Abstract
Thymic epithelial cells (TECs) make up the thymic microenvironments that support the generation of a functionally competent and self-tolerant T-cell repertoire. Cortical (c)TECs, present in the cortex, are essential for early thymocyte development including selection of thymocytes expressing functional TCRs (positive selection). Medullary (m)TECs, located in the medulla, play a key role in late thymocyte development, including depletion of self-reactive T cells (negative selection) and selection of regulatory T cells. In recent years, transcriptomic analysis by single-cell (sc)RNA sequencing (Seq) has revealed TEC heterogeneity previously masked by population-level RNA-Seq or phenotypic studies. We summarize the discoveries made possible by scRNA-Seq, including the identification of novel mTEC subsets, advances in understanding mTEC promiscuous gene expression, and TEC alterations from embryonic to adult stages. Whereas pseudotime analyses of scRNA-Seq data can suggest relationships between TEC subsets, experimental methods such as lineage tracing and reaggregate thymic organ culture (RTOC) are required to test these hypotheses. Lineage tracing – namely, of β5t or Aire expressing cells – has exposed progenitor and parent-daughter cellular relationships within TEC.
Keywords: Thymic epithelial cell, scRNA-Seq, mTEC, cTEC, Tuft, Aire, Promiscuous gene expression, Lineage tracing
1. Introduction
The thymus forms during fetal development, differentiating from the third pharyngeal pouch endoderm on embryonic day (E)9 of mouse gestation with lymphoid progenitors arriving by E11 [1, 2]. The development of the TEC lineage and therefore the thymus is dependent on the transcription factor Foxn1 [3, 4]. The thymus is a three-dimensional arrangement where TECs are compartmentalized in the two anatomical regions of the thymus: cortex and medulla. This compartmentalization is the basis for the broadest classification of TECs into cortical (c)TECs and medullary (m)TECs. Initially reliant on histology to distinguish these sublineages, TECs can now be discriminated by flow cytometry. Based on commonly used markers, TECs are defined as CD45− EpCAM+ with the additional expression of CD205 or Ly51 for cTECs and UEA1 for mTECs [5]. cTECs and mTECs are anatomically and functionally distinct niches providing different essential signals for T-cell development. cTECs support early thymocyte development, including T lineage determination and positive selection of T cells, whereas mTECs foster negative selection and late T-cell development. In recent years, the known diversity of TECs at both the functional and phenotypic levels has expanded. We will provide an overall description of TEC biology, heterogeneity, and parent-daughter relationships based on fundamental studies and new insights revealed by scRNA-Seq and lineage tracing methodologies.
2. cTEC Characteristics and Functional Roles
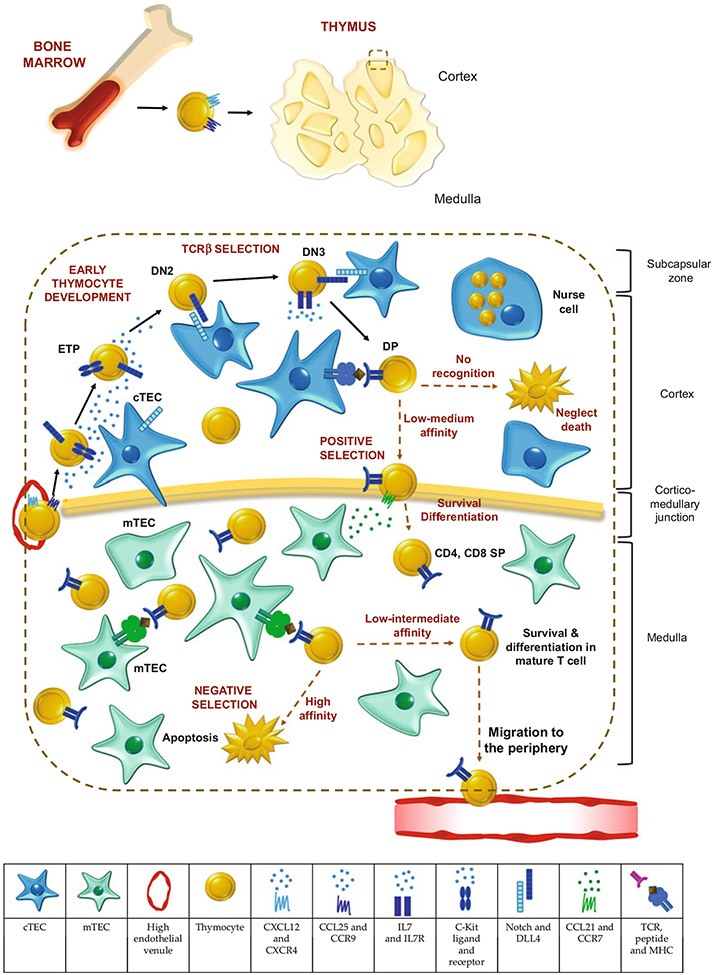
Functionally, cTECs can support diverse processes, including the chemotactic attraction of lymphoid progenitors to the thymus, the development of early thymic progenitors (ETPs) to CD4+CD8+ double-positive (DP) T cells, and functional TCR selection (known as positive selection). These processes are described below and summarized in Fig. 1. Fetal liver progenitors migrate to the thymus before the formation of the vasculature. In contrast, bone marrow-derived progenitors go through the vasculature at the corticomedullary junction to reach the thymus [6]. Once in the thymus, thymocyte migration proceeds through the cortex to the subcapsular zone and back through the cortex into the medulla [7]. Chemokines secreted by cTECs are critical regulators of this cell movement during which thymocytes undergo transitions through multiple developmental stages (DN1/ETP → DN2 → DN3 → DN4 → DP stages). The key events of β-selection and positive selection occur at the DN3 and DP stages, respectively [8-10]. Early thymic progenitors express the chemokine receptors CXCR4 and CCR9 [11-13], whereas cTECs express the corresponding ligands CXCL12 and CCL25 [11, 14,15], thus promoting thymic homing and intrathymic migration. Moreover, cTECs are the source of Kit ligand (also known as stem cell factor – SCF), and signaling through the kit receptor is critical for ETP expansion and development into DN2-stage progenitors [16, 17]. cTECs also express the Notch ligand Delta-like 4 (DLL4), whose signaling is essential for T lineage specification and for the transition of DN3/4 to DP thymocytes [18]. A subset of DLL4+ cTECs expresses high levels of IL-7, which supports the expansion of IL-7R-expressing T-cell precursors. Thymocyte expansion after β-selection enables the generation of TCR repertoire diversity during positive selection [19].
Fig. 1.
Thymic development is supported by thymic epithelial cells (TECs). Early thymic progenitors (ETPs) arrive at the thymus at the corticomedullary junction and then migrate through the cortex. The chemokines CXCL12 and CCL25 secreted by cTECs and the corresponding receptors CXCR4 and CCR9 expressed by thymocytes mediate thymic homing and intrathymic migration. During their movement in the cortex, thymocytes undergo transition through ETP, double-negative (DN)2, DN3, DN4, double-positive (DP), and single-positive (SP) stages. cTECs provide key molecules, such as Kit ligand, Notch ligand Delta-like 4 (DLL4), and IL-7, whose signaling is critical for T lineage specification, expansion, and transition between thymocyte stages. Positive selection by cTECs is enabled by their ability to process and present a wide variety of self-antigens on MHC molecules. Thymic nurse cells are a subset of cTECs that enclose DP thymocytes and provide a microenvironment for a secondary TCR rearrangement. In the medulla, mTECs secrete CCL21 to attract positively selected SP thymocytes. mTECs express an unparalleled number of self-peptides and tissue-restricted antigens – enabling them to mediate negative selection and establish central tolerance. Thymocytes with high affinity to self-peptides are induced to die by apoptosis, while thymocytes with low to intermediate affinity survive and differentiate into mature T cells. Please note that this is a schematic figure showing representative molecules and processes mediating thymocyte development by TECs. However, many other molecules are essential in these tasks
Positive selection is based on recognition of self-peptides presented on MHC class I and class II molecules. During this process, low-medium TCR-peptide/MHC interactions result in TCR signaling that promotes cell survival and differentiation from DP to SP (CD4+ or CD8+) cells. However, T cells that fail to recognize peptide/MHC molecules do not receive TCR signals and die by neglect [20]. In addition, high-affinity interactions result in cell death. The characterization of the peptides that induce positive selection has been extensively addressed. cTECs are the principal source of MHC-associated self-ligands, and they express unique protein degradation machineries crucial for the production of these self-ligands.
The proteasome is the main source of peptides presented by MHC-I. Murata et al. identified β5t, the catalytic subunit of the proteasome exclusively expressed by cTECs [21]. Interestingly, unique β5t-dependent cleavages were identified. Although the specific mechanism remains to be determined, it was shown that β5t-dependent peptides display specific amino acid sequences with C-terminal hydrophobic amino acids that induce low-affinity TCR interactions and promote positive selection [22]. Consistently, β5t-deficient mice display defective development of CD8+ thymocytes [21]. Thus, β5t enables the formation of a highly specialized thymoproteasome that is critical for MHC-I-dependent positive selection.
Similarly, for MHC-II presentation, the inactivation of the thymus-specific serine protease (TSSP), a peptide-producing enzyme localized to the endosomal/lysosomal compartments of cTECs, impaired the positive selection of CD4+ cell [23, 24]. Mice deficient in Prss16 (encoding TSSP) displayed a decreased expression of MHC-II in CDR1+ cTECs and a twofold reduction in the frequency of cTEC-hi (MHC-II hi) [23].
