SUMMARY
In response to antigens, B cells undergo affinity maturation and class switching mediated by activation-induced cytidine deaminase (AID) in germinal centers (GCs) of secondary lymphoid organs, but uncontrolled AID activity can precipitate autoimmunity and cancer. The regulation of GC antibody diversification is of fundamental importance but not well understood. We found that autoimmune regulator (AIRE), the molecule essential for T cell tolerance, is expressed in GC B cells in a CD40-dependent manner, interacts with AID and negatively regulates antibody affinity maturation and class switching by inhibiting AID function. AIRE deficiency in B cells caused altered antibody repertoire, increased somatic hypermutations, elevated autoantibodies to T helper 17 effector cytokines and defective control of skin Candida albicans. These results define a GC B cell checkpoint of humoral immunity and illuminate new approaches of generating high-affinity neutralizing antibodies for immunotherapy.
Keywords: Humoral immunity, checkpoint, B cell, antibody diversification, germinal center, AIRE, AID, immunodeficiency, autoimmunity, immunotherapy
INTRODUCTION
A properly diversified and selected repertoire of antibodies is essential for effective immune defense against diverse pathogens as well as the prevention of autoimmune diseases. After V(D)J recombination in the bone marrow (BM) that generates a primary antibody repertoire, B lymphocytes enter GCs of secondary lymphoid organs, such as the lymph nodes, spleen, tonsils and mucosa-associated lymphoid tissues, to further diversify their antibody repertoire in response to antigens by undergoing class switch recombination (CSR) and somatic hypermutation (SHM), both of which are mediated by the DNA-cytosine deaminase AID (Muramatsu et al., 2000; Revy et al., 2000). SHM introduces point mutations in immunoglobulin (Ig) variable region exons for selection of higher-affinity antibody clones by antigens, whereas CSR replaces Ig constant region exons encoding IgM and IgD with those encoding IgG, IgA, or IgE to provide antibodies with new effector functions (Murphy and Weaver, 2016b). However, aberrant AID activity in B cells can cause mutations in non-Ig loci to precipitate cancer (Casellas et al., 2016), and AID-mediated GC reaction can generate autoreactive antibodies to drive many autoimmune diseases (Vinuesa et al., 2009). The absence of B cell lymphomas and overt autoimmune diseases in healthy individuals amidst ongoing humoral immune responses reflect the existence of physiological mechanisms that restrain AID-mediated antibody diversification in GC B cells. The details of these mechanisms are of fundamental importance but not fully understood.
Humoral immunity is regulated by cell-mediated immunity, in which the molecule AIRE induces the expression of peripheral tissue-specific antigens (TSAs) in medullary epithelial cells (mTECs) and B cells in the thymus to promote the negative selection of self-reactive T cells or their conversion into regulatory T (Treg) cells (Anderson et al., 2002; Malchow et al., 2013; Yamano et al., 2015). AIRE is also expressed in specialized extrathymic cells (eTACs) that can inactivate self-reactive T cells in the periphery (Gardner et al., 2008; Gardner et al., 2013). Loss-of-function mutations in the AIRE gene cause autoimmune polyglandular syndrome type 1 (APS-1) (Finnish-German, 1997; Nagamine et al., 1997) associated with organ-specific autoimmunity, aberrant production of autoantibodies and increased susceptibility to mucocutaneous infection by Candida albicans, an otherwise innocuous commensal microbe in humans. Mysteriously, APS-1 patients can produce high-affinity neutralizing antibodies against T helper 17 (TH17) effector cytokines, which has been suggested to negatively impact anti-fungal immune defense (Kisand et al., 2010; Meyer et al., 2016; Puel et al., 2010). In light of these findings, we sought to determine whether AIRE has a B cell-intrinsic role in regulating peripheral antibody diversification.
Here we show that AIRE is expressed in GC B cells in a CD40-dependent manner, interacts with AID, and negatively regulates AID-mediated peripheral antibody diversification. AIRE-deficient mouse B cells undergo elevated class switching and affinity maturation after antigenic stimulation, which correlates with enhanced generation of genomic uracil, elevated Ig SHM, augmented AID targeting to Ig switch (S) regions and increased interaction of AID with transcriptionally stalled RNA polymerase II (Pol II). In addition, naive B cells of APS-1 patients undergo increased CSR upon stimulation. Mice with AIRE deficiency in B cells have elevated levels of autoantibodies against T helper 17 (TH17) effector cytokines and heightened skin C. albicans burden after infection, which recapitulates the symptoms of APS-1 patients. Our results define a previously unknown but crucial B cell-intrinsic AIRE-mediated GC checkpoint of peripheral antibody diversification that limits autoimmunity.
RESULTS
GC B Cells Express AIRE
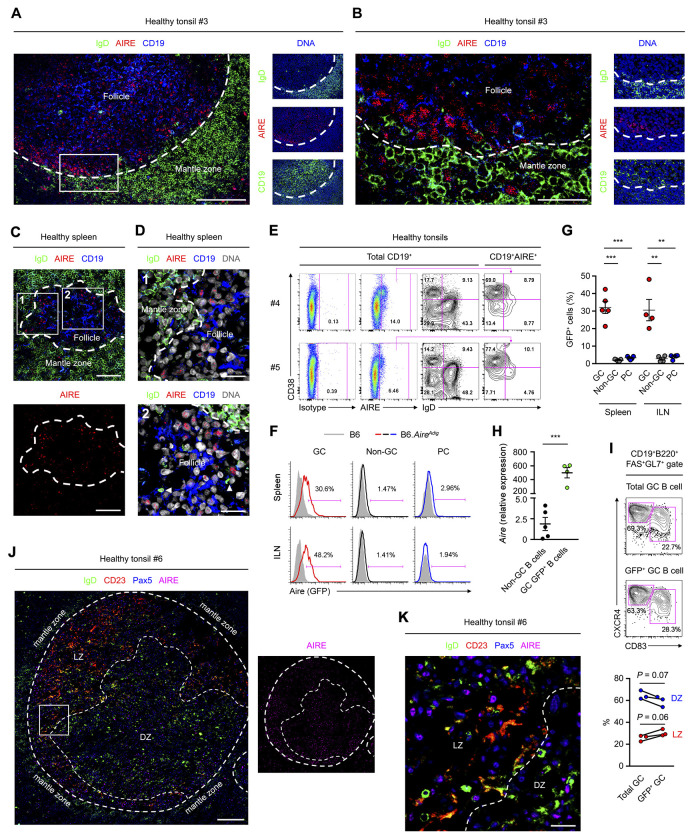
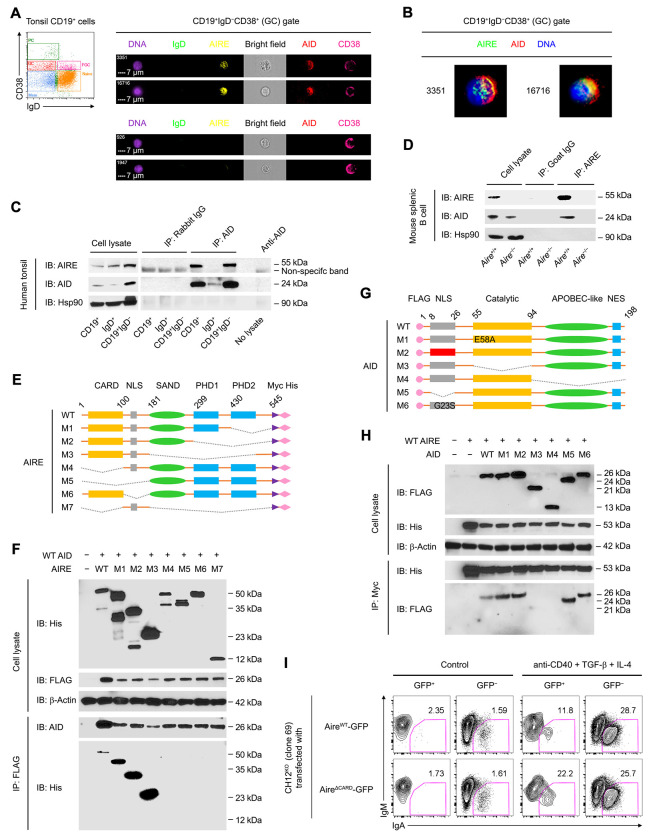
Secondary lymphoid organs are the major sites for peripheral antibody diversification (Murphy and Weaver, 2016a). To determine any roles of AIRE in peripheral antibody diversification, we first examined AIRE expression in B cells of human secondary lymphoid organs using an antibody that detects AIRE in the nuclei of mTECs (Figure S1A). We found many IgD−CD19+ or IgD−Pax5+ B cells inside tonsillar and splenic follicles that harbored nuclear AIRE (Figure 1A–D, Figure S1B and C). Follicular AIRE+ B cells expressed the GC B cell-associated molecule Bcl-6 (Figure S1C). In contrast, tonsillar and splenic IgD+ B cells in the mantle zone and IgD+ plasmablasts in GCs and extrafollicular areas (Chen et al., 2009) expressed little or no AIRE (Figure S1B and Figure 1D). Peripheral blood IgD+CD27− or CD24+CD38lo naive, IgD+CD27+ circulating marginal zone, IgD−CD27+ or CD24hiCD38− memory, IgD−CD27− atypical memory and CD24hiCD38hi transitional B cells as well as CD24−CD38hi plasma cells (PCs) did not express AIRE either (Figure S1D). Consistent with their follicular localization, tonsillar AIRE+ B cells were mostly IgD−CD38+ GC B cells, with a small fraction being IgD+CD38+ founder GC (FGC) or IgD−CD38− memory B cells (Figure 1E).
Figure 1. GC B Cells Express AIRE.
(A and B) Immunofluorescence analysis of the tonsillar tissue of a healthy donor for IgD, CD19, AIRE and DAPI-stained DNA. The dotted line outlines the follicles. Bars: 100 μm (A) or 25 μm (B).
(C and D) Immunofluorescence analysis of tissues of a healthy donor for IgD, AIRE, CD19 and DNA. Arrow heads indicate follicular IgD+ plasmablasts. Dotted lines mark the boundary between follicular mantle zone and the follicle. Bars: 40 μm (C) and 15 μm (D).
(E) Flow cytometric analysis of AIRE expression in tonsillar total CD19+ B cells, IgD+CD38− naive B cells, IgD+CD38+ founder GC (FGC) B cells, IgD−CD38+ GC B cells and IgD−CD38− memory B cells. The data represent 5 donors.
(F and G) Flow cytometric and statistical analyses of Aire (GFP) expression in splenic and ILN viable CD19+B220+FAS+GL7+ GC B cells, CD19+B220+FAS−GL7− non-GC B cells and CD19loB220loCD138+ PCs of B6 mice (shaded histograms, n = 5 for spleen and n = 4 for ILN) or B6.AireAdig mice (colored histograms, n = 5 for spleen and n = 4 for ILN) after 4 i.p. immunizations with NP32-KLH with CFA and subsequently with IFA. Data are represented as mean ± SEM. **P < 0.01, ***P < 0.001, by 1-way ANOVA with Tukey’s post hoc test.
(H) qPCR analysis of Aire transcript levels in CD19+B220+FAS−GL7− non-GC B cells (n = 5) and CD19+B220+FAS+GL7+GFP+ Aire-expressing GC B cells (n = 4) of AireAdig mice after 1 i.p. immunization with SRBC and CFA. Data are represented as mean ± SEM. ***P < 0.001, by 2-tailed unpaired t-test.
(I) Flow cytometric and statistical analyses of CD83+CXCR4lo LZ and CD83−CXCR4hi DZ B cells in splenic total GC and GFP+ GC B cells of immunized B6.AireAdig mice, by 2-tailed paired t-test.
(J and K) Immunofluorescence analysis of the tonsillar tissue of a healthy donor for IgD, CD23, Pax5 and AIRE. The dotted line outlines the follicles and delineates the border between LZ and DZ. Bars: 200 μm (J) or 30 μm (K).
Similar to human B cells, AIRE was found in B cells in the splenic follicles of immunized mice (Figure S1E and F). Consistently, in the AireAdig reporter mice (Gardner et al., 2008) after intraperitoneal immunizations, B cell AIRE expression was detected in FAS+GL7+ GC B cells in the spleen, inguinal lymph nodes (ILNs), mesenteric lymph nodes (MLNs) and Peyer’s patches (PPs) and in thymic B cells, but not in FAS−GL7− non-GC B cells or CD138+ PCs in these tissues, or in peripheral blood or peritoneal B cells (Figure 1F and G, Figure S1G–J). Consistently, Aire transcript level was markedly higher in splenic GFP+ GC B cells than non-GC B cells (Figure 1H). Furthermore, AIRE was detected in both CXCR4+CD83− dark zone (DZ) and CXCR4loCD83+ light zone (LZ) B cells in mouse spleens (Figure 1I) and DZ and LZ B cells in human tonsils (Figure 1J and K). Altogether, these data indicate that AIRE expression in GC B cells is a specific and conserved characteristic of human and mouse secondary lymphoid organs.
Follicular B Cell AIRE Expression Requires CD40 Signaling
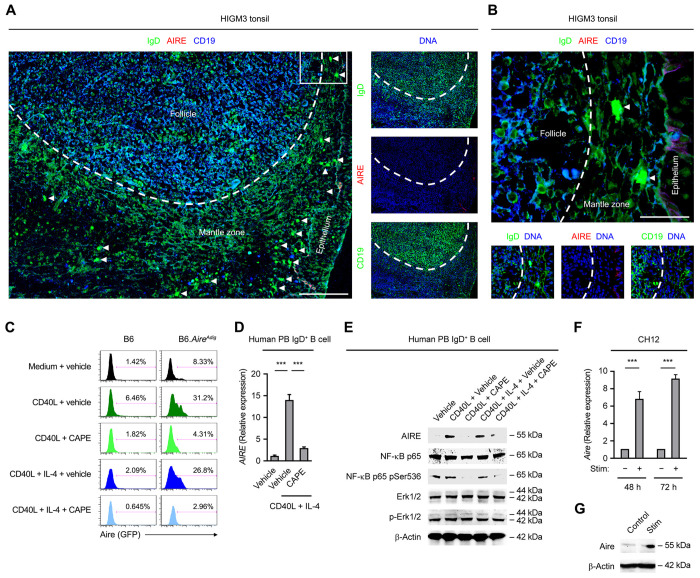
The induction of T cell-dependent GC B cell responses in secondary lymphoid organs involves B cell antigen presentation to primed cognate T helper (TH) cells and clonal proliferation upon receiving TH cell signals, such as CD40-ligand (CD40L) and interleukin (IL)-4 (Liu et al., 1989; Yusuf et al., 2010). CD40 signaling was previously reported to promote AIRE expression by mTECs and thymic B cells (Akiyama et al., 2008; Yamano et al., 2015). To determine the regulation of AIRE expression in follicular B cells, we examined the tonsillar tissue of a patient with the rare primary immunodeficiency (PID) Hyper-IgM Syndrome type 3 (HIGM3), which is caused by loss-of-function mutations in the CD40 gene (Durandy et al., 2005). In contrast to the prominent AIRE expression in tonsillar follicular B cells of healthy subjects (Figure 1A and B, Figure S1B and C), the tonsil of the HIGM3 patient harbored enlarged follicles containing B cells that had no expression of AIRE and failed to downregulate cell surface IgD (Figure 2A and B). Of note, tonsillar IgD plasmablasts were absent from the follicles but still present in the extrafollicular area, which is consistent with the existence of T cell-independent pathways for their generation as we reported previously (Chen et al., 2009), and indicates a specific shutdown of the T cell-dependent GC antibody diversification machinery. These results demonstrate a requirement for CD40 signaling in follicular B cell AIRE expression in vivo.
Figure 2. Follicular B Cell AIRE Expression Requires CD40 Signaling.
(A and B) Immunofluorescence analysis of tonsillar tissues of a HIGM3 patient for IgD, AIRE, CD19 and DNA. The area outlined in A is shown with a higher magnification in B. The dotted line outlines the follicles. Bars: 100 μm (A) or 25 μm (B).
(C) Flow cytometric analysis of Aire (GFP) expression in splenic B cells of a B6 or B6.AireAdig mouse treated for 3 d with medium or CD40L with or without IL-4 in the absence (vehicle) or presence of CAPE. The data represent the results from 3 B6 and 3 B6.AireAdig mice.
(D and E) qRT-PCR and Western Blot analyses of AIRE transcript and protein levels, the protein levels of total and Ser536-phosphorylated NF-κB p65, as well as total and Thr202/Tyr204-phosphorylated Erk1/2 in human peripheral blood IgD+ B cells treated with medium or CD40L, or CD40L and IL-4, in the presence of vehicle or CAPE for 3 d. Data are represented as mean ± SEM. ***P < 0.001, by 2-tailed unpaired t-test.
(F and G) qRT-PCR and Western Blot analyses of Aire transcript and Aire protein levels in mouse CH12 cells treated with anti-CD40, TGF-β and 100 ng/ml IL-4 for 3 d. Data are represented as mean ± SEM. P < 0.001, by 2-tailed unpaired t-test.
