Abstract
Background:
Recent molecular surveillance suggests an unexpectedly high prevalence of non-falciparum malaria in Africa. Malaria control is also challenged by undetected asymptomatic P. falciparum malaria resulting in an undetectable reservoir for potential transmission. Context-specific surveillance of asymptomatic P. falciparum and non-falciparum species is needed to properly inform malaria control programs.
Methods:
We performed quantitative real time PCR for four malaria species in 4,595 primarily adult individuals in Rwanda using the 2014–2015 Demographic Health Survey. We assessed correlates of infection by species to explore attributes associated with each species. Asymptomatic P. ovale spp., P. malariae, and P. falciparum malaria infection had broad spatial distribution across Rwanda. P. vivax infection was rare.
Results:
Overall infection prevalence was 23.6% (95%CI [21.7%, 26.0%]), with P. falciparum and non-falciparum at 17.6% [15.9%, 19.0%] and 8.3% [7.0%, 10.0%], respectively. Parasitemias tended to be low and mixed species infections were common, especially where malaria transmission and overall prevalence was the highest. P. falciparum infection was associated with lower wealth, rural residence and low elevation. Fewer factors were associated with non-falciparum malaria.
Conclusions:
Asymptomatic non-falciparum malaria and P. falciparum malaria are common and widely distributed across Rwanda in adults. Continued molecular monitoring of Plasmodium spp. is needed to strengthen malaria control.
Keywords: Plasmodium, falciparum, asymptomatic, ovale, vivax, malariae
Article Summary
Non-falciparum and asymptomatic Plasmodium falciparum malaria are prevalent across Rwanda, with P. falciparum linked to poverty, rural areas, and low elevation. Mixed infections are frequent. Molecular surveillance is crucial to guide effective malaria control efforts.
Introduction
Despite global efforts in malaria control, malaria prevalence has increased in recent years from 233 million to 249 million cases, with most of this increase coming from countries in the WHO African Region.[1] Malaria infection is caused by five species of the Plasmodium parasite, with most cases and morbidity in Africa attributable to Plasmodium falciparum. Diagnosis of malaria in Africa relies principally on rapid diagnostic tests (RDTs) detecting the antigen P. falciparum histidine rich protein 2 (HRP2), often with a second less sensitive band for pan-species lactate dehydrogenase (LDH).[2] Designed to detect clinical malaria, RDTs for malaria surveillance will often miss low density P. falciparum infections and infections with non-falciparum species. Alternate approaches are needed to define the asymptomatic reservoir.
Molecular detection, such as through real-time PCR, can identify infections at lower parasitemia levels than RDTs can, and better characterize non-falciparum malaria infections. Molecular approaches are particularly important for use in asymptomatic community surveillance.[3] Asymptomatic infections may have significant but underestimated morbidity,[4] and are often followed by symptomatic infection.[5] Thus, characterizing the epidemiology of asymptomatic malaria is important for malaria control and prevention.
Molecular identification of non-falciparum malaria infections (P. malariae, P. ovale curtisi, P. ovale wallikeri, and P. vivax) is critical for addressing morbidity from these parasites and for reaching malaria elimination in settings with falciparum-based diagnostics and treatments. Accurate diagnosis is imperative for malaria control, as the clinical features and treatments differ. In contrast with P. falciparum infection, P. vivax and P. ovale cause relapse through the persistence of hypnozoites (dormant liver stage parasites).[6,7] As hypnozoites do not respond to blood-stage treatment, namely artemisinin-combination therapies (ACTs), the primary treatment for severe malaria in most countries, relapsing infections require radical cure.[8] Thus, their presence may require national malaria control programs to alter therapeutic options in the country. To-date, P. ovale spp. and P. malariae malaria have been more frequently described, due in part to wider implementation of highly sensitive nucleic acid detection, and also true increases in prevalence as P. falciparum is controlled.[9] P. vivax is also being reported more frequently due to wider implementation of molecular diagnostics.[10–12]
Rwanda has historically had strong malaria control, leveraging effective antimalarials, insecticide-treated bed nets, and indoor residual spraying.[13] Malaria cases in Rwanda increased from 48 to 403 per 1,000 between 2012 and 2018, with mortality increasing 41% over the same time.[13] Emerging insecticide resistance, an increase in irrigated agriculture, and insufficient insecticide-treated bed net coverage, among other factors, have been attributed to these increases.[13] From 2018 to 2022, malaria prevalence decreased from an estimated 321 to 76 cases per 1,000, still higher than 2012 estimates,[14] and with detection of non-falciparum species among clinical malaria patients.[15] There has been one recent report of P. vivax commonly occurring in Hue.[15] These inconsistent declines in prevalence suggest the need to characterize the epidemiology of asymptomatic and non-falciparum infections in this period to inform future control efforts.
We used survey, GPS, and biospecimen data collected in the 2014–15 Rwanda Demographic and Health Survey (RDHS), a large population-representative study, to characterize asymptomatic and non-falciparum infections across the country’s adult population.
