Abstract
Importance:
The under-representation of participants with non-European ancestry in genome-wide association studies (GWAS) is a critical issue that has significant implications, including hindering the progress of precision medicine initiatives. This issue is particularly significant in the context of neurodegenerative diseases (NDDs), where current therapeutic approaches have shown limited success. Addressing this under-representation is crucial to harnessing the full potential of genomic medicine in underserved communities and improving outcomes for NDD patients.
Objective:
Our primary objective was to assess the representation of non-European ancestry participants in genetic discovery efforts related to NDDs. We aimed to quantify the extent of inclusion of diverse ancestry groups in NDD studies and determine the number of associated loci identified in more inclusive studies. Specifically, we sought to highlight the disparities in research efforts and outcomes between studies predominantly involving European ancestry participants and those deliberately targeting non-European or multi-ancestry populations across NDDs.
Evidence Review:
We conducted a systematic review utilizing existing GWAS results and publications to assess the inclusion of diverse ancestry groups in neurodegeneration and neurogenetics studies. Our search encompassed studies published up to the end of 2022, with a focus on identifying research that deliberately included non-European or multi-ancestry cohorts. We employed rigorous methods for the inclusion of identified articles and quality assessment.
Findings:
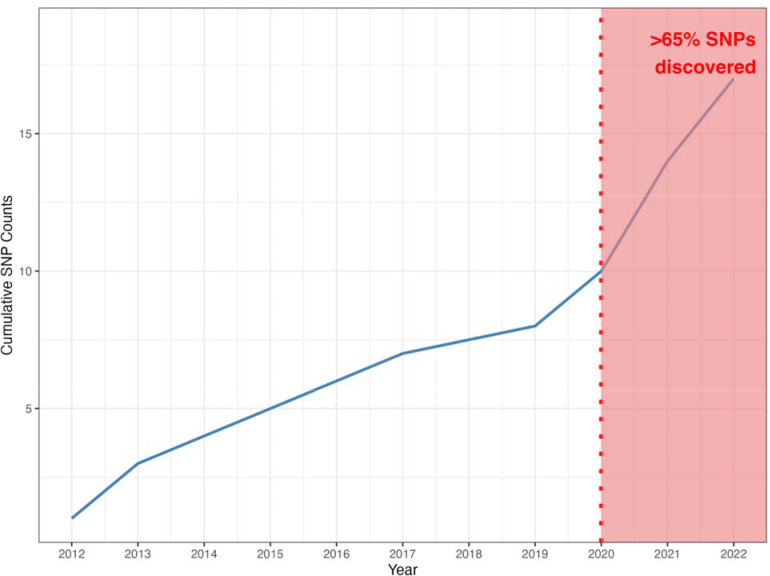
Our review identified a total of 123 NDD GWAS. Strikingly, 82% of these studies predominantly featured participants of European ancestry. Endeavors specifically targeting non-European or multi-ancestry populations across NDDs identified only 52 risk loci. This contrasts with predominantly European studies, which reported over 90 risk loci for a single disease. Encouragingly, over 65% of these discoveries occurred in 2020 or later, indicating a recent increase in studies deliberately including non-European cohorts.
Conclusions and relevance:
Our findings underscore the pressing need for increased diversity in neurodegenerative research. The significant under-representation of non-European ancestry participants in NDD GWAS limits our understanding of the genetic underpinnings of these diseases. To advance the field of neurodegenerative research and develop more effective therapies, it is imperative that future investigations prioritize and harness the genomic diversity present within and across global populations.
Introduction
Genome-wide association studies (GWAS) have shown a significant bias towards individuals of European ancestry, despite comprising only 16% of the global population 1. This underrepresentation issue is particularly salient in the realm of neurodegenerative disease (NDD) studies. For instance, while a recent Alzheimer’s disease (AD) GWAS including ~800,000 individuals of European descent identified 75 disease-associated loci 2, no GWAS studies on AD currently exist for Admixed American or Native American populations. Similarly, Parkinson’s disease research exhibits a glaring imbalance, with black individuals included in just ~4% of published PD studies 3.
The prevalence of NDDs varies significantly among global populations and racial/ethnic groups. This warrants a critical examination of the disparity in genetic research efforts over time. In this manuscript, we present a systematic review spanning from 2012 through 2022, focusing on neurodegenerative disease GWAS research.
Our analysis encompasses common NDDs such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis, as well as less common atypical dementias. Our objective is to quantify the disparity in participant recruitment for genetic studies, shed light on genetic findings in underrepresented populations, and discuss ongoing initiatives aimed at addressing this pervasive issue.
Methods
Search Strategy
The systematic review was conducted in two phases. First, we reviewed the GWAS Catalog; then, since the GWAS Catalog does not include all GWAS studies, we performed a formal literature review in collaboration with the National Library of Medicine (NLM). The keywords used in both searches are included in Supplementary Table 1.
Results from both the GWAS Catalog and the NLM search were uploaded to Covidence 4, a web-based software platform, for further review. We removed duplicate studies and any studies published before 2012 or after 2022.
All studies were filtered to only include genome-wide associations examining neurological disease risk factors, family history of disease, disease progression, age at onset, or survival genome-wide, excluding exome wide studies and those that focused on a targeted set of SNPs or genetic loci. Studies investigating disease subtypes, biomarkers, non-English language studies, and those investigating only rare or structural variation were also excluded.