To further ascertain a role of CD40 signaling in promoting B cell AIRE expression, we treated mouse splenic naive resting B cells and human peripheral blood IgD+ mature naive B cells in vitro with CD40L alone or in combination with IL-4. Consistent with the above in vivo data, these B cells expressed AIRE protein and transcript upon CD40L stimulation (Figure 2C–E). AIRE induction was abrogated by caffeic acid phenethyl ester (CAPE), a selective inhibitor of nuclear factor-kappa B (NF-κB) (Natarajan et al., 1996), a transcription factor activated by CD40 (Figure 2C–E). Furthermore, AIRE transcript and protein were also induced in the mouse B cell line CH12 (Nakamura et al., 1996) upon anti-CD40 stimulation (Figure 2F and G). Collectively, these results show that CD40 signaling promotes AIRE expression in GC B cells in vivo and in B cells and cell lines in vitro.
AIRE in B Cells Inhibits CSR and SHM
As GC is the site of antibody diversification, including CSR and SHM (Murphy and Weaver, 2016b), we sought to determine the B cell-intrinsic function of AIRE in CSR and SHM. We employed several alternative and independent experimental approaches to achieve this goal.
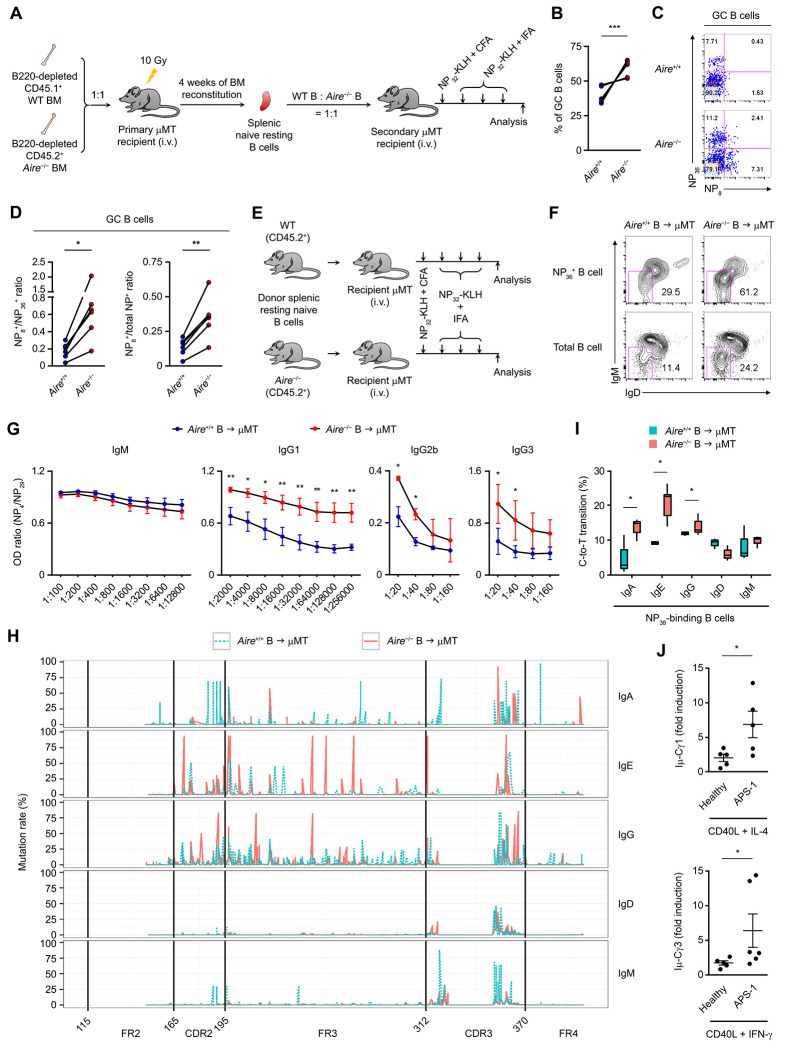
Firstly, we engrafted lethally irradiated B cell-deficient μMT mice with the BM of CD45.1+ AIRE+/+ and CD45.2+ Aire− mice depleted of B220+ cells at a ratio of 1:1 (Figure S2A), allowing the donor B cell compartment to develop in the same environment in which the thymic epithelium is Aire+/+ so that the autoimmune manifestations of AIRE deficiency do not develop. Twenty-eight days later, naive resting splenic unswitched Aire+/+ and Aire−/− B cells from these BM chimeras were adoptively transferred to secondary μMT recipients at a ratio of 1:1 (Figure S2B). These secondary μMT recipients were subsequently immunized with the T cell-dependent antigen (4-hydroxy-3-nitrophenylacetyl)32-Keyhole Limpet Hemocyanin (NP32-KLH) and analyzed the affinity maturation and class switching of NP-specific donor B cells by flow cytometry (Figure 3A). After the immunizations, the splenic GC B cell compartment contained more Aire−/−B cells than Aire+/+ B cells (Figure 3B), and the GC Aire−/− B cells had a higher NP8-to-NP36 binding ratio and a higher percentage of NP8-binding cells in total NP-specific cells than their Aire+/+ counterpart (Figure 3C and D), indicating increased affinity maturation. These data suggest that AIRE inhibits SHM in a B cell-intrinsic manner.
Figure 3. AIRE Inhibits CSR and SHM in B cells.
(A) The generation and immunization of Aire+/+ and Aire−/− BM and B cell chimeric mice.
(B) The percentage of CD45.1+ Aire+/+ and CD45.2+ Aire−/− B cells in the splenic GC B cells of the secondary μMT recipient mice (n = 6) after the immunizations. ***P < 0.001, by 2-tailed paired t-test.
(C and D) Flow cytometric and statistical analyses of the ratio of NP8- to NP36-binding (C) and NP8- to total NP-binding (D) GC B cells after immunizations of the secondary μMT recipient mice (n = 6). **P < 0.05, **P < 0.01, by 2-tailed paired t-test.
(E) The generation and immunization of Aire+/+ and Aire−/− B cell chimeric mice.
(F) Flow cytometric analysis of surface IgD and IgM on NP36-binding B cells in μMT recipients of Aire+/+ or Aire−/− B cells immunized with NP32-KLH. The result represents 3 age- and sex-matched μMT recipients each of B cells from 3–5 age- and sex-matched littermate donor Aire+/+ or Aire−/− mice.
(G) The ratios of the titers of circulating NP4-binding to NP29-binding IgM, IgG1, IgG2b and IgG3 in immunized μMT recipient mice of Aire+/+ or Aire−/− B cells. The results represent 4 experiments, each consisting of B cells from 3–5 age- and sex-matched littermate donor mice and 6–8 age- and sex-matched littermate μMT recipient mice. *P < 0.05, **P < 0.01, by 2-tailed unpaired t-test.
(H) The SHM landscape across IgHV, including FR2, CDR2, FR3, CDR3 and FR4, of NP36-binding IgM−IgD− or IgM+IgD+ Aire+/+ and Aire−/− donor B cells in μMT recipients after immunizations with NP32-KLH. The result represents 3 μMT recipients of Aire+/+ donor B cells and 3 μMT recipients of Aire−/− donor B cells.
(I) Frequencies of C-to-T transitions in SHMs in IgHV of NP-specific IgG+, IgA+ or IgE+ splenic B cells from μMT recipient mice of Aire+/+ or Aire−/− B cells after immunizations with NP32-KLH. Data are represented as median ± upper/lower quartile. *P < 0.05, **P < 0.01, by 1-tailed unpaired t-test.
(J) qRT-PCR analysis of the fold induction of Iμ-Cγ1 and Iμ-Cγ3 post-switch transcript levels in peripheral blood IgD+CD27− naïve B cells from healthy subjects (n = 5) or APS-1 patients (n = 5) stimulated for 3 d with CD40L and IL-4 or IFN-γ over the respective unstimulated control B cells. *P < 0.05, by 1-tailed unpaired t-test (upper panel) or 1-tailed Mann-Whitney U test (lower panel).
Secondly, we adoptively transferred an equal number of naive resting unswitched B cells from CD45.2+ Aire+/+ or CD45.2+ Aire−/− donor mice into μMT recipients and assessed the CSR and SHM of NP-specific donor B cells after the immunizations by flow cytometry, enzyme-linked immunosorbent assay (ELISA) and Ig heavy (IgH) chain variable (V) region mutation profiling with next-generation sequencing (Figure 3E). The Aire+/+ and Aire−/− donor B cells exhibited a similar phenotype and had a comparable NP8-to-NP36 binding ratio before the transfer (Figure S2C–E). Following the immunizations, Aire+/+ and Aire−/− donor B cells entered the GC similarly in their respective recipients (Figure S2F), and GC Aire+/+ and Aire−/− donor B cells exhibited similar expression of major co-stimulatory and co-inhibitory molecules (Figure S2G). μMT recipients of Aire+/+ or Aire−/− B cells had a similar proportion of CXCR5+PD-1+ follicular helper T (TFH) cells (Figure S2H) and Foxp3+CD25+ follicular regulatory T (TFR) cells in the spleen (Figure S2I). However, NP-specific Aire−/− donor B cells exhibited elevated class switching by harboring a much higher fraction of IgM−IgD− cells than NP-specific Aire+/+ donor B cells (Figure 3F), and underwent increased affinity maturation by producing IgG1, IgG2b and IgG3 of higher NP4-to-NP29 binding ratios (Figure 3G). In contrast, a higher NP4-to-NP29 binding ratio was not observed in the IgM compartment (Figure 3G). Increased CSR and SHM of donor Aire−/− B cells were not a secondary effect of differential AIRE expression by donor B cells in the thymus, because we found a negligible number of B cells in the thymus of the μMT recipient mice and the adoptive transfer did not lead to an increase in B cell numbers in the thymic stroma of the recipients at the end of the experiment, indicating that the donor B cells did not enter the thymus of the recipients during the course of our experiment (Figure S2J and K). Analysis of the IgH variable region (IgHV) SHM landscape of NP-specific Aire+/+ and Aire−/− donor B cells sorted from the recipient mice (Figure S2L) showed that Aire−/− NP-specific class-switched IgG and IgE B cells had higher rates of IgHV SHMs in complementarity-determining region 2 (CDR2) and framework region 3 (FR3) than Aire+/+ NP-specific B cells (Figure 3H). Importantly, such an increased SHM profile was not seen in the NP-specific IgM and IgD compartments (Figure 3H). In addition, there was an increased frequency of C-to-T transitions among the SHMs in the IgH V region coding sequences in Aire−/− NP-specific B cells compared to Aire+/+ NP-specific B cells (Figure 3I), which is a molecular signature associated with the action of AID in the IgH V region (Maul et al., 2011). Aire+/+ and Aire−/−splenic B cells exhibited comparable proliferation and apoptosis upon stimulation ex vivo (Figure S3A–D). These data suggest that AIRE suppresses CSR and SHM in AID-experienced B cells.
Thirdly, we sorted IgD+CD27− mature naive B cells from the peripheral blood of healthy subjects and APS-1 patients and compared their CSR in vitro upon stimulation with T cell-dependent stimuli. Naive B cells of APS-1 patients underwent increased CSR compared to those of healthy subjects by expressing higher levels of the post-switch transcript Iμ,-Cγ1 or Iμ,-Cγ3 following stimulation with CD40L and IL-4 or CD40L and interferon (IFN)-γ, respectively (Figure 3J).
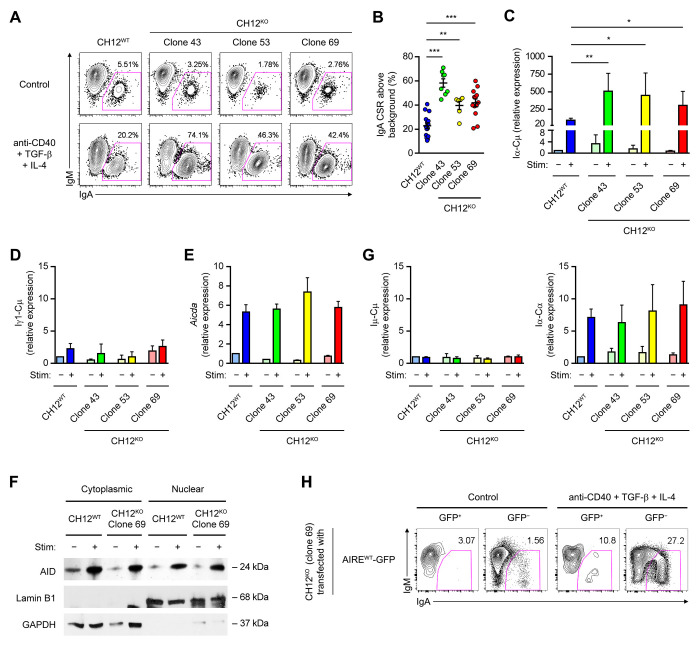
Fourthly, using CRISPR-Cas9-mediated gene editing, we disrupted the Aire gene in the murine B cell line CH12 that class-switches from IgM to IgA upon stimulation with anti-CD40, TGF-β and IL-4 (Nakamura et al., 1996), and identified 3 Aire−/− CH12 clones which were frame-shifted in both Aire alleles (Table S1, and Figure S4A and B), devoid of AIRE protein expression (Figure S4C) and free from detectable CRISPR off-target effects (data not shown). Upon stimulation, these Aire−/− CH12 clones underwent elevated IgA CSR (Figure 4A and B) with concomitantly increased levels of the Iα-Cμ circle transcript compared to their parental Aire+/+ CH12 cells (Figure 4C). The increased CSR was specific to IgA, as Iγ1-Cμ, the circle transcript of IgG1, an isotype that CH12 cells do not switch to, was not affected by Aire deficiency (Figure 4D). Exaggerated IgA CSR in Aire−/− CH12 cells was not a result of increased induction of AID (Figure 4E and F) or germline transcription (Figure 4G), nor a result of increased survival (Figure S4D and E). Remarkably, WT AIRE suppressed cytokine-induced CSR when re-introduced into Aire−/− CH12 cells (Figure 4H). Collectively, the results obtained from these various experimental systems demonstrate the B-cell intrinsic function of AIRE in inhibiting CSR and SHM.
Figure 4. Aire−/− CH12 Cells Undergo Elevated CSR.
(A and B) Flow cytometric and statistical analyses of IgA CSR in WT and Aire−/− CH12 cells treated with medium (Control) or anti-CD40, TGF-β and IL-4 for 3 d. Data in F was determined as the difference between the percentages of IgA+IgM− cells in stimulated samples to unstimulated samples. The results (mean ± SEM) represent or compare 16 experiments involving WT CH12 cells, 8 experiments involving clones 43, 6 experiments involving clone 53, and 13 experiments involving clone 69. **P < 0.01, ***P < 0.001, by 2-tailed unpaired t-test.
(C) qRT-PCR analysis of the Iα-Cμ circle transcript levels (mean ± SEM) in WT and Aire−/− CH12 cells treated with medium (Control) or anti-CD40, TGF-β and IL-4 for 3 d. The results compare 3 experiments. *P < 0.05, **P < 0.01, by 1-tailed unpaired t-test.
(D) qRT-PCR analysis of the Iγ1-Cμ circle transcript level in Aire+/+ CH12 cells and Aire−/− CH12 cell clones 43, 53 and 69 that were either unstimulated or stimulated with anti-CD40, TGF-β and IL-4 for 3 d. The result was normalised using the respective Actb transcript level and expressed as fold of induction (mean ± SEM) relative to unstimulated Aire+/+ CH12 cells.
(E and F) Western Blot analysis of AID in WT and Aire−/− CH12 cells that were either unstimulated or stimulated with anti-CD40, TGF-β and IL-4 for 3 d. Lamin B1 and GAPDH were used as the control for nuclear and cytoplasmic proteins, respectively. The data are presented as mean ± SEM and represent 3 experiments.
(G) qRT-PCR analysis of Aicda and the Iμ-Cμ and Iα-Cα germline transcript levels (mean ± SEM) in Aire+/+ CH12 cells and Aire−/− CH12 cell clones 43, 53 and 69 that were either unstimulated or stimulated with anti-CD40, TGF-β and IL-4 for 3 d.
(H) Flow cytometric analysis of IgA CSR in Aire−/− CH12 cells (clone 69) transfected with a plasmid expressing WT (AIREWT-GFP) AIRE-GFP and treated with medium (Control) or anti-CD40, TGF-β and IL-4 for 3 d. The results represent 3 experiments.