Methods
Study design and population
The 2014–2015 Rwanda DHS studied 12,699 households from 492 GPS-located clusters from all 30 recognized districts. In 50% of households, dried blood spot (DBS) specimens were collected for HIV testing from men aged 15–59 years and women aged 15–49 years, and a subsample of children 0–14 years; in the other 50% of households, rapid malaria diagnostic testing was completed on children aged 6–59 months.[16] Previously, we used these data to estimate clusters that would represent high and low malaria transmission areas.[17] High prevalence clusters had a RDT or microscopy positivity rate of >15%. We included 1,434 samples from these 55 high prevalence clusters in 3 regions. In addition to samples from high prevalence areas, a random subset of 3,161 samples from 402 low prevalence clusters were selected. A total of 4,595 samples from 457 out of 492 DHS clusters were analyzed for four species of malaria infection by real time PCR. (Supplemental Figure 1).
Species-specific real time PCR
DNA from each sample was extracted from three 6 mm DBS punches using Chelex and screened for four species of malaria infection using real-time PCR assays.[18] These assays (Supplemental Table 1) targeted the 18s genes for P. malariae, P. ovale, and P. vivax, and the varATS repeat in P. falciparum.[11,19–21] To allow for quantification of P. falciparum, mock DBS were created using whole blood and cultured 3d7 parasites (MRA-102, BEI Resources, Manasas, VA) and extracted with the same assay used for samples. Controls for non-falciparum species used serial dilutions of plasmid DNA (MRA-180, MRA-179, MRA-178; BEI Resources, Manassas VA), with estimates for parasitemia based on an estimated six 18s rRNA gene copies per parasite.[22] All assays were run for 45 cycles to enable detection of lower density infections. The high cycle number approach has been previously evaluated for P. ovale and P. vivax, where assays were tested against 390 negative controls (human DNA) with no false positives.[23] We had no false positive results in our non-template controls for this study. In addition, we ran each assay to 45 cycles against plasmids of the other species to evaluate for cross-reactivity (Supplemental Table 2). All positive samples were confirmed by manually reviewing the amplification curves in the machine software. Standard curves had a minimum r-squared value of 0.95 across all runs. A positive result for each species was determined using a 45-cycle cutoff, unless otherwise stated. In addition, the assays retain a high specificity at high cycle number.[24]
Spatial & ecological variables
Deidentified survey and geospatial data from the 2014–2015 Rwanda DHS were matched to PCR data using DBS sample barcodes. Clusters with individuals positive for any species of malaria infection were mapped using DHS geospatial coordinates. Individual level covariates assessed for association included sex, age group, wealth quintile, education level, livestock ownership, source of drinking water, bed net ownership, whether the household bed net has been treated with long-lasting insecticide (LLIN = long-lasting insecticide-treated net) and sleeping under a LLIN the night prior to the survey. Cluster level covariates included region, urban/rural status of place of residence, elevation, month of data collection, proportion of a given cluster living in a household with a bed net, proportion of the cluster that slept under an LLIN, land cover, average daily maximum temperature for the current month and precipitation for the prior month. Land cover estimates were taken from the Regional Center for Mapping Resources for Development and SERVIR-Eastern and Southern Africa and cluster-level temperature and precipitation values were obtained from the 2015 Rwanda DHS geospatial covariates. Survey clusters were assigned GPS coordinate values within buffers as described previously in accordance with DHS specifications.[23,25]
Statistical analysis
We estimated species-specific prevalence, non-falciparum prevalence, and overall Plasmodium sp. prevalence, applying HIV sampling weights, inverse propensity for selection weights, and weights to account for selection by cluster transmission intensity and the skewed selection of samples from low and high transmission clusters. [26,27] We estimated bivariate associations between each Plasmodium species and a variety of covariates available in the DHS and investigated in other contexts,[23,28] using the same combination of weights. We report prevalence differences and 95% confidence intervals to assess precision. We analyzed data using the survey (4.2.1), srvyr (v1.2.0), and sf (v1.0–8) packages using R 4.2.1 (R Foundation for Statistical Computing). Shapefiles of Rwanda district boundaries taken from the OCHA Regional Office for Southern and Eastern Africa database.[29]
Results
Study population characteristics
The study population was 40% female, 14% aged 15–24 years, 76% lived in rural areas, and 80% had a primary school education or no education (60% reported primary education, 20% reported preschool/none). Only 3% of the study population was aged 0–15 years, so the results of this survey are only representative of Rwandan adults. Overall, most (83%) individuals reported a household bed net and 68% reported sleeping under a long-lasting insecticide treated net the night before the survey. However, 41% of the study population lived in a household that did not meet the World Health organization’s criteria of at least 1 net per 1.8 household members. At each of our covariates of interest, the study population was comparable to and representative of the overall DHS population (Supplemental Table 3).