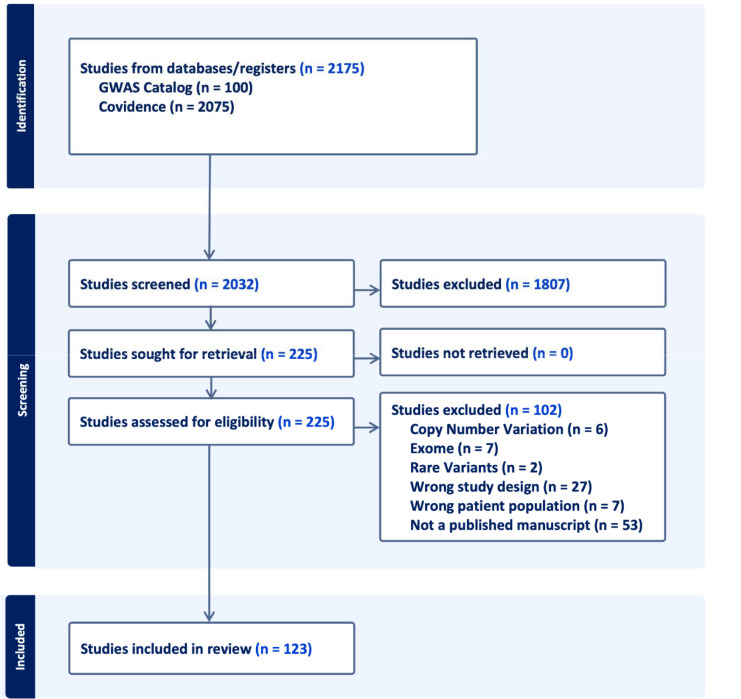
Studies were assessed for eligibility by two independent reviewers and all conflicts were resolved by a third independent reviewer. Publication date, phenotype, and cohort information were extracted from each publication. If multiple phenotypes of interest were analyzed in the same study, information was included in both phenotype categories. The number of samples per ancestry was extracted manually from each study. We looked at seven ancestry groupings: European (EUR), East Asian (EAS), Middle-Eastern (MDE), African (AFR), African American and Caribbean (AAC), Latino and indigenous Americas populations (AMR), and South Asian (SAS). A PRISMA diagram of our filtering process can be found in Figure 1. All 123 studies passing our filters can be found in Supplementary Table 2.
Figure 1: PRISMA flow diagram for systematic review of published GWAS studies and pubmed literature review from NLM.
N in the figure relates to the number of published studies as of April 28th, 2022.
Finally, results were examined manually for all studies passing implemented filtering methods. Novel loci discovered in non-European or multi-ancestry populations, with a p-value below 5E-8, are listed in Table 2.
Table 2: Genome-wide significant (P-value < 5E-8) novel loci nominated in non-European populations or multi-ancestry studies.
We found no diverse or multi-ancestry loci for LBD, FTD, MG, or VaD. P-values are given in parentheses. Nominated loci were determined as the nearest gene or genomic context within 1MB of the significant SNP.
| Disease | Ancestry | Nominated loci |
| AD | AAC | COBL (3.8E-8), SLC10A2 (4.6E-8)11 |
| EAS | RHOBTB3/GLRX (3.07E-19), CTC-278L1.1 (2.49E-23), CTD-2506J14.1 (1.35E-67), CHODL (4.81E-9)15 , FAM47E (5.34E-9)16, CACNA1A (2.49E-8), LRIG1 (1.51E-8)17 | |
| AMR | FBXL7 (6.19E-09)13 | |
| MULTI | OR2B2 (2.14E-8)16, SORL1 (1.04E-8)18, TRANK1 (3.49E-8), VWA5B2 (3.75E-8)19 | |
| PD | EAS | NDN/PWRN4 (3.14E-9)23, RPL3 (2.72E-8) 24 |
| MULTI |
ITGA8 (1.3E-8)26, SV2C (1.17E-10)27 MTF2 (1.15E-10), RP11-360P21.2 (1.65E-10), ADD1 (4.11E-9), SYBU (3.62E-9), IRS2 (2.30E-9), USP8RP11-562A8.5 (6.45E-10), PIGL (2.93E-9), FASN (2.61E-9), MYLK2 (3.86E-9), AJ006998.2 (1.12E-9), Y_RNA (3.81E-9), PPP6R2 (4.09E-10)7, BST1 (4.41E-8)28 |
|
| ALS | EAS | CAMKIG (2.92E-8), CABIN1/SUSD2 (2.35E-9)31, INPP5B (2.24E-8), IQCF5/IQCF1 (2.06E-9), ITGA9 (2.55E-8), PFKP (2.46E-9), MYO18B (2.28E-10), ALCAM (4.00E-8), OPCML (8.43E-9), GPR133 (8.45E-10)32, TYW/CRYZ (2.10E-14), FGD4 (5.19E-9) 33 |
| MULTI | GPX3-TNIP1 (1.3E-8)30, ACSL5-ZDHHC6 (8.3E-9)34, SOD1 (3.5E-18), HLA (3.5E-12), SLC9A8-SPATA2 (3.2E-10), ERGIC1 (5.6E-9), NEK1 (6.9E-9), COG3 (1.2E-8), and PTPRN2 (1.8E-8)36 |
Results
Search results
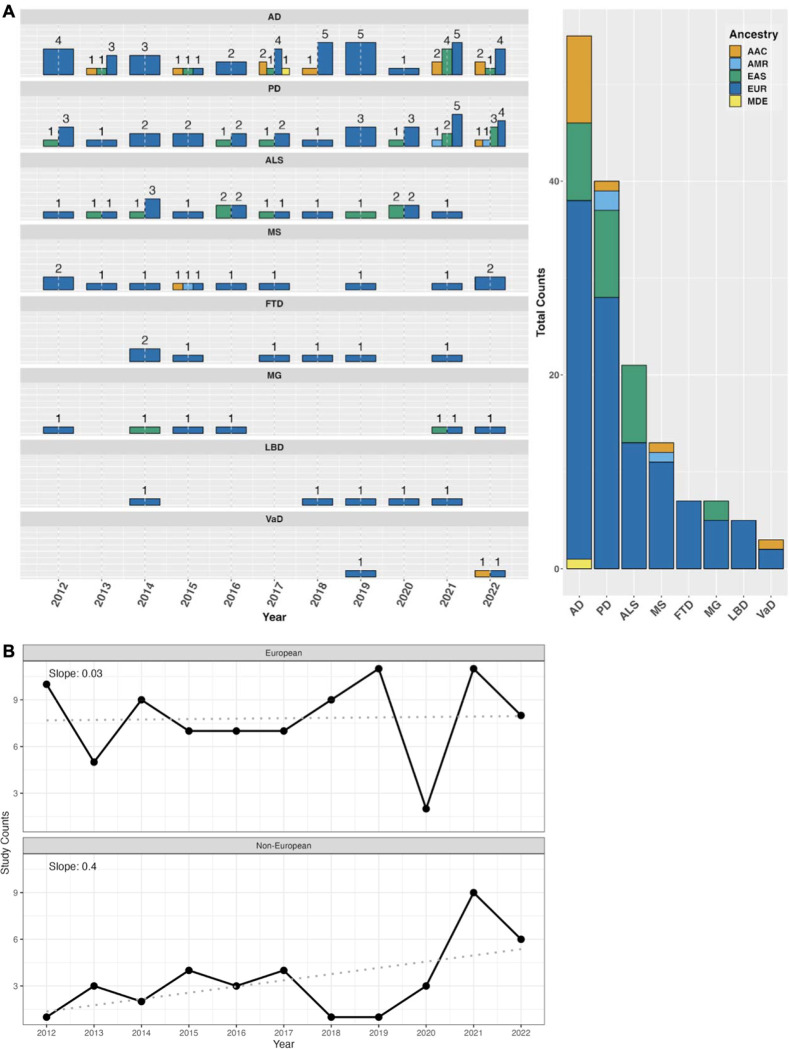
We identified 123 eligible GWAS studies. Unsurprisingly, we found that European populations were overrepresented in GWAS pertaining to NDDs (Figure 2). When non-European populations were included, the sample sizes were on average 15X smaller than the European ancestry samples included in the same disease category. The underrepresentation of non-European populations was particularly evident among the less common NDDs, including Lewy body dementia and frontotemporal dementia, where we did not identify any non-European or multi-ancestry GWAS studies using the outlined search methods. We have summarized the lack of diversity in genetic studies of NDDs in Table 1, and Figure 2.
