignment of biomarkers and chemokine ligand-receptor pairs governing leukocyte recruitment to the skin during CLE.
(A) We queried the Olink dataset for all available chemokine measurements. Significant differences were observed in the levels of CCL8, CCL25, CCL28, CXCL6, CXCL9, and CXCL11 in the Olink interstitial skin fluid analysis for various chemokines (two-way ANOVA with Tukey’s post tests significant as indicated). B) Heatmaps illustrate their cellular sources in the WTA DSP analysis for healthy, SCLE, and DLE samples.
Functional chemotaxis assays recapitulate CXCR3- mediated T cell migration and reveal CXCR1-sensitive CD14+CD16+ and CCR2-sensitive CD14+ populations
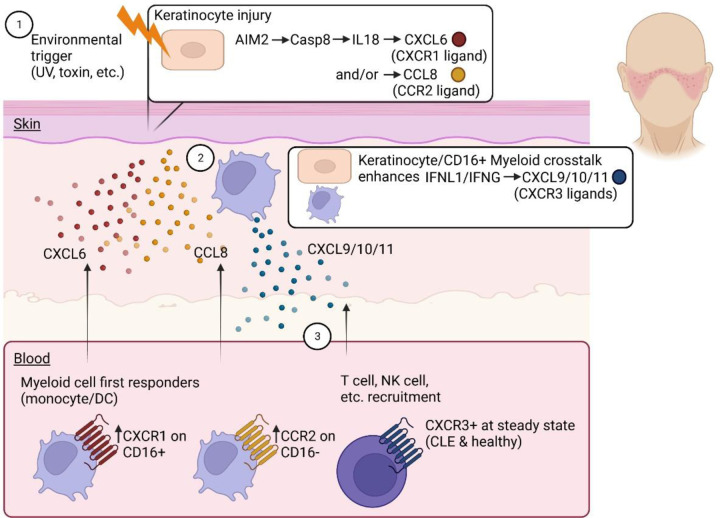
To determine the functional consequences of our data, we drew blood from healthy and CLE donors and performed chemotaxis assays, focusing on the ligands we identified in the Olink dataset (CCL8, CXCL6, CXCL9, CXCL11; Fig 6A & Table S5). We also performed flow cytometry on input cells prior to migration (antibody information & chemokine information in Table S6 & Table S7). Cells were gated on live singlets and were identified with major lineage markers (e.g. CD3+ T cells, CD19+ B cells, CD56+ NK cells). Both lupus and healthy donor T cells expressed CXCR3 (Fig 6B & C) and were able to migrate well towards CXCL9 and CXCL11 (Fig 6D & E, additional donors in Fig S7). However, CXCL9 and CXCL11 are off in healthy skin, and turned on in lupus skin, exemplifying how T cells are poised to respond to IFN-inducible chemokines. We next assessed CXCL6 and its cognate receptor CXCR1 (23), which has been reported to be expressed on skin NK cells (24) and is important for their recruitment into tumors (25). NK cells from both healthy and lupus donors expressed CXCR1 (Fig 6F & G), though there was donor variability for migratory capacity towards the CXCL6 ligand (Fig 6H & I). Like T cells, NK cells migrated to CXCL9 and CXCL11 (Fig 6H & I). CD16+CD14+ myeloid cells were recently identified in nonlesional CLE skin by Billi et al using 10X spatial transcriptomics (26). We gated live, single, CD45+CD3-CD19-CD56- cells (i.e. non-T/B/NK) followed by CD14 and CD16 expression (Fig S8) to confirm these cells are present in CLE blood and skin by flow cytometry (Fig S9). We examined chemokine receptor expression in CD14+CD16-, CD14+CD16+ and CD14-CD16+ myeloid cells and noted significant increases in CXCR1 expression on CD14+CD16+ cells (Fig 6J & K). All CD14+ cells, but particularly CD14+CD16+ cells, migrated towards CXCL6, providing a possible chemokine receptor:ligand pair that mediates their entry into the skin (Fig 6L). In contrast, the CCL8 receptor CCR2 was enriched on CD14+CD16- cells (Fig 6M & N), but not the CCR5 receptor (Fig S10). CD14+CD16- cells migrated best towards CCL8 (Fig 6O). Taken together, these data exemplify how disease states can influence chemokine ligand levels and influence timing and recruitment of specific immune cell populations to the tissue, namely through keratinocytes following injury and subsequently through recruited immune cell populations. We propose the model that keratinocyte injury induces activation of AIM2-related pathways that turn on CXCL6 and CCL8 expression to recruit myeloid cells, which in turn potentiate IFN-related signaling pathways to recruit T cells and other lymphocytes via CXCR3 ligands (Fig 7).
Fig. 6. Functional chemotaxis assays reveal that both healthy and lupus T cells are poised to respond to CXCR3 ligands, whereas CD14+CD16+ myeloid cells are poised to respond to CXCL6 via CXCR1 and CD14+CD16- myeloid cells are poised to respond to CCL8 via CCR2.
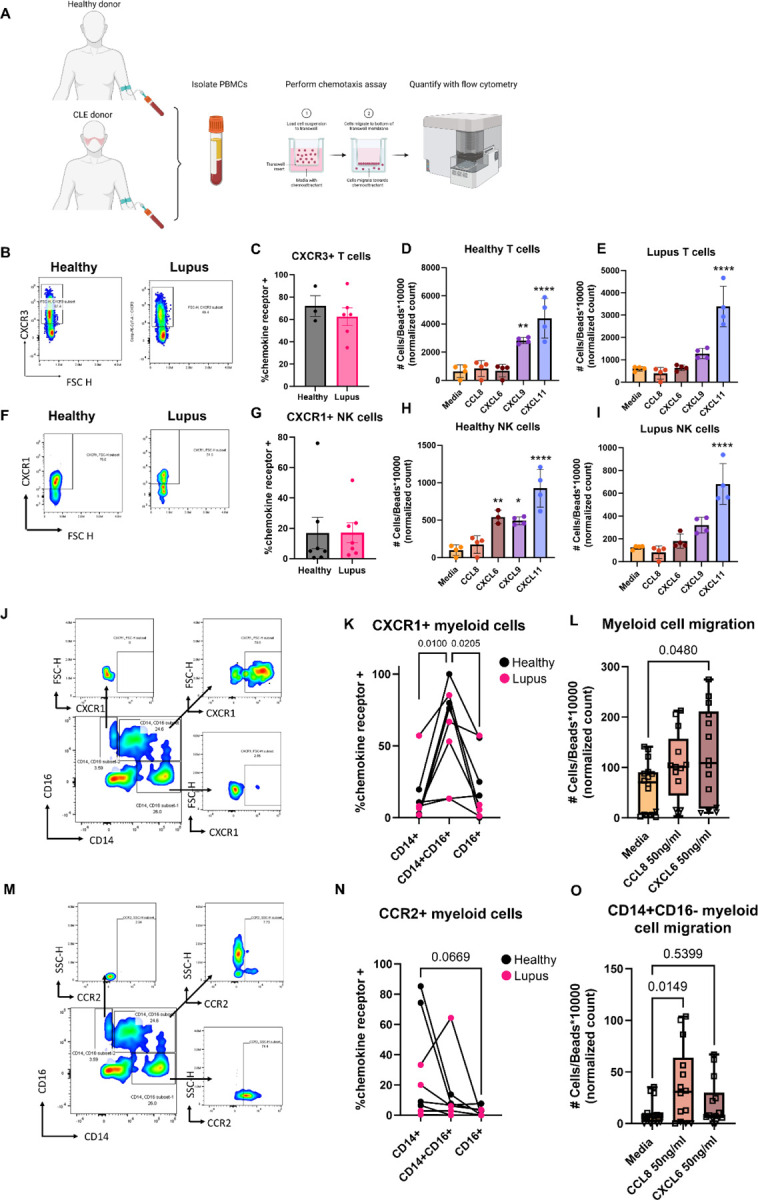
(A) To determine functional output, we performed chemotaxis assays on PBMCs from healthy or CLE donors. Created with Biorender.com. (B) Example flow staining and (C) quantification of CXCR3 on CD3+ T cells (n=3 healthy and n=6 CLE, t test ns). (D) Representative chemotaxis assay for healthy and (E) CLE donor T cells reveals directed migration towards CXCL9 and CXCL11, though the ligands are not turned on in healthy skin (**p<0.01 and ****p<0.0001 by one way ANOVA with Dunnet’s post hoc tests versus media control; donors assayed in quadruplicate; wells were excluded if a bubble was present that precluded migration). (F) Example flow staining and (G) quantification of CXCR1 on CD56+ natural killer (NK) cells from healthy and lupus PBMCs (n=7 healthy and 7 lupus donors, t test ns). (H) Healthy donor and (I) lupus donor directed migration reveals NK cells migrate to both CXCR3 ligands, with donor variable responses towards CXCL6. (**p<0.01 and ****p<0.0001 by one way ANOVA with Dunnet’s post hoc tests versus media control; donors assayed in quadruplicate; wells were excluded if a bubble was present that precluded migration). (J) Example flow staining of myeloid cells demonstrating CXCR1 expression in CD14+, CD14+CD16+ and CD16+ gates. (K) Quantification of CXCR1+ cells demonstrates CD14+CD16+ cells express more CXCR1 than their counterparts. (n=4 healthy and 4 lupus donors; p=0.01 and 0.0205 by repeated measures one way ANOVA matched by donor, with Tukey’s post hoc tests). (L) Chemotaxis assays demonstrate that CD14+ cells migrate towards CXCL6 (n=3 healthy donors denoted by squares and n=1 lupus donor denoted by triangles assayed in triplicate or quadruplicate; one way ANOVA with Dunnet’s post hoc tests compared to media control was significant p=0.048 for CXCL6 but not CCL8. Note that lupus donor chemotactic indices for myeloid cells were lower, but still followed the same trends as healthy donors, likely due to their medications). (M) Example flow staining of myeloid cells demonstrating CCR2 expression in CD14+, CD14+CD16+ and CD16+ gates. (N) Quantification of CXCR1+ cells demonstrates CD14+CD16- cells are trending towards more CCR2 expression than their counterparts (n=4 healthy and 4 lupus donors; p=0.0669 by one way ANOVA with Tukey’s post hoc tests). (O) Chemotaxis assays demonstrate that CD14+CD16- cells migrate towards CCL8. (n=3 healthy donors denoted by squares and n=1 lupus donor denoted by triangles assayed in triplicate or quadruplicate; one way ANOVA with Dunnet’s post hoc tests compared to media control was significant p=0.0149 for CCL8 but not CXCL6).