A fraction of β5t+ cTECs – termed thymic nurse cells (TNCs) – forms complexes with thymocytes that may serve to optimize positive selection. In these complexes, large cTECs enclose multiple DP thymocytes, providing a microenvironment for secondary TCR rearrangement and thereby increasing the chance of successful positive selection [25, 26]. Positively selected SP thymocytes express CCR7 and CCR4 and are attracted to the medulla toward a CCL21/CCL19 and CCL22/CCL17 gradient [27]. Hence, cTECs express several soluble, membrane-associated, and intracellular molecules crucial for early thymocyte migration, development, and selection.
3. mTEC Subset Characteristics and Functional Roles
mTECs are highly specialized antigen-presenting cells that mediate negative selection and establish T-cell self-tolerance, thereby imposing central tolerance. In this process, a wide variety of self-peptides are presented by MHC, and thymocytes with high TCR affinity for those self-peptides are induced to die by apoptosis. Conversely, some high-affinity thymocytes are agonist-selected to differentiate into immunoregulatory Foxp3+CD4+ T regulatory (Treg) cells. To accomplish these tasks, the thymic medulla contains functionally distinct mTEC subsets. The expression levels of MHC-II and CD80 subdivide mTECs into two principal populations: mTEChi (MHC-IIhi, CD80hi) and mTEClo (MHC-IIlo, CD80lo) [28]. An mTEClo subset expresses CCL21 to actively attract positively selected SP thymocytes, which highly express the CCL21 receptor, CCR7 [27, 29]. Promiscuous gene expression (PGE), a distinctive trait of mTEChi, is the ability to express many tissue-restricted antigens (TRAs). Aire is an important transcription factor driving PGE and thus the principal regulator of negative selection. The Aire+ subpopulation constitutes a nondividing, high turnover subset of the mTEChi [30, 31]. Intracellularly, Aire is localized in interchromatin granule clusters, associated with large transcriptional complexes containing many other proteins including RNA pol II, helicases, topoisomerases, transcription initiation and elongation factors, and ribonucleoproteins [32-34]. Notably, Aire induces mTEChi to express XCL1, a chemokine important in the recruitment of intrathymic dendritic cells (DCs) [35], and promotes antigenic transfer to these DCs – a crucial aspect for establishing central tolerance [36]. Some peripheral tissues have been found to express Aire; however, its role in the periphery remains to be understood [37-39]. Deleterious mutations in Aire are associated with autoimmune disorders in humans [40, 41]. Also, Aire-deficient mice exhibit reduced TRA transcription and autoimmune manifestations [42]. Although Aire was considered the unique regulator of PGE for many years, Takaba et al. demonstrated that the transcription factor Fefz2 also regulates TRA expression independently of Aire. Fefz2 has a broader expression profile within mTECs, and its ablation in mice leads to severe autoimmune symptoms [43]. Members of the TNFR superfamily influence the expression of both factors. RANK and CD40 mainly control Aire expression, whereas LTβR signaling regulates Fezf2 expression [43-45 ].
Along with the known role of mTECs in mediating negative selection and late-stage T-cell development, thymocytes participate in the maturation of mTECs and the structural organization of medullary regions. This bidirectional and interdependent communication between thymocytes and thymic stromal cells is known as thymic cross talk. Shores et al. described that mice lacking TCR+ thymocytes presented very few medullary cells that were not well organized into discrete medullary regions [46]. Subsequent studies demonstrated that the expression of functional TCR alpha/beta and CCR7-dependent migration of positively selected thymocytes are each essential for establishment of the medulla [47-49]. In addition, it has been shown that RANKL, CD40L, and LT signaling in mTECs is critical for thymic cross talk [50-55].
4. Single-Cell Assessment of TEC Heterogeneity and Diversity
In recent years, transcriptomic analyses by RNA sequencing (RNA-Seq) have contributed significantly to our understanding of several immune processes, including evolutionary relationships, developmental trajectories, microenvironment complexity, and cell-cell interactions. Additionally, the use of single-cell (sc)RNA-Seq has enhanced the discovery of new immune cell subtypes that were masked by bulk transcriptional approaches and phenotypic studies [56]. Notably, transcriptional studies at single-cell resolution have revealed previously unexpected TEC heterogeneity and novel insights into PGE programming by TECs. This section will describe the main findings on TEC diversity and heterogeneity revealed by scRNA-Seq technologies. A framework of scRNA-Seq is shown in Fig. 2.
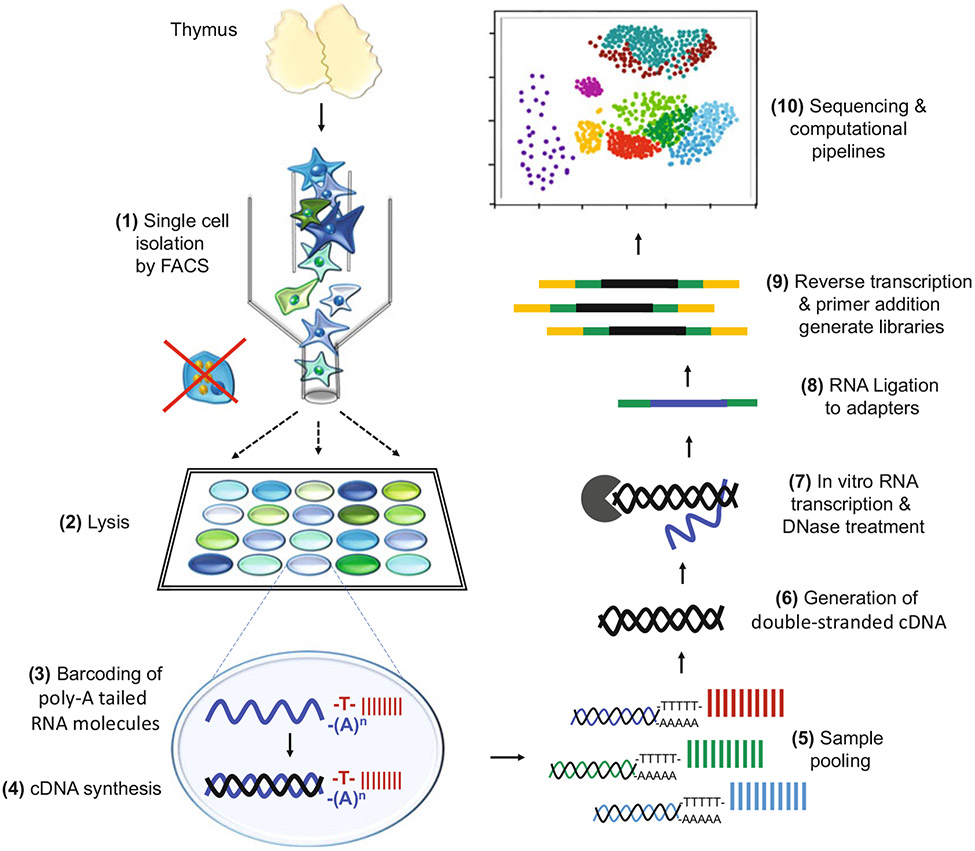
Fig. 2.
Schematic diagram of scRNA-Seq in the TEC scRNA-Seq framework based on the Massively parallel RNA single-cell sequencing framework (MARS-Seq) [93]. Thymic cells are used as an example. Single cells are first isolated by FACS eliminating cell complexes such as nurse cells. Next, cells are lysed in individual wells and, poly-A tailed RNA molecules are barcoded by annealing them to randomized molecular tags (unique molecular identifiers). Molecular tags also contain a T7 promoter (not shown), which will be used later for in vitro transcription. Then, reverse transcription is performed, generating the first cDNA strand. Next, individual cell lysates are pooled, and RNA-cDNA hybrids are converted to double-stranded cDNA (second strand synthesis). Then, in vitro transcription generates single-chain RNA molecules (this constitutes the first round of amplification), which are then fragmented and ligated to sequencing adapters. Finally, reverse transcription and a second round of amplification generate cDNA libraries for sequencing
4.1. Reclassification of the mTEC Compartment into Four Major Groups
As previously mentioned, the mTEC compartment is commonly subdivided into two main groups based on the expression of CD80 and MHC class II – designated mTEChi and mTEClo. However, new single-cell sequencing technologies have provided a more comprehensive atlas of TEC subpopulations and have shown a more complex and diverse molecular mTEC repertoire than was previously appreciated. Independently, Miller et al. [57] and Bornstein et al. [58] used RNA-Seq to subdivide mTEC subsets in postnatal mice. While the results from these groups had significant overlaps, they used distinct approaches and different nomenclatures for their mTEC subgroups. Notably, both groups identified specialized mTEC subpopulations that formed microniches within the medulla. While this section will focus on the results and methods of Bornstein et al., the techniques and conclusions of Miller et al. will be discussed in the lineage tracing section.
The reclassification of mTECs by Bornstein et al. was based on scRNA-Seq data and chromatin profiling (iChIP-Seq and ATAC-Seq). Using these strategies, mTECs were divided into four transcriptionally and epigenetically defined subtypes: mTEC I-IV. These subgroups were retrospectively validated (index sorting strategy) by the de novo identification of surface markers and staining followed by MARS-Seq and qPCR. mTEC I were defined by MHC-IIlo and β4 integrin expression. Concordantly, they exhibited the lowest level of stochastically expressed genes across the TEC populations. In addition, some mTEC I showed high levels of α6 integrin and Sca1, which correspond to the markers of putative TEC progenitors discussed further in the lineage trace section [59]. Finally, mTEC I also contained cells with activating chromatin markers around the Ccl21 locus.