Figure 2: Number of studies over time from 2012 to 2022.
A) Bar plot of study counts by NDD (left) with cumulative counts for each ancestry (right). Data in this figure includes both single and multi ancestry studies. B) Time series of the annual study counts in European and Non-European populations from 2012 to 2022. The slope from a linear regression is also displayed to highlight the rate of change in the number of study counts over time.
Table 1:
Largest GWAS sample size by NDD and ancestry for single and multi ancestry studies.
| NDD | Ancestry | Author | Year | Total samples (% non-European*) | Study |
|---|---|---|---|---|---|
| AD | AAC | Sherva | 2022 | 75,058 | African ancestry GWAS of dementia in a large military cohort identifies significant risk loci |
| EAS | Hirano | 2015 | 17,031 | A genome-wide association study of late-onset Alzheimer’s disease in a Japanese population | |
| EUR | Wightman | 2021 | 1,126,563 | A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease | |
| MULTI | Lake | 2022 | 644,188 (2.83%) | Multi-ancestry meta-analysis and fine-mapping in Alzheimer’s Disease | |
| AMR,MDE | NO STUDIES | ||||
| PD | AMR | Loesch | 2021 | 1,497 | Characterizing the Genetic Architecture of Parkinson’s Disease in Latinos |
| EAS | Foo | 2017 | 14,006 | Genome-wide association study of Parkinson’s disease in East Asians | |
| EUR | Nalls | 2019 | 1,474,097 | Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies | |
| MULTI | Kim | 2022 | 2,525,730 (38.12%) | Multi-ancestry genome-wide meta-analysis in Parkinson’s disease | |
| AAC,MDE | NO STUDIES | ||||
| ALS | EAS | Wei | 2019 | 4,727 | Identification of TYW3/CRYZ and FGD4 as susceptibility genes for amyotrophic lateral sclerosis |
| EUR | vanRheenen | 2016 | 41,398 | Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis | |
| MULTI | van Rheenen | 2021 | 152,268 (12.00%) | Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology | |
| AAC, AMR, MDE | NO STUDIES | ||||
| MS | AAC | Isobe | 2015 | 2319 | An ImmunoChip study of multiple sclerosis risk in African Americans |
| AMR | Ordoñez | 2015 | 161 | Genomewide admixture study in Mexican Mestizos with multiple sclerosis | |
| EUR | Patsopoulos | 2019 | 115,803 | Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility | |
| EAS,MDE,MULTI | NO STUDIES | ||||
| FTD | EUR | Ferrari | 201 4 | 12,928 | Frontotemporal dementia and its subtypes: a genomewide association study |
| AAC, AMR, EAS, MDE, MULTI | NO STUDIES | ||||
| MG | EAS | Na | 2014 | 259 | Whole-genome analysis in Korean patients with autoimmune myasthenia gravis |
| EUR | Chia | 2022 | 45,675 | Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study | |
| MULTI | Sakaue | 2021 | 533,853 (33.48 %) | A cross-population atlas of genetic associations for 220 human phenotypes | |
| AAC,AMR,MDE | NO STUDIES | ||||
| LBD | EUR | Chia | 2021 | 7,372 | Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture |
| AAC,AMR,EAS,MDE,MULTI | NO STUDIES | ||||
| VaD | EUR | Moreno-Grau | 2019 | 4,830 | Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: The GR@ACE project |
| MULTI | Fongang | 2022 | 482,088 (2.40%) | A meta-analysis of genomewide association studies identifies new genetic loci associated with all-cause and vascular dementia | |
| AAC,AMR,EAS,MDE | NO STUDIES | ||||
if applicable
We found 52 novel NDD loci that were identified in non-European or multi-ancestry populations (Table 2). Of these 52 loci, 28 were found in multi-ancestry studies, 21 were found in East Asian studies, and only 3 were found in other populations (AAC and AMR). No loci were discovered in AFR, MDE, or SAS ancestries. Recent studies that combined individuals of multiple ancestries by using standard random-effects and some custom meta-analytic techniques 5 have succeeded in identifying novel disease loci that reach genome-wide significance, including two novel AD loci 6 and 12 novel PD loci 7. However, these studies leverage existing European sample sizes as a backbone for much of the statistical power needed for discovery.
In the following sections, we briefly summarize the results of our systematic review in a disease specific manner. Many of these findings have not been replicated. As datasets become larger and more inclusive, the genetic architecture of these diseases may grow and change.
Alzheimer’s Disease
Largest European GWAS: Wightman 2021
Total samples: 1,126,563
Largest multi-ancestry GWAS: Lake 2022
Total samples: 644,188
Total non-European samples: 18,246
% non-European: 2.83%
Largest non-European GWAS: Sherva 2022
Ancestry: AAC
Total samples: 75,058
The largest Alzheimer’s disease GWAS of European populations included ~1.1 million individuals and identified a total of 38 associated loci 8. Another recent GWAS included ~800,000 individuals of European ancestry and identified a total of 75 loci 2. The discrepancy between identified loci in these studies could be due to many factors, including differences in neuropathological/diagnostic criteria 9.