Fig. 7. Model of chemokine systems governing recruitment of immune cell subsets to form interface dermatitis in cutaneous lupus.
1. Integration of WTA DSP, Olink and flow cytometry data reveals AIM2 pathway activation in keratinocytes proximal to inflammation. 2. Chemokines including CXCL6 and CCL8 can be induced downstream of this pathway. CD14+CD16+ myeloid cells express more CXCR1, and migrate best towards CXCL6, whereas CD14+CD16- myeloid cells express more CCR2 and migrate best towards CCL8. 3. The CXCR3 ligands CXCL9/10/11 are expressed by keratinocytes, but more strongly by CD45+ immune cell ROIs and T cells. Thus, keratinocyte/myeloid crosstalk can reinforce this system to optimally recruit lymphocytes to form interface dermatitis. Created with Biorender.com.
DISCUSSION
In this study, we sought to analyze chemokine orchestrators of T cell rich interface dermatitis in CLE. Chemokines can be released by keratinocytes in response to UV light, infection, or other environmental triggers (27). These chemokines can then recruit immune cells to the skin, which can lead to inflammation. One of the most highly upregulated chemokine families in CLE lesions is the CXCR3 chemokine family(6, 28, 29). CXCR3 binds to its interferon (IFN)-inducible ligands CXCL9, CXCL10 and CXCL11 to mediate leukocyte migration and function(30). IFN signaling has been postulated to drive the pathogenesis of all subtypes CLE(31). Further, CLE is a photosensitive disorder, and UV light induces upregulation of IFN to amplify CXCR3 ligand production in keratinocytes(17, 32, 33), linking the environmental insult to the recruitment of pathogenic immune cells in the context of CLE. The interaction between CXCR3-expressing T-cells and its ligands CXCL9, CXCL10 and CXCL11 have been associated with tissue damage in several CLE subtypes(8). A recent study described CXCL10 as a biomarker that can distinguish CLE patients from systemic lupus patients(34). Based on our data, we would hypothesize that CXCL11 may be a superior biomarker to CXCL10 in predicting disease activity in CLE patients due to both its expression level and chemotactic potency, though this would need to be validated in a larger patient cohort.
Examining chemokine ligand expression in our Olink dataset and in our ROIs, and chemokine receptor expression in our flow cytometry data, confirmed expression of key receptors and also identified potential novel mediators of recruitment in CLE. Based on our Olink and flow cytometry data, we hypothesize that CXCR1 is a key receptor for CXCL6 on proinflammatory CD14+CD16+ APCs, and could be a viable treatment target for CLE, especially given the role of CXCL6 in fibrosis which occurs in DLE and other forms of chronic CLE (35) as well as in renal interstitial fibrosis (36). CXCL6 is a particularly interesting target immunologically, given that it may be upregulated by double stranded RNA or IL1𝝱 (37). This is important, as we know from work by Rodriguez-Pla et al. that IFN priming alone is not sufficient to activate migratory programs in lupus monocytes (38), and from work by Billi et al. that CD14+CD16+ monocyte/dendritic cell populations are among the first cells recruited to the skin in CLE (26). Further studies are needed to understand epigenetic factors that result in increased CXCR1 expression in CD16+ myeloid cells.
We also identified CCL8 as a DEP between lesional and nonlesional blister fluid that preferentially recruits CD16- populations. CCL8 binds multiple receptors (39), of which we noted a trend towards higher expression of CCR2 but not CCR5 in CD14+ classical monocyte/dendritic cell populations. Previous studies have reported that CCR2 mediates basophil recruitment to the skin in CLE(40), and CCR2 is a predicted target for CLE in silico(41). CCR2 deficiency reduces kidney disease and prolongs survival in MRL/lpr mice (42), though it has also been reported to exacerbate a murine model of accelerated nephrotoxic nephritis (43). Elevated CCL8 was also reported to be expressed in the kidney in MRL/lpr mice, where it activated mesenchymal stem cells to produce immunosuppressive factors such as TGF-B1 (44). Future studies examining protective versus pathogenic effects of CCL8/CCR2+ monocyte/dendritic cell populations in the skin during CLE are warranted.
Other potential chemokine ligands and receptors were trending towards significance in our dataset, some of which have been described in the context of lupus and/or skin immunopathogenesis. For example, CCR5 can bind to CCL8 and is upregulated in lupus CD4+ T cells (8). CCR3 was reported to decrease relative to CCR5 in CLE CD4+ T cells (45). CCL25 and its receptor CCR9 have been implicated in Sjogren’s Syndrome pathogenesis (46), and CCL25 blockade prolongs skin allograft survival in mice (47), though the role of this chemokine axis in CLE has not yet been studied. We noted higher CC25 expression in SCLE CD45+ ROIs, compared to more CCL8 expression in DLE CD45+ ROIs, indicating the chemokine context for myeloid recruitment in these 2 clinical subtypes may differ. CCL28 is a ligand for CCR10, and SLE plasmablasts have been reported to exhibit enhanced migration towards CCL28 (48). CCR10 immunohistochemistry staining is stronger in CLE subtypes compared to healthy skin (8), though the functional consequences of the CCL28/CCR10 axis in CLE are unknown.