Prevalence of P. falciparum and non-falciparum infection by real time PCR
A total of 1,231 P. falciparum, 246 P. ovale, 168 P. malariae and 7 P. vivax infections were identified. The overall weighted prevalence of any non-falciparum malaria infection was 8.3% (95% CI: [7.0%, 10.0%]) compared to 17.6% [15.9%, 19.0%] for P. falciparum and 23.6% [21.7%, 26.0%] overall malaria prevalence with HIV sampling, inverse propensity of selection, and transmission intensity correction weights applied. Species specific weighted prevalences were 3.3% [2.7%, 4.0%] and 5.1% [4.0%, 7.0%] for P. malariae and P. ovale spp. Unweighted prevalence for P. vivax was 0.15%, with a manually calculated 95% CI [0.04%, 0.27%]. Unweighted prevalence estimates were calculated for each species based on false-positive rates of high cycle number PCR (Supplemental Table 4). Using a more restrictive cut off of 40 cycles (requiring a higher parasitemia to be positive at approximately 1 parasite per microliter of template DNA), resulted in weighted overall prevalences of 4.3% [3.6%, 5.0%] for any non-falciparum infection, compared to 14.3% [12.7%, 16.0%] for P. falciparum and 17.4% for [15.8%, 19.0%] overall malaria. Species specific weighted prevalences at this cut off were 2.7% [2.2%, 3.0%) and 1.7% [1.2%, 2.0%], and for P. malariae, and P. ovale spp. Unweighted prevalence for P. vivax was 0.09% [0.002%, 0.17%]. The largest difference in estimated prevalence was for P. ovale spp. This is not surprising given the distribution of estimated parasitemia values (Figure 1), showing a lower median parasitemia in the non-falciparum species compared to P. falciparum. District level weighted prevalences and their differences by PCR cut off are shown in Supplemental Table 5 and 6. P. falciparum, P. ovale and P. malariae infections were distributed across the country, while P. vivax infections were more localized (Supplemental Figure 2). District level overall malaria prevalence is shown in Figure 2, while district level prevalences for each species are illustrated in Figure 3. Among P. ovale spp. P. malariae, and P. vivax infections, 45%, 45% and 57% (unweighted counts) were infected with at least one other species of malaria (Supplemental Table 7).
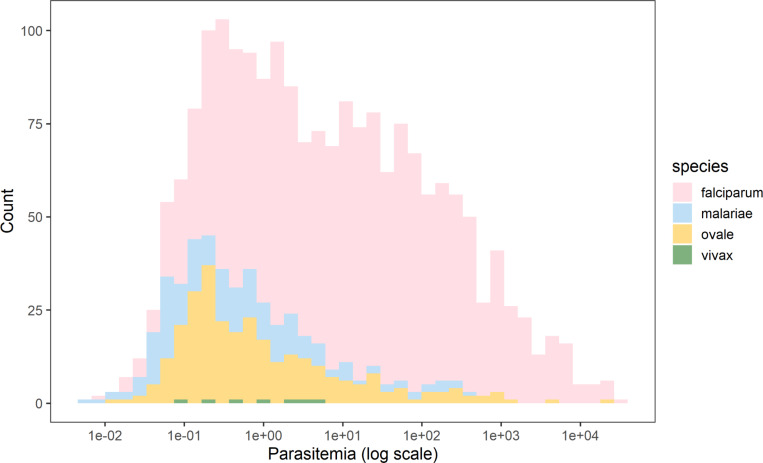
Figure 1. Calculated Parasitemia Estimates for Falciparum and Non-falciparum Infections.
Overall, falciparum had a higher parasite density with a median of 10.90 (IQR 0.96–101.41). The median parasitemia level for P. malariae, P. ovale spp., and P. vivax malaria were 0.35 (IQR: 0.07–1.83), 0.48 (IQR: 0.16–3.25) and 0.94 (IQR: 0.20–3.44), respectively.
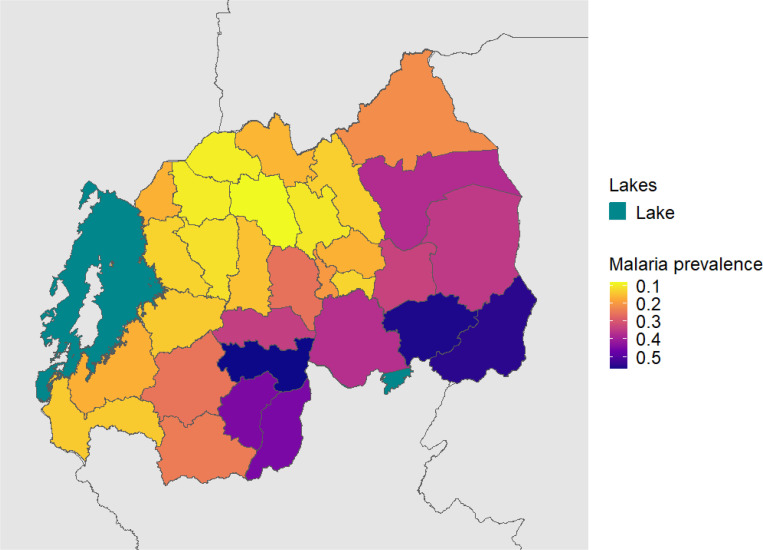
Figure 2. District Level Weighted Overall Malaria Prevalence.
Weighted prevalence of any malaria infection, using HIV sampling, inverse propensity for selection and transmission intensity weights (described above).