A 2013 GWAS conducted in African Americans replicated an association at ABCA7 previously identified in European populations. They found that rs115550680, rare in European populations, was associated with an increased risk for AD in African Americans comparable to the highly pathogenic APOE- 4 variant observed in Europeans 10. A 2017 GWAS in African Americans identified two novel loci at COBL and SLC10A2 11. The most extensive African American GWAS to date, drawing from a military cohort of around 22,000 individuals and a proxy GWAS involving approximately 50,000 individuals, identified significant associations with established AD risk genes such as TREM2, CD2AP, and ABCA7. Notably, distinct lead variants were observed in these loci compared to those found in European cohorts 12.
The only study conducted in Caribbean Hispanic individuals was a 2017 study with 2,451 cases and 2,063 controls. They found a novel and population specific locus near FBXL7 13. The lead SNP, rs75002042, is much more common in individuals with African ancestry compared to individuals of European ancestry, with minor allele frequencies around 20% and 0.009%, respectively. This study also replicated six loci previously reported in European populations, including FRMD4A, CELF1, FERMT2, SLC24A4-RIN3, ABCA7, and CD33 13.
The largest AD study in East Asian populations was conducted in Japanese participants with 1,827 cases and 15,204 controls (discovery + replication) 14, but they did not nominate any genome-wide significant loci. More recent but smaller studies have since been conducted, including a 2021 GWAS in a Chinese cohort that reported four novel loci near RHOBTB3/GLRX, CTC-278L1.1, CTD-2506J14.1, and CHODL 15, a study in Japanese participants that nominated a locus in FAM47E 16, and a study including both Korean and Japanese participants that nominated two novel loci at CACNA1A and LRIG1 17.
Multi-ancestry studies have nominated additional AD loci, however these studies still rely on Europeans as the majority population. SORL1 was first identified as a risk locus for AD in a GWAS that included East Asian and European ancestry populations 18. Other multi-ancestry GWAS identified OR2B2 16, TRANK1 and VWA5B2 19 as novel loci for AD.
While the inclusion of diverse populations in genetic research for AD is arguably better than what is seen for some of the atypical dementias, the largest study size for a non-European population 12 was still only 7% of the total sample size for the largest European AD GWAS.
Parkinson’s disease
Largest European GWAS: Nalls 2019
Total samples: 1,456,306
Largest multi-ancestry GWAS: Kim 2022
Total samples: 2,525,730
Total non-European samples: 962,735
% non-European: 38.12%
Largest non-European GWAS: Foo 2017
Ancestry: EAS
Total samples: 14,006
The largest meta-GWAS of PD risk in individuals of European ancestry found 90 significant risk signals across 78 genomic regions. The 90 nominated risk variants collectively explain roughly 16–36% of the heritable risk of non-monogenic, or complex PD 20.
The largest study in East Asian populations (with exception to a study done in Japan before our review period 21) was conducted with Han Chinese participants, replicating loci previously identified in European populations including SNCA, LRRK2 and MCCC1 in their discovery GWAS of 14,006 participants 22. More recent studies in Chinese populations have nominated a locus on NDN/PWRN4 associated with age at onset and a locus on RPL3 associated with reduced survival 23,24 .
The first and most recent PD GWAS of a South American population was conducted in 2021, replicating an association at SNCA with 1,497 participants 25.
Recently, more multi-ancestry studies have been conducted in PD, nominating novel loci for disease risk and age at onset including ITGA8, SV2C, and BST1 26,27,28. The largest meta-GWAS for PD, which included 4 ancestral populations, identified 12 novel loci: MTF2, RP11–360P21.2, ADD1, SYBU, IRS2, USP8:RP11–562A8.5, PIGL, FASN, MYLK2, AJ006998.2, Y_RNA, and PPP6R2 7.
The largest non-European PD GWAS was in East Asian populations, however, only a few novel loci have been nominated in that ancestry. Multi-ancestry studies have nominated more novel variants in recent studies, but much more work is needed to better understand risk for PD in non-European populations.
Amyotrophic lateral sclerosis
Largest European GWAS: van Rheenen 2016
Total samples: 41,398
Largest multi-ancestry GWAS: van Rheenen 2021
Total samples: 152,268
Total non-European samples: 18,266
% non-European: 12.00%
Largest non-European GWAS: Wei 2019
Ancestry: EAS
Total samples: 4,727
ALS GWAS in European populations have nominated a number of risk loci including C9ORF72, UNC13A, C21orf2, SARM1, MOBP, SCFD1, TBKK1, and KIF5A 29–30.
ALS is the only disease in our review where more genome-wide significant novel loci have been identified in a non-European population than in the largest European-only study. The first GWAS of individuals with Chinese Han ancestry identified CAMKIG and CABIN1/SUSD2 as susceptibility loci for ALS 31. Later studies in the Han Chinese population nominated additional novel loci including INPP5B, IQCF5/IQCF1, ITGA9, PFKP, MYO18B, ALCAM, OPCML, GPR133, TYW/CRYZ and FGD4 32–33. With a total of 12 genome-wide significant loci, East Asian ancestry GWAS for ALS have nominated the most of any single non-European population covered in our review.
Multi-ancestry GWAS for ALS, which typically consist of European and East Asian ancestry populations, have been successful at nominating additional risk loci including GPX3-TNIP1 and ACSL5 30,34,35. The largest ALS GWAS to date was a multi-ancestry study including over 150,000 individuals of European and East Asian ancestry. This study identified a total of 15 risk loci for ALS, replicating 8 previously-identified and nominating 7 novel loci: SOD1, HLA, SLC9A8-SPATA2, ERGIC1, NEK1, COG3, and PTPRN2 36.
Similar to PD, ALS GWAS including or focused on East Asian populations have made more progress than other non-European populations for these diseases. However, much more work is still needed in all populations to progress potential precision medicine initiatives for ALS.
Multiple Sclerosis
Largest European GWAS: Patsopoulos 2019
Total samples: 115,803
There are no multi-ancestry studies in MS.