Our Olink dataset provides insight into novel protein biomarkers of CLE beyond chemokine ligands that have interesting implications in immunopathogenesis, including Flt3L, HGF, CD40 and CEACAM3. Flt3L is a dendritic cell growth and survival factor, and has been previously described in SLE(49). A recent study demonstrated generation of tolerogenic conventional DCs in SLE patients undergoing mesenchymal stem cell therapy(50). Thus, it is possible that the upregulation of Flt3L in skin is a compensatory mechanism. HGF, which as noted above can increase CXCR3 expression on T cells (11), is higher in SLE patients than healthy controls(51) and in patients with higher disease activity(52). HGF administration can prevent lupus nephritis in mice(53), potentially indicating a compensatory mechanism for HGF in CLE. CD40 overexpression causes autoimmune skin disease in mice(54), and increased CD40 expression has been reported in IHC of CLE lesions(55). Genome wide differential expression analysis identified CEACAM3 as a potential gene involved in the pathogenesis of lupus and lupus nephritis(56), though its role in skin disease has yet to be elucidated.
We also found differences in DLE and SCLE that have not previously been appreciated. For example, DLE lesional skin T cells express less B7-H3 than SCLE lesional skin as assessed by protein DSP. B7-H3, also known as CD276, is an immune checkpoint within the B7 molecular family that fine tunes immune responses (57). B7-H3 is abnormally expressed on tumor cells (58) and can also be induced on antigen-presenting cells (APCs), including dendritic cells (DCs) and macrophages (57). B7-H3 plays a pivotal role in mediating the suppressive effects of DCs on T cells and downregulating Th1 responses (59). Therefore, a relative reduction in B7-H3 in DLE as compared to SCLE fits with previously described IFNG signaling nodes identified in DLE by bulk RNA sequencing (9). Also of interest, recent findings by Sun et al. have shown that the serum concentrations of soluble B7-H3 (sB7-H3) in patients with Systemic Lupus Erythematosus (SLE) were notably lower than those in healthy individuals (60). In another study by Zheng et al., B7-H3 knockout (KO) mice exhibited significantly higher serum levels of anti-double-stranded DNA antibodies (anti-dsDNA Abs) when compared to control mice (wild-type C57BL/6 mice) (61). This underscores the pivotal role of B7-H3 in lupus and provides a biochemical foundation for considering the B7-H3 pathway as a potential and effective therapy for SLE and, based on our data, DLE. Further studies are needed to assess the myriad DEGs for DLE vs SCLE ROI comparisons that were identified in the WTA dataset.
While our study provides valuable insights into the immunopathogenesis of CLE and identifies novel chemokine ligand-receptor pairs that contribute to disease progression, it is essential to acknowledge the study’s limitations. First, our research predominantly focuses on a specific subset of chemokine ligand-receptor pairs and immune cell interactions that we were able to measure with current methodologies. CXCR1 is a classical receptor for neutrophil recruitment to inflamed tissues, and ligation of CXCR1 can enhance NET formation(62, 63). We were not able to recover neutrophils from blister fluid, and opted to use Ficoll gradients, which remove the bulk of granulocytes, to isolate PBMCs for chemotaxis assays. Future investigations should broaden the scope to encompass a more comprehensive landscape of potential signaling pathways and interactions that may influence CLE pathogenesis, including methodologies to specifically assess neutrophils and other granulocytes. Moreover, our study relies on a cross-sectional design, which limits our ability to capture the dynamic evolution of CLE over time. A longitudinal approach with serial assessments would offer a more subtle understanding of disease progression. Finally, our findings in different CLE subtypes and patient populations should be examined further with larger patient cohorts: in the future, we plan to expand our study to include a broader array of CLE phenotypes and incorporate larger and more diverse cohorts to provide a more comprehensive understanding of the disease spectrum. Moreover, in-depth mechanistic studies and the development of targeted interventions based on the identified chemokine and other pathways hold promise for improving therapeutic strategies in CLE management.
MATERIALS AND METHODS
Study design & subjects -
The main objective of this study was to characterize chemokine ligand:receptor pairs in CLE to better understand how they orchestrate the interface dermatitis reaction. We set out to answer the question: how are autoreactive T cells recruited to the dermal:epidermal junction in CLE? We hypothesized that, in addition to previously published literature about the CXCR3 chemokine axis in CLE (see for example (6, 8, 28, 29, 32)), additional non-redundant chemokine axes were present in the skin to govern immune cell recruitment. As our study progressed, additional literature was published regarding CD14+CD16+ cells in nonlesional CLE skin (26), which we queried in our datasets and performed functional chemotaxis assays. In using -omics approaches, we also sought to provide unbiased analyses that might yield new insights and targets, to perform dataset concordance analyses, to provide protein-level expression data in addition to previously published RNA-level expression data in the CLE field (see for example (9, 18, 21)), and to answer sub-questions including: can we identify DEGs and/or DEPs in the different major clinical subtypes of CLE?
This study was performed with internal review board (IRB) approved protocols at the UMass Chan Medical School (H-14848 and H00021295 for fresh blood and skin tissue; H00020503 for archival skin tissue), Yale University Institutional Review Board (Human Investigative Committee no. 15010105235 for archival skin tissue), and/or Dartmouth Hitchcock Medical Center (STUDY02001542 for PBMCs), and all samples were obtained with written informed consent and were de-identified before use in experiments. Inclusion/exclusion criteria were as follows: subjects with a diagnosis of cutaneous lupus erythematosus by clinical exam performed by a dermatologist were included. Subjects with recent onset of new lesions and objective clinical signs of activity, specifically erythema and/or hair loss/alopecia, were recruited to represent active disease. Patients on treatment were included in the study, as most lupus patients must remain on maintenance therapy.
Blister biopsies & blood collection -
Suction blisters (1 cm in diameter) were induced on the skin by using the Negative Pressure Instrument Model NP-4 (Electronic Diversities, Finksburg, MD) as previously described(10). Briefly, the suction chambers were applied to the skin with 10–15 mm Hg of negative pressure and a constant temperature of 40C. After blister formation (30–60min), the blister fluid was aspirated using a 1 mL insulin syringe. Cells within the blister fluid were pelleted at 330 x g for 5–10 minutes for cell staining and the supernatant was collected and frozen for future analysis. The cell pellet was resuspended in FACS buffer (1% FBS in PBS; Sigma Aldrich) and transferred to a FACS tube for flow staining. Venipuncture was performed using heparinized tubes, and peripheral blood mononuclear cells (PBMCs) were isolated following a Ficoll density gradient. Cells were washed once in RPMI (Sigma Millipore) and were resuspended in Cell staining buffer (Biolegend) prior to staining and fixation with Fluorofix buffer (Biolegend). These patient samples are noted in Table S1 and in the appropriate results sections, and data are deposited on FlowRepository.
Olink proteomics:
Blister fluid or plasma isolated from heparinized blood was centrifuged at 330 x g for 5 min and aliquoted into 96 well qPCR plates. Samples were shipped to Olink Proteomics Inc. (Cambridge, MA) for analysis in the inflammation and neuroexploratory panels. Blister fluids were analyzed using Olink Inflammation and Neuro Exploratory panels in a high throughput proteomics platform with next generation sequencing readouts. Olink data was analyzed using NPX software,Olink Insights Stat Analysis online tool (https://olinkproteomics.shinyapps.io/OlinkInsightsStatAnalysis/), and OlinkAnalyze package using Rstudio Version 1.3. DEPs are provided in Table S4. Data are deposited on GEO database under accession #GSE182302.