AIRE Interacts with AID in GC B Cells
We next investigated the mechanism by which AIRE inhibits peripheral antibody diversification. Given AID as the obligatory enzyme that mediates CSR and SHM (Muramatsu et al., 2000), we asked whether AIRE inhibits AID function in B cells. To this end, we first examined whether AIRE and AID interact in GC B cells. AIRE and AID co-localized in the nuclei of tonsillar IgD−CD38+ GC B cells (Figure 5A and B, and Figure S5A) but not in IgD+CD38− naive B cells (Figure S5B), IgD−CD38− switched memory B cells (Figure S5C) or IgD−CD38hi switched PCs (Figure S5D), albeit a low level of nuclear AIRE and AID were detected in a small fraction of IgD+CD38+ FGC B cells (Figure S5E). Using an AID antibody validated for immunoprecipitation (IP) and Chromatin IP (ChIP) (Vuong et al., 2009), we found that AIRE interacted with AID in human tonsillar CD19+ and CD19+IgD− cell fractions (Figure 5C). AID and AIRE did not interact via DNA in GC B cells as they still co-immunoprecipitated after DNAse I treatment (Figure S6A). AIRE also co-immunoprecipitated with AID in splenic B cells of immunized WT but not Aire−/− or Aicda−/− mice (Figure 5D and Figure S6B). Based on these data, we conclude that AIRE interacts with AID in GC B cells in vivo.
Figure 5. AIRE Interacts with AID in GC B Cells.
(A and B) Imaging flow cytometric analysis of AIRE and AID in tonsillar IgD−CD38+ GC B cells of a healthy donor. Bars: 7 μm (A). The results represent 3 donors.
(C and D) Co-IP of AIRE and AID in tonsillar CD19+ total, IgD+ naive and FGC and CD19+IgD− GC and memory B cells of a healthy donor, and in splenic CD19+ B cells of a B6 mouse after 3 doses of immunization with SRBCs. The results are representative of tonsils of 4 donors and spleens of 3 mice.
(E) The domain structures of recombinant WT and mutant human AIRE and AID molecules. Dotted lines indicated the deleted regions in the mutant proteins.
(F) Co-IP of WT AID and WT or mutant AIRE in HKB-11 cells 24 h after transfection of plasmid(s) encoding WT AID and WT or mutant AIRE proteins.
(G) The domain structures of recombinant WT and mutant human AID proteins.
(H) Co-IP of WT AIRE and WT or mutant AID in HKB-11 cells 24 h after transfection of plasmid(s) encoding WT AIRE and WT or mutant AID proteins. The results in E and G are representative of 3 experiments.
(I) Flow cytometric analysis of IgA CSR in Aire−/− CH12 cells (clone 69) transfected with a plasmid expressing either WT (AireWT-GFP) or CARD-deficient (AireΔCARD-GFP) AIRE-GFP and treated with medium (Control) or anti-CD40, TGF-β and IL-4 for 3 d. The results represent 3 experiments.
Interaction of AIRE with AID Requires the CARD and NLS domains of AIRE
We subsequently generated a series of deletion mutants of AIRE with C-terminal Myc and His tags to characterize AIRE’s interaction with AID (Figure 5E and Table S2A). AIRE mutants missing the N-terminal caspase activation and recruitment domain (CARD) and/or nuclear localization signal (NLS) lost the ability to interact with AID (Figure 5F), suggesting the requirement for the CARD domain and nuclear localization of AIRE for interaction with AID. Furthermore, using a series of deletion, domain replacement or point mutants of AID with an N-terminal FLAG tag (Figure 5G and Table S2B), we found that the interaction between AID and AIRE required both the catalytic and APOBEC-like domains of AID, although the catalytic activity of AID was not necessary, as the catalytically inactive AIDE58A mutant (Patenaude et al., 2009) still interacted with AIRE (Figure 5H). The AID point mutation G23S, which substantially abrogates the SHM but not much CSR activity of AID (Wei et al., 2011), did not affect the interaction with AIRE (Figure 5H). Echoing the CARD-dependent interaction of AIRE with AID, a CARD domain deletion mutant of AIRE had impaired ability to suppress CSR when introduced into Aire−/− CH12 cells (Figure 5I).
AIRE Inhibits the Function of AID during Antibody Diversification
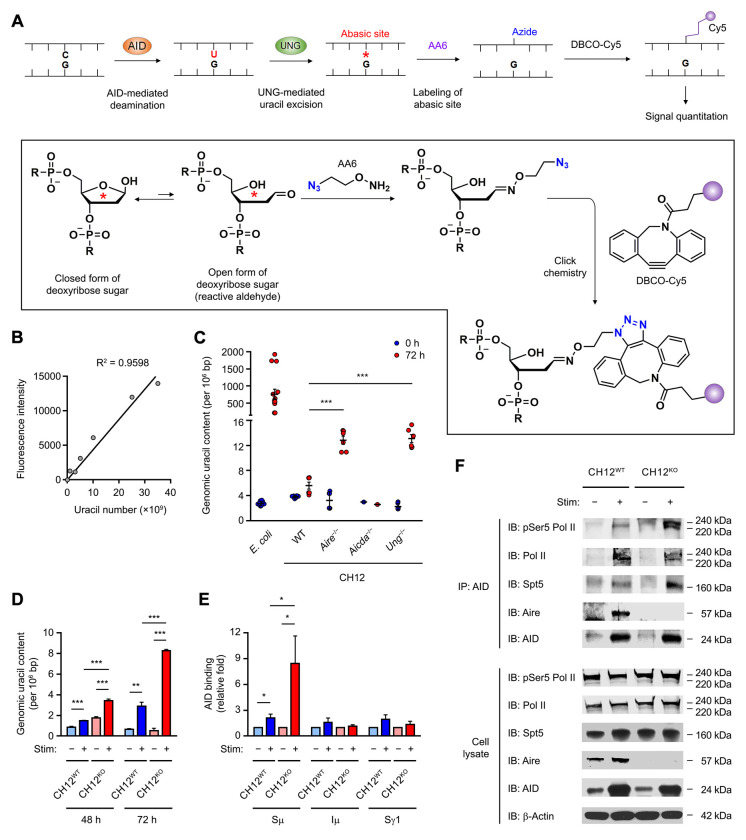
Since AID enzymatically generates uracils in DNA (Bransteitter et al., 2003; Chaudhuri et al., 2003; Sohail et al., 2003), we employed a genomic uracil dot blot assay to directly test the effect of AIRE on the activity of AID (Figure 6A and B). In CH12 cells, increased uracils were detected in the genome upon stimulation with anti-CD40, TGF-β and IL-4 to undergo IgA CSR, which peaked on day 3 (Shalhout et al., 2014), whereas Acida−/− CH12 cells failed to accumulate genomic uracils after stimulation (Figure 6C), indicating that the emergence of genomic uracils was mediated by AID. Upon stimulation, Aire−/− CH12 cells harbored higher levels of genomic uracil than Aire+/+ CH12 cells (Figure 6C and D). This result reflects the inhibition of AID’s enzymatic function by AIRE at a step upstream of the deamination reaction.
Figure 6. AIRE Inhibits AID Function by Interfering with AID Targeting to Its IgH DNA Substrate.
(A and B) The principle, chemistry and calibration of the dot blot assay for the quantitation of genomic uracil content.
(C) The genomic uracil levels in WT, Aire−/−, Aicda−/− or Ung−/− CH12 cells after 72 h of treatment without or with anti-CD40, TGF-β and IL-4. The results are presented as mean ± SEM and represent 3 experiments. ***P < 0.001, by 2-tailed unpaired t-test. Bisulfite-treated E. coli DNA was included as a positive control.
(D) The genomic uracil content in WT and Aire−/− CH12 cells after 48 or 72 h of treatment without or with anti-CD40, TGF-β and IL-4. The results are presented as mean ± SEM and represent 3 experiments. **P < 0.01, ***P < 0.001, by 2-tailed unpaired t-test.
(E) ChIP-qPCR analysis for the interaction of AID with Sμ, Iμ and Sγ1 regions in WT and Aire−/− CH12 cells after 72 h of treatment without or with anti-CD40, TGF-β and IL-4. The results are presented as mean ± SEM and represent 3 experiments. *P < 0.05, by 2-tailed unpaired t-test.
(F) Co-IP of AID with pSer5-Pol II, total Pol II, Spt5 and Aire in WT and Aire−/− CH12 cells after 72 h of treatment without or with anti-CD40, TGF-β and IL-4. The results represent 3 experiments.
We further investigated how AIRE may inhibit AID’s enzymatic activity. Upstream of DNA deamination by AID is the proper targeting of AID to the IgH S regions at sites of Pol II stalling (Chaudhuri et al., 2003; Pavri et al., 2010). We hypothesized that AIRE may interfere with the targeting of AID to IgH S regions during CSR. Using ChIP and IP assays, we found increased AID binding to the Sμ, but not Iμ or Sγ1, region (Figure 6E) and increased interaction of AID with serine-5 phosphorylated Pol II and its associated factor Spt5 at a promoter-proximal pause site (Peterlin and Price, 2006) in stimulated Aire−/− CH12 cells compared to stimulated Aire+/+ CH12 cells (Figure 6F and Figure S6C). These data suggest that AIRE inhibits AID function in B cells by interfering with the interaction of AID with transcriptionally paused Pol II and the targeting of AID to its IgH DNA substrate.
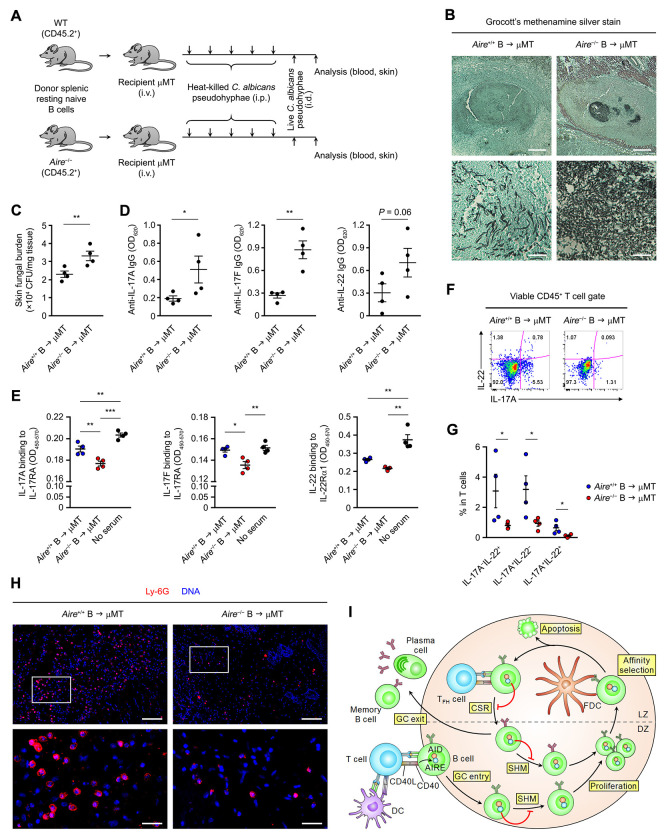
AIRE Deficiency in B Cells Engenders Humoral Autoimmunity and Compromises Skin Candida Immune Defense
Given that the vast majority of APS-1 patients mysteriously develop chronic mucocutaneous candidiasis (CMC) as an early clinical symptom (Kisand and Peterson, 2015), we sought to determine the functional impact of B cell-intrinsic AIRE in humoral immunity and anti-Candida immune defense, and asked whether AIRE deficiency in peripheral B cells could promote APS-1-like CMC. We employed a mouse dermal candidiasis model which allows skin C. albicans infection to be established without the use of immunosuppressive agents (Conti et al., 2014). μMT recipient mice reconstituted with either Aire+/+ or Aire−/− B cells were first exposed to heat-killed C. albicans pseudohyphae and subsequently infected cutaneously with live C. albicans pseudohyphae (Figure 7A). Four days after infection, μMT recipient mice of Aire−/− B cells had heightened fungal burden in the skin (Figure 7B and C) and concomitant elevation of circulating autoantibodies to IL-17A, IL-17F and IL-22 as compared to μMT recipients of Aire+/+ B cells (Figure 7D and Figure S7A). The sera of μMT recipients of Aire−/− B cells contained enhanced blocking activity of the binding of IL-17A, IL-17F and IL-22 to their receptors (Figure 7E). In addition, the dermal infection site of μMT recipients of Aire−/− B cells harbored reduced IL-17A- and IL-22-producing T cells (Figure 7F and G), which was accompanied by diminished neutrophils infiltration into the infected skin tissue (Figure 7H). These results are reminiscent of the aberrant production of class-switched neutralizing autoantibodies against TH17 cytokines in APS-1 patients that may impair anti-C. albicans immunity (Kisand et al., 2010; Meyer et al., 2016; Puel et al., 2010), and collectively show that AIRE deficiency in peripheral B cells compromises cutaneous anti-Candida immune defense and promotes APS-1-like CMC by engendering humoral autoimmunity.
Figure 7. Aire Deficiency in B Cells Promotes Humoral Autoimmunity and Compromises Cutaneous Anti-Candida Defense.
(A) The cutaneous candidiasis infection model.
(B and C) GMS stain of cutaneous C. albicans and skin fungal burden (CFU per mg of tissue) (mean ± SEM) in μMT recipient mice of Aire+/+ or Aire−/− donor B cells 4 d after infection. Bars: 1 mm (B, upper panels) or 100 μm (B, lower panels). **P < 0.01, by 1-tailed unpaired t-test.
(D) ELISA of the levels (mean ± SEM) of autoantibodies binding to IL-17A, IL-17F and IL-22 in the sera of μMT recipient mice of Aire+/+ or Aire−/− donor B cells 4 d after infection. *P < 0.05, **P < 0.01, by 1-tailed unpaired t-test.
(E) ELISA of the levels (mean ± SEM) of blocking activity of IL- 17A, IL- 17F and IL-22 to their receptors in the sera of μMT recipient mice of Aire+/+ or Aire−/− donor B cells 4 d after infection. *P < 0.05, **P < 0.01, ***P < 0.001, by 1-way ANOVA with Tukey’s post hoc test.
(F and G) Flow cytometric and statistical analyses of IL-17A and IL-22 expression in cutaneous T cells of μMT recipient mice of Aire+/+ or Aire−/− donor B cells (n = 4 in each group) 4 d after infection and after ex vivo re-stimulation. The data are represented as mean ± SEM. *P < 0.05, by 1-tailed unpaired t-test.
(H) Immunofluorescence analysis of Ly-6G (red) and DNA (blue) in cutaneous tissues surrounding the C. albicans infection site in μMT recipient mice of Aire+/+ or Aire−/− donor B cells 4 d after infection. The results in B–H represent 2 experiments, with 4 mice per group in each experiment. Bars: 160 μm (upper panels) or 40 μm (lower panels).
(I) A simplified schematic of AIRE-mediated GC checkpoint of antibody diversification in B cells. At the T–B cell border of secondary lymphoid organs, B cells present antigens to and receive co-stimulation from DC-activated T cells, which also induce AIRE expression in B cells via CD40. The activated B cells enter the GC DZ and undergo SHM, proliferation and subsequent affinity selection by interacting with antigens on the surface of follicular dendritic cells (FDCs) in LZ. Low-affinity B cells will undergo apoptosis, whereas high-affinity B cells receive help from T follicular helper (TFH) cells to undergo CSR, and subsequently either re-enter the SHM–proliferation cycle in the DZ or exit the GC as plasma cells or memory B cells. AIRE in B cells limits autoantibody generation by restraining excessive AID activity in the GC.
DISCUSSION
AIRE as A Paradigm of B Cell-Intrinsic Intracellular Immune Checkpoint
The concept and importance of immune checkpoint are now widely appreciated by scientists and clinicians. Immune checkpoints refer to molecules that mediate negative regulation of immune responses and therefore have a crucial role in maintaining immunological self-tolerance and avoiding autoimmunity. Many of the checkpoint molecules can be hijacked by tumors or pathogens for immune evasion and have therefore emerged as therapeutic targets for cancer and infectious diseases. To date, the majority of immune checkpoints under research investigation or clinical development encompass cell surface ligands and receptors that are involved in antigen-presenting cell (APC)–T cell interactions in immune-inductive sites (i.e., secondary lymphoid organs) or immune-effector sites (Sharma and Allison, 2015). Here, we demonstrate AIRE expression in the GC stage of B cell differentiation and its negative regulation on AID-mediated peripheral antibody diversification. Consequently, AIRE deficiency in B cells results in exaggerated antibody diversification and autoimmunity. Importantly, increased SHM of Aire−/− donor B cells was only seen in class-switched isotypes but not the IgM or IgD compartment, indicating an effect of AIRE only in AID-experienced B cells. This is not only consistent with the GC-specific AIRE expression in peripheral B cells, but also suggests that the function of AIRE described here is not a secondary effect of thymic B cells. The result from Aire+/+ and Aire−/− B cell chimeras further corroborates this conclusion and confirms the B cell-intrinsic role of AIRE in limiting antibody diversification. Therefore, AIRE represents a paradigmatic B cell-intrinsic intracellular immune checkpoint of humoral immunity (Figure 7I).