Largest non-European GWAS: Isobe 2015
Ancestry: AAC
Total samples: 2,319
The largest GWAS meta-analysis for MS included 115,803 individuals of European ancestry and found 82 significant genome-wide associations with MS. This study was also the first to identify a risk locus for MS on chromosome X and the identified genetic markers accounted for nearly 50% of the hereditary risk for MS 37.
Studies in non-European populations were more limited in MS than in the previous diseases discussed. The largest GWAS in African Americans was successful at replicating 21 of the loci previously identified in European populations, but did not nominate any new risk loci at a genome-wide significant level 38. The only other study nominated by our review process for MS in non-European populations was conducted in a Mexican population. The study found 4 significant variants, however, these variants had limited regional support and the study was severely underpowered with only 29 cases and 132 controls 39. Due to these limitations, we concluded that the variants identified in this study could not be classified as novel.
Sample sizes for MS GWAS are still relatively small, even for European populations. In addition, we did not find any multi-ancestry studies through our search methods, highlighting a potential opportunity for further discovery for this disease.
Frontotemporal dementia
Largest European GWAS: Ferrari 2014
Total samples: 12,928
There are no multi-ancestry or non-European studies in FTD.
Common risk loci nominated by previous European FTD studies include C9ORF72, GRN, and MAPT 40. The largest FTD GWAS in our review date range included ~13,000 participants of European ancestry and nominated an additional locus in the HLA-DRA/HLA-DRB5 region 41. This study was conducted in 2014, and while more recent GWAS of FTD have been performed, none have surpassed the sample size from the Ferrari study, and many have focused on smaller FTD subtypes 42,43.
No non-European or multi-ancestry GWAS were identified in our systematic review for FTD. Investigation of known genetic risk factors in non-Europeans suggest that C9ORF72 expansions may be quite rare in Chinese populations,44 highlighting the need for further research in this area.
Myasthenia Gravis
Largest European GWAS: Sakaue 2021
Total samples: 355,142
Largest Multi-ancestry GWAS: Sakaue 2021
Total samples: 533,853
Total non-European samples: 178,711
% non-European: 33.47%
Largest non-European GWAS: Na 2014
Ancestry: EAS
Total samples: 259
Known loci for MG include PTPN22, CTLA4, HLA-DQA1, ZBTB10, and TNFRSF11A 45,46, all nominated in European-based GWAS. The most recent GWAS for MG nominated an additional loci at CHRNA1, SFMBT2, and FAM76B, although the latter two did not replicate 47. The largest European and multi-ancestry GWAS for MG to date were both performed in the same study, leveraging 533,853 total samples from Japanese, UK, and Finnish-based biobanks. However, with only 278 cases, the effective sample size (4/(1/ncase+1/ncontrol)) for the meta analysis was insufficiently powered and they did not nominate any new loci for MG 48.
In non-European populations, the literature review identified one Korean GWAS for MG. However, this study was small and did not identify any loci meeting genome-wide significance 49. Other studies have found that there is earlier onset of MG in Asian populations, and higher prevalence of the ocular form in Asian children, highlighting the importance of continued discovery efforts for MG in non-European populations 50.
Lewy body dementia
Largest European GWAS: Chia 2021
Total samples: 7,372
There are no multi-ancestry or non-European studies in LBD.
Previously nominated risk loci for LBD include GBA, APOE, and SNCA 51,52. LBD can be hard to diagnose as there are a number of clinical and genetic overlaps with AD and PD, which may be one of the reasons why there is still limited genetic research for LBD in both European and non-European populations 51,53.
We found no LBD GWAS in any single non-European ancestry populations or any multi-ancestry studies through our search methods. Concrete data on the prevalence of LBD in diverse ancestries is difficult to acquire, showing a potential opportunity for valuable future research.
Vascular dementia
Largest European GWAS: Moreno-Grau 2019
Total samples: 4,830
Largest Multi-ancestry GWAS: Fongang 2022
Total samples: 482,088
Total non-European samples: 11,590
% non-European: 2.40%
There are no non-European studies in VaD.
Despite an approximated prevalence of about 15–20% in all dementia cases 54, vascular dementia remains difficult to study because of the uncertainty of diagnosis. In fact, only two studies on vascular dementia (VaD) passed our criteria and only one of these found genome-wide significant novel loci. The first study was a European GWAS that looked at vascular, mixed, and pure AD phenotypes and nominated loci at ANKRD31 and NDUFAF6 55. The second study that passed our criteria was a multi-ancestry GWAS for all-cause and vascular dementia including participants from European, African, Asian, and Hispanic ancestries, but did not find any significant novel loci 56.
VaD prevalence and risk appears to be higher in South Asian ancestries compared to European or Chinese populations 57,58. Additional studies have suggested that African Americans are most likely to be admitted to inpatient care with a VaD primary diagnosis 59. Despite these findings, there are still limited genetic studies for VaD, and we found no single non-European GWAS, highlighting the need for future research.
Discussion
This review highlights the lack of ancestral diversity in genetic research across neurodegenerative disease GWAS over the past decade. Current research suggests that including non-European populations can improve our understanding of the genetic architecture of disease through novel ancestry-specific discoveries, increased statistical power awarded by studying diverse haplotype structures, and the identification of loci with heterogeneous effects across populations 63.
Additionally, while we looked at seven genetic ancestry groups, these “buckets” do not capture the true diversity of global populations. The African continent is known to have high genetic diversity, yet individuals of African ancestry are routinely grouped into a single category 64 . In fact, we found no studies investigating South Asian (SAS) or continental African (AFR) populations. After investigating the cohorts in our review, we noted that though there were multiple “African” labeled studies, none of them directly investigated individuals in continental Africa, instead looking at African-American or other African-admixed populations. It is critical to mention the reference population used to define the specific population, to prevent the misattribution of genetic features across ancestries. Grouping all participants with any African ancestry into a generalized African category obscures the significant issue of inadequate representation of continental Africans.
In addition, there has been very little research done on admixed populations, and how the combinations of different ancestries affect SNP frequencies and/or gene expression. A GWAS in a Caribbean Hispanic admixed population found that the frequency of a novel locus spanning FBXL7 varied greatly, from 1% in those with European ancestry to 20% in African Americans.13
Furthermore, previous research has shown that the transferability of polygenic risk scores from African Americans to various African populations is highly unreliable 65. The substantial genetic and environmental disparities among individuals of African descent underscore the urgent need to improve diversity in genetic studies.