Histopathological samples:
Archival tissue from Skin biopsies for diagnostic purposes submitted to the pathology laboratory of dermatopathologist (AD) was used for these studies through an institutional review board (IRB)–approved protocol at UMass Chan (H00020503), and clinical features were re-reviewed by a board-certified dermatologist (MR). All samples were de-identified before use in experiments. Skin biopsies from patients with the diagnosis of ‘skin lupus’ including 5 DLE and 4 SCLE obtained between 2005–2013 were chosen. Three control tissues from healthy margins of skin cancer excisions were also selected from the biorepository from age and sex matched subjects. One DLE and one healthy margin sample were unable to be assessed because of being cut-off from the visualization region on the DSP machine, as well as one SCLE sample due to poor staining, with final n’s of 4 DLE biopsies, 4 SCLE biopsies from 3 patients, and 2 healthy margins. Inclusion criteria for CLE samples included interface dermatitis with perivascular lymphocytic infiltrate, increased dermal mucin on histomorphology and clinical findings consistent with CLE. These patient samples are noted in Table S2 and in the appropriate results sections.
Validation dataset analysis for DLE keratinocytes and immune cells:
A separate DSP project was conducted by our collaborators at Yale University School of Medicine. Staining of archived, de-identified human formalin-fixed paraffin-embedded (FFPE) tissues was approved by the Yale University Institutional Review Board (Human Investigative Committee no. 15010105235). This project used the cancer transcriptome atlas (CTA), which includes 1,800+ mRNA probe-based gene targets that cover important aspects of immune response, tumor biology and the microenvironment. We performed comparative analyses between this dataset, comprised of n=3 DLE and n=3 healthy control archival skin tissues, and our DLE keratinocyte and CD3 enriched ROI datasets as a validation cohort using BioVenn (64) to visualize overlapping DEGs. These patient samples are noted in Table S3 and in the appropriate results sections.
Chemotaxis experiments:
The migration of healthy or lupus donor peripheral blood mononuclear cells (PBMCs) was measured with a HTS Transwell® 96-well Permeable chemotaxis plate (Life science, Coring, Arizona, United states) with 5.0-μm pore filters. In brief, PBMCs were obtained from heparinized blood samples using SepMate™ PBMC isolation tubes (STEMCELL Technologies Inc, Massachusetts), according to the manufacturer’s instructions, and were suspended in endotoxin-free RPMI 1640 containing L-glutamine, 5nM HEPES (Corning, Arizona, United States) and 5% Fetal bovine serum (FBS; Sigma Heat Inactivated, US/HI origin). PBMCs were cultured overnight in chemotaxis media containing RPMI and 1% ultrapure BSA prior to use in assays.
The bottom wells were loaded with 200μl of chemotaxis media. We used purified CCL8 (50 ng/ml) CXCL9 (100 ng/ml), CXCL11 (200 ng/ml; all from Biolegend) or CXCL6 (50 ng/ml, R&D Systems), in bottom wells of 5.0-μm plate for chemotaxis of lymphocytes. Chemotaxis medium alone was used as a negative control. The top wells were loaded with 70 μl of PBMC with 1 × 105 cells, and plates were incubated at 37℃ for 2 h. The migrated cells in the bottom chamber of each well were collected and stained with CD3, CD8, CD4, CD14, CD16, CD19, CD56, HLA-DR and Live/dead. The number of each type of cells was normalized to count bright beads (Thermo Fisher) and assessed with flow cytometry (Cytek Aurora). The experiment was performed in triplicate or quadruplicate wells with at least 2 donors per chemokine/condition. In tandem, we performed flow cytometry on the input cells using a chemokine receptor staining panel. Blood donors used for chemotaxis assays and chemokine receptor staining are noted in Table S5 and in the appropriate results sections. Antibody information is provided in Table S6 and chemokine ligand information is provided in Table S7.
Sample Preparation and Digital Spatial Profiling (DSP):
The NanoString DSP technology provides spatial transcriptomics data using a combination of morphology and immunohistochemical staining(65, 66). Tissue sections at 5 µm thickness were placed on Superfrost Plus slides. Slides were deparaffinized and rehydrated by incubating for 3 × 5 min in CitriSolv, 2 × 5 min in 100% ethanol, 2 × 5 min in 95% ethanol, and 2 × 5 min in deionized water. In order to perform antigen retrieval processing, the slides were put in 1X Citrate Buffer (pH 6) into a pressure cooker at high temperature and high pressure for 15 min. After washing in five times in 1X TBS-T, blocking was performed by putting slides in humidity chambers and covering them with Buffer W (NanoString) for 1 h.
Slides were incubated overnight at 4°C overnight in a humidity chamber with 1.25 ug/ml anti-SMA, 5 ug/ml anti-CD45, and 1 ug/ml anti-CD3 antibodies. Slides were washed three times in 1X TBS-T post-staining and then post-fixed in 4% PFA. After two additional washes in 1X TBS-T to remove the fixative, nuclei were stained with 500 nM SYTO83 for 15 min at room temperature followed by one wash in 1X TBS-T before loading onto the GeoMx instrument.
Regions of interest (ROI) using the polygon selection tool were created and cell masking was performed using the morphology markers to define cell type as follows: CD45+CD3+ for T cells, CD45+CD3- for non-T cell-immune cells, CD45- and morphology for keratinocytes, CD45-SMA+ for endothelial cells.
For Whole Transcriptome Atlas (WTA), photocleaved oligos were then collected for sequencing. For protein profiling, we used 62 immune related biomarkers with the following panels: Immune Cell Profiling (18 markers), immuno-oncology (IO) Drug Target (10 markers), Immune Activation Status (8 markers), Immune Cell Typing (7 markers), Cell Death (10 markers), and PI3K/AKT Signaling (9 markers).
Library preparation and sequencing for WTA:
Oligos from each ROI were collected via microcapillary tube inspiration using the DSP platform robotic system and transferred into a microwell plate for use in whole transcriptome atlas (WTA) sequencing, which includes 18,000+ protein encoding mRNA targets that encompasses the whole human transcriptome. Collected oligos were amplified using a forward primer and a reverse primer that serve as Illumina i5/i7 unique dual indexing sequences to index ROI identity. After purifying the PCR products with AMPure XP beads (Beckman Coulter), they were sequenced. Library purity and concentration were measured with DNA Bioanalyzer chip (Agilent). Data are deposited on GEO Database under accession # GSE182825.
nCounter Readout for Protein DSP:
Protein modules were collected and run in the MAX machine at the Comparative Pathobiology and Genomics Shared Resources (“CPGSR”) in Cummings School of Veterinary Medicine at Tufts University.
Data processing and analysis:
Reads after sequencing, were trimmed, merged, and aligned to retrieve the identity of probes. PCR duplicates and duplicate reads were removed and the reads were converted to digital counts. The RNA sequencing saturation was sufficient and above 50%. After removing the outlier probes, the mean of the individual probe counts is considered as the reported count value. using GeoMX software (NanoString), 75% upper quartile (Q3) of the counts per ROI were selected after removing genes with zero counts. The Q3 normalized counts were compared across ROI and disease subtypes using several approaches. Overall differences were determined using linear mixed model analysis in GeoMX software. QC of data was performed using GeoMX software, and normalized counts were uploaded to ROSALIND software for comparison (as in Fig S1, https://rosalind.bio/, (67)). Specific groups of genes of interest, including chemokines/chemokine receptors, were analyzed as groups using GraphPad Prism version 9 with two-way ANOVA and Tukey’s post hoc tests. Median values for groups and ROIs generated in GeoMX software were used to visualize large gene sets as heatmaps using GraphPad Prism.