Functionally Important Levels of AIRE May Have a Broader Expression Pattern
Besides mTECs (Anderson et al., 2002), AIRE protein expression has been previously reported in thymic B cells as well as several normal or malignant cell types of the hematopoietic or non-hematopoietic lineage in the periphery in humans and/or mice (Bianchi et al., 2016; Gardner et al., 2008; Gardner et al., 2013; Hobbs et al., 2015; Lindh et al., 2008; Poliani et al., 2010; Yamano et al., 2015). Intriguingly, little or no AIRE protein was seen in mouse GC B cells in a previous study, which was thought to result from B cell receptor (BCR)-mediated inhibition of AIRE induction; however, mouse B cells still expressed markedly elevated levels (~20 fold) of AIRE transcript when they were stimulated with anti-CD40 in the presence of anti-IgM (Yamano et al., 2015). In contrast, we present evidences obtained using multiple approaches to demonstrate AIRE protein expression in human and mouse GC B cells in vivo, and have also found much higher levels of AIRE transcript in mouse GC B cells than in non-GC B cells (data not shown). Therefore, BCR triggering does not completely abolish AIRE induction in GCs under physiological conditions. Consistent with our finding, BCR signaling in the GCs is reduced due to high phosphatase activity (Khalil et al., 2012) and, in some cases, BCR specificity may not be required for entry into GCs (Silver et al., 2018). Nonetheless, the role of AIRE in inhibiting AID shown here is in line with BCR engagement in facilitating CSR and SHM of antigen-specific B cell clones and their affinity maturation by downregulating AIRE.
We currently do not know if AIRE in GC B cells has a significant impact on TSA expression and Treg induction by these cells as seen in mTECs and B cells in the thymus (Anderson et al., 2002; Malchow et al., 2013; Yamano et al., 2015), or an impact on CD4+ T cell inactivation as seen in eTACs in secondary lymphoid organs (Gardner et al., 2008; Gardner et al., 2013). However, the comparable response of splenic TFR cells in immunized μMT recipients and the similar expression of co-stimulatory and co-inhibitory molecules on donor Aire−/− and Aire−/− B cells would argue against these possibilities. In agreement with others (Gardner et al., 2013; Yamano et al., 2015), we found that the highest level of AIRE expression in mice appeared to be in mTECs and thymic B cells (data now shown). Hence, a lower level of AIRE expression in GC B cells as compared to thymic mTECs and thymic B cells would allow AIRE to perform unique and different functions, and our demonstration of the function of AIRE in regulating antibody diversification suggests that functionally important levels of AIRE have a broader expression pattern than previously appreciated. Lending further credence to this notion is the example of AID, where a much lower expression level in BM B cell precursors compared to that in GC B cells has a profound effect on B cell repertoire as well as both central and peripheral B cell tolerance in humans and mice (Cantaert et al., 2015; Meyers et al., 2011).
AIRE as a Unique Negative Regulator of AID
Many proteins have been reported to interact with and/or regulate AID in B cells, and they are thought to function at various steps of the molecular cascade of antibody diversification ranging from chromatin remodeling, AID targeting, transcriptional regulation, RNA processing, DNA repair and post-translational protein modification (Casellas et al., 2016; Vaidyanathan et al., 2014; Xu et al., 2012). To our knowledge, among the AID-interacting partners identified to date, AIRE is the only one which, when deficient, enhances AID function, whereas all others impair AID function when they are missing. This provides a unique way to up-regulate the activity of AID in B cells. Whether AIRE interacts with AID directly or indirectly via other factors remains to be elucidated. Our results suggest the requirement for the CARD and NLS domains of AIRE in the interaction with AID. CARD is critical for AIRE’s interaction with bromodomain-containing protein 4 (Brd4) which binds acetylated lysines in CARD and bridges AIRE with the positive transcription elongation factor b (P-TEFb) complex to induce ectopic gene expression in mTECs by promoting Pol II elongation (Giraud et al., 2014; Giraud et al., 2012; Oven et al., 2007; Yoshida et al., 2015). The increased interaction of AID with IgH S region and transcriptionally paused Pol II in Aire−/− CH12 cells undergoing CSR is therefore consistent with this body of literature, considering AID is targeted to DNA sites of Pol II pausing (Pavri et al., 2010). The genomic uracil dot blot assay we employed directly detects the product of AID’s enzymatic activity, in contrast to conventional methods such as flow cytometry, ELISA and antibody sequencing that measure the final outcome of CSR and SHM as an indirect readout of AID’s function. Increased generation of genomic uracils in Aire−/− CH12 cells undergoing CSR points to the regulation of AID by AIRE at steps upstream of the deamination reaction, which further agrees with the role of AIRE in restraining AID targeting to its DNA substrate. However, the involvement of other mechanisms upstream or downstream of the deamination reaction, such as the regulation of chromatin availability or DNA repair by AIRE (Abramson et al., 2010; Koh et al., 2018), cannot be discounted.
B Cell-Extrinsic and -Intrinsic AIRE Deficiency May Differentially Contribute to Autoimmunity and Immunodeficiency in APS-1
APS-1 is caused by mutations in a single gene but has emerged as a disease with complex pathogenesis. In fact, it has been classified as a type IV PID, a disease of immune dysregulation (Al-Herz et al., 2011). APS-1-associated CMC amidst the multi-organ autoimmune manifestations indicates the co-occurrence of immunodeficiency and autoimmunity, which are two conditions that are thought to occur usually at the opposite ends of the clinical immune spectrum. Such a seemingly paradoxical feature is now being found in an increasing number of primary and acquired immunodeficiencies and autoimmune diseases, such as selective IgA deficiency (SIgAD), immune dysregulation-polyendocrinopathy-enteropathy-X-linked syndrome (IPEX), severe combined variable immunodeficiency (CVID), acquired immunodeficiency syndrome (AIDS), systemic lupus erythematosus (SLE) and diabetes mellitus (DM) (Bacchetta and Notarangelo, 2013; Grammatikos and Tsokos, 2012; Zandman-Goddard and Shoenfeld, 2002). Our work revealed that, besides the central and peripheral abnormalities in T cell tolerance extrinsic to B cells, APS-1 involves previously unknown B cell-intrinsic dysregulation in peripheral antibody diversification, and this can engender humoral autoimmunity, such as the production of autoreactive antibodies against TH17 effector cytokines. Our findings offer a plausible mechanistic explanation to the clinical observation of the presence of high affinity autoreactive neutralizing antibodies in these patients (Kisand et al., 2010; Meyer et al., 2016; Puel et al., 2010) and the causes of defective anti-Candida immune defense, thereby arguing for the concept that the seemingly contradictory autoimmunity and immunodeficiency can go hand-in-hand if there is an overproduction of autoreactive antibodies that impair protective immunity against pathogens. Of note, B cells were required to cause the multi-organ inflammation in Aire−/− mice not by producing autoantibodies, but by mediating early T cell priming and expansion (Gavanescu et al., 2008). This suggests that the multi-organ autoimmunity and CMC-associated immunodeficiency in APS-1 are differentially contributed by the B cell-extrinsic and -intrinsic consequences of AIRE deficiency. We think that this scenario, if proven to be true, could be a paradigm applicable to understanding many other immunodeficiencies co-presenting with autoimmunity that involve humoral immune dysfunction. Indeed, an interesting example that mirrors this scenario is seen in AID-deficient individuals and mice that suffer from immunodeficiency due to the lack of CSR and SHM in the periphery (Muramatsu et al., 2000; Revy et al., 2000) but also have B cell autoimmunity likely due to missing AID’s potential function in purging autoreactive immature B cells in the BM (Cantaert et al., 2015; Meyers et al., 2011). The insights gained from this study, therefore, could potentially offer a new direction for developing therapies that specifically and actively target the various aspects of pathogenesis of APS-1 and these other diseases.
AIRE Ablation as a Potentially New and Effective Approach of Antibody Generation for Immunotherapy
Besides offering new mechanistic insights into the regulation of the GC antibody diversification machinery and the unique production of high-affinity neutralizing autoantibodies in APS-1, our study underscores the important and emerging idea that immune tolerance mechanisms can be barriers to the generation of effective immunity (Khan et al., 2014; Schroeder et al., 2017), and controlled breaching of peripheral tolerance can permit neutralizing antibody responses that can be therapeutically beneficial. Remarkably, we found increased Ig framework region SHMs in class-switched antigen-specific Aire−/− B cells. Such an observation is reminiscent of the hallmarks of broadly and potently neutralizing antibodies against certain pathogens such as HIV-1 (Klein et al., 2013), the generation of which remains one of the most important mysteries as well as challenges in immunology. Our findings therefore point to a new effective strategy for producing such antibodies by removing a critical B cell-intrinsic brake of AID, which is AIRE. We envisage that further mechanistic elucidation of how B cell-intrinsic AIRE regulates AID function could assist in cracking the elusive molecular underpinning of the generation of broadly neutralizing antibodies and lead to substantial advances in antibody-based immunotherapies against devastating diseases, such as AIDS and cancer.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kang Chen (kang@wayne.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Autoimmune polyglandular syndrome type 1 (APS-1) patients with loss-of-function mutations in the AIRE gene were enrolled in the study with an approved protocol of the Ethics Committee of Medicine of the Hospital District of Helsinki and Uusimaa (HUS), Finland. Hyper-IgM syndrome type 3 (HIGM3) patients with loss-of-function mutations in the CD40 gene were enrolled in the study with an approved Institutional Review Board (IRB) protocol of the Icahn School of Medicine at Mount Sinai. Peripheral blood leukocytes of anonymous healthy donors were obtained from the American Red Cross with a protocol approved by the IRB of Wayne State University (WSU) and the Detroit Medical Centre (DMC). Tonsil, thymus and spleen tissues were obtained after pediatric tonsillectomy, cardiac surgery and splenectomy, respectively, from the Children’s Hospital of Michigan with an IRB protocol approved by WSU and DMC.
Mice
C57BL/6J mice, Aire+/− and μMT were purchased from the Jackson Laboratory. AireAdig mice in C57BL/6 background were previously reported (Gardner et al., 2008). Aicda−/− mice (Muramatsu et al., 2000) were generously provided by Dr. Tasuku Honjo (Kyoto University, Japan). These mice were maintained in the same room at the specific pathogen-free (SPF) facility of the Division of Laboratory Animal Resources (DLAR) at Wayne State University. Aire−/− mice were generated by mating Aire+/− mice. Age- and sex-matched Aire+/+ littermates were randomly assigned to experimental groups or used as controls for ex vivo and in vivo experiments. Prior to any experiment, all Aire and AireAdig mice were genotyped by PCR to ensure the correctness of the genotype. All breeding and experimental protocols were approved by Wayne State University Institutional Animal Care and Use Committee (IACUC).
Primary cell cultures
Peripheral blood IgD+CD27− naive B cells of healthy subjects or APS-1 patients were sorted and stored in RNALater. Primary blood, spleen and tonsil IgD+ B cells were purified by MACS and cultured in RPMI-1640 medium. Mouse cells purified from spleen and lymph nodes were extracted via B cell isolation kit and cultured in RPMI-1640 medium further supplemented with 5% (v/v) NCTC-109 and 50 βM μ-mercaptoethanol.
Cell lines
The human embryonic kidney cell/Burkitt’s lymphoma fusion cell line HKB-11 was cultured in DMEM/F12 supplemented with 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B and 10% FBS. WT, Aire−/−,Acida−/− and Ung−/− CH12 cells (generated in house) were cultured in RPMI-1640 medium further supplemented with 5% (v/v) NCTC-109 and 50 μM β-mercaptoethanol.
Microbe strain
The C. albicans strain SC5314 was purchased from the ATCC, streak plates were made on YPD agar and incubated at 30°C overnight. Plates were then stored at 4°C. For experiments, single colonies were cultured in YPD broth at 30°C for 16 h with shaking at 220 rpm prior to induction of virulent pseudohyphae formation.
METHOD DETAILS
Human blood and tissue sample processing and cell isolation
Peripheral blood mononuclear cells (PBMCs) of APS-1 patients and healthy controls were purified using Ficoll-Paque Plus. Live (7AAD− or Ghost Violet 510−) naive B cells (CD19+IgD+CD27−) and class-switched memory B cells (CD19+IgD−CD27+) were sorted from the PBMCs to a purity of ≥ 99% on a FACSAria II sorter (BD). PBMCs of anonymous healthy donors were isolated using a Histopaque-1077 gradient following the manufacturer’s instruction. Red blood cells (RBCs) were lysed using an ammonium-chloride-potassium (ACK) lysing buffer. Human tonsil and spleen tissues were minced into small pieces, meshed through 100 μm cell strainers, and pelleted at 600 g for 5 min at 4°C. Spleen cells were treated with an ACK buffer to remove erythrocytes and filtered through 40 μm cell strainers. Tonsil and spleen cells were then washed with phosphate-buffered saline (PBS). IgD+ B cells were purified from tonsil cells by magnetic-activated cell sorting (MACS) with a biotinylated goat F(ab’)2 anti-human IgD antibody and anti-biotin magnetic microbeads as previously reported (Chen et al., 2009). CD19+ B cells were similarly obtained using a biotinylated mouse anti-human CD19 (clone HIB19) antibody. The CD19+IgD− fraction was obtained by selecting for CD19+ cells from the IgD− fraction by MACS. Thymic cell suspensions were obtained by mincing human thymus tissues into small pieces and mechanically removing thymocytes followed by 2 rounds of digestion with 0.2% (w/v) Collagenase V and 0.1 mg/ml DNase I in Hank’s Balanced Salt Solution (HBSS) for 45 min at 37°C with shaking. The digested samples were filtered through 70 μm cell strainers and washed with PBS.
Mouse blood and tissue cell isolation
Blood was collected from mice before euthanasia. spleen, inguinal lymph nodes, mesenteric lymph node and Peyer’s patches were collected after euthanasia. Adjacent fat and other tissues were removed before single cells suspensions were prepared, filtered through 100 μm cell strainer. RBCs from blood were removed by centrifugation on Histopaque 1077, and those in spleens were lysed using an ACK buffer. The cells were washed in PBS and counted before cell sorting, flow cytometry or purification by MACS. Resting B cells were isolated from the spleens of age- and sex-matched Aire+/+ or Aire−/− littermates by MACS using a B Cell Isolate Kit. The purity of the isolated B cells ranged from 97–99.6% as determined by flow cytometry based on CD19 and B220 staining.
Mouse immunization
2.5×107 purified Aire+/+ or Aire−/− B cells were introduced via the tail vein into each recipient μMT littermate mouse. One day after the adoptive transfer, each recipient was intraperitoneally (i.p.) immunized with 1 dose of 100 μg NP32-KLH in Complete Freund’s Adjuvant and 3 doses of 100 μg NP32-KLH in Incomplete Freund’s Adjuvant once every week. Four days after the last immunization, mice were sacrificed, and blood and spleens were collected for ELISA, flow cytometry or cell sorting. In some experiments, mice were immunized 3 times, each with 200 μl of 2% sheep red blood cells in CFA for the initial immunization and IFA for the following immunizations. For Figure S1H, mice were immunized i.p. with 4×108 SRBCs in PBS in a final volume of 200 μl/mouse with or without CFA.
BM chimeras and B cell chimeras
BM cells were isolated from the tibias and femurs of age- and sex-matched CD45.1 Aire+/+ and CD45.2 Aire−/− mice by flushing the BM cells with serum-free RPMI-1640 using 27G needles. Following RBC lysis, B220+ BM cells were depleted by MACS after labeling the BM cells with a biotinylated anti-mouse B220 antibody and anti-biotin microbeads. The B220− cells from CD45.1 Aire+/+ and CD45.2 Aire−/− BMs were mixed at the ratio of 1:1 and adoptively transferred via the tail vein into sex-matched primary μMT littermate recipients 1 d after these recipients received 10 Gy total body irradiation, with each recipient receiving a total of 1.5×107 cells. BM reconstitution was allowed to proceed for 28 days. Splenic resting naive B cells were isolated from these primary μMT recipients by MACS using a B Cell Isolation Kit similarly as described above. Purified splenic B cells were counted, adjusted to a ratio of 1:1 and adoptively transferred via the tail vein into sex-matched secondary μMT littermate recipients, with each recipient receiving a total of 1.5×107 cells. Mice were then immunized with NP32-KLH and adjuvants as described above.
Discrimination of intravascular and tissue leukocytes
A published method was used to distinguish intravascular and tissue leukocytes in mice (Anderson et al., 2014). In this method, mice were intravenously administered with 6 μg biotinylated anti-CD45 antibody 2–3 minutes prior to anesthesia and cardiac perfusion with PBS, followed by immediate tissue harvest and digestion. Intravascular cells were subsequently stained with fluorochrome-conjugated streptavidin in combination with other cell surface markers before analysis by flow cytometry.
Culture and stimulation or primary B cells
Peripheral blood IgD+CD27− naive B cells of healthy subjects or APS-1 patients were stimulated with 500 ng/ml soluble CD40L (sCD40L) and 100 ng/ml IL-4 or 100 ng/ml IFN-γ. Purified mouse splenic B cells were stimulated with 500 ng/ml sCD40L with or without 100 ng/ml IL-4, 100 ng/ml IL-21 or 25 μM CAPE. In some experiments, sCD40L was replaced with 5 μg/ml anti-CD40. To determine cell proliferation, the cells were labelled with carboxyfluorescein succinimidyl ester (CFSE) according to the manufacture’s protocol prior to culture. Alternatively, 10 μM 5-ethynyl-2’-deoxyuridine (EdU) was added to the culture medium for 6 hours before EdU incorporation was determined by flow cytometry using a Click-iT EdU Flow Cytometry Assay Kit according to the manufacturer’s protocol.