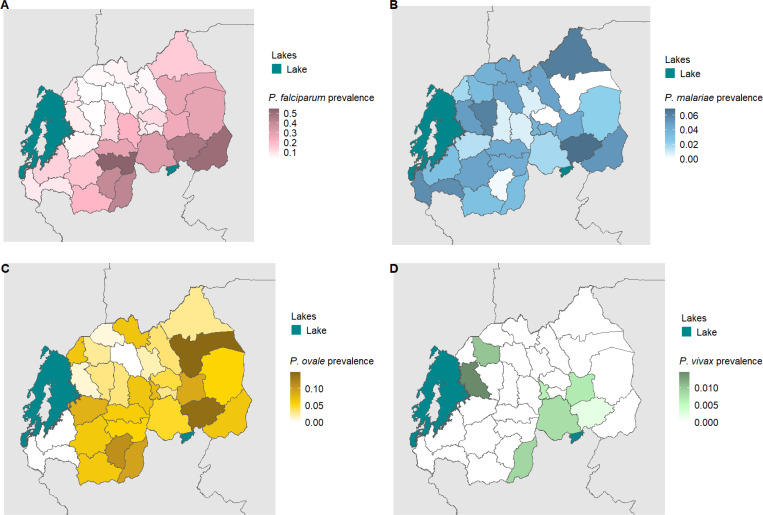
Figure 3. District Level Weighted Prevalence Estimates for Malaria Species.
The weighted prevalence estimate for each species is shown. Panel A, B, C and D represent P. falciparum, P. malariae, P. ovale spp. and P. vivax malaria, respectively.
Bivariate associations for infection
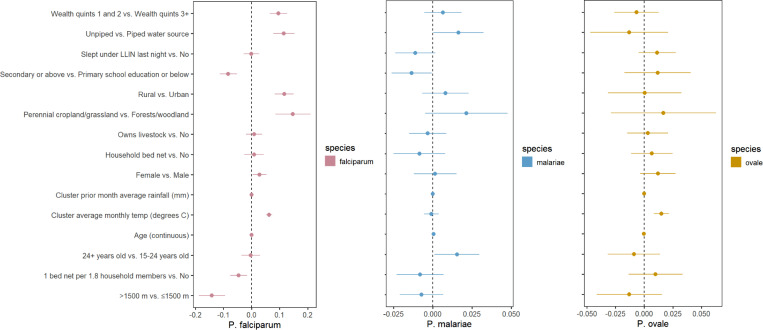
Bivariate regression models using weighted (as previously described) survey data found multiple associations for infection with P. falciparum malaria, but few for non-falciparum malaria (Figure 4). Covariates with more than two categories were dichotomized to reflect interpretable comparisons across the study population. For example, Anopheles mosquito vectors generally do not thrive at elevations over 1,500 m, although this is not a fixed limit. [30] Land cover classifications were dichotomized to compare less vegetation/more human activity to more vegetation/less human activity. No adjusted analyses were conducted as we are only exploring the strength of individual associations based on prevalence measures. Similar to previous work, a higher prevalence of P. falciparum malaria in our dataset was significantly (at a 0.05 confidence level for all associations) associated with multiple study covariates related to socioeconomic status (e.g. lower wealth quintile, lower education status, and unpiped drinking water). Secondary or higher education, residence in a household with at least 1 bed net per 1.8 household members, piped drinking water, continuous increase in prior month’s average rainfall, and higher altitude (>1,500m) were all associated with significantly lower prevalence of P. falciparum malaria. Rural clusters compared to urban, lower wealth index (first and second quintiles compared to the upper three), continuous increase in the cluster’s average monthly temperature, and female participants were associated with higher P. falciparum prevalence. Fewer associations were found for non-falciparum malaria. Lower prevalence of P. malariae infection was significantly associated with piped drinking water and secondary or higher education, and both a 1-year increase in age and participants over 24 (compared to those 15–24 years old) were associated with higher P. malariae prevalence. P. ovale infection was significantly associated with continuous average monthly temperature increase, while lower prevalence of infection was associated with continuous increase in the prior month’s average rainfall. Both significant associations with rainfall increase were smaller than the associations with temperature for the same species, by several orders of magnitude. Associations for P. vivax infection were not attempted due to the limited number of infections in the survey (n=7).
Figure 4. Bivariate associations and between demographic and environmental risk factors and Plasmodium spp. prevalence using weighted survey data.
Models incorporate 2014–15 Rwanda Demographic and Health Survey weights, inverse probability of selection weights, and cluster transmission intensity weights (described in Table 1). Point estimates of prevalence difference are surrounded by confidence intervals. The reference is the second variable listed. Panel A, B and C represent P. falciparum, P. malariae, and P. ovale sp., respectively. Note that each panel has a different scale.
Discussion
In Rwanda, a setting with robust malaria control efforts but inconsistently declining prevalence, we conducted the largest assessment of asymptomatic and non-falciparum malaria to-date, primarily among adults. We found an asymptomatic malaria prevalence of 23.6% using PCR, substantially higher than estimates using RDT and microscopy. Additionally, we observed 8.3% of the population was infected with a non-falciparum species.