Diversity in SNP discovery
While 6 of the 8 NDDs we investigated had non-European representation, only PD had >1,000 cases and >30% non-European samples (Table 1). Additionally, no new significant loci have been identified in diverse population studies for MS, LBD, FTD, VaD, or MG. While the largest non-European cohort in MG included almost 180,000 samples, only 81 MG cases were included. A GWAS with less than 1,000 cases is unlikely to achieve sufficient statistical power for SNP discovery in polygenic diseases where multiple loci with small effect sizes are generally expected 60–62. Alzheimer’s, the most well funded of all the NDDs, has less than 3% diversity among cases in genetic studies. The incorporation of studies that lack statistical power and replicability minimizes the true imbalance between European and non-European studies, maintaining a Eurocentric bias.
In addition, many genetic association studies in East Asian populations did not meet our review criteria because they were not conducted on a genome-wide scale. Instead, these studies often investigated only one or a small group of SNPs that had been previously associated with disease in European populations, potentially missing associations that are specific to non-European populations.
In fact, many loci identified in European populations have heterogeneous effects or ancestry-specific SNP associations. For example, while the APOE alleles account for around a quarter of overall heritability for AD in Europeans 8,16, several studies suggest that the APOE4 allele has a weaker effect in African ancestry 66,67 and Caribbean Hispanic 68 populations. The effect has been found to be greater in Japanese populations 66,67. Heterogeneity of risk at APOE4 has been quantified in a recent multi-ancestry meta-analysis, with an I2 up to 85%, with 50% or more of that risk heterogeneity attributable to genetic ancestry differences 6. We believe that examination of local ancestry at loci with such global differences may help discern whether locus-specific inheritance patterns modulate disease risk.
Similarly, C9ORF72 is one of the most common risk factors for ALS. However, the frequency of the C9ORF72 expansion is lower in Chinese populations (0.3%) as compared to European populations (7%) 30. Recent research suggests that commonly used genetic tests to diagnose ALS may be less accurate in non-European ancestry patients because they are less likely to carry the C9ORF72 structural variant 69 .
Some SNPs with large effect sizes don’t exist or are extremely rare in certain ancestry groups. Variants in ABCA7, for example, increase AD risk more in individuals of African ancestry than in those of European ancestry 70. In fact, ABCA7 has a comparable effect size to APOE in individuals of African ancestry 10. Genetic variants in LRRK2, GBA, and SNCA, which have been associated with increased risk of PD in European ancestry populations, appear to have a negligible effect in individuals from India 71,72,73,74. Without studying diverse populations, researchers would miss the population-specific effects of these loci and potential therapeutic targets which modify their effects.
Looking forward
Despite the inequalities highlighted above, progress is being made. Researchers in AD are taking a strong multi-modal approach to increasing diversity. The Multi-Partner Consortium to Expand Dementia Research in Latin America (ReDLat) is leveraging “on the ground” connections with research communities in Latin America and the Caribbean to grow a diverse database of dementia resources 75,76. The Alzheimer’s Disease NeuroImaging Study (ADNI) is growing more inclusive cell lines and generating partner data for multiple ancestrally diverse samples 77. The NIH’s Center for Alzheimer’s and Related Dementias (CARD) is filling diversity gaps by creating training materials, generating data to complement existing efforts, and providing open science support for researchers in diverse communities.
Multiple efforts are also underway in the PD space. The Genetic Architecture of Parkinson disease in India (GAP-India) plans to develop a large clinical/genomic biobank in India 71. The Latin American Research Consortium on the Genetics of PD (LARGE-PD) aims to address inclusivity and genomic differences within and across Latino populations. Finally, the Global Parkinson’s Genetics Program (GP2) aims to genotype >150,000 individuals from around the world. GP2-funded projects include the Black and African Americans Connections to Parkinson’s Disease Study (BLAACPD), which seeks to assess the genetic architecture of Black and African American individuals with Parkinson’s disease, as well as healthy subjects, from across the United States 78. GP2 is motivated to increase diversity not only just among samples recruited into studies, but also in the investigators making use of the data, providing training and resources to ensure that all researchers are on an open and equal field of play 79.
A list of ongoing efforts for increasing diversity in NDD genetic research, including atypical dementias, can be found in Supplemental Table 3.
These efforts are paying off. More than 65% of the neurodegenerative disease-associated loci discovered in non-European or multi-ancestry populations were identified in the period between 2020 to 2022 (Figure 3). Over the past 10 years, there has been a steady increase in the proportion of non-European samples included in genetic studies (Figure 2B). With the increase in diverse samples in recent years, there has also been a growing interest in the use of multi-ancestry analyses to discover, fine-map and assess heterogeneity at disease risk loci, particularly in AD, PD, and ALS (Figure 2A).
Figure 3: Cumulative count of discovered SNPs from 2012 through 2022.
Notably, more than 65% of the SNPs were identified in the period between 2020 through 2022.
In fact, we are already seeing the benefits of increased diversity on genetic discovery in NDD research. A 2023 GWAS using African and African American samples collected by GP2 and 23andMe, and co-lead by researchers in Nigeria and NIH, found a novel GBA1 locus that is rare in other populations82. We anticipate that in the future, leveraging multiple ancestries will continue to improve fine-mapping resolution to prioritize causal variants5, increase access to and reduce bias in precision medicine practices such as polygenic risk prediction1, and drive many new discoveries in the genetics of NDDs.
Conclusion
Our systematic review highlights a striking disparity in the representation of diverse genetic ancestry populations in NDD research, emphasizing the urgent need for greater inclusivity to advance our understanding of these complex conditions and develop more equitable precision medicine approaches. Efforts to bridge this gap and promote diversity in genetic studies are vital for achieving meaningful progress in the diagnosis, treatment, and prevention of NDDs across global populations.
Supplementary Material
Key Points.
Question:
What is the state of ancestral inclusivity in genetic studies of neurodegenerative diseases?
Findings:
A systematic review of 123 publications on neurodegenerative diseases shows a focus on European populations, with only 18% of studies including any non-European ancestry data. Among 52 novel loci identified in non-European studies, 28 were from multi-ancestry studies (which included Europeans), 21 from East Asian studies, and 3 from other populations.