Generation and validation of Aire−/− CH12 cells
Several clones of Aire−/− CH12 cells were generated by targeting the Aire gene using the CRISPR/Cas9 system as described previously (Ran et al., 2013). Single guide RNAs (sgRNA) targeting exon 1 or exon 3 of mouse Aire gene (GenBank AJ007715.1) were designed using the online tool at http://crispr.mit.edu. Sequences with the highest score for the respective region were selected. To express sgRNAs, pairs of oligonucleotides were synthesized and cloned into pSpCas9(BB)-2A-Puro plasmid as reported (Ran et al., 2013). The sgRNA expression plasmid was then transfected into CH12 cells using electroporation (square wave pulse at 200 V for 30 ms) in serum-free RPMI-1640 with 5 mM glutathione in a 4-mm cuvette. 24 hours after transfection, cells were resuspended in 125 ng/ml puromycin for 48 hours. After a brief expansion in puromycin-free media, single cell clones from transfected cells were screened for loss of the sgRNA targeting site using PCR. Clones with deletions in both alleles were identified by PCR. To determine the exact genomic modifications in each clone, the sgRNA-targeting sites were amplified with primer pairs spanning the targeting sites, and PCR products were sequenced directly using the respective forward primer. In addition, PCR products from clones 43 and 53 were cloned into the pGEM-T Easy vector and sequenced with T7 primer. All three mutant clones used were confirmed to harbor frameshift mutations on both alleles, resulting in termination shortly after the frameshift site. The potential off-target sites in the mouse genome for each guide were identified by the same online tool (http://crispr.mit.edu). Cas9 generally does not tolerate more than 3 mismatches (Hsu et al., 2013). All off-target sites in a potential gene-coding region with non-zero scores (up to 4 mismatches) were verified by sequencing to be intact. The lack of AIRE protein expression in these clones was finally confirmed by Western Blot.
Plasmids
Full-length human AIRE cDNA sequence was cloned into pcDNA3.1(−) with tandem C-terminal Myc and 6-Histidine tag. Sequences coding various domains of AIRE were deleted using a Phusion Site-Directed Mutagenesis Kit using appropriate primers. Briefly, to delete a specific section of AIRE in the vector, a pair of outward primers was designed to amplify the remaining region together with the plasmid backbone. PCR product was then phosphorylated at 5’ end and ligated with Quick T4 ligase to recircularize it. Human AID was obtained by cloning full-length AICDA into pFLAG-CMV2 vector with an N-terminal FLAG tag. Domain-specific deletion mutants and G23S and E58A point mutants of AID were generated using the Phusion Site-Directed Mutagenesis kit using appropriate primers. The full-length Egfp sequence from pcDNA3-eGFP (from Dr. Thilo Hagen, National University of Singapore) was then cloned in frame to the C-terminus of mouse Aire, AireΔNLS, or AireΔCARD using blunt end ligation of PCR-amplified fragments.
Transfection
106 seeded HKB-11 cells were cultured to 70–90% confluence and transfected with 4 μg plasmid DNA using Lipofectamine 3000 in Opti-MEM by following the manufacturer’s instruction. CH12 cells were transfected using the Amaxa cell line nucleofector kit V (Lonza). Cells were pelleted at 100 g for 10 min and resuspended in the electroporation buffer according to the manual. 106 cells in 100 μl were mixed with 2 μg of target plasmids. The mixture was transferred to cuvettes for electroporation with Nucleofactor 2b (Lonza) using the program D-023. After electroporation, complete medium was immediately added to promote recovery. The electroporated cells were subsequently divided equally into 2 wells, with one well left unstimulated and the other stimulated with 1 μg/ml anti-CD40, 1 ng/ml TGF-β1 and 12.5 ng/ml IL-4 for 3 d.
C. albicans culture
A single colony of C. albicans was cultured in YPD broth at 30 °C for 16 h with shaking at 220 rpm. C. albicans existed in the blastospore form after the 16 h culture. The concentration of the culture was quantitated using a hemocytometer. The culture was then diluted 1:10 with fresh YPD broth containing 10% (v/v) heat-inactivated FBS and grown at 37 °C for 3 h with shaking at 220 rpm. An aliquot of the culture was removed and examined under the microscope to ensure that 95% of blastospores switched to the virulent pseudohyphal form. The culture was pelleted by centrifugation at 4,000 rpm for 10 minutes, washed with PBS twice and resuspended in PBS at the concentration of 5×106 CFU per 50 μl based on the quantitation of the culture 3 h ago. The pseudohyphae samples were used for either intradermal infection of mice or the preparation of heat-killed samples by treatment at 95 °C for 2 h followed by 3 rounds of sonication on ice at 30% maximum power for 5 seconds per round using a sonifier.
Cutaneous C. albicans infection
5×107 purified Aire+/+ or Aire−/− B cells were introduced via the tail vein into each recipient μMT mouse littermate. Starting from the day of adoptive transfer, 5 doses each of 106 CFU heat-killed C. albicans pseudohyphae were given intraperitoneally to each recipient mouse every 4 d. Four days after the last injection, mice were infected with 5×106 CFU live C. albicans pseudohyphae in 50 μl PBS per spot at the deep dermis of the shaved dorsal region (Conti et al., 2014). The actual dose of infection was determined by immediately plating serial dilutions of the inoculum on YPD agar in triplicate, incubating the plates at 28 °C for 24 h and colony enumeration. The inoculum size per spot ranged between 3.8–12.3×106 CFU in various experiments. Four days after the infection, blood was obtained prior to sacrificing the mice. The entire dermal injection site was excised for histological evaluation of fungal burden by Grocott’s methenamine silver (GMS) stain or by plating, or for determination of effector T cell response by flow cytometry. For GMS stain, the tissues were immediately fixed in 10% formalin overnight and embedded in paraffin before sectioning. For plating, each tissue was weighed, minced, grounded thoroughly and resuspended in sterile PBS. Serial dilutions of the suspensions were plated on YPD agar in triplicate and incubated at 28 °C for 24 h before colony enumeration. The fungal load was calculated as CFU per mg of tissue. For flow cytometry, the tissues were washed in FBS-free RPMI-1640 twice, minced and digested in FBS-free RPMI-1640 containing 0.7 mg/ml collagenase II, 2 mM EDTA and 25 mM HEPES at 37 °C for 1 h. The digested samples were passed through a 70 μm cell strainer, washed twice with RPMI-1640 containing 10% FBS, 2 mg/ml glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 25 μg/ml amphotericin B. The samples were then cultured in this medium further supplemented with 500 ng/ml PMA, 500 ng/ml ionomycin and 1 μg/ml GolgiPlug at 37 °C for 5 h before being harvested for flow cytometric analysis.
Immunoprecipitation
Cultured cells were harvested, washed with cold PBS twice and lysed with a CelLytic M buffer containing 1× protease inhibitor cocktail and 1× Halt phosphatase Inhibitor for 60 minutes on ice. The lysates were centrifuged at 18,000 g for 15 minutes at 4 °C. Protein concentration in the supernatants was determined by a BCA Protein Assay Kit. Equal amounts of lysate supernatants were used for immunoprecipitation with specific or isotype control antibodies using protein G magnetic beads (Cell Signaling 8740 or Thermo Fisher Scientific 88847) according to the manufacturers’ instructions.
RNA extraction and quantitative real-time polymerase chain reaction
RNA was extracted from cells or tissues other than those from the APS-1 patients using TRIzol. cDNA synthesis was performed using the Superscript III first strand synthesis system in a thermocycler (Bio-Rad T100). qRT-PCR was performed with PowerSYBR Green Master Mix on a StepOnePlus instrument (Applied Biosystems) using pairs of sense and anti-sense primers targeting the genes of interest. For APS-1 patients’ peripheral blood IgD+CD27− B cells, following stimulation, the cells were washed and stored in RNAlater. Prior to RNA isolation, cells were pelleted at 5,000 g for 5 min and the RNAlater was removed. The cells were washed once with ice-cold PBS. RNA was isolated using the lysis and stop solutions in a Cells-to-CT 1-step SYBR Green kit (Thermo Fisher Scientific A25601) and amplified using an iTaq Universal SYBR Green One-Step kit (Bio-Rad 172-5150) on a StepOnePlus instrument using pairs of sense and anti-sense primers targeting the genes of interest. The ACTB (Actb) gene was used as an internal control for normalization.
Chromatin immunoprecipitation and quantitative real-time PCR
ChIP was performed using a ChIP assay kit based on the manufacturer’s instructions with slight modifications. Following 3 d of stimulation of 106 CH12 cells as described above, formaldehyde was added to the culture to the final concentration of 1% and incubated for 10 minutes at 37 °C to crosslink chromatin. The cells were pelleted, washed twice in PBS, resuspended in an SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) for 10 minutes on ice. DNA was sheared by 3 rounds of sonication on ice at 30% maximum power for 3 seconds per round using a sonifier (Thermo Fisher Scientific Q500). After centrifugation at 13,000rpm for 10 minutes, the supernatants were harvested, diluted 10-fold in a ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, 167 mM NaCl, pH 8.1) containing protease inhibitors, and precleared with 50% protein A agarose/salmon sperm DNA slurry for 30 minutes at 4 °C with rotation. After setting aside an aliquot as input, An AID or control antibody was then added and incubated overnight at 4 °C with rotation, followed by the addition of 50% protein A agarose/salmon sperm DNA slurry for 1 h at 4 °C with rotation. The agarose was then pelleted and sequentially washed once with the low salt wash buffer, once with the high salt wash buffer, once with the LiCl wash buffer and twice with TE buffer, all of which were components of the kit. DNA in the bound chromatin was eluted from the beads using an elution buffer (1% SDS, 0.1 M NaHCO3, pH 8.0), reverse-crosslinked from proteins by incubation at 65 °C for 4 h in the presence of 200 mM NaCl, cleaned by 20 μg/ml RNase A treatment for 30 minutes at 37 °C followed by 40 μg/ml proteinase K treatment for 1 h at 45 °C, purified using phenol/chloroform extraction followed by ethanol precipitation with carrier glycogen according to the kit’s manual and resuspended in TE buffer for quantitative real-time PCR analysis using PowerSYBR Green Master Mix on a StepOnePlus instrument (Applied Biosystems). The fold enrichment of DNA was calculated using the ΔΔCT method with control antibody-precipitated samples as an internal reference, and further compared among different CH12 cells and treatments.
Protein extraction and Western Blot
Cells were pelleted and washed twice with ice-cold PBS, lysed with a pH 8.0 protein extraction buffer containing 20 mM Tris-HCl, 150 mM NaCl, 1% IGEPAL CA-630 (NP-40, Sigma-Aldrich I8896), 0.1% sodium dodecyl sulfate (SDS), 1 mM EDTA and protease and phosphatase inhibitor cocktail for 30 minutes on ice. Supernatants were collected after centrifugation, heated at 98 °C in SDS sample buffer with 4% β-mercaptoethanol for 5 minutes to denature proteins. Proteins were resolved in 4–20% Bis-Tris gels or 10% Tris-Glycine gels and transferred to 0.2 μm polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% (w/v) non-fat milk in Tris-buffered saline with Tween-20 for 30 minutes to 1 h, incubated with primary antibodies overnight at 4 °C and subsequently with secondary antibodies conjugated to HRP. Signals were visualized with clarity western-blot ECL substrate and exposed on autoradiograph films.
Genomic uracil quantitation
The uracils in genomic DNA were quantified using AA6 as described previously (Wei et al., 2017; Wei et al., 2015). CH12 cells were harvested by centrifugation and lysed by incubating for 1 h at 37°C in Tris-EDTA buffer (TE) containing 2 μg/ml RNase A (Sigma-Aldrich R6513) and 0.5% SDS, followed by incubation with 100 μg/ml Proteinase K (Qiagen 19131) at 56°C for 3 h. DNA was isolated by phenol:chloroform (1:1) extraction and ethanol precipitation and dissolved in TE. The DNA was then digested with HaeIII (New England BioLabs R0108) and purified as described above. Digested genomic DNA was incubated with 10 mM O-Allylhydroxylamine hydrochloride (Sigma-Aldrich 05983) for 1 h at 37°C to block the pre-existing aldehydic lesions and the DNA was ethanol precipitated. The DNA was then treated with E. coli uracil DNA-glycosylase for 30 min at 37°C to excise uracils and create new abasic sites. The resulting abasic sites were labeled with 2 mM AA6 for 1 hour at 37°C and DNA was re-purified and dissolved in ddH2O. AA6-tagged DNA was labeled with 1.7 μM DBCO-Cy5 (Click Chemistry Tools A130) under Cu-free conditions by shaking the reaction mixture for 2 h at 37°C. Labeled DNA was purified using the DNA Clean and Concentrator kit (Zymo research D4014). Genomic DNA from E. coli CJ236 strain served as the uracil standard. WT and bisulfite-treated E. coli DNA served respectively as the negative and the positive controls. DBCO-Cy5-labeled DNA was spotted onto a positively charged zeta probe membrane (Bio-Rad 1620153) using a vacuum filtration apparatus and the membrane was scanned using a Typhoon 9210 phosphor imager (GE Healthcare). Cy5 fluorescence was analyzed using the ImageQuant software.
Conventional flow cytometry
Cells were incubated with an Fc blocking reagent (Miltenyi Biotec 130-059-901 or Tonbo Biosciences 70-0161) and stained in PBS at 4°C with antibodies to various cell surface antigens. In the experiments that used NP8-FITC and NP36-PE to measure B cell affinity maturation, NP8-FITC was added to the cells and incubated for 15 min before NP36-PE was added. For the staining of intracellular molecules, cells were subsequently fixed and permeabilized using a CytoFix/CytoPerm kit or a Transcription Factor Buffer set. Isotype-matched control antibodies were used to define the baseline staining for the molecules of interest. Cells or beads stained with each fluorochrome were used to establish fluorescent compensation. 7-aminoactinomycin D (7-AAD, Tonbo Biosciences 13-6993-T500 or BD 559925) or Ghost Dye Violet 510 (GV510) was used to identify and exclude non-viable cells from the analysis. Events were acquired on an LSR II or LSR Fortessa flow cytometer (BD) and analyzed using FlowJo version 7 or 10 (Tree Star).
Imaging flow cytometry
CD19+ B cells were purified from tonsillar cell suspensions by MACS with a biotinylated anti-CD19 antibody and anti-biotin microbeads. The cells were then incubated with an Fc blocking reagent and stained at 4°C with antibodies to surface IgD and CD38, fixed and permeabilized, and stained for AID and AIRE or with isotype control antibodies. Nuclei were counter stained with 4’,6-diamidine-2’-phenylindole dihydrochloride (DAPI). Tonsillar cells stained with each fluorochrome were used to establish fluorescent compensation. Cells were imaged on an ImageStream X Mark II imaging flow cytometer (Amnis) and data were analyzed using IDEAS 6.1 (Amnis).
Immunofluorescence analysis
Frozen human tissues were stored at −80°C before 6-7 μm tissue sections were made using a cryostat (Leica CM1950). Sections were fixed with 4% paraformaldehyde, permeabilized in PBS containing 0.2% Triton X-100, incubated with Image-iT FX signal enhancer solution, blocked with PBS containing 1% BSA, 0.1% Triton X-100, 100 μg/ml human IgG (for human tissues only) and 10% serum from the source of the fluorochrome-conjugated antibodies, and stained with various combinations of primary antibodies against the molecules of interest, followed by appropriate fluorochrome-conjugated secondary antibodies. For Bcl-6 staining, the antibody used was not diluted and, instead, a large enough volume was used to cover the tissue section. For AIRE staining, a 1:25 dilution of the Miltenyi Biotec anti-AIRE-APC conjugated antibody was used. Nuclei were visualized with DAPI. Following washing, slides were mounted using a FluoroSave reagent and imaged on a confocal microscope (Zeiss LSM 780 or Leica TCS SP5). Pseudocolor images were processed using Photoshop CS6 (Adobe).