Reanalysis of GSE112943 dataset:
Microarray analysis of bulk RNA was previously performed on matched biobanked samples(18). To compare bulk RNA from spatial transcriptomics, we used Geo2R to download the toptable of significant genes for CCLE/DLE versus healthy and SCLE versus healthy, and compared DEGs with P<0.01 to DEGs from all ROIs pooled from the dataset presented in this manuscript using BioVenn (64).
Statistics -
Statistical analyses were performed using Prism software version 9 (GraphPad) for flow cytometry, NPX software for Olink, and GeoMX software for spatial datasets. For comparisons across disease states and cell types, two-way ANOVAs with main row effects comparisons were performed to identify highly significant biomarkers or cell types for further analysis. Differences were considered significant at a P value of less than 0.05. DSP statistical analyses using Linear Mixed Model (LMM) or unpaired T-tests were performed in GeoMX software with the resulting p-values that were adjusted for multiple comparisons with the Benjamini-Hochberg procedure (FDR, false discovery rate) and considering the size of ROIs and slide scanning batch effect. Box plots and volcano plots were generated in GraphPad Prism version 9. P-value or FDR <0.05 were considered significant, with P<0.01 as highly significant. We also used one-way ANOVA or two-way ANOVA for some comparisons in a group of genes between healthy and disease ROIs to answer specific hypotheses generated by -omics datasets.
Data visualization tools:
Gene Ontology (GO) analysis(68), Gene Set Enrichment Analysis (GSEA)(69) were performed using GeoMX software. The CellPhone Database(70) was queried for cell-cell communication pathway analyses in different ROI types. Venn diagrams were generated with BioVenn software(64), and hierarchical clustering were performed with ClustVis(71) and/or Morpheus software (https://software.broadinstitute.org/morpheus).
Supplementary Material
Fig S1. Quality Control for Digital Spatial Profiling (DSP) Whole Transcriptome Atlas (WTA) spatial transcriptomics.
Fig S2. Quality Control and Pathway Analysis for Regions of Interest (ROIs).
Fig S3. Validation of DSP dataset using historical microarray dataset and a cancer transcriptome atlas (CTA) dataset.
Fig S4. Validation of DLE dataset ROIs as compared to CTA.
Fig S5. Examining CD45+ and epidermal ROIs in DLE vs SCLE reveals pathways and DEGs unique to each CLE subtype.
Fig S6. Stability of chemokines in lesional and nonlesional samples over time.
Fig S7. Additional donors for chemotaxis exhibit similar migratory patterns.
Fig S8. Flow gating strategy for assessing myeloid populations.
Fig S9. Examination of CD14+CD16+ cells in blister biopsies
Fig S10. CCR5 is not enriched on CD14 vs CD16 expressing myeloid cells.
Table S1. Blister biopsy and blood donation patient info & characterization (UMass Chan).
Table S2. Archival CLE biopsies used in WTA Digital Spatial Profiling (UMass Chan).
Table S3. Validation archival CLE biopsies used in CTA Digital Spatial Profiling (Yale).
Table S4. Olink DEPs calculated by NPX software and 2-way ANOVA.
Table S5. Chemotaxis and chemokine receptor staining blood donor information (UMass Chan and Dartmouth Hitchcock).
Table S6. Antibody information & RRIDs.
Table S7. Chemokine information.
Acknowledgments:
We thank those who donated tissue for this study. We thank Celia Hartigan for clinical research support. We thank Alaina Ryan and the Comparative Pathobiology and Genomics Shared Resources (“CPGSR”) in Cummings School of Veterinary Medicine at Tufts University for running the protein DSP plate in the MAX machine. We used Biorender.com for illustrations. SS used ChatGPT to correct English grammar in the paragraphs he wrote for the first draft of the manuscript, and JMR edited for clarity and brevity as a native English speaker.
Funding Statement
Lupus Research Alliance Target Identification in Lupus Award (JMR)
Lupus Research Alliance Diversity Research Supplement Award (JEL)
Dermatology Foundation Women’s Health Career Development Award (JMR)
UMMS CCTS PPP grant (KA, JMR)
National Institute of Arthritis and Musculoskeletal and Skin Diseases Award Numbers 1R21AR079661-01 and 1R01AR08641-01A1 (SSG)
National Institute of Arthritis and Musculoskeletal and Skin Diseases Award K08 AR08777 (MDV)
National Institute of Arthritis and Musculoskeletal and Skin Diseases Award Numbers AR061437 and AR069114 (JEH)
Flow cytometry equipment used for this study is maintained by the UMass Chan Flow Cytometry Core Facility. The UMass Chan Medical School Center for Clinical and Translational Research was responsible for blood and biopsy collection and is supported by NIH Clinical and Translational Sciences Award UL1TR000161. Protein DSP was supported by the Sanderson Center for Optical Experimentation at UMass Chan (University of Massachusetts Medical School - UMass Chan - SCOPE, RRID:SCR_022721) and the CPGSR at Tufts Cummings School of Veterinary Medicine.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests: JMR is an inventor on patent application #63/478,900 “Diagnosis of skin diseases in veterinary and human patients” for CTCL. JEH & JMR are inventors on patent application #62489191, “Diagnosis and Treatment of Vitiligo” which covers targeting IL-15 and Trm for the treatment of vitiligo; and on patent application #15/851,651, “Anti-human CXCR3 antibodies for the Treatment of Vitiligo” which covers targeting CXCR3 for the treatment of vitiligo. JEH holds equity in Rheos Medicines and TeVido BioDevices; is a founder with equity of Villaris Therapeutics, Aldena Therapeutics, NIRA Biosciences, Vimela Therapeutics, and Klirna Therapeutics; has served as a consultant for Pfizer, Sanofi Genzyme, Incyte, Sun Pharmaceuticals, LEO Pharma, Dermavant, Temprian Therapeutics, AbbVie, Janssen, Almirall, Methuselah Health, Pandion, AnaptysBio, Avita, Aclaris Therapeutics, The Expert Institute, BiologicsMD, Boston Pharma, Sonoma Biotherapeutics, Two Biotech, Admirx, Frazier Management, 3rd Rock Ventrures, Gogen Therapeutics, Granular Therapeutics, Platelet Biogenesis, BridgeBio, Merck, Matchpoint Therapeutics, and Klirna; has served as an investigator for Pfizer, Sanofi Genzyme, Incyte, Sun Pharmaceuticals, LEO Pharma, Dermavant, Aclaris Therapeutics, GSK, Celgene, Dermira, and EMD Serono. LZ, ML & YL are employees of NanoString Technologies. MR is principal or co-investigator of studies sponsored by Pfizer, Biogen, AbbVie, Incyte, LEO Pharma, Abeona Therapeutics, Dermavant, and Target RWE; and MR provides consulting for Pfizer, Biogen, Incyte, Takeda, Inzen, ROME Therapeutics, Almirall, Medicxi, Related Sciences, and VisualDx. Remaining authors declare that they have no competing interests.
Data and materials availability:
Datasets related to this article have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (72) and are accessible through GEO Series accession numbers GSE182302 for Olink data, and GSE182825 for spatial transcriptomics. Flow data has been deposited on FlowRepository under accessions FR-FCM-Z4PL, FR-FCM-Z4PM, FR-FCM-Z4PN, FR-FCM-Z4PQ, FR-FCM-Z4PX, FR-FCM-Z6UN, FR-FCM-Z7ZP, FR-FCM-Z7ZQ for blister biopsies and blood immunophenotyping.