The mTEC II subset displayed high levels of the PGE transcription factors, Aire and Fefz2, and antigen presentation associated molecules such as Cd40, H2-Aa, and Cd74. Additionally, epigenetic characterization showed an enrichment of H3K4me2 (marking transcriptionally active genes) in Aire enhancer regulatory elements. Accordantly, mTEC II exhibited the highest levels of stochastically expressed genes but also the broadest TRA expression variability across the TEC subsets.
mTEC III constituted a heterogeneous population that exhibited several genes known to be expressed by previously described post-AIRE cells including Krt10, Sca1, and Pigr [60, 61]. Concordantly, this subset showed low levels of MHC-II and CD80. Furthermore, contrary to mTEC II, mTEC III displayed a more restricted distribution of stochastically expressed genes. This population will be discussed in more detail in the lineage tracing section.
mTEC IV had a gene expression signature associated with intestinal tuft cells. Tuft cells residing in mucosal barriers are distinguished by the presence of apical microvilli harboring tuft-like brush structures and the expression of choline-acetyltransferase (ChAT) and taste receptor signaling members such as TRPM5 [62]. Additionally, they are a major source of IL-25, and their development is dependent on the transcription factor POU2F3. Thymic tuft cells were initially identified by light- and electronmicroscopic immunohistochemistry and were described as a subpopulation of morphologically heterogeneous, medullary epithelial cells [63]. Moreover, a later study by Panneck et al. described them as ChAT-positive cells residing in the medulla. They displayed features associated with tuft intestinal cells such as a lateral microvilli ultrastructure and the expression of taste transduction cascade members [64]. Bornstein et al. and Miller et al. simultaneously described a tuft cell-like mTEC subset via the implementation of scRNA-Seq, bulk RNA-Seq, and lineage tracing [57, 58]. In agreement with the requirement for Pou2f3 by mucosal tuft cells, Pou2f3−/− mice showed severely reduced thymic tuft cells. Notably, the identification of enhancer regulatory elements by H3K4me2 ChIP-Seq and motif enrichment by ATAC-Seq demonstrated enrichment of the Pou2F3 motif specifically in mTEC IV [58]. Aire−/− mice displayed unaltered numbers but molecular alterations (shown by alteration in Gnat3 gene expression) in tuft TECs. This finding suggests that thymic tuft development is Aire-independent but does not rule out the possibility of a nondevelopment role.
As previously mentioned, in the thymus, IL-25 is exclusively expressed by tuft cells. However, the function of thymic tuft cells is incompletely understood [57, 58]. Intestinal tuft cells detect the luminal pathogens which drives IL-25-mediated type 2 immune responses. Concordantly, reduction of the thymic tuft compartment in Pou2f3−/− mice resulted in decreased NKT2 cells [57]. In the intestine, group 2 innate lymphoid cells (ILC2) are activated by tuft cell-derived IL-25 after luminal detection of pathosymbionts. Intriguingly, a significant increase in the thymic ILC2 population was observed in Pou2f3−/− mice. Additionally, thymic transplantation from Pou2f3−/− mice into athymic Foxn1−/− hosts resulted in the generation of higher IL-25 auto-antibody titers compared to C57BL/6 neonatal thymic transplants [57]. Thus, thymic tuft cells could be essential in mediating tolerance to IL-25.
Thymic, and especially TEC, research in humans has also progressed. A recent analysis of human thymus samples from 15 prenatal and 9 postnatal samples by scRNA-Seq comprehensively analyzed thymocyte subsets as well as thymic stromal components, including TECs [65]. Interestingly, early thymic samples were predominantly cTECs, while postnatal samples were predominantly mTECs, which aligns with observations of mouse TEC among which cTEC develop prior to mTECs [28]. TECs were subdivided into eight subtypes: cTEC, mcTEC, mTEC I, mTEC II, mTEC III, mTEC IV, TEC (neuro), and TEC (myo). Although the mcTEC, neuro TEC, and myo TEC subtypes were not found to have direct mouse counterparts, the cTEC and mTEC I-IV designations directly correlated to mouse equivalents and comparison of these populations in human and mouse samples revealed the expression of characteristic genes laid out by Bornstein et al. [58]. As seen in mouse scRNA-Seq samples and detailed below in Subheading 4.3, TECs from different age frames clustered separately. For instance, their mcTEC cluster, an intermediate population between cTECs and mTECs, was almost exclusively present in the late fetal-neonatal time period. Finally, taking advantage of sequencing of all thymic cells, Park et al. analyzed cross-talk interactions by chemokine and chemokine receptor expression and confirmed known relationships at a transcriptional level – verifying that cTECs and DP and early SP T cells use the CCL25/CCR9 axis, whereas mTECs, SP T cells, and activated DCs use the CCL19/CCR7 axis. Interestingly, this study did not recapitulate the use of the XCL1/XCR1 axis documented by mTEC/DC1 in mouse. Instead, XCL1/XCR1 designated interactions between CD1 and CD8aa T cells. The use of this axis by mTECs may have been drowned out by grouping all mTECs together given that XCL1 is specifically produced by Aire+ mTECs [36].
More recently, Bautista et al. profiled human thymus stroma (CD45- EpCam−) and epithelial (CD45- EpCam+) compartments across fetal, postnatal, and adult stages by scRNA-Seq [66]. They identified different clusters corresponding to epithelial, mesenchymal, pericyte, mesothelial, endothelial, immune, and red blood cells. This study subsequently focused on TEC heterogeneity and assessed other stromal populations for their potential to influence TEC development and function. The mesenchyme cluster was found to highly express RSPO3-, SFRP2-, and FRZB-secreted factors directly associated with the Wnt/β-catenin signaling pathway, which has been described as a regulator of TEC development [67-69]. Endothelial cells expressed growth factors TGFβ1 and IGF1 suggesting that they can also contribute to TEC development. The epithelial compartment was further analyzed, and TECs were subdivided into nine subclusters: cTEC-hi, cTEC-lo, immature TEC, mTEC-lo, Aire+ mTEC-hi, corneocyte-like mTEC, neuroendocrine TEC, myoid TECm and myelin+ TEC. To validate these findings, they compared their dataset to the one previously published by Park et al. [65]. Most of the clusters corresponded, but one population of early (fetal) cTEC was found only in the Park dataset, whereas one population of myelin+ cells was only found in the Bautista dataset. Importantly, both works identified immature TEC that did not express any of the functional genes of cTECs and mTECs and that accumulated in the adult thymus. Immature TECs expressed ZBED2, IGFBP5, and low levels of KRT15, among other genes. Notably, knockdown of ZBED2 in human basal keratinocytes induces differentiation, suggesting that this gene maintains an immature state in keratinocytes [70] and may have a similar role in TEC. By re-clustering mTEC and rare TEC populations using a higher resolution analysis, Bautista et al. revealed two additional clusters – tuft/ionocyte and ciliated – and verified their presence in the medulla using immunofluorescence. The tuft chemosensory cells expressed POU2F3, GNAT3, AVIL, PLCB2, and OVOL3, similar to what was already described by Borstein et al. [58] and Miller et al. [57]. However, in the human samples, tuft cells did not express IL-25. This study also found a population of CFRT+ cells localized in the Hassall’s corpuscles or dispersed in the medulla that resembled lung ionocytes and a population of ciliated cells previously observed in the mouse thymus by microscopy, positive for ATHO1, GFI1, LHX3, and FOXJ1. Finally, this analysis showed some markers specific to the myelin+ cells, including SOX10, MPZ, and MBP, therefore resembling Schwann cells.
4.2. TRA Signature, Another Trait of TEC Heterogeneity
Early studies examining TEC heterogeneity at the single-cell level focused on determining the mechanism of PGE. In three early studies, the maximum cellular input was only about 200 mature mTECs. Interestingly, approximately 19,000 genes, including almost 95% of TRA were expressed in this small number of cells [71-73]. How does this small cell subset guarantee a comprehensive representation of the self-antigen repertoire? Interestingly, TECs expressed the highest proportion of genes of any other cell type. Indeed, Sansom et al. [71] used population-level approaches and determined that cTECs and mTECs express 84% and 89%, respectively, of the Ensembl protein-coding genes [74]. Single-cell approaches have confirmed that Aire ensures the promiscuous transcription of an unparalleled number of TRAs. These analyses revealed that Aire-dependent genes are induced in only a minority of mTECs and are expressed in these cells more highly (16-fold) than previously predicted from population-level studies [71]. Moreover, it has been shown that a proportion of Aire-dependent and Aire-independent TRAs are expressed in concert and can be grouped into small but stable clusters. Calculation of the mean genome distance between neighboring genes indicated that numerous genes are clustered densely in specific genomic regions, suggesting that genes for TRAs may be coordinately expressed [72,73].
More recently, Dhalla et al. [75] undertook a large-scale scRNA-Seq analysis of 6894 mTECs taken from Balb/c, C57Bl/6, and Balb/c x C57Bl/6 F1 mice that were either unselected or selected for expression of the TRAs TSPAN8 or GP2. Their goals were to address mTEC heterogeneity with a focus on TRA distribution within mTECs. Their analysis subdivided mTECs into 15 clusters grouped into 8 subtypes: pre-Aire, proliferating, mature, post-Aire, tuft-like, fibroblast-like, ciliated, and GP2-preferred mTECs. The gene expression characteristics of the pre-Aire (Ccl21a), mature (Aire), post-Aire (Krt10, Ivl), and tuft-like (Il25, Dclk1) subtypes matched up with characteristics previously put forward by Bornstein et al. The proliferating cluster was primarily characterized by the expression of G2/M genes such as Mki67. Interestingly, the pre-Aire cluster was subdivided into two clusters – one with higher expression of alpha6 integrin (Itga6) and Sca1(Ly6a) similar to the potential TEC progenitors that will be discussed in the lineage trace section. Pseudotime analysis indicated three trajectories, all beginning in the proliferating mTEC cluster: two continued up through mature and into post-Aire mTEC clusters, whereas the third branched down into the pre-Aire clusters. An alternative analysis, RNA velocity, indicated similar results with the primary difference being a minor population of pre-Aire cells having a trajectory into the proliferating mTECs – thereby connecting pre-Aire and mature mTECs. Probing PGE mechanism, Dhalla et al. characterized modules based off of co-expressed TRAs. Co-expressed TRAs were not grouped by peripheral organ expression or by chromosomal location, arguing against the idea that TRA expression is driven by peripheral tissue programs or chromosomal proximity. The authors instead suggested distinct factors such as TEC maturation stage and/or chromatin proximity may be involved.