While we observed P. falciparum prevalence to be 17.6% in adults, malaria detection by microscopy in the RDHS reported a 2% malaria prevalence among children age 6–59 months and 0.6% among women age 15–49.[31] This high prevalence in adults found using molecular detection is consistent with a study of school-aged individuals in the Huye District in the same year, finding a 22% prevalence of any malaria (19% P. falciparum prevalence) using a combined microscopy and PCR approach.[32] We used an ultra-sensitive assay for P. falciparum to detect what we assumed would be primarily asymptomatic, lower density infections, enhancing the characterization of P. falciparum prevalence. While a higher PCR cycle cut-off may increase concern about false-positive detection, our assays have been run extensively at 45 cycles with little to no evidence of false positives[23], and are consistent with estimates using a lower cut off (40 cycles). The largest change in prevalence occurred with P. ovale spp., where estimated prevalence dropped from 5.1% to 1.7% at different cycle cut offs, reflecting the high number of low density infections detected. The relative decrease in prevalence for each species was not always consistent, with some regions having no decline in P. falciparum prevalence with large declines in P. ovale malaria (e.g. Karongi near Lake Kivu) or the opposite with no decline in non-falciparum but lower P. falciparum prevalence estimates (e.g. Rutsiro and Nyamasheke). With asymptomatic P. falciparum malaria increasing risk of symptomatic disease at one month and contributing to significant morbidity, the high observed prevalence of asymptomatic, low-parasitemia infections offers an important direction for malaria control.[4,5]
Non-falciparum malaria was detected in 8.3% of individuals nationally, a prevalence not previously appreciated in the country. P. ovale spp. and P. malariae were both common in Rwanda (5.1% and 3.3% prevalence, respectively) and distributed in regions of both high and low transmission. A recent household survey estimated that P. falciparum is responsible for 97% of malaria infections in Rwanda, with P. malariae and P. ovale spp. each responsible for 1%–2% of total infections.[33,34] P. vivax is present but remains relatively uncommon (0.15% unweighted prevalence) and sporadic, but consistent with reporting of occasional clinical cases. [33] More recently, a cluster of P. vivax was reported in the Huye District.[15] Given distinct treatment approaches by species, the higher than expected prevalence of P. ovale spp. and occasional P. vivax infection underscores the need for molecular monitoring to guide control efforts towards elimination.
Not surprisingly, mixed species infections were common and widely distributed, but occurred more commonly in clusters in the south and east where malaria transmission was the highest (Supplemental Figure 2). Among P. ovale spp. P. malariae, and P. vivax infections, 44%, 45% and 57% (unweighted) were infected with at least one other species of malaria (Supplemental Table 7). Bivariate associations for mixed infections were largely found in the same direction as those observed for P. falciparum, with lower magnitude and wider confidence intervals attributable to the smaller total counts. Mixed infections are often underappreciated and may lead to severe disease complications. A recent meta-analysis suggested that patients with mixed infections have a higher proportion of pulmonary complications and multiple organ failure than patients with P. falciparum infection alone.[35] The impact of mixed species infections on clinical malaria outcomes in Rwanda is unknown and requires additional evaluation in symptomatic infections, which were not included in this study.
The associations we observed between P. falciparum infection and lower household wealth, no bed net use and lower elevation are consistent with previous studies using these data, with malaria positivity defined by RDT or microscopy.[36,37] We observed malaria prevalence was highest in the South and East as expected, but asymptomatic infection remained common in other areas. Like similar studies, associations of covariates for non-falciparum malaria were few and traditional risk factors for P. falciparum were less strongly associated with non-falciparum malaria.[11,20,23,28] This raises concern for how the control program can target non-falciparum infections without better diagnostics in the community. The reasons for the relative lack of risk factors, especially for P. ovale spp., remains unclear. Relapsing malaria, caused by P. ovale and P. vivax, may not be associated with typical covariates due to the inability to discern between incident or relapse infections in the study.
While this data reflects the epidemiology from nearly a decade ago, it remains important for malaria control. The baseline set here is useful for understanding the interventions that have been used in Rwanda for malaria control if future similar surveys, such as the 2019/20 DHS, Malaria Indicator Surveys, or other broad sampling efforts, are genotyped similarly. The longitudinal data across surveys can be used to understand how malaria interventions impact both asymptomatic malaria and non-falciparum malaria, both of which are not captured by standard DHIS2 data but remain important for malaria elimination.
While this national molecular evaluation offers important perspectives, we are limited by a few key points. As noted above, the cross-sectional nature of a single DHS limits inference regarding transmission and time trends, but use of subsequent (2019–20 DHS) and future DHSs, would allow for ongoing surveillance of these parasites, as has been done in the DRC.[38] We also could not determine if P. ovale spp. and P. vivax infections were newly acquired or the result of relapse from hypnozoites. Detection of these parasites is still important for malaria control programming. Additionally, the relative lack of data for children under 15 makes this survey not representative of the true asymptomatic reservoir and non-falciparum prevalence. We are limited by the sampling framework of the DHS. School aged children are a particularly vulnerable group and often have the highest rate of malaria infection in Africa, including for P. vivax.[39] Without this group, a true population prevalence of asymptomatic infection is difficult to determine. However, we likely underestimate overall population prevalence given the 2017 MIS had an overall prevalence of 14% for children aged 6 months to 14 years of age using a rapid diagnostic test which typically detects 41% of infections compared to PCR.[3,40] In addition, males were overrepresented in the sample. We used an assay that targets 18S ribosomal RNA genes for non-falciparum species, thus could underreport infections by P. vivax that could have been detected with assays that target genes with higher copy numbers in the parasite such as the assay used for falciparum in this study. [41–43] Despite these limitations, the use of existing samples and individual level data from a DHS is a highly informative method to gain insights into national malaria prevalence.