References:
- 1.Garelli C. J., Refat M. A., Nanaware P. P., Ramirez-Ortiz Z. G., Rashighi M., Richmond J. M., Current Insights in Cutaneous Lupus Erythematosus Immunopathogenesis. Front. Immunol. 11, 1353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz H. Y., Dutz J. P., Furukawa F., Goodfield M. J., Kuhn A., Lee L. A., Nyberg F., Szepietowski J. C., Sontheimer R. D., Werth V. P., From pathogenesis, epidemiology, and genetics to definitions, diagnosis, and treatments of cutaneous lupus erythematosus and dermatomyositis: a report from the 3rd International Conference on Cutaneous Lupus Erythematosus (ICCLE) 2013. J. Invest. Dermatol. 135, 7–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reich A., Werth V. P., Furukawa F., Kuhn A., Szczęch J., Samotij D., Szepietowski J. C., Treatment of cutaneous lupus erythematosus: current practice variations. Lupus 25, 964–972 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Chang A. Y., Werth V. P., Treatment of cutaneous lupus. Curr. Rheumatol. Rep. 13, 300–307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stannard J. N., Kahlenberg J. M., Cutaneous lupus erythematosus: updates on pathogenesis and associations with systemic lupus. Curr. Opin. Rheumatol. 28, 453–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel J., Zahn S., Mikus S., Wiechert A., Bieber T., Tüting T., The expression pattern of interferon-inducible proteins reflects the characteristic histological distribution of infiltrating immune cells in different cutaneous lupus erythematosus subsets. Br. J. Dermatol. 157, 752–757 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Wenzel J., Henze S., Wörenkämper E., Basner-Tschakarjan E., Sokolowska-Wojdylo M., Steitz J., Bieber T., Tüting T., Role of the Chemokine Receptor CCR4 and its Ligand Thymus- and Activation-Regulated Chemokine/CCL17 for Lymphocyte Recruitment in Cutaneous Lupus Erythematosus. J. Invest. Dermatol. 124, 1241–1248 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Méndez-Flores S., Hernández-Molina G., Azamar-Llamas D., Zúñiga J., Romero-Díaz J., Furuzawa-Carballeda J., Inflammatory chemokine profiles and their correlations with effector CD4 T cell and regulatory cell subpopulations in cutaneous lupus erythematosus. Cytokine 119, 95–112 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Berthier C. C., Tsoi L. C., Reed T. J., Stannard J. N., Myers E. M., Namas R., Xing X., Lazar S., Lowe L., Kretzler M., Gudjonsson J. E., Kahlenberg J. M., Molecular Profiling of Cutaneous Lupus Lesions Identifies Subgroups Distinct from Clinical Phenotypes. J. Clin. Med. Res. 8 (2019), doi: 10.3390/jcm8081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strassner J. P., Rashighi M., Ahmed Refat M., Richmond J. M., Harris J. E., Suction blistering the lesional skin of vitiligo patients reveals useful biomarkers of disease activity. J. Am. Acad. Dermatol. 76, 847–855.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komarowska I., Coe D., Wang G., Haas R., Mauro C., Kishore M., Cooper D., Nadkarni S., Fu H., Steinbruchel D. A., Pitzalis C., Anderson G., Bucy P., Lombardi G., Breckenridge R., Marelli-Berg F. M., Hepatocyte Growth Factor Receptor c-Met Instructs T Cell Cardiotropism and Promotes T Cell Migration to the Heart via Autocrine Chemokine Release. Immunity 42, 1087–1099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim B., Rothenberg M. E., Sun X., Bachert C., Artis D., Zaheer R., Deniz Y., Rowe P., Cyr S., Neuroimmune interplay during type 2 inflammation: Symptoms, mechanisms, and therapeutic targets in atopic diseases. J. Allergy Clin. Immunol. (2023), doi: 10.1016/j.jaci.2023.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierini R., Juruj C., Perret M., Jones C. L., Mangeot P., Weiss D. S., Henry T., AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 19, 1709–1721 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratsimandresy R. A., Indramohan M., Dorfleutner A., Stehlik C., The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell. Mol. Immunol. 14, 127–142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa Y., Yoshimoto T., Nakanishi K., Contribution of IL-18-induced innate T cell activation to airway inflammation with mucus hypersecretion and airway hyperresponsiveness. Int. Immunol. 18, 847–855 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Struyf S., Proost P., Vandercappellen J., Dempe S., Noyens B., Nelissen S., Gouwy M., Locati M., Opdenakker G., Dinsart C., Van Damme J., Synergistic up-regulation of MCP-2/CCL8 activity is counteracted by chemokine cleavage, limiting its inflammatory and anti-tumoral effects. Eur. J. Immunol. 39, 843–857 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Skopelja-Gardner S., An J., Tai J., Tanaka L., Sun X., The early local and systemic Type I interferon responses to ultraviolet B light exposure are cGAS dependent. Sci. Rep. (2020) (available at https://www.nature.com/articles/s41598-020-64865-w). [DOI] [PMC free article] [PubMed]
- 18.Ko W.-C. C., Li L., Young T. R., McLean-Mandell R. E., Deng A. C., Vanguri V. K., Dresser K., Harris J. E., Gene expression profiling in skin reveals strong similarities between subacute and chronic cutaneous lupus that are distinct from lupus nephritis. J. Invest. Dermatol. (2021), doi: 10.1016/j.jid.2021.04.030. [DOI] [PubMed] [Google Scholar]
- 19.Mande P., Zirak B., Ko W.-C., Taravati K., Bride K. L., Brodeur T. Y., Deng A., Dresser K., Jiang Z., Ettinger R., Fitzgerald K. A., Rosenblum M. D., Harris J. E., Marshak-Rothstein A., Fas ligand promotes an inducible TLR-dependent model of cutaneous lupus-like inflammation. J. Clin. Invest. 128, 2966–2978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garelli C. J., Wong N. B., Piedra-Mora C., Wrijil L. M., Scarglia G., David C. N., Almela R. M., Robinson N. A., Richmond J. M., Shared inflammatory and skin-specific gene signatures reveal common drivers of discoid lupus erythematosus in canines, humans and mice. Current Research in Immunology (2021), doi: 10.1016/j.crimmu.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dey-Rao R., Smith J. R., Chow S., Sinha A. A., Differential gene expression analysis in CCLE lesions provides new insights regarding the genetics basis of skin vs. systemic disease. Genomics 104, 144–155 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Jabbari A., Suárez-Fariñas M., Fuentes-Duculan J., Gonzalez J., Cueto I., Franks A. G., Krueger J. G., Dominant Th1 and Minimal Th17 Skewing in Discoid Lupus Revealed by Transcriptomic Comparison with Psoriasis. J. Invest. Dermatol. 134, 87–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wuyts A., Proost P., Lenaerts J. P., Ben-Baruch A., Van Damme J., Wang J. M., Differential usage of the CXC chemokine receptors 1 and 2 by interleukin-8, granulocyte chemotactic protein-2 and epithelial-cell-derived neutrophil attractant-78. Eur. J. Biochem. 255, 67–73 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Ottaviani C., Nasorri F., Bedini C., de Pità O., Girolomoni G., Cavani A., CD56brightCD16(−) NK cells accumulate in psoriatic skin in response to CXCL10 and CCL5 and exacerbate skin inflammation. Eur. J. Immunol. 