4.3. TEC Transcriptional Changes During Development and Aging
The thymus as a whole and TECs specifically are known to undergo extreme alterations from fetal development to adulthood. Recent work from the Hollander lab has highlighted the dramatic changes in the proportion of TEC subpopulations during aging using flow cytometry and scRNA-Seq analyses on mice from neonatal to 1 year of age. Phenotypically, cTEC populations expand significantly after 4 weeks of age, while mTEC-Lo and mTEC-Hi populations decrease. However, in silico analysis reveals expansion of not only the cTEC population but also a population of TECs expressing genes characteristic of both cTECs (Prss16 and Cxcl12) and mTECs (Ccl21a and Krt5) [76]. scRNA-Seq revealed distinct transcriptional gene expression patterns in TECs throughout life. As with the human samples discussed above, mouse TECs cluster by age instead of by subtype [65, 77, 78]. Kernfeld et al. [78] analyzed mouse TECs from E12.5 through birth and analyzed the mTEC and cTEC compartments by age. Analysis of maturation markers in diffusion maps for pseudotime analysis demonstrated that cTECs upregulate H2-Aa, Cd40, and Enpep (Ly-51), while mTEC upregulate Tnfrsf11a (RANK), Aire, and Cd80. Additionally, ex vivo culturing of E13.5 thymi in FTOC did not interfere with TEC maturation and demonstrated that TECs mature in vitro with the only major difference being a reduced proportion of cTECs with high cell cycle gene expression [78]. Indeed, population RNA-Seq analysis of cTECs and mTECs from E13.5 to 17+ months demonstrated clustering by subtype and by age with age-based clustering for both cTECs and mTECs driven by Myc-target genes including cell cycle and ribosome biogenesis. These results were corroborated in scRNA-Seq and experimental quantification of proliferation, total RNA levels, and Myc expression. Transgenic expression of Myc in TECs drove thymic growth, increasing total thymic size and the proportion of cTECs while maintaining equivalent proportions of mTEC I-IV subtypes. Functionally, transgenic Myc expression enhanced BrdU incorporation in adult TECs, increased the number of active ribosomes in adult TECs, and rescued the ability of adult TECs to successfully engraft after intrathymic transplantation into an irradiated host [77]. A major hurdle for elucidating thymic regeneration moving forward will be identifying upstream controls of Myc as well as the developmental trajectories of TECs and their progenitors in the adult thymus.
5. The Use of Lineage Tracing to Identify TEC Progenitor Populations and Parent-Daughter Relationships
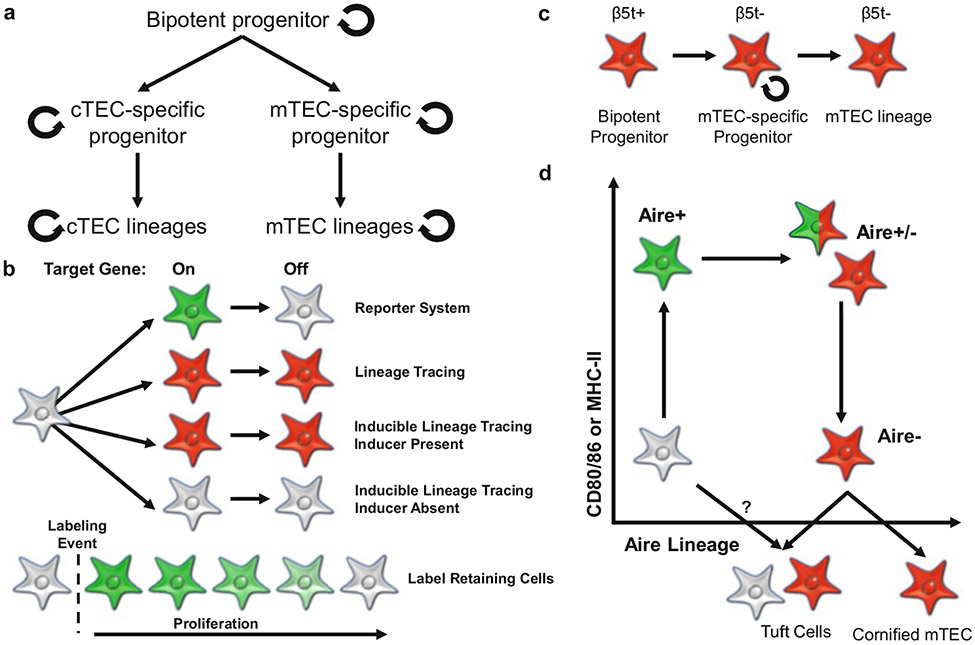
The development and maintenance of TEC populations within the thymus is a crucial research topic for understanding thymic involution and regeneration. Population differentiation and maintenance are generally thought of in a hierarchy with multiple levels of stem or progenitor cells and subsequent differentiation into mature cell types (Fig. 3a). The proliferation required for population maintenance could occur at progenitor stages and/or at mature stages. For TECs, the apex of the hierarchy is presumed to be a bipotential progenitor cell, capable of making both cTEC and mTEC lineages. Although this population is known to exist embryonically, its persistence in adult mice has not been definitively established [79]. The next rung could be cTEC- and mTEC-restricted progenitors followed by development of the cTEC and mTEC mature subtypes. The differentiation of mature TEC subsets is not currently known to be linear or branching or a combination.
Fig. 3.
The use of lineage tracing to ascertain cellular relationships. (a) Potential models for TEC developmental trajectories and levels of maintenance. (b) The fluorescence phenotypes of reporter (green), lineage tracing (red), and inducible lineage tracing (red) models when the target gene turns on and then off. The fluorescence phenotype of label-retaining cell models with details on before and after the labeling event and the impact of proliferation is also shown. (c) β5t lineage tracing of mTEC development indicates that β5t is expressed in bipotent progenitors and that β5t- mTEC-specific progenitors contribute to the maintenance of the mTEC lineage long term. (d) Combination of Aire-reporter (green) and inducible Aire fate mapping (red) reveals multiple stages of mTEC differentiation. Analysis of a short tamoxifen pulse reveals the presence of Aire-reporter+ and Aire-reporter−, lineage-traced mTEC in the mTEChi fraction, as well as Aire-reporter-, lineage-traced mTEC in the mTEClo fraction. Post-Aire mTEClo cells have been subdivided into cornified mTEC and tuft cells. Lineage tracing has not ruled out the potential for an Aire-independent developmental path for tuft cells
While scRNA-Seq has revealed previously unknown heterogeneity within TEC populations and pseudotime analysis tools such as Monocle can hypothesize relationships between these groups, experimental methods are required to test these hypotheses. In contrast to reporter systems, which use a fluorescent protein to indicate when a gene is actively being expressed, lineage tracing (or fate mapping) is a method of permanently marking a cell and its daughter cells – usually with a constitutively expressed fluorophore turned on by Cre recombinase-mediated recombination (Fig. 3b). The converse of this technique is ubiquitous marking followed by an extended chase to identify quiescent progenitor populations (known as label-retaining cells or LRC). There have been two dominant TEC lineage tracing targets: β5t – a cTEC-specific component of the immunoproteasome – and Aire, an mTEC-specific transcription factor. Cre is expressed under the control of specific target genes by inserting the Cre cassette directly into the gene locus or by creating a transgene of the Cre cassette within the target gene locus. Cre activity can be further controlled by creating fusion proteins with inducible domains (namely ERT2, tamoxifen, and Tet, doxycycline). The targeted, transient Cre activity of inducible systems allow for assessment at multiple timepoints (i.e., embryonic vs postnatal vs adult) – an important consideration for TEC given the drastic age-based changes the thymus undergoes [65, 77, 78]. Ultimately, the verification of potential progenitors and parent-daughter relationships must be tested by FACS-based isolation and transplantation into fetal or reaggregate thymic organ cultures (FTOC or RTOC) with the ultimate test being single-cell transfer resulting in reconstitution [79].
In order to search for potential TEC progenitor populations, LRCs were assessed because they retain their label by being quiescent – a common characteristic for stem cells. Osada et al. utilized a tetracycline-off (Tet-Off, tTA) system in keratin 5-expressing TECs to turn off H2B-GFP expression upon doxycycline administration starting at 4 weeks postnatally. After 10 weeks of doxycycline administration, the majority of EpCAM+ LRCs were Sca1+, MHC-IIlo, α6 integrinlo/hi, and β1 integrin+ [80]. Using a 12-day BrdU pulse followed by a 6-month chase, Wong et al. characterized a population of EpCAM+ LRCs with similar cell markers – namely, UEA1−, MHC-IIlo, Sca1hi, and α6 integrinhi – that were able to successfully generate mTEC and cTEC lineages in RTOC [59]. While a putative mTEC-specific progenitor population was also identified within the mTEClo population as Sca1hi and α6 integrinint, RTOCs revealed their ability to develop into mTEChi, but not establish long-term maintenance – although they did persist to a greater extent than their MHC-IIhi mTEC and cTEC counterparts. Dumont-Lagace et al. identified a similar population (UEA1-, MHC-IIlo, Sca1hi, α6 integrinhi, CD24lo) using a Tet-On (rtTA) system that induces H2B-GFP expression in all cells during doxycycline administration [81]. Similarly to Wong et al., a UEA1 + subset of LRCs was also detected, but these cells failed to proliferate in response to induced atrophy, unlike UEA1-LRCs. While the progenitor abilities of the LRC populations (especially the UEA1-populations) identified by these three groups have been tested by ex vivo colony formation, RTOC, and BrdU incorporation during induced atrophy and regeneration, these were done as bulk assays – not definitively ruling out the potential of this population to contain separate lineage-specific precursors for cTECs and mTECs rather than bipotent progenitors. Importantly, all of these models were performed in adult mice – indicating the presence of long-lived, quiescent populations in the adult thymus.