Meaning:
This significant disparity underscores the need for more inclusive research approaches in neurodegenerative diseases, emphasizing multi-ancestry and non-European populations to advance precision medicine and develop treatments effective for diverse populations.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA), National Institutes of Health, Department of Health and Human Services; project number ZO1 AG000535, as well as the National Institute of Neurological Disorders and Stroke. We would also like to thank the National Library of Medicine, including Nancy Terry and Alicia Livinski, for their assistance with the review and Covidence. J.S.Y. is supported by NIH-NIA R01AG062588, R01AG057234, P30AG062422, P01AG019724, U19AG079774; NIH-NINDS U54NS123985; NIH-NIDA 75N95022C00031; the Rainwater Charitable Foundation; the Alzheimer’s Association; the Global Brain Health Institute; and the Mary Oakley Foundation.
Footnotes
Conflict of Interest
C.J, K.S.L., L.J., H.L.L. and M.A.N.’s participation in this project was part of a competitive contract awarded to DataTecnica LLC by the National Institutes of Health to support open science research. M.A.N. also currently serves on the scientific advisory board for Character Bio Inc. and is a scientific founder at Neuron23 Inc. J.S.Y. serves on the scientific advisory board for the Epstein Family Alzheimer’s Research Collaboration.
References
- 1.Martin A. R. et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellenguez C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 1–25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumacher-Schuh A. F. et al. Diversity in Parkinson’s disease genetics research: current landscape and future directions. bioRxiv (2021) doi: 10.1101/2021.12.07.21266995. [DOI] [Google Scholar]
- 4.Innovation V. H. Covidence systematic review software. Preprint at (2019).
- 5.Mägi R. et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet. 26, 3639–3650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lake J. et al. Multi-ancestry meta-analysis and fine-mapping in Alzheimer’s disease. Mol. Psychiatry (2023) doi: 10.1038/s41380-023-02089-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J. J. et al. Multi-ancestry genome-wide meta-analysis in Parkinson’s disease. bioRxiv (2022) doi: 10.1101/2022.08.04.22278432. [DOI] [Google Scholar]
- 8.Wightman D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beach T. G., Monsell S. E., Phillips L. E. & Kukull W. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005–2010. J. Neuropathol. Exp. Neurol. 71, 266–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reitz C. et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. JAMA 309, 1483–1492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mez J. et al. Two novel loci, COBL and SLC10A2, for Alzheimer’s disease in African Americans. Alzheimers. Dement. 13, 119–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherva R. et al. African ancestry GWAS of dementia in a large military cohort identifies significant risk loci. bioRxiv (2022) doi: 10.1101/2022.05.25.22275553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosto G. et al. F box/ LRR repeat protein 7 is genetically associated with Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2, 810–820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirano A. et al. A genome-wide association study of late-onset Alzheimer’s disease in a Japanese population. Psychiatr. Genet. 25, 139–146 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Jia L. et al. Prediction of Alzheimer’s disease using multi-variants from a Chinese genome-wide association study. Brain 144, 924–937 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shigemizu D. et al. Ethnic and trans-ethnic genome-wide association studies identify new loci influencing Japanese Alzheimer’s disease risk. Transl. Psychiatry 11, 151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang S. et al. Potential Novel Genes for Late-Onset Alzheimer’s Disease in East-Asian Descent Identified by APOE-Stratified Genome-Wide Association Study. J. Alzheimers. Dis. 82, 1451–1460 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyashita A. et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One 8, e58618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake J. et al. Multi-ancestry meta-analysis and fine-mapping in Alzheimer’s Disease. bioRxiv (2022) doi: 10.1101/2022.08.04.22278442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nalls M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satake W. et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 41, 1303–1307 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Foo J. N. et al. Genome-wide association study of Parkinson’s disease in East Asians. Hum. Mol. Genet. 26, 226–232 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Li C. et al. Genetic Modifiers of Age at Onset for Parkinson’s Disease in Asians: A Genome-Wide Association Study. Mov. Disord. 36, 2077–2084 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Li C. et al. Genetic Determinants of Survival in Parkinson’s Disease in the Asian Population. Mov. Disord. (2022) doi: 10.1002/mds.29069. [DOI] [PubMed] [Google Scholar]
- 25.Loesch D. P. et al. Characterizing the Genetic Architecture of Parkinson’s Disease in Latinos. Ann. Neurol. 90, 353–365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lill C. M. et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 8, e1002548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foo J. N. et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans: A Genome-Wide Association Study. JAMA Neurol. 77, 746–754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grover S. et al. Genome-wide Association and Meta-analysis of Age at Onset in Parkinson Disease: Evidence From the COURAGE-PD Consortium. Neurology 99, e698–e710 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolas A. et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97, 1268–1283.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benyamin B. et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat. Commun. 8, 611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng M. et al. Genome-wide association analyses in Han Chinese identify two new susceptibility loci for amyotrophic lateral sclerosis. Nat. Genet. 45, 697–700 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Xie T. et al. Genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol. Aging 35, 1778.e9–1778.e23 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wei L. et al. Identification of TYW3/CRYZ and FGD4 as susceptibility genes for amyotrophic lateral sclerosis. Neurol Genet 5, e375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iacoangeli A. et al. Genome-wide Meta-analysis Finds the ACSL5-ZDHHC6 Locus Is Associated with ALS and Links Weight Loss to the Disease Genetics. Cell Rep. 33, 108323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura R. et al. A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Communications Biology 3, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rheenen W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 53, 1636–1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isobe N. et al. An ImmunoChip study of multiple sclerosis risk in African Americans. Brain 138, 1518–1530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ordoñez G. et al. Genomewide admixture study in Mexican Mestizos with multiple sclerosis. Clin. Neurol. Neurosurg. 130, 55–60 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Sirkis D. W., Geier E. G., Bonham L. W., Karch C. M. & Yokoyama J. S. Recent advances in the genetics of frontotemporal dementia. Curr. Genet. Med. Rep. 7, 41–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrari R. et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 13, 686–699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reus L. M. et al. Genome-wide association study of frontotemporal dementia identifies a C9ORF72 haplotype with a median of 12-G4C2 repeats that predisposes to pathological repeat expansions. Transl. Psychiatry 11, 451 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pottier C. et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 17, 548–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Che X.