This study represents the first national investigation of asymptomatic P. falciparum malaria and non-falciparum malaria infection nationally in Rwanda using molecular methods. The prevalence of asymptomatic P. falciparum malaria in adults was significantly higher than estimates with RDT and microscopy in children.[31] P. ovale spp. and P. malariae were found across the country; however, few covariates were found to be significantly associated with non-falciparum infection. P. vivax was found, but infrequently. Most non-falciparum infections had low-density parasitemias and coinfection with P. falciparum was common, especially for P. ovale spp. and P. vivax. The prevalence of P. ovale and P. malariae infection was higher than expected, with few or no discernable risk factors, indicating the need to develop diagnostic plans for these species in communities where their prevalence is increasing. The data from this study is critical for national malaria control goals, given asymptomatic individuals comprise a large reservoir of P. falciparum infections and a high rate of relapsing malaria infection that requires radical cure. Ongoing molecular monitoring, including from the 2019–20 DHS, is imperative to characterize malaria prevalence over time, to guide efforts toward elimination.
Supplementary Material
Acknowledgements:
The following reagent was obtained through BEI Resources, NIAID, NIH: Plasmodium falciparum, Strain 3D7, MRA-102, contributed by Daniel J. Carucci. The following reagent was obtained through BEI Resources, NIAID, NIH: Diagnostic Plasmid Containing the Small Subunit Ribosomal RNA Gene (18S) from Plasmodium vivax, MRA-178, contributed by Peter A. Zimmerman. The following reagent was obtained through BEI Resources, NIAID, NIH: Diagnostic Plasmid Containing the Small Subunit Ribosomal RNA Gene (18S) from Plasmodium malariae, MRA-179, contributed by Peter A. Zimmerman. The following reagent was obtained through BEI Resources, NIAID, NIH: Diagnostic Plasmid Containing the Small Subunit Ribosomal RNA Gene (18S) from Plasmodium ovale, MRA-180, contributed by Peter A. Zimmerman.
Funding:
This work was funded by the National Institutes for Health (R01AI156267 to JAB, JBM and JJJ and K24AI134990 to JJJ). The funding for the DHS was provided by the government of Rwanda, the United States Agency for International Development (USAID), the One United Nations (One UN), the Global Fund to Fight AIDS, Tuberculosis and Malaria (Global Fund), World Vision International, the Swiss Agency for Development and Cooperation (SDC), and the Partners in Health (PIH). ICF International provided technical assistance through The DHS Program, a USAID-funded project providing support and technical assistance in the implementation of population and health surveys in countries worldwide. The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Footnotes
Conflict of Interest/Ethics:
Authors do not declare conflicts of interest associated with this work. Dried Blood Spots (DBS) were provided by the Ministry of Health- Rwanda. This research was reviewed and approved by the Rwanda National Ethics Committee (reference 102/RNEC/2023). Clinical and offset GPS data was downloaded from DHS-MEASURE. The University of North Carolina and Brown University IRBs deemed this non-human subjects research.
Data Availability:
Clinical and spatial data are available through the DHS MEASURE website. PCR data is available upon request with written approval of use of DHS data from DHS MEASURE. Code used for analysis is available at: https://github.com/claudiagaither.