Fate mapping of β5t-expressing cells revealed unexpected results. Murata et al. demonstrated that 80% of cTECs express β5t using mice with the reporter fluorophore Venus substituted into one allele of Psmb11 [21]. Intriguingly, more than 95% of both cTECs and mTECs in 6- to 7-week-old mice are lineage traced in β5t-Cre/LoxP-GFP mice – indicating that both cTECs and mTECs develop from β5t-expressing progenitors. Indeed, immunohistochemical analysis of thymic tissue from E12.75 verified that all TEC were either co-expressing β5t and GFP or were already lineage traced by β5t [82]. This corresponds with prior data from β5t-Venus reporter mice that β5t is detectable by E12.5 [83]. In order to determine if β5t-expressing cells continued to give rise to mTECs postnatally, a Tet-On system (β5t-rtTA; TRE-Cre; LoxP-GFP or ZsGreen) was employed. In accordance with prior results, doxycycline dosing from conception to 4 weeks postnatal resulted in 83% of mTECs and 95% of cTECs being labeled. However, altering the timing of induction to start 1-week postnatal significantly reduced the labeling of mTECs in 4-week-old mice to less than 10%. Interestingly, cTEC labeling also decreased to 40% when doxycycline was withheld until 2 weeks of postnatal life [84]. Replacement of the LoxP-GFP with the confetti cassette (which randomly expresses one of four fluorophores upon Cre-mediated recombination) allowed assessment of single-cell contributions to mTEC and cTEC compartments by immunofluorescence. Doxycycline administration at 1 week of age resulted in clusters of fluorophore-labeled cells that extend into the cortex and medulla – positioning these β5t-expressing bipotential progenitor cells at the cortico-medullary junction [85].
Long term maintenance of TEC has been experimentally probed by time courses as well as by induction of ablation or atrophy and regeneration. In accordance with the above results, cTEC-specific ablation by administration of diphtheria toxin (DT) to mice expressing the DT receptor (DTR) under the control of the cTEC-specific gene Ccr-ckr1 revealed that neonatal cTECs rapidly recover while adult thymic recovery is stunted [86]. Similarly, embryonic ablation of cTECs by expression of nitroreductase (NTR) under the control of Prss16 or Psmb11 (encoding TSSP and β5t, respectively) and administration of the pro-form of the cytotoxic agent corroborated the efficient recovery capacity of the cTEC compartment early in life [87]. Despite the fact that these ablation experiments were not designed to result in complete removal of cTECs, thymic cellularity plummeted in all three instances – highlighting the importance of cTECs for the thymocyte population. However, the impact on cTEC and mTEC subsets was not directly assessed in these studies. Using the inducible β5t-cre mice, a 45-week time course indicated that cells labeled embryonically (E0-birth) and neonatally (D0-1 week) contribute to mTEC and cTEC populations long term with 50–60% of mTEC and cTEC fate mapped at 45 weeks of age [84, 85]. In agreement, the percentage of labeled mTECs in adults remained consistently low after induction of atrophy by irradiation or poly I/C treatment and subsequent regeneration – indicating repopulation by previously labeled mTEC-specific progenitors rather than the creation of new progenitors. Interestingly, doxycycline administration during the regeneration time course labeled approximately 5% of adult mTECs with or without induction of atrophy [84]. Collectively, these results indicate that (1) β5t-expressing cells contribute to both cTEC and mTEC lineages, (2) the majority of mTEC-specific progenitors are created embryonically and neonatally, and (3) mTEC-specific progenitors provide long-term maintenance for the mTEC compartment (Fig. 3c).
While β5t lineage tracing has provided invaluable information of progenitor characteristics, Aire lineage tracing has provided insights into the differentiation and development of mature mTEC subsets. Unlike β5t models which knock in the fluorophore, Cre recombinase, or rtTA cassette into the β5t locus, Aire models have relied on the random incorporation of bacterial artificial chromosome (BAC) transgenes encoding the Cre recombinase or reporter fluorophore within the Aire locus [21, 82, 84, 88, 89]. The first Aire lineage tracing experiments generated multiple transgenic mouse lines, and their characterization revealed that Aire is broadly expressed during early embryogenesis. However, a mouse line with mTEC-specific expression specifically labeled Aire + cells found in the CD80hi, MHC-IIhi compartment, but also indicated the existence of lineage-traced, Aire- cells in the CD80lo, MHC-IIlo compartment [88]. By altering the Cre to a tamoxifen-inducible ERT2-Cre and crossing with Aire +/GFP or −/GFP mice, Nishikawa et al. validated the presence of Aire-, fate-mapped mTECs in the CD80lo, MHC-IIlo subset. They further demonstrated that lineage-traced, Aire-deficient (Aire −/GFP) mTECs failed to downregulate CD80 [88, 89]. Simultaneously, Metzger et al. [90] also created Aire expression-restricted ERT2-Cre mice and combined them with Aire-reporter mice. Time course analysis of the resulting mTECs after a single tamoxifen dose revealed that Aire+ mTECs begin to downregulate Aire within 7 days of induction (becoming reporter-negative, but remaining lineage traced) with about 50% losing Aire expression by 10 days post-induction (Fig. 3d). In 6- to 8-week old mice, the percentage of lineage-traced mTECs increases from D1 to D7 post-induction (climaxing at 35%) but quickly decreases with only 6% of cells lineage traced by D35 post-induction – indicating that although proliferation does occur during mTEC maturation, these populations are not being maintained long term unlike the β5t lineage-traced mTEC populations. Additionally, lineage-traced, reporter-positive cells remain steadfastly MHC-IIhi, while lineage-traced, reporter-negative cells are both MHC-IIhi and MHC-IIlo with MHC-IIlo cells expressing increased terminal differentiation markers such as involucrin and decreased mTEChi markers such as Aire and Xcl1 [90].
The Anderson lab has subsequently used these lineage trace mice to characterize post-Aire cells. Bulk RNA-Seq of cells 7 days post-induction isolated by MHC-II expression levels and lineage trace into pre-Aire (MHC-II-lo, trace−), early-Aire (MHC-IIhi, trace−), late-Aire (MHC-IIhi, trace+), and post-Aire (MHC-IIlo, trace+) revealed two gene signatures: cornified epithelium characterized by Ivl and Krt10 and intestinal tuft cells exemplified by high expression of Trpm5 and Dclk1. Thymic tuft cells can be identified by flow cytometry using DCLK1 and IL-25-reporter expression [57, 58]. The progenitor-successor relationship of tuft cells was assessed using a 10-day tamoxifen pulse that resulted in approximately 50% of IL-25-reporter+ tuft cells being lineage traced by Aire [57]. Indeed, ablation of Aire+ cells using a DTR system (as described above) that co-expresses GFP revealed that while single-dose ablation efficiently eliminates GFP+ mTEChi, extended ablation results in significant reductions in mTEClo populations – including Dclk1+ tuft cells [57]. Bornstein et al. employed fate mapping of Csnb (also known as Csn2) to ascertain tuft cell development. Although Csnb is expressed in mTEC II/III (Aire and post-Aire), but not in mTEC I/IV (pre-Aire/tuft), tuft cells were lineage traced at a frequency of 55%. However, only 40–50% of mTEC II and III were fate mapped – indicating either poor efficiency of the Cre or variable expression of Csnb [58]. Together, these results indicate the potential of Aire-dependent and Aire-independent tuft cell development pathways; however, a longer tamoxifen pulse or a time course has not been performed to ascertain whether all tuft cells naturally proceed through an Aire+ stage or to determine the longevity of this population.
Combination of the Aire fate mapping system with RANK-L blockade and scRNA-Seq analysis revealed further nuances within mTEC subset relationships [91]. In addition to the four subsets identified previously (here referred to as Ccl21a-high, Aire-positive, late-Aire, and tuft), a fifth population termed transit-amplifying cell (TAC) TEC was identified and characterized by active cycling. Pseudotime analysis suggested that TAC-TEC gives rise to both Ccl21a-high and Aire-positive TEC – advocating for a branched mTEC lineage tree. After a 10-day induction, Aire expression lineage traced approximately 90% of Aire-positive and late-Aire clusters and 52% of TAC-TECs. In contrast to the Aire-positive population, 18% of TAC-TECs expressed Aire without being fate-mapped – indicating recent Aire expression. Sequencing of TECs over a 10-week time course following RANK-L blockade, which ablates MHC-II-hi, Aire-positive mTECs, agreed with the predicted pseudotime with TAC-TECs recovering first, followed by Aire-positive and subsequently late-Aire populations. The TAC-TEC to Ccl21a-high branch of the pseudotime has yet to be experimentally tested.