-Q. et al. Genetic Features of MAPT, GRN, C9orf72 and CHCHD10 Gene Mutations in Chinese Patients with Frontotemporal Dementia. Curr. Alzheimer Res. 14, 1102–1108 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Renton A. E. et al. A genome-wide association study of myasthenia gravis. JAMA Neurol. 72, 396–404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seldin M. F. et al. Genome-Wide Association Study of Late-Onset Myasthenia Gravis: Confirmation of TNFRSF11A and Identification of ZBTB10 and Three Distinct HLA Associations. Mol. Med. 21, 769–781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chia R. et al. Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study. Proc. Natl. Acad. Sci. U. S. A. 119, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakaue S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Na S.-J. et al. Whole-genome analysis in Korean patients with autoimmune myasthenia gravis. Yonsei Med. J. 55, 660–668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murai H. et al. Characteristics of myasthenia gravis according to onset-age: Japanese nationwide survey. J. Neurol. Sci. 305, 97–102 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Rongve A. et al. GBA and APOE ε4 associate with sporadic dementia with Lewy bodies in European genome wide association study. Sci. Rep. 9, 7013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chia R. et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat. Genet. 53, 294–303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guerreiro R. et al. Investigating the genetic architecture of dementia with Lewy bodies: a two-stage genome-wide association study. Lancet Neurol. 17, 64–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrijvers E. M. C. et al. Genome-wide association study of vascular dementia. Stroke 43, 315–319 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Moreno-Grau S. et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: The GR@ACE project. Alzheimers. Dement. 15, 1333–1347 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Fongang B. et al. A meta-analysis of genome-wide association studies identifies new genetic loci associated with all-cause and vascular dementia. Alzheimers. Dement. 17 Suppl 3, e056081 (2021). [Google Scholar]
- 57.Singh V., Dhamoon M. S. & Alladi S. Stroke Risk and Vascular Dementia in South Asians. Curr. Atheroscler. Rep. 20, 43 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Sahadevan S. et al. Ethnic differences in Singapore’s dementia prevalence: the stroke, Parkinson’s disease, epilepsy, and dementia in Singapore study. J. Am. Geriatr. Soc. 56, 2061–2068 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Simpkins A. N. Impact of Race-Ethnic and Economic Disparities on Rates of Vascular Dementia in the National Inpatient Sample Database from 2006–2014. J. Stroke Cerebrovasc. Dis. 29, 104731 (2020). [Google Scholar]
- 60.Hindorff L. A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 106, 9362–9367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wray N. R., Goddard M. E. & Visscher P. M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 17, 1520–1528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sham P. C. & Purcell S. M. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet. 15, 335–346 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Manolio T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell M. C. & Tishkoff S. A. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 9, 403–433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamiza A. B. et al. Transferability of genetic risk scores in African populations. Nat. Med. 28, 1163–1166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naslavsky M. S. et al. Global and local ancestry modulate APOE association with Alzheimer’s neuropathology and cognitive outcomes in an admixed sample. bioRxiv (2022) doi: 10.1101/2022.02.02.22270331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jun G. R. et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers. Dement. 13, 727–738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blue E. E., Horimoto A. R. V. R., Mukherjee S., Wijsman E. M. & Thornton T. A. Local ancestry at APOE modifies Alzheimer’s disease risk in Caribbean Hispanics. Alzheimers. Dement. 15, 1524–1532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mesaros M. Investigating the genetic profile of amyotrophic lateral sclerosis in patients of diverse race, ethnicity, and ancestry (REA). (The Ohio State University, 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stepler K. E. et al. ABCA7, a Genetic Risk Factor Associated with Alzheimer’s Disease Risk in African Americans. J. Alzheimers. Dis. (2022) doi: 10.3233/JAD-215306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rajan R. et al. Genetic Architecture of Parkinson’s Disease in the Indian Population: Harnessing Genetic Diversity to Address Critical Gaps in Parkinson’s Disease Research. Front. Neurol. 11, 524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Punia S. et al. Absence/rarity of commonly reported LRRK2 mutations in Indian Parkinson’s disease patients. Neurosci. Lett. 409, 83–88 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Vijayan B., Gopala S. & Kishore A. LRRK2 G2019S mutation does not contribute to Parkinson’s disease in South India. Neurol. India 59, 157–160 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Vishwanathan Padmaja M., Jayaraman M., Srinivasan A. V., Srikumari Srisailapathy C. R. & Ramesh A. The SNCA (A53T, A30P, E46K) and LRRK2 (G2019S) mutations are rare cause of Parkinson’s disease in South Indian patients. Parkinsonism Relat. Disord. 18, 801–802 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Ibanez A. et al. The Multi-Partner Consortium to Expand Dementia Research in Latin America (ReDLat): Driving Multicentric Research and Implementation Science. Front. Neurol. 12, 631722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ibanez A., Parra M. A., Butler C. & Latin America and the Caribbean Consortium on Dementia (LAC-CD). The Latin America and the Caribbean Consortium on Dementia (LAC-CD): From Networking to Research to Implementation Science. J. Alzheimers. Dis. 82, S379–S394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crane P. K. et al. Cognitive assessments in ADNI: Lessons learned from the ADNI psychometrics project. Alzheimer’s & Dementia vol. 17 Preprint at 10.1002/alz.056474 (2021). [DOI] [Google Scholar]
- 78.Bandres-Ciga S. Black and African American Connections to Parkinson’s Disease Study: Addressing Missing Diversity in Parkinson’s Disease Genetics. Mov. Disord. 37, 1559–1561 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Program T. G. P. G. & The Global Parkinson’s Genetics Program. GP2 : The Global Parkinson’s Genetics Program. Movement Disorders vol. 36 842–851 Preprint at 10.1002/mds.28494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borda M. G. et al. Colombian consortium for the study of Lewy body dementia COL-DLB. J. Neurol. Sci. 412, 116807 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Oppedal K. et al. European DLB consortium: diagnostic and prognostic biomarkers in dementia with Lewy bodies, a multicenter international initiative. Neurodegener. Dis. Manag. 9, 247–250 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Rizig M. et al. Identification of genetic risk loci and causal insights associated with Parkinson’s disease in African and African admixed populations: a genome-wide association study. Lancet Neurol. 22, 1015–1025 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.