References
- 1.World malaria report 2022 [Internet]. [cited 2023 Sep 15]. Available from: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022
- 2.Boyce MR, O’Meara WP. Use of malaria RDTs in various health contexts across sub-Saharan Africa: a systematic review. BMC Public Health [Internet]. 2017; 17(1):470. Available from: 10.1186/s12889-017-4398-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, van den Hoogen LL, H Slater, et al. Comparison of diagnostics for the detection of asymptomatic Plasmodium falciparum infections to inform control and elimination strategies. Nature [Internet]. Nature Publishing Group; 2015. [cited 2023 Sep 15]; 528(7580):S86–S93. Available from: https://www.nature.com/articles/nature16039 [DOI] [PubMed] [Google Scholar]
- 4.Chen I, Clarke SE, Gosling R, et al. “Asymptomatic” Malaria: A Chronic and Debilitating Infection That Should Be Treated. PLoS Med [Internet]. PLOS; 2016. [cited 2023 Sep 15]; 13(1). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4718522/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sumner KM, Mangeni JN, Obala AA, et al. Impact of asymptomatic Plasmodium falciparum infection on the risk of subsequent symptomatic malaria in a longitudinal cohort in Kenya. eLife Sciences Publications Limited; 2021. [cited 2023 Sep 15]; . Available from: https://elifesciences.org/articles/68812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter J, Franken G, Mehlhorn H, Labisch A, Häussinger D. What is the evidence for the existence of Plasmodium ovale hypnozoites? Parasitol Res [Internet]. 2010; 107(6):1285–1290. Available from: 10.1007/s00436-010-2071-z [DOI] [PubMed] [Google Scholar]
- 7.Mueller I, Galinski MR, Baird JK, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis [Internet]. 2009; 9(9):555–566. Available from: 10.1016/S1473-3099(09)70177-X [DOI] [PubMed] [Google Scholar]
- 8.Wells TNC, Burrows JN, Baird JK. Targeting the hypnozoite reservoir of Plasmodium vivax: the hidden obstacle to malaria elimination. Trends Parasitol [Internet]. 2010; 26(3):145–151. Available from: 10.1016/j.pt.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 9.Yman V, Wandell G, Mutemi DD, et al. Persistent transmission of Plasmodium malariae and Plasmodium ovale species in an area of declining Plasmodium falciparum transmission in eastern Tanzania. PLoS Negl Trop Dis [Internet]. Public Library of Science; 2019. [cited 2023 Sep 15]; 13(5):e0007414. Available from: https://journals.plos.org/plosntds/article/file?id=10.1371/journal.pntd.0007414&type=printable [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mapping the global endemicity and clinical burden of Plasmodium vivax, 2000–17: a spatial and temporal modelling study. Lancet [Internet]. Elsevier; 2019. [cited 2023 Sep 15]; 394(10195):332–343. Available from: 10.1016/S0140-6736(19)31096-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brazeau NF, Mitchell CL, Morgan AP, et al. The epidemiology of Plasmodium vivax among adults in the Democratic Republic of the Congo. Nat Commun [Internet]. 2021; 12(1):4169. Available from: 10.1038/s41467-021-24216-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twohig KA, Pfeffer DA, Baird JK, et al. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl Trop Dis [Internet]. 2019; 13(1):e0007140. Available from: 10.1371/journal.pntd.0007140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karema C, Wen S, Sidibe A, et al. History of malaria control in Rwanda: implications for future elimination in Rwanda and other malaria-endemic countries. Malar J [Internet]. BMC; 2020. [cited 2023 Sep 15]; 19. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7539391/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.[No title] [Internet]. [cited 2024 Apr 29]. Available from: https://www.rbc.gov.rw/fileadmin/user_upload/report23/Rwanda%20Aide_Memoire_MTR_9%20_March_2023_Final.pdf [Google Scholar]
- 15.van Loon W, Oliveira R, Bergmann C, et al. Plasmodium vivax Malaria in Duffy-Positive Patients in Rwanda. Am J Trop Med Hyg [Internet]. Am J Trop Med Hyg; 2023. [cited 2023 Sep 15]; 109(3). Available from: https://pubmed.ncbi.nlm.nih.gov/37549894/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demographic ICF. health surveys standard recode manual for dhs7. The Demographic and Health Surveys Program. [Google Scholar]
- 17.Kirby R, Giesbrecht D, Karema C, et al. Examining the Early Distribution of the Artemisinin-Resistant R561H Mutation in Areas of Higher Transmission in Rwanda. Open Forum Infect Dis [Internet]. 2023; 10(4):ofad149. Available from: 10.1093/ofid/ofad149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh PS, Metzger DA, Higushi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 10(4): 506–13 (April 1991). Biotechniques [Internet]. 2013; 54(3):134–139. Available from: 10.2144/000114018 [DOI] [PubMed] [Google Scholar]
- 19.Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, Jaton K. Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol [Internet]. 2004; 42(12):5636–5643. Available from: 10.1128/JCM.42.12.5636-5643.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell CL, Brazeau NF, Keeler C, et al. Under the Radar: Epidemiology of Plasmodium ovale in the Democratic Republic of the Congo. J Infect Dis [Internet]. J Infect Dis; 2021. [cited 2023 Sep 15]; 223(6). Available from: https://pubmed.ncbi.nlm.nih.gov/32766832/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med [Internet]. 2015; 12(3):e1001788. Available from: 10.1371/journal.pmed.1001788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercereau-Puijalon O, Barale J-C, Bischoff E. Three multigene families in Plasmodium parasites: facts and questions. Int J Parasitol [Internet]. 2002; 32(11):1323–1344. Available from: 10.1016/s0020-7519(02)00111-x [DOI] [PubMed] [Google Scholar]
- 23.Gumbo A, Topazian HM, Mwanza A, et al. Occurrence and Distribution of Nonfalciparum Malaria Parasite Species Among Adolescents and Adults in Malawi. J Infect Dis [Internet]. J Infect Dis; 2022. [cited 2023 Sep 15]; 225(2). Available from: https://pubmed.ncbi.nlm.nih.gov/34244739/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sendor R, Banek K, Kashamuka MM, et al. Epidemiology of Plasmodium malariae and Plasmodium ovale spp. in Kinshasa Province, Democratic Republic of Congo. Nat Commun [Internet]. 2023; 14(1):6618. Available from: 10.1038/s41467-023-42190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.[No title] [Internet]. [cited 2023 Nov 2]. Available from: https://spatialdata.dhsprogram.com/references/DHS%20Covariates%20Extract%20Data%20Description%202.pdf
- 26.Guide to DHS Statistics [Internet]. [cited 2023 Nov 2]. Available from: https://dhsprogram.com/data/Guide-to-DHS-Statistics/index.cfm
- 27.Rosenbaum PR, Rubin DB. The Central Role of the Propensity Score in Observational Studies for Causal Effects [Internet]. 1982. Available from: https://books.google.com/books/about/The_Central_Role_of_the_Propensity_Score.html?hl=&id=_GQ5GwAACAAJ
- 28.Mitchell CL, Topazian HM, Brazeau NF, et al. Household Prevalence of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale in the Democratic Republic of the Congo, 2013–2014. Clin Infect Dis [Internet]. 2021; 73(11):e3966–e3969. Available from: 10.1093/cid/ciaa1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rwanda - Subnational Administrative Boundaries [Internet]. [cited 2023 Sep 15]. Available from: https://data.humdata.org/dataset/cod-ab-rwa?