While these lineage tracing experiments have provided valuable insight into TEC development, differentiation, and maintenance, there are still many questions to be addressed. Both β5t and Aire lineage tracing have predominantly focused on the mTEC lineage. The β5t lineage tracing model holds promise for elucidation of cTEC lineage questions as well. Indeed, the decrease of fate-mapped cTECs to 40% when induced 2 weeks postnatal indicates that not all cTEC express β5t [84]. Similarly, the 30% decrease in embryonically and neonatally labeled cTECs over the 45-week time course suggests that new cTEC-specific progenitors are made in adult life that may or may not come from β5t-expressing progenitors [84]. Combined with the observation that new mTEC progenitors also arise from β5t-expressing cells in adulthood (albeit fewer), these results imply the presence of active bipotent progenitors in the adult thymus. The putative bipotent progenitors identified by LRC techniques do not express β5t [59] – although it is possible that β5t is turned on only when the cells are triggered to divide and differentiate. These putative progenitor populations have not been assessed in the models of DTR and NTR induced cTEC ablation and regeneration [86]. Additionally, the use of Sox10 to fate map neural crest (NC)-derived cells in the thymus mapped approximately 10% of TECs. It has yet to be determined whether these cells are actually neural crest derived or if Sox10 is co-opted by a subset of TECs – however, it is interesting that an equivalent proportion of both cTECs and mTECs are traced by it. The functionality or long-term contribution of Sox10-traced TECs has not been established [57]. Thus, while significant progress has been made on mTEC lineages, many questions remain. A key step in moving forward will be to reconcile the RNA-Seq signatures of TEC subsets with flow cytometric phenotypic markers (using lineage tracing, reporter systems, and antibodies against surface proteins) to be able to accurately isolate and assess TEC subsets. The segregation of mTEC subsets by definitive reporter systems – such as Aire, Ccl21a, and IL-25 – may help identify specific cell surface markers for these groups [92].
6. Conclusions
The intricate mysteries of the thymus are starting to be unraveled. While foundational research into the functional characteristics of cTECs and mTECs has provided broad characteristics and classifications of TECs, the implementation of scRNA-Seq and lineage tracing has greatly enhanced our knowledge of TEC biology and provided directions for future research. Specifically, ingenious experiments incorporating lineage tracing and scRNA-Seq have improved our ability to distinguish mTEC subsets and verify progenitor-successor relationships. However, even with acknowledgement of these expanded subsets, it is known that certain subsets are likely to be further heterogeneous. For instance, the mTEC I subset is hypothesized to be heterogenous for mature CCL21-producing cells as well as progenitors for Aire+ mTEC [58, 59, 92]. cTEC heterogeneity has been an especially difficult area of study. Even the application of scRNA-Seq has had difficulty with subdividing cTEC groups despite the evidence and hypotheses that bipotent progenitors may reside within cTEC groupings [59]. Indeed, the fact that these progenitors are predicted to be present in minute quantities in the adult thymus combined with the notorious difficultly of cTEC isolation from adult thymi could be inhibiting their ability to group separately in a scRNA-Seq analysis. Additionally, cTEC heterogeneity is underestimated by scRNA-Seq as single-cell sorting inherently eliminates cell complexes such as thymic nurse cells. The combination of scATAC-Seq with scRNA-Seq is expected to reveal further intricacies and hypotheses into TEC developmental trajectories. However, a significant hurdle to the validation of these hypotheses (and a current area of interest) is the identification of reliable and specific surface markers for TEC subsets. We expect that future research into TEC heterogeneity and lineage development will rely upon a combination of new surface markers, the implementation of new reporter and fate mapping models, the development of barcoding models, single-cell analyses, and functional assessment of putative progenitors.
Support Statement
AMS is supported by the patronage of the Children’s Hospital of Mexico.
SCS, JEC and AB are supported by the NIH Intramural Cancer Research Training Award to postdoctoral fellows.
References
- 1.Owen JJ, Ritter MA (1969) Tissue interaction in the development of thymus lymphocytes. J Exp Med 129(2):431–442. 10.1084/jem.129.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon J, Wilson VA, Blair NF, Sheridan J, Farley A, Wilson L, Manley NR, Blackburn CC (2004) Functional evidence for a single endodermal origin for the thymic epithelium. Nat Immunol 5(5):546–553. 10.1038/ni1064 [DOI] [PubMed] [Google Scholar]
- 3.Pantelouris EM (1968) Absence of thymus in a mouse mutant. Nature 217(5126):370–371. 10.1038/217370a0 [DOI] [PubMed] [Google Scholar]
- 4.Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ, Boehm T (1996) Two genetically separable steps in the differentiation of thymic epithelium. Science 272(5263):886–889. 10.1126/science.272.5263.886 [DOI] [PubMed] [Google Scholar]
- 5.Anderson G, Takahama Y (2012) Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol 33(6):256–263. 10.1016/j.it.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 6.Kyewski BA (1987) Seeding of thymic microenvironments defined by distinct thymocytestromal cell interactions is developmentally controlled. J Exp Med 166(2):520–538. 10.1084/jem.166.2.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Love PE, Bhandoola A (2011) Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol 11(7):469–477. 10.1038/nri2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gossens K, Naus S, Corbel SY, Lin S, Rossi FM, Kast J, Ziltener HJ (2009) Thymic progenitor homing and lymphocyte homeostasis are linked via S1P-controlled expression of thymic P-selectin/CCL25. J Exp Med 206(4):761–778. 10.1084/jem.20082502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkinson WE, Rossi SW, Parnell SM, Agace WW, Takahama Y, Jenkinson EJ, Anderson G (2007) Chemokine receptor expression defines heterogeneity in the earliest thymic migrants. Eur J Immunol 37(8):2090–2096. 10.1002/eji.200737212 [DOI] [PubMed] [Google Scholar]
- 10.Rossi FM, Corbel SY, Merzaban JS, Carlow DA, Gossens K, Duenas J, So L, Yi L, Ziltener HJ (2005) Recruitment of adult thymic progenitors is regulated by P-selectin and its ligand PSGL-1. Nat Immunol 6(6):626–634. 10.1038/ni1203 [DOI] [PubMed] [Google Scholar]
- 11.Wurbel MA, Philippe JM, Nguyen C, Victorero G, Freeman T, Wooding P, Miazek A, Mattei MG, Malissen M, Jordan BR, Malissen B, Carrier A, Naquet P (2000) The chemokine TECK is expressed by thymic and intestinal epithelial cells and attracts double- and single-positive thymocytes expressing the TECK receptor CCR9. Eur J Immunol 30(1):262–271. [DOI] [PubMed] [Google Scholar]
- 12.Uehara S, Grinberg A, Farber JM, Love PE (2002) A role for CCR9 in T lymphocyte development and migration. J Immunol 168(6):2811–2819. 10.4049/jimmunol.168.6.2811 [DOI] [PubMed] [Google Scholar]
- 13.Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A (2010) CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood 115(10):1897–1905. 10.1182/blood-2009-08-237784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas B, White AJ, Parnell SM, Henley PM, Jenkinson WE, Anderson G (2017) Progressive changes in CXCR4 expression that define thymocyte positive selection are dispensable for both innate and conventional alphabetaT-cell development. Sci Rep 7(1):5068. 10.1038/s41598-017-05182-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadakia T, Tai X, Kruhlak M, Wisniewski J, Hwang IY, Roy S, Guinter TI, Alag A, Kehrl JH, Zhuang Y, Singer A (2019) E-protein-regulated expression of CXCR4 adheres preselection thymocytes to the thymic cortex. J Exp Med 216(8):1749–1761. 10.1084/jem.20182285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buono M, Facchini R, Matsuoka S, Thongjuea S, Waithe D, Luis TC, Giustacchini A, Besmer P, Mead AJ, Jacobsen SE, Nerlov C(2016)A dynamic niche provides Kit ligand in a stage-specific manner to the earliest thymocyte progenitors. Nat Cell Biol 18(2):157–167. 10.1038/ncb3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodewald HR, Kretzschmar K, Swat W, Takeda S (1995) Intrathymically expressed c-kit ligand (stem cell factor) is a major factor driving expansion of very immature thymocytes in vivo. Immunity 3(3):313–319. 10.1016/1074-7613(95)90116-7 [DOI] [PubMed] [Google Scholar]
- 18.Deftos ML, Bevan MJ (2000) Notch signaling in T cell development. Curr Opin Immunol 12(2):166–172. 10.1016/s0952-7915(99)00067-9 [DOI] [PubMed] [Google Scholar]
- 19.Ribeiro AR, Rodrigues PM, Meireles C, Di Santo JP, Alves NL (2013) Thymocyte selection regulates the homeostasis of IL-7-expressing thymic cortical epithelial cells in vivo. J Immunol 191(3):1200–1209. 10.4049/jimmunol.1203042 [DOI] [PubMed] [Google Scholar]
- 20.Szondy Z, Garabuczi E, Toth K, Kiss B, Koroskenyi K (2012) Thymocyte death by neglect: contribution of engulfing macrophages. Eur J Immunol 42(7):1662–1667. 10.1002/eji.201142338 [DOI] [PubMed] [Google Scholar]
- 21.Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316(5829):1349–1353. 10.1126/science.1141915 [DOI] [PubMed] [Google Scholar]