ELISA
ELISA to determine NP-specific antibody affinity maturation was performed as previously described (Ballon et al., 2011) with minor modifications in the reagents. Briefly, each serum sample was titrated on both NP29-BSA- and NP4-BSA-coated microtiter plates. The ratio of binding to NP4-BSA and NP29-BSA is an indicator of relative Ig affinity maturation. Bound antibodies were detected using horseradish peroxidase (HRP)-conjugated goat-anti-mouse IgG1, IgG2b or IgG. The colorimetric reaction was terminated with the addition of an equal volume of 1 M H2SO4 and quantitated on a microplate reader (BioTek Epoch) at 450 nm. ELISA to determine IgG1 and IgA secretion by ex vivo stimulated mouse B cells was performed using a mouse IgG1 or IgA quantitation set. Anti-IL-17A, IL-17F and IL-22 autoantibodies in the mouse sera were measured using microtiters plates coated with 1 μg/ml recombinant murine IL-17A, IL-17F or IL-22. The plates were blocked with 10% BSA in PBS, washed, incubated with mouse serum samples, washed and then incubated with an alkaline phosphatase (ALP)-conjugated horse-anti-mouse IgG antibody (1:500). Following washing, the colorimetric reaction was developed using the BluePhos phosphatase substrate system and quantitated on a microplate reader (BioTek Epoch) at 620 nm. For the measurement of autoantibodies that block the binding of IL-17A, IL-17F and IL-22 to their receptors, mouse sera were incubated at 4°C overnight with HRP-conjugated IL-17A, IL-17F or IL-22 of the optimized concentrations, and the mixtures were added to microtiter plates coated with IL-17RA homodimer or IL-22Rα1 after the plates were blocked with 10% BSA in PBS. The plates were then incubated at room temperature for 1 hour and washed. The colorimetric reaction was developed and terminated with the addition of an equal volume of 1 M H2SO4 and quantitated on a microplate reader at 450 nm.
IgHV repertoire and mutation analysis
Live (7-AAD−) unswitched (IgM+IgD+) or switched (IgM−IgD−) NP-specific B cells (CD19+B220+NP36+) in the spleens of immunized μMT recipients were sorted using a SONY SH800 cell sorter (SONY Biotechnology) and resuspended in RNAProtect solution. High-throughput IgHV repertoire profiling by RNA-Seq was performed iRepertoire, Inc. (Huntsville, AL, USA). The raw sequences were processed and analyzed using the IMonitor 1.1.0 pipeline (Zhang et al., 2015). With this pipeline tool, each sequence was mapped to the Mus musculus germline V-D-J sequences (IMGT, http://www.imgt.org/vquest/refseqh.html) to identify the V, D and J gene segments and the CDRs, which were also used for clonal clustering. The sequences observed only once in a sample were filtered off to reduce the sequencing error. Subsequently, the sequences were normalized according to the number of cells in each sample. By comparing the sequence of each clone with the germline sequence, the mismatches of nucleotides were regarded as potential mutations. To eliminate PCR noise and sequencing errors, the first 25 bp of the sequences corresponding to the primer-binding site were excluded from the analysis, and the sequences were filtered if 3 successive mismatches were observed in them. Finally, the mutation rate for each IMGT position in the IgHV was calculated if the sequencing depth for that position was ≥10, and the frequency of each type of nucleotide substitution at these mutated positions were computed for each Ig isotype.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analyses
All statistical analyses were performed using Excel or Prism. Graphs represent data from at least 2 independent experiments, each consisting of at least 3 biological replicates. The exact number of the biological replicates (n) in the presented data set are indicated in the figure legends.
Results are expressed as mean ± SEM. Pair-wise statistical difference was assessed by the parametric t-test or the non-parametric Mann-Whitney U test depending on the distribution of the data, which is stated in the figure legends. Multiple group comparisons were performed using 1-way ANOVA with Tukey’s post hoc test. Differences were considered significant when P values were < 0.05. P values > 0.05 are either not shown, marked with NS (not significant), or indicated in the figure if they are close to 0.05. *P < 0.05, **P < 0.01, ***P < 0.001.
DATA AND SOFTWARE AVAILABILITY
Software Availability
All software used for the data analysis in this study is commercially or freely available. The IMonitor software used for immune repertoire analysis has been published (Zhang et al., 2015) and can be downloaded at https://github.com/zhangwei2015/IMonitor.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies against human antigens | ||
| Rat monoclonal anti-AID, unconjugated (mAID-2) | eBioscience | Cat#14-5959 |
| Rabbit polyclonal anti-AID, unconjugated | Jayanta Chaudhuri | (Vuong et al., 2009) |
| Rat monoclonal anti-AID, AF647 (EK2-5G9) | BD | Cat#565785 |
| Mouse monoclonal anti-AIRE, unconjugated (C-2) | Santa Cruz | Cat#sc-373703 |
| Human recombinant anti-AIRE, APC (REA352) | Miltenyi Biotec | Cat#130-105-401 |
| Rat monoclonal anti-AIRE, eF570 (TM-724) | eBioscience | Cat#41-9534 |
| Human recombinant anti-AIRE, APC (REA352) | Miltenyi Biotec | Cat#130-105-359 |
| Mouse monoclonal anti-Bcl6, unconjugated (GI191E/A8) | Ventana | Cat# 227M |
| Mouse monoclonal anti-CD19, Biotin (HIB19) | Biolegend | Cat#302204 |
| Mouse monoclonal anti-CD19, eF450 (HIB19) | Tonbo | Cat#75-0199-T100 |
| Mouse monoclonal anti-CD19, PE (4G7) | BD | Cat#349209 |
| Mouse monoclonal anti-CD19, PE (HIB19) | BD | Cat#555413 |
| Mouse monoclonal anti-CD19, PE-CF594 (HIB19) | BD | Cat#562321 |
| Mouse monoclonal anti-CD23, FITC (9P25) | Beckman | Cat#IM0529 |
| Mouse monoclonal anti-CD19, PE-Cy7 (HIB19) | eBioscience | Cat#25-0199 |
| Mouse monoclonal anti-CD19, QDot655 (SJ25C1) | Thermo Fisher | Cat#Q10179 |
| Mouse monoclonal anti-CD19, BV786 (SJ25C1) | BD | Cat#563326 |
| Mouse monoclonal anti-CD24, APC-eF780 (eBioSN3) | eBioscience | Cat#47-0247 |
| Mouse monoclonal anti-CD27, AF647 (O323) | Biolegend | Cat#302812 |
| Mouse monoclonal anti-CD38, APC (HIT2) | Biolegend | Cat#303510 |
| Mouse monoclonal anti-CD38, PE-Cy7 (HIT2) | Biolegend | Cat#303516 |
| Mouse monoclonal anti-CD45, eF450 (2D1) | eBioscience | Cat#48-9459 |
| Mouse monoclonal anti-CD45, PE-Cy7 (HI30) | Tonbo | Cat#60-0459 |
| Mouse monoclonal anti-EpCAM, AF488 (9C4) | Biolegend | Cat#324210 |
| Rabbit monoclonal anti-Hsp90, unconjugated (C45G5) | Cell Signaling | Cat#4877 |
| Goat polyclonal F(ab)2 anti-IgD, Biotin | Southern Biotech | Cat#2032-08 |
| Goat polyclonal F(ab)2 anti-IgD, FITC | Southern Biotech | Cat#2032-02 |
| Mouse monoclonal anti-IgD, FITC (IA6-2) | BD | Cat#555778 |
| Rabbit polyclonal anti-Lamin B1 unconjugated (H-90) | Santa Cruz | Cat#sc-20682 |
| Rat monoclonal anti-Pax5, AF594 (1H9) | BioLegend | Cat#649711 |
| Mouse monoclonal anti-β-Actin, unconjugated (AC-15) | Sigma-Aldrich | Cat#A5441 |
| Antibodies against mouse antigens | ||
| Rat monoclonal anti-AID, unconjugated (mAID-2) | eBioscience | Cat#14-5959 |
| Rabbit polyclonal anti-AID, unconjugated | Jayanta Chaudhuri | (Vuong et al., 2009) |
| Rabbit polyclonal anti-AIRE, unconjugated (H-300) | Santa Cruz | Cat#sc-33188 |
| Goat polyclonal anti-AIRE, unconjugated (D-17) | Santa Cruz | Cat#sc-17986 |
| Rat monoclonal anti-AIRE, unconjugated (5H12) | eBioscience | Cat#14-5934 |
| Rat monoclonal anti-AIRE, eF660 (5H12) | eBioscience | Cat#50-5934 |
| Human recombinant anti-AIRE, APC (REA352) | Miltenyi Biotec | Cat#130-105-359 |
| Rat monoclonal anti-B220, APC-Cy7 (RA3-6B2) | Biolegend | Cat#103224 |
| Rat monoclonal anti-B220, Biotin (RA3-6B2) | Biolegend | Cat#103204 |
| Rat monoclonal anti-BAFF-R, APC (eBio7H22-E16) | eBioscience | Cat#17-5943 |
| Rat monoclonal anti-CD138, Biotin (281-2) | Biolegend | Cat#142514 |
| Rat monoclonal anti-CD138, PE-Cy7 (281-2) | Biolegend | Cat#142511 |
| Rat monoclonal anti-CD16/CD32, unconjugated (2.4G2) | Tonbo | Cat#70-0161 |
| Rat monoclonal anti-CD16/CD32, unconjugated (2.4G2) | BD | Cat#553141 |
| Rat monoclonal anti-CD19, Biotin (1D3) | BD | Cat#553784 |
| Rat monoclonal anti-CD19, BV650 (6D5) | Biolegend | Cat#115541 |
| Rat monoclonal anti-CD19, FITC (1D3) | Tonbo | Cat#35-0193 |
| Rat monoclonal anti-CD19, Pacific Blue (6D5) | Biolegend | Cat#115523 |
| Rat monoclonal anti-CD19, PE-CF594 (1D3) | BD | Cat#562291 |
| Rat monoclonal anti-CD21, APC (7G6) | BD | Cat#558658 |
| Rat monoclonal anti-CD23, PE (B3B4) | BD | Cat#553139 |
| Rat monoclonal anti-CD25, APC (PC61.5) | Tonbo | Cat#20-0251 |
| Rat monoclonal anti-CD3, APC-Cy7 (17A2) | Tonbo | Cat#25-0032 |
| Rat monoclonal anti-CD38, PE-Cy7 (90) | Biolegend | Cat#102717 |
| Rat monoclonal anti-CD4, unconjugated (GK1.5) | Biolegend | Cat#100401 |
| Rat monoclonal anti-CD4, FITC (GK1.5) | Biolegend | Cat#100406 |
| Rat monoclonal anti-CD4, FITC (GK1.5) | BD | Cat#553729 |
| Rat monoclonal anti-CD4, PE (GK1.5) | Biolegend | Cat#100408 |
| Rat monoclonal anti-CD4, PerCP-Cy5.5 (GK1.5) | Biolegend | Cat#100434 |
| Hamster monoclonal anti-CD40, unconjugated (HM40-3) | eBioscience | Cat#16-0402 |
| Hamster monoclonal anti-CD40, FITC (H40-3) | BD | Cat#553723 |
| Rat monoclonal anti-CD45, violetFluor450 (30-F11) | Tonbo | Cat#75-0451 |
| Rat monoclonal anti-CD45, biotin (30-F11) | eBioscience | Cat#13-0451-85 |
| Rat monoclonal anti-CD62L, PE-Cy7 (MEL-14) | BD | Cat#560516 |
| Hamster monoclonal anti-CD80, PerCP-Cy5.5 (16-10A1) | BD | Cat#560526 |
| Rat monoclonal anti-CD83, BV650 (Michel-19) | Biolegend | Cat#121515 |
| Rat monoclonal anti-CD86, APC (GL-1) | BD | Cat#558703 |
| Rat monoclonal anti-CD93, PE (AA4.1) | Biolegend | Cat#136503 |
| Rat monoclonal anti-CXCR4, Biotin (2B11) | eBioscience | Cat#13-9991 |
| Rat monoclonal anti-CXCR5, PE (L138D7) | Biolegend | Cat#145504 |
| Hamster monoclonal anti-FAS, PE (Jo2) | BD | Cat#554258 |
| Rat monoclonal anti-Foxp3, V450 (MF23) | BD | Cat#561293 |
| Rabbit monoclonal anti-GAPDH, unconjugated (14C10) | Cell Signaling | Cat#2188 |
| Rat monoclonal anti-GL7, AF647 (GL7) | BD | Cat#561529 |
| Rabbit monoclonal anti-Hsp90, unconjugated (C45G5) | Cell Signaling | Cat#4877 |
| Rat monoclonal anti-I-Ab, PerCP-Cy5.5 (AF6-120.1) | Biolegend | Cat#116416 |
| Rat monoclonal anti-ICOSL, PE (HK5.3) | Biolegend | Cat#107405 |
| Rat monoclonal anti-IgA, FITC (C10-3) | BD | Cat#559354 |
| Goat polyclonal anti-IgA, HRP | Bethyl | Cat#A90-103P |
| Rat monoclonal anti-IgD, PE-Cy7 (11-26c.2a) | Biolegend | Cat#405727 |
| Rat monoclonal anti-IgD, PE-Cy7 (11-26c) | eBioscience | Cat#25-5993 |
| Horse polyclonal anti-IgG, ALP | Vector Laboratories | Cat#AP-2000 |
| Goat polyclonal anti-IgG1, HRP | Jackson Immuno | Cat#115-035-205 |
| Rat monoclonal anti-IgG1, PE-CF594 (A85-1) | BD | Cat#562559 |
| Goat polyclonal anti-IgG2b, HRP | Jackson Immuno | Cat#115-035-207 |
| Goat polyclonal anti-IgG3, HRP | Jackson Immuno | Cat#115-035-209 |
| Goat polyclonal F(ab)2 anti-IgM, unconjugated | Southern Biotech | Cat#1022-01 |
| Rat monoclonal anti-IgM, APC (RMM-1) | Biolegend | Cat#406509 |
| Goat polyclonal anti-IgM, HRP | Bethyl | Cat#A90-101P |
| Rat monoclonal anti-IL-17A, BV650 (TC11-18H10) | Biolegend | Cat#506929 |
| Goat polyclonal anti-IL-22, AF647 (Poly5164) | Biolegend | Cat#516406 |
| Rat monoclonal anti-Ly6G, AF647 (1A8) | Biolegend | Cat#127610 |
| Hamster monoclonal anti-PD-1, FITC (J43) | eBioscience | Cat#11-9985 |
| Hamster monoclonal anti-PD-1, PE-Cy7 (J43) | eBioscience | Cat#25-9985 |
| Rat monoclonal anti-PD-L1, PE-Cy7 (10F.9G2) | Biolegend | Cat#124314 |
| Rat monoclonal anti-PD-L2, PE (TY25) | BD | Cat#557796 |
| Goat polyclonal anti-Pol II, unconjugated | Bethyl | Cat#A303-835A |
| Rabbit polyclonal anti-Pol II Ser5, unconjugated | Bethyl | Cat#A304-408A |
| Rabbit polyclonal anti-Spt5, unconjugated | Santa Cruz | Cat#sc-28678 |
| Hamster monoclonal anti-TCRβ, PerCP-Cy5.5 (H57-597) | Biolegend | Cat#109227 |
| Hamster monoclonal anti-TCRγδ, BV421 (GL3) | Biolegend | Cat#118119 |
| Mouse monoclonal anti-β-Actin, unconjugated (AC-15) | Sigma-Aldrich | Cat#A5441 |
| Mouse monoclonal anti-IL17A, unconjugated (eBioMM17F3) | eBioscience | Cat#16-7173-81 |
| Mouse monoclonal anti-IL17F, unconjugated (RN17) | eBioscience | Cat#16-7473-82 |
| Mouse monoclonal anti-IL22, unconjugated (IL22JOP) | eBioscience | Cat#16-7222-82 |
| Antibodies against protein tags | ||
| Mouse monoclonal anti-6xHis, unconjugated (J099B12) | Biolegend | Cat#652502 |
| Mouse monoclonal anti-c-Myc, unconjugated (9E10) | Biolegend | Cat#626802 |
| Rabbit polyclonal anti-FLAG, unconjugated | Cell Signaling | Cat#2368 |
| Mouse monoclonal anti-FLAG, magnetic beads (M2) | Sigma-Aldrich | Cat#F3165 |
| Isotype control antibodies | ||
| Goat polyclonal IgG, unconjugated | Santa Cruz | Cat#sc-2028 |
| Goat polyclonal IgG, AF647 (Poly24030) | Biolegend | Cat#403006 |
| Goat polyclonal F(ab)2 IgG, Biotin | Southern Biotech | Cat#0110-08 |
| Goat polyclonal F(ab)2 IgG, FITC | Southern Biotech | Cat#0110-02 |
| Hamster monoclonal IgG2, PE (B81-3) | BD | Cat#550085 |
| Hamster monoclonal IgG2, PerCP-Cy5.5 (B81-3) | BD | Cat#560562 |
| Hamster monoclonal IgM, FITC (G235-1) | BD | Cat#553960 |
| Mouse monoclonal IgG1, AF647 (MOPC-21) | Biolegend | Cat#400155 |
| Mouse monoclonal IgG1, APC (MOPC-21) | BD | Cat#555751 |
| Mouse monoclonal IgG1, APC (MOPC-21) | Biolegend | Cat#400120 |
| Mouse monoclonal IgG1, Biotin (MOPC-21) | Biolegend | Cat#400103 |
| Mouse monoclonal IgG1, PE (MOPC-21) | BD | Cat#555749 |