36, 118–128 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ng Y. Y., Tay J. C. K., Wang S., CXCR1 Expression to Improve Anti-Cancer Efficacy of Intravenously Injected CAR-NK Cells in Mice with Peritoneal Xenografts. Mol Ther Oncolytics 16, 75–85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billi A. C., Ma F., Plazyo O., Gharaee-Kermani M., Wasikowski R., Hile G. A., Xing X., Yee C. M., Rizvi S. M., Maz M. P., Berthier C. C., Wen F., Tsoi L. C., Pellegrini M., Modlin R. L., Gudjonsson J. E., Kahlenberg J. M., Nonlesional lupus skin contributes to inflammatory education of myeloid cells and primes for cutaneous inflammation. Sci. Transl. Med. 14, eabn2263 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y., Tsoi L. C., Billi A. C., Ward N. L., Harms P. W., Zeng C., Maverakis E., Kahlenberg J. M., Gudjonsson J. E., Cytokinocytes: the diverse contribution of keratinocytes to immune responses in skin. JCI Insight 5 (2020), doi: 10.1172/jci.insight.142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flier J., Boorsma D. M., van Beek P. J., Nieboer C., Stoof T. J., Willemze R., Tensen C. P., Differential expression of CXCR3 targeting chemokines CXCL10, CXCL9, and CXCL11 in different types of skin inflammation. J. Pathol. 194, 398–405 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Wenzel J., Wörenkämper E., Freutel S., Henze S., Haller O., Bieber T., Tüting T., Enhanced type I interferon signalling promotes Th1-biased inflammation in cutaneous lupus erythematosus. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland 205, 435–442 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Groom J. R., Luster A. D., CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 89, 207–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson E. S., Werth V. P., The role of cytokines in the pathogenesis of cutaneous lupus erythematosus. Cytokine 73, 326–334 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Meller S., Winterberg F., Gilliet M., Müller A., Lauceviciute I., Rieker J., Neumann N. J., Kubitza R., Gombert M., Bünemann E., Others, Ultraviolet radiation--induced injury, chemokines, and leukocyte recruitment: an amplification cycle triggering cutaneous lupus erythematosus. Arthritis & Rheumatism 52, 1504–1516 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Scholtissek B., Zahn S., Maier J., Klaeschen S., Braegelmann C., Hoelzel M., Bieber T., Barchet W., Wenzel J., Immunostimulatory Endogenous Nucleic Acids Drive the Lesional Inflammation in Cutaneous Lupus Erythematosus. J. Invest. Dermatol. 137, 1484–1492 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zhu J. L., O’Brien J. C., Barber G., Saxena R., Chong B. F., Elevated serum levels of CXC motif chemokine ligand 10 can distinguish systemic lupus erythematosus patients from cutaneous lupus erythematosus patients. J. Am. Acad. Dermatol. (2021) (available at https://www.sciencedirect.com/science/article/pii/S0190962221008355/pdf?md5=0cec5469b9182262b9aad38462ef9b59&pid=1-s2.0-S0190962221008355-main.pdf). [DOI] [PMC free article] [PubMed]
- 35.Dai C.-L., Yang H.-X., Liu Q.-P., Rahman K., Zhang H., CXCL6: A potential therapeutic target for inflammation and cancer. Clin. Exp. Med. (2023), doi: 10.1007/s10238-023-01152-8. [DOI] [PubMed] [Google Scholar]
- 36.Sun M.-Y., Wang S.-J., Li X.-Q., Shen Y.-L., Lu J.-R., Tian X.-H., Rahman K., Zhang L.-J., Nian H., Zhang H., CXCL6 Promotes Renal Interstitial Fibrosis in Diabetic Nephropathy by Activating JAK/STAT3 Signaling Pathway. Front. Pharmacol. 10, 224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Wuyts A., Struyf S., Gijsbers K., Schutyser E., Put W., Conings R., Lenaerts J.-P., Geboes K., Opdenakker G., Menten P., Proost P., Van Damme J., The CXC Chemokine GCP-2/CXCL6 Is Predominantly Induced in Mesenchymal Cells by Interleukin-1β and Is Down-Regulated by Interferon-γ: Comparison with Interleukin-8/CXCL8. Lab. Invest. 83, 23–34 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Pla A., Patel P., Maecker H. T., Rossello-Urgell J., Baldwin N., Bennett L., Cantrell V., Baisch J., Punaro M., Gotte A., Nassi L., Wright T., Palucka A. K., Banchereau J., Pascual V., IFN priming is necessary but not sufficient to turn on a migratory dendritic cell program in lupus monocytes. J. Immunol. 192, 5586–5598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong X., Gong W., Kuhns D. B., Ben-Baruch A., Howard O. M., Wang J. M., Monocyte chemotactic protein-2 (MCP-2) uses CCR1 and CCR2B as its functional receptors. J. Biol. Chem. 272, 11682–11685 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Pan Q., Feng Y., Peng Y., Zhou H., Deng Z., Li L., Han H., Lin J., Shi L., Wang S., An N., Yang C., Liu H.-F., Basophil Recruitment to Skin Lesions of Patients with Systemic Lupus Erythematosus Mediated by CCR1 and CCR2. Cell. Physiol. Biochem. 43, 832–839 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Dey-Rao R., Sinha A. A., In silico Analyses of Skin and Peripheral Blood Transcriptional Data in Cutaneous Lupus Reveals CCR2-A Novel Potential Therapeutic Target. Front. Immunol. 10, 640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Lema G. P., Maier H., Franz T. J., Escribese M., Chilla S., Segerer S., Camarasa N., Schmid H., Banas B., Kalaydjiev S., Busch D. H., Pfeffer K., Mampaso F., Schlo D. [Combining Diaeresis]ndorff, B. Luckow, Chemokine Receptor Ccr2 Deficiency Reduces Renal Disease and Prolongs Survival in MRL/lpr Lupus-Prone Mice. J. Am. Soc. Nephrol. 16, 3592 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Bird J. E., Giancarli M. R., Kurihara T., Kowala M. C., Valentine M. T., Gitlitz P. H., Pandya D. G., French M. H., Durham S. K., Increased severity of glomerulonephritis in C-C chemokine receptor 2 knockout mice. Kidney Int. 57, 129–136 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Kim H. S., Lee H. K., Kim K., Ahn G. B., Kim M. S., Lee T. Y., Son D. J., Kim Y., Hong J. T., Han S.-B., Mesenchymal stem cells enhance CCL8 expression by podocytes in lupus-prone MRL.Faslpr mice. Sci. Rep. 13, 1–11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freutel S., Gaffal E., Zahn S., Bieber T., Tüting T., Wenzel J., Enhanced CCR5+/CCR3+ T helper cell ratio in patients with active cutaneous lupus erythematosus. Lupus 20, 1300–1304 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Blokland S., Innovative techniques to investigate salivary gland immunopathology and exploration of the CCL25 CCR9 T helper cell axis in Sjögren’s syndrome (Utrecht University, 2018; https://dspace.library.uu.nl/handle/1874/371371). [Google Scholar]
- 47.Li J., Xiong T., Xiao R., Xiong A., Chen J., Altaf E., Zheng Y., Zhu G., He Y., Tan J., Anti-CCL25 antibody prolongs skin allograft survival by blocking CCR9 expression and impairing splenic T-cell function. Arch. Immunol. Ther. Exp. 61, 237–244 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Mei H. E., Hahne S., Redlin A., Hoyer B. F., Wu K., Baganz L., Lisney A. R., Alexander T., Rudolph B., Dörner T., Plasmablasts With a Mucosal Phenotype Contribute to Plasmacytosis in Systemic Lupus Erythematosus. Arthritis Rheumatol 69, 2018–2028 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Gill M. A., Blanco P., Arce E., Pascual V., Banchereau J., Palucka A. K., Blood dendritic cells and DC-poietins in systemic lupus erythematosus. Hum. Immunol. 63, 1172–1180 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Yuan X., Qin X., Wang D., Zhang Z., Tang X., Gao X., Chen W., Sun L., Mesenchymal stem cell therapy induces FLT3L and CD1c+ dendritic cells in systemic lupus erythematosus patients. Nat. Commun. 10, 2498 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robak E., Woźniacka A., Sysa-Jedrzejowska A., Stepień H., Robak T., Serum levels of angiogenic cytokines in systemic lupus erythematosus and their correlation with disease activity. Eur. Cytokine Netw. 12, 445–452 (2001). [PubMed] [Google Scholar]