- 30.Carlson CJ, Bannon E, Mendenhall E, Newfield T, Bansal S. Rapid range shifts in African Anopheles mosquitoes over the last century. Biol Lett [Internet]. 2023; 19(2):20220365. Available from: 10.1098/rsbl.2022.0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.[No title] [Internet]. [cited 2023 Sep 15]. Available from: https://dhsprogram.com/pubs/pdf/FR316/FR316.pdf
- 32.Sifft KC, Geus D, Mukampunga C, et al. Asymptomatic only at first sight: malaria infection among schoolchildren in highland Rwanda. Malar J [Internet]. Malar J; 2016. [cited 2023 Sep 15]; 15(1). Available from: https://pubmed.ncbi.nlm.nih.gov/27842542/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCaffery JN, Munyaneza T, Uwimana A, et al. Symptomatic Infection in Rwanda. Open Forum Infect Dis [Internet]. 2022; 9(3):ofac025. Available from: 10.1093/ofid/ofac025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uwimana A. Rwandan National Malaria Control Program (unpublished data). 2018. [Google Scholar]
- 35.Kotepui M, Kotepui KU, De Jesus Milanez G, Masangkay FR. Plasmodium spp. mixed infection leading to severe malaria: a systematic review and meta-analysis. Sci Rep [Internet]. 2020; 10(1):11068. Available from: 10.1038/s41598-020-68082-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubana E, Munyaneza A, Sande S, et al. “A comparative analysis of risk factors of malaria” case study Gisagara and Bugesera District of Rwanda. RDHS 2014/2015. A retrospective study. BMC Public Health [Internet]. BioMed Central; 2023. [cited 2023 Sep 15]; 23(1):1–9. Available from: https://bmcpublichealth.biomedcentral.com/articles/10.1186/s12889-023-15104-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudasingwa G, Cho S-I. Determinants of the persistence of malaria in Rwanda. Malar J [Internet]. BioMed Central; 2020. [cited 2023 Sep 15]; 19(1):1–9. Available from: https://malariajournal.biomedcentral.com/articles/10.1186/s12936-020-3117-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deutsch-Feldman M, Aydemir O, Carrel M, et al. The changing landscape of Plasmodium falciparum drug resistance in the Democratic Republic of Congo. BMC Infect Dis [Internet]. 2019; 19(1):872. Available from: 10.1186/s12879-019-4523-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sendor R, Mitchell CL, Chacky F, et al. Similar Prevalence of Plasmodium falciparum and Non-P. falciparum Malaria Infections among Schoolchildren, Tanzania. Emerg Infect Dis [Internet]. 2023; 29(6):1143–1153. Available from: 10.3201/eid2906.221016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Habyarimana F, Ramroop S. Prevalence and Risk Factors Associated with Malaria among Children Aged Six Months to 14 Years Old in Rwanda: Evidence from 2017 Rwanda Malaria Indicator Survey. International Journal of Environmental Research and Public Health [Internet]. 2020. [cited 2024 Oct 30]; 17(21):7975. Available from: https://pmc.ncbi.nlm.nih.gov/articles/PMC7672573/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann NE, Gruenberg M, Nate E, et al. Assessment of ultra-sensitive malaria diagnosis versus standard molecular diagnostics for malaria elimination: an in-depth molecular community cross-sectional study. Lancet Infect Dis [Internet]. 2018; 18(10):1108–1116. Available from: 10.1016/S1473-3099(18)30411-0 [DOI] [PubMed] [Google Scholar]
- 42.Gruenberg M, Moniz CA, Hofmann NE, et al. Plasmodium vivax molecular diagnostics in community surveys: pitfalls and solutions. Malar J [Internet]. 2018; 17(1):55. Available from: 10.1186/s12936-018-2201-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gruenberg M, Moniz CA, Hofmann NE, et al. Utility of ultra-sensitive qPCR to detect Plasmodium falciparum and Plasmodium vivax infections under different transmission intensities. Malar J [Internet]. 2020; 19(1):319. Available from: 10.1186/s12936-020-03374-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Clinical and spatial data are available through the DHS MEASURE website. PCR data is available upon request with written approval of use of DHS data from DHS MEASURE. Code used for analysis is available at: https://github.com/claudiagaither.