| Mouse monoclonal IgG1, PE-Cy7 (MOPC-21) | BD | Cat#555872 |
| Mouse monoclonal IgG1, PE-Cy7 (MOPC-21) | Biolegend | Cat#400126 |
| Mouse monoclonal IgG2a, FITC (X39) | BD | Cat#349051 |
| Rabbit polyclonal IgG, unconjugated | Santa Cruz | Cat#sc-2027 |
| Rabbit polyclonal IgG, unconjugated | R&D | Cat#AF-008 |
| Rat monoclonal IgG1, BV650 (RTK2071) | Biolegend | Cat#400437 |
| Rat monoclonal IgG2a, unconjugated (RTK2758) | Biolegend | Cat#400501 |
| Rat monoclonal IgG2a, APC (RTK2758) | Biolegend | Cat#400511 |
| Rat monoclonal IgG2a, Biotin (eBR2a) | eBioscience | Cat#13-4321 |
| Rat monoclonal IgG2a, eF570 (eBR2a) | eBioscience | Cat#41-4321 |
| Rat monoclonal IgG2a, PE (eBR2a) | eBioscience | Cat#12-4321 |
| Rat monoclonal IgG2a, PerCP-Cy5.5 (RTK2758) | Biolegend | Cat#400531 |
| Rat monoclonal IgG2a, PE-Cy7 (RTK2758) | Biolegend | Cat#400521 |
| Rat monoclonal IgG2b, unconjugated (eB149/10H5) | eBioscience | Cat#14-4031 |
| Rat monoclonal IgG2b, AF647 (A95-1) | BD | Cat#557691 |
| Rat monoclonal IgG2b, Biotin (RTK4530) | Biolegend | Cat#400603 |
| Rat monoclonal IgG2b, eF660 (eB149/10H5) | eBioscience | Cat#50-4031 |
| Rat monoclonal IgG2b, PE (A95-1) | BD | Cat#553989 |
| Rat monoclonal IgG2b, PE-Cy7 (RTK4530) | Biolegend | Cat#400617 |
| Human recombinant IgG, APC (REA293) | Miltenyi Biotec | Cat#130-104-615 |
| Human recombinant IgG, PE (REA293) | Miltenyi Biotec | Cat#130-104-613 |
| Secondary antibodies | ||
| Mouse monoclonal anti-Biotin, magnetic microbeads | Miltenyi Biotec | Cat#130-090-485 |
| Donkey polyclonal anti-goat IgG, HRP | Santa Cruz | Cat#sc-2020 |
| Donkey polyclonal anti-mouse IgG, AF546 | Thermo Fisher | Cat#A10036 |
| Donkey polyclonal anti-mouse IgG, CF647 | Sigma-Aldrich | Cat#SAB4600176 |
| Donkey polyclonal anti-mouse IgG, HRP | Santa Cruz | Cat#sc-2318 |
| Donkey polyclonal F(ab’)2 anti-rat IgG, HRP | Jackson Immuno | Cat#712-036-153 |
| Goat polyclonal F(ab’)2 anti-mouse IgG, Cy3 | Jackson Immuno | Cat#115-166-006 |
| Goat polyclonal F(ab’)2 anti-mouse IgG, FITC | Southern Biotech | Cat#1032-02 |
| Goat polyclonal F(ab’)2 anti-rabbit IgG, Cy3 | Jackson Immuno | Cat#111-166-047 |
| Goat polyclonal anti-rabbit IgG, HRP | Santa Cruz | Cat#sc-2004 |
| Bacterial, Fungal and Viral Strains | ||
| Candida albicans | ATCC | Cat#MYA-2876 |
| Heat Inactivated C. albicans pseudohyphae | Made in house | This paper |
| Live C. albicans pseudohyphae | Made in house | This paper |
| Biological Samples | ||
| Human blood | American Red Cross | This paper |
| Human spleen | Children’s Hospital of Michigan | This paper |
| Human thymus | Children’s Hospital of Michigan | This paper |
| Human tonsil, healthy | Children’s Hospital of Michigan | This paper |
| Human tonsil, HIGM3 | Mount Sinai Medical Center | This paper |
| Sheep red blood cell | Innovative | Cat#IC100-0210 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| NP8, FITC | BioSearch | Cat#N-5050F |
| NP36, PE | BioSearch | Cat#N-5070-1 |
| NP32, KLH | BioSearch | Cat#N-5060 |
| NP4, BSA | BioSearch | Cat#N-5050L-10 |
| NP29, BSA | BioSearch | Cat#N-5050H-10 |
| UEA-1, Biotin | Vector Laboratories | Cat#B-1065 |
| Streptavidin, AF488 | Thermo Fisher | Cat#S11223 |
| Streptavidin, AF546 | Thermo Fisher | Cat#S11225 |
| Streptavidin, PerCP-Cy5.5 | BD | Cat#551419 |
| Streptavidin, BV605 | Biolegend | Cat#405229 |
| Streptavidin, BV785 | Biolegend | Cat#405249 |
| Streptavidin, Qdot605 | Thermo Fisher | Cat#Q10101MP |
| Aldehyde Reactive Probe | Cayman Chemical | Cat#10009350 |
| RNase A | Sigma | Cat# R6513 |
| Proteinase K | Qiagen | Cat# 9133 |
| HaeIII | New England Biolabs | Cat# R0108S |
| O-Allylhydroxylamine hydrochloride (AA7) | Sigma | Cat# 05983 |
| Methoxyamine hydrochloride | Sigma | Cat# 226904 |
| Alkoxyamine-6 (AA6) | Ashok Bhagwat | (Wei et al., 2017) |
| Alkoxyamine-3 (AA3) | Ashok Bhagwat | (Wei et al., 2015) |
| DBCO -Cy5 | Click Chemistry Tools | Cat# A130-1 |
| Cy5 Azide | Lumiprobe | Cat# A3330 |
| Copper bromide | Sigma | Cat# 254185 |
| TBTA Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine | Sigma | Cat# 678937 |
| DNA Clean and Concentrator kit | Zymo research | Cat# D4014 |
| SYBR Gold nucleic acid gel stain | Invitrogen | Cat# S11494 |
| Zeta probe membrane | Bio-Rad | Cat# 1620165 |
| Complete Freund’s Adjuvant | Thermo Fisher | Cat#77140 |
| Incomplete Freund’s Adjuvant | Thermo Fisher | Cat#77145 |
| Histopaque-1077 | Sigma-Aldrich | Cat#10771 |
| Ammonium-Chloride-Potassium (ACK) Lysis Buffer | Thermo Fisher | Cat#A1049201 |
| Collagenase II | Worthington | Cat#LS004177 |
| Collagenase V | Sigma-Aldrich | Cat#C9263-1G |
| DNase I | Roche | Cat#11284932001 |
| CD40L (Human) | Peprotech | Cat#310-02 |
| IL-4 (Human) | Peprotech | Cat#200-21 |
| IFN-β (Human) | Peprotech | Cat#300-02 |
| CD40L (Mouse) | Peprotech | Cat#315-15 |
| IL-4 (Mouse) | Peprotech | Cat#214-14 |
| IL-4 (Mouse) | R&D | Cat#404-ML |
| IL-17A (Mouse) | Peprotech | Cat#210-17 |
| IL-17F (Mouse) | Peprotech | Cat#210-17F |
| IL-17RA homodimer (Mouse) | R&D | Cat# 4481-MR-100 |
| IL-21 (Mouse) | Peprotech | Cat#210-21 |
| IL-22 (Mouse) | Peprotech | Cat#210-22 |
| IL-22Rα1 (Mouse) | R&D | Cat# 4294-MR-050 |
| TGF-β1 (Mouse) | R&D | Cat#7666-MB/CF |
| AIRE and AID WT and mutant proteins, Table S2 | Kang Chen | This paper |
| Caffeic Acid Phenethyl Ester (CAPE) | Cayman | Cat#70750 |
| Carboxyfluorescein Succinimidyl Ester (CFSE) | Biolegend | Cat#422701 |
| Lipofectamine 3000 | Thermo Fisher | Cat#L3000015 |
| Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#P8340 |
| TRIzol | Thermo Fisher | Cat#15596026 |
| RNALater | Thermo Fisher | Cat#AM7020 |
| RNAProtect | QIAGEN | Cat#76526 |
| NP-40 | Sigma-Aldrich | Cat#I8896 |
| 4-20% Bis-Tris Gel | GeneScript | Cat#M42012 |
| 10% Tris-Glycine Gel | Bio-Rad | Cat#4561034 |
| Polyvinylidene fluoride (PVDF) Membrane | Bio-Rad | Cat#1620177 |
| Nitrocellulose Membrane | Bio-Rad | Cat#1620115 |
| ECL Substrate | Bio-Rad | Cat#170-5061 |
| Western Blotting Chemiluminescence Luminol Reagent | Santa Cruz | Cat#sc-2048 |
| Fc Blocking Reagent | Miltenyi Biotec | Cat#130-059-901 |
| Fc Blocking Reagent | Tonbo Biosciences | Cat#70-0161 |
| CytoFix/CytoPerm | BD | Cat#554722 |
| Transcription Factor Buffer | BD | Cat#562725 |
| 7-aminoactinomycin D (7AAD) | Tonbo Biosciences | Cat#13-6993-T500 |
| 7-aminoactinomycin D (7AAD) | BD | Cat#559925 |
| Ghost Dye Violet 510 (GV510) | Tonbo Biosciences | Cat#13-0870-T500 |
| Anti-Biotin Magnetic Microbeads | Miltenyi Biotec | Cat#130-090-485 |
| 4’,6-diamidine-2’-phenylindole dihydrochloride (DAPI) | Sigma-Aldrich | Cat#D9542 |
| BluePhos Phosphate Substrate | KPL | Cat#50-88-02 |
| BbsI restriction enzyme | Thermo Fisher | Cat#ER1011 |
| Ficoll-Paque | GE Healthcare | Cat#17-1440-03 |
| Golgi Plug | BD | Cat#55029 |
| PMA | Sigma-Aldrich | Cat#P1585-1MG |
| Ionomycin | Thermo Fisher | Cat#I24222 |
| Quick T4 Ligase | New Engl. Biolabs | Cat#M2200L |
| Opti-MEM | Thermo Fisher | Cat#31985070 |
| Protein G Beads | Cell Signaling | Cat#8740 |
| Protein G Beads | Thermo Fisher | Cat#88847 |
| CelLytic M Buffer | Sigma-Aldrich | Cat#C2978 |
| Halt Phosphatase Inhibitor | Thermo Fisher | Cat#78426 |
| FluorSave mounting medium | EMD Millipore | Cat#345789 |
| HRP conjugation kit | Abcam | Cat#Ab102890 |
| Image-iT FX Signal Enhancer | Thermo Fisher | Cat#I36933 |
| Critical Commercial Assays | ||
| Mouse B Cell Isolation kit | Miltenyi Biotec | Cat#130-090-862 |
| Click-iT EdU Flow Cytometry Assay kit | Thermo Fisher | Cat#C10418 |
| Phusion Site-Directed Mutagenesis kit | Thermo Fisher | Cat#F541 |
| Amaxa Cell Line Nucleofector Kit V | Lonza | Cat#VCA-1003 |
| BCA Protein Assay kit | Thermo Fisher | Cat#23225 |
| iTaq Universal SYBR Green One-Step kit | Bio-Rad | Cat#172-5150 |
| Cells-to-CT 1-step SYBR Green kit | Thermo Fisher | Cat#A25601 |
| ChIP Assay kit | EMD Millipore | Cat#17-295 |
| Mouse IgA ELISA | Bethyl | Cat#E90-103 |
| Superscript III First Strand Synthesis | Thermo Fisher | Cat#18808051 |
| PowerSYBR Green Master Mix | Thermo Fisher | Cat#4367660 |
| Experimental Models: Cell Lines | ||
| CH12F3, WT | Tasuku Honjo | (Nakamura et al., 1996) |
| CH12F3, Aire−/− (clones 43, 53 and 69) | Kang Chen | This Paper |
| CH12F3, Acida−/− | Kefei Yu | This Paper |
| CH12F3, Ung−/− | Ashok Bhagwat | This Paper |
| HKB-11 | ATCC | Cat#12568 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | Jackson | Cat#000664 |
| Aire+/− (B6.129S2-Airetm1.1Doi/J) | Jackson | Cat#004743 |
| μMT (B6.129S2-Ighmtm1Cgn/J) | Jackson | Cat#002288 |
| B6.AireAdig reporter | Mark Anderson | (Gardner et al., 2008) |
| Aicda −/− | Tasuku Honjo | (Muramatsu et al., 2000) |
| CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) | Jackson | Cat#002014 |
| Oligonucleotides | ||
| A) qRT-PCR Primers | ||
| Human Primers: | ||
|
ACTB: _F: AGAGCTACGAGCTGCCTGAC _R: AGCACTGTGTTGGCGTACAG |
Sigma-Aldrich | (Wang et al., 2013) |
| Iμ_F: GTGATTAAGGAGAAACACTTTGAT | Sigma-Aldrich | (Chen et al., 2009) |
| Cγ1_R: CCAGGGCTGCTGTGCCCCCA | Sigma-Aldrich | This Paper |
| Cγ3_R: CCAGGGCCGCTGTGCCCCCA | Sigma-Aldrich | This Paper |
|
AIRE: _F: CCAGGCTCTCAACTGAAGGC _R: GAATCCCGTTCCCGAGTGG |
Sigma-Aldrich | (Dudakovic et al., 2015) |
| Mouse Primers: | ||
|
Actb _F: TGCGTGACATCAAAGAGAAG _R: CGGATGTCAACGTCACACTT |
Sigma-Aldrich | (Steuerwald et al., 2000) |
|
Aicda _F: GAAAGTCACGCTGGAG _R: TCTCATGCCGTCCCTT |
Sigma-Aldrich | (Xu et al., 2015) |
|
Aire (B6.129S2-Airetm1.1Doi/J): _F: GACAAGTTCCAGGAGACGCT _R: AAGCCGTCCAGGATGCTATG |
Sigma-Aldrich | This Paper |
| Iα-Cβ circle transcript: Iα_F: CCAGGCATGGTTGAGATAGAGATAG Cβ_R: AATGGTGCTGGGCAGGAAGT |
Sigma-Aldrich | (Cao et al., 2015) |
| Iγ1-Cμ circle transcript: Iγ1_F: GGCCCTTCCAGATCTTTGAG Cμ_R: AATGGTGCTGGGCAGGAAGT |
Sigma-Aldrich | (Doi et al., 2008) |
| Iμ-Cμ germline transcript: Iμ_F: CTCTGGCCCTGCTTATTGTTG Cμ’_R: GAAGACATTTGGGAAGGACTG |
Sigma-Aldrich | (Muramatsu et al., 2000) |
| Iα-Cα germline transcript: Iα_F: CCTGGCTGTTCCCCTATGAA Cα_R: GAGCTCGTGGGAGTGTCAGTG |
Sigma-Aldrich | (Boersma et al., 2015) |
| Sμ (after ChIP): Sμ_F: TAGTAAGCGAGGCTCTAAAAAGCAT Sμ_R: AGAACAGTCCAGTGTAGGCAGTAGA |
Sigma-Aldrich | (Nowak et al., 2011) |
| Iμ (after ChIP): Iμ_F: GCTCAGCCTGGACTTTCGGTTTGGT Iμ_R: GGAGTCAAGATGGCCGATCAGAACC |
Sigma-Aldrich | (Nowak et al., 2011) |
| Sγ1 (after ChIP): Sγ1_F: TATGATGGAAAGAGGGTAGCATTCACC Sγ1_R: CTCCTTCCCAATCTCCCGTG |
Sigma-Aldrich | (Nowak et al., 2011) |
| sgRNA (CH12 mutant 69): GCACCGCACCGAGATCGCGG (PAM:TGG) |
Feng Zhang | http://crispr.mit.edu |
| sgRNA (CH12 mutants 43 and 53): ACCTAAACCAGTCCCGGAAA (PAM:GGG) |
Feng Zhang | http://crispr.mit.edu |
| B) Primers for screening CRISPR mutants | ||
| CH12 Mutant 69: _F: CTTTCCCGCTTCCTCTATCC _R: ACTGTCTATGGCCACCGC |
Sigma-Aldrich | This paper |
| CH12 Mutant 43: _F1: ACCTAAACCAGTCCCGGAAA _F2 (Second Mutation): CCATTGTTCCTGCCCCTG _R: ACCGTTTCCAAGAGGAAGGT |
Sigma-Aldrich | This paper |
| CH12 Mutant 53: _F: ACCTAAACCAGTCCCGGAAA _R: ACCGTTTCCAAGAGGAAGGT |
Sigma-Aldrich | This paper |
| C) Primers used to generate AIRE and AICDA mutants, Table S3 | Sigma-Aldrich | This paper |
| D) Oligos used to clone sgRNA for CH12 CRISPR mutants, Table S3 | Sigma-Aldrich | This paper |
| E) Primers to amplify β-Actin gDNA after DNAse I treatment | ||
|
ACTB_gDNA:_F: CAAGGCCAACCGCGAGAAGA _R: TGTGCTGGGGTCTTGGGATG |
Sigma-Aldrich | This paper |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-Puro | Addgene | Cat#48139 |
| pFLAG-CMV2 | Sigma-Aldrich | Cat#E7033 |
| pcDNA3.1(-) | Thermo Fisher | Cat#V38520 |
| pcDNA3-eGFP | Thilo Hagen | N/A |
| pCMV-Tag1 | Agilent Technologies | Cat#211170 |
| Software and Algorithms | ||
| FlowJo 7 and 10 | Tree Star | N/A |
| Adobe Photoshop CS6 | Adobe | N/A |
| Prism 6 | GraphPad | N/A |
| StepOne RT-PCR software | Applied Biosystems | N/A |
| IDEAS 6.1 | Amnis | N/A |
| VectorNTI 10 | Thermo Fisher | N/A |
| ImageQuant TL 8.1 | GE Healthcare | N/A |
| IMonitor 1.1.0 | Wei Zhang | (Zhang et al., 2015) |
ACKNOWLEDGMENTS
We are grateful to D. Mathis and C. Benoist (Harvard Medical School, USA) for AIRE cDNA plasmids, T. Honjo (Kyoto University, Japan) for Aicda−/− mice and CH12 cells, M. Anderson (University of California-San Francisco, USA) for AireAdig mice and discussions, T. Hagen (National University of Singapore) for the pcDNA3-eGFP vector, J.X. Ang, G. Lee and Q. Peng (Republic Polytechnic, Singapore) for assistance in creating and validating Aire−/− CH12 cells, C. Lim (Republic Polytechnic, Singapore) for assistance in constructing Aire mutant plasmids, E. van Buren and J. Back (Microscopy and Flow Cytometry Core of the Barbara Ann Karmanos Cancer Institute, USA) for assistance in cell sorting and imaging flow cytometry, and S. Dzinic, K. White and J. Kushner (Animal Model and Therapeutics Evaluation Core of the Barbara Ann Karmanos Cancer Institute, USA) for assistance in some animal experiments. The Chen laboratory is supported by the US National Institutes of Health (R21AI122256, U01AI95776 Mucosal Immunology Studies Team Young Investigator Award), Burroughs Wellcome Fund (1013738 Preterm Birth Initiative), American Congress of Obstetricians and Gynecologists (Industry Grant in Immunization), China Jiangsu Province Department of Health (Expert of Medical Research Program) and Wayne State University (Office of the Vice President for Research, Perinatal Research Initiative). The Bhagwat laboratory was supported by the National Institutes of Health grant R01GM057200 and Wayne State University Bridge award. S.Z. is supported by the Lee Kuan Yew Postdoctoral Fellowship.