- 52.Liu Y., Zheng M., Yin W.-H., Zhang B., Relationship of serum levels of HGF and MMP-9 with disease activity of patients with systemic lupus erythematosus. Zhejiang Da Xue Xue Bao Yi Xue Ban 33, 340–3, 348 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Kuroiwa T., Iwasaki T., Imado T., Sekiguchi M., Fujimoto J., Sano H., Hepatocyte growth factor prevents lupus nephritis in a murine lupus model of chronic graft-versus-host disease. Arthritis Res. Ther. 8, R123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehling A., Loser K., Varga G., Metze D., Luger T. A., Schwarz T., Grabbe S., Beissert S., Overexpression of CD40 ligand in murine epidermis results in chronic skin inflammation and systemic autoimmunity. J. Exp. Med. 194, 615–628 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caproni M., Torchia D., Antiga E., Giomi B., Mercuri M., Volpi W., Fabbri P., The CD40/CD40 ligand system in the skin of patients with subacute cutaneous lupus erythematosus. J. Rheumatol. 34, 2412–2416 (2007). [PubMed] [Google Scholar]
- 56.AlFadhli S., Ghanem A. A. M., Nizam R., Genome-wide differential expression reveals candidate genes involved in the pathogenesis of lupus and lupus nephritis. Int. J. Rheum. Dis. 19, 55–64 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Yi K. H., Chen L., Fine tuning the immune response through B7-H3 and B7-H4. Immunol. Rev. 229, 145–151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castellanos J. R., Purvis I. J., Labak C. M., Guda M. R., Tsung A. J., Velpula K. K., Asuthkar S., B7-H3 role in the immune landscape of cancer. Am. J. Clin. Exp. Immunol. 6, 66–75 (2017). [PMC free article] [PubMed] [Google Scholar]
- 59.Suh W.-K., Gajewska B. U., Okada H., Gronski M. A., Bertram E. M., Dawicki W., Duncan G. S., Bukczynski J., Plyte S., Elia A., Wakeham A., Itie A., Chung S., Da Costa J., Arya S., Horan T., Campbell P., Gaida K., Ohashi P. S., Watts T. H., Yoshinaga S. K., Bray M. R., Jordana M., Mak T. W., The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol. 4, 899–906 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Sun J., Lai H., Shen D., Wu P., Yang J., Sun Z., Guo Y., Reduced sB7-H3 Expression in the Peripheral Blood of Systemic Lupus Erythematosus Patients. J Immunol Res 2017, 5728512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng X., Xiao Z. X., Hu L., Fang X., Luo L., Chen L., Dendritic cell-associated B7-H3 suppresses the production of autoantibodies and renal inflammation in a mouse model of systemic lupus erythematosus. Cell Death Dis. 10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teijeira Á., Garasa S., Gato M., Alfaro C., Migueliz I., Cirella A., de Andrea C., Ochoa M. C., Otano I., Etxeberria I., Andueza M. P., Nieto C. P., Resano L., Azpilikueta A., Allegretti M., de Pizzol M., Ponz-Sarvisé M., Rouzaut A., Sanmamed M. F., Schalper K., Carleton M., Mellado M., Rodriguez-Ruiz M. E., Berraondo P., Perez-Gracia J. L., Melero I., CXCR1 and CXCR2 Chemokine Receptor Agonists Produced by Tumors Induce Neutrophil Extracellular Traps that Interfere with Immune Cytotoxicity. Immunity 52, 856–871.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Florey O. J., Johns M., Esho O. O., Mason J. C., Haskard D. O., Antiendothelial cell antibodies mediate enhanced leukocyte adhesion to cytokine-activated endothelial cells through a novel mechanism requiring cooperation between FcγRIIa and CXCR1/2. Blood 109, 3881–3889 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Hulsen T., de Vlieg J., Alkema W., BioVenn--a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 1–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toki M. I., Merritt C. R., Wong P. F., Smithy J. W., High-Plex Predictive Marker Discovery for Melanoma Immunotherapy–Treated Patients Using Digital Spatial Profiling. Clin. Cancer Res. (2019) (available at https://clincancerres.aacrjournals.org/content/25/18/5503.abstract?casa_token=-0FBFJL_rYwAAAAA:-CU2EZjsJeFHL5ZpArZYsvJH0WTQTqbHWN8brtTlrWeaSzNu8PrqP1ZN3B7l7hpBRsNuSVsBZUuUZD8N). [DOI] [PMC free article] [PubMed]
- 66.Merritt C. R., Ong G. T., Church S. E., Barker K., Danaher P., Geiss G., Hoang M., Jung J., Liang Y., McKay-Fleisch J., Nguyen K., Norgaard Z., Sorg K., Sprague I., Warren C., Warren S., Webster P. J., Zhou Z., Zollinger D. R., Dunaway D. L., Mills G. B., Beechem J. M., Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Perkins J. R., Dawes J. M., McMahon S. B., Bennett D. L. H., Orengo C., Kohl M., ReadqPCR and NormqPCR: R packages for the reading, quality checking and normalisation of RT-qPCR quantification cycle (Cq) data. BMC Genomics 13, 296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin D., Brun C., Remy E., Mouren P., Thieffry D., Jacq B., GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 5, R101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efremova M., Vento-Tormo M., Teichmann S. A., Vento-Tormo R., CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Metsalu T., Vilo J., ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43, W566–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Quality Control for Digital Spatial Profiling (DSP) Whole Transcriptome Atlas (WTA) spatial transcriptomics.
Fig S2. Quality Control and Pathway Analysis for Regions of Interest (ROIs).
Fig S3. Validation of DSP dataset using historical microarray dataset and a cancer transcriptome atlas (CTA) dataset.
Fig S4. Validation of DLE dataset ROIs as compared to CTA.
Fig S5. Examining CD45+ and epidermal ROIs in DLE vs SCLE reveals pathways and DEGs unique to each CLE subtype.
Fig S6. Stability of chemokines in lesional and nonlesional samples over time.
Fig S7. Additional donors for chemotaxis exhibit similar migratory patterns.
Fig S8. Flow gating strategy for assessing myeloid populations.
Fig S9. Examination of CD14+CD16+ cells in blister biopsies
Fig S10. CCR5 is not enriched on CD14 vs CD16 expressing myeloid cells.
Table S1. Blister biopsy and blood donation patient info & characterization (UMass Chan).
Table S2. Archival CLE biopsies used in WTA Digital Spatial Profiling (UMass Chan).
Table S3. Validation archival CLE biopsies used in CTA Digital Spatial Profiling (Yale).
Table S4. Olink DEPs calculated by NPX software and 2-way ANOVA.
Table S5. Chemotaxis and chemokine receptor staining blood donor information (UMass Chan and Dartmouth Hitchcock).
Table S6. Antibody information & RRIDs.
Table S7. Chemokine information.
Data Availability Statement
Datasets related to this article have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (72) and are accessible through GEO Series accession numbers GSE182302 for Olink data, and GSE182825 for spatial transcriptomics. Flow data has been deposited on FlowRepository under accessions FR-FCM-Z4PL, FR-FCM-Z4PM, FR-FCM-Z4PN, FR-FCM-Z4PQ, FR-FCM-Z4PX, FR-FCM-Z6UN, FR-FCM-Z7ZP, FR-FCM-Z7ZQ for blister biopsies and blood immunophenotyping.