- 22.Sasaki K, Takada K, Ohte Y, Kondo H, Sorimachi H, Tanaka K, Takahama Y, Murata S (2015) Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat Commun 6:7484. 10.1038/ncomms8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gommeaux J, Gregoire C, Nguessan P, Richelme M, Malissen M, Guerder S, Malissen B, Carrier A (2009) Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol 39(4):956–964. 10.1002/eji.200839175 [DOI] [PubMed] [Google Scholar]
- 24.Carrier A, Nguyen C, Victorero G, Granjeaud S, Rocha D, Bernard K, Miazek A, Ferrier P, Malissen M, Naquet P, Malissen B, Jordan BR(1999) Differential gene expression in CD3epsilon- and RAG1-deficient thymuses: definition of a set of genes potentially involved in thymocyte maturation. Immunogenetics 50(5–6):255–270. 10.1007/s002510050601 [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa Y, Ohigashi I, Nitta T, Sakata M, Tanaka K, Murata S, Kanagawa O, Takahama Y (2012) Thymic nurse cells provide microenvironment for secondary T cell receptor alpha rearrangement in cortical thymocytes. Proc Natl Acad Sci U S A 109(50):20572–20577. 10.1073/pnas.1213069109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wekerle H, Ketelsen UP (1980) Thymic nurse cells–Ia-bearing epithelium involved in T-lymphocyte differentiation? Nature 283(5745):402–404. 10.1038/283402a0 [DOI] [PubMed] [Google Scholar]
- 27.Ueno T, Saito F, Gray DH, Kuse S, Hieshima K, Nakano H, Kakiuchi T, Lipp M, Boyd RL, Takahama Y (2004) CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med 200(4):493–505. 10.1084/jem.20040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray DH, Seach N, Ueno T, Milton MK, Liston A, Lew AM, Goodnow CC, Boyd RL (2006) Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood 108(12):3777–3785. 10.1182/blood-2006-02-004531 [DOI] [PubMed] [Google Scholar]
- 29.Lkhagvasuren E, Sakata M, Ohigashi I, Takahama Y (2013) Lymphotoxin beta receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J Immunol 190(10):5110–5117. 10.4049/jimmunol.1203203 [DOI] [PubMed] [Google Scholar]
- 30.Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M, Minato N (2007) Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol 8(3):304–311. 10.1038/ni1438 [DOI] [PubMed] [Google Scholar]
- 31.Gray D, Abramson J, Benoist C, Mathis D (2007) Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med 204(11):2521–2528. 10.1084/jem.20070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bansal K, Yoshida H, Benoist C, Mathis D (2017) The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol 18(3):263–273. 10.1038/ni.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramson J, Giraud M, Benoist C, Mathis D (2010) Aire’s partners in the molecular control of immunological tolerance. Cell 140(1):123–135. 10.1016/j.cell.2009.12.030 [DOI] [PubMed] [Google Scholar]
- 34.Derbinski J, Schulte A, Kyewski B, Klein L (2001) Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol 2(11):1032–1039. 10.1038/ni723 [DOI] [PubMed] [Google Scholar]
- 35.Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT, Bosl MR, Hollander GA, Hayashi Y, Malefyt Rde W, Nitta T, Takahama Y (2011) Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med 208(2):383–394. 10.1084/jem.20102327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancaster JN, Thyagarajan HM, Srinivasan J, Li Y, Hu Z, Ehrlich LIR (2019) Live-cell imaging reveals the relative contributions of antigen-presenting cell subsets to thymic central tolerance. Nat Commun 10(1):2220. 10.1038/s41467-019-09727-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klamp T, Sahin U, Kyewski B, Schwendemann J, Dhaene K, Tureci O (2006) Expression profiling of autoimmune regulator AIRE mRNA in a comprehensive set of human normal and neoplastic tissues. Immunol Lett 106(2):172–179. 10.1016/j.imlet.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 38.Kogawa K, Nagafuchi S, Katsuta H, Kudoh J, Tamiya S, Sakai Y, Shimizu N, Harada M (2002) Expression of AIRE gene in peripheral monocyte/dendritic cell lineage. Immunol Lett 80(3):195–198. 10.1016/s0165-2478(01)00314-5 [DOI] [PubMed] [Google Scholar]
- 39.Schaller CE, Wang CL, Beck-Engeser G, Goss L, Scott HS, Anderson MS, Wabl M (2008) Expression of Aire and the early wave of apoptosis in spermatogenesis. J Immunol 180(3):1338–1343. 10.4049/jimmunol.180.3.1338 [DOI] [PubMed] [Google Scholar]
- 40.Finnish-German AC (1997) An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet 17(4):399–403. 10.1038/ng1297-399 [DOI] [PubMed] [Google Scholar]
- 41.Scott HS, Heino M, Peterson P, Mittaz L, Lalioti MD, Betterle C, Cohen A, Seri M, Lerone M, Romeo G, Collin P, Salo M, Metcalfe R, Weetman A, Papasavvas MP, Rossier C, Nagamine K, Kudoh J, Shimizu N, Krohn KJ, Antonarakis SE (1998) Common mutations in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients of different origins. Mol Endocrinol 12(8):1112–1119. 10.1210/mend.12.8.0143 [DOI] [PubMed] [Google Scholar]
- 42.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D (2002) Projection of an immunological self shadow within the thymus by the aire protein. Science 298(5597):1395–1401. 10.1126/science.1075958 [DOI] [PubMed] [Google Scholar]
- 43.Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, Kodama T, Takayanagi H (2015) Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell 163(4):975–987. 10.1016/j.cell.2015.10.013 [DOI] [PubMed] [Google Scholar]
- 44.Aaltonen J, Horelli-Kuitunen N, Fan JB, Bjorses P, Perheentupa J, Myers R, Palotie A, Peltonen L (1997) High-resolution physical and transcriptional mapping of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy locus on chromosome 21q22.3 by FISH. Genome Res 7(8):820–829. 10.1101/gr.7.8.820 [DOI] [PubMed] [Google Scholar]
- 45.Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, Krohn KJ, Lalioti MD, Mullis PE, Antonarakis SE, Kawasaki K, Asakawa S, Ito F, Shimizu N (1997) Positional cloning of the APECED gene. Nat Genet 17(4):393–398. 10.1038/ng1297-393 [DOI] [PubMed] [Google Scholar]
- 46.Shores EW, Van Ewijk W, Singer A (1991) Disorganization and restoration of thymic medullary epithelial cells in T cell receptor-negative scid mice: evidence that receptor-bearing lymphocytes influence maturation of the thymic microenvironment. Eur J Immunol 21(7):1657–1661. 10.1002/eji.1830210711 [DOI] [PubMed] [Google Scholar]
- 47.Palmer DB, Viney JL, Ritter MA, Hayday AC, Owen MJ (1993) Expression of the alpha beta T-cell receptor is necessary for the generation of the thymic medulla. Dev Immunol 3(3):175–179. 10.1155/1993/56290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Negishi I, Motoyama N, Nakayama K, Nakayama K, Senju S, Hatakeyama S, Zhang Q, Chan AC, Loh DY (1995) Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature 376(6539):435–438. 10.1038/376435a0 [DOI] [PubMed] [Google Scholar]
- 49.Kurobe H, Liu C, Ueno T, Saito F, Ohigashi I, Seach N, Arakaki R, Hayashi Y, Kitagawa T, Lipp M, Boyd RL, Takahama Y (2006) CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity 24(2):165–177. 10.1016/j.immuni.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 50.Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, Hayashi Y, Matsumoto M, Matsuo K, Penninger JM, Takayanagi H, Yokota Y, Yamada H, Yoshikai Y, Inoue J, Akiyama T, Takahama Y (2008) The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity 29(3):438–450. 10.1016/j.immuni.2008.06.018 [DOI] [PubMed] [Google Scholar]
- 51.Rossi SW, Kim MY, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, McConnell FM, Scott HS, Penninger JM, Jenkinson EJ, Lane PJ, Anderson G (2007) RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med 204(6):1267–1272. 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akiyama T, Shimo Y, Yanai H, Qin J, Ohshima D, Maruyama Y, Asaumi Y, Kitazawa J, Takayanagi H, Penninger JM, Matsumoto M, Nitta T, Takahama Y, Inoue J (2008) The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity 29(3):423–437. 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 53.Dunn RJ, Luedecker CJ, Haugen HS, Clegg CH, Farr AG (1997) Thymic overexpression of CD40 ligand disrupts normal thymic epithelial organization. J Histochem Cytochem 45(1):129–141. 10.1177/002215549704500116 [DOI] [PubMed] [Google Scholar]
- 54.Venanzi ES, Gray DH, Benoist C, Mathis D (2007) Lymphotoxin pathway and Aire influences on thymic medullary epithelial cells are unconnected. J Immunol 179(9):5693–5700. 10.4049/jimmunol.179.9.5693 [DOI] [PubMed] [Google Scholar]
- 55.Zhu M, Chin RK, Tumanov AV, Liu X, Fu YX (2007) Lymphotoxin beta receptor is required for the migration and selection of autoreactive T cells in thymic medulla. J Immunol 179(12):8069–8075. 10.4049/jimmunol.179.12.8069 [DOI] [PubMed] [Google Scholar]
- 56.Chen H, Ye F, Guo G (2019) Revolutionizing immunology with single-cell RNA sequencing. Cell Mol Immunol 16(3):242–249. 10.1038/s41423-019-0214-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, Parent AV, Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS (2018) Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development. Nature 559(7715):627–631. 10.1038/s41586-018-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Toth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann VS, Abramson J, Amit I (2018) Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559(7715):622–626. 10.1038/s41586-018-0346-1 [DOI] [PubMed] [Google Scholar]