REFERENCES
- Abramson J., Giraud M., Benoist C., and Mathis D. (2010). Aire’s partners in the molecular control of immunological tolerance. Cell 140, 123–135. [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., et al. (2008). The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity 29, 423–437. [DOI] [PubMed] [Google Scholar]
- Al-Herz W., Bousfiha A., Casanova J.L., Chapel H., Conley M.E., Cunningham-Rundles C., Etzioni A., Fischer A., Franco J.L., Geha R.S., et al. (2011). Primary immunodeficiency diseases: an update on the classification from the international union of immunological societies expert committee for primary immunodeficiency. Frontiers in immunology 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., et al. (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J., von Boehmer H., Bronson R., Dierich A., Benoist C., et al. (2002). Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Bacchetta R., and Notarangelo L.D. (2013). Immunodeficiency with autoimmunity: beyond the paradox. Frontiers in immunology 4, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballon G., Chen K., Perez R., Tam W., and Cesarman E. (2011). Kaposi sarcoma herpesvirus (KSHV) vFLIP oncoprotein induces B cell transdifferentiation and tumorigenesis in mice. The Journal of clinical investigation 121, 1141–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi F., Sommariva M., De Cecco L., Triulzi T., Romero-Cordoba S., Tagliabue E., Sfondrini L., and Balsari A. (2016). Expression and prognostic significance of the autoimmune regulator gene in breast cancer cells. Cell Cycle 15, 3220–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma V., Moatti N., Segura-Bayona S., Peuscher M.H., van der Torre J., Wevers B.A., Orthwein A., Durocher D., and Jacobs J.J.L. (2015). MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5’ end resection. Nature 521, 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Scharff M.D., and Goodman M.F. (2003). Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proceedings of the National Academy of Sciences of the United States of America 100, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantaert T., Schickel J.N., Bannock J.M., Ng Y.S., Massad C., Oe T., Wu R., Lavoie A., Walter J.E., Notarangelo L.D., et al. (2015). Activation-Induced Cytidine Deaminase Expression in Human B Cell Precursors Is Essential for Central B Cell Tolerance. Immunity 43, 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao A T., Yao S., Gong B., Nurieva R.I., Elson C.O., andCong Y. (2015). Interleukin (IL)-21 promotes intestinal IgA response to microbiota. Mucosal immunology 8, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., and Di Noia J.M. (2016). Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nature reviews Immunology 16, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., and Alt F.W. (2003). Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730. [DOI] [PubMed] [Google Scholar]
- Chen K., Xu W., Wilson M., He B., Miller N.W., Bengten E., Edholm E.S., Santini P.A., Rath P., Chiu A., et al. (2009). Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nature immunology 10, 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti H.R., Huppler A.R., Whibley N., and Gaffen S.L. (2014). Animal models for candidiasis. Curr Protoc Immunol 105, 19.16.11–19.16.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi T., Obayashi K., Kadowaki T., Fujii H., and Koyasu S. (2008). PI3K is a negative regulator of IgE production. International immunology 20, 499–508. [DOI] [PubMed] [Google Scholar]
- Dudakovic A., Camilleri E.T., Xu F., Riester S.M., McGee-Lawrence M.E., Bradley E.W., Paradise C.R., Lewallen E.A., Thaler R., Deyle D.R., et al. (2015). Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. J Biol Chem 290, 27604–27617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandy A., Revy P., Imai K., and Fischer A. (2005). Hyper-immunoglobulin M syndromes caused by intrinsic B-lymphocyte defects. Immunological reviews 203, 67–79. [DOI] [PubMed] [Google Scholar]
- Finnish-German A.C. (1997). An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nature genetics 17, 399–403. [DOI] [PubMed] [Google Scholar]
- Gardner J.M., Devoss J.J., Friedman R.S., Wong D.J., Tan Y.X., Zhou X., Johannes K.P., Su M.A., Chang H.Y., Krummel M.F., et al. (2008). Deletional tolerance mediated by extrathymic Aire-expressing cells. Science 321, 843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J.M., Metzger T.C., McMahon E.J., Au-Yeung B.B., Krawisz A.K., Lu W., Price J.D., Johannes K.P., Satpathy A.T., Murphy K.M., et al. (2013). Extrathymic Aire-expressing cells are a distinct bone marrow-derived population that induce functional inactivation of CD4(+) T cells. Immunity 39, 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavanescu I., Benoist C., and Mathis D. (2008). B cells are required for Aire-deficient mice to develop multi-organ autoinflammation: A therapeutic approach for APECED patients. Proceedings of the National Academy of Sciences of the United States of America 105, 13009–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud M., Jmari N., Du L., Carallis F., Nieland T.J., Perez-Campo F.M., Bensaude O., Root D.E.,Hacohen N., Mathis D., et al. (2014). An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proceedings of the National Academy of Sciences of the United States of America 111, 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud M., Yoshida H., Abramson J., Rahl P.B., Young R.A., Mathis D., and Benoist C. (2012). Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proceedings of the National Academy of Sciences of the United States of America 109, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikos A.P., and Tsokos G.C. (2012). Immunodeficiency and autoimmunity: lessons from systemic lupus erythematosus. Trends in molecular medicine 18, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R.P., DePianto D.J., Jacob J.T., Han M.C., Chung B.M., Batazzi AS., Poll B.G., Guo Y., Han J., Ong S., et al. (2015). Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes. Nature genetics 47, 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.M., Cambier J.C., and Shlomchik M.J. (2012). B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science 336, 1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I.S., Mouchess M.L., Zhu M.L., Conley B., Fasano K.J., Hou Y., Fong L., Su M.A., and Anderson M.S. (2014). Enhancement of an anti-tumor immune response by transient blockade of central T cell tolerance. The Journal of experimental medicine 211, 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisand K., Boe Wolff A.S., Podkrajsek K.T., Tserel L., Link M., Kisand K.V., Ersvaer E., Perheentupa J., Erichsen M.M., Bratanic N., et al. (2010). Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. The Journal of experimental medicine 207, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisand K., and Peterson P. (2015). Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy. Journal of clinical immunology 35, 463–478. [DOI] [PubMed] [Google Scholar]
- Klein F., Diskin R., Scheid J.F., Gaebler C., Mouquet H., Georgiev I.S., Pancera M., Zhou T., Incesu R.B., Fu B.Z., et al. (2013). Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153, 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh A.S., Miller E.L., Buenrostro J.D., Moskowitz D.M., Wang J., Greenleaf W.J., Chang H.Y., and Crabtree G.R. (2018). Rapid chromatin repression by Aire provides precise control of immune tolerance. Nature immunology 19, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh E., Lind S.M., Lindmark E., Hassler S., Perheentupa J., Peltonen L., Winqvist O., and Karlsson M.C. (2008). AIRE regulates T-cell-independent B-cell responses through BAFF. Proceedings of the National Academy of Sciences of the United States of America 105, 18466–18471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.J., Joshua D.E., Williams G.T., Smith C.A., Gordon J., and MacLennan I.C. (1989). Mechanism of antigen-driven selection in germinal centres. Nature 342, 929–931. [DOI] [PubMed] [Google Scholar]
- Malchow S., Leventhal D.S., Nishi S., Fischer B.I., Shen L., Paner G.P., Amit A.S., Kang C., Geddes J.E., Allison J.P., et al. (2013). Aire-dependent thymic development of tumor-associated regulatory T cells. Science 339, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul R.W., Saribasak H., Martomo S.A., McClure R.L., Yang W., Vaisman A., Gramlich H.S., Schatz D.G., Woodgate R., Wilson D.M. 3rd, et al. (2011). Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nature immunology 12, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Woodward M., Hertel C., Vlaicu P., Haque Y., Karner J., Macagno A., Onuoha S.C., Fishman D., Peterson H., et al. (2016). AIRE-Deficient Patients Harbor Unique High-Affinity Disease-Ameliorating Autoantibodies. Cell 166, 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G., Ng Y.S., Bannock J.M., Lavoie A., Walter J.E., Notarangelo L.D., Kilic S.S., Aksu G., Debre M., Rieux-Laucat F., et al. (2011). Activation-induced cytidine deaminase (AID) is required for B-cell tolerance in humans. Proceedings of the National Academy of Sciences of the United States of America 108, 11554–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., and Honjo T. (2000). Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563. [DOI] [PubMed] [Google Scholar]
- Murphy K.M., and Weaver C. (2016a). Basic Concepts in Immunology. In Janeway’s Immunobiology (Garland Science), pp. 1–36. [Google Scholar]
- Murphy K.M., and Weaver C. (2016b). The Humoral Immune Response. In Janeway’s Immunobiology (Garland Science), pp. 399–441. [Google Scholar]
- Nagamine K., Peterson P., Scott H.S., Kudoh J., Minoshima S., Heino M., Krohn K.J., Lalioti M.D., Mullis P.E., Antonarakis S.E., et al. (1997). Positional cloning of the APECED gene. Nature genetics 17, 393–398. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Kondo S., Sugai M., Nazarea M., Imamura S., and Honjo T. (1996). High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. International immunology 8, 193–201. [DOI] [PubMed] [Google Scholar]
- Natarajan K., Singh S., Burke T.R. Jr., Grunberger D., and Aggarwal B.B. (1996). Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proceedings of the National Academy of Sciences of the United States of America 93, 9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak U., Matthews A.J., Zheng S., and Chaudhuri J. (2011). The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nature immunology 12, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oven I., Brdickova N., Kohoutek J., Vaupotic T., Narat M., and Peterlin B.M. (2007). AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol 27, 8815–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude A.M., Orthwein A., Hu Y., Campo V.A., Kavli B., Buschiazzo A., and Di Noia J.M. (2009). Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol 16, 517–527. [DOI] [PubMed] [Google Scholar]
- Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., et al. (2010). Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143, 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B.M., and Price D.H. (2006). Controlling the elongation phase of transcription with P-TEFb. Molecular cell 23, 297–305. [DOI] [PubMed] [Google Scholar]
- Poliani P.L., Kisand K., Marrella V., Ravanini M., Notarangelo L.D., Villa A., Peterson P., and Facchetti F. (2010). Human peripheral lymphoid tissues contain autoimmune regulator-expressing dendritic cells. The American journal of pathology 176, 1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A., Doffinger R., Natividad A., Chrabieh M., Barcenas-Morales G., Picard C., Cobat A., Ouachee-Chardin M., Toulon A., Bustamante J., et al. (2010). Autoantibodies against IL- 17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. The Journal of experimental medicine 207, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. (2000). Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell 102, 565–575. [DOI] [PubMed] [Google Scholar]
- Schroeder K.M.S., Agazio A., Strauch P.J., Jones S.T., Thompson S.B., Harper M.S., Pelanda R., Santiago M.L., and Torres R.M. (2017). Breaching peripheral tolerance promotes the production of HIV-1-neutralizing antibodies. The Journal of experimental medicine 214, 2283–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalhout S., Haddad D., Sosin A., Holland T.C., Al-Katib A., Martin A., and Bhagwat A.S. (2014). Genomic uracil homeostasis during normal B cell maturation and loss of this balance during B cell cancer development. Mol Cell Biol 34, 4019–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., and Allison J.P. (2015). The future of immune checkpoint therapy. Science 348, 56–61. [DOI] [PubMed] [Google Scholar]
- Silver J., Zuo T., Chaudhary N., Kumari R., Tong P., Giguere S., Granato A., Donthula R., Devereaux C., and Wesemann D.R. (2018). Stochasticity enables BCR-independent germinal center initiation and antibody affinity maturation. The Journal of experimental medicine 215, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A., Klapacz J., Samaranayake M., Ullah A., and Bhagwat A.S. (2003). Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic acids research 31, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerwald N., Cohen J., Herrera R.J., and Brenner C.A. (2000). Quantification of mRNA in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT-PCR. Mol Hum Reprod 6, 448–453. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan B., Yen W.F., Pucella J.N., and Chaudhuri J. (2014). AIDing Chromatin and Transcription-Coupled Orchestration of Immunoglobulin Class-Switch Recombination. Frontiers in immunology 5, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Sanz I., and Cook M.C. (2009). Dysregulation of germinal centres in autoimmune disease. Nature reviews Immunology 9, 845–857. [DOI] [PubMed] [Google Scholar]
- Vuong B.Q., Lee M., Kabir S., Irimia C., Macchiarulo S., McKnight G.S., and Chaudhuri J. (2009). Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nature immunology 10, 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Huang J., Lyu H., Lee C.K., Tan J., Wang J., and Liu B. (2013). Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis 4, e556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Shinkura R., Doi Y., Maruya M., Fagarasan S., and Honjo T. (2011). Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nature immunology 12, 264–270. [DOI] [PubMed] [Google Scholar]
- Wei S., Perera M.L.W., Sakhtemani R., and Bhagwat A.S. (2017). A novel class of chemicals that react with abasic sites in DNA and specifically kill B cell cancers. PLoS One 12, e0185010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S., Shalhout S., Ahn Y.H., and Bhagwat A.S. (2015). A versatile new tool to quantify abasic sites in DNA and inhibit base excision repair. DNA Repair (Amst) 27, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Chapman J.R., Brandsma I., Yuan J., Mistrik M., Bouwman P., Bartkova J., Gogola E., Warmerdam D., Barazas M., et al. (2015). REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zan H., Pone E.J., Mai T., and Casali P. (2012). Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nature reviews Immunology 12, 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Nedjic J., Hinterberger M., Steinert M., Koser S., Pinto S., Gerdes N., Lutgens E., Ishimaru N., Busslinger M., et al. (2015). Thymic B Cells Are Licensed to Present Self Antigens for Central T Cell Tolerance Induction. Immunity 42, 1048–1061. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Bansal K., Schaefer U., Chapman T., Rioja I., Proekt I., Anderson M.S., Prinjha R.K., Tarakhovsky A., Benoist C., et al. (2015). Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proceedings of the National Academy of Sciences of the United States of America 112, E4448–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I., Kageyama R., Monticelli L., Johnston R.J., Ditoro D., Hansen K., Barnett B., and Crotty S. (2010). Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). Journal of immunology 185, 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandman-Goddard G., and Shoenfeld Y. (2002). HIV and autoimmunity. Autoimmunity reviews 1, 329–337. [DOI] [PubMed] [Google Scholar]
- Zhang W., Du Y., Su Z., Wang C., Zeng X., Zhang R., Hong X., Nie C., Wu J., Cao H., et al. (2015). IMonitor: A Robust Pipeline for TCR and BCR Repertoire Analysis. Genetics 201, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Software Availability
All software used for the data analysis in this study is commercially or freely available. The IMonitor software used for immune repertoire analysis has been published (Zhang et al., 2015) and can be downloaded at https://github.com/zhangwei2015/IMonitor.