- 59.Wong K, Lister NL, Barsanti M, Lim JM, Hammett MV, Khong DM, Siatskas C, Gray DH, Boyd RL, Chidgey AP (2014) Multilineage potential and self-renewal define an epithelial progenitor cell population in the adult thymus. Cell Rep 8(4):1198–1209. 10.1016/j.celrep.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Laan M, Bichele R, Kisand K, Scott HS, Peterson P (2012) Post-Aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front Immunol 3(March):19. 10.3389/fimmu.2012.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hale LP, Markert ML (2004) Corticosteroids regulate epithelial cell differentiation and Hassall body formation in the human thymus. J Immunol 172(1):617–624. 10.4049/jimmunol.172.1.617 [DOI] [PubMed] [Google Scholar]
- 62.Schneider C, O’Leary CE, Locksley RM (2019) Regulation of immune responses by tuft cells. Nat Rev Immunol 19(9):584–593. 10.1038/s41577-019-0176-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farr A, Nelson A, Hosier S, Kim A (1993) A novel cytokine-responsive cell surface glyco-protein defines a subset of medullary thymic epithelium in situ. J Immunol 150(4):1160–1171 [PubMed] [Google Scholar]
- 64.Panneck AR, Rafiq A, Schutz B, Soultanova A, Deckmann K, Chubanov V, Gudermann T, Weihe E, Krasteva-Christ G, Grau V, del Rey A, Kummer W (2014) Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res 358(3):737–748. 10.1007/s00441-014-2002-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, Wilbrey-Clark A, Roberts K, Kedlian VR, Ferdinand JR, He X, Webb S, Maunder D, Vandamme N, Mahbubani KT, Polanski K, Mamanova L, Bolt L, Crossland D, de Rita F, Fuller A, Filby A, Reynolds G, Dixon D, Saeb-Parsy K, Lisgo S, Henderson D, Vento-Tormo R, Bayraktar OA, Barker RA, Meyer KB, Saeys Y, Bonfanti P, Behjati S, Clatworthy MR, Taghon T, Haniffa M, Teichmann SA (2020) A cell atlas of human thymic development defines T cell repertoire formation. Science 367(6480):eaay3224. 10.1126/science.aay3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bautista JL, Cramer NT, Miller CN, Chavez J, Berrios DI, Byrnes LE, Germino J, Ntranos V, Sneddon JB, Burt TD, Gardner JM, Ye CJ, Anderson MS, Parent AV (2021) Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat Commun 12(1):1096. 10.1038/s41467-021-21346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lepletier A, Hun ML, Hammett MV, Wong K, Naeem H, Hedger M, Loveland K, Chidgey AP (2019) Interplay between follistatin, activin a, and BMP4 signaling regulates postnatal thymic epithelial progenitor cell differentiation during aging. Cell Rep 27(13):3887–3901. e3884. 10.1016/j.celrep.2019.05.045 [DOI] [PubMed] [Google Scholar]
- 68.Wertheimer T, Velardi E, Tsai J, Cooper K, Xiao S, Kloss CC, Ottmuller KJ, Mokhtari Z, Brede C, deRoos P, Kinsella S, Palikuqi B, Ginsberg M, Young LF, Kreines F, Lieberman SR, Lazrak A, Guo P, Malard F, Smith OM, Shono Y, Jenq RR, Hanash AM, Nolan DJ, Butler JM, Beilhack A, Manley NR, Rafii S, Dudakov JA, van den Brink MRM (2018) Production of BMP4 by endothelial cells is crucial for endogenous thymic regeneration. Sci Immunol 3(19):eaal2736. 10.1126/sciimmunol.aal2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balciunaite G, Keller MP, Balciunaite E, Piali L, Zuklys S, Mathieu YD, Gill J, Boyd R, Sussman DJ, Hollander GA (2002) Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat Immunol 3(11):1102–1108. 10.1038/ni850 [DOI] [PubMed] [Google Scholar]
- 70.Finnegan A, Cho RJ, Luu A, Harirchian P, Lee J, Cheng JB, Song JS (2019) Single-cell transcriptomics reveals spatial and temporal turnover of keratinocyte differentiation regulators. Front Genet 10:775. 10.3389/fgene.2019.00775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, Heger A, Ponting CP, Hollander GA (2014) Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res 24(12):1918–1931. 10.1101/gr.171645.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meredith M, Zemmour D, Mathis D, Benoist C (2015) Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol 16(9):942–949. 10.1038/ni.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Kuchler R, Huber W, Kyewski B, Steinmetz LM (2015) Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol 16(9):933–941. 10.1038/ni.3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM (2013) Ensembl 2013. Nucleic Acids Res 41 (Database issue):D48–D55. 10.1093/nar/gks1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dhalla F, Baran-Gale J, Maio S, Chappell L, Hollander GA, Ponting CP (2020) Biologically indeterminate yet ordered promiscuous gene expression in single medullary thymic epithelial cells. EMBO J 39(1):e101828. 10.15252/embj.2019101828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baran-Gale J, Morgan MD, Maio S, Dhalla F, Calvo-Asensio I, Deadman ME, Handel AE, Maynard A, Chen S, Green F, Sit RV, Neff NF, Darmanis S, Tan W, May AP, Marioni JC, Ponting CP, Hollander GA (2020) Ageing compromises mouse thymus function and remodels epithelial cell differentiation. Elife 9: e56221. 10.7554/eLife.56221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cowan JE, Malin J, Zhao Y, Seedhom MO, Harly C, Ohigashi I, Kelly M, Takahama Y, Yewdell JW, Cam M, Bhandoola A (2019) Myc controls a distinct transcriptional program in fetal thymic epithelial cells that determines thymus growth. Nat Commun 10(1):5498. 10.1038/s41467-019-13465-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R (2018) A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity 48(6):1258–1270. e1256. 10.1016/j.immuni.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ (2006) Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature 441(7096):988–991. 10.1038/nature04813 [DOI] [PubMed] [Google Scholar]
- 80.Osada M, Singh VJ, Wu K, Sant’Angelo DB, Pezzano M (2013) Label retention identifies a multipotent mesenchymal stem cell-like population in the postnatal thymus. PLoS One 8(12):e83024. 10.1371/journal.pone.0083024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dumont-Lagace M, Gerbe H, Daouda T, Laverdure JP, Brochu S, Lemieux S, Gagnon E, Perreault C (2017) Detection of quiescent radioresistant epithelial progenitors in the adult thymus. Front Immunol 8:1717. 10.3389/fimmu.2017.01717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohigashi I, Zuklys S, Sakata M, Mayer CE, Zhanybekova S, Murata S, Tanaka K, Hollander GA, Takahama Y (2013) Aire-expressing thymic medullary epithelial cells originate from beta5t-expressing progenitor cells. Proc Natl Acad Sci U S A 110(24):9885–9890. 10.1073/pnas.1301799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kitaoka M, Tsuruda Y, Tanaka Y, Goto M, Mitsumori M, Hayashi K, Hiraishi Y, Miyawaki K, Noji S, Kamiya N (2011) Transglutaminase-mediated synthesis of a DNA-(enzyme)n probe for highly sensitive DNA detection. Chemistry 17(19):5387–5392. 10.1002/chem.201003744 [DOI] [PubMed] [Google Scholar]
- 84.Ohigashi I, Zuklys S, Sakata M, Mayer CE, Hamazaki Y, Minato N, Hollander GA, Takahama Y (2015) Adult thymic medullary epithelium is maintained and regenerated by lineage-restricted cells rather than bipotent progenitors. Cell Rep 13(7):1432–1443. 10.1016/j.celrep.2015.10.012 [DOI] [PubMed] [Google Scholar]
- 85.Mayer CE, Zuklys S, Zhanybekova S, Ohigashi I, Teh HY, Sansom SN, Shikama-Dorn N, Hafen K, Macaulay IC, Deadman ME, Ponting CP, Takahama Y, Hollander GA (2016) Dynamic spatio-temporal contribution of single beta5t+ cortical epithelial precursors to the thymus medulla. Eur J Immunol 46(4):846–856. 10.1002/eji.201545995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rode I, Boehm T (2012) Regenerative capacity of adult cortical thymic epithelial cells. Proc Natl Acad Sci U S A 109(9):3463–3468. 10.1073/pnas.1118823109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hirakawa M, Nagakubo D, Kanzler B, Avilov S, Krauth B, Happe C, Swann JB, Nusser A, Boehm T (2018) Fundamental parameters of the developing thymic epithelium in the mouse. Sci Rep 8(1):11095. 10.1038/s41598-018-29460-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nishikawa Y, Hirota F, Yano M, Kitajima H, Miyazaki J, Kawamoto H, Mouri Y, Matsumoto M (2010) Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J Exp Med 207(5):963–971. 10.1084/jem.20092144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nishikawa Y, Nishijima H, Matsumoto M, Morimoto J, Hirota F, Takahashi S, Luche H, Fehling HJ, Mouri Y, Matsumoto M (2014) Temporal lineage tracing of Aire-expressing cells reveals a requirement for Aire in their maturation program. J Immunol 192(6):2585–2592. 10.4049/jimmunol.1302786 [DOI] [PubMed] [Google Scholar]
- 90.Metzger TC, Khan IS, Gardner JM, Mouchess ML, Johannes KP, Krawisz AK, Skrzypczynska KM, Anderson MS (2013) Lineage tracing and cell ablation identify a post-Aire-expressing thymic epithelial cell population. Cell Rep 5(1):166–179. 10.1016/j.celrep.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wells KL, Miller CN, Gschwind AR, Wei W, Phipps JD, Anderson MS, Steinmetz LM (2020) Combined transient ablation and single-cell RNA-sequencing reveals the development of medullary thymic epithelial cells. Elife 9:e60188. 10.7554/eLife.60188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kozai M, Kubo Y, Katakai T, Kondo H, Kiyonari H, Schaeuble K, Luther SA, Ishimaru N, Ohigashi I, Takahama Y (2017) Essential role of CCL21 in establishment of central self-tolerance in T cells. J Exp Med 214(7):1925–1935. 10.1084/jem.20161864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I (2014) Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343(6172):776–779. 10.1126/science.1247651 [DOI] [PMC free article] [PubMed] [Google Scholar]